Abstract
Background
Although Methicillin resistant Staphylococcus aureus (MRSA) is one of the major pathogens of healthcare associated infections, we had only sporadic cases in our intensive care unit (ICU) for years.
Aims
To investigate the sudden increase in the number of MRSA cases in ICU.
Study design
Descriptive study.
Methods
From the 5th December 2016 to 26th January 2017, we detected 11 new MRSA cases in ICU. Screening of 73 ICU healthcare workers (HCWs) and screening of 13 patients was performed for outbreak investigation. Nine clinical isolates available in stocks and eight screening MRSA isolates were included in molecular studies. PFGE, spa-mecA-mecC-PVL in-house multiplex PCR assay and spa typing, SCCmec typing were performed for all isolates. Sequence type of the representative strain was determined by Multi-Locus Sequence typing (MLST).
Results
All strains were mecA positive, PVL negative, and have the same antimicrobial susceptibility pattern except for two strains. All clinical, two patient screening and three nasal isolates of HCWs showed the same pulsotype, named clone A. The spa type of outbreak isolates is t030 and the SCCmec type is SCCmecIII; the MLST type of representative strain is ST239 (PFGE pulsotype A, ST239-SCCmecIII-t030). Unrelated three isolates had PFGE pulsotype B-SCCmecI-t030, PFGE pulsotype C-SCCmecIII-t459, PFGE pulsotype D-SCCmecIII.
Conclusion
Molecular typing techniques are the cornerstones for the investigation of outbreaks. Infection control measures, such as enhancing cleaning procedures, promoting hand hygiene, should be enforced in the ICU unit. All patients, including those who have already been discharged to other departments, must be put on contact isolation. HCWs carrying the MRSA strains could be offered decolonization.
Methicillin-resistant Staphylococcus aureus (MRSA) was described for the first time in1961, after the use of methicillin in 1959. MRSA is a causative agent of a broad spectrum of infectious diseases and considered as a frequent pathogen of health care-associated infections (1, 2). Methicillin resistance in S aureus is encoded by mecA or mecC genes that are carried on a mobile genetic element known as staphylococcal cassette chromosome (SCC) (1, 3). MRSA outbreaks are serious situations that can result in significant mortality and morbidity (4). Rapid diagnostic tests are needed to detect infections caused by MRSA strains and to limit the spread of these strains (3). Genotyping of outbreak strains to detect the possible source of the outbreak and immediate implementation of control measures are necessary. Frequently used molecular methods for typing purposes are pulsed-field gel electrophoresis (PFGE), spa typing, SCCmec typing, and multilocus sequence typing (MLST).
In this study, we aimed to investigate new MRSA cases in the intensive care unit (ICU). We screened patients and health care workers (HCWs) and performed spa typing, PFGE, SCCmec typing, and MLST on MRSA strains to decide whether any transmission was occurring and measures of infection prevention were mandatory.
MATERIALS AND METHODS
Ethical Considerations
Ethics committee approval was received for this study from the Local Ethics Committe of Şişli Hamidiye Etfal Training and Research Hospital (date: 30.04.2019; number: 2377).
Outbreak description and bacterial isolates
From December 5, 2016, to January 26, 2017, we detected 11 new MRSA cases in the adult ICU. On December 20, 23, and 24, 2016, the Clinical Microbiology Department noticed 3 consecutive MRSA isolates, 2 from the blood and 1 from tracheal aspirate culture, having the same antibiotic susceptibility pattern belonging to 3 different patients in the adult ICU. We searched the Laboratory Information System for MRSA growths starting from October; we found 3 more isolates and 2 more cases who had already been discharged from the ICU to the neurology and general surgery wards. The first isolate was from a blood culture dated December 5 and belonged to a male patient who had a positive blood culture on December 20. We determined him as the index case. The index case was a morbidly obese patient hospitalized in the ICU because of respiratory failure owing to pneumonia and left ventricular dysfunction.
Nasal screening of 74 ICU HCWs and nasal, throat, axilla, groin, and rectum screening of 13 patients were performed for outbreak investigation. Identification and antimicrobial susceptibility test (AST) were done by MALDI-TOF MS (Bruker Daltonik GmbH, Bremen, Germany) and Phoenix100 ID/AST system (Becton Dickinson, Sparks, MD, USA), respectively.
