Abstract
Grain size, grain number per panicle, and grain weight are crucial determinants of yield-related traits in cereals. Understanding the genetic basis of grain yield-related traits has been the main research object and nodal in crop science. Sorghum and maize, as very close C4 crops with high photosynthetic rates, stress tolerance and large biomass characteristics, are extensively used to produce food, feed, and biofuels worldwide. In this review, we comprehensively summarize a large number of quantitative trait loci (QTLs) associated with grain yield in sorghum and maize. We placed great emphasis on discussing 22 fine-mapped QTLs and 30 functionally characterized genes, which greatly hinders our deep understanding at the molecular mechanism level. This review provides a general overview of the comprehensive findings on grain yield QTLs and discusses the emerging trend in molecular marker-assisted breeding with these QTLs.
Keywords: QTLs, grain yield, sorghum, maize, marker-assisted breeding
1. Introduction
Increasing world population growth, which is expected to reach 9 billion, will require increasing food production to be doubled by 2050 [1,2]. Facing limited cultivated land, a lack of water resources and extreme environmental deterioration, humans have been seeking sustainable ways to maximize crop production to feed the soaring population [3]. Since world food production is mainly expected from crops, one way to meet this challenge is to improve the unit yield of crops [4]. Sorghum (Sorghum bicolor L. Moench), as the fifth largest food crop, guarantees food security for ~5 million people living in drought, saline-alkali, and barren areas worldwide [5,6]. Maize (Zea mays L.), as one of the three largest crops, feeds one-third of the world’s population [7]. Currently, sorghum and maize have also become available for feed, fodder, and emerging bioenergy [8,9]. Both sorghum and maize are the closest and relatively recent domesticated crops from their wild progenitor, which diverged from each other approximately 11.9 million years ago and distantly diverged approximately 50 Mya from their rice ancestor [10].
Grain yield is one of the most complex quantitative features with a varied genetic basis and is affected by multiple genetic components and environmental factors [11]. Grain weight is a grain yield component with high heritability. Grain size (grain length, grain width, and grain thickness) also significantly contributes to grain yield [12,13,14]. Grain yield is also determined by grain number per panicle in sorghum, maize, and other cereals [15,16,17,18]. In addition, the grain-filling rate and multiple-ovary development at anthesis also affect grain weight and grain number per panicle, respectively [19].
The complex genetic architecture of grain yield-related traits is often controlled by multiple genes known as quantitative trait loci (QTLs), which are genetic loci responsible for phenotypic variation of a quantitative trait. QTLs generally refers to the genomic position of genes that control quantitative traits, which are located by flanking genetic markers. Researchers found one or more QTLs next to genetic markers by looking for the relationship between genetic markers and quantitative traits of interest. In other words, QTLs and flanking markers should be linked. QTL mapping refers to locating QTLs on the genetic map by using several methods, such as interval mapping (IM) and composite interval mapping (CIM). The Kosambi function is used to determine the distance between QTLs and genetic markers. A QTL with LOD value more than 2.5 usually represents a credibly genetic locus for a trait. WinQTLCart (version 2.5) is one of the most used linkage mapping software [20] and the results obtained are more relatively accurate when compared to others.
Map-based cloning and genome-wide association studies (GWASs) are two effective ways to dissect complex quantitative traits and identify candidate loci across the whole genome [21,22]. The discovery of major QTLs for grain yield-related traits is a key goal of crop genetic research. The fast development of DNA markers and the sorghum and maize genome sequencing projects make QTL detection for grain yield traits easier. In particular, in the last two decades, numerous studies have used a genetic linkage mapping method to map QTLs associated with grain yield-related traits in sorghum and maize [20,23]. However, cloned genes related to sorghum and maize yield-related traits are not well studied, maybe due to complex polygenic control and variable environmental effects. This review summarizes current knowledge of initial and fine-mapped QTLs and functionally characterized genes associated with sorghum and maize grain yield-related traits to provide a more in-depth understanding of genetic networks in crop floret development and grain yield improvement.
2. Increased Grain Yield through Crop Domestication
Crop domestication is a complex combination of various selection pressures, with frequent fluctuations in introgression levels throughout human history [24]. Archaeological and genetic studies suggested that sorghum [25] and maize [26] were domesticated in different regions from their wild ancestors. Sorghum originated from four prominent wild sorghum (Sorghum bicolor ssp. verticilliflorum (L.) Moench) races, notably aethiopicum, verticilliflorum, virgatum, and arundinaceum [27], in Ethiopia and Sudan 8000 years ago [25] and then swiftly disseminated to Southern Africa and Asia. Maize was domesticated 9000 years ago in southwest Mexico from the wild progenitor teosinte (Zea mays ssp. Parviglumis) [26,28]. According to recent comparative genomics techniques, maize and sorghum have tight genetic collinearity and distantly share a common ancestor, which was closely related to 11.9 MYA [10,29]. They also hypothesized that both share a highly common identity between duplicated loci of maize and the corresponding orthologous region of sorghum [10]. A current study also shows high homologous similarity associated with seed protein quality based on phylogenetic analysis of 511 single-copy homologs between sorghum kafirins and maize zeins [29,30].
During crop domestication, symbolic phenotypic modifications, such as increased grain size and grain weight, were key targets by creating genetic variety from wild species [31]. Wild species generally have small grains to avoid being found and eaten by animals to ensure their own transmission and reproduction [32]. However, they were gradually domesticated into large and heavy seeds to meet the demand for human food resources [33]. Genetic changes in other grain yield-related traits have also accumulated to feed the world. A case in point is that grain number per panicle of both sorghum and maize cultivars increased more than their wild progenitors [34,35,36]. Bommert et al. reported that grain number per panicle was negatively correlated with grain size and some other inflorescence architectures [37]. It is difficult to greatly improve current sorghum and maize production due to the bottleneck effect of a few favorable alleles [36,38].
Nodal positioning, simple pathways, minimal pleiotropic effects, and selection on standing genetic variations from wild species are main factors for promoting convergent molecular domestication in crops [39]. Convergent increased grain yield in crop evolution is generally caused by genetic mutations within orthologous genes. For example, rice GS3, located at a key nodal position of the G-protein pathway, was first cloned to be a major gene for grain length and grain weight [40,41]. Then, ZmGS3 [42] in maize and SbGS3 [43] in sorghum were also reported to be associated with grain weight in diverse varieties. Naturally, rare and excellent mutations originating from wild progenitors have accidently increased grain size, grain weight, and grain number. These beneficial alleles were consciously selected and broadly used in cultivar breeding by humans. For example, eight QTLs for kernel weight and four QTLs for kernel row number were identified through a segregated population derived from teosinte and maize [44]. In sorghum, nine QTLs have been detected for grain weight and grain width derived from wild Sorghum Virgatum and domesticated Sorghum bicolor [45]. In a recent study, fifteen significant QTLs for grain yield-related traits were identified from a population derived from wild Sorghum halepense and Sorghum bicolor [46].
3. Genetic Dissection of Grain Yield-Related Traits in Sorghum and Maize
3.1. Sorghum
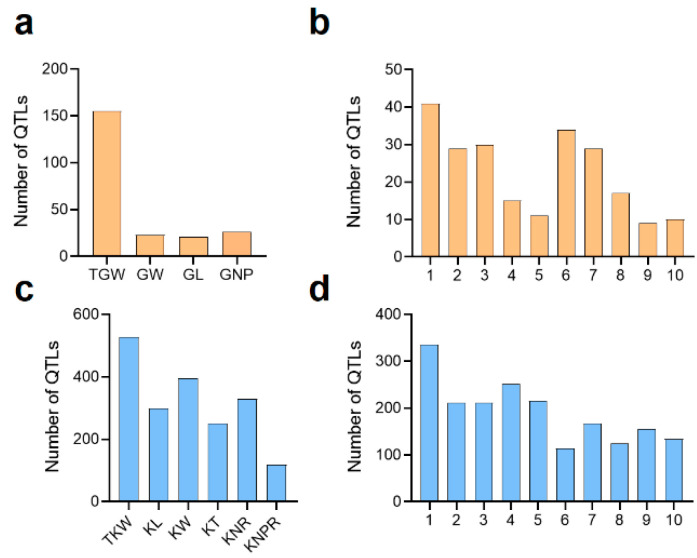
Grain yield-related traits have been extensively investigated in sorghum over the last two decades, and a large number of QTLs for grain weight and grain size are now mapped. Sorghum grain weight has shown to be highly inversely related to grain number per panicle [47,48] and significantly correlated with grain length and grain width [43]. To date, over 168 primarily mapped QTLs associated with grain size have been reported in sorghum. Among them, 155 QTLs located on ten chromosomes have been mapped for thousand grain weight (TGW) in various independent studies published between 1995 and 2021 [13,34,43,45,46,47,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66] (Figure 1a and Table S1). Few QTLs related to grain size (21 QTLs for grain length, 23 QTLs for grain width, and 26 QTLs for grain number per panicle) have been identified. Most grain size QTLs contribute minor phenotypic effects [43,45] and are unevenly distributed among ten chromosomes (Figure 1b).
Figure 1.
Number and distribution of quantitative trait loci (QTLs) for grain yield-related traits in sorghum and maize. (a) Number of QTLs associated with thousand-grain weight (TGW), grain width (GW), grain length (GL), and grain number per panicle (GNP) in sorghum. (b) Distribution of QTLs for grain size on ten sorghum chromosomes. (c) Number of QTLs associated with 1000-kernel weight (TKW), grain length (GL), grain width (GW), grain thickness (GT), kernel row number (KNR), and kernel number per row (KNPR) in maize. (d) Distribution of QTLs for grain yield-related traits on ten maize chromosomes.
Because of the quantitative nature, sensitivity to environmental effects, and complex genetic networks of grain yield-related traits, inadequate works have fine-mapped these QTLs in sorghum (Table 1). To date, few reports have attempted to predict candidate genes associated with sorghum grain weight and grain size QTLs. qTGW1a was recently defined as a 33-kb region on the long arm of chromosome 1 by flanking markers SM010165 and SM010171, which contain three candidate genes. One of them, SORBI_3001G341700, encodes an atypical G-protein γ subunit, and as a causative gene of qTGW1a, it negatively controls grain weight in sorghum. The wild-type qTGW1a allele showed decreased grain size, plant height and grain yield in transgenic overexpression rice lines [43]. This locus is homologous to rice GS3 and maize ZmGS3, which have also been shown to be negative regulators of grain length and grain weight [41,42,67,68]. Tao et al. [69] also confirmed a major QTL for grain size, which harbors the SbGS3 gene in a diverse sorghum panel. Two previously identified QTLs, QKwt.uga-C [65] and qtl1bGW [51], were also reported to be colocalized with qTGW1a [43]. GS3 can explain up to ~70% of the phenotypic variation in grain length among 180 rice varieties, while sorghum qTGW1a and maize ZmGS3 can explain 4–10% and 8% of the phenotypic variation in grain weight, respectively [42,43]. These results suggest that both sorghum and maize Gγ-like proteins cause minor effects on grain yield despite rice Gγ-like proteins.
Table 1.
Fine-mapped QTLs associated with grain yield-related traits in sorghum and maize.
| Crop | Trait a | QTL | Chr b | Marker Interval | Distance c | Candi d | Reference |
|---|---|---|---|---|---|---|---|
| Sorghum | GW | qGW1 | 1 | SB00037–SB00219 | 101 kb | 13 | [13] |
| GW | qTGW1a | 1 | SM010165–SM010171 | 33 kb | 3 | [43] | |
| Maize | KRN | qKRN8 | 8 | umc2571–umc2593 | 520 kb | 6 | [70] |
| KRN | qkrnw4 | 4 | Ch4.200–Ch4.K-2 | 33 kb | 2 | [71] | |
| KW | qKW-9.2 | 9 | FSR6–MSR36 | 630 kb | 28 | [72] | |
| KW | qKW7 | 7 | 7H-16–7F-5 | 647 kb | 4 | [73] | |
| KNPR | qKNR6 | 6 | M6–M8 | 110 kb | 2 | [74] | |
| KNPR | qKNPR6 | 6 | N6M19–umc1257 | 198 kb | 6 | [75] | |
| KNPR | qKN | 10 | bnlg1360–umc1645 | 480 kb | 1 | [76] | |
| KRN | KRN1.4 | 1 | umc1737–cic001 | 203 kb | 7 | [77] | |
| KRN | qKRN5b | 5 | umc1365–umc2512 | 147.2 kb | 3 | [78] | |
| KRN | krn1 | 1 | SNP1–SNP2 | 6.6 kb | 1 | [79] | |
| KRN | KRN4 | 4 | M6–M8 | 3 kb | 2 | [80] | |
| KL | qKL1.07 | 1 | ML194–ML162 | 1.6 Mb | 1 | [81] | |
| KL | qKL9 | 9 | C9-54–C9-58 | 942 kb | 24 | [82] | |
| KL | qKL-2 | 9 | mk3106–mk3114 | 1.95 Mb | 40 | [83] | |
| KW | qKW7b | 7 | M115.8–M116.7 | 59 kb | 1 | [84] | |
| KRN | KNE4 | 4 | umc1086–M5 | 440 kb | 14 | [85] | |
| GW | qGW4.05 | 4 | ND16–ND19 | 279.6 kb | 2 | [86] | |
| GW | qhkw5-3 | 5 | InYM20–InYM36 | 125.3 kb | 6 | [87] | |
| GW | qGW1.05 | 1 | umc1601–umc1754 | 1.11 Mb | 30 | [88] | |
| GW | qKW9 | 9 | M3484–M3506 | 20 kb | 3 | [89] |
a GW: grain weight; KRNL: kernel row number; KW: kernel width; KL: kernel length; KNPR: kernel number per row. b Chromosome. c physical distance (kb or Mb). d Number of candidate genes.
