Abstract
Zinc finger protein 36 like 1 (ZFP36L1) enhances the turnover of mRNAs containing AU-rich elements (AREs) in their 3'-untranslated regions (3′UTR). The physiological and pathological functions of ZFP36L1 in liver, however, remain largely unknown. Liver-specific ZFP36L1-deficient (Zfp36l1flox/flox/Cre+; L1LKO) mice were generated to investigate the role of ZFP36L1 in liver physiology and pathology. Under normal conditions, the L1LKO mice and their littermate controls (Zfp36l1flox/flox/Cre−; L1FLX) appeared normal. When fed a Lieber-DeCarli liquid diet containing alcohol, L1LKO mice were significantly protected from developing alcohol-induced hepatic steatosis, injury, and inflammation compared with L1FLX mice. Most importantly, fibroblast growth factor 21 (Fgf21) mRNA was significantly increased in the livers of alcohol diet–fed L1LKO mice compared with the alcohol diet–fed L1FLX group. The Fgf21 mRNA contains three AREs in its 3′UTR, and Fgf21 3′UTR was directly regulated by ZFP36L1 in luciferase reporter assays. Steady-state levels of Fgf21 mRNA were significantly decreased by wild-type ZFP36L1, but not by a non-binding zinc finger ZFP36L1 mutant. Finally, wild-type ZFP36L1, but not the ZFP36L1 mutant, bound to the Fgf21 3′UTR ARE RNA probe. These results demonstrate that ZFP36L1 inactivation protects against alcohol-induced hepatic steatosis and liver injury and inflammation, possibly by stabilizing Fgf21 mRNA. These findings suggest that the modulation of ZFP36L1 may be beneficial in the prevention or treatment of human alcoholic liver disease.
The mouse zinc finger protein 36 (ZFP36) family consists of four RNA binding proteins: zinc finger protein 36, alias tristetraprolin (TTP), zinc finger protein 36 like 1 (ZFP36L1), zinc finger protein 36 like 2 (ZFP36L2), and zinc finger protein 36 like 3 (ZFP36L3). All four of these proteins regulate mRNA stability by binding to AU-rich elements (AREs) within the 3′-untranslated regions (3′UTRs) of target mRNAs, resulting in ARE-mediated mRNA degradation.1,2 Despite their common mode of biochemical action, the ZFP36 family members appear to play roles in different physiological processes. This is apparent from the fact that germline deletion of all four ZFP36 family members in mice has resulted in varied phenotypes.3, 4, 5, 6 ZFP36, or TTP, is the best-studied family member and plays an essential role in controlling inflammation.7,8 However, the roles of the other three members of the ZFP36 family remain poorly understood.
Recent studies have found that ZFP36L1 is involved in the regulation of mRNAs associated with metabolic functions. For example, ZFP36L1 has been shown to post-transcriptionally regulate the levels of the low-density lipoprotein receptor, a protein that is highly expressed in the liver and is a major determinant of circulating levels of low-density lipoprotein, linking ZFP36L1 to cholesterol metabolism.9 ZFP36L1 has also been shown to post-transcriptionally regulate uncoupling protein 1 (UCP1), which plays an important role in thermogenesis.10 Similarly, ZFP36L1 was recently shown to regulate liver bile acid metabolism.11 These findings indicate that ZFP36L1 may play critical roles in regulating liver metabolic homeostasis. However, the detailed physiological functions of ZFP36L1 remain particularly elusive, partly due to the embryonic lethal phenotype that is observed following germline deletion of ZFP36L1 in mice.4
This study sought to understand the physiological roles of ZFP36L1 in the liver. It was hypothesized that the removal of ZFP36L1 could alter certain pathological processes associated with liver. To test this, Cre-Lox technology was employed to delete Zfp36l1 in hepatocytes and cholangiocytes.12 Liver-specific ZFP36L1-deficient (L1LKO) mice and their littermate control mice (L1FLX) were then subjected to a model of alcoholic steatohepatitis.13 The mice were analyzed for histologic, biochemical, and transcriptional alterations associated with the development of alcoholic steatosis, liver injury, and inflammation. The results show a novel protective effect of liver-specific ablation of ZFP36L1 against alcohol-induced steatosis and liver inflammation, and identified Fgf21 mRNA as a likely target of ZFP36L1.
Materials and Methods
Mice
The floxed mice carrying loxP-flanked exon 2 of Zfp36l1 alleles were generated by gene targeting in C57BL/6 embryonic stem cells at Xenogen Biosciences (Cranbury, NJ). Liver-specific Zfp36l1 knockout (Zfp36l1flox/flox/Cre+; L1LKO) mice were generated by crossing the homozygous floxed mice (Zfp36lflox/flox) with albumin-Cre (Alb-Cre)14 mice (Jackson Laboratory, Bar Harbor, ME). Genotype status of the progeny was determined by PCR using primer 1 (P1), 5′-CTCTTGCTGGTCACTACCGTCGCT-3′ and primer 2 (P2), 5′-TCAATGTAAGCCAGCAAGTGCAGC-3′, which amplified a 484-bp endogenous Zfp36l1 wild-type (WT) allele and a 618-bp mutant Zfp36l1 floxed allele. PCR conditions were 93°C for 3 minutes followed by 30 cycles of 93°C for 30 seconds, 58°C for 30 seconds, and 72°C for 1 minute, with a final extension of 72°C for 10 minutes. Eight- to 12-week–old female L1LKO and littermate control (Zfp36l1flox/flox/Cre−; L1FLX) mice were used in all studies. All the animal experiments were performed in accordance with principles and procedures outlined in the NIH Guide for the Care and Use of Laboratory Animals15 and were approved by the Louisiana State University Animal Care and Use Committee.
Lieber-DeCarli Liquid Diet Model of Chronic-Plus-Binge Model of Alcoholic Liver Disease
Eight- to 12-week–old female L1LKO or L1FLX mice were subjected to alcoholic liver disease as described by Bertola et al.13 Briefly, mice were first acclimatized for 5 days to tube feeding and liquid diet ad libitum. Following acclimatization, mice were maintained for 10 days on a fresh liquid diet (free access) containing 5% ethanol (v/v) (Lieber-DeCarli ‘82 Shake and Pour ethanol liquid diet; product number F1258SP; Bio-Serv, Flemington, NJ) or pair-fed with a control isocaloric diet (Lieber-DeCarli ’82, Shake and Pour control liquid diet; product number F1259SP; Bio-Serv) in which the same number of calories was obtained from dextrin maltose. The freshly constituted liquid diet was stored in the refrigerator and used within 3 days. The refrigerated diet was brought to room temperature, and a calculated volume of fresh ethanol was added daily. On day 11, a single intragastric (oral gavage) administration of ethanol (5-g ethanol/kg body weight; 31.5% ethanol) or an isocaloric dose of maltose-dextrin (control group) was administered between 8 AM and 9 AM. Mice were humanely euthanized 9 hours after gavage feeding, and serum and liver were collected.
Necropsy and Tissue Collection
Mice were humanely euthanized, midline laparotomy was performed, and blood was then collected by cardiac puncture for serum. The livers were removed and weighed, and the left lateral lobe was dissected into three parts, of which the first part was stored in RNAlater (Life Technologies, Carlsbad, CA), the second part was snap-frozen and stored at −80°C, and the third part was fixed in 10% neutral buffered formalin. The remaining liver lobes were fixed in 10% neutral buffered formalin.
Histology
The 5-μm sections of liver were stained with hematoxylin and eosin for routine histologic analyses. Quantification of steatosis was performed by stereological point counting.16 Briefly, the histologic microscopic images were overlaid by a regular grid of points. The grid contained approximately 300 intersections within the tissue boundary that were counted as either fat deposits or no fat deposits depending on whether they covered the fat droplets or not. The fractional area of fat droplets was determined by dividing the number of points covering fat droplets to the total number of points.
Oil Red O Staining
Oil Red O staining was used to confirm hepatic steatosis. Briefly, freshly collected liver tissue was embedded in optimal cutting temperature compound (Sakura Finetek, Torrance, CA), frozen on dry ice, and then stored at −80°C. Later, 10 µm sections were obtained with a cryostat and fixed with 10% neutral buffered formalin, washed thoroughly with running tap water for 5 minutes, and then stained in Oil Red O solution (Electron Microscopy Sciences, Hatfield, PA) for 8 minutes followed by rinsing in 60% isopropanol and counterstaining with Harris hematoxylin for 1 minute. Sections were then mounted with glycerol jelly medium.
Immunohistochemistry
Immunohistochemical stainings for myeloperoxidase (MPO) and F4/80 were performed on formalin-fixed, paraffin-embedded liver sections. Briefly, liver sections were first deparaffinized with Citrisolv (Decon Labs Inc., King of Prussia, PA) (2 × 5 minutes each) and were rehydrated through descending grades of ethanol (100%, 95%, 70%, 30%, distilled water; 3 minutes each). Antigen retrieval was performed using a proteinase K–mediated antigen retrieval method. Endogenous peroxides were quenched with 3% hydrogen peroxide solution for 10 minutes at room temperature. Sections were blocked for 20 minutes with bocking serum followed by incubation in the primary antibodies [rabbit anti-mouse myeloperoxidase monoclonal antibody (ab208670; Abcam, Waltham, MA) and rat anti-mouse F4/80 antibody (MCA497G; Bio-Rad, Hercules, CA)] at room temperature for 1 hour. Sections were washed and incubated with respective biotinylated secondary antibodies for 1 hour at room temperature. The sections were then rinsed in deionized water (2 × 5 minutes each) and processed using VECTASTAIN Elite ABC HRP Kits (PK-6101/6104; Vector Laboratories, Burlingame, CA), followed by chromogenic substrate conversion using ImmPACT NovaRED HRP Substrate Kit (SK-4800; Vector Laboratories). Sections were counterstained with Gill’s Hematoxylin-I (EMD Millipore Corporation, Burlington, MA) for 30 seconds, rinsed in deionized water, dehydrated with ascending grades of ethanol, and coverslipped with VectaMount mounting media (H-5000; Vector Laboratories).
Liver Triglyceride Assay
Liver triglyceride levels were measured using Triglyceride Assay Kit (ab65336; Abcam) per the manufacturer’s recommendations. Briefly, 30 mg liver tissue samples were heated in 400 µL of 5% NP-40 solution (VWR chemicals, Solon, OH) at 90°C for 5 minutes to dissolve triglycerides. The heating cycles were repeated until a clear lysate was obtained by centrifugation. The triglyceride levels were then determined in liver lysates in duplicates and quantified at 570 nm using Spark Multimode Microplate Reader (Tecan Trading AG, Mannedorf, Switzerland). Liver triglyceride values were calculated and expressed as mg/g of the liver tissue.
Serum Enzymes and Lipids Analyses
Analyses for alanine transaminase (ALT), aspartate transaminase (AST), cholesterol, and triglycerides were performed on a Beckman Coulter AU680 chemistry analyzer using Beckman Coulter reagents (Beckman-Coulter, Irving, TX) at the clinical pathology core at Louisiana State University.
Enzyme-Linked Immunosorbent Assay
Serum levels of fibroblast growth factor (FGF21) were determined using an enzyme-linked immunosorbent assay kit (R&D systems, Minneapolis, MN) according to the manufacturer's instructions. The optical density was measured at 450 nm and 570 nm using Spark Multimode Microplate Reader (Tecan Trading AG, Mannedorf, Switzerland). The optical density readings at 570 nm were subtracted from those at 450 nm to correct for optical imperfections in the plate.
RNA Extraction
Total RNA was extracted using the GE Healthcare Illustra RNAspin MiniRNA isolation kit, according to the manufacturer's instructions (GE Healthcare, Little Chalfont, UK). The RNA content and purity were determined by measuring absorbance at 260 nm and 260/280 ratio, respectively, on a NanoDrop 8000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA). The quality of the RNA was determined using an Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA).
