Abstract
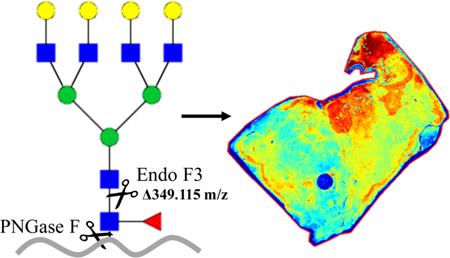
Specific alterations in N-linked glycans, such as core fucosylation, are associated with many cancers and other disease states. Because of the many possible anomeric linkages associated with fucosylated N-glycans, determination of specific anomeric linkages and the site of fucosylation (i.e., core vs outer arm) can be difficult to elucidate. A new MALDI mass spectrometry imaging workflow in formalin-fixed clinical tissues is described using recombinant endoglycosidase F3 (Endo F3), an enzyme with a specific preference for cleaving core-fucosylated N-glycans attached to glycoproteins. In contrast to the broader substrate enzyme peptide-N-glycosidase F (PNGaseF), Endo F3 cleaves between the two core N-acetylglucosamine residues at the protein attachment site. On tissues, this results in a mass shift of 349.137 a.m.u. for core-fucosylated N-glycans when compared to N-glycans released with standard PNGaseF. Endo F3 can be used singly and in combination with PNGaseF digestion of the same tissue sections. Initial results in liver and prostate tissues indicate core-fucosylated glycans associated to specific tissue regions while still demonstrating a diverse mix of core- and outer arm-fucosylated glycans throughout all regions of tissue. By determining these specific linkages while preserving localization, more targeted diagnostic biomarkers for disease states are possible without the need for microdissection or solubilization of the tissue.
Keywords: MALDI, mass apectrometry imaging, endoglycosidase F3, Endo F3, PNGase F, dual Enzyme, N-glycans, N-glycosylation
Graphical Abstract
INTRODUCTION
It is well-established that many aspects of the molecular development and progression of cancer are directly linked to changes in glycosylation.1–10 In most cases, glycan analysis has been done with serum and not directly from the cancer tissue itself.11–27 Serum is often used as it is easily obtained, but it is limited, in that it comprises dilute levels of tumor-derived material. Thus, direct tissue analysis is preferred. However, the mixing of different cell types and the loss of proteins while processing complicate glycan analysis of tissue often lead to misleading data and misrepresentation of tumor-specific analysis. To combat this, we have developed a method of tissue-based glycan imaging that allows for both qualitative and quantitative in situ N-linked glycan analysis on tissues using matrix-assisted laser desorption/ionization imaging mass spectrometry (MALDI IMS).28 This method was codeveloped in 2013 by Drake and Mehta laboratories and has continued to evolve29–31 to allow for better analysis of sialylated glycans32 and for the simultaneous analysis of glycans and proteins.33 However, a major limitation of the MALDI-TOF imaging methods is the inability to obtain true structural and linkage information of a PNGase F-released glycan. To address this limitation, we began to examine other enzymes that may allow for more structural information via IMS.
In 1982, a novel glycosidase preparation from Flavobacterium meningosepticum, designated endo-β-N-acetylglucosaminidase F, was described34 and was found to include three distinct endoglycosidase activities, termed Endo F1, Endo F2, and Endo F3.35 These three endoglycosidases cleave the β(1−4) link between the two core GlcNAcs of asparagine-linked glycans, but have specificities for distinct oligosaccharide structures.36 For example, Endo F1 cleaves high mannose and hybrid structures, but not complex oligosaccharides, and core fucosylation of hybrid structures reduces the rate of cleavage by 50-fold. Endo F2 cleaves primarily complex glycans with core fucosylation having little impact on glycan cleavage. In contrast, Endo F3 has no activity on oligomannose and hybrid molecules; it has a reported 400-fold increase in activity toward core-fucosylated structures as compared to triantennary structures at a pH of 4.5, thus reducing the amount of nonspecific N-glycan cleavage.37 With this in mind, Endo F3 was applied to different MALDI IMS workflows alone or in conjunction with PNGase F. This workflow would allow for the structural characterization of core-fucosylated glycans in tissues, while maintaining the localization of N-glycans in tissues.
Fucosylatransferase 8 (FUT8), the only known enzyme responsible for core fucosylation, has been implicated in a variety of settings including non-small-cell lung cancer, melanoma, and hepatocellular carcinoma, demonstrating an increase in invasion and metastasis for patients with elevated levels of FUT8 or core-fucosylated N-glycans.38–40 These previous studies show the importance of determining core fucosylation as opposed to outer arm fucosylation in terms of N-linked glycosylation and the clinical relevance of this methodology described below.
EXPERIMENTAL SECTION
Cloning, Expression, and Purification of Endo F3
The cDNA fragment encoding the Endo F3 gene was amplified by polymerase chain reaction from the genomic DNA of Elizabethkingia meningoseptica (UniProtKB—P36913) without the N-terminus signal sequence. Additionally, a His tag (×10) was added to its C-terminus. Amplified DNA fragments were cloned into pQE-60 by NcoI/BlpI (Genscript, Piscataway, NJ). The constructed plasmid, pQE-60-Endo F3–10xHis, was transformed into BL21 (DE3). The transformants were cultured in Luria−Bertani broth supplemented with 100 μg/ mL ampicillin. Cultures were grown at 37 °C until the cells reached an A600nm of about 0.5, and 0.5 mM IPTG was added to the culture to induce protein overproduction at 20 °C. The next day, the cells were harvested by centrifugation. The cell pellets were resuspended in phosphate-buffered salinewith added Pierce protease inhibitor tablets (Thermo Fisher Scientific, Waltham, MA), which are stored at −20 °C. OmniCleave endonuclease (Lucigen Corporation, Middleton, WI) and MgCl2 were added to the thawed cell suspension. The cell suspension was incubated at room temperature for at least 1 h with rocking. The cells were lysed using a French press (GlenMills Inc., Clifton, NJ) as per the manufacturer’s instructions. The cell lysis was applied to HisTrap FF (GE Healthcare, Pittsburgh, PA) and washed using 20 mM sodium phosphate, 0.5 M NaCl, and 20 mM imidazole (pH 7.4). The bound His-tagged protein was eluted with a gradient from 150 to 500 mM imidazole in 20 mM sodium phosphate and 0.5 M NaCl (pH 7.4). The purified Endo F3 was desalted and concentrated using 20 mM Tris-HCl and 50 mM NaCl (pH 7.5), using a Spin-X UF concentrator (10 kDa; Corning). The protein purity was confirmed using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
In-Solution Digestion by Endo F3
Human Fetuin-A (Assaypro, St. Charles, MO) or RNase B (New England BioLabs) was incubated with Endo F3 at an enzyme-to-protein-ratio of 1:5 (w/w) at 37 °C for 3 h. For our purposes, 1 μg of Endo F3 was added to 5 μg of the protein at a pH of 4.5.