The other major QTL for sorghum grain weight, qGW1, was fine mapped into a 101 kb targeted region on the short arm of chromosome 1 flanking by markers SB00037 and SB00219. qGW1 could explain 20–40% of phenotypic variations across multiple genetic backgrounds and various environments. However, among the 13 putative candidate genes in this region, none of them had homologs to previously reported grain weight-related genes in other plants [13]. Even so, we compared reported genes that were domesticated in sorghum evolution [90] and found one possible gene (Sobic.001G038900) with a strong selection signal located in the fine-mapped region. This hypothesized gene encodes a DUF567 protein with high expression at the anthesis stage, and it should be further confirmed by researchers. Moreover, a novel QTL within a 5 Mb genomic region on chromosome 5 for TGW was recently discovered by Boyles et al. [50]. A closer look revealed that the highest-ranking marker was linked with a remorin protein (SORBI _3005G188400), which was primarily expressed in early young panicle and developing embryo tissues [91].
3.2. Maize
Kernel weight is confirmed to be significantly correlated with kernel length in maize [92,93,94]. Kernel size (same as sorghum grain size) also positively contributes to the end-use quality of maize [95]. Compared to sorghum, much more improvements have been made to identify major QTLs or genes for grain yield-related traits in maize. Currently, over 1920 QTLs associated with kernel size and kernel weight reported from 1994 to 2021 have been identified [11,12,44,53,70,71,72,73,83,92,93,94,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126,127,128,129,130,131]. Among these loci, 528 QTLs related to hundred kernel weight (HKW), 299 QTLs associated with kernel length, 386 QTLs associated with kernel width, 250 QTLs related to kernel thickness, 118 QTLs associated with kernel number per row (KNPR), and 329 QTLs related to kernel row number (KRN) were detected (Figure 1c and Table S2). All these QTLs were unevenly distributed among the ten chromosomes of maize (Figure 1d).
A total of 20 loci for grain yield-related traits were fine mapped in maize (Table 1). GW4.05, qhkw5-3, qGW1.05, and qKW9 are four major QTLs for kernel weight that have mapped on chromosomes 4, 5, 1, and 9, respectively. qGW4.05 mapped into a 279.6 kb interval between markers ND16 and ND19 that explained 23.9% of phenotypic variation and contained two annotated genes. GRMZM2G039934 is predicted to be a candidate for qGW4.05, which encodes a putative leucine-rich repeat receptor-like protein kinase. Six polymorphic sites in the CDS were significantly associated with kernel weight and size between parental lines (HZS and LV28). qGW4.05 exhibits the best pleiotropic effect on kernel weight, length, and width [86]. The other major QTL for kernel weight, qhkw5-3, was recently fine mapped into a 125.3 kb physical region between markers InYM20 and InYM36. Within this locus, six genes have been annotated [87]. qGW1.05 was narrowed down to a 1.11 Mb target region flanking the SSR markers umcl601 and umcl754, which explains 20.94% of the phenotypic variation in KW and contains 30 predicted genes [88]. qKW9 was recently narrowed down to a 20 kb target region between markers M3484 and M3506, with 3 annotated genes. One of them, Zm00001d048451, which encodes a PLS-DYW-type PPR protein, is the causal gene for kernel weight [89].
qKL1.07, qKL9, and qKL-2 are three major QTLs for kernel length. qKL1.07 on chromosome 1 was delimited into a 1.6 Mb genomic fragment flanking markers ML194 and ML162, which harbors 5 genes. qKL1.07, GRMZM2G348452 (ZmCKX10), encodes a cytokinin oxidase that explains 11% of phenotypic variations in kernel length [81]. qKL9 is the other major QTL on chromosome 9, which was identified to increase kernel length and HKW [82]. Liu et al. [94] previously reported that qKL9 was highly associated with kernel length in maize in multiple environments by using F2:3 families. This was repeatedly confirmed by mapping the BC2F2 and BC3F1 populations with 16.09% phenotypic variance in kernel length. This locus was eventually delimited into a 942 kb region between markers C9-54 and C9-58, which contained 24 annotated genes. Among them, Zm00001d046723, which encodes an expansin-A20 protein, is a potential candidate. Three indel variations in the 5′UTR of this gene induce significant differences in expression levels between near-isogenic lines (NILs) McqKL9−A and Mc, which are also associated with kernel length [82]. Another recent QTL, qKL-2, was physically mapped to a 1.95 Mb interval between markers mk3106 and mk3114 on chromosome 9 containing 40 genes. Only one gene, GRMZM2G006080, is supposed to be a putative candidate for qKL-2, which encodes the receptor-like protein kinase FERONIA [83]. The receptor kinase FERONIA has also demonstrated itself as a signaling pathway that negatively regulates the elongation of integument cells and then controls seed size in A. thaliana [132].
Two major QTLs, qKW7 and qKW9.2 for kernel width, were fine mapped on chromosomes 7 and 9, respectively. A major QTL, qKW7, was narrowed down to a 647 kb target region between markers 7H-16 and 7F-5, which contains 4 annotated genes. Among these kinases, GRMZM2G114706 encodes an ankyrin protein kinase that positively regulates kernel weight and kernel width in maize [73]. A major locus qKW7 for kernel width was recently fine-mapped using a series of backcross populations derived from a cross between YE478 and Huangzaosi. It was divided into two tightly linked intervals (qKW7a and qKW7b) with opposite phenotypic effects. qKW7a had a minor additive effect and was highly influenced by the environment. However, qKW7b had a high LOD value with additive effects and a larger kernel width due to harboring the YE478 allele, which is a promising locus for kernel width. The locus was eventually narrowed down to a 59 kb region, which contains a single gene. Zm00001d020460 encodes a putative zinc finger homeodomain (ZF-HD) transcription factor whose gene expression level is associated with kernel development [84]. qKW-9.2 for kernel width was mapped into a 630 kb region between markers FSR6 and MSR36 on chromosome 9, which explained 5.23–11.26% of phenotypic variances in 4 different locations [72].
KNE4, qKNR6, qKNPR6, and qKN are four major QTLs for KNPR that have been fine mapped on chromosomes 4, 6, and 10. KNE4 was localized to a 440 kb genomic region flanking umc1667 and umc2135. Only Zm00001d052399, which encodes a long-chain acyl-CoA synthetase and explains 26% of phenotypic variation in KNPR, is supposed to be a candidate gene [85]. qKNR6 was recently mapped into a 110 kb target region flanking markers M6 and M8, which harbors two candidate genes. Among these genes, Zm00001d036602 is a causative gene for qKNR6, which encodes a serine/threonine protein kinase that regulates KNPR variation at different gene expression levels [74]. qKNPR6 was narrowed into a 198 kb target region on chromosome 6 between markers N6M19 and umc1257, which contains 6 candidate genes. qKNPR6 explains 56.3% of KNPR variation and exhibits pleiotropic effects on ear length and kernel weight [75]. qKN was identified as a 480 kb region between markers bnlg1360 and umc1645 [76]. A favorable allele derived from parent 178 increased the KNPR by 6.08–10.76% [76].
The other major QTLs, KRN1.4, krn1, qkrnw4, KRN4, qKRN5b, and qKRN8, for KRN were fine-mapped on chromosomes 1, 4, 5, and 8, respectively. KRN1.4, a major locus that explained 50.48% of the phenotypic variance for KRN, was narrowed into a 203 kb region containing 7 predicted candidate genes. Among them, Indeterminate spikelet1 (ids1) is a probable candidate gene that encodes an APETALA2 (AP2)-like transcription factor that regulates inflorescence branching, floral meristem determinacy, and spikelet meristem determinacy [77]. qKRN5, as a major QTL for KRN, was previously reported on chromosome 5. Currently, through fine mapping in some advanced backcross populations, qKRN5 was dissected into two tightly linked loci, qKRN5a and qKRN5b. qKRN5b was a major QTL with a higher additive effect than qKRN5a and subsequently fine mapped into a 147.2 kb target region flanked by markers SC3603d1 and SC14631, which harbored three putative candidate genes. Among them, Zm00001d013603 encodes an endonuclease/exonuclease/phosphatase family protein that can hydrolyze phosphatidylinositol diphosphates and was identified as the causal gene of qKRN5b based on expression analysis and sequence variation between the two parental lines [78]. The other major QTL of krn1 has been delimited into a 6.6 kb genomic region on chromosome 1 flanked by SNP1 and SNP2 markers. A single gene located in krn1, Zm00001d034629, which encodes an (AP2) transcription factor, controls KRN in maize [79]. KRN4 was fine-mapped into a 3 kb region between markers M6 and M8. Within this region, a 1.2 kb transposon-containing insertion was present in the parental line H21NX531, which may function in increased KRN since this indel was strongly associated with KRN variation in diverse maize inbred lines [80]. qkrnw4 was located at a 33 kb interval between the Ch4.200-1 and Ch4. K-2 target region on chromosome 4, which contains two possible candidate genes. Zm00001d052910 encodes a putative protein and was supposed to be a candidate for KRN based on its gene expression and bioinformatics analysis [71]. Recently, qKRN8, a novel QTL for KRN in maize, was fine-mapped into a 520 kb target region and localized an interval between markers umc2571 and umc2593 on chromosome 8, which harbors six annotated genes. Among these genes, Zm00001d010007 encodes a START domain-containing protein as a causal gene for qKRN8 since the differential expression pattern was found in immature ears of NILs qKRN8Lian87 and qKRN8V54 [70].
4. Functionally Characterized Genes Associated with Grain Yield-Related Traits in Sorghum and Maize
In the last two decades, genome sequencing and DNA markers have led to rapid progress in cloning genes underlying grain yield-related traits. To date, a total of 30 genes have been cloned with grain yield-related traits in sorghum and maize (Table 2). Zou et al. [43] recently reported the only cloned gene, qTGW1a, for sorghum grain weight. qTGW1a (SORBI_3001G341700), the ortholog to rice GS3 [40,67] and maize ZmGS3 [42], encodes a G-protein γ subunit located at the N-terminus [43]. Overexpression of the sorghum qTGW1a gene was shown to be a negative regulator of grain weight in sorghum. A 5-bp insertion in the fifth exon of qTGW1a in the parental line LTR108 with a large grain weight results in a 61-amino-acid truncation in the C-terminal domain [43]. However, the authors showed that the natural truncated qTGW1a version caused heavier grain weight, which is inconsistent with the fact that rice truncated GS3-4 caused shorter grain length. This should be further carefully confirmed by the authors since the C-terminal domain can inhibit the negative regulatory function of the N-terminal domain [133]. GS3 is the first identified gene for determining grain size on chromosome 3 in rice, and it has been shown to explain up to 72% of phenotypic variance [40,67]. Mutation at the second exon changes a cysteine codon (TGC) to a stop codon (TGA) in the large rice grain varieties, while ZmGS3 has 5 exons encoding a protein with 198 amino acids. ZmGS3 encodes a putative transmembrane protein that has been demonstrated to play a function in maize kernel formation through methods that differ from rice GS3 [42,67]. Furthermore, qTGW1a is a minor gene for variations in sorghum grain weight due to its 4–10% phenotypic explanation [43].
Table 2.
