Abstract
While meropenem MICs were strongly influenced by the presence or absence of the MexAB-OprM efflux pump in both OprD-proficient and -deficient strain backgrounds, MICs of imipenem and of ER-35786 remained unchanged, demonstrating that meropenem is a substrate of MexAB-OprM but not imipenem and ER-35786. In vitro, all three carbapenems selected loss of OprD as a first mechanism of resistance. However, in an OprD-deficient background, meropenem was able to select MexAB-OprM overproducers as a secondary resistance mechanism, while ER-35786 selected a mutant cross-resistant to sparfloxacin and cefpirome.
In Pseudomonas aeruginosa, the potency of β-lactam molecules is limited by several barriers. First, the bacterium’s rather impermeable outer membrane (1, 24) significantly decreases the access of the mostly hydrophilic β-lactams to their targets, the penicillin-binding proteins. Second, chromosomal and plasmid-carried β-lactamases (2, 15) enzymatically hydrolyze β-lactam molecules in the periplasmic space. Finally, active efflux systems extrude β-lactams (16, 17). Indeed, the constitutively expressed MexAB-OprM efflux system (9, 10) includes most β-lactams in its broad-substrate spectrum, while the MexCD-OprJ system (19), when derepressed, extrudes only cephems (4, 11). The MexEF-OprN system (7) does not contribute to β-lactam efflux; however, its overexpression indirectly affects the efficacy of carbapenems through a concomitant reduction (7, 12) of the carbapenem-specific OprD porin protein.
Masuda and Ohya (12) showed that mutants overexpressing MexAB-OprM are more resistant to meropenem but not to imipenem or panipenem compared to wild type. This finding led to the suggestion (9) that meropenem behaves as a substrate of this pump because of the presence of a hydrophobic side chain at position 2, whereas imipenem or panipenem, containing strongly charged, hydrophilic side-chains, cannot become a substrate. However, the correlation between resistance and efflux may not be simple, because the influx of carbapenems is affected by the levels of OprD.
In the present study, we therefore examined the activity of the three carbapenems, imipenem, meropenem, and ER-35786 (Fig. 1), in the presence and absence of OprD, and determined the mechanisms of resistance selected in vitro by the three antibiotics.
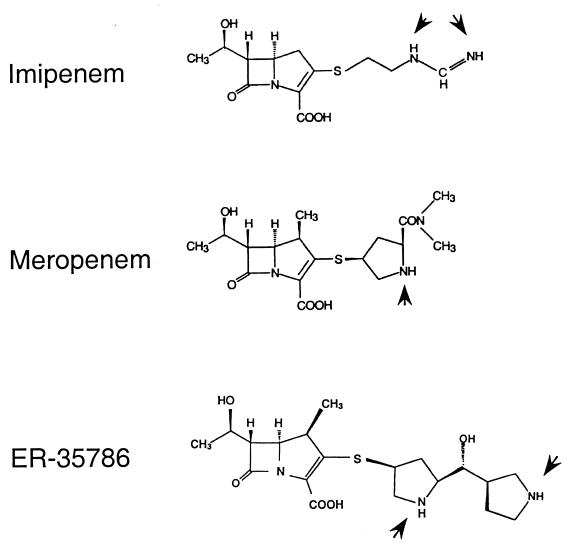
FIG. 1.
Chemical structures of the three carbapenems studied. Arrowheads indicate nitrogen atoms which can be charged positively.
Activity of carbapenems against mutants with well-defined resistance mechanisms.
Derivatives of PAO1 with all possible combinations of OprD (influx) and MexAB-OprM (efflux) expression were constructed (Table 1). The oprD::ΩTc knockout mutant PASE1 (2a) was transduced with phage E79tv2 (5) grown on the oprM::ΩHg mutant K613 (20) to generate the defined oprD-oprM double mutant PA1425. A nalB-type derivative of strain PASE1, called PA1426 and overexpressing the mexAB-oprM operon, was obtained by plating PASE1 on Luria-Bertani (LB) agar containing carbenicillin (SmithKline Beecham Pharmaceuticals, Worthing, Great Britain) at a concentration of 100 μg/ml. Western blot analysis with a rabbit anti-OprD antibody (2a) confirmed the absence of OprD in strains PASE1, PA1425, and PA1426. By using an anti-OprM antibody (25), OprM was determined to be undetectable in PAO1T and PA1425 but was overexpressed in PA1423 and PA1426 (data not shown). None of the strains produced detectable β-lactamase activities under noninducing conditions, thereby excluding fortuitous derepression of β-lactamases during the construction of the strains.
