Abstract
Twelve SHV-type extended-spectrum β-lactamase (ESBL)-producing Escherichia coli lac mutant isolates were recovered in October 1997 from 11 patients of the neonatal ward in a Warsaw hospital. The outbreak was clonal; however, some of the isolates expressed a much higher level of resistance to several β-lactam antibiotics, including expanded-spectrum cephalosporins. This phenotype has been attributed to β-lactamase hyperproduction correlating with the multiplication of ESBL gene copies, as was demonstrated for representative isolates.
The growing use of newer β-lactam antibiotics in recent years in Polish hospitals has created a risk of the efficient spread of extended-spectrum β-lactamases (ESBLs). A 1995 survey involving 10 hospitals demonstrated that 20.1% of Klebsiella pneumoniae isolates (of 189 collected) and 4.9% of Escherichia coli isolates (of 502 collected) were resistant to ceftazidime (9). Several different ESBLs belonging to the TEM, SHV, and CTX-M families, some of which may be endemic for Polish nosocomial environments, have been identified in subsequent analyses (6, 7). In this work, we describe a Warsaw hospital outbreak caused by ESBL-expressing E. coli which, probably during the outbreak, acquired genetic changes resulting in hyperproduction of the enzyme and a substantial increase in the level of resistance.
Twelve ESBL-producing E. coli lac mutant isolates were collected between 3 and 30 October 1997 from infected or colonized neonates at the Praski Hospital in Warsaw. Two of the first three isolates were recovered from a 5-day-old child with septic shock who was transferred from the neonatal ward to the intensive-care unit; one was from a urine sample (isolate 1657/97), and the other was cultured from a tracheal tube swab (isolate 1658/97). The remaining isolate was from a throat swab specimen from an asymptomatic child in the neonatal ward. One of the nine subsequent isolates was recovered from a blood sample obtained from another neonate with sepsis, whereas eight were cultured from throat swabs of different colonized children. Identification to the species was done in the hospital microbiology laboratory by the ID32E ATB test (bioMérieux), routine susceptibility testing was performed by the disc diffusion method in accordance with National Committee for Clinical Laboratory Standards (NCCLS) guidelines (13), and ESBL production was detected by the double-disc test (10). All of the isolates were subjected to a detailed epidemiological analysis in which the ESBL-producing E. coli lac mutant strain 173/97, isolated in December 1996 from a patient in the internal medicine ward of the Praski Hospital, was included.
The initial susceptibility testing, performed in the hospital laboratory, suggested that the outbreak under analysis could have been caused by two ESBL-producing E. coli lac mutant strains differing in their levels of resistance to antimicrobial agents. The susceptibility analysis was repeated by evaluation of MICs of different antibiotics. MICs were determined by the agar dilution method in accordance with NCCLS guidelines (13); antimicrobial standards were supplied by their corresponding manufacturers. Results of the analysis (Table 1) confirmed the preliminary observations, and two groups of isolates were distinguished within the analyzed material. One of the groups, consisting of seven isolates (the “susceptible” group), was characterized by relatively low MICs and expressed resistance (in terms of NCCLS criteria) only to ampicillin of the β-lactams tested. The remaining group of five isolates (the “resistant” group) was defined by substantially higher MICs of all β-lactams studied than the susceptible group, except for cefoxitin, imipenem, and β-lactam–β-lactamase inhibitor combinations. These isolates were resistant to ampicillin, piperacillin, ceftazidime, and aztreonam and exhibited an intermediate level of resistance to cefotaxime and cefepime. The greatest difference observed concerned ceftazidime and cefotaxime, the MICs of which were 32 times higher for the resistant isolates than for the susceptible ones. Both groups of isolates were characterized by having the same MICs of aminoglycosides tested and exhibited intermediate levels of resistance to gentamicin and susceptibility to amikacin. Of the two isolates recovered from a single infected patient at the beginning of the outbreak, one belonged to the resistant group (isolate 1657/97) and the other belonged to the susceptible group (isolate 1658/97). The MICs characterizing E. coli isolate 173/97 were very similar to those for the susceptible isolates.
