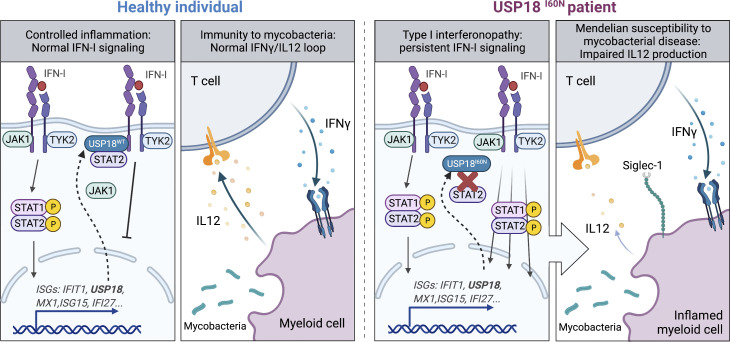
Martin-Fernandez et al. describe patients with partial USP18 deficiency, which underlies both type I interferonopathy and Mendelian susceptibility to mycobacterial disease (MSMD). This work delineates the lack of negative regulation of the IFN-I signaling pathway leading to depression of the IFN-γ–IL12 loop as a cause of MSMD.
Abstract
Human USP18 is an interferon (IFN)-stimulated gene product and a negative regulator of type I IFN (IFN-I) signaling. It also removes covalently linked ISG15 from proteins, in a process called deISGylation. In turn, ISG15 prevents USP18 from being degraded by the proteasome. Autosomal recessive complete USP18 deficiency is life-threatening in infancy owing to uncontrolled IFN-I–mediated autoinflammation. We report three Moroccan siblings with autoinflammation and mycobacterial disease who are homozygous for a new USP18 variant. We demonstrate that the mutant USP18 (p.I60N) is normally stabilized by ISG15 and efficient for deISGylation but interacts poorly with the receptor-anchoring STAT2 and is impaired in negative regulation of IFN-I signaling. We also show that IFN-γ–dependent induction of IL-12 and IL-23 is reduced owing to IFN-I–mediated impairment of myeloid cells to produce both cytokines. Thus, insufficient negative regulation of IFN-I signaling by USP18-I60N underlies a specific type I interferonopathy, which impairs IL-12 and IL-23 production by myeloid cells, thereby explaining predisposition to mycobacterial disease.
Graphical Abstract
Introduction
Type I IFNs (IFN-Is) play a central role in protective immunity against viral infections. However, overabundance or excessive duration of IFN-I signaling can be pathogenic, including the development of type I interferonopathies. These monogenic disorders cover a wide phenotypic clinical spectrum, varying from severe neurological symptoms and death to mild cutaneous disease, systemic inflammation, and asymptomatic intracranial calcifications (Rodero and Crow, 2016; Taft and Bogunovic, 2018). Despite differences in their clinical phenotypic features, all type I interferonopathies are characterized by persistent IFN-I–mediated inflammation and an ensuing increase in the expression of IFN-stimulated genes (ISGs), such as IFIT1, MX1, RSAD2, and IFI27 (Crow, 2011). Type I interferonopathies include Aicardi-Goutières syndrome, familial chilblain lupus, spondyloenchondrodysplasia, and some monogenic forms of systemic lupus erythematosus. In recent years, an increasing number of genetic disorders presenting with perturbations in the regulation of IFN-I responses have been discovered and included in the larger type I interferonopathy family (Tangye et al., 2020). These include proteasome associated autoinflammatory syndromes (Ohmura, 2019; Torrelo et al., 2010), Singleton-Merten syndrome and its atypical presentation (Jang et al., 2015; Rutsch et al., 2015; Pettersson et al., 2017), stimulator of IFN genes–associated vasculopathy with onset in infancy (Liu et al., 2014), STAT1 gain-of-function (GOF; Liu et al., 2011; Toubiana et al., 2016), JAK1 GOF (Gruber et al., 2020b), STAT2 GOF (Gruber et al., 2020a; Duncan et al., 2019; Kozlova et al., 2021), ISG15 deficiency (Bogunovic et al., 2012; Zhang et al., 2015; Hermann and Bogunovic, 2017; Martin-Fernandez et al., 2020), and ubiquitin-specific peptidase 18 (USP18) complete deficiency, presenting as pseudo-TORCH (toxoplasmosis, other [syphilis, varicella, mumps, parvovirus and HIV], rubella, cytomegalovirus, and herpes simplex) syndrome (Meuwissen et al., 2016; Alsohime et al., 2020).
Most of the deleterious mutations in type I interferonopathies occur in genes involved in the metabolism of nonself and self nucleic acids or in their recognition machinery (i.e., TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, ADAR1, and IFIH1), which result in the augmented production of IFN-I (Crow and Rehwinkel, 2009; Rice et al., 2013). Alternatively, STAT1 and JAK1 GOF deficiencies result in enhanced cellular responses to various IFNs including IFN-Is. The disease-causing mechanism in ISG15 and USP18 deficiencies lies in a defective negative feedback control of IFN-I signaling. In response to IFN-I stimulation, USP18 is upregulated and is responsible for the dampening of this pathway to prevent overt IFN-I inflammation (Malakhova et al., 2006; Taft and Bogunovic, 2018). Intracellular ISG15, also an ISG, maintains adequate levels of this negative feedback regulator (Zhang et al., 2015). Therefore, in these deficiencies, the high levels of ISGs and the related symptoms are due to dysregulation of the response to IFN-I, as opposed to dysregulation of IFN-I induction, as is the case in other type I interferonopathies. ISG15-deficient patients also suffer from Mendelian susceptibility to mycobacterial disease (MSMD), owing to the lack of secreted ISG15, which triggers production of IFN-γ by T and natural killer (NK) cells (Bogunovic et al., 2012). In contrast, autosomal recessive (AR) complete USP18 deficiency, as described to date, is lethal at birth in the absence of adequate treatment, owing to strong IFN-I signaling, and has not been associated with susceptibility to infection (Meuwissen et al., 2016; Alsohime et al., 2020). Here we studied three patients from Morocco with type I interferonopathy and susceptibility to bacillus Calmette-Guerin (BCG) vaccine, mimicking AR ISG15 deficiency.
Results
Rare homozygous variant of USP18 in three patients with central nervous system inflammation and bacterial disease
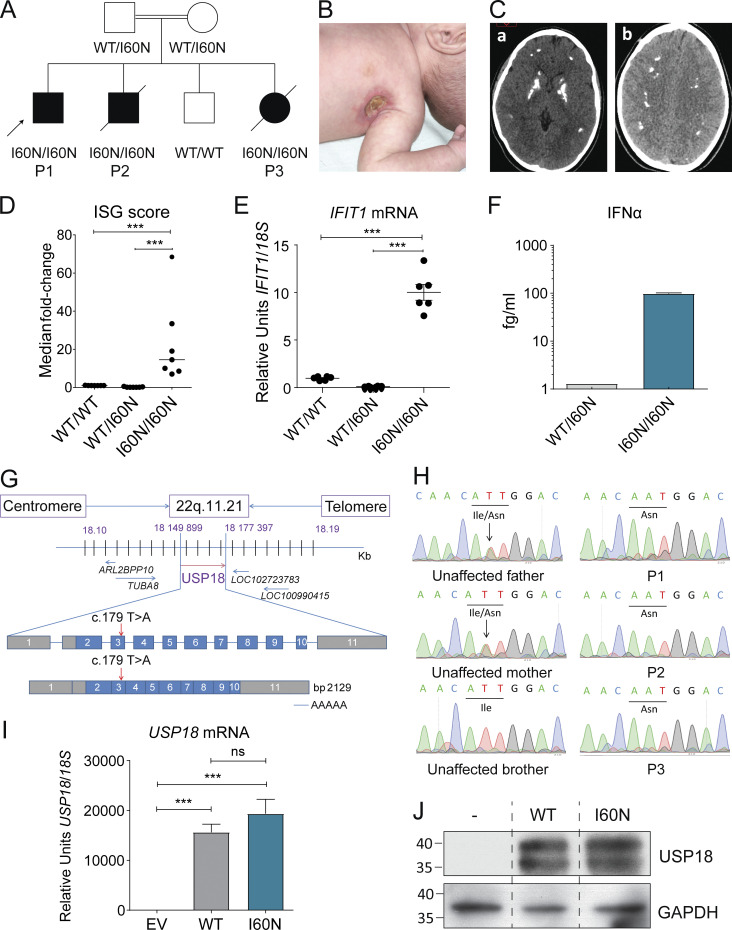
We studied a consanguineous family from Morocco with four children (Fig. 1 A). At ∼1 mo of age, three of these children (P1, P2, and P3) presented with localized fistulizing lymphadenopathies after BCG vaccination (BCG-itis; Fig. 1 B). P1 is currently 23 yr old and doing well. P2 died at the age of 4 mo of systemic inflammation after initial hospitalization for a staphylococcal infection. P3 had severe learning and motor disabilities and died at the age of 9 yr from excessive inflammation consequent to respiratory infection. Additionally, computed tomography scans of P1 revealed pronounced intracranial calcifications in the basal ganglia (Fig. 1 C). We assessed ex vivo mRNA levels of several ISGs (IFIT1, MX1, IFI27, RSAD2, IFIT2, and ISG15) in blood cells of P1. Accordingly, IFN score (and individual ISGs) was significantly higher in P1 compared with healthy controls (including a healthy sibling and mother; Fig 1, D and E; and Fig. S1, A–E), suggesting that the patient suffered from classic type I interferonopathy. The plasma level of IFN-α was significantly elevated in P1 compared with the mother (Fig. 1 F). Collectively, the three affected individuals in this family presented early with MSMD and later with severe systemic inflammation resulting in either compatibility with life or early death. Given these unusual clinical presentations, we performed whole-exome sequencing (WES) of these patients (P1, P2, and P3) and searched for candidate genetic variants based on the AR inheritance model given familial consanguinity (Table S1). The homozygosity rates inferred from WES were 5.2%, 4.3%, and 3.2% respectively (Fig. S1 F), confirming parental consanguinity. A homozygous variant, c.179T>A, was found in exon 3 of the USP18 gene, resulting in the substitution of the isoleucine at position 60 by an asparagine (p.I60N; Fig. 1 G). No other MSMD or type I interferonopathy causing allele variants was detected (Table S1). Sanger sequencing confirmed the homozygous mutation (c.179T>A/c.179T>A) in three patients (P1, P2, and P3) consistent with complete penetrance for both phenotypes (Fig. 1 H). This variant was also detected in cDNA from Epstein–Barr immortalized B lymphocytes (EBV-B) cells from P2 and human telomerase reverse transcriptase (hTERT)–immortalized fibroblasts of P1 (Fig. S1 G). Both parents are heterozygous and the healthy sibling is WT, indicating a familial segregation consistent with AR inheritance. These findings suggested that the patients had AR USP18 deficiency.
Figure 1.
Identification of a rare variant in exon 3 of USP18 in the offspring of a consanguineous family. (A) Familial segregation of the USP18-I60N allele. The kindred includes three affected siblings (P1, P2, and P3) and one unaffected (WT homozygous) sibling. (B) Axillary lymph node fistulation in P2. (C) Axial view of cerebral computed tomography scans of P1. Two images from the same patient are shown (a and b). (D) IFN score in the patient and controls, including healthy control (WT/WT) and the heterozygous mother (WT/I60N) calculated from the median fold-change in relative quantification value for a panel of six IFN-stimulated genes (Fig. 1 E and Fig. S1, A–E; Livingston et al., 2014). Each dot represents the mean of different replicates of each gene. Data analysis was performed with unpaired t tests. ***, P < 0.0001. (E) mRNA expression of a representative ISG (IFIT1) measured from whole-blood RNA isolated from WT healthy control (WT/WT), heterozygous mother (WT/I60N), or P1 (I60N/I60N) as assessed by quantitative RT-PCR. Bars represent the mean ± SD; each dot represents a replicate for each gene. Data analysis was performed with unpaired t tests. ***, P < 0.0001. (F) Quantification of circulating IFN-α by digital ELISA (SiMoA) in plasma from the heterozygous mother (WT/I60N) and P1 (I60N/I60N). Bars represent the mean ± SD. (G) Schematic localization of the USP18 variant in the genomic DNA and mRNA, with coding regions in blue, untranslated regions in gray, and the mutation indicated by red arrows. (H) Electropherogram showing the USP18 homozygous c.179T>A; p.I60N variant in P1, P2, and P3; and the heterozygous state in the unaffected parents. The unaffected brother shows WT sequence. (I) 293T cells were transfected with pTRIP expressing USP18: EV, WT USP18 (WT), or USP18 c.179T>A variant (I60N). Relative mRNA levels for USP18 assessed by quantitative RT-PCR and performed three times each with technical triplicates; representative experiment shown. Data analysis was performed with unpaired t tests. ns, P > 0.05; ***, P < 0.0001. (J) 293T cells were transfected with pTRIP expressing USP18: EV, WT USP18 (WT), or USP18 c.179T>A variant (I60N). Cell lysates were analyzed by Western blot for levels of USP18 protein; representative experiment shown. The molecular weight markers (in kD) are shown on the left. All results in the figure are representative of at least two independent experiments.