A total of 9 clinical isolates available in stocks (7 blood, 1 tracheal aspirate, 1 abscess; 1 isolate per patient) and 8 screening MRSA isolates (5 HCWs and 3 patients) were included in molecular studies.
Evaluation of mecA, mecC, spa, and PVL genes by multiplex polymerase chain reaction
In-house multiplex polymerase chain reaction (PCR) assay was performed as described by Stegger et al. (5). Instagene Matrix (Bio-Rad) was used for DNA extraction. Confirmation of methicillin resistance and identification of isolates were done by amplification of mecA/mecC genes and spa gene, respectively. We also searched for the Panton-Valentine leukocidin (PVL) gene.
spa typing
spa gene amplicons were purified and sequenced. The open-source spa typing program at http://spatyper.fortinbras.us/ was used for spa sequence analysis.
Pulsed-field gel electrophoresis
PFGE typing of S aureus strains was performed following the protocol of Yetkin et al. (6). Briefly, bacterial cells were embedded into low melting agarose including lysostaphin (5 U/mL). Cells in plugs were digested with proteinase K. After washing the plugs, genomic DNA in the plugs was restricted by 30 U of SmaI (Promega Corporation, Durham, NC, USA) for 24 hours at 25°C in a water bath. DNA fragments were separated on 1% agarose gels run in 0.5 × Tris-borate-ethylenediaminetetraacetic acid buffer using a CHEF-DR III system (Bio-Rad Laboratories, Nazareth, Belgium). The electrophoresis conditions were 14°C at 6 V/cm2 for 20 hours. The initial and final switch times were 5.3 and 34.9 seconds, respectively. The gel was stained with ethidium bromide (5 μg/mL) for 20 minutes and destained with distilled water for 30 minutes. The DNA band profiles were visualized under ultraviolet light, photographed using a Gel Logic 2200 Imaging System (Kodak Co, Rochester, NY, USA), and analyzed by GelCompar software (version 7.0; Applied Maths, Sint-Martens-Latem, Belgium). A 1% band tolerance was used for the comparison of DNA profiles. The clonal relationship among isolates was evaluated using the criteria of Tenover et al. (7).
SCCmec typing
SCCmec typing was performed using in-house multiplex PCR according to the procedure described by McClure-Warnier et al. (8). A total of 9 primer pairs were used for the screening of major SCCmec types and subtypes I to V.
Multilocus sequence typing
The allelic profile of the outbreak MRSA strain was obtained by sequencing internal fragments of 7 house-keeping genes as described at http://saureus.mlst.net/misc/info.asp. (9): carbamate kinase (arc), Shikimate dehydrogenase (aro), Glycerol kinase (glp), Guanylate kinase (gmk), Phosphate acetyltransferase (pta), triosephosphate isomerase (tpi), and acetyl coenzyme A acetyltransferase (yqi). The DNA sequence of each allele analyzed using data analysis software at the http://saureus.mlst.net/sql/singlelocus.asp website and allele numbers of 7 amplicons were found out. By entering all allelic numbers of the strain in a program at the http://saureus.mlst.net/sql/allelicprofilechoice.as website, the sequence type (ST) of the outbreak strain was determined.
RESULTS
Patient and HCW demographics, clonal characteristics, and antibiotic susceptibilities of MRSA isolates and molecular typing test results are presented in Table 1.
TABLE 1.
