Abstract
The circular RNA (circRNA) circLAMA3 is significantly downregulated in bladder cancer tissues and cell lines. However, its function in bladder cancer has not yet been explored, and further research is needed. In this study, functional experiments demonstrated that circLAMA3 significantly inhibited the proliferation, migration, and invasion of bladder cancer cells and inhibited bladder cancer growth in vivo. Mechanistically, circLAMA3 directly binds to and promotes the degradation of MYCN mRNA, thereby reducing the MYCN protein expression in bladder cancer cells. Decreased expression of the MYCN protein inhibits the promoter activity and expression of CDK6. Ultimately, circLAMA3 affects DNA replication by downregulating CDK6, resulting in G0/G1 phase arrest and inhibition of bladder cancer proliferation. In summary, we report a potential novel regulatory mechanism via which a circRNA directly binds an mRNA and thereby regulates its fate. Moreover, circLAMA3 significantly affects the progression of bladder cancer and has potential as a diagnostic biomarker and therapeutic target for bladder cancer.
Keywords: bladder cancer, circLAMA3, proliferation, MYCN, CDK6, mRNA degradation
Graphical abstract

The research reports a potential novel regulatory mechanism via which a circRNA directly binds an mRNA and thereby regulates its fate. Moreover, circLAMA3 significantly affects the progression of bladder cancer and has potential as a diagnostic biomarker and therapeutic target for bladder cancer.
Introduction
Bladder cancer is the sixth most common cancer in the male population, and its incidence has gradually increased in recent decades. Approximately 549,000 new bladder cancer diagnoses and approximately 200,000 deaths occur worldwide annually. The morbidity and mortality rates of bladder cancer in men are approximately 4 times higher than those in women,1 and approximately 70% of bladder cancers are diagnosed at an early stage and are usually removed by urethral bladder tumor resection.2 Approximately 60% of bladder cancers recur after transurethral cystectomy. Due to the recurrence and resistance of bladder cancer, the five-year survival rate of patients after surgery remains low, at approximately 50%.3 With the exception of surgical resection and triple adjuvant chemotherapy, effective specific drugs and treatment methods for bladder cancer are still lacking. Exploring the mechanisms related to the occurrence and development of bladder cancer and identifying key genes and new targets with high specificity are important for improving the early diagnosis, prognosis, and accurate treatment of bladder cancer.
Recent rapid progress in the field of noncoding RNA has provided a new direction for the treatment of tumors. Previous studies have demonstrated that a variety of noncoding RNAs are involved in the occurrence,4 development,5 invasion, and metastasis6 of various human malignancies. These tumor-related biological behaviors play important roles in the process of cancer development. Therefore, an in-depth study of the relationship between noncoding RNAs and tumors may provide new options for cancer diagnosis and treatment. As a new type of noncoding RNA, circular RNAs (circRNAs) have gradually entered the field of bladder cancer research. Unlike traditional noncoding RNAs, circRNAs have high stability, tissue specificity, and peak expression patterns due to their unique circular structure.7 A previous study even showed that the hundreds of circRNAs in the blood were detected at higher levels than their precursor mRNAs.8 Multiple studies have shown that circRNAs play an important biological role in a variety of malignant tumors, such as colorectal cancer,9 lung cancer,10 breast cancer,11 and gastric cancer.11 Gastric cancer studies have revealed that hsa_circ_0001649 is significantly downregulated in gastric cancer tissues12 and significantly upregulated after therapeutic surgery. Li et al. also found that circRNA expression levels are significantly correlated with the tumor stage and lymph node metastasis of gastric cancer.13 In addition, hsa_circ_0004018 is abnormally expressed in liver cancer; its expression level is related to the level of alpha fetoprotein (AFP), tumor diameter, and differentiation and differs from those in other chronic liver diseases.14 Considering the high stability and abundance of circRNAs, exploring their potential as disease biomarkers will be valuable.
Many studies have been conducted to explore the mechanism by which circRNAs affect tumor progression. Current research shows that circRNAs function mainly by interacting with other molecules to alter their original functions. The most widely studied theory is that circRNA acts as miRNA sponges,15 participating in the competing endogenous RNA (ceRNA) network, and indirectly affects the expression of downstream mRNAs, thereby ultimately regulating cancer progression. In addition to miRNAs, circRNAs can also interact with other molecules, such as proteins. We previously confirmed that circNOL10 interacts with SCML1 and inhibits its ubiquitination, thereby affecting cell mitochondrial function and lung cancer processes.16 CircFOXO3 enhances the sensitivity of breast cancer cells to cisplatin and doxorubicin by binding to p53 and MDM2.17 CircRNAs are known to bind miRNAs via complementary pairing to RNA-RNA base sequences. However, whether circRNAs also bind mRNAs and affect their fate via this RNA-RNA binding mechanism remains unknown.
In this study, we explored the function and related mechanisms of circRNAs in bladder cancer by analyzing a series of research data. Gene chip technology was used to detect the expression of circRNAs in human bladder cancer cells and human urothelial cells, and circLAMA3 was shown to be significantly downregulated in bladder cancer and to have potential diagnostic value. In vitro and in vivo studies confirmed that circLAMA3 participates in bladder cancer development, and bioinformatics analysis revealed that circLAMA3 encodes no proteins but is potentially involved in multiple life processes at the nucleic acid level. Further research found that circLAMA3 directly targets MYCN and promotes the degradation of MYCN mRNA, thereby inhibiting the promoter activity of CDK6 and eventually leading to G0/G1 phase arrest and bladder cancer cell apoptosis. We herein confirmed for the first time that a circRNA can directly bind mRNA through sequence complementation and cause its degradation, which broadens our understanding of how circRNA exerts their biological functions. This new regulatory mechanism of circRNAs may play roles in various physiological and pathological processes. In conclusion, circLAMA3 has the potential to become a new target for the diagnosis and treatment of bladder cancer.
Results
Circular RNA circLAMA3 is significantly downregulated in bladder cancer
With the rapid development and clinical application of chip technology, many noncoding RNAs have been identified and gradually emerged in clinical treatment strategies. To explore the role of circRNAs in bladder cancer, we conducted gene chip analyses of human urothelial immortalized SV-HUC-1 cells as a control group and human bladder cancer UM-UC-3 cells as an experimental group (Figure S1A and Table S2). We screened the eight circRNAs that were most significantly downregulated in the circRNA differential expression profile of bladder cancer cells. The original expression profile data are available in the Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/gds) under the following accession number: GSE159239. Hsa_circ_0047253 was most significantly downregulated in bladder cancer cells (Figure 1A). To further detect the expression of hsa_circ_0047253 in bladder cancer cell lines, we assessed its expression in multiple bladder cancer cell lines, revealing that it was significantly decreased in all five bladder cancer cell lines and expressed at the lowest level in T24 and J82 cells (Figure 1B). To determine the expression trend of hsa_circ_0047253 in bladder cancer tissues, we conducted qPCR analysis of bladder cancer tissues and adjacent tissues. Hsa_circ_0047253 was significantly decreased in bladder cancer tissues, confirming its low expression trend in bladder cancer (Figure 1C). Receiver operating characteristic (ROC) curve analysis revealed that the area under the curve (AUC) value of hsa_circ_0047253 was 0.7578 in patients with low-grade bladder cancer and 0.8099 in patients with high-grade bladder cancer, suggesting that hsa_circ_0047253 has diagnostic value in bladder cancer (Figures 1D and 1E). In addition, hsa_circ_0047253 showed a low-surface trend in colorectal cancer tissues (Figure S1B). The decreased hsa_circ_0047253 expression in bladder cancer tissues was significantly correlated with higher grade and T stage in bladder cancer patients (Figures S1C and S1D). Hsa_circ_0047253 was shown to be located on the LAMA3 gene of chromosome 18 and was named circLAMA3 based on its gene symbol. CircLAMA3 is 391 bp in length and is formed by the circularization of exons 45, 46, and 47 (Figure 1F). To determine the circular structure of circLAMA3, we designed polymerized and dispersed primers and conducted agarose gel electrophoresis. CircLAMA3 was resistant to RNase digestion, confirming its circular structure (Figures 1G and 1H). Ta clone sequencing was performed on the circLAMA3 qPCR product to visualize the circularization cleavage site (Figure S1E). To determine the role of circLAMA3 in the progression of bladder cancer, we conducted bioinformatics analysis of circLAMA3 using DAVID:https://david.ncifcrf.gov/, and the results suggested that circLAMA3 affects cell proliferation, invasion, and other functions through multiple signaling pathways, further affecting the bladder cancer developmental process (Figure 1I and Table S3).