Quantitative PCR
RT-qPCR for the quantification of Tnf, Mcp1, Il10, and Cd36 transcripts was performed using the SYBR Green method. Briefly, 1 μg of total RNA was used for cDNA synthesis using an iScript cDNA synthesis kit (Bio-Rad). Quantitative real-time RT-PCR was performed on the ABI Prism 7900 Sequence Detection System (Applied Biosystems, Waltham, MA). Primer sequences were: Tnf, forward 5′-CTTCTGTCTACTGAACTTCGGG-3′, reverse 5′-CAGGCTTGTCACTCGAATTTTG-3; Mcp1, forward 5′-GTCCCTGTCATGCTTCTGG-3′, reverse 5′-GCTCTCCAGCCTACTCATTG-3′; Il10, forward 5′-AGCCGGGAAGACAATAACTG-3′, reverse 5′-GGAGTCGGTTAGCAGTATGTTG-3′; Cd36, forward 5′-GCGACATGATTAATGGCACAG-3′, reverse 5′-GATCCGAACACAGCGTAGATAG-3′.
The CT values were normalized to Gapdh internal controls. The expression levels were calculated according to the method. Quantitative PCR for Fgf21 and Zfp36l1 was performed using Taqman assays (Fgf21:hs00173927; Zfp36l1: hs00245183).
Library Preparation and RNA Deep Sequencing
Library preparation and RNA sequencing was performed at Novogene Corporation Inc. (Sacramento, CA). Briefly, 1 μg of RNA was used for cDNA library construction using an NEBNext Ultra II RNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, MA) according to the manufacturer's protocol. mRNA was enriched using oligo (dT) beads followed by two rounds of purification and random fragmentation. The first-strand cDNA was synthesized using random hexamers followed by generation of the second strand. After a series of terminal repair, polyadenylation, and sequencing adaptor ligation, the double-stranded cDNA library was completed following size selection and PCR enrichment. The resulting 250- to 350-bp insert libraries were quantified using a Qubit 2.0 fluorometer (Thermo Fisher Scientific) and quantitative PCR. Size distribution was analyzed using an Agilent 2100 Bioanalyzer (Agilent Technologies). Qualified libraries were sequenced on an Illumina NovaSeq 6000 Platform (Illumina, Inc., San Diego, CA) using a paired-end 150 run (2 × 150 bases). Approximately 20 M raw reads were generated from each library.
Immunoblotting
Cells/tissues were lysed using Pierce RIPA buffer (Thermo Fisher Scientific) supplemented with Pierce protease inhibitor mixture (Thermo Fisher Scientific) and phosphatase inhibitors (10 mM sodium fluoride and 1 mM sodium orthovanadate). Tissues were mechanically homogenized using a homogenizer (Fisherbrand 150 handheld homogenizer; Thermo Fisher Scientific). Tissue lysates were centrifuged (13,000 × g, 10 minutes, 4°C) to remove insoluble material, and protein concentrations of the supernatants were measured using the Bradford assay (Bio-Rad Laboratories). Denatured proteins were separated on a 4% to 12% Bis-Tris plus gel (Invitrogen, Carlsbad, CA), transferred onto polyvinylidene difluoride membranes (Invitrogen), and probed with a 1:1000 dilution of BRF1/2 primary antibody (Cell Signaling Technology, Danvers, MA), followed by a 1:3000 dilution of horseradish peroxidase–conjugated goat anti-rabbit IgG. Signals were determined either on a ChemiDoc Imaging System (Bio-Rad Laboratories) or on X-ray film.
Luciferase Reporter Assay
HEK293 cells were procured from ATCC (Manassas, VA). Cells were cultured in EMEM with 10% FBS. pcDNA 3.1+/C-(K)-DYK Zfp36l1, pcDNA 3.1+/C-(K)-DYK Zfp36l1-zinc finger mutant C135R, C173R (two amino-acid mutations within the two zinc fingers at positions 135 and 173), and pcDNA 3.1+/C-(K)-DYK empty vector were obtained from GenScript Biotech (Piscataway, NJ). Ambion pMIR-REPORT Luciferase expression reporter vector was obtained from Thermo Fisher Scientific, and the 3′UTR of Ffg21 mRNA was cloned into a pMIR-REPORT luciferase reporter vector. Transfections were done with Lipofectamine reagent (Invitrogen). Luminescence was determined using a Promega Luciferase Reporter Assay Kit (Promega, Madison, WI), according to the manufacturer's instructions, on a Spark multimode microplate reader (Tecan Trading AG, Mannedorf, Switzerland). Briefly, cells were seeded onto 6-well plates and transfected with the respective constructs (0.5 μg) when they reached a confluency of approximately 60% to 65%. At 48 hours after transfection, the growth medium was removed, and cells were gently rinsed with phosphate-buffered saline, followed by addition of 1× reporter lysis buffer to lyse the cells. To ensure complete lysis, cells were subjected to a single freeze–thaw cycle. Lysed cells were scraped, transferred into microcentrifuge tubes, and then centrifuged at 12,000 × g for 15 seconds at room temperature. The supernatants were transferred into new microcentrifuge tubes. A total of 20 μL of cell lysate was used per well of a 96-well plate for luciferase assays. The Spark multimode microplate reader was programed to add 100 μL of luciferase assay reagent per well with the injector. Luminescence was determined immediately (10 seconds). Luminescence was normalized to beta-galactosidase measurements.
Cotransfection Assay
For cotransfection assays, HEK293 cells were cotransfected with pcDNA 3.1+/C-(K)-DYK empty vector alone, the pcDNA 3.1+/C-(K)-DYK Zfp36l1-WT plasmid or the zinc finger mutant, pcDNA 3.1+/C-(K)-DYK Zfp36l1-C135R, C173R, and the full-length pcDNA 3.1+/C-(K)-DYK-Fgf21plasmid, which contained an intact 3′UTR (GenScript Biotech). Two concentrations of Zfp36l1 (WT and mutant) plasmids were used, 0.25 μg and 0.50 μg, whereas 0.5 μg of the Fgf21 plasmid was transfected in all. The amount of total transfected plasmid was kept constant for all the plates, and normalized by the addition of vector control plasmid. The cells were harvested after 48 hours, RNA was isolated, cDNA was prepared using iScript Reverse Transcription Supermix (Bio-Rad Laboratories), and then the levels of Fgf21 and Zfp36l1 mRNAs were assessed by quantitative real-time PCR.
Electrophoretic Mobility Shift Assays
RNA-ARE probes were designed from the Fgf21-3′UTR ARE sequence and synthesized with 5′-biotin–end labelling (Integrated DNA Technologies, Coralville, IA). HEK293 cells were transfected with the ZFP36L1 expression plasmid (pcDNA 3.1+/C-(K)-DYK Zfp36l1) or its zinc finger mutant (pcDNA 3.1+/C-(K)-DYK Zfp36l1-C135R, C173R), and cytosolic cell extracts were prepared. Briefly, the transfection mixture was removed, and cells were rinsed with ice-cold Ca2+- and Mg2+-free Dulbecco's phosphate-buffered saline. The cells were then scraped and centrifuged at 600 × g for 3 minutes at 4°C. The supernatants were removed, and the pellets were rinsed with ice-cold diethyl-pyrocarbonate–treated water containing 8 μg/mL leupeptin, 0.2 mM phenylmethylsulfonyl fluoride, and 1 μg/mL pepstatin A. The cells were then lysed in a hypotonic cell lysis buffer containing 10 mM HEPES (pH 7.6), 5 mM KCl, 5% (v/v) glycerol, 0.25% (v/v) Nonidet P-40, 1 μg/mL pepstatin A, 0.1 mM phenylmethylsulfonyl fluoride, and 8 μg/mL leupeptin. Lysates were centrifuged for 15 minutes at 15,000 × g at 4°C. The supernatant was transferred to a fresh tube, and potassium chloride and glycerol were added to final concentrations of 40 mM and 15% (v/v), respectively. Cell extracts were aliquoted and stored at −80°C until ready to use. For the electrophoretic mobility shift assay, approximately 8 μg of cytosolic extracts prepared from HEK293 cells transfected with either the vector alone, or with the WT or mutant ZFP36L1 expression constructs, were incubated with 300 mM 5′-biotin–end-labelled ARE probes at room temperature for 30 minutes. The total reaction volume was 20 μL and consisted of 10 mM HEPES (pH 7.6), 40 mM KCl, 2.5% (v/v) glycerol, 3 mM MgCl2, 2.5 μg/μL heparin, and 50 ng/μL yeast tRNA. Following incubation, the reaction mixture was loaded onto 6% to 8% nondenaturing acrylamide gels and subjected to electrophoresis (120 V for 60 minutes in 0.5× Tris/borate/EDTA buffer). Gels were transferred onto Biodyne B nylon membranes (0.45 μm; Thermo Fisher Scientific) in 0.5× Tris/borate/EDTA buffer (60 V for 30 minutes). RNA–protein complexes and the unbound RNA probe were detected using the Chemiluminescent Nucleic Acid Detection Module (Thermo Fisher Scientific), following the manufacturer's instructions.
Statistical Analysis
Statistical significance between two groups was determined by t-test assuming unequal variance. Significant differences among groups were determined by one-way analysis of variance followed by Tukey's post hoc test for multiple comparisons. All data are expressed as means ± SEM. P < 0.05 is considered statistically significant. Statistical analyses were performed using GraphPad Prism software version 7.0 (GraphPad Software, La Jolla, CA).
Nucleotide Sequence Accession Number
The RNA-Seq data have been deposited in the NCBI Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo; accession number GSE163444).
Results
Generation and Characterization of ZFP36L1 Liver-Specific Knockout Mice
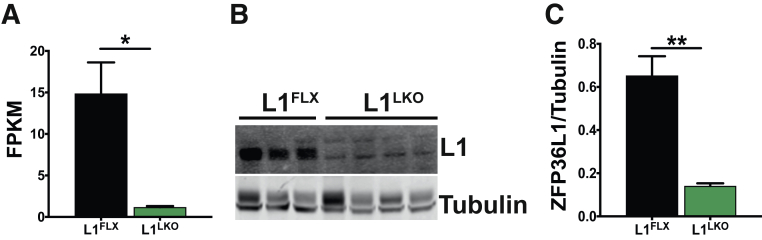
To investigate the role of ZFP36L1 in liver, liver-specific Zfp36l1-deficient mice (L1LKO) were generated using the Cre-LoxP recombination approach. RNA-Seq analysis of liver RNA showed at least a fivefold reduction in the mRNA levels of Zfp36l1 in L1LKO mice compared with littermate control L1FLX mice (Figure 1A). Western blot analysis of liver tissue lysates showed minimal to no detectable expression of ZFP36L1 in L1LKO mice compared with littermate control L1FLX mice (Figure 1B). Quantitatively, the expression of ZFP36L1 was significantly reduced in L1LKO mice compared with littermate control L1FLX mice (Figure 1C). The L1LKO mice developed normally and did not exhibit any growth defects or anatomic or histologic abnormalities (data not shown).
Figure 1.
mRNA and protein expression of ZFP36L1 in control (L1FLX) versus ZFP36L1 liver-specific knockout (L1LKO) mice. A: FPKM (fragments per kilobase of transcripts per million mapped reads) values from liver RNA-Seq data showing the expression of Zfp36l1 mRNA in liver homogenates from ZFP36L1-sufficient (L1FLX) and liver-specific ZFP36L1-deficient (L1LKO) mice. B: An immunoblot showing expression of ZFP36L1 in liver homogenates from ZFP36L1-sufficient (L1FLX) and liver-specific ZFP36L1-deficient (L1LKO) mice. C: Quantification of immunoblots shown in panel B was performed using ImageJ version 1.53i (NIH, Bethesda, MD; https://imagej.nih.gov/ij). Data are expressed as means ± SEM. n = 4 mice (A); n = 3 L1FLX mice (B); n = 4 L1LKO mice (B). ∗P < 0.05, ∗∗P < 0.01 (unpaired t-test).
