Abstract
We have previously suggested a central role for mitochondria in the observed sex differences in metabolic traits. However, the mechanisms by which sex differences affect adipose mitochondrial function and metabolic syndrome are unclear. Here we show that in both mice and humans, adipose mitochondrial functions are elevated in females and are strongly associated with adiposity, insulin resistance and plasma lipids. Using a panel of diverse inbred strains of mice, we identify a genetic locus on mouse chromosome 17 that controls mitochondrial mass and function in adipose tissue in a sex- and tissue-specific manner. This locus contains Ndufv2 and regulates the expression of at least 89 mitochondrial genes in females, including oxidative phosphorylation genes and those related to mitochondrial DNA content. Overexpression studies indicate that Ndufv2 mediates these effects by regulating supercomplex assembly and elevating mitochondrial reactive oxygen species production, which generates a signal that increases mitochondrial biogenesis.
To address the complexity of common metabolic disorders such as obesity, insulin resistance and dyslipidaemias, we have employed a ‘systems genetics’ approach in which high-throughput ‘omics’ technologies, such as transcriptomics, are used quantitatively to assess the many molecular differences that occur among individuals in a population and then to relate these to physiological functions or disease states1. Mouse models offer important advantages for such studies as environmental factors, particularly the diet, can be controlled, relevant tissues can be examined and experimental perturbations can be performed. We previously developed and characterized a systems genetics population resource, termed the hybrid mouse diversity panel (HMDP), that consists of a panel of approximately 100 genetically diverse inbred mouse strains2. Important advantages of this resource include the ability to carry out high-resolution association mapping, analogous to human genome-wide association studies (GWAS) and, as the mice are members of inbred strains, to replicate data and examine different environments on identical genetic backgrounds. During the past 10 years, we have characterized both male and female HMDP mice for many metabolic and cardiovascular traits3–13.
In the present study, we used this resource to determine how genetic variations and sex differences affect mitochondria and their functions, and how these in turn affect metabolic phenotypes. Mitochondria clearly play major roles in cardiometabolic diseases14–18, but the causal relationships of mitochondrial function to these traits are not well understood. Similarly, sex differences in metabolic phenotypes have been amply described in mice, humans and other species, with females generally exhibiting more beneficial metabolic profiles, but the underlying mechanisms remain unclear4,19–22. Notably, most detailed studies have employed a single genetic background (strain C57BL/6 J) with no regard to genetic variation. Recently, using the HMDP resource, we reported that in mice, female adipose is enriched for mitochondria-related gene networks and that mitochondrial functions exhibit gene-by-sex interactions11.
Mitochondria are unique organelles made up of proteins encoded by two genomes. The mitochondrial genome, composed of a circular DNA genome (mitochondrial DNA, ∼16.5 kb in vertebrates) encoding essential subunits of respiratory complexes I, III, IV and V, as well as two ribosomal and 22 transfer RNAs, is required for translation by mitoribosomes within the matrix. However, the vast majority of mitochondrial proteins, approximately 1200 in all, are encoded by the nuclear genome. Mitochondria not only provide energy for the cells through ATP production, but they also generate metabolites for biosynthesis, protein modification and signal transduction. In addition, in certain cell types such as brown adipocytes and white adipocytes that have undergone ‘browning’, mitochondria produce heat by futile cycling23. Therefore, an understanding of the sex differences regulating these transcripts that in turn control metabolic traits could not only reveal targets for treatment but would also be helpful for diagnosis and personalized medicine.
We now report that tissue-specific sex differences in mitochondria occur in adipose tissues in both mice and humans, and that they are strongly associated with metabolic syndrome traits in both species. We carried out mapping studies in the HMDP to identify the basis of the sex-specific differences in mitochondrial gene expression and uncovered a female-specific genetic locus in adipose that significantly controls ~10% of the approximately 1,200 mitochondria-related transcriptomes, with one-third belonging to oxidative phosphorylation (OXPHOS). Furthermore, this genetic locus was significantly associated with obesity-related traits in a female-specific manner. Using two different inbred strains, we demonstrate that adipose Ndufv2 is the causal candidate gene for this locus in regulating obesity via promoting highly functional mitochondria and/or transcripts in a strain-by-sex manner. Our study supports a causal relationship between adipose mitochondrial function and cardiometabolic traits.
Results
Sex-specific adipose mitochondrial expression in both mice and humans.
Using three common inbred strains, we recently reported that female mice had higher mitochondrial function in their adipose tissue and that gene-by-sex interactions affecting adipose mitochondrial functions contributed majorly to the sex differences in metabolic traits11. Mitochondria are best known for their role in ATP production via OXPHOS. Therefore, to explore the functional relevance of these gene-by-sex interactions, we focused on the gene expression profiles of all nuclear-encoded adipose OXPHOS genes (91 genes, 134 probes) between the two sexes of HMDP. We observed that, in general, female HMDP mice had higher adipose expression of OXPHOS genes (75 genes, 110 probes; 82% of OXPHOS genes) compared to males (Fig. 1a). In contrast, no female-specific enrichments were observed in liver expression of OXPHOS genes (Fig. 1b).
Fig. 1 |. Sex and tissue-specific expression profiles of oXPHoS genes in both mice and humans.
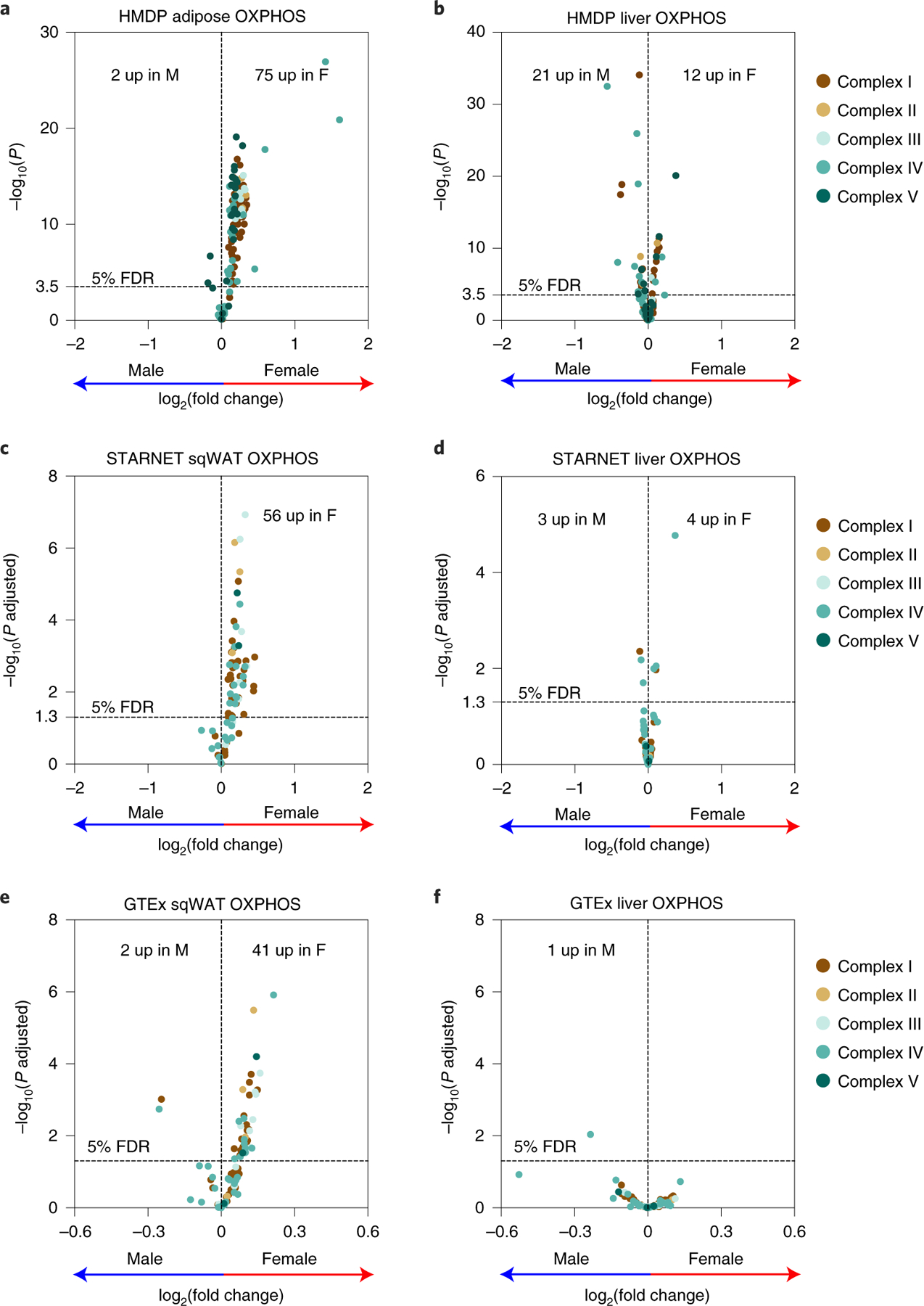
a–f, Volcano plots showing sex differences in the expression profiles of OXPHOS genes in a, adipose (n = 98 sex-matched strains) and b, liver tissues (n = 97 sex-matched strains) isolated from HMDP. To check for translational relevance, we examined the sex differences in the expression profiles of OXPHOS genes in the c, subcutaneous adipose and d, liver tissues from the STARNET cohort (n = 600 patients) or e, subcutaneous adipose and f, liver tissues from the human GTEx cohort (n = 663 adipose or 226 liver donors), respectively. Genes corresponding to individual OXPHOS complexes are colour coded. Horizontal dotted lines represent 5% false discovery rate (FDR)-corrected significance threshold. Data are presented as log2(fold change) between sex. P values were calculated using DESeq2 Bioconductor
To examine the relevance in humans of our sex and tissue-specific observations in mice, we examined the expression of OXPHOS-related genes in 600 human patient samples from the Stockholm-Tartu Atherosclerosis Reverse Networks Engineering Task (STARNET) study24. We observed similar tissue-by-sex interactions in human samples, with female subcutaneous adipose having higher expression of OXPHOS genes (56 genes; 67% of OXPHOS genes) compared to males (Fig. 1c) but not in liver tissues (Fig. 1d). We also independently examined both the human subcutaneous adipose and liver tissues from the GTEx Project data25 and saw similar observations with only the adipose having higher OXPHOS gene expression (41 genes; 44% of OXPHOS genes) in females (Fig. 1e) but no differences between sexes in the liver tissues (Fig. 1f). Taken together, we conclude that female adipose tissues have increased expression of mitochondrial OXPHOS genes in both mice and humans.
Adipose mitochondria levels strongly predict metabolic syndrome traits in both mice and humans.
To understand further whether this sex and tissue-specific regulation translates to metabolic traits, we used adipose mtDNA copy number, as a surrogate measure for mitochondrial mass26, across the HMDP. We quantified mtDNA content from 90 female and 104 male strains (216 female and 260 male mice) of HMDP mice and observed that females were significantly enriched with mtDNA compared to males (Fig. 2a). We also observed that mtDNA levels were strongly associated with metabolic traits (Supplementary Table 1); for instance, adipose mtDNA copy number was negatively correlated to both body weight (Fig. 2b) and homoeostatic model assessment for insulin resistance (HOMA-IR) (Fig. 2c) in both sexes, albeit more strongly in females.
Fig. 2 |. Adipose mitochondria levels strongly predict metabolic traits in both mice and humans.

a, Sex differences in the adipose mtDNA levels isolated from HMDP (n = 90 female and 104 male strains). Bicorrelation plots of female or male adipose mtDNA levels and b, body weight or c, HOMA-IR levels in HMDP. d, Distribution of mtDNA levels isolated from adipose (n = 483) and blood (n = 179) from the human Metabolic Syndrome in Men (METSIM) cohort. Bicorrelation plots of adipose or blood mtDNA levels and e, body mass index or f, HOMA-IR levels in METSIM. g, List of bicorrelations between adipose or blood mtDNA levels and multiple metabolic traits in the METSIM cohort. Red and blue dots represent female and male HMDP datapoints, respectively. Yellow and grey dots (or bars) represent adipose and blood from METSIM, respectively. Data are presented as median and interquartile range (boxplots). P values were calculated using (a) unpaired two-tailed Student’s t-test; (b, c, e–g) BicorAndPvalue function of the WGCNA R-package that uses unpaired two-tailed Student’s t-test. See also Supplementary Tables 1 and 2.
To examine the translational relevance of our mouse adipose mitochondrial associations, we quantitated the mtDNA content in both adipose and blood tissues collected from a well-characterized human cohort of the Finnish population, the METSIM study27 (Fig. 2d). Using biweight midcorrelation (bicor), we observed that human adipose mtDNA copy number was negatively correlated to body mass index (P = 2.72 × 10−6) (Fig. 2e), HOMA-IR (P = 1.49 × 10−7) (Fig. 2f) and plasma triglycerides (P = 8.54 × 10−8) and positively associated with high-density lipoprotein cholesterol levels (P = 1.68 × 10−9) (Fig. 2g). In contrast, the human blood mtDNA copy number was not correlated with any of these traits (Fig. 2g). All possible associations are listed in Supplementary Table 2. As a coordinated upregulation of mtDNA content and OXPHOS gene expression is a strong indication of elevated mitochondrial biogenesis, we conclude that adipose tissue mitochondrial mass is a strong predictor of metabolic traits in both mice and humans.
Sex and tissue-specific trans-regulatory hotspot in adipose mitochondria.
