Abstract
Background
Viral load (VL) testing in people living with HIV (PLHIV) helps to monitor antiretroviral therapy (ART). VL is still largely tested using central laboratory‐based platforms, which have long test turnaround times and involve sophisticated equipment. VL tests with point‐of‐care (POC) platforms capable of being used near the patient are potentially easy to use, give quick results, are cost‐effective, and could replace central or reference VL testing platforms.
Objectives
To estimate the diagnostic accuracy of POC tests to detect high viral load levels in PLHIV attending healthcare facilities.
Search methods
We searched eight electronic databases using standard, extensive Cochrane search methods, and did not use any language, document type, or publication status limitations. We also searched the reference lists of included studies and relevant systematic reviews, and consulted an expert in the field from the World Health Organization (WHO) HIV Department for potentially relevant studies. The latest search was 23 November 2020.
Selection criteria
We included any primary study that compared the results of a VL test with a POC platform to that of a central laboratory‐based reference test to detect high viral load in PLHIV on HIV/AIDS care or follow‐up. We included all forms of POC tests for VL as defined by study authors, regardless of the healthcare facility in which the test was conducted. We excluded diagnostic case‐control studies with healthy controls and studies that did not provide sufficient data to create the 2 × 2 tables to calculate sensitivity and specificity. We did not limit our study inclusion to age, gender, or geographical setting.
Data collection and analysis
Two review authors independently screened the titles, abstracts, and full texts of the search results to identify eligible articles. They also independently extracted data using a standardized data extraction form and conducted risk of bias assessment using the Quality Assessment of Diagnostic Accuracy Studies (QUADAS‐2) tool. Using participants as the unit of analysis, we fitted simplified univariable models for sensitivity and specificity separately, employing a random‐effects model to estimate the summary sensitivity and specificity at the current and commonly reported World Health Organization (WHO) threshold (≥ 1000 copies/mL). The bivariate models did not converge to give a model estimate.
Main results
We identified 18 studies (24 evaluations, 10,034 participants) defining high viral loads at main thresholds ≥ 1000 copies/mL (n = 20), ≥ 5000 copies/mL (n = 1), and ≥ 40 copies/mL (n = 3). All evaluations were done on samples from PLHIV retrieved from routine HIV/AIDS care centres or health facilities. For clinical applicability, we included 14 studies (20 evaluations, 8659 participants) assessing high viral load at the clinical threshold of ≥ 1000 copies/mL in the meta‐analyses. Of these, sub‐Saharan Africa, Europe, and Asia contributed 16, three, and one evaluation respectively. All included participants were on ART in only nine evaluations; in the other 11 evaluations the proportion of participants on ART was either partial or not clearly stated. Thirteen evaluations included adults only (n = 13), five mixed populations of adults and children, whilst in the remaining two the age of included populations was not clearly stated. The majority of evaluations included commercially available tests (n = 18). Ten evaluations were POC VL tests conducted near the patient in a peripheral or onsite laboratory, whilst the other 10 were evaluations of POC VL tests in a central or reference laboratory setting. The test types evaluated as POC VL tests included Xpert HIV‐1 Viral Load test (n = 8), SAMBA HIV‐1 Semi‐Q Test (n = 9), Alere Q NAT prototype assay for HIV‐1 (n = 2) and m‐PIMA HIV‐1/2 Viral Load test (n = 1). The majority of evaluations (n = 17) used plasma samples, whilst the rest (n = 3) utilized whole blood samples.
Pooled sensitivity (95% confidence interval (CI)) of POC VL at a threshold of ≥ 1000 copies/mL was 96.6% (94.8 to 97.8) (20 evaluations, 2522 participants), and pooled specificity (95% CI) was 95.7% (90.8 to 98.0) (20 evaluations, 6137 participants). Median prevalence for high viral load (≥ 1000 copies/mL) (n = 20) was 33.4% (range 6.9% to 88.5%).
Limitations
The risk of bias was mostly assessed as unclear across the four domains due to incomplete reporting.
Authors' conclusions
We found POC VL to have high sensitivity and high specificity for the diagnosis of high HIV viral load in PLHIV attending healthcare facilities at a clinical threshold of ≥ 1000 copies/mL.
Plain language summary
Point‐of‐care tests for detecting high viral load in people living with HIV attending healthcare facilities
Why is improving the diagnosis of high HIV viral load infection important?
It helps to monitor the HIV virus levels in people living with HIV (PLHIV) who are receiving antiretroviral therapy (ART). High virus levels indicate that the medications are failing to suppress the virus, a condition known as ART treatment failure, which has a risk of severe illness and death. Rapid diagnostic tests that detect high HIV virus levels quickly near the patient (point‐of‐care) can increase access to early changes in ART.
What is the aim of this review?
To determine the accuracy of point‐of‐care (POC) tests for diagnosing high HIV virus levels in PLHIV attending healthcare facilities.
What was studied in this review?
Point‐of‐care tests for viral load detection with results measured against central laboratory tests (reference test). We included all forms of tests with POC platforms for VL regardless of the healthcare facility in which the test was conducted.
What are the main results in this review?
Fourteen studies that completed 20 evaluations involving 8659 participants compared molecular POC tests for diagnosing high virus levels at the clinically recommended positivity threshold of ≥ 1000 copies/mL.
What are the strengths and limitations of this review?
The review included sufficient studies done on samples from PLHIV retrieved from routine HIV/AIDS care centres or health facilities, but it was unclear if all included participants were on ART. Also, none of the included tests was a true POC test conducted at the patient's side: half of the included studies (n = 10) evaluated POC tests in onsite laboratories near the patient, and the other half were tests with POC platforms evaluated in a central or reference laboratory (n = 10).
To whom do the results of this review apply?
PLHIV with suspected high viral loads attending healthcare facilities.
What are the implications of this review?
In theory, for a population of 1000 PLHIV where 100 have high virus levels, 136 people would receive a positive result with the molecular POC test; of these, 39 will not have high viral levels (false‐positive result) and would be incorrectly identified as not responding to ART treatment, possibly leading to unnecessary testing or further treatment; and 864 would receive a negative test result with the molecular POC test; of these, three will actually have high virus levels (false‐negative result) and would be missed whilst failing ART treatment.
How up‐to‐date is this review?
The evidence is current to 23 November 2020.
Summary of findings
Summary of findings 1. Summary of findings table.
| Question | What is the diagnostic accuracy of point‐of‐care tests to detect high viral load levels in people living with HIV? | ||||||
| Population | People living with either HIV‐1 or HIV‐2 with suspected high viral loads attending health facilities | ||||||
| Index test | Tests with point‐of‐care platforms for detecting HIV viral load (POC VL) | ||||||
| Comparator test | None | ||||||
| Target condition | High viral load | ||||||
| Reference test | Central laboratory testing for HIV viral load | ||||||
| Role | If accurate, index test results will be used to monitor viral load to decide on change of drug therapy. This will replace the reference standard of laboratory testing. | ||||||
| Limitations | TEST: POC VL THRESHOLD: ≥ 1000 copies/mL defined as treatment failure | ||||||
| Risk of bias |
Mostly unclear risk of bias Method of recruitment in most studies (except two studies) largely unclear. Blinding of index and reference tests was not well‐reported, but is unlikely to have introduced bias. Interval between index and reference tests not well‐reported, but is unlikely to have introduced bias. |
||||||
| Applicability of evidence to question | Patient selection: all evaluations were done on samples from PLHIV retrieved from routine HIV/AIDS care centres or health facilities. Nearly half of the included studies had ART‐exclusive populations, whilst in the other studies the ART status was either unclear or mixed, comprising both ART‐experienced and ART‐naive participants. Nonetheless, this is reflective of routine care settings where mixed populations of ART‐experienced, ‐naive, and ‐non‐adherent are present due to barriers in ART initiation and adherence. Index test: none of the evaluations was done at the patient's side (not true point‐of‐care tests). About half of the included POC VL tests were evaluated onsite in the health facility laboratory or in a peripheral laboratory near the patient. The other half were evaluated in a central or reference laboratory setting and not near the patient.a This is reflective of many resource‐limited settings where testing locations for POC tests are often blurred. |
||||||
| Findings | TEST: POC VL THRESHOLD: ≥ 1000 copies/mL defined as the clinical threshold for treatment failure | ||||||
| Quantity of evidence | Number of evaluations N = 20 | Total participants N = 8659 | Total with target condition N = 2522 | Medianprevalence33.4% | |||
| Accuracy | Test consequences | Effect per 1000 patients tested at different prevalenceb settings | |||||
| 2.5% | 10% | 30% | 40% | ||||
| Sensitivity | 96.6% (94.8 to 97.8) | True‐positives (patients with high viral load or treatment failure) | Will receive appropriate change in drug treatment | 24 (24 to 24) | 97 (95 to 98) | 290 (284 to 293) | 386 (379 to 391) |
| False‐negatives (patients incorrectly classified as not having high viral load or treatment failure) | Will not receive required change in drug treatment |
1 (1 to 1) | 3 (2 to 5) | 10 (7 to 16) | 14 (9 to 21) | ||
| Specificity | 95.7% (90.8 to 98.0) | True‐negatives (patients without high viral load or treatment failure) |
Appropriately do not change drug treatment |
933 (885 to 956) | 861 (817 to 882) | 670 (636 to 686) | 574 (545 to 588) |
| False‐positives (patients incorrectly classified as having high viral load or treatment failure) | Will receive unnecessary change in drug treatment |
42 (19 to 90) | 39 (18 to 83) | 30 (14 to 64) | 26 (12 to 55) | ||
| Consistency | Minimal heterogeneity for sensitivity between studies, but heterogeneity present for specificity, especially for laboratory evaluations of POC VL test | ||||||
| Indirect test comparisonsc | |||||||
| Included tests | Xpert HIV‐1, n = 8 | SAMBA HIV‐1 Semi‐Q Test, n = 9 | Alere q prototype for HIV‐1, n = 2 | m‐PIMA HIV‐1/2, n = 1 | |||
| Sensitivity | No statistically significant difference between Xpert (97%) and SAMBA (95%); P = 0.21 | ‐ | ‐ | ||||
| Specificity | No statistically significant difference between Xpert (96%) and SAMBA (97%); P = 0.43 | ‐ | ‐ | ||||
Abbreviations: ART: antiretroviral therapy; PLHIV: people living with HIV; POC: point of care; VL: viral load
a'Near the patient' implies that testing was done onsite in the health facility laboratory or decentralized peripheral laboratory. bValues of prevalence chosen to represent rates of detecting treatment failure on a single test, for low (2.5%), medium (10%), and high (30% and 40%) prevalence scenarios. cIndirect test comparisons only possible where data were sufficient (i.e. Xpert versus SAMBA).
Background
It is estimated that in 2019 there were about 38 million people living with HIV globally, of whom 25.4 million (67%) people living with HIV (PLHIV) were on antiretroviral therapy (ART) (UNAIDS 2020). In sub‐Saharan Africa in 2019, there were about 25.6 million PLHIV, of which about 17 million (69%) were on ART (UNAIDS 2020). In order to effectively sustain treatment for people on ART, it is essential to know the HIV viral load (VL) levels in those undergoing treatment. VL (the number of HIV viral ribonucleic acid (RNA) particles per millilitre of blood) is the recommended monitoring approach to diagnose and confirm ART treatment failure (WHO 2016). VL is usually measured in plasma; however, some technologies use whole blood (UNITAID 2015). In Africa, it is estimated that less than 20% of people on ART received routine VL testing in 2013 (ASLM 2013). This could be partly be explained by poor access to VL testing services. Currently, VL testing is largely done on central laboratory‐based platforms that involve sophisticated equipment requiring dedicated laboratory space, substantial financial resources, and trained laboratory technicians. These laboratory tests require venous blood collection, cold chain storage of collected samples, and instrument‐based sample processing techniques. With transport shortcomings being a common challenge in resource‐limited settings, delays in transporting samples to the laboratory and relaying test results back to the health centre lead to delays in changing therapy in cases of treatment failure. To overcome this challenge, point‐of‐care tests are increasingly being developed because they are potentially easy to use, cost‐effective, and require less laboratory infrastructure. They could also potentially reduce patient waiting time and therefore reduce loss to follow‐up cases (UNITAID 2014; UNITAID 2015; WHO 2014).
Target condition being diagnosed
The target condition of this review is high HIV VL levels in blood or plasma of people living with either HIV‐1 or HIV‐2 on HIV/AIDS care or follow‐up in health facilities. The World Health Organization (WHO) recommends a policy of initiating ART on all PLHIV regardless of immunological status (WHO 2015). The main objective of ART is to reduce HIV VL to undetectable levels, meaning that the concentration HIV RNA cannot be detectable by a test. In PLHIV, it is therefore essential to monitor VL levels especially after ART initiation. The higher the VL, the higher the increased risk of transmission when VL is detectable and the faster the CD4 cells and body's immune system are destroyed. Detectable VL can be a reflection of poor adherence to treatment or treatment failure once poor adherence is ruled out. Intermittent low‐level viraemia (50 copies/mL to 1000 copies/mL) not associated with treatment failure may also occur during effective treatment (Havlir 2001). Current WHO guidelines on ART define a high or detectable VL level as 1000 copies/mL or greater and treatment failure as a persistently high VL concentration (1000 copies/mL or greater) in two consecutive measurements (with adherence support between measurements) (WHO 2016). Treatment failure should trigger evaluation or changing of the antiretroviral drugs included in ART. Delayed detection of treatment failure may therefore lead to progression of HIV infection to AIDS or the resistance of the infection to ART, or increase the risk of HIV transmission (UNITAID 2015; WHO 2013). Analysed data of 9200 adults on ART for at least four months from population‐based surveys from five Southern African countries conducted between 2015 and 2017 revealed that 11.2% had non‐suppressed viral loads (≥ 1000 copies/mL) including 8.2% who experienced virological failure (on ART and viral load ≥ 1000 copies/mL) (Haas 2020). In addition, the proportion of those with non‐suppressed viral load was about 35% in the Eastern and Southern Africa region, and about 55% in the Western and Central Africa region (UNAIDS 2020).
Index test(s)
In this Cochrane Review, we estimated the accuracy of molecular tests with point‐of‐care (POC) platforms in detecting high VL levels (POC VL) on PLHIV. Molecular POC VL include semi‐quantitative and quantitative tests that quantify the copies of HIV virus in plasma or whole blood (UNITAID 2014; UNITAID 2015). Results are reported as HIV copies in a millilitre (copies/mL). There is no established optimal threshold for detecting VL concentration or defining virological failure (Fox 2012; Ritchie 2014; WHO 2013; WHO 2016). In 2013, the WHO lowered the threshold for detecting high VL levels from 5000 copies/mL to 1000 copies/mL based on evidence that below 1000 copies/mL, intermittent low‐level viraemia (50 copies/mL to 1000 copies/mL) not associated with treatment failure can occur during effective treatment (Ritchie 2014; WHO 2013). Also, the risk of HIV transmission and progression of disease is minimal when VL concentration is less than 1000 copies/mL. Nonetheless, the lower limit of VL detection depends on the test and sample used. For example, a capillary sample from a finger prick may not accurately detect a VL level below 5000 copies/mL (ASLM 2013; UNITAID 2015).
Ideally, true POC tests are conducted on patient samples next to the patient or at the bedside in settings with minimaI laboratory and training requirements (Level 1 facilities). However, in resource‐limited settings testing locations are often blurred, as tests designed with POC platforms have been evaluated and implemented across a variety of healthcare and laboratory settings ranging from primary level next to patients (Level 1 facilities) to district (Level 2) and provincial levels (Level 3) (UNITAID 2015). To this end, various definitions of POC testing have been proposed with no universally accepted definition (Drain 2014; UNITAID 2015). For example, some definitions consider technical characteristics of the test (rapid test with minimal infrastructure requirements) (Wu 2012), or its effect on management (linking to decision making at the same patient visit) (Pai 2012), or its location (at the patient site or near the treatment facility) (Drain 2014). Another definition of a POC test would be a diagnostic test that is administered near the patient or at a health facility, with a fast turnaround time, leading to a change in patient management (Schito 2012). WHO developed the ASSURED (Affordable, Sensitive, Specific, User‐friendly, Robust & Rapid, Equipment free, and Deliverable to end‐users) criteria for the ideal rapid test for resource‐limited settings (Wu 2012). In order to maximize the utility of our review, we considered all forms of tests designed with POC platforms for VL regardless of the health facility setting in which the test was conducted.
Clinical pathway
The role of POC VL for monitoring response to ART will be to act as a replacement for laboratory‐based VL testing platforms in the current testing algorithms outlined in Figure 1.
1.
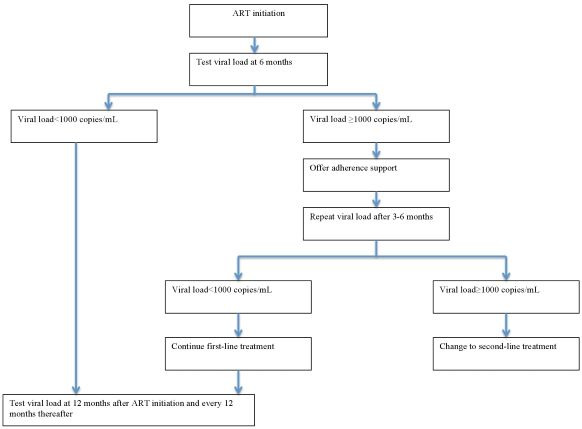
Routine viral load testing algorithm. Abbreviations: ART: antiretroviral therapy.
In routine care, current WHO guidelines recommend that VL testing be done at six and 12 months after initiation of ART and repeated every 12 months thereafter. If the VL is detectable at any time (1000 copies/mL or greater), it is recommended that a patient undergo intensive adherence support and repeat VL testing three to six months later. If the VL is still detectable and non‐adherence can be ruled out, a clinician may then decide to change to second‐line therapy (WHO 2013; WHO 2016).
In this review, we focused on the accuracy of a single POC VL test done at one time point in PLHIV attending healthcare facilities.
Alternative test(s)
Alternative HIV VL tests include non‐nucleic acid tests (non‐molecular tests) that detect HIV viral enzymes (reverse transcriptase) and HIV viral proteins (p24 antigen), markers that can be correlated to HIV RNA. These tests indirectly reflect VL concentration and are currently not commonly used (UNITAID 2015).
Alternative methods for monitoring response to ART include immunological monitoring through CD4 testing and clinical monitoring through WHO clinical staging. For example, in adults, a persistent CD4 count less than 100 cells/mm3 or a new or recurrent clinical condition indicative of WHO clinical stage 4 after six months of treatment is regarded as treatment failure. However, these methods are less sensitive and specific than VL testing and are not recommended as the first‐line approach for monitoring response to ART (Rutherford 2014). This may lead to delayed detection of treatment failure or to unnecessary therapy switches. In addition, the WHO revised its guidelines in 2013 to recommend that all PLHIV be started on ART regardless of CD4 count and clinical status (WHO 2013). In this regard, using these criteria to monitor response to therapy will not be an accurate measure of treatment failure. Nonetheless, these alternative tests may still be used in areas that do not have access to VL testing (WHO 2013).
Rationale
In 2014, the Joint United Nations Programme on HIV/AIDS (UNAIDS) declared the 90‐90‐90 target: it aimed to have at least 90% of HIV‐positive people diagnosed, at least 90% of those diagnosed receiving ART, and at least 90% of those receiving ART having suppressed viral replication by 2020 (WHO 2016). POC VL tests being developed to detect HIV RNA and treatment failure in HIV‐positive people on ART in resource‐limited settings will be instrumental in checking if the third target will be met effectively. If these POC VL tests have a high level of accuracy, they can replace or complement central laboratory‐based testing platforms because they are quicker to use and may minimize delays in initiating therapy or changing therapy in cases of treatment failure (UNITAID 2014; UNITAID 2015). A high sensitivity is required because false‐negative results will lead to a delay in detecting treatment failure or adherence concerns related to treatment, which will ultimately lead to progression to AIDS and mortality. A high specificity is also required because false‐positive results will lead to unnecessary switching to costly second‐line therapy. A test with an optimal combination of sensitivity and specificity is thus needed.
Objectives
To estimate the diagnostic accuracy of POC tests to detect high viral load levels in PLHIV attending healthcare facilities.
Secondary objectives
To investigate sources of heterogeneity in test accuracy estimates including age (children versus adults), test type (commercially available versus in‐house assays), sample type (whole blood versus plasma), test threshold (1000 copies/mL or greater versus other thresholds), location of testing (near patient versus central laboratory evaluations), geographical location (sub‐Saharan Africa versus other regions), and methodological quality (high versus low risk of bias).
Methods
Criteria for considering studies for this review
Types of studies
We included any primary study that compared the results of the POC VL index test to that of a central laboratory‐based reference standard (cross‐sectional, prospective, and retrospective study designs or diagnostic accuracy studies performed within randomized trials) and that provided sufficient data to create the 2 × 2 table to calculate sensitivity, specificity, and negative and positive predictive values. We excluded ecological studies and diagnostic case‐control studies in which the test performance was compared in participants with the target condition versus healthy controls, as specificity will be overestimated (Macaskill 2013). We excluded studies without a reference standard, case reports and case‐series studies, animal or laboratory studies, reviews, discussion papers, non‐research letters, commentaries, or editorials.
Participants
People infected with either HIV‐1 or HIV‐2 irrespective of age and gender, undergoing HIV/AIDS care or follow‐up from any healthcare facility or geographical setting.
