Abstract
Lung cancer (LC) is a commonly diagnosed cancer with an unsatisfactory prognosis. Extracellular vesicles (EVs) are lipid bilayer-delimited particles that mediate cell-cell communication by transporting various biomacromolecules, such as nucleic acids, proteins, and lipids. Noncoding RNAs (ncRNAs), including microRNAs, circular RNAs, and long noncoding RNAs, are important noncoding transcripts that play critical roles in a variety of physiological and pathological processes, especially in cancer. ncRNAs have been verified to be packaged into EVs and transported between LC cells and stromal cells, regulating multiple LC malignant phenotypes, such as proliferation, migration, invasion, epithelial-mesenchymal transition, metastasis, and treatment resistance. Additionally, EVs can be detected in various body fluids and are associated with the stage, grade, and metastasis of LC. Herein, we summarize the biological characteristics and functions of EV ncRNAs in the biological processes of LC, focusing on their potential to serve as diagnostic and prognostic biomarkers of LC as well as their probable role in the clinical treatment of LC. EV ncRNAs provide a new perspective for understanding the mechanism underlying LC pathogenesis and development, which might benefit numerous LC patients in the future.
Keywords: extracellular vesicle, noncoding RNA, lung cancer progression, prospective biomarkers, clinical values
Graphical Abstract
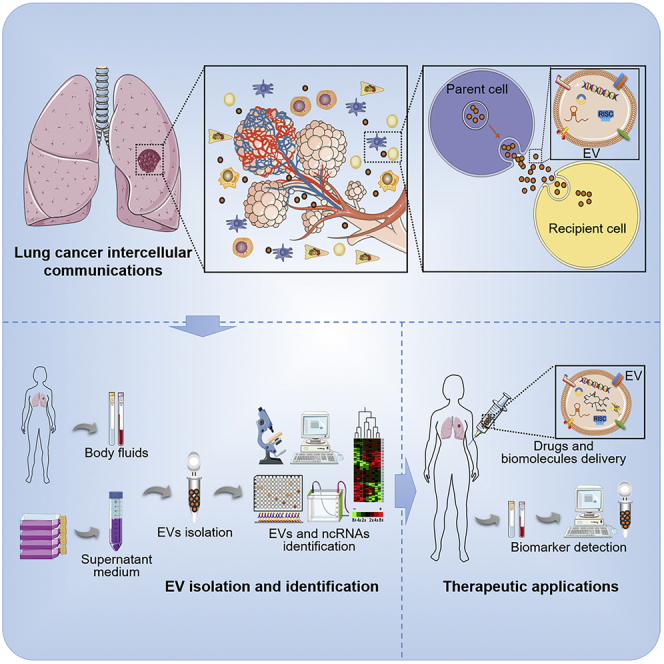
Shan et al. summarize the current knowledge of the biogenesis and functions of extracellular vesicle (EV) noncoding RNAs (ncRNAs) in lung cancer biological processes. The crucial roles of EV ncRNAs provide a novel insight for lung cancer pathogenesis and development, suggesting noninvasive biomarkers and potential targets for anti-lung cancer therapy.
Introduction
Lung cancer (LC), a commonly diagnosed and lethal cancer, accounts for 11.4% of the total cases and 18% of the total cancer-related deaths worldwide.1 LC is a highly aggressive, rapidly metastatic, and highly heterogeneous cancer that can occur in many different parts of the bronchial tree, therefore exhibiting highly different symptoms and signs.2 Even worse, LC metastasizes via blood vessels as well as the lymphatic system, which usually results in an increased rate of recurrence and distant metastasis, as well as shortened survival of patients.3 Despite advances in treatments such as surgery, radiotherapy, chemotherapy, and targeted therapy, 70% of LC patients are already at an advanced stage when diagnosed.2 The 5-year survival of patients with LC remains poor.4 The poor prognosis of LC patients also stems from insufficient understanding of its molecular pathogenesis, lack of a timely diagnosis, and a lack of sensitive monitoring tools. Therefore, it is crucial to clarify the molecular mechanism of LC occurrence and development and to identify reliable biomarkers.
The tumor microenvironment (TME) has been considered to be associated with cancer progression and high mortality.5,6 The LC TME relies on complex interactions between tumor cells and adjacent noncancerous cells, such as fibroblasts, endothelial cells, and immune cells.7 Cancer cells can directly interact with neighboring cells through membrane receptors and their ligands or interact with distant cells by releasing cytokines, chemokines, and metabolites into the circulatory system.8 In recent decades, extracellular vesicles (EVs) have been considered a new type of intercellular communication and have received much attention.9,10 EVs can realize the molecular exchange between cancer cells and stromal cells, reshape the local TME, and stimulate the occurrence, development, and distant metastasis of cancer.11, 12, 13 Among the EV cargoes, noncoding RNAs (ncRNAs), such as microRNAs (miRNAs), circular RNAs (circRNAs), and long noncoding RNAs (lncRNAs), have become crucial regulators due to their roles in the initiation and progression of various cancers.14,15 In this review, we summarize the functions of EV ncRNAs in LC, with a focus on their potential as biomarkers and therapeutic targets for LC.
Biogenesis and characteristics of EVs
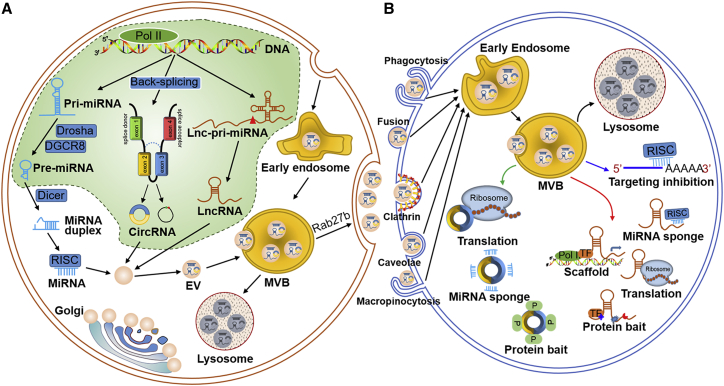
EVs, with a diameter of 30–150 nm, are small EVs secreted by cells that exist in almost all body fluids, such as the blood, cerebrospinal fluid, saliva, urine, breast milk, amniotic fluid, and ascites.16, 17, 18 The biosynthesis of EVs begins with early endosomes formed by endocytosis of the plasma membrane (Figure 1A). Then, the early endosomes become late endosomes, which then mature to form multivesicular bodies (MVBs) that package EVs and their cargoes. Finally, the MVBs can be phagocytosed and degraded by lysosomes or released outside the cell.19
Figure 1.
Biology of EV ncRNAs
(A) The biogenesis of ncRNAs and EVs. miRNA genes are transcribed into pri-miRNAs by RNA polymerase II (Pol II), and further cleaved by Drosha and DGCR8 to generate pre-miRNAs. After exporting into cytoplasm, pre-miRNAs are cleaved by Dicer to produce a miRNA duplex. Then the miRNA duplex loads on AGO. One strand of the duplex is selectively anchored into the AGO to form the RISC complex, thereby regulating the expression of target mRNA. circRNAs get their closed loop structures through back-splicing in nucleus. circRNAs are composed of extrons and/or introns depending on the methods of lariat-driven circularization, intron-pairing-driven circularization, and intron cyclization. lncRNA genes are transcribed through the mediation of Pol I/II to form lnc-pri-miRNAs, and are processed by RNase P/H to generate mature lncRNAs and pre-miRNAs. Mature ncRNAs are then sorted into MVBs. The MVBs are finally released as EVs in a Rab27b-dependent manner, or transported into lysosomes for degradation. (B) EVs can be internalized by recipient cells in different ways, such as phagocytosis, membrane fusion, clathrin-dependent endocytosis, caveolae-mediated endocytosis, and micropinocytosis. Once entering the recipient cells, EV ncRNAs exert their biological functions by acting as miRNA sponges, protein baits, protein scaffolds, and encoding proteins.
Accumulating evidence has revealed that both the endosomal sorting complex required for transport (ESCRT)-dependent and ESCRT-independent pathways are involved in the process of membrane shaping and scission of EVs, depending on the types of EV cargoes and the cellular context, and can be affected by signals and stimuli that the cells received. The ESCRT machinery consists of four subcomplexes as ESCRT-0, Ⅰ, Ⅱ, and Ⅲ.20 It was reported that these subcomplexes act in a stepwise manner. ESCRT-0 assembles on membranes and is responsible for capturing polyubiquitylated cargoes. Then ESCRT-0 recruits ESCRT-Ⅰ and hands over ubiquitylated cargoes. ESCRT-Ⅰ recruits ESCRT-Ⅱ and co-mediates endosome membrane invagination, into which cargoes are sorted. ESCRT-Ⅲ takes charge of fission and scission of vesicles. Moreover, the ESCRT-dependent pathway could intersect with the syndecan-syntenin-ALIX pathway, which supports cargo sequestration, the budding process, and the biogenesis of EVs.21 However, EVs could still form once the ESCRTs were exhausted, suggesting the existence of an ESCRT-independent pathway.22 Neural sphingomyelinase 2 (nSMase 2) and proteins of the tetraspanin family (like CD63, CD81, CD82, and CD9) are involved in the ESCRT-independent EV cargo sorting processes.23, 24, 25, 26 In contrast, EV cargoes' sortation could also modulate EV biogenesis. The abnormal expression of certain cargoes, such as the major histocompatibility complex (MHC) class Ⅱ, might recruit sorting machineries, thereby promoting MVB formation and the subsequent secretion of EVs.26
During EV biogenesis, multiple biomolecules, such as proteins, nucleic acids, and lipids, are packaged into EVs. Among the numerous EV cargoes, the sorting mechanism of EV miRNAs is relatively well studied. Increasing evidence has confirmed that miRNAs can be selectively sorted into EVs rather than a passive process. According to the current knowledge, there are four mechanistic hypotheses for the selective sorting of miRNAs into EVs. (1) The nSMase 2-dependent pathway. nSMase 2 is a rate-limiting enzyme in ceramide biosynthesis, while ceramide is involved in EV biogenesis.27 nSMase 2 is the first regulatory proteins that is related to EV miRNAs secretion.28 Kosaka et al.27 found that the inhibition of nSMase 2 reduces the sorting of miR-16 and miR-146a into EVs, while nSMase 2 overexpression enhanced EV miR-16 and miR-146a levels with no changes in cellular miRNAs. (2) The heterogeneous nuclear ribonucleoproteins (hnRNPs) family-dependent pathway. The hnRNPA2B1 protein was found to be sumoylated and to bind with miR-198 and miR-601 through their GGAG/UGCA motifs, thereby controlling their loading into EVs.29 In separate research, Lee et al.30,31 demonstrated that hnRNPA2B1 mediates the sorting into EVs of miR-17 and miR-93, which possess AGG/UAG motifs. HnRNPA1 is another member of the hnRNP family, which could be sumoylated and regulate the loading process of miR-198 into EVs.32 In addition, hnRNP-Q was confirmed to bind with the GGCU sequences in miR-3470a and miR-194-2-3p, which are highly enriched in EVs.33 The N-terminal unit for the RNA recognition domain of hnRNP-Q is critical for EV loading.34 (3) The 3′ end of miRNA-dependent pathway. A study by Koppers-Lalic et al.35 revealed that post-transcriptional modifications, especially 3′ end adenylation and uridylation of miRNAs, could affect miRNA sorting into EVs. Notably, 3′-end-adenylated miRNAs tended to be enriched in cells, whereas 3′-end-uridylated forms were more likely to be detected in EVs isolated from human urine samples. (4) The RNA-induced silencing complex (RISC)-dependent pathway. The RISC, which consists of miRNA, Argonaute protein (AGO), and GW182, is responsible for the translational repression of the mRNAs.36 It was demonstrated that the core components of RISC were co-localized with MVB proteins.37 Meanwhile, blocking lysosomal degradation of MVB results in RISC accumulation.38 Besides, the expression level of the target gene can also affect the number of miRNAs sorted into EVs.39 In sum, specific miRNA sequences may guide their sorting into EVs, and certain RBPs also participate in the regulation of EV miRNA sorting.
Once the MVBs move to and fuse with the plasma membrane, the internal vesicles are then released to the extracellular substance as EVs. Small GTPases of the Ral and Rab families are crucial factors in EV secretion. Rab27a was found to affect the size of MVBs, while Rab27b affects the distribution of MVBs in cells.40 After secretion, EVs participate in the regulation of cell behavior and intercellular communication through various mechanisms. On the one hand, the EV cargoes directly interact with the surface molecules of the recipient cell to achieve signal transduction through ligand-receptor interactions.41 On the other hand, EV cargoes can fuse with the plasma membrane and directly enter the recipient cell (Figure 1B).42
Several decades ago, Johnstone et al.43,44 considered EVs as a mechanism of shedding surface membrane components, and the formation of EVs might be a major route for externalization of superfluous membrane proteins during reticulocyte maturation. Currently, extensive research has found that EVs are crucial regulators of various physiological and pathological processes, including cancer.45 EVs might affect cancer initiation and progression in various ways. Cancer-derived EVs can be internalized by stromal cells, thus creating a suitable environment for cancer occurrence and development.46 Meanwhile, EVs secreted by cancer-associated fibroblasts (CAFs) contain complete amino acids and lipids, which can meet the requirements of tumor growth under conditions of nutrient stress.47 Increasing evidence shows that EVs play a role in regulating tumorigenesis by regulating angiogenesis, immunity, metastasis, and treatment resistance.48 The potential of tumor EVs as biomarkers and novel anti-tumor targets has attracted great attention.
Characteristics and biological functions of EV ncRNAs
Various kinds of biomacromolecules, such as proteins, lipids, and nucleic acids, have been assessed in EVs where they can exist stably and exert their important biological functions.49 In recent years, the identification of EV ncRNAs has substantially advanced due to high-throughput sequencing and other technologies used to detect low-abundance RNA samples. These ncRNA populations include miRNAs, circRNAs, lncRNAs, transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), Y RNAs, and piwi-interacting RNAs (piRNAs), which can be transferred from parent cells to recipient cells or communicate in the TME, thus regulating various normal physiological and pathological processes, especially in cancer.50, 51, 52, 53, 54 Herein, we focus on the characteristics and biological roles of EV ncRNAs (Figure 1).
miRNAs in EVs
miRNAs are endogenous small RNAs consisting of approximately 20–24 nucleotides that recognize target genes through complementary base pairing to mRNA 3′ untranslated regions (UTRs), thus disrupting their stability and reducing the expression of those genes.55 Mechanistically, the generation of miRNAs starts from the formation of pri-miRNAs in the nucleus, and they are further cleaved by Drosha and DGCR8 to generate pre-miRNAs. Then, the RNase III enzyme Dicer cleaves the terminal loop of the pre-miRNA to generate a 20–22-nucleotide miRNA duplex, which is then loaded on AGO. After that, the passenger strand is degraded, while the mature miRNA is loaded into AGO to form the RISC, and guides RISC to the target mRNA to perform post-transcriptional silencing functions. The miRNA may originate from the 5′ side of the pre-miRNA (referred to as the 5p strand) or the 3′ side of the pre-miRNA (referred to as the 3p strand).56 Usually only one miRNA strand dominates. However, in some cases, both strands of the duplex generate the functional mature miRNA to regulate gene expression (Figure 1A).57,58 To date, approximately 2,000 miRNAs have been identified in the human genome, and the number is still growing.59 In the past few decades, identification of the crucial roles of miRNAs in cancer initiation, progression, and anticancer therapy has gained momentum.60,61 Functionally, miRNAs might be oncomiRNAs as well as tumor-suppressive miRNAs in different cellular contexts, considering that a single miRNA might have several target mRNAs.62
Among various types of ncRNAs, miRNAs were first identified in EVs.63 Peng et al.64 identified 706 miRNAs from non-small cell lung cancer (NSCLC) patients' plasma EVs, among which 155 EV miRNAs were differently expressed between NSCLC patients and healthy persons. In another study, Jin's group65 classified the EV miRNA profiles in early-stage NSCLC patients' plasma using next-generation sequencing, and identified several differently expressed EV miRNAs as highly sensitive, noninvasive biomarkers for early NSCLC diagnosis. Accumulating evidence has verified that EV miRNAs regulate numerous cancer-related life events through the participation of cell-cell communication.50 Importantly, the high abundance of EVs in body fluids, as well as the relevance of miRNAs to cancer progression and prognosis, have made EV miRNAs potential biomarkers and ideal therapeutic targets for cancer.66
circRNAs in EVs
circRNAs are new types of ncRNAs with covalently linked circular structures.62,67 In contrast to linear RNAs, circRNAs are thought to be the result of exon-skipping events through mechanisms of intron-pairing-driven circularization and lariat-driven circularization (Figure 1A).68,69 Existing studies have described the characteristics of circRNAs: diversity and high abundance in various tissues, stability and conservation, and tissue and spatial-temporal specificity.62 circRNAs might possess various functions in vital pathophysiological processes, such as serving as miRNA sponges, binding to functional proteins, encoding proteins, regulating pre-RNA splicing, and managing parental gene expression. Importantly, increasing evidence has verified that circRNAs participate in the occurrence and development of many kinds of diseases, especially cancer.70
EV circRNAs were first identified in 2015, and related studies have flourished since then.71 Yang et al.72 investigated the expression profile of EV circRNAs in NSCLC patients' serum by using a high-throughput circRNA microarray, and found that circRNA_102481 is highly expressed in serum EVs of EGFR-TKI-resistant patients. Additionally, Wang and colleagues73 analyzed the circRNAs expression profile in plasma EVs of lung squamous cell carcinoma patients, and 252 differently expressed EV circRNAs were identified, among which hsa_circ_0014235 and hsa_circ_0025580 were significantly upregulated. Compared with producer cells, circRNAs are more stable and enriched in EVs, and can be transferred into recipient cells and the TME, thus regulating the behavior of recipient cells and promoting cancer progression (Figure 1B).45 Specifically, EVs circRNAs have been recognized as crucial modulators of numerous physiological and pathological processes, and might become readily noninvasive biomarkers for the prediction, diagnosis, and prognostic evaluation of diseases, especially cancers.
lncRNAs in EVs
lncRNAs are a class of endogenous ncRNAs >200 nucleotides in length that were identified approximately three decades ago.74 In mammalian genomes, 4%–9% of sequences produce transcripts that are lncRNAs (1%–2% for mRNAs).75 With the development of high-throughput technologies, the number of lncRNAs in humans has been certified to be more than 160,000.76 Increasing evidence has suggested that lncRNAs are not transcriptional “noise” but instead have various physiological functions, such as X chromosome inactivation, genomic imprinting, chromatin modification, transcriptional regulation, mRNA turnover, and protein translation.77,78 Importantly, lncRNAs are abundantly expressed and are widely relevant to various kinds of cancers, and their aberrant expression and mutations are associated with tumorigenesis, tumor progression, and metastasis.79, 80, 81
Likewise, lncRNAs can be sorted into EVs and act as mediators in cell-cell communication (Figure 1).82 Recently, EV lncRNAs have been reported to be involved in the regulation of proliferation, angiogenesis, metastasis, and drug resistance in LC.83, 84, 85 Moreover, the dysregulation of EV lncRNAs has been found to affect cellular behavior and cell-cell communication in the LC TME.86 Furthermore, EV lncRNAs have been demonstrated to be closely related to clinicopathological characteristics and treatment outcomes, indicating the possibility of EV lncRNAs serving as diagnostic and prognostic biomarkers for LC patients.87,88 lncRNAs have gained increasing attention in EV investigations.
