Abstract
Cocaine addiction remains a serious problem lacking an effective pharmacological treatment. Thus, we have developed a high-affinity anti-cocaine monoclonal antibody (mAb), h2E2, for the treatment of cocaine use disorders. We show that selective tryptophan (Trp) oxidation by 2,2′-azobis(2-amidinopropane) dihydrochloride (AAPH) resulted in a loss of high-affinity binding of cocaine to this mAb. The newly developed use of excess methionine (Met) to protect mAb met residues from AAPH oxidation did not substantially attenuate the effects of oxidation on cocaine binding but greatly decreased the modification of met residues in the mAb. Similar large decreases in ligand affinity (5000–10,000-fold) upon oxidation were observed using cocaine and two cocaine metabolites, cocaethylene and benzoylecgonine, which also bind with nanomolar affinity to this h2E2 mAb. The decrease in binding affinity was accompanied by a decrease of approximately 50% in Trp fluorescence, and increases in mAb 310 to 370 nm absorbance were consistent with the presence of oxidized forms of Trp. Finally, mass spectral analysis of peptides derived from control and AAPH-oxidized mAb indicated that excess free met did effectively protect mAb met residues from oxidation, and that AAPH-oxidized mAb heavy-chain Trp33 and light-chain Trp91 residues are important for cocaine binding, consistent with a recently derived h2E2 Fab fragment crystal structure containing bound benzoylecgonine. Thus, protection of the anti-cocaine h2E2 mAb from Trp oxidation prior to its clinical administration is critical for its proposed therapeutic use in the treatment of cocaine use disorders.
Keywords: tryptophan, oxidation, monoclonal antibody, cocaine, ligand affinity, addiction therapy
Abbreviations: AAPH, 2,2′-azobis(2-amidinopropane) dihydrochloride; apo h2E2, unliganded h2E2; BE, benzoylecgonine; CE, cocaethylene; DASPMI, 4-(4-(dimethylamino)styryl)-N-methylpyridinium iodide; DSF, differential scanning fluorimetry; FA, formic acid; h2E2, humanized anticocaine monoclonal antibody; HC, heavy chain; LC, light chain; mAb, monoclonal antibody; Met, l-methionine; RBA, relative binding affinity; RTI, Research Triangle Institute; TmB, Boltzmann fitting–derived melting temperature; TmD, derivative-derived melting temperature; Trp, l-tryptophan
Monoclonal antibodies (mAbs) have been increasingly employed as therapeutic drugs for treating many different conditions and diseases. There are currently no Food and Drug Administration–approved therapies for the treatment of cocaine use disorders (1). Thus, we have developed and characterized a high-affinity anti-cocaine mAb, h2E2, which also binds the active metabolite of cocaine, cocaethylene (CE), and one of the inactive metabolites, benzoylecgonine (BE), with high (nanomolar) affinity (2, 3, 4, 5). We have published several ways to characterize the function (cocaine and cocaine metabolite binding) of this mAb, which include ligand-induced intrinsic (tyrosine and tryptophan [Trp]) mAb fluorescence quenching (6) and extrinsic fluorescent dyes and differential scanning fluorimetry (DSF) (7) and dye absorbance (5) to quantitate ligand binding to this mAb. We also recently published a crystal structure of the Fab fragment of this h2E2 mAb (8). Although this crystal structure was consistent with several tyrosine and Trp residues being important for BE binding (the metabolite of cocaine that was cocrystalized with the Fab fragment), it did not explain the observed different affinities of the mAb for CE, cocaine, and BE. Also, it did not suggest which, if any, of these amino acid residues are important for any possible loss of therapeutic function (loss of high-affinity cocaine binding) because of spontaneous oxidation that could occur during production, formulation, storage, or clinical usage of this mAb.
In this study, we used the oxidant, 2,2′-azobis(2-amidinopropane) dihydrochloride (AAPH) as an indicator of mAb oxidation susceptibility (9), both in the absence and presence of a high concentration of methionine (Met; 260 mM), to protect met mAb residues from oxidation (10, 11). Thus, the effects of Trp-selective oxidation on mAb high-affinity binding of cocaine and metabolites were determined, followed by mass spectral analysis of proteolytic peptides to identify residues that were oxidized, and whose oxidation resulted in loss of high-affinity binding. We employed several recently developed methodologies in order to characterize the effects of oxidation on h2E2 mAb ligand binding and stability, including 4-(4-(dimethylamino)styryl)-N-methylpyridinium iodide (DASPMI) dye–based DSF (12), DASPMI-based quantitation of the number of high-affinity binding sites (5), and nonreducing SDS-PAGE after mild heating of mAb samples. The methods explore possible oxidation-induced differences in susceptibility of mAb domains to SDS-induced unfolding of protein structure (12). We identified heavy-chain (HC) and light-chain (LC) complementarity-determining region Trp residues whose oxidation corresponded to a loss of high-affinity cocaine and cocaine metabolite binding. Specifically, oxidized HC Trp33 and LC Trp91 (both in the cocaine-binding site (8)) could be detected and quantified using liquid chromatography–mass spectrometry after AAPH oxidation both with and without met. The results indicated that excess Met did indeed protect oxidation-susceptible Met residues from AAPH oxidation, whereas not substantially reducing the extent of oxidation of two mAb variable region Trp residues, HC Trp33 and LC Trp91, in h2E2 mAb samples exhibiting a loss of high-affinity cocaine binding upon mild oxidation.
Results
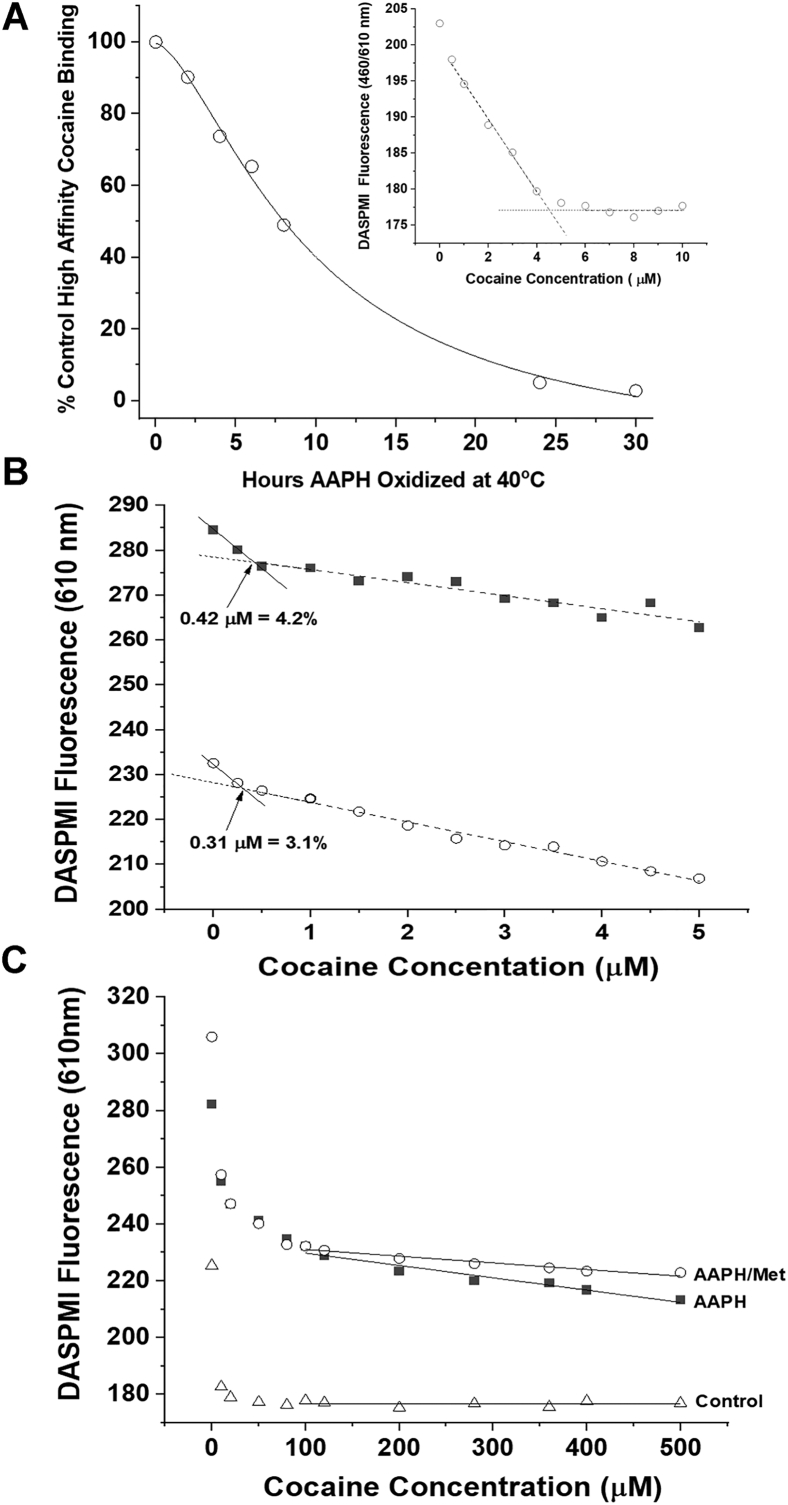
Earlier published results utilizing the anti-cocaine h2E2 mAb suggested that Trp residues are important for cocaine and high-affinity cocaine metabolite binding to this mAb (6, 8), so forced oxidation of Trp by AAPH was utilized, both in the absence and presence of a high concentration of free Met (260 mM, to protect mAb Met from oxidation (11)). It should be noted that to make these experiments most relevant to any possible oxidation that may occur during production, storage, or clinical utilization of the anti-cocaine h2E2 mAb, the oxidations were performed in the buffer used for its clinical formulation, which is 10 mM histidine, 10% sucrose, and 0.01% polysorbate 80, pH = 6.0. Thus, it is expected in the samples with added Met, that both Met and histidine should be at least partly protected from possible AAPH oxidation by the presence of those free amino acids. Oxidation of 19 mg/ml (≈130 μM) h2E2 mAb in the formulation buffer utilizing 5 mM AAPH at 40 °C resulted in a time-dependent decrease in high-affinity cocaine binding, as assessed by a newly developed assay using the fluorescence of the DASPMI reporter dye to measure high-affinity cocaine binding (Fig. 1A, and the inset of Fig. 1A; (13)). As seen in Figure 1B, nearly all high-affinity cocaine binding is abolished (96–97% reduction) after 24 h of AAPH treatment, both in the absence (filled square symbols) and the presence of 260 mM Met (open circles). Using control (untreated or sham processed) mAb, this assay results in a straight line with a slope of zero at saturating cocaine concentrations (see 2.5 μM of mAb control data in the inset of Fig. 1, A and C). Thus, the nonzero ligand–saturated slopes seen in the DASPMI binding assays shown in Figure 1, B and C suggest that there is low-affinity cocaine binding remaining in the AAPH-oxidized samples, and that this binding affinity might be in the high micromolar range, since the nonzero slope continues out to at least 500 μM cocaine for both the AAPH only and the AAPH/Met-oxidized mAb (Fig. 1C).