Alcohol Diet–Fed L1LKO Mice Exhibit Significantly Attenuated Hepatic Steatosis, Injury, and Inflammation Compared with Alcohol Diet–Fed Littermate Control L1FLX Mice
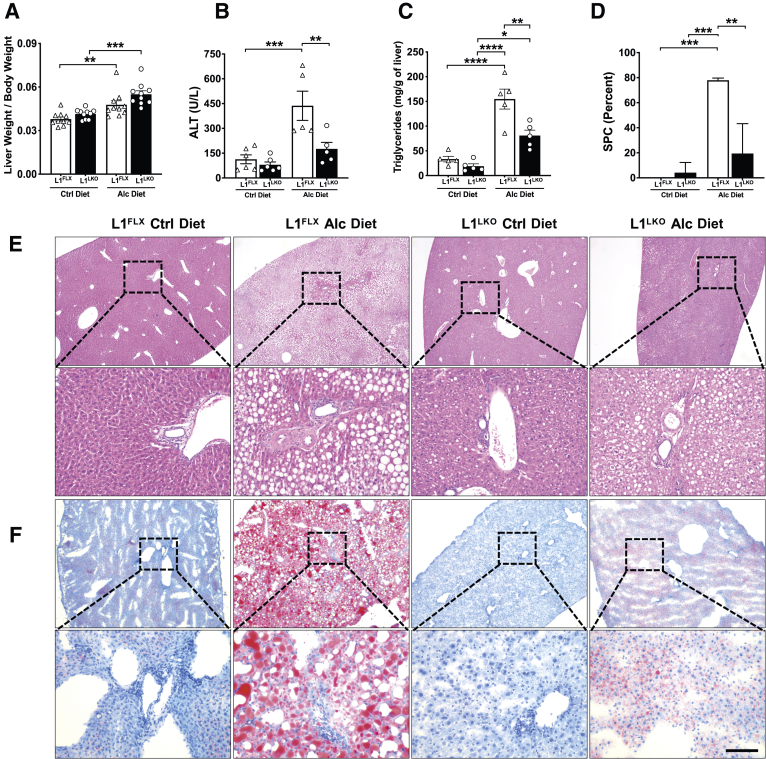
In order to determine the effect of ZFP36L1 deletion in stressed liver cells, L1FLX and L1LKO mice were subjected to alcohol-induced liver injury and steatosis by feeding them the alcohol-containing Lieber-DeCarli diet (alcohol group). The weights of both the L1FLX and L1LKO mice did not change significantly after alcohol ingestion over a period of 11 days compared with their initial body weights (Supplemental Figure S1A). Alcohol significantly increased the liver weight to body weight ratio after alcohol-diet feeding in both L1FLX and L1LKO mice, but the difference was not significant between the alcohol diet–fed L1FLX and L1LKO groups (Figure 2A).
Figure 2.
ZFP36L1 LKO mice are protected from alcohol-induced liver injury. A: Liver weight to body weight ratios of alcohol diet–fed ZFP36L1-sufficient (L1FLX) and liver-specific ZFP36L1-deficient (L1LKO) mice and those that were pair-fed with control diet. B: Serum levels of alanine transaminase (ALT) in alcohol diet–fed ZFP36L1-sufficient (L1FLX) and liver-specific ZFP36L1-deficient (L1LKO) mice and the control diet–fed ZFP36L1-sufficient (L1FLX) and liver-specific ZFP36L1-deficient (L1LKO). C: Liver triglyceride levels in alcohol diet–fed ZFP36L1-sufficient (L1FLX) and liver-specific ZFP36L1-deficient (L1LKO) mice and the control diet–fed ZFP36L1-sufficient (L1FLX) and liver-specific ZFP36L1-deficient (L1LKO) mice. D: Quantification of hepatic steatosis of images shown in E. E: Representative liver histology images from ZFP36L1-sufficient (L1FLX) and liver-specific ZFP36L1-deficient (L1LKO) mice that were fed with alcohol diet versus those that were pair-fed with control diet are shown. F: Oil Red O–stained liver sections from ZFP36L1-sufficient (L1FLX) and liver-specific ZFP36L1-deficient (L1LKO) mice that were fed with alcohol diet versus those that were pair-fed with control diet are shown. Representative images from three independent experiments are shown. Boxed areas in E and F are shown at higher magnification below. Data are expressed as means ± SEM. n = 10 mice in each group (A); n = 5 mice in the alcohol-diet group (B); n = 6 mice in the control diet group (B); n = 5 mice (C); n = 3–4 (D). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, and ∗∗∗∗P < 0.0001 (one-way analysis of variance followed by Tukey's correction for multiple comparisons). Scale bar = 50 μm. Alc, alcohol; Ctrl, control.
Alcohol results in hepatic lesions, particularly steatosis, hepatic injury, and hepatic inflammation.17 Serum levels of ALT, a marker of liver injury, were significantly increased in alcohol diet–fed L1FLX mice compared with control diet–fed (pair-fed) L1FLX mice. However, alcohol ingestion did not increase the levels of ALT in L1LKO mice (Figure 2B). The levels of AST were also increased in alcohol diet–fed mice, but the difference was not statistically significant between the L1FLX and L1LKO groups (Supplemental Figure S1B). Liver triglycerides were significantly increased in alcohol diet–fed L1FLX and L1LKO mice compared with their respective control diet–fed (pair-fed) counterparts. However, the levels of liver triglycerides were significantly less in alcohol diet–fed L1LKO compared with alcohol diet–fed L1FLX mice (Figure 2C). The serum levels of triglycerides and cholesterol were not significantly increased after alcohol diet feeding compared with control diet feeding in either of the two genotype groups (Supplemental Figure S1, C and D).
Steatosis is the earliest response of the liver to alcohol ingestion and is characterized by the accumulation of lipid droplets within the hepatocytes, starting at the centrilobular areas and progressing toward the mid-lobular and periportal regions in later stages.17,18 Consistent with these reports, the alcohol diet significantly increased microvesicular and macrovesicular steatosis in L1FLX mice compared with control diet–fed L1FLX mice. Remarkably, alcohol diet–fed L1LKO mice exhibited markedly diminished hepatic steatosis compared with the alcohol diet–fed L1FLX mice (Figure 2, D–F).
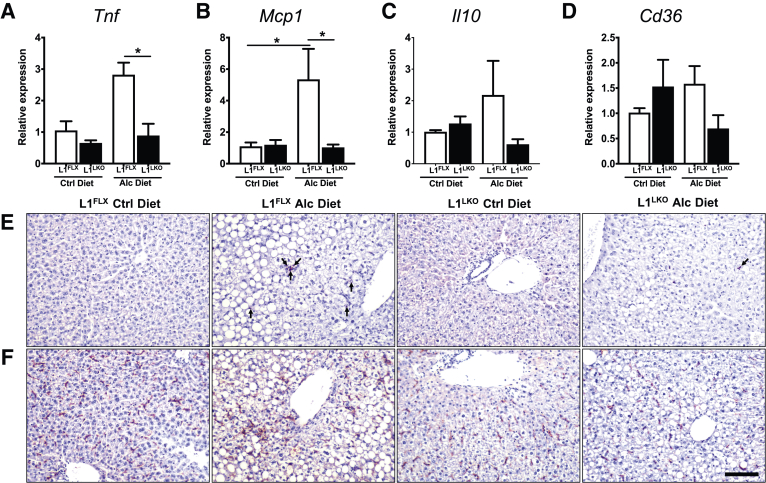
Alcohol-induced steatosis is usually accompanied by hepatic inflammation.17 Certain proinflammatory mediators, particularly tumor necrosis factor (TNF) and MCP1, have been implicated in alcoholic liver inflammation.19,20 Similarly, IL-10 and CD36, a fatty acid translocase, have also been shown to play a role in alcoholic liver disease.21,22 Therefore, mRNA expression levels for these proinflammatory cytokines and mediators were assessed in alcohol diet–fed L1FLX and L1LKO mice. The mRNA expression levels of both Tnf and Mcp1 were significantly increased in alcohol diet–fed L1FLX mice compared with control diet–fed L1FLX mice (Figure 3, A and B). However, the increase in expression of Tnf and Mcp1 was significantly attenuated in alcohol diet–fed L1LKO mice compared with alcohol diet–fed L1FLX mice (Figure 3, A and B). The mRNA expression levels of Il10 and Cd36 also showed similar trends, but the differences were not statistically significant between alcohol diet–fed L1LKO and alcohol diet–fed L1FLX mice (Figure 3, C and D). Increased inflammation in alcohol diet–fed L1FLX mice was also evident from increased staining for neutrophils [MPO-positive] in this group compared with control diet–fed L1FLX mice. The alcohol diet–fed L1LKO mice had fewer numbers of neutrophils in tissue sections compared with alcohol diet–fed L1FLX mice (Figure 3E). There were no differences in the numbers of F4/80-positive macrophages within the liver tissue sections (Figure 3F). Taken together, these data demonstrate that liver-specific deletion of ZFP36L1 confers significant protection against hepatic steatosis, hepatic injury, and hepatic inflammation in alcohol diet–fed mice.
Figure 3.
ZFP36L1 LKO mice were protected from alcohol-induced liver inflammation. Relative quantification of Tnf (A), Mcp1 (B), Il10 (C), and Cd36 (D) mRNA expression in the liver homogenates from alcohol diet–fed ZFP36L1-sufficient (L1FLX) and liver-specific ZFP36L1-deficient (L1LKO) mice, and those that were pair-fed with control diet. Representative photomicrographs of myeloperoxidase (MPO) (E) and F4/80 (F) stained liver sections from alcohol diet–fed ZFP36L1-sufficient (L1FLX) and liver-specific ZFP36L1-deficient (L1LKO) mice and those that were pair-fed with control diet. Arrows depict cells that stained positive for MPO. Data are expressed as means ± SEM. n = 3 to 4 mice. ∗P < 0.05 (one-way analysis of variance followed by Tukey's correction for multiple comparisons). Scale bar = 50 μm. Alc, alcohol; Cd36, cluster of differentiation 36; Ctrl, control; Il10, interleukin-10; Mcp1, monocyte chemoattractant protein-1; Tnf, tumor necrosis factor.
ZFP36L1 Deficiency Results in Reduced Numbers of Differentially Expressed Transcripts in Alcohol Diet–Fed Mice
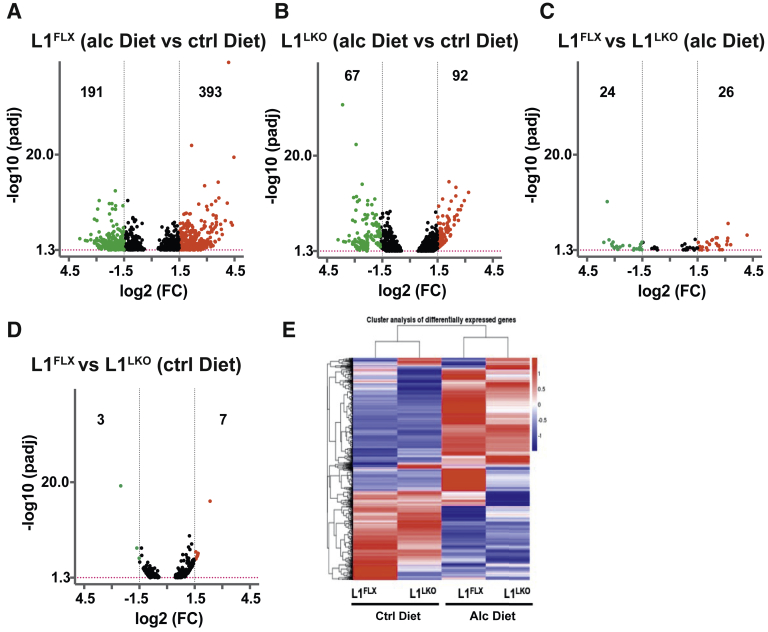
In order to identify transcriptomic changes through which ZFP36L1 modulates alcoholic hepatic steatosis, hepatic injury, and hepatic inflammation, RNA-Seq analysis of total liver RNA obtained from the four groups: control diet–fed and alcohol diet–fed L1FLX and L1LKO mice was performed. The RNA-Seq data are expressed as fragments per kilobase per million mapped reads. The inclusion criteria for increase or decrease were set as greater than or less than 1.5-fold (log2FC), with adjusted P < 0.05. Interestingly, although 584 transcripts were significantly differentially expressed (393 up-regulated; 191 down-regulated) in the alcohol diet–fed L1FLX mice compared with control diet–fed L1FLX mice, only 159 transcripts were differentially expressed (92 up-regulated; 67 down-regulated) in the alcohol diet–fed L1LKO mice compared with control diet–fed L1LKO mice (Figure 4, A and B). Fifty transcripts were differentially expressed (26 up-regulated; 24 down-regulated) between alcohol diet–fed L1LKO and alcohol diet–fed L1FLX mice (Figure 4C), whereas only 10 transcripts were differentially expressed (7 up-regulated; 3 down-regulated) between control diet–fed L1LKO and control diet–fed L1FLX mice (Figure 4D). Figure 4E shows a heat map comparing differentially expressed transcripts in the four groups. These data indicate that, in line with the increased steatosis, liver injury, and inflammation seen in the alcohol diet–fed L1FLX mice, there were larger numbers of transcriptomic changes, whereas minimal to no steatosis and significantly attenuated liver injury and inflammation were associated with comparatively lesser numbers of transcriptomic changes in alcohol diet–fed L1LKO mice.