Patient and healthcare workers demographics, isolation sites, clonal characteristics, and antibiotic susceptibilities of MRSA isolate
| Isolate identifier | Age | Sex | Diagnosis | Isolation site | Therapy | Outcome | PFGE | spa type | SCCmec type | Cefoxitin | Ciprofloxacin | Clindamycin | Daptomycin | Erythromycin | Fusidic acid | Gentamicin | Levofloxacin | Linezolid | Rifampicin | Tetracycline | TMP-SXT | Tobramycin | Vancomycin |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patients’ isolates | |||||||||||||||||||||||
| CI-1 | 48 | M | Pneumonia | Blood | PIP-TZP 4.5 g IV TID; Daptomycin 500 mg IV daily | Died | A | t030 | Type III | R | R | R | S | R | S | R | R | S | R | R | S | R | S |
| CI-2 | 79 | F | Cerebrovascular disease | Trachea | Linezolid 600 mg IV BID | Discharged | A | t030 | Type III | R | R | R | S | R | S | R | R | S | R | R | S | R | S |
| CI-3 | 74 | F | Meninx benign neoplasm | Trachea | Vancomycin 1 g IV daily | Died | A | t030 | Type III | R | R | R | S | R | S | R | R | S | R | R | S | R | S |
| CI-4 | 68 | M | Colon cancer | Blood | Daptomycin 350 mg IV daily | Discharged | A | t030 | Type III | R | R | R | S | R | S | R | R | S | R | R | S | R | S |
| CI-5 | 46 | F | Falling from high | Trachea | Tigecycline 50 mg IV BID; Meropenem 1 g IV TID | Discharged | A | t030 | Type III | R | R | R | S | R | S | R | R | S | R | R | S | R | S |
| CI-6 | 49 | F | Over cancer | Blood | Vancomycin 1 g IV BID | Discharged | A | t030 | Type III | R | R | R | S | R | S | R | R | S | R | R | S | R | S |
| CI-7 | 88 | M | Colon cancer | Abscess | Tigecycline 50 mg IV BID; Meropenem 1 g IV TID | Discharged | A | t030 | Type III | R | R | R | S | R | S | R | R | S | R | R | S | R | S |
| CI-8 | 23 | M | Drug user, intoxication | Blood | Tigecycline 50 mg IV BID; Meropenem 1 g IV TID | Discharged | A | t030 | Type I | R | R | S | S | S | S | S | R | S | R | R | S | S | S |
| CI-9 | 86 | M | Fournier’s gangrene | Blood | Linezolid 600 mg BID | Died | A | t030 | Type III | R | R | S | S | S | S | S | R | S | R | R | S | S | S |
| Screening cultures isolates | |||||||||||||||||||||||
| Pt-S-1 | 63 | K | Cerebellar tumor, epilepsy | Throat, nasal | Vancomycin 1 g IV BID | Died | C | t459 | Type III | R | R | R | S | R | S | R | R | S | R | R | S | R | S |
| Pt-S-2* | 74 | F | Meninx benign neoplasm | Nasal, groin | Vancomycin 1 g IV daily | Died | A | t030 | Type III | R | R | R | S | R | S | R | R | S | R | R | S | R | S |
| Pt-S-3** | 86 | M | Fournier’s gangrene | Throat, nasal, groin, rectum | Linezolid 600 mg IV BID | Died | A | t030 | Type III | R | R | R | S | R | S | R | R | S | R | R | S | R | S |
| HCW-1 | 44 | E | Nasal | TMP-SXT 160/800 mg PO BID | A | t030 | Type III | R | R | R | S | R | S | R | R | S | R | R | S | R | S | ||
| HCW-2 | 23 | E | Nasal | TMP-SXT 160/800 mg PO BID | D | ND | Type III | R | R | R | S | R | S | R | R | S | R | R | S | R | S | ||
| HCW-3 | 28 | E | Nasal | TMP-SXT 160/800 mg PO BID | A | t030 | Type V | R | R | R | S | R | S | R | R | S | R | R | S | R | S | ||
| HCW-4 | 30 | K | Nasal | Not decolonized | A | t030 | Type III | R | R | S | S | S | S | S | R | S | R | R | S | S | S | ||
| HCW-5 | 24 | K | Nasal | TMP-SXT 160/800 mg PO BID | B | t030 | Type I | R | R | S | S | S | S | S | R | S | R | R | S | S | S |
BID: 2 times a day, CI: clinical isolate, F: female, HCW: healthcare worker, IV: intravenous, M: male, MRSA: methicillin-resistant Staphylococcus aureus, PIP-TZP: piperacillin/tazobactam, PO: by mouth, Pt-S: patient screening, R: resistant, S: susceptible, TID: 3 times a day, TMP-SXT: trimethoprim/sulfamethoxazole
Pt-S-2 and CI-3 belonged to the same patient;
Pt-S-3 and CI-9 belonged to the same patient
Screening cultures
Screening was performed in the ICU on December 24. Of the 13 patients, 5 were carrying MRSA strains, in multiple sites of their body; 2 of 5 sharing the same antibiotic susceptibility profile were shown to carry the outbreak strain by molecular typing methods (15%). Of these 2 colonized patients, one carried the outbreak strain at nasal and groin sites, and the other one at the throat, nasal, groin, and rectum sites. Rectum-colonized patient was an 86-year-old man with a Fournier’s gangrene; 18 of 73 HCWs were S aureus positive for their nasal swab cultures (10 methicillin-susceptible Staphylococcus aureus [MSSA]; 8 MRSA), and molecular typing was done for 5 MRSA sharing the same or similar antibiotic susceptibility pattern with outbreak strain (Table 1). Moreover, 3 of HCWs’ nasal isolates were the outbreak strain (4%).
