Abstract
Simple Summary
Non-Small-Cell Lung Cancer (NSCLC) is the primary cause of cancer-related death worldwide. Patients carrying Epidermal Growth Factor Receptor (EGFR) mutations usually benefit from targeted therapy treatment. Nonetheless, primary or acquired resistance mechanisms lead to treatment discontinuation and disease progression. Tumor protein 53 (TP53) mutations are the most common mutations in NSCLC, and several reports highlighted a role for these mutations in influencing prognosis and responsiveness to EGFR targeted therapy. In this review, we discuss the emerging data about the role of TP53 in predicting EGFR mutated NSCLC patients’ prognosis and responsiveness to targeted therapy.
Abstract
Non-Small-Cell Lung Cancer (NSCLC) is the primary cause of cancer-related death worldwide. Oncogene-addicted patients usually benefit from targeted therapy, but primary and acquired resistance mechanisms inevitably occur. Tumor protein 53 (TP53) gene is the most frequently mutated gene in cancer, including NSCLC. TP53 mutations are able to induce carcinogenesis, tumor development and resistance to therapy, influencing patient prognosis and responsiveness to therapy. TP53 mutants present in different forms, suggesting that different gene alterations confer specific acquired protein functions. In recent years, many associations between different TP53 mutations and responses to Epidermal Growth Factor Receptor (EGFR) targeted therapy in NSCLC patients have been found. In this review, we discuss the current landscape concerning the role of TP53 mutants to guide primary and acquired resistance to Tyrosine-Kinase Inhibitors (TKIs) EGFR-directed, investigating the possible mechanisms of TP53 mutants within the cellular compartments. We also discuss the role of the TP53 mutations in predicting the response to targeted therapy with EGFR-TKIs, as a possible biomarker to guide patient stratification for treatment.
Keywords: Non-Small-Cell Lung Cancer (NSCLC), Epidermal Growth Factor Receptor (EGFR), Tumor protein 53 (TP53), targeted therapy, resistance mechanisms
1. Introduction
Lung cancer (LC) is the main cause of cancer-related death worldwide [1]. Non-Small Cell Lung Cancer (NSCLC), the most common LC histology, is a heterogeneous malignancy comprising molecular subtypes for which targeted agents are available in clinical practice [2]. Epidermal Growth Factor Receptor (EGFR) is the most common altered targetable gene in NSCLC, and its mutations (mainly exon 19 deletions and exon 21 L8585R mutations) represent the predictive biomarker for first-, second- and third-generation EGFR-Tyrosine Kinase Inhibitors (TKIs). Nonetheless, resistance to EGFR targeted therapy inevitably occurs, and molecular mechanisms at the basis of primary and acquired resistance to TKIs still have to be elucidated [3].
Tumor protein 53 (TP53) gene encodes for p53 protein, a transcription factor recognized as a master regulator of a wide range of cellular processes such as proliferation, differentiation, apoptosis, metabolism and DNA repair [4]. Gene alterations affecting TP53 are the most commons across all cancers, and are involved in cancer onset, development, progression and response to therapies, thus also affecting the patient’s prognosis [4].
To date, several TP53 mutations have been highlighted, with relative peculiar protein associated functions; more than 70% of these alterations are represented by missense mutations along the DNA-binding domain (DBD), resulting in different consequences at a cellular, organismal and clinical level [4]. Gene alterations affecting TP53 are proved to be a strong prognostic factor for NSCLC [5], and recent reports indicate a role for these mutations in predicting EGFR-mutated NSCLC patients responsiveness to TKIs. In this review, we focus on the relation between TP53 mutations and EGFR-mutated NSCLC subtype, discussing the achieved results on the role of such mutations in predicting responsiveness to TKIs. We also explore possible cellular mechanisms that TP53 mutants activate to guide the resistance to therapy and discuss the emerging data on the role of these gene mutations for a possible patient stratification for EGFR-mutated patients. Moreover, we discuss the classification systems proposed to date, as it has been demonstrated that different mutations confer different characteristics to the cancer cell.
2. EGFR-Mutated NSCLC
LC is the main cause of cancer-related mortality worldwide (18.4%), while NSCLC accounts for 80–85% of lung cancers [1]. In patients affected by metastatic NSCLC, clinical guidelines recommend the testing of activating mutations, the majority of which are linked with an activation of EGFR that occurs in 10–20% of Caucasian and 50% of Asian patients [1,6].
The EGFR gene is located in the short arm of chromosome 7 (7p11.2) [7]. EGFR belongs to the HER/ErbB2 family, a group of receptor tyrosine kinases that include epidermal receptor tyrosine kinases 1 (EGFR, ERBB1), HER2/ERBB2, HER3/ERBB3 and HER4/ERBB4 [8]. These receptors share a similar structure and are composed of three regions: the intracellular, the extracellular, and the transmembrane regions [9]. When the receptor is activated by the ligand, it dimerizes and autophosphorylates the tyrosine residues in the cytoplasmic domain. This step consequently allows the triggering of the intracellular signaling and gene transcription process [9]. The intracellular signaling cascade is mediated through the following pathways: RAS/RAF/MEK/MAPK, PI3K-AKT, JAK/STAT, Src kinase, the Endocytic pathway [10]. Downstream EGFR signaling influences gene expression, apoptosis inhibition, proliferation, angiogenesis, cell motility, and metastasis [6,7,9,11].
The EGFR activity can be dysregulated by several activating mutations that occur within the exons from 18 to 21 (encoding the kinase domain) [8]. Exon 19 deletions (ex19Del) of amino acids 747–750 account for 45% of all EGFR mutations, while exon 21 mutations account for 40–45% and are characterized by the substitution of leucine for arginine (L858R) [11]. The remaining 10% are uncommon mutations affecting exons 18 and 20. Among these, the most frequent (4–8%) is the exon 20 insertion, which is also associated with resistance to the three generations of EGFR inhibitors [6]. Ex19Del and L858R are responsible for a constitutional activation of the receptor: ex19Del shortens the activation loop and prevents the rotation of the alpha helix causing a destabilization of the inactive conformation. L858R causes the interaction between the N-lobe and the C-lobe in the inactive conformation. L858R causes steric hindrance and leads to a constitutive active conformation [6].
2.1. EGFR+ Clinical Management: Clinical Trials Demonstrating First-, Second- and Third-Generation TKIs Efficacy
2.1.1. First-Generation of EGFR TKIs
Gefitinib and erlotinib are first-generation reversible EGFR inhibitors. The activity of gefitinib and erlotinib were initially evaluated in unselected NSCLC patients with poor results [12]. Despite these disappointing results, a retrospective analysis of responders to these treatments allowed the researchers to identify EGFR mutations as predictive biomarkers for responsiveness to EGFR TKIs.
The IPASS trial was the first randomized phase III trial evaluating the efficacy of gefitinib versus chemotherapy (carboplatin/docetaxel) in 1217 treatment-naïve EGFR-mutated NSCLC patients. This study met its progression-free survival (PFS) primary endpoint and objective response rate (ORR) endpoint [13]. Indeed, the PFS (9.5 months vs. 6.3 months; HR 0.48, p < 0.001) and ORR (71.2% vs. 47.3%) in the gefitinib arm was superior to those of the chemotherapy group. It is of note that this study confirmed that EGFR mutations are the strongest predictive biomarker for response to front-line gefitinib. The phase III NEJ002 study confirmed the superiority in terms of PFS gefitinib compared to carboplatin and paclitaxel (10.8 months vs. 5.4 months; HR 0.3; p < 0.001) [14]. Similarly, in the phase III WJT0G3405 trial, PFS was higher in the gefitinib arm than in the carboplatin/docetaxel arm (9.2 vs. 6.3 months, HR 0.489, p < 0.0001) [15]. Moreover, two phase III trials compared erlotinib to chemotherapy confirming the superiority of this EGFR TKI in terms of PFS [16,17]. As shown in Table 1, a total of six large phase III trials showed a strong benefit of EGFR TKIs versus chemotherapy as a first-line treatment in terms of PFS and ORR in patients with EGFR-mutated NSCLC [13,14,15,16,17,18,19]. However, these studies did not demonstrate a significant benefit in Overall Survival (OS), probably due to the high cross-over rate between the Gefitinib or Erlotinib arm and the chemotherapy arm [20].
Table 1.
Phase III trials comparing EGFR inhibitors to chemotherapy in first-line treatment.
| Study | Treatment Arms | mPFS (Months) | mOS (Months) | ORR (%) | Ref. |
|---|---|---|---|---|---|
| IPASS | Gefitinib vs. carboplatin/paclitaxel | 9.8 vs. 6.4 p < 0.001 |
21.6 vs. 21.9 p = 0.99 |
71.2 vs. 47.3 | [15] |
| NEJ002 | Gefitinib vs. carboplatin/paclitaxel | 10.8 vs. 5.4 p < 0.001 |
27.7 vs. 26.6 p = 0.48 |
73.7 vs. 30.7 | [13] |
| WJT0G3405 | Gefitinib vs. cisplatin/docetaxel | 9.2 vs. 6.3 p < 0.0001 |
36.0 vs. 39.0 | 62.1 vs. 32.2 | [16] |
| OPTIMAL | Erlotinib vs. carboplatin/gemcitabine | 13.1 vs. 4.6 p < 0.0001 |
22.8 vs. 27.2 p = 0.27 |
83.0 vs. 36.0 | [17] |
| EURTAC | Erlotinib vs. cisplatin/docetaxel | 9.7 vs. 5.2 p < 0.0001 |
19.3 vs. 19.5 p = 0.87 |
64.0 vs. 18.0 | [18] |
| ENSURE | Erlotinib vs. cisplatin/gemcitabine | 11 vs. 5.6 | 26.3 vs. 25.5 | 62.7 vs. 33.6 | [20] |
| CONVINCE | Icotinib vs. cisplatin/pemetrexed | 11.2 vs. 7.9 p = 0.006 |
30.5 vs. 32.1 p = 0.89 |
NR | [21] |
| LUX-Lung 3 | Afatinib vs. cisplatin/Pemetrexed | 11.1 vs. 6.9 p = 0.001 |
28.2 vs. 28.2 p = 0.39 |
56.1 vs. 22.6 | [22] |
| LUX-Lung 6 | Afatinib vs. cisplatin/gemcitabine | 11.0 vs. 5.6 p < 0.0001 |
23.1 vs. 23.5 p = 0.61 |
66.9 vs. 23.0 | [23] |
| AURA 3 | Osimertinib vs. platinum/pemetrexed | 10.1 vs. 4.4 p < 0.001 |
NA | 26.8 vs. 22.5 | [24] |
| CTONG | Erlotinib vs. gefitinib | 13.2 vs. 11.1 | 22.4 vs. 20.7 | NR | [25] |
| LUX-Lung 7 | Afatinib vs. gefitinib | 13.7 vs. 11.5 p = 0.007 |
27.9 vs. 24.5 | 70 vs. 56 | [26] |
| FLAURA | Osimertinib vs. erlotinib or gefitinib | 18.9 vs. 10.2 p < 0.001 |
NR | 80.0 vs. 76 | [27] |
mPFS: median Progression-Free Survival. mOS: median Overall Survival. ORR: Objective Response Rate. NR: Not Reported.
Based on these results, gefitinib was approved by the FDA (US Food and Drug Administration) in 2015 and erlotinib in 2016 for the treatment of patients with metastatic NSCLC whose tumors have EGFR exon 19 deletions or exon 21 (L858R) substitution mutations.
The efficacy of another first-generation EGFR-TKI, namely icotinib, has been evaluated in a phase III CONVINCE trial. In this first-line study, icotinib showed a higher PFS in comparison with the chemotherapy arm (11.2 months vs. 7.9 months; HR 0.61; p = 0.06) [21]. Therefore, based on CONVINCE results, icotinib was approved by the China Food and Drug Administration (CFDA) in June 2011 as a first-line treatment, enriching the EGFR-mutated NSCLC therapeutic armamentarium.
The evolution of the EGFR TKIs in the therapeutic landscape entailed the need to define a better first-line strategy treatment in this patient setting. Therefore, the phase III CTONG 0901 trial was conducted to compare the efficacy and safety of gefitinib with that of erlotinib in patients with metastatic NSCLC characterized by EGFR exon 19 or 21 mutations [28]. The results of this comparison demonstrated that the PFS and OS of the erlotinib and gefitinib arms were 13.2 months vs. 11.1 months (HR 0.80; p = 0.108) and 22.4 vs. 20.7 months (HR 0.98; p = 0.902) respectively. Even though this study did not meet its primary endpoint, the subgroup analyses demonstrated that patients with EGFR exon 19 mutations had a significantly higher RR (62.2% vs. 43.5%, p = 0.003) and superior median OS (22.9 vs. 17.8 months, p = 0.022) than those with exon 21 mutations treated with erlotinib or gefitinib.
The ICOGEN trial is another randomized, double-blind, phase III trial that was conducted to evaluate the safety and efficacy of icotinib and gefitinib in advanced NSCLC patients previously treated with chemotherapy [29]. In this study, the PFS (7.8 months for icotinib vs. 5.3 months for gefitinib, p = 0.32, and OS 20.9 vs. 20.2; p = 0.76) were similar between the two arms, but the toxicity of the icotinib arm was lower than that of the gefitinib arm (60.5% vs. 70.4%). Thus, compared with the results of these two head-to-head studies, the three first-generation EGFR-TKIs do not seem to be significantly different in terms of the PFS and safety profile.
2.1.2. First-Generation EGFR TKIs Combined with Other Agents
Based on the evidence of a link between EGFR stimulation and increased angiogenesis, first-generation EGFR TKIs were tested in combination with anti-angiogenic compounds. An anti-vascular endothelial growth factor (anti-VEGF) + erlotinib combination has been investigated in patients with untreated, advanced, EGFR mutated NSCLC in the phase II JO25567 trial, which evaluated the efficacy of erlotinib and bevacizumab compared with erlotinib alone in patients with EGFR mutation-positive NSCLC. The PFS benefit was more consistent for the erlotinib + bevacizumab arm than in the erlotinib arm (median, 16.0 vs. 9.7 months; HR, 0.54; 95% CI, 0.36–0.79; p = 0.0015), leading to EMA approval for this combination [30].