TABLE 1.
MICs for the outbreak isolates and the E. coli 173/97 strain evaluated by the agar dilution methoda
| Antibiotic(s) | MIC (μg/ml) for bacterial strain(s):
|
|||
|---|---|---|---|---|
| Resistant group (n = 5) | Susceptible group (n = 7) | E. coli 173/97b(n = 1) | E. coli ATCC 25922 | |
| Ampicillin | >512 | 256 | 256 | 4 |
| Piperacillin | 256 | 16 | 32 | 2 |
| Piperacillin + tazobactamc | 2 | 1 | 1 | 2 |
| Ceftazidime | 128 | 4 | 4 | 0.25 |
| Ceftazidime + clavulanated | 0.125 | 0.06 | 0.06 | 0.125 |
| Cefotaxime | 32 | 1 | 1 | 0.06 |
| Cefotaxime + clavulanate | ≤0.03 | ≤0.03 | ≤0.03 | ≤0.03 |
| Cefepime | 16 | 1 | 0.5 | 0.06 |
| Cefepime + clavulanate | 0.06 | 0.06 | 0.06 | 0.06 |
| Ceftibuten | 8 | 0.5 | 0.5 | 0.25 |
| Ceftibuten + clavulanate | 0.125 | 0.125 | 0.125 | 0.25 |
| Aztreonam | 256 | 8 | 8 | 0.125 |
| Aztreonam + clavulanate | ≤0.03 | ≤0.03 | ≤0.03 | 0.06 |
| Cefoxitin | 8 | 4 | 2 | 2 |
| Imipenem | 0.06 | 0.06 | 0.125 | 0.06 |
| Gentamicin | 8 | 8 | 8 | 1 |
| Amikacin | 16 | 16 | 16 | 1 |
This method is described in reference 13.
E. coli 173/97 was isolated in December 1996 (10 months before the outbreak).
The concentration of tazobactam was 4 μg/ml.
The concentration of clavulanate was constant at 2 μg/ml in all combinations.
All of the isolates were typed by the randomly amplified polymorphic DNA (RAPD) and genomic DNA restriction fragment length polymorphism (RFLP) approaches. The RAPD analysis was performed with the RAPD-7 primer (19) as described previously (6); the RFLP typing was carried out according to the method of Struelens et al. (18), using a CHEF DR II apparatus for pulsed-field gel electrophoresis (Bio-Rad). The RAPD method could not be used to differentiate the outbreak isolates, and it suggested that they were closely related to E. coli isolate 173/97 (a single band difference was evident [results not shown]). All of the outbreak isolates produced nearly identical XbaI RFLP patterns, with the only difference being a single band distinguishing the resistant and susceptible groups of isolates. E. coli isolate 173/97 was characterized by an RFLP pattern distinct from but similar to those of the outbreak isolates (results not shown).
Crude protein extracts of all of the isolates were subjected to isoelectric focusing (IEF) according to the procedure of Matthew et al. (12), using a model 111 Mini IEF Cell (Bio-Rad). After IEF, ceftazidimase activity was assigned to specific β-lactamase bands by the bioassay approach of Bauernfeind et al. (1). A single major β-lactamase band with a pI of 8.2 was detected in extracts of all outbreak isolates, as well as E. coli isolate 173/97, and subsequently demonstrated to possess ceftazidime-hydrolyzing activity (results not shown). Total DNA of the isolates was purified and used in PCRs with SHV-A and SHV-B primers specific for genes encoding SHV β-lactamases (2), as previously described (6). PCR products of the expected size of about 300 bp were found for all of the analyzed isolates (results not shown). The pI value of 8.2 characterizes SHV-5 (2), SHV-9 (16), and SHV-12 (14) ESBLs, and genes coding for these enzymes can be amplified by the use of SHV-A and SHV-B primers. Therefore, the studied isolates produced either one of the β-lactamases mentioned above or another, related SHV-type enzyme.