Antibiotic susceptibilities
We performed the AST using the recommended methods and interpretation of the European Committee on Antimicrobial Susceptibility Testing. The outbreak strain was resistant to ciprofloxacin (CIP), levofloxacin (LEV), clindamycin (DA), erythromycin (E), tetracycline (TE), gentamicin (GN), and tobramycin (TOB) and susceptible to trimethoprim-sulfamethoxazole (TMP-SXT), daptomycin (DPC), fusidic acid (FD), linezolid (LZD), and vancomycin (VA) except clinical isolate 8 and clinical isolate 9. Clinical isolate 8 and clinical isolate 9 were susceptible to DA, E, GN, and TOB.
Decolonization of HCWs
All of the HCWs, except 1 pregnant nurse, were decolonized by TMP-SXT tablets (800/160 mg twice a day for 10 days). We checked the colonization status of HCWs 1 week after the completion of treatment. They were all cleared of the MRSA.
Evaluation of mecA-mecC-SPAPVL multiplex PCR
All strains were positive for the mecA gene and negative for the PVL gene.
Molecular typing
All clinical isolates and 2 patient screening and 3 nasal isolates of HCWs showed the same PFGE pulsotype (PT) that was named as clone A (Figure 1). The spa ST of these isolates was t030, and the SCCmec type was SCCmecIII. The MLST result was ST239. Clinical isolate 8 and HCW-3 have different SCCmec types, SCCmecI and SCCmecV, respectively. Unrelated 3 isolates had PFGE PT B-SCCmecI-t030, PFGE PT C-SCCmecIII-t459, and PFGE PT D-SCCmecIII (Figure 2).
FIG. 1.
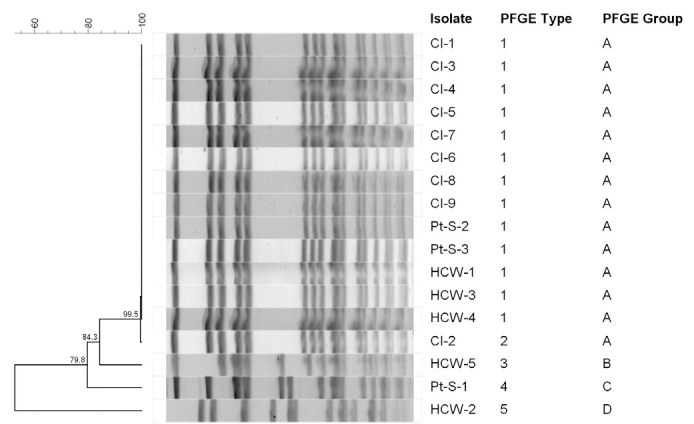
PFGE smaI restriction patterns of MRSA clinical isolates clone A. Lanes 1 and 15, Salmonella braenderup molecular size marker; lanes 4 to 7, CI-1, CI-2, Cl-3, and CI-4; and lanes 9 to 13, CI-5, CI-6, CI-8, CI-7, and CI-9, respectively; lanes 2 to 4 and 14, CIs that were unrelated to the outbreak; lane 8, empty
CI: clinical isolate, MRSA: methicillin-resistant Staphylococcus aureus, PFGF: pulsed-field gel electrophoresis
FIG. 2.