Glycan Sequencing
Human Fetuin-A was run on SDS-PAGE gel, stained, and cut out. The gel pieces were alkylated in the dark for 30 min with iodoacetamide, fixed in a solution of 10% methanol and 7% acetic acid for 1 h, washed in acetonitrile, followed by subsequent steps of washing in 20 mM ammonium bicarbonate (pH 7.0) and acetonitrile before being dried in a SpeedVac. PNGase F (PNGASE F Prime, N-Zyme Scientifics, Doyles-town, PA) or Endo F3 was diluted with corresponding buffer and allowed to absorb into and cover the gel pieces and then incubated overnight at 37 °C. The glycans were eluted from the gel pieces by sonication in Milli-Q water, dried down, and labeled with a 2AB dye as previously described.41 The labeled glycans were subsequently enriched from free 2AB dye using paper chromatography and filtered using a poly-(tetrafluoroethylene) syringe filter unit. Fluorescently labeled glycans were then separated on a normal-phase Waters Alliance high-performance liquid chromatography (HPLC) system as previously described.41 Samples were further digested with sialidase for calculation of the glucose unit (GU) value and compared with the GlycoStore database.42
On-Slide Tissue Preparation
Multiple formalin-fixed paraffin-embedded (FFPE) blocks of tissues were obtained for optimization and analysis. Tissue microarray (TMA) slides were purchased from US Biomax (Rockville, MD), while all other tissue blocks (prostate, cervix, and liver) were provided by the Medical University of South Carolina Biorepository and Tissue Analysis Shared Resource (Charleston, SC). The FFPE blocks were sectioned onto the slides at 5 μm and then prepped for imaging as previously described.43 In brief, the slides were washed and deparaffinized by heating at 60 °C for 1 h and then washed sequentially in xylene, a dilution of ethanol, and water. The slides then underwent antigen retrieval using citraconic anhydride and were placed in a steam chamber for 30 min. Finally, buffer exchange was performed and the slides were desiccated. The enzyme was then applied to the slides using an M3 TM-Sprayer Tissue MALDI Sample Preparation System (HTX Technologies, LLC) at 0.1 μg/μL. PNGase F was sprayed in HPLC water, while Endo F3 was sprayed in a solution of 87 μM acetic acid (pH 4.43) for better efficiency. The slides were then placed in a humidity chamber and incubated at 37 °C for 2 h and then desiccated. Finally, the matrix was applied (α-cyano-4-hydroxycinnamic acid, 0.042 g CHCA in 6 mL of 50% acetonitrile/49.9% water/0.1% trifluoroacetic acid) using the same M3 TM-Sprayer.
N-Glycan Imaging via MALDI IMS
As previously described, tissues were analyzed via imaging N-glycans using a MALDI FTICR mass spectrometer (SolariX Dual Source, 7T, Bruker Daltonics, m/z 500−5000). The data were then analyzed and visualized using FlexImaging 5.0 and SCiLS Lab 2017b (Bruker Daltonics). Finally, glycans were built and validated against the database in GlycoWorkbench, as well as built for graphical interpretation.28,44
N-Glycan Removal
In cases where F3 was applied first, glycans were collected from the slide and analyzed as previously described.45 In brief, the slides were placed in 100% ethanol for removal of matrix and then placed in a series of dilutions of ethanol (95 and 70%). Next, the slides were placed in a high-pH cleaning solution (10 mM Tris, pH 8.98), HPLC grade water, then a low-pH cleaning solution (citraconic buffer, pH 3), and then HPLC grade water again. The slides were then desiccated and dried. Following the cleaning, the tissues were then prepped for PNGase F application by following the same tissue preparation and glycan imaging protocol as previously described;40 however, the dewaxing and antigen retrieval steps were omitted, beginning with enzyme application on the tissue.
RESULTS
In-Solution Analysis of Endo F3 Activity on N-Linked Glycans
The deglycosylation activity of the purified recombinant Endo F3 was tested initially using two well-characterized glyco-proteins, RNase B and Fetuin-A, to confirm the activity of Endo F3 acting on core-fucosylated glycans only (Figure 1). Human Fetuin-A is a circulating plasma glycoprotein with two N-linked and three O-linked carbohydrate side chains.46 The heterogeneity of Fetuin-A is mainly due to extensive modification with variable amounts of sialic acids; some less abundant glycoforms were found to be core-fucosylated.47 RNase B is a well-characterized glycoprotein from bovine pancreas that only contains noncore-fucosylated high mannose N-glycans attached to a single N-linked glycosylation site.48 As shown by SDS-PAGE, the recombinant Endo F3 will cleave Fetuin-A but not RNase B as shown by the band shift on the gel, which is consistent with Endo F3-reported sensitivity and specificity. In contrast, treatment with PNGase F leads to a band shift of RNase B. This supports the claim that we can differentially cleave glycans on proteins based on the composition of the glycans attached to them, specifically ignoring high-mannose glycans that do not contain a core fucose modification.
Figure 1.
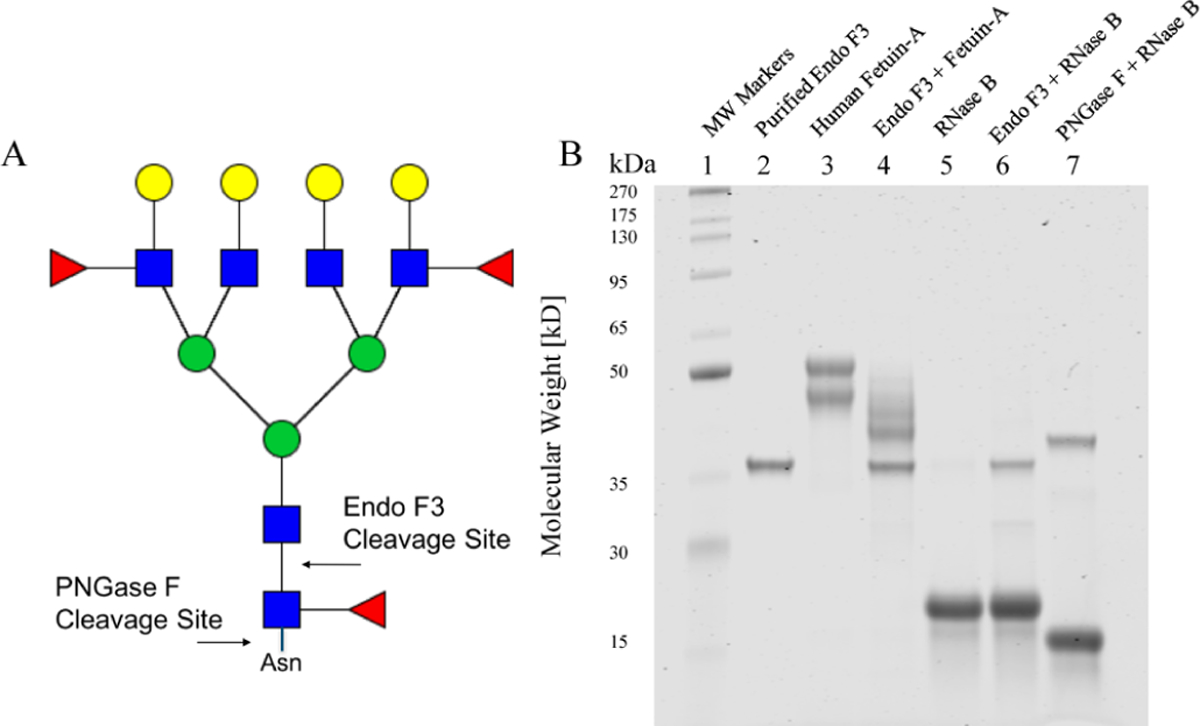
SDS-PAGE analysis of N-linked glycans cleaved by PNGase F or Endo F3. (A) Cartoon description of Endo F3 vs PNGase F cleavage on core-fucosylated N-linked glycans. For glycans, red triangles represent fucose, blue squares represent N-acetylglucosamine, green circles represent mannose, and yellow circles represent galactose. (B) SDS-PAGE analysis of Endo F3 and PNGase F digestion of human Fetuin-A or RNase B
The glycan profile of Fetuin-A was also investigated by normal-phase HPLC. The chromatograms are shown in Figure 2. A standard curve using the homopolymer dextran was used to convert the elution time into glucose units and is shown at the bottom of the figure. Among PNGase F-released glycans, sialic acid removal simplified the profiles, and further treatment with bovine kidney fucosidase (result not shown) removed peaks at GU 7.70, representing a biantennary glycan with a core α−1,6-linked fucose (F(6)A2G2) that only contributed to 2.2% of the total glycan profile. On the other hand, the three major glycans released by Endo F3 are all core-fucosylated biantennary with variable amounts of sialic acids that represented 83.0% of the total glycan profile; with the removal of sialic acids, the three species combined into one peak at GU 7.20 which is F(6)A2G2*, considering that with Endo F3 digestion, one GlcNAc and the core α−1,6-linked fucose were left on the protein as opposed to the cleavage at the asparagine residue for PNGase F (Figure 1A).
Figure 2.