Figure 1.
The effect of AAPH oxidation on cocaine binding to h2E2 mAb.A, the time course of AAPH oxidation induced loss of high-affinity cocaine binding to h2E2 mAb at 40 °C in formulation buffer (plus 260 mM methionine). Inset shows an example of the DASPMI high-affinity cocaine-binding fluorescence assay used to obtain the data, using the control (0 h at 40 °C) mAb as example data. B, DASPMI-binding assay used to determine the percentage of high-affinity cocaine-binding sites remaining, comparing data obtained after 24 h of oxidation at 40 °C with AAPH, in the absence (top) and presence of 260 mM methionine to attenuate methionine oxidation. C, binding of the AAPH-oxidized mAb samples from B titrated to very high concentrations of cocaine, compared with the control h2E2 mAb over the same cocaine concentration range. Straight line fits to data are shown in A (inset), B, and C. Note the nonzero slopes of the straight line fits at the high concentrations of cocaine in the oxidized samples in B and C, indicating that low-affinity cocaine binding is likely present in these oxidized mAb samples. AAPH, 2,2′-azobis(2-amidinopropane) dihydrochloride; DASPMI, 4-(4-(dimethylamino)styryl)-N-methylpyridinium iodide; mAb, monoclonal antibody
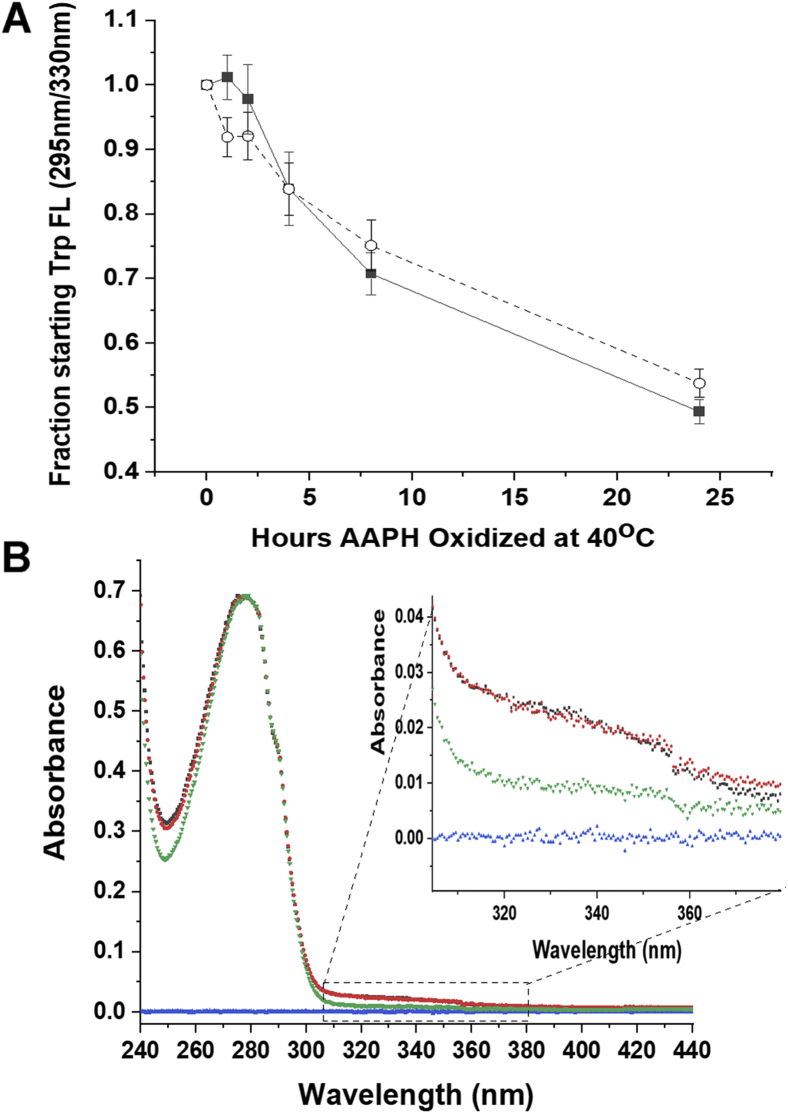
The effect of AAPH oxidation on mAb Trp fluorescence is fairly dramatic, decreasing this fluorescence about 50% both in the absence and presence of Met, after 24 h of oxidation at 40 °C (Fig. 2A, Trp fluorescence measured using excitation at 295 nm and emission at 330 nm). We note that no change in the maximal Trp emission wavelength (330 nm) was observed after AAPH oxidation (not shown). The effects of AAPH oxidation on the UV absorbance of the mAb after 24 h of oxidation at 40 °C are shown in Figure 2B (measured after separation of the mAb from excess reagents). There was also little, if any, change in the 280 nm absorbance caused by AAPH oxidation as a function of time (data not shown). However, there was an increase in absorbance in the 310 to 370 nm range (Fig. 2B, expanded inset), suggestive of a small amount of Trp oxidation products (because of the production of kynurenine and N′-formylkynurenine, which absorb at about 365 and 325 nm, respectively (14)). In addition, an increase in the mAb absorbance at the minimum of about 250 nm caused by oxidation was also noted (Fig. 2B).
Figure 2.
Spectroscopic effects of AAPH oxidation on the h2E2 mAb.A, the time course of the decrease in tryptophan fluorescence (excitation at 295 nm, emission at 330 nm) upon oxidation with 5 mM AAPH, in the absence (filled squares) and the presence of 260 mM methionine (open circles). Error bars denote the SEMs for three independent experiments. B, the effects of 24 h of 5 mM AAPH oxidation at 40 °C on the absorbance spectrum of the h2E2 mAb. Shown are the buffer blank (blue), untreated mAb (green), AAPH-oxidized mAb (black), and AAPH/260 mM methionine-oxidized mAb (red). The AAPH-oxidized samples were separated from excess reagents using a 40 kDa MWCO spin filter, prior to 40-fold dilution into 20 mM Mops/200 mM NaCl, pH = 7.4 buffer, and spectral measurements. Very little, if any, change in absorbance at 280 nm was observed, so the mAb spectra were normalized to yield the same absorbance at 280 nm for easier visual comparison of spectra. Note the increase in absorbance at 310 to 370 nm (see inset) as well as the increase in the absorbance minimum at about 250 nm because of AAPH oxidation. AAPH, 2,2′-azobis(2-amidinopropane) dihydrochloride; mAb, monoclonal antibody; MWCO, molecular weight cutoff.