Figure S1.
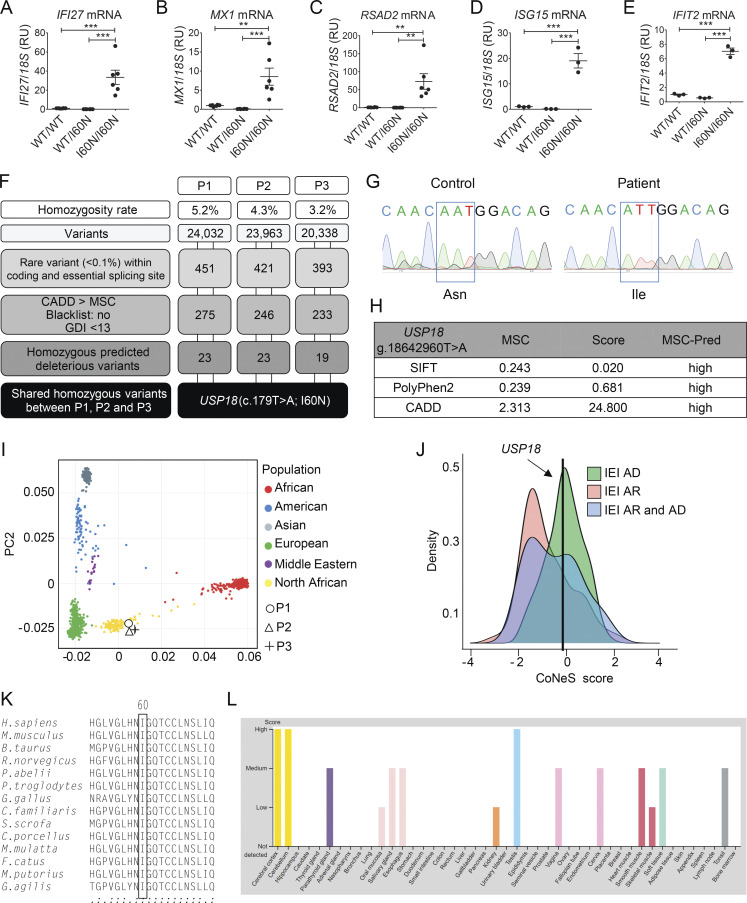
Characterization of the USP18 I60N allele. (A–E) Relative mRNA levels for IFI27, IFIT2, RSAD2, ISG15, and MX1 in peripheral blood from a healthy control (WT/WT), heterozygous mother (WT/I60N), or P1 (I60N/I60N) as assessed by quantitative RT-PCR. Bars represent the mean of two independent experiments; data analysis was performed with unpaired t tests. **, P < 0.001; ***, P < 0.0001. (F) Schematic demonstrating variant analysis pipeline and results from WES. GDI, gene damage index. (G) Electropherograms showing the WT USP18 sequence of control or homozygote c.179T>A, p.I60N mutation in P2 cDNA extracted from EBV-B cells. (H) Predictive (Pred) effects of I60N mutation determined by using PolyPhen-2, Sorting Intolerant from Tolerant (SIFT), and CADD scores. (I) Principal component analysis showing the origins of P1, P2, and P3 plotted on main ethnic origins extracted from the 1,000 Genomes database and our own WES database. (J) CoNeS (consensus negative selection) for USP18 and its distribution for genes causing inborn errors of immunity (IEI), according to disease mode of inheritance. AD, autosomal dominant. (K) Multiple sequence alignment of human USP18 and its orthologues. The Ile60 residue of human USP18 (top row) and the corresponding residues in the other species are boxed. (L) Levels of expression of USP18 in different tissues according to the Human Atlas website (https://www.proteinatlas.org/ENSG00000184979-USP18/tissue). RU, relative units.
The I60N USP18 variant is rare and hypomorphic
The I60N variant was absent in our in-house database of >15,000 exomes and in the The Greater Middle East Variomedatabase; however, it was found at a very low frequency of 3.98 × 10−6 in the gnomAD 2.1.1 database. In silico analysis revealed that the variant was predicted to be protein-damaging by Combined Annotation Dependent Depletion (CADD; score = 24.8), with a mutation significance cutoff (MSC) of 2.313 (Fig. S1 H). The ancestries of these patients were confirmed by principal component analysis (Fig. S1 I). The consensus negative selection score of USP18 was low, suggesting that the genetic variants in this gene are under negative selection (Fig. S1 J). Combined with the familial segregation, the rarity of the allele, the absence of other candidate genes, and the potential damaging impact of the substitution suggested that the c.179T>A variant could be a disease-causing mutation.
USP18 protein (372 aa) has the three-domain architecture of cysteine proteases with a typical catalytic triad (Basters et al., 2017). The Ile60 residue is highly conserved across species (Fig. S1 K). The expression of USP18 is variable across different tissues and organs (Fig. S1 L). To functionally characterize the I60N allele, we first compared levels of cloned USP18-WT and USP18-I60N transiently transfected in 293T cells. As shown in Fig. 1, I and J, the I60N substitution did not impair expression.
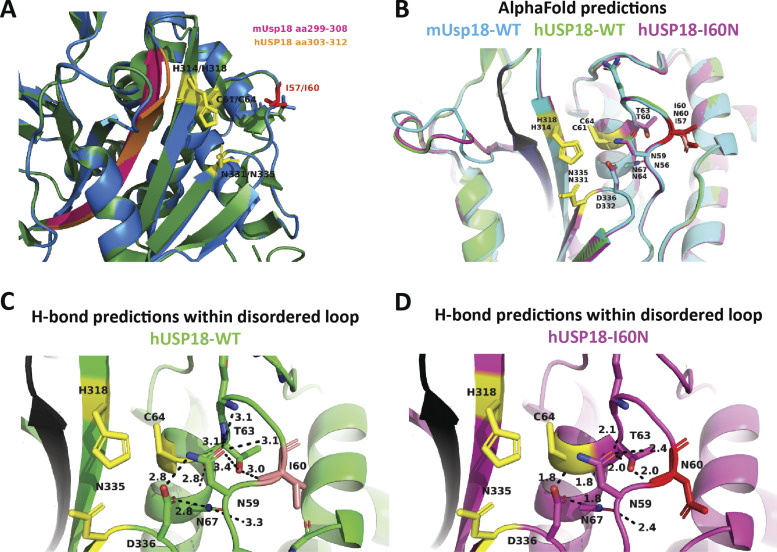
The crystal structure of the murine Usp18 (mUsp18) has been solved (Basters et al., 2017). I60 corresponds to I57 in mUsp18. This residue is located in a disordered loop above the catalytic triad (C61, H314, and N331; Fig. 2 A). All residues in this disordered loop are conserved between the human and murine orthologs. Of note, the side chain of I57 points toward an α-helix, while the side chains of the neighboring N56, G58, and Q59 residues orient toward the catalytic triad (Fig. 2 A).
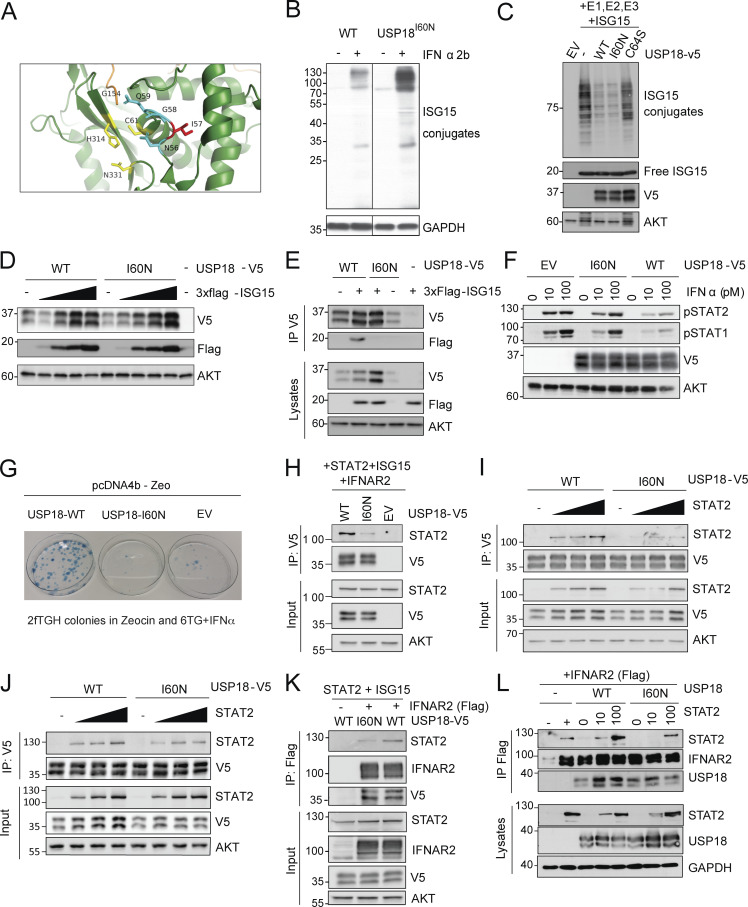
Figure 2.
Characterization of the enzymatic and regulatory functions of USP18 allele. (A) Close-up view of the catalytic cleft of mUsp18 (green, chain A) complexed to mIsg15 (orange; Basters et al., 2017; PDB entry: 5CHV). Residues of the catalytic triad (C61, H314, and N331) are shown in yellow. I57 (I60 in hUSP18) is located in a disordered loop above the catalytic triad. I57 is shown in red, and the neighboring residues of the loop (N56, G58, and Q59) in cyan. Of note, the side chain of I57 points toward an α-helix (aa 117–126), while the side chains of the neighboring N56, G58, and Q59 residues orient toward the catalytic triad. Figure made using PyMOL. (B) hTERT-immortalized fibroblasts from a control donor (WT) or P1 (USP18I60N) were treated with 1,000 IU/ml IFN-α2b for 24 h. Cell lysates were analyzed by Western blot with the indicated antibodies; representative experiment shown. (C) 293T cells were cotransfected with UbE1L (E1), UbCH8 (E2), HerC5 (E3), and 3xFlag-ISG15 in combination with USP18-V5 (WT), the c.179T>A variant (I60N), or the catalytically inactive mutant C64S. Cell lysates were analyzed by Western blot as indicated. ISG15 (conjugates and free) was detected with anti-ISG15 antibodies (a gift of E.J. Borden). In this and all panels below, USP18 was detected using V5 antibodies. (D) 293T cells were cotransfected with USP18-V5 WT or I60N and increasing amounts of 3xFlag-ISG15. Cell lysates were analyzed by Western blot as indicated. (E) 293T cells were cotransfected with USP18-V5 WT or I60N with 3xFlag-ISG15, as indicated. Immunoprecipitation (IP) of USP18 was performed using V5 antibodies. Immunoprecipitates were analyzed by Western blot. (F) 293T cells were transfected with EV, USP18-V5, WT, or I60N. Cells were then treated with the indicated doses of IFN-α2 for 20 min. Fresh lysates were analyzed by Western blot as indicated. (G) 2fTGH cells were transfected with empty pcDNA4b vector (EV), USP18-WT, or -I60N. Transfected cells were seeded in Zeocin-containing medium. 11 d later, fresh medium with Zeocin and 6TG plus IFN-α2b was added. Colonies were fixed and stained 7 d later. Only cells not responding to IFN, i.e., expressing functional USP18, survive. (H) 293T cells were cotransfected with USP18-V5, WT, or I60N or EV together with STAT2, IFNAR2-Flag, and ISG15. Immunoprecipitation of USP18 was performed using V5 antibodies. (I) 293T cells (p60 dish) were cotransfected with 650 ng of USP18-V5 WT or I60N and increasing amounts of STAT2 (210, 420, and 650 ng). 24 h after transfection, USP18 was immunoprecipitated using V5 antibodies. Immunoprecipitates were analyzed by Western blot with STAT2 and V5 antibodies. (J) Experiment as described in I, except that cells were harvested 48 h after transfection. In the immunoprecipitate, USP18 was revealed using USP18 antibodies. (K) 293T cells were cotransfected with USP18-V5, WT, or I60N together with STAT2, IFNAR2-Flag, and ISG15. Immunoprecipitation of IFNAR2 was performed using Flag antibodies. (L) 293T cells were cotransfected with USP18-V5, WT, or I60N together with IFNAR2-Flag and increasing amounts of STAT2 (10 and 100 ng). Immunoprecipitation of IFNAR2 (∼90–100 kD) was performed using Flag antibodies. All results in the figure are representative of at least two independent experiments. The molecular weight markers (kD) are shown on the left.