Figure 1.
CircLAMA3 expression was significantly decreased in bladder cancer
(A) Microarray analysis of human immortalized urothelial SV-HUC-1 and UMUC3 cells by circRNA chip. A total of 1,188 circRNAs were differentially expressed. CircRNA PCR verification was performed on the eight most obvious circRNAs in the microarray results. Hsa_circ_0047253 exhibited the most obvious downregulation and was named circLAMA3 according to its gene symbol. An asterisk (∗) indicates a significant difference (p < 0.05). (B) The relative expression of circLAMA3 in human bladder cancer cells and normal cells as determined by qPCR. An asterisk (∗) indicates a significant difference (p < 0.05). (C) qPCR analysis of the relative expression levels of circLAMA3 in 50 pairs of fully matched human bladder cancer clinical samples collected by our team. N: normal tissue; T: tumor tissue. An asterisk (∗) indicates a significant difference (p < 0.05). (D and E) ROC curve analysis was used to evaluate the diagnostic value of circLAMA3. In low-grade bladder cancer tissues, the AUC value of circLAMA3 was 0.7578. In high-grade bladder cancer tissues, the AUC value of circLAMA3 was 0.8099 (p < 0.05). (F) circLAMA3 is transcribed from the parent gene pre-LAMA3 and is formed by the circularization of exons 45, 46, and 47. (G and H) Scattered and polymerized primers were designed for PCR-agarose gel electrophoresis to verify the circular structure of circLAMA3. PCR primers spanned the splice site to ensure detection specificity. (I) Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of circLAMA3 using OriginPro. The Y axis represents the KEGG pathway, the size of the dots represents the number of differentially expressed genes, and the color of the dots represents the range of p values.
Circular RNA circLAMA3 significantly inhibits the proliferation and invasion of bladder cancer
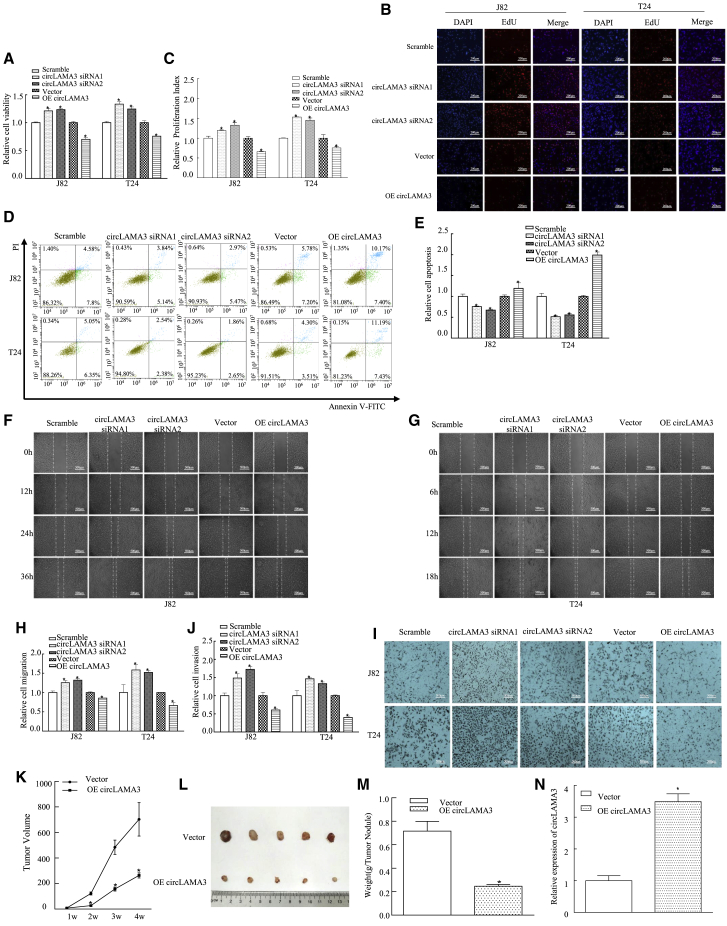
According to the bioinformatics analysis results, circLAMA3 may play an important biological role in the progression of bladder cancer. To explore this further, we designed specific small interfering RNAs (siRNAs) at the circRNA circularization site to silence its expression in J82 and T24 cells and used circularized vectors (Figure S1F) to overexpress circLAMA3 in bladder cancer cells. qPCR revealed that the silencing efficiency of circLAMA3 in J82 and T24 cells was greater than 50% compared with the control group cells. The overexpression level of circLAMA3 also exceeded the baseline level by more than 10-fold (Figure S1G). In addition, the silencing and overexpression of circLAMA3 did not affect LAMA3 (Figure S1H). We next assessed the in vitro function of circLAMA3 by transfecting circLAMA3-specific siRNA and overexpression plasmids into cells. siRNA1 and siRNA2 targeting circLAMA3 significantly increased the viability, inhibited the apoptosis, and increased the migration and invasion of J82/T24 cells compared with the values in the scramble group. Compared with the vector group, the circLAMA3 overexpression group showed the opposite effect of that in the siRNA group, and the difference was statistically significant (Figures 2A–2I). These results indicated that circLAMA3 inhibited the proliferation of bladder cancer in vitro.
Figure 2.
CircLAMA3 significantly inhibited the proliferation and migration of bladder cancer cells in vitro and in vivo
(A) The effect of circLAMA3 on cell viability was determined by the CCK-8 assay. An asterisk (∗) indicates a significant difference (p < 0.05). (B and C) The effect of circLAMA3 on cell proliferation was measured by the EdU assay. Nuclei were stained with DAPI, and cell proliferation activity was determined by the EdU/DAPI ratio. An asterisk (∗) indicates a significant difference (p < 0.05). (D and E) Effect of circLAMA3 on apoptosis as determined by Annexin V-FITC/PI staining and FCM detection. (F–H) A wound-healing test was performed to assess the effect of circLAMA3 on cell migration. (I and J) Transwell experiments verified the effect of circLAMA3 on cell invasion. (K) The widths and lengths (ab) of subcutaneous tumors in nude mice were measured weekly, and the tumor volume was calculated as V = 1/2ab2. An asterisk (∗) indicates a significant difference (p < 0.05). (L and M) Subcutaneous tumor tissue was removed from nude mice on day 28 after the subcutaneous injection of cells. The subcutaneous tumor tissues from the two groups were photographed and weighed. An asterisk (∗) indicates a significant difference (p < 0.05). (N) qPCR was performed to detect the expression of circLAMA3 in subcutaneous tumor tissues from nude mice. An asterisk (∗) indicates a significant difference (p < 0.05).