A phase III trial evaluated the efficacy of the addition of ramucirumab (anti-VEGF receptor inhibitor) to erlotinib, finding a significant benefit in PFS (19.4 vs. 12.4 months, HR: 0.59, p < 0.0001) [31]. Based on these results, despite the fact that the data of OS are immature, this combination has been approved by the FDA (US Food and Drug Administration) for the first-line treatment of patients with metastatic NSCLC harboring EGFR exon 19 deletions or exon 21 mutations. Interestingly, a biomarker analysis from the RELAY trial highlighted that p53 and EGFR co-mutations were associated with prolonged PFS in the experimental arm, both in exon 19 and 21 mutated patients [32]. The addition of chemotherapy to first-generation TKIs was also investigated. A recent meta-analysis of randomized controlled trials that compared EGFR-TKI monotherapy with the combination of EGFR-TKI and chemotherapy showed that there was a benefit in terms of ORR, PFS and OS in favor of the combination arm, with an acceptable toxicity profile. These data are of interest and the combination of chemotherapy with EGFR TKIs could be a potential first-line treatment in selective patients [22].
2.1.3. Second-Generation of EGFR TKIs
Afatinib and dacomitinib are second-generation irreversible EGFR TKIs with a similar structure with that of gefitinib or erlotinib but with a side chain that binds covalently to cysteine-797 of EGFR with a subsequent irreversible EGFR inhibition [33,34].
Afatinib was evaluated as a first-line treatment in comparison with cisplatin and pemetrexed in the phase III LUX-Lung 3 trial, demonstrating superiority in terms of PFS compared to chemotherapy (11.1 months vs. 6.9 months; HR 0.58; p = 0.001). In this trial, only patients with common EGFR mutations (exon 19 deletions and L858R) were considered, with a reported increase in PFS of 13.6 months for the experimental arm vs. 6.9 months for the chemotherapy arm (HR = 0.47; 95% CI, 0.34–0.65; p = 0.001). Interestingly, the PFS result was superior in patients with tumours harboring an exon 19 deletion with respect to the L858R mutation [35]. On the basis of this clinical trial result, afatinib was approved as a treatment for treatment-naïve patients affected by advanced EGFR-mutated NSCLC. In the LUX-Lung 6 trial, patients were randomized to receive afatinib or gemcitabine/cisplatin chemotherapy. The final results of this study showed a statistically significant benefit in terms of the PFS for the afatinib arm (11.0 vs. 5.6 months; HR 0.28; p < 0.0001) [36]. Despite the PFS benefit for afatinib in these two trials, no difference in terms of survival benefit in the overall population was reported. However, in a prespecific EGFR exon 19 subgroup analysis, afatinib demonstrated a significant improvement in OS compared with the chemotherapy group, in both trials. On the other hand, in the LUX-Lung 7 study, a head-to-head comparison between afatinib and gefitinib, no significant differences in terms of PFS were found (11.0 months for afatinib vs. 10.9 months for gefitinib; HR 0.73; p = 0.017) [24].
Another second-generation EGFR inhibitor, dacomitinib, was evaluated in the phase III ARCHER-1050 trial [37]. A total of 452 patients with EGFR-mutant NSCLC were randomized to receive as a first-line treatment dacomitinib 45 mg/day or gefitinib 250 mg/day. The results of this trial demonstrated that the PFS of the dacomitinib was statistically longer compared with the gefitinib arm (14.7 months vs. 9.2 months HR: 0.59; 95% CI: 0.47–0.74). In responders, the duration of response was longer in the dacomitinib arm vs. the gefitinib arm in both EGFR mutation subgroups. However, a dose reduction of dacomitinib was still needed in 66.5% of the trial patients due to a more significant toxicity profile of dacomitinib. Moreover, mOS was significantly longer in the dacomitinib arm than in the gefitinib arm (34.1 months vs. 26.8 months, respectively; HR 0.76; 95% CI, 0.582 to 0.993; p = 0.0438). In the updated OS analysis, a significant improvement in survival was confirmed for the dacomitinib group in patients with exon 21 (L858R) substitution mutations (32.5 months vs. 23.2 months; HR 0.67; 95% CI: 0.47–0.94; p = 0.02); whereas, there was no survival benefit in patients with the exon 19 deletion mutation (36.7 months for dacomitinib vs. 30.8 months for gefitinib, HR 0.85; p = 0.30) [23].
2.1.4. Third-Generation of EGFR TKIs
One of the most common mechanisms of acquired resistance during the treatment with first- and second-generation EGFR TKIs is represented by the mutation of T790M (EGFR exon 20). Therefore, clinical development was focused on second-line treatments with the subsequent development of third-generation EGFR TKIs. Osimertinib is an oral, third-generation, irreversible EGFR TKI designed to have activity against T790M mutations as well as activity in the central nervous system [27]. The phase III AURA3 trial evaluated the effects of osimertinib or cisplatin/pemetrexed in randomized EGFR-positive patients with T790M resistance [38]. This study demonstrated a significant improvement in PFS, with a median of 10.1 months in the osimertinib arm compared to 4.4 months with chemotherapy (p < 0.001). Furthermore, osimertinib was demonstrated to be superior to chemotherapy in terms of ORR (71% vs. 31%) and with an intracranial ORR of 70%. Based on the results of this study, osimertinib was approved as a second-line treatment of EGFR T790M-mutated NSCLC patients.
A sequent phase III FLAURA study assessed the efficacy and safety of osimertinib in patients with previously untreated EGFR mutated advanced NSCLC compared with the standard first-generation EGFR-TKIs (gefitinib or erlotinib) [39]. The results of the FLAURA trial showed that osimertinib resulted in significantly longer PFS (18.9 months vs. 10.2 months; HR 0.46; p < 0.001) with a better safety profile with respect to standard EGFR-TKIs. Despite crossover by 31% of the patients in the control arm, osimertinib confirmed its superiority in terms of OS with a median OS of 38.6 months (95.05% CI, 34.5–41.8) vs. 31.8 months (95.05% CI, 26.6, 36.0) for standard EGFR TKI treatment, respectively (HR 0.799; 95.05% CI, 0.641–0.997; p = 0.0462). Based on these practice-changing results, osimertinib was approved as a first-line treatment for patients with metastatic NSCLC harboring EGFR exon 19 deletions or exon 21 L858R mutations.
2.2. EGFR TKIs for the Treatment of Uncommon EGFR Mutations
Approximately 10–15% of EGFR-mutated tumors present uncommon somatic EGFR mutations such as exon 18 nucleotide alterations, exon 19 in frame insertions, exon 20 alterations, and exon 21 mutation L861Q. The knowledge concerning the efficacy of EGFR TKIs in patients affected by NSCLC carrying uncommon EGFR mutations is limited to small clinical investigations or case reports. A post hoc analysis of the LUX-Lung 2, LUX-Lung 3, and LUX-Lung 6 trials demonstrated that patients with L861Q, G719X and S768I responded well to afatinib treatment, whereas afatinib did not show any efficacy in patients with an exon 20 insertion mutation [40,41]. Another retrospective study confirmed the minor efficacy of first-generation EGFR TKIs in patients with G718X and L861Q [42]. Encouraging results come from the clinical activity of amivantamab, a bispecific antibody directed against EGFR and MET tyrosine kinase receptors, and against tumors with EGFR exon 20ins mutations [43]. However, we have to wait for the conclusion of ongoing phase III studies to better define its role in this patient setting. Moreover, osimertinib efficacy was tested in a phase II trial of enrolled patients with uncommon EGFR mutations, demonstrating a 50% objective response rate and 8.2 median PFS [44].
3. Common Resistance Mechanisms to EGFR-TKIs
Despite the known and confirmed efficacy of EGFR-TKIs in the treatment of EGFR mutated NSCLC, about 5–25% of patients, who receive target therapies, show no benefits from this treatment [45]. The cause of this resistance is the presence of mechanisms that, generally, could be grouped into on-target EGFR-dependent and off-target EGFR-independent. Moreover, primary, or secondary resistance mechanisms could be observed, depending on whether they occurred from the beginning of the treatment with EGFR-TKI or after an initial period of response or stability.
3.1. On-Target Mechanisms of Resistance
On-target mechanisms of resistance occur mostly with the use of first- and second-generation EGFR-TKIs at about 50%, compared to 10–15% for third-generation TKI used as a first-line and 20% as a second-line [3]. One of the most known on-target mutations is T790M, characterized by a threonine substituted with a methionine in the 720 position of exon 20. T790M mostly develops as a resistance mechanism to first- and second-generation EGFR-TKIs used as a first-line treatment, occurring in about 49–63% of patients [46,47,48]. Finding this mutation as a diagnosis of a primary mechanism of resistance is rare. In this case, a possible germinal origin of the mutation should be considered [46]. Other reported rare alterations conferring resistance to first- and second-generation EGFR-TKIs are the missense mutations D761Y, L747S and T854A, while EGFR amplification occurs in 8–10% of cases.
On the other hand, some on-target missense mutations conferring resistance to osimertinib have been identified. Exon 20 C797S is able to avoid the binding of osimertinib to EGFR, as well as other third-generation TKIs (e.g., rociletinib, narzatinib, olmutinib) [49,50]. This mutation is the most common tertiary mechanism of resistance and accounts for about 10–26% of cases resistant to osimertinib when used as a second-line treatment; although, when it is used as a first-line therapy, its prevalence is 7% lower, though it remains the second most frequent event behind MET amplification [38,51]. The same effect occurs with mutations affecting L792. On the other hand, it was demonstrated that this resistance mutation to third-generation TKIs is sensible to first-generation gefitinib in vitro [46,50,51]. Another mutation interfering with the binding of osimertinib is the substitution in the 718 residue (L718Q, L718V), placed in the adenosine triphosphate (ATP) binding site. Usually, these mutations confer independent resistance to osimertinib, not coexisting with the C797 mutation [52,53]. Anecdotal and rare mutations interfering with osimertinib activity include C797G and mutations in the C796 residue (C796R, G796S, G796D) in exon 20, near C797 [49,52,54,55,56,57]. In the same protein domain, the G719A mutation has been reported in association with osimertinib resistance. Similarly, other exon 20 mutations (G724S and SV768IL) induce resistance to third-generation TKIs [50,53,56,57]. Finally, the amplification of EGFR and a deletion in exon 19 have been described as additional mechanisms of resistance to osimertinib in the II line setting [58,59]. Lazertinib, a third-generation TKI targeting activating and T790M EGFR mutations, showed important clinical activity in EGFR-mutated NSCLC patients as a second-line treatment after progression to a first- or second-generation TKI [60] and its efficacy, in combination with amivantamab after progression to osimertinib treatment, was demonstrated [61].
3.2. Off-Target Mechanisms of Resistance
Off-target resistance mechanisms induced by TKI treatment include alterations affecting pathways that bypass signaling activation, in an EGFR-independent manner. As TP53 mutations involved in the process of histologic transformation to SCLC are a resistance mechanism to EGFR-TKIs, this issue is discussed later.
3.2.1. MET Amplification
MET amplification is one of the most frequent mechanisms of resistance to all generations of EGFR-TKIs, used as both a first- and second-line of treatment.
It was reported in 5–22% of cases treated with first- and second-generation TKIs, mostly in association with an EGFR exon 19 deletion, while it is the most common cause of resistance to osimertinib used in II line [62,63,64]. This data has been observed in the AURA3 trial, where about 19% of the cfDNA samples at the progression showed MET amplification, though the percentage was lower in tumor tissue [53,62,64]. In this setting, MET amplification occurs with or without the loss of T790M, and in 7% of cases could be present with a C797S mutation [63,65].
3.2.2. HER2
The tyrosine kinase receptor Erb2, encoded by HER2, stimulates the activation of the MAPK and PI3K pathways, mediating resistance to EGFR-TKIs. HER2 amplification has been described in 12% of tumor samples of patients treated with first-generation TKI [66], in 5% of cases treated with osimertinib in II line, and in 2% of cases treated in I line [65,66,67]. This last percentage has been found when the cfDNA was analyzed, and no HER2 amplification was detected in the tumor tissue.
3.2.3. KRAS, BRAF and PI3K
KRAS mutation as a mechanism of resistance to EGFR-TKIs is very rare. G12S mutation has been described as resistant to osimertinib in II line; other KRAS mutations, such as G12D, G13D, Q61R and Q61K, have been found to confer resistance to osimertinib as well (reported in less than 1% of cases) [63,65,68,69,70]. In the FLAURA study, mutations of KRAS, like G12D and A146T, were described in the cfDNA of 3% of cases [51]; in the study conducted by Schoenfeld on tumor tissue, KRAS mutation G12A was recorded in one case (4%) [54]. Another mechanism of resistance observed is the BRAF V600F; after treatment with osimertinib in II line, it was observed in 3% of the cfDNA samples [65]. The same frequency was also reported in the cfDNA analyzed after progression with osimertinib used in I line [54]. At progression after osimertinib is used as a second-line treatment, the PI3KCA mutations E454K, E452K, R88Q, N345K, and E418K, have been identified as mechanisms of resistance in 4–11% of cases [62,64,68,70]; in another study, the percentage of these mutations noted on tumor tissue was 17% [71]. Moreover, PI3KCA amplification has been detected, using NGS, in the AURA3 study in 4% of the cases [65]. The main clinical trials investigating new therapeutic strategies for patients with progressive disease after treatment with Osimertinib are resumed in Table 2.
Table 2.