Figure 4.
Differentially expressed transcripts in L1LKO mice and L1FLX mice following control diet and alcohol diet feeding. Volcano plots depict differentially expressed transcripts (red: up-regulated; green: down-regulated) in alcohol diet–fed versus control diet–fed L1FLX mice (A); alcohol diet–fed versus control diet–fed L1LKO mice (B); alcohol diet–fed L1FLX versus L1LKO mice (C); and control diet–fed L1FLX versus L1LKO mice (D). Black vertical dashed lines indicate a log2fold-change cutoff of 1.5. The red horizontal dashed line indicates −log10 (P value) cutoff of 1.3 (P = 0.05). Heat map of differentially expressed transcripts from alcohol diet–fed ZFP36L1-sufficient (L1FLX) and liver-specific ZFP36L1-deficient (L1LKO) mice, and control diet–fed ZFP36L1-sufficient (L1FLX) and liver-specific ZFP36L1-deficient (L1LKO) mice (E). Alc, alcohol; Ctrl, control; FC, fold change.
Next, the expression levels of known proinflammatory mRNAs between the two groups were compared to investigate whether the large numbers of gene expression changes in the alcohol diet–fed L1FLX versus alcohol diet–fed L1LKO group could be accounted for by proinflammatory mRNAs. A large proportion of the known inflammatory genes were significantly up-regulated in the L1FLX group (alcohol diet–fed L1FLX mice versus control diet–fed L1FLX mice), but not in the L1LKO group (alcohol diet–fed L1LKO mice versus control diet–fed L1LKO mice) (Table 1). Key inflammatory genes included toll-like 4-receptor Cd14, macrophage scavenger receptor, Marco, interleukin receptor, Il3ra, chemokines including Ccl2, Ccr1, Cxcl2, Cxcl14, and Ccrl2, TNF receptor superfamily proteins, Tnfrsf10b and Tnfrsf21, NFΚB family proteins Ikbip and Nfkbib, MAP kinase 4 (Mapk4), complement component, C3ar1, cell surface receptors, Cd34, Cd68, and Cd44, protein kinase substrate protein, Marcksl1, and metalloelastases, Mmp12 and Timp. Interestingly, few of the mRNAs that achieved statistical significance in the alcohol diet–fed L1LKO group were up-regulated by a comparatively lower-fold change when compared with the alcohol diet–fed L1FLX group. Feeding alcohol resulted in significantly increased liver inflammation in L1FLX mice, but not in L1LKO mice.
Table 1.
Inflammatory Genes
| Gene symbol | Gene name | L1FLX alcohol diet vs ctrl diet (log2FC) | L1FLX alcohol diet vs ctrl diet (padj) | L1LKO alcohol diet vs ctrl diet (log2FC) | L1LKO alcohol diet vs ctrl diet (padj) |
|---|---|---|---|---|---|
| TLR4 receptor | |||||
| Cd14 | CD14 antigen | 3.0 | 6.53−10 | 1.1 | NA |
| Macrophage receptor | |||||
| Marco | Macrophage receptor with collagenous structure | 1.6 | 1.81−05 | 0.1 | 0.89227 |
| Interleukin receptor | |||||
| Il3ra | Interleukin 3 receptor alpha chain | 2.4 | 0.01761 | -0.5 | 0.24085 |
| Chemokines | |||||
| Ccl2 | Chemokine (C-C_motif) ligand 2 | 3.0 | 1.76−05 | 0.6 | NA |
| Ccr1 | Chemokine (C-C_motif) receptor 1 | 2.9 | 0.00231 | 0.6 | 0.38265 |
| Cxcl2 | Chemokine (C-X-C_motif) ligand 2 | 1.9 | 0.04027 | 0.5 | NA |
| Cxcl14 | Chemokine (C-X-C_motif) ligand 14 | 1.5 | 0.01405 | 0.6 | 0.33444 |
| Ccrl2 | Chemokine (C-C_motif) receptor-like 2 | 1.5 | 0.00243 | 0.4 | 0.55244 |
| Ccl9 | Chemokine (C-C_motif) ligand 9 | 1.4 | 0.00053 | 0.8 | 0.05965 |
| TNF superfamily | |||||
| Tnfrsf12a | Tumor necrosis factor receptor superfamily member 12a | 2.5 | 2.12−05 | 1.8 | 0.00100 |
| Tnfrsf10b | Tumor necrosis factor receptor superfamily member 10b | 2.3 | 0.03836 | 0.8 | 0.25032 |
| Tnfrsf23 | Tumor necrosis factor receptor superfamily member 23 | 2.1 | 0.00451 | 1.2 | 0.03907 |
| Tnfrsf21 | Tumor necrosis factor receptor superfamily member 21 | 0.8 | 0.00636 | 0.2 | 0.76364 |
| Tnfaip1 | Tumor necrosis factor alpha-induced protein 1 (endothelial) | 0.8 | 8.79−06 | 0.6 | 0.08765 |
| NFkB family | |||||
| Relb | Avian reticuloendotheliosis viral (v-rel) oncogene related B | 1.4 | 0.00535 | 1.1 | 0.05787 |
| Rell1 | RELT-like 1 | 1.2 | 0.02623 | 1.1 | 0.00801 |
| Ikbip | IKBKB interacting protein | 1.0 | 0.00159 | 0.7 | 0.18132 |
| Nfkbib | Nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor beta | 0.6 | 0.04846 | 0.4 | 0.36545 |
| MAP kinase family | |||||
| Mapk4 | Mitogen activated protein kinase 4 | 2.2 | 0.01868 | 0.9 | 0.15484 |
| Map3k6 | Mitogen activated protein kinase kinase kinase 6 | 1.9 | 0.04403 | 0.9 | 0.09448 |
| Complement components | |||||
| C5ar1 | Complement component 5a receptor 1 | 2.7 | 4.26−06 | 1.5 | 0.00133 |
| C3ar1 | Complement component 3a receptor 1 | 1.5 | 0.00041 | 0.7 | 0.24271 |
| Cell-surface proteins | |||||
| Cd63 | CD63 antigen | 2.9 | 1.10−06 | 1.4 | 0.00210 |
| Cd34 | CD34 antigen | 1.3 | 0.03684 | 0.5 | 0.45291 |
| Cd68 | CD68 antigen | 1.1 | 0.01152 | -0.1 | 0.86209 |
| Cd44 | CD44 antigen | 0.9 | 0.03224 | 0.2 | 0.67557 |
| Protein kinase substrate protein | |||||
| Marcksl1 | MARCKS-like_1 | 0.7 | 0.00503 | 0.3 | 0.47187 |
| Metalloelastases | |||||
| Mmp12 | Matrix metallopeptidase_12 | 3.4 | 2.95−08 | 1.1 | NA |
| Timp1 | Tissue inhibitor of metalloproteinase 1 | 2.9 | 0.00165 | 0.6 | 0.44009 |
Differentially expressed inflammatory genes in alcohol-diet–fed ZFP36L1-sufficient (L1FLX) and liver-specific ZFP36L1-deficient (L1LKO) mice and control diet–fed ZFP36L1-sufficient (L1FLX) and liver-specific ZFP36L1-deficient (L1LKO) mice are shown. Bold values depict significant differences.
Comparison of Expression of Alcohol-Regulated Metabolic mRNAs in Alcohol Diet–Fed L1LKO versus the Alcohol Diet–Fed L1FLX Mice
Alcohol ingestion results in metabolic changes including increased lipogenesis, decreased fatty acid oxidation, mobilization of fat from various body deposits into the liver, and impaired very low-density lipoprotein secretion, all resulting in the development of hepatic steatosis.17 A large number of metabolic genes are involved in this process. To delineate the mechanisms for differential responses of the L1FLX versus the L1LKO mice to alcohol feeding, the RNA-Seq data set was queried for changes in known metabolic transcripts. Sirtuin 1 (Sirt1), fatty acid synthase (Fasn), stearoyl-coenzyme A desaturase 1 (Scd1), acyl-coenzyme A dehydrogenase family member 8 (Acad8), and cytochrome P450 family 2 subfamily e polypeptide 1 (Cyp2e1) were significantly down-regulated in alcohol diet–fed L1FLX group but were unchanged in the alcohol diet–fed L1LKO group, whereas Lipin-3 and lipoprotein lipase (Lpl) were significantly up-regulated in alcohol diet–fed L1FLX group but were unchanged in the alcohol diet–fed L1LKO group. Peroxisome proliferator activated receptor alpha (Ppara) and sterol regulatory element binding transcription factor 1 (Srebf1) were significantly down-regulated in both the L1FLX and L1LKO groups following alcohol ingestion, although the fold change was higher in the L1FLX group (Table 2).
Table 2.
Metabolic Genes
| Gene symbol | Gene name | L1FLX alcohol diet vs ctrl diet (log2FC) | L1FLX alcohol diet vs ctrl diet (padj) | L1LKO alcohol diet vs ctrl diet (log2FC) | L1LKO alcohol diet vs ctrl diet (padj) |
|---|---|---|---|---|---|
| Metabolic regulator genes | |||||
| Sirt1 | Sirtuin 1 | −0.93 | 0.01 | −0.08 | 0.88 |
| Sirt3 | Sirtuin 3 | −1.32 | 0.02 | −0.90 | 0.001 |
| Lipid metabolism | |||||
| Lpin1 | Lipin 1 | −1.12 | 0.07 | −1.28 | 0.01 |
| Lpin2 | Lipin 2 | −0.31 | 0.75 | −0.90 | 0.008 |
| Lpin3 | Lipin 3 | 1.33 | 0.02 | 0.25 | 0.67 |
| Lpl | Lipoprotein lipase | 2.3 | 0.001 | 0.3 | 0.55 |
| Lipogenesis | |||||
| Srebf1 | Sterol regulatory element binding transcription factor 1 | −2.28 | 5.99−05 | −1.68 | 6.15−07 |
| Fasn | Fatty acid synthase | −1.78 | 0.05 | −0.64 | 0.32 |
| Scd1 | Stearoyl coenzyme A desaturase 1 | −1.89 | 0.03 | −0.54 | 0.48 |
| Fatty acid oxidation | |||||
| Ppara | Peroxisome proliferator activated receptor alpha | −1.19 | 0.01 | −0.98 | 0.04 |
| Acad8 | Acyl-coenzyme A dehydrogenase family member 8 | −0.65 | 0.03 | −0.29 | 0.36 |
| Cpt1a | Carnitine palmitoyltransferase 1a liver | −0.39 | 0.71 | −0.94 | 0.01 |
| Cd36 | CD36 antigen | −0.02 | 0.98 | 0.27 | 0.75 |
| Fgf21 | Fibroblast growth factor 21 | 0.7 | 0.60 | 2.0 | 5.92−06 |
| Alcohol metabolism | |||||
| Aldh1a1 | Aldehyde dehydrogenase family 1 subfamily A1 | −0.75 | 0.28 | 0.21 | 0.68 |
| Cyp2e1 | Cytochrome P450 family 2 subfamily e polypeptide 1 | −1.54 | 0.003 | −0.74 | 0.14 |
Differentially expressed metabolic genes in alcohol-diet–fed ZFP36L1-sufficient (L1FLX) and liver-specific ZFP36L1-deficient (L1LKO) mice and control diet–fed ZFP36L1-sufficient (L1FLX) and liver-specific ZFP36L1-deficient (L1LKO) mice are shown. Bold values depict significant differences.