Circular RNA circLAMA3 significantly inhibits the proliferation of bladder cancer cells in vivo
The above experiments indicated that circLAMA3 can inhibit the proliferation of bladder cancer in vitro. To determine whether circLAMA3 has the same biological function in animals, we built a subcutaneous tumor formation model using nude mice. Using a GFP-labeled lentiviral packaging vector (Figure S2A), we constructed a stable T24 cell line to overexpress (OE) circLAMA3 and a stable control cell line, and qPCR revealed that the stable cell line overexpressing circLAMA3 was successfully constructed (Figure S2B). The tumor growth rates differed between the experimental and control groups, as subcutaneous tumors appeared later (Figure 2K) and tissue volumes and weights were decreased in the OE circLAMA3 group compared with the vector control (Figures 2L and 2M). This result confirmed that circLAMA3 could inhibit the proliferation of bladder cancer cells in vivo. After extracting RNA from tumor tissue and performing reverse transcription, qPCR revealed higher circLAMA3 expression in OE circLAMA3 tumor tissues than in the control group (Figure 2N). Together, the above results demonstrated that circLAMA3 inhibited the proliferation of bladder cancer in vitro and in vivo, indicating that decreased circLAMA3 expression plays a role in bladder cancer proliferation.
Circular RNA circLAMA3 inhibits CDK6 expression and cell-cycle progression in bladder cancer
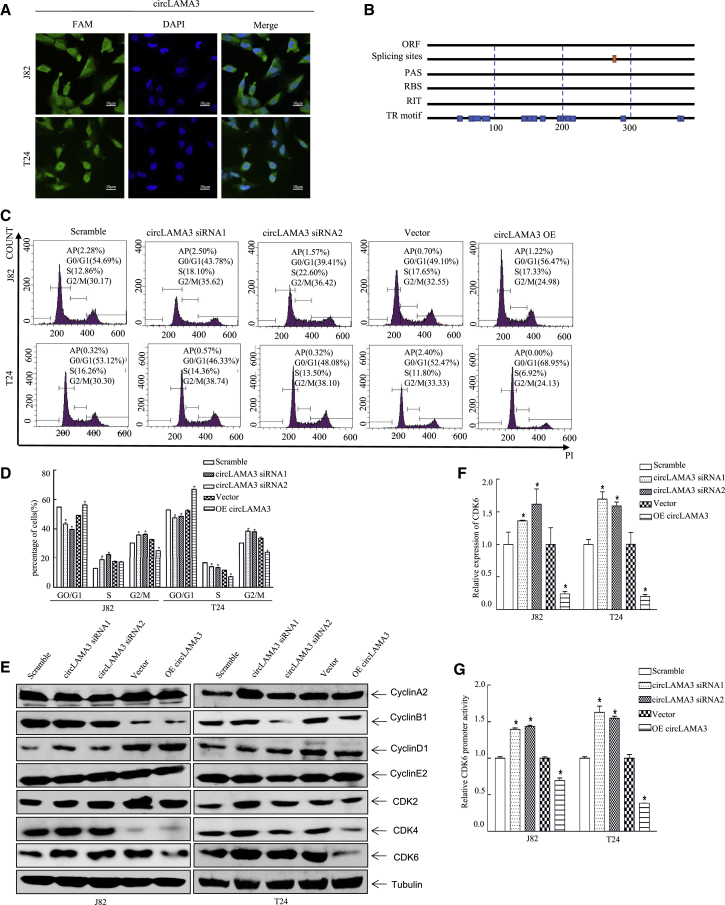
To explore the relevant mechanism by which circLAMA3 inhibits the proliferation of bladder cancer cells, we determined its localization in J82 and T24 cells using fluorescence in situ hybridization (FISH). CircLAMA3 was distributed in both the cytoplasm and nucleus but mainly localized in the nucleus (Figure 3A). Bioinformatics analysis demonstrated that circLAMA3 lacked open reading frame (ORF) regions, polyadenylation sites, ribosome binding sites, and Rho-dependent terminators but had transcriptional regulatory motifs and splice sites, suggesting that it lacks protein coding ability and is involved in transcriptional regulation in cells (Figure 3B). Cell-cycle detection via flow cytometry (FCM) showed that compared with those in the control group, the numbers of J82 and T24 cells in the G0/G1 phase were significantly reduced after transfection with circLAMA3 siRNA. After transfection of the circLAMA3 overexpression plasmid, the cells were arrested at the G0/G1 phase (Figures 3C and 3D). Therefore, circLAMA3 may inhibit bladder cancer cell proliferation by inducing G0/G1 phase arrest.
Figure 3.
Overexpression of circLAMA3-induced G0/G1 phase arrest in bladder cancer cells
(A) The distribution of circLAMA3 in T24 and J82 cells as determined by FISH. Nuclei were stained with DAPI, and circLAMA3 was labeled with a specific probe containing GFP. The map shows the intracellular distribution of circLAMA3. (B) The circLAMA3 nucleic acid sequence indicates an open reading frame (ORF), splice site, polyadenylation site (PAS), ribosome binding site (RBS), Rho-independent terminator (RIT), transcription regulation motif (TR motif), and codons. Two splice sites and several transcriptional regulatory motifs were observed. This structure suggests that circLAMA3 likely does not encode a protein. (C and D) FCM was performed to assess the role of circLAMA3 in the cell cycle. CircRNA circLAMA3 was transiently silenced and overexpressed in J82/T24 cells; compared with the scramble group; the transfection of circLAMA3 siRNA reduced the G0/G1 phase arrest and accelerated the cell-cycle process. Compared with the vector group, transfection of the circLMA3 overexpression plasmid promoted the G0/G1 phase arrest of J82/T24 cells and decelerated the cell-cycle process. An asterisk (∗) indicates a significant difference (p < 0.05). (E) circLAMA3 inhibits cell-cycle progression by inhibiting the expression of CDK6. Western blotting was performed to assess the effect of circLAMA3 on G0/G1 phase kinesins. (F) Determination of the effects of circLAMA3 transient silencing and overexpression on CDK6 mRNA levels. An asterisk (∗) indicates a significant difference (p < 0.05). (G) The luciferase reporter gene driven by the CDK6 promoter and TK reporter gene were co-transfected into T24 (vector and OE circLAMA3) and J82 (vector and OE circLAMA3) cells. An asterisk (∗) indicates a significant difference (p < 0.05).
To clarify the mechanism by which circLAMA3 prohibits J82/T24 cells from exiting the G0/G1 phase, we consulted the literature and verified our results by performing western blot (WB) analysis of classic G0/G1 phase kinesins. Studies have demonstrated that CDK4/CDK6 interact with cyclin D to promote the phosphorylation of the pRb protein, thereby activating DNA replication during the S phase.18 CDK2 promotes the G1/S transition by combining with cyclin E during the G1 phase,19 and CDK2 participates in S phase replication initiation by combining with cyclin A.20 In addition, cyclin B has been reported to be active throughout the mitotic stage.21 Considering the above experimental results, we used WB to detect changes in the expression levels of cyclin A, cyclin B, cyclin D, cyclin E, CDK2, CDK4, and CDK6 after overexpression of circLAMA3 in J82 and T24 cells. Compared with that in the scramble group, CDK6 expression was increased in cells transfected with circLAMA3 siRNA and decreased in cells transfected with OE circLAMA3 (Figure 3E). Therefore, decreased CDK6 expression may play a key role in circLAMA3-induced G0/G1 phase arrest.
To assess the relationship between circLAMA3 and CDK6, we measured the mRNA expression of CDK6 and found that circLAMA3 inhibited the protein level of CDK6 in an mRNA-dependent manner (Figure 3F). We also explored the interaction between circLAMA3 and CDK6 using a luciferase reporter assay. The luciferase reporter gene driven by the CDK6 promoter was co-transfected with the TK reporter gene into T24 and J82 (vector and OE circLAMA3) cells, and the transcriptional activity of the CDK6 promoter was examined 24 h after transfection. Compared with that in the control group, the fluorescence intensity was significantly reduced in the circLAMA3 overexpression group (Figure 3G). These results indicate that decreased circLAMA3 expression in bladder cancer weakens its inhibitory effect on CDK6 promoter activity, enhances CDK6 expression, promotes the processes occurring the G0/G1 phase, and ultimately promotes the proliferation of bladder cancer cells.
