Abstract
Gene expression manipulation of specific metabolic pathways can be used to obtain bioaccumulation of valuable molecules and desired quality traits in plants. A single-gene approach to impact different traits would be greatly desirable in agrospace applications, where several aspects of plant physiology can be affected, influencing growth. In this work, MicroTom hairy root cultures expressing a MYB-like transcription factor that regulates the biosynthesis of anthocyanins in Petunia hybrida (PhAN4), were considered as a testbed for bio-fortified tomato whole plants aimed at agrospace applications. Ectopic expression of PhAN4 promoted biosynthesis of anthocyanins, allowing to profile 5 major derivatives of delphinidin and petunidin together with pelargonidin and malvidin-based anthocyanins, unusual in tomato. Consistent with PhAN4 features, transcriptomic profiling indicated upregulation of genes correlated to anthocyanin biosynthesis. Interestingly, a transcriptome reprogramming oriented to positive regulation of cell response to biotic, abiotic, and redox stimuli was evidenced. PhAN4 hairy root cultures showed the significant capability to counteract reactive oxygen species (ROS) accumulation and protein misfolding upon high-dose gamma irradiation, which is among the most potent pro-oxidant stress that can be encountered in space. These results may have significance in the engineering of whole tomato plants that can benefit space agriculture.
Keywords: MicroTom, hairy root cultures, agrospace, biofortification, anthocyanins, gamma radiation
Introduction
Anthocyanins are valuable water-soluble plant pigments. They accumulate in the vacuole of specialized cells and play a crucial role in pigmentation of flowers and fruits, pollinators attraction, plant-pathogen interaction, protection against ultraviolet (UV) light, and modulation of reactive oxygen species (ROS)-signaling both in reproductive and in vegetative tissues (Brunetti et al., 2013). As plant-derived molecules, anthocyanins are naturally present in the human diet where they are predominantly represented as glycosides, in a multitude of fruits and vegetables, among which berries and grapes have the highest content (Bognar et al., 2013). Anthocyanins act as health-promoting and chronic-diseases-preventing molecules, due to antioxidant, anti-inflammatory, anti-proliferative and anti-neurodegenerative functions (Blesso, 2019; Krga and Milenkovic, 2019; Tian et al., 2019; Bendokas et al., 2020; Kalt et al., 2020). Due to these properties, anthocyanins have increasing applications in the food sector and there is also growing interested in the design of food crops with improved levels and composition of these antioxidant nutraceuticals. In particular, the research involved in the definition of plants intended for ‘agrospace’ applications, will have to tackle the issue to provide fresh and healthy food for space crews in the context of a harmful ionizing irradiated environment, and to cope with cultivation areas subjected to possible biotic contaminations, as well (Amalfitano et al., 2020; Bijlani et al., 2021). Agrospace crops are, therefore, candidates for the application of technologies aiming to improve both their content in antioxidant nutraceuticals and resistance to biotic and abiotic stresses (Zabel et al., 2015, 2016; Massa et al., 2016; Khodadad et al., 2020).
Tomato (Solanum lycopersicum L.) is a rich source of bioactive molecules such as carotenoids (in particular, lycopene), polyphenols and flavonoids, ascorbic acid, and other vitamins (Gerszberg et al., 2015; Martí et al., 2016). However, anthocyanins are poorly accumulated in cultivated tomatoes and even the fruits of cultivar harboring natural mutants for the Abg (Aubergine), Aft (Anthocyanin fruit), and Atv (Atroviolaceum) loci, only contain these molecules in the peel (Mes et al., 2008; Gonzali et al., 2009; Povero et al., 2011). Accumulation in fruit flesh and other organs upon genetic engineering indicates that tomatoes can be manipulated to this end (Zhang et al., 2014; Lloyd et al., 2017). By breeding, tomato lines were generated that combine the dominant Atv allele with Aft or Abg. These lines accumulated up to 0.1% (in fresh weight) of the anthocyanin petunidin-3-(p-coumaryl)-rutinoside-5-glucoside in the fruit epidermis (Mes et al., 2008; Povero et al., 2011). Gene and pathway engineering are powerful approaches to enhance the biosynthesis of anthocyanins in plants, and they have been successfully applied in food staples (Garg et al., 2018). Transcription factors, regulating the expression of structural biosynthetic genes, control the activity of the anthocyanin pathway in all plant species (Gonzalez et al., 2008). In particular, proteins belonging to specific clades of R2R3-MYB, bHLH, WDR, and WRKY have been shown to regulate anthocyanins biosynthesis combined in the MBWW transcription complex, as shown for a multitude of plant species among which tomato (Ramsay and Glover, 2005; Zhang et al., 2014; Gao et al., 2018). The combined expression of a MYB and bHLH regulators of the anthocyanin pathway from Antirrhinum majus (Delila and Rosea1, respectively) under a fruit-specific promoter, resulted in the production of anthocyanins in fruits peel and flesh of tomato (Butelli et al., 2008). Studies have demonstrated that pathway engineering approaches implying the sole use of MYB factors belonging to the SG6 clade are sufficient to restore the biosynthesis of anthocyanins by promotion of the transcription of their bHLH partners and, therefore, by reconstituting the MBWW (Mehrtens et al., 2005; Takos et al., 2006; Zhang et al., 2019). The tomato ANT1 gene encodes a MYB transcription factor belonging to the SG6 clade, highly homologous to the Antirrhinum Rosea. It has been demonstrated that ectopic expression of ANT1 from a tomato wild relative (S. chilense), induces purple spotting on the epidermis of tomatoes (Mathews et al., 2003; Schreiber et al., 2012). The 35S promoter-driven expression of either the Solanum lycopersicum ANT1 or AN2 (another SG6 MYB), has been shown to induce anthocyanins production in the flesh and peel of the fruit and different organs of tomato plants (Kiferle et al., 2015). Upon overexpression of SlAN2, together with anthocyanins accumulation in fruits, flower organs, and vegetative parts, an enhancement of the emission of volatile molecules contributing to the aroma of fruits was found, as well (Jian et al., 2019). In addition, SlAN2 has been related to the variation of levels of specialized metabolites other than anthocyanins, and of fruit softening (Meng et al., 2015). These findings seem to confirm that MYB transcription factors of the SG6 clade can regulate various, sometimes unrelated, processes in tomatoes, as well (Stracke et al., 2001; Zimmermann et al., 2004; Zhang et al., 2019). Therefore, a SG6 MYB-based approach may be considered suitable to affect multiple pathways in tomatoes.
The Anthocyanin4 gene of Petunia hybrida (PhAN4) is a SG6 member of a small family of genes encoding very similar MYBs phylogenetically related to the snapdragon AmROSEA, the tomato SlANT1, and other anthocyanin-regulating MYBs from a multitude of plant species. All these petunia MYBs are involved in the induction of anthocyanins accumulation in different plant parts and response to different stimuli (Povero, 2011). In the present study, we performed Agrobacterium rhizogenes-mediated transfer of a construct for the expression of PhAN4 into the miniature tomato genotype MicroTom to generate hairy root cultures (HRCs). HRCs were intended as a testbed for whole plant engineering strategies able to improve traits for space cultivation. We previously reported about improved in vivo response to space-mimicking conditions (i.e., static magnetic fields and X and gamma rays) of PhAN4-engineered HRC (Villani et al., 2017; Desiderio et al., 2019), confirming that this plant-based expression system, used over the last 30 years to produce various specialized metabolites and recombinant proteins of pharmaceutical value (Gutierrez-Valdes et al., 2020; Häkkinen et al., 2020), is useful in studies on the adaptation of plants to extraterrestrial conditions, as well. Recently, HRCs served in several plant species as handier and faster biotechnology tools, compared to whole plant transformation, to gain biological insights in gene function, spatial and temporal gene expression studies, and signaling pathways in plant cell response to a changing environment (Ron et al., 2014).
Anthocyanin biosynthesis engineering in HRCs was reported in a few species (Sharma et al., 2013; Li et al., 2016; Thwe et al., 2016; Hou et al., 2017), and, to date, never in tomatoes. In this study, PhAN4 gene expression resulted in anthocyanins accumulation in tomato HRCs. Transcript profiling showed that several genes encoding enzymes and transcription factors involved in anthocyanins biosynthesis were upregulated. Interestingly, also genes correlated to cell response to biotic and abiotic stress, including redox stimuli, resulted in transcriptionally upregulated.
In addition, we report about the antioxidant properties and diminished generation of ROS in HRCs expressing PhAN4 exposed to ionizing gamma radiation. PhAN4 HRCs were endowed with a ninefold enhanced antioxidant capacity per se compared to controls by 2,2-diphenyl-1-picrylhydrazyl (DPPH) assay. Furthermore, ROS accumulation was counteracted after gamma radiation, as shown by Electron Spin Resonance (ESR) Spectroscopy. Both UV-VIS spectra and photoluminescence analysis demonstrated that polyphenols content and stability of soluble protein folding were not significantly affected by high dose gamma irradiation in PhAN4-engineered HRCs compared to control.
In conclusion, MicroTom HRCs represented a simplified model that allowed to rapidly test PhAN4 expression effects on tomato cells, possibly opening the way to the application of the strategy to the engineering of whole plants intended for cultivation in harsh environments like future space outposts.
Materials and Methods
Gene and Constructs
Anthocyanin4 (PhAN4) complementary DNA (cDNA) from petals of Petunia × hybrida cultivar Violet 30 (GenBank: HQ428105.1) was amplified with primers containing AttB sites and recombined into pDONR221 (RU Ghent) to produce an entry clone. This was then recombined with pKGW,0 (RU Ghent) to produce the 35S:AN4 construct and in pK7FWG2 (RU Ghent) to yield the 35S:GFP-PhAN4 construct (where the GFP gene fusion was adopted to possibly stabilize the PhAN4 transcription factor).
Hairy Root Cultures Generation
Solanum lycopersicum (cv. MicroTom) clonal hairy root lines were obtained from wild-type leaf explants by infection with A. rhizogenes A4 (ATCC, 43057™) harboring either the 35S:PhAN4 or the 35S:GFP-PhAN4 or no additional construct. Bacteria were grown in a YEB medium with 50 μg/ml rifampicin and 50 μg/ml kanamycin at 28°C and 220 rpm to OD600 = 0.6. Bacteria were centrifuged at 3,000 × g for 15 min and resuspended at OD600 = 1 in Murashige and Skoog medium (MS, Duchefa) with 30 g/l sucrose and 200 μM acetosyringone, pH 5.8. Leaves from 3-week-old MicroTom plants were harvested, sterilized in 0.1% (v/v) sodium hypochlorite solution for 15 min, and aseptically cut into explants of 1 cm × 1 cm. Explants were immersed in the recombinant A. rhizogenes suspension for 15 min, in a rotary shaker at the minimum speed, and in the dark. Explants were dried onto sterilized tissue paper and transferred on their adaxial side, on MS agar medium co-culture plates with 100 μM acetosyringone and incubated in the dark for 4 days. Explants were then blotted and transferred to MS medium supplemented with 250 μg/ml cefotaxime (Cef) at 25°C. Fresh growing hairy roots were obtained after 8–10 days. Emerging roots of 1 cm in length were excised and transferred to new plates. A. rhizogenes was eliminated with decreasing Cef concentrations (0.25, 0.125, and 0.05 μg/ml) until no antibiotic was added. HRCs were screened for pigmentation under a dissecting microscope. Growth was estimated by the increase in fresh weight at different time points after subculture over a 28-day culture period recorded for three biological replicates for chosen hairy root clones. Hairy root biomass harvested for analysis was carefully handled, pulverized in liquid nitrogen, and immediately stored at −80°C. For metabolite content and antioxidant properties analysis, HRCs were lyophilized in a freeze-dry system (FreeZone Labconco, Kansas City, MO, United States).
Polymerase Chain Reaction Assays
Standard polymerase chain reaction (PCR) assays were performed on genomic DNA of kanamycin-resistant hairy root clones (extracted with NucleoSpin Plant II Kit; Macherey-Nagel; Duren, Germany) with primers specific for PhAN4, rol B, rol C, virC1, respectively (Supplementary Table 1), to select hairy root lines carrying PhAN4 transgene clean from A. rhizogenes in the tissue culture. In selected HRC clones, SYBR Green real-time PCR was used to determine the PhAN4 copy number. The tomato actin 41 gene (NCBI Reference Sequence: NM_001330119.1) served as an endogenous gene reference. For qPCR (i-Cycler iQ detection system; BioRad Laboratories Inc., Milan, Italy) Kapa SYBR Fast 2× qPCR Master Mix (KAPA Biosystems, Milano, Italy) was used, according to the manufacturer’s instructions. Samples were amplified at 95°C for 3 min, followed by 40 cycles of denaturation at 95°C for 15 s, annealing, and extension at 60°C for 30 s.
Total RNA was isolated using the RNeasy Plant Mini Kit (Qiagen; Valencia, CA, United States) and then treated with amplification grade DNaseI (Invitrogen, Cambridge, MA, United States). cDNA was synthesized using the iScript™ cDNA Synthesis Kit (BioRad Laboratories Inc., Milan, Italy) and used as a template for real time-PCR analysis (Kapa SYBR Fast 2× qPCR Master Mix; KAPA Biosystems, Milan, Italy) in iCycler iQ detection system (BioRad Laboratories Inc., Milan, Italy). The actin 41 gene was used as the reference gene. Primers are listed in Supplementary Table 1. Relative gene expression levels were obtained using the 2–ΔCT formula (Livak and Schmittgen, 2001).
Phenylpropanoids Identification
Anthocyanin profile was carried out on representative PhAN4 and GFP-PhAN4 HRCs by liquid chromatography coupled to high-resolution mass spectrometry (LC-HRMS) as reported before (Diretto et al., 2019; Carmona et al., 2021) with slight modifications. Briefly, 3 mg (dried weight) of ground hairy roots were re-suspended in 600 μl of 85:15 MeOH: 1N HCl, vortexed, shaken in Mixer Mill (MM) for 15′ at 20 Hz frequency and gently mixed at 4°C O.N. Samples were then centrifuged at 20,000 × g for 20 min, the supernatant recovered, completely dried and re-suspended in 600 μl of spiked (with 0.5 μg/ml formononetin, as internal standard) 75% MeOH + 0.1% formic acid. Samples were then centrifuged 10 min at 20,000 × g at RT, and the supernatant was transferred to HPLC vials for MS analysis with a Q-Exactive mass spectrometer (Thermo Fisher Scientific, Cambridge, MA, United States), coupled to a HPLC system equipped with a photodiode array detector (Dionex, Califiornia, United States). LC separation of anthocyanins was performed injecting 5 μl of sample on a C18 Luna reverse-phase column (100 × 2.1 mm, 2.5 μm; Phenomenex, Torrance, CA, United States), using as mobile phase water + 0.1% formic acid (A) and acetonitrile + 0.1% formic acid (B) at a total flow rate of 250 μl/min. The separation was developed using 5% B for 0.5 min, followed by a 24 min linear gradient to 75% B. The ionization was performed using heated electrospray ionization (HESI) source, with nitrogen used as sheath and auxiliary gas, and set to 35 and 10 units, respectively. The vaporizer temperature was 250°C, the capillary temperature was 30°C, the spray voltage was set to 3.5 kV, the probe heater temperature was 390°C, and the S-lens RF level was set at 50. The acquisition was performed in the mass range 110/1,600 m/z both in positive and in negative ion mode with the following parameters: resolution 70,000, microscan 1, AGC target 1e6, maximum injection time 50. UV-VIS detection was continuous from 220 to 700 nm. All solvents used were LC-MS grade (Merck Millipore, Burlington, MA, United States). Identification was achieved based on accurate masses and by comparison with authentic reference substances. The ion peak areas were normalized to the ion peak area of the internal standard (formononetin).
Total anthocyanins content was measured by spectrophotometric analysis, as described in the study of Brito et al. (2014), using the extinction coefficient of the most abundant anthocyanin (petunidin-3-(p-coumaroyl)-rutinoside-5-glucoside).
Determination of the Total Phenolics Accumulation Level
Total phenolic content of representative PhAN4 and GFP-PhAN4 MicroTom HRC was estimated by colorimetric assay with modified (Inglett et al., 2010) Folin–Ciocalteu reagent (Merk, Germany) (Şensoy et al., 2006). Briefly, 4.25 ml of de-ionized water was mixed with 0.25 ml of ethanolic extract diluted 1:5 with 80% (v/v) ethanol and 0.25 ml of Folin-Ciocalteu reagent. After 7 min incubation in the dark at room temperature, 0.5 ml of saturated sodium carbonate solution (20%) was added, and the mixture was incubated for 40 min in the dark at room temperature. Absorbance was measured at 725 nm using a UV-VIS spectrophotometer (PerkinElmer, Waltham, MA, United States). A standard curve was prepared with gallic acid, as the reference standard. Final values were obtained by interpolating the absorbance values recorded for tomato hairy root extracts with the gallic acid calibration curve. The total phenolic content was expressed as μg of gallic acid equivalents (GAE)/g of dry weight. Each analysis consisted of triplicate measurements of each sample and data were averaged over the three measurements.
1,1-Diphenyl-2-Picrylhydrazyl Radical Scavenging Activity Assay
Phenolic compounds accumulating in representative PhAN4 and GFP-PhAN4 HRCs were extracted (Morishita et al., 2007) to apply the DPPH free radical scavenging method in order to establish their antioxidant properties. Ground, freeze-dried tomato HRC (30 mg) were extracted with 0.6 ml EtOH 80% (v/v), shaken in a water bath at 80°C for 40 min, and then centrifuged at 3,000 rpm for 15 min. The recovered supernatant was filtered through polytetrafluoroethylene membrane (0.45 μm) and stored at −20°C. The antioxidant capacity of the ethanolic extracts was spectrophotometrically tested by DPPH (1,1-diphenyl-2-picrylhydrazyl) (Li et al., 2010), with some modifications. Briefly, 0.25 ml of diluted ethanolic extract was added to 2.9 ml of 0.06 mM DPPH working solution. The mixture was shaken and allowed to stand at room temperature, in the dark, for 30 min. Absorbance was measured at 515 nm using a UV-VIS spectrophotometer. Lower absorbance values of the reaction mixture indicated higher free radical scavenging activity. The inhibition of free radical DPPH was expressed as DPPH scavenging effect (% inhibition) I% = {(A0 − A1)/A0) × 100}, where A1 and A0 are the absorbance values of blank and of tested samples, respectively. Trolox (vitamin E equivalent antioxidant) was used as the reference standard. Each analysis consisted of triplicate measurements of each sample and data were averaged over the three measurements.
Complementary DNA Library Construction and Sequencing for Transcriptomic Analysis
Total RNA from wild type and PhAN4-1 HRC was extracted using the RNeasy Plant Mini Kit (Qiagen; Valencia, CA, United States). RNA was quantified using the Qubit ® fluorometer (Thermo Fisher Scientific, Waltham, MA, United States) and assayed through Agilent 2100 Bioanalyzer ® (Agilent Technologies, Santa Clara, CA, United States) for quality and Integrity Number (RIN) evaluation. Samples with 8 ≤ RIN ≤ 10 were considered. For each sample, equal amounts of RNA (2 μg) extracted from three biological replicates were pooled for cDNA library construction. First, the poly-A mRNA in the total RNA was pulled down using poly-T oligo-attached magnetic beads. After purification, the mRNA was fragmented at 95°C for 2 min along with RT primer and first-strand buffer. The fragmented RNA was used for the synthesis of the first-strand cDNA by adding DTT, dNTPs, Rnase Inhibitor, and SMARTScribe. All chemicals were incubated for 2 h at 42°C. This was followed by second-strand cDNA synthesis using template-switching oligonucleotide and an additional 1 μl of SMARTScribe. Subsequently, the cDNAs were purified using two rounds of AMPure beads. The samples were enriched by using PCR to create the final library. A final fragment size purification step was performed using the Blue Pippin system selecting fragments between 250 and 500 bp. The libraries were confirmed by electrophoresis on a 1% TAE agarose gel and the Agilent BioAnalyzer 2100 ® after purification by AMPure bead. The average read size was estimated at 250–350 bp. The libraries were sequenced at Novogene Corporation using two lanes of an Illumina HiSeq4000 system with a pair-end run of 2 × 150 bp (Illumina, San Diego, CA, United States).
Sequencing Read Mapping and Identification of Differentially Expressed Genes
Raw RNA-seq libraries were analyzed according to the bioinformatic in-house pipeline (University of Amsterdam, Netherlands)1. The raw reads (in FASTQ format) generated from sequencing were cleaned using Trimmomatic version 0.36 (Institut Pasteur, France) (Cock et al., 2009) by removing adaptor-polluted reads, reads with unknown sequences “N” accounting for more than 5% and low-quality reads (with a mass value less than 10 and proportion of a total number of bases in the reads greater than 20%). The clean reads were mapped to the S. lycopersicum reference genome sequence version ITAG4 (Consortium et al., 2012) downloaded from the Sol Genomics Network database (Fernandez-Pozo et al., 2015). Two programs were used for this purpose: Hisat2 version 2.1 (Institut Pasteur, France) (Kim et al., 2015) and STAR version 2.5.2b (Institut Pasteur, France) (Dobin et al., 2013). The mapping results were compared with the Picard Tools CollectAlignmentSummaryMetrics version 1.138 (Broad Institute, Cambridge, MA, United States)2. The STAR mapping results were selected for further analysis. BAM files were transformed to a subread matrix file using the Rsubread version 1.34.4 R package (Bioconductor, open source) (Liao et al., 2019). Differential expression analysis was performed with the DESeq2 R package version 1.22.2 (Bioconductor, open source) (Love et al., 2014). Genes with an adjusted P-value of ≤0.001 and a log2 fold change of ≥2 were defined as differentially expressed (Benjamini and Hochberg, 1995).
Functional Annotation and Enrichment Pathway Analysis of Differentially Expressed Genes and Identification of Tomato Genes
The list of DEGs (Cluster 1 – PhAN4/WT < 1 and Cluster 2 – PhAN4/WT > 1) was analyzed using the g:Profiler (Reimand et al., 2007) with the default parameters using Organism Solanum lycopersicum. No terms were statistically significant under the “Measure underrepresentation” option. The results were exported as CSV and uploaded into R Studio where they were plotted with ggplot23. The gene functional annotation was performed by sequence homology search with different protein data sets using BLASTP and Protein domains search using InterPro Scan.
Measurements of Tomato Hairy Root Cultures pH
Measurement of both control and PhAN4 HRCs pH was accomplished as described by Verweij et al. (2008). Briefly, 10 mg of hairy root material were ground in 2 ml distilled water and immediately measured with a pH electrode (edge ® Multiparameter pH Meter – Hanna Instruments, Italy).
