Abstract
The nasopharyngeal swab is a gold standard for detecting SARS-CoV-2. However, the inconvenience of this method compelled us to compare its efficiency with saliva and gargle samples, which we collected sequentially from 229 individuals. Saliva outperformed gargle samples, constituting a reliable RNA viral source with similar performance to nasopharyngeal samples.
Keywords: SARS-CoV-2, RT-PCR, Detection, Saliva, Gargle
Nasopharyngeal and oropharyngeal swabs have been used as the gold standard for detecting SARS-COV-2 infection [1]. However, the high number of testings have put the supply chain of swabs, viral transport medium, molecular kits, and personal protection equipment under pressure. Furthermore, swab collections are painful and cause discomfort, contributing to inadequate sampling and false-negative results [2].
Saliva and gargle have been alternatives to overcome limitations of increasing SARS-CoV-2 testing using swabs. They are painless, easy to collect, have a low risk of nosocomial transmission, and can be applied in large-scale testing; therefore, they are suitable for SARS-CoV-2 screening of students/staff, professionals and travelers, among others [3]. However, the performance of molecular tests using these samples has been contradictory since it can be influenced by the time between collection and onset of symptoms, type of RNA isolation and RT-PCR kits, and number of samples evaluated. To evaluate the feasibility and reliability of using these alternative samples, as in a previous study of our group [4], we compared the performance of the RT-PCR test for SARS-CoV-2 diagnosis in nasopharyngeal swabs (NS), saliva, and gargle sequentially obtained in a cohort of 229 health care workers referred to Occupational Health due to symptoms or exposure to a COVID-19 case.
The study was performed at Complexo Hospital de Clínicas/Universidade Federal do Paraná (CHC/UFPR), a tertiary public hospital in Curitiba, Brazil, between August 2020 and November 2020. The Institutional Review Board of CHC/UFPR approved the study (No. 31687620.2.0000.0096). Three sequential samples were collected from each participant: (1) NS in viral transport medium, (2) whole oral fluid, and (3) saline gargle. The Supplementary Methods illustrates the methodology in detail.
A total of 229 participants were included, 177 (77%) of which reported symptoms, 35 (15%) were asymptomatic, and 17 did not complete the form. The clinical and epidemiological characteristics of participants by SARS-CoV-2 RT-PCR NS positive results are in Supplementary Table 1. Epidemiological factors were not statistically significant with SARS-CoV-2 infection.
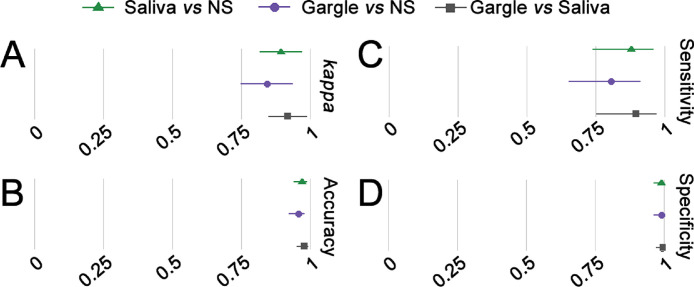
We compared the samples of saliva and gargle with NS to estimate the sensitivity, specificity, and accuracy of the tests evaluated, and all of them had a good performance. The kappa index was 0.89 for NS x saliva and 0.84 for NS x gargle (Fig. 1 ). The saliva samples had 87.80% sensitivity, 98.94% specificity, and 96.94% accuracy, whereas gargle samples had 80.49% sensitivity, 98.94% specificity, and 95.63% accuracy.
Fig. 1.
Forest plot for accuracy measures of SARS-CoV-2 detection by RT-PCR using saliva and gargle compared with NS. Cohen's kappa (A), accuracy (B), sensitivity (C), and specificity (D) of SARS-CoV-2 detection by RT-PCR comparing saliva x NS (green), gargle x NS (purple), and gargle x saliva (gray). Lines represent a 95% confidence interval. NS = nasopharyngeal swab.