MYCN mediates the regulatory effect of circLAMA3 on CDK6 transcription
Based on the results of the above experiments, we performed bioinformatics analysis and revealed that circLAMA3 and CDK6 lack binding sites for interaction. Therefore, we speculated that circLAMA3 regulates the promoter activity of CDK6 through an intermediary molecule. A variety of transcription factors can bind the CDK6 promoter region to promote CDK6 transcription. For example, the bHLH region of the MYC protein family can be combined with the E box of the CDK6 promoter region,22 the E2F protein family,18 P53,23 and others. In addition, bioinformatics analysis revealed that circLAMA3 can directly bind MYCN and E2F1 mRNA. To determine whether MYCN and E2F1 participate in the circLAMA3 inhibition of CDK6 transcription, we used WB to determine the expression levels of MYCN and E2F1 after silencing and overexpressing circLAMA3. Compared with that in the scramble group, the expression of MYCN was increased after the transfection of circLAMA3 siRNA and decreased after the transfection of OE circLAMA3 (Figure 4A). Studies have shown that MYCN can bind to the CDK6 promoter region at nucleotides 138 and 884 to mediate its transcriptional activation, thereby accelerating the cell-cycle process and promoting cell proliferation.22 To further assess the effect of the transcription factor MYCN on CDK6, we overexpressed MYCN in T24 (OE circLAMA3) and J82 (OE circLAMA3) cells and their corresponding controls to evaluate the expression of CDK6. WB confirmed that MYCN was successfully overexpressed (Figure 4B) and the overexpression efficiency (Figure S2C). Ectopic expression of MYCN significantly increased the expression of CDK6 (Figure 4B), indicating that MYCN is a key factor that mediates the transcript expression of CDK6. In addition, a luciferase reporter gene encoding the CDK6 promoter was co-transfected with the TK reporter gene into T24 and J82 cells overexpressing MYCN and its control plasmid, and the transcriptional activity of the CDK6 promoter was detected 24 h later. Compared with that in the control group, the promoter activity of CDK6 was significantly increased in the MYCN overexpression group (Figures 4C and 4D), demonstrating that MYCN plays a key role in mediating the inhibitory effect of circLAMA3 on CDK6 transcription.
Figure 4.
Downregulation of MYCN mediated the inhibition of CDK6 transcription by circLAMA3
(A) Semiquantitative detection of the MYCN and E2F1 proteins in J82/T24 cells by western blotting. (B) Western blotting was performed to detect the expression of CDK6 in T24 (vector and OE circLAMA3) and J82 (vector and OE circLAMA3) cells after overexpression of MYCN. (C and D) The luciferase reporter gene driven by the CDK6 promoter was co-transfected with the TK reporter gene into T24/J82 cells overexpressing MYCN and its control plasmid, and the transcriptional activity of the CDK6 promoter was detected 24 h after transfection. The induction multiple was normalized using pRL-TK as an internal control. An asterisk (∗) indicates a significant difference (p < 0.05). (E and F) The cell proliferation ability of T24 OE circLAMA3 cells ectopically expressing MYCN as determined by the CCK-8 assay. An asterisk (∗) indicates a significant difference (p < 0.05). (G and H) PI staining and FCM detection of the T24 (OE circLAMA3) and J82 (OE circLAMA3) cell cycles after the ectopic expression of MYCN. An asterisk (∗) indicates a significant difference (p < 0.05). (I and J) The proliferation ability of T24 (OE circLAMA3) and J82 (OE circLAMA3) cells ectopically expressing MYCN was determined by the EdU assay; nuclei were stained with DAPI, and the cell proliferation activity was determined by the EdU/DAPI ratio. An asterisk (∗) indicates a significant difference (p < 0.05). (K) Immunohistochemistry detection of the MYCN protein in nude mice.
Finally, to assess the role of MYCN in the circLAMA3 inhibition of bladder cancer proliferation, we ectopically expressed the MYCN plasmid in T24 (OE circLAMA3) and J82 (OE circLAMA3) cells. Compared with that in the control group, the proliferative capacity of bladder cancer cells was significantly improved in cells ectopically expressing MYCN (Figures 4E–4J). In addition, immunohistochemistry analysis revealed high protein expression of MYCN in the subcutaneous tumor tissues of nude mice (Figure 4K). Thus, the transcription factor MYCN may mediate the inhibitory effect of circLAMA3 on the transcription and expression of CDK6 in bladder cancer cells, leading to G0/G1 phase arrest and ultimately affecting the proliferation of bladder cancer.
Circular RNA circLAMA3 directly promotes the degradation of MYCN mRNA and inhibits the expression of CDK6
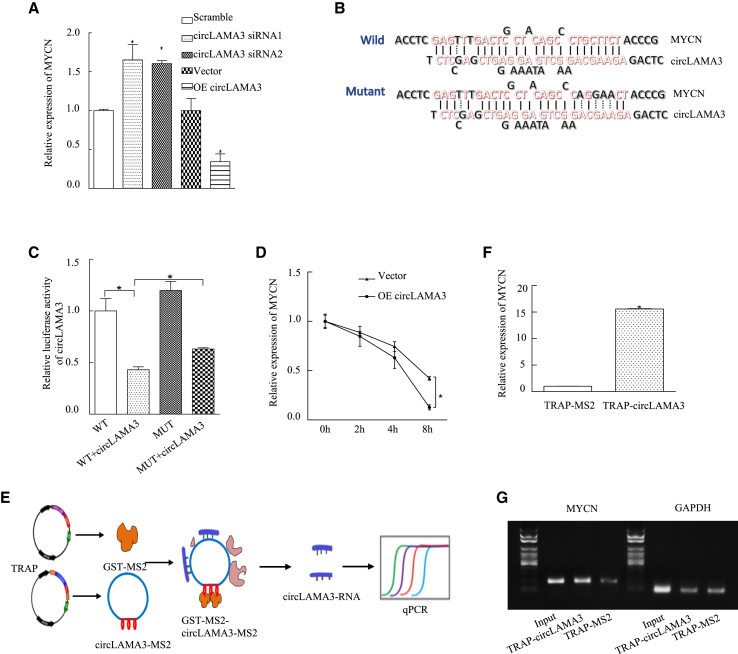
MYCN is an oncogene, and previous studies have shown that its expression level is correlated with many types of cancer. To explore the specific mechanism by which circLAMA3 downregulates the protein expression of MYCN, we first performed qPCR and found that the MYCN mRNA level was increased significantly after transfection of circLAMA3 siRNA compared with that in the scramble group; compared with that in the vector group, the MYCN mRNA level was decreased significantly after transfection of OE circLAMA3. This result demonstrates that circLAMA3 may regulate MYCN at the mRNA level (Figure 5A).
Figure 5.
CircLAMA3 can directly bind an mRNA and inhibit its stability
(A) circLAMA3 downregulated the mRNA levels of MYCN. qPCR was performed to detect the mRNA expression of MYCN in T24 and J82 cells after the silencing and overexpression of circLAMA3. An asterisk (∗) indicates a significant difference (p < 0.05). (B) IntaRNA analyses of the potential binding regions of circLAMA3 and MYCN mRNA. (C) The binding site between circLAMA3 and MYCN was mutated and detected using a dual luciferase reporter gene. An asterisk (∗) indicates a significant difference (p < 0.05). (D) T24 (vector and OE circLAMA3) cells were treated with ActD for 0, 2, 4, and 8 h. RNA was then extracted, and qPCR was performed to detect the MYCN mRNA levels. An asterisk (∗) indicates a significant difference (p < 0.05). (E) Schematic of the TRAP experiment. (F and G) The TRAP test was performed to assess the direct binding effect of circLAMA3 and MYCN. An asterisk (∗) indicates a significant difference (p < 0.05).