Index tests
We included studies evaluating the accuracy of molecular VL tests designed with POC platforms that could be used near the patient regardless of the health facility setting in which the test was conducted. In resource‐limited settings, however, testing locations are often blurred, as POC tests have been evaluated and implemented across a variety of healthcare and laboratory settings (UNITAID 2015). We considered the current WHO‐recommended threshold (1000 copies/mL or greater) as the main threshold to define test positivity (WHO 2013; WHO 2016). We also considered the previous WHO‐recommended threshold (5000 copies/mL or greater) (WHO 2010), and other thresholds that may have been used for test evaluations in subgroup analyses.
Examples of POC VL tests include semi‐quantitative tests or quantitative tests as shown below (Drain 2019):
Xpert HIV‐1 Viral Load (Cepheid);
SAMBA I HIV‐1 Semi‐Quantitative Test;
SAMBA II HIV‐1 Semi‐Quantitative Test;
m‐PIMA (formerly Alere q HIV‐1/2 assay (quantitative whole blood assay);
Truelab Real Time micro PCR system (Molbio HIV‐1);
Savanna RealTime HIV‐1 Viral Load assay (Quidel);
cobas Liat Analyzer (Roche) (production postponed, not currently available);
Xpert HIV‐1 Viral Load (Cepheid);
ZIVA (Cavidi);
Liat Analyzer (IQuum Inc);
EOSCAPE HIV Rapid RNA Assay System;
Truelab Real Time micro PCR system (Molbio);
RT CPA HIV‐1 viral load.
Of all these tests, only Xpert HIV‐1, SAMBA I & II, m‐PIMA (formerly Alere), and Molbio are currently available. In addition, only Xpert HIV‐1 VL assay and m‐PIMA test are WHO prequalified.
Semi‐quantitative tests provide output as either positive or negative with assay results being read as lines on the lateral flow strips. For SAMBA Semi‐Q test, for example, the presence of test line indicates a viral load > 1000 copies/mL, and the absence of a test line indicates a viral load < 1000 copies/mL (Ritchie 2014). On the other hand, results of quantitative tests are expressed as copies/mL.
Target conditions
A high HIV VL level in people living with HIV‐1 or HIV‐2.
Reference standards
Laboratory‐based testing platforms to detect high VL levels taken at the same time (within 24 hours) as the sample for POC VL tests. Most laboratory‐based VL platforms are designed to detect the HIV virus in plasma that is extracted from a venous blood sample though centrifugation. Typical laboratories for VL technologies involve sophisticated equipment and have three rooms for sample extraction, reagent preparation, and amplification (and detection) of the HIV virus (UNITAID 2015). Examples of laboratory‐based platforms for VL are nucleic acid‐based tests (NAT), including five commercially available reverse transcriptase polymerase chain reaction (RT‐PCR)‐based VL assays:
COBAS AmpliPrep/COBAS TaqMan version 2.0 (CAP/CTM v2.0) (Roche);
RealTime HIV‐1 (Abbott);
VERSANT HIV RNA 1.0 (kPCR) (Siemens);
Artus HIV‐1 QS‐RGQ (QIAGEN);
RT‐TMA technology for Panther system (Hologic).
Current and previous WHO‐recommended thresholds to detect high HIV VL levels in plasma and classify a patient as having treatment failure include 1000 copies/mL or greater (WHO 2013; WHO 2016), and 5000 copies/mL or greater (WHO 2010). We included data where the threshold of 1000 copies/mL were presented but also collected data of the 5000 copies/mL threshold.
Where studies used a tie‐breaker approach (where a second test/PCR for discordant results), we included results for the first test/PCR only in the 2 × 2 tables to avoid inflation of sensitivity and specificity (Ritchie 2014). Some included evaluations used a tie‐breaker approach (Goel 2017a; Goel 2017b; Goel 2017c; Goel 2017d; Ritchie 2014b; Ritchie 2014c). We mostly included results of the first reference in the analysis, but made an exception for Goel 2017c. This evaluation used Roche CAP/CTM v2.0 assay as the first reference, and Abbott RealTime HIV‐1 assay as the second reference test to handle discrepant results. There were seven discrepant results using original Roche testing, and six discrepant results were concordant/similar with tie‐breaker testing (Abbot = Roche); it was challenging getting the exact 2 x 2 table with the original results, hence results of the reference test were based on tie‐breaker results. This is unlikely to have introduced bias, as it was only one differing result.
Search methods for identification of studies
Electronic searches
We searched the following electronic databases with no language, document type, or publication status limitations.
Cochrane Central Register of Controlled Trials (CENTRAL) in the Cochrane Library (Issue 11 of 12, November 2020)
MEDLINE Ovid (1946 to 16 November 2020)
Embase Ovid (1947 to 16 November 2020)
LILACS (Latin American and Caribbean Health Sciences Literature database) (searched 22 November 2020)
World Health Organization (WHO) International Clinical Trials Registry Platform (ICTRP) (www.who.int/clinical-trials-registry-platform) (searched 22 November 2020)
WHO Global Index Medicus (www.globalindexmedicus.net/) (searched 22 November 2020)
ClinicalTrials.gov (www.clinicaltrials.gov/) (searched 22 November 2020)
Web of Science (Core Collection, includes Science Citation Index Expanded (SCI‐EXPANDED)/Conference Proceedings Citation Index‐Science (CPCI‐S)) (1990 to 23 November 2020)
Search resources and strategies are presented in Appendix 1.
Searching other resources
We searched the reference lists of included studies, relevant systematic reviews, and conference proceedings (Conference on Retroviruses and Opportunistic Infections, International AIDS Society Conference, and African Society for Laboratory Medicine). We consulted experts in the field such as the WHO HIV Department for potentially relevant studies.
Data collection and analysis
Selection of studies
We de‐duplicated search results in EndNote X7 (EndNote 2016). Two review authors (EAO and EEO) independently screened the titles and abstracts of the search results to identify potentially eligible articles. Reports that were obviously not relevant based on title and abstract and duplicates were removed. The two review authors (EAO and EEO) then independently assessed the full texts of journal articles or conference proceedings for eligibility based on our a priori inclusion criteria. Any disagreements were resolved by consensus or by consulting a third review author (SM or JD). We documented our justifications for excluding articles from the review in the 'Characteristics of excluded studies' table. Details of the included studies are presented in the 'Characteristics of included studies' table, and the study selection process is illustrated in a PRISMA flow diagram.
Data extraction and management
We extracted the following information on study characteristics: study design; demographic and participant characteristics; methods of collecting and preparing blood specimen; time point at which VL testing is done after ART initiation; index test and reference standard characteristics; test cut‐off and performance; main outcome data or results; number of true‐positive, false‐positive, false‐negative, and true‐negative results (Appendix 2).
Two review authors (EAO and EEO) independently extracted data, resolving any disagreements by discussion or by consulting a third review author (SM or JD).
Assessment of methodological quality
We used the QUADAS‐2 (Quality Assessment of Diagnostic Accuracy Studies) tool to assess the risk of bias and applicability concerns of the included studies (Whiting 2011). We tailored the tool in line with the context of our review question (Appendix 3). Two review authors (EAO and EEO) independently assessed the included studies using the tool outlined in Appendix 3. Any disagreements were resolved by consensus or by consulting a third review author (SM or JD).
Statistical analysis and data synthesis
Our unit of analysis was the participant. For each study, we identified the threshold(s) used to define test positivity and constructed 2 × 2 tables (true‐positive, false‐positive, false‐negative, true‐negative) at the presented thresholds. We performed the main analysis with study data using the current WHO‐recommended threshold (1000 copies/mL or greater) definition of test positivity (WHO 2016). We undertook subgroup analyses separately at other commonly presented thresholds. We conducted preliminary exploratory analyses on diagnostic accuracy by plotting estimates of sensitivity and specificity from each study on forest plots and in receiver operating characteristic (ROC) space. These analyses enabled visual assessment of the variation between studies, and also facilitated investigations of heterogeneity for exploring the effect of certain characteristics on test performance.
To estimate the summary sensitivity and specificity at the current WHO threshold (≥ 1000 copies/mL) for the main meta‐analysis, investigate sources of heterogeneity, and compare the accuracy of two or more tests, we fitted simplified univariable models for sensitivity and specificity separately, using a random‐effects model (Takwoingi 2017). The bivariate model with random effects accounts for within‐study variability, and correlation of sensitivity and specificity did not converge to give a model estimate (Macaskill 2013; Reitsma 2005). We therefore fitted simplified models, using univariable models for sensitivity and specificity separately, employing a random‐effects model (Takwoingi 2017). There were two reasons for model convergence problems. Firstly, three studies reported specificity values that were very different from most studies (19%, 45%, and 48% compared to the rest of studies, within range of 92% to 100%). This caused instability in model fitting. Secondly, in analyses not including outlier specificity values, most values of specificity were close to 100%, meaning that there was no correlation between sensitivity and specificity, so bivariate models did not converge.
For comparisons between tests that had sufficient data, we included all studies in the analysis (indirect comparison). We performed analyses using Review Manager 5 (RevMan 5) (Review Manager 2020), and the meta‐analysis using Stata (Stata 2017).
Investigations of heterogeneity
Where there were sufficient data, we investigated sources of heterogeneity in estimates of test accuracy. We added the following covariates to the univariate model to assess the influence on test performance: manufacturer test type (Xpert versus SAMBA) and location of testing (near patient versus central laboratory).
Sensitivity analyses
Where there were sufficient data, we used sensitivity analyses to explore the effect of other test thresholds, ART status, geographical setting, and study quality. We estimated sensitivity and specificity at other commonly used thresholds (≥ 40 copies/mL). We restricted the analysis to studies that exclusively included participants on ART, and to studies conducted in sub‐Saharan Africa. Our risk of bias assessment was mostly unclear for the included studies, and most studies had either high concerns for applicability for participant selection, index and reference test. We therefore did not conduct sensitivity analyses for studies at low risk of bias for participant selection or high applicability for index test conduct. In addition, the proportion of children included was unclear, therefore we conducted a sensitivity analysis by restricting analysis to studies that included only adults. Limited data precluded a comparison of commercial tests to in‐house test and whole blood to plasma blood samples. We instead restricted the analysis to studies that included commercial tests and those that used plasma samples.
Assessment of reporting bias
We did not assess reporting bias due to various methodological shortcomings associated with assessing reporting bias in diagnostic accuracy studies (Macaskill 2013).
Assessment of the strength of the evidence
We summarized the main findings of the review, reporting the numbers of true‐positives, true‐negatives, false‐positives, and false‐negatives per 1000 people tested in the summary of findings table. There are some methodological challenges with GRADE for diagnostic test accuracy reviews (Gopalakrishna 2014; Gopalakrishna 2016), therefore rather than following any formal process for downgrading the evidence, we described the following concepts, which constitute an assessment of strength of the evidence.
Precision of the study estimates.
Heterogeneity in study findings.
Risk of bias.
Concerns about applicability.
Indirect comparisons between tests.
These issues cover the key domains of GRADE (GRADE 2013), except publication bias, which cannot be assessed, and would permit inclusion of the evidence in a GRADE assessment should a guideline developer wish to do so.
Results
Results of the search
A summary of search results is provided in Figure 2. Our search yielded 2555 potentially eligible articles, of which 11 were found through additional searches. We screened 2178 titles and abstracts and retrieved the full texts for 110 articles. We assessed the full texts, and excluded 92 articles, and included 18 studies in the systematic review and 14 studies in the meta‐analyses. The meta‐analyses included studies that assessed accuracy of POC VL at a threshold of ≥ 1000 copies/mL.
2.
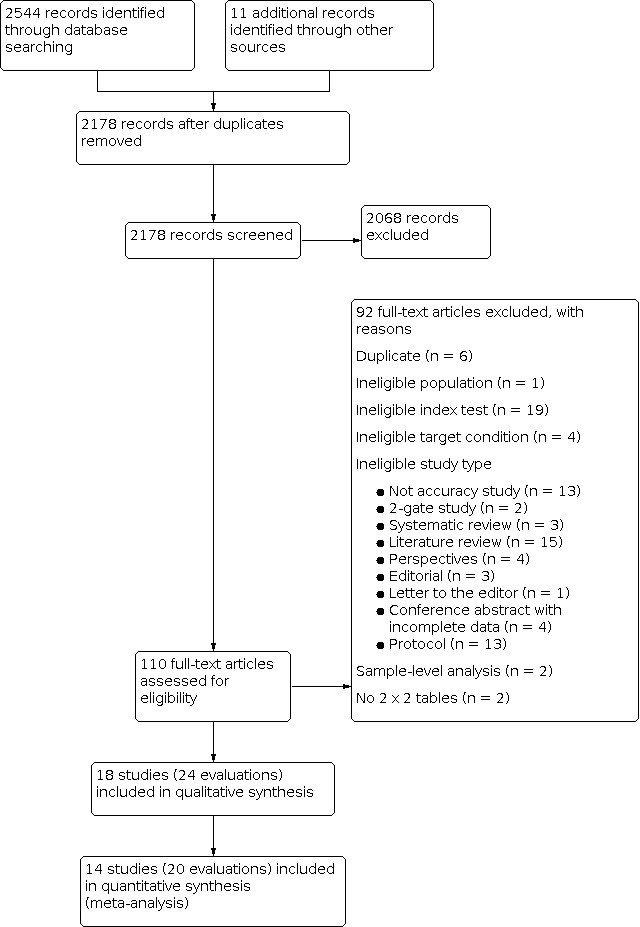
Study flow diagram.
Included studies
We identified a total of 18 studies (24 evaluations, 10,034 participants) defining high viral loads at main thresholds ≥ 1000 copies/mL (n = 20), ≥ 5000 copies/mL (n = 1), and ≥ 40 copies/mL (n = 3). All evaluations were done on samples from PLHIV retrieved from routine HIV/AIDS care centres or health facilities. Twenty evaluations had a cross‐sectional design, three had cohort‐like designs, and the design for one study was unclear. Five evaluations reported a random‐sampling strategy, whilst the rest (n = 19) had an unclear sampling strategy. Full details of the included studies are provided in the Characteristics of included studies section. For clinical applicability, we included 14 studies (20 evaluations, 8659 participants) assessing high viral load at the clinical threshold of ≥ 1000 copies/mL in the meta‐analyses. Of these evaluations, 17 had a cross‐sectional design, and three had cohort‐like designs. Also, four evaluations reported a random‐sampling strategy, whilst the rest (n = 16) had an unclear sampling strategy. Half (n = 10) of the samples retrieved from patients were tested near the patient (in the health facility laboratory or decentralized or peripheral laboratory), and the other half (n = 10) away from the patient at a central or reference laboratory. Most evaluations used plasma samples (n = 17), except for three evaluations, which utilized whole blood samples.
Excluded studies
We excluded 92 articles after critically reading the full texts. Full details of the excluded studies are provided in the Characteristics of excluded studies section. In summary, six were duplicates, one was a primary study with an ineligible population (exclusively ART‐naive population retrieved from a household community survey, not from an HIV/AIDS care centre) (Moyo 2016), 19 included ineligible index tests, four studies had ineligible target conditions, and 58 were ineligible study types including reviews, editorials, perspectives, protocols, conference abstracts with incomplete data, and non‐accuracy studies. Two studies evaluated the accuracy of the tests at sample level, and we could not construct 2 x 2 tables for two studies.
Methodological quality of included studies
In Figure 3 and Figure 4, we have summarized the results of quality appraisal for 24 evaluations included in the systematic review that defined high viral loads across three main thresholds: ≥ 1000 copies/mL (n = 20), ≥ 5000 copies/mL (n = 1), and ≥ 40 copies/mL (n = 3). We evaluated these studies for risk of bias based on the following QUADAS‐2 domains (Whiting 2011): participant selection, index test, reference standard, and participant flow. The risk of bias was mostly assessed as unclear across the four domains due to incomplete reporting. We assessed about 90% of evaluations in the patient selection domain as unclear due to mostly poor reporting of patient sampling method or inappropriate exclusions. For the index test and reference tests domains about 55% and 65% of evaluations, respectively, were judged as unclear due to poor reporting of blinding of the test results. Lastly, about 70% of evaluations in the flow‐and‐timing domain unclearly reported the interval between the index and reference tests or whether all test results were included in the final analysis. The included studies had some concerns for applicability across two domains: patient selection and index test. Viral load monitoring is mostly essential for patients who have initiated ART. For patient selection, about 30% of evaluations included ART‐naive populations in the samples. Also, 30% did not clearly report ART status of the included populations, though the samples were retrieved from routine HIV/AIDS care centres. For the index test domain, about 50% of evaluations had concerns for applicability because they were conducted in central or reference laboratories.
3.
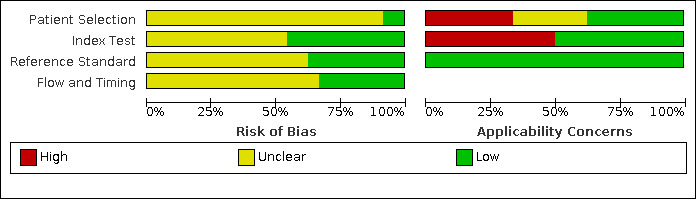
Risk of bias and applicability concerns graph: review authors' judgements about each domain presented as percentages across included studies.
4.
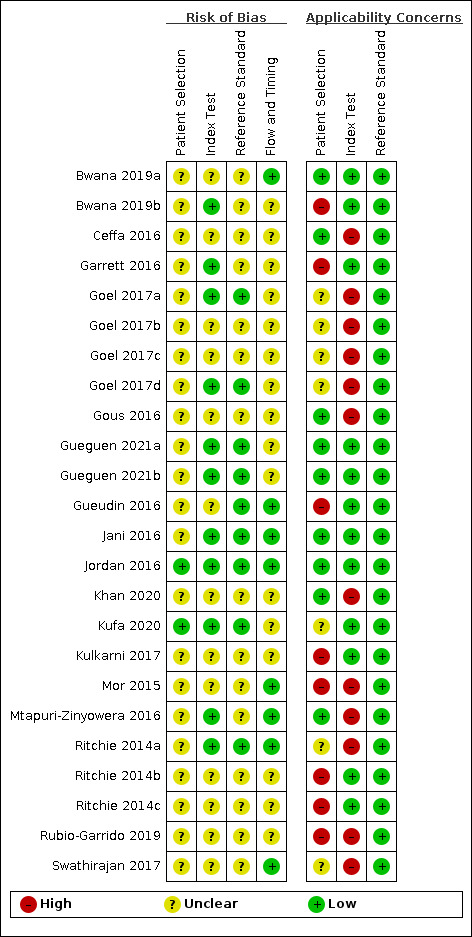
Risk of bias and applicability concerns summary: review authors' judgements about each domain for each included study.
Findings
A summary of the main findings is provided in Table 1.
We identified a total of 18 studies (24 evaluations, 10,034 participants) defining high viral loads at main thresholds ≥ 1000 copies/mL (n = 20), ≥ 5000 copies/mL (n = 1), and ≥ 40 copies/mL (n = 3). All evaluations were done on samples from PLHIV retrieved from routine HIV/AIDS care centres or health facilities. None of the tests was a true POC test done at the patient's side; all were conducted in laboratories, either in onsite laboratories near the patient (n = 12) or at a central or reference laboratory (n = 12).
For clinical applicability, we focused on and included 14 studies (20 evaluations, 8659 participants) assessing high VL at the clinical threshold of ≥ 1000 copies/mL in the meta‐analyses. Of these, sub‐Saharan Africa, Europe, and Asia contributed 16, three, and one evaluation respectively. All included participants were on ART in only nine evaluations; in the other 11 the proportion of participants on ART was either partial or unclearly stated. Thirteen evaluations included adults only (n = 13), five mixed populations of adults and children, and two did not clearly state the age of populations included. The majority of evaluations included commercially available tests (n = 18). Ten evaluations were POC VL tests conducted near the patient in a peripheral or onsite laboratory, whilst the other 10 were evaluations of POC VL tests in a central or reference laboratory setting. The test types evaluated as POC VL tests included Xpert HIV‐1 Viral Load test (n = 8), SAMBA HIV‐1 Semi‐Q Test (n = 9), Alere Q NAT prototype assay for HIV‐1 (n = 2), and m‐PIMA HIV‐1/2 Viral Load test (n = 1). The majority of evaluations (n = 17) used plasma samples, whilst the rest (n = 3) utilized whole blood samples.
The reference tests used in the included 20 evaluations (≥ 1000 copies/mL) varied. Some evaluations only used one type of reference test, as follows: Roche COBAS AmpliPrep/COBAS TaqMan (CAP/CTM) HIV‐1 (n = 5), Abbott (n = 5), and NUCLISENS (n = 1). Other evaluations used a combination: Roche and Abbott (n = 6), NUCLISENS and Abbott (n = 1), and Abbott QIAGEN (n = 1). The reference test was unclearly reported in one evaluation.
For POC VL evaluations with the threshold ≥ 1000 copies/mL only, the forest plot in Figure 5 and summary receiver operating characteristic (SROC) plot in Figure 6 reveal some heterogeneity for estimates of sensitivity (range 89% to 100%) and more heterogeneity for estimates of specificity (range 19% to 100%).
5.
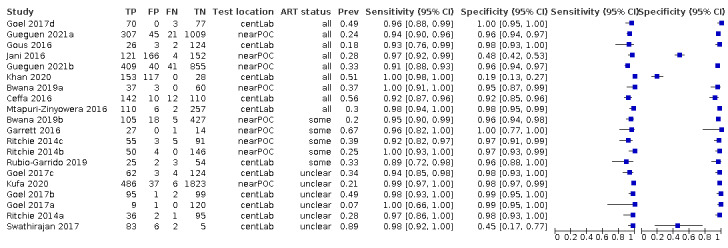
Forest plot of POC VL evaluations at clinical threshold ≥ 1000 copies/mL. Abbreviations: centLab (central laboratory), nearPOC (near point of care or near patient site in the field).