Functionally characterized genes associated with grain yield-related traits in sorghum and maize.
| Crop | Gene | Trait a | Annotation | Variations | Ref b |
|---|---|---|---|---|---|
| Sorghum | MSD1 | GNP | TCP-domain TF | Missense mutation in msd1-1/2 | [16] |
| MSD2 | GNP | lipoxygenase (LOX) | Nonsense mutation in msd2-1, missense mutation in msd2-2, nonsense mutation in msd2-3 | [18] | |
| MSD3 | GNP | ω-3 fatty acid desaturase enzyme | in msd3-2, nonsense mutation in msd3-3, alternative splicing in msd3-1 and msd3-4 | [17] | |
| qTGW1a | GW | G-protein γ subunit | 5 bp insertion, frame shift | [43] | |
| Maize | ZmCEP1 | KS | C-terminal encoded peptide | Two frameshift mutations (1 bp insertion, 1 bp deletion) in zmcep1 | [134] |
| KNR6 | KNPR | Protein kinase | substitution mutations | [74] | |
| ZmNPF7.9 | KS | Nitrate transporter | Single nucleotide mutation (G to A) | [135] | |
| ids1/Ts6 | KRN | AP2-domain TF | 5 kb indel | [79] | |
| ZmSWEET4c | SF | Hexose transporter | Insertion in zmsweet4c | [136] | |
| ZmBAM1d | KW | CLV1/BAM receptor kinase | Insertion in zmbam1d | [137] | |
| qKW9 | KW | DYW-PPR protein | Deletion in qkw9 | [89] | |
| PPR2263 | KS | DYW-PPR protein | Insertion in ppr2263 | [138] | |
| Emp4 | KS | PLS- PPR proteins | Insertions in the emp4 | [139] | |
| Emp5 | KS | PLS- PPR proteins | 1.4 kb insertion in emp5 | [140] | |
| Emp7 | KS | PLS- PPR proteins | Insertion in emp7 | [141] | |
| Emp10 | KS | P-type PPR protein | 431 bp deletion in emp10 | [142] | |
| Emp9 | KS | P-type PPR protein | Insertion in emp9 | [143] | |
| Emp11 | KS | P-type PPR protein | Insertion in emp11 | [144] | |
| Emp12 | KS | PPR protein | Insertion in emp12 | [145] | |
| Emp16 | KS | P-type PPR protein | Insertion in emp16 | [146] | |
| Dek2 | KS | P-type PPR protein | Insertion in dek2 | [147] | |
| Zmsmk9 | KS | P-type PPR protein | Frameshift mutation in zmsmk9 | [148] | |
| Dek10 | KS | E-subgroup PPR protein | 5 bp insertion in dek10 | [149] | |
| Dek35 | KS | P-type PPR protein | Insertion in dek35 | [150] | |
| Dek36 | KS | E+ subgroup PPR | Insertion in dek36 | [151] | |
| Dek37 | KS | P-type PPR protein | Insertion in dek37 | [152] | |
| Dek39 | KS | PLS-PPR protein | Nonsense mutation in dek39 | [153] | |
| Smk1 | KS | PPR-E class protein | Missense, insertions in smk1 | [154] | |
| FEA2 | KRN | Leucine-rich repeat (LRR) receptor-like protein | Expression differences in fea2 | [155] | |
| UB3 | KRN | SBP-box TF | 2 kb transposon-containing indel | [80] |
a GNP: grain number per panicle; GW: grain weight; KS: kernel size; KNPR: kernel number per row; KRN: kernel row number; SF: seed filling; KW: kernel. b Reference. TF: transcription factor.
Multiseeded (MSD) genes (MSD1, MSD2, and MSD3) regulate a critical pathway involved in sorghum grain number per panicle [16,17,18]. Compared to wild-type BTx623, identified mutants with multiseeded panicles can produce up to over 200% of grain number per panicle. The MSD1 gene, SORBI_3007G021140 (SbTCP16), encodes a plant-specific teosinte branched/cycloidea/proliferating cell nuclear antigen-domain (TCP) transcription factor that is involved in jasmonic acid (JA) biosynthesis. Even though the msd1 mutant has a smaller grain weight, it compensates by producing more grains per panicle than the WT [156]. In addition, SORBI_3001G121600 (SbTCP2), a class II TCP-domain protein of the CYC/B1 family and homolog to maize TB1 and barley VRS5/HvTB1, also affects grain number in sorghum [157,158]. MSD2 (SORBI_3004G078600), an ortholog of the maize tassel seed 2 (TS2) gene, encodes a lipoxygenase (LOX), which can catalyze the conversion of free α-linolenic acid (18:3) to 13(S)-hydroperoxylinolenic acid, which acts as the first committed step of JA biosynthesis [18]. JAs play an important role in regulating inflorescence development of plants, such as pollen development in Arabidopsis [159], embryo development in tomatos [160,161], and spikelet formation in rice [162]. Both tassel seed 1 (TS1)- and TS2-mediated sex determination in maize were identified [163,164,165]. Ts1 encodes a plastid-targeted 13-lipooxygenase that catalyzes the first committed step in JA biosynthesis. In addition, SbTs1 (Ts1 ortholog of sorghum) is also highly correlated with the MSD phenotype in sorghum, which suggests a similar JA-mediated MSD pathway in crops [165]. Currently, new findings have reported that a six-row barley mutant vrs2 shows an MSD-like feature by Hv36160 and encodes the SHORT INTERNODES (SHI) transcriptional regulator, which is regulated by cross impacts of auxin, gibberellin and cytokinin [166]. Furthermore, MSD1 can regulate the gene expression of MSD2 by directly binding to its promoter [18]. Because JA biosynthesis is blocked in the msd1 and msd2 mutants, the pedicellate spikelet continues to develop into viable grains with complete spikelet fertility in both sessile spikelets (SSs) and pedicellate spikelets (PSs). Recently, a report showed that PS, as a functional organ, contributes to seed weight by translocating its photosynthetic products to the SSs in 10029 sorghums [167]. Restoring the fertility of PS ultimately causes a twofold increase in grain number per panicle through lost functions of MSD1 and MSD2 genes. [16,18]. MSD3, SORBI_3001G407600, which encodes a plastidial Ꙍ-3 fatty acid desaturase that acid desaturase enhances grain number by lowering JA levels. It catalyzes the conversion of linoleic acid (18:2) to linolenic acid (18:3), emphasizing the relevance of the JA regulatory module(s) in the control of the fertility of PS in sorghum. Loss-of-function mutations in MSD3 possess dramatically reduced linolenic acid (18:3), which lowers the level of endogenous JA [17]. The msd1, msd2, and msd3 mutant phenotypes can be recovered to sterile spikelets as the wild type by using methyl-JA treatment [16,17,18].
Although many QTLs for maize grain yield-related traits have already been identified, there have been few efforts to characterize the causative genes. However, several genes that regulate maize kernel development have been identified and functionally characterized. For example, Emp16, Emp10, Emp11, Emp12, Dek2, Dek35, Dek37, and Zmsmk9, which encode P-type pentatricopeptide repeat (PPR) proteins, are involved in the alternative splicing of mitochondrial genes to regulate maize kernel development. Loss of function of these genes can damage mitochondrial function and cause delayed kernel development, resulting in undersized kernels and decreased kernel yield [142,144,145,146,147,148,150,152]. The PLS-type pentatricopeptide repeat (PPR) genes Emp4, Emp5, Emp7, Emp9, Dek36, Dek10, Dek39, PPR2263, and Smk1 encode proteins that function in the C-to-U editing of transcripts in mitochondria and chloroplasts [138,139,140,141,143,149,151,153,154]. Loss of function of these distinguished genes potentially delays embryo and endosperm development, leading to insufficient kernel yield [168].
ZmSWEET4c, ZmNPF7.9, ZmCEP1, ZmBAM1d, qKW9, FEA2, KNR6, ids1/Ts6, and UB3 have also been responsible for grain yield-related traits in different manners. ZmSWEET4c, an ortholog of rice OsSWEET4, encodes a hexose transporter protein. It functions in sugar transport across the basal endosperm transfer cell layer (BETL), resulting in an increase in seed filling. Loss of function of both ZmSWEET4c and OsSWEET4 is defective in seed filling, including a dramatic loss of endosperm. The locus also shows a strong selection signature in ZmSWEET4c during maize domestication [136]. Recent reports show that the nitrate transporter ZmNPF7.9/NRT1.5 is involved in maize kernel development [169]. ZmNPF7.9 is highly expressed in the cells of BETL. Loss of function of ZmNPF7.9 results in faulty seed growth and aberrant starch deposition, resulting in a significant reduction in grain weight [135]. ZmCEP1, an ortholog of OsCEP6.1, as a short C-terminal encoded peptide (CEP), functions as a negative regulator of kernel weight in maize [134]. Compared to the wild type, overexpression of ZmCEP1 significantly reduces plant height, kernel size and 100-kernel weight, while knockout of ZmCEP1 by CRISPR/Cas9 noticeably enhances kernel yield-related traits [170]. The authors also showed that ZmCEP1 participates in nitrogen metabolism, nitrate, sugar transport and auxin response pathways to affect kernel development [134].
CLV1/BAM family genes were reported to control shoot meristem development [171], floret number and fruit size in plants [172,173]. Currently, a major QTL, qHKW1, which encodes a CLAVATA1 (CLV1)/BARELY ANY MERISTEM (BAM)-related receptor kinase, accounts for 18.4% of phenotypic variation in maize HKW. Sequence variation revealed a positive selection signal with low DNA methylation in the promoter region of ZmBAM1d, which enhances its gene expression and increases HKW during maize evolution [137]. Furthermore, overexpression of the retromer complex subunit ZmVPS29 results in a slenderer kernel morphology with a lower KW, a greater KNPR, and a larger grain yield per plant (YPP). ZmVPS29 also played a crucial role in improving maize yield during domestication [174]. Another major gene, qKW9, encodes a DYW-PPR protein and is essential for C-to-U editing ndhB, a subunit of the chloroplast NADH dehydrogenase-like complex. A 13 bp deletion in the coding sequence of qKW9 reduced photosynthetic activity, resulting in less maternal photosynthetic availability during the grain-filling process and a lower kernel weight [89]. FASCIATED EAR2 (FEA2), an ortholog of Arabidopsis CLAVATA2 (CLV2), encodes a leucine-rich repeat (LRR) receptor-like protein and regulates KRN by transferring signals from ZmCLV3 (the homolog to Arabidopsis CLAVATA3) peptide ligand to WUSCHEL (WUS) homeodomain transcription factor [175,176]. The loss function of FEA2 has irregular kernel rows, increased KNPR, and a much higher KRN than the wild type. The fea2-null mutant also shows a lower number of total kernels due to massive proliferation and resource competition [155]. ids1/Ts6 and Zm00001d034629 are orthologs of the wheat q gene, which encodes an AP2 domain protein. Overexpression of Ts6/ids1 has been demonstrated to produce more SPMs and higher KRN [79]. Ts6/ids1 can affect the transcripts of the downstream fea3, fea4, and ra3 genes involved in SPM development in maize [79]. Genes, such as kernel row number 4 (KRN4) [80], FEA2 [155], and GRF-interacting factor 1 (GIF1) [177], also control the development of maize ears. Among them, KRN4 can enhance kernel yield by increasing KNPR. KRN4, a 3-kb indel fragment located 60 kb downstream of the SBP-box gene Unbranched3 (UB3), is responsible for variations in KRN by regulating UB3 transcripts [80]. qKNR6, Zm00001d036602, encodes a serine/threonine protein kinase. Overexpression of KNR6 increases kernel yield, while the presence of transposable elements and long terminal repeat retrotransposons in the regulatory region of KNR6 decreases kernel yield in maize. KNR6 can phosphorylate Arf GTPase-activating protein (AGAP) to also affect ear length and kernel number [74].
5. Conclusions and Prospective
The past two decades have identified hundreds of QTLs for grain yield-related traits in sorghum and maize (Figure 1 and Tables S1 and S2). Most of the fine-mapped QTLs and functionally characterized genes related to grain weight, grain length, grain width, and grain number per panicle in sorghum and maize are shown in Table 1 and Table 2 and Tables S1 and S2. Grain yield-related traits are generally controlled by a few major and multiple minor loci due to its quantitative nature and environmental factors. We should focus more on QTLs that steadily occur in various environments to promote fast-track discovery of candidate genes and further gene functions for grain yield-related traits. Highly efficient map-based cloning mainly depends on the number of recombinants and marker density. Once a target QTL is narrowed into a small region, to clone the candidate genes accurately, it is better to select the recombinants with extremely recessive phenotypes. When the number of candidate genes are mapped to single digits, expression pattern analysis, genetic variation detection, and functional prediction of coding proteins between the two alleles are three available approaches to obtain the final candidate. Genetic scholars currently also use BSA-Seq, SNP chip, and GWAS strategies to obtain the major genes quickly and accurately by high-throughput genotyping technologies, such as the low-cost Specific locus amplified fragment sequencing (SLAF-seq) [178,179,180,181].