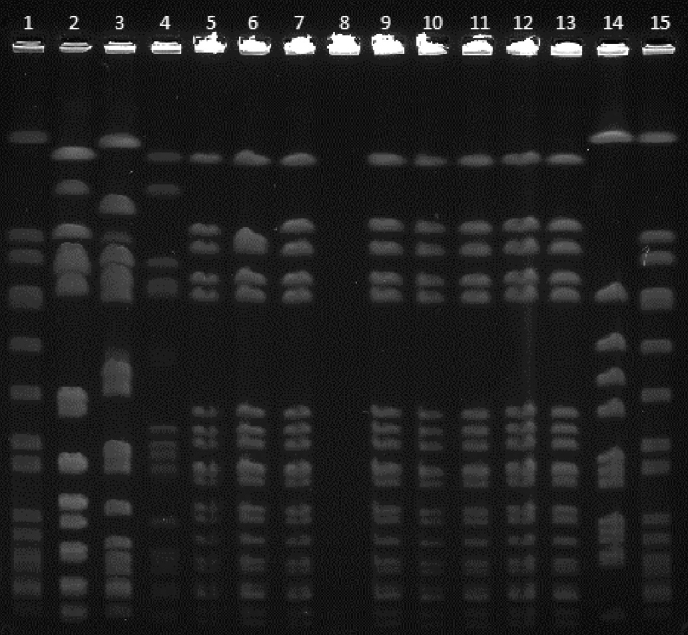
PFGE dendrogram showing different PTs seen over the duration of outbreak (A to D). PT A belongs to outbreak strain
PFGE: pulsed-field gel electrophoresis, PT: pulsotype
DISCUSSION
In this study, we report our experience about the MRSA outbreak in an adult ICU. The outbreak was noticed in the middle of December 2016; by that time, 4 patients had already been discharged from the ICU to other clinical wards. We assume that our index case was a male patient who stayed in the ICU for more than a month. Few HCWs were colonized with the outbreak strain, and we assume HCWs might have transmitted the outbreak strain to other patients during the medical care as reported in the literature before (10).
To interrupt the MRSA transmission, various infection control measures were established immediately. Nasal screening of HCWs and nasal, throat, axilla, groin, and rectal screening of ICU patients were performed (11). All of HCWs carrying the MRSA strains except 1 pregnant employee were treated with co-trimoxazole as a decolonization procedure because mupirocin ointment for nasal usage is not available in the market in our country (12). We put our infected and colonized patients in contact isolation, including those who had already been discharged to other wards (13). Infection control nurses gave educational sessions to HCWs on promoting hand hygiene and enhancing cleaning procedures. We screened patients a month later and there were still patients colonized with the outbreak strain, but no new MRSA cases occurred. Therefore, we declared the outbreak was over and decided to follow up on the situation. Identification of an outbreak is usually based on molecular typing, which is a significant tool in infection control. Molecular typing methods show the nosocomial transmission of the successful clone and elucidate its transmission routes and sources in an outbreak setting (14). Different molecular methods are available for typing of S aureus isolates, including PFGE, multilocus variable number of tandem repeat analysis, SCCmec typing, spa typing, MLST, and full-genome sequencing (1, 3, 15). PFGE is still accepted as the “gold standard” for molecular typing of MRSA, and SmaI is the enzyme of the choice for macrorestriction (16). The major drawback of PFGE is the insufficient comparability of results between laboratories, which might be overcome by using standardized protocols nationally (2, 16). In contrast, results of MLST are comparable between laboratories, but its routine use in infection control is restricted by high cost, labor intensity, and availability of DNA sequencer (16). Recently, it has been reported that spa typing and MLST typing have comparable performance in a macroepidemiologic study (15). Moreover, spa typing has a higher discriminatory power over MLST, so 1 ST type can harbor several spa types, but they remain within the same clonal complex (17). In this study, we used several typing methods, PFGE, SCCmec typing, spa typing, and MLST, simultaneously, for the characterization of the outbreak strain. The outbreak isolates showed the same PFGE PT named as clone A and were ST239-SCCmecIII-t030 strains. ST239-SCCmecIII-t030 has previously been reported as the most frequent hospital-associated (HA) MRSA clone in Turkey (18) and named as TR09. Other studies from Turkey also reported the dominance of MLST ST239 and spa type t030 (19–21) characterized MRSA clones. Oksuz et al. (21) found Vienna/Hungarian/Brazilian clone (ST239-MRSA-III) as