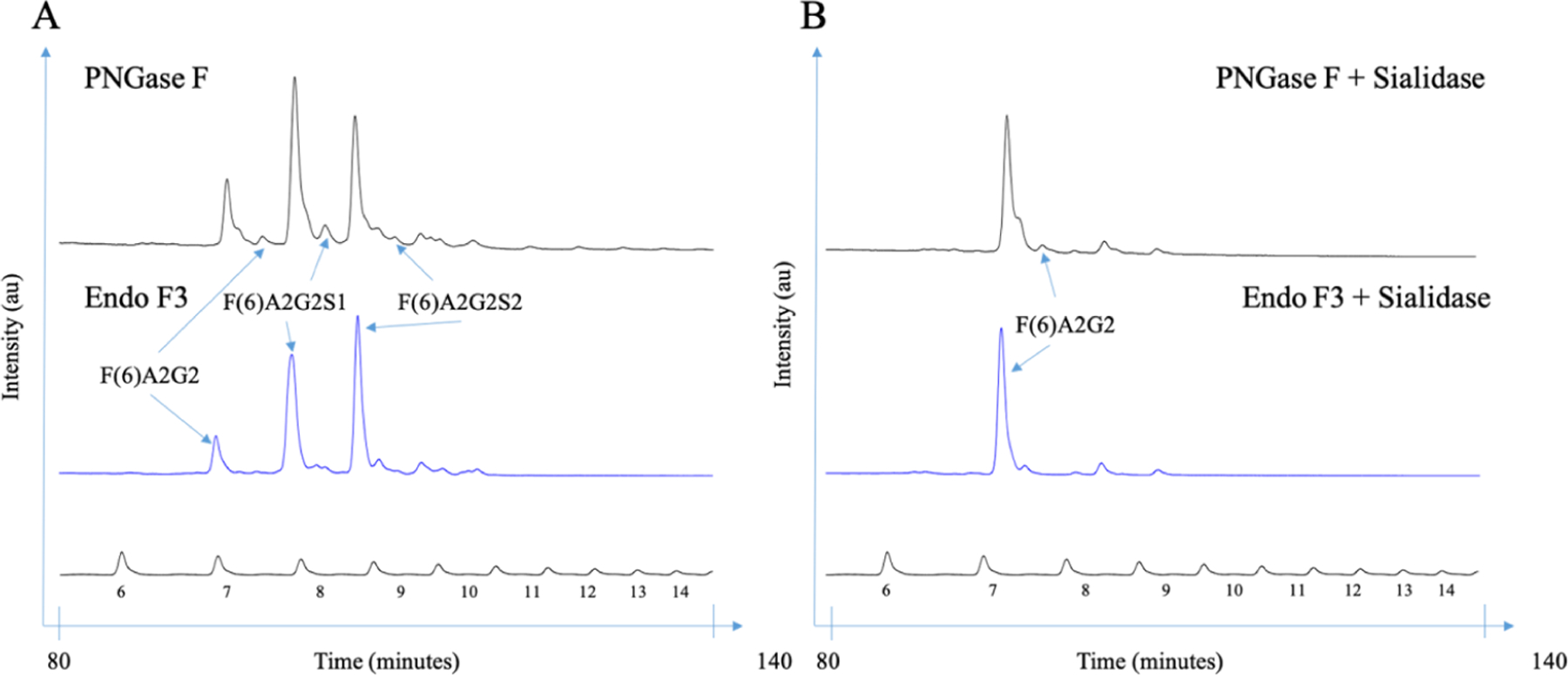
HPLC analysis of human Fetuin A N-linked glycans following Endo F3 and sialidase digestions. (A) N-linked glycans sequencing of human Fetuin-A released by PNGase F or Endo F3 and (B) followed by sequential sialidase digestion.
On-Tissue Analysis of Endo F3 Digestion Using MALDI IMS
Keeping the conserved GlcNAc and fucose residues in mind, we then applied the enzyme to the well-established tissue imaging protocol as described above (Figure 3). With the differential cleavage of Endo F3 as compared to PNGase F, we saw a mass shift of 349.137 m/z for core-fucosylated glycans. When applied, we saw the downward shift in the mass spectra of core-fucosylated N-glycans, while effectively prohibiting cleavage of N-glycans that do not contain a core fucose residue, similar to what was observed via HPLC (Figure 4). The benefit of tissue imaging is the conservation of spatial localization for the analytes without the need for microdissection or solubilization, and this work maintains this advantage as shown in Figure 5. Following analysis of Endo F3 application on tissues, we found over 30 N-linked glycans to be core-fucosylated (Supporting Information, Tables S1 and S2), and the main N-glycans found to be core-fucosylated are demonstrated in Table 1. These N-glycans also showed localization to specific regions of the tissue. As shown in Figure 5, a prostate cancer tissue section (Supporting Information, Figure S1) underwent a variety of treatments, where the first column of images represents masses for the tissue following a general PNGase F digestion, the second column represents an Endo F3 digestion, and finally, the last column represents an Endo F3 digestion, wash, and sequential PNGase F digestion as described above. As shown in the first row of Figure 5, we see the distribution of the N-glycan A2G2F (1809.6393 predicted m/z) undergoing PNGase F digestion (Figure 5A), a serial tissue section with Endo F3 digestion (Figure 5B) and the same tissue section washed, and a sequential PNGase F digestion applied (Figure 5C). These results show that we do not achieve any PNGase F cleavage activity on the glycans with our Endo F3 digestion, but are still able to achieve the same spatial distribution of the glycans following an Endo F3 digestion, albeit at a lower overall intensity relative to the initial PNGase F digestion. The second row of Figure 5 shows the truncated N-glycan F(6)A2G2 (1460.5023 predicted m/z) that underwent the same treatments. Figure 5D shows that we do not observe this mass following PNGase F digestion; however, in Figure 5E, we see this mass following the Endo F3 digestion as expected. We are also able to efficiently remove the Endo F3-cleaved glycans following washing and PNGase F application as shown by Figure 5F. Finally, in the third row of Figure 5, we show the distribution of the N-glycan Man8 (1743.5810 predicted m/z) which should never contain a core fucose. Again, we see a similar situation as observed in the first row, with PNGase F cleavage (Figure 5G), no cleavage with Endo F3 (Figure 5H), and a less efficient salvage with a sequential PNGase F digestion (Figure 5I). To be certain that the effectiveness of the Endo F3 digestion was not tissue-specific, we also performed similar digestions on multiple tissue types (Supporting Information, Figure S2). To further explore the process of the dual-enzyme cleavage, initial experiments were conducted to determine the possibility of mixing both PNGase F and Endo F3 in one spray. The enzymes were initially combined at a 3:1, 1:1, or 1:3 ratio of Endo F3 and PNGase F, and it was found that the lower concentration of Endo F3 was better suited for cleaving both core- and noncore-fucosylated N-glycans (data not shown). Therefore, further experiments were done to obtain a 1:20 ratio of Endo F3 to PNGase F, and this demonstrated the best spectra regarding efficient cleavage of all N-glycans of interest (Supporting Information, Figure S3). Efficiency and control experiments are still ongoing; however, this is a promising start to further optimize the dual enzymatic workflow.
Figure 3.