Gamma Irradiation Tests
Irradiation tests were performed at the Calliope facility, a pool-type irradiation plant equipped with a 60Co gamma source in a high volume (7 m × 6 m × 3.9 m) shielded cell at ENEA (Casaccia Research Centre, Rome, Italy). The source emits radiations consisting of two gamma photons with a mean energy of 1.25 MeV (Baccaro et al., 2019). Fricke dosimetric system was employed for the determination of the absorbed dose during the irradiation tests. HRCs-derived samples were irradiated at room temperature, at three different absorbed doses (0.5, 1, and 2 kGy), and a dose rate of 1.8 kGywater/h.
Electron Spin Resonance Spectroscopy Before and After Gamma Irradiation
The molecular species accumulating upon the ectopic expression of PhAN4 were investigated for possible efficient maintenance of the ability to counteract the generation of reactive oxygen species upon strong ROS inducers such as ionizing radiations. Gamma rays were used to generate peroxyl radicals (which are proportional to the number of paramagnetic species present in the samples) in lyophilized HRC powder. We explored ROS formation before and after 0.5, 1, and 2 kGy absorbed dose in AN4-1 and control HRCs sample sets by ESR Spectroscopy measurements. Not irradiated sample sets were used as references. Each set consisted of two replicates. For each ESR analysis, 8 ± 0.1 mg of HRC lyophilized powder was split into two PT-Capillaries (NOX-A.8.1-PT NOXYGEN, Holland) that were then inserted in a conventional quartz sample tube (o.d./i.d. of 4/3 mm) closed by a plastic lid. Irradiated samples were analyzed straight after the end of irradiation and ESR signals and were normalized to the sample mass. ESR measurements were acquired using an ESR e-scan spectrometer (Bruker, Billerica, MA, United States) operating in the X-band frequency (9.4 GHz) with a field modulation frequency of 86 kHz and modulation amplitude of 5.152 G. The ESR spectra were recorded at a central magnetic field of 3466 Gauss with a sweep width of 160 G, microwave power of 0.14 mW, microwave frequency of 9.75 GHz. The ESR spectra reported in this work derived from the accumulation of four scans. Bruker WinEPR data processing software (Bruker, Billerica, MA, United States) was used for data elaboration.
Ultraviolet-Visible Absorbance Spectra Analysis Before and After Gamma Irradiation
Ultraviolet-visible (UV-VIS) spectra were obtained from crude extracts of AN4-1 and control HRCs. Briefly, lyophilized HRCs were ground in liquid nitrogen and the resulting powder was finely homogenized using an Ultraturrax homogenizer (IKA, Germany) in water:HCl (100:1, v/v). Samples were incubated at 500 rpm for 1 h at R.T., clarified by centrifugation at 11,000 × g for 30 min. The resulting supernatants were examined at 280–600 nm, by a UV-spectrophotometer (Lambda 950, Perkin Elmer, Waltham, MA, United States) at R.T. with a slit width of 2 nm, using a 10 mm cell.
Photoluminescence Analysis Before and After Gamma Irradiation
Samples were finely ground in liquid nitrogen with mortar and pestle, resuspended, and homogenized in phosphate-buffered saline pH 7.2 (PBS, 1:3 w/v) containing a protease inhibitor cocktail (Complete™; Roche, Mannheim, Germany) to extract soluble proteins. Photoluminescence emission spectra of extracts were determined before and straight after 2 kGy absorbed dose in AN4-1 and control HRC dried biomass sample sets. Two replicates per set were poured into quartz cuvettes with an optical path length of 1 cm (104F-QS, Hellma, Germany). The emission spectra were recorded using the Edinburgh Instruments FS 5 spectrometer in the range 300–800 nm with 280 nm excitation wavelength. The recorded spectra were mass-normalized and corrected for background scattering (reference: extraction buffer).
Statistical Analysis
All data (HRCs growth, PhAN4 gene expression, total phenolic content, DPPH antioxidant capacity, total anthocyanin content) were subjected to one-way ANOVA with Tukey’s post-test to determine the differences in average of all tested parameters ± SD. A p-value less than 0.05 was considered statistically significant. GraphPad Prism version 8.0.2 for Windows (GraphPad Software, San Diego, CA, United States) was used for graphical and statistical data processing.
Results
Hairy Roots Generation and Screening
Both 35S:PhAN4 and 35S:GFP-PhAN4 constructs were independently transferred into the miniature tomato genotype MicroTom to generate hairy root cultures. Control HRCs were obtained by transformation with A. rhizogenes not containing PhAN4. HRCs were collected from independent explants. While control HRCs grew as unpigmented organ cultures (Figure 1A, left), HRCs generated by 35S:PhAN4 and 35S:GFP-PhAN4 showed purple pigmentation (Figure 1A, right) that may vary among clones. Purple pigmentation was present on primary and secondary branches and was maintained on kanamycin selection (Supplementary Figure 1). HRCs showed typical abundant secondary branching. Three clones for each construct were selected and further analyzed. No statistical difference was found in the growth rate of PhAN4, GFP-PhAN4, and control HRCs (Figure 1B).
FIGURE 1.
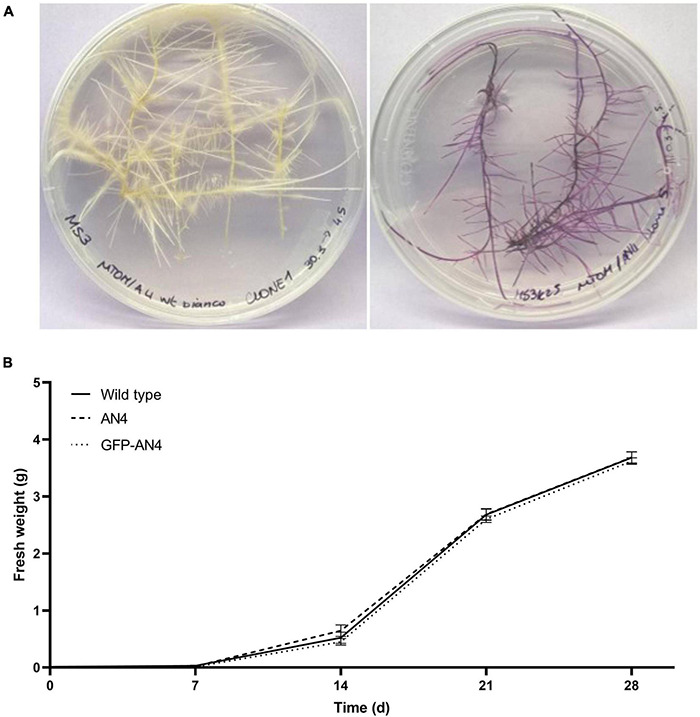
Control (left) and transformed (35S:AN4, right) MicroTom hairy roots (A). Hairy root culture (HRC) growth estimation by an increase in fresh weight along 28 days of cultures in MS medium (one-way ANOVA analysis of variance with Tukey’s post-hoc test) by GraphPad Prism (B).
Polymerase chain reaction (PCR) screening showed amplification of the expected fragments from genomic DNA, confirming integration of the necessary root-inducing genes from A. rhizogenes and of the PhAN4 transgene (Supplementary Figure 2A). No transgene loss was observed over time (Supplementary Figure 2B). Control HRCs were negative for PhAN4 amplification, as expected. Integration of PhAN4 was estimated at copy numbers ranging from 4 to 8 copies, depending on the selected clone analyzed (Supplementary Figure 3). PhAN4 transcripts were detected in both HRCs harboring either 35S:PhAN4 or 35S:GFP-PhAN4 constructs, while they were absent in the control, as expected (Supplementary Figure 3).
Liquid Chromatography Coupled to High-Resolution Mass Spectrometry Analysis of Phenylpropanoids
Total anthocyanins content was measured. AN4-1 and AN4-4 HRCs showed the highest anthocyanins concentrations, equal to 37 and 36.6 μg/gdried weight (Figure 2A), respectively. The anthocyanins profile was determined by LC-HRMS and compared to control HRCs. Identification was achieved by m/z ion reconstruction starting with the aglycon (delphinidin, petunidin, pelargonidin, malvidin), followed by the recognition of all the conjugated sugar and phenolic moieties. Subsequently, absolute quantification was performed as previously described (Diretto et al., 2019; Carmona et al., 2021) and by interpolating anthocyanin signal intensities in the roots compared to the ones of external calibration curves of the Pelargonidin-3-glucoside and Delphinidin 3,5-O-diglucoside standards. In our experimental conditions, anthocyanins were virtually undetectable in control HRCs. Petunidin-3-(p-coumaroyl)-rutinoside-5-glucoside1 and Delphinidin 3,5-O-diglucoside were the most abundant anthocyanins in the PhAN4 roots, followed by a second group including Petunidin-3-(p-coumaroyl)-rutinoside-5-glucoside2, Petunidin-3-feruloyl-rutinoside-5-glucoside and Delphinidin-3-(p-coumaroyl)-rutinoside-5-glucoside. Two additional anthocyanins, Pelargonidin-3-glucoside and Malvidin-3-O-(4”’coumaroyl)-rutinose-5-O-glucose, were detected, although at low levels (Figure 2B; Su et al., 2016). All lines show about the same relative amount of the different anthocyanin species.
FIGURE 2.
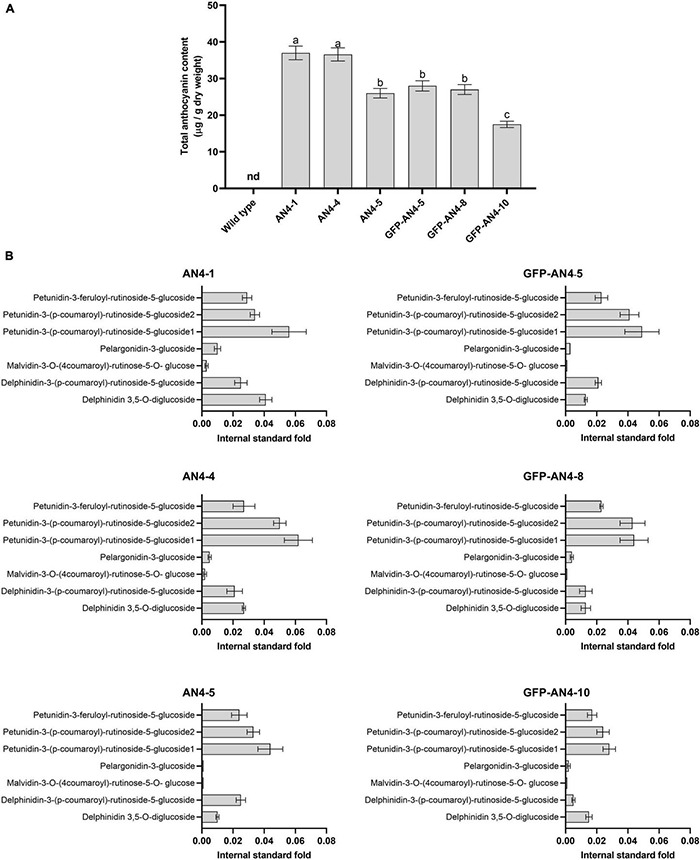
Anthocyanins in PhAN4 HRC clones. Total anthocyanins content (A) and amount of specific anthocyanins (B) of six representative PhAN4-expressing hairy root clones. Columns report average values ± SD (n = 3).
To evaluate the effect of the accumulation of anthocyanins on their precursors and the final balance on flavonoids accumulation, a detailed analysis of phenylpropanoids was carried out by LC-HRMS on the engineered HRCs. A graphical representation of the accumulation levels of anthocyanins precursors is shown for the best anthocyanin-accumulating AN4-1 HRC (Figures 3, 4) and the other engineered HRC (Supplementary Files 1, 2). A series of phenolic acids and their derivatives (e.g., dicaffeoylquinic, 5-caffeoyl-quinic, and 4-caffeoyl-quinic acids) were accumulated at a significantly lower level compared to control. This finding might be ascribed to the role of these compounds as flavanones and flavonols precursors of their sugar-decorated derivatives and of anthocyanins that, in turn, resulted to be enhanced in accumulation in engineered HRCs. As a consequence of the lower accumulation of phenolic acids, hydroxycinnamic acid accumulated at reduced levels in engineered HRCs, with respect to control. Interestingly, the hydroxybenzoic acid level was not significantly different. Significantly higher levels of other valuable phenolics such as coumaric acid and caffeic acid derivatives were found in AN4-1 HRC compared to control (Figure 3).
FIGURE 3.
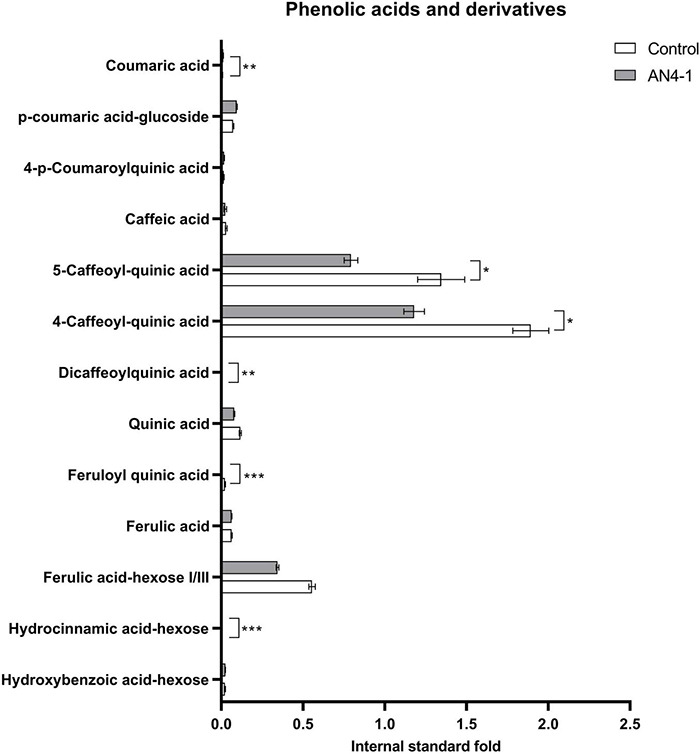
Graph representing phenolic compounds and their derivatives expressed as internal standard (formononetin) fold of control and AN4-1 HRCs. Columns report average values ± SD (n = 3). *p-value ≤ 0.05, **p-value ≤ 0.01 and ***p-value ≤ 0.001.
FIGURE 4.
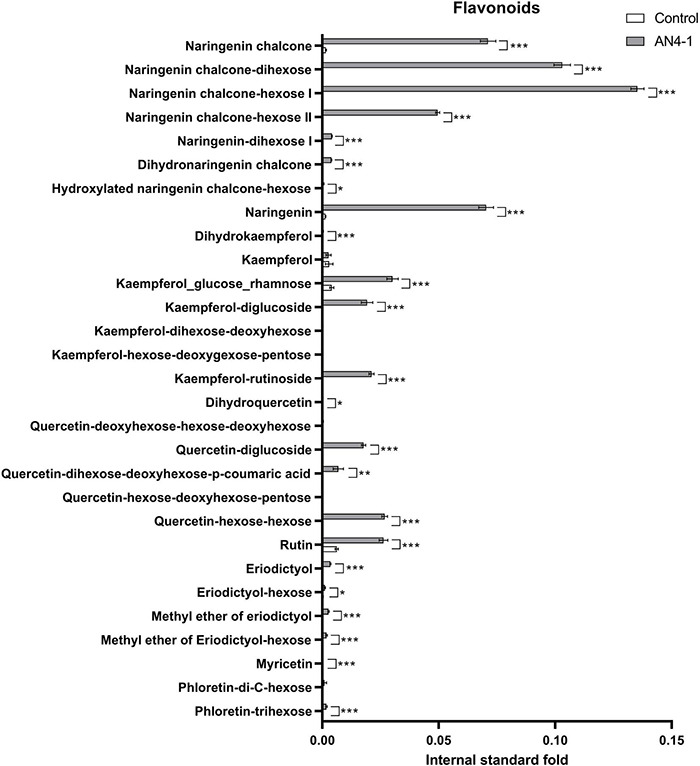
Flavonoids are expressed as internal standard (formononetin) fold of control and AN4-1 HRCs. Columns report average values ± SD (n = 3). *p-value ≤ 0.05, **p-value ≤ 0.01 and ***p-value ≤ 0.001.
Accordingly, among flavonoids, both flavanones (i.e., naringenin chalcone and its sugar-decorated derivatives, naringenin, eriodictyol) and flavonols (i.e., dihydrokaempferol, sugar-decorated kaempferol, dihydroquercetin, quercetin derivatives, rutin, and myricetin) resulted in an overall significantly higher accumulation in engineered HRCs compared to control (Figure 4 and Supplementary File 2). Notably, and coherently with phenolic acids precursor function, the observed fold change levels on most flavonoid groups (from naringenin and kaempferol derivatives to quercetin derivatives and rutin) displayed a much larger extent (192.82 ± 39.1 for naringenin dihexose I) compared to the phenolic acid precursor (0.623 ± 0.069 for 4-caffeoyl-quinic acids).
AN4-1 HRC was chosen as the candidate to perform the subsequent transcriptome analysis due to the higher content of the different anthocyanin and flavonoid species compared to the remaining clones.
Transcriptome Analysis of Tomato PhAN4 Hairy Roots
The response to the constitutive expression of PhAN4 in tomato HRCs were analyzed transcriptome-wide by RNAseq analysis and compared to control HRCs. AN4-1 showed a total of 442 differentially expressed genes (DEGs), of which 331 were upregulated and 111 were downregulated (Table 1 and Supplementary File 3). After Gene Ontology Enrichment (GOE) analysis, 38 upregulated and 8 downregulated genes were termed (Supplementary File 4). For upregulated DEGs, 3 GO categories were assigned: Molecular Function (MF), Biological Process (BP), and Cellular Component (CC) (Figure 5).
TABLE 1.
List of the more representative and significant DEGs obtained from the GOE and functional annotation analyses in relation to agrospace application.