TABLE 1.
Bacterial strains
| P. aeruginosa strain | Relevant characteristics | Source or reference |
|---|---|---|
| PAO1 | Wild type | Laboratory collection |
| PA1423 | Overexpressing MexAB-OprM | 8 |
| PAO1T | oprM::ΩHg | 14 |
| PASE1 | oprD::ΩTc | S. F. Epp et al. |
| PA1425 | oprD::ΩTc oprM::ΩHg | This study |
| PA1426 | oprD::ΩTc, overexpressing MexAB-OprM, selected on carbenicillin | This study |
| PA1433 | oprD::ΩTc, decreased expression of 55-kDa protein, selected on meropenem | This study |
| PA1434 | oprD::ΩTc, overexpressing MexAB-OprM, selected on meropenem | This study |
| PA1436 | oprD::ΩTc, decreased expression of 55-kDa protein, selected on ER-35786 | This study |
Susceptibility to antimicrobial agents was assayed by the microdilution method with Mueller-Hinton broth (6). In an OprD-sufficient background, the OprM-deficient strain PAO1T was hypersusceptible to all antibiotics tested except imipenem (Merck-Sharp and Dohme-Chibret, Zurich, Switzerland) and ER-35786 (Eisai Co., Ltd., Tsukuba, Japan) (Table 2). By contrast, strain PA1423, overexpressing the MexAB-OprM system, showed increased resistance to all the antibiotics tested, again with the exception of imipenem and ER-35786. Parallel MIC changes were also observed in an OprD-negative background, where both MexAB-OprM deficiency (PA1425) and MexAB-OprM overexpression (PA1426) altered the MICs of meropenem (Imperial Chemical Industries, Macclesfild, Great Britain), without changing those of imipenem and ER-35786. These results strongly suggest that meropenem is a substrate of the MexAB-OprM system, while imipenem and ER-35786 are not.
TABLE 2.
Effects of OprM and OprD expression levels on susceptibilities of carbapenems and other antimicrobial agents
| Antibiotic | MICs (μg/ml) for strains with indicated phenotypea
|
|||||
|---|---|---|---|---|---|---|
| PAO1 OprM+ OprD+ | PAO1T OprM− OprD+ | PA1423 OprM++ OprD+ | PASE1 OprM+ OprD− | PA1425 OprM− OprD− | PA1426 OprM++ OprD− | |
| Imipenem | 1 | 1 | 1 | 16 | 16 | 16 |
| Meropenem | 0.5 | 0.125 | 4 | 4 | 0.5 | 16 |
| ER-35786 | 0.25 | 0.25 | 0.25 | 1 | 0.5 | 0.5 |
| Carbenicillin | 64 | 1 | 256 | 64 | 2 | 256 |
| Cefpirome | 2 | 0.25 | 4 | 2 | 0.25 | 8 |
| Sparfloxacin | 0.5 | 0.125 | 2 | 0.5 | 0.125 | 2 |
+, wild-type expression level; −, knockout mutation; ++, overexpression.
One possible explanation for the differential behavior of imipenem and meropenem regarding the MexAB-OprM system is their different permeation rates. Based on liposome swelling assays, imipenem (736 nm/s) penetrates about 10 times more rapidly through OprD than meropenem (73 nm/s), while the penetration coefficients for imipenem (6 nm/s) and meropenem (5.5 nm/s) are comparable in OprD-deficient strains (23). Therefore, the rapid imipenem influx through OprD could saturate the efflux pump such that increased MexAB-OprM expression would not affect imipenem MICs. However, since in the OprD-deficient mutant imipenem activity is not influenced by the levels of MexAB-OprM expression, efflux pump saturation is not a valid explanation.