USP18 specifically cleaves (deISGylates) the small ubiquitin-like ISG15 from ISGylated proteins (Malakhov et al., 2002). Interestingly, patient-derived dermal fibroblasts that were stimulated with IFN exhibited higher levels of global ISGylation than control fibroblasts (Fig. 2 B). Because the components of the ISGylation machinery are all encoded by ISGs, the high ISGylation observed in patients’ cells could be due to lack of negative regulation. Alternatively, it could reflect a defect in deISGylation. To discriminate between these two possibilities, we compared the enzymatic activity of USP18-WT, USP18-I60N, and the catalytically dead USP18-C64S in 293T cells cotransfected with ISG15 and the ISGylation machinery (UbE1L, UbCH8, and HerC5). As shown in Fig. 2 C, the level of global ISGylation was comparable in cells expressing USP18-WT or USP18-I60N and lower than in cells expressing the C64S mutant. This result indicates that USP18-I60N is catalytically competent.
We previously showed that free ISG15 protects USP18 from SKP2-mediated proteasomal targeting (Zhang et al., 2015; Speer et al., 2016; Vuillier et al., 2019). Hence, in cells from ISG15-deficient patients, the level of IFN-induced USP18 is abnormally low but can be restored by ectopic ISG15 (Zhang et al., 2015; Vuillier et al., 2019). On this basis, we evaluated whether ISG15 stabilized USP18-I60N. For this, cells were cotransfected with USP18-WT or -I60N along with increasing amounts of ISG15, and protein levels were monitored. As shown in Fig. 2 D, USP18-WT and -I60N were both stabilized by ISG15. Next, we used a coimmunoprecipitation assay to monitor noncovalent binding of USP18-WT and -I60N with ISG15. Importantly, as opposed to the WT protein, USP18-I60N did not complex with ISG15 (Fig. 2 E), confirming that USP18 stabilization does not require ISG15 binding (Vuillier et al., 2019).
Lastly, we assessed the ability of USP18-I60N to negatively regulate IFN-I signaling. First, we transiently expressed empty vector (EV), USP18-WT, or -I60N in 293T cells and measured STAT1/2 activation in response to a 30-min pulse of IFN-α2. The extent of STAT1/2 phosphorylation was strongly reduced only in cells expressing USP18-WT, suggesting impaired function of I60N (Fig. 2 F). The ability of USP18-I60N to negatively regulate IFN-I signaling was also evaluated in a stringent survival assay using 2fTGH cells (Pellegrini et al., 1989). These cells contain an IFN-I–inducible bacterial enzyme (guanine-phosphoribosyl transferase) and die in medium supplemented with 6-thioguanine and IFN-I. 2fTGH cells transfected with USP18-WT survived in this medium, because of efficient blockage of IFN-I signaling, whereas cells transfected with USP18-I60N or EV did not yield surviving colonies (Fig. 2 G).
Impaired regulation of IFN-I signaling by the complex USP18-I60N/IFNAR2/STAT2
STAT2 is known to contribute to the negative regulation of IFN-I signaling through recruitment of USP18 to the receptor subunit IFNAR2 (Arimoto et al., 2017). USP18 is a highly flexible protein and adopts distinct conformational changes, as shown upon binding to ISG15 (Basters et al., 2017). The N- and C-terminal regions of USP18 are involved in the interaction with IFNAR2, and two adjacent regions are critical for binding to STAT2 (Arimoto et al., 2017). One such conserved region (aa 303–312 in human (h)USP18; aa 299–308 in mUsp18) folds in a β-sheet, which is located on the opposite side of the catalytic cleft with respect to the disordered loop containing I60 (Figs. 2 A and S2 A). Structural alignment of AlphaFold-predicted mUsp18, hUSP18, and hUSP18-I60N proteins revealed high degree of similarity in the disordered loop containing I60 (Fig. S2 B). Interestingly, slight differences between hUSP18-WT and hUSP18-I60N were observed at the level of hydrogen bonds formed by residues around I60 (Fig. S2, C and D). Hence, it is plausible that changes in bond strength may decrease the flexibility of the variant protein and affect the β-sheet involved in STAT2 binding.
Figure S2.
Prediction modeling of human USP18-WT and I60N variant by SWISS-MODEL and AlphaFold. (A) Overlay of unbound mUsp18 (green) and predicted structure of hUSP18 (blue) modeled by SWISS-MODEL. Residues of the catalytic cleft are shown in yellow, I57/I60 highlighted in red, and residues of β-sheet essential for STAT2 binding highlighted in pink and orange for murine and human USP18, respectively. PDB entry: 5CHT. (B) Alignment of AlphaFold-predicted structures of mUsp18 (cyan), hUSP18 (green), and hUSP18-I60N (magenta) in non–ISG15-bound state. Residue numbering corresponds to human/murine residues in USP18. Residues of the catalytic cleft are shown in yellow. I60/I57 highlighted in salmon/red. Potential STAT2 binding β-sheet is conserved and highlighted in black. (C and D) Polar residue contact prediction within and around the disordered loop. H-bonds formed within the disordered loop of human USP18-WT (C) and USP18-I60N (D) set at a cutoff at 3.5 Å. Figure made using PyMOL.
We thus examined the interaction between USP18-I60N and STAT2 in 293T cells. First, USP18-WT or -I60N was cotransfected with STAT2. To somehow mimic native conditions, IFNAR2 and ISG15 were added as well. 24 h after transfection, USP18 was immunoprecipitated and STAT2 was analyzed by Western blot. The amount of STAT2 brought down by USP18-I60N was considerably less than the amount brought down by USP18-WT (Fig. 2 H). Next, we sought to evaluate whether binding of USP18-I60N could be reinstated by increasing the amount of STAT2. Thus, USP18 (WT or I60N) was cotransfected with increasing doses of STAT2 in the absence of IFNAR2 and ISG15. USP18 was immunoprecipitated and STAT2 analyzed 24 or 48 h after transfection. USP18-I60N was found to be less competent than USP18-WT in binding STAT2 at 24 h, even at a high STAT2 dose (Fig. 2 I), whereas this difference was largely lost in an independent experiment performed 48 h after transfection (Fig. 2 J).
Using a similar experimental setting as in Fig. 2 H, IFNAR2 was immunoprecipitated to measure the partners. We found that the levels of USP18-WT and -I60N bound to IFNAR2 were equivalent. STAT2 was found to be bound to IFNAR2 but clearly reduced in cells expressing USP18-I60N (Fig. 2 K). We performed similar experiments in a STAT2 dose-dependent manner in the absence of ISG15. The results show a defective interaction of STAT2 at the receptor level in presence of USP18-I60N. In agreement, when increasing doses of STAT2 were transfected, we detected interaction of STAT2 with IFNAR2 even in presence of USP18-I60N (Fig. 2 L), suggesting that accumulation of STAT2 ultimately can allow for the formation of the trimeric complex and a late negative regulation (albeit hypomorphic for I60N) of the IFN-I signaling cascade.
Overall, the above data demonstrate that the USP18-I60N protein is correctly expressed, is sustained by ISG15, and retains normal deISGylating activity but is hypomorphic in terms of its ability to negatively regulate IFN-I signaling, owing to its impaired binding to STAT2 and disturbed formation of the IFNAR2/STAT2/USP18 complex.
Enhanced IFN-I responses in fibroblasts of patient with I60N USP18 mutation
We monitored the level of ISG transcripts after continuous stimulation of patient fibroblasts with IFN-α2b. We observed normal peak responses (8 h) but persistent ISG transcripts at late time points (24 h; Fig. 3, A–D; and Fig. S3, A–D). We also monitored the basal and IFN-I–induced levels of three ISG transcripts after stimulating fibroblasts for 12 h with IFN-α2b, washing with PBS, and leaving to rest for 36 h to simulate the resolution of the IFN response. Normal basal ISG levels but decreased resolution of all analyzed ISGs was observed in P1 fibroblasts with respect to fibroblasts from the heterozygous mother, the healthy brother, or a control donor (Fig. S3, E–G). Levels of ISG proteins were higher in cells of P1 stimulated with IFN-I with respect to control cells (Fig. 3 E and Fig. S3, H and I). Since USP18 is itself an ISG, we also detected increased levels of USP18 in P1, which, evidently, is insufficient to maintain adequate negative regulation over this time period. We compared the expression levels of ISGs in P1 ISG15-deficient and complete USP18-deficient fibroblasts after 12-h IFN-α2b priming and a 36-h rest. Fibroblasts from ISG15-deficient patients are unable to properly control IFN-I response, and when primed with IFN-α2b, their ISG transcripts are more abundant than in control cells (Zhang et al., 2015). This increase is further enhanced in complete USP18-deficient fibroblasts, which in part explains the stark phenotypic difference. Interestingly, ISG levels in P1 fibroblasts were similar to those detected in complete USP18-deficient fibroblasts, and significantly higher than those in ISG15-deficient cells (Fig. 3, F–H). This observation was especially puzzling for P1, as he is currently 23 and attending college (his genotype-sharing siblings lived 4 mo and 9 yr, respectively), while all five completely USP18-deficient children died perinatally (Meuwissen et al., 2016). Because I60N-expressing cells and USP18-null cells respond similarly to a first IFN-I stimulation (priming), we hypothesized that the I60N allele provides some protection from subsequent IFN-I challenge. It is well established that IFN-I–induced USP18 desensitizes cells to a second challenge with IFN-I (Francois-Newton et al., 2012). We thus primed cells with IFN-α2b for 12 h followed by a resting period of 36 h, and then assessed STAT phosphorylation in response to a second short challenge with IFN-α2b. Unlike USP18-null cells, WT cells were refractory to the second IFN-α2b challenge in terms of STAT1 and STAT2 phosphorylation (Fig. 3 I). Importantly, P1 fibroblasts showed an intermediate or partially refractory phenotype, suggesting that over time, USP18 I60N protein accumulates to high levels, which allows partial protection from secondary challenge with IFN-α2b (Fig. 3 I). Similar results were obtained when we repeated experiments using different doses of IFN-α2b (Fig. S3 J). Combined, this set of experiments suggests that, unlike complete deficiency, P1 cells were capable of establishing partial protection from secondary IFN-I challenge. This difference likely underlies the vastly different clinical presentation of disease in comparison to complete USP18 deficiency.
Figure 3.