Bioinformatics analysis revealed a region in which circLAMA3 and MYCN interact (Figures 5B and S2D), and destroying these mutual binding sites weakened their binding ability (Figure 5C). We speculate that circLAMA3 impacts the stability of MYCN mRNA, and we thus performed an mRNA stability experiment to further assess this possibility. T24 (OE circLAMA3) cells and control group cells were treated with actinomycin D (ActD), and the half-life of MYCN was used to set the detection time point. The results are shown in Figure 5D. After overexpression of circLAMA3, the degradation rate of MYCN mRNA was significantly accelerated, indicating that circLAMA3 may regulate the MYCN expression level by promoting the degradation of its mRNA. To assess the direct interaction between circLAMA3 and MYCN, we transfected the GST-MS2 fusion expression vector and circLAMA3-MS2 stem-loop structure tandem repeat vector into T24 cells to obtain the GST-MS2 fusion protein, and circLAMA3-MS2 fusion RNA was used to form the GST-MS2-circLAMA3-MS2 complex in cells. The cells were lysed at 48 h after transfection, and the target RNA directly bound to circLAMA3 was obtained using glutathione affinity agarose beads. The tagged RNA affinity purification (TRAP) experimental results indicated that circLAMA3 could directly bind to MYCN mRNA. At the same time, we assessed the colocalization of circLAMA3 and MYCN using the FISH assay (Figure S2E). Together, these experimental results demonstrate that the overexpression of circLAMA3 reduces its stability by directly binding MYCN mRNA and decreasing the protein expression of MYCN. These phenomena decrease the expression of CDK6, ultimately inducing cell-cycle arrest and restricting cell proliferation.
Discussion
Increasing evidence has demonstrated that circRNAs play an important role in a variety of human cancers. CircRNA-based diagnostic strategies may provide new insights into cancer. CircPRKCI promotes lung adenocarcinoma proliferation and is one of the most common genomic aberrations in many tumors.24 CiRS-7 is significantly related to AFP, and its high abundance promotes liver microvascular infiltration and plays a role in the progression of liver cancer.25 CircMAN2B2 significantly promotes the proliferation and migration of non-small-cell lung cancer (NSCLC) cells and has excellent diagnostic accuracy.26 In this study, we assessed the differential expression profile of circRNAs in bladder cancer using gene chip technology; and bioinformatics, cell modeling, and clinical sample analyses identified circLAMA3 as the most representative marker that was significantly expressed in multiple of types of bladder cancer. The levels of circLAMA3 were shown to be significantly reduced in both bladder cancer cells and bladder cancer tissues. At the same time, circLAMA3 was correlated with higher grade and T stage in bladder cancer patients. Our study identified circLAMA3 as a potential diagnostic biomarker for bladder cancer, and our bioinformatics analysis showed that circLAMA3 may function via cancer-related pathways. Subsequently, we conducted in vivo and in vitro functional experiments to confirm that low expression of circLAMA3 significantly promotes the development of bladder cancer. This is also the first indication that circLAMA3 inhibits cancer.
CircRNAs are characterized by richness, high stability, and evolutionary conservation among species, highlighting their unique characteristics and biological functions.7,27,28 CircRNAs were first reported to biologically function as miRNA sponges in previous studies. Researchers found overlapping localization of ciRS-7 and miR-7 by performing gene in situ hybridization analysis of the mouse brain, and miRNA activity was found to be decreased in tissues co-expressing ciRS-7 and miR-7.29 In addition, hsa_circ_001783 was shown to significantly promote the proliferation and colony-forming ability of breast cancer cells via the absorption of miR-200c-3p.30 Hsa_circ_0007142 targets miR-103a-2-5p in colorectal cancer cells31 and plays a role in the poor differentiation and lymphatic metastasis of colorectal cancer through the circRNA-miRNA-mRNA network. MiRNAs can be combined with miRNA response elements on mRNAs, circRNAs, and long noncoding RNAS (lncRNAs) by complementary matching in seed sequences,11 forming an endogenous ceRNA network to regulate mRNA transcription.32 As an indispensable component of the ceRNA mechanism, mRNAs may feasibly have circRNA response elements that can interact with circRNAs and mediate their biological functions, but this has not yet been verified. Herein, RegRNA and IntaRNA analyses led to the prediction that circLAMA3 has an MYCN binding site. Therefore, by performing a series of experiments, such as mRNA stability, dual luciferase reporter gene, and TRAP assays, we confirmed that circLAMA3 can directly bind MYCN mRNA. By binding to MYCN mRNA, circLAMA3 induces its degradation and thereby reduces the expression of the MYCN protein, which plays a vital role in the human bladder cancer proliferation process. This is also the first proposal and verification of the theory that circRNAs directly target mRNA sand induce their degradation. Next, we will focus on exploring the molecular mechanism underlying circRNA-induced mRNA degradation.
Our study found that MYCN is upregulated in bladder cancer and involved in the regulation of bladder cancer cell proliferation via circLAMA3. MYCN, an oncogene with DNA-binding transcription factor activity,33 regulates transcription by specifically binding to the bHLH domain of the E box promoter region. The abnormal expression of MYCN is related to the occurrence, metastasis, and treatment of various tumors.34,35 Studies have found that MYCN regulates the proliferation and apoptosis of neuroblastoma by interacting with the transcription target NLRR1,36 and MYCN has been reported to mediate NSCLC chemoresistance by binding to the HES1 promoter, thereby inhibiting apoptosis. High expression of MYCN is closely related to advanced stages of NSCLC and short survival times of these patients.37 In prostate cancer, MYCN can transcriptionally activate PARP1 and BRCA1 to promote malignant cell activities.38 These results indicate that the abnormal expression of MYCN is closely related to tumor progression.39 The results of this study indicate that MYCN mediates the transcriptional activation of CDK6 and participates in the circLAMA3-induced inhibition of bladder cancer cell proliferation. Subsequent rescue experiments further confirmed that ectopic expression of MYCN could significantly restore the inhibitory effect of circLAMA3 on the proliferation of bladder cancer.
Our study found that the upregulation of MYCN increased the promoter activity of CDK6 in bladder cancer cells, indicating that MYCN is a key regulator of CDK6. The CDK6 protein, which is key for classic cell-cycle regulation, binds to cyclin D to thereby promote the phosphorylation of pRb, inducing the release of the transcription factor E2F and initiating DNA replication in the S phase.40 CDK6 is essential for the G0/G1 phase of the cell cycle, and the loss of cell-cycle regulation is the first step in tumor progression. To date, the mechanism by which circRNAs promote CDK6-induced bladder cancer is unclear. The results of this study indicate that circLAMA3 interacts with MYCN and inhibits the expression of CDK6, potentially representing a novel mechanism by which the circLAMA3/MYCN/CDK6 axis regulates the progression of bladder cancer. This study may provide new inspiration for the clinical treatment of bladder cancer.