Other ncRNAs in EVs
tRNAs, rRNAs, Y RNAs, and piRNAs are also members of the ncRNA family. Increasing evidence shows that these ncRNAs can be encapsulated into EVs, and they participate in the initiation and development of many diseases, including cancer.
tRNAs are small nucleic molecules consisting of 73–95 nucleotides characterized by their evolutionarily conserved structures terminate with the CCA trinucleotide at the 3′ end and the three hairpins.89,90 The main and well-known function of tRNAs is carrying amino acids to the ribosome for protein synthesis. In recent years, tRNAs have been thought to act as primers for reverse transcription in viruses, inhibit protein translation, or regulate cell proliferation and DNA damage.91, 92, 93 Likewise, full-length mature tRNAs and tRNA-derived small RNAs (tsRNAs) have been identified in EVs. Moreover, the abundance of tRNAs in EVs varies in different cell types. Approximately 50% of total small RNAs in adipose mesenchymal stem cell-derived EVs are tRNAs, while in bone marrow mesenchymal stem cell (BMSC)-derived EVs, only 23%–35% are tRNAs.94 Surprisingly, in the newest study by Zhu's group,53 they demonstrated the ubiquitous presence of tsRNAs in EVs extract from liver cancer cell culture medium and liver patients' plasma. Compared with healthy controls, tRNA-ValTAC-3, tRNA-GlyTCC-5, tRNA-ValAAC-5, and tRNA-GluCTC-5 were verified to be upregulated in liver cancer patients' plasma EVs, suggesting the biomarker potential of EV tsRNAs in liver cancer. The clinical application value of EV tRNAs in cancer is worthy of further study.
rRNA is the major component of the ribosomes and is answerable for peptide bond formation during protein biosynthesis.95,96 rRNAs are single stranded, but a wide double-stranded region is formed by spatial folding.97 In recent years, research has suggested that the processing of rRNA is the prominent function of nuclear RNA EVs.98 Schuller et al.99 investigated the structure of the ribosomal subunit within RNA EVs, and found that the rRNA spacer sequences internal transcribed spacer 2 is removed from rRNA precursors by a complex of 10 proteins in EVs, thereby forming the matured 60S rRNA. Specifically, several rRNAs have been detected in EVs of cancer cells. A high-throughput sequencing of breast cancer cells revealed that about 80% of EV RNA sequencing (RNA-seq) reads match to rRNAs.100 In another separate study, Fiskaa et al.101 found that about 43% of breast cancer cell EV-derived small RNA reads map to rRNAs, and present a high abundance of 5′ 28S rRNAs in EVs compared with parent cells. In contrast, endothelial and immune cell-derived EVs did not include these 5′ 28S rRNA fragments. In total, these results suggest that rRNAs are specifically secreted into EVs under pathological conditions, and might be associated with cancer progression.
Y RNAs are a class of transcripts by RNA polymerase III (Pol III) that consist of 83–112 nucleotides.102 The biological function of Y RNAs is not clear, although several hypotheses are proposed. Ro60 is an RNA-binding protein that binds with misfolded and aberrant noncoding RNAs and channels them into a decay pathway for unfolding and degradation.103 Specifically, since Y RNA and rRNA possess overlapped binding sites to Ro60, a possible function of Y RNAs is to spatially prevent misfolded rRNAs from entering the Ro60 active pocket, thus participating in rRNA quality control.103,104 Existing data from bacterial and animal systems support the function of Ro60-Y RNA interaction in reducing cellular stress for survival.105 Moreover, Y RNAs and Y RNA-derived fragments have been verified to play a role in carcinogenesis due to their effects in regulating cell proliferation and inflammation, and have shown potential as diagnostic and prognostic biomarkers in cancer.106 In addition, deep RNA sequencing demonstrated that Y RNA is a highly abundant ncRNA species enriched in EVs in cancer. In chronic lymphocytic leukemia (CLL), Y RNAs are identified as the most abundant significantly enriched RNA in EVs from patients' plasma, which is notably different from the cellular Y RNA distribution.107 Transference of CLL-derived EV Y RNA conferred monocytes CLL-associated phenotypes, suggesting the establishment of tumor-supportive microenvironment in CLL. Likewise, Y RNA fragments were found to be massively packaged into the EVs of anaplastic large cell lymphoma patients that are more aggressive and advanced.108 Y RNA fragments were also identified in EVs secreted by melanoma cells.109 Research on the correlation between EV Y RNA and cancer progression may provide clues for the development of new anticancer strategies.
piRNAs constitute a type of small ncRNAs containing 24–31 nucleotides with a 10th adenosine or 5′-terminal uridine bias, lacking characteristic secondary structures.110 The biosynthesis of piRNA is to first produce piRNA intermediates from the piRNA cluster, and then generate mature piRNA through the cleavage of Zucchini riboendonuclease.111 Importantly, piRNAs interact with piwi protein to form a piRNA/piwi complex, which participates in the regulation of spermiogenesis, germline stem cell maintenance, genomic rearrangement, transposon silencing, and epigenetic modification.112 So far, piRNAs have been identified in germinal tissues as well as somatic tissues, such as brain tissue, heart tissue, and plasma.113,114 Several studies have dissected EV piRNA profiling in a number of diseases, such as heart failure, Alzheimer disease, and cancers, which contribute to the distinguishment of health volunteers from patients, revealing the potential of EV piRNAs as diagnostic biomarkers.54,115,116 Dysregulation of piRNAs and piwi proteins is also found in cancers, and some of them are involved in tumorigenesis, cancer diagnosis, and prognosis.112,117 Recently, piRNAs were identified in EVs of human semen, plasma, and cultured human cancer cell lines.54,116,118 It has been confirmed that piRNAs could be delivered by EVs and affect the behavior of recipient cells. For example, De Luca et al.110 reported that umbilical cord blood CD34+ stem (UCB-CD34+) cells become less differentiated and more viable after exposure to BMSC-derived EVs. Further analysis revealed that it is the miRNAs and piRNAs in EVs that regulate the differentiation and apoptosis of UCB-CD34+ cells. However, functions of piRNAs in EVs have not been thoroughly examined. Thus, further experimental and clinical studies are required.
The involvement of EV ncRNAs in LC biology and therapy
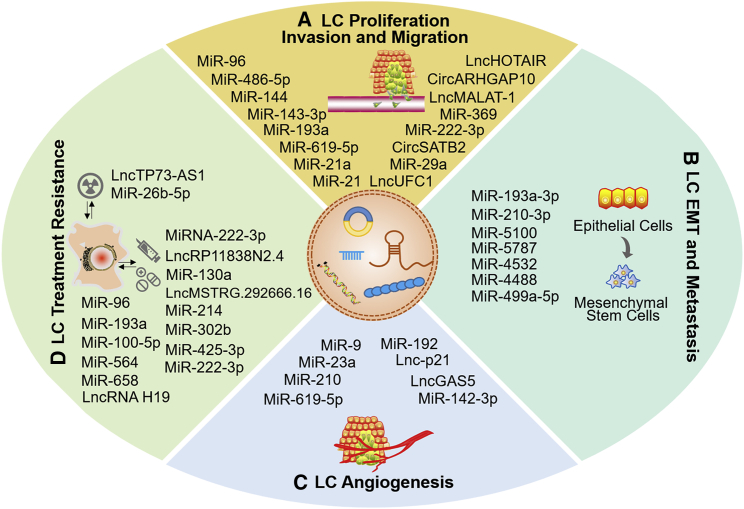
LC is the most common and deadly cancer, with the characteristics of rapid proliferation, high malignancy, late detection, aggressive metastasis, strong drug resistance, and a poor prognosis.119 It is urgent to clarify the underlying mechanism of LC occurrence and development and to bring new insight to clinical therapy for LC. Recently, a growing number of studies have focused on EV cargoes and their usage in LC genesis and progression.120 In this section, we pay attention to the roles of EV ncRNAs in LC proliferation and survival, angiogenesis, invasion, metastasis, epithelial-mesenchymal transition (EMT), and therapy resistance (Table 1; Figure 2).
Table 1.
The biological functions of EV ncRNAs in LC
| ncRNAs | Dysregulation | Target | Experiment type (in vitro/in vivo) | Biological functions | Reference |
|---|---|---|---|---|---|
| miR-9 | – | SOCS5 | in vitro (NSCLC cells→ endothelial cells) | promote endothelial cell migration and angiogenesis | Zhuang et al.121 |
| miR-21 | upregulated | PDCD4 | in vitro (LAC cells→ osteoclasts) | promote tumor osteolytic metastasis | Xu et al.122 |
| miR-21/miR-29a | upregulated | TLR8 | in vitro (LLCs→ macrophages); in vivo (mouse LC model, intravenously injected with LLC-secreted EVs) | trigger pre-metastatic inflammatory response | Fabbri et al.123 |
| miR-21a | upregulated | PDCD4 | in vitro (LLCs→ MDSCs); in vivo (mouse LC model, intravenously injected with LLC-secreted EVs) | promote MDSCs expansion and accumulation, promote cancer cell growth | Zhang et al.124 |
| miR-23a | upregulated | ZO-1 | in vitro (hypoxic LC cells → endothelial cells); in vivo (human, serum EVs) | promote angiogenesis, increase migration | Hsu et al.125 |
| miR-31-5p | upregulated | SATB2 | in vitro (hypoxic LC cells → endothelial cells); in vivo (human, plasma EVs) | promote migration, invasion, and metastasis | Yu et al.126 |
| miR-96 | upregulated | LMO7 | in vitro (NSCLC cells → NSCLC cells); in vivo (human, serum EVs) | promote cell proliferation, migration, and DDP resistance | Wu et al.127 |
| miR-100-5p | Downregulated | – | in vitro (A549/DDP cells → A549 cells); in vivo (mouse LC model, intratumorally injected with A549/DDP cell-secreted EVs) | alter other LC cells' sensitivity to DDP | Qin et al.128 |
| miR-103a | upregulated | PTEN | in vitro (hypoxic LC cells → M2 macrophages) | increase M2 macrophage polarization | Hsu et al.129 |
| miR-130a | – | PUM2 | in vitro (CAFs → DDP-sensitive NSCLC cells) | promote survival of DDP-sensitive NSCLC cells | Zhang et al.130 |
| miR-142-3p | upregulated | TGFbetaR1 | in vitro (LAC cells → endothelial and fibroblast cells) | promote angiogenesis and cancer-associated fibroblast phenotype | Lawson et al.131 |
| miR-143-3p | upregulated | ITM2B | in vitro (MDSCs →LC cells); in vivo (mouse LC model, intratumorally injected with MDSC-secreted EVs) | promote proliferation | Zhou et al.132 |
| miR-144 | Downregulated | CCNE1/2 | in vitro (BMSCs →NSCLC cells); in vivo (mouse LC model, intravenously injected with BMSC-secreted EVs) | inhibit proliferation | Liang et al.133 |
| miR-192 | Downregulated | – | in vitro (NSCLC cells → osteoclasts); in vivo (mouse LC model, intravenously injected with NSCLC cell-secreted EVs) | inhibit angiogenesis and osteoclastogenesis | Valencia et al.134 |
| miR-193a | – | LRRC1 | in vitro (BMSCs → NSCLC cells); in vivo (mouse LC model, intravenously injected with BMSC-secreted EVs) | promote proliferation, migration, invasion, and DDP-resistant cell apoptosis | Wu et al.135 |
| miR-193a-3p, miR-210-3p, and miR-5100 | upregulated | – | in vitro (hypoxic BMSCs → Epithelial cancer cells); in vivo (human, plasma EVs; mouse LC model, intravenously injected with hypoxic BMSC-secreted EVs) | promote invasion and EMT | Zhang et al.136 |
| miR-210 | upregulated | TET2 | in vitro (LC cells →fibroblasts); in vivo (human, plasma EVs; mouse angiogenesis model, fibroblasts mixed with LC cell-secreted EVs) | promote fibroblasts transfer into CAFs and angiogenesis | Fan et al.137 |
| miR-222-3p | – | SOCS3 | in vitro (A549/GR cells → A549 cells); in vivo (human, plasma EVs; mouse LC model, injected with A549/GR cell-secreted EVs) | promote proliferation, migration, invasion, and gemcitabine resistance | Wei et al.138 |
| miR-214 | upregulated | – | in vitro (PC-9/GR cells → PC-9 cells); in vivo (mouse LC model, hypodermic injected with PC-9/GR cell-secreted EVs) | promote gefitinib resistance | Zhang et al.139 |
| miR-302b | upregulated | TGFbetaRII | in vitro (lowly metastatic LC cells → highly metastatic LAC cells) | suppress LC cell proliferation and migration | Li et al.140 |
| miR-369 | upregulated | NF1 | in vitro (CAFs → LAC cells); in vivo (mouse LC model, hypodermic injected with CAF-secreted EVs) | promote proliferation, migration, and invasion | Guo et al.141 |
| miR-425-3p | upregulated | AKT | in vitro (DDP-resistant NSCLC cells → DDP-sensitive NSCLC cells); in vivo (human, serum EVs) | facilitated autophagic activation and chemoresistance | Ma et al.142 |
| miR-486-5p | Downregulated | NEK2 | in vitro (NSCLC cells → NSCLC cells); in vivo (human, serum EVs) | induce cell-cycle arrest, inhibit cell proliferation, and metastasis | Hu et al.143 |
| miR-499a-5p | upregulated | mTOR | in vitro (highly metastatic LAC cells → LAC cells) | promote cell proliferation, migration, and EMT | He et al.144 |
| miR-564 and miR-658 | upregulated | – | in vitro (PC-9/gefitinib cells → PC-9 cells) | induce drug resistance in sensitive cells | Azuma et al.145 |
| miR-619-5p | upregulated | RCAN1.4 | in vitro (NSCLC cells → endothelial cells); in vivo (human, plasma EVs) | promote proliferation, metastasis, and angiogenesis | Kim et al.146 |
| miR-5787, miR-4532, and miR-4488 | upregulated | – | in vitro (mesenchymal cells → epithelial cells) | promote migration, invasion, and EMT | Tang et al.147 |
| circSATB2 | upregulated | miR-326 | in vitro (NSCLC cells → NSCLC cells); in vivo (human, serum EVs) | promote cell proliferation, migration, and invasion | Zhang et al.148 |
| circARHGAP10 | upregulated | miR-638 | in vitro (NSCLC cells → NSCLC cells); in vivo (human, serum EVs) | promote proliferation, migration, invasion, and glycolysis | Fang et al.149 |
| circUSP7 | upregulated | miR-934 | in vitro (NSCLC cells → CD8+ T cells); in vivo (human, plasma EVs) | inhibits CD8+ T cell function, promote resistance to immunotherapy | Chen et al.150 |
| lncHOTAIR | upregulated | miR-203 | in vitro (NSCLC cells → NSCLC cells); in vivo (human, serum EVs) | promote proliferation, migration, and invasion | Zhang et al.83 |
| lncMALAT-1 | upregulated | – | in vitro (NSCLC cells → NSCLC cells); in vivo (human, serum EVs) | promote cell growth and migration, inhibit apoptosis | Zhang et al.151 |
| lncUFC1 | upregulated | EZH2 | in vitro (NSCLC cells → NSCLC cells); in vivo (human, serum EVs) | promote proliferation, migration, and invasion | Zang et al.152 |
| lncRNA-p21 | upregulated | – | in vitro (LC cells → HUVECs); in vivo (human, plasma EVs) | promote tube formation and enhance tumor cell adhesion to endothelial cells | Castellano et al.153 |
| lncGAS5 | Downregulated | miR-29-3p | in vitro (LC cells →HUVECs); in vivo (mouse LC model, serum EVs) | promote proliferation and tube formation, inhibit apoptosis | Cheng et al.154 |
| lncAGAP2-AS1 | upregulated | miR-296 | in vitro (M2 macrophage → NSCLC cells) | promote the radiotherapy immunity | Zhang et al.155 |
| lncPCAT-1 | upregulated | miR-182/miR-217 | in vivo (human, serum EVs) | promote pre-metastatic niche formation and tumor metastasis; induce G0/G1 cell-cycle arrest; promote chemoresistance and tumor growth | Domvri et al.156 |
| lncRNA MSTRG.292666.16 | upregulated | – | in vitro (osimertinib-resistant cells → osimertinib-sensitive cells); in vivo (human, plasma EVs) | associated with asimertinib resistance | Deng et al.86 |
| lncRNA RP11838N2.4 | upregulated | – | in vitro (erlotinib-resistant cells → erlotinib-sensitive cells); in vivo (human, serum EVs) | promote erlotinib resistance | Zhang et al.157 |
| lncRNA H19 | upregulated | – | in vitro (gefitinib-resistant NSCLC cells → gefitinib-sensitive NSCLC cells) | promote gefitinib resistance | Lei et al.158 |
NSCLC, non-small cell lung cancer; LAC, lung adenocarcinoma; LLC, Lewis LAC; LC, lung cancer; EV, extracellular vesicle; MDSC, myeloid-derived suppressor cell; DDP, cisplatin; CAF, cancer-associated fibroblast; BMSC, bone marrow mesenchymal stem cell; GR, gemcitabine resistance; EMT, epithelial-mesenchymal transition; HUVEC, human umbilical vein endothelial cell.