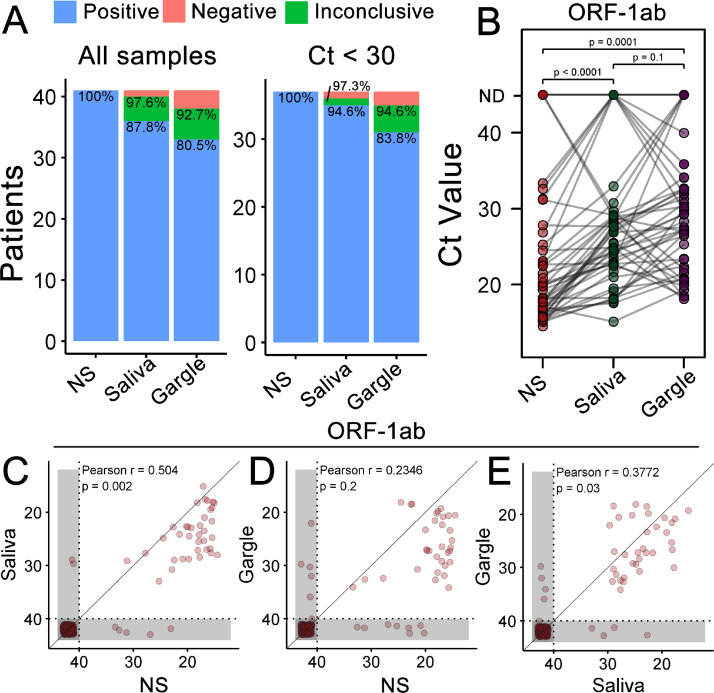
Among the 229 patients evaluated, 41 (17.9%) were positive for SARS-CoV-2 in NS tests (Fig. 2 A), 36 (87.8%) of which were also confirmed using saliva, and 33 (80.5%) using gargle samples. Due to inconclusive findings when only the N viral target was detected, we also evaluated performance, considering those cases as positive. As a result, the true positive rate increased to 40 (97.6%) for saliva and 36 (92.7%) in gargle samples (Fig. 2A). The sample input, measured as the cycle threshold (Ct) of the RNaseP human target (Supplementary Fig. 1A), was the same for NS and saliva samples (P = 0.5) and lower for gargle than for NS and saliva samples (P < 0.0001). Remarkably, two patients were positive only for saliva and gargle samples, with negative results for NS.
Fig. 2.
Detection of SARS-CoV-2 using saliva and gargle, compared to the NS. (A) True-positive rate (TPR) of saliva and gargle samples in comparison to NS. Two positive patients in saliva and gargle but negative in the NS were removed from this plot. Percentages are related to positive (below) or positive + inconclusive (above) TPR values. Inconclusive results, according to kit manufacturer instructions, render only amplification of the N gene target. (B) Paired ORF-1ab target comparison between NS, saliva, and gargle. P-values derived from paired t test. Correlation between ORF-1ab Ct values from NS and saliva (C), NS and gargle (D), and gargle and saliva (E). Gray areas represent undetermined Ct values, which are the values represented in both axes. Pearson's correlation and derived P-values were calculated excluding undetermined samples (lower-left corner in shaded area). Ct = cycle threshold; NS = nasopharyngeal swab.
Comparing the Ct values of the ORF-1ab target gene in the three samples from each patient (Fig. 2B), we observed a higher viral load, denoted as lower Ct values, in NS samples than in saliva (P < 0.0001) and gargle (P = 0.0001) samples. In contrast, we found no difference between saliva and gargle samples (P = 0.1). Similar results were obtained for the nucleocapsid gene (Supplementary Fig. 1B), although saliva outperformed gargle for this target, showing lower Ct values (P = 0.003).
We also tested the correlation of Ct values between distinct sample sources (Fig. 2C–E). ORF-1ab detection in saliva showed better correlation with NS (Pearson r = 0.504, P = 0.002) than gargle (Pearson r = 0.2346, P = 0.2). Notably, correlation between saliva and gargle samples (Pearson r = 0.3772, P = 0.03, Fig. 2E) did not surpass saliva and NS. However, the same best performance of saliva when compared to gargle was obtained with nucleocapsid targets, with higher correlations for all comparisons (Supplementary Fig. 1C–E).
We detected better concordance and sensitivity between saliva and NS than between gargle and NS; however, specificity and accuracy were similar for both comparisons. There have been contradictory reports on SARS-CoV-2 detection by RT-PCR in saliva and gargle samples. However, a recent meta-analysis with 5,922 patients from 16 studies showed a detection sensitivity for saliva of 83.2% [5], suggesting a similar accuracy for saliva and NS samples, especially in the ambulatory setting [5]. We highlight that the contradictory results reported in this meta-analysis may also be due to the different extraction and amplification kits used during the tests. We found that saliva and gargle had the best results in patients with higher viral loads in the nasopharynx. However, in the work of Yee and cols [6]., ten cases of negative samples for the nasopharynx were positive for saliva (mean Ct = 32.4).
Gargle samples are less studied than saliva samples, despite being more suitable for automation due to higher fluidity. In contrast to our observations, a recent report described a higher sensitivity for gargle (98%, 39/40) than for saliva samples (79%, 26/33) [7]. Such differences may be related to the extraction method used for each sample since no extensive optimization was performed to improve detection. Besides, not every subject provided the three samples in that study [7], which may have affected the availability of nucleic acids. Because gargle was the last sample collected from our participants, we could not exclude the hypothesis that a limited target-RNA remains after saliva sampling, thus compromising virus detection.
Since saliva can be self-collected, it may prove to be a substitute for SARS-CoV-2 surveillance, particularly in home environments, to test individuals in quarantine. Other advantages of using saliva samples include reducing risk exposure to health care workers and decreasing use of supplies, such as swabs and personal protective equipment.
