Malin et al. show that the homophilic adhesion molecule Sidekick interacts alternately with the WAVE regulatory complex and with Polychaetoid/Zonula occludence-1 at tricellular adherens junctions to dynamically rebalance opposing protrusive and contractile forces that repeatedly expand and contract cell contacts to maintain cell–cell contacts and ensure proper epithelial remodeling.
Abstract
Contractile actomyosin and protrusive branched F-actin networks interact in a dynamic balance, repeatedly contracting and expanding apical cell contacts to organize the epithelium of the developing fly retina. Previously we showed that the immunoglobulin superfamily protein Sidekick (Sdk) contributes to contraction by recruiting the actin binding protein Polychaetoid (Pyd) to vertices. Here we show that as tension increases during contraction, Sdk progressively accumulates at vertices, where it toggles to recruit the WAVE regulatory complex (WRC) to promote actin branching and protrusion. Sdk alternately interacts with the WRC and Pyd using the same C-terminal motif. With increasing protrusion, levels of Sdk and the WRC decrease at vertices while levels of Pyd increase paving the way for another round of contraction. Thus, by virtue of dynamic association with vertices and interchangeable associations with contractile and protrusive effectors, Sdk is central to controlling the balance between contraction and expansion that shapes this epithelium.
Introduction
Epithelial cells coordinately move, change shape, exchange neighbors, proliferate, and die to reshape epithelial layers during development (Pinheiro and Bellaiche, 2018). Tensile forces generated by actomyosin networks drive many of these cell behaviors (Levayer and Lecuit, 2012). By coupling mechanically to the cell cortex through linker proteins, the contraction of actomyosin networks can deform the cell surface, alter cell shape and influence the shape of neighboring cells (Charras and Yap, 2018). Isometric activation of actomyosin networks can uniformly decrease the apical cell perimeter to drive processes such as apical constriction and tissue bending, while polarized activation can promote the polarized cell shape changes that drive processes such as cell intercalation and tissue elongation (Lecuit et al., 2011).
In addition to tensile forces, cells can generate protrusive forces that can push on and expand the cell surface to direct epithelial remodeling (Pollard and Cooper, 1986). Protrusive forces are generated by branching F-actin networks and control a range of fundamental morphogenetic processes (Pollitt and Insall, 2009; Tajiri et al., 2011; Takenawa and Suetsugu, 2007). Protrusive forces are primarily regulated by the assembly and activation of the pentameric WAVE regulatory complex (WRC) at the cell surface (Chen et al., 2014b; Chen et al., 2010; Eden et al., 2002; Ismail et al., 2009; Koronakis et al., 2011; Lebensohn and Kirschner, 2009; Mendoza, 2013; Oikawa et al., 2004). The WRC is targeted to the cell surface, at least in part, by binding to a short sequence motif known as the WRC interacting receptor sequence (WIRS), which is found in functionally diverse transmembrane proteins (Chen et al., 2014a).
In some developmental processes, one force, either tensile or protrusive, will dominate. In others, it is likely that they exist in a dynamic balance. One such process is the development of the Drosophila compound eye, which elaborates 800 nearly identical ommatidia (Cagan, 2009; Carthew, 2007; Johnson, 2021). After cell specification and rearrangement generate the initial arrangement of cell types in ommatidia, the asymmetric distribution of tensile and protrusive mechanical forces in the epithelium directs cell shape changes that produce the final ordered structure (Fig. 1 A; Blackie et al., 2020; Chan et al., 2017; Del Signore et al., 2018; Galy et al., 2011; Hayashi and Carthew, 2004; Letizia et al., 2019; Zallen et al., 2002). Actomyosin networks are cyclically activated at specific contacts by accumulation of nonmuscle MyoII. The WRC and branched F-actin also cyclically accumulate at the same contacts, but with inverse temporal dynamics. If F-actin branching is inhibited, the amplitude of contact lengthening is reduced and the coordination of contractile with protrusive dynamics is impaired. Consequently, the ommatidia fail to develop correctly. Thus, in this system, F-actin branching and actomyosin contractility are working together to repeatedly push and pull on cell contacts. This balancing act reorders and regularizes the packing geometries of ommatidial cells (Del Signore et al., 2018).
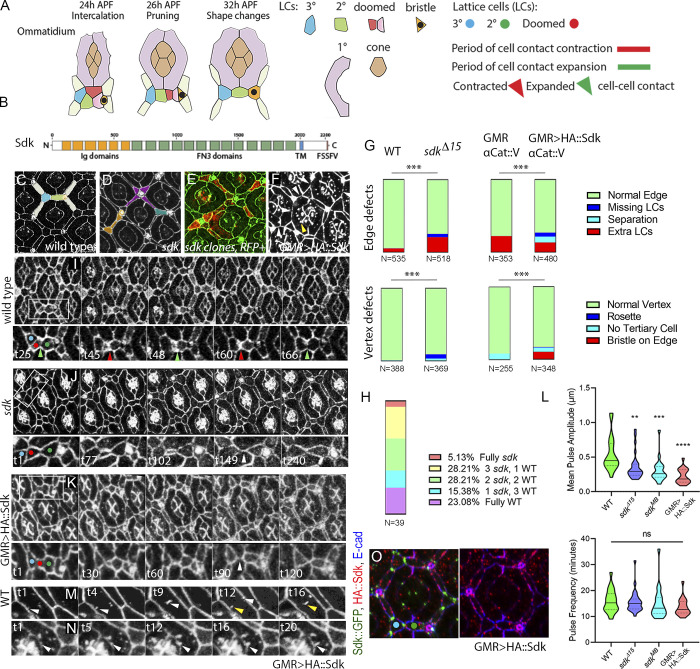
Figure 1.
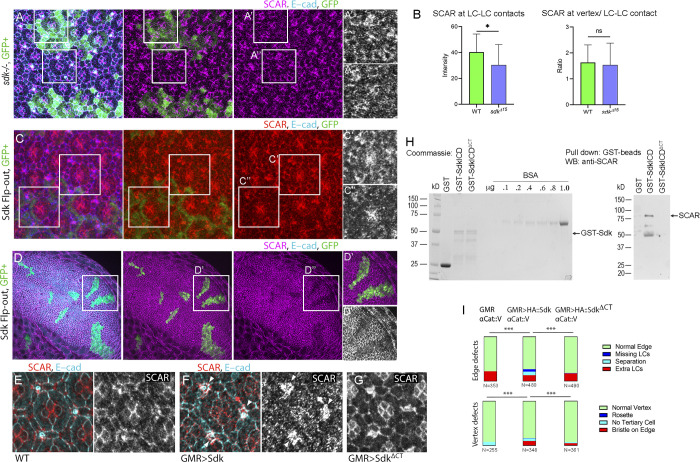
Sdk controls epithelial remodeling and fluctuations of cell contact length. (Refers to Videos 1 and 2 and Table S1.) (A) Left: Schematics of major steps in lattice remodeling. Right: Types of LCs, periods of expansion and contraction of LC–LC contacts, and contracted or expanded cell–cell contacts in this and subsequent figures. (B) Sdk consists of 6 immunoglobulin (Ig) domains, 13 Fibronectin type III domains (FN3), a transmembrane (TM) domain and a predicted WIRS motif. (C–H) Cellular defects in sdk mutants and HA::Sdk-expressing eyes compared with WT. (C) WT; each bristle cell, and 3° LC, connects to three 2° LCs as in colored overlay. (D) sdk mutant; common defects include cellular rosettes (magenta), missing LCs (blue), and extra LCs (brown; Fig. S1). (E) sdk mutant clones; cellular rosettes can consist of mutant and/or WT cells, see quantification in H. (F) GMR>HA::Sdk; LC–LC contacts are lost and replaced with 1°–1° contacts (white arrowheads) and large elongated vesicles persist at the cell surface (yellow arrowhead). (G) Quantification of the frequency of ommatidia vertices associated with intercalation defects and the frequency of edges associated with extra cells or missing cells and misplaced bristles in sdk mutants and GMR>HA::Sdk eyes compared with WT. χ2 test; ***, P < 0.001. (H) Quantification of the mutant and WT cell composition in 39 rosettes in 5 eyes containing sdk MARCM clones. (I–L) Cellular and contact dynamics in sdk mutant and GMR>HA::Sdk eyes compared with WT, 26–28 h APF (Video 1). (I) WT; contacts expand and contract cyclically. (J) sdk mutants; a subset of contacts transiently separate. (K) GMR>HA::Sdk; contacts separate either transiently or permanently. (L) Quantification of the amplitude of contact expansion and frequency of contact pulsing along horizontal edges. WT, n = 20 contacts in two retinas; sdkMB05054, n = 20 contacts in two retinas; sdkΔ15, n = 30 contacts in three retinas; GMR>HA::Sdk, n = 20 contacts in two retinas. Kruskal–Wallis test with Dunn’s multiple comparison test for pulse amplitudes, overall P < 0.0001; sdkΔ15 versus WT, P = 0.0189; sdkMB05054 versus WT, P = 0.0037; GMR>Sdk::HA versus WT, P < 0.0001, for pulse frequencies, overall P = .0923, sdkΔ15 vs. WT P > .9999, sdkMB05054 vs. WT P = .3918, GMR>HA::Sdk vs. WT P = .1341. (M) WT; vesicles bud from cone–1° contacts and move to 1°–LC contacts. Arrowheads trace vesicle movement. (N) GMR>HA::Sdk causes a delay of vesicle budding and their enlargement to form tubules. Arrowhead traces a vesicle arrested at budding (Video 2). (O) GMR>HA::Sdk expression in Sdk::GFP eyes. 36 h APF; HA::Sdk is broadly distributed along cell contacts and is excluded from vertices at this and earlier stages. Scale bar, 5 μm.
While mechanisms that regulate actomyosin contractility have been extensively studied in several systems, the mechanisms that regulate protrusive F-actin branching during the remodeling of adherens junctions (AJs) remain poorly understood. It is unclear how the WRC localizes to specific cell–cell contacts, how it is cyclically activated at these contacts, and how the assembly of branched F-actin networks and contractile actomyosin networks are coordinated. Here we test the hypothesis that a protein containing a WIRS motif is involved in this process, and we identify a key role for the Sidekick (Sdk) protein. Previously we showed that Sdk promotes actomyosin contraction by binding to Polychaetoid (Pyd; Letizia et al., 2019). Here we show that Sdk uses the same cytoplasmic C-terminal motif to recruit the WRC and thus promote expansion. Our findings highlight the role of Sdk in coordinating competing forces to regulate cell contact length and consequently epithelial morphogenesis.
Results
Sdk contains a putative WIRS motif
Ommatidia in the Drosophila eye are composed of lattice cells (LCs) surrounding central primary (1°) and cone cells (Fig. 1 A). During lattice remodeling, the WRC accumulates dynamically along LC–LC contacts at the plane of AJs (Del Signore et al., 2018). We hypothesized that a protein containing a WIRS motif would recruit and preferentially localize the WRC to the LC–LC contacts. The WIRS is a weak five- to six-residue consensus motif (Chen et al., 2014a). We therefore searched for proteins predicted to contain a WIRS motif that had been previously implicated in eye epithelial remodeling. The immunoglobulin superfamily protein Sdk fulfilled these two criteria (Fig. 1 B). Sdk is a homophilic adhesion protein expressed widely in nervous and epithelial tissues (Nguyen et al., 1997; Yamagata et al., 2002). Sdk’s five C-terminal amino acids, FSSFV, are conserved and were selected computationally as a possible WIRS motif (Chen et al., 2014a; Yamagata et al., 2002). Moreover, the missing LCs and irregularly shaped ommatidia in sdk mutants implicate sdk in the mechanics of lattice remodeling (Letizia et al., 2019; Nguyen et al., 1997). We therefore investigated whether Sdk could localize the WRC.
sdk controls cell behavior and contact dynamics during epithelial remodeling
Mutations in the WRC subunits SCAR or abi or the Arp2/3 complex subunit arpc2 cause strong defects in cell intercalation, cell number, and connectivity between LCs (Del Signore et al., 2018). To determine whether sdk mutants cause similar defects, we stained developing sdk mutant eyes for E-cadherin (E-cad) to examine epithelial structure. In WT, three secondary (2°) LCs connect to each mechanosensory bristle and tertiary (3°) LC (Fig. 1 C). In sdk mutants, a subset of mechanosensory bristles connected with four or more 2° LCs, forming cellular rosettes, a phenotype that can arise from defects in cell intercalation (Fig. 1 D; Letizia et al., 2019). The formation of rosettes reduced the number of edges in ommatidia from six to five or four and increased the number of LCs along some edges. However, at other edges, LCs were missing and a subset of the LC–LC contacts were short (Figs. 1, D and G, and S1; and Table S1). Likewise, analysis of sdk mutant clones revealed defects in cell intercalation and connectivity between LCs (Fig. 1, E and H). Some cell clusters that failed to intercalate consisted of both mutant and WT cells. A small subset of rosettes that formed adjacent to patches of sdk mutant cells comprised only WT cells. These cell-autonomous and nonautonomous defects are consistent with a role for sdk in transmitting forces between cells that are necessary to resolve T1 transitions and reorder the epithelium (Finegan et al., 2019; Letizia et al., 2019; Uechi and Kuranaga, 2019).
Figure S1.