To understand further how natural genetic variations control sex-specific regulation of mitochondrial biogenesis and their contribution to metabolic traits, we focused on the expression profiles of nuclear-encoded mitochondrial genes, listed in MitoCarta2.028 in adipose tissues extracted from both sexes of approximately 100 strains of HMDP fed a high fat, high sucrose (HF/HS) diet. Out of 1,158 genes listed in MitoCarta2.0, we retrieved expression data for 911 genes (1,312 probes) and then mapped their respective expression abundance levels to the HMDP genotypes. We refer to the genetic variants that alter expression levels of nearby genes (<1 Mb) as cis (local) expression quantitative trait loci (eQTLs) and the remainder as trans (distal) eQTLs. When a trans-eQTL locus affects the expression of multiple genes, we defined it as a trans-eQTL hotspot. Based on these criteria, we identified both cis and trans regulated nuclear-encoded mitochondrial genes in a sex-specific manner (Fig. 3a,b), with a prominent trans-eQTL hotspot located on chromosome (chr) 17 in female adipose (Fig. 3b). P value distribution of trans associations between mitochondria genes (1,312 probes) and the lead expression single nucleotide polymorphism (eSNP) (rs48062344) of chr17 trans-eQTL hotspot revealed strong enrichment of low P values only in female but not male adipose. This enrichment suggested that the chr17 locus is a female-specific master regulator of adipose mitochondrial biogenesis (Fig. 3c). In contrast, when we examined the eQTL networks in HMDP liver tissues, we found no such associations (Extended Data Fig. 1), thereby revealing a sex and tissue-specific regulation. When we focused on the trans-eQTL hub located on chr17, we observed that it significantly controlled the expression of 89 genes (P < 1 × 10−6) in female adipose tissue, representing approximately 10% of the available mitochondrial genes (Fig. 3d). Among these, 28 trans-genes (one-third) belong to nuclear-encoded OXPHOS genes.
Fig. 3 |. Sex-specific genetic architecture of adipose mitochondrial gene expression.

High-resolution association mapping of 911 (1,312 probes) mitochondrial gene abundance levels from a, male and b, female adipose tissues isolated from ~100 HMDP strains to identify eQTL networks. The x and y axis represent SNP and gene position on the mouse genome, respectively. Each dot represents a significant association. Associations between gene expression and genetic variants located within 1 Mb of the respective gene location were considered as cis-eQTLs (P < 1 × 10−5) and are shown along the diagonal axis and the rest were considered as trans-eQTLs (P < 1 × 10−6). Based on these criteria, we uncovered a distinct female-specific trans-eQTL hotspot located on chr17 that significantly controls 89 adipose genes (red arrow). c, Frequency distributions of P values of association between the chr17 trans-eQTL lead eSNP (rs48062344) and mitochondria-related transcripts in the male and female adipose tissues. d, Circos plot representation of the female-specific chr17 trans-eQTL hotspot and the genomic location of 89 target adipose mitochondrial genes (P < 1 × 10−6). P values of association (a–d) were calculated using factored spectrally transformed linear mixed models (FaST-LMM) that uses the likelihood ratio test. See also Extended Data Fig. 1.
Chr17 trans-eQTL controls metabolic syndrome traits in HMDP.
To understand further the sex and tissue-specific regulation mediated by the chr17 lead eSNP (rs48062344), we focused on the expression profiles of all nuclear-encoded adipose OXPHOS genes. We observed that only female cohorts harbouring TT genotypes had higher adipose expression of OXPHOS genes (46 genes, 65 probes; 51% of OXPHOS genes) compared to cohorts harbouring CC genotypes (Fig. 4a). In contrast, no such genetic associations were observed in the expression profiles of OXPHOS genes in male adipose or liver from both sexes (Extended Data Fig. 2). In addition to gene expression, our analyses also show that among the females, cohorts that harbour TT genotype had more mtDNA compared to CC genotype (Fig. 4b). Again, we did not observe any associations in male HMDP (Fig. 4f). Furthermore, the chr17 locus appears to control mtDNA copy number, as the peak SNP (rs48062344) exhibited a strong association (P = 6.06 × 10−5) in females, but not males (P = 0.57) (Extended Data Fig. 3).
Fig. 4 |. Chr17 trans-eQTL controls metabolic syndrome traits in HMDP.

a, Volcano plots showing genetic (lead SNP of chr17 locus) differences in the expression profiles of OXPHOS genes in the female adipose (n = 98 female strains; CC: 56 and TT: 42). Genotype distribution plots of mtDNA levels in the b, female and c, male adipose at chr17 trans-eQTL lead SNP, respectively (n = 89 female (CC: 51 and TT: 38) and 104 male (CC: 57 and TT: 47) strains). d, Sex differences in the variance explained by the chr17 trans-eQTL lead SNP (rs48062344) for multiple metabolic traits in HMDP. Genotype distribution plots of gonadal white adipose tissue (gWAT) weights in the e, female and f, male HMDP at chr17 trans-eQTL lead SNP, respectively (n = 98 sex-matched strains; CC: 56 and TT: 42). g, The Ndufv2 gene locus is shown on mouse chromosome 17 from 66 to 68 Mb. Linkage disequilibrium (LD), calculated by R2 to gonadal gWAT s48062344 (green vertical line), which was the chr17 trans-eQTL lead SNP as well as peak cis-eQTL for Ndufv2, is shown in the upper triangular heatmap. Red pixels indicate complete LD between pairwise genomic segments. The LD block from 66 to 67.5 Mb was subjected to motif mutation analysis for the ERE motif and the top two variants, rs3713670 and rs46900561, whose reference (C57BL6/J) alleles decrease ERE motif scores are highlighted by red and yellow arrows, respectively. The ERE motif position weight matrix is shown beneath each sequence alignment. h, Genotype distribution plots of gWAT weights in the female and male HMDP at the top variant, rs3713670 (n = 102 female (AA: 49 and TT: 53) and 111 male (AA: 54 and GG: 57) strains). Red and blue dots (or bars) represent female and male HMDP datapoints (or associations), respectively. Data are presented as median and interquartile range (boxplots). P values were calculated using DESeq2 Bioconductor package that uses Wald test (a) or unpaired two-tailed Student’s t-test (b, c, e, f and h). See also Extended Data Figs. 2 and 3 and Supplementary Table 3.
Combining the previous two results, we hypothesized that the female cohorts harbouring TT genotype had higher mitochondrial biogenesis in their adipose tissues resulting in lower metabolic burden. To test this, we performed genotype-by-phenotype associations (GWAS) in HMDP. We report that the trans-acting lead eSNP (rs48062344) explains about 30–40% of the variance of several metabolic traits in a female-specific manner, with gWAT weight shown as an example (Fig. 4d). Among the females, gWAT weights were significantly lower in cohorts harbouring TT genotype (Fig. 4e). No such genotype differences were observed in males (Fig. 4f).
Adipose Ndufv2 mediates Chr17 trans-eQTL effects in a sex-specific manner.
To identify the potential cis-effector gene mediating the effects of chr17 trans-eQTL control, we used three criteria: (1) the trans-acting lead eSNP must show cis-regulation; (2) the association occurs only in female adipose tissues; and (3) it has strong associations with metabolic syndrome traits and adipose mtDNA content. Based on this, we identified adipose Ndufv2 as a potential cis-effector gene as it satisfied all these criteria. Secondary candidates included Ralbp1 (not associated with adipose mtDNA levels (P = 0.07)) and Ptprm (not cis-regulated (P = 3.70 × 10−5)).
We reasoned that the genetic variants affecting the oestrogen receptor (ER) binding element (ERE) might underlie the sex-specific regulation of Ndufv2. Therefore, we identified ERE in the region in LD with the trans-eQTL lead eSNP (rs48062344) at chr17:66–67.5 Mb (Fig. 4g). By comparing ERE motif scores between genome sequence with ‘alternate’, non-C57BL6/J alleles, relative to reference alleles, we identified ten SNPs in which alternative sequence matched an ERE motif and scored higher than the reference sequence (Supplementary Table 3). The top two SNPs with greatest predicted consequence on binding affinity are shown in Fig. 4g. In support of these results, when we compared the gWAT weights within female and male HMDP cohorts harbouring differential genotypes at the top SNP (rs3713670), we observed significant genotype differences only in females (Fig. 4h). Thus, these data identify candidate functional SNPs that regulate Ndufv2 transcription via ER genomic binding.
High-resolution association mapping of female adipose Ndufv2 expression revealed that the lead eSNP (rs48062344) is the same as the trans-acting lead eSNP (Fig. 5a). In contrast, Ndufv2 expression in male adipose or in liver of either sex is not genetically cis-regulated (Fig. 5b and Extended Data Fig. 4a,b). When we analysed the expression data of Ndufv2 in 98 sex-matched HMDP strains, female HMDP mice tended to have higher expression compared to males (Fig. 5c), and among the females, cohorts harbouring the TT genotype had higher expression (Fig. 5d). Again, no such associations were observed in male adipose (Fig. 5e) or liver tissues (Extended Data Fig. 4c–e). Further analyses using gonadectomized mice that were fed either a chow or HF/HS diet revealed that Ndufv2 expression is hormonally regulated in females irrespective of the diet (Fig. 5f). In contrast, minor sex-by-diet interactions exist in male adipose (Fig. 5g), while no such hormonal regulations were observed in liver tissues (Fig. 5h,i).
Fig. 5 |. Adipose Ndufv2 is the causal regulator of chr17 trans-eQTL locus.
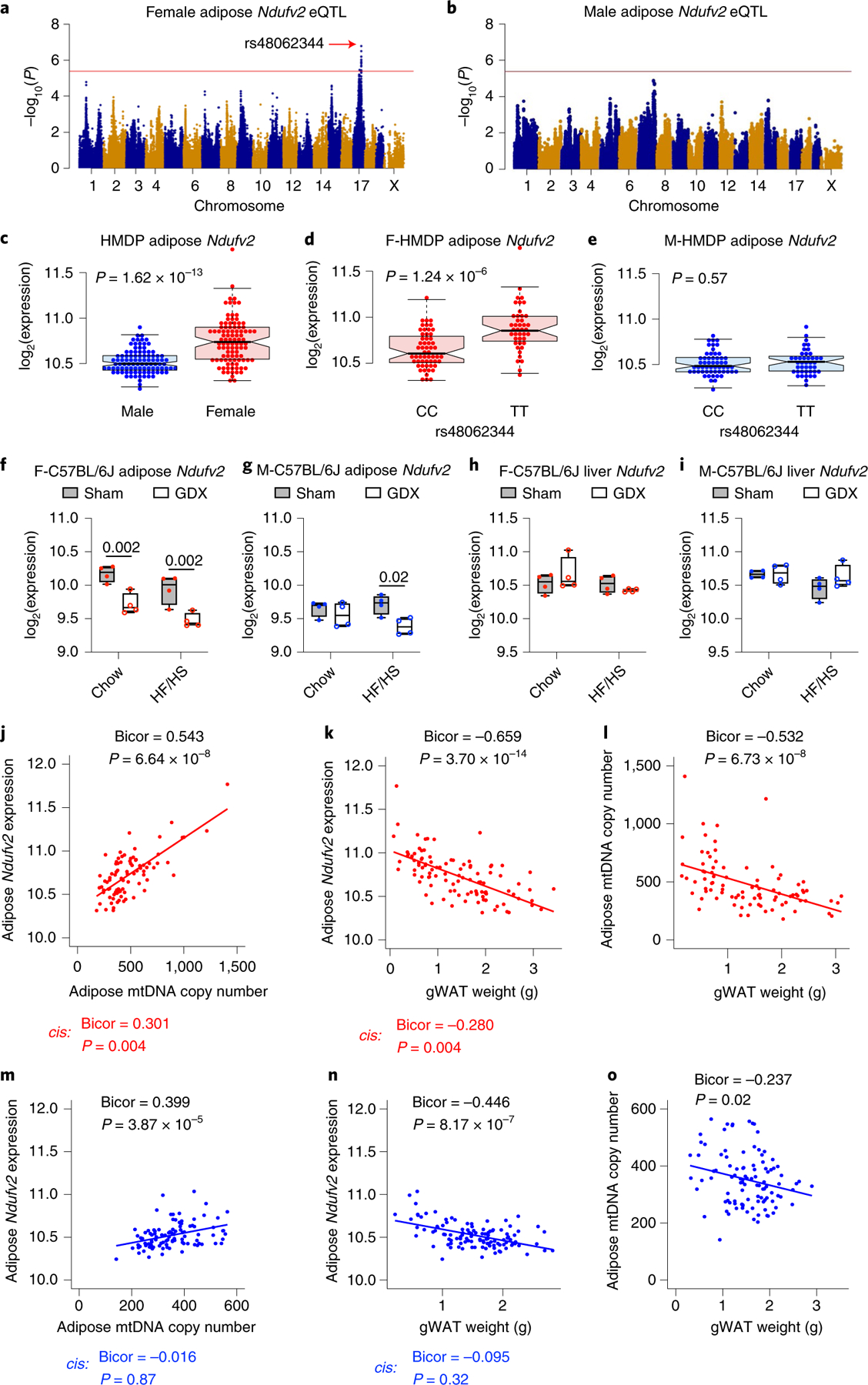
Association mapping of a, female and b, male adipose Ndufv2 expression from HMDP. The red line represents the significance threshold (P = 4.1 × 10−6). Adipose Ndufv2 expression between c, sex or genotypes (lead SNP of chr17 locus) within d, female or e, male HMDP (n = 98 sex-matched strains; CC: 56 and TT: 42). Average Ndufv2 expression in f, female and g, male adipose or h, female and i, male liver isolated from intact or gonadectomized C57BL/6 J mice of both sexes (n = 4 per group). Bicorrelation plots between adipose Ndufv2 and j, mtDNA levels or k, gWAT weights, and l, between adipose mtDNA levels and gWAT weights from female HMDP (n = 86–103 female strains). Similarly, bicorrelation plots between adipose Ndufv2 and m, mtDNA levels or n, gWAT weights, and o, between adipose mtDNA levels and gWAT weights from male HMDP (n = 100–112 male strains). Red and blue dots represent female and male HMDP datapoints, respectively. Data are presented as median and interquartile range (boxplots). P values were calculated using FaST-LMM that uses the likelihood ratio test (a, b), unpaired two-tailed Student’s t-test (c–e), two-factor analysis of variance (ANOVA) corrected by post-hoc Holm–Sidak’s multiple comparisons test (f–i) or BicorAndPvalue function of the WGCNA R-package that uses unpaired two-tailed Student’s t-test (j–o). See also Extended Data Fig. 4.