6.
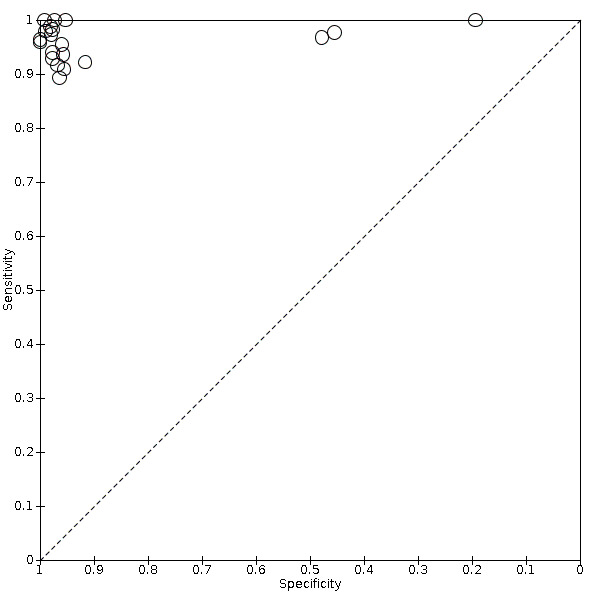
Summary ROC plot of POC VL evaluations at clinical threshold ≥ 1000 copies/mL.
A. Primary analysis, POC VL for detection of treatment failure (high viral load ≥ 1000 copies/mL)
The primary meta‐analysis was limited to 20 evaluations that reported a threshold of ≥ 1000 copies/mL, the current WHO‐recommended clinical threshold for treatment failure. Median prevalence for high viral load (≥ 1000 copies/mL) (n = 20) was 33.4% (range 6.9% to 88.5%). Ten were evaluations of the POC VL tests in the field or at point of care, and 10 were evaluations of the POC VL tests in a laboratory setting.
For these 20 evaluations, sensitivity estimates ranged from 89% to 100% (Figure 5). Specificity estimates ranged from 19% to 100%. Notably, three studies had low specificity results (19% in Khan 2020, 45% in Swathirajan 2017, and 48% in Jani 2016). Of these, two studies where specificity results were low (19% and 48%, Khan 2020; Jani 2016) used whole blood samples, in contrast to the majority of studies where plasma samples were used.
Khan 2020 (specificity 19%) was a laboratory‐based cross‐sectional study evaluating a prototype assay, Alere q (Alere Technologies, Jena, Germany), performed using a prototype cartridge on routinely collected whole blood samples from ART clinics from mostly adult PLHIV (93%). An additional study also used whole blood samples; Jani 2016 was a field evaluation of Alere q (Alere Technologies, Jena, Germany) on routinely collected whole blood samples from adult PLHIV on ART from a peri‐urban primary health centre in Mozambique. Swathirajan 2017 was an evaluation of Xpert HIV‐1 Viral Load assay in a tertiary AIDS care and research centre in India. The demographics of the samples in this study were unclear.
POC VL pooled sensitivity and specificity (95% confidence interval (CI)) against laboratory‐based assays at a threshold ≥ 1000 copies/mL were 96.6% (94.8 to 97.8) (20 evaluations, 2522 participants) and 95.7% (90.8 to 98.0) (20 evaluations, 6137 participants).
B. Investigating sources of heterogeneity
A summary of the variation in sensitivity and specificity is provided in Table 2.
1. Sources of variation in accuracy estimates.
| Sensitivity (%) | Specificity (%) | Comparisons | ||
| Main meta‐analysisa | @ ≥ 1000 copies/mL n = 20 | 96.6 (94.8 to 97.8) | 95.7 (90.8 to 98.0) | ‐ |
| Subgroup analysisa | ||||
| Test type | Xpert HIV‐1 Viral Load test (n = 8) | 96.9 (94.0 to 98.4) | 95.6 (89.4 to 98.2) | Difference in sensitivity Xpert versus Sambab 2.1% (−1.2 to 5.3) |
| SAMBA HIV‐1 Semi‐Q Test (n = 9) | 94.8 (91.6 to 96.9) | 97.2 (95.3 to 98.4) | Difference in specificity Xpert versus Sambab −1.7% (−5.9 to 2.5) |
|
| Location | Central lab (n = 10) | 96.5 (93.7 to 98.1) | 95.8 (84.0 to 99.0) | Difference in sensitivity Lab versus near patientb,c −0.1% (−3.0 to 2.7) |
| Near patientc (n = 10) | 96.7 (94.1 to 98.2) | 95.6 (90.8 to 98.0) | Difference in specificity Lab versus near patientb,c 0.2% (−6.5 to 6.9) |
|
| Sensitivity analysisa | ||||
| ART status | All on ART (n = 9) | 96.5 (92.6 to 98.4) | 90.1 (71.6 to 97.0) | ‐ |
| Region | Africa (n = 16) | 95.3 (94.4 to 96.1) | 92.1 (91.4 to 92.8) | ‐ |
| Age | Adults only (n = 13) | 97.2 (95.6 to 98.2) | 97.4 (94.3 to 98.8) | ‐ |
| Test group | Commercial assay (n = 18) | 96.1 (94.2 to 97.4) | 96.9 (95.2 to 98.1) | ‐ |
| Sample type | Plasma (n = 17) | 96.0 (94.0 to 97.3) | 97.0 (96.1 to 97.8) | ‐ |
| Threshold | Threshold @ ≥ 40 copies/mL (n = 7) | 85.6 (74.9 to 92.2) | 95.9 (90.7 to 98.2) | ‐ |
Abbreviations: ART: antiretroviral therapy
aWe fitted simplified univariable models for sensitivity and specificity separately, using a random‐effects model when the bivariate models did not converge to give a model estimate. bIndirect test comparisons were conducted. c'Near the patient' implies that testing was done onsite in the health facility laboratory or decentralized peripheral laboratory.
Subgroup analysis
Guided by the availability of sufficient data, we conducted subgroup analysis for the following covariates: location (field or near‐point of care versus central lab), test type (Xpert versus SAMBA), and threshold (at ≥ 40 copies/mL) (Table 2). For POC tests conducted near the patient (n = 10), pooled sensitivity (95% CI) was 96.7% (94.1 to 98.2), and specificity was (95% CI) 95.6% (90.8 to 98.0). For POC tests conducted in the central laboratory (n = 10), pooled sensitivity (95% CI) was 96.5% (93.7 to 98.1), and specificity was (95% CI) 95.8% (84.0 to 99.0). There was no statistically significant difference in the sensitivity (‐0.1% (−3.0 to 2.7), P = 0.92) and specificity (0.2% (−6.5 to 6.9), P = 0.95) of POC tests conducted in the central laboratory compared to those conducted near the patient.
The pooled sensitivity (95% CI) of Xpert Viral Load test (n = 8) was 96.9% (94.0 to 98.4), and specificity (95% CI) was 95.6% (89.4 to 98.2). The pooled sensitivity (95% CI) of SAMBA (n = 9) was 94.8% (91.6 to 96.9), and specificity was 97.2% (95.3 to 98.4). There was no statistically significant difference in the sensitivity (2.1% (−1.2 to 5.3), P = 0.21) and specificity (−1.7% (−5.9 to 2.5), P = 0.43) of Xpert VL test compared to SAMBA.
Pooled sensitivity (95% CI) for one other reported threshold (≥ 40 copies/mL) was 85.6% (74.9 to 92.2), and pooled specificity was 95.9% (90.7 to 98.2). A lower threshold for viral load may have been more difficult to detect compared to using the higher threshold, and more cases may have been missed.
Sensitivity analysis
When only studies with clearly reported ART‐exclusive populations were included (n = 9), pooled sensitivity and specificity (95% CI) against laboratory tests were 96.5% (92.6 to 98.4) and 90.1% (71.6 to 97.0), respectively. When only studies done in sub‐Saharan Africa were included (n = 16), pooled sensitivity and specificity (95% CI) against laboratory tests were 95.3% (94.4 to 96.1) and 92.1% (91.4 to 92.8). Restricting the analysis to adults (n = 13) yielded a sensitivity (95% CI) of 97.2% (95.6 to 98.2) and specificity (95% CI) of 97.4% (94.3 to 98.8). Restricting the analysis to commercial assays (n = 18) yielded a sensitivity (95% CI) of 96.1% (94.2 to 97.4) and specificity (95% CI) of 96.9% (95.2 to 98.1). Restricting the analysis to plasma samples (n = 17) yielded a sensitivity (95% CI) of 96.0% (94.0 to 97.3) and specificity (95% CI) of 97.0% (96.1 to 97.8). We did not restrict to studies with low risk of bias because no studies were judged as high risk of bias.
Apart from the main threshold ≥ 1000 copies/mL, various other thresholds were reported in the studies, including: ≥ 40 copies/mL, ≥ 200 copies/mL, ≥ 300 copies/mL, ≥ 3000 copies/mL, ≥ 4000 copies/mL, ≥ 5000 copies/mL, and ≥ 10,000 copies/mL. Some studies reported more than one threshold. Data were insufficient to pool accuracy estimates at one other threshold of ≥ 40 copies/mL. At the threshold ≥ 40 copies/mL (n = 7, 2288 participants), pooled sensitivity (95% CI) was 85.6% (74.9 to 92.2), and pooled specificity (95% CI) was 95.9% (90.7 to 98.2). These evaluations were conducted using the following tests: Xpert HIV‐1 Viral Load assay (n = 5), Alere Q prototype assay (n = 1), and m‐PIMA HIV‐1/2 assay (n = 1).
Discussion
This review evaluated the diagnostic accuracy of POC VL tests in detecting high viral loads in PLHIV in comparison with central laboratory testing as the reference standard, from 18 studies published between the years 2014 and 2020 (24 evaluations). To assess the diagnostic accuracy of POC VL tests to detect high HIV viral load at the WHO clinically recommended threshold of ≥ 1000 copies/mL, estimates from 20 evaluations were statistically pooled in the meta‐analysis.
Summary of main results
We included 14 studies (20 evaluations, 8659 participants) assessing high HIV viral load at the clinical threshold of ≥ 1000 copies/mL in the meta‐analyses. Of these, sub‐Saharan Africa, Europe, and Asia contributed 16, three, and one evaluation respectively. All evaluations were done on samples from PLHIV retrieved from routine HIV/AIDS care centres or health facilities. All included participants were on ART in only nine evaluations; in the other 11 the proportion of participants on ART was either partial or unclearly stated. For this, median prevalence for high viral load (≥ 1000 copies/mL) (n = 20) was 33.4% (range 6.9% to 88.5%). Thirteen evaluations included adults only (n = 13), five mixed populations of adults and children, and two evaluations did not clearly state the age of included populations. The majority of evaluations included commercially available tests (n = 18). None of the tests was a true POC test done at the patient's side; all were conducted in laboratories, either in onsite laboratories near the patient (n = 10) or at a central or reference laboratory (n = 10). The test types evaluated as POC VL tests included Xpert HIV‐1 Viral Load test (n = 8), SAMBA HIV‐1 Semi‐Q Test (n = 9), Alere Q NAT prototype assay for HIV‐1 (n = 2), and m‐PIMA HIV‐1/2 Viral Load test (n = 1). The majority of evaluations (n = 17) used plasma samples, whilst the rest (n = 3) utilized whole blood samples.
For these 20 evaluations, sensitivity estimates ranged from 89% to 100% and specificity estimates from 19% to 100%. Noting the three studies with low specificity results, two were unusual in using whole blood rather plasma samples (Jani 2016; Khan 2020), and one was a smaller study (Swathirajan 2017). POC VL pooled sensitivity and specificity (95% CI) against laboratory tests at a threshold ≥ 1000 copies/mL were 96.6% (94.8 to 97.8) (20 evaluations, 2522 participants) and 95.7% (90.8 to 98.0) (20 evaluations, 6137 participants). For POC VL tests conducted in the central laboratory (n = 10), pooled sensitivity (95% CI) was 96.5% (93.7 to 98.1), and specificity was 95.8% (84.0 to 99.0); for POC VL conducted in the field, sensitivity and specificity estimates were similar at 96.7% (94.1 to 98.2) and 95.6% (90.8 to 98.0), respectively. When the analysis was restricted to studies with clearly reported ART‐exclusive populations (n = 9), pooled sensitivity was similar to the overall analysis (96.5% versus 96.6%), and specificity was lower (90.1% versus 95.7%).
Risk of bias assessment was mostly unclear due to poor reporting. The included studies had some concerns for applicability for patient selection and index test domains. Not all included participants were on ART, and some tests with POC platforms were conducted in a laboratory setting rather than in the field near the patient.
In a hypothetical cohort of 1000 PLHIV, where 100 have high viral load, 136 people would receive a positive result with the molecular POC test; of these 39 will not have high viral loads (false‐positive result) and would be incorrectly identified as not responding to ART treatment, possibly leading to unnecessary testing or further treatment; and 864 people would receive a negative test result with the molecular POC test; of these three will actually have high virus levels (false‐negative results) and would be missed whilst failing ART treatment.
Strengths and weaknesses of the review
We searched multiple databases and literature sources and contacted experts for additional studies. We also contacted authors for additional information. A similar meta‐analysis was recently published evaluating the performance of Cepheid Xpert HIV‐1 Viral Load plasma assay to accurately detect treatment failure (Sacks 2019). Whereas that study focused on Cepheid Xpert viral load assay for HIV‐1, our review included other index tests including SAMBA for HIV‐1, Alere for HIV‐1, and m‐PIMA HIV‐1/2. The sensitivity (95% CI) and specificity for Xpert in the review by Sacks and colleagues at 1000 copies/mL were 96.47% (95.1 to 97.5) and 96.59% (92.9 to 98.4). Our review revealed similar estimates for sensitivity (96.9% (94.0 to 98.4)), and slightly lower estimates for specificity (95.6% (89.4 to 98.2)) than those in Sacks 2019.
Our review included samples from PLHIV retrieved from routine HIV/AIDS care centres or health facilities, but not all included PLHIV were on ART, as some studies had mixed populations of patients on ART (reported proportions ranging from 52% to 80%) and those not on ART. This could be a reflection of the barriers to initiating ART in those newly diagnosed with HIV in HIV/AIDS care centres or health facilities (Loeliger 2016; Moges 2020; Patel 2016). Also, in other studies, the ART status of included participants was not reported, but samples were from HIV/AIDS care centres or hospitals. Considering the WHO recommendation that all HIV‐infected individuals be on ART regardless of immunological status, we assumed that a sizeable proportion of participants in the unclearly reported studies were on ART (WHO 2015). Enriching the sample with those not yet on ART may introduce bias if the viral load measures in this group are higher (and thus easier to detect) than those in individuals on ART who are experiencing a treatment failure. An overestimation of sensitivity would be expected, but when we restricted the analysis to studies with participants on ART exclusively, the sensitivity was similar (96.5% versus 96.6%), but specificity was lower (90.1% versus 95.7%) compared to the overall pooled analysis.
Secondly, all included index tests were laboratory evaluations of the POC VL tests, with some conducted in the field near the patient in onsite laboratories and others conducted in central laboratories. However, our review found no statistically significant differences in the sensitivity and specificity of the POC tests conducted in the central laboratory versus those conducted in the field near the patient. Of note, there is often a blur with regard to the definition of POC tests, as tests designed with POC platforms are conducted near the patient in peripheral laboratories or even in central laboratory settings.
Thirdly, our overall meta‐analysis included three studies with outlier specificity results (19% to 48%) (Jani 2016; Khan 2020; Swathirajan 2017), compared to the rest of the included studies, whose specificity results ranged from 92% to 100%. Jani 2016 and Khan 2020 included samples from PLHIV on ART from peri‐urban and urban health centres in Mozambique and South Africa, respectively. They both evaluated Alere Q NAT, a prototype RNA amplification assay on whole blood samples, which measures both plasma‐ and cell‐associated RNA (total RNA). The cell‐associated RNA in the whole blood samples can lead to higher viral load measurements (hence higher false‐positive results) when coupled with detection methodology limitations in the test (Jani 2016; Khan 2020). Swathirajan and colleagues, on the other hand, evaluated Xpert HIV‐1 Viral Load test on a sample set that predominantly had viral load measurements greater than 1000 copies/mL. Only 11 out of 103 specimens had viral load measurements of less than 1000 copies/mL. Indeed, this could have contributed to the higher viral load quantification levels (85%) detected by Xpert VL test compared to the reference standard Abbott RealTime PCR assay. This study included samples from HIV‐1 patients undergoing care at a tertiary AIDS research and care centre (Swathirajan 2017).
Lastly, limitations in the reporting of included studies limited our investigations of all possible sources of variation. The median prevalence for high viral load (≥ 1000 copies/mL) (n = 20) was 33.4% (range 6.9% to 88.5%) in the studies included in our review. Well‐reported estimates of adherence to ART would have helped explain high prevalence of viral load estimates better. Also, some evaluations used two reference tests to handle discrepant results. We aimed to consider only the result of the first reference test in the analysis where discrepant results were retested with a second reference test. However, we made an exception for Goel 2017c, where results of the resolution made by the second test were included in the analysis. There were seven discrepant results using original Roche testing, and six discrepant results were concordant/similar with tie‐breaker testing. This was unlikely to have introduced bias, as it was only one differing result. In addition, data were insufficient to pool results at other reported thresholds (≥ 200 copies/mL, ≥ 400 copies/mL, ≥ 3000 copies/mL, ≥ 5000 copies/mL, ≥ 10,000 copies/mL). Newer POC HIV viral load assays should achieve a lower limit of quantification, such as 200 copies/mL, given the availability of newer medications with greater efficacy for maintaining viral suppression (Drain 2019).
Applicability of findings to the review question
The findings of this review had some concerns for applicability to the review question with regard to the population included and index test. Our review included samples from PLHIV retrieved from routine HIV/AIDS care facilities or hospitals, but not all of the participants included in the review were on ART. Some studies included a mixture of ART‐naive and ART‐experienced participants, and in some studies ART status was not reported. Some studies evaluated tests with rapid POC platforms in central laboratory settings instead of at or near the patient's side, though this is reflective of what occurs in many resource‐limited settings. In resource‐limited settings it is often unclear what defines a true POC test, as tests with POC platforms have been evaluated and implemented across a wide range of healthcare and laboratory facilities (UNITAID 2015).
Authors' conclusions
Implications for practice.
The point‐of‐care viral load (POC VL) tests have a high sensitivity and high specificity to detect or exclude high viral loads at ≥ 1000 copies/mL in people living with HIV (PLHIV) compared to central laboratory‐based assays. About half of included evaluations of the POC VL tests were conducted in a central laboratory setting and not near the patient, but there was no statically significant difference in accuracy between settings. These tests may complement or replace traditional central laboratory‐based viral assays. Also, in resource‐poor settings where patients have limited access to health facilities and would otherwise exceed the recommended time for a POC VL, field or near POC VL testing may be useful as an initial screening test to ensure these cohorts of patients are not left completely unmonitored. The World Health Organization has recommended a policy of initiating antiretroviral therapy (ART) in all PLHIV regardless of immunological status (WHO 2015). Though all of the included studies retrieved samples from routine HIV/AIDS care centres, not all included samples were from patients on ART. In health facilities and HIV care centres, barriers and delays to initiating ART in PLHIV need to be investigated and reasons acted upon such as providing counselling beyond initial diagnosis and following up patients. For example, a qualitative study amongst newly diagnosed HIV‐positive patients in Ethiopia cited patient disbelief in test results, having no symptoms, and preference for spiritual healing as barriers to the initiation of ART on the same day or at next visit (Moges 2020), and a qualitative study amongst HIV‐discordant couples in Kenya found that barriers to ART initiation included denial of diagnosis, stigma, challenges in obtaining refills, and perceived side effects of ART (Patel 2016). In addition, a qualitative study seeking perspectives of community health workers in South Africa highlighted ART initiation barriers, including: inadequate patient education and social support, fear of lifelong therapy amongst patients, preference for alternative medicines, patient dissatisfaction with health services, and low socio‐economic status (Loeliger 2016).
Implications for research.
Estimates of adherence to ART need to be investigated and reported in future studies evaluating the accuracy and impact of POC VL. This would help better explain the accuracy of POC VL in the context of high prevalence of VL in those on ART. Also, research into the development and evaluation of true POC tests on exclusively ART‐experienced populations conducted near or at the patient's side are needed. More clinical trials evaluating the effect of these POC tests compared to laboratory standard‐of‐care tests on people‐important outcomes such as time to change in treatment, emotional effects (stigma), morbidity and mortality will be useful in gauging the utility of these tests in different settings. For example, Drain and colleagues conducted an open‐label, non‐inferiority, randomized controlled trial to evaluate the effectiveness of POC HIV VL testing with task shifting on treatment and care outcomes (combined viral suppression (< 200 copies/mL) and retention at 12 months after enrolment) for adults on ART when compared with standard laboratory VL testing in South Africa (Drain 2020). This trial demonstrated that POC VL testing combined with task shifting significantly improved viral suppression and retention in HIV care in a public clinic in Durban, South Africa. Diagnostic accuracy is considered as indirect evidence on people‐important outcomes. With the availability of direct evidence regarding the effect or clinical impact of HIV POC diagnostics on people‐important outcomes (Drain 2020), it is preferable to base decisions on the existing direct evidence and the certainty of that evidence.
History
Protocol first published: Issue 11, 2018
Acknowledgements
The CIDG Contact Editor is Dr Nathan Ford, and the DTA Contact Editor is Dr Danielle van der Windt.
The editorial base of the Cochrane Infectious Diseases Group is funded by UK aid from the UK government for the benefit of low‐ and middle‐income countries (project number 300342‐104). The views expressed do not necessarily reflect the UK government’s official policies.