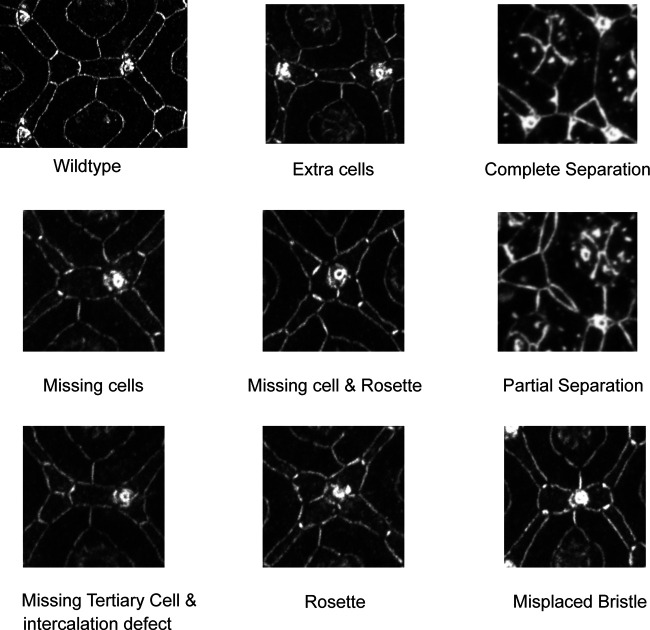
Common phenotypes in sdk mutant and HA::Sdk-expressing eyes. (Corresponds to Figs. 1 and 4.) Representative examples of cellular defects in sdk mutant eyes and HA::Sdk-expressing eyes that were scored in Figs. 1 G and 4 H and in Fig. 4I. Common defects include missing and extra LCs, misplaced bristles, rosettes, and other intercalation defects and separation of LC–LC contacts that lead to a cell contact forming between 1° cells of adjacent ommatidia.
After cell intercalation, the LC–LC contacts expand and contract dynamically, and new contacts form between LCs immediately after pruning. To test how sdk affects epithelial organization in this later stage, we live imaged sdk mutant eyes with an α-Catenin::GFP (α-Cat::GFP) protein trap to examine epithelial cell dynamics (Fig. 1, I and J and Video 1). In sdk mutants, the amplitudes of contact expansion decreased, and the formation of new LC–LC contacts after pruning was delayed. This resulted in transient separation of LC–LC contacts and formation of aberrant contacts between 1° cells of adjacent ommatidia (Fig. 1, J and L and Video 1). While the amplitude of contact pulsing decreased, the frequency was not affected (Fig. 1 L).
Video 1.
Cellular dynamics in WT compared with sdk mutant and HA::Sdk expressing eyes. (Corresponds to Fig. 1.) Pulsing contacts and pruned cells are demarcated with yellow and white rectangles, respectively. Left panel: a WT eye showing normal dynamics of repeated contraction and expansion of LC–LC contacts during lattice remodeling. Contacts between LCs are reestablished immediately following pruning of intervening LCs. Middle panel: sdk mutant eyes showing abnormal narrowing of LC–LC contacts and transient separation of a subset of these contacts. Right panel: GMR>HA::Sdk expressing eyes showing abnormal narrowing and separation of LC–LC contacts.
To determine if an excess of Sdk can affect epithelial development, we overexpressed a full-length HA-tagged Sdk transgene (HA::Sdk) and examined phenotypes in fixed and live tissue using the approach described above. HA::Sdk overexpression induced similar but stronger phenotypes compared with sdk mutants including defects in cell intercalation and loss of LCs and LC–LC contacts (Fig. 1, F, G, and K; and Video 1). Cells that formed rosettes at earlier stages were subsequently pruned from the lattice. Analysis of time-lapse videos showed decreased amplitudes of LC–LC contact expansion in HA::Sdk eyes compared with WT (Fig. 1, K and L). HA::Sdk also caused prolonged association of enlarged endocytic vesicles and tubular structures with the cell surface, a phenotype not observed in sdk mutants (Fig. 1, M and N; and Video 2). These types of endocytic defects are typically observed in mutants affecting the Arp2/3 complex and F-actin branching, suggesting that Sdk affects F-actin branching, possibly by regulating the WRC (Georgiou et al., 2008; Harris and Tepass, 2008; Leibfried et al., 2008). While endogenous Sdk localized preferentially to vertices at 36 h after puparium formation (APF) as previously described (Letizia et al., 2019), the overexpressed HA::Sdk protein localized broadly to the cell surface and was excluded from vertices (Fig. 1 O). This mislocalization of Sdk protein may result in defects in epithelial remodeling. The sdk and HA::Sdk phenotypes were similar to those in mutants for WRC and Arp2/3 complex subunits (Del Signore et al., 2018), consistent with a functional interaction between Sdk and these effectors that controls epithelial remodeling.
Video 2.
Trafficking defects in HA::Sdk-expressing eyes. (Corresponds to Fig. 1.) Left: In WT, AJs are trafficked preferentially from cone cells toward the LCs. After endocytosis, vesicles bud off from cone–1° cell edges, move toward the LCs in straight lines, and fuse with the 1°–LC edges. In HA::Sdk-expressing cells, vesicles form on cone–1° cell edges but fail to bud off and move in a timely manner. These vesicles linger and form enlarged tubular structures.
Sdk affects F-actin and Rho kinase (Rok) dynamics at LC–LC contacts
The above results suggested that Sdk affects the contractile-protrusive force dynamics at LC–LC contacts. In WT, an increase in F-actin levels correlates with contact lengthening, while increased MyoII and Rok, which phosphorylates and activates MyoII, correlates with contact shortening (Del Signore et al., 2018). To determine whether sdk affects F-actin and Rok dynamics, we live imaged F-actin and Rok in WT, sdk mutant, and HA::Sdk-expressing eyes (Fig. 2, A–C). We used the actin-binding protein Lifeact tagged with Ruby (Lifeact::Ruby) and Rok tagged with GFP (Rok::GFP) to follow F-actin and Rok dynamics, respectively (Video 3). We then measured the length of the LC–LC contacts and the signal intensities of F-actin and Rok along these contacts over time. Finally, we computed the temporal cross-correlation between either F-actin or Rok and contact length to determine whether sdk loss or HA::Sdk expression disrupted the temporal relationships between actin, Rok, and contact length.
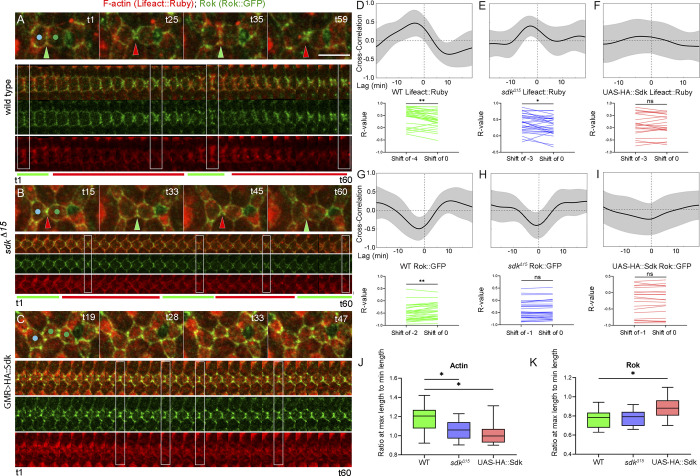
Figure 2.
Sdk affects F-actin and Rok dynamics at LC–LC contacts. (Refers to Table S2 and Video 3.) (A–C) Actin and Rok dynamics in WT (A), sdkΔ15 mutant (B), and GMR>HA::Sdk eyes (C). Top: Snapshots from a time-lapse video. Bottom: Kymographs of a cell contact. Boxes correspond to time points shown in top panel. (D–F) Time-shifted Pearson correlation plots for contact length versus F-actin. In this and subsequent plots, black line is the mean correlation, gray band the SD. Length positively correlates with F-actin in WT (D) at a shift of −4 min, and in sdkΔ15 (E) at a shift of −3 min, but no correlation is detected in GMR>HA::Sdk (F). (G–I) Pearson correlation plots for contact length versus Rok. Length negatively correlates with Rok in WT (G) at a shift of −2 min and in sdkΔ15 (H) at a shift of −1 min. (I) GMR>HA::Sdk: length has a weak negative correlation with Rok at a shift of −1 min. (J and K) Levels of F-actin (J) and Rok (K) in contacts with maximal length relative to contacts with minimal length in WT, sdk mutants, and GMR>HA::Sdk eyes. Kruskal–Wallis test of F-actin ratios, P = 0.0090. Dunn’s multiple comparisons test: WT versus sdkΔ15, P = 0.0327; WT versus GMR>HA::Sdk, P = 0.0146. Rok ratios, P = 0.0435; WT versus GMR>HA::Sdk, P = 0.0293; WT versus sdkΔ15, P > 0.9999. Scale bar, 5 μm.
Video 3.
Cytoskeletal dynamic defects in sdk mutant and HA::Sdk expressing eyes. (Corresponds to Fig. 2). Time-lapse videos of F-actin (Lifeact::Ruby) and Rok (Rok::GFP) in WT, sdkΔ15 mutant, and HA::Sdk expressing eyes. Note robust, inversely coordinated pulsing of actin and Rok in WT, reduced amplitude of contact pulsing in sdkΔ15, and sustained accumulation of Rok and loss of actin dynamics in HA::Sdk eyes.
In WT, F-actin levels correlated with contact length, peaking before the maximal increase of contact length. In sdk mutant eyes, this correlation decreased but not significantly. However, in HA::Sdk eyes, changes in contact length and F-actin were both reduced, and as a result, this correlation was completely lost (Fig. 2, D–F; and Table S2). Rok levels in WT peaked before the maximal decrease of contact length. In sdk mutants, the negative correlation between Rok and contact length decreased significantly, while in HA::Sdk eyes it was completely lost (Fig. 2, G–I; and Table S2). These results provide evidence that Sdk affects the assembly and/or dynamics of protrusive branched F-actin, contractile actomyosin or both networks, with Sdk overexpression having stronger effects on network behavior than sdk loss.
To determine whether Sdk influences the pulse amplitude of F-actin and Rok accumulation, we generated kymographs of individual cell contacts in the time-lapse videos. We then measured the signal intensities of F-actin and Rok at maximal expansion relative to maximal contraction. We found that F-actin accumulation during contact expansion was less intense in both sdk mutants and HA::Sdk eyes compared with WT (Fig. 2 J). Additionally, the increase in Rok levels during contraction was comparable to WT in sdk mutants but significantly less intense in HA::Sdk (Fig. 2 K). Taken together, these data provide evidence that Sdk influences the pulsed assembly of branched F-actin networks and Rok-regulated contractile actomyosin networks, either directly or indirectly.
Sdk accumulates preferentially with F-actin and the WRC at LC–LC contacts
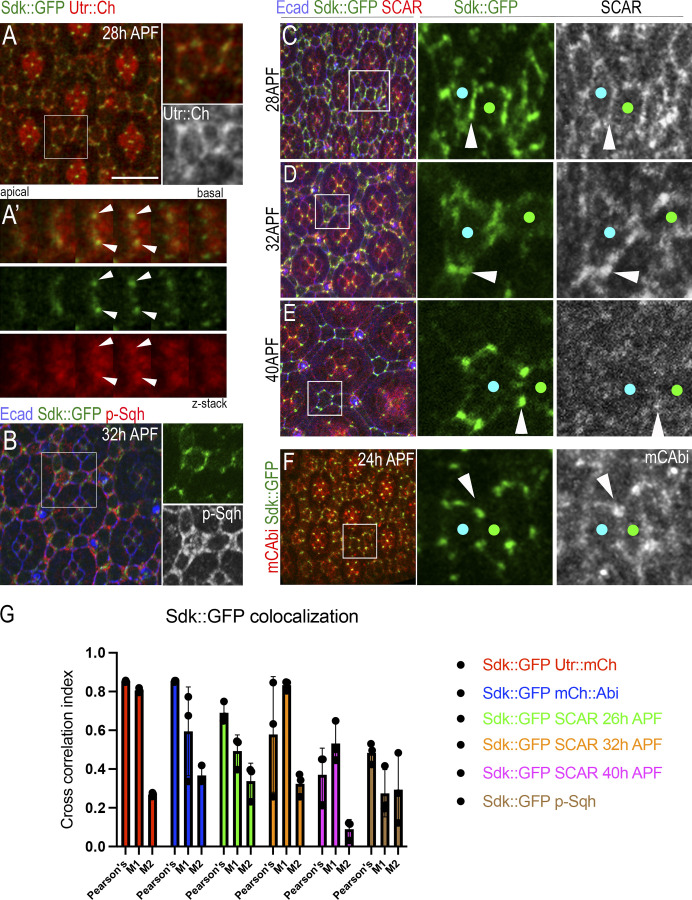
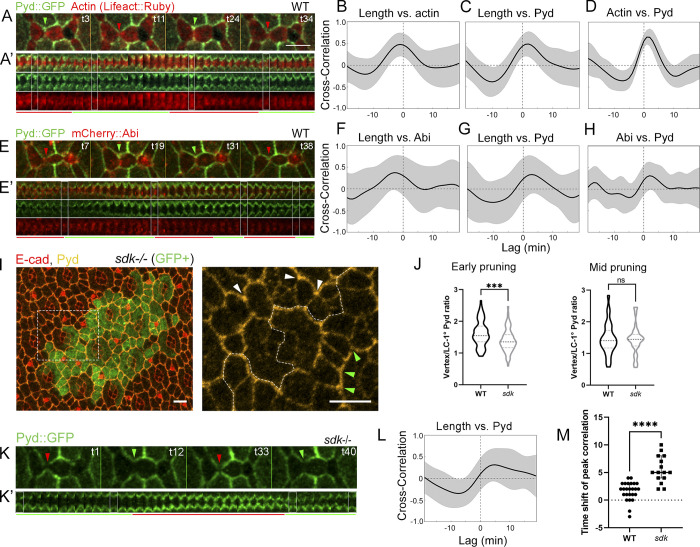
To test how Sdk could affect these dynamics, we examined Sdk protein localization relative to protrusive F-actin and contractile actomyosin networks. We used both a Sdk::GFP protein trap (Venken et al., 2011) and antibodies to detect Sdk. In fixed tissue, Sdk colocalized strongly with actin and weakly with the active phosphorylated form of MyoII (p-MyoII) that flanked the LC–LC contacts (Fig. 3, A, B, and G). To test whether Sdk could physically target the WRC to vertices and LC–LC contacts, we examined Sdk subcellular distribution relative to the WRC subunits SCAR and Abi. We used antibodies to detect SCAR and an mCherry::Abi (mC::Abi) reporter to detect Abi. We found that Sdk localized to the AJs and preferentially to vertices throughout development. Interestingly, Sdk distribution evolved over time. During lattice remodeling and the fluctuations of LC–LC contact length (24–32 h APF), Sdk accumulated strongly both at vertices and along the LC–LC contacts (Fig. 3 C), while at later stages, 32–40 h APF, as the cells assumed their stable shapes, Sdk became progressively restricted to vertices (Fig. 3, D and E). Colocalization analysis revealed that part of the SCAR pool strongly colocalized with Sdk at LC–LC contacts and vertices at earlier stages (Fig. 3, C, D, and G, 28–32 h APF) and colocalization decreased at later stages (Fig. 3, E and G; 40 h APF). Inspection of Sdk relative to mC::Abi by live imaging and colocalization analysis confirmed that the two proteins strongly colocalize during contact remodeling (Fig. 3, F and G). Taken together, the colocalization of Sdk and SCAR/Abi at vertices and LC–LC contacts, the assembly of F-actin at sites of Sdk accumulation, and the decrease in F-actin dynamics in sdk mutant and HA::Sdk-expressing eyes during pulsing reinforce the idea that sdk interacts with the WRC to affect protrusive F-actin dynamics and the contractile-protrusive force balance affecting tissue remodeling.