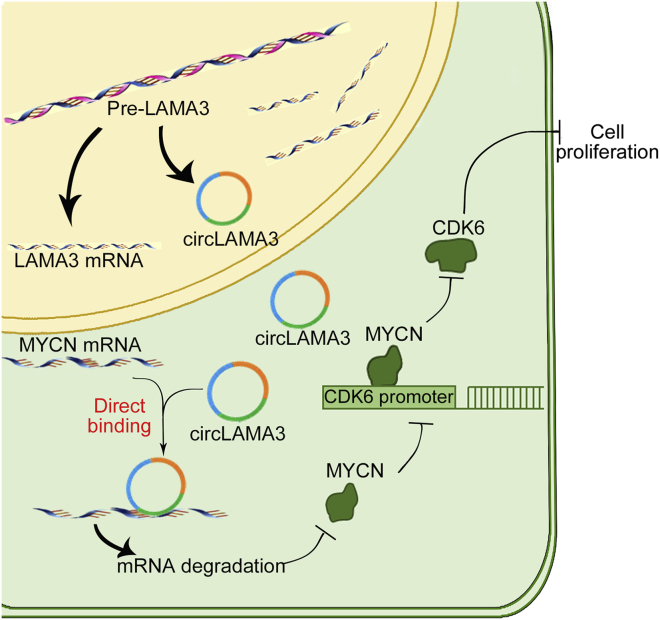
In conclusion, this study is the first to demonstrate the low expression of circLAMA3 in bladder cancer tissues and cells and to propose a new mechanism by which circRNAs directly target mRNA stability (Figure 6). In bladder cancer, as the expression of circLAMA3 decreases, the stability of its direct binding to the target gene MYCN weakens, leading to the increased expression of MYCN. MYCN enhances the activation of CDK6 transcription, promotes the expression of CDK6, and accelerates the G0/G1 phase. This process eventually leads to increased bladder cancer proliferation. These results show that low circLAMA3 expression has potential as a therapeutic target for bladder cancer. In addition, overexpression of circLAMA3 can significantly inhibit the proliferation of bladder cancer.
Figure 6.
Schematic diagram of the molecular mechanism by which circLAMA3 inhibits the proliferation of human bladder cancer cells
Materials and methods
Tissue samples and experimental animals
From 2017 to 2019, 50 pairs of bladder cancer tumor tissues and adjacent tissues were obtained from patients who were diagnosed with bladder cancer and underwent surgery at the First Affiliated Hospital of Wenzhou Medical University. The adjacent tissue was located ≥3 cm from the edge of the bladder cancer tissue. All of the tissue samples were frozen in liquid nitrogen and stored at −80°C. All of the organizational uses were approved by the ethics committee of Wenzhou Medical University, and the patients provided informed consent.
Four-week-old female athymic nude mice were purchased from the Shanghai Slack Experimental Animal Center (experimental animal certificate number, SCXK (Su) 201904657). The mice were raised in the SPF-level experimental area of the Experimental Animal Center of Wenzhou Medical University. All animal experiments were approved by the Animal Research Committee of Wenzhou Medical University, and animal research was conducted in accordance with international guidelines.
Cell culture and transfection
Human normal urothelial SV-HUC-1 cells and human bladder cancer cell lines (RT4, T24, J82, UM-UC-3, and TCCSUP) were used. The SV-HUC-1 cell line was obtained from American Type Culture Collection (ATCC), and the RT4, T24, J82, UM-UC-3, and TCCSUP cell lines were obtained from the National Collection of Authenticated Cell Cultures. SV-HUC-1 cells were cultured in F-12K medium (21127022, Gibco, New York, USA) supplemented with 10% fetal bovine serum (FBS; 1750114, Gibco). J82 and TCCSUP cells were cultured in Minimum Essential Medium (MEM) (11095080, Gibco) containing 10% FBS. T24 cells were cultured in DMEM-F12 medium (10565018, Gibco) containing 5% FBS. UM-UC-3 and RT4 cells were cultured in DMEM (11995065, Gibco) containing 10% FBS. The cells were cultured in a humid environment (5% CO2 and 95% O2) at 37°C. Plasmids were transfected in vitro using PolyJet Transfection Reagent (SignaGen Laboratories, Gaithersburg, MD, USA) according to the manufacturer's instructions, and RiboFect CP (riboBio, Guangzhou, China) was used to transfect siRNA (GenePharma, Shanghai, China). All of the siRNA sequences are shown in Table S1.
RNA extraction, reverse transcription, and qPCR
Total RNA was isolated from the tissues and cells manually using TRIzol reagent (Invitrogen, Grand Island, NY, USA), and cDNA was synthesized using a SuperScript IV First-Strand Synthesis System (Invitrogen) and random primers (Invitrogen). Real-time quantitative PCR (qPCR) analysis was performed on a QuantStudio 6 (Q6) Real-Time qPCR System (Thermo Fisher Scientific, Waltham, USA), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as the internal control. All of the primer sequences are shown in Table S1.
CCK-8 assay of cell viability
A Cell Counting Kit-8 (CCK-8) (Beyotime Biotechnology, Shanghai, China) was used to assess cell viability. The cells were collected and seeded in a 96-well plate at 2,000 cells/100 μL/well. The cells were cultured at 5% CO2 at 37°C for 12 h before being transfected. The fresh medium was provided, and the culturing continued for 48 h. A total of 10 μL of CCK-8 reagent was added to each well, and the absorbance was detected at 450 nm after 2 h of incubation.
EdU assay of cell proliferation
A Cell-Light EdU Apollo 567 in vitro imaging kit (riboBio) was used according to the manufacturer's instructions. The main steps included cell culture, 5-ethynyl-2'-deoxyuridine (EdU) labeling, cell immobilization, Apollo staining, DNA staining, and fluorescence microscope photography. The data were analyzed using ImageJ software.
FCM assays of the cell cycle and apoptosis
The transfected cells were stained with propidium iodide (KGA512, KeyGen Biotech, Nanjing, China), and the cell cycle was assessed using CytoFLEX FCM (Beckman Coulter, Brea, CA, USA). The percentages of cells in the G0/G1, S, and G2 phases were determined and compared. For apoptosis detection, the cells were stained with an Annexin V-FITC/PI KGA106 Apoptosis Detection Kit (Keygen Biotech), and apoptosis was detected using CytoFLEX FCM. The ratio of early apoptotic cells to late apoptotic cells was calculated.
Wound-healing assay of cell migration
A total of 5 × 105 cells were seeded in 6-well plates and reached 95% confluence after 48 h of transfection. They were washed twice with PBS, after which fresh medium was provided, and a scratch of uniform width was made with a sterile pipette tip. The same portion of the scratch was photographed at specified times to assess the scratch width, and the scratch area was measured. The experiment was repeated three times.
Transwell assay of cell invasion
A total of 3 × 104 cells in 0.1% FBS medium were seeded in a Transwell chamber (353097, Corning, NY, USA) and allowed to migrate for 24 h. Invasion was assessed according to the manufacturer's instructions (354480, Corning BioCoat Matrigel invasion chambers). A total of 3 × 104 cells were seeded in 400 μL of 0.1% medium in the upper chamber, and 700 μL of complete medium was placed in the lower chamber. The cells were allowed to invade the chambers for 24 h. The cells were fixed with 3.7% formalin for 5 min at room temperature, washed twice with PBS, and transferred to 100% methanol for 20 min. They were then washed twice with PBS and stained with Giemsa (1:1 dilution in PBS) for 15 min in the dark at room temperature. They were washed twice with PBS, and cells were scraped from the upper surface of the membrane with a cotton swab. Five areas were randomly selected for counting the cells on the membrane under a light microscope (DMi1).
FISH assay of subcellular localization
A total of 4 × 105 cells were seeded into 12-well plates and reached 80% confluence after 24 h of transfection. The following assay was performed according to the manufacturer's instructions (BersinBio, Guangzhou, China). The main steps included cell fixation, cell permeabilization, cell dehydration, hybridization, DAPI staining, and fluorescence microscopy photography. The data were analyzed using ImageJ software.