We acknowledge Information Specialists Anel Schoonees and Vittoria Lutje, who assisted in developing the search terms and search strategy and in conducting the searches.
We acknowledge the contribution of Dr Karla Soares‐Weiser and Artemisia Kakourou in developing the protocol and writing the initial final report that was submitted to the WHO ART Guideline Committee in 2015.
Eleanor A Ochodo is supported by a grant from the UK MRC/DFID African Research Leader grant scheme (grant number: T008768). The UK MRC and DFID has no role in the design, conduct, and interpretation of this protocol and review. EO is also partly supported by the Research, Evidence and Development Initiative (READ‐It). READ‐It (project number 300342‐104) is funded by UK aid from the UK government; however, the views expressed do not necessarily reflect the UK government’s official policies.
Appendices
Appendix 1. Search resources and strategies
Search strategy as per updated search done on 16 to 23 November 2020
Medline (Ovid)
Ovid MEDLINE(R) and Epub Ahead of Print, In‐Process & Other Non‐Indexed Citations, Daily and Versions(R) <January 2020 to November 16, 2020>
Search date: 16 November 2020
1 exp HIV/ or exp HIV Infections/ or Acquired Immunodeficiency Syndrome/ or (Acquired Immunodeficiency Syndrome? or Acquired Immunologic Deficiency Syndrome? or Acquired Immun? Deficiency Syndrome? or Human Immunodeficiency Virus$ or Human T Cell Lymphotropic Virus$ or Human T Lymphotropic Virus$ or Human T Cell Leukemia Virus$ or LAV HTLV III or Lymphadenopathy Associated Virus$ or HIV or HIV 1 or HIV 2 or HIV/aids or HIV I or LAV 2 or LAV HTLV III or HIV II or HTLV III or HTLV IV or SBL 6669 or AIDS).ti,ab.
2 Viral Load/ or exp Nucleic Acid Amplification Techniques/ or nucleic acid hybridization/ or self‐sustained sequence replication/ or polymerase chain reaction/ or reverse transcriptase polymerase chain reaction/ or Branched DNA signal amplification assay/ or (Viral Load$ or Virus$ Load$ or Viral Burden? or Virus$ Burden? or Virus Titer$ or Viral Titer$ or VL$ or NAT or NATs or NAAT or NAATs or Nucleic Acid Amplif$ or DNA Amplif$ or RNA Amplif$ or nucleic acid sequence based amplification or NASBA or nucleic acid hybridization or nucleic acid hybridization or nucleic acid test$ or nucleic acid based test$ or transcription‐mediated amplification or self‐sustained sequence replication or polymerase chain reaction or PCR or RT‐PCR or RTPCR or bDNA or b‐ DNA or branched DNA or branched‐chain DNA).ti,ab.
3 Point‐of‐Care Systems/ or (Point of Care or Care Technolog$ Point$ or Bedside Test$ or Bedside Comput$ or Bedside Technolog$ or Rapid Test$ or Rapid Diagnos$ or RDT).ti,ab. (39098)
4 1 and 2 and 3
5 limit 4 to yr="1990 ‐Current"
6 (animals not (humans and animals)).sh.
7 5 not 6
Embase (Ovid)
Embase January 2020 ‐Present, updated daily
Search date: 16 November 2020
1 exp Human immunodeficiency virus/ or exp acquired immune deficiency syndrome/ or exp human immunodeficiency virus infection/ or exp human immunodeficiency virus 1/ or exp human immunodeficiency virus 2/ or (Acquired Immunodeficiency Syndrome? or Acquired Immunologic Deficiency Syndrome? or Acquired Immun? Deficiency Syndrome? or Human Immunodeficiency Virus$ or Human T Cell Lymphotropic Virus$ or Human T Lymphotropic Virus$ or Human T Cell Leukemia Virus$ or LAV HTLV III or Lymphadenopathy Associated Virus$ or HIV or HIV 1 or HIV 2 or HIV/aids or HIV I or LAV 2 or LAV HTLV III or HIV II or HTLV III or HTLV IV or SBL 6669 or AIDS).ti,ab.
2 Viral Load/ or nucleic acid amplification/ or nucleic acid hybridization/ or nucleic acid sequence based amplification/ or polymerase chain reaction/ or reverse transcription polymerase chain reaction/ or branched DNA signal amplification assay/ or (Viral Load$ or Virus$ Load$ or Viral Burden? or Virus$ Burden? or Virus Titer$ or Viral Titer$ or VL$ or NAT or NATs or NAAT or NAATs or Nucleic Acid Amplif$ or DNA Amplif$ or RNA Amplif$ or nucleic acid sequence based amplification or NASBA or nucleic acid hybridization or nucleic acid hybridization or nucleic acid test$ or nucleic acid based test$ or transcription‐mediated amplification or self‐sustained sequence replication or polymerase chain reaction or PCR or RT‐PCR or RTPCR or bDNA or b‐ DNA or branched DNA or branched‐chain DNA).ti,ab.
3 Point of care testing/ or exp rapid test/ or (Point of Care or Care Technolog$ Point$ or Bedside Test$ or Bedside Comput$ or Bedside Technolog$ or Rapid Test$ or Rapid Diagnos$ or RDT).ti,ab.
4 exp animals/ or exp invertebrate/ or animal experiment/ or animal model/ or animal tissue/ or animal cell/ or nonhuman/
5 human/ or normal human/ or human cell/
6 4 and 5
7 4 not 6
8 1 and 2 and 3
9 8 not 7
10 limit 9 to yr="1990 ‐Current"
11 limit 10 to exclude medline journals
ClinicalTrials.gov
Date of search: 22 November 2020
Advanced search
(Viral Load* or Virus* Load* or Viral Burden* or Virus* Burden* or Virus Titer* or Viral Titer* or VL* or Point of Care OR Care Technolog* Point* OR Bedside Test* OR Bedside Comput* OR Bedside Technolog* OR Rapid Test* OR Rapid Diagnos* OR RDT) | (Acquired Immunodeficiency Syndrome* OR Acquired Immunologic Deficiency Syndrome* OR Acquired Immun* Deficiency Syndrome* OR Human Immunodeficiency Virus* OR HIV* OR AIDS*)
Web of Science Core Collection
Includes: Science Citation Index Expanded (SCI‐EXPANDED)/ and Conference Proceedings Citation Index‐ Science (CPCI‐S).
Date of search: 22 November 2020
TITLE: ((Acquired Immunodeficiency Syndrome* OR Acquired Immunologic Deficiency Syndrome* OR Acquired Immun* Deficiency Syndrome* OR Human Immunodeficiency Virus* OR Human T Cell Lymphotropic Virus* OR Human T Lymphotropic Virus* OR Human T Cell Leukemia Virus* OR LAV HTLV III OR Lymphadenopathy Associated Virus* OR HIV OR HIV 1 OR HIV 2 OR HIV/AIDS OR HIV I OR LAV 2 OR LAV HTLV III OR HIV II OR HTLV III OR HTLV IV OR SBL 6669 OR AIDS)) ANDTITLE: ((NAT OR NATs OR NAAT OR NAATs OR Nucleic Acid Amplif* OR DNA Amplif* OR RNA Amplif* OR nucleic acid sequence based amplification OR NASBA OR nucleic acid hybridization OR nucleic acid hybridization OR nucleic acid test* OR nucleic acid based test*OR transcription‐mediated amplification OR self‐sustained sequence replication OR polymerase chain reaction OR PCR OR RT‐PCR OR RTPCR OR bDNA OR b‐DNA OR branched DNA OR branched‐chain DNA)) ANDTITLE: ((Viral Load* or Virus* Load* or Viral Burden* or Virus* Burden* or Virus Titer* or Viral Titer* or VL or Point of Care OR Care Technolog* Point* OR Bedside Test* OR Bedside Comput* OR Bedside Technolog* OR Rapid Test* OR Rapid Diagnos* OR RDT))
LILACS (Virtual Health Library)
Date of search: 22 November 2020
Words: (Acquired Immunodeficiency Syndrome$ OR Acquired Immunologic Deficiency Syndrome$ OR Acquired Immun$ Deficiency Syndrome$ OR Human Immunodeficiency Virus$ OR Human T Cell Lymphotropic Virus$ OR Human T Lymphotropic Virus$ OR Human T Cell Leukemia Virus$ OR LAV HTLV III OR Lymphadenopathy Associated Virus$ OR HIV OR HIV 1 OR HIV 2 OR HIV/AIDS OR HIV I OR LAV 2 OR LAV HTLV III OR HIV II OR HTLV III OR HTLV IV OR SBL 6669 OR AIDS) AND
Words: (NAT OR NATs OR NAAT OR NAATs OR Nucleic Acid Amplif$ OR DNA Amplif$ OR RNA Amplif$ OR nucleic acid sequence based amplification OR NASBA OR nucleic acid hybridization OR nucleic acid hybridization OR nucleic acid test$ OR nucleic acid based test$ OR transcription‐mediated amplification OR self‐sustained sequence replication OR polymerase chain reaction OR PCR OR RT‐PCR OR RTPCR OR bDNA OR b‐DNA OR branched DNA OR branched‐chain DNA) AND
Words: (Viral Load$ or Virus$ Load$ or Viral Burden$ or Virus$ Burden$ or Virus Titer$ or Viral Titer$ or VL or Point of Care OR Care Technolog$ Point$ OR Bedside Test$ OR Bedside Comput$ OR Bedside Technolog$ OR Rapid Test$ OR Rapid Diagnos$ OR RDT)
WHO Global Index Medicus
Search date: 22 November 2020
https://www.globalindexmedicus.net/
Searched in Title, Abstract, Subject:
(tw:((Acquired Immunodeficiency Syndrome$) OR (Acquired Immunologic Deficiency Syndrome$) OR (Acquired Immun$ Deficiency Syndrome$) OR (Human Immunodeficiency Virus$) OR (HIV) OR (HIV/AIDS) OR (AIDS))) AND (tw:((viral load$) OR (virus load$) OR (viral burden$) OR (virus$ burden$) OR (virus titer$) OR (viral titer$) OR (VL$) OR (point of care) OR (care technolog$ Point$) OR (bedside test$) OR (bedside comput$) OR (bedside Technolog$) OR (rapid test$) OR (rapid diagnos$) OR (RDT))) AND (tw:((NAT) OR (NATs) OR (NAAT) OR (NAATs) OR (nucleic acid amplif$) OR (DNA Amplif$) OR (RNA Amplif$) OR (nucleic acid sequence based amplification) OR (NASBA) OR (nucleic acid hybridization) OR (nucleic acid hybridisation) OR (nucleic acid test$) OR (transcription‐mediated amplification) OR (self‐sustained sequence replication) OR (polymerase chain reaction) OR (PCR) OR (RT‐PCR) OR (RTPCR) OR (bDNA) OR (b‐DNA) OR (branched DNA) OR (branched‐chain DNA) OR (branched chain DNA)))
World Health Organization International Clinical Trials Registry Platform (WHO ICTRP)
http://apps.who.int/trialsearch/
Date: 23 November 2020
(Acquired Immunodeficiency Syndrome* OR Acquired Immunologic Deficiency Syndrome* OR Acquired Immun* Deficiency Syndrome* OR Human Immunodeficiency Virus* OR HIV* OR AIDS*) in the Condition
AND
(Viral Load* or Virus* Load* or Viral Burden* or Virus* Burden* or Virus Titer* or Viral Titer* or VL* or Point of Care OR Care Technolog* Point* OR Bedside Test* OR Bedside Comput* OR Bedside Technolog* OR Rapid Test* OR Rapid Diagnos* OR RDT) in the Intervention
Recruitment status: ALL
World Health Organization International Clinical Trials Registry Platform (WHO ICTRP)
https://www.who.int/clinical‐trials‐registry‐platform
Date: 23 November 2020
(HIV* and point of care)
CENTRAL in Cochrane Library
Date of search: 23 November 2020
#1 MeSH descriptor: [HIV] explode all trees
#2 MeSH descriptor: [HIV Infections] explode all trees
#3 ((Acquired Immunodeficiency Syndrome* or Acquired Immunologic Deficiency Syndrome* or Acquired Immun* Deficiency Syndrome* or Human Immunodeficiency Virus* or Human T Cell Lymphotropic Virus* or Human T Lymphotropic Virus* or Human T Cell Leukemia Virus* or LAV HTLV III or Lymphadenopathy Associated Virus* or HIV or “HIV 1” or “HIV 2” or “HIVAIDS” or HIV I or “LAV 2” or LAV HTLV III or HIV II or HTLV III or HTLV IV or “SBL 6669” or AIDS)):ti,ab,kw
#4 #1 or #2 or #3
#5 MeSH descriptor: [Viral Load] explode all trees
#6 ((Viral Load* or Virus* Load* or Viral Burden* or Virus* Burden* or Virus Titer* or Viral Titer* or VL*)):ti,ab,kw
#7 MeSH descriptor: [Nucleic Acid Amplification Techniques] explode all trees
#8 MeSH descriptor: [Nucleic Acid Hybridization] explode all trees
#9 MeSH descriptor: [Self‐Sustained Sequence Replication] explode all trees
#10 MeSH descriptor: [Polymerase Chain Reaction] explode all trees
#11 MeSH descriptor: [Reverse Transcriptase Polymerase Chain Reaction] explode all trees
#12 MeSH descriptor: [Branched DNA Signal Amplification Assay] explode all trees
#13 ((NAT or NATs or NAAT or NAATs or Nucleic Acid Amplif* or DNA Amplif* or RNA Amplif* or nucleic acid sequence based amplification or NASBA or nucleic acid hybridization or nucleic acid hybridization or nucleic acid test* or nucleic acid based test* or transcription‐mediated amplification or self‐sustained sequence replication or polymerase chain reaction or PCR or RT‐PCR or RTPCR or bDNA or b‐DNA or branched DNA or branched‐chain DNA)):ti,ab,kw
#14 #5 OR #6 OR #7 OR #8 OR #9 OR #10 OR #11 OR #12 OR #13
#15 MeSH descriptor: [Point‐of‐Care Systems] explode all trees
#16 ((Point of Care or Care Technolog* Point* or Bedside Test* or Bedside Comput* or Bedside Technolog* or Rapid Test* or Rapid Diagnos* or RDT)):ti,ab,kw
#17 #15 or #16
#18 #4 and #14 and #17 with Cochrane Library publication date from Jan 1990 to present, in Trials
Appendix 2. Data to be extracted
We will extract the following information for cross‐sectional, cohort, and case‐control studies.
Study ID: studies by the name of the first author and the year in which the study was first published.
Eligibility: study design, population, HIV status, details of antiretroviral therapy used.
Study details: aim/objective of the study, inclusion and exclusion criteria, study design, prospective/retrospective, whether study was restricted to a subgroup of a larger cohort, how sample size was determined, region and country, setting (inpatients, outpatients), study start and end dates, duration of follow‐up, and sponsor/source of funding.
Study population: description of the participants included in the study (age, gender), predefined inclusion or exclusion criteria (or both), special populations, number of participants recruited/included in the study, how participants were allocated to groups, ART used (first or second line).
Interventions: details of POC VL test used, manufacturer/brand name, conduct of the test, test cut‐off and performance, staff performing test, specimens or sample type, time point at which VL testing was done after ART initiation.
Accuracy estimates: true‐positives, false‐positives, false‐negatives, true‐negatives.
Study aim and comments: short description of the overall aim of the study, and any additional comments on the study.
Appendix 3. QUADAS‐2; list of signalling questions, risk of bias, and applicability
| Domain | Participant selection | Index test (IT) | Reference standard (RS) | Flow and timing |
| Description | Methods of participant selection | How IT was conducted and reported | How RS was conducted and reported | Describe participants who did not receive and time interval between IT or RS |
| Signalling questions (yes, no, unclear) | Consecutive or random sample of participants? Yes: when the authors reported random participant sampling or consecutive enrolment. No: when participants were selected, for example, based on previous (reference or index) test results. Unclear: if there was insufficient information on study sampling. |
IT results interpreted without knowledge of the results of RS? Yes: when study reported that results of the ITs were interpreted without knowledge of RS results or when ITs were done before the RS. No: when study reported that results of the ITs were interpreted with knowledge of RS results or in cases when RS were used before the index tests. Unclear: when there was insufficient information on when the IT and RS were interpreted. |
RS likely to correctly classify the target condition? Yes: if the RS threshold was clearly reported as > 1000 copies/mL or > 5000 copies/mL. No: if the RS threshold was not reported or if other thresholds were used without justification. Unclear: if there was insufficient information to make a judgement. |
Appropriate interval between IT and RS? Yes: if samples tested by both the RS and IT were taken at the same time or within 24 hours. No: if samples tested by both the RS and IT were taken at the same time or within 24 hours. Unclear: when there was no or insufficient information on time period. |
| Was a case‐control design avoided? Yes: if a case‐control design was not used. No: if a case‐control design was used. Unclear: if there was insufficient information on study design. |
Prespecified threshold used? Yes: when the authors reported the use of one prespecified cut‐off value. A prespecified threshold also included statements such as 'the test was scored according to manufacturer’s instructions'. No: when multiple cut‐off values were tested and the best one chosen afterwards. Unclear: when only one cut‐off value was used, but this was not explicitly stated in the methods section. |
RS results interpreted without knowledge of the results of IT? Yes: when study reported that results of the RS were interpreted without knowledge of IT results, or in cases when RS were used before the IT. No: when study reported that results of the RS were interpreted with knowledge of the IT results in cases when IT were used before the RS. Unclear: when there was insufficient information on when the IT and RS were interpreted. |
Number of participants receiving a RS, and included in the analysis? Yes: when the whole sample or a random selection of the sample or a selection of the sample with consecutive series received verification using an RS. No: when a part of the sample that was non‐randomly or non‐consecutively selected receives verification with the RS. Unclear: when there was no or insufficient information to ascertain if the whole sample or a random selection of the sample received verification with an RS. |
|
| Did the study avoid inappropriate exclusions? Yes: no participants were excluded after inclusion. No: for example, when specific participants were excluded (e.g. those with mild disease because they are more difficult to detect). Unclear: if there was insufficient information on inclusion/exclusion criteria. |
Number of participants receiving same RS, and included in the analysis? Yes: when study participants were tested with the same reference standard RS regardless of index test result. No: when different RS were used. Unclear: when there was no or insufficient information the different RS used. |
|||
| Were all participants included in the analysis? Yes: when the participants included in the study were also included in the analysis. No: when some participants/results were missing. Unclear: when there was no or insufficient information to make a judgement. | ||||
| Risk of bias (high, low, unclear) | Could the selection of participants have introduced bias? | Could the conduct or interpretation of the IT have introduced bias? | Could the RS, its conduct, or its interpretation have introduced bias? | Could the participant flow have introduced bias? |
| Applicability concerns (high, low, unclear) | Are there concerns that the included participants do not match the review question? High: if some included participants were not on ART. Low: if all participants were on ART. Unclear: if there was insufficient information to make a judgement. |
Are there concerns that the IT, its conduct, or interpretation differs from the review question? High: if IT was not a true POC, i.e. required ancillary laboratory equipment or staff or testing done on frozen samples, or if IT was not commercially available (a prototype). Low: if IT was a true POC and commercially available. Unclear: if there was insufficient information to make a judgement. |
Are there concerns that the target condition as defined by the RS does not match the review question? High: if the RS threshold was not reported, or if other thresholds were used without justification. Low: if the RS threshold was clearly reported as > 1000 copies/mL or > 5000 copies/mL. Unclear: if there was insufficient information to make a judgement. |
‐ |
Scoring risk of bias assessment.
Abbreviations: IT: index test; POC: point of care; RS: reference standard. | ||||
Data
Presented below are all the data for all of the tests entered into the review.