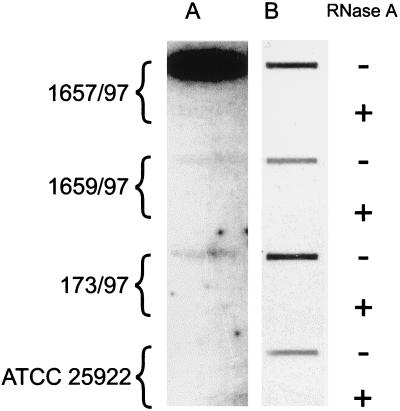
Protein extracts of the 1657/97 isolate (representative of the resistant group), the 1658/97 isolate (representative of the susceptible group), and the E. coli isolate 173/97 were tested for β-lactamase activity spectrophotometrically by the method of O’Callaghan et al. (15). The rates of nitrocefin hydrolysis were 2.1 μmol/min/mg of protein for the 1657/97 isolate, 0.2 μmol/min/mg of protein for the 1658/97 isolate, and 0.3 μmol/min/mg of protein for E. coli isolate 173/97. To reveal whether the differences in the β-lactamase specific activities of the susceptible and resistant isolates could be due to quantitative differences in ESBL-encoding gene expression, the SHV mRNA levels in isolates representing both groups were compared. Total RNA was purified from cells of the 1657/97 isolate (resistant), the 1658/97 isolate (susceptible), E. coli isolate 173/97, and E. coli ATCC 25922 (negative control). RNA was extracted with the TRI reagent (Molecular Research Center, Inc., Cincinnati, Ohio) in accordance with the manufacturer’s recommendations. The resulting preparations were treated with RQ1 RNase-free DNase (Promega) and split into halves. One-half of each preparation was digested with DNase-free RNase A (Boehringer Mannheim); the final preparations were blotted onto a nylon membrane (Boehringer Mannheim) by using a Bio-Dot slot format apparatus (Bio-Rad). The 300-bp PCR product, representing part of the blaSHV gene (see above), was gel purified and used as a hybridization probe (the SHV probe). The probe was labeled with [α-32P]dATP and [α-32P]dCTP (Amersham) by using the Megaprime DNA labeling system (Amersham). Following hybridization and exposure, the probe was washed out of the membrane, which was then rehybridized with the 16S plus 23S rRNA gene (rDNA) probe. The rDNA probe was obtained by PCR as previously described (7). Probe labeling, hybridization, and signal detection were performed with the ECL direct nucleic acid labeling and detection systems (Amersham). The results of the analysis are shown in Fig. 1. The resistant isolate 1657/97 was found to express a much higher level of the SHV mRNA than the susceptible isolate 1658/97 or the E. coli isolate 173/97. This effect could not be explained by the slight quantitative differences in RNA which were demonstrated by rRNA hybridization. The specificity of the SHV mRNA detection was confirmed by the lack of any signal with the RNA of E. coli ATCC 25922. The results could not be due to the presence of DNA in the RNA preparations, as evidenced by the lack of hybridization in RNase-treated controls.
FIG. 1.
RNA hybridization analysis. The level of mRNA encoding the SHV-type β-lactamase was found to be substantially higher in cells of the resistant 1657/97 isolate than in the susceptible 1658/97 and 173/97 strains (A), and these differences could not be explained by differences in the total amounts of RNA blotted onto the membrane (B), as determined by hybridization with the rDNA probe. −, not treated with DNase-free RNase; +, treated with DNase-free RNase.