Ongoing clinical trials in Osimertinib-resistant EGFR-mutated NSCLC.
| Clinical Trial Number | Phase | Treatment Arms | Primary Endpoint | Link to the Clinical Trial |
|---|---|---|---|---|
| NCT03515837 | III | Pembrolizumab + pemetrexed + chemo vs. placebo + pemetrexed + chemo |
PFS, OS | https://clinicaltrials.gov/ct2/show/NCT03515837 (accessed on 28 January 2022) |
| NCT03778229 | II | Osimertinib + savolitinib | ORR | https://www.clinicaltrials.gov/ct2/show/NCT03778229 (accessed on 28 January 2022) |
| NCT03944772 | II | Osimertinib + savolitinib vs. osimertinib + gefitinib vs. osimertinib + necitumumab vs. durvalumab + carboplatin + pemetrexed |
ORR | https://www.clinicaltrials.gov/ct2/show/NCT03944772 (accessed on 28 January 2022) |
| NCT03940703 | II | Tepotinib + osimertinib | Safety, ORR | https://clinicaltrials.gov/ct2/show/NCT03940703 (accessed on 28 January 2022) |
| NCT04136535 | II | Pemetrexed and carboplatin with or without anlotinib | PFS | https://www.clinicaltrials.gov/ct2/show/NCT4136535 (accessed on 28 January 2022) |
| NCT03532698 | II | Osimertinib + aspirin | ORR | https://www.clinicaltrials.gov/ct2/show/NCT03532698 (accessed on 28 January 2022) |
| NCT04316351 | II | Toripalimab + pemetrexed + anlotinib | ORR | https://www.clinicaltrials.gov/ct2/show/NCT04316351 (accessed on 28 January 2022) |
| NCT03784599 | I/II | Trastuzumab emtansine and osimertinib | Safety, ORR | https://www.clinicaltrials.gov/ct2/show/NCT03784599 (accessed on 28 January 2022) |
| NCT03891615 | I | Osimertinib + Niraparib | MTD | https://www.clinicaltrials.gov/ct2/show/NCT03891615 (accessed on 28 January 2022) |
| NCT03516214 | I | Nazartinib and trametinib | MTD; RP2D | https://www.clinicaltrials.gov/ct2/show/NCT03516214 (accessed on 28 January 2022) |
EGFR: Epidermal Growth Factor Receptor. NSCLC: Non-Small-Cell Lung Cancer. MDT: maximum tolerated dose. OS: Overall Survival. PFS: Progression-Free Survival. RP2D: recommended phase II dose. ORR: Objective response rate.
4. TP53: The Master Regulator of the Genome
The 53 KD protein p53, encoded by the Tumor protein P53 (TP53) gene is a master regulator of cell fate and a powerful antiproliferative transcription factor that controls the epigenetic program and dictates the expression of a plethora of target genes in response to multiple external stresses [72]. In a balanced homeostasis cell state, p53 is maintained at low cytoplasmic levels and kept mostly inactivated by the regulatory action of the E3 ubiquitin-protein ligase, MDM2; various cellular stimuli, including DNA damage and replication induced by oncogenic deregulation, releases MDM2-mediated degradation of p53 and promotes p53 activation by its phosphorylation [73].
The best understood function of p53 focuses on its DNA-binding ability to induce cell cycle arrest and promote apoptosis [74]. However, p53 also plays a pivotal role in controlling the overall integrity of the genome, as it is often addressed as the “guardian of the genome”. Upon DNA damage, multiple signaling pathways converge to activate p53, either promoting the repair of the damaged DNA sequence or blocking the DNA replication fork, and in doing so, the propagation of genomic instability and mutations [75]. In addition, p53 can limit chromosomal rearrangements by blocking centromere duplication and telomeric dysfunctions that lead to aberrant mitosis [76]. Mainly for this reason, the absence or inactivation of p53 permits cell survival and facilitates aneuploidy, which is a common step towards further accumulation of oncogenic abnormalities. In addition, p53 suppresses shattering genomic rearrangements such as chromothripsis that typically occur when cells have bypassed replicative senescence [77]. Although the extent of chromothripsis contribution to oncogenesis is still open to debate, this phenomenon is significantly more recurrent when TP53 is deleted or mutated [78]. Another important way by which p53 contributes to maintaining genomic integrity is via the suppression of retrotransposon reactivation and mobilization that can lead to mutagenesis throughout the genome [79]. Specifically, p53 binding to long interspersed nuclear element (LINE) elements promotes LINE epigenetic silencing and might protect the cell from transposon-associated mutagenesis [80].
The control of genome integrity by p53 extends to multiple layers that are far beyond its well-known function as a transcription factor [72]. Several genome-wide studies have shown that p53 possesses repressive functions that directly depend on the DNA-binding capacity of p53 [81,82]. ChIP-Seq genome scanning revealed the presence of up to 10,000 possible positions in the genome that p53 potentially binds, which are widespread and do not always represent the binding preferences of p53 as a transcription factor [83]. Furthermore, many of the transcriptionally active promoters bound by p53 do not display direct p53-dependent regulation, suggesting the existence of different dynamics through which p53 might regulate the chromatin status and the overall stability of the genome [81]. For example, p53 can bind with high affinity to regions of chromatin in a closed status [84]. These findings may also raise the hypothesis that it is the nucleosomal structure rather than the DNA sequence affinity that dictates the genomic binding pattern of p53. A comparison between normal and cancer cells suggested that enrichment in CpG islands and hypomethylation of the DNA may drive p53 binding, which likely arise from the overall altered structure of chromatin during oncogenic transformation [85]. In a context of DNA damage, p53 can form a complex with the remodeling and splicing factor 1 (RSF1) forming a complex with the histone acetyltransferase p300 [86].
How the interplay between the transcription factor and the chromatin remodeling activity of p53, in a normal cellular context or upon the induction of DNA damage, affects the cell fate and is a question currently awaiting answers; it might have important implications when thinking about cancer therapy. In this concern, p53 inactivation that occurs in cancer may be unique in its ability to both favor genomic instability and sustain survival by downgrading p53 activity as a transcriptional repressor.
TP53 Mutations in Human Cancer
The TP53 gene has long been recognized as the most frequently mutated gene in human cancer [87]. It is, in fact, well accepted that various mutations in the TP53 gene are the most common genetic lesions found in cancer cells, and mutational dysfunction of the p53 protein is a major contributor to cancer development, progression, metastasis, and resistance to therapy. Further, the presence of mutations that abrogate p53 functionality could also predispose a patient to resistance to cancer therapy [88]. Still, no effective medication that can block the oncogenic derangements derived from p53 inactivation has been approved for use in clinic. The inadequacy in restoring tumor suppressor activity of p53 mutants might also depend on the variety of effects that the different p53 mutations have on the cell [4]. Nevertheless, the precise characterization, in terms of functionality and pathological consequences, of the various p53 mutations is particularly relevant for their use as clinical biomarkers and the optimization of therapeutic options.
Most frequently, TP53 mutations in cancer cells occur in one allele, while the other allele has been lost or deleted following major chromosomal rearrangements, leading to a loss of heterozygosity (LOH) that results in the expression of the sole mutated TP53 allele [89]. However, a good fraction of tumors do not present with LOH for TP53, indicating that mutations of the gene might not be necessarily the primary driver of oncogenesis, but might occur at later stages and be just one among many critical pathological events that accumulate during a cancer cell’s life. Typical alterations of TP53 include frameshifts (deletions and insertions), nonsense, silent, and missense mutations that may occur throughout the entire gene sequence [4]. Missense mutations are by far the most common alterations of TP53 (>70%), and normally cluster in the DNA-binding domain (DBD, exons 5–8). Minor hotspot mutations may occur in other coding regions of the gene that are still associated with amino acid residues responsible for the interaction of the protein with DNA. Typically, all of these TP53 mutations have been collectively considered equally for their ability to interfere with the tumor suppressor activity of p53, but there are actually profound differences in the classes of mutations, which may produce distinct outcomes [4].
The main function of p53 as a tumor suppressor is linked to its ability to induce cell death or to put the cell into a permanent senescent status. However, it is now quite established that several gain of function (GOF) mutations may happen in TP53, and sustains the notion that cancer cells may actually be addicted to mutated p53 [90]. A depletion of mutated p53 leads to cancer cell death, while ectopic expression of mutant p53 promotes survival via increased genomic instability, angiogenesis, and invasion [91,92,93]. Moreover, recent discoveries unveiled the capacity of p53, in certain contexts, to promote cell survival by also sustaining metabolism and maintaining the balance between glycolysis and oxidative stress, thus, limiting the production of reactive oxygen species [94]. Specifically, chances of p53 levels over time rather than its absolute levels are pivotal in driving cell fate and determining how the cell can respond to perturbations [95]. For example, during treatment with the chemotherapeutic agent Cisplatin, the mRNA stability of p53 target genes, which respond to p53 temporal regulation, is the main determinant in deciding whether the cell will survive to treatment [96]. This notion contributes to the question that multiple levels of p53 regulation may exist, and deciphering the complexity of p53 function relies upon the integration of the tumor suppressive activity of p53 and the understanding that deregulation of some elements of the p53 pathway might also provide the tumour with a survival advantage. Metabolic status, the overall mutational profile, and the epigenetic state of the cell are all determinants of how the tumor suppressive function of intact p53 might be restored during cancer treatment.
From a clinical perspective, targeting GOF mutations of p53 may have direct effects on the proliferation and survival of cancer cells that are addicted to p53. However, drugs that target the mutant form of p53, either to block GOF activity or to restore the tumor suppressive activity of p53, should have little interference on the wild-type p53. In particular, restoration of the wild-type function of p53 has been heavily pursued [97]. After almost four decades of studying, p53 is still considered undraggable, especially for the numerous off-target effects that many compounds, initially found as being able to target specifically p53 mutants, actually have. In particular, drugs that promote degradation of mutant p53, have adverse effects on many ubiquitous cellular pathways [98]. Similarly, use of non-selective anti-MDM2 inhibitors that should act in enhancing wild-type p53 activity have been revealed to be problematic due to many adverse side effects in patients [98].
This negative trend might, however, soon change, thanks to the availability of novel FDA-approved drugs, already being tested in clinical trials, that are more specific for individual p53 mutations that stratify with patient characteristics [4]. The evolving understanding of the specific functions of the different p53 mutant types might open the door, in the near future, to effective mutant p53 directed therapeutic strategies that will benefit over 50% of cancer patients, particularly those patients with TP53 mutations.
5. TP53 Mutations in NSCLC
As with the majority of cancers, TP53 is the most common mutated gene in NSCLC, also [99,100,101] showing a predominant clonal expression [102]. Data analysis from The Cancer Genome Atlas (TCGA) database highlights that TP53 mutations occur in the exons 4–8 of the gene in 44.8% of cases, confirming that DBD represents the hotspot of the protein. The authors showed that TP53 mutations are able to affect the prognosis of NSCLC patients (OS 27 vs. 19 months, p < 0.001); moreover, different mutations result in different survival times, suggesting that different mutations play different roles at a molecular level [5]. Several studies identified that mutations affecting TP53 are the most influencing prognostic factor, both in early and advanced NSCLC [103,104,105,106]. Moreover, it has been recently reported that activation of TP53 is involved in the EGFR-signaling pathway and in the apoptosis process induced by platinum-based chemotherapy [107]. This evidence, together with the fact that TP53 mutations are more frequent in EGFR-mutated patients, suggest that some of these oncogene-addicted tumors could possess an underlying biology and molecular mechanisms based on two main biomarkers to guide cancer progression [5,108]. On the other hand, in an attempt to find predictive biomarkers for a neo-adjuvant therapy for stage II–III EGFR-mutated NSCLC, exon 4/5 TP53 missense mutations have been found to be a stratification factor for OS and treatment [109]; another study based on the IALT trial case series, found that TP53 mutations play a role in predicting the efficacy of adjuvant platinum-based chemotherapy [110]. In the next paragraph, we discuss the emerging role of TP53 in EGFR-mutated patients, both in terms of prognostic impact and resistance to targeted therapy.