| Gene ID | Log2Fc | Gene name | Function | References |
| Solyc03g020080.3.1 | 2.017930333 | SlPI11 | Pin-II type proteinase inhibitor/biotic (herbivorous and insects resistance), abiotic (drought/heat) | Fan et al., 2020 |
| Solyc03g020030.3.1 | 10.72904412 | SlPI16 | Pin-II type proteinase inhibitor/biotic (wound stress, insect resistance), abiotic (heat) | Fan et al., 2020 |
| Solyc11g020960.2.1 | 2.362943887 | SlPI51 | Proteinase inhibitor II/biotic (wound stress) | Fan et al., 2020 |
| Solyc08g080630.3.1 | 2.33279266 | SlPI31 | Proteinase inhibitor 1/biotic (TSWV infection), abiotic (drought) | Fan et al., 2020 |
| Solyc10g086090.2.1 | 2.247200225 | SlPI47 | Trypsin inhibitor 1/abiotic (drought) | Fan et al., 2020 |
| Solyc10g086100.2.1 | 2.406338004 | SlPI48 | Proteinase inhibitor/abiotic (heat) | Fan et al., 2020 |
| Solyc01g059965.1.1 | 3.476158885 | SlGluB | Beta-1,3-glucanase/biotic (defense against pathogens: C. fulvum, P. infestans) | van Kan et al., 1992; Fan et al., 2021 |
| Solyc09g091510.3.1 | 12.25514456 | CHS | Chalcone synthase 1/phenylpropanoids biosynthesis | Zhang et al., 2019 |
| Solyc05g053550.3.1 | 9.83030415 | CHS | Chalcone synthase 2/phenylpropanoids biosynthesis | Kalt et al., 2020 |
| Solyc02g083860.3.1 | 6.299607501 | F3H | Flavonoid-3-hydroxylase/phenylpropanoids biosynthesis | Aoki et al., 2010; Zhang et al., 2019 |
| Solyc02g085020.3.1 | 10.09460014 | DFR | Dihydroflavonol reductase/phenylpropanoids biosynthesis | Bongue-Bartelsman et al., 1994 |
| Solyc01g106650.3.1 | 3.003680242 | – | Xyloglucan endotransglucosylase/cell elongation | Li P. et al., 2020 |
| Solyc11g011210.2.1 | 2.189943987 | RSI-1 | RSI-1 precursor/lateral root initiation | Taylor and Scheuring, 1994 |
| Solyc03g093390.3.1 | 2.390710296 | LeEXPB2 | Expansin-B15-like/sexual reproduction | Sundaresan et al., 2016 |
| Solyc08g077910.3.1 | 3.227703527 | – | Expansin-like B1/sexual reproduction | Nveawiah-Yoho et al., 2013 |
| Solyc05g052245.1.1 | 2.186779111 | – | Expansin A8-like/cell wall organization | – |
| Solyc09g010860.3.1 | 2.630126243 | EXPA4 | Expansin 4-like/cell wall organization | – |
| Solyc02g062510.3.1 | 2.392338587 | – | Peroxidase 72-like/phenylpropanoids biosynthesis | Nveawiah-Yoho et al., 2013 |
| Solyc04g080760.3.1 | 2.42913063 | – | Peroxidase 9/abiotic stress (hypoxia tolerance) | Andolfo et al., 2014 |
| Solyc02g077300.2.1 | 7.236518572 | – | Peroxidase 19/phenylpropanoids biosynthesis, biotic (ToMV infection) | Andolfo et al., 2014; Safavi-Rizi et al., 2020 |
| Solyc06g054320.1.1 | 2.690758759 | – | Dirigent protein/lignin biosynthesis | Paniagua et al., 2017 |
| Solyc10g055190.1.1 | 5.216357243 | – | Dirigent protein/lignin biosynthesis | Paniagua et al., 2017 |
| Solyc10g055200.1.1 | 2.06850204 | – | Dirigent protein/lignin biosynthesis | Paniagua et al., 2017 |
| Solyc04g010270.1.1 | 2.350516375 | – | Dirigent protein/lignin biosynthesis | Paniagua et al., 2017 |
| Solyc02g076710.3.1 | 2.307987898 | CathB | Cathepsin B-like cysteine/biotic (hypersensitive response) | McLellan et al., 2009 |
| Solyc05g053890.2.1 | −2.032569214 | – | UDP-GT-like/flavonoids glycosylation | Fernandez-Moreno et al., 2016 |
| Solyc01g096560.2.1 | 4.109312617 | TomLOXD | Subtilisin-like protease/biotic (wound stress, resistance to insects and necrotrophic pathogens) | Yan et al., 2013 |
| Solyc07g054840.3.1 | 3.341226163 | AtMYB41 | Transcription factor 41/abiotic (salt tolerance) | Hoang et al., 2012 |
| Solyc03g095810.3.1 | 6.923791313 | – | Trichome birefringence-like/xylan acetylation (resistance against micro-organisms, cold and drought) | Zhang et al., 2020 |
| Solyc10g007970.2.1 | 6.961895177 | – | Transcription factor 77/abiotic (water deficit) | Asins et al., 2021 |
| Solyc07g043690.2.1 | 2.800732902 | SlNPR1 | 3-Hydroxyisobutyryl-CoA hydrolase/abiotic (drought stress) | Li J. et al., 2019 |
| Solyc03g096460.3.1 | 2.302142017 | – | Wound signaling | Scranton et al., 2013 |
| Solyc02g080790.3.1 | 3.317661852 | SlDHS | Deoxyhypusine synthase/osmotic stress and chilling injury | Wang et al., 2001; Gupta et al., 2013 |
| Solyc10g081300.1.1 | 2.564715 | SlMC8 | Metacaspase 9/biotic (apoptosis induction by pests), abiotic (drought, cold and salt) | Liu et al., 2016 |
| Solyc10g080690.2.1 | 2.304387313 | – | Patatin/abiotic (flooding) | de Ollas et al., 2021 |
| Solyc06g073760.3.1 | 4.071580028 | BGL2 | Beta-glucosidase/sugar/organic acid ratio tomato fruits/resistance against different pathogens/softening | Liu et al., 2016 |
| Solyc12g035225.1.1 | 9.263765052 | RICESLEEPER1 | BED zinc-finger/abiotic (salt tolerance) | Kashyap et al., 2020 |
| Solyc12g010500.2.1 | 5.358159809 | – | E3 ubiquitin protein/biotic (resistance to X. perforans) | Shi and Panthee, 2020 |
| Solyc12g010670.1.1 | 5.364030417 | – | E3 ubiquitin protein/biotic (resistance to X. perforans) | Shi and Panthee, 2020 |
| Solyc12g009630.2.1 | 5.183567126 | SlCaM3 | Calcium-binding protein/biotic (B. cinerea), abiotic (mechanical wounding, salt and cold stress) | Peng et al., 2014; Shi and Du, 2020 |
| Solyc02g067750.3.1 | 6.915338794 | CA1 | Carbonic anhydrase/abiotic (drought stress) | Li X. et al., 2020 |
| Solyc08g075705.1.1 | 4.258074963 | pTRX y2 | Thioredoxin Y2/ROS detoxification, redox signaling network regulation | Serrato et al., 2013 |
| Solyc09g007190.3.1 | −2.218315202 | PRXL2A | Peroxiredoxin-like 2A/redox regulatory protein | – |
| Solyc03g098760.2.1 | −5.109469505 | – | I3 Kunitz-type trypsin inhibitor/biotic (infection Tetranychus species infection) | Islam et al., 2015; Schimmel et al., 2018 |
| Solyc09g089490.3.1 | −5.211378167 | SlPI40 | Proteinase inhibitor/abiotic (drought, salt) biotic (B. cinerea, TSWV infection) | Fan et al., 2020 |
| Solyc06g008760.1.1 | −3.056115855 | – | Glutaredoxin-C13-like/upregulated in tomato rin mutants | Kumar et al., 2016 |
| Solyc07g055610.2.1 | −2.055605009 | PR1 | Resistance protein R1/biotic (resistance to late blight and F. oxysporum infection) | Bournival et al., 1989; Pan et al., 2000 |
| Solyc01g010480.3.1 | −2.123574853 | KAT1 | K+ channel KAT1/abiotic (tolerance to potassium deficiency) | Zhao et al., 2018 |
| Solyc01g102610.3.1 | −3.607916378 | FRO6 | Ferric reduction oxidase 6/biotic (P. solani infection) | Buoso et al., 2019 |
| Solyc05g010320.3.1 | 4.308610241 | CHI | Chalcone isomerase/phenylpropanoids biosynthesis | Morita et al., 2014; Li Z. et al., 2019 |
| Solyc05g052240.3.1 | 8.452278685 | CHI | Chalcone isomerase/phenylpropanoids biosynthesis | Ron et al., 2014; Hou et al., 2017 |
| Solyc11g066580.2.1 | 16.35234434 | F3′5′H | Flavonoid-3′,5′-hydroxylase/phenylpropanoids biosynthesis | Shi and Panthee, 2020 |
| Solyc08g080040.3.1 | 8.861102345 | ANS | Anthocyanin synthase/phenylpropanoids biosynthesis | Shi and Panthee, 2020 |
| Solyc04g078140.3.1 | 8.35041607 | DilFl | Cytochrome B5/phenylpropanoids biosynthesis (essential for full activity of F3′5′H) | de Vetten et al., 1999 |
| Solyc09g082660.3.1 | 15.05027118 | OMT | Caffeoyl-CoA-O-methyltransferase/phenylpropanoids biosynthesis | Roldan et al., 2014; Shi and Panthee, 2020 |
| Solyc02g062975.1.1 | 7.081891304 | 3UFGT | UDP-glucose flavonoid 3-O-glucosyl transferases/phenylpropanoids biosynthesis | Hu et al., 2011; Tohge et al., 2020 |
| Solyc10g083440.1.1 | 9.972475187 | 3UFGT | UDP-glucose flavonoid 3-O-glucosyl transferases/phenylpropanoids biosynthesis | Hu et al., 2011; Tohge et al., 2017 |
| Solyc12g098590.2.1 | 16.55105848 | 3UFGT | UDP-glucose flavonoid 3-O-glucosyl transferases/phenylpropanoids biosynthesis | Hu et al., 2011; Tohge et al., 2020 |
| Solyc09g059170.2.1 | 8.326262972 | 3RT | Anthocyanidin-3-O-glucoside rhamnosyltransferase/phenylpropanoids biosynthesis | Tohge et al., 2020 |
| Solyc12g088170.2.1 | 13.21347105 | AAT | Flavonoid-3-O-rutinoside-4”’-O-phenylacyltransferase/phenylpropanoids biosynthesis | Florio et al., 2021 |
| Solyc03g025190.4.1 | 16.80814437 | MTP77 | Transparent testa 2-like/toxic compound extrusion, regulation of cell turgescence | dos Santos et al., 2017 |
| Solyc10g006120.2.1 | 7.357873128 | LDOX | Leucoanthocyanidin dioxygenase/phenylpropanoids biosynthesis | Pelletier et al., 1997 |
| Solyc07g052490.3.1 | 7.173073383 | Atv | Myb-like transcription factor Atv/anthocyanin biosynthesis | Cao et al., 2017 |
| Solyc10g086290.2.1 | 8.651275639 | SlAN2 | AN2-like transcription factor/anthocyanin biosynthesis | Sun C. et al., 2019 |
| Solyc12g005800.2.1 | 7.517786731 | SlMYBATV-like | R3-MYB repressor/anthocyanin biosynthesis | Cao et al., 2017 |
| Solyc09g065100.2.1 | 14.18672027 | AN1 | AN1-like transcription factor/anthocyanin biosynthesis | Qiu et al., 2016 |
| Solyc10g084380.1.1 | 3.439413134 | PH3 | WRKY transcription factor/anthocyanin biosynthesis | Verweij et al., 2016 |
| Solyc10g083900.2.1 | 3.542474125 | Myb27 | MYB transcription factor/inhibitor of anthocyanin biosynthesis | Albert et al., 2011, 2014 |
| Solyc01g095640.2.1 | 4.928760848 | SlTRY | Trichome initiation factor ECT3/anthocyanin biosynthesis | Tominaga-Wada et al., 2013; Kim et al., 2017 |
| Solyc01g105880.4.1 | 3.469067651 | TPS4 | Monoterpenoid synthase/terpene biosynthesis | Velázquez-Márquez et al., 2021 |
| Solyc09g092470.2.1 | 5.695783056 | TPS14 | Sesquiterpene synthase/terpene biosynthesis, biotic (F. oxysporum resistance) | Velázquez-Márquez et al., 2021 |
| Solyc11g017240.2.1 | 3.78371117 | SlCM2 | Chorismate mutase/volatile compounds biosynthesis, abiotic (drought stress) | Tzin et al., 2015; Filiz et al., 2019 |
| Solyc08g008630.3.1 | −3.340796451 | Dwarf27 | Beta-carotene isomerase D27/strigolactone and beta-carotene biosynthesis, biotic (psyllid resistance) | Harrison et al., 2021 |
| Solyc04g050930.3.1 | −2.072953851 | VDE | Violaxanthin de-epoxidase VDE/carotenoid biosynthesis, biotic (P. syringae infection), abiotic (anoxia) | Yang Y. X. et al., 2015 |
| Solyc05g010180.3.1 | −2.221215961 | CRTISO | Carotenoid isomerase/carotenoid biosynthesis | Isaacson et al., 2002 |
| Solyc12g006140.2.1 | 4.364619823 | – | Chlorophyll a/b-binding protein/fruits ripening | Zouari et al., 2014 |
| Solyc02g065220.3.1 | −2.141082227 | – | Cytochrome P450/fruits ripening (extended shelf-life | Gao et al., 2018 |
| Solyc09g066150.1.1 | −4.574546484 | – | Cytochrome P450/fruits ripening (extended shelf-life) | Gao et al., 2018 |
| Solyc07g006570.3.1 | 4.221848981 | – | Ribonuclease 3-like/RNA biogenesis | Consortium et al., 2012 |
| Solyc02g065230.3.1 | 3.170387722 | IMT7 | Cytochrome P450 | – |
| Solyc02g091440.2.1 | 2.129583909 | bHLH83-like | bHLH/enhance root hair initiation, promote flowering in short day and maintain the iron balance | Qian et al., 2021 |
| Solyc12g088130.2.1 | 4.327213562 | bHLH93-like | bHLH/enhance root hair initiation, promote flowering in short day | – |
| Solyc04g077780.3.1 | 2.019567752 | – | LIM transcription factor/cytoskeleton organization | – |
| Solyc12g013850.2.1 | −2.036803744 | – | Glycosyltransferase/regulates anther development and male-sterility | Omidvar et al., 2015 |
| Solyc02g081340.3.1 | 10.53476424 | GST | Glutathione-S-transferase/anthocyanin biosynthesis | Alfenito et al., 1998 |
| Solyc01g058030.2.1 | 8.08401203 | – | Gibberellin 2-beta-dioxygenase/determines dwarf phenotype with shorter internodes | Sun X. et al., 2019 |
| Solyc05g054360.3.1 | 6.837513657 | – | Pectin methylesterase/flower initiation | Wen et al., 2020 |
| Solyc12g010500.2.1 | 5.358159809 | – | U-box protein/biotic (resistance to X. perforans | Shi and Panthee, 2020 |
The function was provided by SolGenomics Network annotation and by specific literature (in bold).
FIGURE 5.
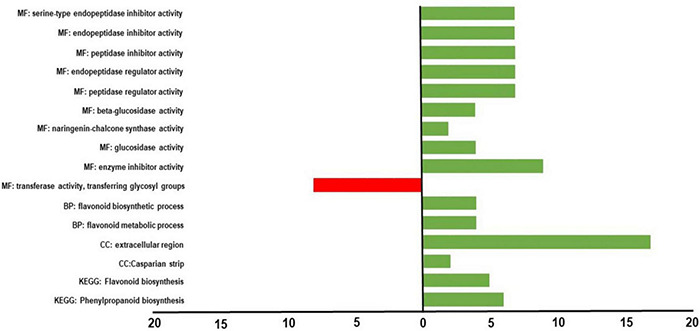
GOs distribution of differentially expressed genes (DEGs). Gene Ontology (GO) terms associated with upregulated (green bars) and downregulated (red bars) genes in the PhAN4 HRCs compared to control HRCs, based on “Molecular Functions” (MF), “Biological Process” (BP), “Cellular Component” (CC) and KEGG ontological domain.
Gene Ontology Enrichment Analysis
Molecular function (MF) resulted in the most abundantly represented GO category, with 9 GO terms and an intersection size of 15 genes over a query size of 165 genes. The most enriched terms were related to ‘enzyme inhibitory activity,’ with nine upregulated DEGs, seven of which are classified as serine-type endopeptidase inhibitors in tomatoes. Among them, Solyc03g020080.3.1, Solyc03g020030.3.1, Solyc11g020960.2.1, Solyc08g080630.3.1, Solyc10g086090.2.1, Solyc10g086100.2.1 encode specific Pin-II type proteinase inhibitors or protease inhibitors that have been already demonstrated to be players of the defense against wounding, pathogens, and pests and of response to abiotic stress, especially UV, drought, and heat stress in tomato (Bergey et al., 1996; Conconi et al., 1996; Fan et al., 2020). Within MF also ‘beta-glucosidase’ and ‘glucosidase activity’ terms were represented with four upregulated DEGs. Among them, Solyc01g059965.1.1 has been demonstrated to be implicated in the defense of tomato plants against pathogens (Murata et al., 2019), and its expression has been found to be modulated in tomatoes upon infection with Cladosporium fulvum and Phytophthora infestans (van Kan et al., 1992; Fan et al., 2021). Upregulation of Solyc06g073760.3.1 has been correlated to increased softening of transgenic tomato overexpressing a MADS-box transcription factor, affecting fruit development and ripening (Huang et al., 2017).
Within the BP category, coherently with secondary metabolites analysis, the ‘flavonoid biosynthetic process’ term was found. In particular, GOE analysis highlighted the upregulation of four genes: Solyc05g053550.3.1, Solyc09g091510.3.1, Solyc02g083860.3.1, Solyc02g085020.3.1. Solyc09g091510.3.1, and Solyc05g053550.3.1 are tomato chalcone synthase (CHS) I and II, respectively, that are early pathway genes that determine the accumulation of the naringenin chalcone precursor of anthocyanins and other flavonoids (Zhang et al., 2019). Solyc02g083860.3.1 is tomato Flavonoid-3-hydroxylase (F3H) that leads to the accumulation of dihydromyricetin, dihydrokaempferol, and dihydroquercetin precursors of anthocyanins and of other flavonoids in tomatoes (Aoki et al., 2010; Zhang et al., 2019). Solyc02g085020.3.1 is tomato dihydroflavonol reductase (DFR) that leads to the accumulation of leucoanthocyanidins in tomatoes (Bongue-Bartelsman et al., 1994).
Within the CC GO category, ‘Extracellular Region’ and ‘Casparian Strip’ terms were categorized. Seventeen genes over a query size of 115, were found to be upregulated within these terms. These genes, in many cases, are specifically correlated to lignin biosynthesis, cell wall organization, and resistance to abiotic and biotic stress functions in tomatoes. In particular, Solyc05g052245.1.1 and Solyc09g010860.3.1 are the tomato expansins A8-like and EXPA4, involved in cell wall organization (Consortium et al., 2012). Solyc02g062510.3.1, which shares 86% homology with the S. tuberosum Peroxidase 72-like gene, and Solyc02g077300.2.1 (tomato Peroxidase 19) are involved in the phenylpropanoids biosynthesis (Mei et al., 2009). In addition, also Solyc04g080760.3.1 (tomato Peroxidase 9) plays a role in hypoxia tolerance and maintaining the iron balance in tomato (Safavi-Rizi et al., 2020). Peroxidase 9 was also demonstrated to be a pathogenesis-related protein upregulated upon Tomato Mosaic Virus infection (Andolfo et al., 2014). Solyc06g054320.1.1, Solyc10g055190.1.1, Solyc10g055200.1.1, and Solyc04g010270.1.1 represent tomato dirigent proteins that contribute to the dimerization of conyferil alcohol, a crucial step toward lignin biosynthesis, modulating cell wall metabolism during abiotic and biotic stress exposure in tomato (Paniagua et al., 2017). Solyc01g110110.3.1 and Solyc02g076710.3.1 are tomato cysteine proteinases. In particular, Solyc02g076710.3.1 shares 90% homology with N. benthamiana cathepsin B-like cysteine proteases that are involved in the hypersensitive response (McLellan et al., 2009). Solyc08g080630.3.1, already highlighted as an upregulated DEG within the MF category GOE, is the tomato SlPI31 protease inhibitor. This gene has been demonstrated to be upregulated under Tomato Spotted Wilt Virus infection in tomato roots and leaves and has been shown to be upregulated in drought-tolerant tomato lines and drought-sensitive varieties under drought conditions (Fan et al., 2020).
Gene ontology enrichment (GOE) analysis revealed only one downregulated DEG (Supplementary File 4) belonging to the Molecular Function term ‘glycosyl transferase.’ Solyc05g053890.2.1 represents the complete sequence of the tomato UDP-GT-like which is probably related to glycosylation of flavonoids prior to their transport to the vacuole (Solyc04g016200.1.1, Solyc04g016210.3.1, Solyc05g053890.2.1, Solyc01g095760.3.1 are incomplete sequences of tomato UDP-GT-like) (Fernandez-Moreno et al., 2016).
Differentially Expressed Genes Analysis
Despite no GOE being found in the Response to stimulus category, a relatively high number of DEGs was found that could be associated with response to abiotic and biotic stress response. Fifteen upregulated and seven downregulated DEGs were found to be correlated with such response. Among upregulated DEGs involved in early signals of defense responses against environmental cues, Solyc01g096560.2.1 (Log2Fc = 4,10; TomLOXD; Supplementary File 3), was retrieved. TomLOXD encodes a lipoxygenase that has been demonstrated to elevate wound-induced jasmonate response, upregulation of wound-induced genes, and enhanced resistance to insects and necrotrophic pathogens in tomatoes (Yan et al., 2013). Solyc07g054840.3.1 (Log2Fc = 3.34; tomato transcription factor 41) shares the best homology with Myb41 of A. thaliana where it functions as a Map-kinase involved in several signaling pathways that control plant development and salt stress tolerance (Hoang et al., 2012). Solyc03g095810.3.1 (Log2Fc = 6.92; tomato Trichome birefringence-like protein) upregulation mediates xylan acetylation and has been demonstrated essential in tomatoes for invading microorganism resistance and against environmental stress like cold and drought (Zhang et al., 2020). Solyc10g007970.2.1 (Log2Fc = 6.96; tomato WRKY transcription factor 77) has been demonstrated to be involved in the signaling to water deficit in tomatoes (Asins et al., 2021). Solyc07g043690.2.1 (Log2Fc = 2.80; tomato SlNPR1) has been suggested to regulate tomato plant drought response (Li J. et al., 2019). Solyc03g096460.3.1 (Log2Fc = 2.3) is a known modulator of wound signaling in tomatoes (Scranton et al., 2013). Solyc02g080790.3.1 (Log2Fc = 3.31; tomato deoxypusine synthase SlDHS) has been shown to be upregulated during osmotic stress and chilling injury (Wang et al., 2001; Gupta et al., 2013). Solyc10g081300.1.1 (Log2Fc = 2.56; SlMC8), encodes a metacaspase that is upregulated during apoptosis induction by pests and regulated by drought, cold, and salt in tomatoes (Liu et al., 2016). Solyc10g080690.2.1 (Log2Fc = 2.3; tomato patatin defense protein) has been demonstrated to be upregulated upon soil flooding in tomatoes (de Ollas et al., 2021). Solyc06g073760.3.1 (Log2Fc = 4.07; tomato β 1,3-glucanase 2 BGL2) has been found to be negatively correlated with the sugar/organic acid ratio of tomato fruits (Li et al., 2021). The simultaneous upregulation of BGL2 and PR1, the marker genes of the salicylic acid (SA) pathway, is a hallmark of systemic resistance induced in tomato plants against different pathogens and can be followed by accumulation of SA at high levels (Peng et al., 2004; Fahim et al., 2016; Hanan et al., 2020). Solyc12g035225.1.1 (Log2Fc = 9.26; tomato putative zinc-finger domain-containing protein) shares 75% homology with rice RICESLEEPER1 that was found to be upregulated in S. chilense in relation to transcription factors for salt tolerance (Kashyap et al., 2020). Solyc12g010500.2.1 and Solyc12g010670.1.1 (Log2Fc = 5.35 and 5.36, respectively) are categorized as E3 ubiquitin proteins that have been found to be upregulated in tomato genotypes resistant to Xanthomonas perforans (Shi and Panthee, 2020). Solyc12g009630.2.1 (Log2Fc = 5.18; tomato calmodulin SlCaM3), encodes an important calcium-binding protein that has been found to be upregulated in tomato stem and roots upon Botrytis cinerea infection and mechanical wounding (Peng et al., 2014). In addition, it has been reported that SlCaM3 is strongly expressed under salt and cold stress in tomatoes (Shi and Du, 2020). Solyc02g067750.3.1 (Log2Fc = 6.91) shares 80% identity with N. benthamiana CA1. During drought stress CA1 proteins gradually diminish within the chloroplast and are accumulated in the cytosol, suggesting that they could be translocated from chloroplasts to the cytosol and act as a signal messenger from the chloroplast in tomato (Li P. et al., 2020). Importantly, in view of the possible improvement of resistance to abiotic stresses relevant to space, Solyc08g075705.1.1 (Log2Fc = 4.25; tomato plastidial thioredoxin Y2, pTRX y2) was found to be upregulated. PTRX y2 together with pTRX y1 and x-type TRXs is mostly involved in ROS detoxification and takes part in the complex redox signaling network regulating tomato plant development (Serrato et al., 2013).
Among downregulated DEGs correlated with resistance functions, Solyc09g007190.3.1 (Log2Fc = −2.21; tomato peroxiredoxin-like 2A PRXL2A) redox regulatory protein was found. Solyc03g098760.2.1 (Log2Fc = −5.1; tomato proteinase inhibitor I3 Kunitz-type trypsin inhibitor) has been demonstrated to be upregulated upon the cell-content feeding mite Tetranychus species infection (Schimmel et al., 2018) and it is told to protect seeds from predators (Islam et al., 2015). Solyc09g089490.3.1 (Log2Fc = −5.21; tomato SlPI40 protease inhibitor) can be induced by abiotic (drought and salt) and biotic (Botrytis cinerea and Tomato Spotted Wilt Virus) stress (Fan et al., 2020). Solyc06g008760.1.1 (Log2Fc = −3.05; tomato Glutaredoxin-C13-like) has been demonstrated to be upregulated in ripening inhibitor (rin) mutants of S. lycopersicum (Kumar et al., 2016). Solyc07g055610.2.1 (Log2Fc = −2.05); tomato Resistance protein R1) has been related to resistance to late blight (Bournival et al., 1989) and it has been shown to be linked to proteinase inhibitor I3 upon Fusarium oxysporum infection (Bournival et al., 1989; Pan et al., 2000). Solyc01g010480 (Log2Fc = −2.2; tomato K+ channel KAT1) was demonstrated to be highly upregulated in low K tolerant tomato genotypes upon potassium deficiency (Zhao et al., 2018). Solyc01g102610.3.1 (Log2Fc = −3.60; tomato FRO6) is involved in nutrient transport in phloem and was observed to be downregulated under Phytoplasma solani infection of tomato (Buoso et al., 2019).