In conclusion, viral detection using saliva and gargle samples is viable. Furthermore, they offer a lower transmission risk during collection and are cheaper than swabs. Therefore, they can be a good alternative for high throughput screening of asymptomatic populations.
Authors’ contributions
Gustavo Genelhoud: Investigation, Writing - Original Draft. Douglas Adamoski: Conceptualization, Methodology, Investigation, Visualization, Writing - Original Draft, Supervision. Regiane Nogueira Spalanzani: Investigation. Lucas Bochnia-Bueno: Investigation. Jaqueline Carvalho de Oliveira: Investigation, Writing - Review & Editing. Daniela Fiori Gradia: Funding acquisition, Investigation, Writing - Review & Editing. Ana Cláudia Bonatto: Investigation. Roseli Wassem: Investigation. Sonia Mara Raboni: Resources, Writing - Original Draft. Meri Bordignon Nogueira: Conceptualization, Writing - Original Draft. Patricia Savio de Araujo-Souza: Conceptualization, Methodology, Visualization, Writing - Original Draft, Project administration.
Availability of data and material
Upon request.
Code availability
Not applicable.
Ethics approval
Approval was obtained from the ethics committee of CHC/UFPR. The procedures used in this study adhere to the tenets of the Declaration of Helsinki.
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Consent for publication
All authors read and approved the final manuscript.
Acknowledgments
We are grateful to Dr Maria da Graça Bicalho, Dr Emanuel Maltempi de Souza, and to the team of volunteers for all their support. We also thank Dr Daniel Pacheco Bruschi and the Instituto de Biologia Molecular do Paraná (IBMP) for generous gifts of RNA isolation reagent and RT-PCR kits, respectively.
Funding
This study was supported by the Pro-Reitoria de Planejamento, Orçamento e Finanças (PROPLAN)/ Setor de Ciências da Saúde/ Universidade Federal do Paraná/ Brazil, with resources from Ministério da Educação e Cultura (MEC), for the execution of specific support actions to fight COVID-19; Laboratórios de Campanha – Rede vírus/Ministério de Ciência, Tecnologia e Inovação (MCTI) - Brazil. It was also financially supported by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES)/PROAP—Finance Code 001 from Ministério da Educação (MEC) - Brazil.
Declaration of competing interest
The authors have no relevant financial or non-financial interests to disclose.
Footnotes
Abbreviations: CHC-UFPR, Complexo Hospital de Clínicas/Universidade Federal do Paraná; Ct, cycle threshold; NS, nasopharyngeal swab.
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.diagmicrobio.2022.115678.
Appendix. Supplementary materials
References
- 1.Hanson KE, Caliendo AM, Arias CA, Hayden MK, Englund JA, Lee MJ, et al. Infectious diseases Society of America Guidelines on the Diagnosis of Coronavirus Disease. 2019. 2021 doi: 10.1093/cid/ciaa760. , article number 2021.ciaa760. [DOI] [Google Scholar]
- 2.Kinloch NN, Ritchie G, Brumme CJ, Dong W, Dong W, Lawson T, et al. Suboptimal biological sampling as a probable cause of false-negative COVID-19 diagnostic test results. J Infect Dis. 2021;222(6):899–902. doi: 10.1093/infdis/jiaa370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun Q, Li J, Ren H, Pastor L, Loginova Y, Madej R, et al. Saliva as a testing specimen with or without pooling for SARS-CoV-2 detection by multiplex RT-PCR test. PLoS One. 2021 doi: 10.1371/journal.pone.0243183. , article number e0243183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adamoski D, de Oliveira JC, Bonatto AC, Wassem R, Nogueira MB, Raboni SM, et al. Large-scale screening of asymptomatic persons for SARS-CoV-2 variants of concern and gamma takeover, Brazil. Emerg Infect Dis. 2021;27(12):3124–3127. doi: 10.3201/eid2712.211326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butler-Laporte G, Lawandi A, Schiller I, Yao M, Dendukuri N, McDonald EG, et al. Comparison of saliva and nasopharyngeal swab nucleic acid amplification testing for detection of SARS-CoV-2: a systematic review and meta-analysis. JAMA Intern Med. 2021;181(3):353–360. doi: 10.1001/jamainternmed.2020.8876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yee R, Truong TT, Pannaraj PS, Eubanks N, Gai E, Jumarang J, et al. Saliva is a promising alternative specimen for the detection of SARS-CoV-2 in children and adults. J Clin Microbiol. 2021 doi: 10.1128/JCM.02686-20. , article number e02686-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldfarb DM, Tilley P, Al-Rawahi GN, Srigley JA, Ford G, Pedersen H, et al. Self-collected saline gargle samples as an alternative to health care worker-collected nasopharyngeal swabs for COVID-19 diagnosis in outpatients. J Clin Microbiol. 2021 doi: 10.1128/JCM.02427-20. , article number e02427-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Upon request.