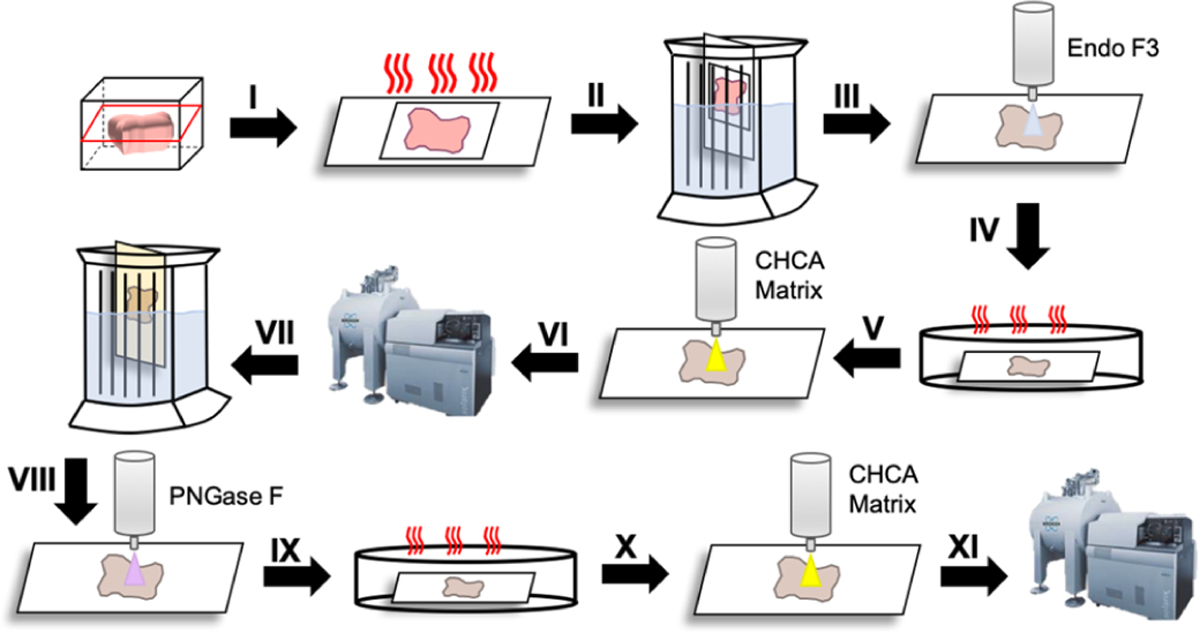
Generalized workflow of Endo F3 and PNGase F treatments on a tissue. Begin by cutting the tissue from the FFPE block at 5 μm onto the slide; (I) heat the slide at 60 °C for 1 h; (II) dewax in the series of xylene, ethanol dilutions, and water, antigen retrieval in citraconic buffer; (III) apply Endo F3 to the tissue; (IV) incubate in a humidity chamber at 37 °C for 2 h; (V) apply the CHCA matrix to the tissue; (VI) Image on MALDI-FT-ICR; (VII) Clear the matrix and glycans with ethanol dilutions, high-pH and low-pH washes; (VII) apply PNGase F to the tissue; (IX) incubate in the humidity chamber at 37 °C for 2 h; (X) Apply the CHCA matrix to the tissue; (XI) Image on MALDI-FT-ICR.
Figure 4.
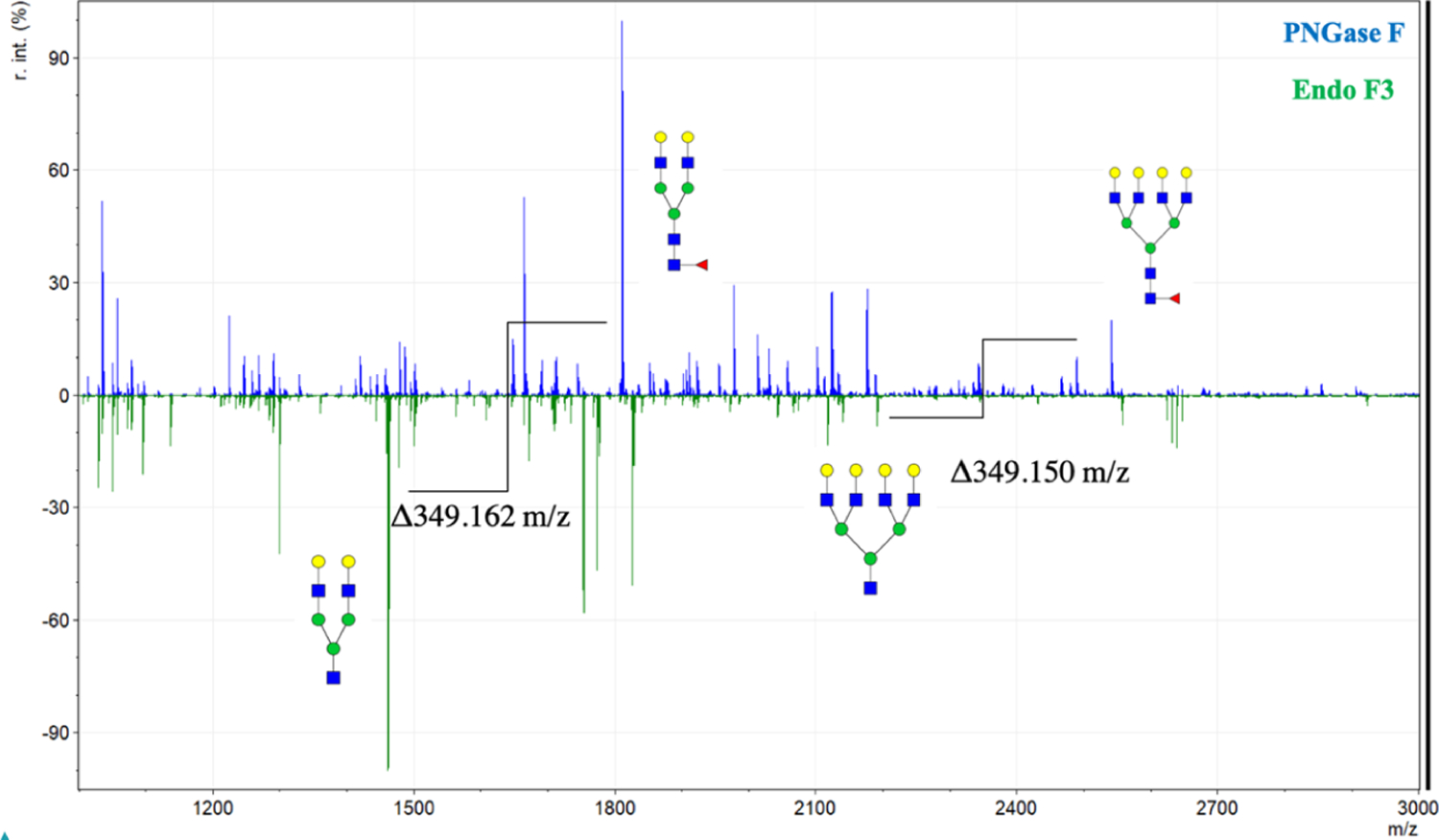
Full mass spectra for prostate cancer tissues treated with Endo F3 and PNGase F. Full example mass spectra are represented for PNGase F (blue, top) and for Endo F3 (green, bottom) applied to prostate cancer tissues. Two major PNGase F fucosylated glycans and their Endo F3 counterparts are highlighted along with the observed corresponding mass shift. Further N-glycan identification from the corresponding mass spectra peaks can be found in the Supporting Information Table S1 for PNGase F and Supporting Information Table S2 for Endo F3.
Figure 5.