Despite GOE retrieving only four genes among the ‘flavonoid biosynthetic process’ term, many other upregulated DEGs resulted in the analysis that is associated with biosynthesis of anthocyanins and other specialized metabolites (Figure 6). Twelve additional structural genes and six transcription factors were identified as upregulated DEGs. In addition to CHS, F3H and DFR, DEGs analysis revealed also tomato naringenin-chalcone isomerase (CHI) (Solyc05g010320.3.1; Log2Fc = 4.30 and Solyc05g052240.3.1; Log2Fc = 8.45), tomato flavonoid-3′,5′-hydroxylase F3′5′H (Solyc11g066580.2.1; Log2Fc = 16.35), tomato anthocyanin synthase ANS (Solyc08g080040.3.1, Log2Fc = 8.86) were found. Interestingly, also Solyc04g078140.3.1, which shares the best homology with Cytochrome b5 of P. hybrida, where it is essential for full activity of F3′5′H, was upregulated (Log2Fc = 8.35) (de Vetten et al., 1999). In addition, tomato caffeoyl-CoA-O-methyltransferase Solyc09g082660.3.1; Log2Fc = 15.0503), already identified as the prime candidate gene responsible for anthocyanin methylation in tomatoes due to significant correlation of expression with ANS, DFR, and F3′5′H in Rosea1 and Delila fruits (Roldan et al., 2014), resulted upregulated. It shares the best homology with P. hybrida O-methyltransferase (OMT). Interestingly, it was found to have a significant correlation with abiotic stress in tomatoes (Shi and Panthee, 2020). Three tomato UDP-glucose flavonoid 3-O-glucosyl transferases (3UFGT) (Solyc02g062975.1.1, Log2Fc = 7.08; Solyc10g083440.1.1, Log2Fc = 9.97; Solyc12g098590.2.1, Log2Fc = 16.55) catalyzing the transfer of the glucosyl moiety from UDP-glucose to the 3-hydroxyl group of anthocyanidins in tomato were found among upregulated DEGs, as well (Hu et al., 2011; Tohge et al., 2020). Furthermore, anthocyanidin-3-O-glucoside rhamnosyltransferase (3RT) (Solyc09g059170.2.1, Log2Fc = 8.3262), which in tomato controls the conversion of anthocyanidin-3-glucosides to anthocyanidin-3-rutinosides by the UDP rhamnose, was found among upregulated DEGs (Tohge et al., 2020). Tomato anthocyanin acyltransferase (AAT, or Flavonoid-3-O-rutinoside-4”’-O-phenylacyltransferase; Solyc12g088170.2.1, Log2Fc = 13.21) resulted upregulated, as well. Solyc02g081340.3.1 (Log2Fc = 10.53) resulted among upregulated DEGs. In tomato, it encodes a putative Glutathione S-Transferase that shares 84% homology with P. hybrida GST that is responsible for anthocyanin sequestration in the vacuole (Alfenito et al., 1998). Solyc03g025190.4.1 (Log2Fc = 16.80; tomato MTP77) resulted highly upregulated among DEGs. This gene belongs to clade 1 of the multidrug and toxic compound extrusion (MATE) family member (Transparent testa 2-like), which in tomato has been associated with vacuolar chloride channels related to the regulation of cell turgescence. In Micro-Tom, many MATE belonging to clade 1 have been functionally related to the transport of secondary metabolites (dos Santos et al., 2017). Solyc10g006120.2.1 (Log2Fc = 7) was found to be upregulated, as well. This gene shares 59% sequence identity with leucoanthocyanidin dioxygenase (LDOX) from P. hybrida and 61% with S. tuberosum FLS (Pelletier et al., 1997). LDOX has been demonstrated to be a bi-functional enzyme being able both to convert leucoanthocyanidins into anthocyanidins and to catalyze the in planta formation of flavonols in fls1-2 mutants of A. thaliana (Preuß et al., 2009).
FIGURE 6.
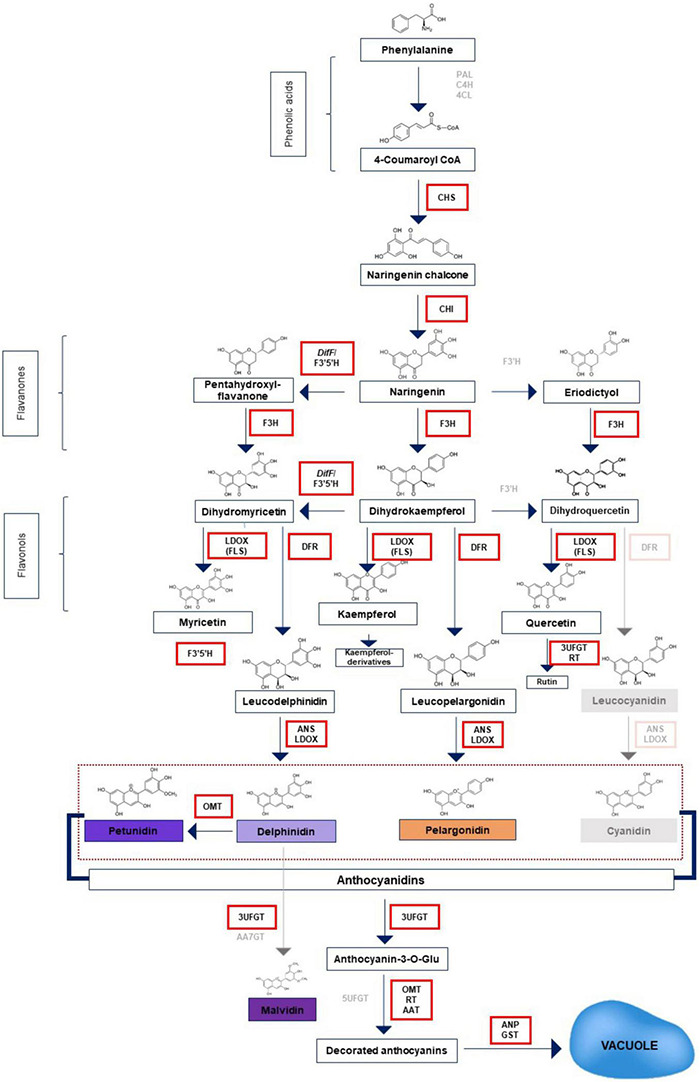
Schematic diagram of the anthocyanin biosynthetic pathway. The structural genes upregulated by ectopic expression of PhAN4 in MicroTom HRCs and leading to accumulation of compounds are in black and marked by red boxes. PAL, phenylalanine ammonia-lyase; C4H, cinnamate-4-hydroxylase; 4CL, 4-coumarate CoA ligase; CHS, chalcone synthase; CHI, chalcone isomerase; F3′H, flavonoid-3′-hydroxylase; F3′5′H, flavonoid-3′,5′-hydroxylase; DFR, dihydroflavonol reductase; ANS, anthocyanin synthase; OMT, O-methyltransferase; AAT, anthocyanin acyltransferase; RT, rhamnosyltransferase; UFGT, UDP-glucose flavonoid 3-O-glucosyl transferase; AA7GT, cyanidin-3-O-glucoside-7-O-glucosyltransferase; ANP, anthocyanin permease; GST, glutathione-S-transferase.
Together with the regulation of structural genes, PhAN4 positively modulated also transcription factors involved in anthocyanin biosynthesis. Solyc07g052490.3.1 (Log2Fc = 7.17, tomato Myb-like transcription factor Atv) is involved in anthocyanin biosynthesis (Cao et al., 2017). Also, Solyc10g086290.2.1 (Log2Fc = 8.65), which shares 83% homology with Solanum tuberosum AN2-like, is a R2R3Myb involved in anthocyanin biosynthesis, was found (Sun C. et al., 2019; Colanero et al., 2020; Yan et al., 2020). Solyc09g065100.2.1 (Log2Fc = 14.18) maps the Hoffman’s anthocyaninless (AH) locus and encodes a bHLH factor (SlAN1) that positively regulates anthocyanin biosynthesis in tomatoes (Qiu et al., 2016). No effects of PhAN4 expression were found on another known regulator of anthocyanins biosynthesis, AN11. Additionally, Solyc10g084380.1.1 was upregulated (Log2Fc = 3.43). In Solanum lycopersicum, this gene encodes a WRKY that shares 76% homology with PH3 from Petunia hybrida where it regulates vacuolar acidification and boosts anthocyanin biosynthesis, as well (Verweij et al., 2016).
Among upregulated DEGs, putative repressors of anthocyanins biosynthesis were found. The upregulated Solyc12g005800.2.1 (Log2Fc = 7.51) is the tomato SlMYBATV-like (Cao et al., 2017), a R3-MYB repressor. Solyc10g083900.2.1 (Log2Fc = 3.54; tomato R2R3Myb transcription factor 27) shares the best homology with Myb27 from P. hybrida, where it is a repressor of the synthesis of anthocyanins (Albert et al., 2011, 2014) and Solyc01g095640.2.1 (Log2Fc = 4.92; tomato ETC3 or SlTRY) orthologous in A. thaliana acts a repressor of anthocyanins accumulation, as well (Tominaga-Wada et al., 2013).
To complete the description related to the modulation of genes related to specialized metabolites, three DEGs resulted to be upregulated and three downregulated. Among upregulated DEGS, Solyc01g105880.4.1 and Solyc09g092470.2.1 (Log2Fc = 3.46 and 5.69, respectively; tomato monoterpenoid synthases 2 TPS4 and TPS14), are involved in the biosynthesis of monoterpenes and sesquiterpenes, respectively. TPS4 has been found to be upregulated in tomato cultivars resistant to Fusarium oxysporum (Velázquez-Márquez et al., 2021). Solyc11g017240.2.1 (Log2Fc = 3.78; tomato chorismate mutase 2 SlCM2), catalyzes the first step of the shikimate pathway from phenylalanine to the volatile compounds responsible for tomato fruit aroma and quality and defense from biotic and abiotic stress response (Tzin et al., 2015). In tomatoes, SlCM2 has been also shown to be upregulated under drought stress (Filiz et al., 2019). Among downregulated genes, Solyc08g008630.3.1 (Log2Fc = −3.34; tomato chloroplastic beta-carotene isomerase D27 Dwarf27) is involved in strigolactone and beta-carotene biosynthesis and has been found to be downregulated in psyllid-infested tomato plants (Harrison et al., 2021). Solyc04g050930.3.1 (Log2Fc = −2.07; tomato violaxanthin de-epoxidase VDE) is involved in the carotenoid biosynthesis. Suppression of VDE can induce the photo-inhibition of the PSII and, at the same time, it results in an accumulation of fucoxanthin that functions as an efficient anti-oxidant in anoxia conditions (Tohge et al., 2020). Moreover, it has been shown that VDE is downregulated in tomato plants upon Pseudomonas syringae pv. tomato DC3000 infection (Yang Y. X. et al., 2015). Solyc05g010180.3.1 (Log2Fc = −2.22; tomato carotenoid isomerase CRTISO) is involved in carotenoids biosynthesis. In MicroTom fruits, the downregulation of carotenoid isomerase has been demonstrated to induce an accumulation of zeta-carotene and cis-prolycopene (dos Santos et al., 2017; Florio et al., 2021), both elevating and modifying carotenoid profiles toward more bioavailable forms compared to wild-type (Pelletier et al., 1997).
To complete the description related to DEGs that were modulated upon expression of PhAN4 and that are related to plant physiology, in addition to Solyc06g073760.3.1 (already described in GOE analysis in the MF category), Solyc12g006140.2.1 (Log2Fc = 4.36; tomato chloroplastic light-harvesting chlorophyll a/b-binding protein 37) was found to be upregulated. Its upregulation was demonstrated to improve photosynthesis and in extending the shelf life in tomato plants (Zouari et al., 2014). Among downregulated DEGs, Solyc02g065220.3.1 (Log2Fc = −2.14; tomato cytochrome P450) was shown to have significantly different transcript levels between purple and red sectors of VIGs Del/Ros tomatoes late ripening. Its downregulation has been associated with extended shelf life (Gao et al., 2018). Solyc09g066150.1.1 (Log2Fc = −4.57; putative tomato cytochrome P450) suppression has been found in transgenic tomato plants unable to perform DNA methylation and its upregulation has been associated with fruit ripening (Zhao et al., 2018).
Phenolic Content and Antioxidant (2,2-Diphenyl-1-Picrylhydrazyl) Activity
To verify whether PhAN4 expression was associated with increased antioxidant activity, in vitro scavenging activity of the hydroalcoholic soluble fraction of HRCs was assessed by non-enzymatic 2,2-diphenyl-1-picrylhydrazyl (DPPH) assay (de Torre et al., 2019). The total phenolic content (expressed as μg of gallic acid equivalents (GAE)/g of dry weight, DW) was found to be ninefold and eightfold compared to control (18.2 × 103 μg GAE/g DW in AN4-1 and 16.4 × 103 μg GAE/g DW AN4-GFP-8 HRCs (Figure 7A). The trolox equivalent antioxidant capacity of the hydroalcoholic soluble fraction (containing anthocyanins) in purple HRCs appears to be thirty (in the AN4-1 clone) and twenty (in the AN4-GFP-8 clone) times higher than in control HRCs (Figure 7B).
FIGURE 7.
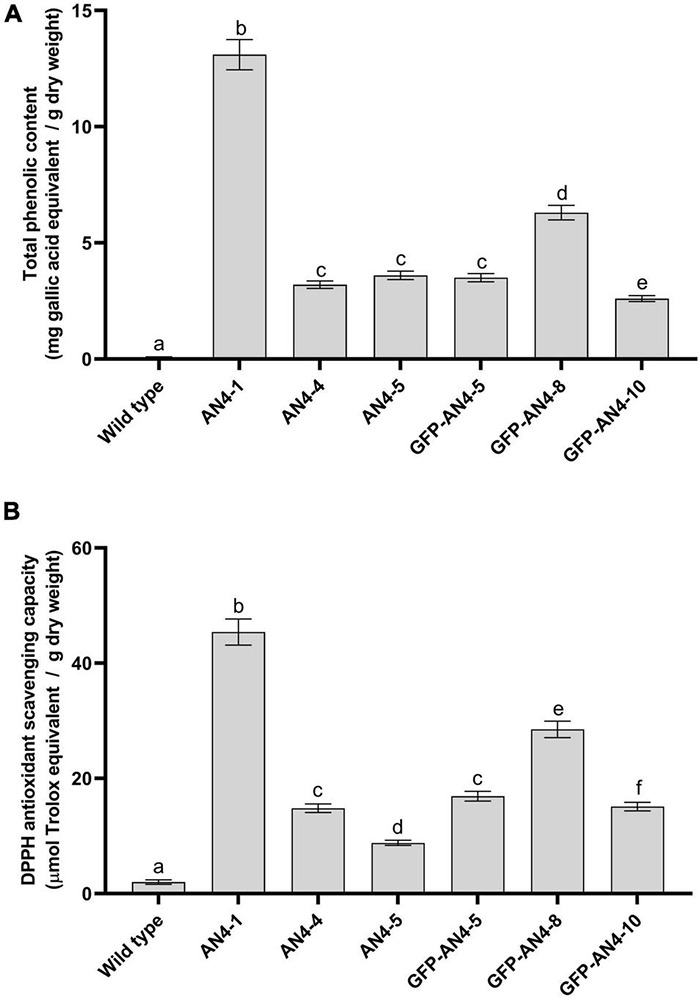
Total phenolic content in MicroTom transgenic HRCs reported in mg of gallic acid equivalents/g of dry weight (A). Analysis of antioxidant activity (DPPH) in MicroTom transgenic HRCs reported as μg Trolox equivalents/g dry weight (DW) (B). Each analysis consisted of triplicate measurements of each sample and data were averaged over the three measurements. A univariate statistical analysis based on Student’s t-test and one way-ANOVA (P ≤ 0.05) was carried out.
Electron Spin Resonance Analysis
To further characterize the effect of the ectopic expression of PhAN4, MicroTom HRC lyophilized material was investigated for its ability to counteract the generation of reactive oxygen species (ROS) and for the maintenance of this feature after high dose gamma radiation. The AN4-1 HRC was chosen as the best candidate to evaluate resistance to radiation by Electron Spin Resonance (ESR). This analysis assessed the amount of peroxyl radicals in lyophilized HRCs. The HRCs ESR signal is characterized by a singlet signal at 3454 Gauss, correspondent to peroxyl radicals induced by gamma irradiation (Andersen et al., 2000; Ichikawa et al., 2001; Nagata et al., 2003; Rossetto et al., 2007; Esatbeyoglu et al., 2014; Faure et al., 2014). Negligible satellite peaks below 3,440 and above 3,480 Gauss are present as well and are related to several different contributions (such as cellulose-like molecules and anthocyanins) (Andersen et al., 2000; Tuner and Korkmaz, 2007; Petrisor et al., 2008; Esatbeyoglu et al., 2014; Faure et al., 2014). Typical ESR spectra of HRCs irradiated at 2,000 Gy absorbed dose, are shown in Figure 8A.
FIGURE 8.
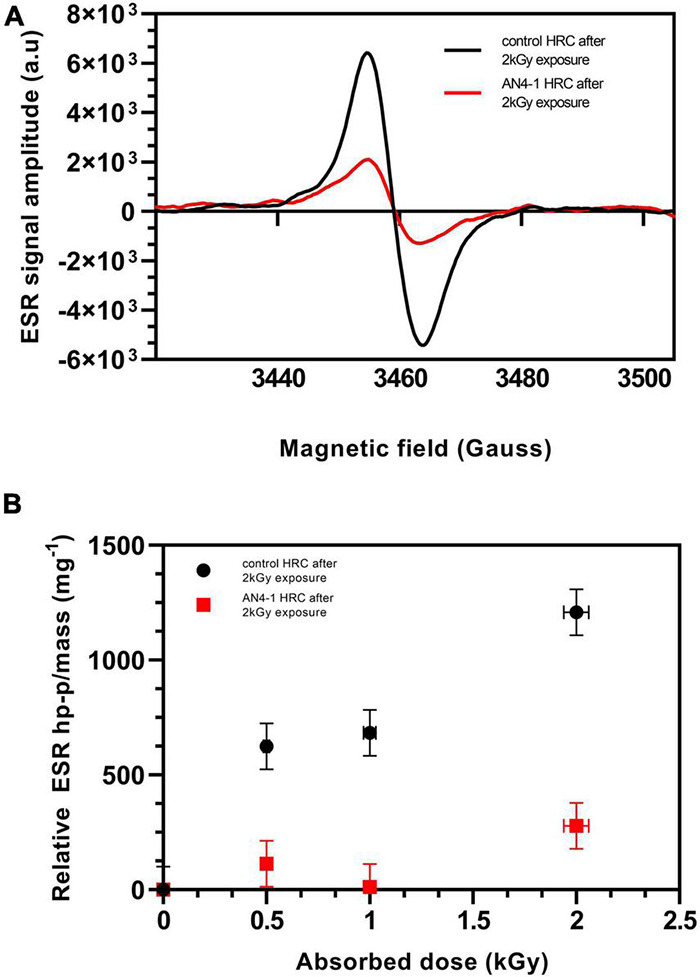
ESR spectra of AN4-1 and control HRCs gamma-irradiated at 2 kGy absorbed dose (dose rate = 1.8) (A). ESR kGy water/h signal intensity of white and purple HRCs gamma are reported as a function of the absorbed dose (up to 2 kGy absorbed dose; dose rate = 1.8 kGy water/h) (B). The intensity of each signal is expressed as the peak-to-peak height normalized for mass units and by subtracting the intensities of the HRCs signals before irradiation.
The AN4-1 and control spectra before gamma radiation exposure do not show significant peaks (data not shown). After irradiation (0.5, 1, and 2 kGy), the control shows a signal intensity significantly higher than AN4-1 (Figures 8A,B). In particular, AN4-1 shows only a negligible signal increase after irradiation from the lowest to the highest absorbed dose. On the contrary, control HRC shows a constant and nearly linear increase of singlet intensity, already evident at low absorbed doses.
Ultraviolet-Visible Absorbance Spectra
We have used UV-VIS analysis of HRC extracts to characterize the resistance to gamma irradiation of the phenolic compounds accumulated in the AN4-1 line. Before irradiation, both control and AN4-1 acidic extracts revealed the main peak at 280 nm, followed by another peak at 315–320 nm (Figure 9).
FIGURE 9.
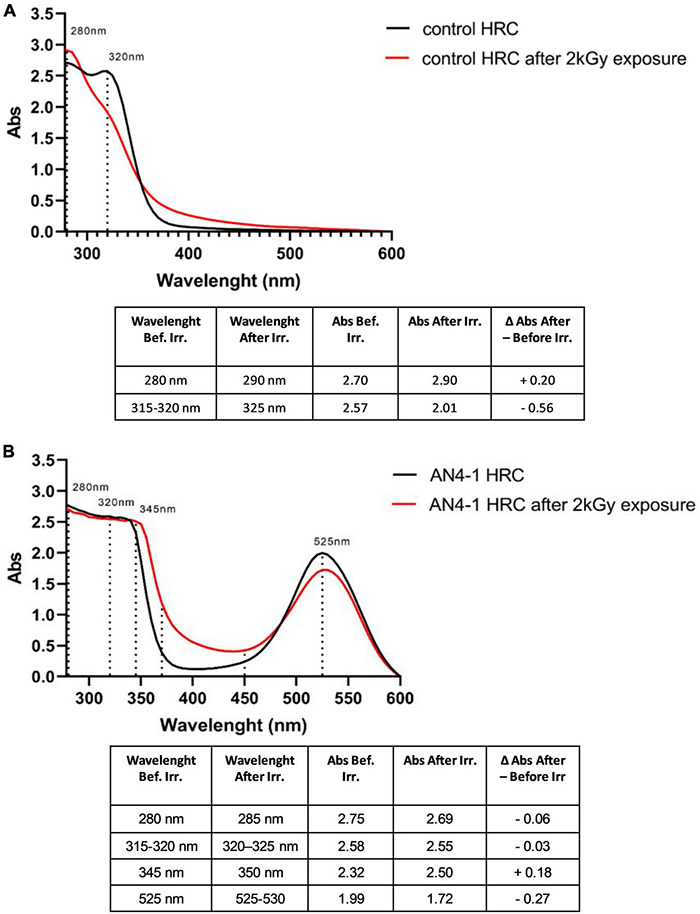
UV-VIS spectra of MicroTom HRCs before and after 2 kGy absorbed dose. In control, peaks at 280 nm and 320 nm indicate the presence of flavonols, hydroxycinnamic acids, tannins, and flavanols (A). In AN4-1, the additional peak at 345 nm completes the flavonols group profile compared to control, while the peak at 525 nm represents anthocyanins. The curve in the region between 400 and 450 nm refers to possible glycosylation, precipitation, complexation of anthocyanins (B).