Western blotting
The cells were collected, and protein was extracted from the cell lysate with lysis buffer (10 mM tris(hydroxymethyl)aminomethane hydrochloride [Tris-HCl] [pH 7.4], 1% SDS, 1 mM Na3VO4, and proteasome inhibitor). A NanoDrop 2000 instrument (Thermo Fisher Scientific, Holtsville, NY, USA) was used to determine the protein concentration. WB bands were detected using an ECF substrate kit (RPN5787, GE Healthcare, Butler, PA, USA), and images were acquired using a Typhoon FLA 7000 imager (GE Healthcare).
Double luciferase reporter assay
The CDK6 promoter luciferase reporter vector was co-transfected with the TK plasmid into T24/J82 (OE circLAMA3) cells using the Dual-Luciferase Reporter Assay System (Promega, Fitchburg, WI, USA) in accordance with the manufacturer's instructions. After 24 h, the cells were lysed with PLB lysate for 15 min. A total of 20 μL of lysate was pipetted into a 96-well plate. After the addition of luciferase reagent (LAR II), the first value was detected by chemiluminescence (Centro LB 960, Berthold Technologies, Germany) and recorded as the luciferin activity. After Stop & Glo solution was added, the machine test recorded the second time value as the TK value. The first test results were corrected with the TK values.
mRNA stability test
Cells were plated on a 6-well plate at a certain density. When the cell density reached approximately 90%, ActD (10 μg/mL) was added for 0, 2, 4, and 8 h. The cells were then collected and washed with PBS, after which 1 mL of TRIzol reagent was added to lyse the cells. Cellular RNA was extracted, and the mRNA degradation rate was determined by qPCR.
Tagged RNA affinity purification experiment
A TRAP kit (Bes5106, BersinBio, Guangzhou, China) was performed in accordance with the manufacturer's instructions. The main steps included cell lysate preparation, pulldown, RNA purification, and RNA analysis. For TRAP-RNA, qPCR was conducted using a reverse transcription kit and a qPCR kit. A total of 3–5 μL of qPCR products was used to perform agarose gel electrophoresis to assess the amplification.
Statistical analysis
All of the experimental data are expressed as the mean ± SD. Data processing was performed using SPSS 18.0 statistical software. GraphPad Prism 5.0 software (GraphPad Software, San Diego, CA, USA) was used to statistically analyze the data; the t test was used to determine the statistical significance between the treated and untreated groups. The experimental results were obtained from at least three independent experiments and are expressed as the mean ± SD. A value of p < 0.05 was considered to indicate statistical significance between the experimental group and the control group.
Acknowledgments
We thank doctor Dapang Rao of the Second Affiliated Hospital and Yuying Children's Hospital of Wenzhou Medical University, Wenzhou, Zhejiang 325027, China, for critical technical support. This work was partially supported by the Natural Science Foundation of China (NSFC81903356).
Author contributions
Aruo Nan conceived and designed the study. Shuilian Wu and Haotian Xu performed the experiments and manuscript writing. Ruirui Zhang, Xin Wang, Jialei Yang, Sixian Chen, Xiaofei Li, and Wanting He collected the clinical samples, analyzed the data, and revised manuscript. All authors read and approved the final manuscript.
Declaration of interests
The authors have no conflicts of interest to declare.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omto.2022.02.020.
Supplemental information
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Keck B., Stoehr R., Wach S., Rogler A., Hofstaedter F., Lehmann J., Montironi R., Sibonye M., Fritsche H.M., Lopez-Beltran A., et al. The plasmacytoid carcinoma of the bladder--rare variant of aggressive urothelial carcinoma. Int. J. Cancer. 2011;129:346–354. doi: 10.1002/ijc.25700. [DOI] [PubMed] [Google Scholar]
- 3.Grayson M. Bladder cancer. Nature. 2017;551:S33. doi: 10.1038/551S33a. [DOI] [PubMed] [Google Scholar]
- 4.Xiao T., Xue J., Shi M., Chen C., Luo F., Xu H., Chen X., Sun B., Sun Q., Yang Q., et al. Circ008913, via miR-889 regulation of DAB2IP/ZEB1, is involved in the arsenite-induced acquisition of CSC-like properties by human keratinocytes in carcinogenesis. Metallomics. 2018;10:1328–1338. doi: 10.1039/c8mt00207j. [DOI] [PubMed] [Google Scholar]
- 5.Berrout J., Kyriakopoulou E., Moparthi L., Hogea A.S., Berrout L., Ivan C., Lorger M., Boyle J., Peers C., Muench S., et al. TRPA1-FGFR2 binding event is a regulatory oncogenic driver modulated by miRNA-142-3p. Nat. Commun. 2017;8:947. doi: 10.1038/s41467-017-00983-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiao Y., Liu G., Sun Y., Gao Y., Ouyang X., Chang C., Gong L., Yeh S. Targeting the estrogen receptor alpha (ERalpha)-mediated circ-SMG1.72/miR-141-3p/Gelsolin signaling to better suppress the HCC cell invasion. Oncogene. 2020;39:2493–2508. doi: 10.1038/s41388-019-1150-6. [DOI] [PubMed] [Google Scholar]
- 7.Kristensen L.S., Hansen T.B., Veno M.T., Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37:555–565. doi: 10.1038/onc.2017.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meng S., Zhou H., Feng Z., Xu Z., Tang Y., Li P., Wu M. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol. Cancer. 2017;16:94. doi: 10.1186/s12943-017-0663-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J., Yin J., Wang X., Liu H., Hu Y., Yan X., Zhuang B., Yu Z., Han S. Changing expression profiles of mRNA, lncRNA, circRNA, and miRNA in lung tissue reveal the pathophysiological of bronchopulmonary dysplasia (BPD) in mouse model. J. Cell. Biochem. 2019;120:9369–9380. doi: 10.1002/jcb.28212. [DOI] [PubMed] [Google Scholar]
- 10.Jian X., He H., Zhu J., Zhang Q., Zheng Z., Liang X., Chen L., Yang M., Peng K., Zhang Z., et al. Hsa_circ_001680 affects the proliferation and migration of CRC and mediates its chemoresistance by regulating BMI1 through miR-340. Mol. Cancer. 2020;19:20. doi: 10.1186/s12943-020-1134-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guan Y.J., Ma J.Y., Song W. Identification of circRNA-miRNA-mRNA regulatory network in gastric cancer by analysis of microarray data. Cancer Cell Int. 2019;19:183. doi: 10.1186/s12935-019-0905-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li W.H., Song Y.C., Zhang H., Zhou Z.J., Xie X., Zeng Q.N., Guo K., Wang T., Xia P., Chang D.M. Decreased expression of Hsa_circ_00001649 in gastric cancer and its clinical significance. Dis. Markers. 2017;2017:4587698. doi: 10.1155/2017/4587698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li P., Chen S., Chen H., Mo X., Li T., Shao Y., Xiao B., Guo J. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin. Chim. Acta. 2015;444:132–136. doi: 10.1016/j.cca.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 14.Fu L., Yao T., Chen Q., Mo X., Hu Y., Guo J. Screening differential circular RNA expression profiles reveals hsa_circ_0004018 is associated with hepatocellular carcinoma. Oncotarget. 