Figure 2.
The participation of EV ncRNAs in lung cancer biology
EV ncRNAs play important roles in lung cancer progression, such as cell proliferation, invasion, and migration (A), EMT and metastasis (B), angiogenesis (C), and treatment resistance (D).
EV ncRNAs and cell proliferation, invasion, and migration in LC
Uncontrolled proliferation and enhanced invasion and migration ability are typical malignant phenotypes of LC cells, which ultimately lead to poor prognosis of LC patients. Therefore, discovering the molecular mechanisms and key regulatory factors of LC cellular proliferation, invasion, and migration is vital for understanding the pathogenesis of LC and for selecting appropriate treatment options. Mounting evidence has revealed that EV ncRNAs might be important mediators in the regulation of LC cell proliferation, invasion, and migration (Figure 2A).
MiR-96 is upregulated in various cancers and is regarded as an oncomiRNA.159 In LC-derived EVs, miR-96 is found at significantly higher levels than in the EVs of healthy donors.127 Importantly, EV miR-96 is positively associated with the LC grade. Higher EV miR-96 levels can be detected in LC cells with higher invasiveness. Functionally, miR-96 promotes cell proliferation, migration, and cisplatin (DDP) resistance in A549 cells by directly inhibiting LMO7 expression. In another separate study, Hu and colleagues143 demonstrated that EV miR-486-5p regulates the cell cycle and proliferation by targeting NEK2 in lung adenocarcinoma (LAC). EV miR-486-5p might be a potential biomarker for LAC diagnosis and a target for LAC treatment. Moreover, miR-let-7e was found to be downregulated in NSCLC tissues and serum-derived EVs, which is relevant to the low survival rate of NSCLC patients.160 Functionally, serum EV miR-let-7e suppresses NSCLC cellular viability, invasion, and migration in vitro and in vivo by targeting SUV39H2.
In addition to miRNAs, EV circRNAs are involved in the regulation of proliferation in LC. Zhang et al.148 reported that circSATB2 could be packaged in EVs and promote the proliferation, migration, and invasion of NSCLC cells, as well as induce abnormal proliferation of bronchial epithelial cells. Importantly, circSATB2 is highly expressed in the serum EVs of LC patients compared with healthy donors, suggesting its possible use as a noninvasive diagnostic biomarker of NSCLC. In another study, circARHGAP10 was reported to be upregulated in EVs of NSCLC cells and tissues, as well as in patients' serum-derived EVs.149 Functionally, circARHGAP10 promotes the proliferation, invasion, and migration of NSCLC cells and facilitates glycolysis. MiR-638 is the direct target of circARHGAP10, which inhibits the downstream oncogene FAM83F to repress the progression of NSCLC. The circARHGAP10-miR-638-FAM83F axis might be a potential target for the investigation of novel anticancer strategies in NSCLC.
In addition, some lncRNAs have been clarified to be selectively sorted in EVs and associated with the regulation of proliferation of LC. Zhang et al.83 demonstrated that the expression of lncHOTAIR is increased in the serum EVs of LC patients compared with healthy donors. Additionally, EV lncHOTAIR enhanced the proliferation, migration, and invasion of A549 and H1299 cells, which might be mediated by sponging miR-203. In another separate study, Zhang's group151 identified a highly expressed EV lncRNA, lncMALAT-1, from the serum of LC patients. More importantly, the expression of EV lncMALAT-1 is positively correlated with the tumor stage. Accumulation of serum-derived EV lncMALAT-1 could promote cell growth and migration and prevent apoptosis in LC cell lines. Furthermore, Zang and colleagues152 demonstrated that the expression of lncUFC1 is elevated in tissues, serum, and serum EVs in NSCLC patients. NSCLC cell-derived EV promotes proliferation, migration, and invasion through transferring lncUFC1 in A549 cells. Specifically, lncUFC1 could downregulate PTEN expression and activate the AKT pathway by binding to EZH2, thus promoting the tumorigenesis of NSCLC. In brief, ncRNAs can be selectively packaged into EVs and participate in the regulation of proliferation, invasion, and migration in LC.
EV ncRNAs stimulate EMT and metastasis in LC
EMT is a process by which epithelial cells lose epithelial polarity and cell-cell adhesion to transform into mesenchymal stem cells (MSCs), granting the cells the ability to invade and migrate, therefore participating in multiple physiological and pathological processes, such as individual development, tissue healing, organ fibrosis, and cancer.161 Importantly, cancer cells in the primary site lose their adhesion ability during the EMT process; at the same time, the expression of E-cadherin decreases, the expression of mesenchymal markers and fibronectin rises, and the cell motility and invasion ability are enhanced to invade the extracellular matrix.162 In recent years, a growing body of research has revealed that EVs largely contribute to EMT and the subsequent metastasis in various cancers.163 More importantly, EV cargoes, such as ncRNAs, could be transferred and internalized by certain recipient cells, thus regulating the behavior of these cells and the process of EMT, subsequently affecting cancer development and metastasis (Figure 2B).
Zhang et al.136 reported that BMSC-delivered miR-193a-3p, miR-210-3p, and miR-5100 can be transferred to epithelial cancer cells, promoting the metastasis of LC cells through activating STAT3-dependent EMT. Moreover, Tang's group147 recently found that EV miR-5787, miR-4532, and miR-4488 secreted by MSCs contribute to signal transduction in the EMT process and the stimulation of mesenchymal marker expression in epithelial cells, therefore promoting the metastasis of LC. In another separate study, He and colleagues144 clarified the effect of miR-499a-5p on promoting the migration, EMT, and metastasis of LC cells. MiR-499a-5p-containing EVs derived from highly metastatic cells activate the mammalian target of rapamycin (mTOR) signaling pathway to facilitate cellular proliferation, migration, and EMT, which provides new insight into modulating LC metastasis. In total, these results demonstrated that EV ncRNAs could affect LC progression and metastasis through regulating the EMT process.
EV ncRNAs induce LC angiogenesis
Angiogenesis, namely the formation of new blood vessels from pre-existing vessels, is pivotal for tumor growth and hematogenous metastasis of various solid tumors, including LC.164 Increasing evidence has demonstrated that ncRNAs play important roles in tumor angiogenesis. Vascular endothelial cells capture EVs released by stromal cells, tumor cells, and MSCs, regulating the formation of new blood vessels. Extensive studies have revealed that EV ncRNAs participate in the regulation of tumor angiogenesis (Figure 2C). Zhuang et al.121 reported that miR-9 could be secreted from NSCLC cells via EVs and then be internalized by endothelial cells. MiR-9 reduced SOCS5 levels to activate the JAK-STAT pathway to promote angiogenesis, further promoting tumor cell growth. In addition, miR-23a was demonstrated to be upregulated in EVs in LC under hypoxic conditions.125 EV miR-23a causes the accumulation of HIF-1α in endothelial cells by suppressing PHD1/2, thus leading to neovascularization in LC. Specifically, EV miR-23a inhibits ZO-1 to increase vascular permeability. Therefore, EV miR-23a could be an intercellular communication regulator involved in LC angiogenesis. Other studies have found that CAFs possess the ability to promote tumor angiogenesis. Fan's group137 proved that miR-210 activates the JAK2/STAT3 pathway by targeting TET2 in CAFs. Specifically, EVs containing overexpressed miR-210 could stimulate the transferring of fibroblasts into CAFs, resulting in the promotion of angiogenesis in LC. In another study, miR-619-5p was shown to be upregulated in NSCLC patients' plasma-released EVs, and it has dual functions in tumor progression: promoting angiogenesis in endothelial cells and increasing proliferation and metastasis in NSCLC cells.146
For EV lncRNAs, EV lncRNA-p21 was demonstrated to be upregulated in NSCLC patient plasma.153 Functionally, tumor-derived EVs containing lncRNA-p21 promote tube formation and tumor cell adhesion to endothelial cells by targeting miRNAs related to angiogenesis and metastasis. In addition, Cheng's group154 reported that the low expression of EV lncGAS5 derived from LC tissues is positively correlated with human umbilical vein endothelial cell (HUVEC) proliferation and tube formation. Lentivirus-mediated lncGAS5 overexpression reversed the effects of LC cell-derived EVs on HUVECs. EV lncGAS5 might be a novel target for inhibiting LC angiogenesis. Taken together, these results suggest that ncRNAs selectively sorted in EVs play important roles in regulating LC angiogenesis.
EV ncRNAs remodel the TME of LC
The TME is a complex scaffold that consists of diverse cell populations, structural molecules, and signaling factors that interact with tumor cells and support tumorigenesis and development.165 In the TME, EV ncRNAs can transfer between cancer cells and stromal cells, thus regulating the behaviors of recipient cells and participating in various prominent biological processes involved in the TME, such as immune escape, metabolism alternation, hypoxia, and chronic inflammation.
Organisms are monitored by the immune system, which recognizes and eliminates the incipient cancer cells so as to restrain nascent tumors.166 However, most solid tumors are able to avoid the monitoring and elimination of the immune system, leading to the occurrence and development of cancer. Accumulating evidence has revealed that the secretion of tumor-associated immunosuppressive factors and the development of tumor-antagonizing immune cells could be influenced by EV ncRNAs. For instance, transforming growth factor (TGF) is the immunosuppressive central to the TME.167 In the recent study by Li et al.,140 they reported that miR-302b was significantly upregulated in LC cell-derived EVs. Overexpression of miR-302b inhibits LC cell proliferation and migration accompanied by the decreased expression of TGFβRII. In addition, the dysfunction of tumor-antagonizing immune cells, CD8+ T cells, has been verified in NSCLC. Chen's group150 demonstrated the existence of circUSP7 in LC patients' plasma EVs. CircUSP7 increased SHP2 expression by sponging miR-934, thereby inhibiting IFN-γ, TNF-α, granzyme-B, and Perforin secretion by CD8+ T cells. Moreover, Domvri and colleagues156 verified that EV lncPCAT-1 was highly expressed KRAS-mutant LC patients' serum. LncPCAT-1 increased the expression of miR-182/miR-217, thus promoting pre-metastatic niche formation and LC immunosuppressive metastasis. LncPCAT-1 could also regulate fibroblast differentiation and CAF-mediated stromal activation, thereby affecting chemoresistance and tumor growth.
Cancer cells undergo dramatic changes in the metabolism of glucose, amino acids, and fatty acids to survive in the stressful TME.168 Aerobic glycolysis, also known as Warburg effect, is the major way through which cancer cells get energy and generate biomolecules under diminished nutrient supply conditions.169 A growing body of evidence has demonstrated that EV ncRNAs could modulate the glycolysis metabolism of LC. For example, EV circRNAs, such as circ-MEMO1, circ_0008928, hsa_circRNA_0002130, and circARHGAP10 were found to facilitate glycolysis metabolism and pathological process of NSCLC through acting as miRNA sponges, which providing a perspective for further understanding the pathogenesis of NSCLC.149,170, 171, 172
As the rapid and uncontrolled proliferation of cancer cells exceeds the oxygen supply, hypoxia is one of the characteristics of the TME in almost all solid tumors.173 Cancer cells, once they acclimate to the hypoxic conditions, lead to changes in gene expression and proteomics, resulting in a more aggressive and therapy-resistant phenotype. It was demonstrated that hypoxia enhanced the release of EVs into the TME.174 EV ncRNAs derived from hypoxic LC cells or stromal cells participate in the regulation of biological processes such as the growth of new vasculatures, EMT, and metastasis, so as to guarantee the survival of cancer cells and promote LC development. MiR-193a-3p, miR-210-3p, and miR-5100 carried by hypoxic BMSCs EVs were confirmed to promote LC metastasis by directly targeting STAT3 in vitro and in vivo.136 Especially, EV miR-193a-3p was able to distinguish LC patients from healthy controls, or metastatic LC patients from non-metastatic LC patients, revealing the diagnostic accuracy of EV miR-193a. In other research, hypoxic LC cell-secreted EV miR-23a, miR-103a, and miR-31-5p were reported to promote angiogenesis and affect LC malignant phenotypes, respectively.125,126,129
Chronic inflammation is one of the characteristics of cancer.166 Inflammatory cells and inflammatory factors within the TME promote the activation of inflammatory pathways, thereby stimulating the malignant transformation, and the occurrence and development of cancer. Emerging evidence suggests that EV ncRNAs could target and modulate the key components and processes of chronic inflammation. Macrophages are the most studied inflammatory cells in LC and participate in inflammation by secreting inflammatory factors and chemokines, thereby regulating LC progression.175 It was demonstrated that M2 macrophage-derived EV ncRNAs, such as miR-501-3p, miR-942, miR-155, miR-196-5p, and lncAGAP2-AS1, were able to promote LC cell growth, migration, invasion, EMT, and angiogenesis.155,176, 177, 178 Moreover, inflammation-related signal pathways in LC could be affected by EV ncRNAs. For example, Ni et al.85 found that LC cell-derived EV lncRNA-SOX2OT promotes cell invasion, migration, and bone metastasis by targeting the TGF-β/pTHrP/RANKL pathway. In the studies by Fan's and Du's groups, EV miR-3473b and miR-210 were verified to promote intrapulmonary colonization and angiogenesis by modulating nuclear factor (NF)-κB and STAT3 signaling, respectively.137,179
In general, the TME is a complex environment composed of a variety of cellular and acellular components. EV ncRNAs can be secreted and transmitted between donor cells and recipient cells and regulate the behavior of recipient cells in the TME, thereby affecting the occurrence, development, and metastasis of LC.
EV ncRNAs interfere with LC therapy
Approximately 60% of LC patients need radiotherapy during the course of their cancer treatment. EV ncRNAs interfere with the resistance and sensitivity of LC cells to radiotherapy, as well as the effect of treatment (Figure 2D). Zheng's group180 reported that EV miR-96 is highly expressed in NSCLC patients' plasma. Especially, EV miR-96 was correlated with radioresistance, vascular invasion, and poor overall survival (OS) rate in LC, suggesting the potential of EV miR-96 as a diagnostic and prognostic biomarker for LC radioresistance. In addition, Han et al.181 demonstrated that LC cell-derived EVs containing miR-26b-5p could be internalized by radioresistant LC cells, promoting DNA damage, apoptosis, and radiosensitivity by inhibiting ATF2. Specifically, miR-26b-5p can be detected in serum EVs, revealing the potential of EV miR-26b-5p as a noninvasive biomarker for LC radiotherapy monitoring.
Chemotherapy is one of the most important approaches in LC therapy. However, chemotherapy resistance often appears in clinical practice and becomes an insurmountable obstacle in the treatment of LC. Therefore, an in-depth study of the molecular mechanism of chemotherapy resistance is crucial to find more effective treatment strategies for LC. Recent studies have shown that EV ncRNAs secreted by stromal cells regulate chemotherapy resistance in LC (Figure 2D). Zhang et al.130 found that NSCLC-derived CAFs are innately resistant to DDP. EV miR-130a transferred from CAFs significantly improves the survival rate of NSCLC cells during DDP treatment. Furthermore, Wu's group135 demonstrated that human BMSC-derived EVs that are transfected with miR-193a suppress cellular viability, invasion, and migration and promote the apoptosis of DDP-resistant NSCLC cells by targeting and downregulating LRRC1. These results suggest that stromal cell-derived EV ncRNAs might be potential therapeutic targets for DDP resistance in NSCLC.
Importantly, EVs originated from drug-resistant LC cells can be transferred to and internalized by chemotherapy-sensitive LC cells, thus spreading drug resistance, revealing the potential participation of EV ncRNAs in these processes. For instance, gefitinib-resistant LC cell-derived EVs containing miR-214, miR-302b, miR-564, and miR-658 could confer gefitinib resistance to normal LC cells.139,140,145 Moreover, DDP-resistant LC cell-secreted EVs carrying miR-425-3p and miR-100-5p could be internalized by DDP-sensitive LC cells, providing chemotherapy resistance to recipient cells in c-myc/AKT1- and mTOR-dependent manners, respectively128,142. Furthermore, there are other EV ncRNAs that are transmitted from drug-resistant cells to drug-sensitive cells and affect chemotherapy resistance, such as lncMSTRG.292666.16 to osimertinib, lncRP11838N2.4 to erlotinib, and miRNA-222-3p to gemcitabine.86,138,157 Overall, EV ncRNAs, as important molecules in the regulation of radioresistance and chemoresistance in LC, are of great significance for improving the clinical therapeutic effect.
Diagnostic, prognostic, and therapeutic applications of EV ncRNAs in LC
EV ncRNAs serve as promising biomarkers in LC
EVs contain a variety of functional molecules, such as ncRNAs, that reflect the complexity and heterogeneity of tumors.101 Increasing research has implicated the important roles of EV ncRNAs in the diagnosis and prognosis of LC, as well as anti-LC therapeutic applications.153,182, 183, 184 Importantly, EV ncRNAs are readily available from almost all types of human fluids, providing a liquid tool for noninvasive clinical detection (Table 2).9
Table 2.