In accordance with metabolomic data, in the spectra obtained in absence of irradiation, peaks around 280 nm and 320 nm indicate the presence of flavonols, hydroxycinnamic acids, tannins, and flavanols (Solìs-Oviedo and de La Cruz Pech-Canul, 2019). The shoulder at 345 nm together with that at 280 nm further defines the UV-VIS spectra of the flavonols in AN4-1 (Saha et al., 2021). Anthocyanins and anthocyanins associated with phenolic acids also produce peaks around 280 nm and 320 nm, respectively (Solìs-Oviedo and de La Cruz Pech-Canul, 2019), contributing to the profile of AN4-1 HRCs in Figure 7B. Anthocyanins result in an additional characteristic peak at 525 nm (Vivar-Quintana et al., 2002; da Silva et al., 2007; Fedenko et al., 2017), which, as expected, is observed in AN4-1 and not in control HRCs. Gamma irradiation determined a slight absorbance increase at 280 nm and a significant decrease at 320 nm in controls. On the contrary, these peaks remained unchanged in AN4-1. In AN4-1, the absorbance at 400–450 nm is possibly an indication of glycosylation, precipitation, complexation with tannins of anthocyanins upon irradiation (Saha et al., 2021). Gamma irradiation determined a slight decrease of the absorbance at 525 nm in AN4-1, as confirmed by a slight discoloration of the root material after irradiation.
Photoluminescence Analysis
Photoluminescence emission spectra were analyzed to determine the resistance to misfolding and oxidation of MicroTom HRCs soluble proteins after gamma irradiation. Photoluminescence spectra of extracts containing soluble proteins from control (Figure 10A) and AN4-1 HRCs (Figure 10B) were produced. The mass-normalized emission spectra of not irradiated soluble protein samples are similar in controls and AN4-1 and are characterized by high peaks mainly corresponding to tryptophan (370 nm) (Yang H. et al., 2015; Hilaire et al., 2017) and its metabolic products such as kynurenine (470 nm) and 3-hydroxykynurenine (439 nm) (Lohmann et al., 1988; Daly et al., 2009; Gakamsky et al., 2017). In addition, a peak at 470 nm is indicative of the presence of carbamate anions (Pan et al., 2013). In control HRC, the 2 kGy absorbed dose caused a decrease of fluorescence emission intensity for all three peaks. In particular, the Δ% photoluminescence intensity after and before the irradiation shows that the peak at 370 nm is reduced by 67.87%, the peaks at 439 nm and 470 nm by 37.5%, and 28.57%, respectively (Figure 8A). In AN4-1 only the 370 nm peak decreased (−27.64%), while a slight increase (+18.57%) of the kynurenine peak (470 nm) was present. No significant modifications were observed at 439 nm (Figure 10B).
FIGURE 10.
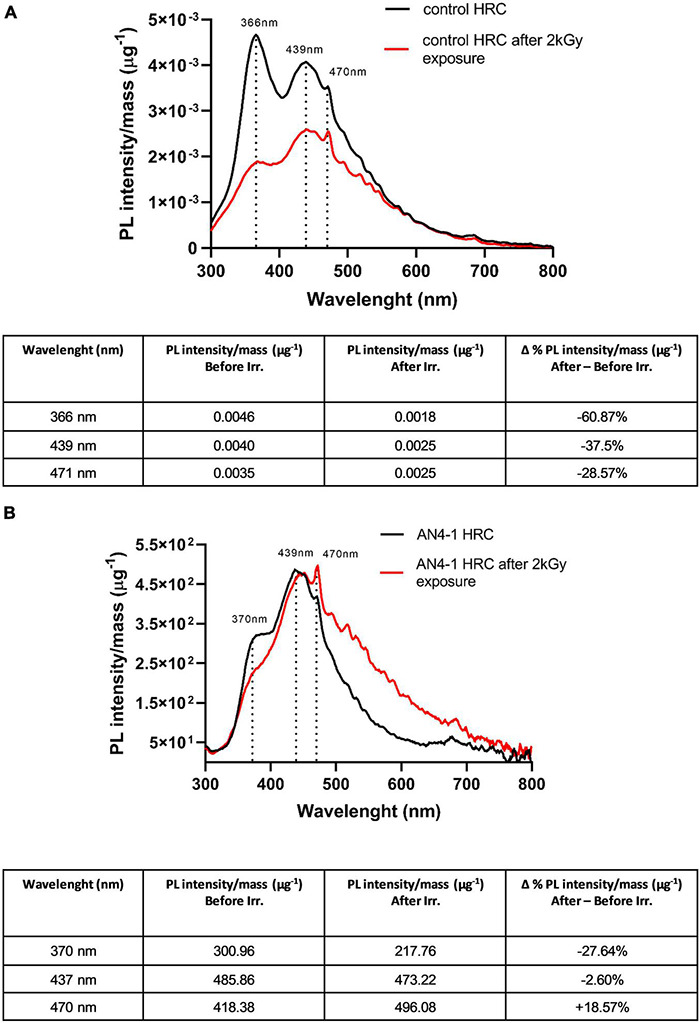
Photoluminescence spectra of control (A) and AN4-1 (B) HRCs and related mass-normalized emission intensities (tables) before and after 2 kGy gamma irradiation. Main peaks refer to tryptophan (370 nm), its degradation products kynurenine and 3-hydroxykynurenine (439 and 470 nm), and carbamate anion (470 nm).
Discussion
Anthocyanins are well known for their antioxidant and health-protection properties. Tomato has been already subjected to genetic manipulation for improved levels and composition of these high-value compounds (Schauer et al., 2006; Klee and Tieman, 2013; Giovannoni, 2018; Wang et al., 2019). Nutraceutical improvement of tomato is expected to provide a nourishing food source for future long-term manned space missions such as NASA’s “Artemis” lunar exploration program. The Artemis initiative includes sending a suite of new technology demonstrators to establish a sustained human presence by 2028 (DeZwart and Henderson, 2021). Low-power systems to grow plants will have the role to provide fresh and nutritious food to supplement astronauts’ diet and provide psychological benefits. The experiments conducted in the VEGGIE module onboard the International Space Station over the last few years have pioneered this scenario (Wolff et al., 2014; Fu et al., 2016; Massa et al., 2016; Zabel et al., 2016; Imhof et al., 2018).
The space environment, totally unusual for plants, can affect their metabolic processes and, consequently, growth, due to high-energy ionizing radiation, microgravity, magnetic field, and ultra-vacuum (Williams et al., 2004; Lotito and Frei, 2006; Moghaddam et al., 2011; Thamaphat et al., 2015; van Hoeck et al., 2017). Among abiotic stresses that plants will have to cope with in extraterrestrial environments artificially adapted to space agriculture, pro-oxidant stimuli are, indeed, the most prominent, due to cosmic ionizing radiation. Acting directly and indirectly to delay oxidative damage, enzymatic players, and metabolites activating endogenous antioxidant defense systems may help plant growth in the space environment. ROS attack plant cells causing loss of their structure and function (Vandenhove et al., 2010; Moghaddam et al., 2011; Marcu et al., 2013a; Vardhan and Shukla, 2017; Gudkov et al., 2019), rapidly reacting with almost all structural and functional organic molecules in the plant cells and causing irreversible oxidative damage on DNA, lipids, and proteins (Scandalios, 2002; Sewelam et al., 2016). The issue of counteracting overproduction of free radicals generated by harmful ionizing radiation is crucial for human survival in space outposts, as well, and opens the way to the ideal ‘anti-oxidant space fresh food.’ Indeed, the oral intake of clinically tested chemical radioprotectants (i.e., thiols, aminothiols, thiadiazoles, and benzothiazoles) is limited due to toxicity (Copp et al., 2013). Therefore, there is great interest in the development of natural antioxidants possibly produced by plants and introduced with the diet (Gómez et al., 2021). Together with the above-mentioned abiotic ‘macro-stresses,’ plants intended for space agriculture will have to cope with a plethora of abiotic ‘micro-stresses’ related to the different cultivation environment/systems that will be adopted. Furthermore, plants will have to share with humans future crewed space habitats (spaceflights, planetary outposts, and life-support systems) where stringent microbial quality requirements may not be achieved (Amalfitano et al., 2020).
Plants have an intrinsic potential for adaptation as the heritage of ancestors that survived in the harsh initial terrestrial environments (de Vries and Archibald, 2018). The ‘design’ of plant ideotypes suitable to these environments may be achieved by further refinement of existing biochemical and physiological features, such as specialized metabolites, that plants use to survive stress. Novel genotypes boosting biosynthetic pathways for the production of specialized metabolites can be generated by manipulation of their regulators. Among regulators, there are MYB transcription factors, known for their contribution to the increased complexity of land plants and to be regulators of plant responses to the environment (Lloyd et al., 2017; Gao et al., 2018; Kashyap et al., 2020; Shi and Panthee, 2020). Members of this large family of regulators are key players in the modulation, among others, of the biosynthesis of flavonoids, like anthocyanins, in different plant parts and response to all kinds of stimuli (Ambawat et al., 2013; Liu et al., 2015; Roy, 2016). MicroTom is extremely compact, early yielding, and would be suitable for agriculture in the confined area of a crewed space module. Here, we described the ectopic expression of the anthocyanin R2R3-MYB regulator PhAN4 from Petunia hybrida in MicroTom hairy roots as a testbed for future engineered whole plants. As known, anthocyanin production is regulated by R2R3-MYB, bHLH, WDR, and WRKY cooperation (MBWW transcriptional complex) (Spelt et al., 2000; Koes et al., 2005). In this complex, the R2R3-MYB transcription factor is probably the limiting factor, in that the WDR and bHLH proteins are thought to be constitutively expressed (Zhang et al., 2013, 2014). When specific MYB proteins are expressed in plants, this induces anthocyanin accumulation. Since some specific MYB transcription factors can function in regulating not only the biosynthesis of anthocyanins but also other traits (Meng et al., 2015; Vardhan and Shukla, 2017; Jian et al., 2019), we investigated if PhAN4 may affect, together with accumulation of anthocyanins, features associated with improved traits related to survival in harsh and confined environments such as future space outposts.
Differently from the wealth of literature available on whole transgenic plants and cell culture systems, data concerning anthocyanins production are limited for hairy roots. To date, no data are available on anthocyanin accumulation in tomato hairy roots by metabolic engineering to make comparisons with our work. Although not comparable to levels achieved in transgenic plants (Butelli et al., 2008; Povero et al., 2011; Kiferle et al., 2015), accumulation of anthocyanins occurred in MicroTom HRCs upon PhAN4 transformation, as expected due to the nature of the transgene. Importantly, no significant variation of transformed HRCs growth rates was found compared to controls. This gives a clue about the non-detrimental activity of PhAN4 toward primary metabolism in this system.
In accordance with literature related to transgenic tomato plants (Butelli et al., 2008; Su et al., 2016; Jian et al., 2019), mass spectrometry allowed us to identify glycosylated and acylated delphinidin and petunidin as the most abundant anthocyanins in PhAN4 HRCs. Interestingly, two additional anthocyanins, unusual in tomato, pelargonidin-3-glucoside, and malvidin-3-O-(4”’coumaroyl)-rutinose-5-O-glucose, were detected, as well. Malvidin and Pelargonidin derivatives have already been detected in Del and Ros1 and LC and C1 tomato plants, respectively (Bovy et al., 2002; Su et al., 2016). Interestingly, pelargonidin plays a major role in reducing genotoxic stress induced by environmental toxicants in plants (Khandelwal and Abraham, 2014). In addition, rather than other anthocyanins, pelargonidin-3-glucoside, that was accumulated in engineered MicroTom HRCs, has high bioavailability, being absorbed in an intact form into the gastrointestinal wall, undergoing first-pass metabolism and entering the systemic circulation as 4-hydroxybenzoic acid, a stable metabolite that is considered one of the main players of the systemic health effects of anthocyanins (Fang, 2014). In order to achieve a better understanding of PhAN4 in the tomato hairy root phenylpropanoid pathway, we measured 14 phenolic acids and derivatives and 29 flavonoids: notably, a reduction in the content of some members of the former, and a massive increase in most of the compounds in the latter were observed, thus proving PhAN4 is able to trigger the metabolic flux at both flavonoid and anthocyanin levels.
Data on the accumulation of anthocyanins and phenylpropanoids obtained by mass spectrometry was supported by transcriptome-wide RNA-seq analysis. RNA-seq, in complex, depicted a reprogramming oriented not only to anthocyanin accumulation, as expected, but also to positive regulation of cell response to biotic and abiotic stress and, possibly, to fruit quality-related traits.
Expression of PhAN4 increased the transcript levels of almost all of the genes encoding enzymes required for anthocyanin biosynthesis with the exception of PAL, C4H, 4CL, F3′H, and 5UFGT. To facilitate comparisons, transcriptomic research on tomato plants for anthocyanins enrichment has been summarized in Table 2. CHS and CHI had been already found to be upregulated by overexpression of SlAN2 in tomatoes, but not by ectopic expression of Del/Ros that, in turn, were able to upregulate F3H (Butelli et al., 2008; Jian et al., 2019). DFR had been already shown to be upregulated in the stem and leaf of tomato plants overexpressing SlAN2 and SlAN1 R2R3-MYBs (Kiferle et al., 2015). Interestingly, despite FLS was not found among upregulated DEGs in PhAN4 HRCs, LDOX, a known bi-functional enzyme being able not only the conversion of leucoanthocyanidins in anthocyanidins but also to catalyze the formation of flavonols, resulted to be differentially upregulated in PhAN4 HRCs. This may explain the accumulation of quercetin, kaempferol derivatives, and myricetin together with anthocyanidins. The flavonol biosynthetic activity of the upregulated LDOX is counterbalanced by the upregulated DFR. In tomatoes, DFR has a substantial preference for dihydromyricetin, which can be also derived by dihydrokaempferol and dihydroquercetin by F3′5′H activity. Interestingly, DilFl a tomato homolog of cytochrome b5 of P. hybrida, where it is essential for full activity of F3′5′H, was found to be upregulated in PhAN4 HRCs. These findings may explain the differential accumulation of delphinidin and petunidin compared to pelargonidin, malvidin, and cyanidin (the latter was not detected at all). Among late biosynthetic genes, upregulation of ANS had been already shown upon SlAN2 over-expression and Del/Ros expression in tomatoes. OMT and AAT transferases have already been described to be targets of a MYB in tomatoes, as well (Zhang et al., 2019). Their upregulation in PhAN4 HRCs, together with that of GST, PH3, OMT, 3UFGT, RT, and AAT may explain the accumulation of the glycosylated and acylated forms of anthocyanidins. In particular, the function of the upregulated PH3 (Solyc10g084380.1.1) in PhAN4 HRC is rather to be associated with anthocyanin biosynthesis boosting than with acidification of pH, as tomato lacks PH1 and PH5 (i.e., the genes encoding the vacuolar P-ATPases which cooperate with PH3 and PH4 in determining flower color by hyperacidification of petal cell vacuoles in Petunia). Nevertheless, preliminary evaluation of pH of PhAN4 and control HRC homogenates (Verweij et al., 2008) does not allow to exclude that the acidification function may occur (Supplementary Figure 5), given that pH differences between PhAN4 and control HRC seem to be in accordance with variations recorded between vacuolar P-ATPases-defective mutants and wild type Petunia hybrida (Faraco et al., 2014). The upregulation of anthocyanin biosynthesis-relevant transcription factors further expands the influence of PhAN4 MYB in tomato gene expression regulation. Among those, the upregulation of SlAN1, SlAN2, and WD40 are coherent with already proposed models of anthocyanin biosynthesis regulation (Koes et al., 2005; Liu et al., 2018).
TABLE 2.
Summary of results from the main research on tomato plants enrichment with anthocyanins obtained by both conventional breeding and genetic engineering approaches.
| Tomato line | Origin | Main overexpressed genes | Main anthocyanins detected | Anthocyanin concentration | References |
| V118 | Breeding | – | Pet-3-(p-coumaryl)-rut-5-glc; Pet-3-caeoyl-rut-5-glc; Mal-3(p-coumaryl)-rut-5-glc |
50.18 mg 100 g–1 DW 9.04 mg 100 g–1 DW 13.09 mg 100 g–1 DW |
Li et al., 2011 |
| Aft/Aft × atv/atv | Breeding | – | Pet-3-(p-coumaroyl)-rut-5-glc; Del-3-rut |
Peel: 116.11 mg 100 g–1 FW | Mes et al., 2008 |
| Sun Black (Aft/Aft × atv/atv) | Breeding | – | Pet-3-(trans-p-coumaroyl)-rut-5-glc; Mal-3-(trans-p-coumaroyl)-rut-5-glc |
More than 1 mg g–1 DW | Mazzucato et al., 2013 |
| Blue Japan Indigo tomato (Aft/Aft × atv/atv) |
Breeding | — | Pet + p-coumaroyl + rut + glyc; Mal + p-coumaroyl + rut; Del |
Peel: 17 mg g–1 DW Pulp: 0.1 mg g–1 DW |
Ooe et al., 2016 |
| Aft/Aft × atv/atv × hp2/hp2 | Breeding | – | Pet-(p-coumaroyl)-rut-hex; Del-3-(p-coumaroyl)-rut-glyc; Pet-(p-coumaroyl)-rut-hex; Pet-3-(caffeoyl)-rut-5-glyc; Mal-3-(p-coumaroyl)-rut-5-glyc; Cya-3-O-rut; |
Peel: 90.91 mg 100 g–1 FW | da Silva Souza et al., 2020 |
| ANT1 from S. chilense | Genetic engineering | CHS, DFR, 3-GT, 5-GT, GST, ANP | Del-3-rut-5-glc; Del-3-(p-coumaroyl)-rut-glyc; Del-3-(caffeoyl)-rut-5-glyc; Pet-3-rut-5-glc; Pet-3-(p-coumaroyl)-rut-5-glyc; Pet-3-(caffeoyl)-rut-5-glyc; Mal-3-rut-5-glc; Mal-3-(p-coumaryl)-rut-5-glyc; Mal-3-(caffeoyl)-rut-5-glyc |
3.574 mg g–1 FW | Mathews et al., 2003 |
| Del/Ros1 | Genetic engineering | PAL, C3H, CHI, F3′5′H, DFR, ANS, 3-GT, 5-GT, RT, AAC, GST, ANP | Pet-3-(trans-p-coumaroyl)-rut-5-glc; Del-3-(trans-p-coumaroyl)-rut-5-glc; Pet-3-(feruloyl)-rut-5-glc; Del-3-(feruloyl)-rut-5-glc |
2.835 ± 0.456 mg g–1 FW | Butelli et al., 2008 |
| Del/Ros1 | Genetic engineering | – | Del-3-(trans-p-coumaroyl)-rut-5-glc; Pet-3-(trans-p-coumaroyl)-rut-5-glc; Mal-3-(p-coumaroyl)-rut-5-glc; Mal-3-(feruloyl)-rut-5-glc |
Peel: 5.1 ± 0.5 g kg–1 DW Flesh: 5.8 ± 0.3 g kg–1 DW Whole fruit: 5.2 ± 0.5 g Peo-3-glc equivalent kg–1 DW, or 0.5% of DW |
Su et al., 2016 |
| ANT1 from S. lycopersicum | Genetic engineering | CHI, F3H, DFR, ANS, 3-GT | Pet Mal Del |
– | Schreiber et al., 2012 |
| SlANT1 and SlAN2 | Genetic engineering | SlAN2, SlANT1, SlAN1, SlAN11, SlJAF13, SlDFR | – | – | Kiferle et al., 2015 |
| Del/Ros1 × AtMYB12 | Genetic engineering | PAL, 4CL, CHS, CHI, F3H, FLS, DFR, ANP, 3-GT, C3H | Del-3-(trans-p-coumaroyl)-rut-5-glc; Pet-3-(trans-p-coumaroyl)-rut-5-glc; Pet-3-(feruloyl)-rut-5-glc; Mal-3-(p-coumaroyl)-rut-5-glc |
1.154 ± 0.011 mg g–1 FW 2.857 ± 0.218 mg g–1 FW 0.922 ± 0.102 mg g–1 FW 0.598 ± 0.011 mg g–1 FW |
Zhang et al., 2015 |
| SlMYB75 | Genetic engineering | PAL, CHI, CHS, AAC, ANS, 3-RT, LDOX | – | ∼2 mg g–1 FW | Jian et al., 2019 |
| PhAN4 | Genetic engineering (transformed tomato hairy root cultures) | CHS, CHI, F3′5′H, F3H LDOX, DFR, ANS, 3-GT, RT, OMT, AAT, ANP, GST | Pet-3-(feruloyl)-rut-5-glc; Pet-3-(p-coumaroyl)-rut-5-glc; Pel-3glc; Mal-3-(4-coumaroyl)-rut-5-glc; Del-3-(p-coumaroyl)-rut-5-glc; Del-3-5-glc |
37 μg g–1 DW |
The main overexpressed genes and anthocyanins content are shown where available.
Del, delphinidin; Pet, petunidin; Mal, malvidin; Cya, cyanidin; Peo, peonidin; Pel, pelargonidin; rut, rutinoside; glc, glucoside; glyc, glycoside; hex, hexoside; FW, fresh weight; DW, dry weight.
Interestingly, the PhAN4-related transcriptomic enhancement was found to involve more than 30 genes specifically related to response to biotic/abiotic and oxidative stress, as well. Indeed, GOE analysis was associated with proteinase inhibitors, beta-glucosidases, glucanases, glycosylases, xylan acetylases, resistance proteins, and other classes of enzymes that are players of defense against pathogens and pests and of response to abiotic stresses specifically associated with wounding, cold, heat, drought, hypoxia, and to UV exposure in tomato (Fan et al., 2020). Among those, the upregulated beta-glucosidases, together with the dirigent proteins and the monoterpenoid and sesquiterpenoid synthases that were found in the present work, are involved in the formation of required intermediates for cell wall lignification. The positive regulation of trichome birefringence-like protein mediating xylan acetylation also is another tool in possible protection against environmental stresses among which there is cold and excess of minerals in the soil. Acetylation of wall polymers is, indeed, vital for plant growth and adaptation to various environments, and is required for the structural integrity of the leaf surface exerting a global impact on plant stress responses (Gao et al., 2017). In relation to the positive regulation of beta-1,3-glucanases, beta-glucosidases, and xylan acetylases, together with expansions, it has to be added that these genes are related to fruit softening or improved emission of volatiles, as well (Tzin et al., 2015; Li P. et al., 2020). In this sense, the positive regulation of chlorophyll a/b binding protein (that is normally upregulated during fruit ripening) and of one gene related to volatile compounds biosynthesis (chorismate mutase) was found in PhAN4 HRCs. At the same time, negative regulation of other genes, which down expression is involved in the extended shelf life of fruits, was found. The evaluation of these aspects might be important in view of the development/design of whole transgenic tomato plants in relation to the quality traits of fruits.