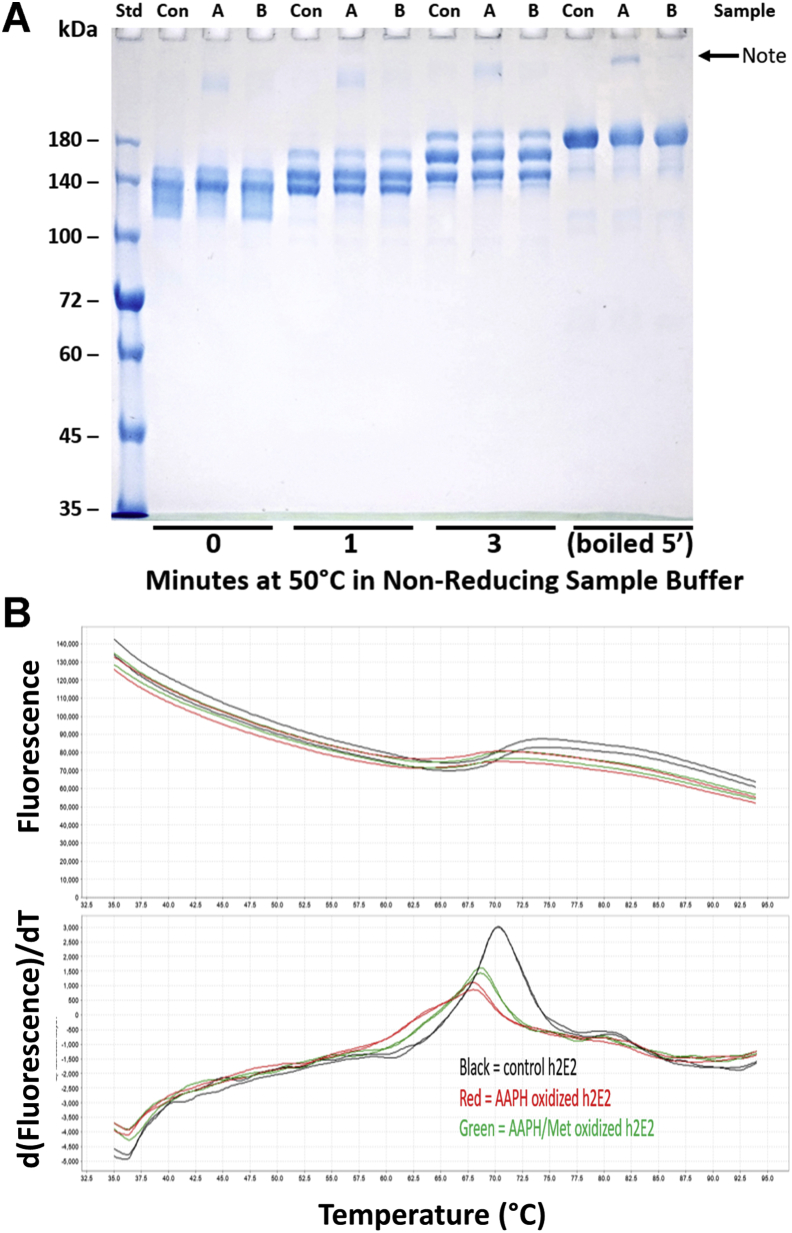
The effects of AAPH oxidation on the stability of the unfolding domains of the mAb were assessed by both nonreducing SDS-PAGE with no or mild sample heating prior to electrophoresis (12, 15) and by DSF using the rotor DASPMI dye (16) shown to report on cocaine binding and to not bind to the Fc part of the mAb (7). As shown in Figure 3A, the susceptibility to partial domain unfolding induced by no or mild heating (heated at 50 °C for a few minutes) in 5% SDS Laemmli sample buffer (containing no reductant) is not changed by AAPH oxidation either without (“A” lanes) or with Met present during oxidation (“B” lanes) from the control mAb (“Con” lanes). There is a small amount of high molecular weight aggregate present in the AAPH-only samples (“A” lanes), which is not evident in either the control or the AAPH/Met-oxidized samples, suggesting that this small amount of aggregate is mediated by Met oxidation of the mAb. Using the more sensitive technique of DSF and DASPMI rotor dye (7) to assess mAb stability, there is a destabilization in the portion of the mAb variable region that binds the dye and reports on the thermal stabilization induced by cocaine binding, as indicated by the decrease in the melting temperatures (TmDs) for the oxidized mAbs, with the TmD being slightly lower for the 24 h, 40 °C 5 mM AAPH-oxidized sample without added Met (the red traces in Fig. 3B, see also Table 1).
Figure 3.
Comparison of the susceptibility to SDS and thermally induced denaturation for control and AAPH-oxidized mAb.A, time course of thermally induced domain unfolding at 50 °C for untreated (Con), 5 mM AAPH-oxidized (“A”), and 5 mM AAPH/260 mM Met-oxidized mAb (oxidation at 40 °C for 24 h) in a 5% SDS containing Laemmli sample buffer, run on a nonreducing Laemmli gel. Aliquots of these samples were also boiled to completely denature the mAbs (last three lanes). Note that although no obvious differential susceptibility to SDS-induced domain unfolding because of AAPH oxidation is observed, there is a small amount of large molecular weight aggregate evident in the AAPH-treated mAb, which is virtually absent in the AAPH/Met-treated and control mAb. B, the dye fluorescence and first derivative of the dye fluorescence changes observed using differential scanning fluorimetry (DSF) employing 50 μM DASPMI as the reporter dye in PBS buffer. As evident from the decreased temperatures for the derivative plot peak maximums, the oxidized mAb samples (treated for 24 h at 40 °C with 5 mM AAPH, AAPH-mAb in red, and AAPH/Met-mAb in green) are less thermally stable than the control mAb (control mAb is shown in black traces, duplicates of all three samples are shown in B). AAPH, 2,2′-azobis(2-amidinopropane) dihydrochloride; DASPMI, 4-(4-(dimethylamino)styryl)-N-methylpyridinium iodide; mAb, monoclonal antibody.
Table 1.
Summary of 24 h, 40 °C oxidations of h2E2 mAb in formulation buffer, using 5 mM AAPH or 5 mM AAPH plus 260 mM Met
| Protein | Ligand added | Apo TmD (°C) | Apo TmB (°C) | Kd at TmB (μM) | RBA | Oxidation-induced increase in Kd |
|---|---|---|---|---|---|---|
| Control h2E2 mAb | None | 70.33 ± 0.05 | 70.41 ± 0.03 | |||
| Control h2E2 mAb | CE | 0.0106 ± 0.00596 | 0.366 ± 0.065 | — | ||
| Control h2E2 mAb | Cocaine | 0.0228 ± 0.0132 | (1.00) | — | ||
| Control h2E2 mAb | BE | 0.1508 ± 0.07686 | 5.30 ± 1.30 | — | ||
| AAPH-h2E2 mAb | None | 67.97 ± 0.03 | 67.70 ± 0.03 | |||
| AAPH-h2E2 mAb | CE | 79.7 ± 20.6 | 0.325 ± 0.05 | 7519 X | ||
| AAPH-h2E2 mAb | Cocaine | 242 ± 32 | (1.00) | 10,614 X | ||
| AAPH-h2E2 mAb | BE | 946 ± 373 | 3.81 ± 1.13 | 6273 X | ||
| AAPH/Met-h2E2 mAb | None | 68.60 ± 0.03 | 68.37 ± 0.05 | |||
| AAPH/Met-h2E2 mAb | CE | 72.1 ± 9.0 | 0.310 ± 0.011 | 6802 X | ||
| AAPH/Met-h2E2 mAb | Cocaine | 233 ± 32 | (1.00) | 10,219 X | ||
| AAPH/Met-h2E2 mAb | BE | 926 ± 144 | 4.00 ±0.64 | 6140 X |
mAb samples were analyzed by differential scanning fluorimetry (DSF) in PBS buffer, using the fluorescent DASPMI reporting dye. Data are from three independent experiments, and the values reported are means ± standard deviations. “Apo” denotes no ligand (cocaine or cocaine metabolite) added to the mAb. RBA is the relative binding affinity, assigning the binding affinity for cocaine as 1.0 for each set of samples analyzed using three ligands. TmD is the temperature at the peak maximum for the derivative of the fluorescence DSF curves, and TmB is the temperature at the middle of the unfolding transition determined by Boltzmann fitting of the DSF fluorescence data. Affinities (Kd) at the TmB for each sample are calculated from the DSF data via ΔG values derived from the DSF data, as previously reported (7). The oxidation-induced fold increases in Kd (decrease in affinity) are calculated by dividing the reported average Kd for the oxidized mAb by the average Kd for the control mAb for each of the three ligands, CE, cocaine, and BE.