Accumulation of USP18 I60N protects from subsequent IFN challenges. (A–D) hTERT-immortalized fibroblasts from control donor (C1 WT/WT), P1 (I60N/I60N), heterozygous parent (WT/I60N), or healthy brother (C2 WT/WT) were treated with 1,000 IU/ml IFN-α2b for 8 or 24 h. Relative mRNA levels were assessed for IFIT1, MX1, RSAD2, and USP18 performed three times each with technical triplicates. The results are represented relative to 8 h as the peak of ISG induction. A representative experiment is shown. Data analysis was performed with unpaired t tests. ns, P > 0.05; *, P < 0.01; **, P < 0.001; ***, P < 0.0001. (E) hTert-immortalized fibroblasts from a control donor (Control) or P1 were treated with 1,000 IU/ml IFN-α2b for 24 h. Cell lysates were analyzed by Western blot for the indicated antibodies; representative experiment shown. (F–H) hTERT-immortalized fibroblasts from a control donor (C1 WT/WT), ISG15-deficient donor (ISG15−/−), USP18-deficient donor (USP18−/−), P1 (I60N/I60N), heterozygous parent (WT/I60N), or healthy brother (C2 WT/WT) treated with 1,000 IU/ml IFN-α2b for 12 h, washed with PBS, and left to rest for 36 h. Relative mRNA levels were assessed for RSAD2, IFIT1, and MX1, performed three times each with technical triplicates; representative experiment shown. Bars represent the mean ± SEM. Statistical analysis performed by one-way ANOVA. **, P < 0.001; ****, P <0.00001. (I) hTERT-immortalized fibroblasts from control donor (Control), USP8-deficient donor (USP18−/−), and P1 (USP18I60N) were primed with 1,000 IU/ml IFN-α2b for 12 h, washed, left to rest for 36 h, and restimulated with increasing amounts of IFN-α2b for 30 min. Cell lysates were analyzed by Western blot for the indicated antibodies; representative experiment shown. (J) hTert-immortalized fibroblasts from a control donor (Control) or P1 were mock-transduced (−) or transduced with Luc-RFP (Luc) or WT USP18 (WT), sorted, treated with the indicated doses of IFN-α2b for 12 h, washed with PBS, and left to rest for 36 h, after which relative mRNA levels were assessed for RSAD2, performed three times each with technical triplicates; representative experiment shown. Bars represent the mean ± SD. (K) hTert-immortalized fibroblasts from control donor (Control), P1 transduced with Luc-RFP (Patient-Luc), or P1 transduced with WT USP18 (Patient-WT) were primed with 1,000 IU/ml IFN-α2b for 12 h, washed, left to rest for 36 h, and restimulated with IFN-α2b for 20 min. Cell lysates were analyzed by Western blot with the indicated antibodies. All results in the figure are representative of at least two independent experiments. The molecular weight markers (kD) are shown on the left. RU, relative units.
Figure S3.
The I60N mutation leads to increased ISG mRNA and protein levels. (A–D) hTert-immortalized fibroblasts from control donor or P1 were treated with 1,000 IU/ml IFN-α2b for 8 or 24 h. Relative mRNA levels were assessed for IFIT1, MX1, RSAD2, and USP18, performed three times each with technical triplicates. A representative experiment is shown. Data analysis was performed with unpaired t tests. ns, P > 0.05; *, P < 0.01; **, P < 0.001; ***, P < 0.0001. (E–G) hTert-immortalized fibroblasts from control donor (C1 WT/WT), P1 (I60N/I60N), mother (WT/I60N), and healthy brother (C2 WT/WT) were treated with 1,000 IU/ml IFN-α2b for 12 h, washed, and left to rest for 36 h. Relative mRNA levels were assessed for IFIT1, MX1, and RSAD2, performed three times each with technical triplicates; representative experiment shown. Statistical analysis performed by one-way ANOVA. ***, P < 0.0001. (H and I) hTert-immortalized fibroblasts from a control donor (Control), P1 (I60N), or USP18-deficient patient (P2) were treated with IFN-α2b (100 pM) for the indicated times. Cell lysates were analyzed by Western blot for the indicated antibodies; representative experiment shown. (J) hTERT-immortalized fibroblasts from a control donor (Control), P1, or P1 lentivirally transduced with WT-USP18 were primed with IFN-α2b (500 pM) for 8 h, washed, left to rest for 36 h, and restimulated with the indicated doses of IFN-α2b for 20 min. Cell lysates were analyzed by Western blot with the indicated antibodies; a representative experiment is shown. (K) hTert-immortalized fibroblasts from a control donor (Control), P1, or P1 lentivirally transduced with WT-USP18 were treated with IFN-α2b (1,000 U/ml) for the indicated times. Cell lysates were analyzed by Western blot for the indicated antibodies; representative experiment shown. (L–N) hTert-immortalized fibroblasts from a control donor (Control) or P1 were mock-transduced (−) or transduced with Luc-RFP (Luc) or WT USP18 (WT), sorted, treated with the indicated doses of IFN-α2b for 12 h, washed with PBS, and left to rest for 36 h, after which relative mRNA levels were assessed for the genes indicated, performed three times each with technical triplicates; representative experiment shown. Bars represent the mean ± SD. (O) hTert-immortalized fibroblasts from Control untransduced (−) or USP18-deficient patient untransduced (−) or transduced with Luc or with USP18-WT or -I60N and sorted. Cells were treated with the indicated doses of IFN-α2b for 12 h, washed with PBS, and left to rest for 36 h, after which relative mRNA levels were assessed for MX1, performed three times each with technical triplicates; representative experiment shown. Bars represent the mean ± SD. R.U., relative units. All results are representative of at least two independent experiments. The molecular weight markers (kD) are shown on the left.
USP18 rescues the P1 cellular phenotype in vitro
Finally, we transduced control and P1 fibroblasts with lentiviral particles expressing luciferase (Luc) or USP18. USP18-WT transduction almost completely rescued the patient's phenotype, normalizing the ISGylation profile (Fig. S3 K) and ISG mRNA (RSAD2, MX1, and IFIT1; Fig. 3 J and Fig. S3, L–N). Additionally, cells transduced with USP18-WT (but not Luc) regained the ability to be desensitized to a secondary challenge with IFN (Fig. 3 K). To further confirm these results, we also transduced USP18-deficient fibroblasts with USP18-WT or -I60N lentiviral constructs. USP18-WT normalized the IFN-I response, as ISG mRNA levels were similar to control fibroblasts (Meuwissen et al., 2016). However, USP18-I60N failed to rescue USP18 deficiency and mimicked P1’s phenotype (Fig. S3 O). These reconstitution experiments indicate that the P1 USP18-I60N variant is responsible for the abnormal activation of IFN-I signaling and is causative of the patients’ IFN-I–mediated clinical presentations. The study of the individual alleles and patient cells established that the patients had a partial, as opposed to complete (loss-of-expression/loss-of-function), form of USP18 deficiency.
Mycobacterial disease is driven by defects in IFN-γ–mediated immunity
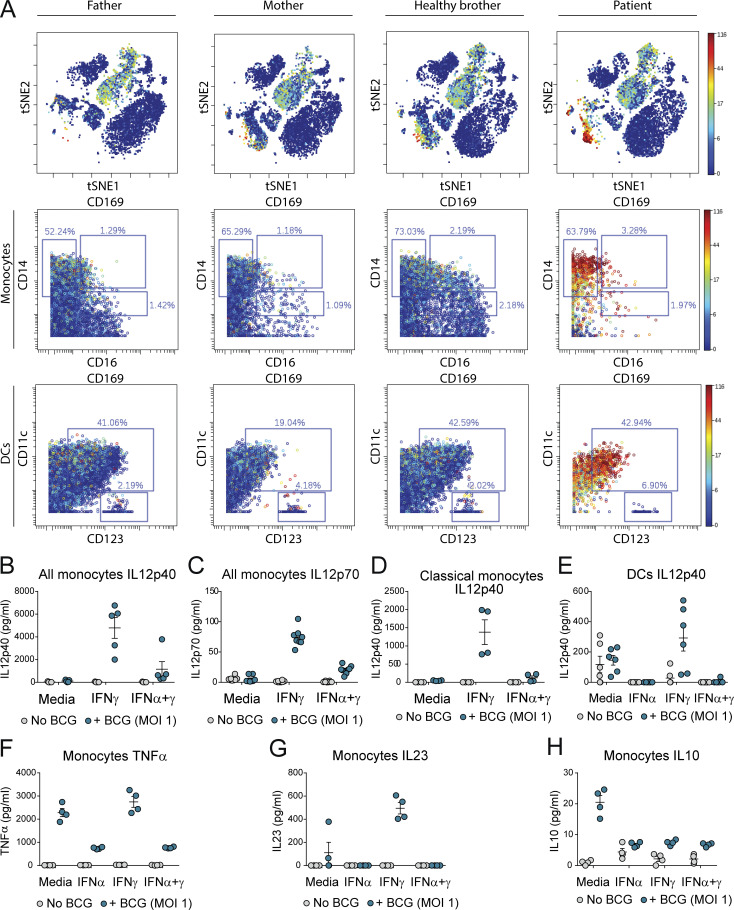
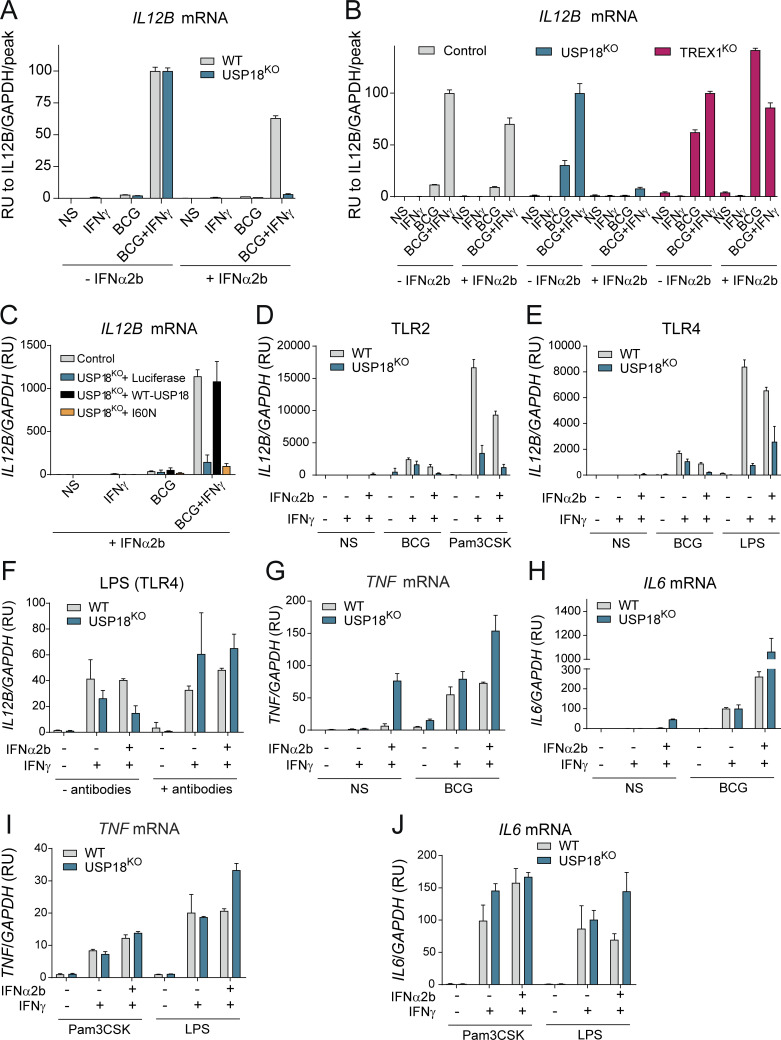
MSMD caused by BCG vaccination in ISG15-deficient patients results from a decrease in the production of IFN-γ by lymphocytes and NK cells (Bogunovic et al., 2012; Swaim et al., 2017). Given that the three patients with USP18-I60N mutations developed BCG-itis, we addressed the possibility of a defect in the IFN-γ axis. However, no impairment in the secretion of IFN-γ or IL-12 in whole blood activation was observed for P1 and P2 (Fig. S4, A–C). We also evaluated the ability of P1 CD4+ T cells to produce IL-22, IL17A, IL-17F, and IFN-γ in response to PMA/ionomycin and detected no defect compared with WT or heterozygous controls (Fig. S4, D–G). These findings are in line with normal secretion of ISG15 by P1 (Fig. S4 H). MSMD can also be due to mutations in Cytochrome B-245 β chain (CYBB) affecting macrophage-specific oxidative burst (Bustamante et al., 2011) or caused by mutations leading to loss of specific blood cell subtypes, like CDc1+ dendritic cells in IRF8 (Hambleton et al., 2011) and SPPL2A (Kong et al., 2018) deficiencies. The former possibility did not seem etiologic in USP18 I60N, as respiratory burst on neutrophils and monocytes was normal upon phorbol ester stimulation (Fig. S4 I). Similarly, stimulation of neutrophils from P1 with TNF, N-formyl-methionyl-leucyl-phenylalanine (fMLP), or their combination resulted in phenotypes comparable to controls (Fig. S4 J). We then assessed peripheral lymphoid and myeloid cells. We reviewed the complete blood counts of P1 and P2, which were both unremarkable (Table 1). In-depth immunophenotyping by conventional flow cytometry of P1 blood cells did not reveal abnormalities in the different subsets of T cells tested (Table 2). This was confirmed by cytometry by time of flight (CyTOF) analysis for all cell compartments (Table 3), with the exception of mucosal-associated invariant T cells (MAIT