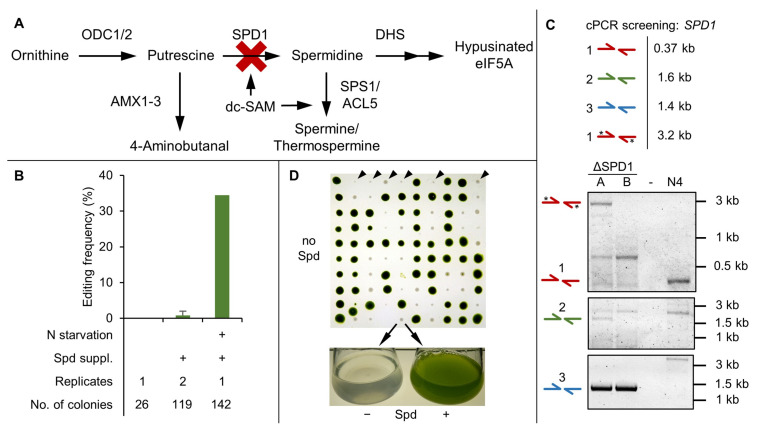
Figure 3.
SPD1 knockout study using Cas9-RNP-based gene editing. (A) Schematic of the spermidine metabolic pathway. Functional knockout of the SPD1 gene (red cross) should disrupt spermidine formation. (B) Editing frequency of the SPD1 locus depending on pre-culture treatment and spermidine supplementation (0.1 mM) as determined by colony PCR and spermidine starvation assay. The number of individual transformations and screened colonies are indicated. Error bars indicate the standard deviation of replicates. (C) Representative cPCR results of two independent mutants using the indicated primer pairs (compare Figure 2). Asterisks (*) indicate a secondary PCR product derived from amplification of the repair template. Sequencing results and uncropped gel images are available in Supplementary Figures S6 and S7 respectively. (D) Phenotypical screening via spermidine starvation assay. Colonies emerging after transformation were transferred to a spermidine-less plate. Colonies grew slower and bleached after 10 to 14 days, confirming the spermidine auxotrophy of the ΔSPD1 mutants (black triangles in the first row). Colonies were also grown in liquid medium with and without spermidine (lower panel). (N4: N-UVM4 parental strain, ODC: Ornithine decarboxylase, SPD1: Spermidine synthase, DHS: Deoxyhypusine synthase, SPS1: Spermine synthase, ACL5: Thermospermine synthase, AMX: Amine oxidase, dc-SAM: decarboxylated S-adenosylmethionine, eIF5A: eukaryotic translation factor 5A, Spd: Spermidine).