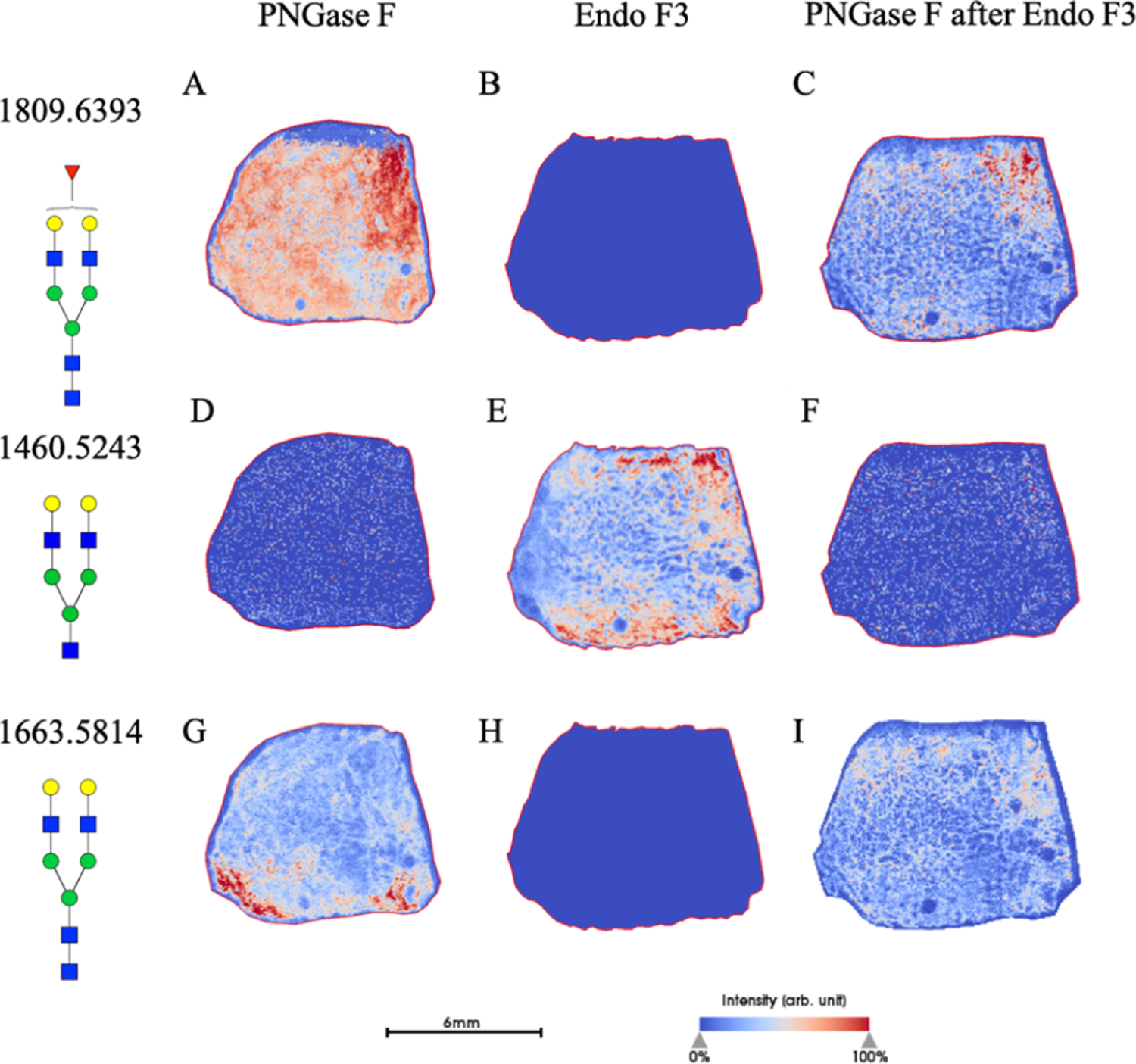
Prostate cancer tissues analyzed upon multiple enzymatic digestions. A prostate cancer tissue section undergoing PNGase F treatment (A,D,G), Endo F3 treatment (B,E,H) or sequential PNGase F treatment following a wash of the Endo F3-treated tissue (C,F,I). A known core-fucosylated glycan, A2G2F distribution is shown for the PNGase F mass shift of 1809.6393 m/z (A−C) and for the Endo F3-treated, a mass shift of 1460.5023 m/z (D−F). Finally, high-mannose glycan Man8 (1743.5810 m/z) distribution is shown (G−I). Scale bar and intensity bar are included.
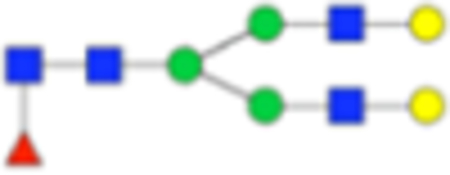
Table 1.
Main Core-Fucosylated N-Glycans Found in the Prostate Tissuea
| PNGase F Composition | PNGase F Structure | PNGase F m/z | Endo F3 Compositon | Endo F3 m/z | Endo F3 Structure |
|---|---|---|---|---|---|
| Hex5dHex1HexNAc4 + 1Na |
|
1809.6393 | Hex5HexNAc3 + 1NA | 1460.5023 |
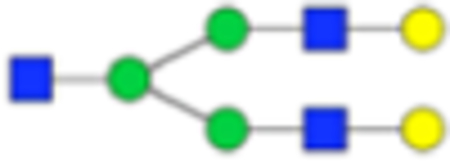
|
| Hex5dHex1HexNAc5 + 1Na |
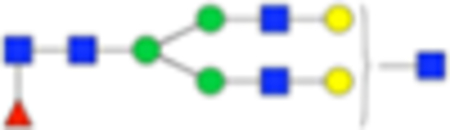
|
2012.7187 | Hex5HexNAc4 + 1NA | 1663.5817 |

|
| Hex5dHex1HexNAc4 NeuAc1 + 1Na |
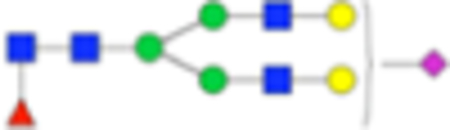
|
2100.7347 | Hex5HexNAc3 NeuAc1 + 1NA | 1751.5977 |

|
| Hex6dHex1HexNAc5 + 1Na |

|
2174.7715 | Hex6HexNAc4 + 1Na | 1825.6345 |

|
| Hex6dHex1HexNAc5 NeuAc1 + 1Na |
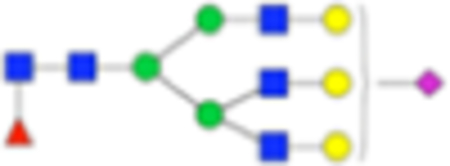
|
2465.8669 | Hex6HexNAc4 NeuAc1 + 1Na | 2116.7299 |
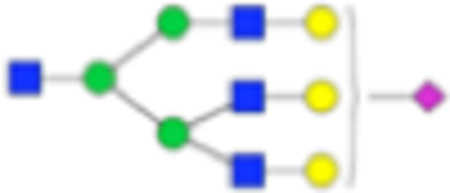
|
| Hex7dHex1HexNAc6 + 1Na |
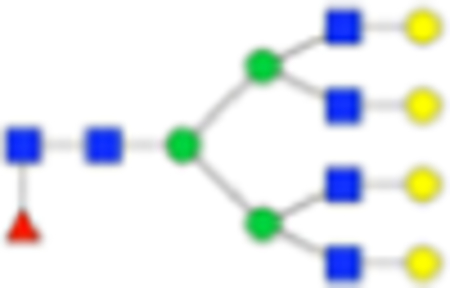
|
2539.9307 | Hex7HexNAc5 + 1Na | 2190.7667 |
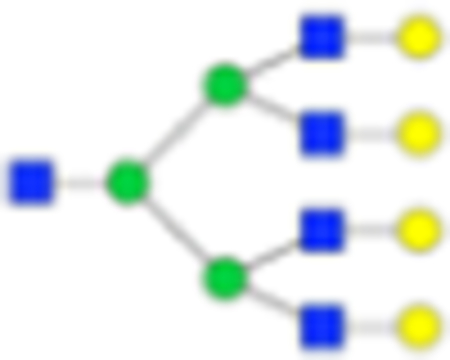
|
| Hex9dHex1HexNAc8 + 1Na |
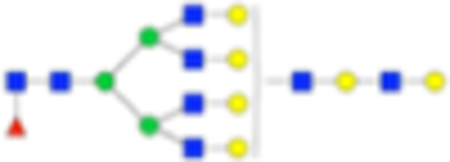
|
3270.1681 | Hex9HexNAc7 + 1Na | 2921.0311 |
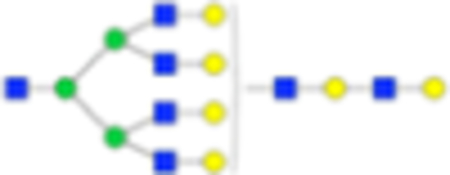
|
Six main N-glycans found to be core-fucosylated in the prostate tissue, showing the mass-to-charge ratio, composition, and structure for both the PNGase F- and Endo F3-cleaved glycoforms.
Endo F3 Application to Patient Tumor Microarrays
With the ability to determine core versus outer arm fucosylation, we then wanted to apply this technique to patient samples to determine the relevancy of this technique for determining clinically relevant factors. As previously described, core fucose is implicated in many cancer progressions; so we applied the Endo F3 by following the PNGase F protocol to a purchased hepatocellular carcinoma TMA set (US Biomax) as previously analyzed by our group.49 In Figure 6, we see two different fucosylated glycans implicated in the paper, A2G2F and A4G4F (1809.6393 and 2539.9037 predicted m/z, respectively). Figures 6A,B represent F(6)A2G2 and F(6)A4G4 in their reduced forms following Endo F3 digestion (1460.5023 and 2190.7667 predicted m/z, respectively), while Figure 6C,D represent the sequential wash and PNGase F digestion for noncore-fucosylated A2G2F and A4G4F. On examining the results, we see that there are TMA cores that contain relatively more of the core-fucosylated versions of the glycans, while some contain relatively more noncore-fucosylated glycans. While this is not an absolute quantitation, and more direct analysis will be required to determine the abundance of core versus outer arm fucosylation, this work shows promise that we can further parse out the underlying mechanisms and difference resulting from the tumor heterogeneity among patients. For example, in previously published results involving this tumor microarray, it was shown that survival probability is decreased in patients with elevated levels of A4G4F2 (m/z 2685.968);49 however, when examined under the dual-enzymatic conditions described above, elevated levels of the glycan mentioned above involving no core fucosylation show no significant difference in survival probability (Supporting Information, Figure S4), thus demonstrating the effectiveness and relevance of the dual-enzymatic approach. Further studies are needed and ongoing regarding patient outcomes and tumor grading and scoring in terms of core versus outer arm fucosylation.