One player of ROS detoxification and regulator of the redox signaling network of tomato, plastidial thioredoxin Y2, was found to be highly upregulated, as well. Moreover, the analysis demonstrated the downregulation of violaxanthin de-epoxidase (VDE) and carotenoid isomerase (CRTISO), two genes involved in carotenoid biosynthesis. Despite being involved in the photo-inhibition of the PSII, suppression of VDE can induce, at the same time, an accumulation of fucoxanthin that functions as an efficient anti-oxidant in anoxia conditions (Mikami and Hosokawa, 2013). The downregulation of carotenoid isomerase was demonstrated to induce an accumulation of zeta-carotene and cis-prolycopene in tomato fruits (Isaacson et al., 2002; Pinheiro et al., 2019), both elevating and modifying carotenoid profiles toward more bioavailable forms compared to wild-type (Cooperstone et al., 2015). Therefore, the ROS counteracting potential of PhAN4 HRC material may be related not only to anthocyanins but also to other accumulating anti-oxidant specialized metabolites.
In view of the future development of natural anti-oxidants produced by engineered plants and possibly administered with the diet, and to define whether the significantly higher DPPH antioxidant capacity of specialized metabolites accumulating in PhAN4 HRCs may be efficiently maintained after ionizing radiation, we exposed lyophilized biomass from AN4-1 HRC to high dose 60Co gamma radiation. Gamma rays are a component of cosmic ionizing radiation and induce the formation of free radical species (i.e., paramagnetic species). When carried out in an air atmosphere, gamma rays induce the formation of ROS, such as superoxide, peroxide, and hydroxyl radicals, that are responsible for several oxidative processes in biological systems (Marcu et al., 2013b; Schreurs et al., 2016). Gamma rays are perfect ROS inducers both directly through water radiolysis, and indirectly via the activation of a broad range of signaling processes (e.g., damages to the mitochondria or cell microenvironment) (Riley, 1994; Azzam et al., 2012; Buttarelli et al., 2019). ESR Spectroscopy was used as a sensitive tool to identify the entity of the paramagnetic species generated in the HCRs samples after gamma irradiation (Aleksieva et al., 2009; Hawkins and Davies, 2014; D’Errico et al., 2018). ESR analyses revealed a significant difference in the overall amounts of radical species accumulated in the two HRC molecular backgrounds. At the same absorbed dose, control generates a very intense peroxyl radical signal, while a considerably lower level of the singlet intensity is generated in AN4-1. This result reveals that the molecular set formed upon PhAN4 expression provides very effective free radical scavengers efficiently counteracting oxidative stress upon gamma radiation.
The UV-VIS analysis allowed us to confirm the results obtained by metabolomic data and to characterize the effect of ionizing radiation on the antioxidant compounds. Gamma irradiation, via releasing free radicals in solution, may alter these plant constituents (Lateef and Al-Nimer, 2009; Lalande et al., 2019). Compared to the substantially unchanged spectra of AN4-1, the absorbance decrease of the 320 nm peak (i.e., flavonols, hydroxycinnamic acids, tannins, flavonols, and anthocyanins associated to caffeic and coumaric acids) controls upon irradiation, suggests that the anthocyanins accumulated in PhAN4 HRCs are particularly stable to gamma high absorbed dose and probably have protective effects on other biomolecules. Indirectly, this result shows that these molecules may give a major contribution to the ROS buffering capacity under radiation shown by ESR and to the overall antioxidant potential. As a confirmation of the possible significance of this result in vivo, it was reported that, even at higher absorbed doses than that used in this work, X rays (e.g., another type of ionizing radiation) leave the level of flavonoids in an aqueous solution unchanged due to the radiolysis-mediated formation of depsides that, in addition, maintain good anti-oxidant properties, as well (Kozlowski et al., 2007; Lateef and Al-Nimer, 2009).
For proteins to be functional within a cell requires coordinated folding processes to obtain a correct 3D shape. Disruptions to protein folding, that can occur in space due to ionizing radiation, can have profound biological implications for all organisms, including plant cells, leading to dysfunctions (Blanco et al., 2018; Lalande et al., 2019). For this reason, investigations were conducted through photoluminescence analysis on HRCs’ total soluble protein extracts. When a protein is exposed to the wavelength of 280 nm, mainly the tryptophan and tyrosine residues get excited, which would reflect upon its tertiary structure (Yang H. et al., 2015). The maximum and emission peak position reflects upon secondary, tertiary, and quaternary structures (Hilaire et al., 2017). Moreover, the extent of protein oxidation is measured by determining the loss of specific tryptophan fluorescence. The emission spectra of HRCs showed an overall decrease in maximum emission intensities only in the case of irradiated controls, revealing that the structure of soluble proteins is partially lost upon irradiation in those samples. This result demonstrates a low capacity of proteins of control HRCs to counteract unfolding, binding to hydrophobic pockets, and aggregation, witnessed by the loss of fluorescence due to the burial of tryptophan residues after radiation. In particular, the Δ% photoluminescence intensity/mass units after and before irradiation indicated that soluble proteins from control undergo a doubled oxidative stress compared to AN4-1 samples. Being polyphenols emitting at 280 and 320 nm extracted by PBS together with total soluble proteins (anthocyanins are not efficiently extracted, Supplementary Figure 4), the contribution of these classes of molecules may be hypothesized in stabilizing proteins, as proofs of the interaction of anthocyanins with proteins seem to suggest (Sui et al., 2018). Possible additional contributions of enzymatic players to mitigation of misfolding and aggregation in vivo may be the object of future studies.
In conclusion, plant biotechnology methods may thus be exploited for the generation of plants capable of dealing with harsh conditions such as those typical of space outposts. MicroTom HRC allowed to rapidly test PhAN4 expression effects on tomato cells, possibly opening the way to apply to the engineering of whole plants able to perform in a suitable and predictable manner in those environments.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://dataview.ncbi.nlm.nih.gov/object/PRJNA794337?reviewer=estprpllasrt3f3libdoij8rco.
Author Contributions
SM planned and designed the project, undertook tomato transformation experiments, PCR screening and maintenance of HRCs, and sample preparation for subsequent analysis, as well as wrote the manuscript. RP handled maintenance of HRCs, contributed to sample preparation and transcriptomic data retrieving and interpretation, and contributed to the writing of the manuscript and the preparation of figures. AB performed sequencing read mapping, identification of DEGs and functional annotation, and enrichment pathway analysis of DEGs. HP undertook cDNA library construction and sequencing for transcriptomic analysis. MB contributed to the identification of DEGs and functional annotation of genes. AC and IS performed gamma irradiation experiments, ESR, UV-VIS, and photoluminescence analysis and contributed to writing the manuscript. GD and OD performed MS analysis, anthocyanin identification/quantification, and contributed to writing the manuscript. FP conducted the qPCR analysis. AD and PD performed the Trolox assay and the quantification of total polyphenol content and contributed to writing the manuscript. FQ, RK, and CS assembled and kindly provided the PhAN4 constructs used in this work. ElB contributed to the maintenance of HRCs. EuB reviewed and contributed to the writing of the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are deeply grateful to Stefania Baccaro (Fusion and Nuclear Safety Technologies Department, Casaccia Research Center, Rome, Italy) for enthusiastic support and for sharing precious advice concerning radiation experiments, although premature death did not allow her to see the end of the work. We thank Alessia Fiore (ENEA, BIOTEC) for help in the qualitative analysis of RNAs extracted from HRCs and Giulio Metelli (University of Viterbo/ENEA BIOTEC) for support in collecting data on HRCs growth.
Footnotes
Funding
This work was supported by the ENEA/ASI (Italian Space Agency) BIOExTREME and HORTSPACE Projects (ASI n. 2014-007-R.0, ASI n. 2017-11-H.0).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.830931/full#supplementary-material
Primer identification (id.) as described in “Materials and Methods” for HRCs PCR screening (for, forward; rev, reverse).
List of figures representing the level of different phenolic compounds and their derivatives in control and PhAN4 HRCs.
List of figures representing the level of different flavonoids in control and PhAN4 HRCs.
Complete list of DEGs.
GOE analysis results.
References
- Albert N. W., Davies K. M., Lewis D. H., Zhang H., Montefiori M., Brendolise C., et al. (2014). A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in Eudicots. Plant Cell 26 962–980. 10.1105/tpc.113.122069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert N. W., Lewis D. H., Zhang H., Schwinn K. E., Jameson P. E., Davies K. M. (2011). Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. Plant J. 65 771–784. 10.1111/j.1365-313X.2010.04465.x [DOI] [PubMed] [Google Scholar]
- Aleksieva K., Georgieva L., Tzvetkova E., Yordanov N. D. (2009). EPR study on tomatoes before and after gamma-irradiation. Radiat. Phys. Chem. 78 823–825. 10.1016/j.radphyschem.2009.05.013 [DOI] [Google Scholar]
- Alfenito M. R., Souer E., Goodman C. D., Buell R., Mol J., Koes R., et al. (1998). Functional complementation of anthocyanin sequestration in the vacuole by widely divergent glutathione S-transferases. Plant Cell 10 1135–1149. 10.1105/tpc.10.7.1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amalfitano S., Levantesi C., Copetti D., Stefani F., Locantore I., Guarnieri V., et al. (2020). Water and microbial monitoring technologies towards the near future space exploration. Water Res. 177:115787. 10.1016/j.watres.2020.115787 [DOI] [PubMed] [Google Scholar]
- Ambawat S., Sharma P., Yadav N. R., Yadav R. C. (2013). MYB transcription factor genes as regulators for plant responses: an overview. Phys. Mol. Biol. Plants 19 307–321. 10.1007/s12298-013-0179-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen M. L., Outtrup H., Skibsted L. H. (2000). Potential antioxidants in beer assessed by ESR spin trapping. J. Agric. Food Chem. 48 3106–3111. 10.1021/jf000354+ [DOI] [PubMed] [Google Scholar]
- Andolfo G., Ferriello F., Tardella L., Ferrarini A., Sigillo L., Frusciante L., et al. (2014). Tomato genome-wide transcriptional responses to Fusarium wilt and Tomato Mosaic Virus. PLoS One 9:e94963. 10.1371/journal.pone.0094963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki K., Yano K., Suzuki A., Kawamura S., Sakurai N., Suda K., et al. (2010). Large-scale analysis of full-length cDNA from the tomato (Solanum lycopersicum) cultivar Micro-Tom, a reference system for the Solanaceae genomics. BMC Genomics 11:210. 10.1186/1471-2164-11-210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asins M. J., Albacete A., Martínez-Andújar C., Celiktopuz E., Solmaz İ, Sarı N., et al. (2021). Genetic analysis of root-to-shoot signaling and rootstock-mediated tolerance to water deficit in tomato. Genes 12 1–25. 10.3390/genes12010010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzam E. I., Jay-Gerin J.-P., Pain D. (2012). Ionizing radiation-induced metabolic oxidative stress and prolonged cell injury. Cancer Lett. 327 48–60. 10.1016/j.canlet.2011.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccaro S., Cemmi A., Sarcina I. D. (2019). Calliope 60 Co gamma irradiation facility for space qualification at ENEA-Casaccia research centre. Phys. Astron. Int. J. 3 94–100. 10.15406/paij.2019.03.00164 [DOI] [Google Scholar]
- Bendokas V., Skemiene K., Trumbeckaite S., Stanys V., Passamonti S., Borutaite V., et al. (2020). Anthocyanins: from plant pigments to health benefits at mitochondrial level. Crit. Rev. Food Sci. Nutr. 60 3352–3365. 10.1080/10408398.2019.1687421 [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the False Discovery Rate: a Practical and Powerful Approach to Multiple Testing. J. R. Statist. Soc. Ser. B 57 289–300. 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- Bergey D. R., Howe G. A., Ryan C. A. (1996). Polypeptide signaling for plant defensive genes exhibits analogies to defense signaling in animals. Proc. Natl. Acad. Sci. U. S. A. 93 12053–12058. 10.1073/pnas.93.22.12053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijlani S., Stephens E., Singh N. K., Venkateswaran K., Wang C. C. C. (2021). Advances in space microbiology. Iscience 24:102395. 10.1016/j.isci.2021.102395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco Y., de Diego-Castilla G., Viúdez-Moreiras D., Cavalcante-Silva E., Rodríguez-Manfredi J. A., Davila A. F., et al. (2018). Effects of Gamma and Electron Radiation on the Structural Integrity of Organic Molecules and Macromolecular Biomarkers Measured by Microarray Immunoassays and Their Astrobiological Implications. Astrobiology 18 1497–1516. 10.1089/ast.2016.1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blesso C. N. (2019). Dietary Anthocyanins and Human Health. Nutrients 11 10–13. 10.3390/nu11092107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bognar E., Sarszegi Z., Szabo A., Debreceni B., Kalman N., Tucsek Z., et al. (2013). Antioxidant and Anti-Inflammatory Effects in RAW264.7 Macrophages of Malvidin, a Major Red Wine Polyphenol. PLoS One 8:e65355. 10.1371/journal.pone.0065355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongue-Bartelsman M., O’Neill S. D., Yusen T., Yoder J. I. (1994). Characterization of the gene encoding dihydroflavonol 4-reductase in tomato. Gene 138 153–157. 10.1016/0378-1119(94)90799-4 [DOI] [PubMed] [Google Scholar]
- Bournival B. L., Scott J. W., Vallejos C. E. (1989). An isozyme marker for resistance to race 3 of Fusarium oxysporum f. sp. lycopersici in tomato. Theor. Appl. Genet. 78 489–494. 10.1007/BF00290832 [DOI] [PubMed] [Google Scholar]
- Bovy A., de Vos R., Kemper M., Schijlen E., Almenar Pertejo M., Muir S., et al. (2002). High-flavonol tomatoes resulting from the heterologous expression of the maize transcription factor genes LC and C1. Plant Cell 14 2509–2526. 10.1105/tpc.004218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito A., Areche C., Sepúlveda B., Kennelly E. J., Simirgiotis M. J. (2014). Anthocyanin characterization, total phenolic quantification and antioxidant features of some chilean edible berry extracts. Molecules 19 10936–10955. 10.3390/molecules190810936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunetti C., di Ferdinando M., Fini A., Pollastri S., Tattini M. (2013). Flavonoids as antioxidants and developmental regulators: relative significance in plants and humans. Int. J. Mol. Sci. 14 3540–3555. 10.3390/ijms14023540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buoso S., Pagliari L., Musetti R., Martini M., Marroni F., Schmidt W., et al. (2019). “Candidatus phytoplasma solani” interferes with the distribution and uptake of iron in tomato. BMC Genomics 20:703. 10.1186/s12864-019-6062-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butelli E., Titta L., Giorgio M., Mock H. P., Matros A., Peterek S., et al. (2008). Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 26 1301–1308. 10.1038/nbt.1506 [DOI] [PubMed] [Google Scholar]
- Buttarelli M., Babini G., Raspaglio G., Filippetti F., Battaglia A., Ciucci A., et al. (2019). A combined ANXA2-NDRG1-STAT1 gene signature predicts response to chemoradiotherapy in cervical cancer. J. Exp. Clin. Cancer Res. 38:279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X., Qiu Z., Wang X., van Giang T., Liu X., Wang J., et al. (2017). A putative R3 MYB repressor is the candidate gene underlying atroviolacium, a locus for anthocyanin pigmentation in tomato fruit. J. Exp. Bot. 68 5745–5758. 10.1093/jxb/erx382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona L., Alquézar B., Diretto G., Sevi F., Malara T., Lafuente M. T., et al. (2021). Curing and low-temperature combined post-harvest storage enhances anthocyanin biosynthesis in blood oranges. Food Chem. 342:128334. 10.1016/j.foodchem.2020.128334 [DOI] [PubMed] [Google Scholar]
- Cock P. J. A., Fields C. J., Goto N., Heuer M. L., Rice P. M. (2009). The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 38 1767–1771. 10.1093/nar/gkp1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colanero S., Perata P., Gonzali S. (2020). What’s behind Purple Tomatoes? Insight into the Mechanisms of Anthocyanin Synthesis in Tomato Fruits. Plant Physiol. 182 1841–1853. 10.1104/pp.19.01530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conconi A., Smerdon M. J., Howe G. A., Ryan C. A. (1996). The octadecanoid signalling pathway in plants mediates Response To Ultraviolet Radiation. Nature 383 826–829. [DOI] [PubMed] [Google Scholar]
- Consortium T. T. G., Sato S., Tabata S., Hirakawa H., Asamizu E., Shirasawa K., et al. (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485:635. 10.1038/nature11119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooperstone J. L., Ralston R. A., Riedl K. M., Haufe T. C., Ralf M. (2015). Enhanced bioavailability of lycopene when consumed as cis-isomers from tangerine compared to red tomato juice, a randomized, cross-over clinical trial. Mol. Nutr. Food Res. 59 658–669. 10.1002/mnfr.201400658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copp R. R., Peebles D. D., Soref C. M., Fahl W. E. (2013). Radioprotective efficacy and toxicity of a new family of aminothiol analogs. Int. J. Radiat. Biol. 89 485–492. 10.3109/09553002.2013.770579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva F. L., Escribano-Bailón M. T., Pérez Alonso J. J., Rivas-Gonzalo J. C., Santos-Buelga C. (2007). Anthocyanin pigments in strawberry. LWT Food Sci. Technol. 40 374–382. 10.1016/j.lwt.2005.09.018 [DOI] [Google Scholar]
- da Silva Souza M. A., Peres L. E. P., Freschi J. R., Purgatto E., Lajolo F. M., Hassimotto N. M. A. (2020). Changes in flavonoid and carotenoid profiles alter volatile organic compounds in purple and orange cherry tomatoes obtained by allele introgression. J. Sci. Food Agric. 100 1662–1670. 10.1002/jsfa.10180 [DOI] [PubMed] [Google Scholar]
- Daly S., Bianchini R., Polefka T., Jumbelic L., Jachowicz J. (2009). Fluorescence and coloration of grey hair. Int. J. Cosmet. Sci. 31 347–359. 10.1111/j.1468-2494.2009.00500.x [DOI] [PubMed] [Google Scholar]
- de Ollas C., González-Guzmán M., Pitarch Z., Matus J. T., Candela H., Rambla J. L., et al. (2021). Identification of ABA-Mediated Genetic and Metabolic Responses to Soil Flooding in Tomato (Solanum lycopersicum L. Mill). Front. Plant Sci. 12:613059. 10.3389/fpls.2021.613059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Torre M. P., Cavero R. Y., Calvo M. I., Vizmanos J. L. W. (2019). A simple and a reliable method to quantify antioxidant activity in vivo. Antioxidants 8 1–11. 10.3390/antiox8050142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vetten N., ter Horst J., van Schaik H. P., de Boer A., Mol J., Koes R. (1999). A cytochrome b5 is required for full activity of flavonoid 3′,5′-hydroxylase, a cytochrome P450 involved in the formation of blue flower colors. Proc. Natl. Acad. Sci. U. S. A. 96 778–783. 10.1073/pnas.96.2.778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries J., Archibald J. M. (2018). Plant evolution: landmarks on the path to terrestrial life. New Phytol. 217 1428–1434. 10.1111/nph.14975 [DOI] [PubMed] [Google Scholar]
- D’Errico G., Vitiello G., de Tommaso G., Abdel-Gawad F. K., Brundo M. V., Ferrante M., et al. (2018). Electron Spin Resonance (ESR) for the study of Reactive Oxygen Species (ROS) on the isolated frog skin (Pelophylax bergeri): a non-invasive method for environmental monitoring. Environ. Res. 165 11–18. 10.1016/j.envres.2018.03.044 [DOI] [PubMed] [Google Scholar]
- Desiderio A., Salzano A. M., Scaloni A., Massa S., Pimpinella M., de Coste V., et al. (2019). Effects of Simulated Space Radiations on the Tomato Root Proteome. Front. Plant Sci. 10:1334. 10.3389/fpls.2019.01334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeZwart M., Henderson S. (2021). Commercial and Military Uses of Outer Space. Germany: Springer. [Google Scholar]
- Diretto G., Jin X., Capell T., Zhu C., Gomez-Gomez L. (2019). Differential accumulation of pelargonidin glycosides in petals at three different developmental stages of the orange-flowered gentian (Gentiana lutea L. var. aurantiaca). PLoS One 14:e0212062. 10.1371/journal.pone.0212062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Santos A. L., Chaves-Silva S., Yang L., Maia L. G. S., Chalfun-Júnior A., Sinharoy S., et al. (2017). Global analysis of the MATE gene family of metabolite transporters in tomato. BMC Plant Biol. 17:185. 10.1186/s12870-017-1115-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esatbeyoglu T., Wagner A. E., Motafakkerazad R., Nakajima Y., Matsugo S., Rimbach G. (2014). Free radical scavenging and antioxidant activity of betanin: electron spin resonance spectroscopy studies and studies in cultured cells. Food Chem. Toxicol. 73 119–126. 10.1016/j.fct.2014.08.007 [DOI] [PubMed] [Google Scholar]
- Fahim S., Hussein W., Awad H. (2016). Systemic Resistance Induction of Tomato Plants against ToMV Virus by Surfactin Produced from Bacillus subtilis BMG02. Am. J. of Microbiol. Res. 4 153–158. 10.12691/ajmr-4-5-5 [DOI] [Google Scholar]