EV ncRNAs as diagnosis and prognostic biomarkers in LC
| NcRNAs | Source | Cases and samples | Biomarker potential | AUC, sensitivity, and specificity | Reference |
|---|---|---|---|---|---|
| miR-1-3p, miR-144-5p, and miR-150-5p | pleural lavage | 46 LC patients and 25 healthy controls | promising biomarkers of LC diagnosis | AUC = 0.899; sensitivity = 80.25%; specificity = 92.31% | Roman-Canal et al.185 |
| miR-17-5p | serum | 100 NSCLC patients and 90 healthy controls | potential diagnostic biomarker for NSCLC patients | AUC = 0.86; sensitivity = 63%; specificity = 93.3% | Zhang et al.186 |
| miR-20b-5p and miR-3187-5p | serum | 276 NSCLC patients and 282 healthy controls | diagnosis biomarkers for early-stage NSCLC | AUC = 0.848 | Zhang et al.187 |
| miR-21 | pleural lavage | 144 primary LAC patients and 55 healthy controls | prognostic biomarker in primary LAC | – | Watabe et al.188 |
| miR-21 and miR-4257 | plasma | 195 NSCLC patients | predictive biomarker for recurrence in NSCLC patients | – | Dejima et al.189 |
| miR-126 | BAL fluid | 13 LAC patients and 15 healthy controls | diagnostic biomarkers in early-stage LAC | – | Kim et al.190 |
| miR-126 | serum | 45 NSCLC patients | diagnostics biomarker and therapeutic target for NSCLC | AUC = 0.875 and 0.835 for stage Ⅰ/Ⅱ and Ⅲ/Ⅳ NSCLC, respectively | Grimolizzi et al.191 |
| miR-146-5p | serum | six DDP-resistant NSCLC patients and six DDP-sensitive NSCLC patients | biomarker predicting DDP efficacy and real-time monitoring DDP resistance for NSCLC | – | Yuwen et al.192 |
| miR-193a-3p, miR-210-3p, and miR-5100 | plasma | 21 metastasis LC patients and 20 non-metastasis LC patients | biomarkers for LC metastasis | AUC = 0.8600, 0.8369 and 0.8016, respectively | Zhang et al.136 |
| miR-1290 | serum | 70 LAC patients and 40 healthy controls | potential biomarker for the diagnosis and prognosis of LAC | AUC = 0.937; sensitivity = 80%; specificity = 96.7% | Wu et al.193 |
| miR-216b | serum | 105 NSCLC patients and 60 healthy controls | potential diagnostic and prognostic biomarker for NSCLC | AUC = 0.84; sensitivity = 86.7%; specificity = 75% | Liu et al.194 |
| miR-342-5p and miR-574-5p | plasma | seven early-stage LAC patients and seven healthy controls | diagnostic biomarkers for early-stage LAC | AUC = 0.813; sensitivity = 80%; specificity = 73.2% | Han et al.195 |
| miR-378a, miR-379, miR-139-5p, and miR-200b-5p | plasma | 30 LC patients | biomarkers for screening and diagnose LC | AUC = 0.76; sensitivity = 96%; specificity = 60% | Cazzoli et al.196 |
| miR-1269a | serum | 147 NSCLC patients and 149 healthy controls | diagnostic biomarker for NSCLC | AUC = 0.915; sensitivity = 77%; specificity = 89% | Wang et al.197 |
| miR-5864 and miR-125b-5p | serum | 330 NSCLC patients and 312 healthy controls | diagnostic and prognostic biomarkers for NSCLC | AUC = 0.733; sensitivity = 62.4%; specificity = 70% | Zhang et al.183 |
| miR-7977 | serum | 65 LAC patients | LC biomarker | AUC = 0.787 | Chen et al.182 |
| circRNA-002178 | plasma | 120 LAC patients and 30 healthy controls | biomarkers for LAC early diagnosis | AUC = 0.9956 | Wang et al.198 |
| circRNA_0056616 | plasma | 42 LAC patients with lymph node metastasis and 48 without lymph node metastasis | potential biomarker for lymph node metastasis in LAC | AUC = 0.812; sensitivity = 79.2%; specificity = 81% | He et al.199 |
| circ_0047921, circ_0056285 and circ_0007761 | serum | 30 NSCLC patients and 45 healthy controls | biomarkers for NSCLC diagnosis in Chinese population | AUC = 0.926 | Xian et al.200 |
| hsa_circ_0,002,130 | serum | 28 osimertinib-resistant NSCLC and 32 osimertinib-sensitive NSCLC patients | biomarker for osimertinib resistance in LC | AUC = 0.792 | Ma et al.172 |
| hsa_circ_0014235 and hsa_circ_0025580 | plasma | 30 pairs of LUSC patients and healthy controls | diagnostic biomarkers for LUSC | AUC = 0.8254 and 0.8003, respectively | Wang et al.73 |
| circ-MEMO1 | serum | 52 pairs of NSCLC tissue sample and adjacent normal tissues | biomarker for the early diagnosis for NSCLC | AUC = 0.76; sensitivity = 56.67%; specificity = 96% | Ding et al.170 |
| lncGAS5 | serum | 64 NSCLC patients | single diagnostic biomarker for NSCLC | AUC = 0.929; sensitivity = 85.94%; specificity = 70% | Li et al.201 |
| lncTBILA and lncAGAP2-AS1 | serum | 150 NSCLC patients | potential diagnostic biomarkers for NSCLC | AUC = 0.853 | Tao et al.202 |
| lncSOX2-OT | plasma | 75 LSCC patients and 79 healthy controls | noninvasive biomarker for LSCC | AUC = 0.815; sensitivity = 76%; specificity = 73.17% | Teng et al.88 |
| lncMALAT-1 | serum | 77 NSCLC patients | biomarker and therapeutic target for NSCLC | AUC = 0.73; sensitivity = 60.1%; specificity = 80.9% | Zhang et al.151 |
| lncRNA-p21 | plasma | 56 NSCLC patients | prognostic biomarker for NSCLC | AUC = 0.639 | Castellano et al.153 |
AUC, area under the curve; BAL, bronchoalveolar lavage; LUSC, lung squamous cell carcinoma; LSCC, lung squamous cell carcinoma.
Kim et al.190 investigated the expression level of miRNAs in EVs isolated from bronchoalveolar lavage (BAL) fluid, comparing early-stage LAC patients and healthy controls. MiR-126 was upregulated in the BAL fluid of LAC patients compared with healthy people, which makes it a potential diagnostic biomarker for early-stage LAC. Specifically, miR-126 could also be detected in serum EVs.191 Downregulation of miR-126 was detected in the serum of advanced-stage NSCLC patients compared with healthy controls. EV miR-126 might serve as a potential diagnostic biomarker and personalized therapeutic target in LC. Moreover, Roman-Canal et al.185 performed pleural lavage on LC patients and analyzed the expression of EV miRNAs in the lavage fluid. Fourteen miRNAs were found to be dysregulated between cancer and control cases, among which miR-1-3p, miR-144-5p, and miR-150-5p were the three most significantly dysregulated with a high area under the curve (AUC) value, sensitivity, and specificity. These results suggest that EV miR-1-3p, miR-144-5p, and miR-150-5p are promising noninvasive biomarkers for LC diagnosis.
The expression of circRNAs and lncRNAs in EVs has also been investigated. In the work of Xian et al.,200 three EV circRNAs (circ_0047921, circ_0056285, and circ_0007761) showed diagnostic value in NSCLC patients (AUC = 0.926). Additionally, circ_0047921 could distinguish NSCLC patients from chronic obstructive pulmonary disease controls (AUC = 0.890). However, circ_0056285-circ_0007761 combinations could distinguish NSCLC patients from tuberculosis controls (AUC = 0.820). The expression level of circ_0056285 was related to clinical stages and lymph node metastasis of NSCLC. These three EV circRNAs might serve as diagnostic and prognostic biomarkers of NSCLC in the Chinese population. According to the research of Castellano et al.,153 patients with high expression levels of EV lncRNA-p21 have a shorter time to relapse and a shorter OS. In addition, they generated receiver operating characteristic (ROC) curves according to the survival time and lncRNA-p21 expression. The AUCs for 6-, 12-, 24-, and 36-month survival were 0.639, 0.737, 0.686, and 0.632, respectively.
EV ncRNAs might also be potential biomarkers for monitoring the outcomes of treatment. Yuwen's group192 found that advanced NSCLC patients with low serum EV miR-146a-5p possessed a higher recurrence risk than those with high serum EV miR-146a-5p levels. Additionally, in the process of DDP resistance, miR-146a-5p expression is decreased in either serum EVs or NSCLC cells, suggesting that overexpression of miR-146a-5p might reverse DDP resistance in NSCLC. Moreover, Ma et al.172 demonstrated that hsa_circ_0002130 is upregulated in osimertinib-resistant NSCLC cells and regulates glycolysis by targeting miR-498. Importantly, hsa_circ_0002130 was highly expressed in osimertinib-resistant NSCLC patient serum EVs compared with osimertinib-sensitive NSCLC patients (AUC = 0.792). Serum EV hsa_circ_0002130 might be a noninvasive biomarker monitoring osimertinib resistance in NSCLC. In summary, EVs regulate the progression of LC by mediating the communication between neighboring cells and distant cells. EV ncRNAs in body fluids possess pivotal clinical value as biomarkers for LC diagnosis, prognosis, and treatment monitoring, and their clinical application deserves further investigation.
Potential applications of EV ncRNAs in anti-LC therapy
Currently, an increasing number of studies have demonstrated that EVs are ideal delivery vehicles. Lai et al.203 created a sensitive reporter for EV imaging and expression monitoring in organs and body fluids. Importantly, they found that injected EVs can be delivered to tumor tissues 1 h after injection. Additionally, due to the peptides and ligands on the surface, EVs are able to deliver their cargoes to recipient cells directly. Moreover, many candidate ligands (such as drugs and biomacromolecules) are unstable in vivo, which is a huge challenge for their clinical applications. Since the lipid bilayer membrane structure of EVs can sequester drug enzymes, nucleases, and proteases, encapsulation of these ligands into EVs can enhance their stability and delivery efficiency. Besides, since EVs are intracellular components, they possess low immunogenicity and high biocompatibility in vivo. Therefore, EV cargoes might provide a novel nanobiomedical therapeutic approach for cancers.
Increasing evidence has confirmed the anti-LC potency of exosomal ncRNAs. Nie and colleagues184 demonstrated that EVs derived from breast cancer cells could be internalized by NSCLC cells through an interaction between integrin β4 on EVs and surfactant protein C on A549 cells. Especially, systemic administration of miR-126-loaded EVs significantly suppressed proliferation and migration in A549 cells by interrupting the PTEN/PI3K/AKT pathway. In a lung metastasis mouse model, miR-126-loaded EVs also had an efficacious anticancer effect. In another study, Jeong et al.204 investigated the anti-angiogenic and anti-growth effects of miR-497-loaded engineered EVs on NSCLC cells cultured in 2D and 3D microfluidic devices. MiR-497-loaded EVs effectively inhibit the expression of YAP1, VEGF-A, and CCNE1 and further suppress the angiogenesis and migration of tumors compared with the control. Altogether, these studies demonstrated that EVs could be vehicles for ncRNA delivery, and EV ncRNAs can be used as predictive, cost-efficient tools for the development of targeted LC therapy.
In addition to ncRNAs, other kinds of biomacromolecules, such as peptides, lipids, and even exogenous DNAs and drugs, can be loaded into EVs and transferred to recipient cells to exert anti-tumor effects, providing broad application prospects for EV-based delivery systems.205 However, many basic studies are required before the extensive application of EV ncRNAs in the clinical practice of LC treatment.
Isolation and identification of EVs
Methods for EV isolation
EV ncRNAs have been proved to have great application potential. Therefore, it is imperative to isolate and identify EVs from wide range of interfering substances and cellular fragments. With the development of the biomedical industry, several kinds of techniques for EVs isolation have been widely adopted, such as centrifugation techniques, polymer precipitation, size-based isolation techniques, immunoaffinity capture techniques, and microfluidic techniques.206
The centrifugation technique is the most commonly used method and is the gold standard for EV separation.207 This method usually requires a series of centrifugal forces with different rotation speeds and a certain amount of time. Although the technique is simple to operate and the amount of EVs obtained is large, the recovery rate is unstable and the purity is challenged. In addition, polyethylene glycol (PEG) is often used in polymer precipitation techniques to combine with water molecules, resulting in the decrease of solubility and precipitation of EVs.208 In recent years, several PEG-based commercial EV extraction kits, such as ExoQuick, Pure-Exo, and Total EV Isolation, have appeared. Nevertheless, lipoproteins and polymers may be co-precipitated and affect the purity of EVs.
Moreover, there are two types of size-based EV isolation techniques: size exclusion chromatography (SEC) and ultrafiltration.209,210 In SEC, the components with small hydrodynamic radius, such as proteins and oligonucleotides, can pass through the pore of the stationary phase, resulting in a longer elution time, while EVs possess relatively large hydrodynamic radius, leading to an early elution. SEC has been shown to be superior to other techniques in the purity of isolated EVs due to the amelioration of protein contamination. However, it is worth noting that SEC cannot distinguish microvesicles with the same size.208 If different EV subtypes need to be identified, a combination of SEC and immunocapture methods can be used. Ultrafiltration relies on the use of membranes with specified pore diameters to isolate particles of a pre-determined size range.211 Since the volume of EVs is larger than biological macromolecules such as proteins and nucleic acids, ultrafiltration membranes with different cutoff molecular weight can be used for EV separation. This method is simple to operate and does not require special equipment and reagents.
Immunoaffinity capture techniques is a method that is based on the interaction between the antibodies fixed on the chromatographic stationary phase and magnetic beads and EV surface biomarkers, through which the specific enrichment of EVs can be achieved. Membrane proteins expressed on the EV surface, such as CD9, EpCAM, CD63, and RAB5, can be used as specific markers for EV separation.212 Immunoaffinity capture can be used not only for the isolation of EVs with same surface markers but also for the qualitative and quantitative identification of EVs.213 However, this method is not suitable for separating EVs from a large quantity of samples, and non-specific adsorption might cause impurities mixed into obtained EVs.
The microfluidic technique is a promising attempt at EV isolation in recent years.214 Based on the biological, physical, and chemical properties of EVs (such as size, density, and immunoaffinity), the microfluidic technique can achieve precise separation of EVs. Additionally, innovative EV separation methods that combine electrophoresis, acoustics, and electromagnetics are also under rapid development.215,216 Nevertheless, because of the lack of standardization and large-scale clinical sample testing, as well as the lack of relevant method verification, the microfluidic technique currently is mainly used in disease diagnosis. In general, efficient and economical separation and purification of a sufficient number of pure EVs is the main bottleneck of current research; therefore, it is urgent to develop novel techniques for EV isolation.
Methods for EV identification
Basic research and clinical applications require pure and uniform EVs. Therefore, it is urgent to identify and control the quality of the isolated EVs. Some molecular, biochemical, and biophysical properties of EVs, such as size, morphology, concentration, membrane potential, and surface markers, as well as the inside cargoes, can be utilized for EVs identification.217 In this section, we summarize the current methods available for EV characterization.
Electron microscopy (EM), dynamic light scattering (DLS), and nanoparticle tracking analysis (NTA) are effective methods for the determination of EV size. EM is a technique that is widely used to visualize and characterize biological samples.218 Three types of EM, transmission electron microscopy (TEM), scanning electron microscope (SEM), and cryoelectron microscopy (cryo-EM), are commonly employed to represent the microstructures of EVs.217 However, the requirement of complex and multistep sample preparations before EM detection, like dehydration or fixation, might induce alteration of EV morphology. In addition, the electron beam may also damage biological samples in some cases.219 EVs possess obvious membrane boundaries under the EM, and form cup-shaped structures with varying sizes of 30–150 nm. DLS, also known as photon correlation spectroscopy, is a method that uses a monochromatic coherent laser beam to pass through suspended particles, and the size of the particles in the solution can be estimated by observing the time-dependent fluctuations in scattering intensity.220 This method is simple and convenient to operate, and can be used to measure particles ranging from 1 nm to 6 μm in size. The effectiveness of DLS in measuring EVs' size and distribution has been confirmed in many studies.221,222 However, the measurement of DLS is easily disturbed by the physical and chemical properties of the samples, such as color, electrical properties, and magnetic properties, and is sensitive to dust and impurities.220 Therefore, DLS requires higher sample purity. In recent years, NTA has been used as one of the methods for characterization and identification of EVs.223 Compared with other characterization methods, the NTA technique can maintain the original state of EVs through simple sample processing, and can quickly detect the average size, modal value, and size distribution of EVs. Specifically, fluorescence intensity can be detected by the NTA system.217 Therefore, NTA can be used in combination with immunological methods to detect antigens on EVs by using fluorophore-labelled antibodies, which can be utilized for real-time monitoring of diseases.224
In addition, zeta potential is a physical property of EVs that affects the stability of EVs.225 Zeta potential can be assessed by measuring the magnitude of electrostatic or charge repulsion or attraction between EVs. The ZetaView and Zetasizer Nano series are newly launched instruments that are capable of characterizing size and zeta potential of particles. Studies have employed them for the identification and characterization of EVs. For example, Jeong et al.226 found that EVs from murine embryonic stem cells are 60–120 nm in diameter, with a zeta potential of −14.54 ± 1.31 mV. Danilo's group225 measured the size and zeta potential of neuroblastoma cell-derived vesicles by using the Zetasizer Nano instrument. The diameter of EVs was about 70 nm, and the zeta potential ranged from −14.8 ± 1.55 to −12 ± 0.15 mV. Commonly, particles stably exist at absolute values of the zeta potential less than −30 mV or more than 30 mV.227 Therefore, EVs are unstable, and tend to settle and aggregate automatically.
Moreover, western blot and flow cytometry are traditional identification and screening techniques that are based on EV surface markers (CD9, CD63, CD81, HSP90, etc.).228 High-quality monoclonal antibodies are key factors in improving the reliability of detection. In a recent study by Zhou et al.,229 they identified the EVs isolated from LC cells through detecting the surface markers CD9 and CD63 by western blot assays. In comparison, flow cytometry, with faster detection speed, is suitable for high-throughput screening, at the same time analyzing the size and volume of EV particles. The widely used enzyme-linked immunosorbent assays can be also utilized for EV identification. The quantitative detection of EV surface markers can be performed using specific antibodies.230 In addition to these methods, some new technologies for EV identification have emerged based on EV surface markers. Yoshioka et al.231 performed the ExoScreen method for detecting EVs isolated from colorectal cancer patients' serum. This method was described as an amplified luminescent proximity homogeneous assay that need streptavidin-coated donor beads and photosensitizer acceptor beads. The emitted fluorescent signal will appear only if the acceptor beads are within about 200 nm from donor beads, thereby avoiding the interference of proteins and large vesicles.