To relate functionally the trans-acting lead eSNP (rs48062344), adipose Ndufv2 expression, adipose mtDNA content and gWAT weights, we computed bicor between these traits and calculated how the cis component of adipose Ndufv2 expression contributed to these associations. We observed that adipose Ndufv2 expression is positively correlated to adipose mtDNA content (Fig. 5j) and negatively correlated to gWAT weights (Fig. 5k) in females, with the cis component of the expression strongly contributing to both these associations. Finally, we observed that adipose mtDNA copy number is negatively correlated to gWAT weights (Fig. 5l). In contrast, although we observed correlations (not as strong as in females) between male adipose Ndufv2 expression, adipose mtDNA copy number and gWAT weights, respectively, the cis component of the male adipose Ndufv2 expression did not contribute to these associations. In addition, we did not observe strong correlations between adipose mtDNA copy number and gWAT weights in males (Fig. 5m–o). Taken together, we propose that the trans-eSNP effect on female adipose Ndufv2 expression affects weight-related metabolic traits through its effect on mitochondrial biogenesis.
Adipose Ndufv2 is causal for regulating adiposity in a strain-by-sex manner.
To test our hypothesis that Ndufv2 is the causal gene for mediating the trans-SNP effects in regulating fat accumulation, we overexpressed either GFP or NDUFV2 in an adipose-specific manner using adeno-associated viral serotype-8 (AAV8) vectors that contain both adiponectin promoter/enhancer elements for adipose specificity and miR122 target sequences for liver exclusion (AAV8-hAdp-GFP-miR122T or AAV-hAdp-NDUFV2-miR122T). To exclude host genetic bias, we used two different inbred strains harbouring alternate genotypes at chr17 locus lead eSNP (rs48062344), namely, C57BL/6 J (CC genotype) and A/J (TT genotype). Despite the fact that both C57BL/6 J and A/J females overexpressing NDUFV2 ate the same amount of food, C57BL/6 J females only showed a minor decrease, whereas A/J females showed a more pronounced decrease in total body weight (Fig. 6a,f, Extended Data Fig. 5a,c) that was explained by a reduction in body fat mass (Fig. 6b,g and Extended Data Fig. 5a,c). The quantitative PCR (qPCR) analyses in gWAT confirmed increased adipose Ndufv2 expression both in C57BL/6 J and A/J (Fig. 6c,h). To determine whether increased lipolysis was associated with decreased fat mass, we measured the levels of the lipolysis product glycerol in plasma. We observed a trend to a decrease in glycerol levels in C57BL/6 J females overexpressing NDUFV2 (Fig. 6d), which was significant in A/J females overexpressing NDUFV2 (Fig. 6i). Individual tissue weights revealed that gWAT weights were significantly reduced both in C57BL/6 J and A/J females overexpressing NDUFV2 (Fig. 6e,j). As decreased body fat mass is strongly associated with improved insulin sensitivity, we subjected a cohort of A/J mice overexpressing GFP or NDUFV2 to insulin tolerance tests (ITTs). We observed that A/J females overexpressing NDUFV2 had reduced basal/fasting glycaemia when normalized to GFP glycaemia after insulin injection (Extended Data Fig. 5e). That the reduction in fasting glucose was a result of improved insulin sensitivity was confirmed by a decrease in HOMA-IR values and fasting insulinaemia in A/J females overexpressing NDUFV2 (Extended Data Figs. 5f,g). The selective decrease in insulin resistance during fasting is consistent with decreased white adipose tissue (WAT) lipolysis induced by NDFUV2 overexpression29. We observed that C57BL/6 J males overexpressing NDUFV2 had a trend toward increased weight gain, specifically fat mass (Extended Data Figs. 5b and 6a–e). In contrast, A/J males overexpressing NDUFV2 had lower body weight, total fat mass and gWAT weights (Extended Data Figs. 5d and 6f–j). However, NDUFV2 overexpression in A/J males did not change the ITT curves but increased HOMA-IR and fasting insulinaemia (Extended Data Fig. 5h–j), the latter suggesting that NDUFV2 was increasing lipolysis during fasting in A/J males. Taken together, these data support the conclusion that NDUFV2 regulates adipose mass and insulin sensitivity in a sex-by-strain dependent manner, with its gain in function being more beneficial in females of an A/J rather than C57BL/6 J background. Additional genetic considerations might also explain the metabolic differences between A/J and C57BL/6 J, such as the sex-specific regulation of mitochondrial function independently of chromosome 17 and the lack of super assembly complex factor I (SCAFI) in C57BL/6 J30. Interestingly, it has been demonstrated that supercomplex formation can alter lipid metabolism to optimize metabolic resources31.
Fig. 6 |. Adipose Ndufv2 overexpression regulated adiposity in a strain-by-sex manner.

Eight-week-old females of a–e, C57BL/6 J or f–j, A/J mice were injected with AAV vectors expressing either GFP or NDUFV2 in an adipose-specific manner and fed a HF/HS diet for 8 additional weeks. Metabolic traits such as a, total mass and food intake; b, fat and lean mass were monitored over 8 weeks. Comparisons of c, adipose Ndufv2 expression; d, plasma free glycerol; e, tissue weights from C57BL/6 J females overexpressing GFP or NDUFV2, respectively. Similarly, f, total mass and food intake; g, fat and lean mass; h, adipose Ndufv2 expression; i, plasma free glycerol and j, tissue weights were measured from A/J females overexpressing GFP or NDUFV2, respectively. Data are presented as mean ± s.e.m. (n = 7 per group). P values were calculated using repeated measures two-factor (a, f) or three-factor (b, g) or two-factor (e, j) ANOVA corrected by post-hoc Holm–Sidak’s multiple comparisons test, or unpaired two-tailed Student’s t-test (c, d, h, i). See also Extended Data Figs. 5– and 6 and Supplementary Fig. 1.
To determine whether the decrease in fat mass induced by NDUFV2 overexpression was caused by increased energy expenditure (EE), we analysed whole body energy balance in mice overexpressing NDUFV2 in mice using indirect calorimetry. We observed a trend of increased EE and oxygen consumption rate (VO2) in A/J females overexpressing NDUFV2 that did not reach statistical significance by analysis of covariance (ANCOVA) (Supplementary Fig. 1A–J). There were no significant changes between A/J males overexpressing GFP or NDUFV2 (Supplementary Fig. 1K–T). As we observed lower lean mass in A/J females overexpressing NDUFV2, which by itself decreases EE, we considered the possibility that the effect on lean mass was masking an increase in EE in WAT induced by NDUFV2 overexpression. Thus, to test whether an increase in EE in WAT explained decreased body fat, we analysed the effects of NDUFV2 overexpression on mitochondrial biogenesis and EE in isolated WAT.
Adipose Ndufv2 increases WAT mitochondrial biogenesis in a strain-by-sex manner.
Increased OXPHOS transcripts content are strong indicators of elevated mitochondrial biogenesis. Thus, we determined whether NDUFV2 overexpression in WAT was sufficient to change the content of OXPHOS transcripts. We observed that NDUFV2 overexpression induced a robust expression of mitochondrial OXPHOS genes in both sexes and strains, but more pronounced in the A/J strain (Fig. 7a–d). To confirm these data further, we performed RNA sequencing on gonadal adipose tissues and focused on OXPHOS genes. Differential expression analyses revealed that a significant proportion of OXPHOS genes were upregulated in NDUFV2 overexpressing animals (Supplementary Fig. 2A,B). To gain further insight into the mechanisms behind increased mitochondrial biogenesis, we measured Ppargc1a and Ucp1 expression in both C57BL/6 J and A/J females. We studied these genes as important markers of an engagement of the transcriptional programme increasing mitochondrial biogenesis in WAT32–34. UCP1 is a marker of brown/beige adipocytes located within WAT, which have higher mitochondrial content, and Ppargc1a is a transcriptional co-activator that increases mitochondrial biogenesis in multiple tissues. We observed that both Ppargc1a and Ucp1 expression were only increased by NDUFV2 overexpression in A/J but not C57BL/6 J females (Supplementary Fig. 2C–F). Immunoblots of mitochondria OXPHOS proteins corroborated the relevance of the transcriptional upregulation induced by NDUFV2 (Fig. 7e). Altogether, these data were either suggesting that white adipocytes had more mitochondria or that there was an increase in beige adipocytes. To determine whether beiging occurred, we examined NDUFV2 overexpressing WAT by histology. We found that NDUFV2 overexpression in females of both strains shifted the distribution towards smaller adipocytes (Extended Data Fig. 7a,c). Despite being smaller, the adipocytes were unilocular, which advocated against an increase in beige adipocyte recruitment induced by NDUFV2 expression in A/J females. In contrast, NDUFV2 overexpression in C57BL/6 J males shifted the distribution toward larger adipocytes (Extended Data Fig. 7b), while in A/J males there was a subtle shift towards smaller adipocytes (Extended Data Fig. 7d). An analysis of their average sizes yielded similar conclusions (insets of Extended Data Fig. 7).
Fig. 7 |. Adipose Ndufv2 overexpression regulated mitochondria in a strain-by-sex manner.

Relative normalized expression values of Ndufv2 (complex I), Sdhc (complex II), Atp5a1 (complex V) and Cpt1a (FAO) in gonadal adipose tissues from a, female and b, male C57BL/6 J or c, female and d, male A/J mice overexpressing GFP or NDUFV2, respectively (C57BL/6 J n = 5 per group; A/J n = 6 per group). e, Representative immunoblots probed for OXPHOS proteins (CI to CV) and vinculin as a loading control, and their corresponding quantifications (females n = 4 per group; males n = 3 per group). Comparisons of f, mitochondrial RCR and g, coupling efficiency in the gonadal adipose tissues from female C57BL/6 J mice overexpressing GFP or NDUFV2, respectively (n = 4 per group). Similarly, h, RCR and i, coupling efficiency were measured in the gonadal adipose tissues from female A/J mice overexpressing GFP or NDUFV2, respectively (n = 4 per group). Red and blue dots (or bars) represent female and male datapoints, respectively. Data are presented as mean ± s.e.m. P values were calculated using unpaired two-tailed Student’s t-test. See also Extended Data Figs. 7–9 and Supplementary Fig. 2.
Adipose Ndufv2 increases mitochondrial coupling and total mitochondria EE in WAT in a strain-by-sex manner.
We next determined the effects of NDUFV2 expression on mitochondrial EE and uncoupling. Respirometry analyses on isolated mitochondria from WAT with pyruvate/malate as a substrate showed an increased respiratory control ratio (RCR) (state 3/state 4o) (Fig. 7f) and an increased coupling efficiency ([state 3–state 4o]/[state 3–AA]) (Fig. 7g) in C57BL/6 J females overexpressing NDUFV2. The increase in mitochondrial ATP synthesis efficiency was much more pronounced in A/J females (Fig. 7h–i); in contrast, males of both the C57BL/6 J (Extended Data Fig. 8a,b) and A/J (Extended Data Fig. 8c,d) strains exhibited unaltered mitochondrial activities. Elevated coupled respiration suggests that UCP1 is not the major effector of NDUFV2 expression on mitochondrial respiratory function, as indeed UCP1 decreases coupling. Consequently, elevated coupled respiration is a functional demonstration that NDUFV2 mediates a change in WAT mitochondria without inducing beiging35. Moreover, the respirometry data show that the electron transport chain capacity per unit of mitochondria is not increased by NDUFV2. Indeed, the opposite effect was observed, as respiration rates were decreased in all states in both C57BL/6 J and A/J females overexpressing NDUFV2 in WAT (Extended Data Fig. 8e–h). Consequently, an increase in the specific electron transport chain activity per mitochondria or uncoupling cannot explain the decrease in adipose mass. However, isolated mitochondria measurements will miss an increase in total WAT EE licensed by an increase in mitochondrial mass/biogenesis. Therefore, we analysed the effects of NDUFV2 overexpression on mitochondria mass in WAT by staining total WAT lysates with the mitochondrial dye (MitoTracker Deep Red FM (MTDR)), as published36. We note that the MTDR dye can be used to stain mitochondria in fixed cells and frozen samples and therefore can still specifically stain mitochondria that do not have an active membrane potential36. We detected more mitochondria in NDUFV2 overexpressing female A/J WAT lysates, normalized to total protein content (Fig. 8a). In contrast, NDUFV2 overexpression did not affect complex activities or mitochondria content in A/J males (Fig. 8c–d). To obtain a functional confirmation of this increase in mitochondrial mass induced by NDUFV2, we measured total OXPHOS complex activities in these WAT lysates. We observed increased complex-IV activity in total WAT lysates from both A/J and C57BL/6 J mice (Fig. 8b,e), with the effect being larger in A/J and reaching statistical significance for complexes II and IV (P = 0.05) (Fig. 8b). Moreover, immunoblot analyses revealed that fatty acid oxidation proteins, CPT1a and LCAD, were increased in female C57BL/6 J mice, with no changes in UCP1 content (Fig. 8f). In contrast, NDUFV2 overexpressing male C57BL/6 J did not show any significant changes in complex activities or protein levels (Fig. 8g,h). Thus, these data support the conclusion that NDUFV2 induces an increase in mitochondrial biogenesis to support elevated EE in a sex- and tissue-specific manner.
Fig. 8 |. Adipose Ndufv2 overexpression increased mitochondrial biogenesis and protein levels via RoS generation by altering supercomplex composition in a strain-by-sex manner.