the most prevalent clone (53.9%) in a university hospital for more than 5 years, and the rest of the MRSA clones were largely diverse. Geographic distribution of major HA MRSA clones is known worldwide, but sometimes shifts of these MRSA clones have occured over time in countries, in a region, or even in a single hospital (22). MRSA evolved from MSSA in 1961 by the acquisition of SCCmec (1). Since then, a limited number of clones with a certain genetic background and SCCmec type have spread worldwide (22). MRSA ST239 lineage is a globally disseminated HA MRSA consisting of more than 5 clades, such as Asia, North America, South America, Europe, and Australia (23). ST239 is a successful clone in terms of infiltration and adaptation in hospital settings (24). Several studies are being conducted to identify the reasons behind this success. Recently, Hong et al. (24) reported that a higher expression of staphylococcal protein A (SpA) in HA MRSA ST239 helps for durable colonization and immune evasion of the clone. Furthermore, they stipulated that SpA may be a significant factor for the adaptation and persistence of the ST239 clone in hospital environments. Moreover, 2 colonization factors, sasX protein first described in ST239 clone during an outbreak in London and arginine catabolic mobile element (ACME), were found to be associated with the epidemiologic success of the MRSA. ACME was first described in CA-MRSA USA300 and has spread horizontally to HA sequence types such as ST5 and ST239 (25, 26). Finally, enteric carriage of HA MRSA has been shown for ST228 and was considered responsible for maintaining a long-term outbreak in a tertiary hospital (27). In our patient group, we found out that 1 patient had enteric colonization with the outbreak strain. However, he was not the index case.
The outbreak strain was resistant to CIP, LEV, DA, E, TE, GN, and TOB and susceptible to TMP-SXT, DPC, FD, LZD, and VA. This resistance pattern was very similar to that indicated in previous reports (21, 23, 28). PVL was not detected on the outbreak strain.
In conclusion, we described an MRSA outbreak in the ICU owing to the clonal spread of the specific strain. Infection control measures were put in practice immediately upon the detection of clustering. Through the microbiologic typing methods, outbreak strain was described in detail and the index case was identified. The outbreak was successfully controlled through universal contact precautions and treatment of infected patients and decolonization of HCWs for nasal carriage.
Footnotes
Ethics Committee Approval: Ethics committee approval was received for this study from the Local Ethics Committe of Şişli Hamidiye Etfal Training and Research Hospital (date: 30.04.2019; number: 2377).
Patient Consent for Publication: The authors did not take any informed consent from patient due to study design.
Data-sharing Statement: N/A.
Author Contributions: Concept - B.Bayraktar.; Design - B.Bayraktar, A.G.; Supervision - B.Bayraktar, B.Bozdağan.; Resources - B.Bayraktar, B.Bozdağan, R.D., N.U.; Materials - B.Bayraktar, B.Bozdağan, R.D.; Data Collection and/or Processing - L.T., E.O., S.Ş., D.E.; Analysis and/or Interpretation - B.Bayraktar, E.O., A.G., E.A.; Literature Search - B.Bayraktar, L.T.; Writing Manuscript - B.Bayraktar.; Critical Review - B.Bayraktar, A.G.
Conflict of Interest: The authors have no conflicts of interest to declare.
Funding: The authors declared that this study had received no financial support. All expenses belong to the authors.