Figure 3.
Sdk protein colocalizes with F-actin and the WRC at vertices and LC–LC contacts. (A–F) Sdk::GFP localization relative to F-actin (A–A′), p-MyoII (B), SCAR (C–E), and mC::Abi (F). Sdk::GFP protein trap (green), SCAR (red), and E-cad (blue). (G) Cross-correlation analysis of pixels between Sdk::GFP and putative interacting proteins. Sdk::GFP shows the highest degree of colocalization with Actin, Abi, and SCAR at both 26 and 32 h APF. The lowest colocalizations are with SCAR at 40 h APF and with p-MyoII. The relative magnitudes of the Manders’ colocalization coefficients M1 and M2 show that Actin, Abi, and SCAR are more widely distributed relative to Sdk. n = 30 ommatidia in three eyes per experiment. Error bars represent SD. Scale bar, 10 μm.
Sdk affects the subcellular distribution of the WRC subunit SCAR
To test if Sdk and the WRC interact functionally, we examined SCAR accumulation at LC–LC contacts in genetically marked sdk mutant clones compared with nearby WT cells. We found a statistically significant decrease of SCAR levels in patches of sdk mutant cells compared with the adjacent WT cells (Fig. 4, A and B). Our prior work showed that WRC levels increase in expanding contacts and decrease in contracting contacts (Del Signore et al., 2018). Therefore, these measurements in fixed tissue are likely to underestimate the effect of sdk because they cannot distinguish between expanding and contracting contacts. That said, the observation that SCAR levels are not lost but just reduced suggests that other proteins in addition to Sdk localize the WRC to cell contacts. Likewise, the clonal expression of HA::Sdk decreased SCAR accumulation cell-autonomously in the retina and in the developing wing epithelium (Fig. 4, C and D). Additionally, broad expression of HA::Sdk in the retina using GMR-GAL4 decreased SCAR accumulation in all epithelial cells but not in the mechanosensory bristles that do not express this driver (Fig. 4, E and F). These results support the idea that Sdk targets the WRC to LC–LC contacts either directly or indirectly to control actin dynamics and lattice remodeling. They further suggest that HA::Sdk overexpression promotes dispersion of WRC subunits from cell contacts.
Figure 4.
Sdk binds and regulates WRC localization and activity via its conserved C-terminal motif. (A) sdk mutant MARCM clones stained for SCAR, 26 h APF. (B) SCAR levels decrease cell-autonomously in the clones. SCAR levels at contacts: Mann-Whitney U test, P = .0356; SCAR levels at vertices/full contacts: Mann-Whitney U test, P = .1753; n = 30 WT contacts and 40 sdkΔ15contacts across three retinas each. (C and D) HA::Sdk expressing clones in retina, 26 h APF (C) and third instar wing imaginal disc (D). SCAR levels decrease cell-autonomously in the clones. (E–G) GMR-GAL4 (E), GMR>HA::Sdk (F), and GMR>HA::SdkΔCT (G) retinas stained for SCAR, 32 h APF. SCAR levels decrease broadly in the GMR>HA::Sdk eyes but are normal in HA::SdkΔCT eyes. Mechanosensory bristles do not express GMR-GAL4 (arrowheads). (H) Detection of Sdk–SCAR interaction using a GST pulldown assay. Left: Expression of GST alone, GST fused to the Sdk ICD (GST-SdkICD), and GST fused to the Sdk ICD with a deleted C-terminal motif (GST-SdkICDΔCT) were assessed using Coomassie-stained gels. Right: Equal levels of GST, GST-SdkICD, and GST-SdkICDΔCT were used to pull down SCAR from S2 cell lysates. SCAR formed a complex with SdkICD but not with the SdkICDΔCT deletion mutant. The experiment was repeated three times, and one representative result is shown. (I) Quantification of eye phenotypes induced by HA::Sdk and HA::SdkICDΔCT. Deletion of the Sdk C-terminal sequence motif reversed HA::Sdk induced phenotypes. χ2 test; **, P < 0.01; ***, P = 0.0001; ****, P < 0.0001. Scale bar, 10 μm. Source data are available for this figure: SourceData F4.
Sdk physically interacts with SCAR via its cytoplasmic C-terminal motif
Next, we asked if Sdk targets the WRC to the cell surface via its conserved cytoplasmic C-terminal motif. We used the Sdk intracellular domain (ICD) and a deletion mutant lacking this motif (SdkΔCT), each fused to GST (Astigarraga et al., 2018; Letizia et al., 2019), to pull down endogenous SCAR from Schneider 2 (S2) cell lysates. We found strong SCAR binding to the Sdk ICD. However, removing the Sdk C-terminal motif abolished this binding (Fig. 4 H). We next compared the dominant-negative effects induced by full-length HA::Sdk transgene to those of a deletion mutant lacking the C-terminal motif. We used upstream activating sequence (UAS) transgenes that were inserted at the same genomic locus by site-specific recombination (Astigarraga et al., 2018). We found that deleting the C-terminal motif from HA::Sdk nearly abolished its strong dominant-negative effects (Fig. 4 I) and its ability to disperse SCAR from cell contacts (Fig. 4 G). These results indicate that the C-terminal motif of Sdk functions in part as a WIRS motif that targets the WRC to cell contacts.
Cytoskeletal dynamics regulate pulsed Sdk dynamics at vertices
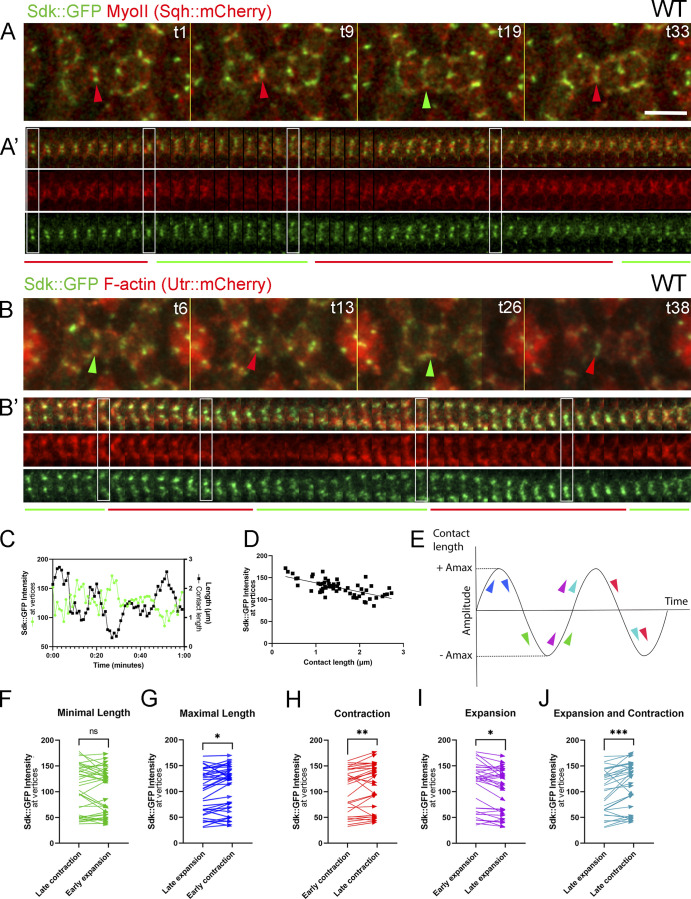
To determine whether the mechanical forces that alter contact length influence Sdk distribution, we examined Sdk dynamics by live imaging Sdk::GFP relative to F-actin and separately relative to MyoII using Utr::mCherry and Sqh::mCherry reporters, respectively. We found that Sdk was enriched at vertices but redistributed dynamically between vertices and LC–LC contacts during the fluctuations in contact length (Fig. 5, A–B’; and Video 4). During contractions, Sdk::GFP levels increased at vertices. During expansions, Sdk levels were high during the early and mid-phase of expansion. However, during the late phase of expansion and at maximal expansion, Sdk levels decreased at vertices and Sdk dispersed along the LC–LC contacts (Fig. 5, A–B’). Traces of Sdk::GFP levels relative to contact length and correlation plots between Sdk::GFP levels and contact length illustrate this relationship (Fig. 5, C and D).
Figure 5.
Sdk localization to vertices and cell contacts is dynamic. (Refers to Video 4.) (A–B′) Dynamics of Sdk::GFP localization relative to MyoII (A and A′) and F-actin (B and B′). (A and B) Upper panels: Snapshots from a video at 28 h APF. (A′ and B′) Lower panels: Kymographs of the contacts shown above. Note increased Sdk::GFP accumulation at vertices during contraction and decreased accumulation and dispersal along LC–LC contacts during expansion. (C) Trace of Sdk::GFP signal intensity at vertices relative to contact length in a single LC–LC contact. Sdk::GFP intensity increases as length decreases and vice versa. (D) Correlation between Sdk::GFP intensity and contact length. r = −0.68, P < 0.0001. (E) Schematics of contact length fluctuations and measurement scheme of Sdk::GFP levels relative to the fluctuations in contact length. A, amplitude. (F–J) The intensity values of Sdk::GFP between the different phases of contact length fluctuations were compared using Wilcoxon tests. Sdk::GFP is higher in shorter contacts with the largest difference in intensities when comparing late expansion versus late contraction. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Scale bar, 5 μm.
Video 4.
Sdk::GFP dynamics relative to F-actin and MyoII. (Corresponds to Fig. 5.) Time-lapse videos of Sdk::GFP relative to F-actin (left) and MyoII (right). Note that Sdk::GFP levels at vertices increase during contraction. During expansion, Sdk levels decrease at vertices and disperse along LC–LC contacts.
To refine the analysis, we measured Sdk::GFP levels in time-lapse videos at vertices of fully contracted and fully expanded contacts. We also measured Sdk::GFP levels before and after peak contraction and before and after peak expansion (Fig. 5 E). We then quantified the relative levels of Sdk between the different phases (Fig. 5, F–J). During contraction, Sdk levels were lower at the onset of contraction and were significantly enriched near peak contraction. During expansion, Sdk levels were higher at the onset of expansion and decreased near peak expansion. Finally, we quantified the temporal cross-correlation between Sdk::GFP signal intensity and LC–LC contact length. We found that Sdk::GFP levels peaked on average 2–4 min before the maximal contraction of the LC–LC contacts (Fig. 6, E–E’; and Table S2). The high Sdk levels at vertices at the onset of expansion are consistent with a role for Sdk in controlling F-actin branching that lengthens the LC–LC contacts and decreases bond tension.
Figure 6.