Comparisons of a, MTDR staining of the mitochondria and b, frozen respirometry quantifications in frozen gonadal adipose homogenates between GFP and NDUFV2 overexpressing female A/J mice (GFP n = 6; NDUFV2 n = 8). Similarly, c, MTDR staining of the mitochondria and d, frozen respirometry quantifications were measured between GFP and NDUFV2 overexpressing male A/J mice (GFP n = 7; NDUFV2 n = 8). Likewise, e, frozen respirometry quantifications and f, immunoblot analyses of CPT1a, LCAD, UCP1 and vinculin (loading control) levels and their corresponding quantifications from female C57BL/6 J mice (n = 7 per group except UCP1 n = 4 per group) and g, frozen respirometry quantifications and h, immunoblot analyses of CPT1a, LCAD, UCP1 and vinculin (loading control) levels and their corresponding quantifications from male C57BL/6 J mice (n = 8 per group except UCP1 n = 4 per group), respectively, are shown. Finally, i, immunoblot staining of NDUFV2 overexpression; j, oxygen consumption rates before and after the addition of N-acetylcysteine (NAC) (n = 10 replicates per group), and k, blue native gel electrophoreses followed by immunoblotting for NDUFA9 (complex I), Core2 (complex III) and SDHA (complex II) for supercomplex visualization in stably transduced differentiated 3T3-L1 cells are shown. Data are presented as mean ± s.e.m. P values were calculated using two-factor ANOVA corrected by post-hoc Holm–Sidak’s multiple comparisons test (b, d, h, j) or unpaired two-tailed Student’s t-test (a, c, j). See also Extended Data Fig. 10 and Supplementary Fig. 3.
To discern whether these effects on mitochondrial function were upstream or downstream of decreased fat mass, we designed our next experiment such that the GFP and NDUFV2 mice had similar body fat mass. For this, we subjected the C57BL/6 J females to a HF/HS diet for the first 6 weeks without any interventions. After 6 weeks, the animals were randomly injected with AAV carrying either GFP or NDUFV2 and were continued on the diet. Further, to exclude cage effects, all cages had mice that received either of the two viruses. Post-AAV injection, body weight and composition were regularly monitored, and the experiment was ended (week 12) before the animals showed any noticeable differences in their body weight. As designed, we did not observe any significant differences in body weight (Extended Data Fig. 9a) or body composition (Extended Data Fig. 9b) or organ weights (Extended Data Fig. 9c) between the two groups, despite Ndufv2 overexpression (Extended Data Fig. 9d). Yet, the bioenergetic analyses revealed that NDUFV2 females had an increased RCR (Extended Data Fig. 9e) and coupling efficiency (Extended Data Fig. 9f) with reduced respiration rates (Extended Data Fig. 9g). These data support the conclusion that the effects of NDUFV2 on mitochondrial function are not a consequence of decreased fat mass.
To determine whether NDUFV2 overexpression will have similar effects on mitochondria from other tissues, we overexpressed GFP or NDUFV2 in a liver-specific manner using AAV8 vectors that contain thyroxine binding globulin (Tbg) promoter for liver specificity (AAV8-Tbg-GFP or AAV8-Tbg-NDUFV2). Although we did not observe dramatic changes in body weight or WAT due to liver NDUFV2 overexpression, we did observe a similar increase in mitochondrial transcripts, in both females and males, as seen in adipose overexpression (Supplementary Fig. 3). We note that the liver-specific NDUFV2 expression was several fold greater than adipose expression.
NDUFV2 overexpression changes supercomplex assembly and ROS-dependent mitochondrial respiration.
Mitochondrial reactive oxygen species (ROS) were previously demonstrated to decrease mitochondrial respiration acutely in WAT37, with these ROS known to transduce a compensatory signal to increase mitochondrial biogenesis38,39. To test whether ROS was responsible for the effects of NDUFV2 on mitochondria, we determined whether the antioxidant NAC could reverse the actions of NDUFV2. To restrict NAC actions to white adipocytes, we moved to a cell culture model. We overexpressed NDUFV2 in AML12 (liver) and differentiated 3T3-L1 (adipose) cells using plasmid transfection and lentiviral transduction, respectively. The qPCR and immunoblot analyses confirmed NDUFV2 overexpression in AML12 (Extended Data Fig. 10a) and differentiated 3T3-L1 (Fig. 8i), respectively. Similar to our in vivo analyses, our in vitro experiments revealed that NDUFV2 overexpression increased coupled respiration, measured by higher RCR values on different complex-I substrates, in both AML12 and differentiated 3T3-L1 cells (Extended Data Fig. 10). Nevertheless and similar to our in vivo experiments (Extended Data Figs. 8e,g and 9g), we observed lower oxygen consumption rates in both AML12 and differentiated 3T3-L1 cells expressing NDUFV2 (Extended Data Fig. 10). After validating our in vitro system, we added NAC to scavenge ROS and observed that only NDUFV2 overexpressing 3T3-L1 cells increased mitochondrial respiration in response to NAC (Fig. 8j). Moreover, we observed NDUFV2 overexpression increased OXPHOS genes in AML12 cells (Extended Data Fig. 10n). Nevertheless, we were unable to see an increase in mitochondrial mass in vitro possibly due to altered ROS-dependent transcriptional programmes, as previously published40, resulting from the loss of important regulatory mechanisms in 3T3 cells as compared to WAT or the hyperoxic and high glucose environment of cells in culture.
NDUFV2 is a core subunit of complex I, located in the region of NADH binding. Complex I assembles with complex III and IV of the electron transport chain to form supercomplexes, which can increase the rates of electron transfer and decrease ROS production. Thus, one could hypothesize that an excess of NDUFV2 subunits could change complex I or supercomplex assembly to increase ROS production. As evidence exists that complex III dimers are known to reduce ROS production via intermonomer electron transfer41 and that ROS generated from complex III regulates adipocyte differentiation42, we sought to determine whether NDUFV2 affects the supercomplex composition by performing blue native gel electrophoresis on 3T3-L1 cell lysates overexpressing NDUFV2. We observed that NDUFV2 overexpression decreased the number of complex III dimers in differentiated 3T3-L1 cells, while preserving the content of supercomplexes I–III2–IV2 (Fig. 8k). Such a change can be explained by a decrease in complex III dimerization or by an increase in the proportion of complex III dedicated to the respirasome43,44. However, the increase in CI and CIII super assembly reduces ROS generation, supporting the conclusion that the decrease in complex III dimers we observed is reflecting a decrease in complex III dimerization45. Alternatively, it is possible that having a decrease in the availability of free complex III dimers causes the electrons entering via complex II to execute reverse electron transport, namely providing the electrons to complex I instead of complex III. Indeed, reverse electron transport results in increased ROS production from complex I. Taken together, we conclude that mitochondrial ROS generation mediated by NDUFV2 overexpression, via modifying complex III dimerization and distribution between complexes and supercomplexes, is likely to be the signal initiating the increase in mitochondrial biogenesis.
In summary, our in vivo validation studies revealed that adipose Ndufv2 directly regulates adiposity in a strain-by-sex manner via modulation of mitochondrial transcript levels and that it increases mitochondrial ROS by directly affecting the supercomplex composition. It is clear, however, that this is not the only genetic factor influencing mitochondrial functions and sex differences in metabolic traits because its effects are in part dependent on sex and genetic background.
Discussion
Our initial discovery relating adipose mitochondrial function to sex differences in cardiometabolic traits came from systems genetics analyses in the HMDP population11. These studies strongly suggested a central role of mitochondria, and this was confirmed by isolating the mitochondria and showing that adipose tissues of females of most strains exhibited increased respiration11. In the present study, we used the HMDP along with two human cohorts (STARNET and METSIM) to investigate the role of sex and genetics in regulating both mitochondrial content and metabolic syndrome traits. Several conclusions have emerged. First, in both mice and humans (STARNET and GTEx cohort), we identified sex- and tissue-specific regulation of mitochondrial transcripts. Second, we observed a strong relationship between the mitochondrial DNA copy number in adipose and metabolic traits in both mice and humans (METSIM cohort). Third, using systems genetics, we identified a mouse locus that controls the increased mitochondrial function in adipose in a sex-specific manner. Fourth, our mouse GWAS analyses identified a strong candidate gene, Ndufv2, which is regulated by both genetic variation and sex hormones in a tissue-specific fashion that controls, in trans, many nuclear-encoded mitochondrial transcripts in the adipose. Finally, we performed overexpression studies in mice and reported that Ndufv2 can regulate obesity by controlling mitochondrial genes or function in a strain-by-sex manner. We discuss these points in turn below.
An important question is whether sex differences similar to those observed in mice also occur in human populations. There is a paucity of data on sex differences in adipose gene expression, but at least one study found a large sex difference in the expression of UCP1, involved in proton futile cycling, in white adipose, with females having much higher levels than males46. Here, we observed significantly higher levels of transcripts for mitochondrial electron transport chain proteins in females of the STARNET cohort of about 600 WATs from patients that have undergone open chest surgery24. We found that mitochondrial DNA content in adipose is strikingly associated with cardiometabolic traits in the Finnish METSIM cohort. Thus, some of our sex-specific and tissue-specific observations are not unique to mice but are translatable to humans. We have thus far examined only gonadal and subcutaneous fat depots, and the regulation of mitochondria may differ in other white adipose depots and in brown fat.
Mapping of the loci controlling gene expression revealed that about 10% of nuclear-encoded mitochondrial genes are significantly controlled (P < 1 × 10−6) in trans by a genetic locus in chr17 in a sex and tissue-specific manner (many other mitochondrial genes show associations but not attaining significance when multiple testing comparisons are applied). Further, we demonstrated that adipose gene NDUFV2, a core subunit of complex I, mediates the effects of the genetic locus in a strain-by-sex manner. Mitochondrial complex I is the largest among the electron transport chain and is composed of approximately 45 subunits, of which 7 are encoded by the mitochondrial genome47–49. Complex I deficiency is the most common mitochondrial pathology and has been associated with common mitochondrial pathology and has been associated with a wide range of disorders, including neurological, cancer and cardiovascular diseases15,50–52. NDUFV2 is part of the matrix-exposed N-module, which oxidizes NADH and is also the primary site of ROS production, thus being a damage-prone area in need for constant turnover and replacement of damaged subunits53. On this note, Lazarou et al. proposed that complex I biogenesis involves two mechanisms: (1) de novo synthesis and assembly of both mtDNA and nuclear-encoded subunits, and (2) exchange of pre-existing subunits with freshly synthesized ones. They also demonstrated that NDUFV2 is one of the subunits that can be imported into the mitochondria to replace existing subunits54. Recently, Szczepanowska et al. reported the existence of a complex I salvage pathway mediated by mitochondrial membrane protease (CLPXP) that selectively degrades damaged N-module subunits, enabling exchange of freshly made N-module subunits55. They also identified NDUFV2 as a putative substrate for CLPXP activity. Interestingly, our chr17 trans-eQTL lead SNP was associated with the protease gene, Clpp (P = 2.10 × 10−6). Our results suggest that NDUFV2 levels may act as a regulator of mitochondrial genes encoded by both nuclear and mitochondrial genomes, probably via the mitochondrial ROS generation. We do, however, note that the NDUFV2 locus is not significantly associated with metabolic traits in human populations based on searches of commonly used public databases. Whether this is due to genetic drift or differences in selection during the evolution of the two species or a lack of power in the human studies is unclear.
Mitochondrial ROS has both physiological and pathophysiological roles in adipose tissues. Physiological levels of ROS have been shown to be involved in regulating mitochondrial respiration37,56,57, lipolysis56,58 and differentiation42 in adipose, and increasing mitochondrial biogenesis38–40. However, pathological increases in ROS production during obesity lead to disorders such as insulin resistance and diabetes59. Here, we showed that NDUFV2-mediated physiological ROS production, possibly via reduced dimeric complex III content, appears to be contributing to its functional role in regulating mitochondrial biogenesis and function. In this regard, increased mitochondrial coupling revealed by higher RCR values in females expressing increased NDUFV2 indicates a tightly coupled mitochondrial pool, suggesting that ROS generation induced by NDUFV2 is not damaging the mitochondrial membranes to induce proton leak. Also, we showed that NDUFV2 overexpression may increase mitochondrial biogenesis via complex III-mediated ROS generation, as complex III dimerization was decreased. It should be noted that as these are effects that only occur at low physiological concentrations in ROS, acting as a signalling molecule, there are no methodologies sensitive enough to detect these transient increases in ROS in WAT in vivo. Most of the ROS estimations done in vivo are in liver, heart and muscle, cell types with 10–100 times more mitochondria per cell than WAT. We are aware that this is a limitation of our study. Taken together, these findings suggest that increased NDUFV2 overexpression in females results in an increased number of coupled mitochondria to meet the increased ATP demands to prevent lipotoxicity in obesity.
It is noteworthy that reduced mitochondrial function is associated with the same set of metabolic syndrome traits in mice and humans, including obesity, impaired glucose tolerance, hyperinsulinaemia, reduced high-density lipoprotein cholesterol and increased triglycerides60. Since Reaven noted in 1988 that insulin resistance could underlie much of the clustering of these traits61, a large body of work has supported the concept that metabolic syndrome traits exhibit causal interactions and common aetiologies62. Our present results in mice and humans now show that mtDNA content and mitochondrial function in WAT also strongly associate with these traits. Moreover, our experiments with NDUFV2 in mice support the concept that mitochondrial function is not only associated with but also is causal for the set of metabolic syndrome traits. The evolutionary explanation for increased adipose mitochondrial function in females is as yet unclear. It may be related to differences in physical characteristics such as body mass or differences in requirements for thermoregulation, such as during pregnancy63.
In conclusion, we show that adipose mitochondrial function is regulated in a tissue-specific and sex-specific manner and that this has profound effects on a variety of metabolic traits related to type 2 diabetes and cardiovascular diseases in both mice and humans. We have identified one pathway, involving sex-specific regulation of the complex I protein NDUFV2, that contributes to the sex differences. However, there are clearly additional genetic or environmental factors that contribute to the variation in adipose mitochondrial functions.
Methods
Mice.