Alternatively, the physicochemical properties of the three carbapenems could be responsible. Each side chain attached at position 2 of the molecule contains nitrogen atoms which can be protonated (Fig. 1). While imipenem and meropenem contain basic groups with measured pKa values of 9.91 (21) and 7.4 (22), respectively, ER-35786 contains 2 basic centers in the pyrrolidine rings with pKa values of >10. This means that at physiological pH, >99% of the C2 side chains of imipenem and ER-35786 are positively charged, against 50% of the meropenem side chains. If membrane insertion, as suggested previously (9, 17), is a prerequisite for subsequent extrusion by an efflux mechanism, then the difference in “amphiphilicity” could explain the differential behavior of the three carbapenems with respect to efflux systems. Interestingly, panipenem, which has a similar imine radical as imipenem at the C2-substituent, also remains unaffected in its activity by efflux systems (13).
In vitro selection of carbapenem resistance.
Another way to look at the contribution of efflux and OprD to carbapenem activity is to analyze the mechanisms of resistance after carbapenem exposure. For this purpose, 109 to 1010 CFUs of wild-type PAO1 were exposed on LB agar plates containing carbapenems at concentrations of two, three, four, or eight times the MIC. Spontaneous resistant colonies appeared after incubation at 37°C for 24 to 48 h with similar frequencies for the three compounds. At two to four times the MICs, frequencies varied from 3 × 10−8 to 1 × 10−9 and were less than 10−9 at 8 times the MIC. All colonies tested (eight from each selection) displayed increased resistance only to the three carbapenems. Total lysates of five spontaneous mutants obtained on each carbapenem were analysed by Western blotting for the presence of OprD. None of them showed OprD reactivity, demonstrating loss of OprD as the first mechanism of resistance.
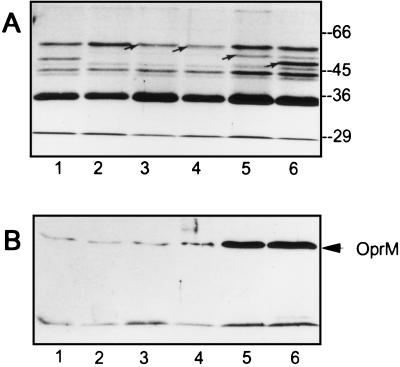
Interesting differences between the three carbapenems were seen when the selection was performed with the OprD-deficient strain PASE1. Again 109 to 1010 CFUs were spread on agar plates containing carbapenem concentrations ranging from 3 to 16 times the MICs. After 48 h of incubation, no colonies grew on imipenem (48 μg/ml) containing agar. At 16 μg of meropenem/ml (4 × MIC) and at 8 μg of ER-35786/ml (8 × MIC) resistant colonies were obtained at a frequency of 2 × 10−8. Analysis of the antibiotic resistance profile of seven colonies obtained on meropenem revealed that six were nalB-type mutants. Indeed, one representative clone, called PA1434, showed a strong outer membrane protein band hybridizing with anti-OprM antibody after Western blotting, confirming overexpression of the MexAB-OprM system (Fig. 2B). Compared to the parental strain PASE1, the remaining mutant, termed PA1433, showed slightly increased MICs of meropenem, ER-35786, and sparfloxacin (Rhône-Poulenc, Paris, France) but not of imipenem (Table 3) and expressed wild-type levels of OprM (Fig. 2B). Spontaneous mutants obtained on ER-35786 were found to have very similar antibiotic resistance profiles. One representative mutant, called PA1436, was characterized by increases in the MICs of ER-35786 (eightfold), sparfloxacin (fourfold), and cefpirome (twofold) but showed unchanged imipenem MICs (Table 3). Careful examination of outer membrane fractions (14) by sodium dodecyl sulfate-polyacrylamide gel electrophoresis revealed decreased expression of a protein band of approximately 55 kDa in mutants PA1433 and PA1436 (Fig. 2A, lanes 3 and 4) compared to the parental strain PASE1 (Fig. 2A, lane 2). This protein could represent a new porin protein. Indeed, alternative ports of entry have been proposed for meropenem (18) and for the synthetic carbapenem BMS-181139 (3). Interestingly, analysis of preliminary sequence data from the P. aeruginosa genome (http://www.pseudomonas.com/) suggests the existence of 14 open reading frames sharing significant homology with either OprD or OprE porins (http://www.interchg.ubc.ca/bobh/).
FIG. 2.