Grain weight is a key component of grain yield-related traits in sorghum and maize. It is mainly determined by genetic factors and is also affected by environmental stress, to a certain extent. The abiotic stress, such as drought, salt, alkali, and barren, could cause indirect yield loss by producing ionic stress, osmotic stress, and oxidative stress [182]. For example, compared to Sorghum propinquum, there are beneficial alleles of at least ten QTLs of Sorghum bicolor that could increase total aboveground biomass and grain yield under salt stress conditions [183]. When drought occurs in maize during the flowering stage, asynchronism occurs in the anthesis and silking interval (ASI), which leads to serious yield losses. Researchers found the genetic manipulation of ZmEXPA4 in developing maize ears could significantly reduce the ASI under drought conditions [184]. In addition to these factors, Murry et al. studied the genetic potential of sugar content in grain and stems of sorghum and found that the increase of stem sugar restricted the increase of grain yield. Thus, we can also use the QTLs related to sugar content to design grain sorghum (low-sugar stem) and sweet sorghum (high-sugar stem) [185].
Overall, this review summarizes noteworthy contributions to update the genetic basis of grain yield-related traits in sorghum and maize. The functionally characterized genes related to maize kernel yield may provide possible causal candidates for numerous collinear QTLs of sorghum, but further investigation should be given. With a more robust knowledge of these QTLs and their gene functions, we believe that genetic researchers and breeders could make good use of these molecular markers and beneficial alleles and remove linked harmful mutations to raise crop production by marker-assist breeding system (MAS). Furthermore, CRISPR-based genome editing [186], a new kind of revolutionary “5G-plant” technology and a transgene (DNA)-free approach to develop genetically modified organism (GMO), can rapidly allow for an optimum combination of various grain yield-related traits for designing ideal crops by directly, artificially, and accurately editing of target sequence in the future.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/ijms23052405/s1.
Author Contributions
Q.X., P.X. and W.B. designed and developed the framework; W.B. and P.X. wrote the manuscript; Q.X. and P.X. checked the literature review; Q.X., P.X. and W.B. contributed to the critical review and editing of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Key R&D Program of China (2019YFD1002701), Na-tional Natural Science Foundation of China (U1906204), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA24010306) and the Agricultural Breeding Program in NingXia Province (2019NYYZ04).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Charles H.G.J., Beddington J.R., Crute I.R., Haddad L., Lawrence D., Muir J.F., Pretty J., Robinson S., Thomas S.M., Toulmin C. Food Security, The Challenge of Feeding 9 Billion People. Science. 2010;327:812. doi: 10.1126/science.1185383. [DOI] [PubMed] [Google Scholar]
- 2.Schütz H.J.M., Verhoff M. A How to Feed the World in 2050. Arch. Kriminol. 2011;228:151–159. [PubMed] [Google Scholar]
- 3.Foley J.A., Ramankutty N., Brauman K.A., Cassidy E.S., Gerber J.S., Johnston M., Mueller N.D., O’Connell C., Ray D.K., West P.C., et al. Solutions for a cultivated planet. Nature. 2011;478:337–342. doi: 10.1038/nature10452. [DOI] [PubMed] [Google Scholar]
- 4.Herridge R.P., Day R.C., Baldwin S., Macknight R.C. Rapid analysis of seed size in Arabidopsis for mutant and QTL discovery. Plant Methods. 2011;7:3. doi: 10.1186/1746-4811-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morris G.P., Ramu P., Deshpande S.P., Hash C.T., Shah T., Upadhyaya H.D., Riera-Lizarazu O., Brown P.J., Acharya C.B., Mitchell S.E., et al. Population genomic and genome-wide association studies of agroclimatic traits in sorghum. Proc. Natl. Acad. Sci. USA. 2013;110:453–458. doi: 10.1073/pnas.1215985110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paterson A.H. Genomics of sorghum. Int. J. Plant Genom. 2008;2008:362451. doi: 10.1155/2008/362451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ranum P., Pena-Rosas J.P., Garcia-Casal M.N. Global maize production, utilization, and consumption. Ann. N. Y. Acad. Sci. 2014;1312:105–112. doi: 10.1111/nyas.12396. [DOI] [PubMed] [Google Scholar]
- 8.Paterson A.H., Bowers J.E., Bruggmann R., Dubchak I., Grimwood J., Gundlach H., Haberer G., Hellsten U., Mitros T., Poliakov A., et al. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457:551–556. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- 9.Ray D.K., Mueller N.D., West P.C., Foley J.A. Yield Trends Are Insufficient to Double Global Crop Production by 2050. PLoS ONE. 2013;8:e66428. doi: 10.1371/journal.pone.0066428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swigonova Z., Lai J., Ma J., Ramakrishna W., Llaca V., Bennetzen J.L., Messing J. Close split of sorghum and maize genome progenitors. Genome Res. 2004;14:1916–1923. doi: 10.1101/gr.2332504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Messmer R., Fracheboud Y., Banziger M., Vargas M., Stamp P., Ribaut J.M. Drought stress and tropical maize: QTL-by-environment interactions and stability of QTLs across environments for yield components and secondary traits. Theor. Appl. Genet. 2009;119:913–930. doi: 10.1007/s00122-009-1099-x. [DOI] [PubMed] [Google Scholar]
- 12.Li C., Li Y., Sun B., Peng B., Liu C., Liu Z., Yang Z., Li Q., Tan W., Zhang Y., et al. Quantitative trait loci mapping for yield components and kernel-related traits in multiple connected RIL populations in maize. Euphytica. 2013;193:303–316. doi: 10.1007/s10681-013-0901-7. [DOI] [Google Scholar]
- 13.Han L., Chen J., Mace E.S., Liu Y., Zhu M., Yuyama N., Jordan D.R., Cai H. Fine mapping of qGW1, a major QTL for grain weight in sorghum. Theor. Appl. Genet. 2015;128:1813–1825. doi: 10.1007/s00122-015-2549-2. [DOI] [PubMed] [Google Scholar]
- 14.Liu C., Zhou Q., Dong L., Wang H., Liu F., Weng J., Li X., Xie C. Genetic architecture of the maize kernel row number revealed by combining QTL mapping using a high-density genetic map and bulked segregant RNA sequencing. BMC Genom. 2016;17:915. doi: 10.1186/s12864-016-3240-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boyles R.E., Cooper E.A., Myers M.T., Brenton Z., Rauh B.L., Morris G.P., Kresovich S. Genome-Wide Association Studies of Grain Yield Components in Diverse Sorghum Germplasm. Plant Genome. 2016;9 doi: 10.3835/plantgenome2015.09.0091. [DOI] [PubMed] [Google Scholar]
- 16.Jiao Y., Lee Y.K., Gladman N., Chopra R., Christensen S.A., Regulski M., Burow G., Hayes C., Burke J., Ware D., et al. MSD1 regulates pedicellate spikelet fertility in sorghum through the jasmonic acid pathway. Nat. Commun. 2018;9:822. doi: 10.1038/s41467-018-03238-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dampanaboina L., Jiao Y., Chen J., Gladman N., Chopra R., Burow G., Hayes C., Christensen S.A., Burke J., Ware D., et al. Sorghum MSD3 Encodes an omega-3 Fatty Acid Desaturase that Increases Grain Number by Reducing Jasmonic Acid Levels. Int. J. Mol. Sci. 2019;20:5359. doi: 10.3390/ijms20215359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gladman N., Jiao Y., Lee Y.K., Zhang L., Chopra R., Regulski M., Burow G., Hayes C., Christensen S.A., Dampanaboina L., et al. Fertility of Pedicellate Spikelets in Sorghum Is Controlled by a Jasmonic Acid Regulatory Module. Int. J. Mol. Sci. 2019;20:4951. doi: 10.3390/ijms20194951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Z., van Oosterom E.J., Jordan D.R., Hammer G.L. Pre-anthesis ovary development determines genotypic differences in potential kernel weight in sorghum. J. Exp. Bot. 2009;60:1399–1408. doi: 10.1093/jxb/erp019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silva Lda C., Wang S., Zeng Z.B. Composite interval mapping and multiple interval mapping: Procedures and guidelines for using Windows QTL Cartographer. Methods Mol. Biol. 2012;871:75–119. doi: 10.1007/978-1-61779-785-9_6. [DOI] [PubMed] [Google Scholar]
- 21.Mace E., Innes D., Hunt C., Wang X., Tao Y., Baxter J., Hassall M., Hathorn A., Jordan D. The Sorghum QTL Atlas: A powerful tool for trait dissection, comparative genomics and crop improvement. Theor. Appl. Genet. 2019;132:751–766. doi: 10.1007/s00122-018-3212-5. [DOI] [PubMed] [Google Scholar]
- 22.Schaid D.J., Chen W., Larson N.B. From genome-wide associations to candidate causal variants by statistical fine-mapping. Nat. Rev. Genet. 2018;19:491–504. doi: 10.1038/s41576-018-0016-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mace E.S., Jordan D.R. Integrating sorghum whole genome sequence information with a compendium of sorghum QTL studies reveals uneven distribution of QTL and of gene-rich regions with significant implications for crop improvement. Theor. Appl. Genet. 2011;123:169–191. doi: 10.1007/s00122-011-1575-y. [DOI] [PubMed] [Google Scholar]
- 24.Hufford M.B., Lubinksy P., Pyhajarvi T., Devengenzo M.T., Ellstrand N.C., Ross-Ibarra J. The genomic signature of crop-wild introgression in maize. PLoS Genet. 2013;9:e1003477. doi: 10.1371/annotation/2eef7b5b-29b2-412f-8472-8fd7f9bd65ab. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wendorf F., Close A.E., Schild R., Wasylikowa K., Housley R.A., Harlan J.R., Królik H. Saharan exploitation of plants 8000 years BP. Nature. 1992;359:721–724. doi: 10.1038/359721a0. [DOI] [Google Scholar]
- 26.van Heerwaarden J., Doebley J., Briggs W.H., Glaubitz J.C., Goodman M.M., de Jesus Sanchez Gonzalez J., Ross-Ibarra J. Genetic signals of origin, spread, and introgression in a large sample of maize landraces. Proc. Natl. Acad. Sci. USA. 2011;108:1088–1092. doi: 10.1073/pnas.1013011108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harlan J.R., De Wet J.M.J., Stemler A., editors. Origins of African Plant Domestication. De Gruyter Mouton; The Hague, Paris: 1976. [DOI] [Google Scholar]
- 28.Matsuoka Y.V.Y., Goodman M.M., Sanchez J.G., Buckler E., Doebley J. A single domestication for maize shown. Proc. Natl. Acad. Sci. USA. 2002;99:6080–6084. doi: 10.1073/pnas.052125199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zou C., Li L., Miki D., Li D., Tang Q., Xiao L., Rajput S., Deng P., Peng L., Jia W., et al. The genome of broomcorn millet. Nat. Commun. 2019;10:436. doi: 10.1038/s41467-019-08409-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu J.H., Messing J. Amplification of prolamin storage protein genes in different subfamilies of the Poaceae. Theor. Appl. Genet. 2009;119:1397–1412. doi: 10.1007/s00122-009-1143-x. [DOI] [PubMed] [Google Scholar]