For characterization of the constituents and functional properties of EV cargoes, multi-omics analysis is needed. The obtainment of EV hallmarks requires evaluation at several levels, including transcriptome, proteome, epigenome, and metabolome. In the past few decades, researchers have made many attempts to reveal the structures, compositions, and biological functions of EVs through single-omics analysis, such as searching the abnormal expression of EV ncRNAs and proteins through transcriptomics and proteomics analysis, respectively.232, 233, 234 However, these single-omics analyses often have limitations. It is difficult to establish a dynamic relationship between the expression changes of EV cargoes and the disease phenotypes. In contrast, multi-omics analysis obtains large-scale omics data from different molecular levels. The integrative analysis of multiple omics information is conducive to in-depth understanding of the regulation and causality between EV cargoes, thereby resolving the molecular mechanism and genetic basis of biological and pathological processes. Multi-omics analysis of EVs has been widely employed in cancer research. For instance, Luo et al.235 reported the transcriptomics and proteomics landscape of EVs derived from human LAC stem-like cells. They obtained a variety of mRNAs, circRNAs, and lncRNAs enriched in EVs through transcriptomics analysis. Importantly, integrative analysis of transcriptomics and proteomics data suggested the association of EV RNAs and proteins to tumorigenesis, revealing the capacity to serve as diagnosis biomarkers for LC.
In total, as an important basis for investigating the biological characteristics and clinical application of EVs, the isolation and identification technology of EVs still needs to be continuously improved.
Conclusions and prospects
According to the Cancer Report 2020, LC is the most common malignancy in humans worldwide.1 Due to the complexity of LC occurrence and development, as well as the great heterogeneity among patients, there are few universal treatment strategies for LC. In recent years, the morbidity and mortality of LC have been on the rise, and therapeutic resistance often occurs. In view of this, it is urgent to investigate the underlying mechanisms of LC and to discover effective strategies for LC precision therapy. ncRNAs, including miRNAs, circRNAs, and lncRNAs, provide a treasure house for discovering pathogenic mechanisms and finding therapeutic targets for LC. EVs are a novel method of intercellular communication and are enriched with cargoes, including ncRNAs.15 Increasing evidence suggests that ncRNAs can be selectively taken up into EVs, secreted, and transferred between specific donor and recipient cells, regulating numerous malignant phenotypes of LC, such as proliferation, migration, EMT, metastasis, angiogenesis, and treatment resistance.139,144,148,154 Many studies have confirmed that EV ncRNAs have the potential to be used as diagnostic and prognostic biomarkers in LC, and they are promising therapeutic targets and reoccurrence risk monitoring tools.87,236
However, some challenges should not be ignored before EV ncRNAs can be used in the clinic. EVs in body fluids are often derived from a combination of complex sources, and it is necessary to explore their sources to confirm the direct correlation between certain EV ncRNAs and LC. In addition, due to the low content of EVs in body fluids, it is necessary to optimize the separation and detection methods of EVs and to establish a standardized and quantitative evaluation system. Current EV extraction methods, such as ultracentrifugation, precipitation, ultrafiltration, and immunoaffinity capture, are time consuming, expensive, low yield, and easy to contaminate, bringing inconvenience to follow-up studies.237 A more convenient and accurate extraction method for EVs needs to be discovered. Moreover, EVs mediate cell-cell communication by transferring biomolecules between cells. Understanding how cargoes are selectively packaged into EVs is an urgent research question. Furthermore, it has been reported that engineered EVs can be used as delivery vehicles for ncRNAs, peptides, lipids, exogenous DNAs, and even drugs.184,205 However, how EVs mediate the recognition of target cells and the release of their cargoes remains unknown.
In summary, EV ncRNAs are promising biomarkers and potential therapeutic targets, as well as prospective drug delivery vehicles for LC. Further study is required to elucidate the properties and regulatory mechanisms underlying EV ncRNAs, benefiting additional LC patients in the near future.
Acknowledgments
This work was supported by China Postdoctoral Science Foundation Funded Project (project no. 2018M632612) and the National Natural Science Foundation of China (project no. 51803098).
Author contributions
C.S. and Y.L. contributed equally to this work. C.S. and Y.L. drafted and wrote the manuscript. H.J.C. and F.W. collected the related paper. X.Z.C. and Q.K.Y. revised the manuscript. K.W. and Y.W. participated in the design of the review and helped to draft and revise the manuscript. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
References
- 1.Wild C.P., Weiderpass E., Stewart B.W. IARC Press; 2020. World Cancer Report 2020: Cancer Research for Cancer Prevention. [Google Scholar]
- 2.Lemjabbar-Alaoui H., Hassan O.U., Yang Y.W., Buchanan P. Lung cancer: biology and treatment options. Biochim. Biophys. Acta. 2015;1856:189–210. doi: 10.1016/j.bbcan.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gabor S., Renner H., Popper H., Anegg U., Sankin O., Matzi V., Lindenmann J., Smolle Juttner F.M. Invasion of blood vessels as significant prognostic factor in radically resected T1-3N0M0 non-small-cell lung cancer. Eur. J. Cardiothorac. Surg. 2004;25:439–442. doi: 10.1016/j.ejcts.2003.11.033. [DOI] [PubMed] [Google Scholar]
- 4.Hirsch F.R., Scagliotti G.V., Mulshine J.L., Kwon R., Curran W.J., Jr., Wu Y.L., Paz-Ares L. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389:299–311. doi: 10.1016/S0140-6736(16)30958-8. [DOI] [PubMed] [Google Scholar]
- 5.Yan J., Zhang Z., Zhan X., Chen K., Pu Y., Liang Y., He B. In situ injection of dual-delivery PEG based MMP-2 sensitive hydrogels for enhanced tumor penetration and chemo-immune combination therapy. Nanoscale. 2021;13:9577–9589. doi: 10.1039/d1nr01155c. [DOI] [PubMed] [Google Scholar]
- 6.Xia H., Liang Y., Chen K., Guo C., Wang M., Cao J., Han S., Ma Q., Sun Y., He B. Reduction-sensitive polymeric micelles as amplifying oxidative stress vehicles for enhanced antitumor therapy. Colloids Surf. B Biointerfaces. 2021;203:111733. doi: 10.1016/j.colsurfb.2021.111733. [DOI] [PubMed] [Google Scholar]
- 7.Wang L., Zhu B., Zhang M., Wang X. Roles of immune microenvironment heterogeneity in therapy-associated biomarkers in lung cancer. Semin. Cell Dev. Biol. 2017;64:90–97. doi: 10.1016/j.semcdb.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 8.Chiodoni C., Di Martino M.T., Zazzeroni F., Caraglia M., Donadelli M., Meschini S., Leonetti C., Scotlandi K. Cell communication and signaling: how to turn bad language into positive one. J. Exp. Clin. Cancer Res. 2019;38:128. doi: 10.1186/s13046-019-1122-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raposo G., Stahl P.D. Extracellular vesicles: a new communication paradigm? Nat. Rev. Mol. Cell Biol. 2019;20:509–510. doi: 10.1038/s41580-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 10.Tkach M., Thery C. Communication by extracellular vesicles: where we are and where we need to go. Cell. 2016;164:1226–1232. doi: 10.1016/j.cell.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 11.Lafitte M., Lecointre C., Roche S. Roles of exosomes in metastatic colorectal cancer. Am. J. Physiol. Cell Physiol. 2019;317:C869–C880. doi: 10.1152/ajpcell.00218.2019. [DOI] [PubMed] [Google Scholar]
- 12.Morad G., Carman C.V., Hagedorn E.J., Perlin J.R., Zon L.I., Mustafaoglu N., Park T.E., Ingber D.E., Daisy C.C., Moses M.A. Tumor-derived extracellular vesicles breach the intact blood-brain barrier via transcytosis. ACS Nano. 2019;13:13853–13865. doi: 10.1021/acsnano.9b04397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wortzel I., Dror S., Kenific C.M., Lyden D. Exosome-mediated metastasis: communication from a distance. Dev. Cell. 2019;49:347–360. doi: 10.1016/j.devcel.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 14.Fan Q., Yang L., Zhang X., Peng X., Wei S., Su D., Zhai Z., Hua X., Li H. The emerging role of exosome-derived non-coding RNAs in cancer biology. Cancer Lett. 2018;414:107–115. doi: 10.1016/j.canlet.2017.10.040. [DOI] [PubMed] [Google Scholar]
- 15.Xie Y., Dang W., Zhang S., Yue W., Yang L., Zhai X., Yan Q., Lu J. The role of exosomal noncoding RNAs in cancer. Mol. Cancer. 2019;18:37. doi: 10.1186/s12943-019-0984-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boriachek K., Islam M.N., Moller A., Salomon C., Nguyen N.T., Hossain M.S.A., Yamauchi Y., Shiddiky M.J.A. Biological functions and current advances in isolation and detection strategies for exosome nanovesicles. Small. 2018;14 doi: 10.1002/smll.201702153. [DOI] [PubMed] [Google Scholar]
- 17.Kowal J., Tkach M., Thery C. Biogenesis and secretion of exosomes. Curr. Opin. Cell Biol. 2014;29:116–125. doi: 10.1016/j.ceb.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 18.Jiang L., Gu Y., Du Y., Liu J. Exosomes: diagnostic biomarkers and therapeutic delivery vehicles for cancer. Mol. Pharm. 2019;16:3333–3349. doi: 10.1021/acs.molpharmaceut.9b00409. [DOI] [PubMed] [Google Scholar]
- 19.Kalluri R., LeBleu V.S. The biology, function, and biomedical applications of exosomes. Science. 2020;367:eaau6977. doi: 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vietri M., Radulovic M., Stenmark H. The many functions of ESCRTs. Nat. Rev. Mol. Cell Biol. 2020;21:25–42. doi: 10.1038/s41580-019-0177-4. [DOI] [PubMed] [Google Scholar]
- 21.Baietti M.F., Zhang Z., Mortier E., Melchior A., Degeest G., Geeraerts A., Ivarsson Y., Depoortere F., Coomans C., Vermeiren E., et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 2012;14:677–685. doi: 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- 22.Stuffers S., Sem Wegner C., Stenmark H., Brech A. Multivesicular endosome biogenesis in the absence of ESCRTs. Traffic. 2009;10:925–937. doi: 10.1111/j.1600-0854.2009.00920.x. [DOI] [PubMed] [Google Scholar]
- 23.Trajkovic K., Hsu C., Chiantia S., Rajendran L., Wenzel D., Wieland F., Schwille P., Brugger B., Simons M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 24.van Niel G., Charrin S., Simoes S., Romao M., Rochin L., Saftig P., Marks M.S., Rubinstein E., Raposo G. The tetraspanin CD63 regulates ESCRT-independent and -dependent endosomal sorting during melanogenesis. Dev. Cell. 2011;21:708–721. doi: 10.1016/j.devcel.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chairoungdua A., Smith D.L., Pochard P., Hull M., Caplan M.J. Exosome release of beta-catenin: a novel mechanism that antagonizes Wnt signaling. J. Cell Biol. 2010;190:1079–1091. doi: 10.1083/jcb.201002049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buschow S.I., Nolte-'t Hoen E.N., van Niel G., Pols M.S., ten Broeke T., Lauwen M., Ossendorp F., Melief C.J., Raposo G., Wubbolts R., et al. MHC II in dendritic cells is targeted to lysosomes or T cell-induced exosomes via distinct multivesicular body pathways. Traffic. 2009;10:1528–1542. doi: 10.1111/j.1600-0854.2009.00963.x. [DOI] [PubMed] [Google Scholar]
- 27.Kosaka N., Iguchi H., Yoshioka Y., Takeshita F., Matsuki Y., Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J. Biol. Chem. 2010;285:17442–17452. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kosaka N., Iguchi H., Hagiwara K., Yoshioka Y., Takeshita F., Ochiya T. Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J. Biol. Chem. 2013;288:10849–10859. doi: 10.1074/jbc.M112.446831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Villarroya-Beltri C., Gutierrez-Vazquez C., Sanchez-Cabo F., Perez-Hernandez D., Vazquez J., Martin-Cofreces N., Martinez-Herrera D.J., Pascual-Montano A., Mittelbrunn M., Sanchez-Madrid F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu B., Su S., Patil D.P., Liu H., Gan J., Jaffrey S.R., Ma J. Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1. Nat. Commun. 2018;9:420. doi: 10.1038/s41467-017-02770-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee H., Li C., Zhang Y., Zhang D., Otterbein L.E., Jin Y. Caveolin-1 selectively regulates microRNA sorting into microvesicles after noxious stimuli. J. Exp. Med. 2019;216:2202–2220. doi: 10.1084/jem.20182313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li T., Evdokimov E., Shen R.F., Chao C.C., Tekle E., Wang T., Stadtman E.R., Yang D.C., Chock P.B. Sumoylation of heterogeneous nuclear ribonucleoproteins, zinc finger proteins, and nuclear pore complex proteins: a proteomic analysis. Proc. Natl. Acad. Sci. U S A. 2004;101:8551–8556. doi: 10.1073/pnas.0402889101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Santangelo L., Giurato G., Cicchini C., Montaldo C., Mancone C., Tarallo R., Battistelli C., Alonzi T., Weisz A., Tripodi M. The RNA-binding protein SYNCRIP is a component of the hepatocyte exosomal machinery controlling microRNA sorting. Cell Rep. 2016;17:799–808. doi: 10.1016/j.celrep.2016.09.031. [DOI] [PubMed] [Google Scholar]
- 34.Hobor F., Dallmann A., Ball N.J., Cicchini C., Battistelli C., Ogrodowicz R.W., Christodoulou E., Martin S.R., Castello A., Tripodi M., et al. A cryptic RNA-binding domain mediates Syncrip recognition and exosomal partitioning of miRNA targets. Nat. Commun. 2018;9:831. doi: 10.1038/s41467-018-03182-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koppers-Lalic D., Hackenberg M., Bijnsdorp I.V., van Eijndhoven M.A.J., Sadek P., Sie D., Zini N., Middeldorp J.M., Ylstra B., de Menezes R.X., et al. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep. 2014;8:1649–1658. doi: 10.1016/j.celrep.2014.08.027. [DOI] [PubMed] [Google Scholar]
- 36.Yao B., La L.B., Chen Y.C., Chang L.J., Chan E.K. Defining a new role of GW182 in maintaining miRNA stability. EMBO Rep. 2012;13:1102–1108. doi: 10.1038/embor.2012.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gibbings D.J., Ciaudo C., Erhardt M., Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat. Cell Biol. 2009;11:1143–1149. doi: 10.1038/ncb1929. [DOI] [PubMed] [Google Scholar]
- 38.Lee Y.S., Pressman S., Andress A.P., Kim K., White J.L., Cassidy J.J., Li X., Lubell K., Lim D.H., Cho I.S., et al. Silencing by small RNAs is linked to endosomal trafficking. Nat. Cell Biol. 2009;11:1150–1156. doi: 10.1038/ncb1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Squadrito M.L., Baer C., Burdet F., Maderna C., Gilfillan G.D., Lyle R., Ibberson M., De Palma M. Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell Rep. 2014;8:1432–1446. doi: 10.1016/j.celrep.2014.07.035. [DOI] [PubMed] [Google Scholar]
- 40.Ostrowski M., Carmo N.B., Krumeich S., Fanget I., Raposo G., Savina A., Moita C.F., Schauer K., Hume A.N., Freitas R.P., et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 2010;12:19–30. doi: 10.1038/ncb2000. sup pp 11–13. [DOI] [PubMed] [Google Scholar]
- 41.Gu J., Qian H., Shen L., Zhang X., Zhu W., Huang L., Yan Y., Mao F., Zhao C., Shi Y., et al. Gastric cancer exosomes trigger differentiation of umbilical cord derived mesenchymal stem cells to carcinoma-associated fibroblasts through TGF-beta/Smad pathway. PLoS One. 2012;7:e52465. doi: 10.1371/journal.pone.0052465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nawaz M., Camussi G., Valadi H., Nazarenko I., Ekstrom K., Wang X., Principe S., Shah N., Ashraf N.M., Fatima F., et al. The emerging role of extracellular vesicles as biomarkers for urogenital cancers. Nat. Rev. Urol. 2014;11:688–701. doi: 10.1038/nrurol.2014.301. [DOI] [PubMed] [Google Scholar]
- 43.Johnstone R.M., Adam M., Hammond J.R., Orr L., Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes) J. Biol. Chem. 1987;262:9412–9420. [PubMed] [Google Scholar]
- 44.Johnstone R.M. The Jeanne Manery-Fisher Memorial Lecture 1991. Maturation of reticulocytes: formation of exosomes as a mechanism for shedding membrane proteins. Biochem. Cell Biol. 1992;70:179–190. doi: 10.1139/o92-028. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y., Liu J., Ma J., Sun T., Zhou Q., Wang W., Wang G., Wu P., Wang H., Jiang L., et al. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol. Cancer. 2019;18:116. doi: 10.1186/s12943-019-1041-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van der Vos K.E., Abels E.R., Zhang X., Lai C., Carrizosa E., Oakley D., Prabhakar S., Mardini O., Crommentuijn M.H., Skog J., et al. Directly visualized glioblastoma-derived extracellular vesicles transfer RNA to microglia/macrophages in the brain. Neuro Oncol. 2016;18:58–69. doi: 10.1093/neuonc/nov244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao H., Yang L., Baddour J., Achreja A., Bernard V., Moss T., Marini J.C., Tudawe T., Seviour E.G., San Lucas F.A., et al. Tumor microenvironment derived exosomes pleiotropically modulate cancer cell metabolism. Elife. 2016;5:e10250. doi: 10.7554/eLife.10250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kalluri R. The biology and function of exosomes in cancer. J. Clin. Invest. 2016;126:1208–1215. doi: 10.1172/JCI81135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pegtel D.M., Gould S.J. Exosomes. Annu. Rev. Biochem. 2019;88:487–514. doi: 10.1146/annurev-biochem-013118-111902. [DOI] [PubMed] [Google Scholar]