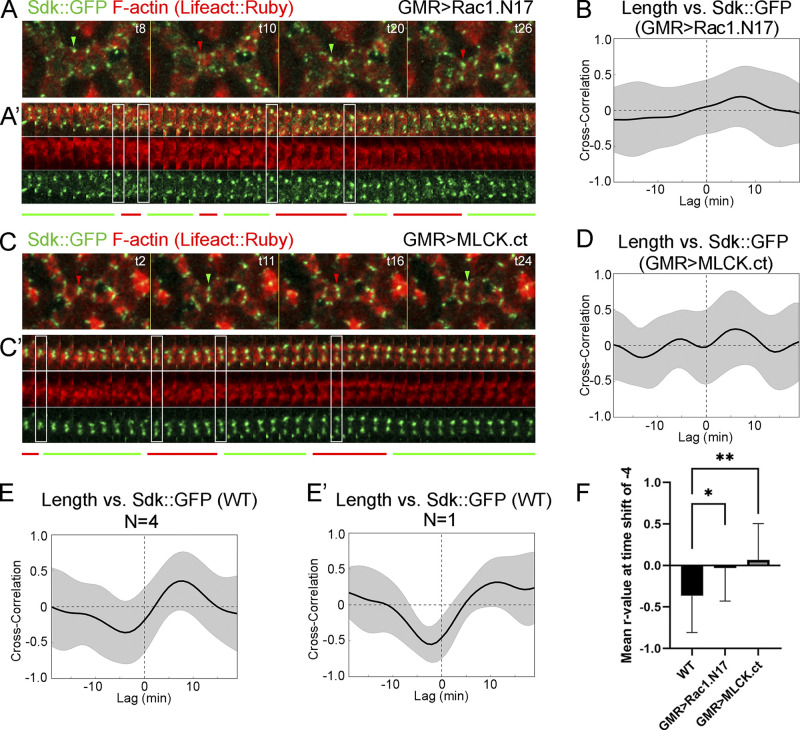
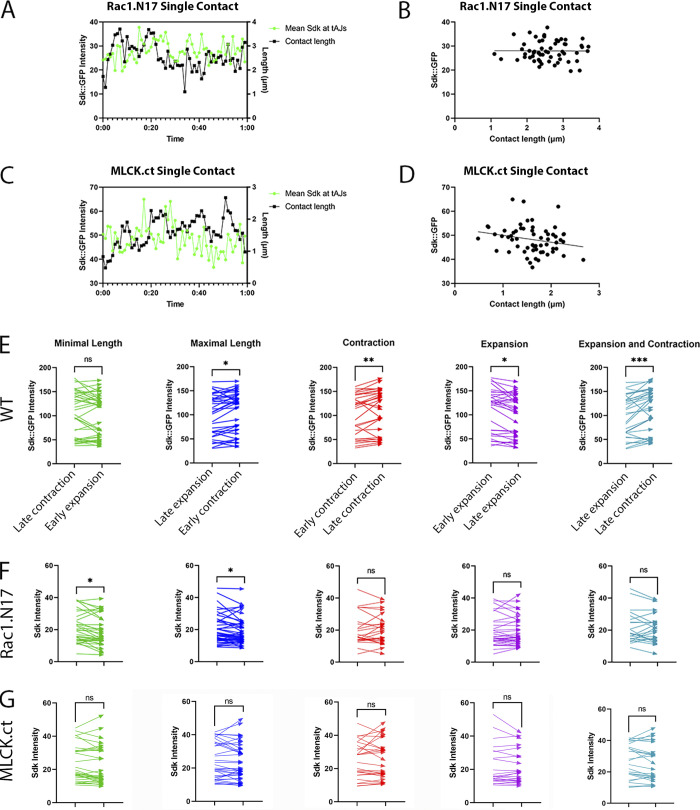
Tension affects Sdk dynamic localization to vertices. (Refers to Table S2 and Video 5.) (A–D) Sdk::GFP dynamics relative to actin and contact length in eyes expressing dominant-negative Rac1.N17 (A and B) and constitutively active MLCK.ct (C and D) compared with WT (E and E′) at 28 h APF. (A and C) Snapshots of a dynamic contact from a time-lapse video. (A′ and C′) Kymographs of the LC–LC contacts shown above. The regular patterns of expansion and contraction seen in WT are disrupted. (B and D–E′) Pearson correlation plots between Sdk levels at vertices and contact length in Rac1.N17 (B), MLCK.ct (D) and WT (E and E′) from four eyes (E) or a single eye (E′). (F) The mean peak correlation for WT contacts is significantly stronger than the correlation at the same time-shift for Rac1.N17 and MLCK.ct contacts (ANOVA, P = 0.0018; Dunn’s multiple comparisons test; *, P < 0.05; **, P < 0.01). Scale bar, 5 μm.
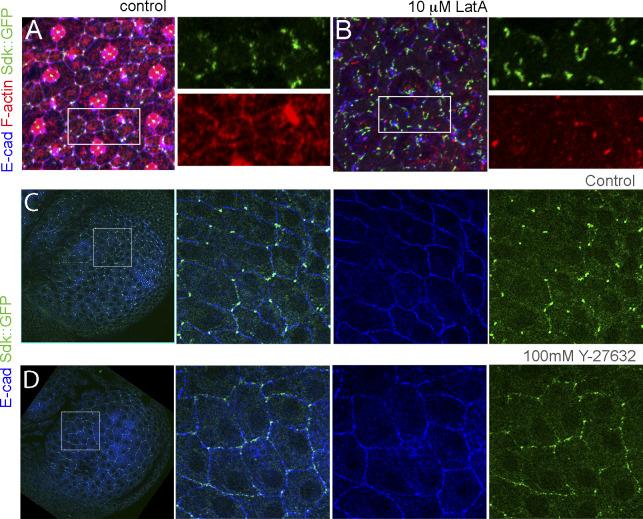
The increase in Sdk levels during contraction provides evidence that Sdk is recruited to vertices in response to tension (Letizia et al., 2019). To further test this model, we asked if the assembly of actin filaments is required. We found that treatment of dissected eyes with latrunculin A, a toxin that inhibits actin polymerization, decreased the abundance of F-actin and disrupted the continuity of AJs. In these eyes, Sdk was detected in elongated and string-like structures and not in spots as in WT (Fig. S2, A and B). We then asked if decreasing tension directly with the Rok inhibitor Y-27632 affected Sdk enrichment at vertices. The treatment led to the dispersion of Sdk from vertices to cell edges (Fig. S2, C and D). This dispersal was most striking in the wing peripodial epithelium.
Figure S2.
F-actin assembly and tension enhance Sdk localization to vertices. (Corresponds to Figs. 5 and 6.) (A and B) Dissected pupal eyes expressing Sdk::GFP were incubated in M3 medium and treated with DMSO (A; control) or the actin-monomer-binding toxin Latrunculin A in DMSO (B) and stained for E-cad (blue) and F-actin (red). (C and D) Dissected wing imaginal discs expressing Sdk::GFP were incubated in M3 medium and treated with deionized water (C; control) or the Rok inhibitor Y-27632 (D) and stained for E-cad (blue). Scale bar, 5 μm in A–D.
We then examined the effects of cytoskeletal mechanics on Sdk::GFP dynamics. First, we generated eyes expressing dominant-negative Rac1.N17, which was shown to likely disrupt both Rac and Rho signaling by sequestering upstream guanine nucleotide exchange factors and thereby decrease F-actin polymerization and decouple actomyosin fluctuations from contact length pulsing (Del Signore et al., 2018; Warner and Longmore, 2009a, 2009b). In these eyes, although Sdk still accumulated at vertices, it frequently spread away from vertices (Fig. 6, A–A’; and Video 5). Temporal cross-correlation analyses showed that the negative correlation between Sdk::GFP levels and contact length observed in WT was lost in Rac1.N17 eyes (Fig. 6, A, B, and F; and Table S2). The disruption of Sdk::GFP dynamics was also observed in traces of Sdk::GFP levels relative to contact length, in correlation plots between Sdk::GFP levels and contact length, and in the comparative analysis of Sdk::GFP levels during the different phases of contact pulsing (Fig. S3, A, B, and F). Second, we generated eyes that overexpress an activated form of myosin light chain kinase (MLCK), MLCK.ct, to increase tension (Jordan and Karess, 1997; Kim et al., 2002). In these eyes, Sdk levels were high and in abnormal aggregates along LC–LC contacts, and contact length fluctuations were not accompanied by the predictable changes in Sdk levels, as shown by cross-correlation analyses carried out using the approaches described above (Figs. 6, C, D, and F; and Fig. S3, C, D, and G; Video 5; and Table S2).
Video 5.
Sdk::GFP pulsatile dynamics in Rac1.N17 and MLCK.ct-expressing eyes. (Corresponds to Fig. 6.) Time-lapse videos of Sdk::GFP relative to F-actin in Rac1.N17 (left panels) and MLCK.ct (right panel) eyes. Note that the pulsatile Sdk::GFP accumulation is disrupted in these eyes.
Figure S3.
Pulse analysis of Sdk::GFP in Rac1.N17 and MLCK.ct-expressing eyes compared with WT (corresponds to Fig. 6). (A and C) A trace of Sdk::GFP levels relative to contact length at vertices at a developmental stage equivalent to 28 h APF in eyes expressing dominant-negative Rac1.N17 (A) and constitutively active MLCK.ct (C). (B and D) Correlation of Sdk::GFP levels at individual contacts relative to contact lengths in Rac1.N17 (r = −0.01; B) and MLCK.ct eyes (r = −0.22; D). (E) Paired comparisons of relative levels of Sdk::GFP at the different phases of contact pulsing in WT eyes. All statistics performed are Wilcoxon tests. Minimal length: P = 0.0929, n = 39 pairs from 17 contacts in 3 eyes; maximal length: P = 0.0229, n = 36 pairs from 17 contacts in 3 eyes; contraction: P = 0.0063, n = 30 pairs from 17 contacts in 3 eyes; expansion: P = 0.0105, n = 32 pairs from 16 contacts in 3 eyes; expansion and contraction: P = 0.0002, n = 31 pairs from 17 contacts in 3 eyes. (F) Paired comparisons of relative levels of Sdk at different time points in pulses in Rac1.N17-expressing eyes. All statistics performed are Wilcoxon tests. Minimal length: P = 0.0166, n = 37 pairs from 22 contacts in 5 eyes; maximal length: P = 0.0436, n = 44 pairs from 23 contacts in 5 eyes; contraction: P > 0.9999, n = 25 pairs from 18 contacts in 5 eyes; expansion: P = 0.9193, n = 34 pairs from 23 contacts in 5 eyes; expansion and contraction: P = 0.3786, n = 24 pairs from 19 contacts in 5 eyes. (G) Paired comparisons of relative levels of Sdk at different time points in pulses in MLCK.ct-expressing eyes. All statistics performed are Wilcoxon tests. Minimal length: P = 0.2129, n = 30 pairs from 12 contacts in 2 eyes; maximal length: P = 0.3364, n = 34 pairs from 12 contacts in 2 eyes; contraction: P = 0.8368, n = 26 pairs from 11 contacts in 2 eyes; expansion: P = 0.7442, n = 22 pairs from 12 contacts in 2 eyes; expansion and contraction: P = 0.3632, n = 26 pairs from 11 contacts in 2 eyes.
Overall, we provide evidence that Sdk dynamics at vertices is mechanosensitive and is regulated by actin assembly and actomyosin contractility. Thus, Sdk affects both the assembly of F-actin networks and their coupling to the cell surface and is itself regulated by these networks. This feedback may contribute to the pulsing of contractile and protrusive F-actin networks and the pulsatile changes in cell contact length that underlie lattice remodeling.
Sdk controls the timing of pulsatile Pyd accumulation at vertices
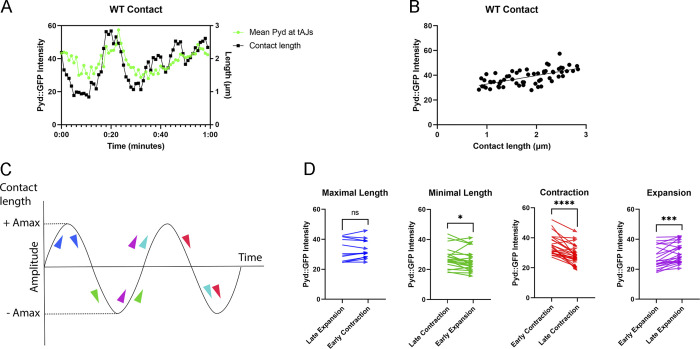
Pyd links surface proteins to actin filaments to transmit tensile forces (Choi et al., 2011; Letizia et al., 2019; Manning et al., 2019). Pyd binds the C-terminal motif of Sdk and accumulates at vertices in the wing imaginal disc epithelium in a Sdk-dependent manner (Letizia et al., 2019), and Pyd depletion mimics the sdk mutant eye phenotype (Seppa et al., 2008). To investigate how Sdk can control both protrusive and contractile forces, we characterized the dynamics of a Pyd::GFP protein trap relative to F-actin and mC::Abi (Figs. 7, A–H; and S4). In time-lapse videos, Pyd was enriched at vertices of highly expanded LC–LC contacts. However, unlike Abi and F-actin levels, which peaked before maximal contact expansion Fig. 7, B and F; Video 6; Del Signore et al., 2018), Pyd levels peaked 2–3 min after maximal expansion (Fig. 7, C and G). Within individual contacts, contact length, F-actin levels, and Abi levels all peaked 1–3 min before Pyd levels peaked (Table S2). These dynamics suggest that Pyd accumulates in response to contact expansion and the corresponding drop in bond tension to promote bond tension and contact contraction.
Figure 7.