- Fan K. T., Hsu Y., Yeh C. F., Chang C. H., Chang W. H., Chen Y. R. (2021). Quantitative proteomics reveals the dynamic regulation of the tomato proteome in response to phytophthora infestans. Int. J. Mol. Sci. 22:4174. 10.3390/ijms22084174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y., Yang W., Yan Q., Chen C., Li J. (2020). Genome-Wide Identification and Expression Analysis. Genes 11:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J. (2014). Some anthocyanins could be efficiently absorbed across the gastrointestinal mucosa: extensive presystemic metabolism reduces apparent bioavailability. J. Agric. Food Chem. 62 3904–3911. 10.1021/jf405356b [DOI] [PubMed] [Google Scholar]
- Faraco M., Spelt C., Bliek M., Verweij W., Hoshino A., Espen L., et al. (2014). Hyperacidification of Vacuoles by the Combined Action of Two Different P-ATPases in the Tonoplast Determines Flower Color. Cell Rep. 6 32–43. 10.1016/j.celrep.2013.12.009 [DOI] [PubMed] [Google Scholar]
- Faure A. M., Sánchez-Ferrer A., Zabara A., Andersen M. L., Nyström L. (2014). Modulating the structural properties of β-d-glucan degradation products by alternative reaction pathways. Carbohydrate Polym. 99 679–686. 10.1016/j.carbpol.2013.08.022 [DOI] [PubMed] [Google Scholar]
- Fedenko V. S., Shemet S. A., Landi M. (2017). UV–vis spectroscopy and colorimetric models for detecting anthocyanin-metal complexes in plants: an overview of in vitro and in vivo techniques. J. Plant Physiol. 212 13–28. 10.1016/j.jplph.2017.02.001 [DOI] [PubMed] [Google Scholar]
- Fernandez-Moreno J.-P., Tzfadia O., Forment J., Presa S., Rogachev I., Meir S., et al. (2016). Characterization of a New Pink-Fruited Tomato Mutant Results in the Identification of a Null Allele of the SlMYB12 Transcription Factor. Plant Physiol. 171 1821–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Pozo N., Menda N., Edwards J. D., Saha S., Tecle I. Y., Strickler S. R., et al. (2015). The Sol Genomics Network (SGN)-from genotype to phenotype to breeding. Nucleic Acids Res. 43 D1036–D1041. 10.1093/nar/gku1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filiz E., Cetin D., Akbudak M. A. (2019). Aromatic amino acids biosynthesis genes identification and expression analysis under salt and drought stresses in Solanum lycopersicum L. Sci. Hortic. 250 127–137. 10.1016/j.scienta.2019.02.044 [DOI] [Google Scholar]
- Florio F. E., Gattolin S., Toppino L., Bassolino L., Fibiani M., Scalzo R. L., et al. (2021). A SmelAAT Acyltransferase Variant Causes a Major Difference in Eggplant (Solanum melongena L.) Peel Anthocyanin Composition. Int. J. Mol. Sci. 22:9174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Li L., Xie B., Dong C., Wang M., Jia B., et al. (2016). How to establish a bioregenerative life support system for long-term crewed missions to the moon or mars. Astrobiology 16 925–936. 10.1089/ast.2016.1477 [DOI] [PubMed] [Google Scholar]
- Gakamsky A., Duncan R. R., Howarth N. M., Dhillon B., Buttenschön K. K., Daly D. J., et al. (2017). Tryptophan and Non-Tryptophan Fluorescence of the Eye Lens Proteins Provides Diagnostics of Cataract at the Molecular Level. Sci. Rep. 7 1–15. 10.1038/srep40375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., He C., Zhang D., Liu X., Xu Z., Tian Y., et al. (2017). Two trichome birefringence-like proteins mediate xylan acetylation, which is essential for leaf blight resistance in rice. Plant Physiol. 173 470–481. 10.1104/pp.16.01618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Liu J., Chen Y., Tang H., Wang Y., He Y., et al. (2018). Tomato SlAN11 regulates flavonoid biosynthesis and seed dormancy by interaction with bHLH proteins but not with MYB proteins. Hortic. Res. 5:27. 10.1038/s41438-018-0032-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg M., Sharma N., Sharma S., Kapoor P., Kumar A., Chunduri V., et al. (2018). Biofortified Crops Generated by Breeding, Agronomy, and Transgenic Approaches Are Improving Lives of Millions of People around the World. Front. Nutr. 5:12. 10.3389/fnut.2018.00012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerszberg A., Hnatuszko-Konka K., Kowalczyk T., Kononowicz A. K. (2015). Tomato (Solanum lycopersicum L.) in the service of biotechnology. Plant Cell Tissue Organ Cult. 120 881–902. 10.1007/s11240-014-0664-4 [DOI] [Google Scholar]
- Giovannoni J. (2018). Tomato Multiomics Reveals Consequences of Crop Domestication and Improvement. Cell 172 6–8. 10.1016/j.cell.2017.12.036 [DOI] [PubMed] [Google Scholar]
- Gómez X., Sanon S., Zambrano K., Asquel S., Bassantes M., Morales J. E., et al. (2021). Key points for the development of antioxidant cocktails to prevent cellular stress and damage caused by reactive oxygen species (ROS) during manned space missions. NPJ Microgravity 7 35. 10.1038/s41526-021-00162-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez A., Zhao M., Leavitt J. M., Lloyd A. M. (2008). Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53 814–827. 10.1111/j.1365-313X.2007.03373.x [DOI] [PubMed] [Google Scholar]
- Gonzali S., Mazzucato A., Perata P. (2009). Purple as a tomato: towards high anthocyanin tomatoes. Trends Plant Sci. 14 237–241. 10.1016/j.tplants.2009.02.001 [DOI] [PubMed] [Google Scholar]
- Gudkov S. V., Grinberg M. A., Sukhov V., Vodeneev V. (2019). Effect of ionizing radiation on physiological and molecular processes in plants. J. Environ. Radioact. 202 8–24. 10.1016/j.jenvrad.2019.02.001 [DOI] [PubMed] [Google Scholar]
- Gupta S., Shi X., Lindquist I. E., Devitt N., Mudge J., Rashotte A. M. (2013). Transcriptome profiling of cytokinin and auxin regulation in tomato root. J. Exp. Bot. 64 695–704. 10.1093/jxb/ers365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Valdes N., Häkkinen S. T., Lemasson C., Guillet M., Oksman-Caldentey K. M., Ritala A., et al. (2020). Hairy Root Cultures—A Versatile Tool With Multiple Applications. Front. Plant Sci. 11:33. 10.3389/fpls.2020.00033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häkkinen S. T., Nygren H., Nohynek L., Puupponen-Pimiä R., Heiniö R. L., Maiorova N., et al. (2020). Plant cell cultures as food—aspects of sustainability and safety. Plant Cell Rep. 39 1655–1668. 10.1007/s00299-020-02592-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanan A., Basit A., Nazir T., Zeeshan Majeed M., Qiu D. (2020). Anti-insect activity of a partially purified protein derived from the entomopathogenic fungus Lecanicillium lecanii (Zimmermann) and its putative role in a tomato defense mechanism against green peach aphid. J. Inverteb. Pathol. 11:574. 10.1016/j.jip.2019.107282 [DOI] [PubMed] [Google Scholar]
- Harrison K., Mendoza-Herrera A., Levy J. G., Tamborindeguy C. (2021). Lasting consequences of psyllid (Bactericera cockerelli L.) infestation on tomato defense, gene expression, and growth. BMC Plant Biol. 21:114. 10.1186/s12870-021-02876-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins C. L., Davies M. J. (2014). Detection and characterisation of radicals in biological materials using EPR methodology. Biochim. Biophys. Acta 1840 708–721. 10.1016/j.bbagen.2013.03.034 [DOI] [PubMed] [Google Scholar]
- Hilaire M. R., Ahmed I. A., Lin C. W., Jo H., DeGrado W. F., Gai F. (2017). Blue fluorescent amino acid for biological spectroscopy and microscopy. Proc. Natl. Acad. Sci. U. S. A. 114 6005–6009. 10.1073/pnas.1705586114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang M. H. T., Nguyen X. C., Lee K., Kwon Y. S., Pham H. T. T., Park H. C., et al. (2012). Phosphorylation by AtMPK6 is required for the biological function of AtMYB41 in Arabidopsis. Biochem. Biophys. Res. Commun. 422 181–186. 10.1016/j.bbrc.2012.04.137 [DOI] [PubMed] [Google Scholar]
- Hou X. J., Li J. M., Liu B. L., Wei L. (2017). Co-expression of basic helix–loop–helix protein (bHLH) and transcriptional activator-Myb genes induced anthocyanin biosynthesis in hairy root culture of Nicotiana tabacum L and Ipomea tricolor. Acta Physiol. Plantar. 39 1–7. 10.1007/s11738-017-2362-4 [DOI] [Google Scholar]
- Hu C., Gong Y., Jin S., Zhu Q. (2011). Molecular analysis of a UDP-glucose: flavonoid 3-O-glucosyltransferase (UFGT) gene from purple potato (Solanum tuberosum). Mol. Biol. Rep. 38 561–567. 10.1007/s11033-010-0141-z [DOI] [PubMed] [Google Scholar]
- Huang B., Routaboul J. M., Liu M., Deng W., Maza E., Mila I., et al. (2017). Overexpression of the class D MADS-box gene Sl-AGL11 impacts fleshy tissue differentiation and structure in tomato fruits. J. Exp. Bot. 68 4869–4884. 10.1093/jxb/erx303 [DOI] [PubMed] [Google Scholar]
- Ichikawa H., Ichiyanagi T., Xu B., Yoshii Y., Nakajima M., Konishi T. (2001). Antioxidant activity of anthocyanin extract from purple black rice. J. Med. Food 4 211–218. 10.1089/10966200152744481 [DOI] [PubMed] [Google Scholar]
- Imhof B., Schlacht I. L., Waclavicek R., Schubert D., Zeidler C., Vrakking V., et al. (2018). “Eden Iss – A Simulation Testbed To An Advanced Exploration Design Concept For A Greenhouse For Moon And Mars” in 69th International Astronautical Congress. Germany: Institute of Space Systems. 1–5. [Google Scholar]
- Inglett G. E., Rose D. J., Chen D., Stevenson D. G., Biswas A. (2010). Phenolic content and antioxidant activity of extracts from whole buckwheat (Fagopyrum esculentum Möench) with or without microwave irradiation. Food Chem. 119 1216–1219. 10.1016/j.foodchem.2009.07.041 [DOI] [Google Scholar]
- Isaacson T., Ronen G., Zamir D., Hirschberg J. (2002). Cloning of tangerine from tomato reveals a Carotenoid isomerase essential for the production of β-carotene and xanthophylls in plants. Plant Cell 14 333–342. 10.1105/tpc.010303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam A., Mercer C. F., Leung S., Dijkwel P. P., McManus M. T. (2015). Transcription of biotic stress associated genes in white clover (Trifolium repens L.) differs in response to cyst and root-knot nematode infection. PLoS One 10:e0137981. 10.1371/journal.pone.0137981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian W., Cao H., Yuan S., Liu Y., Lu J., Lu W., et al. (2019). SlMYB75, an MYB-type transcription factor, promotes anthocyanin accumulation and enhances volatile aroma production in tomato fruits. Hortic. Res. 6:22. 10.1038/s41438-018-0098-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalt W., Cassidy A., Howard L. R., Krikorian R., Stull A. J., Tremblay F., et al. (2020). Recent Research on the Health Benefits of Blueberries and Their Anthocyanins. Adv. Nutr. 11 224–236. 10.1093/advances/nmz065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap S. P., Prasanna H. C., Kumari N., Mishra P., Singh B. (2020). Understanding salt tolerance mechanism using transcriptome profiling and de novo assembly of wild tomato Solanum chilense. Sci. Rep. 10 1–20. 10.1038/s41598-020-72474-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandelwal N., Abraham S. K. (2014). Intake of anthocyanidins pelargonidin and cyanidin reduces genotoxic stress in mice induced by diepoxybutane, urethane and endogenous nitrosation. Environ. Toxicol. Pharmacol. 37 837–843. 10.1016/j.etap.2014.02.012 [DOI] [PubMed] [Google Scholar]
- Khodadad C. L. M., Hummerick M. E., Spencer L. S. E., Dixit A. R., Richards J. T., Romeyn M. W., et al. (2020). Microbiological and Nutritional Analysis of Lettuce Crops Grown on the International Space Station. Front. Plant Sci. 11:492. 10.3389/fpls.2020.00199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiferle C., Fantini E., Bassolino L., Povero G., Spelt C., Buti S., et al. (2015). Tomato R2R3-MYB proteins SlANT1 and SlAN2: same protein activity, different roles. PLoS One 10:e0136365. 10.1371/journal.pone.0136365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S. L. (2015). HISAT: a fast spliced aligner with low memory requirements Daehwan HHS Public Access. Nat. Methods 12 357–360. 10.1038/nmeth.3317.HISAT [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Park J., Yeom S. I., Kim Y. M., Seo E., Kim K. T., et al. (2017). New reference genome sequences of hot pepper reveal the massive evolution of plant disease-resistance genes by retroduplication. Genome Biol. 18 1–11. 10.1186/s13059-017-1341-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee H. J., Tieman D. M. (2013). Genetic challenges of flavor improvement in tomato. Trends Genet. 29 257–262. 10.1016/j.tig.2012.12.003 [DOI] [PubMed] [Google Scholar]
- Koes R., Verweij W., Quattrocchio F. (2005). Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 10 236–242. 10.1016/j.tplants.2005.03.002 [DOI] [PubMed] [Google Scholar]
- Kozlowski D., Marsal P., Steel M., Mokrini R., Duroux J. L., Lazzaroni R., et al. (2007). Theoretical investigation of the formation of a new series of antioxidant depsides from the radiolysis of flavonoid compounds. Radiat. Res. 168 243–252. 10.1667/RR0824.1 [DOI] [PubMed] [Google Scholar]
- Krga I., Milenkovic D. (2019). Anthocyanins: from Sources and Bioavailability to Cardiovascular-Health Benefits and Molecular Mechanisms of Action. J. Agric. Food Chem. 67 1771–1783. 10.1021/acs.jafc.8b06737 [DOI] [PubMed] [Google Scholar]
- Kumar V., Irfan M., Ghosh S., Chakraborty N., Chakraborty S., Datta A. (2016). Fruit ripening mutants reveal cell metabolism and redox state during ripening. Protoplasma 253 581–594. 10.1007/s00709-015-0836-z [DOI] [PubMed] [Google Scholar]
- Lalande M., Schwob L., Vizcaino V., Chirot F., Dugourd P., Schlathölter T., et al. (2019). Direct Radiation Effects on the Structure and Stability of Collagen and Other Proteins. ChemBioChem 20 2972–2980. 10.1002/cbic.201900202 [DOI] [PubMed] [Google Scholar]
- Lateef Z. W. A., Al-Nimer M. S. M. (2009). X-Rays Irradiation Produced Dual Effects on the Constituents of Medicinal Plants Extracts. J. Biol. Sci. 9 872–877. [Google Scholar]
- Li D., Li X., Ding X. (2010). Composition and antioxidative properties of the flavonoid-rich fractions from tartary buckwheat grains. Food Sci. Biotechnol. 19 711–716. 10.1007/s10068-010-0100-4 [DOI] [Google Scholar]
- Li H., Deng Z., Liu R., Young J. C., Zhu H., Loewen S., et al. (2011). Characterization of Phytochemicals and Antioxidant Activities of a Purple Tomato (Solanum lycopersicum L.). J. Agric. Food Chem. 59 11803–11811. 10.1021/jf202364v [DOI] [PubMed] [Google Scholar]
- Li J., Han G., Sun C., Sui N. (2019). Research advances of MYB transcription factors in plant stress resistance and breeding. Plant Signal. Behav. 14 1–9. 10.1080/15592324.2019.1613131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Vickrey T. L., McNally M. G., Sato S. J., Clemente T. E., Mower J. P. (2019). Assessing anthocyanin biosynthesis in solanaceae as a model pathway for secondary metabolism. Genes 10:559. 10.3390/genes10080559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Wang J., Wang B., Huang S., Hu J., Yang T., et al. (2021). Identification of the Carbohydrate and Organic Acid Metabolism Genes Responsible for Brix in Tomato Fruit by Transcriptome and Metabolome Analysis. Front. Genet. 12:714942. 10.3389/fgene.2021.714942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Chen B., Zhang G., Chen L., Dong Q., Wen J., et al. (2016). Regulation of anthocyanin and proanthocyanidin biosynthesis by Medicago truncatula bHLH transcription factor MtTT8. New Phytol. 210 905–921. 10.1111/nph.13816 [DOI] [PubMed] [Google Scholar]
- Li P., Liu H., Yang H., Pu X., Li C., Huo H., et al. (2020). Translocation of Drought-Responsive Proteins from the Chloroplasts. Cells 9 1–17. 10.3390/cells9010259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Tieman D., Liu Z., Chen K., Klee H. J. (2020). Identification of a lipase gene with a role in tomato fruit short-chain fatty acid-derived flavor volatiles by genome-wide association. Plant J. 104 631–644. 10.1111/tpj.14951 [DOI] [PubMed] [Google Scholar]
- Liao Y., Smyth G. K., Shi W. (2019). The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 47:e47. 10.1093/nar/gkz114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Liu J., Wei Y. (2016). Identification and analysis of the metacaspase gene family in tomato. Biochem. Biophys. Res. Commun. 479 523–529. 10.1016/j.bbrc.2016.09.103 [DOI] [PubMed] [Google Scholar]
- Liu J., Osbourn A., Ma P. (2015). MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol. Plant 8 689–708. 10.1016/j.molp.2015.03.012 [DOI] [PubMed] [Google Scholar]
- Liu Y., Tikunov Y., Schouten R. E., Marcelis L. F. M., Visser R. G. F., Bovy A. (2018). Anthocyanin biosynthesis and degradation mechanisms in Solanaceous vegetables: a review. Front. Chem. 6:52. 10.3389/fchem.2018.00052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lloyd A., Brockman A., Aguirre L., Campbell A., Bean A., Cantero A., et al. (2017). Advances in the MYB-bHLH-WD Repeat (MBW) pigment regulatory model: addition of a WRKY factor and co-option of an anthocyanin MYB for betalain regulation. Plant Cell Physiol 58 1431–1441. 10.1093/pcp/pcx075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann W., Lohmann C., Ibrahim M. (1988). Fluorescence spectra of NADH/NAD, kynurenine, tryptophan, and tyrosine. Naturwissenschaften 75 141–142. 10.1007/BF00405305 [DOI] [PubMed] [Google Scholar]
- Lotito S. B., Frei B. (2006). Consumption of flavonoid-rich foods and increased plasma antioxidant capacity in humans: cause, consequence, or epiphenomenon? Free Radic. Biol. Med. 41 1727–1746. 10.1016/j.freeradbiomed.2006.04.033 [DOI] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15 1–21. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcu D., Cristea V., Daraban L. (2013a). Dose-dependent effects of gamma radiation on lettuce (Lactuca sativa var. capitata) seedlings. Int. J. Radiat. Biol. 89 219–223. 10.3109/09553002.2013.734946 [DOI] [PubMed] [Google Scholar]
- Marcu D., Damian G., Cosma C. (2013b). Gamma radiation effects on seed germination, growth and pigment content, and ESR study of induced free radicals in maize (Zea mays). J. Biol. Phys. 39 625–634. 10.1007/s10867-013-9322-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Yang D., Li X., Zhao S., Sui N., Meng Q. (2015). Physiological changes in fruit ripening caused by overexpression of tomato SlAN2, an R2R3-MYB factor. Plant Physiol. Biochem. 89 24–30. 10.1016/j.plaphy.2015.02.005 [DOI] [PubMed] [Google Scholar]
- Martí R., Roselló S., Cebolla-Cornejo J. (2016). Tomato as a source of carotenoids and polyphenols targeted to cancer prevention. Cancers 8 1–28. 10.3390/cancers8060058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massa G. D., Wheeler R. M., Morrow R. C., Levine H. G. (2016). Growth chambers on the International Space Station for large plants. Acta Hortic. 1134 215–221. 10.17660/ActaHortic.2016.1134.29 34854763 [DOI] [Google Scholar]
- Mathews H., Clendennen S. K., Caldwell C. G., Liu X. L., Connors K., Matheis N., et al. (2003). Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification, and transport. Plant Cell 15 1689–1703. 10.1105/tpc.012963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzucato A., Willems D., Bernini R., Picarella M. E., Santangelo E., Ruiu F., et al. (2013). Novel phenotypes related to the breeding of purple-fruited tomatoes and effect of peel extracts on human cancer cell proliferation. Plant Physiol. Biochem. 72 125–133. 10.1016/J.PLAPHY.2013.05.012 [DOI] [PubMed] [Google Scholar]
- McLellan H., Gilroy E. M., Yun B. W., Birch P. R. J., Loake G. J. (2009). Functional redundancy in the Arabidopsis Cathepsin B gene family contributes to basal defence, the hypersensitive response and senescence. New Phytol. 183 408–418. 10.1111/j.1469-8137.2009.02865.x [DOI] [PubMed] [Google Scholar]
- Mehrtens F., Kranz H., Bednarek P., Weisshaar B. (2005). The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol. 138 1083–1096. 10.1104/pp.104.058032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei W., Qin Y., Song W., Li J., Zhu Y. (2009). Cotton GhPOX1 encoding plant class III peroxidase may be responsible for the high level of reactive oxygen species production that is related to cotton fiber elongation. J. Genet. Genomics 36 141–150. 10.1016/S1673-8527(08)60101-0 [DOI] [PubMed] [Google Scholar]
- Mes P. J., Boches P., Myers J. R., Durst R. (2008). Characterization of tomatoes expressing anthocyanin in the fruit. J. Am. Soc. Hortic. Sci. 133 262–269. 10.21273/jashs.133.2.262 [DOI] [Google Scholar]
- Mikami K., Hosokawa M. (2013). Biosynthetic pathway and health benefits of fucoxanthin, an algae-specific xanthophyll in brown seaweeds. Int. J. Mol. Sci. 14 13763–13781. 10.3390/ijms140713763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghaddam S. S., Jaafar H., Ibrahim R., Rahmat A., Aziz M. A., Philip E. (2011). Effects of acute gamma irradiation on physiological traits and flavonoid accumulation of Centella asiatica. Molecules 16 4994–5007. 10.3390/molecules16064994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita T., Yamaguchi H., Degi K. (2007). The contribution of polyphenols to antioxidative activity in common buckwheat and Tartary buckwheat grain. Plant Prod. Sci. 10 99–104. 10.1626/pps.10.99 34921734 [DOI] [Google Scholar]
- Morita Y., Takagi K., Fukuchi-Mizutani M., Ishiguro K., Tanaka Y., Nitasaka E., et al. (2014). A chalcone isomerase-like protein enhances flavonoid production and flower pigmentation. Plant J. 78 294–304. 10.1111/tpj.12469 [DOI] [PubMed] [Google Scholar]
- Murata M., Nakai Y., Kawazu K., Ishizaka M., Kajiwara H., Ab H., et al. (2019). Loliolide, a carotenoid metabolite, is a potential endogenous inducer of herbivore resistance. Plant Physiol. 179 1822–1833. 10.1104/pp.18.00837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata T., Todoriki S., Masumizu T., Suda I., Furuta S., Du Z., et al. (2003). Levels of active oxygen species are controlled by ascorbic acid and anthocyanin in Arabidopsis. J. Agric. Food Chem. 51 2992–2999. 10.1021/jf026179+ [DOI] [PubMed] [Google Scholar]