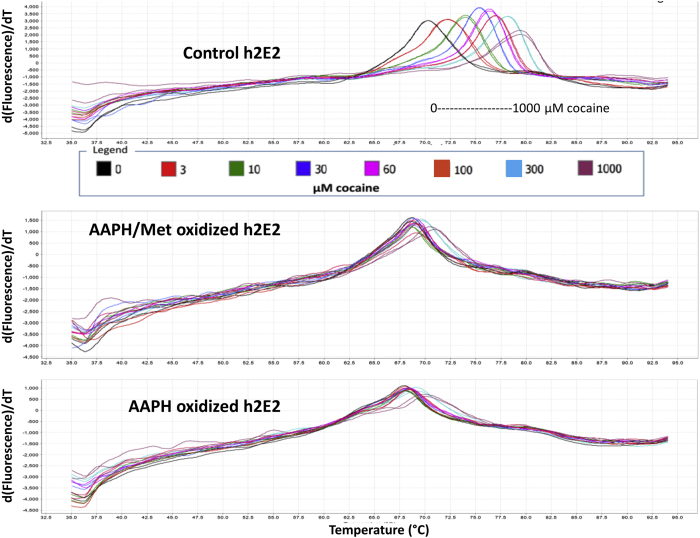
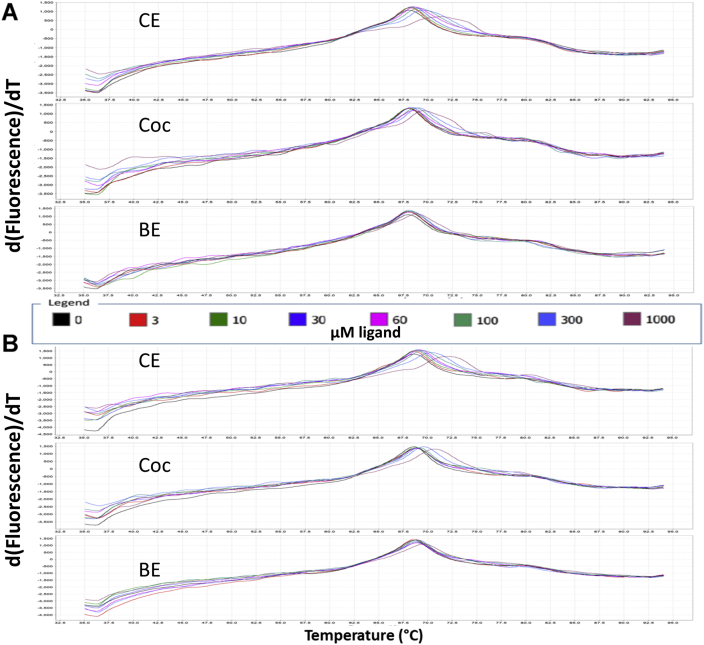
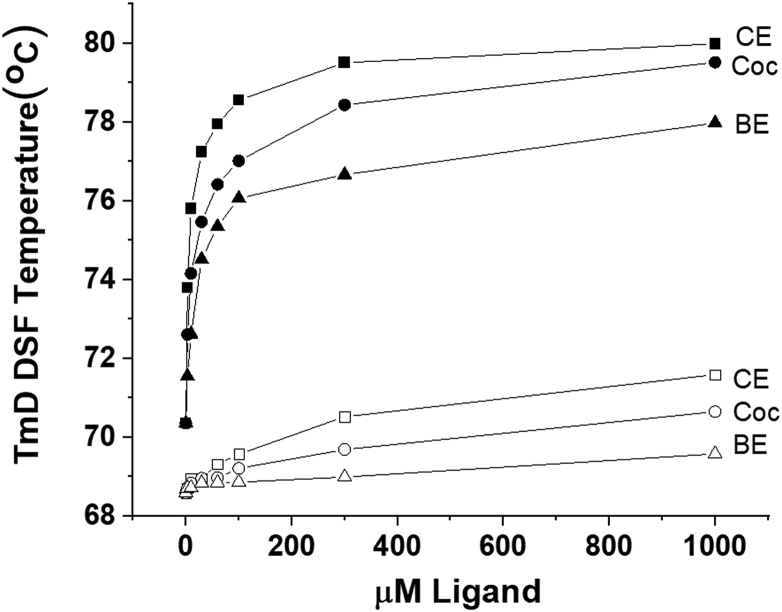
DSF was also used to measure the effect of 24 h AAPH oxidation on the binding of cocaine, as quantitated by the cocaine-induced mAb stabilization, leading to the increase in the temperature at which the thermal unfolding of the ligand-binding domain of the mAb takes place. Thus, control, AAPH/Met, and AAPH-only oxidized samples are compared in Figure 4 regarding the stabilizing effects of concentrations of cocaine, titrating from 1 to 1000 μM cocaine. Obvious shifts in the derivative plot TmD peaks are seen at all concentrations of cocaine used for the control mAb (top), in contrast to both the AAPH-oxidized samples, where TmD shifts are only visually obvious above 100 μM cocaine (Fig. 4). This phenomenon was also observed for the two high-affinity cocaine metabolites, CE and BE, as shown for both the AAPH-only oxidized mAb (Fig. 5A) and the AAPH/Met-oxidized mAb (Fig. 5B). Also evident in Figure 5 are the relative binding affinities (RBAs) of these three ligands, with larger ligand-induced thermal stabilization shifts noted for the higher affinity ligands, with TmD shifts in the rank order CE > cocaine > BE [the Kds for these ligands for control h2E2 mAb at 20 °C are 1, 4, and 20 nM, respectively, for CE, cocaine, and BE, as measured by ligand-induced intrinsic mAb fluorescence quenching at 22 °C (6)]. The TmD DSF values as a function of ligand concentration for a typical such experiment are shown in Figure 6, comparing the control mAb data (filled symbols) with the AAPH/Met-oxidized mAb data (open symbols) for these three ligands.
Figure 4.
DASPMI dye DSF derivative fluorescence data for 2.5 μM h2E2 in PBS buffer: control, AAPH-treated, and AAPH/Met-treated (24 h at 40 °C) mAb as a function of cocaine ligand added (0–1000 μM).Top panel, displays control mAb data, middle panel is data obtained using the AAPH/Met-mAb, whereas the bottom panel is AAPH-treated mAb data (both treated for 24 h at 40 °C). All samples were analyzed in duplicate, using the concentrations of cocaine specified by the legend and the trace colors in the figure. The concentrations of ligands (0–1000 μM) for each data trace are specified by the legend embedded in the figure. AAPH, 2,2′-azobis(2-amidinopropane) dihydrochloride; DASPMI, 4-(4-(dimethylamino)styryl)-N-methylpyridinium iodide; DSF, differential scanning fluorimetry; mAb, monoclonal antibody.
Figure 5.
DASPMI dye DSF derivative fluorescence data for 2.5 μM h2E2 in PBS buffer: 5 mM AAPH-treated and AAPH/Met-treated (24 h at 40 °C) mAb as a function of ligand added (0–1000 μM) for cocaineand two mAb high-affinity cocaine metabolites, cocaethylene (CE) and benzoylecgonine (BE).A, top to bottom, CE, cocaine, and BE ligand titrations for the AAPH-treated mAb. B, top to bottom, CE, cocaine, and BE ligand titrations for the AAPH/Met-treated mAb. The concentrations of ligands (0–1000 μM) for each data trace are specified by the legend embedded in the figure. AAPH, 2,2′-azobis(2-amidinopropane) dihydrochloride; DASPMI, 4-(4-(dimethylamino)styryl)-N-methylpyridinium iodide; DSF, differential scanning fluorimetry; mAb, monoclonal antibody.
Figure 6.
DASPMI DSF derivative peak temperature maximums (TmDs) for control and 24 h, 40 °C 5 mM AAPH/260 mM Met-treated 2.5 μM h2E2 mAb as a function of ligand concentration.Filled symbols are control mAb data, whereas open symbols are the AAPH/Met mAb-oxidized samples, titrated with 0 to 1000 μM CE, cocaine, or BE ligands. TmB values for each sample are all close to TmD values at each ligand concentration, and thus TmB values are not shown to increase the visual clarity of presentation. AAPH, 2,2′-azobis(2-amidinopropane) dihydrochloride; BE, benzoylecgonine; CE, cocaethylene; DASPMI, 4-(4-(dimethylamino)styryl)-N-methylpyridinium iodide; DSF, differential scanning fluorimetry.