- 50.Sun Z., Shi K., Yang S., Liu J., Zhou Q., Wang G., Song J., Li Z., Zhang Z., Yuan W. Effect of exosomal miRNA on cancer biology and clinical applications. Mol. Cancer. 2018;17:147. doi: 10.1186/s12943-018-0897-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang X.Y., Huang Z.L., Huang J., Xu B., Huang X.Y., Xu Y.H., Zhou J., Tang Z.Y. Exosomal circRNA-100338 promotes hepatocellular carcinoma metastasis via enhancing invasiveness and angiogenesis. J. Exp. Clin. Cancer Res. 2020;39:20. doi: 10.1186/s13046-020-1529-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin L.Y., Yang L., Zeng Q., Wang L., Chen M.L., Zhao Z.H., Ye G.D., Luo Q.C., Lv P.Y., Guo Q.W., et al. Tumor-originated exosomal lncUEGC1 as a circulating biomarker for early-stage gastric cancer. Mol. Cancer. 2018;17:84. doi: 10.1186/s12943-018-0834-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu L., Li J., Gong Y., Wu Q., Tan S., Sun D., Xu X., Zuo Y., Zhao Y., Wei Y.Q., et al. Exosomal tRNA-derived small RNA as a promising biomarker for cancer diagnosis. Mol. Cancer. 2019;18:74. doi: 10.1186/s12943-019-1000-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang J., Xue F.T., Li Y.Y., Liu W., Zhang S. Exosomal piRNA sequencing reveals differences between heart failure and healthy patients. Eur. Rev. Med. Pharmacol. Sci. 2018;22:7952–7961. doi: 10.26355/eurrev_201811_16423. [DOI] [PubMed] [Google Scholar]
- 55.Peng Y., Croce C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016;1:15004. doi: 10.1038/sigtrans.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Medley J.C., Panzade G., Zinovyeva A.Y. microRNA strand selection: unwinding the rules. Wiley Interdiscip. Rev. RNA. 2021;12:e1627. doi: 10.1002/wrna.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Okamura K., Liu N., Lai E.C. Distinct mechanisms for microRNA strand selection by Drosophila Argonautes. Mol. Cell. 2009;36:431–444. doi: 10.1016/j.molcel.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ghildiyal M., Xu J., Seitz H., Weng Z., Zamore P.D. Sorting of Drosophila small silencing RNAs partitions microRNA∗ strands into the RNA interference pathway. RNA. 2010;16:43–56. doi: 10.1261/rna.1972910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kozomara A., Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee Y.S., Dutta A. MicroRNAs in cancer. Annu. Rev. Pathol. 2009;4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hayes J., Peruzzi P.P., Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol. Med. 2014;20:460–469. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 62.Shan C., Zhang Y., Hao X., Gao J., Chen X., Wang K. Biogenesis, functions and clinical significance of circRNAs in gastric cancer. Mol. Cancer. 2019;18:136. doi: 10.1186/s12943-019-1069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Valadi H., Ekstrom K., Bossios A., Sjostrand M., Lee J.J., Lotvall J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 64.Peng X.X., Yu R., Wu X., Wu S.Y., Pi C., Chen Z.H., Zhang X.C., Gao C.Y., Shao Y.W., Liu L., et al. Correlation of plasma exosomal microRNAs with the efficacy of immunotherapy in EGFR/ALK wild-type advanced non-small cell lung cancer. J. Immunother. Cancer. 2020;8:e000376. doi: 10.1136/jitc-2019-000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jin X., Chen Y., Chen H., Fei S., Chen D., Cai X., Liu L., Lin B., Su H., Zhao L., et al. Evaluation of tumor-derived exosomal miRNA as potential diagnostic biomarkers for early-stage non-small cell lung cancer using next-generation sequencing. Clin. Cancer Res. 2017;23:5311–5319. doi: 10.1158/1078-0432.CCR-17-0577. [DOI] [PubMed] [Google Scholar]
- 66.Li Y., Yin Z., Fan J., Zhang S., Yang W. The roles of exosomal miRNAs and lncRNAs in lung diseases. Signal Transduct. Target. Ther. 2019;4:47. doi: 10.1038/s41392-019-0080-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang Y., Jia D.D., Zhang Y.F., Cheng M.D., Zhu W.X., Li P.F., Zhang Y.F. The emerging function and clinical significance of circRNAs in thyroid cancer and autoimmune thyroid diseases. Int. J. Biol. Sci. 2021;17:1731–1741. doi: 10.7150/ijbs.55381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kelly S., Greenman C., Cook P.R., Papantonis A. Exon skipping is correlated with exon circularization. J. Mol. Biol. 2015;427:2414–2417. doi: 10.1016/j.jmb.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 70.Han B., Chao J., Yao H. Circular RNA and its mechanisms in disease: from the bench to the clinic. Pharmacol. Ther. 2018;187:31–44. doi: 10.1016/j.pharmthera.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 71.Li Y., Zheng Q., Bao C., Li S., Guo W., Zhao J., Chen D., Gu J., He X., Huang S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang B., Teng F., Chang L., Wang J., Liu D.L., Cui Y.S., Li G.H. Tumor-derived exosomal circRNA_102481 contributes to EGFR-TKIs resistance via the miR-30a-5p/ROR1 axis in non-small cell lung cancer. Aging (Albany NY) 2021;13:13264–13286. doi: 10.18632/aging.203011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Y., Zhang H., Wang J., Li B., Wang X. Circular RNA expression profile of lung squamous cell carcinoma: identification of potential biomarkers and therapeutic targets. Biosci. Rep. 2020;40 doi: 10.1042/BSR20194512. BSR20194512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pachnis V., Brannan C.I., Tilghman S.M. The structure and expression of a novel gene activated in early mouse embryogenesis. EMBO J. 1988;7:673–681. doi: 10.1002/j.1460-2075.1988.tb02862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kapranov P., Willingham A.T., Gingeras T.R. Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 76.Zhao Y., Li H., Fang S., Kang Y., Wu W., Hao Y., Li Z., Bu D., Sun N., Zhang M.Q., et al. NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic Acids Res. 2016;44:D203–D208. doi: 10.1093/nar/gkv1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chew C.L., Conos S.A., Unal B., Tergaonkar V. Noncoding RNAs: master regulators of inflammatory signaling. Trends Mol. Med. 2018;24:66–84. doi: 10.1016/j.molmed.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 78.Kopp F., Mendell J.T. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vitiello M., Tuccoli A., Poliseno L. Long non-coding RNAs in cancer: implications for personalized therapy. Cell. Oncol. (Dordr) 2015;38:17–28. doi: 10.1007/s13402-014-0180-x. [DOI] [PubMed] [Google Scholar]
- 80.Kornfeld J.W., Bruning J.C. Regulation of metabolism by long, non-coding RNAs. Front. Genet. 2014;5:57. doi: 10.3389/fgene.2014.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bartonicek N., Maag J.L., Dinger M.E. Long noncoding RNAs in cancer: mechanisms of action and technological advancements. Mol. Cancer. 2016;15:43. doi: 10.1186/s12943-016-0530-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sun Z., Yang S., Zhou Q., Wang G., Song J., Li Z., Zhang Z., Xu J., Xia K., Chang Y., et al. Emerging role of exosome-derived long non-coding RNAs in tumor microenvironment. Mol. Cancer. 2018;17:82. doi: 10.1186/s12943-018-0831-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang C., Xu L., Deng G., Ding Y., Bi K., Jin H., Shu J., Yang J., Deng H., Wang Z., et al. Exosomal HOTAIR promotes proliferation, migration and invasion of lung cancer by sponging miR-203. Sci. China Life Sci. 2020;63:1265–1268. doi: 10.1007/s11427-019-1579-x. [DOI] [PubMed] [Google Scholar]
- 84.Chen X., Wang Z., Tong F., Dong X., Wu G., Zhang R. lncRNA UCA1 promotes gefitinib resistance as a ceRNA to target FOSL2 by sponging miR-143 in non-small cell lung cancer. Mol. Ther. Nucleic Acids. 2020;19:643–653. doi: 10.1016/j.omtn.2019.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 85.Ni J., Zhang X., Li J., Zheng Z., Zhang J., Zhao W., Liu L. Tumour-derived exosomal lncRNA-SOX2OT promotes bone metastasis of non-small cell lung cancer by targeting the miRNA-194-5p/RAC1 signalling axis in osteoclasts. Cell Death Dis. 2021;12:662. doi: 10.1038/s41419-021-03928-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Deng Q., Fang Q., Xie B., Sun H., Bao Y., Zhou S. Exosomal long non-coding RNA MSTRG.292666.16 is associated with osimertinib (AZD9291) resistance in non-small cell lung cancer. Aging (Albany NY) 2020;12:8001–8015. doi: 10.18632/aging.103119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang X., Guo H., Bao Y., Yu H., Xie D., Wang X. Exosomal long non-coding RNA DLX6-AS1 as a potential diagnostic biomarker for non-small cell lung cancer. Oncol. Lett. 2019;18:5197–5204. doi: 10.3892/ol.2019.10892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Teng Y., Kang H., Chu Y. Identification of an exosomal long noncoding RNA SOX2-OT in plasma as a promising biomarker for lung squamous cell carcinoma. Genet. Test. Mol. Biomark. 2019;23:235–240. doi: 10.1089/gtmb.2018.0103. [DOI] [PubMed] [Google Scholar]
- 89.Anderson P., Ivanov P. tRNA fragments in human health and disease. FEBS Lett. 2014;588:4297–4304. doi: 10.1016/j.febslet.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schimmel P. The emerging complexity of the tRNA world: mammalian tRNAs beyond protein synthesis. Nat. Rev. Mol. Cell Biol. 2018;19:45–58. doi: 10.1038/nrm.2017.77. [DOI] [PubMed] [Google Scholar]
- 91.Yeung M.L., Bennasser Y., Watashi K., Le S.Y., Houzet L., Jeang K.T. Pyrosequencing of small non-coding RNAs in HIV-1 infected cells: evidence for the processing of a viral-cellular double-stranded RNA hybrid. Nucleic Acids Res. 2009;37:6575–6586. doi: 10.1093/nar/gkp707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sobala A., Hutvagner G. Small RNAs derived from the 5' end of tRNA can inhibit protein translation in human cells. RNA Biol. 2013;10:553–563. doi: 10.4161/rna.24285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Maute R.L., Schneider C., Sumazin P., Holmes A., Califano A., Basso K., Dalla-Favera R. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc. Natl. Acad. Sci. U S A. 2013;110:1404–1409. doi: 10.1073/pnas.1206761110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Baglio S.R., Rooijers K., Koppers-Lalic D., Verweij F.J., Perez Lanzon M., Zini N., Naaijkens B., Perut F., Niessen H.W., Baldini N., et al. Human bone marrow- and adipose-mesenchymal stem cells secrete exosomes enriched in distinctive miRNA and tRNA species. Stem Cell Res. Ther. 2015;6:127. doi: 10.1186/s13287-015-0116-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Steitz T.A. A structural understanding of the dynamic ribosome machine. Nat. Rev. Mol. Cell Biol. 2008;9:242–253. doi: 10.1038/nrm2352. [DOI] [PubMed] [Google Scholar]
- 96.Ramakrishnan V. Ribosome structure and the mechanism of translation. Cell. 2002;108:557–572. doi: 10.1016/s0092-8674(02)00619-0. [DOI] [PubMed] [Google Scholar]
- 97.Xie Q., Li C., Song X., Wu L., Jiang Q., Qiu Z., Cao H., Yu K., Wan C., Li J., et al. Folate deficiency facilitates recruitment of upstream binding factor to hot spots of DNA double-strand breaks of rRNA genes and promotes its transcription. Nucleic Acids Res. 2017;45:2472–2489. doi: 10.1093/nar/gkw1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kilchert C., Wittmann S., Vasiljeva L. The regulation and functions of the nuclear RNA exosome complex. Nat. Rev. Mol. Cell Biol. 2016;17:227–239. doi: 10.1038/nrm.2015.15. [DOI] [PubMed] [Google Scholar]
- 99.Schuller J.M., Falk S., Fromm L., Hurt E., Conti E. Structure of the nuclear exosome captured on a maturing preribosome. Science. 2018;360:219–222. doi: 10.1126/science.aar5428. [DOI] [PubMed] [Google Scholar]
- 100.Jenjaroenpun P., Kremenska Y., Nair V.M., Kremenskoy M., Joseph B., Kurochkin I.V. Characterization of RNA in exosomes secreted by human breast cancer cell lines using next-generation sequencing. PeerJ. 2013;1:e201. doi: 10.7717/peerj.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fiskaa T., Knutsen E., Nikolaisen M.A., Jorgensen T.E., Johansen S.D., Perander M., Seternes O.M. Distinct small RNA signatures in extracellular vesicles derived from breast cancer cell lines. PLoS One. 2016;11:e0161824. doi: 10.1371/journal.pone.0161824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kowalski M.P., Krude T. Functional roles of non-coding Y RNAs. Int. J. Biochem. Cell Biol. 2015;66:20–29. doi: 10.1016/j.biocel.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chen X., Taylor D.W., Fowler C.C., Galan J.E., Wang H.W., Wolin S.L. An RNA degradation machine sculpted by Ro autoantigen and noncoding RNA. Cell. 2013;153:166–177. doi: 10.1016/j.cell.2013.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen X., Wurtmann E.J., Van Batavia J., Zybailov B., Washburn M.P., Wolin S.L. An ortholog of the Ro autoantigen functions in 23S rRNA maturation in D. radiodurans. Genes Dev. 2007;21:1328–1339. doi: 10.1101/gad.1548207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Boccitto M., Wolin S.L. Ro60 and Y RNAs: structure, functions, and roles in autoimmunity. Crit. Rev. Biochem. Mol. Biol. 2019;54:133–152. doi: 10.1080/10409238.2019.1608902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Guglas K., Kolodziejczak I., Kolenda T., Kopczynska M., Teresiak A., Sobocinska J., Blizniak R., Lamperska K. YRNAs and YRNA-derived fragments as new players in cancer research and their potential role in diagnostics. Int. J. Mol. Sci. 2020;21:5682. doi: 10.3390/ijms21165682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Haderk F., Schulz R., Iskar M., Cid L.L., Worst T., Willmund K.V., Schulz A., Warnken U., Seiler J., Benner A., et al. Tumor-derived exosomes modulate PD-L1 expression in monocytes. Sci. Immunol. 2017;2:eaah5509. doi: 10.1126/sciimmunol.aah5509. [DOI] [PubMed] [Google Scholar]
- 108.Lovisa F., Di Battista P., Gaffo E., Damanti C.C., Garbin A., Gallingani I., Carraro E., Pillon M., Biffi A., Bortoluzzi S., et al. RNY4 in circulating exosomes of patients with pediatric anaplastic large cell lymphoma: an active player? Front. Oncol. 2020;10:238. doi: 10.3389/fonc.2020.00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lunavat T.R., Cheng L., Kim D.K., Bhadury J., Jang S.C., Lasser C., Sharples R.A., Lopez M.D., Nilsson J., Gho Y.S., et al. Small RNA deep sequencing discriminates subsets of extracellular vesicles released by melanoma cells–evidence of unique microRNA cargos. RNA Biol. 2015;12:810–823. doi: 10.1080/15476286.2015.1056975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.De Luca L., Trino S., Laurenzana I., Simeon V., Calice G., Raimondo S., Podesta M., Santodirocco M., Di Mauro L., La Rocca F., et al. MiRNAs and piRNAs from bone marrow mesenchymal stem cell extracellular vesicles induce cell survival and inhibit cell differentiation of cord blood hematopoietic stem cells: a new insight in transplantation. Oncotarget. 2016;7:6676–6692. doi: 10.18632/oncotarget.6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Czech B., Hannon G.J. One loop to rule them all: the ping-pong cycle and piRNA-guided silencing. Trends Biochem. Sci. 2016;41:324–337. doi: 10.1016/j.tibs.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Han Y.N., Li Y., Xia S.Q., Zhang Y.Y., Zheng J.H., Li W. PIWI proteins and PIWI-interacting RNA: emerging roles in cancer. Cell. Physiol. Biochem. 2017;44:1–20. doi: 10.1159/000484541. [DOI] [PubMed] [Google Scholar]
- 113.Esposito T., Magliocca S., Formicola D., Gianfrancesco F. piR_015520 belongs to Piwi-associated RNAs regulates expression of the human melatonin receptor 1A gene. PLoS One. 2011;6:e22727. doi: 10.1371/journal.pone.0022727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Freedman J.E., Gerstein M., Mick E., Rozowsky J., Levy D., Kitchen R., Das S., Shah R., Danielson K., Beaulieu L., et al. Diverse human extracellular RNAs are widely detected in human plasma. Nat. Commun. 2016;7:11106. doi: 10.1038/ncomms11106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jain G., Stuendl A., Rao P., Berulava T., Pena Centeno T., Kaurani L., Burkhardt S., Delalle I., Kornhuber J., Hull M., et al. A combined miRNA-piRNA signature to detect Alzheimer's disease. Transl. Psychiatry. 2019;9:250. doi: 10.1038/s41398-019-0579-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gu X., Wang C., Deng H., Qing C., Liu R., Liu S., Xue X. Exosomal piRNA profiling revealed unique circulating piRNA signatures of cholangiocarcinoma and gallbladder carcinoma. Acta Biochim. Biophys. Sin. (Shanghai) 2020;52:475–484. doi: 10.1093/abbs/gmaa028. [DOI] [PubMed] [Google Scholar]