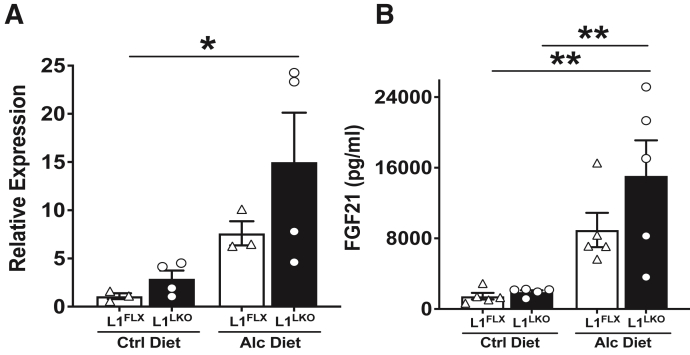
FGF21, a Peptide Metabolic Hormone, Is Significantly Increased in the Alcohol Diet–Fed L1LKO Group Compared with the Alcohol Diet–Fed L1FLX Group
FGF21 is known to be up-regulated following alcohol feeding and has been reported to have a protective role in alcoholic steatohepatitis, both in mice and humans.23,24 RNA-Seq analyses revealed that Fgf21 was significantly up-regulated by 4.2-fold (Log2FC = 2.0; padj = 5.92E06) in alcohol diet–fed L1LKO mice versus the control diet–fed L1LKO mice but was unchanged in alcohol diet–fed L1FLX mice versus the control diet–fed L1FLX (padj = 0.60) (Table 2). Consistent with the RNA-Seq data, quantitative PCR also indicated significant up-regulation of Fgf21 mRNA in the alcohol diet–fed L1LKO group (Figure 5A). Next, to test whether secretory levels of FGF21 in the serum were affected in alcohol diet–fed L1LKO mice, an enzyme-linked immunosorbent assay was performed on serum samples collected from alcohol diet–fed and control diet–fed L1FLX and L1LKO mice. The results showed that FGF21 was significantly increased in alcohol diet–fed L1LKO mice, but not in the alcohol diet–fed L1FLX mice (Figure 5B). These data indicated that Fgf21 may be regulated by ZFP36L1 in this model.
Figure 5.
FGF21 is significantly increased in alcohol diet–fed ZFP36L1 LKO mice. A: Relative expression of Fgf21 mRNA in the liver homogenate from alcohol diet–fed ZFP36L1-sufficient (L1FLX) and liver-specific ZFP36L1-deficient (L1LKO) mice and control diet–fed ZFP36L1-sufficient (L1FLX) and liver-specific ZFP36L1-deficient (L1LKO) mice. B: Secreted levels of FGF21 (picograms per milliliter) in the serum of alcohol diet–fed ZFP36L1-sufficient (L1FLX) and liver-specific ZFP36L1-deficient (L1LKO) mice, and control diet–fed ZFP36L1-sufficient (L1FLX) and liver-specific ZFP36L1-deficient (L1LKO) mice. Data are expressed as means ± SEM. n = 3 to 4 mice (A); n = 5 mice (B). ∗P < 0.05, ∗∗P < 0.01 (one-way analysis of variance followed by Tukey's correction for multiple comparisons). Alc, alcohol; Ctrl, control; FGF21, fibroblast growth factor 21.
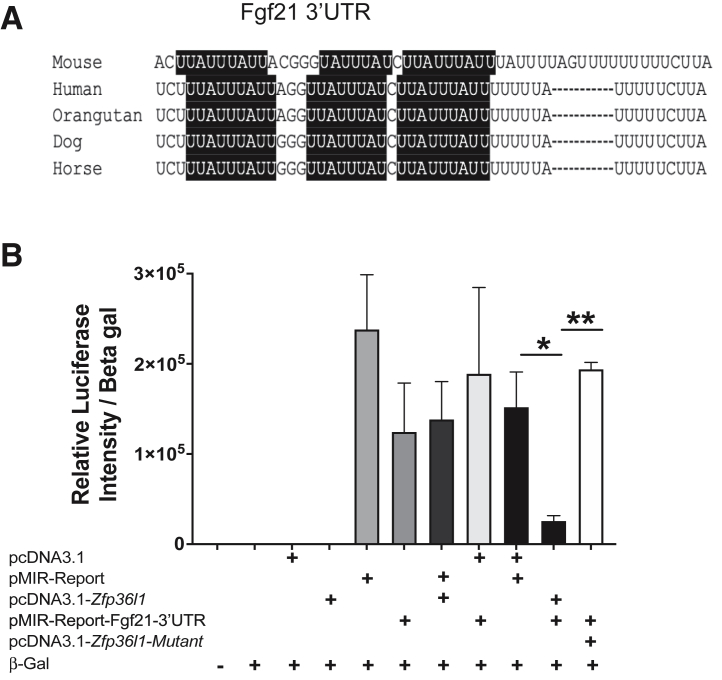
ZFP36L1 Regulates Fgf21 mRNA Turnover through AREs in Its 3′UTR
Because ZFP36 family members regulate their target mRNAs by binding to their 3′UTR AREs,2 whether Fgf21 mRNA contains AREs in its 3′UTR and whether those could be potential ZFP36L1 binding sites was investigated. As expected, the 3′UTR of Fgf21 mRNA contains at least three highly conserved potential ZFP36L1 binding sites (AREs) within its 3′UTR (Figure 6A). Next, to test whether ZFP36L1 regulates Fgf21 mRNA, the complete 3′UTR of Fgf21 mRNA was cloned (a total of 163 bp were cloned that also included 37 bp of the protein coding region) into the pMIR-REPORT luciferase reporter construct. Whether ZFP36L1 can regulate the stability of Fgf21 mRNA 3′UTR in luciferase reporter assays was tested. As shown in Figure 6B, the untransfected control (bar 1) or the transfection of beta-galactosidase alone (bar 2) did not result in detectable luciferase activity/luminescence. Similarly, transfection of an empty pcDNA3.1 vector alone (bar 3) or a plasmid expressing WT ZFP36L1 (pcDNA3.1-Zfp36l1) did not result in detectable luciferase activity. Luciferase activity was reported from cells transfected with the pMIR-REPORT (luciferase reporter expression vector) (bar 5), the pMIR-REPORT containing Fgf21-3′UTR (bar 6), a combination of pMIR-REPORT and pcDNA3.1 plasmid expressing WT ZFP36L1 (bar 7), a combination of pMIR-REPORT expressing Fgf21-3′UTR and pcDNA 3.1 empty vector (bar 8), and a combination of two empty plasmids (pMIR-REPORT + pcDNA 3.1) (bar 9). Luciferase activity was significantly inhibited (bar 10) when WT ZFP36L1 and the pMIR-Fgf21-3′UTR were present together, indicating that ZFP36L1 decreased the stability of Fgf21-3′UTR. In comparison to this, the luciferase activity was significantly rescued when the ZFP36L1 zinc finger mutant (C135R, C173R) was present (bar 11), indicating that the two intact zinc fingers are essentially required for this effect. These data show that ZFP36L1 directly regulates Fgf21-3′UTR and that this effect is mediated through the two zinc finger domains of ZFP36L1
Figure 6.
ZFP36L1 regulates the 3′UTR of Fgf21 mRNA. A: At least three evolutionarily conserved AU-rich elements (AREs) are present in the 3′-untranslated region (3′UTR) of the Fgf21 transcript. The AREs are highlighted in black. B: HEK293 cells were transfected with empty vector (pcDNA 3.1), luciferase reporter (pMIR-REPORT), WT-human ZFP36L1 expression plasmid (pcDNA 3.1-Zfp36l1), ZFP36L1 expression plasmid with C135R, C173R zinc finger mutation (pcDNA 3.1-Zfp36l1-mutant), and pMIR-REPORT vector expressing the 3′UTR of Fgf21 (pMIR-REPORT-Fgf21-3′UTR) in various combinations. Beta-galactosidase (Beta gal) was transfected as an internal control, and data are represented as relative luciferase activity normalized to beta-galactosidase activity. Bar 1: untransfected; bar 2: beta-galactosidase only; bar 3: pcDNA 3.1 empty vector only; bar 4: pcDNA 3.1-Zfp36l1-wt; bar 5; pMIR-REPORT empty vector only; bar 6: pMIR-REPORT-Fgf21-3′UTR-reporter; bar 7: pMIR-REPORT empty vector + pcDNA 3.1 Zfp36l1-wt; bar 8: pcDNA 3.1 empty vector + pMIR-REPORT Fgf21-3′UTR-reporter; bar 9: pcDNA 3.1 empty vector + pMIR-REPORT empty vector; bar 10: pcDNA 3.1 Zfp36l1-wt + pMIR-REPORT-Fgf21-3′UTR; bar 11: pcDNA 3.1 Zfp36l1-mutant + pMIR-REPORT Fgf21-3′UTR. Data are expressed as means ± SEM. n = 3. ∗P < 0.05, ∗∗P < 0.01 (one-way analysis of variance followed by Tukey's correction for multiple comparisons).
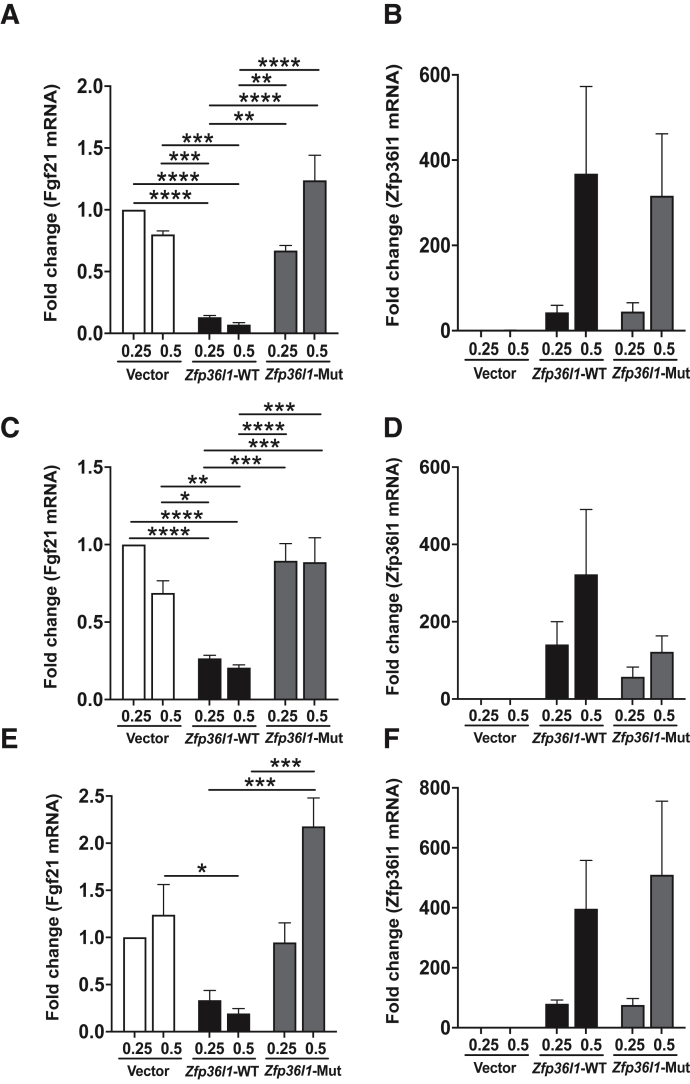
ZFP36L1 Promotes the Decay of Full-Length Fgf21 mRNA via the Zinc Finger Motif
The ability of ZFP36L1 to destabilize a full-length transcript of Fgf21 through cotransfection assays in HEK293 cells was tested next. When a full-length Fgf21-mRNA was coexpressed with ZFP36L1-WT protein, the steady-state levels of Fgf21 mRNA were decreased at both 0.25 μg and 0.5 μg concentrations of ZFP36L1 (Figure 7, A, C, and E). This effect was statistically significant at both 12 and 24 hours for 0.5 and 0.25 μg (Figure 7, A and C) and at 48 hours for 0.5 μg (Figure 7E). However, when a full-length Fgf21-mRNA was coexpressed with zinc finger ZFP36L1-mutant (C135R, C173R), the levels of Fgf21 mRNA did not decrease at either the low (0.25 μg) or the high (0.5 μg) concentration of ZFP36L1-mutant plasmid (Figure 7, A, C, and E). Figures 7, B, D, and F show the dose-dependent increase in the expression of ZFP36L1-WT and ZFP36L1-mutant in this experiment at 12 (Figure 7B), 24 (Figure 7D), and 48 (Figure 7F) hours, respectively. This assay suggested that ZFP36L1 promotes the decay of Fgf21 mRNA and that the zinc finger motif of ZFP36L1 is required for this effect.