A summary of all the DSF data is given in Table 1, which reports the means and standard deviations for the Tms of the apo (i.e., unliganded) mAb as well as the Kd values (calculated at the apo TmB temperature for each sample, as described previously (7)). Also shown in Table 1 are the RBAs for each sample, which compare the Kd values for the three ligands, normalizing the Kd for cocaine to 1.00, for each sample analyzed. In addition, the last column in Table 1 reports the average fold increase in the Kd (at the apo TmB temperature) for all three ligands for the AAPH-oxidized samples, to assess whether the oxidation had any differential effects on the binding of these three ligands. The results in Table 1 indicate that AAPH oxidation destabilizes the ligand-binding domain of the apo mAb (as evidenced by the decreases in the apo TmB and apo TmD values); that the effects on ligand binding of both AAPH oxidation and AAPH/Met oxidation are very similar for all three ligands, with the Kd values for all three ligands being increased by oxidation (denoting a lowering of affinities) by about 5000-fold to 10,000-fold (from nanomolar to micromolar affinities); and that the RBA of these three mAb ligands after AAPH oxidation are not changed.
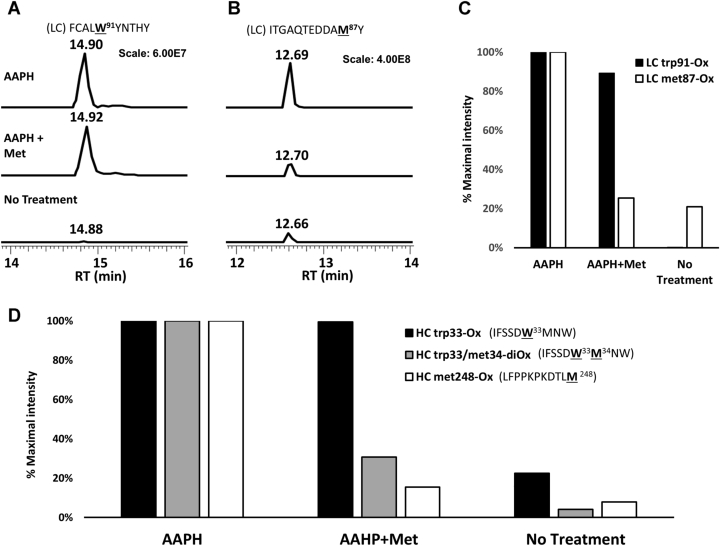
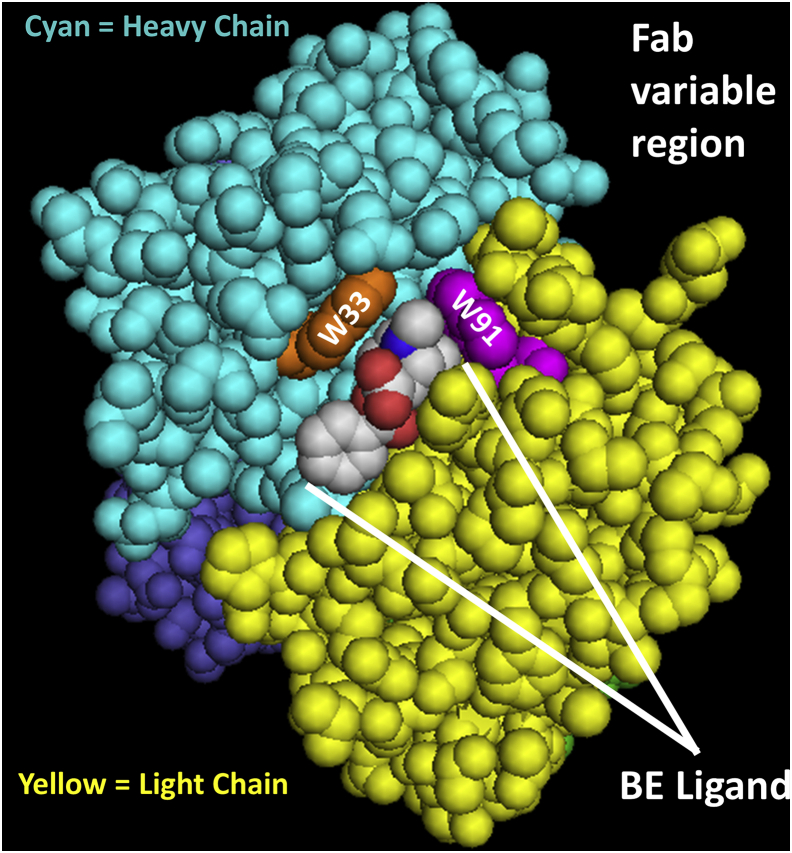
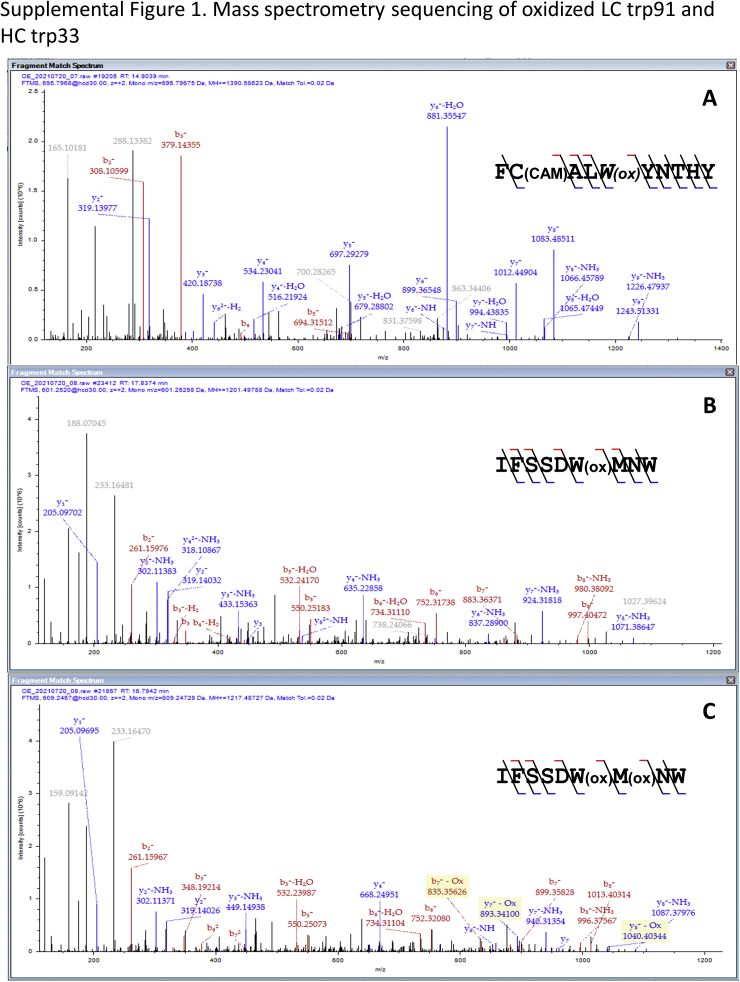
Validation of the changes in oxidation status in the cocaine-binding site Trps (LC-Trp91 and HC-Trp33) and Met residues was accomplished using thermolysin digestion and liquid chromatography–mass spectrometry of the AAPH-treated h2E2 mAb samples (see Table S1 for a complete list of thermolysin peptides identified at >95% confidence). As expected, and demonstrated by data in Figure 7, AAPH treatment of h2E2 produced robust Trp oxidation of LC Trp91 (Figs. 7, A and C and S1A) as well as HC Trp33 oxidation and was accompanied by dioxidations at met34 as well (Figs. 7D and S1, B and C), even in the presence of excess Met during AAPH oxidation. In addition, the robust AAPH oxidation of most Met residues could be controlled by adding excess Met to the AAPH reaction, as judged by the peptide containing LC met87 (7B and 7C) and HC met248 (7D). In both the LC and HC, the addition of excess Met to the AAPH reaction brought the Met oxidation level down to nearly the no treatment baseline, while maintaining high levels of oxidized Trp. The location of the two AAPH-oxidized Trp residues in relation to the BE metabolite in the published Fab fragment crystal structure of this h2E2 mAb (Protein Data Bank code: 6NFN) is shown in Figure 8.