- 117.Ng K.W., Anderson C., Marshall E.A., Minatel B.C., Enfield K.S., Saprunoff H.L., Lam W.L., Martinez V.D. Piwi-interacting RNAs in cancer: emerging functions and clinical utility. Mol. Cancer. 2016;15:5. doi: 10.1186/s12943-016-0491-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Vojtech L., Woo S., Hughes S., Levy C., Ballweber L., Sauteraud R.P., Strobl J., Westerberg K., Gottardo R., Tewari M., et al. Exosomes in human semen carry a distinctive repertoire of small non-coding RNAs with potential regulatory functions. Nucleic Acids Res. 2014;42:7290–7304. doi: 10.1093/nar/gku347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bade B.C., Dela Cruz C.S. Lung cancer 2020: epidemiology, etiology, and prevention. Clin. Chest Med. 2020;41:1–24. doi: 10.1016/j.ccm.2019.10.001. [DOI] [PubMed] [Google Scholar]
- 120.Chen R., Xu X., Qian Z., Zhang C., Niu Y., Wang Z., Sun J., Zhang X., Yu Y. The biological functions and clinical applications of exosomes in lung cancer. Cell. Mol. Life Sci. 2019;76:4613–4633. doi: 10.1007/s00018-019-03233-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhuang G., Wu X., Jiang Z., Kasman I., Yao J., Guan Y., Oeh J., Modrusan Z., Bais C., Sampath D., et al. Tumour-secreted miR-9 promotes endothelial cell migration and angiogenesis by activating the JAK-STAT pathway. EMBO J. 2012;31:3513–3523. doi: 10.1038/emboj.2012.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Xu Z., Liu X., Wang H., Li J., Dai L., Li J., Dong C. Lung adenocarcinoma cell-derived exosomal miR-21 facilitates osteoclastogenesis. Gene. 2018;666:116–122. doi: 10.1016/j.gene.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 123.Fabbri M., Paone A., Calore F., Galli R., Gaudio E., Santhanam R., Lovat F., Fadda P., Mao C., Nuovo G.J., et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Natl. Acad. Sci. U S A. 2012;109:E2110–E2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang X., Li F., Tang Y., Ren Q., Xiao B., Wan Y., Jiang S. miR-21a in exosomes from Lewis lung carcinoma cells accelerates tumor growth through targeting PDCD4 to enhance expansion of myeloid-derived suppressor cells. Oncogene. 2020;39:6354–6369. doi: 10.1038/s41388-020-01406-9. [DOI] [PubMed] [Google Scholar]
- 125.Hsu Y.L., Hung J.Y., Chang W.A., Lin Y.S., Pan Y.C., Tsai P.H., Wu C.Y., Kuo P.L. Hypoxic lung cancer-secreted exosomal miR-23a increased angiogenesis and vascular permeability by targeting prolyl hydroxylase and tight junction protein ZO-1. Oncogene. 2017;36:4929–4942. doi: 10.1038/onc.2017.105. [DOI] [PubMed] [Google Scholar]
- 126.Yu F., Liang M., Huang Y., Wu W., Zheng B., Chen C. Hypoxic tumor-derived exosomal miR-31-5p promotes lung adenocarcinoma metastasis by negatively regulating SATB2-reversed EMT and activating MEK/ERK signaling. J. Exp. Clin. Cancer Res. 2021;40:179. doi: 10.1186/s13046-021-01979-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wu H., Zhou J., Mei S., Wu D., Mu Z., Chen B., Xie Y., Ye Y., Liu J. Circulating exosomal microRNA-96 promotes cell proliferation, migration and drug resistance by targeting LMO7. J. Cell. Mol. Med. 2017;21:1228–1236. doi: 10.1111/jcmm.13056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Qin X., Yu S., Zhou L., Shi M., Hu Y., Xu X., Shen B., Liu S., Yan D., Feng J. Cisplatin-resistant lung cancer cell-derived exosomes increase cisplatin resistance of recipient cells in exosomal miR-100-5p-dependent manner. Int. J. Nanomed. 2017;12:3721–3733. doi: 10.2147/IJN.S131516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hsu Y.L., Hung J.Y., Chang W.A., Jian S.F., Lin Y.S., Pan Y.C., Wu C.Y., Kuo P.L. Hypoxic lung-cancer-derived extracellular vesicle MicroRNA-103a increases the oncogenic effects of macrophages by targeting PTEN. Mol. Ther. 2018;26:568–581. doi: 10.1016/j.ymthe.2017.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhang T., Zhang P., Li H.X. CAFs-derived exosomal miRNA-130a confers cisplatin resistance of NSCLC cells through PUM2-dependent packaging. Int. J. Nanomed. 2021;16:561–577. doi: 10.2147/IJN.S271976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lawson J., Dickman C., Towle R., Jabalee J., Javer A., Garnis C. Extracellular vesicle secretion of miR-142-3p from lung adenocarcinoma cells induces tumor promoting changes in the stroma through cell-cell communication. Mol. Carcinog. 2019;58:376–387. doi: 10.1002/mc.22935. [DOI] [PubMed] [Google Scholar]
- 132.Zhou J.H., Yao Z.X., Zheng Z., Yang J., Wang R., Fu S.J., Pan X.F., Liu Z.H., Wu K. G-MDSCs-derived exosomal miRNA-143-3p promotes proliferation via targeting of ITM2B in lung cancer. Onco Targets Ther. 2020;13:9701–9719. doi: 10.2147/OTT.S256378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Liang Y., Zhang D., Li L., Xin T., Zhao Y., Ma R., Du J. Exosomal microRNA-144 from bone marrow-derived mesenchymal stem cells inhibits the progression of non-small cell lung cancer by targeting CCNE1 and CCNE2. Stem Cell Res. Ther. 2020;11:87. doi: 10.1186/s13287-020-1580-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Valencia K., Luis-Ravelo D., Bovy N., Anton I., Martinez-Canarias S., Zandueta C., Ormazabal C., Struman I., Tabruyn S., Rebmann V., et al. miRNA cargo within exosome-like vesicle transfer influences metastatic bone colonization. Mol. Oncol. 2014;8:689–703. doi: 10.1016/j.molonc.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wu H., Mu X., Liu L., Wu H., Hu X., Chen L., Liu J., Mu Y., Yuan F., Liu W., et al. Bone marrow mesenchymal stem cells-derived exosomal microRNA-193a reduces cisplatin resistance of non-small cell lung cancer cells via targeting LRRC1. Cell Death Dis. 2020;11:801. doi: 10.1038/s41419-020-02962-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhang X., Sai B., Wang F., Wang L., Wang Y., Zheng L., Li G., Tang J., Xiang J. Hypoxic BMSC-derived exosomal miRNAs promote metastasis of lung cancer cells via STAT3-induced EMT. Mol. Cancer. 2019;18:40. doi: 10.1186/s12943-019-0959-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Fan J., Xu G., Chang Z., Zhu L., Yao J. miR-210 transferred by lung cancer cell-derived exosomes may act as proangiogenic factor in cancer-associated fibroblasts by modulating JAK2/STAT3 pathway. Clin. Sci. (Lond) 2020;134:807–825. doi: 10.1042/CS20200039. [DOI] [PubMed] [Google Scholar]
- 138.Wei F., Ma C., Zhou T., Dong X., Luo Q., Geng L., Ding L., Zhang Y., Zhang L., Li N., et al. Exosomes derived from gemcitabine-resistant cells transfer malignant phenotypic traits via delivery of miRNA-222-3p. Mol. Cancer. 2017;16:132. doi: 10.1186/s12943-017-0694-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhang Y., Li M., Hu C. Exosomal transfer of miR-214 mediates gefitinib resistance in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2018;507:457–464. doi: 10.1016/j.bbrc.2018.11.061. [DOI] [PubMed] [Google Scholar]
- 140.Li J., Yu J., Zhang H., Wang B., Guo H., Bai J., Wang J., Dong Y., Zhao Y., Wang Y. Exosomes-derived MiR-302b suppresses lung cancer cell proliferation and migration via TGFbetaRII inhibition. Cell. Physiol. Biochem. 2016;38:1715–1726. doi: 10.1159/000443111. [DOI] [PubMed] [Google Scholar]
- 141.Guo L., Li B., Yang J., Shen J., Ji J., Miao M. Fibroblastderived exosomal microRNA369 potentiates migration and invasion of lung squamous cell carcinoma cells via NF1mediated MAPK signaling pathway. Int. J. Mol. Med. 2020;46:595–608. doi: 10.3892/ijmm.2020.4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ma Y., Yuwen D., Chen J., Zheng B., Gao J., Fan M., Xue W., Wang Y., Li W., Shu Y., et al. Exosomal transfer of cisplatin-induced miR-425-3p confers cisplatin resistance in NSCLC through activating autophagy. Int. J. Nanomed. 2019;14:8121–8132. doi: 10.2147/IJN.S221383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hu H., Xu H., Lu F., Zhang J., Xu L., Xu S., Jiang H., Zeng Q., Chen E., He Z. Exosome-derived miR-486-5p regulates cell cycle, proliferation and metastasis in lung adenocarcinoma via targeting NEK2. Front. Bioeng. Biotechnol. 2020;8:259. doi: 10.3389/fbioe.2020.00259. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 144.He S., Li Z., Yu Y., Zeng Q., Cheng Y., Ji W., Xia W., Lu S. Exosomal miR-499a-5p promotes cell proliferation, migration and EMT via mTOR signaling pathway in lung adenocarcinoma. Exp. Cell Res. 2019;379:203–213. doi: 10.1016/j.yexcr.2019.03.035. [DOI] [PubMed] [Google Scholar]
- 145.Azuma Y., Yokobori T., Mogi A., Yajima T., Kosaka T., Iijima M., Shimizu K., Shirabe K., Kuwano H. Cancer exosomal microRNAs from gefitinib-resistant lung cancer cells cause therapeutic resistance in gefitinib-sensitive cells. Surg. Today. 2020;50:1099–1106. doi: 10.1007/s00595-020-01976-x. [DOI] [PubMed] [Google Scholar]
- 146.Kim D.H., Park S., Kim H., Choi Y.J., Kim S.Y., Sung K.J., Sung Y.H., Choi C.M., Yun M., Yi Y.S., et al. Tumor-derived exosomal miR-619-5p promotes tumor angiogenesis and metastasis through the inhibition of RCAN1.4. Cancer Lett. 2020;475:2–13. doi: 10.1016/j.canlet.2020.01.023. [DOI] [PubMed] [Google Scholar]
- 147.Tang Y.T., Huang Y.Y., Li J.H., Qin S.H., Xu Y., An T.X., Liu C.C., Wang Q., Zheng L. Alterations in exosomal miRNA profile upon epithelial-mesenchymal transition in human lung cancer cell lines. BMC Genomics. 2018;19:802. doi: 10.1186/s12864-018-5143-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhang N., Nan A., Chen L., Li X., Jia Y., Qiu M., Dai X., Zhou H., Zhu J., Zhang H., et al. Circular RNA circSATB2 promotes progression of non-small cell lung cancer cells. Mol. Cancer. 2020;19:101. doi: 10.1186/s12943-020-01221-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Fang K., Chen X., Qiu F., Xu J., Xiong H., Zhang Z. Serum-derived exosomes-mediated circular RNA ARHGAP10 modulates the progression of non-small-cell lung cancer through the miR-638/FAM83F axis. Cancer Biother. Radiopharm. 2020 doi: 10.1089/cbr.2019.3534. [DOI] [PubMed] [Google Scholar]
- 150.Chen S.W., Zhu S.Q., Pei X., Qiu B.Q., Xiong D., Long X., Lin K., Lu F., Xu J.J., Wu Y.B. Cancer cell-derived exosomal circUSP7 induces CD8(+) T cell dysfunction and anti-PD1 resistance by regulating the miR-934/SHP2 axis in NSCLC. Mol. Cancer. 2021;20:144. doi: 10.1186/s12943-021-01448-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zhang R., Xia Y., Wang Z., Zheng J., Chen Y., Li X., Wang Y., Ming H. Serum long non coding RNA MALAT-1 protected by exosomes is up-regulated and promotes cell proliferation and migration in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2017;490:406–414. doi: 10.1016/j.bbrc.2017.06.055. [DOI] [PubMed] [Google Scholar]
- 152.Zang X., Gu J., Zhang J., Shi H., Hou S., Xu X., Chen Y., Zhang Y., Mao F., Qian H., et al. Exosome-transmitted lncRNA UFC1 promotes non-small-cell lung cancer progression by EZH2-mediated epigenetic silencing of PTEN expression. Cell Death Dis. 2020;11:215. doi: 10.1038/s41419-020-2409-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Castellano J.J., Marrades R.M., Molins L., Vinolas N., Moises J., Canals J., Han B., Li Y., Martinez D., Monzo M., et al. Extracellular vesicle lincRNA-p21 expression in tumor-draining pulmonary vein defines prognosis in NSCLC and modulates endothelial cell behavior. Cancers (Basel) 2020;12:734. doi: 10.3390/cancers12030734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Cheng Y., Dai X., Yang T., Zhang N., Liu Z., Jiang Y. Low long noncoding RNA growth arrest-specific transcript 5 expression in the exosomes of lung cancer cells promotes tumor angiogenesis. J. Oncol. 2019;2019:2476175. doi: 10.1155/2019/2476175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zhang F., Sang Y., Chen D., Wu X., Wang X., Yang W., Chen Y. M2 macrophage-derived exosomal long non-coding RNA AGAP2-AS1 enhances radiotherapy immunity in lung cancer by reducing microRNA-296 and elevating NOTCH2. Cell Death Dis. 2021;12:467. doi: 10.1038/s41419-021-03700-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Domvri K., Petanidis S., Anestakis D., Porpodis K., Bai C., Zarogoulidis P., Freitag L., Hohenforst-Schmidt W., Katopodi T. Exosomal lncRNA PCAT-1 promotes Kras-associated chemoresistance via immunosuppressive miR-182/miR-217 signaling and p27/CDK6 regulation. Oncotarget. 2020;11:2847–2862. doi: 10.18632/oncotarget.27675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zhang W., Cai X., Yu J., Lu X., Qian Q., Qian W. Exosome-mediated transfer of lncRNA RP11838N2.4 promotes erlotinib resistance in non-small cell lung cancer. Int. J. Oncol. 2018;53:527–538. doi: 10.3892/ijo.2018.4412. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 158.Lei Y., Guo W., Chen B., Chen L., Gong J., Li W. Tumorreleased lncRNA H19 promotes gefitinib resistance via packaging into exosomes in nonsmall cell lung cancer. Oncol. Rep. 2018;40:3438–3446. doi: 10.3892/or.2018.6762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 159.Yue C., Chen J., Li Z., Li L., Chen J., Guo Y. microRNA-96 promotes occurrence and pro0gression of colorectal cancer via regulation of the AMPKalpha2-FTO-m6A/MYC axis. J. Exp. Clin. Cancer Res. 2020;39:240. doi: 10.1186/s13046-020-01731-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Xu S., Zheng L., Kang L., Xu H., Gao L. microRNA-let-7e in serum-derived exosomes inhibits the metastasis of non-small-cell lung cancer in a SUV39H2/LSD1/CDH1-dependent manner. Cancer Gene Ther. 2020;28:250–264. doi: 10.1038/s41417-020-00216-1. [DOI] [PubMed] [Google Scholar]
- 161.De Craene B., Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat. Rev. Cancer. 2013;13:97–110. doi: 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- 162.Bakir B., Chiarella A.M., Pitarresi J.R., Rustgi A.K. EMT, MET, plasticity, and tumor metastasis. Trends Cell Biol. 2020;30:764–776. doi: 10.1016/j.tcb.2020.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Syn N., Wang L., Sethi G., Thiery J.P., Goh B.C. Exosome-mediated metastasis: from epithelial-mesenchymal transition to escape from immunosurveillance. Trends Pharmacol. Sci. 2016;37:606–617. doi: 10.1016/j.tips.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 164.Folkman J. What is the evidence that tumors are angiogenesis dependent? J. Natl. Cancer Inst. 1990;82:4–6. doi: 10.1093/jnci/82.1.4. [DOI] [PubMed] [Google Scholar]
- 165.Lin E.W., Karakasheva T.A., Hicks P.D., Bass A.J., Rustgi A.K. The tumor microenvironment in esophageal cancer. Oncogene. 2016;35:5337–5349. doi: 10.1038/onc.2016.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 167.Batlle E., Massague J. Transforming growth factor-beta signaling in immunity and cancer. Immunity. 2019;50:924–940. doi: 10.1016/j.immuni.2019.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Luengo A., Gui D.Y., Vander Heiden M.G. Targeting metabolism for cancer therapy. Cell Chem. Biol. 2017;24:1161–1180. doi: 10.1016/j.chembiol.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Elia I., Haigis M.C. Metabolites and the tumour microenvironment: from cellular mechanisms to systemic metabolism. Nat. Metab. 2021;3:21–32. doi: 10.1038/s42255-020-00317-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ding C., Xi G., Wang G., Cui D., Zhang B., Wang H., Jiang G., Song J., Xu G., Wang J. Exosomal circ-MEMO1 promotes the progression and aerobic glycolysis of non-small cell lung cancer through targeting MiR-101-3p/KRAS Axis. Front. Genet. 2020;11:962. doi: 10.3389/fgene.2020.00962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Shi Q., Ji T., Ma Z., Tan Q., Liang J. Serum exosomes-based biomarker circ_0008928 regulates cisplatin sensitivity, tumor progression, and glycolysis metabolism by miR-488/HK2 axis in cisplatin-resistant nonsmall cell lung carcinoma. Cancer Biother. Radiopharm. 2021 doi: 10.1089/cbr.2020.4490. [DOI] [PubMed] [Google Scholar]
- 172.Ma J., Qi G., Li L. A novel serum exosomes-based biomarker hsa_circ_0002130 facilitates osimertinib-resistance in non-small cell lung cancer by sponging miR-498. Onco Targets Ther. 2020;13:5293–5307. doi: 10.2147/OTT.S243214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Pouyssegur J., Dayan F., Mazure N.M. Hypoxia signalling in cancer and approaches to enforce tumour regression. Nature. 2006;441:437–443. doi: 10.1038/nature04871. [DOI] [PubMed] [Google Scholar]
- 174.King H.W., Michael M.Z., Gleadle J.M. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer. 2012;12:421. doi: 10.1186/1471-2407-12-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Tan Z., Xue H., Sun Y., Zhang C., Song Y., Qi Y. The role of tumor inflammatory microenvironment in lung cancer. Front. Pharmacol. 2021;12:688625. doi: 10.3389/fphar.2021.688625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lei J., Chen P., Zhang F., Zhang N., Zhu J., Wang X., Jiang T. M2 macrophages-derived exosomal microRNA-501-3p promotes the progression of lung cancer via targeting WD repeat domain 82. Cancer Cell Int. 2021;21:91. doi: 10.1186/s12935-021-01783-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Wei K., Ma Z., Yang F., Zhao X., Jiang W., Pan C., Li Z., Pan X., He Z., Xu J., et al. M2 macrophage-derived exosomes promote lung adenocarcinoma progression by delivering miR-942. Cancer Lett. 2022;526:205–216. doi: 10.1016/j.canlet.2021.10.045. [DOI] [PubMed] [Google Scholar]