Sdk controls the timing of Pyd localization to vertices. (Refers to Fig. S4 and Video 6.) (A–H) Pyd::GFP dynamics relative to F-actin (A–D) and mC::Abi (E–H) at 28 h APF. (A and E) Snapshots of a dynamic cell contact. (A′ and C′) Kymographs of the LC–LC contacts shown above. Pyd::GFP at vertices increases during expansion and decreases during contraction. (B–D) Pearson correlation charts for contact length, F-actin, and Pyd in WT. Contact length correlates positively with F-actin at a shift of −1 min (B) and with Pyd at a shift of +2 min (C). (D) Actin correlates positively with Pyd at a shift of +1 min. (F–H) Pearson correlation charts for contact length, Abi, and Pyd in WT eyes. (F and G) Contact length correlates positively with Abi at a shift of −3 min and with Pyd at a shift of +3 min. (H) Abi correlates positively with Pyd at a shift of +2 min. (I) 26 h APF; a sdk mutant MARCM clone stained for Pyd and E-cad. Pyd levels decrease at vertices in the sdk clone. Representative vertices are marked with arrowheads. (J) Pyd accumulation at vertices is reduced in sdk clones at 26 h APF (unpaired t test, n = 77 sdk and 73 WT contacts in 2 eyes, P = 0.0001; left) but not at 28 h APF (unpaired t test, n = 30 sdk and 30 WT contacts in 2 eyes, P = 0.8824; right). (K) Snapshots of Pyd::GFP localization in a sdk mutant eyes. (K′) Kymograph of the above contact. Pyd accumulation during expansion is delayed compared with WT. (L) Pearson correlation for contact length versus Pyd in sdk eyes shows peak correlation at a shift of +5 min. (M) The time shift of the peak r values for individual contacts is significantly higher in sdk mutants compared with WT (Mann–Whitney U test, P < 0.0001, n = 24 WT and 15 sdk contacts across 3 retinas each). Scale bars, 5 μm.
Figure S4.
Pyd::GFP pulse analyses. (Corresponds to Fig. 7.) (A) Trace of Pyd::GFP levels relative to contact length in a single LC–LC contact at 28 h APF. (B) Correlation between Pyd::GFP levels and contact length in A. (C) Schematics of contact length fluctuations and the scheme for measuring Pyd::GFP levels relative to the fluctuations in contact length. (D) Paired comparisons of relative levels of Pyd::GFP at the different phases of contact pulsing in WT eyes. Maximal length: n = 12, P = 0.2620; minimal length: n = 22, P = 0.0391; contraction: n = 26, P < 0.0001; expansion: n = 22, P = 0.0006. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. All statistics performed are Wilcoxon tests.
Video 6.
Pyd::GFP dynamics in a sdk mutant eye compared with WT. (Corresponds to Fig. 7.) Time-lapse videos of Pyd::GFP relative to F-actin in a WT eye (left) and Pyd::GFP alone in a sdk mutant eye (right). In WT, Pyd::GFP accumulates during contact expansion, while in sdk mutants. Pyd::GFP accumulates during contraction.
To determine if Pyd localization depends on Sdk, we stained eyes with sdk mutant clones for Pyd (Fig. 7 I). At ∼26 h APF, Pyd levels were lower at vertices in the absence of sdk. However, at later stages, Pyd levels at vertices were similar between mutant and WT cells (Fig. 7 J), indicating that additional mechanisms can target Pyd to cell contacts. To determine if Sdk affects Pyd dynamics at the later stage, we live-imaged Pyd::GFP in eyes fully mutant for sdk and measured the temporal cross-correlation between Pyd levels at vertices and contact length (Fig. 7, K–M; Video 6; and Table S2). This correlation was slightly reduced from WT but still present. However, the time shift of the peak correlation increased from an average of 2 min after maximal contact expansion in WT to an average of 5 min in sdk mutants. This result provides evidence that Sdk controls the timing of Pyd recruitment to vertices and thus Pyd’s participation in contact pulsing during the switching between expansion and contraction.
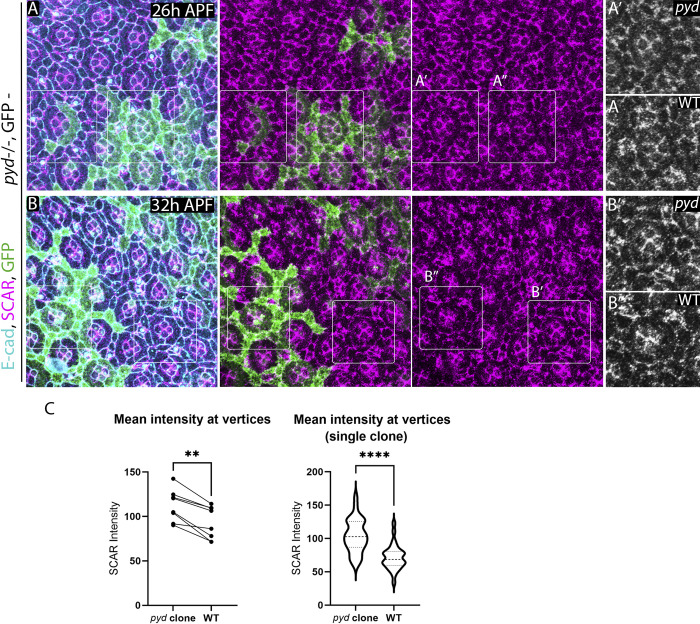
Since Pyd and the WRC both bind to the C-terminus of Sdk, we tested whether the presence of Pyd affects the Sdk-dependent accumulation of SCAR at cell contacts by examining SCAR localization in pyd-null mutant clones. We found that SCAR levels increased significantly at vertices in pyd mutant clones compared with WT (Fig. 8). This finding supports the idea that the same region of Sdk can bind to either the WRC or Pyd to control contact pulsing.
Figure 8.
pyd affects SCAR localization to vertices. (A and B) pyd mutant clones stained for SCAR (A) during 26 h APF and after pruning (32 h APF; B). SCAR levels increase in pyd clones compared with WT cells. (C) Paired comparisons of relative levels of SCAR at vertices of LCs in mutant compared with WT cells in multiple clones (left) and within a single clone (right). Wilcoxon test, n = 8 clones and adjacent WT regions from 3 eyes, 10–30 vertices per region, P = 0.0078. Single clone: unpaired t test, n = 30 vertices per region, P < 0.0001.
Discussion
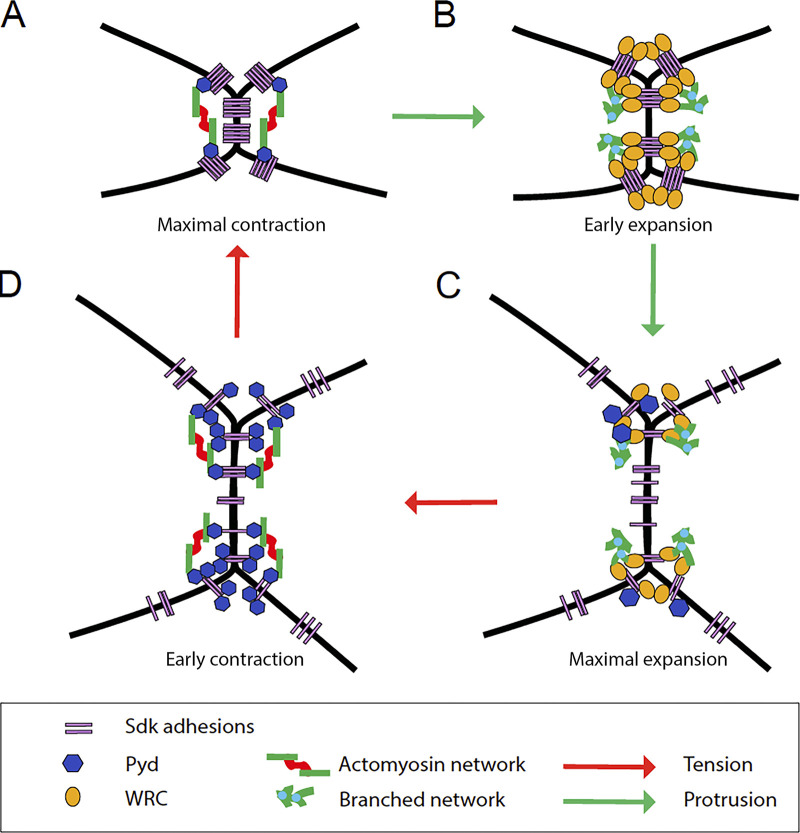
Tensile forces contract cell contacts and lead to recruitment of Sdk, which promotes contraction by interacting with Pyd via its C-terminal motif. However, at a critical point in the contraction phase, Sdk function shifts to promoting protrusion by targeting the WRC to apical junctions via the same sequence motif, which also functions as a WIRS. With increased protrusion counterbalancing contraction, Sdk levels ebb and Pyd levels increase, paving the way for the next contraction (Fig. 9). Thus, Sdk is central to coordinating the interplay between protrusive and contractile forces that control lattice remodeling. Sdk proteins, the WRC, and Pyd are evolutionarily conserved, suggesting that their interaction here may be prototypical.
Figure 9.
Sdk interacts alternately with Pyd and the WRC to control contact length pulsing. By virtue of tension-dependent localization to vertices and interchangeable interactions with the WRC and Pyd, through the same C-terminal motif, Sdk controls the balance between contraction and expansion of cell contacts that shapes the epithelium of the fly retina. (A) With increasing contraction and tension, Sdk progressively accumulates at vertices. (B) In contracted contacts, actomyosin networks disassemble and Sdk recruits the WRC to promote actin branching and protrusion. (C) With increasing protrusion and contact expansion, levels of Sdk and the WRC decrease at vertices. (D) In expanded contacts, Sdk toggles to interact with Pyd to promote the next contraction. (A) In contracted contacts, Pyd levels decrease again, paving the way for the next expansion. Thus, owing to a dynamic association with vertices and alternating associations with contractile and protrusive effectors, Sdk acts as a tension sensor, transducer, and feedback regulator to modulate cell contact length that shapes the epithelium.
Regulation of WRC function at LC–LC contacts
F-actin branching counterbalances and regulates contractile forces at the level of AJs to remodel the epithelium of the fly retina. Branched F-actin accumulates during contact expansion and inversely correlates with contact contraction. Indeed, protrusive forces dynamically balance contractile forces, as perturbing F-actin branching severely disrupts the pulsed dynamics of both protrusive and contractile networks (Del Signore et al., 2018). Despite the key role of protrusive forces, the mechanism that targets the WRC to the appropriate membranes to promote actin branching was unknown. Here we provide evidence that Sdk plays a key role in targeting the WRC to the appropriate membrane domains to control contact pulsing. sdk mutant phenotypes are weaker than in mutants for the WRC subunit SCAR, consistent with the finding that in sdk mutants the localization of the WRC to LC–LC contacts is reduced but not fully abolished. Thus, Sdk may not be the sole protein that controls F-actin branching at LC–LC contacts. Sdk overexpression induces stronger defects in epithelial remodeling and a strong decrease in SCAR localization and F-actin dynamics at LC–LC contacts. Overexpressed HA::Sdk localizes broadly and is excluded from vertices, likely leading to WRC mislocalization.
The insight that Sdk is involved in regulating protrusive forces modifies our understanding of how Sdk functions more broadly in shaping epithelia. We recently reported that Sdk promotes the accumulation of Pyd and its partner Canoe (Cno) to vertices (Letizia et al., 2019). Pyd and Cno have been proposed to link Sdk to the actin cytoskeleton to enable vertices to transmit tensile forces during epithelial remodeling (Choi et al., 2011; Letizia et al., 2019; Manning et al., 2019; Sawyer et al., 2011; Sawyer et al., 2009; Yu and Zallen, 2020). Supporting this idea, Sdk binds Pyd and promotes accumulation of Pyd, Cno, and F-actin at vertices (Letizia et al., 2019). Here we examined Pyd dynamics in WT and sdk mutant eyes. We found that Pyd accumulates in expanding contacts and peaks on average 2 min after maximal contact expansion. In sdk mutants, peak Pyd accumulation is delayed, providing evidence that Sdk targets Pyd to expanding contacts to initiate the next cycle of contraction. The same C-terminal motif of Sdk can interact with either Pyd to transmit tensile forces or the WRC to generate protrusive forces. However, the temporal characteristics of the binding argue against a simple competitive mechanism and suggest the possible involvement of other factors such as tension-regulated conformational changes or clustering. By repeatedly engaging and disengaging these opposing actin regulators, Sdk fine-tunes the levels of tensile and protrusive forces that pull and push vertices and the transition between these two mechanical states. Sdk thus toggles the system between expansion and contraction to dynamically reorder the epithelium while preventing the separation of cell contacts.
Moreover, we show that the level of tension impacts Sdk accumulation. Thus, Sdk functions as a tension sensor. When tension is high and Sdk becomes enriched, it recruits the WRC, and when tension is lower, Sdk levels ebb as it functionally shifts to recruiting Pyd. Thus, Sdk also functions as a tension transducer, alternately interacting with different effectors depending on the level of tension. Finally, the ability of Sdk to recruit either contractile or protrusive force effectors reveals the role for Sdk in dynamic feedback regulation of the level of tension itself. Thus, with respect to tension, Sdk has three distinct roles. It is a sensor, transducer, and feedback regulator. The enriched localization of Sdk at vertices, the geometric points that define the epithelial structure, enables Sdk to efficiently remodel this structure.