cells), which were absent from P1 peripheral blood mononuclear cells (PBMCs). MAIT cells are innate-like T cells defined by their semi-invariant αβ TCR. They are very sensitive to stimuli including IL-12, IL-18, and IFN-α and have the capacity to migrate to inflamed tissues. Patients with IL-12Rβ2, IL-23R, IL-12Rβ1, and ROR-γT deficiencies have a low frequency of MAIT cells (Martínez-Barricarte et al., 2018; Okada et al., 2015). We speculate that the reduction of circulating MAIT cells may result from the recruitment of these cells to the inflamed tissues. To eliminate the possible contribution of this cell type to the infectious phenotype, we performed an assay in which we depleted MAIT cells from healthy donor PBMCs and then stimulated them with BCG or BCG plus IL-12. IFN-γ production was not affected in the depleted sample. These results together with evidence from others (Okada et al., 2015) suggest that the absence of the MAIT subset observed in P1 PBMCs is not responsible for a mycobacterial disease (Fig. S4, K and L). Interestingly, CyTOF analysis revealed that the monocytic compartment (classic and nonclassic) and dendritic cells (CDc1+ and CD141+) specifically expressed higher levels of Siglec-1 (CD169), a marker of IFN-I response (Fig. 4 A). To test whether IFN-I–inflamed myeloid cells were functionally defective in terms of responsiveness to BCG or IFN-γ, we sorted monocytes (all three subtypes combined, or just classic CD14+CD16−) and CD11c+ dendritic cells from healthy controls, stimulated them with IFN-I, and then challenged them with BCG or BCG plus IFN-γ. Our results demonstrate that IFN-I priming reduces the ability of the tested myeloid cells to produce IL-12p40, IL-12p70, IL-23, and TNF, all necessary to control mycobacteria. Interestingly, this was not the case for IL-10 (Fig. 4, B–H). Furthermore, this appears to be transcriptionally controlled, as we detected reduced levels of IL12B and TNF mRNA (Fig. S4, M and N). To test this in a USP18-null inflammatory context, we used WT and USP18KO THP-1 cells. As detailed previously, cells were primed with IFN-I to create the inflammatory environment and then stimulated with BCG or BCG plus IFN-γ. We observed a more profound inhibition of IL-12 expression in the USP18KO primed cells compared with their WT isogenic controls (Fig. 5 A). In this scenario, we wanted to test why the high IFN-I levels in patients with other type I interferonopathies do not cause MSMD (i.e., AGS1-7), with the exception of ISG15 deficiency and recessive STAT2 GOF mutations (Bogunovic et al., 2012; Gruber et al., 2020a; Kozlova et al., 2021). To explain this, we tested TREXKO THP1 cells. TREX1-deficient patients develop type I interferonopathy caused by the accumulation of nucleic acids within the cytoplasm and the resulting overt IFN-I production (as opposed to inability to negatively regulate the IFN-α/β receptor (IFNAR), as in ISG15 and USP18 deficiencies, and STAT2 GOF). Our results showed no defect in the TREX1KO cells compared with WT cells (Fig. 5 B). This suggests that persistent IFN-I signaling results in suppression of IL-12 induction and the ensuing MSMD, while mere high IFN-I production and normal IFN-I negative regulation do not affect IL-12 production, and explains the lack of MSMD in these type I interferonopathies. Probably, MSMD in ISG15-deficient patients is thus due not only to the inability to produce IFN-γ (Bogunovic et al., 2012) but also to concomitant suppression of IL-12 production in some subsets of myeloid cells.
Figure S4.
Characterization of the MSMD phenotype caused by the USP18 I60N variant. (A–C) Cytokine production in the supernatants of whole-blood cells from local controls (n = 28), travel controls (n = 22), P1, P2, and mother, left unstimulated or stimulated with BCG alone or BCG plus cytokine (indicated), as detected by ELISA. (D–G) The ability to produce IL-22, IL-17A, IL-17F, and IFNγ by CD4+ T cells was assayed in Control and USP18 I60N patient. (H) Control and P1 fibroblasts were stimulated with 1,000 U/ml of IFN-α2b for 24 h. The presence of secreted ISG15 was analyzed by immunoblotting of the supernatant proteins. The molecular weight markers (kD) are shown on the left.(I) Dihydrorhodamine 123 (DHR) fluorescence was assayed using samples from healthy control and I60N patient neutrophils and monocytes treated with or without PMA. (J) DHR fluorescence was assayed using samples from healthy control, I60N patient, and CGD patient neutrophils treated with the indicated stimuli. MFI, mean fluorescence intensity. (K) Gating strategy for the quantification and identification of MAIT cells in PBMCs from father, mother, healthy brother, and P1 analyzed by CyTOF. (L) IFN-γ production was measured by ELISA in the supernatants of total PBMCs and MAIT-depleted PBMCs that were left unstimulated, stimulated with BCG alone or BCG plus IL-12, or stimulated with PMA/ionomycin. (M and N) Control monocytes were treated with 1,000 IU/ml IFN-α2b and 1,000 U/ml IFN-γ overnight, washed, and stimulated with BCG for 18 h, followed by measurement of mRNA for IL12B and TNF. All results are representative of at least two independent experiments. MOI, multiplicity of infection; RU, relative units.
Table 1.
Hemograms for USP18-deficient patients
| Component | P1 | Normal range | P2 | Normal range |
|---|---|---|---|---|
| Red cells (× 109/liter) | 4.5 | 4.5–4.9 | 3.89 | 3.9–4.6 |
| Hemoglobin (g/dl) | 11.9 | 13–14.5 | 9.30 | 11.5–12 |
| Hematocrit (%) | 35 | 37–43 | 29.40 | 34–37 |
| VGM (fl) | 79.7 | 78–88 | 75.60 | 75–81 |
| Leucocytes (× 109/liter) | 4.1 | 4.5–13.5 | 5.0 | 6–17 |
| Neutrophils (× 109/liter) | 1.0 | 1.8–8.0 | 2.90 | 1.5–8.5 |
| Eosinophils (× 109/liter) | 0.4 | 0–0.3 | 0 | 0–0.3 |
| Basophils (× 109/liter) | 0 | 0–0.2 | 0 | 0–0.2 |
| Lymphocytes (× 109/liter) | 2.1 | 1.2–5.2 | 1.40 | 2–8 |
| Monocytes (× 109/liter) | 0.6 | 0.2–1 | 0.60 | 0.2–1 |
| Platelets (× 109/liter) | 212 | 150–450 | 344 | 175–200 |
VGM, average globular volume.
Table 2.
Patient immunological phenotypes from the analysis of whole blood samples
| Phenotype | P1 | Normal range | P2 | Normal range |
|---|---|---|---|---|
| Lymphocytes/μl | 1,864 | 2,100 | ||
| T cells | ||||
| CD3+ | ||||
| % | 77 | 56–84 | 66 | 56–75 |
| /μl | 1,435 | 1,000–2,200 | 1,386 | 1,400–3,700 |
| CD4+ | ||||
| % | 26 | 31–52 | 32 | 28–47 |
| /mm3 | 485 | 530–1,300 | 672 | 700–2,200 |
| CD8+ | ||||
| % | 39 | 18–35 | 29 | 16–30 |
| /mm3 | 727 | 330–920 | 609 | 490–1,300 |
| CD45RA+/CD45+ (%) | 72 | 58–70 | 79 | 73–86 |
| CD31+CD45RA+/CD4+ (%) | 53 | 43–55 | 41 | 57–65 |
| CCR7+CD45RA+/CD8+ (%) | 65 | 52–68 | ND | |
| CCR7+CD45RA−/CD8+ (%) | 2 | 3–4 | ND | |
| CCR7−CD45RA−/CD8+ (%) | 12 | 11–20 | ND | |
| CCR7−CD45RA+/CD8+ (%) | 21 | 16–28 | ND | |
| B cells | ||||
| CD19+ | ||||
| % | 17 | 3.5–24 | 12 | 14–33 |
| /μl | 317 | 193–628 | 252 | 390–1,400 |
| CD27+/CD19+ (%) | 5 | 7–29 | ND | |
| CD27−IgD+/CD19+ (%) | 90 | 61.6–87.4 | ND | |
| CD27+IgD+/CD19+ (%) | 2 | 2.6–13.4 | ND | |
| CD27+IgD−/CD19+ (%) | 3 | 4–21.2 | ND | |
| CD24++CD38++CD27−IgD+/CD19+ (%) | 4 | 3.9–7.8 | ND | |
| CD24−CD38++/CD19+ (%) | 1 | 0.3–1.7 | ND | |
| CD21lowCD38low/CD19+ (%) | 2 | 0.9–3.3 | ND | |
| NK cells | ||||
| CD16+CD56+ | ||||
| % | 6 | 3–22 | 16 | |
| /μl | 112 | 70–480 | 336 |
ND, not done.
Table 3.
CyTOF profiling of P1 and his family members
| Percentage of total | Father | Mother | Brother | Patient | ||
|---|---|---|---|---|---|---|
| pDCs (calculated) | 0.46 | 1.05 | 0.37 | 0.77 | ||
| mDCs | CD1c+ | 0.46 | 0.75 | 0.52 | 0.33 | |
| CD141+ | 0.03 | 0.02 | 0.01 | 0.06 | ||
| Total | 2.63 | 1.30 | 0.96 | 0.98 | ||
| Monocytes | Mo1 | 9.71 | 14.42 | 12.15 | 6.82 | |
| Mo2 | 0.26 | 0.26 | 0.37 | 0.21 | ||
| Mo3 | 0.25 | 0.29 | 0.38 | 0.30 | ||
| NK cells | NK1 | 2.81 | 1.83 | 3.26 | 1.00 | |
| NK2 | 0.24 | 0.08 | 0.24 | 0.32 | ||
| NK3 | 24.32 | 28.33 | 20.60 | 13.87 | ||
| T cells | ||||||
| CD8+ | Naive | 3.97 | 12.04 | 12.27 | 20.82 | |
| EMRA | 2.73 | 2.36 | 9.06 | 9.38 | ||
| CM | 1.41 | 2.78 | 1.30 | 2.12 | ||
| EM | 10.08 | 4.66 | 6.65 | 7.38 | ||
| CD4+ | Naive | 16.73 | 16.88 | 15.86 | 20.93 | |
| EMRA | 2.61 | 1.80 | 2.26 | 2.05 | ||
| CM | 9.40 | 6.11 | 2.49 | 4.27 | ||
| EM | 4.85 | 2.31 | 2.15 | 2.06 | ||
| NKT cells | 0.00 | 0.14 | 0.11 | 0.05 | ||
| CD4+ | Th1 | 2.57 | 1.03 | 0.93 | 1.20 | |
| Th2 | 29.70 | 24.50 | 20.83 | 27.28 | ||
| Th17 | 0.86 | 1.22 | 0.83 | 0.51 | ||
| MAIT | 0.245 | 0.852 | 0.316 | 0.036 | ||
| ILCs | ILC1 | 0.032 | 0.077 | 0.076 | 0.045 | |
| ILC2 | 0.017 | 0.080 | 0.085 | 0.03 | ||
| ILC3 | 0.016 | 0.034 | 0.058 | 0.018 | ||
| Total | 0.026 | 0.111 | 0.132 | 0.027 | ||
CM, central memory; EM, effector memory; EMRA, effector memory cells re-expressing CD45RA; mDC, myeloid dendritic cell; pDC, plasmacytoid dendritic cell.
Figure 4.