5.1. TP53 Mutations in EGFR-Positive NSCLC: Clinical Significance
The role of TP53 mutations in EGFR-mutated NSCLC has been widely investigated in recent years. One of the first attempts to establish the role of TP53 mutations in this subset of patients was performed by Molina-Vila and colleagues, who explored the prognosis of 125 wild-type (wt) EGFR and 193 (training cohort) and 64 (validation cohort) mutated EGFR NSCLC patients. The authors categorized TP53 mutations as disruptive and non-disruptive ones. Disruptive mutations were identified in stop codons along all the protein structures and non-conservative mutations within the DBD, while non-disruptive mutations were conservative alterations, as well as non-conservative mutations outside the DBD, apart from the stop codons. Their results showed that TP53 non-disruptive mutations are an EGFR- and KRAS-independent prognostic factor (OS 13.3 months vs. 24.6 months, p < 0.001); they also showed that non-disruptive mutations are a prognostic but not a predictive factor in the subgroup of EGFR-mutated patients (median OS 17.8 months vs. 28.4 months in the training cohort, p = 0.004; median OS 18.1 months vs. 37.8 months in the validation cohort, p = 0.006), highlighting a slight trend in the PFS in a multivariate analysis of erlotinib-treated patients (PFS 11.0 vs. 15.0 months, p = 0.14) [111]. Similar results were recently achieved by Aggarwal and colleagues, who highlighted a worse OS for 114 EGFR and TP53 co-mutated patients treated with first-line TKIs with respect to sole EGFR patients (median OS 33.3 months vs. 53.6 months; p = 0.021), with a trend when the data were adjusted for age, smoking status, and performance status [112]. A large cohort study showed a trend for TP53 mutations in predicting a worse OS of EGFR-mutated patients receiving targeted therapy with respect to wt TP53 patients (2.9 years vs. NR, p = 0.06); the trend became more evident when categorizing TP53 mutations as disruptive and non-disruptive (p = 0.055), reaching statistical significance when pooling data from patients with targetable alterations in EGFR, ALK and ROS1 genes (mOS 2.6 years vs. NR, p = 0.009) [113]. In the attempt to find predictive biomarkers for EGFR-mutated NSCLC patients, our group highlighted that mutations in TP53 are able to influence responsiveness to first-line first- and second-generation TKIs in a case series of 136 EGFR-mutated NSCLC patients. In particular, TP53 exon 8 mutations were able to identify a subset of patients with worse DCR with respect to wt exon 8 patients (41.7% vs. 87.3%, p < 0.001) PFS (4.2 vs. 16.8 months, p < 0.001) and OS (7.6 months vs. NR, p = 0.006), with respect to TP53 wt and mutated TP53 in other exon patients [114]. Later, we confirmed our results in an independent case series of EGFR-mutated patients (HR for PFS 3.16, 95% CI 1.59–6.28, p = 0.001) [115]. These results were confirmed in a larger case series of NSCLC patients, where TP53 exon 8 mutations were able to predict the prognosis of EGFR-mutated patients, independently of the received treatment [116]. These results were not confirmed by the abovementioned study, which found that TP53 in exon 8 were not predictors for PFS compared with mutations in other exons (13 months vs. 13.1 months, p = 0.2) [112]. A study by Labbè and colleagues in a cohort of 60 mutant EGFR patients found that TP53 mutations were associated with worse PFS to first-line TKIs only when considering missense mutations (HR 1.91, CI 1.01–3.60, p = 0.04) [117]. These studies underline the importance of a categorization of TP53 mutations; considering that, when taken together, TP53 mutations were not able to reach significant associations with clinical outcome in advanced or early stage NSCLC, even in patients treated with third-generation TKIs [106,111,114,115,117,118,119,120]. The same observation was also highlighted in EGFR-mutated NSCLC patients by Jin et al., who found that TP53 had the higher co-mutation rate with respect to other genic alterations (72.5%), but showed no associations with survival [121]. On the other hand, studies on Asian patients found that TP53 mutations are able to predict PFS of EGFR-mutated patients treated with both first- or second- and third-generation EGFR-TKIs (HR: 2.02; 95% CI: 1.04–3.93, p = 0.038 and HR: 2.23, 95% CI 1.16–4.29, p = 0.017, respectively) [122]. The same results were achieved in a small case series of gefitinib-treated patients, where TP53 mutations affected exclusively short- or intermediate responders (66.7% vs. 0, p = 0.009), and by a study by Yu and colleagues, who found that TP53 mutations in pre-treated patients predicted shorter time-to-progression (HR 1.7, p = 0.006) [123,124]. An interesting study by Roeper and colleagues found that TP53 mutations have a strong negative impact on the clinical outcome of EGFR-mutated patients (ORR, PFS and OS) whether considered as disruptive or non-disruptive; pathogenic or non-pathogenic; or exon 8 or non-exon 8 [125]. Another study found that TP53 mutations are associated with early resistance to EGFR-TKIs, influencing the prognosis of short-responders to TKIs (<6 months, TP53 mutations found in 87.5% of short-responders and in 16.7% of long-responders, p = 0.0002) [126]; these results were later confirmed by another study, which found that TP53 mutations influences the prognosis of short-responders (<3 months, HR = 1.87, 95% CI 1.06–3.29, p = 0.03) and short-survivors (<6 months, HR = 2.73, 95% CI 1.20–6.21, p = 0.017) [127], and by another study, which found that 100% of non-responders to gefitinib were TP53 mutated, with respect to 39% of responders (p < 0.001) [128]. It has recently been demonstrated that TP53 mutations are independently associated with PFS in both first-, second-, and third-generation EGFR-TKIs (HR: 2.02; 95% CI: 1.04–3.93, p = 0.038 and HR: 2.23, 95% CI 1.16–4.29, p = 0.017, respectively) [122]. An interesting study by Tsui and colleagues explored the dynamics of resistance mechanisms to EGFR-TKI treatment through circulating DNA, finding that pretreatment of TP53 mutations predicted a worse OS of patients (HR 0.43, 95% CI 0.2.0.97, p = 0.035), and that TP53 mutation was observed in a patient experiencing progressive disease and without T790M [129]. Results from positive studies demonstrating a role of TP53 mutations in predicting the clinical outcome of EGFR-mutated patients are resumed in Table 3.
Table 3.
Studies that found that TP53 mutations are prognostic for EGFR-mutated NSCLC patients treated with EGFR-TKIs.
| TP53 Status | Number of Patients (Generation of TKI) | Result | Ref. |
|---|---|---|---|
| Non-disruptive mutations | 193 (I) | OS | [105] |
| Any mutation | 131 (I–II) | OS | [106] |
| Any mutation | 116 (I–II) | OS | [107] |
| Exon 8 mutations | 123 (I–II) | DCR, PFS, OS | [108] |
| Exon 8 mutations | 136 (I–II) | OS | [109] |
| Exon 8 mutations | 379 (I–II) | OS | [110] |
| Missense mutations | 60 (I–II) | PFS | [111] |
| Any mutation | 75; 82 (I–II; III) | PFS, PFS | [116] |
| Any mutation | 18 (I) | TTP | [117] |
| Any mutation | 374 (I–II) | TTP | [118] |
| Any mutation | 28 (I) | TTP | [119] |
| Any mutation | 132 (I) | PFS, OS | [121] |
| Any mutation | 71 (I) | TTP, OS | [120] |
| Any mutation | 50 (I) | OS | [122] |
| Any mutationExon 6, 7 mutations | 368 (I) | PFS, OS | [129] |
| Exon 4, 7 mutations | 256 (I) | PFS, OS | [130] |
EGFR: Epidermal Growth Factor Receptor; TKI: Tyrosine Kinase Inhibitor; OS: Overall Survival; DCR: Disease Control Rate; PFS: Progression-Free Survival; TTP: Time-to-progression.
It has been highlighted that the different methodologies used to detect TP53 mutations have different sensitivities, as next-generation sequence technologies have led to an assessment of a greater rate of mutations in the case series; moreover, massive parallel sequencing has led to the sequencing of the entire TP53 gene, highlighting mutations out of the DNA binding domain.
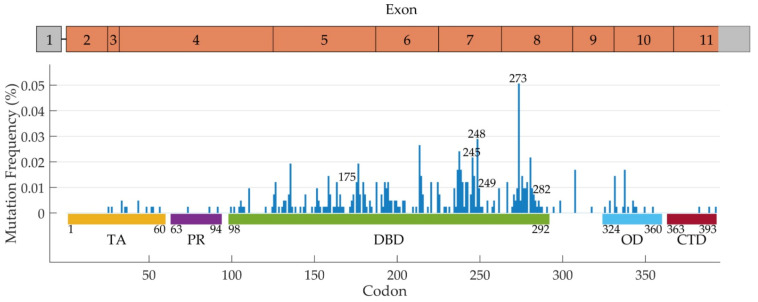
Two meta-analyses were also conducted, to better elucidate the role of TP53 in mutant EGFR patients. The pooled results from 11 studies (1049 EGFR-mutated patients) highlighted that patients with concomitant TP53 mutations have worse PFS (HR 1.76, 95% CI 1.44–2.16, p < 0.001) and OS (HR 1.83, 95% CI 1.47–2.29, p < 0.001). To better estimate the role of such mutations in predicting the response to EGR-TKIs, the authors performed a subgroup analysis of TKI-treated patients. The results remained significant both for patients receiving a TKI as a first-line therapy (HR for PFS 1.69, 95% CI 1.25–2.27, p = 0.001; HR for OS 1.94, 95% CI 1.36–2.76, p < 0.001) and for patients receiving TKIs in an all lines setting, for PFS and OS (HR for PFS 1.99, 95% CI 1.62–2.44, p = 0.001; HR for OS 1.93, 95% CI 1.45–2.58, p < 0.001) [130]. Another meta-analysis based on 2979 EGFR-mutated patients confirmed a worse OS for TP53 mutated patients versus wt TP53 patients (HR 1.73, 95% CI 1.22–2.44, p = 0.002). Interestingly, in a subgroup analysis, patients treated with TKIs had a worse PFS compared to other patients (HR 2.18, 95% CI 1.42–3.36, p < 0.001), even though this data was not confirmed by ORR analysis (RR 1.15, 95% CI 0.92–1.44, p = 0.212) [131]. Even though, in these studies, a categorization of TP53 mutations was not performed, a clear role for TP53 mutations as a negative factor for PFS and OS in EGFR-mutated patients treated with EGFR-TKIs was highlighted. In a study based on TCGA data, Wang et al., found that patients carrying TP53 and EGFR co-mutations had a worse OS with respect to wt/wt patients (38.4 months vs. 51.9 months, p = 0.023) [132]. Other indications highlight that TP53 mutations are able to influence the response to TKIs depending on the type of EGFR alterations. Considering that patients with EGFR exon 19 deletions usually have a major benefit from first-line EGFR-TKIs compared to other EGFR mutations, it has been reported that TP53 mutations are able to mainly affect the ORR and PFS of this subgroup of patients [114,133,134,135]. On the other hand, a recent study from the BENEFIT trial cohort investigated the TP53 mutations in liquid biopsies of EGFR-mutated patients and found that exon 19 deletions and TP53 co-mutations were predictors of better PFS and OS compared to exon 21 L858R/TP53 co-mutations (HR for PFS 1.53, p = 0.02; HR for OS 0.77, p = 0.37). The same study also demonstrated a shorter PFS and OS for TP53 mutated patients vs. TP53 wt ones (HR for PFS 0.66, 95% CI 0.48–0.89, p = 0.007; HR for PFS 0.54, 95% CI 0.40–0.74, p < 0.001), with TP53 mutations affecting the PFS and OS of patients with EGFR L858R mutations more than those with an exon 19 deletion. Interestingly, the authors also highlighted that patients with TP53 exon 6 and 7 mutations experienced worse PFS and OS, with a role in predicting prognosis for exon 5 TP53 mutations when categorized as disruptive and non-disruptive (OS HR = 2.04, 95% CI 0.99–4.19, p < 0.005) [25]. Recent results based on the CTONG 0901 trial identified that patients with TP53 mutations in exons 4 or 7 experience worse PFS and OS with respect to patients with mutations affecting other exons of the gene or wild-type TP53 (PFS 9.4, 11.0, and 14.5 months, respectively p = 0.009; OS 15.8, 20.0, and 26.1 months, respectively p = 0.004) [136]. The frequencies of TP53 mutations in EGFR-mutated patients are reported in Figure 1.
Figure 1.
Frequency of TP53 mutations in EGFR-mutated NSCLC patients, extracted by cBioPortal (https://www.cbioportal.org/, accessed on 28 January 2022). The percentage of TP53 codon mutations along all gene exons, and the most frequently mutated codons are reported. TA: transactivation domain; PR: proline rich domain; DBD: DNA binding domain; OD: oligomerization domain; CTD: carboxy-terminal domain.
5.2. TP53 Mutations in EGFR-Positive NSCLC: Phenotypic Changes
TP53 mutations also play a pivotal role in the histologic transformation to small-cell lung cancer (SCLC), a mechanism of resistance to first- and second-generation EGFR-TKIs known since 2011 [62]. The inactivation of genes like RB1 and TP53 play a crucial role in this transformation, as RB1 and TP53 co-alterations present in patients’ tissues at baseline are more likely to predict histologic transformation, with respect to the presence of only one mutation, and the presence of both mutations is a significant prognostic factor [137,138,139]. In this setting, it seems that TP53 mutation confers genomic instability to cancer cells resulting in a facilitated cell plasticity and phenotype reprogramming. The frequency of this evolution is about 3–5% and, usually, EGFR mutation is maintained [139,140,141].
Transformation to SCLC has also been observed as a mechanism of resistance to third-generation EGFR-TKIs used as a second-line treatment, with a frequency between 2 and 15%. This mechanism has been noticed with loss of T790M, but also when the resistance mutation is maintained, suggesting a focal and clonal tumor evolution [69,142]. When osimertinib is used as a first-line of treatment, transformation to SCLC has occurred in 4% of cases, as indicated by Schoenfeld and colleagues [54]. Transformation toward squamous cell carcinoma has been identified after treatment with osimertinib in II or later line and in I line, with a percentage of 9% and 7% respectively [54].
Recent results highlight that different TP53 mutations influence, in diverse ways, the response to EGFR-TKIs in cell lines. In particular, some mutations are associated with primary resistance to EGFR-TKIs, while others can induce epithelial-mesenchymal transition (EMT) as an acquired resistance mechanism [141]. Considering that TP53 participates in the regulation of the EGFR pathway [107], and that different mutations induce different mechanisms in different cell line models, it could be interesting to better investigate the cellular functions of the different TP53 mutants. In this regard, a classification based on the disturbance grade of the p53 protein has been proposed [111], and several studies have confirmed that TP53 mutations have different roles at a cellular level, and that some types of mutations are associated with oncogenic GOF [142]. Even though many studies of different case series brought interesting results based on this classification, which mutations and through which cellular mechanisms remain unanswered questions. Moreover, a study by Wei and colleagues highlights that TP53 mutations are generally involved in resistance and primary and metastatic relapse, but different TP53 exon alterations are involved in different mechanisms [143], and there is evidence that TP53 mutations are found more frequently in association with EGFR mutations [144,145,146], suggesting that a subset of EGFR-NSCLC could have a “double-oncogene” addition, for the role that such mutations display.
6. Conclusions
NSCLC is the primary cause of cancer-related deaths. Several pieces of evidence suggest that TP53 mutations are able to be used to identify a subset of patients with worse prognosis and a worse response to EGFR-TKIs; thus, identifying the different mutations associated with different outcomes. Given that TP53 exerts its functions through a wide range of cellular pathways, it would be necessary not only to understand the function of a single mutation, but to consider how this mutation could affect which pathway, though which cellular and molecular mechanisms lie at the base of these processes still has to be elucidated. For this reason, the more that is revealed about the role of these gene mutations in NSCLC, the better we will understand the role in primary or acquired resistance to EGFR-TKIs in this malignancy. What we can assume so far is that, as observed in other tumors, TP53 is a prognostic factor; however, after many reports concerning its effect on the efficacy of EGFR-TKIs, we now need to understand some of the molecular processes that link such mutations and resistance to EGFR-targeted therapy, as this could guide resistance to therapy. The link between TP53 mutations and targeted therapy response should be considered a starting point for new investigations, and further studies are needed to investigate these mechanisms to effectively predict responsiveness and survival; thus, better tailoring targeted therapy for EGFR-NSCLC patients.