2017;8:58405–58416. doi: 10.18632/oncotarget.16881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng Q., Bao C., Guo W., Li S., Chen J., Chen B., Luo Y., Lyu D., Li Y., Shi G., et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nan A., Chen L., Zhang N., Jia Y., Li X., Zhou H., Ling Y., Wang Z., Yang C., Liu S., Jiang Y. Circular RNA circNOL10 inhibits lung cancer development by promoting SCLM1-mediated transcriptional regulation of the humanin polypeptide family. Adv. Sci. (Weinh) 2019;6:1800654. doi: 10.1002/advs.201800654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu W.Y. Roles of the circular RNA circ-Foxo3 in breast cancer progression. Cell Cycle. 2017;16:589–590. doi: 10.1080/15384101.2017.1278935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khleif S.N., DeGregori J., Yee C.L., Otterson G.A., Kaye F.J., Nevins J.R., Howley P.M. Inhibition of cyclin D-CDK4/CDK6 activity is associated with an E2F-mediated induction of cyclin kinase inhibitor activity. Proc. Natl. Acad. Sci. U S A. 1996;93:4350–4354. doi: 10.1073/pnas.93.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barrios N., Campuzano S. Expanding the Iroquois genes repertoire: a non-transcriptional function in cell cycle progression. Fly (Austin) 2015;9:126–131. doi: 10.1080/19336934.2016.1139654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H.L., Ma Y., Ma Y., Li Y., Chen X.B., Dong W.L., Wang R.L. The design of novel inhibitors for treating cancer by targeting CDC25B through disruption of CDC25B-CDK2/Cyclin A interaction using computational approaches. Oncotarget. 2017;8:33225–33240. doi: 10.18632/oncotarget.16600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun X., Zhangyuan G., Shi L., Wang Y., Sun B., Ding Q. Prognostic and clinicopathological significance of cyclin B expression in patients with breast cancer: a meta-analysis. Medicine (Baltimore) 2017;96:e6860. doi: 10.1097/MD.0000000000006860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woo C.W., Tan F., Cassano H., Lee J., Lee K.C., Thiele C.J. Use of RNA interference to elucidate the effect of MYCN on cell cycle in neuroblastoma. Pediatr. Blood Cancer. 2008;50:208–212. doi: 10.1002/pbc.21195. [DOI] [PubMed] [Google Scholar]
- 23.Yang M., Lin X., Rowe A., Rognes T., Eide L., Bjoras M. Transcriptome analysis of human OXR1 depleted cells reveals its role in regulating the p53 signaling pathway. Sci. Rep. 2015;5:17409. doi: 10.1038/srep17409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qiu M., Xia W., Chen R., Wang S., Xu Y., Ma Z., Xu W., Zhang E., Wang J., Fang T., et al. The circular RNA circPRKCI promotes tumor growth in lung adenocarcinoma. Cancer Res. 2018;78:2839–2851. doi: 10.1158/0008-5472.CAN-17-2808. [DOI] [PubMed] [Google Scholar]
- 25.Xue J., Liu Y., Luo F., Lu X., Xu H., Liu X., Lu L., Yang Q., Chen C., Fan W., Liu Q. Circ100284, via miR-217 regulation of EZH2, is involved in the arsenite-accelerated cell cycle of human keratinocytes in carcinogenesis. Biochim. Biophys. Acta Mol. Basis Dis. 2017;1863:753–763. doi: 10.1016/j.bbadis.2016.12.018. [DOI] [PubMed] [Google Scholar]
- 26.Ma X., Yang X., Bao W., Li S., Liang S., Sun Y., Zhao Y., Wang J., Zhao C. Circular RNA circMAN2B2 facilitates lung cancer cell proliferation and invasion via miR-1275/FOXK1 axis. Biochem. Biophys. Res. Commun. 2018;498:1009–1015. doi: 10.1016/j.bbrc.2018.03.105. [DOI] [PubMed] [Google Scholar]
- 27.Zhang H.D., Jiang L.H., Sun D.W., Hou J.C., Ji Z.L. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25:1–7. doi: 10.1007/s12282-017-0793-9. [DOI] [PubMed] [Google Scholar]
- 28.Feng J., Chen K., Dong X., Xu X., Jin Y., Zhang X., Chen W., Han Y., Shao L., Gao Y., He C. Genome-wide identification of cancer-specific alternative splicing in circRNA. Mol. Cancer. 2019;18:35. doi: 10.1186/s12943-019-0996-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hansen T.B., Kjems J., Damgaard C.K. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73:5609–5612. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]
- 30.Liu Z., Zhou Y., Liang G., Ling Y., Tan W., Tan L., Andrews R., Zhong W., Zhang X., Song E., Gong C. Circular RNA hsa_circ_001783 regulates breast cancer progression via sponging miR-200c-3p. Cell Death Dis. 2019;10:55. doi: 10.1038/s41419-018-1287-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu C.L., Sha X., Wang Y., Li J., Zhang M.Y., Guo Z.Y., Sun S.A., He J.D. Circular RNA hsa_circ_0007142 is upregulated and targets miR-103a-2-5p in colorectal cancer. J. Oncol. 2019;2019:9836819. doi: 10.1155/2019/9836819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghizoni J.S., Nichele R., de Oliveira M.T., Pamato S., Pereira J.R. The utilization of saliva as an early diagnostic tool for oral cancer: microRNA as a biomarker. Clin. Transl. Oncol. 2020;22:804–812. doi: 10.1007/s12094-019-02210-y. [DOI] [PubMed] [Google Scholar]
- 33.Chipumuro E., Marco E., Christensen C.L., Kwiatkowski N., Zhang T., Hatheway C.M., Abraham B.J., Sharma B., Yeung C., Altabef A., et al. CDK7 inhibition suppresses super-enhancer-linked oncogenic transcription in MYCN-driven cancer. Cell. 2014;159:1126–1139. doi: 10.1016/j.cell.2014.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang H., Hong B., Li X., Deng K., Li H., Yan Lui V.W., Lin W. JQ1 synergizes with the Bcl-2 inhibitor ABT-263 against MYCN-amplified small cell lung cancer. Oncotarget. 2017;8:86312–86324. doi: 10.18632/oncotarget.21146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qin X.Y., Suzuki H., Honda M., Okada H., Kaneko S., Inoue I., Ebisui E., Hashimoto K., Carninci P., Kanki K., et al. Prevention of hepatocellular carcinoma by targeting MYCN-positive liver cancer stem cells with acyclic retinoid. Proc. Natl. Acad. Sci. U S A. 2018;115:4969–4974. doi: 10.1073/pnas.1802279115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hossain S., Takatori A., Nakamura Y., Suenaga Y., Kamijo T., Nakagawara A. NLRR1 enhances EGF-mediated MYCN induction in neuroblastoma and accelerates tumor growth in vivo. Cancer Res. 2012;72:4587–4596. doi: 10.1158/0008-5472.CAN-12-0943. [DOI] [PubMed] [Google Scholar]
- 37.Tong Q., Ouyang S., Chen R., Huang J., Guo L. MYCN-mediated regulation of the HES1 promoter enhances the chemoresistance of small-cell lung cancer by modulating apoptosis. Am. J. Cancer Res. 2019;9:1938–1956. [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang W., Liu B., Wu W., Li L., Broom B.M., Basourakos S.P., Korentzelos D., Luan Y., Wang J., Yang G., et al. Targeting the MYCN-PARP-DNA damage response pathway in neuroendocrine prostate cancer. Clin. Cancer Res. 2018;24:696–707. doi: 10.1158/1078-0432.CCR-17-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hansel D.E., Swain E., Dreicer R., Tubbs R.R. HER2 overexpression and amplification in urothelial carcinoma of the bladder is associated with MYC coamplification in a subset of cases. Am. J. Clin. Pathol. 2008;130:274–281. doi: 10.1309/41VLTFX3YPP1HF6F. [DOI] [PubMed] [Google Scholar]
- 40.Sarma U., Biswas I., Das A., Das G.C., Saikia C., Sarma B. p16INK4a expression in cervical lesions correlates with histologic grading - a tertiary level medical facility based retrospective study. Asian Pac. J. Cancer Prev. 2017;18:2643–2647. doi: 10.22034/APJCP.2017.18.10.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.