MSMD phenotype caused by the USP18 I60N variant is due to impaired IL-12 production by monocytes. (A) t-distributed stochastic neighbor embedding (tSNE) plots obtained from CyTOF analysis of PBMCs from father, mother, healthy brother, and P1. CD169 (SIGLEC-1) expression is highlighted. Backgating of CD169 positive cells tSNE plots showing the localization of CD14+ cells. DCs, dendritic cells. (B–H) Control FACS-sorted monocytes (bulk or classic) or dendritic cells were treated with 1,000 IU/ml IFN-α2b and 1,000 IU/ml IFN-γ overnight, washed, and stimulated with BCG for 24 h, followed by measurement of IL-12p40 and other cytokines by ELISA. A representative figure of at least two independent experiments is shown. MOI, multiplicity of infection.
Figure 5.
Impairment in IL-12 production by monocytes in USP18 I60N–carrying patients is due to uncontrolled IFN-I inflammation. (A) WT or USP18KO THP1 cells were primed with 1,000 IU/ml IFN-α2b for 12 h and stimulated with BCG or BCG + IFN-γ. 24 h later, RNA was isolated, and relative IL12B mRNA levels were assessed. The results are calculated relative to the peak of induction (BCG + IFN-γ). A representative experiment is shown. (B) WT, USP18KO, or TREX1KO THP1 cells were primed with 1,000 IU/ml IFN-α2b for 12 h and stimulated with BCG or BCG + IFN-γ. 24 h later, RNA was isolated, and relative IL12B mRNA levels were assessed. The results are calculated relative to the peak of induction (BCG + IFN-γ). A representative experiment is shown. (C) USP18KO THP1 cells were transduced with Luc-RFP, WT USP18, or USP18 I60N, sorted, treated with 1,000 IU/ml IFN-α2b for 12 h, and stimulated with BCG or BCG + IFN-γ. 24 h later, RNA was isolated, and relative IL12B mRNA levels were assessed. A representative experiment is shown. (D) WT or USP18KO THP1 cells were primed with 1,000 IU/ml IFN-α2b for 12 h and stimulated with BCG or Pam3CSK, with or without IFN-γ. 24 h later, RNA was isolated, and the relative IL12B mRNA levels were assessed. A representative experiment is shown. (E) WT or USP18KO THP1 cells were primed with 1,000 IU/ml IFN-α2b for 12 h and stimulated with BCG or LPS, with or without IFN-γ. 24 h later, RNA was isolated, and relative IL12B mRNA levels were assessed. A representative experiment is shown. (F) WT or USP18KO THP1 cells were pretreated with blocking antibodies for IFNAR2 and IFN-β for 12 h, primed with 1,000 IU/ml IFN-α2b for 12 h, and stimulated with LPS or LPS + IFN-γ. 24 h later, RNA was isolated, and relative IL12B mRNA levels were assessed. A representative experiment is shown. (G) WT or USP18KO THP1 cells were primed with 1,000 IU/ml IFN-α2b for 12 h and stimulated with BCG or BCG + IFN-γ. 24 h later, RNA was isolated, and relative TNF mRNA levels were assessed. A representative experiment is shown. (H) WT or USP18KO THP1 cells were primed with 1,000 IU/ml IFN-α2b for 12 h and stimulated with BCG or BCG + IFN-γ. 24 h later, RNA was isolated, and relative IL6 mRNA levels were assessed. A representative experiment is shown. (I and J) WT or USP18KO THP1 cells were primed with 1,000 IU/ml IFN-α2b for 12 h and stimulated with Pam3CSK or LPS with or without IFN-γ. 24 h later, RNA was isolated, and the relative expression of the indicated genes were assessed. All results in the figure are representative of at least two independent experiments. RU, relative units.
To further confirm our results, we transduced the USP18KO THP1 with USP18-WT, USP18–I60N, or Luc as a control and tested these cells using our in vitro system. Priming with IFN-α2b induced a similar inhibition of IL-12 expression in the USP18KO cells transduced with USP18-I60N and the cells transduced with Luc. On the other hand, transduction of these cells with USP18-WT completely rescued their ability to express IL-12 in presence of IFN-α2b (Fig. 5 C).
BCG contains numerous pathogen-associated molecular patterns (PAMPs; i.e., TLR ligands) responsible for IL12 expression. We tested LPS (TLR4) and Pam3CSK (TLR2) in our in vitro system. USP18KO cells showed lower levels of expression of IL12B in presence of both LPS and Pam3CSK compared with WT (Fig. 5, D and E). As LPS can induce IFN-I, we hypothesized that this IFN inhibits the production of IL12B in absence of the exogenous one in USP18-null cells. Cells pretreated with blocking antibodies for both IFNAR2 and IFN-β restored IL12B expression to levels similar to the WT THP1 (Fig. 5 F), indicating the contribution of TLR4 (and likely other, untested pattern recognition receptors) in the impaired IL12B expression in USP18 mutated cells. Moreover, IL-12 is controlled by NF-κB (Liu et al., 2017). We did not detect inhibition of the expression of TNF or IL6 in cells primed with IFN-I (Fig. 5, G and H). We also tested this in cells stimulated with Pam3CSK or LPS and did not observe inhibition of TNF or IL6 expression upon IFN priming (Fig. 5, I and J). Combined, these data suggest that USP18’s partial deficiency impairs the ability of myeloid cells to produce IL-12/23, accounting for the immunodeficiency of these patients specific to BCG vaccination. The defect observed in the USP18 I60N patients is probably cell type specific and likely dominant in peripheral tissues, thus not readily observed in bulk blood assays, as was the case with patients carrying mutations for autosomal dominant (AD) IRF8, AR SPPL2A, or X-linked recessive CYBB-MSMD deficiency.
Discussion
Mendelian type I interferonopathies result from germline mutations in TMEM173, ADA2, TREX1, RNASEH2A/B/C, SAMHD1, ADAR1, IFIH1, DNASE2, DNASE1L3, ACP5, POLA1, and OAS1 and present with molecular features of persistently augmented ISGs due to dysregulation in the induction of IFN-I (Tangye et al., 2020). Patients with deficiency in ISG15 or USP18, or those with STAT2 GOF (Duncan et al., 2019; Gruber et al., 2020a), exhibit persistent elevated levels of ISGs at steady state in the absence of any infectious agent. The dysregulation is not in the induction of IFN-I cytokines, as in all other type I interferonopathy patients, but rather in the capacity to respond to IFN-I (Meuwissen et al., 2016; Zhang et al., 2015; Gruber et al., 2020a). AR complete USP18 deficiency causes perinatal death due to uncontrolled IFN-I–mediated inflammation (Meuwissen et al., 2016). Here we report a new USP18 variant (p.I60N) in three siblings from a consanguineous family from Morocco that represents the first AR partial USP18 deficiency. The USP18 I60N variant is normally expressed, can be stabilized by ISG15, and retains catalytic activity. However, the I60N variant is hypomorphic in its ability to suppress IFN-I signaling.
STAT2 was recently discovered to recruit USP18 to IFNAR2, where USP18 exerts its negative regulatory function (Arimoto et al., 2017). Patients with STAT2 GOF mutations clinically phenocopy USP18 deficiency owing to impaired STAT2-mediated recruitment of USP18 to IFNAR2 (R148Q) or improper interaction of STAT2 with USP18 (R148W; Gruber et al., 2020a; Duncan et al., 2019; Kozlova et al., 2021). Preliminary in silico prediction analyses suggest that the I60N variant may affect bond strength, possibly decreasing the flexibility of the protein and the interaction with STAT2. Experimentally, we show that USP18-I60N is partially impaired in binding STAT2. Therefore, we propose that type I interferonopathy in these patients is consequent to altered formation and/or stability of the essential negative regulatory complex. The hypomorphic nature of this allele and the persistent ISG induction allows accumulation of USP18-I60N, leading to compensation and partial rescue. Thus, these patients are partially protected from excessive IFN-I inflammation compared with previously described AR complete USP18-deficient patients (Meuwissen et al., 2016). Indeed, considering the cell type specificity and high plasticity features of IFN-I signaling, the extent of desensitization and its functional consequences are likely to vary among cell types. Basal expression of USP18 is variable across different tissues (Fig. S1 L). The presence of basal ganglia calcifications in one of patients suggests the augmented importance of the negative regulation in this tissue per se. Future studies are necessary to determine the pattern of the tissue specificity in USP18-I60N.
MSMD is normally caused by inborn errors of IFN-γ immunity (Bustamante et al., 2014). At times these germline errors can also result in specific cell subset defects such as CD1c+ dendritic cells or macrophages (Bustamante et al., 2011; Hambleton et al., 2011; Kong et al., 2018). MSMD patients usually have a major infectious episode followed by normal immunity to the same pathogen and absence of any persistent inflammation (Bustamante et al., 2014). On the other hand, patients with type I interferonopathies (hundreds of them) do not develop susceptibility to mycobacteria, despite a large number of them being BCG vaccinated (Crow, 2011; Rodero and Crow, 2016). AR complete ISG15 deficiency is an exception. ISG15-deficient patients have impaired production of IFN-γ by lymphocytes and NK cells (albeit mild) owing to lack of extracellular ISG15 and present with persistent IFN-I inflammation (also mild), due to lack of intracellular ISG15. Clinically, just like the vast majority of MSMD patients, individuals deficient in ISG15 presented with only one infectious episode after BCG vaccination. Their immunity to other bacteria (intracellular or otherwise) and viruses is intact (Bogunovic et al., 2012; Hermann and Bogunovic, 2017; Zhang et al., 2015; Martin-Fernandez et al., 2020). In fact, we recently postulated that, given the persistent mild IFN-I inflammation, ISG15-deficient individuals may present with enhanced control of viral infections, which we documented in vitro (Speer et al., 2016). The USP18 hypomorphic allele described here phenocopies ISG15 deficiency, as it causes type I interferonopathy and MSMD. Importantly, unlike ISG15 deficient patients, USP18-I60N individuals exhibit no defects in the IFN-γ/IL-12 loop in ex vivo whole blood assays. However the persistent IFN-I response, notably in myeloid cells, impairs their ability to produce IL-12 and IL-23 (Muglia Amancio et al., 2021). USP18-I60N leads to susceptibility to mycobacteria, not because this gene product intrinsically helps control mycobacterial infection, but rather because this allele cannot control the associated IFN-I inflammation. This suggests that the BCG-associated disease of ISG15-deficient individuals could be due to poor control of IFN-I (and not only lack of soluble ISG15 stimulating IFN-γ production), although this aspect remains to be tested.
Another question refers to the fact that patients with the same USP18-I60N genotype have lived for 4 mo and 9 yr or are currently doing well at 23 yr of age with no therapy. We postulate that differences in abnormal presence of IFN-I (quantity, frequency, or persistence) led to differential pathologic outcomes. While all patients received BCG vaccination at birth, giving rise to their uniform MSMD presentation, the different clinical outcomes may be due to secondary exposure to other common microbes. In P1 these instances were likely spaced apart and not overt at either instance. However, it is conceivable that P2 and P3 experienced more frequent and/or pronounced common childhood infections. This heterogenous clinical presentation of the same genetic defect is not uncommon in other Mendelian interferonopathies. While likely underestimating the in natura complexity, our experiments do suggest that the timing, magnitude, and duration of IFN-I–mediated insults are crucial.
In summary, the findings presented above provide genetic evidence of coexisting immune hyperactivity and immune deficiency. Here, in the instance of USP18 hypomorphic alleles, it appears that a mutation causing autoinflammation is the driver of susceptibility to infectious agents. Hence, as the study of human genetics suggests repeatedly, we must reconsider the conventional wisdom that autoinflammation and susceptibility to infection are opposite ends on the spectrum of immunity.
Materials and methods
Genomic DNA extraction and WES
Genomic DNA was isolated from the whole blood of P1 and P3 and from EBV-B cells from P2 with iPrep instruments from Thermo Fisher Scientific. 3 μg of DNA was used for generation of WES in the three patients (Table S1). The Agilent 50 Mb SureSelect exome kit was used in accordance with the manufacturer’s instructions. Burrows-Wheeler Aligner was used to align the reads with the human reference genome hg19, before recalibration and annotation with GATK, PICARD, and ANNOVAR. Filter of variants was achieved with in-house software. Mutation was verified by Sanger methods (PCR amplification conditions available upon request). PCR products were analyzed by electrophoresis in 1% agarose gels, sequenced with the V3.1 Big Dye terminator cycle sequencing kit, and analyzed on an ABI Prism 3700 machine (Applied Biosystems).