Figure 6.
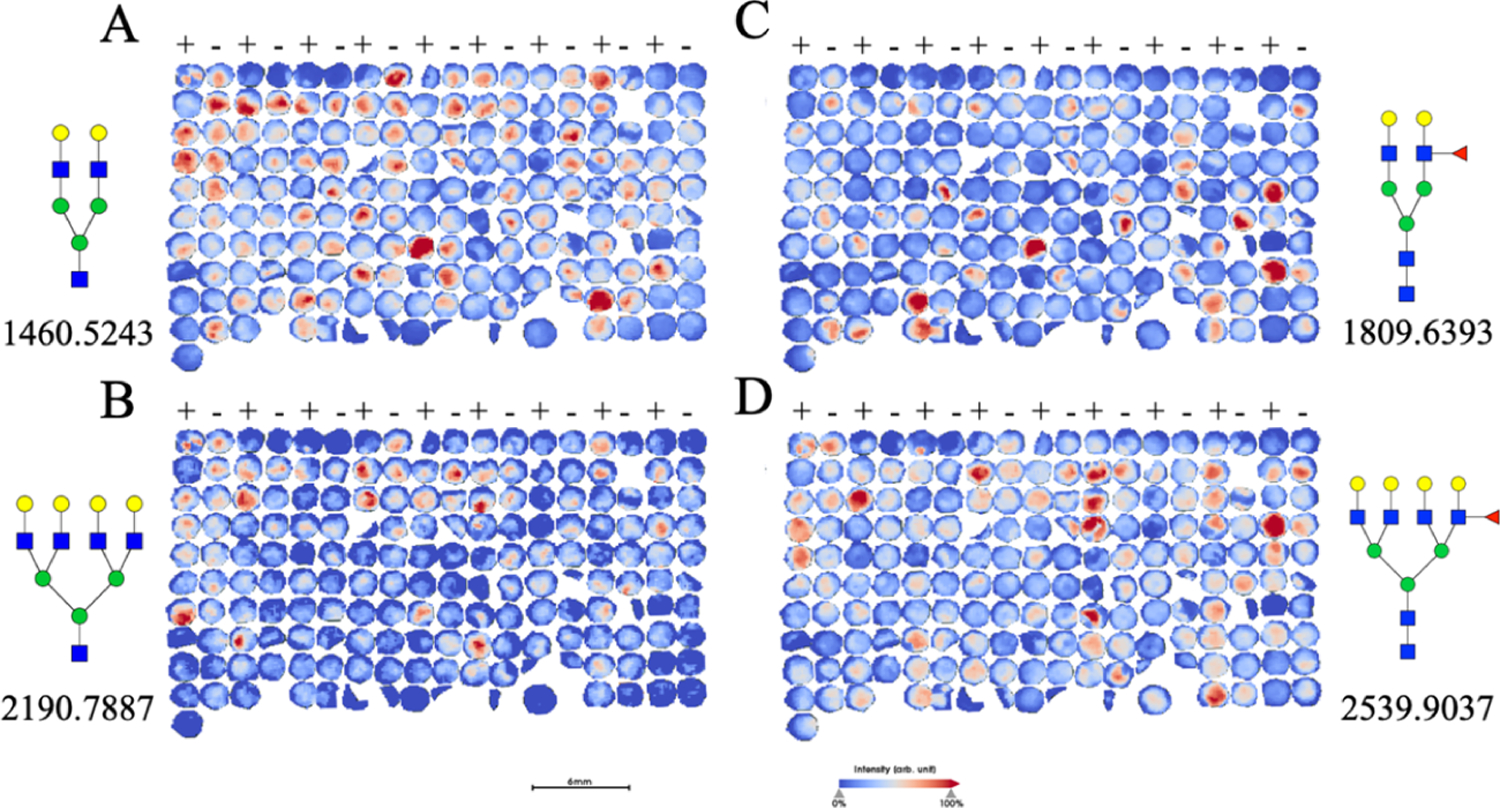
Patient TMA treated with multiple enzymatic digestions. Patient TMAs treated with Endo F3 (A,B), followed by a PNGase F digestion (C,D) are shown with two prominent core-fucosylated glycans of F(6)A2G2 and F(6)A4G4 abundance shown (1809.6393 and 2539.9037 m/z, respectively). (+) indicates cancerous tissue, while (−) indicates normal, untransformed tissue.
DISCUSSION
As we know, fucosylation of N-linked glycans has been associated with several types of cancers,50 especially, changes upon the addition of core α−1,6-linked fucose are associated with the development of hepatocellular carcinoma.51 Compared to PNGase F, Endo F3 works more efficiently and selectively on core α−1,6-linked fucosylated structures. Without the interference and noise of all the other complex glycans released by PNGase F, we can focus on the core α−1,6-linked fucosylated structures while comparing patients’ samples with that of healthy controls. This is demonstrated in Figure 5, showing that core versus outer arm fucosylation does vary from patient to patient, although the underlying mechanism is still unclear.
The most notable benefit of this work is addressing one major drawback of MALDI IMS. With the ability to distinguish between the anomeric linkages of the fucose additions of the glycans, more in-depth analysis of tissue is possible without the use of serial sections or other structural elucidation techniques that lose their spatial localization afforded with imaging, such as proteomic analysis or ion mobility.52
Additionally, this methodology has the potential to improve glycopeptide analysis in the field of proteomics. With the residual HexNAc and Fucose residue being left following Endo F3 cleavage, this could be utilized in proteomic analysis as a more specific precursor ion. When used appropriately, this precursor ion could be indicative of glycopeptides that contained core-fucosylated N-glycans, further elucidating the structural motifs of the attached N-glycans with well-established and easy-to-perform proteomic analyses, such as electron-transfer dissociation.
While this technique can effectively determine core-fucosylated N-glycans, the protocol will still require further optimization to reach efficiency levels similar to that of the PNGase F. As it stands now, PNGase F digestion works on N-glycans substantially more efficiently than Endo F3 digestion, rendering quantitative analysis difficult. However, despite the flaw in quantitative analysis, the qualitative abilities of the data are able to further elucidate the localization and relative abundance of these core-fucosylated glycans. With this information, more distinct patterns and features can be acquired from the tissue, allowing for more comprehensive analysis of tissue imaging and glycosylation as it relates to tumor heterogeneity.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants R01 CA120206 (A.S.M.), U01 CA168856 (A.S.M.), U54 MD010706 (R.R.D.), and the Hollings Cancer Center Predoctoral Fellowship (C.A.W.).
Footnotes
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.jproteome.0c00024
The authors declare no competing financial interest.
Contributor Information
Connor A. West, Department of Cell and Molecular Pharmacology and Experimental Therapeutics, Medical University of South Carolina, Charleston, South Carolina 29425, United States
Hongyan Liang, Department of Cell and Molecular Pharmacology and Experimental Therapeutics, Medical University of South Carolina, Charleston, South Carolina 29425, United States.
Richard R. Drake, Department of Cell and Molecular Pharmacology and Experimental Therapeutics, Medical University of South Carolina, Charleston, South Carolina 29425, United States
Anand S. Mehta, Department of Cell and Molecular Pharmacology and Experimental Therapeutics, Medical University of South Carolina, Charleston, South Carolina 29425, United States.