Regulation of WRC function in epithelial remodeling
The WRC is regulated not only by its selective targeting to a particular location at the cell surface, but also through its activation and inhibition. When activated, the WRC binds and activates the Arp2/3 complex to generate branched F-actin networks that can expand the cell surface (Chen et al., 2014b; Del Signore et al., 2018; Eden et al., 2002; Ismail et al., 2009; Koronakis et al., 2011; Lebensohn and Kirschner, 2009; Mendoza, 2013; Oikawa et al., 2004). We previously showed that the dynamics of the Rac1 RhoGTPase and the phosphoinositide PIP3 coincide with the cyclical activation of F-actin branching. Conversely, the dynamics of the Rho1 RhoGTPase and Rok correlate with the cyclical activation of contractile networks (Del Signore et al., 2018). The identification of Sdk as a WRC receptor therefore suggests that the assembly of branched F-actin networks at AJs is regulated in a two-step process. In the first step, Sdk directly recruits the WRC to vertices and LC–LC contacts by binding to the Sdk intracellular WIRS motif. In the second step, multiple coincidental signals, including Rac1 and PIP3, activate the WRC to promote F-actin branching (Chen et al., 2017; Lebensohn and Kirschner, 2009; Schaks et al., 2018). Branched F-actin network assembly is followed by disassembly, suggesting the involvement of a delayed negative feedback loop that terminates the cycle and enables the next round of contraction, and Sdk itself could regulate this phase as discussed above.
Sdk and the WRC in other systems
The Sdk–WRC interaction identified here may be broadly used. In the epithelium of the renal glomerulus, inappropriate up-regulation of Sdk-1 expression by podocytes has been linked to their dedifferentiation and loss of proper foot-process architecture, leading to collapsed glomeruli and glomerulosclerosis (Kaufman et al., 2007). Our results raise the possibility that the Sdk–WRC interaction could be relevant to this phenomenon, as the WRC and the Arp2/3 complex play critical roles in podocyte protrusion and adhesion (Schell et al., 2018). Likewise, the Sdk–WRC interactions may be relevant to synapse formation, both in the Drosophila visual system, where the single sdk gene controls the sorting of photoreceptor axons into synaptic modules (Astigarraga et al., 2018), and in the vertebrate visual system, where both Sdk-1 and -2 determine the specificity of synapse formation (Krishnaswamy et al., 2015; Yamagata and Sanes, 2010; Yamagata et al., 2002). In addition, previous work has identified the role of sdk in resolving T1 transitions occurring during intercalation of epidermal and tracheal cells in embryos and LCs in the fly retina (Finegan et al., 2019; Letizia et al., 2019; Uechi and Kuranaga, 2019). While one proposed mechanism is through a nonautonomous role in recruiting MyoII to junctions of neighboring cells to pull on and lengthen contacts (Uechi and Kuranaga, 2019), our current findings suggest that the Sdk-WRC interactions may also contribute autonomously to contact lengthening. The effect of Sdk on WRC targeting may be restricted to specific locations. In vertebrate epithelial cells, the immunoglobulin superfamily protein Neogenin utilizes its WIRS motif to target the WRC to AJs. However, Neogenin is localized uniformly at AJs (Lee et al., 2016). In contrast, the preferential localization of Sdk to vertices and LC–LC contacts is consistent with a specialized role in regulating localized forces.
In sum, we show that Sdk accumulation depends on tension. At contracted contacts, it binds and targets the WRC to regulate protrusive branched F-actin dynamics and contact expansion. This enables the system to switch from contraction to expansion. With decreasing tension, Sdk then interacts with Pyd in expanding contacts through the same C-terminal motif. This enables transmission of tensile forces and promotes the switch between expansion and contraction (Fig. 9). By localizing dynamically to vertices and interacting interchangeably with regulators of contractile and protrusive networks, Sdk acts as a tension sensor, transducer, and feedback regulator. Thus, Sdk is central to dynamically tuning the tensile and protrusive forces that remodel the retinal epithelium.
Materials and methods
Contact for reagent and resource sharing
Further information and requests for reagents should be directed to and will be fulfilled by the lead contact, Victor Hatini (victor.hatini@tufts.edu).
Experimental model and subject detail
Fly strains
To examine the sdk loss-of-function phenotypes, we used sdkMB05054, a Minos transposable insertion in a coding exon of sdk (Bellen et al., 2004), and sdkΔ15, containing a deletion of 1,240 bp at the 5′ end of sdk (Astigarraga et al., 2018). Sdk protein was not detected in sdkMB05054 or sdkΔ15 mutant clones, confirming that the mutants are null. To examine the Sdk overexpression phenotypes and the requirement of the Sdk C-terminal motif, we expressed UAS-HA::Sdk and a variant, respectively, with a deleted C-terminal motif, UAS-HA::SdkΔCT. These lines were previously inserted in the same landing site using site-specific recombination (Astigarraga et al., 2018). We used a Sdk::GFP protein trap line, sdkMI01498-GFSTF.1, to examine Sdk protein distribution, dynamics, and interaction with SCAR, sdkMB05059 FRT19A; UAS-lacZ to generate genetically marked mutant clones, sdkMB05045 and sdkΔ15 for live imaging, 82BFRT pydex147 to generate genetically marked mutant clones (Djiane et al., 2011), and a Pyd::GFP protein trap line, pydMI01205-GFSTF.1, to examine Pyd distribution and dynamics.
Fly lines from the Bloomington Drosophila Stock Center: (1) sdkMB05054, (2) sdkMI01498-GFSTF.1, (3) UAS-Lifeact::Ruby, (4) Ubi-Abi::mCherry, (5) sqh-GFP::Rok, (6) sqhAX3/FM7c; sqh-Sqh::GFP, (7) GMR-GAL4, (8) Actin>y+>GAL4, UAS-GFP, (9) α-Cat::GFP, (10) Ubi-mRFP, hsFLP, FRT19A, (11) sqhax3 FRT19A (Karess et al., 1991), (12) SCARΔ37 FRT40A (Zallen et al., 2002), (13) y w hsFLP, UAS-GFPnls, tub-GAL4; FRT82B tub-GAL80, (14) w; FRT82B Ubi-GFP, (15) UAS-MLCK.ct, and (16) UAS-Rac.N17. α-Cat::VenusCPTI002596 was obtained from the Kyoto stock center. Fly lines previously described by Del Signore et al. (2018): (1) GMR-GAL4, α-Cat::Venus, (2) UAS-Lifeact::Ruby; GMR-GAL4, (3) UAS-Lifeact::GFP; sqh-Sqh::mCherry, GMR-GAL4, (4) UAS-Lifeact::Ruby; sqh-Rok::GFP, GMR-GAL4, (5) sqh-UtrABD::GFP; ubi-Abi::mCherry, and (9) GMR-GAL4, UAS-α-Cat::GFP; Abi::mCherry. Additional stocks used: (1) sqh-Sqh::mCherry and (2) sqh-UtrABD::GFP, sqh-Sqh::mCherry (gift of A. Martin); and (3) GMR-GAL4, UAS-α-Cat::GFP (gift of R. Cagan). The following stocks were generated in this study: (1) sdkMI01498-GFSTF.1; mCherry::Abi, (2) sdkMI01498-GFSTF.1; sqh-Sqh::mCherry, (3) sdkMI01498-GFSTF.1; sqh-UtrABD::mCherry, (4) sdkMI01498-GFSTF.1; UAS-Rac.N17, and (5) sdkMI01498-GFSTF.1; UAS-MLCK.ct.
Genetic analysis
GMR-GAL4 lines were used to broadly express UAS transgenes in the eye (Wernet et al., 2003). The FLP/FRT (Xu and Rubin, 1993) and MARCM techniques (Lee and Luo, 2001) were used to generate genetically marked clones by FLP-mediated mitotic recombination. Ubi-mRFP, hsFLP, FRT19A was used to generate sdkMB05049 mutant FLP/FRT clones; y w hsFLP; FRT82B Ubi-GFP/TM6b, Tb to generate pydex147 mutant FLP/FRT clones; eyFLP, tub-GAL80, FRT19A; tub-GAL4, UAS-GFP/CyO, P(Tb, Ubi-RFP) to generate sdkMB05049 MARCM clones. y w hsFLP, UAS-GFPnls, tub-GAL4; FRT82B tub-GAL80 (gift of G. Struhl) was crossed to FRT82B UAS-HA::Sdk to generate MARCM clones expressing the UAS-HA::Sdk transgene. Mitotic clones were induced by heat shock for 30 min at 34°C. There are no known differences in eye morphogenesis between males and females; therefore, sex has not been factored in as a biological variable into the experiments.
Quantitative assessment of eye phenotypes
Images were processed and features were counted using Fiji. We created a region of interest (ROI) in the center of retinas in which to count cellular defects and used the Cell Counter tool to count defects within the area. In addition, we counted the total number of edges, vertices, and ommatidia within the ROI. The following defects were normalized to the number of edges: (1) a missing LC, defined as any edge with only one bristle and one LC, or two LCs and no bristles; (2) a separation, defined as any edge along which 1° cells from two different ommatidia were in direct contact; and (3) an extra LC, defined as any edge with more than three LCs, or two bristles and more than two LCs. The following defects were normalized to the number of vertices: (1) a rosette, defined as a bristle with four LCs contacting it instead of the normal three; (2) a missing 3° LC, defined as a vertex at which a 3° LC was missing and where a contact between multiple secondary cells was formed; and (3) a misplaced bristle, defined as an edge with a bristle in the middle. Statistical differences between conditions were examined using χ2 test. To determine the autonomy of sdk function, we selected rosettes for analysis. Each rosette was scored for how many of the four or more LCs contacting the bristle were sdk mutant and how many were WT.
Drug inhibition
Latrunculin A, a toxin that binds actin monomers and inhibits F-actin polymerization, was dissolved in DMSO and used at a concentration of 2 and 10 μM for 30 min at 25°C in M3 insect medium to treat dissected pupal eyes. Control eyes were treated with DMSO only. A 2-μM Latrunculin A treatment was sufficient to disassemble the medioapical F-actin network, while a 10-μM treatment was sufficient to disassemble both the medioapical and the junctional networks. The Rok inhibitor Y-27632 was dissolved in deionized water and used to treat wing imaginal discs and pupal eyes at a concentration of 10 μM for 1 h at 25°C in M3 insect medium. Control eyes were treated with deionized water only.
Method detail
Immunofluorescence
White prepupae (0 h APF) were selected and aged on glass slides in a humidifying chamber at 25°C. Pupal eyes were dissected in PBS, fixed for 35 min in 4% PFA in PBS, and stained with antibodies in PBS with 3% BSA, 0.3% Triton X-100, and 0.01% sodium azide. Primary antibodies used were rat anti-E-cad (DSHB #DCAD2, 1:100), mouse anti-SCAR (DSHB #P1C1, 1:100), guinea pig anti-Sqh1P (gift of R. Ward; 1:100), guinea pig anti-Sdk (Astigarraga et al., 2018; 1:300), and mouse anti-Pyd (DSHB, PYD2, 1:200). Secondary antibodies conjugated with Alexa Fluor 405 (Calbiochem), Alexa Fluor 488, Alexa Fluor 647 (Molecular Probes), and Cy3 and Cy5 (Jackson ImmunoResearch) were used at 1:150. Actin was stained with Actin-stain 488 and Actin-stain 555 (Cytoskeleton, #PHDG1 and #PHDR1) at 1:150.
Sample preparation for live imaging
Pupae were selected and aged as described above. Pupae expressing UAS-Rac.N17 and UAS-MLCK.ct were aged at 18°C to maintain epithelial integrity and then shifted to 25°C at the pupal stage to increase transgene expression. Image data were collected on an LSM800 laser scanning confocal microscope. Live imaging of pupal eyes was conducted as previously described (Del Signore et al., 2018). Briefly, before imaging, the operculum and surrounding pupal case were peeled carefully to expose the eyes. Intact pupae were inserted in a slit created in an agarose block with eyes facing the coverslip. The agarose block was fitted with a custom-made rubber gasket and capped with custom-built humidified chamber.
Confocal imaging
For analysis of epithelial remodeling and protein dynamics, an image stack was obtained every 1 min unless otherwise noted with optimal pinhole using a 63×, 1.4-NA, plan Apochromat oil-immersion objective, 0.7 μm per optical section with a 10–50% overlap between sections, at a scan speed of 7, averaging of 1, with an overall pixel dwell time of ∼1 s.
Quantification and statistical analysis
Analysis of contact dynamics
We examined contact dynamics at ∼28-h APF at the end of the pruning stage, when some, but not all, ommatidia edges had two or more 2° cells. Individual LC–LC contacts along horizontal edges were traced manually at 1-min intervals for 60 min using a Fiji plugin with a line segment selection producing 40–60 ROIs per contact. A custom Fiji Junction Analyzer script measured the length of each ROI. Then, a Python script operating on the extracted length values calculated the features of the pulsing. The data were smoothed using a Gaussian filter with σ = 2. Pulse amplitudes were taken using the absolute value of the difference between adjacent local maxima and minima. This value was divided by the average contact length to get the normalized amplitude. Pulse frequencies were taken using the number of minutes between maxima. Values for a given contact were averaged to produce an average amplitude or frequency per contact. Statistics were performed on average amplitudes and frequencies per contact using a Kruskal–Wallis test with Dunn’s multiple comparison test in Prism (v8.3.1).