(A) Outer membrane preparations of defined and spontaneous PAO1 derived mutants. Lane 1, PAO1; lane 2, PASE1; lane 3, PA1433; lane 4, PA1436; lane 5, PA1434; lane 6, PA1423. Arrows indicate reduced expression of 55-kDa protein (lanes 3, 4) and presence of OprM (lane 5) and OprD (lane 6). (B) Western blots of total cell lysates obtained from the same strains as in panel A. The blots were revealed with anti-OprM antibody by using a chemiluminescent detection kit.
TABLE 3.
Resistance phenotypes obtained with OprD-deficient mutant PASE1 after exposure to carbapenems
| Antibiotic | MICs (μg/ml)
|
||||
|---|---|---|---|---|---|
| PAO1 | PASE1 | PA1433 | PA1434 | PA1436 | |
| Imipenem | 1 | 16 | 16 | 16 | 16 |
| Meropenem | 0.5 | 4 | 8 | 8 | 2 |
| ER-35786 | 0.25 | 1 | 4 | 1 | 8 |
| Carbenicillin | 64 | 64 | 64 | 256 | 32 |
| Cefpirome | 2 | 2 | 2 | 4 | 4 |
| Gentamicin | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 |
| Chloramphenicol | 64 | 64 | 64 | 128 | 32 |
| Sparfloxacin | 0.5 | 0.5 | 1 | 1 | 2 |
In conclusion, a second step of carbapenem resistance is possible in OprD-deficient strains, affecting this time not only carbapenems but also non-carbapenem antibiotics such as cefpirome and quinolones. The fact that this selection occurred with meropenem and ER-35786 but not after in vitro exposure to imipenem may have some clinical significance.
Acknowledgments
We are grateful to I. Ziha-Zarifi and P. Plésiat (University Hospital Center, Besançon, France) for providing the OprM antiserum and to F. Ohba and N. Watanabe (Eisai Co., Ltd., Tsukuba, Japan) for supplying ER-35786. We thank H. Nikaido for help with the pKa analysis and A. Sasaki for providing references on imipenem and meropenem. We appreciate the skillful technical assistance of L. Kocjancic Curty.
This work was supported by the Fonds National Suisse pour la Recherche Scientifique.
REFERENCES
- 1.Angus B L, Carey A M, Caron D A, Kropinski A M, Hancock R E. Outer membrane permeability in Pseudomonas aeruginosa: comparison of a wild-type with an antibiotic-supersusceptible mutant. Antimicrob Agents Chemother. 1982;21:299–309. doi: 10.1128/aac.21.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen H Y, Yuan M, Livermore D M. Mechanisms of resistance to beta-lactam antibiotics amongst Pseudomonas aeruginosa isolates collected in the UK in 1993. J Med Microbiol. 1995;43:300–309. doi: 10.1099/00222615-43-4-300. [DOI] [PubMed] [Google Scholar]
- 2a.Epp, S. F. Unpublished data.
- 3.Fung-Tomc J C, Gradelski E, Kolek B, Minassian B, Pucci M, Kessler R E, Bonner D P. Activity of carbapenem BMS-181139 against Pseudomonas aeruginosa is not dependent on porin protein D2. Antimicrob Agents Chemother. 1995;39:386–393. doi: 10.1128/aac.39.2.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gotoh N, Tsujimoto H, Tsuda M, Okamoto K, Nomura A, Wada T, Nakahashi M, Nishino T. Characterization of the MexC-MexD-OprJ multidrug efflux system in ΔmexA-mexB-oprM mutants of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1998;42:1938–1943. doi: 10.1128/aac.42.8.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haas D, Holloway B W, Schamböck A, Leisinger T. The genetic organization of arginine biosynthesis in Pseudomonas aeruginosa. Mol Gen Genet. 1977;154:7–22. doi: 10.1007/BF00265571. [DOI] [PubMed] [Google Scholar]
- 6.Jorgensen J H, Ferraro M J, Craig W A, Doern G V, Finegold S M, Fung-Tomc J, Hansen S L, Hindler J, Reller L B, Swenson J M, Tenover F C, Testa R T, Wikler M A. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Villanova, Pa: The National Commitee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 7.Köhler T, Michea-Hamzehpour M, Henze U, Gotoh N, Curty L K, Pechère J C. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol Microbiol. 1997;23:345–354. doi: 10.1046/j.1365-2958.1997.2281594.x. [DOI] [PubMed] [Google Scholar]