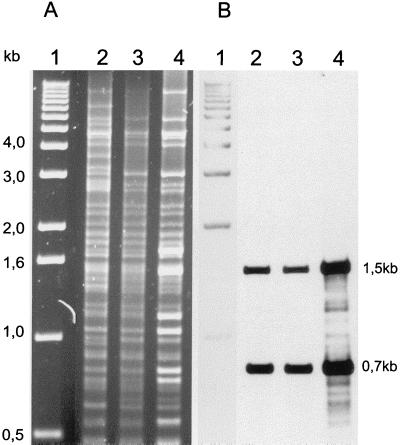
In order to explain the differences in the blaSHV genes transcription levels, we decided to compare plasmid DNA from the 1657/97 and 1658/97 isolates and the E. coli 173/97 strain. Plasmid preparations were purified by using a Qiagen Plasmid Midi kit (Qiagen) as previously described (6). Plasmid DNA was digested with the PstI restriction enzyme (MBI Fermentas), electrophoresed in a 1% agarose gel (Sigma), and blotted onto a nylon membrane (Boehringer Mannheim) for hybridization with the SHV probe (see above). Labeling of the probe and hybridization signal detection were performed with the ECL direct nucleic acid labeling and detection systems (Amersham). The results of this analysis are presented in Fig. 2. The PstI restriction patterns of plasmids from isolates 1657/97 and 1658/97 were identical in terms of sizes of produced bands (within the range of visible bands). However, some of the bands within the pattern of the plasmid from the resistant isolate 1657/97 were nonproportionally more intense, opposite of the situation for the susceptible isolate 1658/97. Two of these bands (DNA fragments of about 1.5 and 0.7 kb) hybridized with the SHV probe. These data suggested that the region(s) containing the blaSHV gene was multiplied in the plasmid from the resistant isolate. Plasmid DNA purified from isolate 173/97 was found to have a restriction pattern very similar to that of the plasmids present in the outbreak isolates, differing only by the presence of some additional PstI bands. The same DNA fragments were shown to hybridize with the SHV probe. These data suggested that the plasmid found in cells of the susceptible isolate 1658/97 may have evolved from the plasmid specific for isolate 173/97 by a DNA deletion event.
FIG. 2.
Plasmid DNA analysis. (A) Results of PstI restriction analysis; (B) results of hybridization of plasmid DNA with the SHV probe. Lane 1, the 1-kb DNA ladder (Gibco BRL); lane 2, E. coli isolate 173/97; lane 3, the susceptible isolate 1658/97; lane 4, the resistant isolate 1657/97. The positions of molecular size markers are indicated on the left (from 0.5 to 4.0 kb).
Data presented in this work document a clonal outbreak, caused by an E. coli lac mutant strain producing an SHV-type ESBL, in the neonatal ward of a large (600-bed) hospital in Warsaw. A high consumption of antibiotics (35.2 kg of cefuroxime, 5.2 kg of ceftriaxone, and 2.9 kg of ceftazidime in 1997) is one of the factors promoting the rapid evolution of ESBL-encoding genes that have spread in the microflora of that environment. In 1996 and 1997, numerous isolates of the family Enterobacteriaceae expressing different types of ESBLs were identified by the hospital microbiology laboratory staff (7). The outbreak analyzed here may have been a direct consequence of a sewage system defect that resulted in contamination of the cloak room used by the personnel of the neonatal ward a week before the first isolation occurred. It is possible that the epidemic strain has evolved from the other E. coli lac mutant strain maintained in the hospital, represented by isolate 173/97. This strain, collected about a year earlier, was shown to produce the same β-lactamase pattern and similar RAPD and RFLP patterns and to contain a clearly related plasmid. E. coli isolate 173/97 was characterized by having antimicrobial-agent MICs almost identical to those of the susceptible fraction of the outbreak isolates, which in the case of β-lactams could be correlated with the comparable levels of the ESBL mRNA expressed by the 173/97 and 1658/97 isolates.
Two groups of isolates, representing two strains strongly differing in their levels of resistance to β-lactams, were identified during the analyzed outbreak. Several lines of evidence suggested that the resistant strain could have evolved from the susceptible one by a genetic event resulting in a dramatic increase in the level of mRNA coding for the SHV-type ESBL. Plasmid DNA purified from the resistant isolate was shown to contain several copies of the blaSHV gene; however, this phenomenon seems to be insufficient to explain the scale of the SHV mRNA level difference observed between the resistant and susceptible isolates. It is possible that the recombination event which led to the gene multiplication created a new local sequence context for the ESBL-encoding gene and exposed at least one of the gene copies to elements enhancing transcription. An elevation of β-lactamase production resulting in an increase in the resistance level has been reported several times to date and has been attributed to gene copy multiplication (17), point promoter-up mutations (3, 4, 11), or insertion of transposable elements in the vicinity of the promoter (8). The single-band difference between the RFLP patterns of the resistant and susceptible isolates reflects a chromosomal DNA rearrangement which occurred in parallel with the plasmid DNA change during the evolution of the resistant strain. It is possible that the resistant strain emerged during the progress of infection of the patient from whom isolates 1657/97 and 1658/97 were recovered; however, it is equally possible that evolved earlier and that the patient was infected by the already mixed population of E. coli strains.