Mice were handled according to approved Institutional Animal Care and Use Committee (IACUC) protocols (no. 92–169) of the University of California at Los Angeles (UCLA) and were housed in an IACUC-approved vivarium with daily monitoring. The HMDP study design was previously described in detail3,4. Briefly, mice were fed ad libitum a chow diet (Ralston Purina Company) until 8 weeks of age and then placed ad libitum on a HF/HS diet (Research Diets-D12266B) with 16.8% kcal protein, 51.4% kcal carbohydrate, 31.8% kcal fat for an additional 8 weeks. The gonadectomy study design was previously described4. Briefly, at 6 weeks of age, female and male C57BL/6 J mice were either gonadectomized or sham-operated under isoflurane anaesthesia and placed on a HF/HS diet around 8 weeks of age. All mice were purchased from the Jackson Laboratory and bred at UCLA. Mice were maintained on a 14-h light/10-h dark cycle (light is on between 6:00 and 20:00) at a temperature of 25 °C and 30–70% humidity. On the day of the experiment, the mice were killed after 4-h fasting.
HMDP adipose gene expression analysis.
Global gene expression of HMDP adipose tissue was analysed using Affymetrix HT_MG430A arrays as previously described2. To remove known batch effects on the gene expression data, the ComBat method from the SVA Bioconductor package64 was used.
Human population cohorts.
Gene expression of human visceral and subcutaneous adipose tissue and liver were obtained from the STARNET study24. Human subcutaneous adipose DNA and metabolically relevant clinical traits were obtained from the METSIM study27.
Association mapping and correction for population structure.
Genotypes for the mice strains were obtained using the mouse diversity array2. After filtering for quality or missing genotypes, about 200,000 remained. Genome-wide association for phenotypes and transcript levels was performed using FaST-LMM, which applies a linear mixed model to correct for population structure65. A cutoff value for genome-wide significance was set at 4.1 × 10−6, as previously determined2. Whereas GWAS results imply causality and therefore correction for population structure is essential, correlation does not imply causality, and to conclude that a relationship is causal requires genetic mapping or experiments. Although our correlations are not corrected for population structure, only a small fraction of correlation appears to be explained by this. Thus, for the metabolic and molecular traits studied here, the correlations observed in the recombinant inbred strains, a subset of the HMDP that do not have significant population structure, exhibited very similar correlations as the entire HMDP.
Mitochondrial DNA content.
Mitochondrial DNA content was measured as previously described66–68. Briefly, total (mitochondrial and nuclear) DNA from gonadal adipose tissue was isolated by phenol/chloroform/isoamyl alcohol extraction. Both mitochondrial and nuclear DNA were amplified by qPCR with 25 ng of total DNA using the primer pairs listed in Supplementary Table 4. mtDNA content, normalized to nuclear DNA, was calculated using the equation 2 × 2−ΔCt (ΔCt = mito Ct – nuclear Ct) and for AAV experiments, reported as relative units of corresponding GFP groups.
Motif mutation analyses within the chr17:66–67.5 Mb genomic region.
To test for potential functional SNPs whose alleles distinguish DNA motifs of the ERE, to which ER binds, sequence variants were retrieved from the Sanger Institute (mgp.v3 SNPs and Indels69) and restricted to genomic region in LD with lead Ndufv2 cis-eQTL SNP rs48062344 (chr17:66–67.5 Mb; mm10 genome build) using vcftools70. The genomic region from 66 to 67.5 Mb on chr17 was binned into 1,000 bp regions and input into MMARGE71 and analysed using the ‘mutation_analysis’ subroutine using the ERE motif and –keep option for reference and alternate alleles (as downloaded from the Sanger vcf files). This resulted in output files demarcating genomic locations where the ERE motif was present, the genomic background (reference or alternate) on which the motif was discovered, as well as the strength with which that genomic sequence matched the motif. Using this information, custom bash and R code were used to match up positions of ERE motifs on either genomic background and ERE motifs of equal position and strength were discarded. The variants that mutated ERE motifs were further restricted to those in which the alternate allele had greater ERE scores (versus reference) and were ranked according to these scores.
Adeno-associated virus generation.
Recombinant adeno-associated virus serotype 8 (AAV8) was generated as described elsewhere12,72 using either mouse Ndufv2 complementary DNA or GFP. For adipose-specific expression, an adiponectin promoter (AAV8-hAdp) with a liver-specific micro RNA target sequence (miR122T) was used72, while a thyroxine binding globulin (AAV8-Tbg) promoter was used for liver-specific expression. The University of Pennsylvania vector core was utilized for complete viral generation.
NDUFV2 overexpression studies.
Eight-week-old female and male C57BL/6 J or A/J mice were injected intravenously with adipose or liver-specific AAV containing NDUFV2 or GFP constructs for overexpression studies (1012 titre in 200 µl saline). A day after the injection, the animals were placed on HF/HS diet for an additional 8 weeks. Body composition (fat and lean mass) was determined using NMR (Brüker Biospin). On the day of euthanasia, the animals were fasted for 4 h. Retro-orbital blood was collected to isolate plasma for analysing glucose, insulin and lipid. Liver, four white adipose depots (gonadal, subcutaneous, mesenteric and retroperitoneal) and brown adipose were collected for weight measurements.
Quantitative polymerase chain reaction.
We performed qPCR following the manufacturer’s protocol. Briefly, total RNA from adipose tissues was first isolated using QIAzol (QiagenD), then first-strand cDNA was made using the high-capacity cDNA reverse transcription kit (Applied Biosystems). Finally, relative quantitative gene expression levels were measured using a Kapa SYBR fast qPCR kit (Kapa Biosystems) on a LightCycler 480 II (Roche). All analyses were done using the Roche LightCycler1.5.0 software. Relative normalized expression was measured using the equation 2-ΔΔCt with the geometric mean of B2M and TBP used to normalize all qPCR targets73. All qPCR primer sequences were listed in Supplementary Table 5.
Bioenergetics of isolated mitochondria.
Isolated mitochondrial respiration from adipose tissue74 was measured as described previously. Briefly, mitochondria were obtained by dual centrifugation and resuspended in respiration buffer74 and kept on ice. Mitochondrial respiration was obtained with an XF24 or XF96 Seahorse bioanalyzer (Agilent). For the complex I respiration, the measures were collected in the presence of 10 mM pyruvate (complex I substrate), 2 mM malate and 4 mM carbonyl cyanide-4 (trifluoromethoxy) phenylhydrazone (FCCP). RCR (state 3/state 4o) is a mitochondrial quality index representing how well the oxygen consumption is coupled to ATP synthesis75. Thus, higher RCR values in females expressing increased NDUFV2 indicate a tightly coupled mitochondrial pool. On the other hand, coupling efficiency ([state 3–state 4o]/[state 3–AA]) is defined as the proportion of mitochondrial oxygen consumption used for ATP synthesis75,76 and an increased coupling efficiency could be a result of lower proton leak that decreases respiration under an oligomycin or higher ADP phosphorylation rate that increases respiration after stimulation with ADP.
Immunoblotting of adipose mitochondria.
Immunoblotting of adipose mitochondria for the different OXPHOS proteins was done as previously described5. Primary antibodies used were Total OXPHOS Rodent WB Antibody Cocktail (1/5,000 dilution, no. ab110413, Abcam), SDHA (1/5,000 dilution, no. 459200, Thermo Fisher Scientific), NDUFB8 (1/5,000 dilution, no. 459210, Thermo Fisher Scientific), LCAD (1/5000 dilution, no. ab129711, Abcam), CPT1a (1/2000 dilution, no. 15184–1-AP, Proteintech), UCP1 (1/5,000 dilution, no. ab10983, Abcam), NDUFV2 (1/5,000 dilution, no. 15301–1-AP, Proteintech), MYC (1/2,000 dilution, no. 05–724, Millipore), GAPDH (1/5,000 dilution, no. GTX100118, GeneTex) and Vinculin (1/5,000 dilution, no. V9131, Sigma). Image contrast was uniformly reduced to enhance visibility. Band densitometry was quantified using ImageJ Gel Plugin (National Institutes of Health (NIH)).
Insulin tolerance test.
For ITT, animals were first fasted for 6 h before being injected intraperitoneally with insulin (1 U kg−1), following which regular plasma glucose measures were done using a glucometer.
Indirect calorimetry analyses.
Oxygen (O2)/carbon dioxide (CO2) levels and physical activity (XYZ, laser beam breaks counts) were determined using Oxymax Comprehensive Lab Animal Monitoring System (CLAMS, Columbus Instruments) at UCLA. Mice were first acclimated for 3 days in CLAMS and then the data were collected the following 2 days and nights. RER was calculated as CO2 consumption (VCO2)/O2 consumption (VO2). EE in cal min−1 was derived from the Lusk equation: (3.815 + 1.232 × RER) × VO2, with VO2 in ml min−1. The system was calibrated with gas of a known O2 and CO2 percentage before every experiment. The ANCOVA was performed with Sigma Plot 14, Systat Software Inc. using the MMPC website: https://www.mmpc.org/shared/regression.aspx.
Frozen respirometry analyses.
Respirometry in previously frozen samples was performed as described elsewhere using total tissue lysates36,77.
Cellular bioenergetics.
AML12 cells previously seeded in 6-well plates were transfected with a plasmid containing either GFP or Ndufv2 cDNA using TransIT-X2 Dynamic Delivery System (Mirus Bio). Ndufv2 overexpression was confirmed by qPCR. After 30 h of transfection, cells were replated on a XF24 plate (25,000 cells per well) with either 25 mM glucose or 25 mM galactose. Coupling assays with PMP permeabilization reagent (Agilent) in the presence of 10 mM pyruvate, 40 µM palmitoyl-carnitine and 10 mM glutamate (individually or together) were run as previously described5. 3T3-L1 pre-adipocytes were cultured and differentiated according to the manufacturer’s instructions (Zen-Bio). Briefly, cells were transduced with either a control or NDUFV2-Lentivirus (Origene) and selected with 2.5 µg ml−1 puromycin. Ndufv2 overexpression was confirmed by western blot using a specific antibody (no. 15301–1-AP, Proteintech). To measure respiration, cells were differentiated in 6-well plates and replated in a XF24 plate (40,000 cells per well) at day 5 and analysed at day 6. Coupling assays with PMP permeabilization reagent (Agilent) in the presence of 10 mM pyruvate, 40 µM palmitoyl-carnitine or 10 mM succinate were run as previously described5. To measure ROS-sensitive respiration, cellular respiration in complete unbuffered Dulbecco’s modified Eagle’s medium was measured before and 100 min after 10 mM NAC injection.
Blue native gel electrophoresis.
Blue native gel electrophoresis was performed in differentiated 3T3 cells. Samples were prepared as previously described78 with slight modifications. Briefly, 2 × 106 adipocytes were resuspended in 200 µl PBS. To permeabilize the plasma membrane, 70 µl of digitonin (8 mg ml−1 freshly prepared) was added to the cell suspension and samples were incubated on ice for 10 min. After this incubation, 1 ml of ice-cold PBS was added and samples were centrifuged at 4 C at 10,000g for 10 min, the pellet was then resuspended in 100 µl of BN sample buffer (500 mM 6-aminohexanoic acid, 50 mM imidazole, 1 mM ethylenediamine tetraacetic acid, pH 7). Mitochondrial supercomplexes were solubilized by adding 10 µl of digitonin 10%. Samples were incubated on ice for 5 min and then centrifuged at 4 C at 20,000g for 30 min. Supernatant containing supercomplexes were mixed with 5% Blue G-250, 5% glycerol in BN sample buffer and loaded on a 4–12% native gel (Invitrogen) following the manufacturer’s recommendations. Gel was transferred to a polyvinylidene difluoride membrane and blotted using the following antibodies: NDUFA9 (1/5,000 dilution, no. ab14713, Abcam); UQCR2 (1/5,000 dilution, no. 14742–1-AP, Proteintech); SDHA (1/5,000 dilution, no. 459200, Thermo Fisher Scientific).
Statistical analysis.
Statistical analyses were performed using Prism v9.2.0 (GraphPad Software). Errors bars plotted on graphs are presented as the mean ± s.e.m. unless reported otherwise. The critical significance value (α) was set at 0.05, and if the P values were less than α, we reported that the observed differences were statistically significant. Correlations were calculated using biweight midcorrelation using the bicorAndPvalue function of the WGCNA package79.
Extended Data
Extended Data Fig. 1 |. Chr17 locus is not associated with liver mitochondria.