- 31.Allaby R.G., Fuller D.Q., Brown T.A. The genetic expectations of a protracted model for the origins of domesticated crops. Proc. Natl. Acad. Sci. USA. 2008;105:13982–13986. doi: 10.1073/pnas.0803780105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Susko D.J., Lovett-Doust L. Patterns of seed mass variation and their effects on seedling traits in Alliaria petiolata (Brassicaceae) Am. J. Bot. 2000;87:56–66. doi: 10.2307/2656685. [DOI] [PubMed] [Google Scholar]
- 33.Flint-Garcia S.A., Bodnar A.L., Scott M.P. Wide variability in kernel composition, seed characteristics, and zein profiles among diverse maize inbreds, landraces, and teosinte. Theor. Appl. Genet. 2009;119:1129–1142. doi: 10.1007/s00122-009-1115-1. [DOI] [PubMed] [Google Scholar]
- 34.Takanashi H., Shichijo M., Sakamoto L., Kajiya-Kanegae H., Iwata H., Sakamoto W., Tsutsumi N. Genetic dissection of QTLs associated with spikelet-related traits and grain size in sorghum. Sci. Rep. 2021;11:9398. doi: 10.1038/s41598-021-88917-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang C.J., Samayoa L.F., Bradbury P.J., Olukolu B.A., Xue W., York A.M., Tuholski M.R., Wang W., Daskalska L.L., Neumeyer M.A., et al. The genetic architecture of teosinte catalyzed and constrained maize domestication. Proc. Natl. Acad. Sci. USA. 2019;116:5643–5652. doi: 10.1073/pnas.1820997116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flint-Garcia S.A. Genetics and consequences of crop domestication. J. Agric. Food Chem. 2013;61:1876–8267. doi: 10.1021/jf305511d. [DOI] [PubMed] [Google Scholar]
- 37.Bommert N.S.-N.P., Jackson D., Hirano H.-Y. Genetics and evolution of inflorescence and flower development in grasses. Plant Cell Physiol. 2005;46:69–78. doi: 10.1093/pcp/pci504. [DOI] [PubMed] [Google Scholar]
- 38.Smith O., Nicholson W.V., Kistler L., Mace E., Clapham A., Rose P., Stevens C., Ware R., Samavedam S., Barker G., et al. A domestication history of dynamic adaptation and genomic deterioration in Sorghum. Nat. Plants. 2019;5:369–379. doi: 10.1038/s41477-019-0397-9. [DOI] [PubMed] [Google Scholar]
- 39.Lenser T., Theissen G. Molecular mechanisms involved in convergent crop domestication. Trends Plant Sci. 2013;18:704–714. doi: 10.1016/j.tplants.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 40.Wang C.F.S.Y.C., Xing Y. A causal C-A mutation in the second exon of GS3 highly associated with rice grain length and validated as a functional marker. Theor. Appl. Genet. 2009;118:465–472. doi: 10.1007/s00122-008-0913-1. [DOI] [PubMed] [Google Scholar]
- 41.Sun S., Wang L., Mao H., Shao L., Li X., Xiao J., Ouyang Y., Zhang Q. A G-protein pathway determines grain size in rice. Nat. Commun. 2018;9:851. doi: 10.1038/s41467-018-03141-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Q., Yang X., Bai G., Warburton M.L., Mahuku G., Gore M., Dai J., Li J., Yan J. Cloning and characterization of a putative GS3 ortholog involved in maize kernel development. Theor. Appl. Genet. 2010;120:753–763. doi: 10.1007/s00122-009-1196-x. [DOI] [PubMed] [Google Scholar]
- 43.Zou G., Zhai G., Yan S., Li S., Zhou L., Ding Y., Liu H., Zhang Z., Zou J., Zhang L., et al. Sorghum qTGW1a encodes a G-protein subunit and acts as a negative regulator of grain size. J. Exp. Bot. 2020;71:5389–5401. doi: 10.1093/jxb/eraa277. [DOI] [PubMed] [Google Scholar]
- 44.Liu Z., Cook J., Melia-Hancock S., Guill K., Bottoms C., Garcia A., Ott O., Nelson R., Recker J., Balint-Kurti P., et al. Expanding Maize Genetic Resources with Predomestication Alleles: Maize-Teosinte Introgression Populations. Plant Genome. 2016;9 doi: 10.3835/plantgenome2015.07.0053. [DOI] [PubMed] [Google Scholar]
- 45.Liu H., Liu H., Zhou L., Lin Z. Genetic Architecture of domestication- and improvement-related traits using a population derived from Sorghum virgatum and Sorghum bicolor. Plant Sci. 2019;283:135–146. doi: 10.1016/j.plantsci.2019.02.013. [DOI] [PubMed] [Google Scholar]
- 46.Nabukalu P., Kong W., Cox T.S., Pierce G.J., Compton R., Tang H., Paterson A.H. Genetic variation underlying kernel size, shape, and color in two interspecific S. bicolor2 × S. halepense subpopulations. Genet. Resour. Crop Evol. 2021;69:1261–1281. doi: 10.1007/s10722-021-01303-4. [DOI] [Google Scholar]
- 47.Tao Y., Mace E., George-Jaeggli B., Hunt C., Cruickshank A., Henzell R., Jordan D. Novel Grain Weight Loci Revealed in a Cross between Cultivated and Wild Sorghum. Plant Genome. 2018;11:170089. doi: 10.3835/plantgenome2017.10.0089. [DOI] [PubMed] [Google Scholar]
- 48.Yang Z., van Oosterom E.J., Jordan D.R., Doherty A., Hammer G.L. Genetic Variation in Potential Kernel Size Affects Kernel Growth and Yield of Sorghum. Crop Sci. 2010;50:685–695. doi: 10.2135/cropsci2009.06.0294. [DOI] [Google Scholar]
- 49.Bai C., Wang C., Wang P., Zhu Z., Cong L., Li D., Liu Y., Zheng W., Lu X. QTL mapping of agronomically important traits in sorghum (Sorghum bicolor L.) Euphytica. 2017;213:285. doi: 10.1007/s10681-017-2075-1. [DOI] [Google Scholar]
- 50.Boyles R.E., Pfeiffer B.K., Cooper E.A., Zielinski K.J., Myers M.T., Rooney W.L., Kresovich S. Quantitative Trait Loci Mapping of Agronomic and Yield Traits in Two Grain Sorghum Biparental Families. Crop Sci. 2017;57:2443–2456. doi: 10.2135/cropsci2016.12.0988. [DOI] [Google Scholar]
- 51.Shehzad T., Okuno K. QTL mapping for yield and yield-contributing traits in sorghum (Sorghum bicolor (L.) Moench) with genome-based SSR markers. Euphytica. 2014;203:17–31. doi: 10.1007/s10681-014-1243-9. [DOI] [Google Scholar]
- 52.Brown P.J., Klein P.E., Bortiri E., Acharya C.B., Rooney W.L., Kresovich S. Inheritance of inflorescence architecture in sorghum. Theor. Appl. Genet. 2006;113:931–942. doi: 10.1007/s00122-006-0352-9. [DOI] [PubMed] [Google Scholar]
- 53.Zhao Y., Su C. Mapping quantitative trait loci for yield-related traits and predicting candidate genes for grain weight in maize. Sci. Rep. 2019;9:16112. doi: 10.1038/s41598-019-52222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Murray S.C., Rooney W.L., Mitchell S.E., Sharma A., Klein P.E., Mullet J.E., Kresovich S. Genetic Improvement of Sorghum as a Biofuel Feedstock: II. QTL for Stem and Leaf Structural Carbohydrates. Crop Sci. 2008;48:2180–2193. doi: 10.2135/cropsci2008.01.0068. [DOI] [Google Scholar]
- 55.Nagaraja Reddy R., Madhusudhana R., Murali Mohan S., Chakravarthi D.V., Mehtre S.P., Seetharama N., Patil J.V. Mapping QTL for grain yield and other agronomic traits in post-rainy sorghum [Sorghum bicolor (L.) Moench] Theor. Appl. Genet. 2013;126:1921–1939. doi: 10.1007/s00122-013-2107-8. [DOI] [PubMed] [Google Scholar]
- 56.Srinivas G., Satish K., Madhusudhana R., Reddy R.N., Mohan S.M., Seetharama N. Identification of quantitative trait loci for agronomically important traits and their association with genic-microsatellite markers in sorghum. Theor. Appl. Genet. 2009;118:1439–1454. doi: 10.1007/s00122-009-0993-6. [DOI] [PubMed] [Google Scholar]
- 57.Fakrudin B., Kavil S.P., Girma Y., Arun S.S., Dadakhalandar D., Gurusiddesh B.H., Patil A.M., Thudi M., Khadi B.M., Kamatar M.Y., et al. Molecular mapping of genomic regions harbouring QTLs for root and yield traits in sorghum (Sorghum bicolor L. Moench) Physiol. Mol. Biol. Plants. 2013;19:409–419. doi: 10.1007/s12298-013-0188-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Phuong N., Stützel H., Uptmoor R. Quantitative trait loci associated to agronomic traits and yield components in a Sorghum bicolor L. Moench RIL population cultivated under pre-flowering drought and well-watered conditions. Agric. Sci. 2013;4:781–791. doi: 10.4236/as.2013.412107. [DOI] [Google Scholar]
- 59.Said A.A., Uptmoor R., El-Soda M. Mapping quantitative trait loci associated with yield and its related traits in Sorghum bicolor. Egypt. J. Agron. 2018;40:251–259. doi: 10.21608/agro.2018.5431.1113. [DOI] [Google Scholar]
- 60.Paterson A.H., Lin Y.R., Li Z., Schertz K.F., Doebley J.F., Pinson S.R., Liu S.-C., Stansel J.W., Irvine J.E. Convergent Domestication of Cereal Crops by Independent Mutations at Corresponding Genetic Loci. Science. 1995;269:1714–1718. doi: 10.1126/science.269.5231.1714. [DOI] [PubMed] [Google Scholar]
- 61.Tuinstra M.R., Grote E.M., Goldsbrough P.B., Ejeta G. Genetic analysis of post-flowering drought tolerance and components of grain development in Sorghum bicolor (L.) Moench. Mol. Breed. 1997;3:439–448. doi: 10.1023/A:1009673126345. [DOI] [Google Scholar]
- 62.Guindo D., Teme N., Vaksmann M., Doumbia M., Vilmus I., Guitton B., Sissoko A., Mestres C., Davrieux F., Fliedel G., et al. Quantitative trait loci for sorghum grain morphology and quality traits: Toward breeding for a traditional food preparation of West-Africa. J. Cereal Sci. 2019;85:256–272. doi: 10.1016/j.jcs.2018.11.012. [DOI] [Google Scholar]
- 63.Rami J.F., Dufour P., Trouche G., Fliedel G., Mestres C., Davrieux F., Blanchard P., Hamon P. Quantitative trait loci for grain quality, productivity, morphological and agronomical traits in sorghum (Sorghum bicolor L. Moench) Theor. Appl. Genet. 1998;97:605–616. doi: 10.1007/s001220050936. [DOI] [Google Scholar]
- 64.Mocoeur A., Zhang Y.M., Liu Z.Q., Shen X., Zhang L.M., Rasmussen S.K., Jing H.C. Stability and genetic control of morphological, biomass and biofuel traits under temperate maritime and continental conditions in sweet sorghum (Sorghum bicolour) Theor. Appl. Genet. 2015;128:1685–1701. doi: 10.1007/s00122-015-2538-5. [DOI] [PubMed] [Google Scholar]
- 65.Feltus F.A., Hart G.E., Schertz K.F., Casa A.M., Kresovich S., Abraham S., Klein P.E., Brown P.J., Paterson A.H. Alignment of Genetic Maps and QTLs between Inter-and Intra-Specific Sorghum Populations. Theor. Appl. Genet. 2006;112:1295–1305. doi: 10.1007/s00122-006-0232-3. [DOI] [PubMed] [Google Scholar]
- 66.Mekonnen T.B., Dong H., Getinet M., Gabizew A., Paterson A., Bantte K. QTL analysis in multiple sorghum mapping populations facilitates dissection of the genetic control of agronomic and yield-related traits in sorghum [Sorghum bicolor (Moench)] Euphytica. 2022;218:24. doi: 10.1007/s10681-022-02968-3. [DOI] [Google Scholar]
- 67.Fan C., Xing Y., Mao H., Lu T., Han B., Xu C., Li X., Zhang Q. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 2006;112:1164–1171. doi: 10.1007/s00122-006-0218-1. [DOI] [PubMed] [Google Scholar]
- 68.Liu H., Li Q., Xing Y. Genes Contributing to Domestication of Rice Seed Traits and Its Global Expansion. Genes. 2018;9:489. doi: 10.3390/genes9100489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tao Y., Zhao X., Wang X., Hathorn A., Hunt C., Cruickshank A.W., Oosterom E.J., Godwin I.D., Mace E.S., Jordan D.R. Large-scale GWAS in sorghum reveals common genetic control of grain size among cereals. Plant Biotechnol. J. 2019;18:1093–1105. doi: 10.1111/pbi.13284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Han X.Q.Y., Sandrine A.M.N., Qiu F. Fine mapping of qKRN8, a QTL for maize kernel row number, and prediction of the candidate gene. Theor. Appl. Genet. 2020;133:3139–3150. doi: 10.1007/s00122-020-03660-7. [DOI] [PubMed] [Google Scholar]