REFERENCES
- (1).Nishimori I; Perini F; Mountjoy KP; Sanderson SD; Johnson N; Cerny RL; Gross ML; Fontenot JD; Hollingsworth MA N-acetylgalactosamine glycosylation of MUC1 tandem repeat peptides by pancreatic tumor cell extracts. Cancer Res 1994, 54, 3738–3744. [PubMed] [Google Scholar]
- (2).Hollingsworth MA; Strawhecker JM; Caffrey TC; Mack DR Expression of MUC1, MUC2, MUC3 and MUC4 mucin mRNAs in human pancreatic and intestinal tumor cell lines. Int. J. Cancer 1994, 57, 198–203. [DOI] [PubMed] [Google Scholar]
- (3).Chambers JA; Hollingsworth MA; Trezise AE; Harris A Developmental expression of mucin genes MUC1 and MUC2. J. Cell Sci 1994, 107, 413–424. [DOI] [PubMed] [Google Scholar]
- (4).Hollingsworth MA; Swanson BJ Mucins in cancer: protection and control of the cell surface. Nat. Rev. Cancer 2004, 4, 45–60. [DOI] [PubMed] [Google Scholar]
- (5).Batra SK; Kern HF; Worlock AJ; Metzgar RS; Hollingsworth MA Transfection of the human Muc 1 mucin gene into a poorly differentiated human pancreatic tumor cell line, Panc1: integration, expression and ultrastructural changes. J. Cell Sci 1991, 100, 841–849. [DOI] [PubMed] [Google Scholar]
- (6).Andrianifahanana M; Moniaux N; Schmied BM; Ringel J; Friess H; Hollingsworth MA; Büchler; Aubert JP; Batra SK Mucin (MUC) gene expression in human pancreatic adenocarcinoma and chronic pancreatitis: a potential role of MUC4 as a tumor marker of diagnostic significance. Clin. Cancer Res 2001, 7, 4033–4040. [PubMed] [Google Scholar]
- (7).Shibahara H; Tamada S; Higashi M; Goto M; Batra SK; Hollingsworth MA; Imai K; Yonezawa S MUC4 is a novel prognostic factor of intrahepatic cholangiocarcinoma-mass forming type. Hepatology 2004, 39, 220–229. [DOI] [PubMed] [Google Scholar]
- (8).Andrianifahanana M; Agrawal A; Singh AP; Moniaux N; van Seuningen I; Aubert J-P; Meza J; Batra SK Synergistic induction of the MUC4 mucin gene by interferon-gamma and retinoic acid in human pancreatic tumour cells involves a reprogramming of signalling pathways. Oncogene 2005, 24, 6143–6154. [DOI] [PubMed] [Google Scholar]
- (9).Singh AP; Chauhan SC; Bafna S; Johansson SL; Smith LM; Moniaux N; Lin M-F; Batra SK Aberrant expression of transmembrane mucins, MUC1 and MUC4, in human prostate carcinomas. Prostate 2006, 66, 421–429. [DOI] [PubMed] [Google Scholar]
- (10).Ludwig JA; Weinstein JN Biomarkers in cancer staging, prognosis and treatment selection. Nat. Rev. Cancer 2005, 5, 845–856. [DOI] [PubMed] [Google Scholar]
- (11).Lai KKY; Kolippakkam D; Beretta L Comprehensive and quantitative proteome profiling of the mouse liver and plasma. Hepatology 2008, 47, 1043–1051. [DOI] [PubMed] [Google Scholar]
- (12).Mato JM; He F; Beretta L The 2006 Human Liver Proteome Project (HLPP) Workshops. Prot. Clin. Appl 2007, 1, 442–445. [DOI] [PubMed] [Google Scholar]
- (13).Beretta L Liver proteomics applied to translational research in liver disease and cancer. Proteonomics Clin. Appl 2010, 4, 359–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Yi X; Luk JM; Lee NP; Peng J; Leng X; Guan X-Y; Lau GK; Beretta L; Fan S-T Association of mortalin (HSPA9) with liver cancer metastasis and prediction for early tumor recurrence. Mol. Cell. Proteomics 2008, 7, 315–325. [DOI] [PubMed] [Google Scholar]
- (15).Goldman R; Ressom HW; Varghese RS; Goldman L; Bascug G; Loffredo CA; Abdel-Hamid M; Gouda I; Ezzat S; Kyselova Z; Mechref Y; Novotny MV Detection of hepatocellular carcinoma using glycomic analysis. Clin. Cancer Res 2009, 15, 1808–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Lattová E; McKenzie EJ; Gruwel MLH; Spicer V; Goldman R; Perreault H Mass spectrometric study of N-glycans from serum of woodchucks with liver cancer. Rapid Commun. Mass Spectrom 2009, 23, 2983–2995. [DOI] [PubMed] [Google Scholar]
- (17).Isailovic D; Kurulugama RT; Plasencia MD; Stokes ST; Kyselova Z; Goldman R; Mechref Y; Novotny MV; Clemmer DE Profiling of human serum glycans associated with liver cancer and cirrhosis by IMS-MS. J. Proteome Res 2008, 7, 1109–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Amano M; Nishimura S-I Large-scale glycomics for discovering cancer-associated N-glycans by integrating glycoblotting and mass spectrometry. Methods Enzymol 2010, 478, 109–125. [DOI] [PubMed] [Google Scholar]
- (19).Fang M; Dewaele S; Zhao Y.-p.; Stärkel P; Vanhooren V; Chen Y.-m.; Ji X; Luo M; Sun B.-m.; Horsmans Y; Dell A; Haslam SM; Grassi P; Libert C; Gao C.-f.; Chen C Serum N-glycome biomarker for monitoring development of DENA-induced hepatocellular carcinoma in rat. Molecular Cancer 2010, 9, 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Liu Y; He J; Li C; Benitez R; Fu S; Marrero J; Lubman DM Identification and confirmation of biomarkers using an integrated platform for quantitative analysis of glycoproteins and their glycosylations. J. Proteome Res 2010, 9, 798–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Nakagawa T; Miyoshi E; Yakushijin T; Hiramatsu N; Igura T; Hayashi N; Taniguchi N; Kondo A Glycomic analysis of alpha-fetoprotein L3 in hepatoma cell lines and hepatocellular carcinoma patients. J. Proteome Res 2008, 7, 2222–2233. [DOI] [PubMed] [Google Scholar]
- (22).Nakagawa T; Takeishi S; Kameyama A; Yagi H; Yoshioka T; Moriwaki K; Masuda T; Matsumoto H; Kato K; Narimatsu H; Taniguchi N; Miyoshi E Glycomic analyses of glycoproteins in bile and serum during rat hepatocarcinogenesis. J. Proteome Res 2010, 9, 4888–4896. [DOI] [PubMed] [Google Scholar]
- (23).An HJ; Lebrilla CB A glycomics approach to the discovery of potential cancer biomarkers. Methods Mol. Biol 2010, 600, 199–213. [DOI] [PubMed] [Google Scholar]
- (24).An HJ; Kronewitter SR; de Leoz MLA; Lebrilla CB Glycomics and disease markers. Curr. Opin. Chem. Biol 2009, 13, 601–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Packer NH; von der Lieth C-W; Aoki-Kinoshita KF; Lebrilla CB; Paulson JC; Raman R; Rudd P; Sasisekharan R; Taniguchi N; York WS Frontiers in glycomics: bioinformatics and biomarkers in disease. An NIH white paper prepared from discussions by the focus groups at a workshop on the NIH campus, Bethesda MD (September 11–13, 2006). Proteomics 2008, 8, 8–20. [DOI] [PubMed] [Google Scholar]
- (26).Kirmiz C; Li B; An HJ; Clowers BH; Chew HK; Lam KS; Ferrige A; Alecio R; Borowsky AD; Sulaimon S; Lebrilla CB; Miyamoto S A serum glycomics approach to breast cancer biomarkers. Mol. Cell. Proteomics 2007, 6, 43–55. [DOI] [PubMed] [Google Scholar]