Figure 7.
ZFP36L1 reduces the steady state levels of Fgf21 mRNA. Wild-type Zfp36l1 or its non-binding mutant (zinc finger mutant) plasmids were cotransfected with plasmid expressing full-length Fgf21 mRNA in HEK293 cells. A, C, and E: Relative expression of Fgf21 mRNA 12 hours (A), 24 hours (C), and 48 hours (E) post-transfection. Bars 1 and 2 show Fgf21 mRNA expression following the transfection of pcDNA 3.1 empty vector at 0.25 and 0.50 μg, respectively; bars 3 and 4 show Fgf21 mRNA expression following the transfection of wild-type Zfp36l1 at 0.25 and 0.50 μg, respectively; and bars 5 and 6 show Fgf21 mRNA expression following the transfection of zinc finger mutant Zfp36l1 at 0.25 and 0.50 μg, respectively. B, D, and F: Relative expression of Zfp36l1 in HEK293 cells transfected with pcDNA 3.1 empty vector, wild-type Zfp36l1 expression plasmid, and zinc finger mutant Zfp36l1 expression plasmid at 12 hours (B), 24 hours (D), and 48 hours (F) post-transfection. Data are expressed as means ± SEM. n = 4 independent experiments. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, and ∗∗∗∗P < 0.0001 (one-way analysis of variance followed by Tukey's correction for multiple comparisons). Mut, mutant; WT, wild-type.
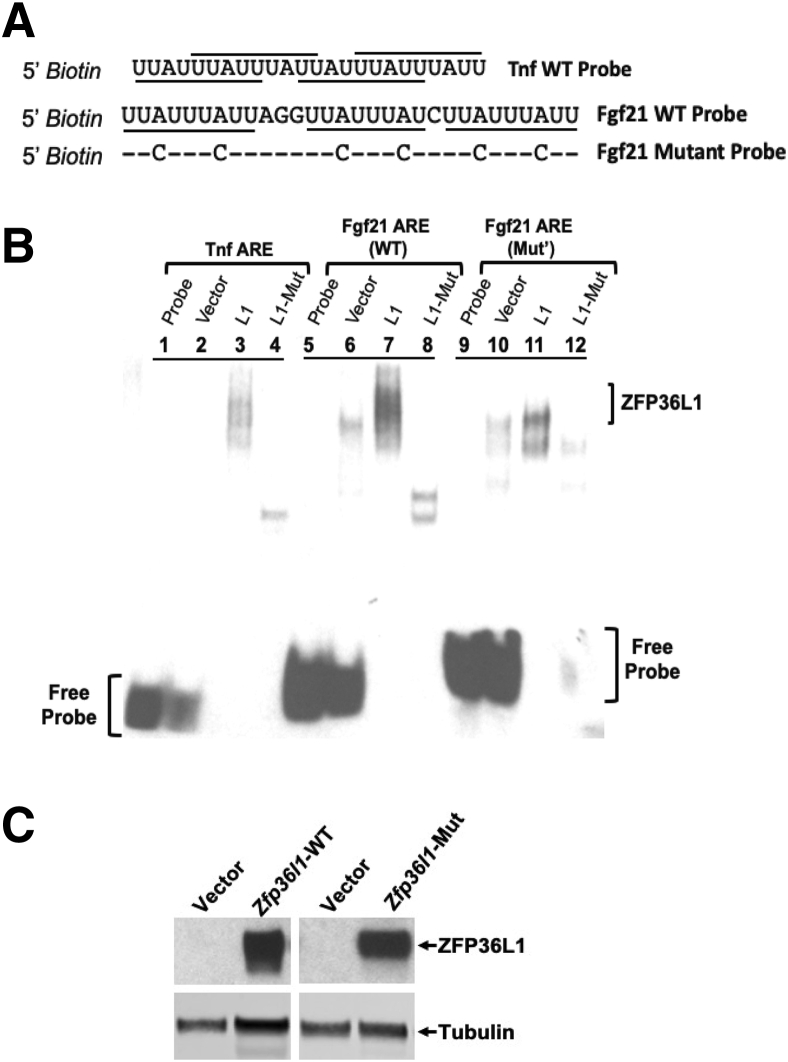
ZFP36L1 Binds to Fgf21-mRNA 3′UTR AU-Rich Region
Finally, whether ZFP36L1 expressed in HEK293 cells can directly bind to AREs within the 3′UTR of Fgf21 mRNA was tested. Biotin 5′-end–labelled RNA probes designed from the 3′UTR of Fgf21 mRNA were synthesized. The probes consisted of a WT Fgf21-ARE and a mutant Fgf21-ARE in which all the six core A residues were mutated to C residues (Figure 8A). Mutation of core A residues to C residues has been previously shown to abrogate the binding of TTP to its target mRNA probes.25 A Tnf 3′UTR-ARE probe was used as a positive control. Cell lysates expressing WT ZFP36L1 or zinc finger mutant ZFP36L1 (C135R, C173R) were incubated with WT Tnf, WT Fgf21, or mutated-Fgf21 mRNA ARE probes, and RNA mobility shift assays were performed. As shown in Figure 8B, cytosolic extracts from cells expressing vector alone (lane 2) did not bind Tnf-ARE. The migration position of the free probe is depicted in lane 1. Tnf-ARE was able to bind to ZFP36L1, as is evident from the slower migration of the probe (lane 3). The Tnf-ARE probe was also able to bind to a small amount of zinc finger mutant-ZFP36L1, but with greatly reduced ability (lane 4) compared with WT ZFP36L1 (lane 3), as is evident from the increased migration (downward shifting of the band) in this case (lane 4). When the WT Fgf21-ARE probe was used, significant binding was evident with the WT ZFP36L1 protein (lane 7). The bound Fgf21-ZFP36L1 complex exhibited significantly reduced migration in the gel, indicating good binding. The binding also appeared to be much better than was observed with the Tnf-ARE, as is evident from the larger size and increased intensity of the band (lane 7). Again, the zinc finger mutant-ZFP36L1 was still able to bind the WT Fgf21-ARE but with markedly reduced ability (lane 8). The position of the WT Fgf21-ARE free probe is shown in lane 5. Lane 6 shows the vector control. Finally, as shown in lane 11, the mutant-Fgf21-ARE probe was able to bind to the WT ZFP36L1 protein; however, its ability to bind was much less than that of the WT Fgf21-ARE probe (lane 7). This is clear from increased migration (downward shifting of the band) and reduced intensity and splitting of the band (lane 11). The mutant-Fgf21-ARE probe was, however, not able to bind to the mutant-ZFP36L1 (lane 12). The position of the mutant-Fgf21-ARE free probe is shown in lane 9. Lane 10 shows the vector control. Figure 8C shows equivalent expression of WT and mutated ZFP36L1 in HEK293 cells.
Figure 8.
ZFP36L1 binds to the ARE from Fgf21 mRNA in electrophoretic mobility shift assay. RNA electrophoretic gel-shift mobility assay with ZFP36L1-WT, ZFP36L1-mutant, WT-Fgf21-ARE, and Mutant-Fgf21-ARE. A: Sequences of 5′-biotin–labelled RNA probes corresponding to the WT Tnf ARE (https://www.ncbi.nlm.nih.gov/nuccore/X02611; accession number X02611), WT Fgf21 ARE (https://www.ncbi.nlm.nih.gov/nuccore/NM_019113.4; accession number NM_019113.4), and mutant human Fgf21 ARE in which six A residues were replaced by C residues. The ARE binding sites (octamer and nonamers) are underlined. The hyphens indicate the sequence identify at that position to the WT-Fgf21 ARE probe. B: HEK293 cells were transfected with vector alone (pcDNA 3.1), WT-human ZFP36L1 expression construct (pcDNA 3.1-Zfp36l1), or the human ZFP36L1 expression construct with C135R, C173R zinc finger mutation (pcDNA 3.1-Zfp36l1-mutant). Equal amounts of these cytosolic extracts were combined with a probe derived from Tnf ARE (lanes 1 to 4), WT-Fgf21 ARE (lanes 5 to 8), and Mutant-Fgf21 ARE (lanes 9 to 12), and gel-shift assay was performed. Lanes 1 and 2 show the position of the free Tnf probe when either free probe only (lane 1) was used or the probe was combined with empty vector-expressing lysates (lane 2). A large complex was formed with the WT-ZFP36L1 (lane 3), but not with the mutant-ZFP36L1 (lane 4). Lanes 5 (probe alone) and 6 (probe with empty vector) show the position of the free Fgf21-WT probe. The amount of probe and the migration position of the complex formed were shifted with WT-ZFP36L1 (lane 7), but not with the mutant-ZFP36L1 (lane 8). Lanes 9 (probe alone) and 10 (probe with empty vector) show the position of the free Fgf21-mutant probe. The amount of probe and the migration positions of the complex formed were partially shifted with WT-ZFP36L1 (lane 11), but not with the mutant-ZFP36L1 (lane 12). C: Immunoblot of the cytosolic extracts from cells transfected with vector (pcDNA 3.1) alone, wild-type human ZFP36L1, or the zinc finger mutant (C135R, C173R) form of ZFP36L1 that were used in the RNA electrophoretic mobility shift assay. The position of ZFP36L1 is indicated to the right. The bottom gels show the loading control, tubulin. ARE, AU-rich element; Mut, mutant; WT, wild-type.
Discussion
Excessive alcohol consumption is a global problem with enormous clinical and economic consequences.26 Liver, the primary site of alcohol metabolism, sustains the greatest tissue injury following alcohol ingestion. However, the pathogenic mechanisms involved in alcoholic hepatic steatosis, injury, and inflammation remain incompletely understood. The current study describes an interesting role of a post-transcriptional regulator of gene expression, that is, ZFP36L1, in the pathogenesis of alcohol-induced changes in the liver.
Liver-specific ablation of ZFP36L1 (L1LKO mice) resulted in significant reduction in hepatic steatosis, liver injury, and inflammation in response to alcohol when compared with mice that contained an intact Zfp36l1 gene (L1FLX mice). These findings suggest that ZFP36L1 promotes instability of mRNAs that confer protection against alcohol-induced liver injury. Along similar lines, liver-specific loss of ZFP36L1 was shown to confer protection against diet-induced obesity and steatosis by altering bile acid metabolism through regulation of cholesterol 7α−hydroxylase (Cyp7a1) mRNA.11 Loss of ZFP36L1 was also shown to protect against the development of experimental osteoarthritis through the post-transcriptional regulation of heat shock protein 70 family members.27 Finally, a recent study showed that ZFP36L1 is required for the maintenance of the marginal zone B-cell compartment, and this effect is dependent on its regulation of the transcription factors KLF2 and IRF8.28 Together, these studies indicate that ZFP36L1 regulates a variety of physiological and pathological responses through its mRNA targets.