REFERENCES
- 1.Liu J, Chen D, Peters BM, Li L, Li B, Xu Z, et al. Staphylococcal chromosomal cassettes mec (SCCmec): A mobile genetic element in methicillin-resistant Staphylococcus aureus. Microb Pathog. 2016;101:56–67. doi: 10.1016/j.micpath.2016.10.028. [DOI] [PubMed] [Google Scholar]
- 2.Simor AE, Ofner-Agostini M, Bryce E, McGeer A, Paton S, Mulvey MR, et al. Laboratory characterization of methicillin-resistant Staphylococcus aureus in Canadian hospitals: Results of 5 years of National Surveillance, 1995–1999. J Infect Dis. 2002;186:652–60. doi: 10.1086/342292. [DOI] [PubMed] [Google Scholar]
- 3.te Witt R, van Belkum A, van Leeuwen WB. Molecular diagnostics and genotyping of MRSA: An update. Expert Rev Mol Diagn. 2010;10:375–80. doi: 10.1586/erm.10.34. [DOI] [PubMed] [Google Scholar]
- 4.Khan A, Lampitoc M, Slaripour M, McKernan P, Devli R, Muller MP. Rapid control of amethicillin reistant Staphylococcus aureus (MRSA) outbreak in a medical surgical intensive care unit (ICU) Can J Infect Control. 2009;24:12–6. [PubMed] [Google Scholar]
- 5.Stegger M, Andersen PS, Kearns A, Pichon B, Holmes MA, Edwards G, et al. Rapid detection, differentiation and typing of methicillin-resistant Staphylococcus aureus harbouring either mecA or the new mecA homologue mecA (LGA251) Clin Microbiol Infect. 2012;18:395–400. doi: 10.1111/j.1469-0691.2011.03715.x. [DOI] [PubMed] [Google Scholar]
- 6.Yetkin GKÇ, Durmaz B, Durmaz R, Çizmeci Z, İşeri L. Molecular typing of methicillin-resistant Staphylococcus aureus isolated from bloodstream infections in a university hospital. Turk J Med Sci. 2009;39:959–68. [Google Scholar]
- 7.Tenover FC, Arbeit RD, Goering RV. How to select and interpret molecular strain typing methods for epidemiological studies of bacterial infections: A review for healthcare epidemiologists. Molecular Typing Working Group of the Society for Healthcare Epidemiology of America. Infect Control Hosp Epidemiol. 1997;18:426–39. doi: 10.2307/30141252. [DOI] [PubMed] [Google Scholar]
- 8.McClure-Warnier JA, Conly JM, Zhang K. Multiplex PCR assay for typing of staphylococcal cassette chromosome mec types I to V in methicillin resistant Staphylococcus aureus. J Vis Exp. 2013;(79):50779. doi: 10.3791/50779. doi: 10.3791/50779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38:1008–15. doi: 10.1128/JCM.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stock NK, Petráš P, Melter O, Kapounová G, Vopalková P, Kubele J, et al. Importance of multifaceted approaches in infection control: A practical experience from an outbreak investigation. PLoS One. 2016;11:e0157981. doi: 10.1371/journal.pone.0157981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chipolombwe J, Török ME, Mbelle N, Nyasulu P. Methicillin resistant Staphylococcus aureus multiple sites surveillance: A systemic review of the literature. Infect Drug Resist. 2016;9:35–42. doi: 10.2147/IDR.S95372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song X, Cheung S, Klontz K, Short B, Campos J, Singh N. A stepwise approach to control an outbreak and ongoing transmission of methicillin-resistant Staphylococcus aureus in a neonatal intensive care unit. Am J Infect Control. 2010;38:607–11. doi: 10.1016/j.ajic.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 13.Jernigan JA, Titus MG, Groschel DH, Getchell-White S, Farr BM. Effectiveness of contact isolation during a hospital outbreak of methicillin-resistant Staphylococcus aureus. Am J Epidemiol. 1996;143:496–504. doi: 10.1093/oxfordjournals.aje.a008770. [DOI] [PubMed] [Google Scholar]
- 14.Mutters NT, Heeg K, Späth I, Henny N, Gunther F. Improvement of infection control management by routine molecular evaluation of pathogen clusters. Diagn Microbiol Infect Dis. 2017;88:82–7. doi: 10.1016/j.diagmicrobio.2017.01.013. [DOI] [PubMed] [Google Scholar]