- (27).Zhu J; Lin Z; Wu J; Yin H; Dai J; Feng Z; Marrero J; Lubman DM Analysis of serum haptoglobin fucosylation in hepatocellular carcinoma and liver cirrhosis of different etiologies. J. Proteome Res 2014, 13, 2986–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Powers TW; Jones EE; Betesh LR; Romano PR; Gao P; Copland JA; Mehta AS; Drake RR Matrix assisted laser desorption ionization imaging mass spectrometry workflow for spatial profiling analysis of N-linked glycan expression in tissues. Anal. Chem 2013, 85, 9799–9806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Powers T; Holst S; Wuhrer M; Mehta A; Drake R Two-Dimensional N-Glycan Distribution Mapping of Hepatocellular Carcinoma Tissues by MALDI-Imaging Mass Spectrometry. Bio-molecules 2015, 5, 2554–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Powers TW; Neely BA; Shao Y; Tang H; Troyer DA; Mehta AS; Haab BB; Drake RR MALDI imaging mass spectrometry profiling of N-glycans in formalin-fixed paraffin embedded clinical tissue blocks and tissue microarrays. PLoS One 2014, 9, No. e106255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Drake RR; Powers TW; Jones EE; Bruner E; Mehta AS; Angel PM MALDI Mass Spectrometry Imaging of N-Linked Glycans in Cancer Tissues. Adv. Canc. Res 2017, 134, 85–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Holst S; Heijs B; de Haan N; van Zeijl RJM; Briaire-de Bruijn IH; van Pelt GW; Mehta AS; Angel PM; Mesker WE; Tollenaar RA; Drake RR; Bovée JVMG; McDonnell LA; Wuhrer M Linkage-Specific in Situ Sialic Acid Derivatization for N-Glycan Mass Spectrometry Imaging of Formalin-Fixed Paraffin-Embedded Tissues. Anal. Chem 2016, 88, 5904–5913. [DOI] [PubMed] [Google Scholar]
- (33).Heijs B; Holst S; Briaire-de Bruijn IH; van Pelt GW; de Ru AH; van Veelen PA; Drake RR; Mehta AS; Mesker WE; Tollenaar RA; Bovée JVMG; Wuhrer M; McDonnell LA Multimodal Mass Spectrometry Imaging of N-Glycans and Proteins from the Same Tissue Section. Anal. Chem 2016, 88, 7745–7753. [DOI] [PubMed] [Google Scholar]
- (34).Elder JH; Alexander S endo-beta-N-acetylglucosaminidase F: endoglycosidase from Flavobacterium meningosepticum that cleaves both high-mannose and complex glycoproteins. Proc. Natl. Acad. Sci. U.S.A 1982, 79, 4540–4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Trimble RB; Tarentino AL Identification of distinct endoglycosidase (endo) activities in Flavobacterium meningosepticum: endo F1, endo F2, and endo F3. Endo F1 and endo H hydrolyze only high mannose and hybrid glycans. J. Biol. Chem 1991, 266, 1646–1651. [PubMed] [Google Scholar]
- (36).Tarentino AL; Quinones G; Changchien LM; Plummer TH Jr. Multiple endoglycosidase F activities expressed by Flavobacterium meningosepticum endoglycosidases F2 and F3. Molecular cloning, primary sequence, and enzyme expression. J. Biol. Chem 1993, 268, 9702–9708. [PubMed] [Google Scholar]
- (37).Plummer TH Jr.; Phelan AW; Tarentino AL Porcine fibrinogen glycopeptides: substrates for detecting endo-beta-N-acetylglucosaminidases F2 and F3(1). Anal. Biochem 1996, 235, 98–101. [DOI] [PubMed] [Google Scholar]
- (38).Agrawal P; Fontanals-Cirera B; Sokolova E; Jacob S; Vaiana CA; Argibay D; Davalos V; McDermott M; Nayak S; Darvishian F; Castillo M; Ueberheide B; Osman I; Fenyö D; Mahal LK; Hernando E A Systems Biology Approach Identifies FUT8 as a Driver of Melanoma Metastasis. Cancer Cell 2017, 31, 804–819.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Chen C-Y; Jan Y-H; Juan Y-H; Yang C-J; Huang M-S; Yu C-J; Yang P-C; Hsiao M; Hsu T-L; Wong C-H Fucosyltransferase 8 as a functional regulator of nonsmall cell lung cancer. Proc. Natl. Acad. Sci. U.S.A 2013, 110, 630–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Wang Y; Fukuda T; Isaji T; Lu J; Im S; Hang Q; Gu W; Hou S; Ohtsubo K; Gu J Loss of α1,6-fucosyltransferase inhibits chemical-induced hepatocellular carcinoma and tumorigenesis by down-regulating several cell signaling pathways. FASEB J 2015, 29, 3217–3227. [DOI] [PubMed] [Google Scholar]
- (41).Norton P; Comunale MA; Herrera H; Wang M; Houser J; Wimmerova M; Romano PR; Mehta A Development and application of a novel recombinant Aleuria aurantia lectin with enhanced core fucose binding for identification of glycoprotein biomarkers of hepatocellular carcinoma. Proteomics 2016, 16, 3126–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Campbell MP; Royle L; Radcliffe CM; Dwek RA; Rudd PM GlycoBase and autoGU: tools for HPLC-based glycan analysis. Bioinformatics 2008, 24, 1214–1216. [DOI] [PubMed] [Google Scholar]
- (43).Powers TW; Neely BA; Shao Y; Tang H; Troyer DA; Mehta AS; Haab BB; Drake RR MALDI imaging mass spectrometry profiling of N-glycans in formalin-fixed paraffin embedded clinical tissue blocks and tissue microarrays. PLoS One 2014, 9, No. e106255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Ceroni A; Maass K; Geyer H; Geyer R; Dell A; Haslam SM GlycoWorkbench: A Tool for the Computer-Assisted Annotation of Mass Spectra of Glycans. J. Proteome Res 2008, 7, 1650–1659. [DOI] [PubMed] [Google Scholar]
- (45).Angel PM; Mehta A; Norris-Caneda K; Drake RR MALDI Imaging Mass Spectrometry of N-glycans and Tryptic Peptides from the Same Formalin-Fixed, Paraffin-Embedded Tissue Section. Methods Mol. Biol 2017, 1788, 225–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Karamessinis PM; Malamitsi-Puchner A; Boutsikou T; Makridakis M; Vougas K; Fountoulakis M; Vlahou A; Chrousos G Marked Defects in the Expression and Glycosylation of 2-HS Glycoprotein/Fetuin-A in Plasma from Neonates with Intrauterine Growth Restriction PROTEOMICS SCREENING AND POTENTIAL CLINICAL IMPLICATIONS*. Mol. Cell. Proteomics 2008, 7, 591–599. [DOI] [PubMed] [Google Scholar]
- (47).Lin Y-H; Franc V; Heck AJR Similar Albeit Not the Same: In-Depth Analysis of Proteoforms of Human Serum, Bovine Serum, and Recombinant Human Fetuin. J. Proteome Res 2018, 17, 2861–2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Prien JM; Ashline DJ; Lapadula AJ; Zhang H; Reinhold VN The high mannose glycans from bovine ribonuclease B isomer characterization by ion trap MS. J. Am. Soc. Mass Spectrom 2009, 20, 539–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).West CA; Wang M; Herrera H; Liang H; Black A; Angel PM; Drake RR; Mehta AS N-Linked Glycan Branching and Fucosylation Are Increased Directly in Hcc Tissue As Determined through in Situ Glycan Imaging. J. Proteome Res 2018, 17, 3454–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Miyoshi E; Noda K; Yamaguchi Y; Inoue S; Ikeda Y; Wang W; Ko JH; Uozumi N; Li W; Taniguchi N The alpha1–6-fucosyltransferase gene and its biological significance. Biochim. Biophys. Acta 1999, 1473, 9–20. [DOI] [PubMed] [Google Scholar]
- (51).Mehta A; Herrera H; Block T Glycosylation and liver cancer. Adv. Canc. Res 2015, 126, 257–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Wongtrakul-Kish K; Walsh I; Sim LC; Mak A; Liau B; Ding V; Hayati N; Wang H; Choo AB-H; Rudd PM; Nguyen-Khuong T Combining Glucose Units, m/z and Collision Cross Section values: Multi-attribute data for increased accuracy in automated glycosphingolipid glycan identifications and its application in Triple Negative Breast Cancer. Anal. Chem 2019, 91, 9078. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