Author Contributions
Ideation: M.C., L.C., G.B. and P.U. Literature search: M.C., K.A., P.C., L.P. and G.B. Paper draft: M.C., K.A. P.C. and L.P. Revisions: M.C., K.A., I.P., P.C., L.P., M.U., A.D., L.C., G.B., P.U. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
The data presented in this study are available in the article.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Ulivi P., Petracci E., Canale M., Priano I., Capelli L., Calistri D., Chiadini E., Cravero P., Rossi A., Delmonte A., et al. Liquid Biopsy for EGFR Mutation Analysis in Advanced Non-Small-Cell Lung Cancer Patients: Thoughts Drawn from a Real-Life Experience. Biomedicines. 2021;9:1299. doi: 10.3390/biomedicines9101299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Passaro A., Jänne P.A., Mok T., Peters S. Overcoming Therapy Resistance in EGFR-Mutant Lung Cancer. Nat. Cancer. 2021;2:377–391. doi: 10.1038/s43018-021-00195-8. [DOI] [PubMed] [Google Scholar]
- 4.Sabapathy K., Lane D. Therapeutic Targeting of p53: All Mutants Are Equal, but Some Mutants Are More Equal than Others. Nat. Rev. Clin. Oncol. 2017;15:13–30. doi: 10.1038/nrclinonc.2017.151. [DOI] [PubMed] [Google Scholar]
- 5.Jiao X.-D., Qin B.-D., You P., Cai J., Zang Y.-S. The Prognostic Value of TP53 and Its Correlation with EGFR Mutation in Advanced Non-Small Cell Lung Cancer, an Analysis Based on cBioPortal Data Base. Lung Cancer. 2018;123:70–75. doi: 10.1016/j.lungcan.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 6.Harrison P.T., Vyse S., Huang P.H. Rare Epidermal Growth Factor Receptor (EGFR) Mutations in Non-Small Cell Lung Cancer. Semin. Cancer Biol. 2019;61:167–179. doi: 10.1016/j.semcancer.2019.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Da Cunha Santos G., Shepherd F.A., Tsao M.S. EGFR Mutations and Lung Cancer. Annu. Rev. Pathol. 2011;6:49–69. doi: 10.1146/annurev-pathol-011110-130206. [DOI] [PubMed] [Google Scholar]
- 8.Castellanos E., Feld E., Horn L. Driven by Mutations: The Predictive Value of Mutation Subtype in EGFR -Mutated Non–Small Cell Lung Cancer. J. Thorac. Oncol. 2017;12:612–623. doi: 10.1016/j.jtho.2016.12.014. [DOI] [PubMed] [Google Scholar]
- 9.Xu M.J., Johnson D.E., Grandis J.R. EGFR-Targeted Therapies in the Post-Genomic Era. Cancer Metastasis Rev. 2017;36:463–473. doi: 10.1007/s10555-017-9687-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Talukdar S., Emdad L., Das S.K., Fisher P.B. EGFR: An Essential Receptor Tyrosine Kinase-Regulator of Cancer Stem Cells. Adv. Cancer Res. 2020;147:161–188. doi: 10.1016/bs.acr.2020.04.003. [DOI] [PubMed] [Google Scholar]
- 11.Gelatti A.C., Drilon A., Santini F.C. Optimizing the Sequencing of Tyrosine Kinase Inhibitors (TKIs) in Epidermal Growth Factor Receptor (EGFR) Mutation-Positive Non-Small Cell Lung Cancer (NSCLC) Lung Cancer. 2019;137:113–122. doi: 10.1016/j.lungcan.2019.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma S.V., Bell D.W., Settleman J., Haber D.A. Epidermal Growth Factor Receptor Mutations in Lung Cancer. Nat. Rev. Cancer. 2007;7:169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 13.Miyauchi E., Inoue A., Kobayashi K., Maemondo M., Sugawara S., Oizumi S., Isobe H., Gemma A., Saijo Y., Yoshizawa H., et al. Efficacy of Chemotherapy after First-Line Gefitinib Therapy in EGFR Mutation-Positive Advanced Non-Small Cell Lung Cancer-Data from a Randomized Phase III Study Comparing Gefitinib with Carboplatin Plus Paclitaxel (NEJ002) Jpn. J. Clin. Oncol. 2015;45:670–676. doi: 10.1093/jjco/hyv054. [DOI] [PubMed] [Google Scholar]
- 14.Lee J.-K., Hahn S., Kim D.-W., Suh K.J., Keam B., Kim T.M., Lee S.-H., Heo D.S. Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitors vs. Conventional Chemotherapy in Non–Small Cell Lung Cancer Harboring Wild-Type Epidermal Growth Factor Receptor. JAMA. 2014;311:1430–1437. doi: 10.1001/jama.2014.3314. [DOI] [PubMed] [Google Scholar]
- 15.Mok T.S., Wu Y.-L., Thongprasert S., Yang C.-H., Chu D.-T., Saijo N., Sunpaweravong P., Han B., Margono B., Ichinose Y., et al. Gefitinib or Carboplatin–Paclitaxel in Pulmonary Adenocarcinoma. N. Engl. J. Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 16.Mitsudomi T., Morita S., Yatabe Y., Negoro S., Okamoto I., Tsurutani J., Seto T., Satouchi M., Tada H., Hirashima T., et al. Gefitinib Versus Cisplatin Plus Docetaxel in Patients with Non-Small-Cell Lung Cancer Harbouring Mutations of the Epidermal Growth Factor Receptor (WJTOG3405): An Open Label, Randomised Phase 3 Trial. Lancet Oncol. 2009;11:121–128. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 17.Zhou C., Wu Y.-L., Chen G., Feng J., Liu X.-Q., Wang C., Zhang S., Wang J., Zhou S., Ren S., et al. Erlotinib Versus Chemotherapy as First-Line Treatment for Patients with Advanced EGFR Mutation-Positive Non-Small-Cell Lung Cancer (OPTIMAL, CTONG-0802): A Multicentre, Open-Label, Randomised, Phase 3 Study. Lancet Oncol. 2011;12:735–742. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 18.Rosell R., Carcereny E., Gervais R., Vergnenegre A., Massuti B., Felip E., Palmero R., Garcia-Gomez R., Pallares C., Sanchez J.M., et al. Erlotinib Versus Standard Chemotherapy as First-Line Treatment for European Patients with Advanced EGFR Mutation-Positive Non-Small-Cell Lung Cancer (EURTAC): A Multicentre, Open-Label, Randomised Phase 3 Trial. Lancet Oncol. 2012;13:239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 19.Maemondo M., Inoue A., Kobayashi K., Sugawara S., Oizumi S., Isobe H., Gemma A., Harada M., Yoshizawa H., Kinoshita I., et al. Gefitinib or Chemotherapy for Non–Small-Cell Lung Cancer with Mutated EGFR. N. Engl. J. Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 20.Wu Y.-L., Zhou C., Liam C.-K., Wu G., Liu X., Zhong Z., Lu S., Cheng Y., Han B., Chen L., et al. First-Line Erlotinib Versus Gemcitabine/Cisplatin in Patients with Advanced EGFR Mutation-Positive Non-Small-Cell Lung Cancer: Analyses from the Phase III, Randomized, Open-Label, ENSURE Study. Ann. Oncol. 2015;26:1883–1889. doi: 10.1093/annonc/mdv270. [DOI] [PubMed] [Google Scholar]
- 21.Lee C.K., Davies L., Wu Y.-L., Mitsudomi T., Inoue A., Rosell R., Zhou C., Nakagawa K., Thongprasert S., Fukuoka M., et al. Gefitinib or Erlotinib vs. Chemotherapy for EGFR Mutation-Positive Lung Cancer: Individual Patient Data Meta-Analysis of Overall Survival. JNCI J. Natl. Cancer Inst. 2017;109:djw279. doi: 10.1093/jnci/djw279. [DOI] [PubMed] [Google Scholar]
- 22.Shi Y.K., Wang L., Han B.H., Li W., Yu P., Liu Y.P., Ding C.M., Song X., Ma Z.Y., Ren X.L., et al. First-Line Icotinib Versus Cisplatin/Pemetrexed Plus Pemetrexed Maintenance Therapy for Patients with Advanced EGFR Mutation-Positive Lung Adenocarcinoma (CONVINCE): A Phase 3, Open-Label, Randomized Study. Ann. Oncol. 2017;28:2443–2450. doi: 10.1093/annonc/mdx359. [DOI] [PubMed] [Google Scholar]
- 23.Yang J.J., Zhou Q., Yan H.H., Zhang X.C., Chen H.J., Tu H.Y., Wang Z., Xu C.R., Su J., Wang B.C., et al. A Phase III Randomised Controlled Trial of Erlotinib vs. Gefitinib in Advanced Non-Small Cell Lung Cancer with EGFR Mutations. Br. J. Cancer. 2017;116:568–574. doi: 10.1038/bjc.2016.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi Y., Zhang L., Liu X., Zhou C., Zhang S., Wang D., Li Q., Qin S., Hu C., Zhang Y., et al. Icotinib Versus Gefitinib in Previously Treated Advanced Non-Small-Cell Lung Cancer (ICOGEN): A Randomised, Double-Blind Phase 3 Non-Inferiority Trial. Lancet Oncol. 2013;14:953–961. doi: 10.1016/S1470-2045(13)70355-3. [DOI] [PubMed] [Google Scholar]
- 25.Yamamoto N., Seto T., Nishio M., Goto K., Okamoto I., Yamanaka T., Tanaka M., Takahashi K., Fukuoka M. Erlotinib Plus Bevacizumab vs. Erlotinib Monotherapy as First-Line Treatment for Advanced EGFR Mutation-Positive Non-Squamous Non-Small-Cell Lung Cancer: Survival Follow-Up Results of the Randomized JO25567 Study. Lung Cancer. 2020;151:20–24. doi: 10.1016/j.lungcan.2020.11.020. [DOI] [PubMed] [Google Scholar]
- 26.Nakagawa K., Garon E.B., Seto T., Nishio M., Aix S.P., Paz-Ares L., Chiu C.-H., Park K., Novello S., Nadal E., et al. Ramucirumab Plus Erlotinib in Patients with Untreated, EGFR-Mutated, Advanced Non-Small-Cell Lung Cancer (RELAY): A Randomised, Double-Blind, Placebo-Controlled, Phase 3 Trial. Lancet Oncol. 2019;20:1655–1669. doi: 10.1016/S1470-2045(19)30634-5. [DOI] [PubMed] [Google Scholar]
- 27.Nakagawa K., Nadal E., Garon E.B., Nishio M., Seto T., Yamamoto N., Park K., Shih J.-Y., Paz-Ares L., Frimodt-Moller B., et al. RELAY Subgroup Analyses by EGFR Ex19del and Ex21L858R Mutations for Ramucirumab Plus Erlotinib in Metastatic Non–Small Cell Lung Cancer. Clin. Cancer Res. 2021;27:5258–5271. doi: 10.1158/1078-0432.CCR-21-0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu Q., Luo W., Li W., Wang T., Huang L., Xu F. First-Generation EGFR-TKI Plus Chemotherapy Versus EGFR-TKI Alone as First-Line Treatment in Advanced NSCLC with EGFR Activating Mutation: A Systematic Review and Meta-Analysis of Randomized Controlled Trials. Front. Oncol. 2021;11:883. doi: 10.3389/fonc.2021.598265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li D., Ambrogio L., Shimamura T., Kubo S., Takahashi M., Chirieac L.R., Padera R.F., Shapiro G.I., Baum A., Himmelsbach F., et al. BIBW2992, an Irreversible EGFR/HER2 Inhibitor Highly Effective in Preclinical Lung Cancer Models. Oncogene. 2008;27:4702–4711. doi: 10.1038/onc.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Engelman J.A., Zejnullahu K., Gale C.-M., Lifshits E., Gonzales A.J., Shimamura T., Zhao F., Vincent P.W., Naumov G.N., Bradner J.E., et al. PF00299804, an Irreversible Pan-ERBB Inhibitor, Is Effective in Lung Cancer Models with EGFR and ERBB2 Mutations that Are Resistant to Gefitinib. Cancer Res. 2007;67:11924–11932. doi: 10.1158/0008-5472.CAN-07-1885. [DOI] [PubMed] [Google Scholar]
- 31.Yang J.C.-H., Hirsh V., Schuler M., Yamamoto N., O’Byrne K.J., Mok T., Zazulina V., Shahidi M., Lungershausen J., Massey D., et al. Symptom Control and Quality of Life in LUX-Lung 3: A Phase III Study of Afatinib or Cisplatin/Pemetrexed in Patients with Advanced Lung Adenocarcinoma with EGFR Mutations. J. Clin. Oncol. 2013;31:3342–3350. doi: 10.1200/JCO.2012.46.1764. [DOI] [PubMed] [Google Scholar]
- 32.Paz-Ares L., Tan E.-H., O’Byrne K., Zhang L., Hirsh V., Boyer M., Yang J.C.-H., Mok T., Lee K.H., Lu S., et al. Afatinib Versus Gefitinib in Patients with EGFR Mutation-Positive Advanced Non-Small-Cell Lung Cancer: Overall Survival Data from the Phase IIb LUX-Lung 7 Trial. Ann. Oncol. 2017;28:270–277. doi: 10.1093/annonc/mdw611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu Y.-L., Cheng Y., Zhou X., Lee K.H., Nakagawa K., Niho S., Tsuji F., Linke R., Rosell R., Corral J., et al. Dacomitinib Versus Gefitinib as First-Line Treatment for Patients with EGFR-Mutation-Positive Non-Small-Cell Lung Cancer (ARCHER 1050): A Randomised, Open-Label, Phase 3 Trial. Lancet Oncol. 2017;18:1454–1466. doi: 10.1016/S1470-2045(17)30608-3. [DOI] [PubMed] [Google Scholar]
- 34.Mok T.S., Cheng Y., Zhou X., Lee K.H., Nakagawa K., Niho S., Chawla A., Rosell R., Corral J., Migliorino M.R., et al. Updated Overall Survival in a Randomized Study Comparing Dacomitinib with Gefitinib as First-Line Treatment in Patients with Advanced Non-Small-Cell Lung Cancer and EGFR-Activating Mutations. Drugs. 2020;81:257–266. doi: 10.1007/s40265-020-01441-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cross D.A.E., Ashton S.E., Ghiorghiu S., Eberlein C., Nebhan C.A., Spitzler P.J., Orme J.P., Finlay M.R.V., Ward R.A., Mellor M.J., et al. AZD9291, an Irreversible EGFR TKI, Overcomes T790M-Mediated Resistance to EGFR Inhibitors in Lung Cancer. Cancer Discov. 2014;4:1046–1061. doi: 10.1158/2159-8290.CD-14-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mok T.S., Wu Y.-L., Ahn M.-J., Garassino M.C., Kim H.R., Ramalingam S.S., Shepherd F.A., He Y., Akamatsu H., Theelen W.S., et al. Osimertinib or Platinum–Pemetrexed in EGFR T790M–Positive Lung Cancer. N. Engl. J. Med. 2017;376:629–640. doi: 10.1056/NEJMoa1612674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soria J.-C., Ohe Y., Vansteenkiste J., Reungwetwattana T., Chewaskulyong B., Lee K.H., Dechaphunkul A., Imamura F., Nogami N., Kurata T., et al. Osimertinib in Untreated EGFR-Mutated Advanced Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2018;378:113–125. doi: 10.1056/NEJMoa1713137. [DOI] [PubMed] [Google Scholar]