- 15.O’Hara FP, Suaya JA, Ray GT, Baxter R, Brown ML, Mera RM, et al. spa typing and multilocus sequence typing show comparable performance in a macroepidemiologic study of staphylococcus aureus in the United States. Microb Drug Resist. 2016;22:88–96. doi: 10.1089/mdr.2014.0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strommenger B, Kettlitz C, Weniger T, Harmsen D, Friedrich AW, Witte W. Assignment of Staphylococcus isolates to groups by spa typing, SmaI macrorestriction analysis, and multilocus sequence typing. J Clin Microbiol. 2006;44:2533–40. doi: 10.1128/JCM.00420-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deurenberg RH, Vink C, Kalenic S, Friedrich AW, Bruggeman CA, Stobberingh EE. The molecular evolution of methicillin-resistant Staphylococcus aureus. Clin Microbiol Infect. 2007;13:222–35. doi: 10.1111/j.1469-0691.2006.01573.x. [DOI] [PubMed] [Google Scholar]
- 18.Bozdoğan B, Yıldız O, Oryasin E, Kırdar S, Gülcü B, Aktepe O, et al. [t030 is the most common spa type among methicillin-resistant Staphylococcus aureus strains isolated from Turkish hospitals]]. Mikrobiyol Bul. 2013;47:571–81. doi: 10.5578/mb.5770. HYPERLINK ". [DOI] [PubMed] [Google Scholar]
- 19.Alp E, Klaassen CHW, Doganay M, Altoparlak U, Aydın K, Engin A, et al. MRSA genotypes in Turkey: Persistence over 10 years of a single clone of ST239. J Infect. 2009;58:433–8. doi: 10.1016/j.jinf.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 20.Kırca Yulmaz S, Acuner IC, Strommenger B, Bek Y, Witte W. [Infectivity-resistotype-genotype clustering of methicillin-resistant Staphylococcus aureus strains in the Central Blacksea Region of Turkey]. Mikrobiyol Bul. 2014;48:14–27. [PubMed] [Google Scholar]
- 21.Oksuz L, Dupieux C, Tristan A, Bes M, Etienne J, Gurler N. The high diversity of MRSA clones detected in a university hospital in İstanbul. Int J Med Sci. 2013;10:1740–5. doi: 10.7150/ijms.6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deurenberg RH, Stobberingh EE. The molecular evolution of hospital- and community-associated methicillin-resistant Staphylococcus aureus. Curr Mol Med. 2009;9:100–15. doi: 10.2174/156652409787581637. [DOI] [PubMed] [Google Scholar]
- 23.Yamamoto T, Takano T, Higuchi W, Iwao Y, Singur O, Reva I, et al. Comparative genomics and drug resistance of a geographic variant of ST239 methicillin-resistant Staphylococcus aureus emerged in Russia. PLoS One. 2012;7:e29187. doi: 10.1371/journal.pone.0029187. doi: 10.1371/journal.pone.0029187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong X, Qin J, Li T, Dai Y, Wang Y, Liu Q, et al. Staphylococcal protein a promotes colonization and immune evasion of the epidemic healthcare-associated MRSA ST239. Front Microbiol. 2016;7:951. doi: 10.3389/fmicb.2016.00951. doi: 10.3389/fmicb.2016.00951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otto M. How colonization factors are linked to outbreaks of methicillin-resistant Staphylococcus aureus: The roles of SasX and ACME. Biomol Concepts. 2013;4:533–7. doi: 10.1515/bmc-2013-0025. [DOI] [PubMed] [Google Scholar]
- 26.Sabat AJ, Köck R, Akkerboom V, Hendrix R, Skov RL, Becker K, et al. Novel organization of the arginine catabolic mobile element and staphylococcal cassette chromosome mec composite island and its horizontal transfer between distinct Staphylococcus aureus genotypes. Antimicrob Agents Chemother. 2013;57:5774–7. doi: 10.1128/AAC.01321-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Senn L, Clerc O, Zanetti G, Basset P, Prod’hom G, Gordon NC, et al. The stealthy superbug: The role of asymptomatic enteric carriage in maintaining a long-term hospital outbreak of ST228 methicillin-resistant Staphylococcus aureus. mBio. 2016;7:e02039–02015. doi: 10.1128/mBio.02039-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teo J, Tan TY, Hon PY, Lee W, Koh TH, Krishnan P, et al. ST22 and ST239 MRSA duopoly in Singaporean hospitals: 2006–2010. Epidemiol Infect. 2013;141:153–7. doi: 10.1017/S0950268812000337. [DOI] [PMC free article] [PubMed] [Google Scholar]