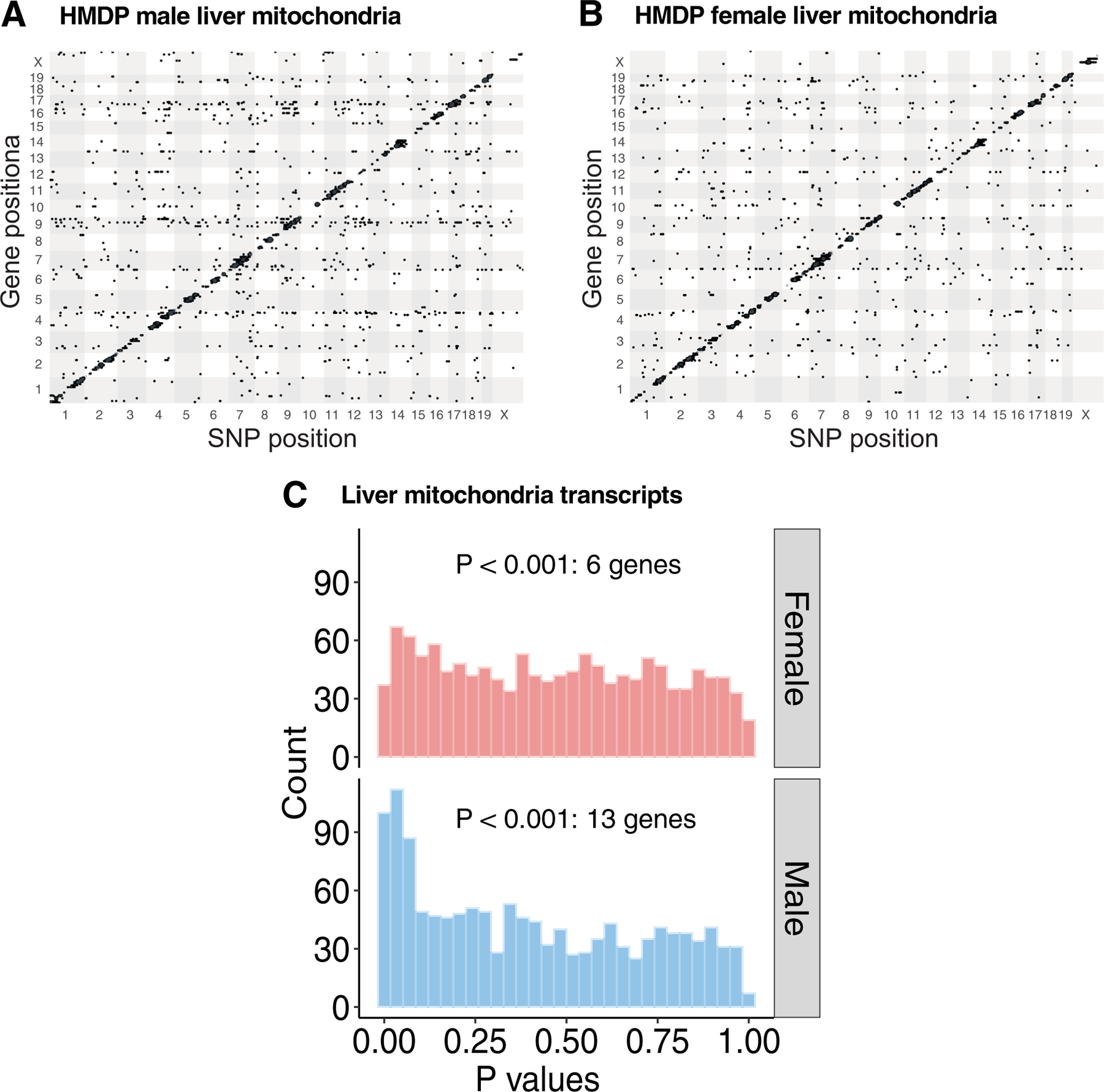
Related to Fig. 3. High-resolution association mapping of 911 (1312 probes) mitochondrial gene abundance levels from (a) male and (b) female liver tissues isolated from ~100 HMDP strains to identify eQTL networks. The X- and Y- axis represent SNP and gene position on the mouse genome, respectively. Each dot represents a significant association. (c) Frequency distributions of P values of association between the chr17 trans-eQTL lead eSNP (rs48062344) and mitochondria-related transcripts in the male and female liver tissues. P values of association were calculated using (A – C) FaST-LMM that uses Likelihood-Ratio test.
Extended Data Fig. 2 |. Chr17 trans-eQTL do not affect the expression profiles of oXPHoS genes in male adipose or both sexes of liver tissues.
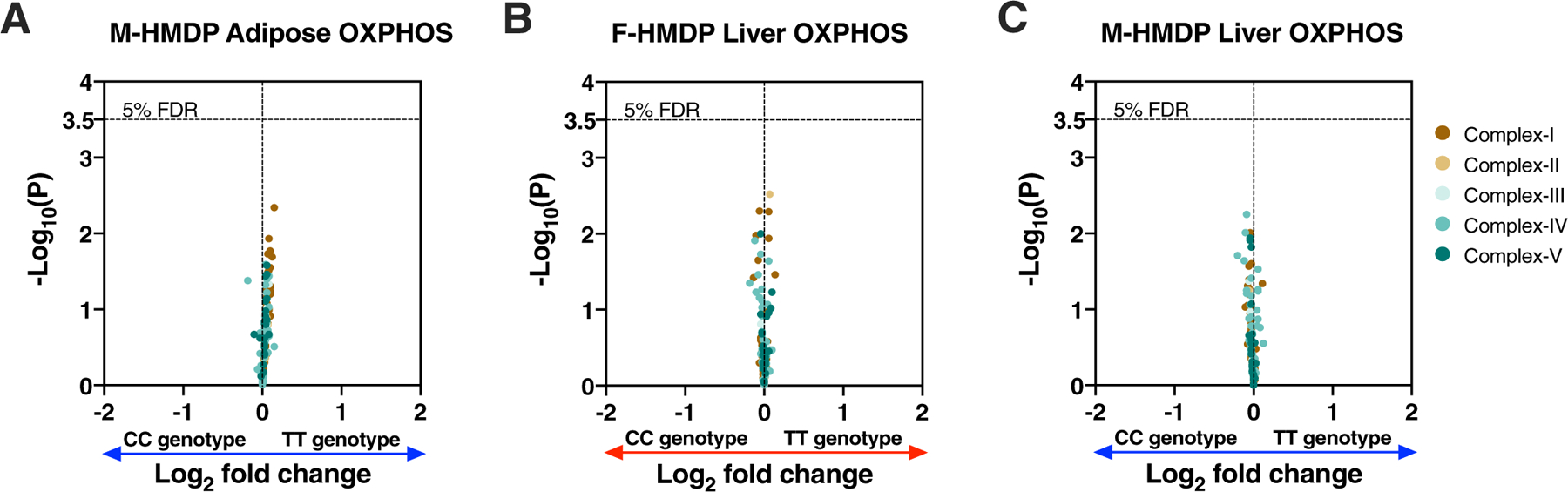
Related to Fig. 4. Volcano plots showing genetic (lead SNP of chr17 locus) differences in the expression profiles of OXPHOS genes in the (a) male adipose (n = 98 male strains; CC: 56 & TT: 42); (b) female and (c) male liver (n = 97 sex-matched strains; CC: 55 & TT: 42) isolated from HMDP. Genes corresponding to individual OXPHOS complexes are color-coded. Horizontal dotted lines represent 5% FDR-corrected significance threshold. Data are presented as log2 fold change between genotype. P values were calculated using (A – C) DESeq2 Bioconductor package that uses Wald test.
Extended Data Fig. 3 |. Association mapping of adipose mtDNA levels in HMDP.

Related to Fig. 4. Association mapping of mtDNA levels from (a) female and (b) male adipose in the HMDP cohort (n = 216 female and 260 male mice) Red line represents significance threshold (HMDP: P = 4.1E-06). P values of association were calculated using (A – B) FaST-LMM that uses Likelihood-Ratio test.
Extended Data Fig. 4 |. Liver Ndufv2 is unaffected by the chr17 trans-eQTL locus.
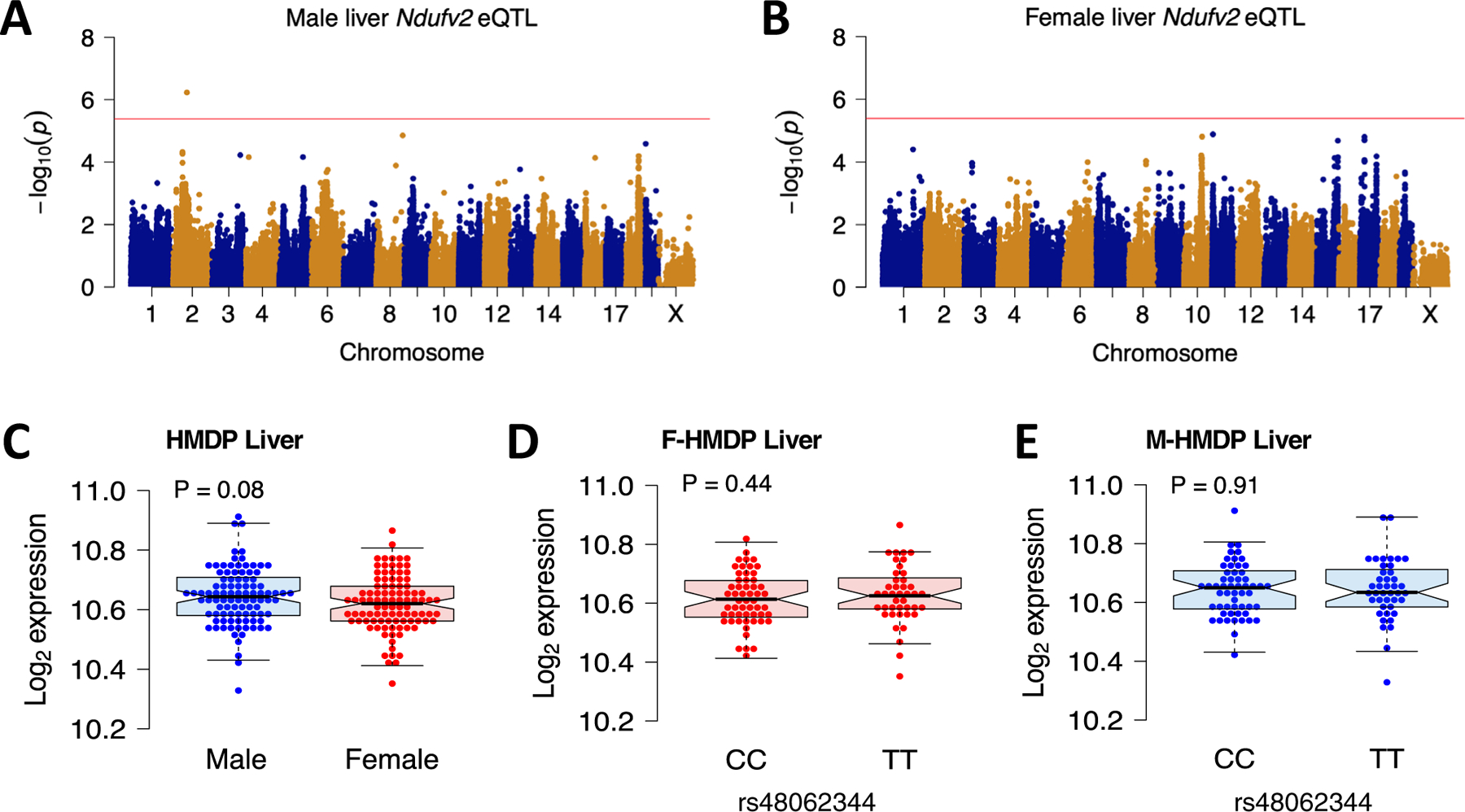
Related to Fig. 5. Association mapping of (a) male and (b) female liver Ndufv2 expression from HMDP. Red line represents significance threshold (P = 4.1E-06). (c – e) Sex and genetic (lead SNP of chr17 locus) differences in the liver Ndufv2 expression from HMDP (n = 97 sex-matched strains; CC: 55 & TT: 42). Data are presented as median and interquartile range (boxplots). P values were calculated using (A – B) FaST-LMM that uses Likelihood-Ratio test; (C – E) Unpaired two-tailed Student’s t test.
Extended Data Fig. 5 |. Changes in body weight and insulin sensitivity mediated by adipose Ndufv2 overexpression.
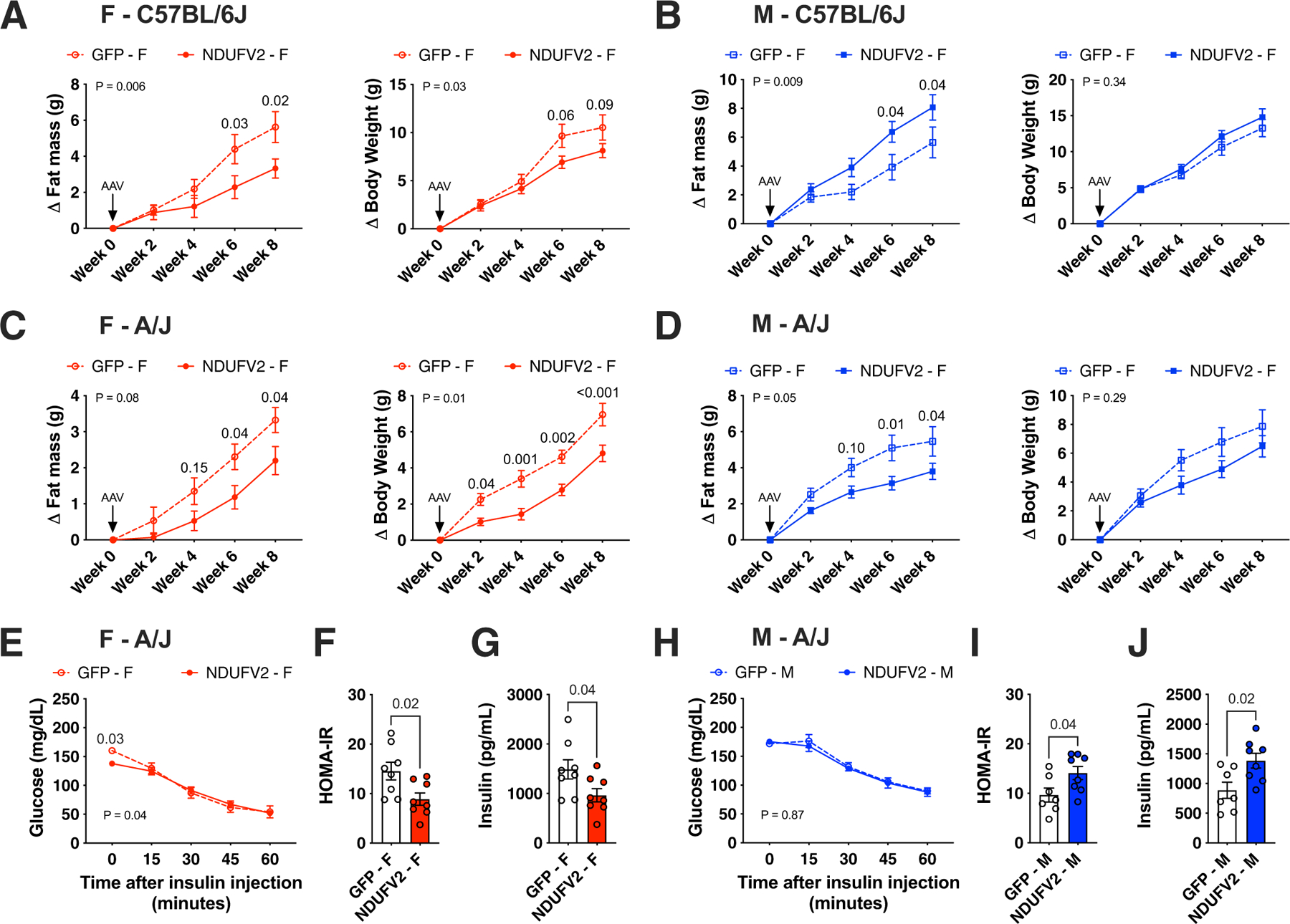
Related to Fig. 6. Eight-week old females or males of (a – b) C57BL/6 J or (c – j) A/J mice were injected with AAV vectors expressing either GFP or NDUFV2 in an adipose-specific manner and fed a HF/HS diet for eight additional weeks. Changes in (A - D) fat mass and total body weight measured every two-weeks (females n = 7 per group; males n = 8 per group), (E and H) ITT, (F and I) end-point HOMA-IR and (G and J) fasting insulin levels (F – A/J n = 8 per group; M – A/J GFP n = 7, NDUFV2 n = 8) are shown. Data are presented as mean ± SEM. P values were calculated using (A – E and H) Repeated measures 2-factor ANOVA corrected by post-hoc ‘Holm-Sidak’s’ multiple comparisons test; (F, G, I and J) Unpaired two-tailed Student’s t test.