- 71.Nie N., Ding X., Chen L., Wu X., An Y., Li C., Song Y., Zhang D., Liu Z., Wang T., et al. Characterization and fine mapping of qkrnw4, a major QTL controlling kernel row number in maize. Theor. Appl. Genet. 2019;132:3321–3331. doi: 10.1007/s00122-019-03427-9. [DOI] [PubMed] [Google Scholar]
- 72.Raihan M.S., Liu J., Huang J., Guo H., Pan Q., Yan J. Multi-environment QTL analysis of grain morphology traits and fine mapping of a kernel-width QTL in Zheng58 x SK maize population. Theor. Appl. Genet. 2016;129:1465–1477. doi: 10.1007/s00122-016-2717-z. [DOI] [PubMed] [Google Scholar]
- 73.Li X., Li Y.-x., Chen L., Wu X., Qin W., Song Y., Zhang D., Wang T., Li Y., Shi Y. Fine mapping of qKW7, a major QTL for kernel weight and kernel width in maize, confirmed by the combined analytic approaches of linkage and association analysis. Euphytica. 2016;210:221–232. doi: 10.1007/s10681-016-1706-2. [DOI] [Google Scholar]
- 74.Jia H., Li M., Li W., Liu L., Jian Y., Yang Z., Shen X., Ning Q., Du Y., Zhao R., et al. A serine/threonine protein kinase encoding gene KERNEL NUMBER PER ROW6 regulates maize grain yield. Nat. Commun. 2020;11:988. doi: 10.1038/s41467-020-14746-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu R., Jia H., Cao X., Huang J., Li F., Tao Y., Qiu F., Zheng Y., Zhang Z. Fine mapping and candidate gene prediction of a pleiotropic quantitative trait locus for yield-related trait in Zea mays. PLoS ONE. 2012;7:e49836. doi: 10.1371/journal.pone.0049836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang G., Wang X., Wang B., Tian Y., Li M., Nie Y., Peng Q., Wang Z. Fine Mapping a Major QTL for Kernel Number per Row under Different Phosphorus Regimes in Maize (Zea mays L.) Theor. Appl. Genet. 2013;126:1545–1553. doi: 10.1007/s00122-013-2072-2. [DOI] [PubMed] [Google Scholar]
- 77.Calderón C.I., Yandell B.S., Doebley J.F. Fine Mapping of a QTL Associated with Kernel Row Number on Chromosome 1 of Maize. PLoS ONE. 2016;11:e0150276. doi: 10.1371/journal.pone.0150276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shen X., Zhao R., Liu L., Zhu C., Li M., Du H., Zhang Z. Identification of a candidate gene underlying qKRN5b for kernel row number in Zea mays L. Theor. Appl. Genet. 2019;132:3439–3448. doi: 10.1007/s00122-019-03436-8. [DOI] [PubMed] [Google Scholar]
- 79.Wang J., Lin Z., Zhang X., Liu H., Zhou L., Zhong S., Li Y., Zhu C., Lin Z. krn1, a major quantitative trait locus for kernel row number in maize. New Phytol. 2019;223:1634–1646. doi: 10.1111/nph.15890. [DOI] [PubMed] [Google Scholar]
- 80.Liu L., Du Y., Shen X., Li M., Sun W., Huang J., Liu Z., Tao Y., Zheng Y., Yan J., et al. KRN4 Controls Quantitative Variation in Maize Kernel Row Number. PLoS Genet. 2015;11:e1005670. doi: 10.1371/journal.pgen.1005670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Qin W., Li Y.-X., Wu X., Li X., Chen L., Shi Y., Song Y., Zhang D., Wang T., Li Y. Fine mapping of qKL1.07, a major QTL for kernel length in maize. Mol. Breed. 2016;36:8. doi: 10.1007/s11032-015-0419-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gong D., Tan Z., Zhao H., Pan Z., Sun Q., Qiu F. Fine mapping of a kernel length-related gene with potential value for maize breeding. Theor. Appl. Genet. 2021;134:1033–1045. doi: 10.1007/s00122-020-03749-z. [DOI] [PubMed] [Google Scholar]
- 83.Wang G., Zhao Y., Mao W., Ma X., Su C. QTL Analysis and Fine Mapping of a Major QTL Conferring Kernel Size in Maize (Zea mays) Front. Genet. 2020;11:603920. doi: 10.3389/fgene.2020.603920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tang B., Li Y., Mu Z., Chen L., Guo H., Chen Z., Li C., Liu X., Zhang D., Shi Y., et al. Fine mapping and candidate gene analysis of qKW7b, a major QTL for kernel width in maize. Mol. Breed. 2020;40:67. doi: 10.1007/s11032-020-01146-4. [DOI] [Google Scholar]
- 85.Zhan J., Wang F., Xing W., Liu J., Fan Z., Tao Y. Fine mapping and candidate gene prediction of a major QTL for kernel number per ear in maize. Mol. Breed. 2018;38:27. doi: 10.1007/s11032-018-0787-0. [DOI] [Google Scholar]
- 86.Chen L., Li Y.-X., Li C., Wu X., Qin W., Li X., Jiao F., Zhang X., Zhang D., Shi Y., et al. Fine-Mapping of QGW4.05, a Major QTL for Kernel Weight and Size in Maize. BMC Plant Biol. 2016;16:81. doi: 10.1186/s12870-016-0768-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li W., Bai Q., Zhan W., Ma C., Wang S., Feng Y., Zhang M., Zhu Y., Cheng M., Xi Z. Fine mapping and candidate gene analysis of qhkw5-3, a major QTL for kernel weight in maize. Theor. Appl. Genet. 2019;132:2579–2589. doi: 10.1007/s00122-019-03372-7. [DOI] [PubMed] [Google Scholar]
- 88.Zhou Q., Dong Y., Shi Q., Zhang L., Chen H., Hu C., Li Y. Verification and fine mapping of qGW1.05, a major QTL for grain weight in maize (Zea mays L.) Mol. Genet. Genom. 2017;292:871–881. doi: 10.1007/s00438-017-1318-0. [DOI] [PubMed] [Google Scholar]
- 89.Huang J., Lu G., Liu L., Raihan M.S., Xu J., Jian L., Zhao L., Tran T.M., Zhang Q., Liu J., et al. The Kernel Size-Related Quantitative Trait Locus qKW9 Encodes a Pentatricopeptide Repeat Protein That Aaffects Photosynthesis and Grain Filling. Plant Physiol. 2020;183:1696–1709. doi: 10.1104/pp.20.00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mace E.S., Tai S., Gilding E.K., Li Y., Prentis P.J., Bian L., Campbell B.C., Hu W., Innes D.J., Han X., et al. Whole-genome sequencing reveals untapped genetic potential in Africa’s indigenous cereal crop sorghum. Nat. Commun. 2013;4:2320. doi: 10.1038/ncomms3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Makita Y., Shimada S., Kawashima M., Kondou-Kuriyama T., Toyoda T., Matsui M. MOROKOSHI: Transcriptome database in Sorghum bicolor. Plant Cell Physiol. 2015;56:e6. doi: 10.1093/pcp/pcu187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li Y.L., Li X.H., Li J.Z., Fu J.F., Wang Y.Z., Wei M.G. Dent corn genetic background influences QTL detection for grain yield and yield components in high-oil maize. Euphytica. 2009;169:273–284. doi: 10.1007/s10681-009-9966-8. [DOI] [Google Scholar]
- 93.Peng B., Li Y., Wang Y., Liu C., Liu Z., Tan W., Zhang Y., Wang D., Shi Y., Li Y., et al. QTL analysis for yield components and kernel-related traits in maize across multi-environments. Theor. Appl. Genet. 2011;122:1305–1320. doi: 10.1007/s00122-011-1532-9. [DOI] [PubMed] [Google Scholar]
- 94.Liu Y., Wang L., Sun C., Zhang Z., Zheng Y., Qiu F. Genetic analysis and major QTL detection for maize kernel size and weight in multi-environments. Theor. Appl. Genet. 2014;127:1019–1037. doi: 10.1007/s00122-014-2276-0. [DOI] [PubMed] [Google Scholar]
- 95.Gupta P.K., Rustgi S., Kumar N. Genetic and molecular basis of grain size and grain number and its relevance to grain productivity in higher plants. Genome. 2006;49:565–571. doi: 10.1139/g06-063. [DOI] [PubMed] [Google Scholar]
- 96.Liu Y., Yi Q., Hou X., Hu Y., Li Y., Yu G., Liu H., Zhang J., Huang Y. Identification of quantitative trait loci for kernel-related traits and the heterosis for these traits in maize (Zea mays L.) Mol. Genet. Genom. 2020;295:121–133. doi: 10.1007/s00438-019-01608-1. [DOI] [PubMed] [Google Scholar]
- 97.Li L., Li X., Li L., Schnable J., Gu R., Wang J. QTL Identification and Epistatic Effect Analysis of Seed Size- and Weight-Related Traits in Zea mays L. Mol. Breed. 2019;39:67. doi: 10.1007/s11032-019-0981-8. [DOI] [Google Scholar]
- 98.Lan T., He K., Chang L., Cui T., Zhao Z., Xue J., Liu J. QTL mapping and genetic analysis for maize kernel size and weight in multi-environments. Euphytica. 2018;214:119. doi: 10.1007/s10681-018-2189-0. [DOI] [Google Scholar]
- 99.Zhang C., Zhou Z., Yong H., Zhang X., Hao Z., Zhang F., Li M., Zhang D., Li X., Wang Z., et al. Analysis of the genetic architecture of maize ear and grain morphological traits by combined linkage and association mapping. Theor. Appl. Genet. 2017;130:1011–1029. doi: 10.1007/s00122-017-2867-7. [DOI] [PubMed] [Google Scholar]
- 100.Shi Z., Song W., Xing J., Duan M., Wang F., Tian H., Xu L., Wang S., Su A., Li C., et al. Molecular mapping of quantitative trait loci for three kernel-related traits in maize using a double haploid population. Mol. Breed. 2017;37:108. doi: 10.1007/s11032-017-0706-9. [DOI] [Google Scholar]
- 101.Su C., Wang W., Gong S., Zuo J., Li S., Xu S. High Density Linkage Map Construction and Mapping of Yield Trait QTLs in Maize (Zea mays) Using the Genotyping-by-Sequencing (GBS) Technology. Front. Plant Sci. 2017;8:706. doi: 10.3389/fpls.2017.00706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yang C., Zhang L., Jia A., Rong T. Identification of QTL for maize grain yield and kernel-related traits. J. Genet. 2016;95:239–247. doi: 10.1007/s12041-016-0628-z. [DOI] [PubMed] [Google Scholar]
- 103.Chen J., Zhang L., Liu S., Li Z., Huang R., Li Y., Cheng H., Li X., Zhou B., Wu S., et al. The Genetic Basis of Natural Variation in Kernel Size and Related Traits Using a Four-Way Cross Population in Maize. PLoS ONE. 2016;11:e0153428. doi: 10.1371/journal.pone.0153428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wei X., Lu X., Zhang Z., Xu M., Mao K., Li W., Wei F., Sun P., Tang J. Genetic analysis of heterosis for maize grain yield and its components in a set of SSSL testcross populations. Euphytica. 2016;210:181–193. doi: 10.1007/s10681-016-1695-1. [DOI] [Google Scholar]
- 105.Huo D., Ning Q., Shen X., Liu L., Zhang Z. QTL Mapping of Kernel Number-Related Traits and Validation of One Major QTL for Ear Length in Maize. PLoS ONE. 2016;11:e0155506. doi: 10.1371/journal.pone.0155506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sa K.J., Park J.Y., Woo S.Y., Ramekar R.V., Jang C.S., Lee J.K. Mapping of QTL traits in corn using a RIL population derived from a cross of dent corn × waxy corn. Genes Genom. 2015;37:1–14. doi: 10.1007/s13258-014-0223-8. [DOI] [Google Scholar]
- 107.Alvarez Prado S., Sadras V.O., Borras L. Independent genetic control of maize (Zea mays L.) kernel weight determination and its phenotypic plasticity. J. Exp. Bot. 2014;65:4479–4487. doi: 10.1093/jxb/eru215. [DOI] [PubMed] [Google Scholar]
- 108.Zhang Z., Liu Z., Hu Y., Li W., Fu Z., Ding D., Li H., Qiao M., Tang J. QTL analysis of Kernel-related traits in maize using an immortalized F2 population. PLoS ONE. 2014;9:e89645. doi: 10.1371/journal.pone.0089645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li F., Jia H.T., Liu L., Zhang C.X., Liu Z.J., Zhang Z.X. Quantitative trait loci mapping for kernel row number using chromosome segment substitution lines in maize. Genet. Mol. Res. 2014;13:1707–1716. doi: 10.4238/2014.January.17.1. [DOI] [PubMed] [Google Scholar]