Time-shifted Pearson’s correlation analysis
Contact dynamics were analyzed by manual tracing with a line segment selection (width = 5 pixels). Vertices were automatically placed at both ends of these line segments as circular ROIs (diameter = 6 pixels). Contact length, pixel mean and total intensity, and background for each channel at each time point were collected using a custom Junction Analyzer Fiji macro. For Lifeact::Ruby Rok::GFP eyes, analysis was based on the mean intensity over the full contact length, and for Lifeact::Ruby Sdk::GFP, Utr::mCh Sdk::GFP, Pyd::GFP Lifeact::Ruby, and Pyd::GFP mCh::Abi eyes, it was based on mean intensity at the vertices. Time-resolved Pearson’s cross-correlations (time windows from ±19 min) were compared between the mean fluorescent marker intensities and contact length using 40–60-min-long videos and calculated using a custom Python script along with packages NumPy and SciPy. Data presented are the average Pearson’s correlations of individual contacts as noted in figure legends and are presented as the mean correlation (R) ± SD. To calculate whether average Pearson r values and time shifts were significantly different from 0, a one-sample t test was used. To calculate whether the peak average r value differed from the average r value at shift of 0, a paired t test was used. For comparisons between groups, ANOVA with post hoc Dunnett’s multiple comparison tests were used to analyze normally distributed data for pulse amplitude. A Kruskal–Wallis test with post-hoc Dunn’s test was used to analyze non–normally distributed data for pulse amplitude, contact length, and cross correlations. For comparisons of peak shifts of length–Pyd correlations between genotypes, the highest local maximum correlation with a time shift within the range of ±10 min was selected. Contacts that did not have a local maximum within this range of time shifts were excluded from the analysis.
Analysis of F-actin and Rok dynamics during fluctuations of contact length
Kymographs of individual contacts were made to verify whether each contact was pulsing. Each contact judged to be pulsing was traced with the line segment selection (width = 15 pixels) at its most expanded and contracted points to measure the mean signal intensity for F-actin and Rok along the contact. For each contact, the mean signal intensity at each maximum was divided by the mean signal intensity at each adjacent minimum to produce a ratio, for a total of two to four ratios per contact. All ratios for a given contact were averaged to create an average ratio for that contact. Average ratios for contacts across different genotypes were compared using a one-way ANOVA with Dunn’s multiple comparison test.
Analysis of Sdk::GFP and Pyd::GFP dynamics during fluctuations of contact length
The Make Montage tool in Fiji was used to generate a kymograph of single contacts that exhibit pulsing behavior. For these contacts, we identified the time points for each pulse at which they were longest and shortest. For Sdk::GFP, we collected data from these time points, as well as from time points 2 min before and after. In the case of minima, for some contacts we selected time points >2 min before and after, selecting only time points at which the tricellular junctions were visibly separated. For Pyd::GFP, we collected data averaged over three time points immediately before and immediately after each extremum. At each time point, we used two circular ROIs (diameter = 5 pixels for Sdk, 9 pixels for Pyd) to measure mean pixel intensity at tricellular junctions and averaged them to generate a mean tricellular junction intensity score. Comparisons between Sdk and Pyd intensities were paired by pulse to avoid effects of pooling eyes with different measured intensities due to biological variation between samples and differences in sample preparation and laser intensities.
Comparing signal intensities between WT and mutant cell clones
LC–LC contacts were selected for analysis if all adjacent cells were mutant or, for the control group, if all adjacent cells were WT. At each contact, circular ROIs were used to measure the signal intensity at a tricellular junction and the center of the bicellular junction between LCs, or the junction between one LC and an adjacent primary cell in Pyd stains. We also took a background measurement at the center of an adjacent LC. In addition, a linear ROI with a width of 5 pixels was used to measure the signal intensity along the entire contact, including tricellular junctions. We calculated the ratio between the measured mean intensity at the tricellular junction and the center of the contact. We compared the results using a t test for normally distributed data and a Mann–Whitney U test for non–normally distributed data. Additionally, we subtracted the background mean intensity from the full-contact mean intensity and compared the results using a Mann–Whitney U test.
Analysis of protein colocalization
Projections of image stacks were converted to binary images using the threshold function for each channel. The threshold levels were initially selected using the JACoP plugin in Fiji, and in some cases manually adjusted to include labeled objects and exclude nonspecific background (Bolte and Cordelières, 2006). Pearson’s coefficient and Manders’ colocalization coefficients M1 and M2 were then calculated using JACoP. To determine the specificity of the colocalization for each pair, we used Van Steensel’s cross-correlation function, which plots Pearson’s correlation coefficient between two channels (on the y axis) as a function displacement of one image relative to the other images (on the x axis). For all the comparisons, the correlation decreased to <0.1 with image displacement, suggesting that each correlation reflected a specific colocalization of the proteins. The cone cells were masked and not included in the analysis.
Protein–protein interaction studies
For GST pulldown assays, WT Sdk ICD (amino acids 2,025–2,224) and mutant ICD lacking the last 6 C-terminal amino acids (amino acids 2,025–2,220) were cloned into a pGEX-4T-1 vector in frame with the GST protein and the thrombin restriction endonuclease site as previously described (Astigarraga et al., 2018). Expression of GST proteins was induced in BL21(DE3) bacterial strain with 0.1 mM IPTG. Bacterial lysates expressing the GST protein only, GST-Sdk-ICD, and GST-Sdk-ICDΔCT were incubated with glutathione-Sepharose 4B beads (GE Life Science) to link the bait protein to the beads. To prepare the lysates, S2 cells were lysed on ice in 400 μl NET buffer (400 mM NaCl, 5 mM EDTA, 50 mM Tris buffer, pH 7.5, and 1% NP-40) supplemented with 1× protein inhibitors cocktail (oComplete; Roche) and 1 mM PMSF for 30 min with vigorous pipetting every 5 min. Lysates were then incubated with glutathione-Sepharose 4B beads alone to clear the lysates. Beads containing equal amounts of GST proteins were then added to the cleared lysates, and the samples were incubated on a nutator for 90 min. Bound beads were washed three times with 1 ml NET buffer, and bound proteins were eluted in 100 μl of 2× sample buffer and processed for Western blot analysis using mouse anti-SCAR antibodies.
Online supplemental material
Fig. S1 shows exles of common cellular defects in sdk mutant eyes and HA::Sdk-expressing eyes. Fig. S2 shows the effects of drug inhibition by Latrunculin A and Y-27632 on Sdk::GFP localization in fixed tissue. Fig. S3 shows traces of Sdk::GFP levels at vertices relative to contact length in eyes expressing dominant-negative Rac1.N17 and constitutively active MLCK.ct, and paired comparisons of relative levels of Sdk::GFP at the different phases of contact pulsing compared with WT. Fig. S4 shows a trace of Pyd::GFP levels relative to contact length, a corresponding Pearson plot, and paired comparisons of Pyd::GFP levels at the different phases of contact pulsing. Table S1 lists the quantification of common cellular phenotypes in sdk mutants and HA::Sdk- and HA::SdkΔCT-expressing eyes compared with WT. Table S2 lists the time-shifted Pearson correlation analyses performed in this study. Video 1 shows cellular dynamics in WT compared with sdk mutant and HA::Sdk-expressing eyes and highlights contact pulsing and formation of new cell–cell contact after cell pruning. Video 2 shows trafficking defects in HA::Sdk-expressing eyes compared with WT. Video 3 shows F-actin and Rok dynamics in sdkΔ15 mutant and HA::Sdk-expressing eyes compared with WT. Video 4 shows Sdk::GFP dynamics relative to F-actin and MyoII in WT eyes. Video 5 shows Sdk::GFP pulsatile dynamics in Rac1.N17 and MLCK.ct-expressing eyes. Video 6 shows Pyd::GFP dynamics in a sdk mutant eye compared with WT.
Supplementary Material
shows quantification of common phenotypes in sdk mutant and HA::Sdk- and HA::SdkΔCT-expressing eyes compared with WT. The data shown correspond to the graphs shown in Figs. 1 G and 4, H and I.
shows time-shifted Pearson correlation analyses to determine temporal relationships between cytoskeletal effectors and the fluctuations in contact length. (Corresponds to Figs. 2, 6, and 7.) Statistics performed on time-shifted Pearson’s correlation data are displayed in Figs. 2, 6, and 7. Each row expresses the mean r value at the time shift nearest to 0 when the correlation between the two values is strongest, whether this mean differs significantly from a hypothetical mean of 0 (P value of one-sample t test), and whether the mean peak r value can be statistically determined to be stronger than the r value at a shift of 0 (P value of paired t tests). Additionally reported are the time shift of this peak and the mean of within-contact differences between the r value at this shift and the r value at a shift of 0 (paired t test).
is the source file for Fig. 4.
Acknowledgments
We thank A. Martin (Massachusetts Institute of Technology, Cambridge, MA), S. Bray (University of Cambridge, Cambridge, UK), and J. Zallen (Memorial Sloan Kettering Cancer Center, New York, NY) for generous gifts of flies; A. Carmena (University Miguel Hernández, Alicante, Spain) and R. Ward (Case Western Reserve University, Cleveland, OH) for generous gifts of antibodies; the Bloomington Stock Center, the Vienna Drosophila Research Center, and the Kyoto Stock Center for fly stocks; and the Developmental Studies Hybridoma Bank for antibodies. We also thank S. Del Signore for his contribution to the early stages of this project. We thank M. Llimargas and S. Del Signore for critical reading of the manuscript and K. G. Commons for editorial suggestions.
This work was supported by grants from the National Institutes of Health to J.E. Treisman (R01 EY025540 and R21 HD102660) and V. Hatini (R01 GM129151).
The authors declare no competing interests.
Author contributions: Conceptualization, V. Hatini; Formal Analysis, J. Malin and V. Hatini; Investigation, J. Malin, C. Rosa Birriel, and V. Hatini; Resources, J.E. Treisman, S. Astigarraga, J. Malin, C. Rosa Birriel, and V. Hatini; Writing—Original Draft, V. Hatini; Writing—Review and Editing, J. Malin, C. Rosa Birriel, J.E. Treisman, and V. Hatini; Supervision, V. Hatini; Funding Acquisition, J.E. Treisman and V. Hatini.