Cells
293T cells, U6A cells, and hTERT-immortalized dermal fibroblasts from P1 were cultured in DMEM (Gibco) supplemented with 10% FBS (Invitrogen), GlutaMAX (350 ng/ml; Gibco), and penicillin/streptomycin (Gibco). EBV-B cells from P2 and THP-1 cells were cultured in RPMI 1640 (Gibco) supplemented with 10% FBS (Invitrogen), GlutaMAX (350 ng/ml; Gibco), and penicillin/streptomycin (Gibco).
Patient and control fibroblasts were stably complemented with USP18-WT, USP18-I60N or Luc by lentiviral transduction. Transduced fibroblasts were sorted on the basis of RFP expression (BD FACS Aria II). USP18 CRISPR KO was performed by transfection of Alt-R S.p. Cas9 Nuclease V3 (IDT 1081058) and Alt-R crRNA gRNAs (IDT) into THP-1 cells with a 4D-Nucleofector (Lonza) using the manufacturer’s optimized protocol. Cells were cultured at 37°C and 10% CO2 and tested for mycoplasma contamination with the MycoAlert PLUS Mycoplasma Detection Kit (Lonza) according to the manufacturer’s instructions.
Plasmids and transfection
293T cells were transiently transfected with Lipofectamine 2000 (Invitrogen) or FuGENE HD (Promega). The variant was introduced using the QuikChange II XL site-directed mutagenesis kit using the pDONOR221-USP18 vector. Then the mutants were flipped into pTRIP expression vector using the Gateway LR Clonase II Enzyme mix. We also used the following constructs: pCAGGS-UbE1l, pCS2+-Herc5, pFlagCMV2-UbcH8, pTRIP-ISG15, pTRIP-STAT2, pMET-IFNAR2-Flag, pcDNA4B-hUSP18-V5, pcDNA4B-hUSP18-I60N-V5, pcDNA3-3xFlag-hISG15 (from J. Huibregtse, University of Texas at Austin, Austin, TX), pRc-CMV-hSTAT2 (from C. Schindler, Columbia University, New York, NY), and pcDNA3.1-hIFNAR2-Flag. In cotransfection experiments, EV, or salmon sperm DNA was added to keep total DNA amount constant.
RNA isolation and quantitative PCR
RNA was extracted from fibroblasts (Qiagen RNeasy) or whole blood (PAXgene Blood RNA Kit) and reverse-transcribed (ABI High Capacity Reverse Transcriptase) according to the instructions provided by the manufacturer. Levels of ISG expression (IFIT1, MX1, IFI27, RSAD2, IFIT2, and ISG15) and IL12B relative to the 18SRNA housekeeper gene were analyzed by Taqman quantitative real-time PCR (TaqMan Universal Master Mix II w/UNG, Roche LightCycler 480 II). Relative expression was calculated by the ΔΔCT method, with comparison with the mean value for mock-treated controls.
Protein analyses
For immunoblotting, cells were lysed in radioimmunoprecipitation assay buffer (Thermo Fisher Scientific), Dithiothreitol, and a protease/phosphatase inhibitor cocktail (Cell Signaling Technology). For coimmunoprecipitation assay, cells were lysed in 50 mM Tris, pH 6.8, 0.5% NP-40, 200 mM NaCl, 10% glycerol, 1 mM EDTA, and 1× protease/phosphatase inhibitor cocktail (Cell Signaling Technology). Cell lysates were incubated with Flag-M2 beads for 2 h at room temperature or with V5 antibodies for 2 h at 4°C, then incubated with Protein A Sepharose 4 Fast Flow and Protein G Sepharose 4 Fast Flow beads (GE Healthcare) for 1 h at 4°C. Immunoprecipitates were subjected to Western blotting. The antibodies used were directed against STAT1 (Santa Cruz Biotechnology), STAT2 (Millipore), phospho-Tyr 689 STAT2 (Millipore), ISG15 (Santa Cruz Biotechnology), mAb cl.2.1, a gift of E.J. Borden, Cleveland Clinic, Cleveland, OH), V5 (Invitrogen), and Flag (Sigma-Aldrich). Antibodies to phospho-Tyr 701 STAT1, USP18, β-Actin, IFIT1, and AKT were all from Cell Signaling Technology. The chemiluminescence detection reagent was Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific) or Western Lightning Plus-ECL (PerkinElmer).
Mass cytometry
Whole blood from P1, mother, father, and healthy brother were processed with Ficoll to collect the mononuclear cell layer. PBMCs were stained and analyzed by mass cytometry (CyTOF) at the Human Immune Monitoring Center of the Icahn School of Medicine at Mt. Sinai. The barcoded cells were combined and stained together with antibodies against selected surface markers for 30 min on ice. Cells were then washed, fixed, resuspended in diH2O containing EQ Four Element Calibration Beads (Fluidigm), and acquired on a CyTOF2 Mass Cytometer (Fluidigm). Data files were normalized by using a bead-based normalization algorithm (CyTOF software, Fluidigm) and de-barcoded using CD45 gating. The gated populations were visualized in lower dimensions using viSNE in Cytobank (https://www.cytobank.org/).
ELISA and Luminex
Plasma collected by Ficoll isolation from heparinized whole blood was clarified by centrifugation. Circulating cytokine levels were determined in magnetic Luminex assays with the Human XL Cyt Disc Premixed Mag Luminex (R&D) according to the manufacturer’s protocol. Samples were quantified on a MAGPIX xMAP Instrument (Luminex). ELISAs for IL12p40, IL12p70, and IL23 (BioLegend) were performed according to the manufacturer’s protocols.
Single-molecule array (SiMoA) digital ELISA
Plasma samples from the patient and healthy donors were isolated by Ficoll gradient and subsequently clarified by centrifugation at high speeds. IFN-α levels were then quantified by digital ELISA using the IFN-α SiMoA Assay Kit (100860; Quanterix) according to the manufacturer’s instructions on a SiMoA HD1 Analyzer.
Ethics statement
This study was conducted in accordance with the Helsinki Declaration, with written informed consent obtained from patients’ families. Experiments were conducted in France and the United States in accordance with local regulations and with the approval of institutional review board of The Rockefeller University, Institut National de la Santé et de la Recherche Médicale (INSERM), and Icahn School of Medicine at Mount Sinai. Icahn School of Medicine institutional review board has approved the study of human subjects (IF2349568).
Online supplemental material
Fig. S1 shows genetic characterization of the USP18 I60N allele. Fig. S2 shows prediction models of human USP18-WT and I60N variant by SWISS-MODEL and AlphaFold. Fig. S3 shows that the I60N mutation leads to increased ISG mRNA and protein levels. Fig. S4 shows results of the characterization of MSMD. Table S1 lists the homozygous rare variants found in WES analysis and shared by P1, P2, and/or P3.
Supplementary Material
lists the homozygous rare variants found in WES analysis and shared by P1, P2, and/or P3.
Acknowledgments
We wish to thank the family members for agreeing to participate in this study. We also thank Yelena Nemirovskaya, Dana Liu, Christine Rivalain, and Lazaro Lorenzo-Diaz for administrative assistance and all members of the laboratory of Human Genetics of Infectious Diseases for helpful discussions.
This research was in part supported by the Division of Intramural Research of the National Institute of Allergy and Infectious Diseases of the National Institutes of Health, the National Institute of Allergy and Infectious Diseases (grant numbers R01AI127372, R01AI148963, R01AI151029, 5R01AI089970, and 5R37AI095983), the National Center for Research Resources and the National Center for Advancing Sciences of the National Institutes of Health (grant number 8UL1TR000043), The Rockefeller University, the St. Giles Foundation, The Yale Center for Mendelian Genomics funded by the National Human Genome Research Institute (UM1HG006504), INSERM, University of Paris, the Integrative Biology of Emerging Infectious Diseases Laboratory of Excellence (ANR-10-LABX-62-IBEID) and the French National Research Agency (ANR) under the “Investments for the future” program (grant number ANR-10-IAHU-01), and GENMSMD (ANR-16-CE17-0005-01 for J. Bustamante). T. LeVoyer is supported by the MD-PhD program of the Imagine Institute with the support of the Bettancourt-Schueller Foundation. Work in Institut Pasteur was supported by grants from Association de la Recherche sur le Cancer (ARC no. 20141201864) and Fondation pour la Recherche Médicale (grant DEQ. 2017033741) and by institutional funding from Institut Pasteur and INSERM. Z. Li was supported by Centre national de la recherche scientifique. L.T. Dynesen was supported by the EU Erasmus+ program. L. Franklin was supported by la Fondation de France (Prix Thérèse Lebrasseur to S. Pellegrini). L. Malle and C. Gruber were supported by National Institute of Child Health and Human Development-Interdisciplinary Training in Systems and Developmental Biology and Birth Defects T32HD075735.
Author contributions: M. Martin-Fernandez designed and performed most of the experiments, analyzed the data, and wrote the manuscript. S. Buta, T. Le Voyer, Z. Li, F. Vuillier, L. Franklin, A. Muglia Amancio, L. Malle, C. Gruber, J. Altman, J. Taft, and L.T. Dynesen performed experiments or generated and analyzed data. F. Ailal, I. Benhsaien, C. Deswarte, M. Roynard, A. Nieto-Patlan, K. Moriya, J. Rosain, N. Boddaert, A. Bousfiha, Y.J. Crow, D. Jankovic, A. Sher, J.-L. Casanova, and S. Pellegrini provided patient samples, analyzed and/or interpreted data, and contributed to the writing of the paper. J. Bustamante and D. Bogunovic helped design the experiments and analyze data, supervised the work, and wrote the manuscript. All authors commented on the manuscript and approved its final version.