Alcoholic hepatitis is a serious consequence of alcoholic liver disease and is accompanied by increased expression of proinflammatory mediators that recruit inflammatory cells into the liver. RNA-sequencing revealed that a large number of inflammatory mRNAs were significantly up-regulated in L1FLX mice, but not in the L1LKO mice, after alcohol administration. For instance, TNFα levels are increased in both human alcoholic liver and in animal models of liver disease.29 Tnf mRNA was significantly up-regulated in L1FLX, but not in the L1LKO group, after alcohol challenge. MCP1, also known as CCL2, levels are increased in the plasma of patients with alcoholic liver disease and have been correlated with liver neutrophil infiltrates after alcohol.30 Further, deficiency of MCP1 has been shown to protect against alcoholic liver injury.19 Mcp1/Ccl2 was significantly up-regulated in the L1FLX, but not in the L1LKO mice, after alcohol administration. Cd14 (a coreceptor for toll-like receptor 4) was specifically up-regulated in the alcohol diet–fed L1FLX group but was unchanged in the alcohol diet–fed L1LKO group. CD14-deficient mice have been shown to exhibit reduced early alcohol-induced liver injury.31 Liver macrophages and monocytes from alcoholic hepatitis patients show increased activation of NFκB.32 Ikbip and Nfkbib, two members of NFκB family, were significantly up-regulated in the alcohol diet–fed L1FLX group but were unchanged in the alcohol diet–fed L1LKO group. Similarly, Mapk4, a member of the MAP kinase signaling family, is up-regulated following alcohol exposure, both in vitro and in vivo.32 Although Mapk4 was significantly up-regulated in the alcohol diet–fed L1FLX group, its expression levels remained unchanged in the alcohol diet–fed L1LKO group. Finally, Mmp12, a metalloprotease involved in extracellular remodeling,33 was significantly up-regulated in the alcohol diet–fed L1FLX group, but not in the alcohol diet–fed L1LKO group. Of note, Mmp12 has been shown to be up-regulated in primary murine macrophages following alcohol exposure.34 These outcomes suggest that the ZFP36L1-deficient liver cells in alcohol-challenged L1LKO mice exhibit mitigated cellular injury that in turn prevents the pronounced inflammation, a response evident in alcohol-challenged L1FLX mice. Second, we speculate that other members of this protein family, particularly ZFP36/TTP, can regulate inflammation, whereas ZFP36L1 regulates metabolic genes within the liver. These possibilities, however, need to be tested further.
FGF21, a liver-specific metabolic hormone, is known to be elevated in the blood of both mice and humans after acute or binge alcohol ingestion.23,35 Interestingly, FGF21 was significantly up-regulated in the alcohol diet–fed L1LKO group, but not in the alcohol diet–fed L1FLX group, suggesting that ZFP36L1 directly or indirectly regulates the expression of Fgf21. This was also indicated by the finding that PPARα, the transcriptional regulator of FGF21, was significantly down-regulated in both the alcohol diet–fed L1FLX and the L1LKO groups. Accordingly, further experiments were conducted to determine whether Fgf21 is a direct target of ZFP36L1. First, luciferase activity assays were used to demonstrate that ZFP36L1 can destabilize Fgf21-3′UTR. Second, cotransfection assays were used to demonstrate that ZFP36L1 can destabilize the full-length Fgf21 mRNA. Third, electrophoretic mobility shift assays were used to demonstrate that ZFP36L1 can directly bind to the 3′UTR ARE region of Fgf21 mRNA. Finally, this effect was shown to be mediated by the two tandem zinc finger domains of ZFP36L1, as indicated by the reappearance of the luciferase activity and the stabilization of Fgf21 mRNA when a zinc finger mutant of ZFP36L1 replaced the WT-ZFP36L1 in the luciferase experiment and the co-expression assays, respectively. These experiments demonstrated that Fgf21 mRNA can be a target of ZFP36L1.
FGF21 has been previously shown to be a target of ZFP36/TTP, another member of the ZFP36 family of RNA binding proteins. For instance, Sawicki et al36 demonstrated that Fgf21 mRNA is post-transcriptionally regulated by ZFP36/TTP, and that liver-specific loss of ZFP36/TTP improves systemic glucose tolerance and insulin sensitivity in mice fed a high-fat diet by up-regulating FGF21. Another study recently demonstrated that CCR4-NOT deadenylase-ZFP36/TTP complex promotes degradation of Fgf21 mRNA, and that ablation of CCR4-NOT deadenylase activity decreases the susceptibility of mice to metabolic disorders, including obesity and steatosis, by increasing FGF21 levels.37 The current data provide evidence of redundancy between ZFP36/TTP and ZFP36L1 in the post-transcriptional regulation of Fgf21 mRNA. However, the relative contribution of ZFP36/TTP versus ZFP36L1 in the post-transcriptional regulation of Fgf21 mRNA remains unexplored and warrants further investigation.
FGF21 has been shown to confer protection from alcohol-induced hepatic steatosis and liver injury in previous studies. In fact, chronic alcohol feeding in the absence of FGF21 results in significant liver injury in mice.23 Mice lacking FGF21 have been shown to exhibit exacerbated alcohol-induced liver steatosis and liver injury through activation of genes involved in lipogenesis and repression of genes involved in beta oxidation of fatty acids.24 Conversely, administration of recombinant FGF21 to WT mice that were chronically fed alcohol was shown to significantly attenuate hepatic steatosis and liver injury.24 Consistent with these reports, up-regulation of FGF21 was shown to align with the hepatoprotective effect in ZFP36L1 liver-specific knockout mice. However, whether the hepatoprotective effect of ZFP36L1 deletion is mediated through the up-regulated expression of FGF21 alone or through the up-regulation and down-regulation of a battery of anti-inflammatory and proinflammatory genes, respectively, remains to be tested.
Previous studies with the ZFP36 RNA binding protein family have suggested that a number of mRNA targets of these proteins are cell-specific. For instance, Tnf has been identified as a myeloid-cell–specific target of ZFP36/TTP.38 Similarly, Notch 1 was identified as a thymocyte-specific target of both ZFP36L1 and ZFP36L2.39 The current study indicates that Fgf21 mRNA is a target of ZFP36L1 in hepatocytes/cholangiocytes. However, it remains unclear whether this relationship is specific to one or both the cell types. Further, whether the deletion of ZFP36L1 in other cell types such as immune cells and hepatic stellate cells within the liver will also result in the up-regulation of Fgf21 also warrants further investigation.
Previous studies with ZFP36 RNA binding protein family have also indicated stimulus specificity for their mRNA targets. For example, myeloid-specific loss of ZFP36/TTP did not result in any abnormalities under basal conditions.40 However, these mice were hypersensitive to lipopolysaccharide challenge that resulted from the stabilization and several-fold up-regulation of the ZFP36/TTP target, Tnf.40 Similarly, TTP deletion in keratinocytes resulted in hypersensitivity to imiquimod-induced skin inflammation in these mice, and this effect was mediated by keratinocyte-specific stabilization and up-regulation of Tnf.41 In a recent study, genetic ablation or silencing of ZFP36L1 significantly protected mice from experimental osteoarthritis by stabilizing members of the heat shock protein 70 family, again indicating stimulus-specific targets and resulting in pathological outcomes.27 Along these lines, Fgf21 also appears to be a stimulus-specific physiologically relevant target of ZFP36L1.
Collectively, the current study reveals a novel post-trans-criptional mechanism of regulation of FGF21 production during alcohol-induced hepatic disease. First, ZFP36L1 deletion in the liver resulted in significant reductions in hepatic steatosis, liver injury, and inflammation in response to alcohol. Second, biochemical studies demonstrated that ZFP36L1 regulates the expression of FGF21, following basic mechanism through which TTP family members operate to regulate their target mRNAs. Of note, FGF21 is considered as a potential treatment of metabolic disease; therefore, various FGF21 analogs have been developed and are undergoing clinical trials for metabolic disorders, including diabetes and obesity.42 The current study suggests that the hepatic ZFP36L1-FGF21 axis is a major signaling route in alcohol-induced hepatic steatosis in mice. This opens up the possibility of silencing ZFP36L1 as a possible method of protection against alcoholic-liver disease.
Acknowledgment
We thank Sherry Ring for histologic tissue processing.
Footnotes
Supported by Louisiana State University (LSU) COBRE (NIH/NIGMS grant # 5P30GM110760 and P20GM130555) and LSUSVM Startup funds (S.P.), and in part by the National Institute of Environmental Health Sciences, NIH, Intramural Research Program (P.J.B.).
Disclosures: None reported.
Supplemental material for this article can be found at http://doi.org/10.1016/j.ajpath.2021.10.017.
Author Contributions
S.P. conceived and designed the study; S.P. and C.S.B. maintained the animal colony. C.S.B. genotyped the animals and performed all the experiments except those listed further. C.S.B. and J.C. performed the luciferase assays. J.C. performed the cotransfection and the electrophoretic mobility shift assays. Y.S. performed the oral gavages. R.K. performed the Oil Red O staining, the immunohistochemisty, and the liver triglyceride assays. P.J.B. provided the Zfp36l1 floxed mice and edited the manuscript; S.P. performed the histopathological studies; S.P. and Y.S. wrote and reviewed the manuscript for intellectual content.
Supplemental Data
A: Change in body weight (initial weight on day 0 was subtracted from final weight on day 11) in both ZFP36L1-sufficient (L1FLX) and liver-specific ZFP36L1-deficient (L1LKO) mice maintained on alcohol diet or control diet. B, C, and D: Levels of AST, triglycerides, and cholesterol in the serum of alcohol diet–fed ZFP36L1-sufficient (L1FLX) and liver-specific ZFP36L1-deficient (L1LKO) mice and the control-diet–fed ZFP36L1-sufficient (L1FLX) and liver-specific ZFP36L1-deficient (L1LKO). n = 10 mice in each group (A); n = 5 mice in the ethanol-diet group (B, C, and D); n = 6 mice in the control-diet group (B, C, and D).