Tests. Data tables by test.
| Test | No. of studies | No. of participants |
|---|---|---|
| 1 POC VL_All | 24 | 10034 |
| 3 POC VL_1000 | 20 | 8659 |
| 4 POC VL_40 | 7 | 2288 |
1. Test.
POC VL_All
3. Test.
POC VL_1000
4. Test.
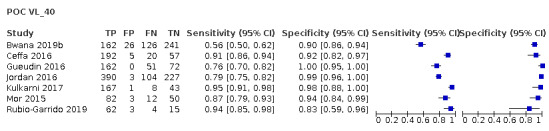
POC VL_40
Characteristics of studies
Characteristics of included studies [ordered by study ID]
Bwana 2019a.
| Study characteristics | |||
| Patient Sampling | In order to assess the performance of the Xpert HIV‐1 quantitative assay on GeneXpert platform, 100 plasma samples were tested on both the platform and on the Abbott m2000 assay and the results were compared. Both the qualitative and the quantitative studies of the performance of the GeneXpert platform were cross‐sectional evaluations of samples obtained from facilities across the country. | ||
| Patient characteristics and setting | HIV‐positive adults on antiretroviral therapy (ART); field in Kenya; contacted author who confirmed that samples were from those on ART. | ||
| Index tests | Xpert HIV‐1 quantitative assay (Cepheid, Sunnyvale, CA, USA); done in peripheral lab on fresh plasma samples from the field. | ||
| Target condition and reference standard(s) | High viral load > 1000 copies/mL; Abbott m2000. | ||
| Flow and timing | In order to assess the performance of the Xpert HIV‐1 quantitative assay on GeneXpert platform, 100 plasma samples were tested on both the platform and on the Abbott m2000 assay and the results were compared. For viral load estimation on the GeneXpert platform, a total of 1.2 mL of plasma was added into the Xpert HIV‐1 Viral Load cartridge using a calibrated pipette. The cartridge was closed and loaded onto the machine. Test results were observed and recorded after 90 minutes. For comparison, viral load estimation was done on the Abbott m2000 using plasma according to manufacturer’s instructions. Test results were observed and recorded after 5 hours. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Unclear | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Bwana 2019b.
| Study characteristics | |||
| Patient Sampling | Cross‐sectional study conducted between June and November 2018. HIV‐positive adults were recruited from Alupe, Nambale, and Matayos Sub County Hospitals as well as Siaya County Referral Hospital. These facilities were selected due to their proximity to the research institute and their high sample volumes. Only consenting participants were enrolled in the study. | ||
| Patient characteristics and setting | HIV‐positive adults (72.89% on ART); selected health facilities in Western Kenya. | ||
| Index tests | m‐PIMA HIV‐1/2 Viral Load; done at point of care (POC) sites on fresh plasma samples. | ||
| Target condition and reference standard(s) | High viral load (viral nucleic acids to HIV‐1/2) Abbott RealTime HIV‐1 assay. | ||
| Flow and timing | For performance evaluation, a venous blood sample was drawn from each participant. At the heath facilities, 4 mL of venous blood was collected from each consenting HIV‐positive adult into an ethylenediaminetetraacetic acid (EDTA) tube using a Vacutainer needle. On the same day, the tubes were centrifuged at 1100 g for 10 min to separate the plasma. Using the provided teat pipette, 25 μL of the resultant plasma was loaded onto the test cartridge; the cartridge was immediately inserted in the m‐PIMA analyser, and the test was left to run until complete. The HIV‐1 results were recorded after 69 min in copies/mL. The remnant samples in the EDTA tubes were shipped to the KEMRI HIV Lab in Alupe at 2 °C to 8 °C. In the reference lab, the remnant samples were received and stored at −30 °C until the next day when they were tested on the comparator. Venous whole blood samples were successfully drawn from 567 participants. 12 samples were excluded from the quantitative data analysis. |
||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Ceffa 2016.
| Study characteristics | |||
| Patient Sampling | To compare viral load results, samples of children ≤ 14 years and adults ≥ 15 years of age who had been taking ART for at least 12 months, and had a viral load test ordered following the clinical flowchart normally in use (viral load test after 12 months of ART treatment), were tested. Participants were enrolled sequentially at the selected sites until the desired sample size was reached. Target enrolment was 200 exposed newborns and 300 adults and/or children, 60 samples with undetectable viral load (< 1.6 log(10) copies/mL), and 60 samples for each range group, as follows: 1.6 to 2.99 log(10) copies/mL), 3 to 3.69 log(10) copies/mL), 3.7 to 3.99 log(10) copies/mL) and > 4 log(10) copies/mL). |
||
| Patient characteristics and setting | Children and adults on ART for at least 12 months; the DREAM laboratory in Blantyre, Malawi, where samples collected at various health centres in different districts were centralized for the analyses. | ||
| Index tests | The Xpert HIV‐1 Viral Load (Cepheid, Sunnyvale, CA, USA) done in central laboratory on frozen plasma samples. | ||
| Target condition and reference standard(s) | High viral load at > 1000 copies/mL and > 40 copies/mL. Abbott RealTime HIV‐1 assay (m2000rt; Abbott Molecular Diagnostics, Mississauga, ON, Canada). | ||
| Flow and timing | EDTA blood samples were collected, and 2 aliquots of plasma from each sample were prepared and stored at −80 °C for a minimum of 1 day and a maximum of 3 months. 1 aliquot was used for routine determination with the Abbott m2000 system, and the second was processed with GeneXpert. The patients received the results of both tests in maximum of 1 month. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Garrett 2016.
| Study characteristics | |||
| Patient Sampling | A total of 42 Xpert HIV‐1 VL assays were performed on plasma samples on whole blood samples collected consecutively from known HIV‐positive South African women who attended for routine study visits in the Centre for the AIDS Programme of Research in South Africa (CAPRISA) 002 study. | ||
| Patient characteristics and setting | HIV‐positive adult women (55% on ART); South Africa. | ||
| Index tests | Xpert HIV‐1 Viral Load were performed in the clinic on fresh (31) plasma samples and frozen (11) plasma samples (field/clinic validation study). | ||
| Target condition and reference standard(s) | High HIV viral load > 1000 copies/mL; Roche TaqMan version 2 assay (Roche Diagnostics, Risch‐Rotkreuz, Switzerland). | ||
| Flow and timing | Samples were collected in 5‐millilitre EDTA tubes. For the Xpert HIV‐1 VL assay, specimens were first centrifuged at 1200 revolutions per minute (rpm) for 10 minutes before transfer of 1 mL of plasma into the assay’s cartridge chamber using a sterile pipette. Assay was then loaded into the GeneXpert System for analysis. HIV viral load testing was performed with the Roche TaqMan version 2 assay (Roche Diagnostics, Risch‐Rotkreuz, Switzerland) as the gold‐standard diagnostic test. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Unclear | ||
| Were all patients included in the analysis? | Unclear | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Goel 2017a.
| Study characteristics | |||
| Patient Sampling | The performance of the SAMBA HIV‐1 Semi‐Q Test for detection of HIV‐1 ribonucleic acid (RNA) at ≥ 1000 copies/mL in HIV‐1 positive patients was evaluated with the SAMBA I platform by testing specimens from consenting adults attending an HIV clinic at St Thomas’ Hospital in London, UK, for routine testing of CD4+ cell count and viral load with the Roche COBAS AmpliPrep/COBAS TaqMan (CAP/CTM) v2.0 assay. Leftover frozen plasma specimens from 130 consecutive patients were tested in a blinded manner with 1 lot of reagents for the SAMBA HIV‐1 Semi‐Q Test at the Diagnostics Development Unit, University of Cambridge, Cambridge, United Kingdom. | ||
| Patient characteristics and setting | Samples from HIV‐1 positive adults attending at HIV clinic at London hospital UK for routine testing and viral load monitoring. | ||
| Index tests | SAMBA HIV‐1 Semi‐Q Test at the Diagnostics Development Unit, University of Cambridge, Cambridge, United Kingdom. Frozen samples tested in central laboratory. | ||
| Target condition and reference standard(s) | Roche COBAS AmpliPrep/COBAS TaqMan (CAP/CTM) v2.0 assay. Abbott HIV‐1 RealTime Assay at the Royal Free Hospital (London, United Kingdom). (Discrepant samples, though original and retested samples similar; unlikely to introduce bias). |
||
| Flow and timing | Leftover frozen plasma specimens from 130 consecutive patients were tested in a blinded manner with 1 lot of reagents for the SAMBA HIV‐1 Semi‐Q Test at the Diagnostics Development Unit, University of Cambridge, Cambridge, United Kingdom. Testing specimens from consenting adults attending an HIV clinic at St Thomas’ Hospital in London, United Kingdom, for routine testing of CD4+ cell count and viral load with the Roche COBAS AmpliPrep/COBAS TaqMan (CAP/CTM) v2.0 assay. | ||
| Comparative | |||
| Notes | 1 discrepant result: Abbott same as Roche. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Unclear | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Low risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Goel 2017b.
| Study characteristics | |||
| Patient Sampling | In Kenya, the SAMBA HIV‐1 Semi‐Q Test as performed on the SAMBA I system by independent trained technicians at the centralized Kenya Medical Research Institute‐Centers for Disease Control and Prevention (KEMRI‐CDC) HIV research laboratory was evaluated with fresh surplus plasma samples obtained from patients attending rural clinics in 6 counties. 200 leftover plasma samples were selected on the basis of 4 viral load categories. | ||
| Patient characteristics and setting | Samples from HIV‐positive patients from rural clinics in Kenya. | ||
| Index tests | SAMBA HIV‐1 Semi‐Q Test on SAMBA I platform, done on fresh plasma samples in central laboratory. | ||
| Target condition and reference standard(s) | High HIV viral load > 1000 copies/mL; Roche CAP/CTM v2.0 assay and the Abbott HIV‐1 RealTime Assay (for discrepant results). Only original reference results included in analysis. | ||
| Flow and timing | All of them tested with same reference standard (main analysis); only a few discrepant results tested with Abbott. Discrepant analysis was performed at an independent laboratory with the Abbott HIV‐1 RealTime Assay. There were 3 discrepant results: Abbott result same as Roche. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Unclear | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Goel 2017c.
| Study characteristics | |||
| Patient Sampling | In Zimbabwe, the performance of the SAMBA HIV‐1 Semi‐Q Test was evaluated by testing plasma specimens from adults attending the Opportunistic Infection Clinic at Harare Hospital. A total of 193 fresh specimens were tested with the SAMBA HIV‐1 Semi‐Q Test as performed on the SAMBA I platform and with the Roche CAP/CTM v2.0 assay at the National Reference Microbiology Laboratory in Harare. | ||
| Patient characteristics and setting | Samples from HIV‐positive adults attending an HIV clinic in Harare, Zimbabwe. | ||
| Index tests | SAMBA HIV‐1 Semi‐Q Test on SAMBA I platform on fresh plasma samples in a central reference laboratory. | ||
| Target condition and reference standard(s) | High HIV viral load > 1000 copies/mL; Roche CAP/CTM v2.0 assay and Abbott HIV‐1 RealTime Assay (discrepant results). 7 discrepant results original Roche testing: 6 discrepant results were concordant with tie‐breaker testing (Abbott = Roche); 1+ Roche and 1‐ SAMBA. Results of the reference test were based on tie‐breaker results but unlikely to introduce bias as it was only one differing result. | ||
| Flow and timing | A total of 193 fresh specimens were tested with the SAMBA HIV‐1 Semi‐Q Test as performed on the SAMBA I platform and with the Roche CAP/CTM v2.0 assay at the National Reference Microbiology Laboratory in Harare. There were 7 discrepant results original Roche testing: 6 discrepant results were concordant with tie‐breaker testing (Abbott = Roche); 1+ Roche and 1‐ SAMBA. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Unclear | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | No | ||
| Were all patients included in the analysis? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Goel 2017d.
| Study characteristics | |||
| Patient Sampling | The performance of the SAMBA HIV‐1 Semi‐Q Test on the SAMBA II system for detection of HIV‐1 RNA at ≥ 1000 copies/mL in HIV‐1–positive patients was evaluated with surplus samples from adults on ART attending the Kiev Regional AIDS Center, Ukraine, for viral load quantification with the Abbott HIV‐1 RealTime Assay. A total of 150 frozen samples were shipped to Cambridge, United Kingdom, and tested with the SAMBA II assay at the Diagnostics Development Unit in a blinded manner. The Abbott HIV‐1 RealTime Assay was performed at the Kiev Regional AIDS Center. Discrepant samples were tested with the QIAGEN artus test by Public Health England, Cambridge. | ||
| Patient characteristics and setting | Surplus samples from HIV‐1 positive adults on ART attending the Kiev Regional AIDS Center, Ukraine; samples tested in laboratory. | ||
| Index tests | SAMBA HIV‐1 Semi‐Q Test on the SAMBA II system tested on frozen plasma samples in central research laboratory. | ||
| Target condition and reference standard(s) | HIV‐1 RNA/viral load > 1000 copies/mL; Abbott HIV‐1 RealTime Assay and QIAGEN artus test (for discrepant results). Only original reference results included in the analysis. | ||
| Flow and timing | A total of 150 frozen samples were shipped to Cambridge, United Kingdom, and tested with the SAMBA II assay at the Diagnostics Development Unit in a blinded manner. The Abbott HIV‐1 RealTime Assay was performed at the Kiev Regional AIDS Center. Discrepant samples were tested with the QIAGEN artus test by Public Health England, Cambridge. In Ukraine, original discordant and retesting with tie breaker yielded similar results. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Unclear | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Low risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | No | ||
| Were all patients included in the analysis? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Gous 2016.
| Study characteristics | |||
| Patient Sampling | Written informed consent was obtained when patients presented at the phlebotomy room of Themba Lethu HIV Clinic, Helen Joseph Hospital, Johannesburg, for routine viral load monitoring. | ||
| Patient characteristics and setting | Adult ART patients in health facilities in Johannesburg, South Africa. | ||
| Index tests | The Xpert HIV‐1 Viral Load assay (Cepheid) on fresh plasma done in lab; University of the Witwatersrand Diagnostics Research testing laboratory. | ||
| Target condition and reference standard(s) | High HIV viral load > 1000 copies/mL; Roche COBAS TaqMan/COBAS Ampliprep version 2 (TaqMan v2) and Roche COBAS. 6800/8800 (Roche Molecular Diagnostics, Branchburg, US) (fresh plasma samples). |
||
| Flow and timing | Following routine blood draw, an additional 4 EDTA.K3 blood tubes were obtained and couriered the same day (approximately 30 minutes) to the University of the Witwatersrand Diagnostics Research testing laboratory. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Unclear | ||
| Were all patients included in the analysis? | Unclear | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Gueguen 2021a.
| Study characteristics | |||
| Patient Sampling | Priority criteria were established for patients on clinical suspicion of virological failure, patients receiving second‐line ART, or third‐line ART from 2015, children and adolescents, and patients who had been receiving ART for more than 4 years. | ||
| Patient characteristics and setting | Patients on ART for between 6 months and 10 years, rural settings in Malawi. SAMBA I‐equipped sites included Chiradzulu District Hospital, together with 4 out of 10 peripheral health centres. | ||
| Index tests | SAMBA I Viral Load (SAMBA I VL) in the field. | ||
| Target condition and reference standard(s) | High viral load reflecting ART failure or efficacy > 1000 copies/mL; NUCLISENS bioMérieux (2013 to 2015); Abbott RealTime HIV‐1 (2016 to 2017). | ||
| Flow and timing | Same‐day point‐of‐care viral load (POC VL) results, but aliquots of the remaining plasma were prepared onsite, frozen in a dedicated freezer on the same day as blood collection, and sent monthly to a reference laboratory in the same country as collection. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Low risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | No | ||
| Were all patients included in the analysis? | Unclear | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Gueguen 2021b.
| Study characteristics | |||
| Patient Sampling | Priority criteria were established for patients on clinical suspicion of virological failure, patients receiving second‐line ART, or third‐line ART from 2015, children and adolescents, and patients who had been receiving ART for more than 4 years. | ||
| Patient characteristics and setting | Patients on ART for between 6 months and 10 years in Uganda; SAMBA I testing was implemented in Arua Regional Referral Hospital. | ||
| Index tests | SAMBA I Viral Load (SAMBA I VL) in field or near patient setting. | ||
| Target condition and reference standard(s) | High viral load > 1000 copies/mL reflecting ART failure or efficacy; Roche COBAS AmpliPrep/COBAS TaqMan HIV‐1 v2.0 (2013 to 2015); Abbott RealTime HIV‐1 (2015 to 2017). | ||
| Flow and timing | Same‐day POC VL results, but aliquots of the remaining plasma were prepared onsite, frozen in a dedicated freezer on the same day as blood collection, and sent monthly to a reference laboratory in the same country as collection. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Low risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | No | ||
| Were all patients included in the analysis? | Unclear | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Gueudin 2016.
| Study characteristics | |||
| Patient Sampling | Clinical performance was evaluated relative to the Abbott RealTime HIV‐1 assay on 285 HIV‐1 seropositive samples selected to cover the assays quantification range (40 copies/mL to 10,000,000 copies/mL), and included RNA undetectable or detected seropositive samples. Samples collected during routine viral load measurements for 285 patients managed at the Charles Nicolle Hospital, Rouen, France. |
||
| Patient characteristics and setting | Samples from 285 HIV‐1 seropositive patients (224 patients on ART and 59 untreated patients; no ART information available for 2 patients); lab setting in France. | ||
| Index tests | Cepheid Xpert HIV‐1 Viral Load assay done on fresh plasma samples in a hospital laboratory in France. | ||
| Target condition and reference standard(s) | High viral load > 40 copies/mL; Abbott RealTime HIV‐1 assay. | ||
| Flow and timing | Fresh samples were stored at 2 °C to 8 °C and tested simultaneously by both techniques within 5 days of collection and separation, and frozen samples were tested simultaneously within the same freeze/thaw cycle. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Unclear | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Low risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Jani 2016.
| Study characteristics | |||
| Patient Sampling | Adult HIV‐positive patients at Polana Caniço Health Centre, Maputo, Mozambique, were invited to participate in the study. Only consenting patients were included in the study. In order to include patients with viral loads throughout all ranges (undetectable, detectable to 10,000 copies/mL, and greater than 10,000 copies/mL), patients were targeted for representation in those 3 ranges based on the following clinical information: on ART for longer than 6 months, on ART for between 4 weeks and 6 months, and on for ART less than 4 weeks, respectively. This clinic was selected based on its proximity to the HIV reference laboratory in Maputo and to facilitate study management and sample logistics. Eligibility criteria included age over 18 years, documented HIV infection, and receipt of ART. Exclusion criteria included any serious medical conditions that could disrupt the accuracy of normal laboratory testing and its interpretation; however, no participants met this criterion. There was no exclusion on grounds of gender, socioeconomic status, race, or ethnicity. | ||
| Patient characteristics and setting | Adult HIV‐positive patients on ART; peri‐urban health centre in Maputo, Mozambique. | ||
| Index tests | Alere Q NAT POC viral load technology (Alere Technologies, Jena, Germany) on fresh whole blood samples (capillary) at point of care. | ||
| Target condition and reference standard(s) | HIV viral load total HIV RNA (HIV‐1/2 RNA); > 1000 copies/mL and > 5000 copies/mL; Roche COBAS AmpliPrep/COBAS TaqMan v2. | ||
| Flow and timing | Viral load testing was performed within 1 week of sample collection. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Low risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Jordan 2016.
| Study characteristics | |||
| Patient Sampling | Participants were enrolled from 4 participating sites, 2 in Europe and 2 in the USA. Fresh plasma samples were tested prospectively, whilst frozen plasma samples were collected prospectively, and tested retrospectively after selection of specimens to cover the assay’s quantification range (40 copies/mL to 10,000,000 copies/mL). Eligibility criteria included a clinician ordered HIV‐1 VL test from a confirmed HIV‐1 positive adult (≥ 18 years) with a known antiviral treatment status. Exclusion criteria included previous enrolment in this study or improper specimen collection. | ||
| Patient characteristics and setting | HIV‐1 positive adults ≥ 18 years of age with known antiviral treatment status; samples from patients from Europe and the USA. | ||
| Index tests | Xpert HIV‐1 Viral Load assay done on fresh and frozen plasma samples in the laboratory. This was a multisite clinical evaluation, implying that Xpert was evaluated in an onsite lab in the 4 sites. | ||
| Target condition and reference standard(s) | High HIV‐1 RNA/viral load > 40 copies/mL and 200 copies/mL; Abbott RealTime HIV‐1 assay done in a reference laboratory. | ||
| Flow and timing | Whole blood was held at 15 °C to 35 °C for up to 6 h or at 2 °C to 8 °C for up to 72 h prior to separating plasma for testing. After centrifugation, fresh plasma was held at 15 °C to 35 °C for up to 8 h or at 2 °C to 8 °C for up to 6 days prior to testing. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Low risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Khan 2020.
| Study characteristics | |||
| Patient Sampling | Routine plasma viral load (VL) and CD4 samples from public sector antiretroviral clinics across the Western Cape Province were used in this evaluation. In order to ensure that most of the VL analytical spectrum was covered, a convenient sampling strategy randomly selected paired CD4/VL EDTA samples where at least 50 samples of plasma VL results were from each of 4 categories: target not detected, < 40 to 1000 copies/mL, 1000 to 10,000 copies/mL, and > 10,000 copies/mL. | ||
| Patient characteristics and setting | Unclear; routine plasma VL and CD4 samples from public sector antiretroviral clinics across the Western Cape Province were used in this evaluation; laboratory‐based cross‐sectional study. | ||
| Index tests | Alere q (Alere Technologies, Jena, Germany) was performed using a prototype cartridge (prototype assay) at Groote Schuur virology laboratory in Cape Town. | ||
| Target condition and reference standard(s) | Abbott RealTime HIV‐1 assay (Abbott Laboratories, Chicago, USA). | ||
| Flow and timing | Once plasma VL and CD4 testing were complete, the remainder of the blood sample from CD4 testing was used for whole blood Alere q test and dried blood spots (DBS) within 72 hours of sample receipt to prevent sample degradation. | ||
| Comparative | |||
| Notes | Final commercially available version known as the m‐PIMA HIV‐1/2 VL test. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Kufa 2020.
| Study characteristics | |||
| Patient Sampling | Prospective study of pregnant or early postpartum women living with HIV (WLHIV) admitted to labour or postdelivery wards and their newborn infants until return of results or discharge, 4 high‐volume tertiary obstetric units (TOUs) in Gauteng, South Africa. All WLHIV admitted to labour or postnatal wards at the 4 TOUs during the study period were offered POC VL or birth polymerase chain reaction (PCR) testing, or both, by routine staff. Specimens were collected by doctors and nurses as part of their routine duties. POC testing was conducted by a POC operator working in a designated POC testing room. | ||
| Patient characteristics and setting | Pregnant or early postpartum WLHIV admitted to labour or postdelivery wards > 95% on ART from 4 high‐volume TOUs in Gauteng, South Africa. | ||
| Index tests | Cepheid’s Xpert HIV‐1 quantitative in the field. | ||
| Target condition and reference standard(s) | High viral load > 1000 copies/mL, central laboratory testing. | ||
| Flow and timing | For both WLHIV and infants, 2 specimens were collected ‐ 1 for POC and the other for central laboratory testing. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Unclear | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Low risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Unclear | ||
| Were all patients included in the analysis? | No | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Kulkarni 2017.