References
- Alsohime, F., Martin-Fernandez M., Temsah M.-H., Alabdulhafid M., Le Voyer T., Alghamdi M., Qiu X., Alotaibi N., Alkahtani A., Buta S., Jouanguy E., et al. 2020. JAK inhibitor therapy in a child with inherited USP18 deficiency. N. Engl. J. Med. 382:256–265. 10.1056/nejmoa1905633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimoto, K.-I., Löchte S., Stoner S.A., Burkart C., Zhang Y., Miyauchi S., Wilmes S., Fan J.-B., Heinisch J.J., Li Z., Yan M., et al. 2017. STAT2 is an essential adaptor in USP18-mediated suppression of type I interferon signaling. Nat. Struct. Mol. Biol. 24:279–289. 10.1038/nsmb.3378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basters, A., Geurink P.P., Röcker A., Witting K.F., Tadayon R., Hess S., Semrau M.S., Storici P., Ovaa H., Knobeloch K.-P., and Fritz G.. 2017. Structural basis of the specificity of USP18 toward ISG15. Nat. Struct. Mol. Biol. 24:270–278. 10.1038/nsmb.3371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogunovic, D., Byun M., Durfee L.A., Abhyankar A., Sanal O., Mansouri D., Salem S., Radovanovic I., V Grant A., Adimi P., Mansouri N., et al. 2012. Mycobacterial disease and impaired IFN-γ immunity in humans with inherited ISG15 deficiency. Science. 337:1684–1688. 10.1126/science.1224026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante, J., Arias A.A., Vogt G., Picard C., Galicia L.B., Prando C., V Grant A., Marchal C.C., Hubeau M., Chapgier A., de Beaucoudrey L., et al. 2011. Germline CYBB mutations that selectively affect macrophages in kindreds with X-linked predisposition to tuberculous mycobacterial disease. Nat. Immunol. 12:213–221. 10.1038/ni.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante, J., Boisson-Dupuis S., Abel L., and Casanova J.-L.. 2014. Mendelian susceptibility to mycobacterial disease: genetic, immunological, and clinical features of inborn errors of IFN-γ immunity. Semin. Immunol. 26:454–470. 10.1016/j.smim.2014.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow, Y.J. 2011. Type I interferonopathies: a novel set of inborn errors of immunity. Ann. NY. Acad. Sci. 1238:91–98. 10.1111/j.1749-6632.2011.06220.x [DOI] [PubMed] [Google Scholar]
- Crow, Y.J., and Rehwinkel J.. 2009. Aicardi-Goutieres syndrome and related phenotypes: linking nucleic acid metabolism with autoimmunity. Hum. Mol. Genet. 18:R130–R136. 10.1093/hmg/ddp293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan, C.J.A., Thompson B.J., Chen R., Rice G.I., Gothe F., Young D.F., Lovell S.C., Shuttleworth V.G., Brocklebank V., Corner B., Skelton A.J., et al. 2019. Severe type I interferonopathy and unrestrained interferon signaling due to a homozygous germline mutation in STAT2. Sci. Immunol. 4:eaav7501. 10.1126/sciimmunol.aav7501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francois-Newton, V., Livingstone M., Payelle-Brogard B., Uzé G., and Pellegrini S.. 2012. USP18 establishes the transcriptional and anti-proliferative interferon α/β differential. Biochem. J. 446:509–516. 10.1042/bj20120541 [DOI] [PubMed] [Google Scholar]
- Gruber, C., Martin-Fernandez M., Ailal F., Qiu X., Taft J., Altman J., Rosain J., Buta S., Bousfiha A., Casanova J.-L., Bustamante J., et al. 2020a. Homozygous STAT2 gain-of-function mutation by loss of USP18 activity in a patient with type I interferonopathy. J. Exp. Med. 217:e20192319. 10.1084/jem.20192319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber, C.N., Calis J.A., Buta S., Evrony G., Martin J.C., Uhl S.A., Caron R., Jarchin L., Dunkin D., Phelps R., Webb B.D., et al. 2020b. Complex autoinflammatory syndrome unveils fundamental principles of JAK1 kinase transcriptional and biochemical function. Immunity. 53:672–684.e11. 10.1016/j.immuni.2020.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hambleton, S., Salem S., Bustamante J., Bigley V., Boisson-Dupuis S., Azevedo J., Fortin A., Haniffa M., Ceron-Gutierrez L., Bacon C.M., Menon G., et al. 2011. IRF8 mutations and human dendritic-cell immunodeficiency. N. Engl. J. Med. 365:127–138. 10.1056/nejmoa1100066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann, M., and Bogunovic D.. 2017. ISG15: in sickness and in Health. Trends Immunol. 38:79–93. 10.1016/j.it.2016.11.001 [DOI] [PubMed] [Google Scholar]
- Jang, M.-A., Kim E.K., Now H., Nguyen N.T.H., Kim W.-J., Yoo J.-Y., Lee J., Jeong Y.-M., Kim C.-H., Kim O.-H., Sohn S., et al. 2015. Mutations in DDX58, which encodes RIG-I, cause atypical Singleton-Merten syndrome. Am. J. Hum. Genet. 96:266–274. 10.1016/j.ajhg.2014.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, X.-F., Martinez-Barricarte R., Kennedy J., Mele F., Lazarov T., Deenick E.K., Ma C.S., Breton G., Lucero K.B., Langlais D., Bousfiha A., et al. 2018. Disruption of an antimycobacterial circuit between dendritic and helper T cells in human SPPL2a deficiency. Nat. Immunol. 19:973–985. 10.1038/s41590-018-0178-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlova, А.L., Leonteva М.Е., Burlakov V.I., Nesterenko Z.А., Laba О.М., Pisareva М.V., Kan N.Y., Khoreva А.L., Roppelt А.А., Yukhacheva D.V., Rodina Y.A., et al. 2021. Clinical case of type I interferonopathy: homozygous STAT2 gain-of-function mutation. Pediatr. Hematol. Immunopathol. 20:132–139. 10.24287/1726-1708-2021-20-3-132-139 [DOI] [Google Scholar]
- Liu, L., Okada S., Kong X.-F., Kreins A.Y., Cypowyj S., Abhyankar A., Toubiana J., Itan Y., Audry M., Nitschke P., Masson C., et al. 2011. Gain-of-function human STAT1 mutations impair IL-17 immunity and underlie chronic mucocutaneous candidiasis. J. Exp. Med. 208:1635–1648. 10.1084/jem.20110958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, T., Zhang L., Joo D., and Sun S.-C.. 2017. NF-κB signaling in inflammation. Signal Transduct. Target. Ther. 2:17023. 10.1038/sigtrans.2017.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Jesus A.A., Marrero B., Yang D., Ramsey S.E., Sanchez G.A.M., Tenbrock K., Wittkowski H., Jones O.Y., Kuehn H.S., Lee C.-C.R., et al. 2014. Activated STING in a vascular and pulmonary syndrome. N. Engl. J. Med. 371:507–518. 10.1056/nejmoa1312625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingston, J.H., Lin J.-P., Dale R.C., Gill D., Brogan P., Munnich A., Kurian M.A., Gonzalez-Martinez V., De Goede C.G.E.L., Falconer A., Forte G., et al. 2014. A type I interferon signature identifies bilateral striatal necrosis due to mutations in ADAR1. J. Med. Genet. 51:76–82. 10.1136/jmedgenet-2013-102038 [DOI] [PubMed] [Google Scholar]
- Malakhov, M.P., Malakhova O.A., Il Kim K., Ritchie K.J., and Zhang D.-E.. 2002. UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J. Biol. Chem. 277:9976–9981. 10.1074/jbc.m109078200 [DOI] [PubMed] [Google Scholar]
- Martin-Fernandez, M., Bravo García-Morato M., Gruber C., Murias Loza S., Malik M.N.H., Alsohime F., Alakeel A., Valdez R., Buta S., Buda G., Marti M.A., et al. 2020. Systemic type I IFN inflammation in human ISG15 deficiency leads to necrotizing skin lesions. Cell Rep. 31:107633. 10.1016/j.celrep.2020.107633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malakhova, O.A., Kim K.I., Luo J.-K., Zou W., Kumar K.G.S., Fuchs S.Y., Shuai K., and Zhang D.-E.. 2006. UBP43 is a novel regulator of interferon signaling independent of its ISG15 isopeptidase activity. EMBO J. 25:2358–2367. 10.1038/sj.emboj.7601149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Barricarte, R., Markle J.G., Ma C.S., Deenick E.K., Ramírez-Alejo N., Mele F., Latorre D., Mahdaviani S.A., Aytekin C., Mansouri D., Bryant V.L., et al. 2018. Human IFN-γ immunity to mycobacteria is governed by both IL-12 and IL-23. Sci. Immunol. 3:eaau6759. 10.1126/sciimmunol.aau6759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen, M.C.E., Schot R., Buta S., Oudesluijs G., Tinschert S., Speer S.D., Li Z., van Unen L., Heijsman D., Goldmann T., Lequin M.H., et al. 2016. Human USP18 deficiency underlies type 1 interferonopathy leading to severe pseudo-TORCH syndrome. J. Exp. Med. 213:1163–1174. 10.1084/jem.20151529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muglia Amancio, A., Mittereder L., Carletti A., Tosh K.W., Green D., Antonelli L.R., Gazzinelli R.T., Sher A., and Jankovic D.. 2021. IFNs reset the differential capacity of human monocyte subsets to produce IL-12 in response to microbial stimulation. J. Immunol. 206:1642–1652. 10.4049/jimmunol.2001194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmura, K. 2019. Nakajo-Nishimura syndrome and related proteasome-associated autoinflammatory syndromes. J. Inflamm. Res. 12:259–265. 10.2147/jir.s194098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada, S., Markle J.G., Deenick E.K., Mele F., Averbuch D., Lagos M., Alzahrani M., Al-Muhsen S., Halwani R., Ma C.S., Wong N., et al. 2015. Impairment of immunity to Candida and Mycobacterium in humans with bi-allelic RORC mutations. Science. 349:606–613. 10.1126/science.aaa4282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini, S., John J., Shearer M., Kerr I.M., and Stark G.R.. 1989. Use of a selectable marker regulated by alpha interferon to obtain mutations in the signaling pathway. Mol. Cell. Biol. 9:4605–4612. 10.1128/mcb.9.11.4605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson, M., Bergendal B., Norderyd J., Nilsson D., Anderlid B.-M., Nordgren A., and Lindstrand A.. 2017. Further evidence for specific IFIH1 mutation as a cause of Singleton-Merten syndrome with phenotypic heterogeneity. Am. J. Med. Genet. A. 173:1396–1399. 10.1002/ajmg.a.38214 [DOI] [PubMed] [Google Scholar]
- Rice, G.I., Forte G.M.A., Szynkiewicz M., Chase D.S., Aeby A., Abdel-Hamid M.S., Ackroyd S., Allcock R., Bailey K.M., Balottin U., Barnerias C., et al. 2013. Assessment of interferon-related biomarkers in Aicardi-Goutières syndrome associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, and ADAR: a case-control study. Lancet Neurol. 12:1159–1169. 10.1016/s1474-4422(13)70258-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodero, M.P., and Crow Y.J.. 2016. Type I interferon–mediated monogenic autoinflammation: the type I interferonopathies, a conceptual overview. J. Exp. Med. 213:2527–2538. 10.1084/jem.20161596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutsch, F., MacDougall M., Lu C., Buers I., Mamaeva O., Nitschke Y., Rice G.I., Erlandsen H., Kehl H.G., Thiele H., Nürnberg P., et al. 2015. A specific IFIH1 gain-of-function mutation causes Singleton-Merten syndrome. Am. J. Hum. Genet. 96:275–282. 10.1016/j.ajhg.2014.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speer, S.D., Li Z., Buta S., Payelle-Brogard B., Qian L., Vigant F., Rubino E., Gardner T.J., Wedeking T., Hermann M., Duehr J., et al. 2016. ISG15 deficiency and increased viral resistance in humans but not mice. Nat. Commun. 7:11496. 10.1038/ncomms11496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaim, C.D., Scott A.F., Canadeo L.A., and Huibregtse J.M.. 2017. Extracellular ISG15 signals cytokine secretion through the LFA-1 integrin receptor. Mol. Cell. 68:581–590.e5. 10.1016/j.molcel.2017.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft, J., and Bogunovic D.. 2018. The Goldilocks zone of type I IFNs: lessons from human genetics. J. Immunol. 201:3479–3485. 10.4049/jimmunol.1800764 [DOI] [PubMed] [Google Scholar]
- Tangye, S.G., Al-Herz W., Bousfiha A., Chatila T., Cunningham-Rundles C., Etzioni A., Franco J.L., Holland S.M., Klein C., Morio T., Ochs H.D., et al. 2020. Human inborn errors of immunity: 2019 update on the classification from the international union of immunological societies expert committee. J. Clin. Immunol. 40:24–64. 10.1007/s10875-019-00737-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrelo, A., Patel S., Colmenero I., Gurbindo D., Lendínez F., Hernández A., López-Robledillo J.C., Dadban A., Requena L., and Paller A.S.. 2010. Chronic atypical neutrophilic dermatosis with lipodystrophy and elevated temperature (CANDLE) syndrome. J. Am. Acad. Dermatol. 62:489–495. 10.1016/j.jaad.2009.04.046 [DOI] [PubMed] [Google Scholar]
- Toubiana, J., Okada S., Hiller J., Oleastro M., Lagos Gomez M., Aldave Becerra J.C., Ouachée-Chardin M., Fouyssac F., Girisha K.M., Etzioni A., Van Montfrans J., et al. 2016. STAT1 Gain-of-Function Study GroupHeterozygous STAT1 gain-of-function mutations underlie an unexpectedly broad clinical phenotype. Blood. 127:3154–3164. 10.1182/blood-2015-11-679902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuillier, F., Li Z., Commere P.H., Dynesen L.T., and Pellegrini S.. 2019. USP18 and ISG15 coordinately impact on SKP2 and cell cycle progression. Sci. Rep. 9:4066. 10.1038/s41598-019-39343-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Bogunovic D., Payelle-Brogard B., Francois-Newton V., Speer S.D., Yuan C., Volpi S., Li Z., Sanal O., Mansouri D., Tezcan I., et al. 2015. Human intracellular ISG15 prevents interferon-α/β over-amplification and auto-inflammation. Nature. 517:89–93. 10.1038/nature13801 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists the homozygous rare variants found in WES analysis and shared by P1, P2, and/or P3.