Figure 7.
Liquid chromatography–mass spectrometry identification of tryptophan residue oxidation in the cocaine-binding site of h2E2 upon oxidation with AAPH.A and B, extracted ion profiles representing the indicated light-chain Trp91 peptide and met87 peptides, respectively, in the oxidized forms after AAPH-only (upper), AAPH + methionine (middle), and no treatment (lower) with 2.5 h thermolysin digestion. Each sample profile is scaled equally to the maximum intensity observed for each of the two peptides, as indicated. C, comparative intensities of the indicated oxidized peptides in A and B, scaled to 100% maximal intensity for each peptide. D, comparative intensities for the heavy-chain Trp33 peptide oxidized at Trp33 alone, Trp33 and met34, and at met248, all scaled to 100% maximal intensity of each peptide. AAPH, 2,2′-azobis(2-amidinopropane) dihydrochloride.
Figure 8.
Crystal structure of the h2E2 mAb Fab fragment (Protein Data Bank code:6NFN) cocrystalized with the benzoylecgonine (BE)cocaine metabolite shown in space filling mode, indicting the location of the light-chain (LC) W91 and the heavy-chain (HC) W33 tryptophan residues shown to be oxidized by AAPH. Only the variable portions of the Fab fragment are shown, with the variable region of the LC in yellow, and the variable portion of the HC in cyan. The BE ligand is shown color coded by element, the LC W91 residue (in the sequence FCALW91YNTHY) is magenta, and the HC W33 residue (in the sequence IFSSDW33M34NW) is orange. Note that these oxidized tryptophan residues are near the tropane ring of the BE ligand, and that this tropane ring is identical in both the cocaine and cocaethylene (CE) structures. The cocaine and CE ligands differ in structure from the BE ligand shown in that the -COOH group on BE, which is directed away from the binding site in this figure, is esterified in cocaine (a methyl ester) and CE (an ethyl ester).
Discussion
There are several lines of evidence indicating that Trp residues in the variable region of the anti-cocaine mAb, h2E2, are important for cocaine and cocaine metabolite binding, including the quenching of intrinsic mAb Trp fluorescence by these high-affinity ligands (6). Recently, the crystal structure of the Fab fragment of this h2E2 mAb was published, both in the apo (nonliganded) state and in the BE cocaine metabolite-bound state. The Fab crystal structure with the BE ligand bound did show several Trp residues in and around the BE-binding site but did not address the importance of possible oxidation of these residues and did not explain the differential ligand affinities noted for cocaine (Kd = 4 nM) and the two other metabolites that bind to this mAb with nanomolar affinity, CE (Kd = 1 nM) and BE (Kd = 20 nM) (see Ref. (6) for Kd values). This is because the carboxylic acid moiety of BE, which is esterified by a methyl group (to form cocaine), or by an ethyl group (to form CE), does not have any direct interactions with the Fab fragment in this structure, since it projects up from the binding site into the solvent.
Thus, in this study, we set out to determine the sensitivity of the function of the mAb to Trp oxidation, the clinically desired function being binding of cocaine with high affinity, sequestering the cocaine in mAb-bound form in the blood, and thus greatly attenuating any central nervous system effects of cocaine. This is useful clinically for prophylactic prevention of cocaine abuse in cocaine use disorders. We also sought to determine if Trp oxidation would have a differential effect on the binding of cocaine and two cocaine metabolites that bind to this h2E2 mAb with nanomolar affinity. We decided that such experiments would be most relevant to the ultimate therapeutic usage of this mAb by performing the oxidation in the buffer in which the mAb is formulated for clinical administration, namely, 10 mM histidine, 10% sucrose, and 0.01% polysorbate 80, pH = 6.0. We used the oxidant, AAPH, as an indicator of mAb oxidation susceptibility (9), both in the absence and presence of a very high concentration of Met (260 mM), to protect Met mAb residues from oxidation (10, 11). The most easily oxidized amino acids in proteins are cysteine, Met, Trp, and histidine. There are no free cysteine residues in this mAb, and histidine and Met oxidation should be largely inhibited by the free amino acid histidine in the formulation buffer and by the addition of a large concentration of the free amino acid Met added in some of the experiments. Thus, we monitored the effects of AAPH Trp-selective oxidation on mAb high-affinity binding of cocaine and metabolites and correlated the changes in binding with the oxidation of specific Trp residues by mass spectral analysis of proteolytic peptides.
The high-affinity binding of cocaine to the mAb was lost over about a day at 40 °C in the formulation buffer using 5 mM AAPH oxidant. This treatment resulted in a time-dependent loss of more than 95% of the high-affinity binding, both in the presence and absence of excess Met (Fig. 1B). Also, from the DASPMI fluorescence assay, there is evidence that there remains some low affinity (micromolar) cocaine binding in the AAPH-oxidized samples (Fig. 1C). This loss of binding correlated with a loss of about 50% of the total Trp fluorescence of this mAb, both with and without added free Met, and the small increase in mAb 310 to 370 nm absorbance suggested the formation of the Trp oxidation products, kynurenine and N′-formylkynurenine, which absorb at about 365 and 325 nm, respectively (14) (Fig. 2).
The effect of AAPH oxidation on the susceptibility of the mAb to mild SDS denaturation measured by nonreducing SDS-PAGE, and on the thermal stability of the cocaine-binding domain of the mAb, as measured by DASPMI dye DSF was examined. No change in the relative thermal denaturation of the various mAb domains in SDS was observed after nonreducing SDS-PAGE, but a small amount of high molecular weight aggregate was observed after oxidation of AAPH in the absence of Met (Fig. 3B). It is possible that at least part of this high molecular weight species is due to a small amount of dityrosine crosslinking, as previously reported by Zheng et al. (17) when using AAPH to oxidize other mAbs, since a small amount of 325 nm excitation/365 nm emission fluorescence characteristic of dityrosine was observed in h2E2 mAb samples after oxidation with AAPH in the absence of Met (data not shown). DSF revealed a relatively small thermal destabilization of the cocaine-binding domain (in the absence of SDS), with this mAb-binding domain being somewhat more destabilized by oxidation in the absence of protective Met (Fig. 3B). This suggests that oxidation of Met in the binding domain measured by DASPMI fluorescence occurs and may somewhat contribute to the destabilization of this domain, but to a much smaller extent than Trp oxidation.
AAPH has an obvious negative effect on the stabilization of the mAb due to the binding of cocaine, both with and without added Met (Fig. 4). This decrease in the affinity of cocaine binding is also observed for CE and BE binding (Fig. 6). A graphical display of these effects, which allows easy visualization, is shown in Figure 6, which displays the TmD values (the maximum temperatures in the first derivative plots of the DASMPI DSF data, both in the absence of AAPH oxidation [filled symbols] and after 24 h of oxidation with 5 mM AAPH in the presence of 260 mM Met [open symbols]). All the DSF data for three independent ligand titration experiments for each of the cocaine-related compounds (CE, cocaine, and BE) are summarized in Table 1. These results indicate that AAPH oxidation destabilizes the ligand-binding domain of the apo mAb, and that the effects on ligand binding of both AAPH oxidation and AAPH/Met oxidation are very similar for all three ligands. The Kd values for all three ligands are increased by oxidation about 5000-fold to 10,000-fold (decreasing the affinities from nanomolar to micromolar), but the RBAs of these three mAb ligands after AAPH oxidation are unchanged. This suggests that the oxidation of the Trp residues by AAPH results in changes in the binding site that are critical for the high-affinity binding of all three ligands.
Mass spectral analyses of peptides derived from thermolysin digests of control and AAPH-oxidized mAb were performed to determine which Trp residues were oxidized by AAPH treatment under these conditions. Thermolysin was used to generate the analyzed peptides since trypsin and trypsin followed by endo Glu-C protease digestion did not result in good peptide mass spectral sequence coverage of the variable regions of the HC and LCs of the mAb, with some of the peptides, including Trp residues derived from the variable regions, not being detected in the mass spectral analysis (data not shown). Analysis of thermolysin-generated peptides following AAPH treatment of h2E2 mAb revealed only oxidation of a few Trp residues, with substantial Trp oxidation of LC Trp91 (Fig. 7, A and C and Table S1). In addition, substantial Trp oxidation of HC Trp33 was observed, which was sometimes accompanied by oxidation at the adjacent met34 residue as well (Fig. 7D). We note that this particular Met residue oxidation occurred even in the presence of 260 mM excess Met. However, in general, AAPH oxidation of mAb Met residues was very much attenuated by adding excess Met to the AAPH oxidation reaction, as judged by the much-reduced Met oxidation of the peptide containing LC met87 (Fig. 7, B and C) and HC met248 (Fig. 7D). This protection of Met oxidation by AAPH is consistent with other literature reports using similar forced oxidation methodologies (10, 11). Susceptibility to AAPH oxidation of the conserved HC met248 residue in the Fc region of the mAb is consistent with several reports, as summarized in a recent review (18), and oxidation of this Met residue can result in decreased antibody stability, increased antibody aggregation tendencies, as well as decreased Fc receptor binding and shorter antibody plasma half-life (18). The addition of excess Met during the AAPH oxidation greatly decreased the oxidation of these Met residues but importantly did not substantially reduce the levels of oxidized Trp. This is consistent with the biochemical results reported in the current study, demonstrating that in terms of the effects of AAPH oxidation on cocaine and cocaine metabolite binding, the presence of this high concentration of Met did not substantially change the negative effect of AAPH oxidation on Trp fluorescence or the decrease in ligand affinities.