A similar example of diversification of an epidemic strain producing ESBL during an outbreak leading to the emergence of a strain overexpressing the enzyme has already been reported by French et al. (5). In their study, however, no differences in the plasmid DNAs of the low- and high-level ESBL-producing isolates of Klebsiella pneumoniae were observed. Both the report of French et al. and the present study demonstrate a very dangerous aspect of ESBL-producing strains, i.e., very efficient increases in their resistance levels. Such events may be promoted by the use of β-lactam antibiotics belonging to the substrate spectrum of ESBLs against the strains which, in spite of ESBL production, are identified in vitro as being susceptible to these antibiotics.
Acknowledgments
We thank Waleria Hryniewicz and Kent Holding for critical reading of the manuscript, Janusz Fiett and Ewa Wasińska for assistance, and Mariola Bisko and Dorota Hoffman-Zacharska for kindly providing radioactive nucleotides.
This work was financed by a grant from the Polish Committee for Scientific Research (KBN) (no. 4 P05D 030 14).
REFERENCES
- 1.Bauernfeind A, Casellas J M, Goldberg M, Holley M, Jungwirth R, Mangold P, Röhnisch T, Schweighart S, Wilhelm R. A new plasmidic cefotaximase from patients infected with Salmonella typhimurium. Infection. 1992;20:158–163. doi: 10.1007/BF01704610. [DOI] [PubMed] [Google Scholar]
- 2.Billot-Klein D, Gutmann L, Collatz E. Nucleotide sequence of the SHV-5 β-lactamase gene of a Klebsiella pneumoniae plasmid. Antimicrob Agents Chemother. 1990;34:2439–2441. doi: 10.1128/aac.34.12.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen S-T, Clowes R C. Two improved promoter sequences for the β-lactamase expression arising from a single base-pair substitution. Nucleic Acids Res. 1984;12:3219–3234. doi: 10.1093/nar/12.7.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen S-T, Clowes R C. Variations between the nucleotide sequences of Tn1, Tn2, and Tn3 and expression of β-lactamase in Pseudomonas aeruginosa and Escherichia coli. J Bacteriol. 1987;169:913–916. doi: 10.1128/jb.169.2.913-916.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.French G L, Shannon K P, Simmons N. Hospital outbreak of Klebsiella pneumoniae resistant to broad-spectrum cephalosporins and β-lactam–β-lactamase inhibitor combinations by hyperproduction of SHV-5 β-lactamase. J Clin Microbiol. 1996;34:358–363. doi: 10.1128/jcm.34.2.358-363.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gniadkowski M, Schneider I, Jungwirth R, Hryniewicz W, Bauernfeind A. Ceftazidime-resistant Enterobacteriaceae isolates from three Polish hospitals: identification of three novel TEM- and SHV-5-type extended-spectrum β-lactamases. Antimicrob Agents Chemother. 1998;42:514–520. doi: 10.1128/aac.42.3.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gniadkowski M, Schneider I, Pałucha A, Jungwirth R, Mikiewicz B, Bauernfeind A. Cefotaxime-resistant Enterobacteriaceae isolates from a hospital in Warsaw, Poland: identification of a new CTX-M-3 cefotaxime-hydrolyzing β-lactamase that is closely related to the CTX-M-1/MEN-1 enzyme. Antimicrob Agents Chemother. 1998;42:827–832. doi: 10.1128/aac.42.4.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goussard S, Sougakoff W, Mabilat C, Bauernfeind A, Courvalin P. An IS1-like element is responsible for high-level synthesis of extended-spectrum β-lactamase TEM-6 in Enterobacteriaceae. J Gen Microbiol. 1991;137:2681–2687. doi: 10.1099/00221287-137-12-2681. [DOI] [PubMed] [Google Scholar]