- 178.Li X., Chen Z., Ni Y., Bian C., Huang J., Chen L., Xie X., Wang J. Tumor-associated macrophages secret exosomal miR-155 and miR-196a-5p to promote metastasis of non-small-cell lung cancer. Transl. Lung Cancer Res. 2021;10:1338–1354. doi: 10.21037/tlcr-20-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Du C., Duan X., Yao X., Wan J., Cheng Y., Wang Y., Yan Y., Zhang L., Zhu L., Ni C., et al. Tumour-derived exosomal miR-3473b promotes lung tumour cell intrapulmonary colonization by activating the nuclear factor-kappaB of local fibroblasts. J. Cell. Mol. Med. 2020;24:7802–7813. doi: 10.1111/jcmm.15411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Zheng Q., Ding H., Wang L., Yan Y., Wan Y., Yi Y., Tao L., Zhu C. Circulating exosomal miR-96 as a novel biomarker for radioresistant non-small-cell lung cancer. J. Oncol. 2021;2021:5893981. doi: 10.1155/2021/5893981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Han F., Huang D., Huang X., Wang W., Yang S., Chen S. Exosomal microRNA-26b-5p down-regulates ATF2 to enhance radiosensitivity of lung adenocarcinoma cells. J. Cell. Mol. Med. 2020;24:7730–7742. doi: 10.1111/jcmm.15402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Chen L., Cao P., Huang C., Wu Q., Chen S., Chen F. Serum exosomal miR-7977 as a novel biomarker for lung adenocarcinoma. J. Cell. Biochem. 2020;121:3382–3391. doi: 10.1002/jcb.29612. [DOI] [PubMed] [Google Scholar]
- 183.Zhang Z., Tang Y., Song X., Xie L., Zhao S., Song X. Tumor-derived exosomal miRNAs as diagnostic biomarkers in non-small cell lung cancer. Front. Oncol. 2020;10:560025. doi: 10.3389/fonc.2020.560025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Nie H., Xie X., Zhang D., Zhou Y., Li B., Li F., Li F., Cheng Y., Mei H., Meng H., et al. Use of lung-specific exosomes for miRNA-126 delivery in non-small cell lung cancer. Nanoscale. 2020;12:877–887. doi: 10.1039/c9nr09011h. [DOI] [PubMed] [Google Scholar]
- 185.Roman-Canal B., Moiola C.P., Gatius S., Bonnin S., Ruiz-Miro M., Gonzalez E., Ojanguren A., Recuero J.L., Gil-Moreno A., Falcon-Perez J.M., et al. EV-associated miRNAs from pleural lavage as potential diagnostic biomarkers in lung cancer. Sci. Rep. 2019;9:15057. doi: 10.1038/s41598-019-51578-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Zhang Y., Zhang Y., Yin Y., Li S. Detection of circulating exosomal miR-17-5p serves as a novel non-invasive diagnostic marker for non-small cell lung cancer patients. Pathol. Res. Pract. 2019;215:152466. doi: 10.1016/j.prp.2019.152466. [DOI] [PubMed] [Google Scholar]
- 187.Zhang Z.J., Song X.G., Xie L., Wang K.Y., Tang Y.Y., Yu M., Feng X.D., Song X.R. Circulating serum exosomal miR-20b-5p and miR-3187-5p as efficient diagnostic biomarkers for early-stage non-small cell lung cancer. Exp. Biol. Med. (Maywood) 2020;245:1428–1436. doi: 10.1177/1535370220945987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Watabe S., Kikuchi Y., Morita S., Komura D., Numakura S., Kumagai-Togashi A., Watanabe M., Matsutani N., Kawamura M., Yasuda M., et al. Clinicopathological significance of microRNA-21 in extracellular vesicles of pleural lavage fluid of lung adenocarcinoma and its functions inducing the mesothelial to mesenchymal transition. Cancer Med. 2020;9:2879–2890. doi: 10.1002/cam4.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Dejima H., Iinuma H., Kanaoka R., Matsutani N., Kawamura M. Exosomal microRNA in plasma as a non-invasive biomarker for the recurrence of non-small cell lung cancer. Oncol. Lett. 2017;13:1256–1263. doi: 10.3892/ol.2017.5569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Kim J.E., Eom J.S., Kim W.Y., Jo E.J., Mok J., Lee K., Kim K.U., Park H.K., Lee M.K., Kim M.H. Diagnostic value of microRNAs derived from exosomes in bronchoalveolar lavage fluid of early-stage lung adenocarcinoma: a pilot study. Thorac. Cancer. 2018;9:911–915. doi: 10.1111/1759-7714.12756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Grimolizzi F., Monaco F., Leoni F., Bracci M., Staffolani S., Bersaglieri C., Gaetani S., Valentino M., Amati M., Rubini C., et al. Exosomal miR-126 as a circulating biomarker in non-small-cell lung cancer regulating cancer progression. Sci. Rep. 2017;7:15277. doi: 10.1038/s41598-017-15475-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Yuwen D.L., Sheng B.B., Liu J., Wenyu W., Shu Y.Q. MiR-146a-5p level in serum exosomes predicts therapeutic effect of cisplatin in non-small cell lung cancer. Eur. Rev. Med. Pharmacol. Sci. 2017;21:2650–2658. [PubMed] [Google Scholar]
- 193.Wu Y., Wei J., Zhang W., Xie M., Wang X., Xu J. Serum exosomal miR-1290 is a potential biomarker for lung adenocarcinoma. Onco Targets Ther. 2020;13:7809–7818. doi: 10.2147/OTT.S263934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Liu W., Liu J., Zhang Q., Wei L. Downregulation of serum exosomal miR-216b predicts unfavorable prognosis in patients with non-small cell lung cancer. Cancer Biomark. 2020;27:113–120. doi: 10.3233/CBM-190914. [DOI] [PubMed] [Google Scholar]
- 195.Han Z., Li Y., Zhang J., Guo C., Li Q., Zhang X., Lan Y., Gu W., Xing Z., Liang L., et al. Tumor-derived circulating exosomal miR-342-5p and miR-574-5p as promising diagnostic biomarkers for early-stage lung adenocarcinoma. Int. J. Med. Sci. 2020;17:1428–1438. doi: 10.7150/ijms.43500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Cazzoli R., Buttitta F., Di Nicola M., Malatesta S., Marchetti A., Rom W.N., Pass H.I. microRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J. Thorac. Oncol. 2013;8:1156–1162. doi: 10.1097/JTO.0b013e318299ac32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Wang X., Jiang X., Li J., Wang J., Binang H., Shi S., Duan W., Zhao Y., Zhang Y. Serum exosomal miR-1269a serves as a diagnostic marker and plays an oncogenic role in non-small cell lung cancer. Thorac. Cancer. 2020;11:3436–3447. doi: 10.1111/1759-7714.13644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Wang J., Zhao X., Wang Y., Ren F., Sun D., Yan Y., Kong X., Bu J., Liu M., Xu S. circRNA-002178 act as a ceRNA to promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis. 2020;11:32. doi: 10.1038/s41419-020-2230-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.He F., Zhong X., Lin Z., Lin J., Qiu M., Li X., Hu Z. Plasma exo-hsa_circRNA_0056616: a potential biomarker for lymph node metastasis in lung adenocarcinoma. J. Cancer. 2020;11:4037–4046. doi: 10.7150/jca.30360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Xian J., Su W., Liu L., Rao B., Lin M., Feng Y., Qiu F., Chen J., Zhou Q., Zhao Z., et al. Identification of three circular RNA cargoes in serum exosomes as diagnostic biomarkers of non-small-cell lung cancer in the Chinese population. J. Mol. Diagn. 2020;22:1096–1108. doi: 10.1016/j.jmoldx.2020.05.011. [DOI] [PubMed] [Google Scholar]
- 201.Li C., Lv Y., Shao C., Chen C., Zhang T., Wei Y., Fan H., Lv T., Liu H., Song Y. Tumor-derived exosomal lncRNA GAS5 as a biomarker for early-stage non-small-cell lung cancer diagnosis. J. Cell. Physiol. 2019;234:20721–20727. doi: 10.1002/jcp.28678. [DOI] [PubMed] [Google Scholar]
- 202.Tao Y., Tang Y., Yang Z., Wu F., Wang L., Yang L., Lei L., Jing Y., Jiang X., Jin H., et al. Exploration of serum exosomal LncRNA TBILA and AGAP2-AS1 as promising biomarkers for diagnosis of non-small cell lung cancer. Int. J. Biol. Sci. 2020;16:471–482. doi: 10.7150/ijbs.39123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Lai C.P., Mardini O., Ericsson M., Prabhakar S., Maguire C., Chen J.W., Tannous B.A., Breakefield X.O. Dynamic biodistribution of extracellular vesicles in vivo using a multimodal imaging reporter. ACS Nano. 2014;8:483–494. doi: 10.1021/nn404945r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Jeong K., Yu Y.J., You J.Y., Rhee W.J., Kim J.A. Exosome-mediated microRNA-497 delivery for anti-cancer therapy in a microfluidic 3D lung cancer model. Lab Chip. 2020;20:548–557. doi: 10.1039/c9lc00958b. [DOI] [PubMed] [Google Scholar]
- 205.Rashed M.H., Bayraktar E., Helal G.K., Abd-Ellah M.F., Amero P., Chavez-Reyes A., Rodriguez-Aguayo C. Exosomes: from garbage bins to promising therapeutic targets. Int. J. Mol. Sci. 2017;18:538. doi: 10.3390/ijms18030538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Yang X.X., Sun C., Wang L., Guo X.L. New insight into isolation, identification techniques and medical applications of exosomes. J. Control. Release. 2019;308:119–129. doi: 10.1016/j.jconrel.2019.07.021. [DOI] [PubMed] [Google Scholar]
- 207.Tiwari S., Kumar V., Randhawa S., Verma S.K. Preparation and characterization of extracellular vesicles. Am. J. Reprod. Immunol. 2021;85:e13367. doi: 10.1111/aji.13367. [DOI] [PubMed] [Google Scholar]
- 208.Sidhom K., Obi P.O., Saleem A. A review of exosomal isolation methods: is size exclusion chromatography the best option? Int. J. Mol. Sci. 2020;21:6466. doi: 10.3390/ijms21186466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Stranska R., Gysbrechts L., Wouters J., Vermeersch P., Bloch K., Dierickx D., Andrei G., Snoeck R. Comparison of membrane affinity-based method with size-exclusion chromatography for isolation of exosome-like vesicles from human plasma. J. Transl. Med. 2018;16:1. doi: 10.1186/s12967-017-1374-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Lobb R.J., Becker M., Wen S.W., Wong C.S., Wiegmans A.P., Leimgruber A., Moller A. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J. Extracell. Vesicles. 2015;4:27031. doi: 10.3402/jev.v4.27031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Konoshenko M.Y., Lekchnov E.A., Vlassov A.V., Laktionov P.P. Isolation of extracellular vesicles: general methodologies and latest trends. Biomed. Res. Int. 2018;2018:8545347. doi: 10.1155/2018/8545347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Keerthikumar S., Chisanga D., Ariyaratne D., Al Saffar H., Anand S., Zhao K., Samuel M., Pathan M., Jois M., Chilamkurti N., et al. ExoCarta: a web-based compendium of exosomal cargo. J. Mol. Biol. 2016;428:688–692. doi: 10.1016/j.jmb.2015.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Taylor D.D., Shah S. Methods of isolating extracellular vesicles impact down-stream analyses of their cargoes. Methods. 2015;87:3–10. doi: 10.1016/j.ymeth.2015.02.019. [DOI] [PubMed] [Google Scholar]
- 214.Contreras-Naranjo J.C., Wu H.J., Ugaz V.M. Microfluidics for exosome isolation and analysis: enabling liquid biopsy for personalized medicine. Lab Chip. 2017;17:3558–3577. doi: 10.1039/c7lc00592j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Davies R.T., Kim J., Jang S.C., Choi E.J., Gho Y.S., Park J. Microfluidic filtration system to isolate extracellular vesicles from blood. Lab Chip. 2012;12:5202–5210. doi: 10.1039/c2lc41006k. [DOI] [PubMed] [Google Scholar]
- 216.Lee K., Shao H., Weissleder R., Lee H. Acoustic purification of extracellular microvesicles. ACS Nano. 2015;9:2321–2327. doi: 10.1021/nn506538f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Szatanek R., Baj-Krzyworzeka M., Zimoch J., Lekka M., Siedlar M., Baran J. The methods of choice for extracellular vesicles (EVs) characterization. Int. J. Mol. Sci. 2017;18:1153. doi: 10.3390/ijms18061153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Deng J.H., Li Z.J., Wang Z.X., Feng J., Huang X.J., Zeng Z.M. Electron microscopy-based comparison and investigation of the morphology of exosomes derived from hepatocellular carcinoma cells isolated at different centrifugal speeds. Microsc. Microanal. 2020;26:310–318. doi: 10.1017/S1431927620000070. [DOI] [PubMed] [Google Scholar]
- 219.Thach R.E., Thach S.S. Damage to biological samples caused by the electron beam during electron microscopy. Biophys. J. 1971;11:204–210. doi: 10.1016/S0006-3495(71)86208-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Gurunathan S., Kang M.H., Jeyaraj M., Qasim M., Kim J.H. Review of the isolation, characterization, biological function, and multifarious therapeutic approaches of exosomes. Cells. 2019;8:307. doi: 10.3390/cells8040307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Lawrie A.S., Albanyan A., Cardigan R.A., Mackie I.J., Harrison P. Microparticle sizing by dynamic light scattering in fresh-frozen plasma. Vox Sang. 2009;96:206–212. doi: 10.1111/j.1423-0410.2008.01151.x. [DOI] [PubMed] [Google Scholar]
- 222.Gercel-Taylor C., Atay S., Tullis R.H., Kesimer M., Taylor D.D. Nanoparticle analysis of circulating cell-derived vesicles in ovarian cancer patients. Anal. Biochem. 2012;428:44–53. doi: 10.1016/j.ab.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 223.Wei H., Chen J., Wang S., Fu F., Zhu X., Wu C., Liu Z., Zhong G., Lin J. A nanodrug consisting of doxorubicin and exosome derived from mesenchymal stem cells for osteosarcoma treatment in vitro. Int. J. Nanomed. 2019;14:8603–8610. doi: 10.2147/IJN.S218988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Zhang W., Ou X., Wu X. Proteomics profiling of plasma exosomes in epithelial ovarian cancer: a potential role in the coagulation cascade, diagnosis and prognosis. Int. J. Oncol. 2019;54:1719–1733. doi: 10.3892/ijo.2019.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Marimpietri D., Petretto A., Raffaghello L., Pezzolo A., Gagliani C., Tacchetti C., Mauri P., Melioli G., Pistoia V. Proteome profiling of neuroblastoma-derived exosomes reveal the expression of proteins potentially involved in tumor progression. PLoS One. 2013;8:e75054. doi: 10.1371/journal.pone.0075054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Jo W., Jeong D., Kim J., Cho S., Jang S.C., Han C., Kang J.Y., Gho Y.S., Park J. Microfluidic fabrication of cell-derived nanovesicles as endogenous RNA carriers. Lab Chip. 2014;14:1261–1269. doi: 10.1039/c3lc50993a. [DOI] [PubMed] [Google Scholar]
- 227.Clogston J.D., Patri A.K. Zeta potential measurement. Methods Mol. Biol. 2011;697:63–70. doi: 10.1007/978-1-60327-198-1_6. [DOI] [PubMed] [Google Scholar]
- 228.Malla R.R., Pandrangi S., Kumari S., Gavara M.M., Badana A.K. Exosomal tetraspanins as regulators of cancer progression and metastasis and novel diagnostic markers. Asia Pac. J. Clin. Oncol. 2018;14:383–391. doi: 10.1111/ajco.12869. [DOI] [PubMed] [Google Scholar]
- 229.Zhou Y., Chen F., Xie X., Nie H., Lian S., Zhong C., Fu C., Shen W., Li B., Ye Y., et al. Tumor-derived exosome promotes metastasis via altering its phenotype and inclusions. J. Cancer. 2021;12:4240–4246. doi: 10.7150/jca.48043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Logozzi M., De Milito A., Lugini L., Borghi M., Calabro L., Spada M., Perdicchio M., Marino M.L., Federici C., Iessi E., et al. High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PLoS One. 2009;4:e5219. doi: 10.1371/journal.pone.0005219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Yoshioka Y., Kosaka N., Konishi Y., Ohta H., Okamoto H., Sonoda H., Nonaka R., Yamamoto H., Ishii H., Mori M., et al. Ultra-sensitive liquid biopsy of circulating extracellular vesicles using ExoScreen. Nat. Commun. 2014;5:3591. doi: 10.1038/ncomms4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Turchinovich A., Drapkina O., Tonevitsky A. Transcriptome of extracellular vesicles: state-of-the-art. Front. Immunol. 2019;10:202. doi: 10.3389/fimmu.2019.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Ghosh S., Bhowmik S., Majumdar S., Goswami A., Chakraborty J., Gupta S., Aggarwal S., Ray S., Chatterjee R., Bhattacharyya S., et al. The exosome encapsulated microRNAs as circulating diagnostic marker for hepatocellular carcinoma with low alpha-fetoprotein. Int. J. Cancer. 2020;147:2934–2947. doi: 10.1002/ijc.33111. [DOI] [PubMed] [Google Scholar]
- 234.Welton J.L., Khanna S., Giles P.J., Brennan P., Brewis I.A., Staffurth J., Mason M.D., Clayton A. Proteomics analysis of bladder cancer exosomes. Mol. Cell. Proteomics. 2010;9:1324–1338. doi: 10.1074/mcp.M000063-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Luo H.T., Zheng Y.Y., Tang J., Shao L.J., Mao Y.H., Yang W., Yang X.F., Li Y., Tian R.J., Li F.R. Dissecting the multi-omics atlas of the exosomes released by human lung adenocarcinoma stem-like cells. NPJ Genom. Med. 2021;6:48. doi: 10.1038/s41525-021-00217-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Zhao X., Li M., Dai X., Yang Y., Peng Y., Xu C., Dai N., Wang D. Downregulation of exosomal miR1273a increases cisplatin resistance of nonsmall cell lung cancer by upregulating the expression of syndecan binding protein. Oncol. Rep. 2020;44:2165–2173. doi: 10.3892/or.2020.7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Gardiner C., Di Vizio D., Sahoo S., Thery C., Witwer K.W., Wauben M., Hill A.F. Techniques used for the isolation and characterization of extracellular vesicles: results of a worldwide survey. J. Extracell. Vesicles. 2016;5:32945. doi: 10.3402/jev.v5.32945. [DOI] [PMC free article] [PubMed] [Google Scholar]