- Nveawiah-Yoho P., Zhou J., Palmer M., Sauve R., Zhou S., Howe K. J., et al. (2013). Identification of Proteins for Salt Tolerance Using a Comparative Proteomics Analysis of Tomato Accessions with Contrasting Salt Tolerance. J. Am. Soc. Hortic. Sci. 138:13. 10.21273/jashs.138.5.382 [DOI] [Google Scholar]
- Omidvar V., Mohorianu I., Dalmay T., Fellner M. (2015). Identification of miRNAs with potential roles in regulation of anther development and male-sterility in 7B-1 male-sterile tomato mutant. BMC Genomics 16:878. 10.1186/s12864-015-2077-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooe E., Ogawa K., Horiuchi T., Tada H., Murase H., Tsuruma K., et al. (2016). Analysis and characterization of anthocyanins and carotenoids in Japanese blue tomato. Biosci. Biotechnol. Biochem. 80 341–349. 10.1080/09168451.2015.1091715 [DOI] [PubMed] [Google Scholar]
- Pan Q., Liu Y. S., Budai-Hadrian O., Sela M., Carmel-Goren L., Zamir D., et al. (2000). Comparative genetics of nucleotide binding site-leucine rich repeat resistance gene homologues in the genomes of two dicotyledons: tomato and arabidopsis. Genetics 155 309–322. 10.1093/genetics/155.1.309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Wang G., Lay C. L., Tan B. H., He C., Liu Y. (2013). Photoluminescence from Amino-Containing Polymer in the Presence of CO 2: carbamato Anion Formed as a Fluorophore. Sci. Rep. 3 1–6. 10.1038/srep02763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paniagua C., Bilkova A., Jackson P., Dabravolski S., Riber W., Didi V., et al. (2017). Dirigent proteins in plants: modulating cell wall metabolism during abiotic and biotic stress exposure. J. Exp. Bot. 68 3287–3301. 10.1093/jxb/erx141 [DOI] [PubMed] [Google Scholar]
- Pelletier K., Murrell J. R., Shirley B. W. (1997). Characterization of flavonol synthase and leucoanthocyanidin dioxygenase genes in Arabidopsis. Further evidence for differential regulation of “early” and “late” genes. Plant Physiol. 113 1437–1445. 10.1104/pp.113.4.1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H., Yang T., Jurick W. M. (2014). Calmodulin gene expression in response to mechanical wounding and Botrytis cinerea infection in tomato fruit. Plants 3 427–441. 10.3390/plants3030427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J., Deng X., Huang J., Jia S., Miao X., Huang Y. (2004). Role of salicylic acid in tomato defense against cotton bollworm, Helicoverpa armigera Hubner. Z. Naturforsch. C J. Biosci. 59 856–862. 10.1515/znc-2004-11-1215 [DOI] [PubMed] [Google Scholar]
- Petrisor D., Damian G., Simon S., Hosu A., Miclaus V. (2008). Antioxidant activity of some types of white wines and juices investigated by EPR spectroscopy. Mod. Phys. Lett. B 22 2689–2698. 10.1142/S0217984908017175 [DOI] [Google Scholar]
- Pinheiro T. T., Peres L. E. P., Purgatto E., Latado R. R., Maniero R. A., Martins M. M., et al. (2019). Citrus carotenoid isomerase gene characterization by complementation of the “Micro-Tom” tangerine mutant. Plant Cell Rep. 38 623–636. 10.1007/s00299-019-02393-2 [DOI] [PubMed] [Google Scholar]
- Povero G. (2011). Physiological and Genetic Control of Anthocyanin Pigmentation in Different Species. Netherlands: Vrije Universiteit Amsterdam. [Google Scholar]
- Povero G., Gonzali S., Bassolino L., Mazzucato A., Perata P. (2011). Transcriptional analysis in high-anthocyanin tomatoes reveals synergistic effect of Aft and atv genes. J. Plant Physiol. 168 270–279. 10.1016/j.jplph.2010.07.022 [DOI] [PubMed] [Google Scholar]
- Preuß A., Stracke R., Weisshaar B., Hillebrecht A., Matern U., Martens S. (2009). Arabidopsis thaliana expresses a second functional flavonol synthase. FEBS Lett. 583 1981–1986. 10.1016/j.febslet.2009.05.006 [DOI] [PubMed] [Google Scholar]
- Qian Y., Zhang T., Yu Y., Gou L., Yang J., Xu J., et al. (2021). Regulatory Mechanisms of bHLH Transcription Factors in Plant Adaptive Responses to Various Abiotic Stresses. Front. Plant Sci. 12:677611. 10.3389/fpls.2021.677611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Z., Wang X., Gao J., Guo Y., Huang Z., Du Y. (2016). The Tomato Hoffman’s Anthocyaninless Gene Encodes a bHLH Transcription Factor Involved in Anthocyanin Biosynthesis That Is Developmentally Regulated and Induced by Low Temperatures. PLoS One 11:e0151067. 10.1371/journal.pone.0151067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay N. A., Glover B. J. (2005). MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 10 63–70. 10.1016/j.tplants.2004.12.011 [DOI] [PubMed] [Google Scholar]
- Reimand J., Kull M., Peterson H., Hansen J., Vilo J. (2007). G:Profiler-a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 35 193–200. 10.1093/nar/gkm226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley P. A. (1994). Free Radicals in Biology: oxidative Stress and the Effects of Ionizing Radiation. Int. J. Radiat. Biol. 65 27–33. [DOI] [PubMed] [Google Scholar]
- Roldan M. V. G., Outchkourov N., van Houwelingen A., Lammers M., de La Fuente I. R., Ziklo N., et al. (2014). An O-methyltransferase modifies accumulation of methylated anthocyanins in seedlings of tomato. Plant J. 80 695–708. 10.1111/tpj.12664 [DOI] [PubMed] [Google Scholar]
- Ron M., Kajala K., Pauluzzi G., Wang D., Reynoso M. A., Zumstein K., et al. (2014). Hairy root transformation using Agrobacterium rhizogenes as a tool for exploring cell type-specific gene expression and function using tomato as a model. Plant Physiol. 166 455–469. 10.1104/pp.114.239392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossetto M., Vanzani P., Lunelli M., Scarpa M., Mattivi F., Rigo A. (2007). Peroxyl radical trapping activity of anthocyanins and generation of free radical intermediates. Free Radic. Res. 41 854–859. 10.1080/10715760701261533 [DOI] [PubMed] [Google Scholar]
- Roy S. (2016). Function of MYB domain transcription factors in abiotic stress and epigenetic control of stress response in plant genome. Plant Signal. Behav. 11:e1117723. 10.1080/15592324.2015.1117723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safavi-Rizi V., Herde M., Stöhr C. (2020). Identification of nitric oxide (NO)-responsive genes under hypoxia in tomato (Solanum lycopersicum L.) root. Sci. Rep. 10 1–20. 10.1038/s41598-020-73613-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S., Singh J., Paul A., Sarkar R., Khan Z., Banerjee K. (2021). Anthocyanin profiling using UV-vis spectroscopy and liquid chromatography mass spectrometry. J. AOAC Int. 103 23–39. 10.5740/jaoacint.19-0201 [DOI] [PubMed] [Google Scholar]
- Scandalios J. G. (2002). Oxidative stress responses - What have genome-scale studies taught us? Genome Biol. 3 1–6. 10.1186/gb-2002-3-7-reviews1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N., Semel Y., Roessner U., Gur A., Balbo I., Carrari F., et al. (2006). Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat. Biotechnol. 24 447–454. 10.1038/nbt1192 [DOI] [PubMed] [Google Scholar]
- Schimmel B. C. J., Alba J. M., Wybouw N., Glas J. J., Meijer T. T., Schuurink R. C., et al. (2018). Distinct signatures of host defense suppression by plant-feeding mites. Int. J. Mol. Sci. 19:3265. 10.3390/ijms19103265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber G., Reuveni M., Evenor D., Oren-Shamir M., Ovadia R., Sapir-Mir M., et al. (2012). ANTHOCYANIN1 from Solanum chilense is more efficient in accumulating anthocyanin metabolites than its Solanum lycopersicum counterpart in association with the ANTHOCYANIN FRUIT phenotype of tomato. Theor. Appl. Genet. 124 295–307. 10.1007/s00122-011-1705-6 [DOI] [PubMed] [Google Scholar]
- Schreurs A. S., Shirazi-Fard Y., Shahnazari M., Alwood J. S., Truong T. A., Tahimic C. G. T., et al. (2016). Dried plum diet protects from bone loss caused by ionizing radiation. Sci. Rep. 6 1–11. 10.1038/srep21343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scranton M. A., Fowler J. H., Girke T., Walling L. L. (2013). Microarray Analysis of Tomato’s Early and Late Wound Response Reveals New Regulatory Targets for Leucine Aminopeptidase A. PLoS One 8:e77889. 10.1371/journal.pone.0077889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Şensoy Í, Rosen R. T., Ho C. T., Karwe M. V. (2006). Effect of processing on buckwheat phenolics and antioxidant activity. Food Chem. 99 388–393. 10.1016/j.foodchem.2005.08.007 [DOI] [Google Scholar]
- Serrato A. J., Fernández-Trijueque J., Barajas-López J., de D., Chueca A., Sahrawy M. (2013). Plastid thioredoxins: a “one-for-all” redox-signaling system in plants. Front. Plant Sci. 4:463. 10.3389/fpls.2013.00463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewelam N., Kazan K., Schenk P. M. (2016). Global plant stress signaling: reactive oxygen species at the cross-road. Front. Plant Sci. 7:187. 10.3389/fpls.2016.00187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P., Padh H., Shrivastava N. (2013). Hairy root cultures: a suitable biological system for studying secondary metabolic pathways in plants. Eng. Life Sci. 13 62–75. 10.1002/elsc.201200030 [DOI] [Google Scholar]
- Shi J., Du X. (2020). Identification, characterization and expression analysis of calmodulin and calmodulin-like proteins in Solanum pennellii. Sci. Rep. 10 1–17. 10.1038/s41598-020-64178-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Panthee D. R. (2020). Transcriptome-based analysis of tomato genotypes resistant to bacterial spot (Xanthomonas perforans) race t4. Int. J. Mol. Sci. 21 1–31. 10.3390/ijms21114070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solìs-Oviedo R. L., de La Cruz Pech-Canul Á. (2019). Frontiers and New Trends in the Science of Fermented Food and Beverages. London: IntechOpen. 10.5772/intechopen.73404 [DOI] [Google Scholar]
- Spelt C., Quattrocchio F., Mol J. N. M., Koes R. (2000). anthocyanin1 of Petunia Encodes a Basic Helix-Loop-Helix Protein That Directly Activates Transcription of Structural Anthocyanin Genes. Plant Cell 12:1619. 10.2307/3871178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R., Werber M., Weisshaar B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4 447–456. 10.1016/S1369-5266(00)00199-0 [DOI] [PubMed] [Google Scholar]
- Su X., Xu J., Rhodes D., Shen Y., Song W., Katz B., et al. (2016). Identification and quantification of anthocyanins in transgenic purple tomato. Food Chem. 202 184–188. 10.1016/j.foodchem.2016.01.128 [DOI] [PubMed] [Google Scholar]
- Sui X., Sun H., Qi B., Zhang M., Li Y., Jiang L. (2018). Functional and conformational changes to soy proteins accompanying anthocyanins: focus on covalent and non-covalent interactions. Food Chem. 245 871–878. 10.1016/j.foodchem.2017.11.090 [DOI] [PubMed] [Google Scholar]
- Sun C., Deng L., Du M., Zhao J., Chen Q., Huang T., et al. (2019). A Transcriptional Network Promotes Anthocyanin Biosynthesis in Tomato Flesh. Mol. Plant 13 42–58. 10.1016/j.molp.2019.10.010 [DOI] [PubMed] [Google Scholar]
- Sun X., Shu J., Mohamed A. M. A., Deng X., Zhi X., Bai J., et al. (2019). Identification and characterization of EI (elongated internode) gene in tomato (solanum lycopersicum). Int. J. Mol. Sci. 20 1–18. 10.3390/ijms20092204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan S., Philosoph-Hadas S., Riov J., Mugasimangalam R., Kuravadi N. A., Kochanek B., et al. (2016). De novo transcriptome sequencing and development of abscission zone-specific microarray as a new molecular tool for analysis of tomato organ abscission. Front. Plant Sci. 6:1258. 10.3389/fpls.2015.01258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takos A. M., Jaffé F. W., Jacob S. R., Bogs J., Robinson S. P., Walker A. R. (2006). Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 142 1216–1232. 10.1104/pp.106.088104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor B. H., Scheuring C. F. (1994). A molecular marker for lateral root initiation: the RSI-1 gene of tomato (Lycopersicon esculentum Mill) is activated in early lateral root primordia. Mol. Gener. Genet. 243 148–157. 10.1007/BF00280311 [DOI] [PubMed] [Google Scholar]
- Thamaphat K., Goodman B. A., Limsuwan P., Smith S. M. (2015). Rapid screening for anthocyanins in cane sugars using ESR spectroscopy. Food Chem. 171 123–127. 10.1016/j.foodchem.2014.08.126 [DOI] [PubMed] [Google Scholar]
- Thwe A., Arasu M. V., Li X., Park C. H., Kim S. J., Al-Dhabi N. A., et al. (2016). Effect of different Agrobacterium rhizogenes strains on hairy root induction and phenylpropanoid biosynthesis in tartary buckwheat (Fagopyrum tataricum Gaertn). Front. Microbiol. 7:318. 10.3389/fmicb.2016.00318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L., Tan Y., Chen G., Wang G., Sun J., Ou S., et al. (2019). Metabolism of anthocyanins and consequent effects on the gut microbiota. Crit. Rev. Food Sci. Nutr. 59 982–991. 10.1080/10408398.2018.1533517 [DOI] [PubMed] [Google Scholar]
- Tohge T., de Souza L. P., Fernie A. R. (2017). Current understanding of the pathways of flavonoid biosynthesis in model and crop plants. J. Exp. Bot. 68 4013–4028. 10.1093/jxb/erx177 [DOI] [PubMed] [Google Scholar]
- Tohge T., Scossa F., Wendenburg R., Frasse P., Balbo I., Watanabe M., et al. (2020). Exploiting Natural Variation in Tomato to Define Pathway Structure and Metabolic Regulation of Fruit Polyphenolics in the Lycopersicum Complex. Mol. Plant 13 1027–1046. 10.1016/j.molp.2020.04.004 [DOI] [PubMed] [Google Scholar]
- Tominaga-Wada R., Nukumizu Y., Wada T. (2013). Tomato (Solanum lycopersicum) homologs of TRIPTYCHON (SLTRY) and GLABRA3 (SLGL3) are involved in anthocyanin accumulation. Plant Signal. Behav. 8 8–10. 10.4161/psb.24575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuner H., Korkmaz M. (2007). Radiostability of butylated hydroxytoluene (BHT): an ESR study. Nuclear Instr. Methods Phys. Res. B Beam Interact. Mater. Atoms 258 388–394. 10.1016/j.nimb.2007.02.098 [DOI] [Google Scholar]
- Tzin V., Rogachev I., Meir S., Moyal Ben Zvi M., Masci T., Vainstein A., et al. (2015). Altered Levels of Aroma and Volatiles by Metabolic Engineering of Shikimate Pathway Genes in Tomato Fruits. AIMS Bioeng. 2 75–92. 10.3934/bioeng.2015.2.75 [DOI] [Google Scholar]
- van Hoeck A., Horemans N., Nauts R., van Hees M., Vandenhove H., Blust R. (2017). Lemna minor plants chronically exposed to ionising radiation: rNA-seq analysis indicates a dose rate dependent shift from acclimation to survival strategies. Plant Sci. 257 84–95. 10.1016/j.plantsci.2017.01.010 [DOI] [PubMed] [Google Scholar]
- van Kan J. A. L., Joosten M. H. A. J., Wagemakers C. A. M., van den Berg-Velthuis G. C. M., de Wit P. J. G. M. (1992). Differential accumulation of mRNAs encoding extracellular and intracellular PR proteins in tomato induced by virulent and avirulent races of Cladosporium fulvum. Plant Mol. Biol. 20 513–527. 10.1007/BF00040610 [DOI] [PubMed] [Google Scholar]
- Vandenhove H., Vanhoudt N., Cuypers A., van Hees M., Wannijn J., Horemans N. (2010). Life-cycle chronic gamma exposure of Arabidopsis thaliana induces growth effects but no discernable effects on oxidative stress pathways. Plant Physiol. Biochem. 48 778–786. 10.1016/j.plaphy.2010.06.006 [DOI] [PubMed] [Google Scholar]
- Vardhan P. V., Shukla L. I. (2017). Gamma irradiation of medicinally important plants and the enhancement of secondary metabolite production. Int. J. Radiat. Biol. 93 967–979. 10.1080/09553002.2017.1344788 [DOI] [PubMed] [Google Scholar]
- Velázquez-Márquez S., De-La-Cruz I. M., Tapia-López R., Núñez-Farfán J. (2021). Tropane alkaloids and terpenes synthase genes of Datura stramonium (Solanaceae). PeerJ. 9:e11466. 10.7717/peerj.11466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verweij W., Spelt C., Di Sansebastiano G. P., Vermeer J., Reale L., Ferranti F., et al. (2008). An H+ P-ATPase on the tonoplast determines vacuolar pH and flower colour. Nat. Cell Biol. 10 1456–1462. 10.1038/ncb1805 [DOI] [PubMed] [Google Scholar]
- Verweij W., Spelt C. E., Bliek M., de Vries M., Wit N., Faraco M., et al. (2016). Functionally similar WRKY proteins regulate vacuolar acidification in petunia and hair development in arabidopsis. Plant Cell 28 786–803. 10.1105/tpc.15.00608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villani M. E., Massa S., Lopresto V., Pinto R., Salzano A. M., Scaloni A., et al. (2017). Effects of high-intensity static magnetic fields on a root-based bioreactor system for space applications. Life Sci. Space Res. 15 79–87. 10.1016/j.lssr.2017.09.002 [DOI] [PubMed] [Google Scholar]
- Vivar-Quintana A. M., Santos-Buelga C., Rivas-Gonzalo J. C. (2002). Anthocyanin-derived pigments and colour of red wines. Anal. Chim. Acta 458 147–155. 10.1016/S0003-2670(01)01619-1 [DOI] [Google Scholar]
- Wang T., Zhang H., Zhu H. (2019). CRISPR technology is revolutionizing the improvement of tomato and other fruit crops. Hortic. Res. 6:77. 10.1038/s41438-019-0159-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T. W., Lu L., Wang D., Thompson J. E. (2001). Isolation and Characterization of Senescence-induced cDNAs Encoding Deoxyhypusine Synthase and Eucaryotic Translation Initiation Factor 5A from Tomato. J. Biol. Chem. 276 17541–17549. 10.1074/jbc.M008544200 [DOI] [PubMed] [Google Scholar]
- Wen B., Zhang F., Wu X., Li H. (2020). Characterization of the Tomato (Solanum lycopersicum) Pectin Methylesterases: evolution, Activity of Isoforms and Expression During Fruit Ripening. Front. Plant Sci. 11:238. 10.3389/fpls.2020.00238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R. J., Spencer J. P. E., Rice-Evans C. (2004). Flavonoids: antioxidants or signalling molecules? Free Radic. Biol. Med. 36 838–849. 10.1016/j.freeradbiomed.2004.01.001 [DOI] [PubMed] [Google Scholar]
- Wolff S. A., Coelho L. H., Karoliussen I., Jost A. I. K. (2014). Effects of the extraterrestrial environment on plants: recommendations for future space experiments for the MELiSSA higher plant compartment. Life 4 189–204. 10.3390/life4020189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Zhai Q., Wei J., Li S., Wang B., Huang T., et al. (2013). Role of Tomato Lipoxygenase D in Wound-Induced Jasmonate Biosynthesis and Plant Immunity to Insect Herbivores. PLoS Genet. 9:e1003964. 10.1371/journal.pgen.1003964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S., Chen N., Huang Z., Li D., Zhi J., Yu B., et al. (2020). Anthocyanin Fruit encodes an R2R3-MYB transcription factor, SlAN2-like, activating the transcription of SlMYBATV to fine-tune anthocyanin content in tomato fruit. New Phytol. 225 2048–2063. [DOI] [PubMed] [Google Scholar]
- Yang H., Xiao X., Zhao X., Wu Y. (2015). “Intrinsic Fluorescence Spectra of Tryptophan. Tyrosine and Phenyloalanine” in Proceedings of the 5th International Conference on Advanced Design and Manufacturing Engineering. (Netherlands: Atlantis Press; ). 224–233. 10.2991/icadme-15.2015.46 [DOI] [Google Scholar]
- Yang Y. X., Wang M. M., Yin Y. L., Onac E., Zhou G. F., Peng S., et al. (2015). RNA-seq analysis reveals the role of red light in resistance against Pseudomonas syringae pv. tomato DC3000 in tomato plants. BMC Genomics 16:120. 10.1186/s12864-015-1228-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabel P., Bamsey M., Schubert D., Tajmar M. (2016). Review and analysis of over 40 years of space plant growth systems. Life Sci. Space Res. 10 1–16. 10.1016/j.lssr.2016.06.004 [DOI] [PubMed] [Google Scholar]
- Zabel P., Bamsey M., Zeidler C., Vrakking V., Johannes B., Rettberg P. (2015). “Introducing EDEN ISS - A European project on advancing plant cultivation technologies and operations,” in 45th International Conference on Environmental Systems. [Google Scholar]
- Zhang H., Koes R., Shang H., Fu Z., Wang L., Dong X., et al. (2019). Identification and functional analysis of three new anthocyanin R2R3-MYB genes in Petunia. Plant Dir 3 1–13. 10.1002/pld3.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Butelli E., Alseekh S., Tohge T., Rallapalli G., Luo J., et al. (2015). Multi-level engineering facilitates the production of phenylpropanoid compounds in tomato. Nat. Commun 6 1–11. 10.1038/ncomms9635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Butelli E., de Stefano R., Schoonbeek H. J., Magusin A., Pagliarani C., et al. (2013). Anthocyanins double the shelf life of tomatoes by delaying overripening and reducing susceptibility to gray mold. Curr. Biol. 23 1094–1100. 10.1016/j.cub.2013.04.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Butelli E., Martin C. (2014). Engineering anthocyanin biosynthesis in plants. Curr. Opin. Plant Biol. 19 81–90. 10.1016/j.pbi.2014.05.011 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Song H., Wang X., Zhou X., Zhang K., Chen X., et al. (2020). The roles of different types of trichomes in tomato resistance to cold, drought, whiteflies, and botrytis. Agronomy 10:411. 10.3390/agronomy10030411 [DOI] [Google Scholar]
- Zhao X., Liu Y., Liu X., Jiang J. (2018). Comparative transcriptome profiling of two tomato genotypes in response to potassium-deficiency stress. Int. J. Mol. Sci. 19:2402. 10.3390/ijms19082402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann I. M., Heim M. A., Weisshaar B., Uhrig J. F. (2004). Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 40 22–34. 10.1111/j.1365-313X.2004.02183.x [DOI] [PubMed] [Google Scholar]
- Zouari I., Salvioli A., Chialva M., Novero M., Miozzi L., Tenore G. C., et al. (2014). From root to fruit: rNA-Seq analysis shows that arbuscular mycorrhizal symbiosis may affect tomato fruit metabolism. BMC Genomics 15:221. 10.1186/1471-2164-15-221 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primer identification (id.) as described in “Materials and Methods” for HRCs PCR screening (for, forward; rev, reverse).
List of figures representing the level of different phenolic compounds and their derivatives in control and PhAN4 HRCs.
List of figures representing the level of different flavonoids in control and PhAN4 HRCs.
Complete list of DEGs.
GOE analysis results.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://dataview.ncbi.nlm.nih.gov/object/PRJNA794337?reviewer=estprpllasrt3f3libdoij8rco.