Extended Data Fig. 6 |. Adipose Ndufv2 overexpression regulated adiposity in a sex-by-strain manner.

Related to Fig. 6. Eight-week old males of (a – e) C57BL/6 J or (f – j) A/J mice were injected with AAV vectors expressing either GFP or NDUFV2 in an adipose-specific manner and fed a HF/HS diet for eight additional weeks. Metabolic traits such as (A and F) total mass and food intake; (B and G) fat and lean mass were monitored over eight weeks. Comparisons of (C and H) adipose Ndufv2 expression; (D and I) plasma free glycerol; (E and J) tissue weights from C57BL/6 J and A/J males, respectively. Data are presented as mean ± SEM (n = 8 per group). P values were calculated using (A and F) Repeated measures 2-factor or (B and G) 3-factor or (E and J) 2-factor ANOVA corrected by post-hoc ‘Holm-Sidak’s’ multiple comparisons test; (C, D, H and I) Unpaired two-tailed Student’s t test.
Extended Data Fig. 7 |. Adipose Ndufv2 overexpression regulated adipocyte size in a sex-by-strain manner.

Related to Fig. 7. Comparisons of adipocyte size distribution in the gonadal adipose tissues from females and males of (a – b) C57BL/6 J or (c – d) A/J mice overexpressing GFP or NDUFV2, respectively. Representative histological sections are shown for each group (scale: 100 µm). Red and blue shades (or bars) represent female and male NDUFV2 datapoints, while brown represents respective GFP datapoints, respectively. Data are presented as frequency distribution of adipocyte sizes or mean ± SEM (F – C57BL/6 J GFP n = 5064 cells from 7 mice, NDUFV2 n = 5249 cells from 7 mice; M – C57BL/6 J GFP n = 11721 cells from 8 mice, NDUFV2 n = 7290 cells from 7 mice; F – A/J GFP n = 5423 cells from 7 mice, NDUFV2 n = 4687 cells from 7 mice; M – A/J GFP n = 4084 cells from 8 mice, NDUFV2 n = 4863 cells from 8 mice). P values were calculated using Unpaired two-tailed Student’s t test.
Extended Data Fig. 8 |. Adipose Ndufv2 overexpression regulated mitochondrial respiration in a sex-by-strain manner.
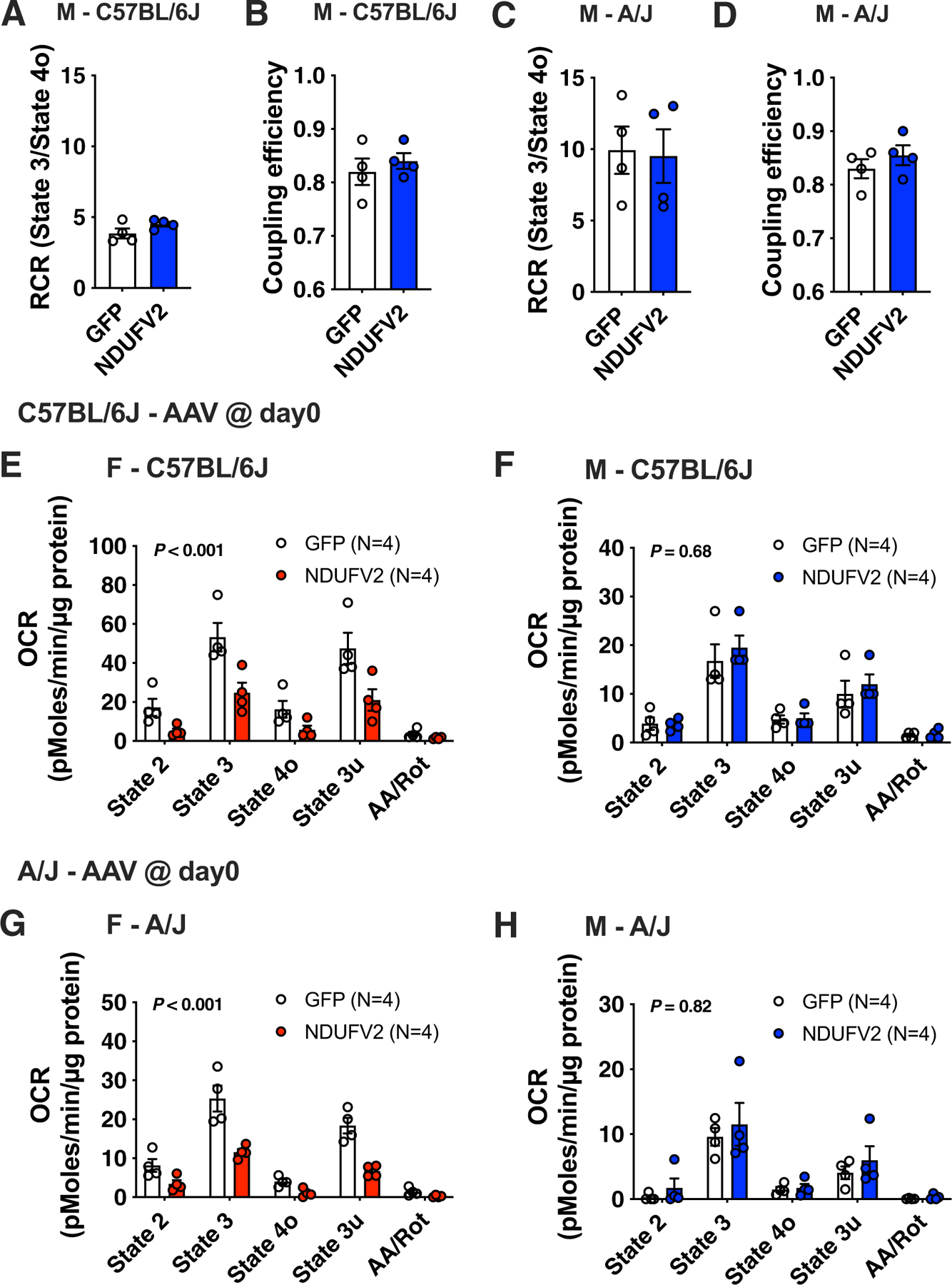
Related to Fig. 7. Comparisons of mitochondrial RCR and coupling efficiency in the gonadal adipose tissues from males of (a – b) C57BL/6 J or (c – d) A/J mice overexpressing GFP or NDUFV2, respectively. Coupling assays of isolated gonadal adipose mitochondria from females and males of (e – f) C57BL/6 J or (g – h) A/J mice overexpressing GFP or NDUFV2, respectively. Data are presented as mean ± SEM (n = 4 per group). P values were calculated using 2-factor ANOVA.
Extended Data Fig. 9 |. Adipose Ndufv2-mediated mitochondrial regulation is not a consequence of body weight.

Related to Fig. 7. Eight-week old female C57BL/6 J mice were fed a HF/HS diet for the first six weeks without any intervention, after which were injected with AAV vectors expressing either GFP or NDUFV2 in an adipose-specific manner and diet continued for six additional weeks. Metabolic traits such as (a) total mass; (b) fat and lean mass were monitored over 12 weeks. Comparisons of (c) tissue weights; (d) adipose Ndufv2 expression; (e) mitochondrial RCR; (f) coupling efficiency and (g) coupling respiration rates between GFP and NDUFV2 animals, respectively. Data are presented as mean ± SEM (n = 6 per group). P values were calculated using (A) Repeated measures 2-factor or (B) 3-factor or (C and G) 2-factor ANOVA corrected by post-hoc ‘Holm-Sidak’s’ multiple comparisons test; (D – F) Unpaired two-tailed Student’s t test.
Extended Data Fig. 10 |. Ndufv2 overexpression regulated mitochondrial function in both AML12 (liver) and differentiated 3T3-L1 (adipose) cells.
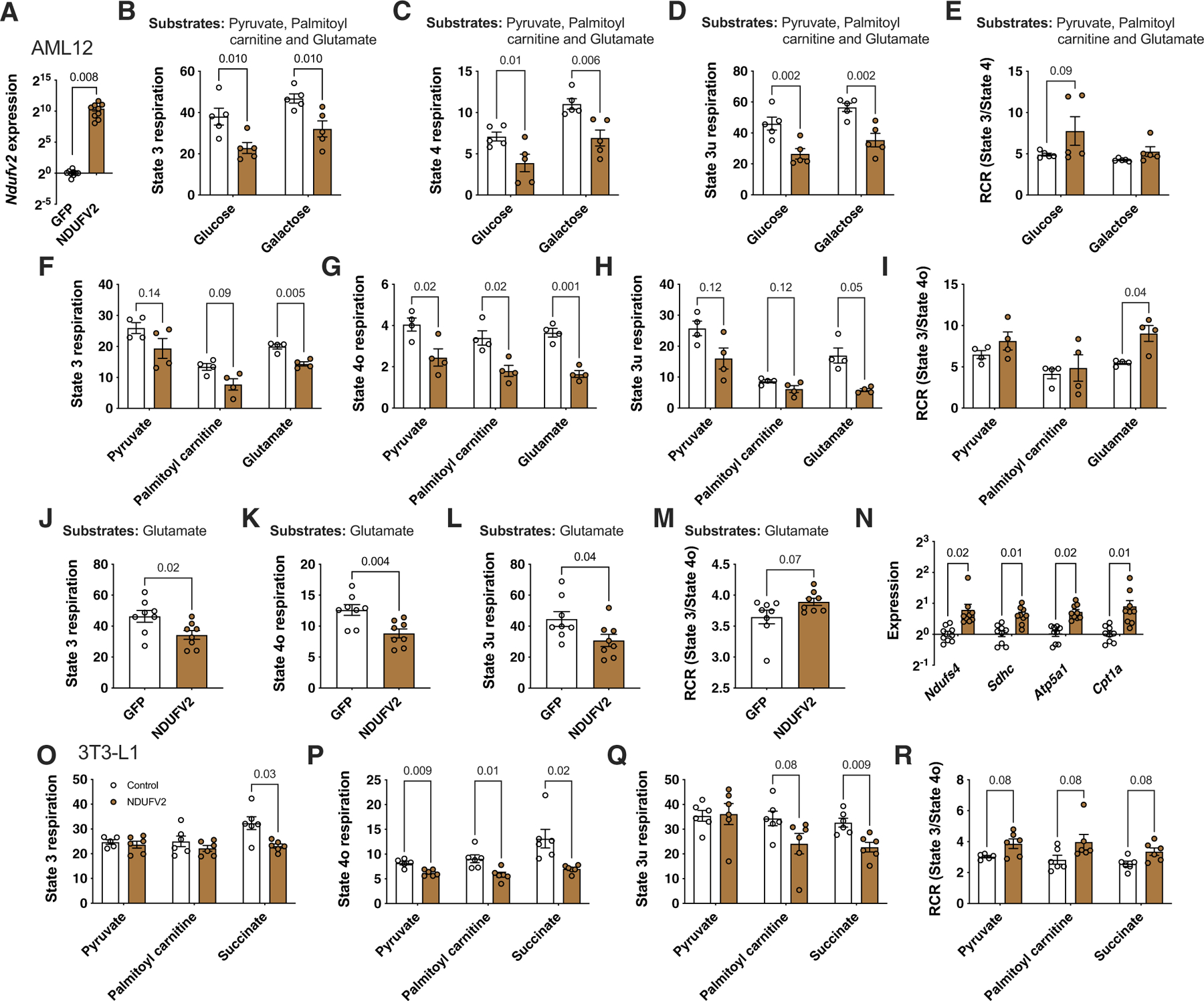
Related to Fig. 8. Comparisons between GFP and NDUFV2 overexpressing AML12 (liver) cells in (a) relative normalized expression values of Ndufv2 (n = 9 per group); Coupling assays and RCR with all three complex I substrates (Pyruvate, Palmitoyl carnitine and Glutamate) either added (b – e) together (n = 5 per group) or (f - m) separately (n = 4 per group except Glutamate, n = 8 per group); and (N) relative normalized expression values of Ndufs4 (complex I), Sdhc (complex II), Atp5a1 (complex V) and Cpt1a (FAO) (n = 9 per group). Similarly, comparisons between control and NDUFV2 overexpressing differentiated 3T3-L1 cells in (O – R) coupling assays and RCR with different substrates (Pyruvate, Palmitoyl carnitine and Succinate) added separately (n = 6 per group). Data are presented as mean ± SEM. P values were calculated using (A and J – N) Unpaired two-tailed Student’s t test; (B – I and O – R) 2-factor ANOVA corrected by post-hoc ‘Holm-Sidak’s’ multiple comparisons test.