- 110.Tian B., Wang J., Wang G., Lübberstedt T. Confirmation of a major QTL on chromosome 10 for maize kernel row number in different environments. Plant Breed. 2014;133:184–188. doi: 10.1111/pbr.12132. [DOI] [Google Scholar]
- 111.Yang G., Li Y., Wang Q., Zhou Y., Zhou Q., Shen B., Zhang F., Liang X. Detection and integration of quantitative trait loci for grain yield components and oil content in two connected recombinant inbred line populations of high-oil maize. Mol. Breed. 2011;29:313–333. doi: 10.1007/s11032-011-9548-z. [DOI] [Google Scholar]
- 112.Cai H., Chu Q., Gu R., Yuan L., Liu J., Zhang X., Chen F., Mi G., Zhang F. Identification of QTLs for plant height, ear height and grain yield in maize (Zea mays L.) in response to nitrogen and phosphorus supply. Plant Breed. 2012;131:502–510. doi: 10.1111/j.1439-0523.2012.01963.x. [DOI] [Google Scholar]
- 113.Lu M., Xie C.-X., Li X.-H., Hao Z.-F., Li M.-S., Weng J.-F., Zhang D.-G., Bai L., Zhang S.-H. Mapping of quantitative trait loci for kernel row number in maize across seven environments. Mol. Breed. 2010;28:143–152. doi: 10.1007/s11032-010-9468-3. [DOI] [Google Scholar]
- 114.Guo J., Chen Z., Liu Z., Wang B., Song W., Li W., Chen J., Dai J., Lai J. Identification of genetic factors affecting plant density response through QTL mapping of yield component traits in maize (Zea mays L.) Euphytica. 2011;182:409. doi: 10.1007/s10681-011-0517-8. [DOI] [Google Scholar]
- 115.Li J.Z., Zhang Z.W., Li Y.L., Wang Q.L., Zhou Y.G. QTL consistency and meta-analysis for grain yield components in three generations in maize. Theor. Appl. Genet. 2011;122:771–782. doi: 10.1007/s00122-010-1485-4. [DOI] [PubMed] [Google Scholar]
- 116.Ding J.Q., Ma J.L., Zhang C.R., Dong H.F., Xi Z.Y., Xia Z.L., Wu J.Y. Mapping for Test Weight by Using F 2:3 Population in Maize. J. Genet. 2011;90:75–80. doi: 10.1007/s12041-011-0036-3. [DOI] [PubMed] [Google Scholar]
- 117.Tang J., Yan J., Ma X., Teng W., Wu W., Dai J., Dhillon B.S., Melchinger A.E., Li J. Dissection of the genetic basis of heterosis in an elite maize hybrid by QTL mapping in an immortalized F2 population. Theor. Appl. Genet. 2010;120:333–340. doi: 10.1007/s00122-009-1213-0. [DOI] [PubMed] [Google Scholar]
- 118.Li Q., Li L., Yang X., Warburton M.L., Bai G., Dai J., Li J., Yan J. Relationship, evolutionary fate and function of two maize co-orthologs of rice GW2 associated with kernel size and weight. BMC Plant Biol. 2010;10:7. doi: 10.1186/1471-2229-10-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zheng H.J., Wu A.Z., Zheng C.C., Wang Y.F., Cai R., Shen X.F., Xu R.R., Liu P., Kong L.J., Dong S.T. QTL mapping of maize (Zea mays) stay-green traits and their relationship to yield. Plant Breed. 2009;128:54–62. doi: 10.1111/j.1439-0523.2008.01529.x. [DOI] [Google Scholar]
- 120.Frascaroli E., Cane M.A., Landi P., Pea G., Gianfranceschi L., Villa M., Morgante M., Pe M.E. Classical genetic and quantitative trait loci analyses of heterosis in a maize hybrid between two elite inbred lines. Genetics. 2007;176:625–644. doi: 10.1534/genetics.106.064493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Coque M., Gallais A. Genomic regions involved in response to grain yield selection at high and low nitrogen fertilization in maize. Theor. Appl. Genet. 2006;112:1205–1220. doi: 10.1007/s00122-006-0222-5. [DOI] [PubMed] [Google Scholar]
- 122.Yan J.-B., Tang H., Huang Y.-Q., Zheng Y.-L., Li J.-S. Quantitative trait loci mapping and epistatic analysis for grain yield and yield components using molecular markers with an elite maize hybrid. Euphytica. 2006;149:121–131. doi: 10.1007/s10681-005-9060-9. [DOI] [Google Scholar]
- 123.Xiao Y.N., Li X.H., George M.L., Li M.S., Zhang S.H., Zheng Y.L. Quantitative Trait Locus Analysis of Drought Tolerance and Yield in Maize in China. Plant Mol. Biol. Rep. 2005;23:155–165. doi: 10.1007/BF02772706. [DOI] [Google Scholar]
- 124.Veldboom L.R., Lee M. Molecular-marker-facilitated studies of morphological traits in maize. II: Determination of QTLs for grain yield and yield components. Theor. Appl. Genet. 1994;89:451–458. doi: 10.1007/BF00225380. [DOI] [PubMed] [Google Scholar]
- 125.Liu Z., Garcia A., McMullen M.D., Flint-Garcia S.A. Genetic Analysis of Kernel Traits in Maize-Teosinte Introgression Populations. G3 Genes Genomes Genet. 2016;6:2523–2530. doi: 10.1534/g3.116.030155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhang X., Ren Z., Luo B., Zhong H., Ma P., Zhang H., Hu H., Wang Y., Liu D., Gao S., et al. Genetic architecture of maize yield traits dissected by QTL mapping and GWAS in maize. Crop J. 2021. in press . [DOI]
- 127.Hao D., Xue L., Zhang Z., Cheng Y., Chen G., Zhou G., Li P., Yang Z., Xu C. Combined linkage and association mapping reveal candidate loci for kernel size and weight in maize. Breed Sci. 2019;69:420–428. doi: 10.1270/jsbbs.18185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Liu J., Huang J., Guo H., Lan L., Wang H., Xu Y., Yang X., Li W., Tong H., Xiao Y., et al. The Conserved and Unique Genetic Architecture of Kernel Size and Weight in Maize and Rice. Plant Physiol. 2017;175:774–785. doi: 10.1104/pp.17.00708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Liu M., Tan X., Yang Y., Liu P., Zhang X., Zhang Y., Wang L., Hu Y., Ma L., Li Z., et al. Analysis of the genetic architecture of maize kernel size traits by combined linkage and association mapping. Plant Biotechnol. J. 2020;18:207–221. doi: 10.1111/pbi.13188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Liu L., Du Y., Huo D., Wang M., Shen X., Yue B., Qiu F., Zheng Y., Yan J., Zhang Z. Genetic architecture of maize kernel row number and whole genome prediction. Theor. Appl. Genet. 2015;128:2243–2254. doi: 10.1007/s00122-015-2581-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wang X., Liao C., Wang X., Yang R., Zhai L., Huang J. Construction of maize–teosinte introgression line population and identification of major quantitative trait loci. Euphytica. 2021;217:179. doi: 10.1007/s10681-021-02912-x. [DOI] [Google Scholar]
- 132.Yu F., Li J., Huang Y., Liu L., Li D., Chen L., Luan S. FERONIA receptor kinase controls seed size in Arabidopsis thaliana. Mol. Plant. 2014;7:920–922. doi: 10.1093/mp/ssu010. [DOI] [PubMed] [Google Scholar]
- 133.Mao H., Sun S., Yao J., Wang C., Yu S., Xu C., Li X., Zhang Q. Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc. Natl. Acad. Sci. USA. 2010;107:19579–19584. doi: 10.1073/pnas.1014419107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Xu R., Li Y., Sui Z., Lan T., Song W., Zhang M., Zhang Y., Xing J. A C-terminal encoded peptide, ZmCEP1, is essential for kernel development in maize. J. Exp. Bot. 2021;72:5390–5406. doi: 10.1093/jxb/erab224. [DOI] [PubMed] [Google Scholar]
- 135.Wei Y.M., Ren Z.J., Wang B.H., Zhang L., Zhao Y.J., Wu J.W., Li L.G., Zhang X.S., Zhao X.Y. A nitrate transporter encoded by ZmNPF7.9 is essential for maize seed development. Plant Sci. 2021;308:110901. doi: 10.1016/j.plantsci.2021.110901. [DOI] [PubMed] [Google Scholar]
- 136.Sosso D., Luo D., Li Q.B., Sasse J., Yang J., Gendrot G., Suzuki M., Koch K.E., McCarty D.R., Chourey P.S., et al. Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport. Nat. Genet. 2015;47:1489–1493. doi: 10.1038/ng.3422. [DOI] [PubMed] [Google Scholar]
- 137.Yang N., Liu J., Gao Q., Gui S., Chen L., Yang L., Huang J., Deng T., Luo J., He L., et al. Genome assembly of a tropical maize inbred line provides insights into structural variation and crop improvement. Nat. Genet. 2019;51:1052–1059. doi: 10.1038/s41588-019-0427-6. [DOI] [PubMed] [Google Scholar]
- 138.Sosso D., Mbelo S., Vernoud V., Gendrot G., Dedieu A., Chambrier P., Dauzat M., Heurtevin L., Guyon V., Takenaka M., et al. PPR2263, a DYW-Subgroup Pentatricopeptide repeat protein, is required for mitochondrial nad5 and cob transcript editing, mitochondrion biogenesis, and maize growth. Plant Cell. 2012;24:676–691. doi: 10.1105/tpc.111.091074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gutierrez-Marcos J.F., Dal Pra M., Giulini A., Costa L.M., Gavazzi G., Cordelier S., Sellam O., Tatout C., Paul W., Perez P., et al. empty pericarp4 encodes a mitochondrion-targeted pentatricopeptide repeat protein necessary for seed development and plant growth in maize. Plant Cell. 2007;19:196–210. doi: 10.1105/tpc.105.039594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Liu Y.J., Xiu Z.H., Meeley R., Tan B.C. Empty pericarp5 encodes a pentatricopeptide repeat protein that is required for mitochondrial RNA editing and seed development in maize. Plant Cell. 2013;25:868–883. doi: 10.1105/tpc.112.106781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sun F., Wang X., Bonnard G., Shen Y., Xiu Z., Li X., Gao D., Zhang Z., Tan B.C. Empty pericarp7 encodes a mitochondrial E-subgroup pentatricopeptide repeat protein that is required for ccmFN editing, mitochondrial function and seed development in maize. Plant J. 2015;84:283–295. doi: 10.1111/tpj.12993. [DOI] [PubMed] [Google Scholar]
- 142.Cai M., Li S., Sun F., Sun Q., Zhao H., Ren X., Zhao Y., Tan B.C., Zhang Z., Qiu F. Emp10 encodes a mitochondrial PPR protein that affects the cis-splicing of nad2 intron 1 and seed development in maize. Plant J. 2017;91:132–144. doi: 10.1111/tpj.13551. [DOI] [PubMed] [Google Scholar]
- 143.Yang Y.Z., Ding S., Wang H.C., Sun F., Huang W.L., Song S., Xu C., Tan B.C. The pentatricopeptide repeat protein EMP9 is required for mitochondrial ccmB and rps4 transcript editing, mitochondrial complex biogenesis and seed development in maize. New Phytol. 2017;214:782–795. doi: 10.1111/nph.14424. [DOI] [PubMed] [Google Scholar]
- 144.Ren X., Pan Z., Zhao H., Zhao J., Cai M., Li J., Zhang Z., Qiu F. EMPTY PERICARP11 serves as a factor for splicing of mitochondrial nad1 intron and is required to ensure proper seed development in maize. J. Exp. Bot. 2017;68:4571–4581. doi: 10.1093/jxb/erx212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sun F., Xiu Z., Jiang R., Liu Y., Zhang X., Yang Y.Z., Li X., Zhang X., Wang Y., Tan B.C. The mitochondrial pentatricopeptide repeat protein EMP12 is involved in the splicing of three nad2 introns and seed development in maize. J. Exp. Bot. 2019;70:963–972. doi: 10.1093/jxb/ery432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Xiu Z., Sun F., Shen Y., Zhang X., Jiang R., Bonnard G., Zhang J., Tan B.C. EMPTY PERICARP16 is required for mitochondrial nad2 intron 4 cis-splicing, complex I assembly and seed development in maize. Plant J. 2016;85:507–519. doi: 10.1111/tpj.13122. [DOI] [PubMed] [Google Scholar]
- 147.Qi W., Yang Y., Feng X., Zhang M., Song R. Mitochondrial Function and Maize Kernel Development Requires Dek2, a Pentatricopeptide Repeat Protein Involved in nad1 mRNA Splicing. Genetics. 2017;205:239–249. doi: 10.1534/genetics.116.196105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Pan Z., Liu M., Xiao Z., Ren X., Zhao H., Gong D., Liang K., Tan Z., Shao Y., Qiu F. ZmSMK9, a pentatricopeptide repeat protein, is involved in the cis-splicing of nad5, kernel development and plant architecture in maize. Plant Sci. 2019;288:110205. doi: 10.1016/j.plantsci.2019.110205. [DOI] [PubMed] [Google Scholar]