- 38.Yang J.C.-H., Wu Y.L., Schuler M., Sebastian M., Popat S., Yamamoto N., Zhou C., Hu C.-P., O’Byrne K., Feng J., et al. Afatinib Versus Cisplatin-Based Chemotherapy for EGFR Mutation-Positive Lung Adenocarcinoma (LUX-Lung 3 and LUX-Lung 6): Analysis of Overall Survival Data from Two Randomised, Phase 3 Trials. Lancet Oncol. 2015;16:141–151. doi: 10.1016/S1470-2045(14)71173-8. [DOI] [PubMed] [Google Scholar]
- 39.Yang J.C.-H., Sequist L.V., Geater S.L., Tsai C.-M., Mok T., Schuler M., Yamamoto N., Yu C.-J., Ou S.-H.I., Zhou C., et al. Clinical Activity of Afatinib in Patients with Advanced Non-Small-Cell Lung Cancer Harbouring Uncommon EGFR Mutations: A Combined Post-Hoc Analysis of LUX-Lung 2, LUX-Lung 3, and LUX-Lung 6. Lancet Oncol. 2015;16:830–838. doi: 10.1016/S1470-2045(15)00026-1. [DOI] [PubMed] [Google Scholar]
- 40.Chiu C.-H., Yang C.-T., Shih J.-Y., Huang M.-S., Su W.-C., Lai R.-S., Wang C.-C., Hsiao S.-H., Lin Y.-C., Ho C.-L., et al. Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitor Treatment Response in Advanced Lung Adenocarcinomas with G719X/L861Q/S768I Mutations. J. Thorac. Oncol. 2015;10:793–799. doi: 10.1097/JTO.0000000000000504. [DOI] [PubMed] [Google Scholar]
- 41.Haura E.B., Cho B.C., Lee J.S., Han J.-Y., Lee K.H., Sanborn R.E., Govindan R., Cho E.K., Kim S.-W., Reckamp K.L., et al. JNJ-61186372 (JNJ-372), an EGFR-cMet Bispecific Antibody, in EGFR-Driven Advanced Non-Small Cell Lung Cancer (NSCLC) J. Clin. Oncol. 2019;37:9009. doi: 10.1200/JCO.2019.37.15_suppl.9009. [DOI] [Google Scholar]
- 42.Cho J.H., Lim S.H., An H.J., Kim K.H., Park K.U., Kang E.J., Choi Y.H., Ahn M.S., Lee M.H., Sun J.-M., et al. Osimertinib for Patients with Non–Small-Cell Lung Cancer Harboring Uncommon EGFR Mutations: A Multicenter, Open-Label, Phase II Trial (KCSG-LU15-09) J. Clin. Oncol. 2020;38:488–495. doi: 10.1200/JCO.19.00931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reita D., Pabst L., Pencreach E., Guérin E., Dano L., Rimelen V., Voegeli A.-C., Vallat L., Mascaux C., Beau-Faller M. Molecular Mechanism of EGFR-TKI Resistance in EGFR-Mutated Non-Small Cell Lung Cancer: Application to Biological Diagnostic and Monitoring. Cancers. 2021;13:4926. doi: 10.3390/cancers13194926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pao W., Girard N. New Driver Mutations in Non-Small-Cell Lung Cancer. Lancet Oncol. 2011;12:175–180. doi: 10.1016/S1470-2045(10)70087-5. [DOI] [PubMed] [Google Scholar]
- 45.Chmielecki J., Foo J., Oxnard G.R., Hutchinson K., Ohashi K., Somwar R., Wang L., Amato K.R., Arcila M., Sos M.L., et al. Optimization of Dosing for EGFR-Mutant Non–Small Cell Lung Cancer with Evolutionary Cancer Modeling. Sci. Transl. Med. 2011;3:90ra59. doi: 10.1126/scitranslmed.3002356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lim S.M., Syn N.L., Cho B.C., Soo R.A. Acquired Resistance to EGFR Targeted Therapy in Non-Small Cell Lung Cancer: Mechanisms and Therapeutic Strategies. Cancer Treat. Rev. 2018;65:1–10. doi: 10.1016/j.ctrv.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 47.Yang Z., Yang N., Ou Q., Xiang Y., Jiang T., Wu X., Bao H., Tong X., Wang X., Shao Y.W., et al. Investigating Novel Resistance Mechanisms to Third-Generation EGFR Tyrosine Kinase Inhibitor Osimertinib in Non–Small Cell Lung Cancer Patients. Clin. Cancer Res. 2018;24:3097–3107. doi: 10.1158/1078-0432.CCR-17-2310. [DOI] [PubMed] [Google Scholar]
- 48.Niederst M.J., Hu H., Mulvey H.E., Lockerman E.L., Garcia A.R., Piotrowska Z., Sequist L.V., Engelman J.A. The Allelic Context of the C797S Mutation Acquired upon Treatment with Third-Generation EGFR Inhibitors Impacts Sensitivity to Subsequent Treatment Strategies. Clin. Cancer Res. 2015;21:3924–3933. doi: 10.1158/1078-0432.CCR-15-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramalingam S.S., Cheng Y., Zhou C., Ohe Y., Imamura F., Cho B.C., Lin M.-C., Majem M., Shah R., Rukazenkov Y., et al. Mechanisms of Acquired Resistance to First-Line Osimertinib: Preliminary Data from the Phase III FLAURA Study. Ann. Oncol. 2018;29:viii740. doi: 10.1093/annonc/mdy424.063. [DOI] [Google Scholar]
- 50.Arulananda S., Do H., Musafer A., Mitchell P., Dobrovic A., John T. Combination Osimertinib and Gefitinib in C797S and T790M EGFR-Mutated Non–Small Cell Lung Cancer. J. Thorac. Oncol. 2017;12:1728–1732. doi: 10.1016/j.jtho.2017.08.006. [DOI] [PubMed] [Google Scholar]
- 51.Ou S.-H.I., Cui J., Schrock A.B., Goldberg M.E., Zhu V.W., Albacker L., Stephens P.J., Miller V.A., Ali S.M. Emergence of Novel and Dominant Acquired EGFR Solvent-Front Mutations at Gly796 (G796S/R) Together with C797S/G and L792F/H Mutations in One EGFR (L858R/T790M) NSCLC Patient Who Progressed on Osimertinib. Lung Cancer. 2017;108:228–231. doi: 10.1016/j.lungcan.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 52.Fang W., Gan J., Huang Y., Zhou H., Zhang L. Acquired EGFR L718V Mutation and Loss of T790M-Mediated Resistance to Osimertinib in a Patient with NSCLC Who Responded to Afatinib. J. Thorac. Oncol. 2019;14:e274–e275. doi: 10.1016/j.jtho.2019.07.018. [DOI] [PubMed] [Google Scholar]
- 53.Schoenfeld A.J., Chan J., Kubota D., Sato H., Rizvi H., Daneshbod Y., Chang J.C., Paik P.K., Offin M., Arcila M.E., et al. Tumor Analyses Reveal Squamous Transformation and Off-Target Alterations as Early Resistance Mechanisms to First-Line Osimertinib in EGFR-Mutant Lung Cancer. Clin. Cancer Res. 2020;26:2654–2663. doi: 10.1158/1078-0432.CCR-19-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Q., Zhang X.-C., Yang J.-J., Yang Z.-F., Bai Y., Su J., Wang Z., Zhang Z., Shao Y., Zhou Q., et al. EGFR L792H and G796R: Two Novel Mutations Mediating Resistance to the Third-Generation EGFR Tyrosine Kinase Inhibitor Osimertinib. J. Thorac. Oncol. 2018;13:1415–1421. doi: 10.1016/j.jtho.2018.05.024. [DOI] [PubMed] [Google Scholar]
- 55.Zheng D., Hu M., Bai Y., Zhu X., Lu X., Wu C., Wang J., Liu L., Wang Z., Ni J., et al. EGFR G796D Mutation Mediates Resistance to Osimertinib. Oncotarget. 2017;8:49671–49679. doi: 10.18632/oncotarget.17913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Y., He B., Zhou D., Li M., Hu C. Newly Emergent Acquired EGFR Exon 18 G724S Mutation after Resistance of a T790M Specific EGFR Inhibitor Osimertinib in Non-Small-Cell Lung Cancer: A Case Report. OncoTargets Ther. 2018;12:51–56. doi: 10.2147/OTT.S188612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oztan A., Fischer S., Schrock A., Erlich R., Lovly C., Stephens P., Ross J., Miller V., Ali S., Ou S.-H.I., et al. Emergence of EGFR G724S Mutation in EGFR-Mutant Lung Adenocarcinoma Post Progression on Osimertinib. Lung Cancer. 2017;111:84–87. doi: 10.1016/j.lungcan.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 58.Nukaga S., Yasuda H., Tsuchihara K., Hamamoto J., Masuzawa K., Kawada I., Naoki K., Matsumoto S., Mimaki S., Ikemura S., et al. Amplification of EGFR Wild-Type Alleles in Non–Small Cell Lung Cancer Cells Confers Acquired Resistance to Mutation-Selective EGFR Tyrosine Kinase Inhibitors. Cancer Res. 2017;77:2078–2089. doi: 10.1158/0008-5472.CAN-16-2359. [DOI] [PubMed] [Google Scholar]
- 59.Shi Y., Xing P., Han X., Wang S., Liu Y., Liu P., Li J., Chang L., Guan Y., Zhang Z., et al. P1.13-18 Exploring the Resistance Mechanism of Osimertinib and Monitoring the Treatment Response Using Plasma ctDNA in Chinese NSCLC Patients. J. Thorac. Oncol. 2018;13:S589. doi: 10.1016/j.jtho.2018.08.875. [DOI] [Google Scholar]
- 60.Graham T.A., McDonald S.A., Wright N.A. Field Cancerization in the GI Tract. Futur. Oncol. 2011;7:981–993. doi: 10.2217/fon.11.70. [DOI] [PubMed] [Google Scholar]
- 61.Sequist L.V., Waltman B.A., Dias-Santagata D., Digumarthy S., Turke A.B., Fidias P., Bergethon K., Shaw A.T., Gettinger S., Cosper A.K., et al. Genotypic and Histological Evolution of Lung Cancers Acquiring Resistance to EGFR Inhibitors. Sci. Transl. Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Le X., Puri S., Negrao M.V., Nilsson M.B., Robichaux J., Boyle T., Hicks J.K., Lovinger K.L., Roarty E., Rinsurongkawong W., et al. Landscape of EGFR-Dependent and -Independent Resistance Mechanisms to Osimertinib and Continuation Therapy Beyond Progression in EGFR-Mutant NSCLC. Clin. Cancer Res. 2018;24:6195–6203. doi: 10.1158/1078-0432.CCR-18-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Engelman J.A., Jänne P.A. Mechanisms of Acquired Resistance to Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitors in Non–Small Cell Lung Cancer: Figure 1. Clin. Cancer Res. 2008;14:2895–2899. doi: 10.1158/1078-0432.CCR-07-2248. [DOI] [PubMed] [Google Scholar]
- 64.Papadimitrakopoulou V.A., Mok T.S., Han J.Y., Ahn M.J., Delmonte A., Ramalingam S.S., Kim S.W., Shepherd F.A., Laskin J., He Y., et al. Osimertinib Versus Platinum–Pemetrexed for Patients with EGFR T790M Advanced NSCLC and Progression on a Prior EGFR-Tyrosine Kinase Inhibitor: AURA3 Overall Survival Analysis. Ann. Oncol. 2020;31:1536–1544. doi: 10.1016/j.annonc.2020.08.2100. [DOI] [PubMed] [Google Scholar]
- 65.Takezawa K., Pirazzoli V., Arcila M.E., Nebhan C.A., Song X., De Stanchina E., Ohashi K., Janjigian Y.Y., Spitzler P.J., Melnick M.A., et al. HER2 Amplification: A Potential Mechanism of Acquired Resistance to EGFR Inhibition in EGFR-Mutant Lung Cancers That Lack the Second-Site EGFRT790M Mutation. Cancer Discov. 2012;2:922–933. doi: 10.1158/2159-8290.CD-12-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Planchard D., Loriot Y., André F., Gobert A., Auger N., Lacroix L., Soria J.C. EGFR-Independent Mechanisms of Acquired Resistance to AZD9291 in EGFR T790M-Positive NSCLC Patients. Ann. Oncol. 2015;26:2073–2078. doi: 10.1093/annonc/mdv319. [DOI] [PubMed] [Google Scholar]
- 67.Jackman D., Pao W., Riely G.J., Engelman J.A., Kris M., Jänne P.A., Lynch T., Johnson B.E., Miller V.A. Clinical Definition of Acquired Resistance to Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitors in Non–Small-Cell Lung Cancer. J. Clin. Oncol. 2010;28:357–360. doi: 10.1200/JCO.2009.24.7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Oxnard G.R., Hu E., Mileham K.F., Husain H., Costa D., Tracy P., Feeney N., Sholl L.M., Dahlberg S.E., Redig A.J., et al. Assessment of Resistance Mechanisms and Clinical Implications in Patients WithEGFRT790M–Positive Lung Cancer and Acquired Resistance to Osimertinib. JAMA Oncol. 2018;4:1527–1534. doi: 10.1001/jamaoncol.2018.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu H.A., Arcila M.E., Rekhtman N., Sima C.S., Zakowski M.F., Pao W., Kris M.G., Miller V.A., Ladanyi M., Riely G.J. Analysis of Tumor Specimens at the Time of Acquired Resistance to EGFR-TKI Therapy in 155 Patients with EGFR-Mutant Lung Cancers. Clin. Cancer Res. 2013;19:2240–2247. doi: 10.1158/1078-0432.CCR-12-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou Y., Wang X.B., Qiu X.P., Shuai Z., Wang C., Zheng F. CDKN2A Promoter Methylation and Hepatocellular Carcinoma Risk: A Meta-Analysis. Clin. Res. Hepatol. Gastroenterol. 2018;42:529–541. doi: 10.1016/j.clinre.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 71.Lee J., Kim H.S., Lee B., Kim H.K., Sun J., Ahn J.S., Ahn M., Park K., Lee S. Genomic Landscape of Acquired Resistance to Third-Generation EGFR Tyrosine Kinase Inhibitors in EGFR T790M-Mutant Non–Small Cell Lung Cancer. Cancer. 2020;126:2704–2712. doi: 10.1002/cncr.32809. [DOI] [PubMed] [Google Scholar]