- 9.Hryniewicz W, Trzciński K, Nowak J, Giffing I. Presented at the 8th International Congress of Bacteriology and Applied Microbiology Division, International Union of Microbiological Societies, Jerusalem, Israel, 18 to 23 September 1996. 1996. National survey of the susceptibility of clinically important bacterial pathogens isolated in Poland in 1995 to piperacillin/tazobactam and other antimicrobial agents. [Google Scholar]
- 10.Jarlier V, Nicolas M, Fournier G, Philippon A. Extended broad-spectrum β-lactamases conferring transferable resistance to newer β-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev Infect Dis. 1988;10:867–878. doi: 10.1093/clinids/10.4.867. [DOI] [PubMed] [Google Scholar]
- 11.Mabilat C, Goussard S, Sougakoff W, Spencer R C, Courvalin P. Direct sequencing of the amplified structural gene and promoter for the extended-broad-spectrum β-lactamase TEM-9 (RHH-1) of Klebsiella pneumoniae. Plasmid. 1990;23:1–8. doi: 10.1016/0147-619x(90)90041-a. [DOI] [PubMed] [Google Scholar]
- 12.Matthew M, Harris A M, Marshall M J, Ross G W. The use of analytical isoelectric focussing for detection and identification of β-lactamases. J Gen Microbiol. 1975;88:169–178. doi: 10.1099/00221287-88-1-169. [DOI] [PubMed] [Google Scholar]
- 13.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 14.Nüesch-Inderbinen M T, Kayser F H, Hächler H. Survey and molecular genetics of SHV β-lactamases in Enterobacteriaceae in Switzerland: two novel enzymes, SHV-11 and SHV-12. Antimicrob Agents Chemother. 1997;41:943–949. doi: 10.1128/aac.41.5.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Callaghan C H, Morris A, Kirby S M, Shingler A H. Novel method for detection of β-lactamases by using a chromogenic cephalosporin substrate. Antimicrob Agents Chemother. 1972;1:283–288. doi: 10.1128/aac.1.4.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prinarakis E E, Tzelepi E, Gazouli M, Mentis A F, Tzouvelekis L S. Characterization of a novel SHV β-lactamase variant that resembles the SHV-5 enzyme. FEMS Microbiol Lett. 1996;139:229–234. doi: 10.1111/j.1574-6968.1996.tb08207.x. [DOI] [PubMed] [Google Scholar]
- 17.Reguera J A, Baquero F, Perez-Diaz J C, Martinez J L. Synergistic effect of dosage and bacterial inoculum in TEM-1 mediated antibiotic resistance. Eur J Clin Microbiol Infect Dis. 1988;7:778–779. doi: 10.1007/BF01975047. [DOI] [PubMed] [Google Scholar]
- 18.Struelens M J, Rost F, Deplano A, Maas A, Schwam V, Serruys E, Cremer M. Pseudomonas aeruginosa and Enterobacteriaceae bacteremia after biliary endoscopy: an outbreak investigation using DNA macrorestriction analysis. Am J Med. 1993;95:489–498. doi: 10.1016/0002-9343(93)90331-i. [DOI] [PubMed] [Google Scholar]
- 19.van Belkum A, Kluytmans J, van Leeuwen W, Bax R, Quint W, Peters E, Fluit A, Vandenbroucke-Grauls C, van den Brule A, Koeleman H, Melchers W, Meis J, Elaichouni A, Vaneechoutte M, Moonens F, Maes N, Struelens M, Tenover F, Verbrugh H. Multicenter evaluation of arbitrary primed PCR for typing of Staphylococcus aureus strains. J Clin Microbiol. 1995;33:1537–1547. doi: 10.1128/jcm.33.6.1537-1547.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]