References
- 1.Wells M.L., Perera L., Blackshear P.J. An ancient family of RNA-binding proteins: still important! Trends Biochem Sci. 2017;42:285–296. doi: 10.1016/j.tibs.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fu M., Blackshear P.J. RNA-binding proteins in immune regulation: a focus on CCCH zinc finger proteins. Nat Rev Immunol. 2017;17:130–143. doi: 10.1038/nri.2016.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stumpo D.J., Broxmeyer H.E., Ward T., Cooper S., Hangoc G., Chung Y.J., Shelley W.C., Richfield E.K., Ray M.K., Yoder M.C., Aplan P.D., Blackshear P.J. Targeted disruption of Zfp36l2, encoding a CCCH tandem zinc finger RNA-binding protein, results in defective hematopoiesis. Blood. 2009;114:2401–2410. doi: 10.1182/blood-2009-04-214619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stumpo D.J., Byrd N.A., Phillips R.S., Ghosh S., Maronpot R.R., Castranio T., Meyers E.N., Mishina Y., Blackshear P.J. Chorioallantoic fusion defects and embryonic lethality resulting from disruption of Zfp36L1, a gene encoding a CCCH tandem zinc finger protein of the Tristetraprolin family. Mol Cell Biol. 2004;24:6445–6455. doi: 10.1128/MCB.24.14.6445-6455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor G.A., Carballo E., Lee D.M., Lai W.S., Thompson M.J., Patel D.D., Schenkman D.I., Gilkeson G.S., Broxmeyer H.E., Haynes B.F., Blackshear P.J. A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity. 1996;4:445–454. doi: 10.1016/s1074-7613(00)80411-2. [DOI] [PubMed] [Google Scholar]
- 6.Stumpo D.J., Trempus C.S., Tucker C.J., Huang W., Li L., Kluckman K., Bortner D.M., Blackshear P.J. Deficiency of the placenta- and yolk sac-specific tristetraprolin family member ZFP36L3 identifies likely mRNA targets and an unexpected link to placental iron metabolism. Development. 2016;143:1424–1433. doi: 10.1242/dev.130369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patial S., Curtis A.D., 2nd, Lai W.S., Stumpo D.J., Hill G.D., Flake G.P., Mannie M.D., Blackshear P.J. Enhanced stability of tristetraprolin mRNA protects mice against immune-mediated inflammatory pathologies. Proc Natl Acad Sci U S A. 2016;113:1865–1870. doi: 10.1073/pnas.1519906113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patial S., Blackshear P.J. Tristetraprolin as a therapeutic target in inflammatory disease. Trends Pharmacol Sci. 2016;37:811–821. doi: 10.1016/j.tips.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adachi S., Homoto M., Tanaka R., Hioki Y., Murakami H., Suga H., Matsumoto M., Nakayama K.I., Hatta T., Iemura S., Natsume T. ZFP36L1 and ZFP36L2 control LDLR mRNA stability via the ERK-RSK pathway. Nucleic Acids Res. 2014;42:10037–10049. doi: 10.1093/nar/gku652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahashi A., Adachi S., Morita M., Tokumasu M., Natsume T., Suzuki T., Yamamoto T. Post-transcriptional stabilization of Ucp1 mRNA protects mice from diet-induced obesity. Cell Rep. 2015;13:2756–2767. doi: 10.1016/j.celrep.2015.11.056. [DOI] [PubMed] [Google Scholar]
- 11.Tarling E.J., Clifford B.L., Cheng J., Morand P., Cheng A., Lester E., Sallam T., Turner M., de Aguiar Vallim T.Q. RNA-binding protein ZFP36L1 maintains posttranscriptional regulation of bile acid metabolism. J Clin Invest. 2017;127:3741–3754. doi: 10.1172/JCI94029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mu X., Pradere J.P., Affo S., Dapito D.H., Friedman R., Lefkovitch J.H., Schwabe R.F. Epithelial transforming growth factor-beta signaling does not contribute to liver fibrosis but protects mice from cholangiocarcinoma. Gastroenterology. 2016;150:720–733. doi: 10.1053/j.gastro.2015.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bertola A., Mathews S., Ki S.H., Wang H., Gao B. Mouse model of chronic and binge ethanol feeding (the NIAAA model) Nat Protoc. 2013;8:627–637. doi: 10.1038/nprot.2013.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Postic C., Shiota M., Niswender K.D., Jetton T.L., Chen Y., Moates J.M., Shelton K.D., Lindner J., Cherrington A.D., Magnuson M.A. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J Biol Chem. 1999;274:305–315. doi: 10.1074/jbc.274.1.305. [DOI] [PubMed] [Google Scholar]
- 15.Committee for the Update of the Guide for the Care and Use of Laboratory Animals; National Research Council . Eighth Edition. National Academies Press; Washington, DC: 2011. Guide for the Care and Use of Laboratory Animals. [Google Scholar]
- 16.Homeyer A., Nasr P., Engel C., Kechagias S., Lundberg P., Ekstedt M., Kost H., Weiss N., Palmer T., Hahn H.K., Treanor D., Lundstrom C. Automated quantification of steatosis: agreement with stereological point counting. Diagn Pathol. 2017;12:80. doi: 10.1186/s13000-017-0671-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Osna N.A., Donohue T.M., Jr., Kharbanda K.K. Alcoholic liver disease: pathogenesis and current management. Alcohol Res. 2017;38:147–161. [PMC free article] [PubMed] [Google Scholar]
- 18.Sakhuja P. Pathology of alcoholic liver disease, can it be differentiated from nonalcoholic steatohepatitis? World J Gastroenterol. 2014;20:16474–16479. doi: 10.3748/wjg.v20.i44.16474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mandrekar P., Ambade A., Lim A., Szabo G., Catalano D. An essential role for monocyte chemoattractant protein-1 in alcoholic liver injury: regulation of proinflammatory cytokines and hepatic steatosis in mice. Hepatology. 2011;54:2185–2197. doi: 10.1002/hep.24599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thakur V., Pritchard M.T., McMullen M.R., Wang Q., Nagy L.E. Chronic ethanol feeding increases activation of NADPH oxidase by lipopolysaccharide in rat Kupffer cells: role of increased reactive oxygen in LPS-stimulated ERK1/2 activation and TNF-alpha production. J Leukoc Biol. 2006;79:1348–1356. doi: 10.1189/jlb.1005613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao B. Hepatoprotective and anti-inflammatory cytokines in alcoholic liver disease. J Gastroenterol Hepatol. 2012;27(Suppl 2):89–93. doi: 10.1111/j.1440-1746.2011.07003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clugston R.D., Jiang H., Lee M.X., Piantedosi R., Yuen J.J., Ramakrishnan R., Lewis M.J., Gottesman M.E., Huang L.S., Goldberg I.J., Berk P.D., Blaner W.S. Altered hepatic lipid metabolism in C57BL/6 mice fed alcohol: a targeted lipidomic and gene expression study. J Lipid Res. 2011;52:2021–2031. doi: 10.1194/jlr.M017368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Desai B.N., Singhal G., Watanabe M., Stevanovic D., Lundasen T., Fisher F.M., Mather M.L., Vardeh H.G., Douris N., Adams A.C., Nasser I.A., FitzGerald G.A., Flier J.S., Skarke C., Maratos-Flier E. Fibroblast growth factor 21 (FGF21) is robustly induced by ethanol and has a protective role in ethanol associated liver injury. Mol Metab. 2017;6:1395–1406. doi: 10.1016/j.molmet.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y., Zhao C., Xiao J., Liu L., Zhang M., Wang C., Wu G., Zheng M.H., Xu L.M., Chen Y.P., Mohammadi M., Chen S.Y., Cave M., McClain C., Li X., Feng W. Fibroblast growth factor 21 deficiency exacerbates chronic alcohol-induced hepatic steatosis and injury. Sci Rep. 2016;6:31026. doi: 10.1038/srep31026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lai W.S., Carrick D.M., Blackshear P.J. Influence of nonameric AU-rich tristetraprolin-binding sites on mRNA deadenylation and turnover. J Biol Chem. 2005;280:34365–34377. doi: 10.1074/jbc.M506757200. [DOI] [PubMed] [Google Scholar]
- 26.Bouchery E.E., Harwood H.J., Sacks J.J., Simon C.J., Brewer R.D. Economic costs of excessive alcohol consumption in the U.S., 2006. Am J Prev Med. 2011;41:516–524. doi: 10.1016/j.amepre.2011.06.045. [DOI] [PubMed] [Google Scholar]
- 27.Son Y.O., Kim H.E., Choi W.S., Chun C.H., Chun J.S. RNA-binding protein ZFP36L1 regulates osteoarthritis by modulating members of the heat shock protein 70 family. Nat Commun. 2019;10:77. doi: 10.1038/s41467-018-08035-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newman R., Ahlfors H., Saveliev A., Galloway A., Hodson D.J., Williams R., Besra G.S., Cook C.N., Cunningham A.F., Bell S.E., Turner M. Maintenance of the marginal-zone B cell compartment specifically requires the RNA-binding protein ZFP36L1. Nat Immunol. 2017;18:683–693. doi: 10.1038/ni.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McClain C.J., Barve S., Barve S., Deaciuc I., Hill D.B. Tumor necrosis factor and alcoholic liver disease. Alcohol Clin Exp Res. 1998;22:248S–252S. doi: 10.1097/00000374-199805001-00006. [DOI] [PubMed] [Google Scholar]
- 30.Degre D., Lemmers A., Gustot T., Ouziel R., Trepo E., Demetter P., Verset L., Quertinmont E., Vercruysse V., Le Moine O., Deviere J., Moreno C. Hepatic expression of CCL2 in alcoholic liver disease is associated with disease severity and neutrophil infiltrates. Clin Exp Immunol. 2012;169:302–310. doi: 10.1111/j.1365-2249.2012.04609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yin M., Bradford B.U., Wheeler M.D., Uesugi T., Froh M., Goyert S.M., Thurman R.G. Reduced early alcohol-induced liver injury in CD14-deficient mice. J Immunol. 2001;166:4737–4742. doi: 10.4049/jimmunol.166.7.4737. [DOI] [PubMed] [Google Scholar]
- 32.Mandrekar P., Szabo G. Signalling pathways in alcohol-induced liver inflammation. J Hepatol. 2009;50:1258–1266. doi: 10.1016/j.jhep.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duarte S., Baber J., Fujii T., Coito A.J. Matrix metalloproteinases in liver injury, repair and fibrosis. Matrix Biol. 2015;44-46:147–156. doi: 10.1016/j.matbio.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim M.J., Nepal S., Lee E.S., Jeong T.C., Kim S.H., Park P.H. Ethanol increases matrix metalloproteinase-12 expression via NADPH oxidase-dependent ROS production in macrophages. Toxicol Appl Pharmacol. 2013;273:77–89. doi: 10.1016/j.taap.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 35.Soberg S., Andersen E.S., Dalsgaard N.B., Jarlhelt I., Hansen N.L., Hoffmann N., Vilsboll T., Chenchar A., Jensen M., Grevengoed T.J., Trammell S.A.J., Knop F.K., Gillum M.P. FGF21, a liver hormone that inhibits alcohol intake in mice, increases in human circulation after acute alcohol ingestion and sustained binge drinking at Oktoberfest. Mol Metab. 2018;11:96–103. doi: 10.1016/j.molmet.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sawicki K.T., Chang H.C., Shapiro J.S., Bayeva M., De Jesus A., Finck B.N., Wertheim J.A., Blackshear P.J., Ardehali H. Hepatic tristetraprolin promotes insulin resistance through RNA destabilization of FGF21. JCI Insight. 2018;3:e95948. doi: 10.1172/jci.insight.95948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morita M., Siddiqui N., Katsumura S., Rouya C., Larsson O., Nagashima T., Hekmatnejad B., Takahashi A., Kiyonari H., Zang M., St-Arnaud R., Oike Y., Giguere V., Topisirovic I., Okada-Hatakeyama M., Yamamoto T., Sonenberg N. Hepatic posttranscriptional network comprised of CCR4-NOT deadenylase and FGF21 maintains systemic metabolic homeostasis. Proc Natl Acad Sci U S A. 2019;116:7973–7981. doi: 10.1073/pnas.1816023116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carballo E., Lai W.S., Blackshear P.J. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 39.Hodson D.J., Janas M.L., Galloway A., Bell S.E., Andrews S., Li C.M., Pannell R., Siebel C.W., MacDonald H.R., De Keersmaecker K., Ferrando A.A., Grutz G., Turner M. Deletion of the RNA-binding proteins ZFP36L1 and ZFP36L2 leads to perturbed thymic development and T lymphoblastic leukemia. Nat Immunol. 2010;11:717–724. doi: 10.1038/ni.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qiu L.Q., Stumpo D.J., Blackshear P.J. Myeloid-specific tristetraprolin deficiency in mice results in extreme lipopolysaccharide sensitivity in an otherwise minimal phenotype. J Immunol. 2012;188:5150–5159. doi: 10.4049/jimmunol.1103700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andrianne M., Assabban A., La C., Mogilenko D., Salle D.S., Fleury S., Doumont G., Van Simaeys G., Nedospasov S.A., Blackshear P.J., Dombrowicz D., Goriely S., Van Maele L. Tristetraprolin expression by keratinocytes controls local and systemic inflammation. JCI Insight. 2017;2:e92979. doi: 10.1172/jci.insight.92979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Struik D., Dommerholt M.B., Jonker J.W. Fibroblast growth factors in control of lipid metabolism: from biological function to clinical application. Curr Opin Lipidol. 2019;30:235–243. doi: 10.1097/MOL.0000000000000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A: Change in body weight (initial weight on day 0 was subtracted from final weight on day 11) in both ZFP36L1-sufficient (L1FLX) and liver-specific ZFP36L1-deficient (L1LKO) mice maintained on alcohol diet or control diet. B, C, and D: Levels of AST, triglycerides, and cholesterol in the serum of alcohol diet–fed ZFP36L1-sufficient (L1FLX) and liver-specific ZFP36L1-deficient (L1LKO) mice and the control-diet–fed ZFP36L1-sufficient (L1FLX) and liver-specific ZFP36L1-deficient (L1LKO). n = 10 mice in each group (A); n = 5 mice in the ethanol-diet group (B, C, and D); n = 6 mice in the control-diet group (B, C, and D).