- 72.Hafner A., Bulyk M.L., Jambhekar A., Lahav G. The Multiple Mechanisms That Regulate P53 Activity and Cell Fate. Nat. Rev. Mol. Cell Biol. 2019;20:199–210. doi: 10.1038/s41580-019-0110-x. [DOI] [PubMed] [Google Scholar]
- 73.Kruiswijk F., Labuschagne C.F., Vousden K.H. p53 in Survival, Death and Metabolic Health: A Lifeguard with a Licence to Kill. Nat. Rev. Mol. Cell Biol. 2015;16:393–405. doi: 10.1038/nrm4007. [DOI] [PubMed] [Google Scholar]
- 74.Kastan M.B., Onyekwere O., Sidransky D., Vogelstein B., Craig R.W. Participation of P53 Protein in the Cellular Response to DNA Damage. Cancer Res. 1991;51:6304–6311. doi: 10.1158/0008-5472.CAN-16-1560. [DOI] [PubMed] [Google Scholar]
- 75.Kastenhuber E.R., Lowe S.W. Putting P53 in Context. Cell. 2017;170:1062–1078. doi: 10.1016/j.cell.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Eischen C.M. Genome Stability Requires P53. Cold Spring Harb. Perspect. Med. 2016;6:a026096. doi: 10.1101/cshperspect.a026096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Maciejowski J., Li Y., Bosco N., Campbell P.J., de Lange T. Chromothripsis and Kataegis Induced by Telomere Crisis. Cell. 2015;163:1641–1654. doi: 10.1016/j.cell.2015.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rausch T., Jones D.T.W., Zapatka M., Stütz A.M., Zichner T., Weischenfeldt J., Jaeger N., Remke M., Shih D.J.H., Northcott P.A., et al. Genome Sequencing of Pediatric Medulloblastoma Links Catastrophic DNA Rearrangements with TP53 Mutations. Cell. 2012;148:59–71. doi: 10.1016/j.cell.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Levine A.J., Ting D., Greenbaum B.D. P53 and the Defenses against Genome Instability Caused by Transposons and Repetitive Elements. BioEssays. 2016;38:508–513. doi: 10.1002/bies.201600031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wylie A., Jones A.E., D’Brot A., Lu W.-J., Kurtz P., Moran J.V., Rakheja D., Chen K.S., Hammer R.E., Comerford S.A., et al. P53 Genes Function to Restrain Mobile Elements. Genes Dev. 2015;30:64–77. doi: 10.1101/gad.266098.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schlereth K., Heyl C., Krampitz A.-M., Mernberger M., Finkernagel F., Scharfe M., Jarek M., Leich E., Rosenwald A., Stiewe T. Characterization of the P53 Cistrome–DNA Binding Cooperativity Dissects P53′s Tumor Suppressor Functions. PLoS Genet. 2013;9:e1003726. doi: 10.1371/journal.pgen.1003726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nikulenkov F., Spinnler C., Li H., Tonelli C., Shi Y., Turunen M., Kivioja T., Ignatiev I., Kel A., Taipale J., et al. Insights into P53 Transcriptional Function via Genome-Wide Chromatin Occupancy and Gene Expression Analysis. Cell Death Differ. 2012;19:1992–2002. doi: 10.1038/cdd.2012.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wei C.-L., Wu Q., Vega V.B., Chiu K.P., Ng P., Zhang T., Shahab A., Yong H.C., Fu Y., Weng Z., et al. A Global Map of P53 Transcription-Factor Binding Sites in the Human Genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 84.Laptenko O., Beckerman R., Freulich E., Prives C. P53 Binding to Nucleosomes within the P21 Promoter in Vivo Leads to Nucleosome Loss and Transcriptional Activation. Proc. Natl. Acad. Sci. USA. 2011;108:10385–10390. doi: 10.1073/pnas.1105680108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Botcheva K., McCorkle S., McCombie W.R., Dunn J.J., Anderson C.W. Distinct P53 Genomic Binding Patterns in Normal and Cancer-Derived Human Cells. Cell Cycle. 2011;10:4237–4249. doi: 10.4161/cc.10.24.18383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Min S., Kim K., Kim S.-G., Cho H., Lee Y. Chromatin-Remodeling Factor, RSF1, Controls P53-Mediated Transcription in Apoptosis Upon DNA Strand Breaks. Cell Death Dis. 2018;9:1–13. doi: 10.1038/s41419-018-1128-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hollstein M., Sidransky D., Vogelstein B., Harris C.C. P53 Mutations in Human Cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 88.Zhou G., Liu Z., Myers J.N. TP53 Mutations in Head and Neck Squamous Cell Carcinoma and Their Impact on Disease Progression and Treatment Response. J. Cell. Biochem. 2016;117:2682–2692. doi: 10.1002/jcb.25592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dearth L.R., Qian H., Wang T., Baroni T.E., Zeng J., Chen S.W., Yi S.Y., Brachmann R.K. Inactive Full-Length P53 Mutants Lacking Dominant Wild-Type P53 Inhibition Highlight Loss of Heterozygosity as an Important Aspect of P53 Status in Human Cancers. Carcinogenesis. 2007;28:289–298. doi: 10.1093/carcin/bgl132. [DOI] [PubMed] [Google Scholar]
- 90.Dittmer D., Pati S., Zambetti G., Chu S., Teresky A.K., Moore M., Finlay C., Levine A.J. Gain of Function Mutations in P53. Nat. Genet. 1993;4:42–46. doi: 10.1038/ng0593-42. [DOI] [PubMed] [Google Scholar]
- 91.Noll J.E., Jeffery J., Al-Ejeh F., Kumar R., Khanna K.K., Callen D., Neilsen P. Mutant P53 Drives Multinucleation and Invasion through a Process That Is Suppressed by ANKRD11. Oncogene. 2011;31:2836–2848. doi: 10.1038/onc.2011.456. [DOI] [PubMed] [Google Scholar]
- 92.El-Hizawi S., Lagowski J.P., Kulesz-Martin M., Albor A. Induction of Gene Amplification as a Gain-of-Function Phenotype of Mutant P53 Proteins. Cancer Res. 2002;62:3264–3270. [PubMed] [Google Scholar]
- 93.Sarig R., Rivlin N., Brosh R., Bornstein C., Kamer I., Ezra O., Molchadsky A., Goldfinger N., Brenner O., Rotter V. Mutant P53 Facilitates Somatic Cell Reprogramming and Augments the Malignant Potential of Reprogrammed Cells. J. Exp. Med. 2010;207:2127–2140. doi: 10.1084/jem.20100797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mantovani F., Collavin L., Del Sal G. Mutant P53 as a Guardian of the Cancer Cell. Cell Death Differ. 2018;26:199–212. doi: 10.1038/s41418-018-0246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Purvis J.E., Karhohs K.W., Mock C., Batchelor E., Loewer A., Lahav G. P53 Dynamics Control Cell Fate. Science. 2012;336:1440–1444. doi: 10.1126/science.1218351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Paek A.L., Liu J.C., Loewer A., Forrester W.C., Lahav G. Cell-to-Cell Variation in P53 Dynamics Leads to Fractional Killing. Cell. 2016;165:631–642. doi: 10.1016/j.cell.2016.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tal P., Eizenberger S., Cohen E., Goldfinger N., Pietrokovski S., Oren M., Rotter V. Cancer Therapeutic Approach Based on Conformational Stabilization of Mutant P53 Protein by Small Peptides. Oncotarget. 2016;7:11817–11837. doi: 10.18632/oncotarget.7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bykov V.N., Wiman K.G. Mutant P53 Reactivation by Small Molecules Makes Its Way to the Clinic. FEBS Lett. 2014;588:2622–2627. doi: 10.1016/j.febslet.2014.04.017. [DOI] [PubMed] [Google Scholar]
- 99.Volckmar A.-L., Leichsenring J., Kirchner M., Christopoulos P., Neumann O., Budczies J., Morais de Oliveira C.M., Rempel E., Buchhalter I., Brandt R., et al. Combined Targeted DNA and RNA Sequencing of Advanced NSCLC in Routine Molecular Diagnostics: Analysis of the First 3000 Heidelberg Cases. Int. J. Cancer. 2019;145:649–661. doi: 10.1002/ijc.32133. [DOI] [PubMed] [Google Scholar]
- 100.Never-smoker N.E.S. The Cancer Genome Atlas Research Network Erratum: Corrigendum: Comprehensive Molecular Profiling of Lung Adenocarcinoma. Nature. 2014;514:262. doi: 10.1038/nature13879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shi J., Hua X., Zhu B., Ravichandran S., Wang M., Nguyen C., Brodie S.A., Palleschi A., Alloisio M., Pariscenti G., et al. Somatic Genomics and Clinical Features of Lung Adenocarcinoma: A Retrospective Study. PLoS Med. 2016;13:e1002162. doi: 10.1371/journal.pmed.1002162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jamal-Hanjani M., Wilson G.A., McGranahan N., Birkbak N.J., Watkins T.B.K., Veeriah S., Shafi S., Johnson D.H., Mitter R., Rosenthal R., et al. Tracking the Evolution of Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2017;376:2109–2121. doi: 10.1056/NEJMoa1616288. [DOI] [PubMed] [Google Scholar]
- 103.Kosaka T., Yatabe Y., Onozato R., Kuwano H., Mitsudomi T. Prognostic Implication of EGFR, KRAS, and TP53 Gene Mutations in a Large Cohort of Japanese Patients with Surgically Treated Lung Adenocarcinoma. J. Thorac. Oncol. 2009;4:22–29. doi: 10.1097/JTO.0b013e3181914111. [DOI] [PubMed] [Google Scholar]
- 104.La Fleur L., Falk-Sörqvist E., Smeds P., Berglund A., Sundström M., Mattsson J.S., Brandén E., Koyi H., Isaksson J., Brunnström H., et al. Mutation Patterns in a Population-Based Non-Small Cell Lung Cancer Cohort and Prognostic Impact of Concomitant Mutations in KRAS and TP53 or STK11. Lung Cancer. 2019;130:50–58. doi: 10.1016/j.lungcan.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 105.Scoccianti C., Vésin A., Martel G., Olivier M., Brambilla E., Timsit J.-F., Tavecchio L.D., Brambilla C., Field J.K., Hainaut P., et al. Prognostic Value ofTP53, KRAS and EGFR Mutations in Non-Small Cell Lung Cancer: The EUELC Cohort. Eur. Respir. J. 2012;40:177–184. doi: 10.1183/09031936.00097311. [DOI] [PubMed] [Google Scholar]
- 106.Tomasini P., Mascaux C., Jao K., Labbe C., Kamel-Reid S., Stockley T., Hwang D.M., Leighl N.B., Liu G., Bradbury P.A., et al. Effect of Coexisting KRAS and TP53 Mutations in Patients Treated with Chemotherapy for Non–Small-Cell Lung Cancer. Clin. Lung Cancer. 2018;20:e338–e345. doi: 10.1016/j.cllc.2018.12.009. [DOI] [PubMed] [Google Scholar]
- 107.Zhang Y., Han C.Y., Duan F.G., Fan X.-X., Yao X.-J., Parks R.J., Tang Y.-J., Wang M.-F., Liu L., Tsang B.K., et al. P53 Sensitizes Chemoresistant Non-Small Cell Lung Cancer via Elevation of Reactive Oxygen Species and Suppression of EGFR/PI3K/AKT Signaling. Cancer Cell Int. 2019;19:1–13. doi: 10.1186/s12935-019-0910-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ma X., Le Teuff G., Lacas B., Tsao M.S., Graziano S., Pignon J.-P., Douillard J.-Y., Le Chevalier T., Seymour L., Filipits M., et al. Prognostic and Predictive Effect of TP53 Mutations in Patients with Non–Small Cell Lung Cancer from Adjuvant Cisplatin–Based Therapy Randomized Trials: A LACE-Bio Pooled Analysis. J. Thorac. Oncol. 2016;11:850–861. doi: 10.1016/j.jtho.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 109.Liu S.-Y., Bao H., Wang Q., Mao W.-M., Chen Y., Tong X., Xu S.-T., Wu L., Wei Y.-C., Liu Y.-Y., et al. Genomic Signatures Define Three Subtypes of EGFR-Mutant Stage II–III Non-Small-Cell Lung Cancer with Distinct Adjuvant Therapy Outcomes. Nat. Commun. 2021;12:1–11. doi: 10.1038/s41467-021-26806-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ma X., Rousseau V., Sun H., Lantuejoul S., Filipits M., Pirker R., Popper H., Mendiboure J., Vataire A.-L., Le Chevalier T., et al. Significance ofTP53 Mutations as Predictive Markers of Adjuvant Cisplatin-Based Chemotherapy in Completely Resected Non-Small-Cell Lung Cancer. Mol. Oncol. 2014;8:555–564. doi: 10.1016/j.molonc.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Molinavila M.A., Bertran-Alamillo J., Gascó A., Mayo-De-Las-Casas C., Sánchez-Ronco M., Pujantell-Pastor L., Bonanno L., Favaretto A., Cardona A.F., Vergnenègre A., et al. Nondisruptive P53 Mutations Are Associated with Shorter Survival in Patients with Advanced Non–Small Cell Lung Cancer. Clin. Cancer Res. 2014;20:4647–4659. doi: 10.1158/1078-0432.CCR-13-2391. [DOI] [PubMed] [Google Scholar]