References
- Astigarraga, S., Douthit J., Tarnogorska D., Creamer M.S., Mano O., Clark D.A., Meinertzhagen I.A., and Treisman J.E.. 2018. Drosophila Sidekick is required in developing photoreceptors to enable visual motion detection. Development. 145:dev158246. 10.1242/dev.158246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen, H.J., Levis R.W., Liao G., He Y., Carlson J.W., Tsang G., Evans-Holm M., Hiesinger P.R., Schulze K.L., Rubin G.M., et al. 2004. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics. 167:761–781. 10.1534/genetics.104.026427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackie, L., Walther R.F., Staddon M.F., Banerjee S., and Pichaud F.. 2020. Cell-type-specific mechanical response and myosin dynamics during retinal lens development in Drosophila. Mol. Biol. Cell. 31:1355–1369. 10.1091/mbc.E19-09-0523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte, S., and Cordelières F.P.. 2006. A guided tour into subcellular colocalization analysis in lightmicroscopy. J. Microsc. 224:213–232. 10.1111/j.1365-2818.2006.01706.x [DOI] [PubMed] [Google Scholar]
- Cagan, R. 2009. Principles of Drosophila eye differentiation. Curr. Top. Dev. Biol. 89:115–135. 10.1016/S0070-2153(09)89005-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew, R.W. 2007. Pattern formation in the Drosophila eye. Curr. Opin. Genet. Dev. 17:309–313. 10.1016/j.gde.2007.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, E.H., Chavadimane Shivakumar P., Clement R., Laugier E., and Lenne P.F.. 2017. Patterned cortical tension mediated by N-cadherin controls cell geometric order in the Drosophila eye. Elife. 6:e22796. 10.7554/eLife.22796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charras, G., and Yap A.S.. 2018. Tensile forces and mechanotransduction at cell-cell junctions. Curr. Biol. 28:R445–R457. 10.1016/j.cub.2018.02.003 [DOI] [PubMed] [Google Scholar]
- Chen, B., Brinkmann K., Chen Z., Pak C.W., Liao Y., Shi S., Henry L., Grishin N.V., Bogdan S., and Rosen M.K.. 2014a. The WAVE regulatory complex links diverse receptors to the actin cytoskeleton. Cell. 156:195–207. 10.1016/j.cell.2013.11.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, B., Chou H.T., Brautigam C.A., Xing W., Yang S., Henry L., Doolittle L.K., Walz T., and Rosen M.K.. 2017. Rac1 GTPase activates the WAVE regulatory complex through two distinct binding sites. Elife. 6:e29795. 10.7554/eLife.29795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X.J., Squarr A.J., Stephan R., Chen B., Higgins T.E., Barry D.J., Martin M.C., Rosen M.K., Bogdan S., and Way M.. 2014b. Ena/VASP proteins cooperate with the WAVE complex to regulate the actin cytoskeleton. Dev. Cell. 30:569–584. 10.1016/j.devcel.2014.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z., Borek D., Padrick S.B., Gomez T.S., Metlagel Z., Ismail A.M., Umetani J., Billadeau D.D., Otwinowski Z., and Rosen M.K.. 2010. Structure and control of the actin regulatory WAVE complex. Nature. 468:533–538. 10.1038/nature09623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, W., Jung K.C., Nelson K.S., Bhat M.A., Beitel G.J., Peifer M., and Fanning A.S.. 2011. The single Drosophila ZO-1 protein Polychaetoid regulates embryonic morphogenesis in coordination with Canoe/afadin and Enabled. Mol. Biol. Cell. 22:2010–2030. 10.1091/mbc.E10-12-1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Signore, S.J., Cilla R., and Hatini V.. 2018. The WAVE regulatory complex and branched F-actin counterbalance contractile force to control cell shape and packing in the Drosophila eye. Dev. Cell. 44:471–483.e4. 10.1016/j.devcel.2017.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djiane, A., Shimizu H., Wilkin M., Mazleyrat S., Jennings M.D., Avis J., Bray S., and Baron M.. 2011. Su(dx) E3 ubiquitin ligase-dependent and -independent functions of polychaetoid, the Drosophila ZO-1 homologue. J. Cell Biol. 192:189–200. 10.1083/jcb.201007023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden, S., Rohatgi R., Podtelejnikov A.V., Mann M., and Kirschner M.W.. 2002. Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature. 418:790–793. 10.1038/nature00859 [DOI] [PubMed] [Google Scholar]
- Finegan, T.M., Hervieux N., Nestor-Bergmann A., Fletcher A.G., Blanchard G.B., and Sanson B.. 2019. The tricellular vertex-specific adhesion molecule Sidekick facilitates polarised cell intercalation during Drosophila axis extension. PLoS Biol. 17:e3000522. 10.1371/journal.pbio.3000522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy, A., Schenck A., Sahin H.B., Qurashi A., Sahel J.A., Diebold C., and Giangrande A.. 2011. CYFIP dependent actin remodeling controls specific aspects of Drosophila eye morphogenesis. Dev. Biol. 359:37–46. 10.1016/j.ydbio.2011.08.009 [DOI] [PubMed] [Google Scholar]
- Georgiou, M., Marinari E., Burden J., and Baum B.. 2008. Cdc42, Par6, and aPKC regulate Arp2/3-mediated endocytosis to control local adherens junction stability. Curr. Biol. 18:1631–1638. 10.1016/j.cub.2008.09.029 [DOI] [PubMed] [Google Scholar]
- Harris, K.P., and Tepass U.. 2008. Cdc42 and Par proteins stabilize dynamic adherens junctions in the Drosophila neuroectoderm through regulation of apical endocytosis. J. Cell Biol. 183:1129–1143. 10.1083/jcb.200807020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi, T., and Carthew R.W.. 2004. Surface mechanics mediate pattern formation in the developing retina. Nature. 431:647–652. 10.1038/nature02952 [DOI] [PubMed] [Google Scholar]
- Ismail, A.M., Padrick S.B., Chen B., Umetani J., and Rosen M.K.. 2009. The WAVE regulatory complex is inhibited. Nat. Struct. Mol. Biol. 16:561–563. 10.1038/nsmb.1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, R.I. 2021. Hexagonal patterning of the Drosophila eye. Dev. Biol. 478:173–182. 10.1016/j.ydbio.2021.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan, P., and Karess R.. 1997. Myosin light chain-activating phosphorylation sites are required for oogenesis in Drosophila. J. Cell Biol. 139:1805–1819. 10.1083/jcb.139.7.1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karess, R.E., Chang X.J., Edwards K.A., Kulkarni S., Aguilera I., and Kiehart D.P.. 1991. The regulatory light chain of nonmuscle myosin is encoded by spaghetti-squash, a gene required for cytokinesis in Drosophila. Cell. 65:1177–1189. 10.1016/0092-8674(91)90013-o [DOI] [PubMed] [Google Scholar]
- Kaufman, L., Yang G., Hayashi K., Ashby J.R., Huang L., Ross M.J., Klotman M.E., and Klotman P.E.. 2007. The homophilic adhesion molecule sidekick-1 contributes to augmented podocyte aggregation in HIV-associated nephropathy. FASEB J. 21:1367–1375. 10.1096/fj.06-7191com [DOI] [PubMed] [Google Scholar]
- Kim, Y.S., Fritz J.L., Seneviratne A.K., and VanBerkum M.F.A.. 2002. Constitutively active myosin light chain kinase alters axon guidance decisions in Drosophila embryos. Dev. Biol. 249:367–381. 10.1006/dbio.2002.0768 [DOI] [PubMed] [Google Scholar]
- Koronakis, V., Hume P.J., Humphreys D., Liu T., Horning O., Jensen O.N., and McGhie E.J.. 2011. WAVE regulatory complex activation by cooperating GTPases Arf and Rac1. Proc. Natl. Acad. Sci. USA. 108:14449–14454. 10.1073/pnas.1107666108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnaswamy, A., Yamagata M., Duan X., Hong Y.K., and Sanes J.R.. 2015. Sidekick 2 directs formation of a retinal circuit that detects differential motion. Nature. 524:466–470. 10.1038/nature14682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebensohn, A.M., and Kirschner M.W.. 2009. Activation of the WAVE complex by coincident signals controls actin assembly. Mol. Cell. 36:512–524. 10.1016/j.molcel.2009.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuit, T., Lenne P.F., and Munro E.. 2011. Force generation, transmission, and integration during cell and tissue morphogenesis. Annu. Rev. Cell Dev. Biol. 27:157–184. 10.1146/annurev-cellbio-100109-104027 [DOI] [PubMed] [Google Scholar]
- Lee, T., and Luo L.. 2001. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 24:251–254. 10.1016/s0166-2236(00)01791-4 [DOI] [PubMed] [Google Scholar]
- Leibfried, A., Fricke R., Morgan M.J., Bogdan S., and Bellaiche Y.. 2008. Drosophila Cip4 and WASp define a branch of the Cdc42-Par6-aPKC pathway regulating E-cadherin endocytosis. Curr. Biol. 18:1639–1648. 10.1016/j.cub.2008.09.063 [DOI] [PubMed] [Google Scholar]
- Letizia, A., He D., Astigarraga S., Colombelli J., Hatini V., Llimargas M., and Treisman J.E.. 2019. Sidekick is a key component of tricellular adherens junctions that acts to resolve cell rearrangements. Dev. Cell. 50:313–326.e5. 10.1016/j.devcel.2019.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levayer, R., and Lecuit T.. 2012. Biomechanical regulation of contractility: spatial control and dynamics. Trends Cell Biol. 22:61–81. 10.1016/j.tcb.2011.10.001 [DOI] [PubMed] [Google Scholar]
- Manning, L.A., Perez-Vale K.Z., Schaefer K.N., Sewell M.T., and Peifer M.. 2019. The Drosophila Afadin and ZO-1 homologues Canoe and Polychaetoid act in parallel to maintain epithelial integrity when challenged by adherens junction remodeling. Mol. Biol. Cell. 30:1938–1960. 10.1091/mbc.E19-04-0209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza, M.C. 2013. Phosphoregulation of the WAVE regulatory complex and signal integration. Semin. Cell Dev. Biol. 24:272–279. 10.1016/j.semcdb.2013.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, D.N., Liu Y., Litsky M.L., and Reinke R.. 1997. The sidekick gene, a member of the immunoglobulin superfamily, is required for pattern formation in the Drosophila eye. Development. 124:3303–3312. 10.1242/dev.124.17.3303 [DOI] [PubMed] [Google Scholar]
- Oikawa, T., Yamaguchi H., Itoh T., Kato M., Ijuin T., Yamazaki D., Suetsugu S., and Takenawa T.. 2004. PtdIns(3,4,5)P3 binding is necessary for WAVE2-induced formation of lamellipodia. Nat. Cell Biol. 6:420–426. 10.1038/ncb1125 [DOI] [PubMed] [Google Scholar]
- Pinheiro, D., and Bellaiche Y.. 2018. Mechanical force-driven adherens junction remodeling and epithelial dynamics. Dev. Cell. 47:3–19. 10.1016/j.devcel.2018.09.014 [DOI] [PubMed] [Google Scholar]
- Pollard, T.D., and Cooper J.A.. 1986. Actin and actin-binding proteins. A critical evaluation of mechanisms and functions. Annu. Rev. Biochem.. 55:987–1035. 10.1146/annurev.bi.55.070186.005011 [DOI] [PubMed] [Google Scholar]
- Pollitt, A.Y., and Insall R.H.. 2009. WASP and SCAR/WAVE proteins: the drivers of actin assembly. J. Cell Sci. 122:2575–2578. 10.1242/jcs.023879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer, J.K., Choi W., Jung K.C., He L., Harris N.J., and Peifer M.. 2011. A contractile actomyosin network linked to adherens junctions by Canoe/afadin helps drive convergent extension. Mol. Biol. Cell. 22:2491–2508. 10.1091/mbc.E11-05-0411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer, J.K., Harris N.J., Slep K.C., Gaul U., and Peifer M.. 2009. The Drosophila afadin homologue Canoe regulates linkage of the actin cytoskeleton to adherens junctions during apical constriction. J. Cell Biol. 186:57–73. 10.1083/jcb.200904001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaks, M., Singh S.P., Kage F., Thomason P., Klunemann T., Steffen A., Blankenfeldt W., Stradal T.E., Insall R.H., and Rottner K.. 2018. Distinct interaction sites of rac GTPase with WAVE regulatory complex have non-redundant functions in vivo. Curr. Biol. 28:3674–3684.e6. 10.1016/j.cub.2018.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell, C., Sabass B., Helmstaedter M., Geist F., Abed A., Yasuda-Yamahara M., Sigle A., Maier J.I., Grahammer F., Siegerist F., et al. 2018. ARP3 Controls the Podocyte Architecture at the Kidney Filtration Barrier. Dev Cell. 47:741–757. 10.1016/j.devcel.2018.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppa, M.J., Johnson R.I., Bao S., and Cagan R.L.. 2008. Polychaetoid controls patterning by modulating adhesion in the Drosophila pupal retina. Dev. Biol. 318:1–16. 10.1016/j.ydbio.2008.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajiri, R., Misaki K., Yonemura S., and Hayashi S.. 2011. Joint morphology in the insect leg: evolutionary history inferred from notch loss-of-function phenotypes in Drosophila. Development. 138:4621–4626. 10.1242/dev.067330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenawa, T., and Suetsugu S.. 2007. The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nat. Rev. Mol. Cell Biol. 8:37–48. 10.1038/nrm2069 [DOI] [PubMed] [Google Scholar]
- Uechi, H., and Kuranaga E.. 2019. The tricellular junction protein sidekick regulates vertex dynamics to promote bicellular junction extension. Dev. Cell. 50:327–338.e5. 10.1016/j.devcel.2019.06.017 [DOI] [PubMed] [Google Scholar]
- Venken, K.J.T., Schulze K.L., Haelterman N.A., Pan H., He Y., Evans-Holm M., Carlson J.W., Levis R.W., Spradling A.C., Hoskins R.A., and Bellen H.J.. 2011. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat. Methods. 8:737–743. 10.1038/nmeth.1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner, S.J., and Longmore G.D.. 2009a. Cdc42 antagonizes Rho1 activity at adherens junctions to limit epithelial cell apical tension. J. Cell Biol. 187:119–133. 10.1083/jcb.200906047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner, S.J., and Longmore G.D.. 2009b. Distinct functions for Rho1 in maintaining adherens junctions and apical tension in remodeling epithelia. J. Cell Biol. 185:1111–1125. 10.1083/jcb.200901029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernet, M.F., Labhart T., Baumann F., Mazzoni E.O., Pichaud F., and Desplan C.. 2003. Homothorax switches function of Drosophila photoreceptors from color to polarized light sensors. Cell. 115:267–279. 10.1016/s0092-8674(03)00848-1 [DOI] [PubMed] [Google Scholar]
- Xu, T., and Rubin G.M.. 1993. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 117:1223–1237. 10.1242/dev.117.4.1223 [DOI] [PubMed] [Google Scholar]
- Yamagata, M., and Sanes J.R.. 2010. Synaptic localization and function of Sidekick recognition molecules require MAGI scaffolding proteins. J. Neurosci. 30:3579–3588. 10.1523/JNEUROSCI.6319-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata, M., Weiner J.A., and Sanes J.R.. 2002. Sidekicks: synaptic adhesion molecules that promote lamina-specific connectivity in the retina. Cell. 110:649–660. 10.1016/s0092-8674(02)00910-8. [DOI] [PubMed] [Google Scholar]
- Yu, H.H., and Zallen J.A.. 2020. Abl and Canoe/Afadin mediate mechanotransduction at tricellular junctions. Science. 370:eaba5528. 10.1126/science.aba5528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallen, J.A., Cohen Y., Hudson A.M., Cooley L., Wieschaus E., and Schejter E.D.. 2002. SCAR is a primary regulator of Arp2/3-dependent morphological events in Drosophila. J. Cell Biol. 156:689–701. 10.1083/jcb.200109057 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows quantification of common phenotypes in sdk mutant and HA::Sdk- and HA::SdkΔCT-expressing eyes compared with WT. The data shown correspond to the graphs shown in Figs. 1 G and 4, H and I.
shows time-shifted Pearson correlation analyses to determine temporal relationships between cytoskeletal effectors and the fluctuations in contact length. (Corresponds to Figs. 2, 6, and 7.) Statistics performed on time-shifted Pearson’s correlation data are displayed in Figs. 2, 6, and 7. Each row expresses the mean r value at the time shift nearest to 0 when the correlation between the two values is strongest, whether this mean differs significantly from a hypothetical mean of 0 (P value of one-sample t test), and whether the mean peak r value can be statistically determined to be stronger than the r value at a shift of 0 (P value of paired t tests). Additionally reported are the time shift of this peak and the mean of within-contact differences between the r value at this shift and the r value at a shift of 0 (paired t test).
is the source file for Fig. 4.