- 8.Köhler T, Michéa-Hamzehpour M, Plésiat P, Kahr A, Pechère J C. Differential selection of multidrug efflux systems by quinolones in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1997;41:2540–2543. doi: 10.1128/aac.41.11.2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li X-Z, Ma D, Livermore D M, Nikaido H. Role of efflux pump(s) in intrinsic resistance of Pseudomonas aeruginosa: active efflux as contributing factor to β-lactam resistance. Antimicrob Agents Chemother. 1994;38:1742–1752. doi: 10.1128/aac.38.8.1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X-Z, Nikaido H, Poole K. Role of MexA-MexB-OprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:1948–1953. doi: 10.1128/aac.39.9.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Masuda N, Gotoh N, Ohya S, Nishino T. Quantitative correlation between susceptibility and OprJ production in NfxB mutants of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1996;40:909–913. doi: 10.1128/aac.40.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masuda N, Ohya S. Cross-resistance to meropenem, cephems, and quinolones in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1992;36:1847–1851. doi: 10.1128/aac.36.9.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masuda N, Sakagawa E, Ohya S. Outer membrane proteins responsible for multiple drug resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:645–649. doi: 10.1128/AAC.39.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michéa-Hamzehpour M, Pechère J-C, Plésiat P, Köhler T. OprK and OprM define two genetically distinct multidrug efflux systems in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:2392–2396. doi: 10.1128/aac.39.11.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minami S, Akama M, Araki H, Watanabe Y, Narita H, Iyobe S, Mitsuhashi S. Imipenem and cephem resistant Pseudomonas aeruginosa carrying plasmids coding for class B beta-lactamase. J Antimicrob Chemother. 1996;37:433–444. doi: 10.1093/jac/37.3.433. [DOI] [PubMed] [Google Scholar]
- 16.Nikaido H. Prevention of drug access to bacterial targets: permeability barrier and active efflux. Science. 1994;264:382–388. doi: 10.1126/science.8153625. [DOI] [PubMed] [Google Scholar]
- 17.Nikaido H. Multidrug efflux pumps of gram-negative bacteria. J Bacteriol. 1996;178:5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perez F J, Gimeno C, Navarro D, Garcia-de-Lomas J. Meropenem permeation through the outer membrane of Pseudomonas aeruginosa can involve pathways other than the OprD porin channel. Chemotherapy. 1996;42:210–214. doi: 10.1159/000239444. [DOI] [PubMed] [Google Scholar]
- 19.Poole K, Gotoh N, Tsujimoto H, Zhao Q, Wada A, Yamasaki T, Neshat S, Yamagishi J, Li X Z, Nishino T. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol Microbiol. 1996;21:713–724. doi: 10.1046/j.1365-2958.1996.281397.x. [DOI] [PubMed] [Google Scholar]
- 20.Poole K, Krebes K, McNally C, Neshat S. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J Bacteriol. 1993;175:7363–7372. doi: 10.1128/jb.175.22.7363-7372.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith G B, Schoenewaldt E F. Stability of N-formimidoylthienamycin in aqueous solution. J Pharm Sci. 1981;70:272–276. doi: 10.1002/jps.2600700312. [DOI] [PubMed] [Google Scholar]
- 22.Takeuchi Y, Sunagawa M, Isobe Y, Hamazume Y, Noguchi T. Stability of a 1 beta-methylcarbapenem antibiotic, meropenem (SM-7338) in aqueous solution. Chem Pharm Bull (Tokyo) 1995;43:689–692. doi: 10.1248/cpb.43.689. [DOI] [PubMed] [Google Scholar]
- 23.Trias J, Nikaido H. Outer membrane protein D2 catalyzes facilitated diffusion of carbapenems and penems through the outer membrane of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1990;34:52–57. doi: 10.1128/aac.34.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshimura F, Nikaido H. Permeability of Pseudomonas aeruginosa outer membrane to hydrophilic solutes. J Bacteriol. 1982;152:636–642. doi: 10.1128/jb.152.2.636-642.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ziha-Zarifi, I., C. Llanes, T. Köhler, J. C. Pechere, and P. Plesiat. In vivo emergence of multidrug-resistant mutants of Pseudomonas aeruginosa overexpressing the active efflux system MexA-MexB-OprM. Antimicrob. Agents Chemother., in press. [DOI] [PMC free article] [PubMed]