| Study characteristics | |||
| Patient Sampling | Study was conducted from June to September 2015 in HIV‐1 positive adults. 314 HIV‐1 seropositive with ART status as follows: (ART naive n = 151, on ART n = 129, suspected ART failure n = 34; individuals were screened to obtain varying viral load ranges) and 20 normal, healthy, HIV seronegative individuals were enrolled at 3 ART centres located in Pune (ART centres: Model Colony; YCM Hospital; Sassoon General Hospital). | ||
| Patient characteristics and setting | HIV‐positive adults (ART naive n = 151, on ART n = 129, suspected ART failure n = 34) (51.9% on ART) from ART centres in health centres in India. | ||
| Index tests | GeneXpert HIV‐1 Quant assay (a point‐of‐care technology) on frozen plasma samples in the laboratory (stored at −70 °C until tested). Testing seems to be at decentralized sites included in the study. (The national ART programme in India was launched in April 2004 in a limited number of hospitals.) | ||
| Target condition and reference standard(s) | HIV viral load at > 5000, > 200, > 40; Abbott HIV‐1 m2000 RealTime PCR | ||
| Flow and timing | The whole blood specimens were collected in 10‐millilitre EDTA Vacutainers (Becton Dickinson, USA), transported to National AIDS Research Institute (NARI), centrifuged at 405 g for 10 min, plasma was separated within 6 h, aliquoted, and stored at −70 °C until tested. Samples collected at same time but tested at different times. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Mor 2015.
| Study characteristics | |||
| Patient Sampling | Study included samples collected between 2013 and 2014 from patients (confirmed HIV‐1 carriers) attending the AIDS clinic of the Sheba Medical Center. Plasma from whole blood samples collected in EDTA‐containing tubes was separated by centrifugation (1100 g for 5 min). Plasma samples (n = 404) spanning the full range of HIV‐1 viral loads were selected based on the NUCLISENS v2.0 results. | ||
| Patient characteristics and setting | Samples from HIV‐1 positive patients (some on ART, others not); Sheba clinic in Israel | ||
| Index tests | The Xpert HIV‐1 Viral Load assay on the GeneXpert platform (Cepheid Inc); frozen plasma samples in a laboratory | ||
| Target condition and reference standard(s) | High HIV viral load results > 40 copies/mL, Abbott RealTime HIV‐1 assay | ||
| Flow and timing | Plasma from whole blood samples collected in EDTA‐containing tubes was separated by centrifugation (1100 g for 5 min). An aliquot (0.5 mL) was initially tested with the NUCLISENS v2.0 assay as part of the regular monitoring of HIV‐1 copy numbers. Separate aliquots of the plasma samples were stored frozen at 20 °C in volumes required for the different assays, with a single freeze‐thaw cycle prior to analysis on the different platforms. On the day of analysis, the aliquots were thawed. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Could the patient flow have introduced bias? | Low risk | ||
Mtapuri‐Zinyowera 2016.
| Study characteristics | |||
| Patient Sampling | Unclear; paired EDTA anticoagulated venous whole blood samples were collected from 375 patients aged ≥ 18 years, on ART for ≥ 3 months, in Zimbabwe. | ||
| Patient characteristics and setting | Adult patients aged ≥ 18 years, on ART for ≥ 3 months, Zimbabwe. | ||
| Index tests | GeneXpert HIV‐1 Quant (Xpert); laboratory evaluation, the National Microbiology Reference Laboratory (NMRL) in Harare, Zimbabwe. | ||
| Target condition and reference standard(s) | High viral load > 1000 copies/mL; bioMérieux NUCLISENS easyMAG/EASYQ v2.0. | ||
| Flow and timing | Paired plasma samples were tested for HIV‐1 viral load on NUCLISENS and Xpert following the manufacturers’ instructions and laboratory standard operating procedures. | ||
| Comparative | |||
| Notes | Conference abstract with 2 x 2 table data. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Ritchie 2014a.
| Study characteristics | |||
| Patient Sampling | Plasma samples from 134 HIV‐1 infected individuals attending The Royal London Hospital (34 patients) or St Thomas’ Hospital (100 patients) in London, UK, were rendered anonymous and provided blinded. | ||
| Patient characteristics and setting | Plasma samples from 134 HIV‐1 infected individuals attending The Royal London Hospital (34 patients) or St Thomas’ Hospital (100 patients) in London, UK (ART status unspecified). | ||
| Index tests | SAMBA I on frozen plasma samples in the laboratory. | ||
| Target condition and reference standard(s) | High viral load > 1000 copies/mL; Roche COBAS AmpliPrep/COBAS TaqMan HIV‐1 test. | ||
| Flow and timing | The plasma samples were stored at −80 °C until tested in‐house with the SAMBA Semi‐Q. Roche TaqMan v2 results and HIV‐1 subtype information (if available) were provided by both hospitals after the SAMBA Semi‐Q testing was completed. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Unclear | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Low risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Ritchie 2014b.
| Study characteristics | |||
| Patient Sampling | A total of 200 samples collected in Chiradzulu, Malawi, from 72 men and 128 women; patients ranged in age from 18 to 61 years. 4 patients were assigned an ID but withdrew from the study with no sample having been collected. | ||
| Patient characteristics and setting | HIV‐positive adults (70 patients (19.8%) were ART naive, and 284 patients (80.2%) had been on ART for a period of 1 month to 10 years at the time of testing) in Malawi. | ||
| Index tests | SAMBA HIV‐1 Semi‐Q Test on fresh plasma samples in the field setting | ||
| Target condition and reference standard(s) | High HIV viral load > 1000 copies/mL; Roche CAP/CTM v2.0 assay and Abbott RealTime; discordant results with Roche were retested with Abbott RealTime | ||
| Flow and timing | The 12 discordant samples (from Malawi and Uganda) were retested with Abbott RealTime at 1 of 2 independent laboratories and in a blinded manner with regard to the SAMBA Semi‐Q and Roche TaqMan v2 results. The Abbott RealTime results were concordant with the Roche TaqMan v2 results for 10 of the 12 samples. This is unlikely to have introduced bias in study estimates, as results were similar. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | No | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | No | ||
| Were all patients included in the analysis? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Ritchie 2014c.
| Study characteristics | |||
| Patient Sampling | 154 samples collected in Arua, Uganda, from 68 men and 86 women; patient age range was from 18 to 71 years. | ||
| Patient characteristics and setting | HIV‐positive adults; 80% on ART in Uganda. | ||
| Index tests | SAMBA HIV‐1 Semi‐Q Test done at point of care on fresh plasma samples. | ||
| Target condition and reference standard(s) | High HIV viral load > 1000 copies/mL; Roche CAP/CTM v2.0 assay and Abbott RealTime; discordant results with Roche were retested with Abbott RealTime. | ||
| Flow and timing | Discrepant results were checked by 2nd reference standard. The 12 discordant samples (from Malawi and Uganda) were retested with Abbott RealTime at 1 of 2 independent laboratories and in a blinded manner with regard to the SAMBA Semi‐Q and Roche TaqMan v2 results. The Abbott RealTime results were concordant with the Roche TaqMan v2 results for 10 of the 12 samples. This is unlikely to have introduced bias in study estimates, as results were similar. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | No | ||
| Were all patients included in the analysis? | Yes | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Rubio‐Garrido 2019.
| Study characteristics | |||
| Patient Sampling | Samples collected in the Democratic Republic of the Congo (DRC) tested in Spain. HIV+ DBS samples tested by viral load assays. From April to November 2016, 160 DBS were collected at Monkole Hospital (Kinshasa, DRC) from 85 children (60 HIV‐non‐infected, 18 HIV‐positive, 7 HIV‐exposed) and 75 HIV‐infected adults (65 treated with clinical suspicion of treatment failure, 10 naive). | ||
| Patient characteristics and setting | 84 (14 children and 70 adults) on ART (n = 69), ART naive (n = 10), ART unknown (n = 5); samples collected from the Democratic Republic of the Congo and tested in a laboratory in Spain. | ||
| Index tests | Xpert HIV‐1 Viral Load (Cepheid) on frozen whole blood DBS samples tested in a lab in Spain. Assays based on real‐time PCR, providing an assay‐specific cycle threshold (Ct), which inversely correlates with the starting concentration of the viral genome in the infected specimen. Ct values were recorded following DBS VL quantification by both Xpert VL and Roche VL platforms using 1 DBS dot in each sample. | ||
| Target condition and reference standard(s) | High HIV viral load > 1000 copies/mL and > 40 copies/mL; Roche COBAS AmpliPrep/COBAS TaqMan HIV‐1 Test v2.0. | ||
| Flow and timing | From April to November 2016, 160 DBS were collected at Monkole Hospital (Kinshasa, Democratic Republic of Congo). They were dried separately on a drying‐rack overnight at room temperature in Monkole Hospital, sealed in a zip‐lock plastic bag with desiccant bags, and stored at −20 °C until transported in dry ice to the laboratories in Madrid and Pamplona, Spain, where children and adult samples, respectively, were stored at −80 °C until further use. HIV diagnosis was firstly performed in DRC using rapid serological tests; in Navarra, Spain, HIV serostatus was confirmed in all adults by two 4th‐generation immunoassays: Elecsys HIV combi PT (Roche) and VIDAS HIV Duo Quick (bioMérieux). HIV‐1 viraemia was quantified using Cepheid Xpert HIV‐1 Viral Load (Xpert VL) 35 and COBAS AmpliPrep/COBAS TaqMan HIV‐1 Test v2.0 (Roche VL) 36 in all HIV+ DBS, both techniques based on real‐time amplification of HIV genome. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Swathirajan 2017.
| Study characteristics | |||
| Patient Sampling | This evaluation study was conducted in specimens collected between July 2015 and June 2016 from HIV‐1 patients attending the Y. R. Gaitonde Centre for AIDS Research and Education (YRG CARE), a tertiary care centre for HIV‐infected individuals in Chennai, Southern India. A total of 103 specimens that were tested by Abbott RealTime PCR as part of patient care services and had remaining samples stored in the freezer at −75 ± 5 °C, were utilized for this validation anonymously without using patient identifiers. | ||
| Patient characteristics and setting | HIV‐1 patients from a tertiary care centre for HIV‐infected individuals in Chennai, Southern India. | ||
| Index tests | Xpert HIV‐1 Viral Load assay on frozen plasma specimens done in a laboratory. | ||
| Target condition and reference standard(s) | High viral load > 1000 copies/mL; Abbott RealTime PCR. | ||
| Flow and timing | Both assays were performed as per the manufacturers’ instructions. A total of 103 specimens that were tested by Abbott RealTime PCR as part of patient care services and had remaining samples stored in the freezer at −75 ± 5 °C, were utilized for this validation anonymously without using patient identifiers. It was ensured that all the specimens were subjected to a single freeze–thaw cycle prior to testing using the Xpert HIV‐1 Viral Load assay. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Unclear | ||
| DOMAIN 2: Index Test (All tests) | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | Unclear risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Abbreviations: ART: antiretroviral therapy; DBS: dried blood spots; EDTA: ethylenediaminetetraacetic acid; PCR: polymerase chain reaction; POC VL: point‐of‐care viral load; RNA: ribonucleic acid; SAMBA: simple amplification‐based assay; Semi‐Q: semi‐quantitative.
Characteristics of excluded studies [ordered by study ID]
| Study | Reason for exclusion |
|---|---|
| Abdissa 2014 | Ineligible index test: not POC VL (PCR laboratory assay) |
| Acharya 2014 | Ineligible index test: not POC VL (HIV‐1 real‐time PCR laboratory assay) |
| ACTRN12618001340224 | Ineligible study type: protocol |
| Afani 2005 | Ineligible index test: not POC VL |
| Agutu 2019 | Ineligible study type: systematic review |
| Aleku 2014 | Ineligible study type: review |
| Amendola 2020 | Ineligible index test: not POC VL (Aptima HIV‐1 real‐time PCR laboratory assay) |
| Anderson 2011 | Ineligible study type: review |
| Anyiwo 2014 | Ineligible study type: review |
| Audu 2015 | Ineligible index test: not POC VL (HIV rapid antibody tests) |
| Avidor 2017 | Sample‐level analysis: 383 samples from 283 patients |
| Avila 2000a | Ineligible index test: not POC VL (branched DNA signal amplification test (bDNA)) |
| Avila 2000b | Duplicate |
| Balachandra 2020 | Ineligible index test: not POC VL (HIV rapid antibody tests) |
| Barbara 2017 | Ineligible study type: review |
| Bastos 2016 | Ineligible study type: conference abstract with incomplete data |
| Bélec 2011 | Ineligible study type: perspective/personal view |
| Berry 2014 | Ineligible index test: not POC VL (IFAST, an RNA extraction test technique) |
| Borysiak 2016 | Ineligible study type: not accuracy study |
| Brook 2018 | Ineligible study type: editorial |
| Bruzzone 2017 | Ineligible study type: 2‐gated design with HIV‐negative controls |
| Chibwesha 2016a | Ineligible study type: protocol |
| Chibwesha 2016b | Duplicate |
| Cogswell 2016 | Ineligible study type: editorial |
| Craik 2016 | Ineligible index test: not POC VL (PCR laboratory assay) |
| Damhorst 2013 | Ineligible study type: review |
| Damond 2001 | Ineligible index test: not POC VL (light cycler real‐time PCR laboratory assay) |
| Désiré 2001 | Ineligible index test: not POC VL (TaqMan real‐time PCR laboratory assay) |
| Dorward 2017 | Ineligible study type: protocol |
| Dorward 2018 | Ineligible study type: perspective/personal view |
| Drain 2017 | Ineligible study type: review |
| Drain 2019 | Ineligible study type: review |
| Drain 2020 | Ineligible study type: not accuracy study |
| Duarte 2017 | Ineligible study type: review |
| Fidler 2017 | Ineligible study type: 2‐gated design with HIV‐negative controls |
| Ganesh 2021 | Ineligible study type: not accuracy study |
| Geretti 2009 | Ineligible study type: review |
| Gurrala 2016 | Ineligible study type: not accuracy study |
| Haleyur Giri Setty 2014 | Ineligible study type: review |
| Harries 2010 | Ineligible study type: perspective/personal view |
| Hopkins 2015 | Ineligible index test: not POC test (Aptima HIV‐1 real‐time PCR laboratory assay) |
| Ibrahim 2017 | Ineligible target condition: detection of HIV‐1 infection |
| ISRCTN12803987(a) | Ineligible study type: protocol |
| ISRCTN12803987(b) | Duplicate |
| Jangam 2013 | Ineligible study type: not accuracy study |
| Kabir 2020 | Ineligible study type: review |
| Kahn 2013 | Ineligible study type: editorial |
| Laursen 2012 | Ineligible study type: perspective/personal view |
| Lee 2010a | Ineligible study type: not accuracy study |
| Lee 2010b | Duplicate |
| Luliano 1995 | Ineligible index test: not POC VL |
| Mani 1999 | Ineligible study type: letter to the editor |
| Manoto 2018 | Ineligible study type: review |
| Mariani 2020 | Sample‐level analysis: 413 samples from 273 patients |
| Masuko 2016 | Ineligible study type: conference abstract with incomplete data |
| Millar 2020 | Ineligible study type: not accuracy study |
| Moyo 2016 | Ineligible population: exclusively ART‐naive population retrieved from a household survey |
| Moyo 2019 | Ineligible study type: conference abstract with incomplete data |
| Moyo 2020 | Ineligible study type: not accuracy study |
| Nacarapa 2019 | Ineligible study type: conference abstract with incomplete data |
| Nash 2017 | Inability to construct 2 x 2 table: correlation study results |
| Nash 2018 | Ineligible study type: systematic review |
| NCT00929604 | Protocol |
| NCT02461576 | Protocol |
| NCT03066128 | Protocol |
| NCT03066128b | Duplicate |
| NCT03187964 | Protocol |
| NCT03288246(a) | Protocol |
| NCT03288246(b) | Duplicate |
| NCT03533868 | Protocol |
| NCT03553693(a) | Protocol |
| NCT03553693(b) | Protocol |
| NCT04517825 | Ineligible study type: protocol |
| Ndlovu 2018 | Ineligible study design: not accuracy study |
| Newman 2020 | Ineligible study type: review |
| Nicholas 2019 | Íneligible study design: not accuracy study |
| Ondiek 2017 | Ineligible target condition |
| Peter 2017 | Ineligible study type: review |
| Phillips 2016 | Ineligible study type: not accuracy study |
| Ritchie 2016 | Ineligible target condition: detection of HIV‐1 subtype |
| Rossetti 2020 | Ineligible index test: not POC VL (Aptima HIV‐1 real‐time PCR laboratory assay) |
| Rowley 2014 | Ineligible study type: review |
| Sacks 2019 | Ineligible study type: systematic review |
| Schalasta 2016 | Ineligible index test: not POC VL (Aptima HIV‐1 real‐time PCR laboratory assay) |
| Schønning 2017 | Ineligible index test: not POC VL (Aptima HIV‐1 real‐time PCR laboratory assay) |
| Scott 2015 | Ineligible index test: Liat HIV Quant plasma (test discontinued, not applicable) |
| Solomon 2016 | Ineligible index test: not POC VL (HIV antibody rapid tests) |
| Tanriverdi 2010 | Ineligible study design: not accuracy study |
| Titchmarsh 2015 | Ineligible index test: accuracy of a method, leukodepletion of a small whole blood volume |
| Vasconcellos 2020 | Inability to construct 2 x 2 tables |
| Villa 2020 | Ineligible study type: not accuracy study |
| Whitlock 2020 | Ineligible target condition: detection of HIV‐1 infection |
Abbreviations: POC VL: point‐of‐care viral load; PCR: polymerase chain reaction; RNA: ribonucleic acid.
Differences between protocol and review
We did not limit inclusion of studies to those that exclusively included antiretroviral therapy (ART) populations because many studies reported mixed populations consisting of both ART‐experienced (range 55% to 80%) and ART‐naive participants. In other studies, the ART status of included participants was not reported. This is reflective of routine care settings where mixed populations of ART experienced, naive, and non‐adherent are present due to barriers in ART initiation and adherence. Given this, we modified the review title to reflect this population in health facilities: 'Point‐of‐care viral load tests to detect high HIV viral load in people living with HIV/AIDS attending health facilities'. The objectives also limit the population to people living with HIV (PLHIV) attending healthcare facilities. Considering that the World Health Organization recommends that all PLHIV be on ART regardless of immunological status, we assumed that a sizeable proportion of participants in the unclearly reported studies were on ART. In Rubio‐Garrido 2019, data were used from only one cohort, due to ambiguities in the 2 x 2 data from the second cohort in the published article. Lastly, we made corrections to the original scoring instructions of low and high risk of bias indicated in the protocol, changing low risk of bias from "if all questions were answered with ‘yes’" to also including "or at least three with yes and the other one with unclear", and high risk of bias from "if one question is answered no" to "if two or more of questions are answered with ‘no’".
Contributions of authors
EAO and EEO were involved in study selection, data extraction, and quality assessment of the included studies.
EAO and SM were involved in the statistical analysis and interpretation of review findings.
EAO wrote the first draft of the review manuscript.
All authors contributed to the revisions and finalization of the manuscript.
Sources of support
Internal sources
Liverpool School of Tropical Medicine, UK
External sources
-
World Health Organization (WHO), Switzerland
The WHO funded the preliminary findings of this review which were presented to the guideline development group meeting in Geneva in June 2015.
-
Foreign, Commonwealth & Development Office (FCDO), UK
Project number 300342‐104
-
UK MRC, UK
This project is funded in part under the UK MRC African Research Leaders award (MR/T008768/1).This award is jointly funded by the UK Medical Research Council (MRC) and the UK Foreign, Commonwealth & Development Office (FCDO) under the MRC/FCDO Concordat agreement and is also part of the EDCTP2 programme supported by the European Union.
Declarations of interest
We presented preliminary findings of this review to the WHO Guideline Meeting Group in Geneva in June 2015.
EAO has no known conflicts of interest.
EEO has no known conflicts of interest.
JD and SM received funding from the World Health Organization to complete the review and present it to the WHO Guideline Meeting Group in 2015.