Supplementary Material
Acknowledgements
We thank Z. Zhou, Y. Meng, S. Charugundla, D. W. Jayasekera, S. Nand and J. Ure for assistance in experiments. This work was supported by NIH grants NIH-P01HL028481, NIH-R01DK117850 and NIH-R01HL144651 (A.J.L.), NIH-R01HL125863 (J.L.M.B.), NIH-R01HL147187 (C.E.R.), NIH-1R01AA026914-01A1 (M. Liesa), NIH-R00DK120875 (K.C.K) and NIH-K99DK120875 (K.C.K); UCLA/UCSD/CTSI grants P30DK41301 (M. Liesa), UL1TR001881 (M. Liesa), P30DK063491 (M. Liesa); the American Heart Association grants A14SFRN20840000 (J.L.M.B.) and 18POST33990256 (K.C.K.); the Academy of Finland 321428 (M. Laakso); the Swedish Research Council 2018-02529 (J.L.M.B); Heart Lung Foundation 20170265 (J.L.M.B.); Astra-Zeneca through ICMC, Karolinska Institutet, Sweden (J.L.M.B.) and the Foundation Leducq 12CVD04 (L.V. and K.R.) and 18CVD02 (J.L.M.B.). The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Competing interests
The authors declare no competing interests.
Additional information
Extended data are available for this paper at https://doi.org/10.1038/s42255-021-00481-w.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s42255-021-00481-w.
Peer review information Primary handling editors: George Caputa and Isabella Samuelson. Nature Metabolism thanks Lawrence Kazak and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Data availability
RNA sequencing raw data can be accessed at the Gene Expression Omnibus under accession GSE64770 (HMDP expression arrays (adipose and liver)) and GSE112947 (Gonadectomized RNA-seq data). GTEx datasets can be found online at the GTEx portal website (https://gtexportal.org/home/datasets). Uncropped scans of blots are available in the source data files. HMDP data from the authors laboratories will be made available on reasonable request. Source data are provided with this paper.
References
- 1.Seldin M, Yang X & Lusis AJ Systems genetics applications in metabolism research. Nat. Metab 1, 1038–1050 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett BJ et al. A high-resolution association mapping panel for the dissection of complex traits in mice. Genome Res 20, 281–290 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parks BW et al. Genetic control of obesity and gut microbiota composition in response to high-fat, high-sucrose diet in mice. Cell Metab 17, 141–152 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parks BW et al. Genetic architecture of insulin resistance in the mouse. Cell Metab 21, 334–347 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chella Krishnan K et al. Sex-specific metabolic functions of adipose Lipocalin-2. Mol. Metab 30, 30–47 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Norheim F et al. Genetic, dietary, and sex-specific regulation of hepatic ceramides and the relationship between hepatic ceramides and IR. J. Lipid Res 59, 1164–1174 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Norheim F et al. Genetic and hormonal control of hepatic steatosis in female and male mice. J. Lipid Res 58, 178–187 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Org E et al. Sex differences and hormonal effects on gut microbiota composition in mice. Gut Microbes 7, 313–322 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chella Krishnan K et al. Integration of multi-omics data from mouse diversity panel highlights mitochondrial dysfunction in non-alcoholic fatty liver disease. Cell Syst 6, 103–115 e107 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hui ST et al. The genetic architecture of diet-Induced hepatic fibrosis in mice. Hepatology 68, 2182–2196 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Norheim F et al. Gene-by-sex interactions in mitochondrial functions and cardio-metabolic traits. Cell Metab 29, 932–949 e934 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seldin MM et al. A strategy for discovery of endocrine interactions with application to whole-body metabolism. Cell Metab 27, 1138–1155 e1136 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seldin MM et al. A systems genetics approach identifies Trp53inp2 as a link between cardiomyocyte glucose utilization and hypertrophic response. Am. J. Physiol. Heart Circ. Physiol 312, H728–H741 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yin X et al. Adipocyte mitochondrial function is reduced in human obesity independent of fat cell size. J. Clin. Endocrinol. Metab 99, E209–E216 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chouchani ET et al. Complex I deficiency due to selective loss of Ndufs4 in the mouse heart results in severe hypertrophic cardiomyopathy. PLoS ONE 9, e94157 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Begriche K, Igoudjil A, Pessayre D & Fromenty B Mitochondrial dysfunction in NASH: causes, consequences and possible means to prevent it. Mitochondrion 6, 1–28 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Sanyal AJ et al. Nonalcoholic steatohepatitis: association of insulin resistance and mitochondrial abnormalities. Gastroenterology 120, 1183–1192 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Kim JA, Wei Y & Sowers JR Role of mitochondrial dysfunction in insulin resistance. Circ. Res 102, 401–414 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mittelstrass K et al. Discovery of sexual dimorphisms in metabolic and genetic biomarkers. PLoS Genet 7, e1002215 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.White UA & Tchoukalova YD Sex dimorphism and depot differences in adipose tissue function. Biochim. Biophys. Acta 1842, 377–392 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang X et al. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res 16, 995–1004 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karp NA et al. Prevalence of sexual dimorphism in mammalian phenotypic traits. Nat. Commun 8, 15475 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wallace DC A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev. Genet 39, 359–407 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Franzen O et al. Cardiometabolic risk loci share downstream cis- and trans-gene regulation across tissues and diseases. Science 353, 827–830 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Consortium GT et al. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robin ED & Wong R Mitochondrial DNA molecules and virtual number of mitochondria per cell in mammalian cells. J. Cell. Physiol 136, 507–513 (1988). [DOI] [PubMed] [Google Scholar]
- 27.Laakso M et al. The Metabolic Syndrome in Men study: a resource for studies of metabolic and cardiovascular diseases. J. Lipid Res 58, 481–493 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Calvo SE, Clauser KR & Mootha VK MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res 44, D1251–D1257 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perry RJ et al. Hepatic acetyl CoA links adipose tissue inflammation to hepatic insulin resistance and type 2 diabetes. Cell 160, 745–758 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lapuente-Brun E et al. Supercomplex assembly determines electron flux in the mitochondrial electron transport chain. Science 340, 1567–1570 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Garcia-Poyatos C et al. Scaf1 promotes respiratory supercomplexes and metabolic efficiency in zebrafish. Embo Rep 21, e50287 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng C-F, Ku H-C & Lin H PGC-1α as a pivotal factor in lipid and metabolic regulation. Int. J. Mol. Sci 19, 3447 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee JH et al. The role of adipose tissue mitochondria: regulation of mitochondrial function for the treatment of metabolic diseases. Int. J. Mol. Sci 20, 4924 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heinonen S, Jokinen R, Rissanen A & Pietilainen KH White adipose tissue mitochondrial metabolism in health and in obesity. Obes. Rev 21, e12958 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Benador IY et al. Mitochondria bound to lipid droplets have unique bioenergetics, composition, and dynamics that support lipid droplet expansion. Cell Metab 27, 869–885.e866 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Acin-Perez R et al. A novel approach to measure mitochondrial respiration in frozen biological samples. EMBO J 39, e104073 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang T et al. Respiration in adipocytes is inhibited by reactive oxygen species. Obesity 18, 1493–1502 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.St-Pierre J et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell 127, 397–408 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Dogan SA et al. Perturbed redox signaling exacerbates a mitochondrial myopathy. Cell Metab 28, 764–775 e765 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moreno-Loshuertos R et al. Differences in reactive oxygen species production explain the phenotypes associated with common mouse mitochondrial DNA variants. Nat. Genet 38, 1261–1268 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Swierczek M et al. An electronic bus bar lies in the core of cytochrome bc1. Science 329, 451–454 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tormos KV et al. Mitochondrial complex III ROS regulate adipocyte differentiation. Cell Metab 14, 537–544 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Acin-Perez R, Fernandez-Silva P, Peleato ML, Perez-Martos A & Enriquez JA Respiratory active mitochondrial supercomplexes. Mol. Cell 32, 529–539 (2008). [DOI] [PubMed] [Google Scholar]
- 44.Acín-Pérez, R. et al. Respiratory complex III is required to maintain complex I in mammalian mitochondria. Mol. Cell 13, 805–815 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maranzana E, Barbero G, Falasca AI, Lenaz G & Genova ML Mitochondrial respiratory supercomplex association limits production of reactive oxygen species from complex I. Antioxid. Redox Signal 19, 1469–1480 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nookaew I et al. Adipose tissue resting energy expenditure and expression of genes involved in mitochondrial function are higher in women than in men. J. Clin. Endocrinol. Metab 98, E370–E378 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hirst J Mitochondrial complex I. Annu. Rev. Biochem 82, 551–575 (2013). [DOI] [PubMed] [Google Scholar]
- 48.Fiedorczuk K et al. Atomic structure of the entire mammalian mitochondrial complex I. Nature 538, 406–410 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu J, Vinothkumar KR & Hirst J Structure of mammalian respiratory complex I. Nature 536, 354–358 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karamanlidis G et al. Mitochondrial complex I deficiency increases protein acetylation and accelerates heart failure. Cell Metab 18, 239–250 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Urra FA, Munoz F, Lovy A & Cardenas C The mitochondrial complex(i)ty of cancer. Front Oncol 7, 118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Papa S & De Rasmo D Complex I deficiencies in neurological disorders. Trends Mol. Med 19, 61–69 (2013). [DOI] [PubMed] [Google Scholar]
- 53.Dröse S & Brandt U Advances in experimental medicine and biology. Adv. Exp. Med Biol 748, 145–169 (2012). [DOI] [PubMed] [Google Scholar]
- 54.Lazarou M, McKenzie M, Ohtake A, Thorburn DR & Ryan MT Analysis of the assembly profiles for mitochondrial- and nuclear-DNA-encoded subunits into complex I. Mol. Cell. Biol 27, 4228–4237 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Szczepanowska K et al. A salvage pathway maintains highly functional respiratory complex I. Nat. Commun 11, 1643 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Latorre-Pellicer A et al. Mitochondrial and nuclear DNA matching shapes metabolism and healthy ageing. Nature 535, 561–565 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Boudina S & Graham TE Mitochondrial function/dysfunction in white adipose tissue. Exp. Physiol 99, 1168–1178 (2014). [DOI] [PubMed] [Google Scholar]
- 58.Krawczyk SA, Haller JF, Ferrante T, Zoeller RA & Corkey BE Reactive oxygen species facilitate translocation of hormone sensitive lipase to the lipid droplet during lipolysis in human differentiated adipocytes. PLoS ONE 7, e34904 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Masschelin PM, Cox AR, Chernis N & Hartig SM The impact of oxidative stress on adipose tissue energy balance. Front Physiol 10, 1638 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Acín-Pérez R et al. Fgr kinase is required for proinflammatory macrophage activation during diet-induced obesity. Nat. Metab 2, 974–988 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reaven GM Banting Lecture 1988. Role of insulin resistance in human disease. Diabetes 37, 1595–1607 (1988). [DOI] [PubMed] [Google Scholar]
- 62.Lusis AJ, Attie AD & Reue K Metabolic syndrome: from epidemiology to systems biology. Nat. Rev. Genet 9, 819–830 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gagnon D & Kenny GP Does sex have an independent effect on thermoeffector responses during exercise in the heat? J. Physiol 590, 5963–5973 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Leek JT, Johnson WE, Parker HS, Jaffe AE & Storey JD The SVA package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lippert C et al. FaST linear mixed models for genome-wide association studies. Nat. Methods 8, 833–835 (2011). [DOI] [PubMed] [Google Scholar]
- 66.Vergnes L, Chin R, Young SG & Reue K Heart-type fatty acid-binding protein is essential for efficient brown adipose tissue fatty acid oxidation and cold tolerance. J. Biol. Chem 286, 380–390 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rooney JP et al. PCR based determination of mitochondrial DNA copy number in multiple species. Methods Mol. Biol 1241, 23–38 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Venegas V & Halberg MC Measurement of mitochondrial DNA copy number. Methods Mol. Biol 837, 327–335 (2012). [DOI] [PubMed] [Google Scholar]
- 69.Keane TM et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477, 289–294 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Danecek P et al. The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Link VM, Romanoski CE, Metzler D & Glass CK MMARGE: Motif Mutation Analysis for Regulatory Genomic Elements. Nucleic Acids Res 46, 7006–7021 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.O’Neill SM et al. Targeting adipose tissue via systemic gene therapy. Gene Ther 21, 653–661 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vandesompele J et al. Accurate normalization of real-time quantitative RT–PCR data by geometric averaging of multiple internal control genes. Genome Biol 3, RESEARCH0034 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rogers GW et al. High throughput microplate respiratory measurements using minimal quantities of isolated mitochondria. PLoS ONE 6, e21746 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brand MD & Nicholls DG Assessing mitochondrial dysfunction in cells. Biochem. J 435, 297–312 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Affourtit C, Jastroch M & Brand MD Uncoupling protein-2 attenuates glucose-stimulated insulin secretion in INS-1E insulinoma cells by lowering mitochondrial reactive oxygen species. Free Radic. Biol. Med 50, 609–616 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Osto C et al. Measuring mitochondrial respiration in previously frozen biological samples. Curr. Protoc. Cell Biol 89, e116 (2020). [DOI] [PubMed] [Google Scholar]
- 78.Garaude J et al. Mitochondrial respiratory-chain adaptations in macrophages contribute to antibacterial host defense. Nat. Immunol 17, 1037–1045 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Langfelder P & Horvath S WGCNA: an R package for weighted correlation network analysis. BMC Bioinf 9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing raw data can be accessed at the Gene Expression Omnibus under accession GSE64770 (HMDP expression arrays (adipose and liver)) and GSE112947 (Gonadectomized RNA-seq data). GTEx datasets can be found online at the GTEx portal website (https://gtexportal.org/home/datasets). Uncropped scans of blots are available in the source data files. HMDP data from the authors laboratories will be made available on reasonable request. Source data are provided with this paper.