- 112.Aggarwal C., Davis C.W., Mick R., Thompson J.C., Ahmed S., Jeffries S., Bagley S., Gabriel P., Evans T.L., Bauml J.M., et al. Influence of TP53 Mutation on Survival in Patients with Advanced EGFR-Mutant Non–Small-Cell Lung Cancer. JCO Precis. Oncol. 2018;2018:1–29. doi: 10.1200/PO.18.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Aisner D.L., Sholl L.M., Berry L.D., Rossi M.R., Chen H., Fujimoto J., Moreira A.L., Ramalingam S.S., Villaruz L.C., Otterson G.A., et al. The Impact of Smoking and TP53 Mutations in Lung Adenocarcinoma Patients with Targetable Mutations—The Lung Cancer Mutation Consortium (LCMC2) Clin. Cancer Res. 2017;24:1038–1047. doi: 10.1158/1078-0432.CCR-17-2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Canale M., Petracci E., Delmonte A., Chiadini E., Dazzi C., Papi M., Capelli L., Casanova C., De Luigi N., Mariotti M., et al. Impact of TP53 Mutations on Outcome in EGFR-Mutated Patients Treated with First-Line Tyrosine Kinase Inhibitors. Clin. Cancer Res. 2016;23:2195–2202. doi: 10.1158/1078-0432.CCR-16-0966. [DOI] [PubMed] [Google Scholar]
- 115.Canale M., Petracci E., Delmonte A., Bronte G., Chiadini E., Ludovini V., Dubini A., Papi M., Baglivo S., De Luigi N., et al. Concomitant TP53 Mutation Confers Worse Prognosis in EGFR-Mutated Non-Small Cell Lung Cancer Patients Treated with TKIs. J. Clin. Med. 2020;9:1047. doi: 10.3390/jcm9041047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liu Y., Xu F., Wang Y., Wu Q., Wang B., Yao Y., Zhang Y., Han-Zhang H., Ye J., Zhang L., et al. Mutations in Exon 8 of TP53 Are Associated with Shorter Survival in Patients with Advanced Lung Cancer. Oncol. Lett. 2019;18:3159–3169. doi: 10.3892/ol.2019.10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Labbé C., Cabanero M., Korpanty G.J., Tomasini P., Doherty M.K., Mascaux C., Jao K., Pitcher B., Wang R., Pintilie M., et al. Prognostic and Predictive Effects of TP53 Co-Mutation in Patients with EGFR-Mutated Non-Small Cell Lung Cancer (NSCLC) Lung Cancer. 2017;111:23–29. doi: 10.1016/j.lungcan.2017.06.014. [DOI] [PubMed] [Google Scholar]
- 118.Ko J.-L., Cheng Y.-W., Chang S.-L., Su J.-M., Chen C.-Y., Lee H. MDM2 mRNA Expression Is a Favorable Prognostic Factor in Non-Small-Cell Lung Cancer. Int. J. Cancer. 2000;89:265–270. doi: 10.1002/1097-0215(20000520)89:3<265::AID-IJC9>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 119.Rachiglio A.M., Fenizia F., Piccirillo M.C., Galetta D., Crinò L., Vincenzi B., Barletta E., Pinto C., Ferraù F., Lambiase M., et al. The Presence of Concomitant Mutations Affects the Activity of EGFR Tyrosine Kinase Inhibitors in EGFR-Mutant Non-Small Cell Lung Cancer (NSCLC) Patients. Cancers. 2019;11:341. doi: 10.3390/cancers11030341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Michels S., Heydt C., Van Veggel B., Deschler-Baier B., Pardo N., Monkhorst K., Rüsseler V., Stratmann J., Griesinger F., Steinhauser S., et al. Genomic Profiling Identifies Outcome-Relevant Mechanisms of Innate and Acquired Resistance to Third-Generation Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitor Therapy in Lung Cancer. JCO Precis. Oncol. 2019;3:1–14. doi: 10.1200/PO.18.00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jin Y., Shi X., Zhao J., He Q., Chen M., Yan J., Ou Q., Wu X., Shao Y.W., Yu X. Mechanisms of Primary Resistance to EGFR Targeted Therapy in Advanced Lung Adenocarcinomas. Lung Cancer. 2018;124:110–116. doi: 10.1016/j.lungcan.2018.07.039. [DOI] [PubMed] [Google Scholar]
- 122.Kim Y., Lee B., Shim J.H., Lee S.-H., Park W.-Y., Choi Y.-L., Sun J.-M., Ahn J.S., Ahn M.-J., Park K. Concurrent Genetic Alterations Predict the Progression to Target Therapy in EGFR-Mutated Advanced NSCLC. J. Thorac. Oncol. 2019;14:193–202. doi: 10.1016/j.jtho.2018.10.150. [DOI] [PubMed] [Google Scholar]
- 123.Bria E., Pilotto S., Amato E., Fassan M., Novello S., Peretti U., Vavalà T., Kinspergher S., Righi L., Santo A., et al. Molecular Heterogeneity Assessment by Next-Generation Sequencing and Response to Gefitinib of EGFR Mutant Advanced Lung Adenocarcinoma. Oncotarget. 2015;6:12783–12795. doi: 10.18632/oncotarget.3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Helena A.Y., Suzawa K., Jordan E.J., Zehir A., Ni A., Kim H.R., Kris M.G., Hellmann M.D., Li B.T., Somwar R., et al. Concurrent Alterations in EGFR-Mutant Lung Cancers Associated with Resistance to EGFR Kinase Inhibitors and Characterization of MTOR as a Mediator of Resistance. Clin. Cancer Res. 2018;24:3108–3118. doi: 10.1158/1078-0432.ccr-17-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Roeper J., Falk M., Chalaris-Rißmann A., Lueers A.C., Ramdani H., Wedeken K., Stropiep U., Diehl L., Tiemann M., Heukamp L.C., et al. TP53 Co-Mutations in EGFR Mutated Patients in NSCLC Stage IV: A Strong Predictive Factor of ORR, PFS and OS in EGFR mt+ NSCLC. Oncotarget. 2020;11:250–264. doi: 10.18632/oncotarget.27430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Xu Y., Tong X., Yan J., Wu X., Shao Y.W., Fan Y. Short-Term Responders of Non–Small Cell Lung Cancer Patients to EGFR Tyrosine Kinase Inhibitors Display High Prevalence of TP53 Mutations and Primary Resistance Mechanisms. Transl. Oncol. 2018;11:1364–1369. doi: 10.1016/j.tranon.2018.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hou H., Qin K., Liang Y., Zhang C., Liu D., Jiang H., Liu K., Zhu J., Lv H., Li T., et al. Concurrent TP53 Mutations Predict Poor Outcomes of EGFR-TKI Treatments in Chinese Patients with Advanced NSCLC. Cancer Manag. Res. 2019;11:5665–5675. doi: 10.2147/CMAR.S201513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lim S.M., Kim H.R., Cho E.K., Min Y.J., Ahn J.S., Ahn M.-J., Park K., Cho B.C., Lee J.-H., Jeong H.C., et al. Targeted Sequencing Identifies Genetic Alterations That Confer Primary Resistance to EGFR Tyrosine Kinase Inhibitor (Korean Lung Cancer Consortium) Oncotarget. 2016;7:36311–36320. doi: 10.18632/oncotarget.8904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Tsui D.W.Y., Murtaza M., Wong A.S.C., Rueda O.M., Smith C.G., Chandrananda D., Soo R.A., Lim H.L., Goh B.C., Caldas C., et al. Dynamics of Multiple Resistance Mechanisms in Plasma DNA during EGFR-Targeted Therapies in Non-Small Cell Lung Cancer. EMBO Mol. Med. 2018;10:e7945. doi: 10.15252/emmm.201707945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Qin K., Hou H., Liang Y., Zhang X. Prognostic Value of TP53 Concurrent Mutations for EGFR-TKIs and ALK-TKIs Based Targeted Therapy in Advanced Non-Small Cell Lung Cancer: A Meta-Analysis. BMC Cancer. 2020;20:1–16. doi: 10.1186/s12885-020-06805-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhang R., Tian P., Chen B., Wang T., Li W. The Prognostic Impact of TP53 Comutation in EGFR Mutant Lung Cancer Patients: A Systematic Review and Meta-Analysis. Postgrad. Med. 2019;131:199–206. doi: 10.1080/00325481.2019.1585690. [DOI] [PubMed] [Google Scholar]
- 132.Wang F., Zhao N., Gao G., Deng H.-B., Wang Z.-H., Deng L.-L., Yang Y., Lu C. Prognostic Value of TP53 Co-Mutation Status Combined with EGFR Mutation in Patients with Lung Adenocarcinoma. J. Cancer Res. Clin. Oncol. 2020;146:2851–2859. doi: 10.1007/s00432-020-03340-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Choi Y.W., Jeon S.Y., Jeong G.S., Lee H.W., Jeong S.H., Kang S.Y., Park J.S., Choi J.-H., Koh Y.W., Han J.H., et al. EGFR Exon 19 Deletion Is Associated with Favorable Overall Survival after First-Line Gefitinib Therapy in Advanced Non–Small Cell Lung Cancer Patients. Am. J. Clin. Oncol. 2018;41:385–390. doi: 10.1097/COC.0000000000000282. [DOI] [PubMed] [Google Scholar]
- 134.Hong W., Wu Q., Zhang J., Zhou Y. Prognostic Value of EGFR 19-del and 21-L858R Mutations in Patients with Non-Small Cell Lung Cancer. Oncol. Lett. 2019;18:3887–3895. doi: 10.3892/ol.2019.10715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.VanderLaan P., Rangachari D., Mockus S.M., Spotlow V., Reddi H.V., Malcolm J., Huberman M.S., Joseph L.J., Kobayashi S.S., Costa D.B. Mutations in TP53, PIK3CA, PTEN and Other Genes in EGFR Mutated Lung Cancers: Correlation with Clinical Outcomes. Lung Cancer. 2017;106:17–21. doi: 10.1016/j.lungcan.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yu R., Bai H., Li T., Gao B., Han J., Chang G., Zhang P., Fei K., He X., Wang J. TP53 Mutations in Circulating Tumor DNA in Advanced Epidermal Growth Factor Receptor-Mutant Lung Adenocarcinoma Patients Treated with Gefitinib. Transl. Oncol. 2021;14:101163. doi: 10.1016/j.tranon.2021.101163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Li X.-M., Li W.-F., Lin J.-T., Yan H.-H., Tu H.-Y., Chen H.-J., Wang B.-C., Wang Z., Zhou Q., Zhang X.-C., et al. Predictive and Prognostic Potential of TP53 in Patients with Advanced Non–Small-Cell Lung Cancer Treated with EGFR-TKI: Analysis of a Phase III Randomized Clinical Trial (CTONG 0901) Clin. Lung Cancer. 2020;22:100–109. doi: 10.1016/j.cllc.2020.11.001. [DOI] [PubMed] [Google Scholar]
- 138.Offin M., Chan J.M., Tenet M., Rizvi H.A., Shen R., Riely G.J., Rekhtman N., Daneshbod Y., Quintanal-Villalonga A., Penson A., et al. Concurrent RB1 and TP53 Alterations Define a Subset of EGFR-Mutant Lung Cancers at Risk for Histologic Transformation and Inferior Clinical Outcomes. J. Thorac. Oncol. 2019;14:1784–1793. doi: 10.1016/j.jtho.2019.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lee J.-K., Lee J., Kim S., Kim S., Youk J., Park S., An Y., Keam B., Kim D.-W., Heo D.S., et al. Clonal History and Genetic Predictors of Transformation into Small-Cell Carcinomas from Lung Adenocarcinomas. J. Clin. Oncol. 2017;35:3065–3074. doi: 10.1200/JCO.2016.71.9096. [DOI] [PubMed] [Google Scholar]
- 140.Niederst M.J., Sequist L.V., Poirier J.T., Mermel C.H., Lockerman E.L., Garcia A.R., Katayama R., Costa C., Ross K.N., Moran T., et al. RB Loss in Resistant EGFR Mutant Lung Adenocarcinomas That Transform to Small-Cell Lung Cancer. Nat. Commun. 2015;6:6377. doi: 10.1038/ncomms7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Piper-Vallillo A.J., Sequist L.V., Piotrowska Z. Emerging Treatment Paradigms for EGFR-Mutant Lung Cancers Progressing on Osimertinib: A Review. J. Clin. Oncol. 2020;38:2926–2936. doi: 10.1200/JCO.19.03123. [DOI] [PubMed] [Google Scholar]
- 142.Clinical Lung Cancer Genome Project The Clinical Lung Cancer Genome Project (CLCGP) and Network Genomic Medicine (NGM) A Genomics-Based Classification of Human Lung Tumors. Sci. Transl. Med. 2013;5:209ra153. doi: 10.1126/scitranslmed.3006802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Jung S., Kim D.H., Choi Y.J., Kim S.Y., Park H., Lee H., Choi C.-M., Sung Y.H., Lee J.C., Rho J.K. Contribution of P53 in Sensitivity to EGFR Tyrosine Kinase Inhibitors in Non-Small Cell Lung Cancer. Sci. Rep. 2021;11:1–10. doi: 10.1038/s41598-021-99267-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Pailler E., Oulhen M., Borget I., Remon J., Ross K., Auger N., Billiot F., Camus M.N., Commo F., Lindsay C.R., et al. Circulating Tumor Cells with Aberrant ALK Copy Number Predict Progression-Free Survival during Crizotinib Treatment in ALK-Rearranged Non–Small Cell Lung Cancer Patients. Cancer Res. 2017;77:2222–2230. doi: 10.1158/0008-5472.CAN-16-3072. [DOI] [PubMed] [Google Scholar]
- 145.Wei Y., Shen K., Lv T., Wang X., Li C., Fan H., Lv Y., Liu H., Song Y. Three New Disease-Progression Modes in NSCLC Patients after EGFR-TKI Treatment by Next-Generation Sequencing Analysis. Lung Cancer. 2018;125:43–50. doi: 10.1016/j.lungcan.2018.08.028. [DOI] [PubMed] [Google Scholar]
- 146.Blakely C.M., Watkins T.B.K., Wu W., Gini B., Chabon J.J., McCoach C.E., McGranahan N., Wilson G.A., Birkbak N., Olivas V.R., et al. Evolution and Clinical Impact of Co-Occurring Genetic Alterations in Advanced-Stage EGFR-Mutant Lung Cancers. Nat. Genet. 2017;49:1693–1704. doi: 10.1038/ng.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available in the article.