New
References
References to studies included in this review
Bwana 2019a {published data only}
- Bwana P, Ageng'o J, Mwau M.Performance and usability of Cepheid GeneXpert HIV-1 qualitative and quantitative assay in Kenya. PLOS ONE 2019;14(3):e0213865. [DOI] [PMC free article] [PubMed] [Google Scholar]
Bwana 2019b {published data only}
- Bwana P, Ageng'o J, Danda J, Mbugua J, Handa A, Mwau M.Performance and usability of mPIMATM HIV 1/2 viral load test in point of care settings in Kenya. Journal of Clinical Virology 2019;121:104202. [DOI] [PubMed] [Google Scholar]
Ceffa 2016 {published data only}
- Ceffa S, Luhanga R, Andreotti M, Brambilla D, Erba F, Jere H, et al.Comparison of the Cepheid GeneXpert and Abbott M2000 HIV-1 real time molecular assays for monitoring HIV-1 viral load and detecting HIV-1 infection. Journal of Virological Methods 2016;229:35-9. [DOI] [PubMed] [Google Scholar]
Garrett 2016 {published data only}
- Garrett NJ, Drain PK, Werner L, Samsunder N, Abdool Karim SS.Diagnostic accuracy of the point-of-care Xpert HIV-1 viral load assay in a South African HIV clinic. Journal of Acquired Immune Deficiency Syndromes 2016;72(2):e45-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Goel 2017a {published data only}
- Goel N, Ritchie AV, Mtapuri-Zinyowera S, Zeh C, Stepchenkova T, Lehga J, et al.Performance of the SAMBA I and II HIV-1 Semi-Q Tests for viral load monitoring at the point-of-care (UK). Journal of Virological Methods 2017;244:39-45. [DOI] [PubMed] [Google Scholar]
Goel 2017b {published data only}
- Goel N, Ritchie AV, Mtapuri-Zinyowera S, Zeh C, Stepchenkova T, Lehga J, et al.Performance of the SAMBA I and II HIV-1 Semi-Q Tests for viral load monitoring at the point-of-care (Kenya). Journal of Virological Methods 2017;244:39-45. [DOI] [PubMed] [Google Scholar]
Goel 2017c {published data only}
- Goel N, Ritchie AV, Mtapuri-Zinyowera S, Zeh C, Stepchenkova T, Lehga J, et al.Performance of the SAMBA I and II HIV-1 Semi-Q Tests for viral load monitoring at the point-of-care (Zimbabwe). Journal of Virological Methods 2017;244:39-45. [DOI] [PubMed] [Google Scholar]
Goel 2017d {published data only}
- Goel N, Ritchie AV, Mtapuri-Zinyowera S, Zeh C, Stepchenkova T, Lehga J, et al.Performance of the SAMBA I and II HIV-1 Semi-Q Tests for viral load monitoring at the point-of-care (Ukraine). Journal of Virological Methods 2017;244:39-45. [DOI] [PubMed] [Google Scholar]
Gous 2016 {published data only}
- Gous N, Scott L, Berrie L, Stevens W.Options to expand HIV viral load testing in South Africa: evaluation of the GeneXpert® HIV-1 viral load assay. PLOS ONE 2016;11(12):e0168244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Gueguen 2021a {published data only}
- Gueguen M, Nicholas S, Poulet E, Schramm B, Szumilin E, Wolters L, et al.Implementation and operational feasibility of SAMBA I HIV-1 semi-quantitative viral load testing at the point-of-care in rural settings in Malawi and Uganda. Tropical Medicine & International Health 2021;26(2):184-94. [DOI] [PubMed] [Google Scholar]
Gueguen 2021b {published data only}
- Gueguen M, Nicholas S, Poulet E, Schramm B, Szumilin E, Wolters L, et al.Implementation and operational feasibility of SAMBA I HIV-1 semi-quantitative viral load testing at the point of care in rural settings in Malawi and Uganda. Tropical Medicine & International Health 2021;26(2):184-94. [DOI] [PubMed] [Google Scholar]
Gueudin 2016 {published data only}
- Gueudin M, Baron A, Alessandri-Gradt E, Lemée V, Mourez T, Etienne M, et al.Performance evaluation of the new HIV-1 quantification assay, Xpert HIV-1 viral load, on a wide panel of HIV-1 variants. Journal of Acquired Immune Deficiency Syndromes 2016;72(5):521-6. [DOI] [PubMed] [Google Scholar]
Jani 2016 {published data only}
- Jani IV, Meggi B, Vubil A, Sitoe NE, Bhatt N, Tobaiwa O, et al.Evaluation of the whole-blood Alere Q NAT point-of-care RNA assay for HIV-1 viral load monitoring in a primary health care setting in Mozambique. Journal of Clinical Microbiology 2016;54(8):2104-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Jordan 2016 {published data only}
- Jordan JA, Plantier JC, Templeton K, Wu AH.Multi-site clinical evaluation of the Xpert HIV-1 viral load assay. Journal of Clinical Virology 2016;80:27-32. [DOI] [PubMed] [Google Scholar]
Khan 2020 {published data only}
- Khan A, Hans L, Hsiao NY.Comparison of Alere q whole blood viral load with DBS and plasma viral load in the classification of HIV virological failure. PLOS ONE 2020;15(5):e0232345. [DOI] [PMC free article] [PubMed] [Google Scholar]
Kufa 2020 {published data only}
- Kufa T, Mazanderani AH, Sherman GG, Mukendi A, Murray T, Moyo F, et al.Point-of-care HIV maternal viral load and early infant diagnosis testing around time of delivery at tertiary obstetric units in South Africa: a prospective study of coverage, results return and turn-around times. Journal of the International AIDS Society 2020;23(4):e25487. [DOI] [PMC free article] [PubMed] [Google Scholar]
Kulkarni 2017 {published data only}
- Kulkarni S, Jadhav S, Khopkar P, Sane S, Londhe R, Chimanpure V, et al.GeneXpert HIV-1 quant assay, a new tool for scale up of viral load monitoring in the success of ART programme in India. BMC Infectious Diseases 2017;17(1):506. [DOI] [PMC free article] [PubMed] [Google Scholar]
Mor 2015 {published data only}
- Mor O, Gozlan Y, Wax M, Mileguir F, Rakovsky A, Noy B, et al.Evaluation of the real time HIV-1, Xpert HIV-1, and Aptima HIV-1 quant dx assays in comparison to the NucliSens EasyQ HIV-1 v2.0 assay for quantification of HIV-1 viral load. Journal of Clinical Microbiology 2015;53(11):3458-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
Mtapuri‐Zinyowera 2016 {published data only}
- Mtapuri-Zinyowera S, Ndlovu Z, Farjardo E, Metcalf C, Kao K, Rumaney M, et al. In:Laboratory evaluation of the GeneXpert HIV-1 Quant viral load assay in Zimbabwe. 23rd Conference on Retroviruses & Opportunistic Infections (CROI); 2016 Feb 22-25; Boston (MA). Boston (MA): Conference on Retroviruses & Opportunistic Infections (CROI), 2016. [Abstract 16-1111] [www.croiconference.org/abstract/laboratory-evaluation-genexpert-hiv-1-viral-load-assay-zimbabwe/]
Ritchie 2014a {published data only}
- Ritchie AV, Ushiro-Lumb I, Edemaga D, Joshi HA, De Ruiter A, Szumilin E, et al.SAMBA HIV semiquantitative test, a new point-of-care viral-load-monitoring assay for resource-limited settings (UK). Journal of Clinical Microbiology 2014;52(9):3377-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
Ritchie 2014b {published data only}
- Ritchie AV, Ushiro-Lumb I, Edemaga D, Joshi HA, De Ruiter A, Szumilin E, et al.SAMBA HIV semiquantitative test, a new point-of-care viral-load-monitoring assay for resource-limited settings (Malawi). Journal of Clinical Microbiology 2014;52(9):3377-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
Ritchie 2014c {published data only}
- Ritchie AV, Ushiro-Lumb I, Edemaga D, Joshi HA, De Ruiter A, Szumilin E, et al.SAMBA HIV semiquantitative test, a new point-of-care viral-load-monitoring assay for resource-limited settings (Uganda). Journal of Clinical Microbiology 2014;52(9):3377-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
Rubio‐Garrido 2019 {published data only}
- Rubio-Garrido M, Ndarabu A, Reina G, Barquin D, Fernandez-Alonso M, Carlos S, et al.Utility of POC Xpert HIV-1 tests for detection-quantification of complex HIV recombinants using dried blood spots from Kinshasa, D. R. Congo. Scientific Reports 2019;9:5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
Swathirajan 2017 {published data only}
- Swathirajan CR, Vignesh R, Boobalan J, Solomon SS, Saravanan S, Balakrishnan P.Performance of point-of-care Xpert HIV-1 plasma viral load assay at a tertiary HIV care centre in Southern India. Journal of Medical Microbiology 2017;66(10):1379-82. [DOI] [PubMed] [Google Scholar]
References to studies excluded from this review
Abdissa 2014 {published data only}
- Abdissa A, Yilma D, Fonager J, Audelin AM, Christensen LH, Olsen MF, et al.Drug resistance in HIV patients with virological failure or slow virological response to antiretroviral therapy in Ethiopia. BMC Infectious Diseases 2014;14:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
Acharya 2014 {published data only}
- Acharya A, Vaniawala S, Shah P, Parekh H, Misra RN, Wani M, et al.A robust HIV-1 viral load detection assay optimized for Indian sub type C specific strains and resource limiting setting. Biological Research 2014;47(1):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
ACTRN12618001340224 {published data only}
- ACTRN12618001340224.A study to examine the tolerability and antiviral activity of switching to Biktarvy tablets taken once daily compared to baseline over 48 weeks in HIV-1 infected antiretroviral therapy (ART) experienced participants aged 55 years and older who are virologically suppressed on a current antiretroviral regimen (CAR). anzctr.org.au/Trial/Registration/TrialReview.aspx?id=375648 (first received 6 August 2018).
Afani 2005 {published data only}
- Afani SA, Ayala CM, Meyer KA, Cabrera CR, Acevedo MW.Resistencia primaria a terapia antirretroviral en pacientes con infección por VIH/SIDA en Chile. Revista Médica de Chile 2005;133(3):295-301. [DOI] [PubMed] [Google Scholar]
Agutu 2019 {published data only}
- Agutu CA, Ngetsa CJ, Price MA, Rinke de Wit TF, Omosa-Manyonyi G, Sanders EJ, et al.Systematic review of the performance and clinical utility of point of care HIV-1 RNA testing for diagnosis and care. PLOS ONE 2019;14(6):e0218369. [DOI] [PMC free article] [PubMed] [Google Scholar]
Aleku 2014 {published data only}
- Aleku GA, Adoga MP, Agwale SM.HIV point-of-care diagnostics: meeting the special needs of sub-Saharan Africa. Journal of Infection in Developing Countries 2014;8(10):1231-43. [DOI] [PubMed] [Google Scholar]
Amendola 2020 {published data only}
- Amendola A, Sberna G, Forbici F, Abbate I, Lorenzini P, Pinnetti C, et al.The dual-target approach in viral HIV-1 viremia testing: an added value to virological monitoring? PLOS ONE 2020;15(2):e0228192. [DOI] [PMC free article] [PubMed] [Google Scholar]
Anderson 2011 {published data only}
- Anderson DA, Crowe SM, Garcia M.Point-of-care testing. Current HIV/AIDS Reports 2011;8(1):31-7. [DOI] [PubMed] [Google Scholar]
Anyiwo 2014 {published data only}
- Anyiwo CE.“Silent” carriers of HIV and the epidemiology of AIDS: a review. Pacific Journal of Medical Sciences 2014;12(2):52-65. [Google Scholar]
Audu 2015 {published data only}
- Audu RA, Okoye RN, Onwuamah CK, Ige FA, Musa AZ, Odunukwe NN, et al.Potential for false-positive HIV test results using rapid HIV testing algorithms. African Journal of Laboratory Medicine 2015;4(1):1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Avidor 2017 {published data only}
- Avidor B, Matus N, Girshengorn S, Achsanov S, Gielman S, Zeldis I, et al.Comparison between Roche and Xpert in HIV-1 RNA quantitation: a high concordance between the two techniques except for a CRF02_AG subtype variant with high viral load titters detected by Roche but undetected by Xpert. Journal of Clinical Virology 2017;93:15-9. [DOI] [PubMed] [Google Scholar]
Avila 2000a {published data only}
- Avila MM, Liberatore D, Martínez Peralta L, Biglione M, Libonatti O, Coll Cárdenas P, et al.Human immunodeficiency virus bDNA assay for pediatric cases. Revista Argentina de Microbiología 2000;32(1):33-8. [PubMed] [Google Scholar]
Avila 2000b {published data only}
- Avila MM, Liberatore D, Martínez Peralta L, Biglione M, Libonatti O, Coll Cárdenas P, et al.Human immunodeficiency virus bDNA assay for pediatric cases. Revista Argentina de Microbiología 2000;32(1):33-8. [PubMed] [Google Scholar]
Balachandra 2020 {published data only}
- Balachandra S, Rogers JH, Ruangtragool L, Radin E, Musuka G, Oboho I, et al.Concurrent advanced HIV disease and viral load suppression in a high-burden setting: findings from the 2015-6 ZIMPHIA survey. PLOS ONE 2020;15(6):e0230205. [DOI] [PMC free article] [PubMed] [Google Scholar]
Barbara 2017 {published data only}
- Barbara C, Reynolds SJ.Optimizing treatment monitoring in resource limited settings in the era of routine viral load monitoring. Current Tropical Medicine Reports 2017;4:1-5. [Google Scholar]
Bastos 2016 {published data only}
- Bastos P, Monteiro F, Tavares G, Amorim A, Ferreira M, Hortelao D, et al.One-step real-time PCR for HIV-2 group A and B RNA plasma viral load in lightcycler 2.0. Journal of the International AIDS Society 2016;19:2. [Google Scholar]
Bélec 2011 {published data only}
- Bélec L, Bonn JP.Challenges in implementing HIV laboratory monitoring in resource-constrained settings: how to do more with less. Future Microbiology 2011;6(11):1251-60. [DOI] [PubMed] [Google Scholar]
Berry 2014 {published data only}
- Berry SM, LaVanway AJ, Pezzi HM, Guckenberger DJ, Anderson MA, Loeb JM, et al.HIV viral RNA extraction in wax immiscible filtration assisted by surface tension (IFAST) devices. Journal of Molecular Diagnostics 2014;16(3):297-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
Borysiak 2016 {published data only}
- Borysiak MD, Bender AT, Boyle DS, Posner JD.Point-of-care HIV-1 diagnostic with integrated nucleic acid extraction and amplification from whole blood. Institute of Electrical and Electronics Engineers (IEEE) 2016;2016:224-7. [DOI: 10.1109/HIC.2016.7797737] [DOI] [Google Scholar]
Brook 2018 {published data only}
- Brook G.HIV viral load point-of-care testing: the what, the whys and the wherefores. Sexually Transmitted Infections 2018;94(6):394-5. [DOI] [PubMed] [Google Scholar]
Bruzzone 2017 {published data only}
- Bruzzone B, Caligiuri P, Nigro N, Arcuri C, Delucis S, Di Biagio A, et al.Xpert HIV-1 viral load assay and VERSANTHIV-1 RNA 1.5 assay: a performance comparison. Journal of Acquired Immune Deficiency Syndrome 2017;74(3):e86-8. [DOI] [PubMed] [Google Scholar]
Chibwesha 2016a {published data only}
- Chibwesha CJ, Ford CE, Mollan KR, Stringer JS.Point-of-care virologic testing to improve outcomes of HIV-infected children in Zambia: a clinical trial protocol. Journal of Acquired Immune Deficiency Syndromes 2016;72(Suppl 2):S197-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Chibwesha 2016b {published data only}
- Chibwesha CJ, Ford CE, Mollan KR, Stringer JSA.Point-of-care virologic testing to improve outcomes of HIV-infected children in Zambia: a clinical trial protocol. Journal of Acquired Immune Deficiency Syndromes 2016;72(Suppl 2):S197-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Cogswell 2016 {published data only}
- Cogswell HA, Ohadi E, Avila C.Viral-load point-of-care technologies to achieve an AIDS-free generation. Future Microbiology 2016;11(1):5-9. [DOI] [PubMed] [Google Scholar]
Craik 2016 {published data only}
- Craik A, Patel P, Patel P, Mallewa J, Malisita K, Bitilinyu-Bangoh J, et al.Challenges with targeted viral load testing for medical inpatients at Queen Elizabeth central hospital in Blantyre, Malawi. Malawi Medical Journal 2016;28(4):179-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
Damhorst 2013 {published data only}
- Damhorst GL, Watkins NN, Bashir R.Micro- and nanotechnology for HIV/AIDS diagnostics in resource-limited settings. IEEE Transactions on Biomedical Engineering 2013;60(3):715-26. [DOI] [PubMed] [Google Scholar]
Damond 2001 {published data only}
- Damond F, Descamps D, Farfara I, Telles JN, Puyeo S, Campa P, et al.Quantification of proviral load of human immunodeficiency virus type 2 subtypes A and B using real-time PCR. Journal of Clinical Microbiology 2001;39(12):4264-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Désiré 2001 {published data only}
- Désiré N, Dehée A, Schneider V, Jacomet C, Goujon C, Girard PM, et al.Quantification of human immunodeficiency virus type 1 proviral load by a TaqMan real-time PCR assay. Journal of Clinical Microbiology 2001;39(4):1303-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Dorward 2017 {published data only}
- Dorward J, Garrett N, Quame-Amaglo J, Samsunder N, Ngobese H, Ngomane N, et al.Protocol for a randomised controlled implementation trial of point-of-care viral load testing and task shifting: the Simplifying HIV TREAtment and Monitoring (STREAM) study. BMJ Open 2017;7(9):e017507. [DOI] [PMC free article] [PubMed] [Google Scholar]
Dorward 2018 {published data only}
- Dorward J, Drain PK, Garrett N.Point-of-care viral load testing and differentiated HIV care. Lancet HIV 2018;5(1):e8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Drain 2017 {published data only}
- Drain PK, Rousseau C.Point-of-care diagnostics: extending the laboratory network to reach the last mile. Current Opinion in HIV and AIDS 2017;12(2):175-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
Drain 2019 {published data only}
- Drain PK, Dorward J, Bender A, Lillis L, Marinucci F, Sacks J, et al.Point-of-care HIV viral load testing: an essential tool for a sustainable global HIV/AIDS response. Clinical Microbiology Reviews 2019;32(3):e00097-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Drain 2020 {published data only}
- Drain PK, Dorward J, Violette LR, Quame-Amaglo J, Thomas KK, Samsunder N, et al.Point-of-care HIV viral load testing combined with task shifting to improve treatment outcomes (STREAM): findings from an open-label, non-inferiority, randomised controlled trial. Lancet HIV 2020;7(4):e229-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Duarte 2017 {published data only}
- Duarte HA, Panpradist N, Beck IA, Lutz B, Lai J, Kanthula RM, et al.Current status of point-of-care testing for human immunodeficiency virus drug resistance. Journal of Infectious Diseases 2017;216(Suppl 9):S824-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Fidler 2017 {published data only}
- Fidler S, Lewis H, Meyerowitz J, Kuldanek K, Thornhill J, Muir D, et al.A pilot evaluation of whole blood finger-prick sampling for point-of-care HIV viral load measurement: the UNICORN study. Scientific Reports 2017;7(1):13658. [DOI] [PMC free article] [PubMed] [Google Scholar]
Ganesh 2021 {published data only}
- Ganesh P, Heller T, Chione B, Gumulira J, Gugsa S, Khan S, et al.Near point of care HIV viral load: targeted testing at large facilities. Journal of Acquired Immune Deficiency Syndromes 2021;86(2):258-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
Geretti 2009 {published data only}
- Geretti AM.HIV testing and monitoring. Medicine 2009;37(7):326-9. [Google Scholar]
Gurrala 2016 {published data only}
- Gurrala R, Lang Z, Shepherd L, Davidson D, Harrison E, McClure M, et al.Novel pH sensing semiconductor for point-of-care detection of HIV-1 viremia. Scientific Reports 2016;6:36000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Haleyur Giri Setty 2014 {published data only}
- Haleyur Giri Setty MK, Hewlett IK.Point of care technologies for HIV. AIDS Research and Treatment 2014;2014:497046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Harries 2010 {published data only}
- Harries AD, Zachariah R, Oosterhout JJ, Reid SD, Hosseinipour MC, Arendt V, et al.Diagnosis and management of antiretroviral-therapy failure in resource-limited settings in sub-Saharan Africa: challenges and perspectives. Lancet Infectious Diseases 2010;10(1):60-5. [DOI] [PubMed] [Google Scholar]
Hopkins 2015 {published data only}
- Hopkins M, Hau S, Tiernan C, Papadimitropoulos A, Chawla A, Beloukas A, et al.Comparative performance of the new Aptima HIV-1 Quant Dx assay with three commercial PCR-based HIV-1 RNA quantitation assays. Journal of Clinical Virology 2015;69:56–62. [DOI] [PubMed] [Google Scholar]
Ibrahim 2017 {published data only}
- Ibrahim M, Moyo S, Mohammed T, Mupfumi L, Gaseitsiwe S, Maswabi K, et al.Brief report: high sensitivity and specificity of the Cepheid Xpert HIV-1 qualitative point-of-care test among newborns in Botswana. Journal of Acquired Immune Deficiency Syndromes 2017;75(5):e128-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
ISRCTN12803987(a) {published data only}
- ISRCTN12803987.Performance evaluation of a point-of-care whole blood viral load test (SAMBA II HIV-1 Semi-Q Whole Blood) to optimise HIV treatment. isrctn.com/ISRCTN12803987 (first received 31 August 2016).
ISRCTN12803987(b) {published data only}
- ISRCTN12803987.Performance evaluation of a point-of-care whole blood viral load test (SAMBA II HIV-1 Semi-Q Whole Blood) to optimise HIV treatment. isrctn.com/ISRCTN12803987 (first received 31 August 2016).