- 149.Qi W., Tian Z., Lu L., Chen X., Chen X., Zhang W., Song R. Editing of Mitochondrial Transcripts nad3 and cox2 by Dek10 Is Essential for Mitochondrial Function and Maize Plant Development. Genetics. 2017;205:1489–1501. doi: 10.1534/genetics.116.199331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Chen X., Feng F., Qi W., Xu L., Yao D., Wang Q., Song R. Dek35 Encodes a PPR Protein that Affects cis-Splicing of Mitochondrial nad4 Intron 1 and Seed Development in Maize. Mol. Plant. 2017;10:427–441. doi: 10.1016/j.molp.2016.08.008. [DOI] [PubMed] [Google Scholar]
- 151.Wang G., Zhong M., Shuai B., Song J., Zhang J., Han L., Ling H., Tang Y., Wang G., Song R. E+ subgroup PPR protein defective kernel 36 is required for multiple mitochondrial transcripts editing and seed development in maize and Arabidopsis. New Phytol. 2017;214:1563–1578. doi: 10.1111/nph.14507. [DOI] [PubMed] [Google Scholar]
- 152.Dai D., Luan S., Chen X., Wang Q., Feng Y., Zhu C., Qi W., Song R. Maize Dek37 Encodes a P-type PPR Protein That Affects cis-Splicing of Mitochondrial nad2 Intron 1 and Seed Development. Genetics. 2018;208:1069–1082. doi: 10.1534/genetics.117.300602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Li X., Gu W., Sun S., Chen Z., Chen J., Song W., Zhao H., Lai J. Defective Kernel 39 encodes a PPR protein required for seed development in maize. J. Integr. Plant Biol. 2018;60:45–64. doi: 10.1111/jipb.12602. [DOI] [PubMed] [Google Scholar]
- 154.Li X.J., Zhang Y.F., Hou M., Sun F., Shen Y., Xiu Z.H., Wang X., Chen Z.L., Sun S.S., Small I., et al. Small kernel 1 encodes a pentatricopeptide repeat protein required for mitochondrial nad7 transcript editing and seed development in maize (Zea mays) and rice (Oryza sativa) Plant J. 2014;79:797–809. doi: 10.1111/tpj.12584. [DOI] [PubMed] [Google Scholar]
- 155.Bommert P., Nagasawa N.S., Jackson D. Quantitative variation in maize kernel row number is controlled by the FASCIATED EAR2 locus. Nat. Genet. 2013;45:334–337. doi: 10.1038/ng.2534. [DOI] [PubMed] [Google Scholar]
- 156.Burow Z.X.G., Hayes C., Burke J. Characterization of a Multiseeded (msd1) Mutant of Sorghum for Increasing Grain Yield. Crop Sci. 2014;54:2030–2037. doi: 10.2135/cropsci2013.08.0566. [DOI] [Google Scholar]
- 157.Francis A., Dhaka N., Bakshi M., Jung K.H., Sharma M.K., Sharma R. Comparative phylogenomic analysis provides insights into TCP gene functions in Sorghum. Sci. Rep. 2016;6:38488. doi: 10.1038/srep38488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ramsay L., Comadran J., Druka A., Marshall D.F., Thomas W.T., Macaulay M., MacKenzie K., Simpson C., Fuller J., Bonar N., et al. INTERMEDIUM-C, a modifier of lateral spikelet fertility in barley, is an ortholog of the maize domestication gene TEOSINTE BRANCHED 1. Nat. Genet. 2011;43:169–172. doi: 10.1038/ng.745. [DOI] [PubMed] [Google Scholar]
- 159.Browse J. The power of mutants for investigating jasmonate biosynthesis and signaling. Phytochemistry. 2009;70:1539–1546. doi: 10.1016/j.phytochem.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 160.Goetz S., Hellwege A., Stenzel I., Kutter C., Hauptmann V., Forner S., McCaig B., Hause G., Miersch O., Wasternack C., et al. Role of cis-12-oxo-phytodienoic acid in tomato embryo development. Plant Physiol. 2012;158:1715–1727. doi: 10.1104/pp.111.192658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Li L., Zhao Y., McCaig B.C., Wingerd B.A., Wang J., Whalon M.E., Pichersky E., Howe G.A. The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell. 2004;16:126–143. doi: 10.1105/tpc.017954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Cai Q., Yuan Z., Chen M., Yin C., Luo Z., Zhao X., Liang W., Hu J., Zhang D. Jasmonic acid regulates spikelet development in rice. Nat. Commun. 2014;5:3476. doi: 10.1038/ncomms4476. [DOI] [PubMed] [Google Scholar]
- 163.DeLong A., Calderon-Urrea A., Dellaporta S.L. Sex determination gene TASSELSEED2 of maize encodes a short-chain alcohol dehydrogenase required for stage.specific floral organ abortion Rapid sequence evolution of the mammalian sex-determining gene SRY Rapid evolution of the sex determining locus in Old World mice and rats. Cell. 1993;74:757–768. doi: 10.1016/0092-8674(93)90522-R. [DOI] [PubMed] [Google Scholar]
- 164.Lunde C., Kimberlin A., Leiboff S., Koo A.J., Hake S. Tasselseed5 overexpresses a wound-inducible enzyme, ZmCYP94B1, that affects jasmonate catabolism, sex determination, and plant architecture in maize. Commun. Biol. 2019;2:114. doi: 10.1038/s42003-019-0354-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Acosta I.F., Laparra H., Romero S.P., Schmelz E., Hamberg M., Mottinger J.P., Moreno M.A., Dellaporta S.L. Tasselseed1 Is a Lipoxygenase Affecting Jasmonic Acid Signaling in Sex Determination of Maize. Science. 2009;323:262–265. doi: 10.1126/science.1164645. [DOI] [PubMed] [Google Scholar]
- 166.Youssef H.M., Eggert K., Koppolu R., Alqudah A.M., Poursarebani N., Fazeli A., Sakuma S., Tagiri A., Rutten T., Govind G., et al. VRS2 regulates hormone-mediated inflorescence patterning in barley. Nat. Genet. 2017;49:157–161. doi: 10.1038/ng.3717. [DOI] [PubMed] [Google Scholar]
- 167.AuBuchon-Elder T., Coneva V., Goad D.M., Jenkins L.M., Yu Y., Allen D.K., Kellogg E.A. Sterile Spikelets Contribute to Yield in Sorghum and Related Grasses. Plant Cell. 2020;32:3500–3518. doi: 10.1105/tpc.20.00424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Doll N.M., Royek S., Fujita S., Okuda S., Chamot S., Stintzi A., Widiez T., Hothorn M., Schaller A., Ingram G. A two-way molecular dialogue between embryo and endosperm is required for seed development. Science. 2020;367:431–435. doi: 10.1126/science.aaz4131. [DOI] [PubMed] [Google Scholar]
- 169.Guan H.-y., Dong Y.-b., Lu S.-p., Liu T.-s., He C.-m., Liu C.-x., Liu Q., Dong R., Wang J., Li Y.-l., et al. Characterization and map-based cloning of miniature2-m1, a gene controlling kernel size in maize. J. Integr. Agric. 2020;19:1961–1973. doi: 10.1016/S2095-3119(19)62797-8. [DOI] [Google Scholar]
- 170.Sui Z., Wang T., Li H., Zhang M., Li Y., Xu R., Xing G., Ni Z., Xin M. Overexpression of Peptide-Encoding OsCEP6.1 Results in Pleiotropic Effects on Growth in Rice (Oryza. sativa) Front. Plant Sci. 2016;7:228. doi: 10.3389/fpls.2016.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Nimchuk Z.L., Zhou Y., Tarr P.T., Peterson B.A., Meyerowitz E.M. Plant stem cell maintenance by transcriptional cross-regulation of related receptor kinases. Development. 2015;142:1043–1049. doi: 10.1242/dev.119677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Somssich M., Je B.I., Simon R., Jackson D. CLAVATA-WUSCHEL signaling in the shoot meristem. Development. 2016;143:3238–3248. doi: 10.1242/dev.133645. [DOI] [PubMed] [Google Scholar]
- 173.Janocha D., Lohmann J.U. From signals to stem cells and back again. Curr. Opin. Plant Biol. 2018;45:136–142. doi: 10.1016/j.pbi.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Chen L., Li Y.X., Li C., Shi Y., Song Y., Zhang D., Wang H., Li Y., Wang T. The retromer protein ZmVPS29 regulates maize kernel morphology likely through an auxin-dependent process(es) Plant Biotechnol. J. 2020;18:1004–1014. doi: 10.1111/pbi.13267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Guo Y., Han L., Hymes M., Denver R., Clark S.E. CLAVATA2 forms a distinct CLE-binding receptor complex regulating Arabidopsis stem cell specification. Plant J. 2010;63:889–900. doi: 10.1111/j.1365-313X.2010.04295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Schoof H., Lenhard M., Haecker A., Mayer K.F., Jürgens G., Laux T. The Stem Cell Population of Arabidopsis Shoot Meristems Is Maintained by a Regulatory Loop between the CLAVATA and WUSCHEL Genes. Cell. 2000;100:635–644. doi: 10.1016/S0092-8674(00)80700-X. [DOI] [PubMed] [Google Scholar]
- 177.Zhang D., Sun W., Singh R., Zheng Y., Cao Z., Li M., Lunde C., Hake S., Zhang Z. GRF-interacting factor1 Regulates Shoot Architecture and Meristem Determinacy in Maize. Plant Cell. 2018;30:360–374. doi: 10.1105/tpc.17.00791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zhang H., Wang X., Pan Q., Li P., Liu Y., Lu X., Zhong W., Li M., Han L., Li J., et al. QTG-Seq Accelerates QTL Fine Mapping through QTL Partitioning and Whole-Genome Sequencing of Bulked Segregant Samples. Mol. Plant. 2019;12:426–437. doi: 10.1016/j.molp.2018.12.018. [DOI] [PubMed] [Google Scholar]
- 179.Takagi H., Abe A., Yoshida K., Kosugi S., Natsume S., Mitsuoka C., Uemura A., Utsushi H., Tamiru M., Takuno S., et al. QTL-seq: Rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J. 2013;74:174–183. doi: 10.1111/tpj.12105. [DOI] [PubMed] [Google Scholar]
- 180.Wu S., Qiu J., Gao Q. QTL-BSA: A Bulked Segregant Analysis and Visualization Pipeline for QTL-seq. Interdiscip. Sci. 2019;11:730–737. doi: 10.1007/s12539-019-00344-9. [DOI] [PubMed] [Google Scholar]
- 181.Zhang S., Hu X., Miao H., Chu Y., Cui F., Yang W., Wang C., Shen Y., Xu T., Zhao L., et al. QTL identification for seed weight and size based on a high-density SLAF-seq genetic map in peanut (Arachis hypogaea L.) BMC Plant Biol. 2019;19:537. doi: 10.1186/s12870-019-2164-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Rankenberg T., Geldhof B., van Veen H., Holsteens K., Van de Poel B., Sasidharan R. Age-Dependent Abiotic Stress Resilience in Plants. Trends Plant Sci. 2021;26:692–705. doi: 10.1016/j.tplants.2020.12.016. [DOI] [PubMed] [Google Scholar]
- 183.Hostetler A.N., Govindarajulu R., Hawkins J.S. QTL mapping in an interspecific sorghum population uncovers candidate regulators of salinity tolerance. Plant Stress. 2021;2:100024. doi: 10.1016/j.stress.2021.100024. [DOI] [Google Scholar]
- 184.Liu B., Zhang B., Yang Z., Liu Y., Yang S., Shi Y., Jiang C., Qin F. Manipulating ZmEXPA4 expression ameliorates the drought-induced prolonged anthesis and silking interval in maize. Plant Cell. 2021;33:2058–2071. doi: 10.1093/plcell/koab083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Murray S.C., Sharma A., Rooney W.L., Klein P.E., Mullet J.E., Mitchell S.E., Kresovich S. Genetic Improvement of Sorghum as a Biofuel Feedstock: I. QTL for Stem Sugar and Grain Nonstructural Carbohydrates. Crop Sci. 2008;48:2165–2179. doi: 10.2135/cropsci2008.01.0016. [DOI] [Google Scholar]
- 186.Mushtaq M., Ahmad Dar A., Skalicky M., Tyagi A., Bhagat N., Basu U., Bhat B.A., Zaid A., Ali S., Dar T.U., et al. CRISPR-Based Genome Editing Tools: Insights into Technological Breakthroughs and Future Challenges. Genes. 2021;12:797. doi: 10.3390/genes12060797. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.