In summary, mild forced Trp oxidation with AAPH resulted in only a few oxidized Trp residues (there are 26 Trp residues present in each h2E2 mAb molecule) at levels that can be detected, as assessed by proteolytic digestion and mass spectrometry analysis. This oxidation greatly decreased the affinity of the mAb for cocaine and cocaine metabolites that bind with nanomolar affinity (increasing the Kds to the micromolar range). However, the decreases in binding affinities are very similar for all three ligands examined and thus do not explain the differences in the binding affinities of these three ligands, which were also not evident upon examination of the BE-Fab crystal structure (8). Protection of Met residues from AAPH oxidation by addition of excess-free Met was effective and allowed Trp-selective oxidation of the intact h2E2 mAb. The two Trp residues identified to be oxidized by AAPH by mass spectral analyses (LC W91 and HC W33) are in, or very near, the BE-binding site in the crystal structure (Fig. 8; Protein Data Bank code: 6NFN). Neither of these Trp residues are in close proximity to the carboxylic acid moiety of the cocrystalized BE ligand, which is esterified to yield cocaine and CE, consistent with the observed lack of change in the RBA observed for these three high-affinity ligands after AAPH oxidation. Oxidation of the anti-cocaine mAb could occur from exposure to oxygen in the air during its production, purification, filtration, packaging, or storage. Data presented in this study do provide some evidence of such “spontaneous” oxidation of both Met87 (Fig. 7C), and to a much lesser extent, Trp33 (Fig. 7D) in control samples, which were not exposed to AAPH (Table S1). Overall, the current study indicates that it will be very important to protect the anti-cocaine h2E2 mAb from oxidation prior to its clinical administration to maintain its high-affinity cocaine binding, which is critical for its proposed therapeutic use in the treatment of cocaine use disorders.
Experimental procedures
Materials
The generation and production of the h2E2 anti-cocaine mAb was previously described (4). The recombinant h2E2 mAb was purified by protein A affinity chromatography and used as supplied by the manufacturer, Catalent. The oxidant, AAPH (catalog no.: 440914-100GPBS), l- Met (catalog no.: M9625-25G), dithiothreitol (catalog no.: D9163-1G), and iodoacetamide (catalog no.: I1149-5G) were purchased from Sigma–Aldrich. PBS buffer concentrate (10×) was purchased from Cambrex (BioWhittaker, without calcium or magnesium; catalog number 17-517Q), and other salts and buffer components were obtained from Fisher. The solid DASPMI dye (4-Di-ASP iodide) was purchased from Thermo Fisher Scientific (catalog no.: D-288; Invitrogen/Life Technologies). Bacillus thermoproteolyticus protease (thermolysin; catalog no.: 161586) was purchased from Boehringer Mannheim. The DASPMI stock dye solution of 20 mM was made by dissolution of the solid dye in dry dimethyl sulfoxide (dye fresh weight = 366.24 g/mol) and stored light protected at −20 °C. The Applied Biosystems 48-well RT–PCR plates (catalog no.: 4375816) and the MicroAmp plate sealing optical adhesive transparent film (catalog no.: 4375928) used in the StepOne RT–PCR used to perform the DSF analyses were purchased from Thermo Fisher Scientific. Forty kilodalton molecular weight cutoff of 500 μl spin filters were used for removal of excess reagents form oxidized mAb (ZebaSpin; catalog no.: PI87767 from Fisher). The 7% Laemmli SDS-PAGE gels were run basically as described by Laemmli (19) and stained for 1 h at room temperature in InstaBlue protein stain (catalog no.: 50-190-5499) purchased from Fisher, prior to photography (without any destaining).
Ten millimolar stock solutions of cocaine and cocaine metabolite ligands were made in distilled water from solids obtained from Research Triangle Institute (RTI): CE, (RTI batch: 9885-1022-125B), cocaine (RTI batch: 14201-12A), and BE (RTI batch: 7474-1022-85A). All liquid chromatography solvents and reagents are of liquid chromatography–mass spectrometry grade from J.T. Baker.
Methods
DASPMI fluorescence assays to determine the number of high-affinity binding sites were performed as described (13), and mAb intrinsic Trp fluorescence was measured using a Hitachi F-2000 spectrofluorometer and a mini cuvette (total sample volume of 0.25 ml). UV spectra were recorded using a Beckman DU800 spectrophotometer and a mini cuvette (total sample volume of 0.1 ml). DSF was performed as described previously (7), using PBS buffer containing 50 μM DASPMI dye, 20 μl sample per well, a heating ramp rate of 0.3% (0.45 °C/min), and recording dye fluorescence of the mAb samples from 35 to 95 °C. The data files obtained for each 48-well plate from the RT–PCR runs were imported into the commercial Protein Thermal Shift version 1.3 software purchased from Applied Biosystems/Life Technologies/Thermo Fisher Scientific. Each sample well was then analyzed with this software, using the single Tm autoanalysis option. The melting temperatures based on the Boltzmann fitting of the fluorescence/temperature raw data (TmB values) and the melting temperatures based on the peak(s) of the first derivative of the fluorescence versus temperature plots (TmD values) were obtained from the software, as well as the best fit Boltzmann parameters for each sample well thermal melting transition, which was used in subsequent quantitative ligand-binding affinity calculations, as described previously (7).
Mapping oxidized Trp and met by thermolysin digestion and mass spectrometry
About 50 μg of each sample (A: AAPH only, B: AAPH + Met, and C: no treatment control) were diluted into 50 mM ammonium bicarbonate buffer, pH = 8.0 (ABC buffer), reduced with dithiothreitol, and alkylated with iodoacetamide as reported previously (20). Each sample was then digested with 2.5 μg of thermolysin at 70 °C for 1, 2.5, 5, and 18 h in a total volume of 100 μl of ABC buffer. The reactions were quenched with 1% formic acid (FA) and dried in a SpeedVac. The samples were reconstituted in 0.1% FA to a concentration of 0.2 μg/μl and centrifuged at 10,000g to remove any particulates prior to injection into the liquid chromatography–mass spectrometry system. Data were collected on an Orbitrap Eclipse mass spectrometer coupled to a Dionex Ultimate 3000 RSLCnano system (Thermo Fisher Scientific). One microgram (5 μl) of the recovered peptides from each sample was injected onto a 5 mm nanoviper μ-Precolumn (i.d. 300 μm, C18 PepMap 100, 5.0 μm, 100 Å) from Thermo Fisher Scientific at 5 μl/min in FA/H2O 0.1/99.9 (v/v) for 5 min to desalt and concentrate the samples. For the chromatographic separation of peptides, the trap column was switched to align with the EASY-Spray column PepMap RSLC C18 with a 150 mm column (i.d. 75 μm, C18, 3.0 μm, 100 Å). The peptides were eluted using a variable mobile phase gradient from 98% phase A (FA/H2O 0.1/99.9, v/v) to 32% phase B (FA/acetonitrile 0.1/99.9, v/v) for 30 min at 300 nl/min. Mass spectrometric 1 data were collected in the Orbitrap (120,000 resolution; maximum injection time 50 ms; automatic gain control 4 × 105). Charge states between two and six were required for mass spectrometry 2 analysis with no dynamic exclusion window used, to maximize the detection of h2E2 mAb peptides. Cycle time was set at 2.5 s. mass spectrometry 2 scans were performed in the ion trap with higher-energy C-trap dissociation fragmentation (isolation window of 0.8 Da; normalized collision energy of 30%; maximum injection time of 40 ms; automatic gain control of 5 × 104). The data were recorded using Xcalibur 4.3 software (Thermo Fisher Scientific). The resulting spectra were searched for thermolysis-digested peptides using a modified Homo sapiens database using Proteome discoverer, version 2.4.1.15 and the Sequest HT search algorithm (Thermo Fisher Scientific). The UniProt (9606) Homo sapiens database was modified by adding the specific sequences of the h2E2 mAb HCs and LCs (2), so that the sequence coverage and the sites of modification could be detected and analyzed as part of normal database search workflow. Variable peptide modifications included in the search parameters were oxidized Met, monooxidized and dioxidized Trp, and kynurenine (an oxidized form of Trp). Full details of the search parameters are provided in Table S2. Relative comparative quantification of specific oxidized peptides for the 2.5 h thermolysin digestion samples was all based on the maximum peak intensity (peak heights) of the indicated peptides from extracted ion chromatograms.