Jangam 2013 {published data only}
- Jangam SR, Agarwal AK, Sur K, Kelso DM.A point-of-care PCR test for HIV-1 detection in resource-limited settings. Biosensors and Bioelectronics 2013;42:69-75. [DOI] [PubMed] [Google Scholar]
Kabir 2020 {published data only}
- Kabir MA, Zilouchian H, Caputi M, Asghar W.Advances in HIV diagnosis and monitoring. Critical Reviews in Biotechnology 2020;40(5):623-38. [DOI] [PubMed] [Google Scholar]
Kahn 2013 {published data only}
- Kahn JG, Marseille EA.Viral load monitoring for antiretroviral therapy in resource-poor settings: an evolving role. AIDS 2013;27(9):1509-11. [DOI] [PubMed] [Google Scholar]
Laursen 2012 {published data only}
- Laursen L.Point-of-care tests poised to alter course of HIV treatment. Nature Medicine 2012;18:1156. [DOI] [PubMed] [Google Scholar]
Lee 2010a {published data only}
- Lee HH, Dineva MA, Chua YL, Ritchie AV, Ushiro-Lumb I, Wisniewski CA.Simple amplification-based assay: a nucleic acid based point-of-care platform for HIV-1 testing. Journal of Infectious Diseases 2010;201(Suppl 1):S65-72. [DOI] [PubMed] [Google Scholar]
Lee 2010b {published data only}
- Lee HH, Dineva MA, Chua YL, Ritchie AV, Ushiro-Lumb I, Wisniewski CA.Simple amplification-based assay: a nucleic acid-based point-of-care platform for HIV-1 testing. Journal of Infectious Diseases 2010;201(Suppl 1):S65-72. [DOI] [PubMed] [Google Scholar]
Luliano 1995 {published data only}
- Luliano R, Forastieri G, Brizzi M, Mazzotta F, Ceccherininelli L.HIV-1 plasma viral load detection by branched DNA signal amplification. Microbiologica 1995;18(3):299-301. [PubMed] [Google Scholar]
Mani 1999 {published data only}
- Mani I, Cao H, Hom D, Johnson JL, Mugerwa RD, Travers KU, et al.Plasma RNA viral load as measured by the branched DNA and nucleic acid sequence-based amplification assays of HIV-1 subtypes A and D in Uganda. Journal of Acquired Immune Deficiency Syndromes 1999;22(2):208-9. [DOI] [PubMed] [Google Scholar]
Manoto 2018 {published data only}
- Manoto SL, Lugongolo M, Govender U, Mthunzi-Kufa P.Point of care diagnostics for HIV in resource limited settings: an overview. Medicina (Kaunas) 2018;54(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Mariani 2020 {published data only}
- Mariani D, Azevedo MC, Vasconcellos I, Ribeiro L, Alves C, Ferreira OC Jr, et al.The performance of a new point-of-care HIV virus load technology to identify patients failing antiretroviral treatment. Journal of Clinical Virology 2020;122:104212. [DOI] [PubMed] [Google Scholar]
Masuko 2016 {published data only}
- Masuko S, Chigiga J, Hodson J, Dedicoat K, Killick P, Osman H, et al.Correlation between GeneXpert, a novel human immunodeficiency virus type 1 (HIV-1) assay for viral load measurement assay with the Abbott m2000 real-time assay. HIV Medicine 2016;17:28-9. [Google Scholar]
Millar 2020 {published data only}
- Millar JR, Bengu N, Fillis R, Sprenger K, Ntlantsana V, Vieira VA, et al.High-frequency failure of combination antiretroviral therapy in paediatric HIV infection is associated with unmet maternal needs causing maternal non-adherence. eClinicalMedicine 2020;22:100344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Moyo 2016 {published data only}
- Moyo S, Mohammed T, Wirth KE, Prague M, Bennett K, Holme MP, et al.Point-of-care Cepheid Xpert HIV-1 Viral Load test in rural African communities is feasible and reliable. Journal of Clinical Microbiology 2016;54(12):3050-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Moyo 2019 {published data only}
- Moyo F, Haeri MA, Murray T, Technau KG, Carmona S, Kufa T, et al.Characterizing viral load burden among HIV-infected women at time of delivery: findings from four tertiary obstetric units in Gauteng, South Africa. Journal of the International AIDS Society 2019;22(Suppl 5).
Moyo 2020 {published data only}
- Moyo F, Haeri Mazanderani A, Murray T, Technau KG, Carmona S, Kufa T, et al.Characterizing viral load burden among HIV-infected women around the time of delivery: findings from four tertiary obstetric units in Gauteng, South Africa. Journal of Acquired Immune Deficiency Syndromes 2020;83(4):390-6. [DOI] [PubMed] [Google Scholar]
Nacarapa 2019 {published data only}
- Nacarapa E, Osorio D, Paredes R.HIV viral suppression in a 16-year cohort (2002-2018), in a rural area, Chokwe, Mozambique. Transactions of the Royal Society of Tropical Medicine and Hygiene 2019;113(Suppl 1):S269-70. [Google Scholar]
Nash 2017 {published data only}
- Nash M, Ramapuram J, Kaiya R, Huddart S, Pai M, Baliga S.Use of the GeneXpert tuberculosis system for HIV viral load testing in India. Lancet 2017;5(8):e754-5. [DOI] [PubMed] [Google Scholar]
Nash 2018 {published data only}
- Nash M, Huddart S, Badar S, Baliga S, Saravu K, Pai M.Performance of the Xpert HIV-1 viral load assay: a systematic review and meta-analysis. Journal of Clinical Microbiology 2018;56(4):e01673-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
NCT00929604 {published data only}
- NCT00929604.HIV viral load monitoring in resource-poor settings [Effectiveness of HIV viral load monitoring of patient outcome in resource-poor settings]. clinicaltrials.gov/ct2/show/NCT00929604 (first received 29 June 2009).
NCT02461576 {published data only}
- NCT02461576.Clinical evaluation of the Xpert® HIV-1 VL [Clinical evaluation of the Xpert® HIV-1 VL: a method comparison study]. clinicaltrials.gov/ct2/show/NCT02461576 (first received 3 June 2015).
NCT03066128 {published data only}
- NCT03066128.Point-of-care viral load testing to enable streamlined care and task shifting for chronic HIV care (STREAM). clinicaltrials.gov/ct2/show/NCT03066128 (first received 28 February 2017).
NCT03066128b {published data only}
- NCT03066128.Point-of-care viral load testing to enable streamlined care and task shifting for chronic HIV care (STREAM). clinicaltrials.gov/ct2/show/NCT03066128 (first received 28 February 2017).
NCT03187964 {published data only}
- NCT03187964.Xpert Ultra and Xpert HIV-VL in people living with HIV [Feasibility, accuracy, and effect of polyvalent point-of-care Xpert MTB/RIF Ultra and Xpert HIV-1 viral load testing in HIV-positive patients initiating ART: a randomised controlled trial]. clinicaltrials.gov/ct2/show/NCT03187964 (first received 15 June 2017).
NCT03288246(a) {published data only}
- NCT03288246.Point-of-care viral load testing among HIV-infected adolescents in Haiti. clinicaltrials.gov/ct2/show/NCT03288246 (first received 20 September 2017).
NCT03288246(b) {published data only}
- NCT03288246.Point-of-care viral load testing among HIV-infected adolescents in Haiti. clinicaltrials.gov/ct2/show/NCT03288246 (first received 20 September 2017).
NCT03533868 {published data only}
- NCT03533868.Reaching 90% HIV suppression: the role of POC viral load monitoring in Nigeria. clinicaltrials.gov/ct2/show/NCT03533868 (first received 23 May 2018).
NCT03553693(a) {published data only}
- NCT03553693.Rapid HIV viral load monitoring in high risk patients in Uganda (RAPID-VL). clinicaltrials.gov/ct2/show/NCT03553693 (first received 12 June 2018).
NCT03553693(b) {published data only}
- NCT03553693.Rapid HIV viral load monitoring in high risk patients in Uganda. clinicaltrials.gov/ct2/show/NCT03553693 (first received 12 June 2018).
NCT04517825 {published data only}
- NCT04517825.More than a machine: make point-of-care HIV-1 viral load testing effective in rural Uganda [IGHID 11920 - more than a machine: exploring the ancillary systems and processes required to make point-of-care HIV-1 viral load testing effective in rural Western Uganda]. clinicaltrials.gov/ct2/show/NCT04517825 (first received 18 August 2020).
Ndlovu 2018 {published data only}
- Ndlovu Z, Fajardo E, Mbofana E, Maparo T, Garone D, Metcalf C, et al.Multidisease testing for HIV and TB using the GeneXpert platform: a feasibility study in rural Zimbabwe. PLOS ONE 2018;13(3):e0193577. [DOI] [PMC free article] [PubMed] [Google Scholar]
Newman 2020 {published data only}
- Newman H, Hardie D.HIV-1 viral load testing in resource-limited settings: challenges and solutions for specimen integrity. Reviews in Medical Virology 2020;31(2):e2165. [DOI] [PubMed] [Google Scholar]
Nicholas 2019 {published data only}
- Nicholas S, Poulet E, Wolters L, Wapling J, Rakesh A, Amoros I, et al.Point-of-care viral load monitoring: outcomes from a decentralized HIV programme in Malawi. Journal of the International AIDS Society 2019;22(8):e25387. [DOI] [PMC free article] [PubMed] [Google Scholar]
Ondiek 2017 {published data only}
- Ondiek J, Namukaya Z, Mtapuri-Zinyowera S, Balkan S, Elbireer A, Ushiro Lumb I, et al.Multicountry validation of SAMBA - a novel molecular point-of-care test for HIV-1 detection in resource-limited setting. Journal of Acquired Immune Deficiency Syndromes 2017;76(2):e52-7. [DOI] [PubMed] [Google Scholar]
Peter 2017 {published data only}
- Peter T, Ellenberger D, Kim AA, Boeras D, Messele T, Roberts T, et al.Early antiretroviral therapy initiation: access and equity of viral load testing for HIV treatment monitoring. Lancet Infectious Diseases 2017;17(1):e26-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Phillips 2016 {published data only}
- Phillips AN, Cambiano V, Nakagawa F, Ford D, Apollo T, Murungu J, et al.Point-of-care viral load testing for sub-Saharan Africa: informing a target product profile. Open Forum Infectious Diseases 2016;3(3):ofw161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Ritchie 2016 {published data only}
- Ritchie AV, Goel N, Sembongi H, Lehga J, Farleigh LE, Edemaga D, et al.Performance evaluation of the point-of-care SAMBA I and II HIV-1 Qual whole blood tests. Journal of Virological Methods 2016;237:143-9. [DOI] [PubMed] [Google Scholar]
Rossetti 2020 {published data only}
- Rossetti R, Smith T, Luo W, Taussig J, Valentine-Graves M, Sullivan P, et al.Performance evaluation of the Aptima HIV-1 RNA Quant assay on the Panther system using the standard and dilution protocols. Journal of Clinical Virology 2020;129:104479. [DOI] [PMC free article] [PubMed] [Google Scholar]
Rowley 2014 {published data only}
- Rowley CF.Developments in CD4 and viral load monitoring in resource-limited settings. Clinical Infectious Diseases 2014;58(3):407-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Sacks 2019 {published data only}
- Sacks JA, Fong Y, Gonzalez MP, Andreotti M, Baliga S, Garrett N, et al.Performance of Cepheid Xpert HIV-1 viral load plasma assay to accurately detect treatment failure. AIDS 2019;33(12):1881-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Schalasta 2016 {published data only}
- Schalasta G, Börner A, Speicher A, Enders M.Comparative evaluation of the Aptima HIV-1 Quant Dx assay and COBAS TaqMan HIV-1 v2.0 assay using the Roche High Pure System for the quantification of HIV-1 RNA in plasma. Clinical Chemistry and Laboratory Medicine 2016;54(3):493–9. [DOI] [PubMed] [Google Scholar]
Schønning 2017 {published data only}
- Schønning K, Johansen K, Landt B, Benfield T, Westh H.Comparison of the Hologic Aptima HIV-1 Quant Dx assay to the Roche COBAS AmpliPrep/COBAS TaqMan HIV-1 Test v2.0 for the quantification of HIV-1 RNA in plasma samples. Journal of Clinical Virology 2017;92:14-9. [DOI] [PubMed] [Google Scholar]
Scott 2015 {published data only}
- Scott L, Gous N, Carmona S, Stevens W.Laboratory evaluation of the Liat HIV Quant (IQuum) whole-blood and plasma HIV-1 viral load assays for point-of-care testing in South Africa. Journal of Clinical Microbiology 2015;53(5):1616-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Solomon 2016 {published data only}
- Solomon SS, Mehta SH, McFall AM, Srikrishnan AK, Saravanan S, Laeyendecker O, et al.Community viral load, antiretroviral therapy coverage, and HIV incidence in India: a cross-sectional, comparative study. Lancet HIV 2016;3(4):e183-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
Tanriverdi 2010 {published data only}
- Tanriverdi S, Chen L, Chen S.A rapid and automated sample-to-result HIV load test for near-patient application. Journal of Infectious Diseases 2010;201(Suppl 1):S52-8. [DOI] [PubMed] [Google Scholar]
Titchmarsh 2015 {published data only}
- Titchmarsh L, Zeh C, Verpoort T, Allain JP, Lee H.Leukodepletion as a point-of-care method for monitoring HIV-1 viral load in whole blood. Journal of Clinical Microbiology 2015;53(4):1080-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Vasconcellos 2020 {published data only}
- Vasconcellos I, Mariani D, Azevedo MC, Ferreira OC, Tanuri A.Development and validation of a simple and rapid way to generate low volume of plasma to be used in point-of-care HIV virus load technologies. Brazilian Journal of Infectious Diseases 2020;24(1):30-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Villa 2020 {published data only}
- Villa G, Abdullahi A, Owusu D, Smith C, Azumah M, Sayeed L, et al.Determining virological suppression and resuppression by point-of-care viral load testing in a HIV care setting in sub-Saharan Africa. eClinicalMedicine 2020;18:100231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Whitlock 2020 {published data only}
- Whitlock G, Nwokolo N.Does qualitative viral load testing shorten the window period for diagnosing HIV in individuals attending for post-exposure prophylaxis? International Journal of STD and AIDS 2020;31(9):816-9. [DOI] [PubMed] [Google Scholar]
Additional references
ASLM 2013
- African Society for Laboratory Medicine.Viral load monitoring in African HIV Treatment Programmes. Expert consultation; Cape Town, South Africa; 2013 Apr 18-20. mfr.osf.io/render?url=https%3A%2F%2Fosf.io%2F56mq8%2Fdownload (accessed prior to 21 February 2022).
Drain 2014
- Drain PK, Hyle EP, Noubary F, Freedberg KA, Wilson D, Bishai W, et al.Evaluating diagnostic point-of-care tests in resource-limited settings. Lancet Infectious Diseases 2014;14(3):239-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
EndNote 2016 [Computer program]
- EndNote.Version X7.7.1. Thomson Reuters, 2016.
Fox 2012
- Fox MP, Cutsem GV, Giddy J, Maskew M, Keiser O, Prozesky H, et al.Rates and predictors of failure of first-line antiretroviral therapy and switch to second-line ART in South Africa. Journal of Acquired Immune Deficiency Syndromes 2012;60(4):428-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Gopalakrishna 2014
- Gopalakrishna G, Mustafa RA, Davenport C, Scholten RJ, Hyde C, Brozek J, et al.Applying Grading of Recommendations Assessment, Development and Evaluation (GRADE) to diagnostic tests was challenging but doable. Journal of Clinical Epidemiology 2014;67(7):760-8. [DOI] [PubMed] [Google Scholar]
Gopalakrishna 2016
- Gopalakrishna G, Leeflang MM, Davenport C, Sanabria AJ, Alonso-Coello P, McCaffery K, et al.Barriers to making recommendations about medical tests: a qualitative study of European guideline developers. BMJ Open 2016;6(9):e010549. [DOI] [PMC free article] [PubMed] [Google Scholar]
GRADE 2013
- Schünemann H, Brożek J, Guyatt G, Oxman A, editor(s).Handbook for grading the quality of evidence and the strength of recommendations using the GRADE approach (updated October 2013). GRADE Working Group, 2013. Available from gdt.guidelinedevelopment.org/app/handbook/handbook.html.
Haas 2020
- Haas AD, Radin E, Hakim AJ, Jahn A, Philip NM, Jonnalagadda S, et al.Prevalence of nonsuppressed viral load and associated factors among HIV-positive adults receiving antiretroviral therapy in Eswatini, Lesotho, Malawi, Zambia and Zimbabwe (2015 to 2017): results from population-based nationally representative surveys. Journal of the International AIDS Society 2020;23(11):e25631. [DOI] [PMC free article] [PubMed] [Google Scholar]
Havlir 2001
- Havlir DV, Bassett R, Levitan D, Gilbert P, Tebas P, Collier AC, et al.Prevalence and predictive value of intermittent viremia with combination HIV therapy. JAMA 2001;286(2):171-9. [DOI] [PubMed] [Google Scholar]
Loeliger 2016
- Loeliger KB, Niccolai LM, Mtungwa LN, Moll A, Shenoi SV.Antiretroviral therapy initiation and adherence in rural South Africa: community health workers' perspectives on barriers and facilitators. AIDS Care 2016;28(8):982-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
Macaskill 2013
- Macaskill P, Gatsonis C, Deeks JJ, Harbord R, Takwoingi Y.Chapter 10: Analysing and presenting results. In: Deeks JJ, Bossuyt PM, Gatsonis C, editor(s). Cochrane Handbook for Systematic Reviews of Diagnostic Test Accuracy Version 1.0.0. The Cochrane Collaboration, 2013. Available from srdta.cochrane.org.
Moges 2020
- Moges NA, Adesina OA, Okunlola MA, Berhane Y.Barriers and facilitators of same-day antiretroviral therapy initiation among people newly diagnosed with HIV in Ethiopia: qualitative study using the transtheoretical model of behavioral change. Journal of Multidisciplinary Healthcare 2020;13:1801-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Pai 2012
- Pai NP, Vadnais C, Denkinger C, Engel N, Pai M.Point-of-care testing for infectious diseases: diversity, complexity, and barriers in low- and middle-income countries. PLOS Medicine 2012;9(9):e1001306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Patel 2016
- Patel RC, Odoyo J, Anand K, Stanford-Moore G, Wakhungu I, Bukusi EA, et al.Facilitators and barriers of antiretroviral therapy initiation among HIV discordant couples in Kenya: qualitative insights from a pre-exposure prophylaxis implementation study. PLOS ONE 2016;11(12):e0168057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Reitsma 2005
- Reitsma JB, Glas AS, Rutjes AW, Scholten RJ, Bossuyt PM, Zwinderman AH.Bivariate analysis of sensitivity and specificity produces informative summary measures in diagnostic reviews. Journal of Clinical Epidemiology 2005;58(10):982-90. [DOI] [PubMed] [Google Scholar]
Review Manager 2020 [Computer program]
- Nordic Cochrane Centre, The Cochrane Collaboration Review Manager 5 (RevMan 5).Version 5.4. Copenhagen: Nordic Cochrane Centre, The Cochrane Collaboration, 2020.
Ritchie 2014
- Ritchie AV, Ushiro-Lumb I, Edemaga D, Joshi HA, De Ruiter A, Szumilin E, et al.SAMBA HIV semiquantitative test, a new point-of-care viral-load-monitoring assay for resource-limited settings. Journal of Clinical Microbiology 2014;52(9):3377-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
Rutherford 2014
- Rutherford GW, Anglemyer A, Easterbroo PJ, Horvath T, Vitoria M, Penazzato M, et al.Predicting treatment failure in adults and children on antiretroviral therapy: a systematic review of the performance characteristics of the 2010 WHO immunologic and clinical criteria for virologic failure. AIDS (London, England) 2014;28(Suppl 2):S161-9. [DOI] [PubMed] [Google Scholar]
Schito 2012
- Schito M, Peter TF, Cavanaugh S, Piatek AS, Young GJ, Alexander H, et al.Opportunities and challenges for cost-efficient implementation of new point-of-care diagnostics for HIV and tuberculosis. Journal of Infectious Diseases 2012;205(Suppl 2):S169-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
Stata 2017 [Computer program]
- Stata.Version 15. College Station, TX, USA: StataCorp, 2017. Available at www.stata.com.
Takwoingi 2017
- Takwoingi Y, Guo B, Riley RD, Deeks JJ.Performance of methods for meta-analysis of diagnostic test accuracy with few studies or sparse data. Statistical Methods in Medical Research 2017;26(4):1896-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
UNAIDS 2020
- UNAIDS.UNAIDS data 2020. unaids.org/en/resources/documents/2020/unaids-data.
UNITAID 2014
- UNITAID.HIV/AIDS diagnostics technology landscape – 4th edition, 2014. unitaid.org/assets/UNITAID-HIV_Diagnostic_Landscape-4th_edition.pdf.
UNITAID 2015
- UNITAID.HIV/AIDS diagnostics technology landscape – 5th edition, 2015. www.unitaid.org/assets/UNITAID_HIV_Nov_2015_Dx_Landscape-1.pdf.
Whiting 2011
- Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al.QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Annals of Internal Medicine 2011;155(8):529-36. [DOI] [PubMed] [Google Scholar]
WHO 2010
- World Health Organization.Antiretroviral therapy for HIV infection in adults and adolescents: recommendations for a public health approach revision, 2010. www.who.int/hiv/pub/arv/adult2010/en. [PubMed]
WHO 2013
- World Health Organization.Consolidated guidelines on the use of antiretroviral drugs for treating and preventing HIV infection. Recommendations for a public health approach, 2013. www.who.int/hiv/pub/guidelines/arv2013/en/. [PubMed]
WHO 2014
- World Health Organization.Technical and operational considerations for implementing HIV viral load testing: interim technical update, 2014. apps.who.int/iris/handle/10665/128121 (accessed prior to 10 September 2018).
WHO 2015
- World Health Organization.Guideline on when to start antiretroviral therapy and on pre-exposure prophylaxis for HIV. September 2015. apps.who.int/iris/bitstream/handle/10665/186275/9789241509565_eng.pdf (accessed prior to 1 November 2021). [PubMed]
WHO 2016
- World Health Organization.Consolidated guidelines on the use of antiretroviral drugs for treating and preventing HIV infection: recommendations for a public health approach – 2nd edition, 2016. www.who.int/hiv/pub/arv/arv-2016/en/. [PubMed]
Wu 2012
- Wu G, Zaman MH.Low-cost tools for diagnosing and monitoring HIV infection in low-resource settings. Bulletin of the World Health Organization 2012;90(12):914-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
References to other published versions of this review
Ochodo 2018
- Ochodo EA, Kakourou A, Mallett S, Deeks JJ.Point-of-care viral load tests to detect high HIV viral load levels in HIV-positive people on antiretroviral therapy. Cochrane Database of Systematic Reviews 2018, Issue 11. Art. No: CD013208. [DOI: 10.1002/14651858.CD013208] [DOI] [PMC free article] [PubMed] [Google Scholar]