Data availability
Data are contained within the article, in Tables S1 and S2 as well as Fig. S1, and the supporting information files.
Supporting information
This article contains supporting information. Twelve raw data files and an Excel file summary table of the files have been uploaded and made available on Figshare with the title, “Kirley_manuscript number JBC-D-21-00934.” The data can be accessed via the link provided below. Once accepted for publication, a DOI can be assigned to include in the article. https://figshare.com/s/1e4e7e4bdd07270310d7.
Conflict of interest
Dr Norman is named as a coinventor on patents for the matter and use of the h2E2 humanized anti-cocaine mAb. A. B. N. has patent nos.: 9,758,593, 9,957,332, and 10,501,556 issued to the University of Cincinnati. All other authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We are grateful to Catalent PharmaSolutions, Inc (Madison, WI) for providing the recombinant humanized h2E2 anti-cocaine mAb protein expressed using their GPEx technology. We are thankful for the use of the StepOne RT–PCR instruments in the laboratory of Dr Guochang Fan (Department of Pharmacology and Systems Physiology) at the University of Cincinnati College of Medicine. Thanks to Michael Wyder in the University of Cincinnati Proteomics laboratory for assistance with the database search process.
Author contributions
T. L. K. conceptualization; T. L. K. and K. D. G. methodology; T. L. K. and K. D. G. formal analysis; T. L. K. and K. D. G. investigation; A. B. N. and K. D. G. resources; T. L. K. writing–original draft; T. L. K., A. B. N., and K. D. G. writing–review & editing; T. L. K. and K. D. G. visualization; T. L. K. supervision; A. B. N. project administration; A. B. N. funding acquisition.
Funding and additional information
This work was supported in part by the National Institutes of Health National Institute on Drug Abuse (grant no.: U01DA050330 [to A. B. N.]). Mass spectrometry data for this study were collected on an Orbitrap mass spectrometer that was obtained in part through a National Institutes of Health high-end instrumentation grant (grant no.: S10OD026717-01; to K. D. G.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Karen Fleming
Supporting information
Supplemental Figure S1.
References
- 1.Vocci F., Ling W. Medications development: Successes and challenges. Pharmacol. Ther. 2005;108:94–108. doi: 10.1016/j.pharmthera.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 2.Wetzel H.N., Webster R.P., Saeed F.O., Kirley T.L., Ball W.J., Norman A.B. Characterization of a recombinant humanized anti-cocaine monoclonal antibody produced from multiple clones for the selection of a master cell bank candidate. Biochem. Biophys. Res. Commun. 2017;487:690–694. doi: 10.1016/j.bbrc.2017.04.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wetzel H.N., Tabet M.R., Ball W.J., Norman A.B. The effects of a humanized recombinant anti-cocaine monoclonal antibody on the disposition of cocaethylene in mice. Int. Immunopharmacol. 2014;23:387–390. doi: 10.1016/j.intimp.2014.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Norman A.B., Gooden F.C., Tabet M.R., Ball W.J. A recombinant humanized anti-cocaine monoclonal antibody inhibits the distribution of cocaine to the brain in rats. Drug Metab. Dispos. 2014;42:1125–1131. doi: 10.1124/dmd.114.057034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kirley T.L., Norman A.B. Ligand binding to a humanized anti-cocaine mAb measured by dye absorption spectroscopy. Biochem. Biophys. Res. Commun. 2021;535:93–98. doi: 10.1016/j.bbrc.2020.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kirley T.L., Norman A.B. Characterization of a recombinant humanized anti-cocaine monoclonal antibody and its Fab fragment. Hum. Vaccin. Immunother. 2015;11:458–467. doi: 10.4161/21645515.2014.990856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kirley T.L., Norman A.B., Wetzel H.N. A novel differential scanning fluorimetry analysis of a humanized anti-cocaine mAb and its ligand binding characteristics. J. Immunol. Methods. 2020;476:112676. doi: 10.1016/j.jim.2019.112676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan K., Zhou M., Ahrendt A.J., Duke N.E.C., Tabaja N., Ball W.J., Kirley T.L., Norman A.B., Joachimiak A., Schiffer M., Wilton R., Pokkuluri P.R. Structural analysis of free and liganded forms of the Fab fragment of a high-affinity anti-cocaine antibody, h2E2. Acta Crystallogr. F Struct. Biol. Commun. 2019;75:697–706. doi: 10.1107/S2053230X19013608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dion M.Z., Wang Y.J., Bregante D., Chan W., Andersen N., Hilderbrand A., Leiske D., Salisbury C.M. The use of a 2,2'-azobis (2-amidinopropane) dihydrochloride stress model as an indicator of oxidation susceptibility for monoclonal antibodies. J. Pharm. Sci. 2018;107:550–558. doi: 10.1016/j.xphs.2017.09.022. [DOI] [PubMed] [Google Scholar]
- 10.Hageman T., Wei H., Kuehne P., Fu J., Ludwig R., Tao L., Leone A., Zocher M., Das T.K. Impact of tryptophan oxidation in complementarity-determining regions of two monoclonal antibodies on structure-function characterized by hydrogen-deuterium exchange mass spectrometry and surface plasmon resonance. Pharm. Res. 2018;36:24. doi: 10.1007/s11095-018-2545-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Folzer E., Diepold K., Bomans K., Finkler C., Schmidt R., Bulau P., Huwyler J., Mahler H.C., Koulov A.V. Selective oxidation of methionine and tryptophan residues in a therapeutic IgG1 molecule. J. Pharm. Sci. 2015;104:2824–2831. doi: 10.1002/jps.24509. [DOI] [PubMed] [Google Scholar]
- 12.Kirley T.L., Norman A.B. Unfolding of IgG domains detected by non-reducing SDS-PAGE. Biochem. Biophys. Res. Commun. 2018;503:944–949. doi: 10.1016/j.bbrc.2018.06.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirley T.L., Norman A.B. Cocaine binding to the Fab fragment of a humanized anti-cocaine mAb quantitated by dye absorption and fluorescence spectroscopy. J. Immunol. Methods. 2021;496:113103. doi: 10.1016/j.jim.2021.113103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukunaga Y., Katsuragi Y., Izumi T., Sakiyama F. Fluorescence characteristics of kynurenine and N'-formylkynurenine. Their use as reporters of the environment of tryptophan 62 in hen egg-white lysozyme. J. Biochem. 1982;92:129–141. doi: 10.1093/oxfordjournals.jbchem.a133909. [DOI] [PubMed] [Google Scholar]
- 15.Kirley T.L., Greis K.D., Norman A.B. Domain unfolding of monoclonal antibody fragments revealed by non-reducing SDS-PAGE. Biochem. Biophys. Rep. 2018;16:138–144. doi: 10.1016/j.bbrep.2018.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong J.J., Wright S.K., Ghozalli I., Mehra R., Furuya K., Katayama D.S. Simultaneous high-throughput conformational and colloidal stability screening using a fluorescent molecular rotor dye, 4-(4-(dimethylamino)styryl)-N-methylpyridinium iodide (DASPMI) J. Biomol. Screen. 2016;21:842–850. doi: 10.1177/1087057116646553. [DOI] [PubMed] [Google Scholar]
- 17.Zheng K., Ren D., Wang Y.J., Lilyestrom W., Scherer T., Hong J.K.Y., Ji J.A. Monoclonal antibody aggregation associated with free radical induced oxidation. Int. J. Mol. Sci. 2021;22:3952. doi: 10.3390/ijms22083952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beck A., Liu H. Macro- and micro-heterogeneity of natural and recombinant IgG antibodies. Antibodies (Basel) 2019;8:18. doi: 10.3390/antib8010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Dwivedi P., Muench D.E., Wagner M., Azam M., Grimes H.L., Greis K.D. Phospho serine and threonine analysis of normal and mutated granulocyte colony stimulating factor receptors. Sci. Data. 2019;6:21. doi: 10.1038/s41597-019-0015-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are contained within the article, in Tables S1 and S2 as well as Fig. S1, and the supporting information files.