Abstract
BACKGROUND AND PURPOSE:
MR imaging provides critical information about fetal brain growth and development. Currently, morphologic analysis primarily relies on manual segmentation, which is time-intensive and has limited repeatability. This work aimed to develop a deep learning–based automatic fetal brain segmentation method that provides improved accuracy and robustness compared with atlas-based methods.
MATERIALS AND METHODS:
A total of 106 fetal MR imaging studies were acquired prospectively from fetuses between 23 and 39 weeks of gestation. We trained a deep learning model on the MR imaging scans of 65 healthy fetuses and compared its performance with a 4D atlas-based segmentation method using the Wilcoxon signed-rank test. The trained model was also evaluated on data from 41 fetuses diagnosed with congenital heart disease.
RESULTS:
The proposed method showed high consistency with the manual segmentation, with an average Dice score of 0.897. It also demonstrated significantly improved performance (P < .001) based on the Dice score and 95% Hausdorff distance in all brain regions compared with the atlas-based method. The performance of the proposed method was consistent across gestational ages. The segmentations of the brains of fetuses with high-risk congenital heart disease were also highly consistent with the manual segmentation, though the Dice score was 7% lower than that of healthy fetuses.
CONCLUSIONS:
The proposed deep learning method provides an efficient and reliable approach for fetal brain segmentation, which outperformed segmentation based on a 4D atlas and has been used in clinical and research settings.
In vivo fetal brain MR imaging has provided critical insight into normal fetal brain development and has led to improved and more accurate diagnoses of brain abnormalities in the high-risk fetus.1 Morphologic fetal MR imaging studies have been used to quantify disturbances in fetal brain development associated with congenital heart disease (CHD).2 However, image segmentation, an essential step in morphologic analysis, is time-consuming and prone to inter-/intraobserver variability.
There are 3 major challenges in fetal MR imaging that affect image quality and reliable anatomic delineation. First, fetal brain anatomy changes rapidly with advancing gestational age (GA), resulting in dramatic morphologic changes in brain tissues. Cortical maturation (ie, gyrification and sulcation) during the second and third trimesters transforms the smooth fetal surface into a highly convoluted structure. Second, changes in water content accompanying active myelination introduce high variations in MR imaging signal intensity and contrast across GAs.3,4 Third, at times, artifacts corrupt fetal images. For example, maternal respiration and irregular fetal movements often result in motion artifacts. Differences in conductivity between amniotic fluid and tissues can cause standing wave artifacts. In addition, the large FOV for the maternal abdomen and limited scan time result in reduced image resolution and partial volume effects, in which a single image voxel may contain mixed tissues.5 These artifacts are more severe in fetal brains than in adult brains. Altogether, these 3 issues hamper fetal brain segmentation.
Because of the limited availability of fetal data, preterm infant brain segmentation is primarily studied as an intermediate approach. Spatiotemporal atlases have been proposed to segment the brain from 28 weeks onward6 and the infant brain at 9–15 months7 and at 0–2 years.8 To address the tissue contrast changes and artifacts, Xue et al4 proposed a modified expectation-maximization method to reduce partial volume effects with subject-specific atlases. Shi et al9 developed a method combining subject-specific characteristics and a conventional atlas with similarity weights. Wang et al10 proposed a patch-based approach based on a subject-specific atlas. We refer you to Devi et al11 for a comprehensive review. Although the fetal brain can be segmented using atlases developed from preterm infants, differences between fetal and preterm brains have been reported, including brain volume12 and neural connectivity13 differences.
In recent years, several groups have developed fetal brain atlases that serve as useful resources for direct segmentation of the fetal brain.14 Habas et al15 developed a 4D atlas based on fetal brain MR images for the mid-second trimester (20–24 weeks). Gholipour et al16 constructed a spatiotemporal atlas for a wider range of GAs between 19 and 39 weeks. However, manual correction is still required after atlas-based segmentation.17 Therefore, it is critical to find an accurate and reliable fetal brain segmentation method that can minimize the intensive work and time involved in manual refinement and, more important, can reduce inter-/intrarater variability, thus improving reproducibility in large-cohort studies.
Deep convolutional neural networks (CNNs) have shown promising performance in fetal medical image analyses, including fetal brain segmentation. In addition to localizing ROIs (eg, SonoNet18), a fully convolutional network has been used to successfully segment the fetal abdomen,19 the whole fetal envelope,20 and the fetal body.21 A multiscale and fine-tuned CNN has been proposed for fetal left ventricle segmentation.22 Additional studies using CNNs focused on fetal brain extraction include the work by Rajchl et al23 called Deepcut, which was based on a CNN and a fully connected conditional random field. P-NET used CNNs with coarse and fine segmentation steps to locate the fetal brain.24 A gradient vector flow network has also been used.25 Likewise, 2D U-Net26 and multistate U-Net27 have been applied to fetal whole-brain extraction. Skull segmentation using a 2-stage CNN, in which the second stage comprises angle incidence and shadow casting maps has also been proposed.28 However, important segmentation that quantifies different brain tissue classes (eg, WM, GM, and CSF) is needed for a more comprehensive volumetric and morphologic assessment of the fetal brain.
A 2D U-Net method was proposed for multitissue fetal brain MR imaging segmentation.29 Khalili et al29 used data augmentation with simulated intensity inhomogeneity artifacts to enhance the robustness of the segmentation. This method, however, was trained on a very small cohort (n = 12). Recently, Payette et al30 evaluated several 2D segmentation methods using the Fetal Tissue Annotation and Segmentation Dataset. Of the deep learning models assessed, the combined IBBM model30 that included information from 3 separate 2D U-Net architectures (ie, axial, coronal, and sagittal) performed the best, suggesting the superiority of using information from 3 planes. 3D U-Net leverages the anatomic information in 3 directions and avoids segmentation failure due to section discontinuity that may arise with 2D models. One of the other models in the study, KispiU, directly compared a 2D with a 3D U-Net. Contrary to expectation, the 2D U-Net performed better; this result was attributed to the reduced number of training samples and the use of nonoverlapping patches in the 3D U-Net.
In this work, we implemented a 3D U-Net for the automatic segmentation of the fetal brain into multiple tissue classes. The proposed method was developed using 65 fetal MR imaging scans from healthy fetuses and was compared with a 4D atlas-based segmentation method. The performance of the 3D U-Net was also evaluated on the brain MR imaging scans of 41 fetuses diagnosed with CHD. We hypothesized that the proposed method would learn fetal brain anatomy in high-order space; thus, this approach could segment brain regions with superior accuracy compared with an atlas-based method. Moreover, we speculated that segmentation performance would be improved across GAs. Last, we hypothesized that the same method can be used to reliably segment the brains of clinically high-risk fetuses, such as those with CHD.
MATERIALS AND METHODS
In this study, MR imaging data were acquired as part of prospective fetal brain longitudinal studies between 2014 and 2017. Pregnant women with healthy or low-risk pregnancies and with fetuses diagnosed with CHD in utero were included in the study. Pregnant women with pregnancy-related complications, multiple pregnancies, known disorders, maternal medications or illicit drug use, claustrophobia, or non-MR imaging–safe implants were excluded. Fetuses with extracardiac anomalies or chromosomal abnormalities were excluded. The study was approved by the institutional review board of Children’s National Medical Center in Washington, DC. Written informed consent was obtained from all volunteers.
MR Imaging Data Acquisition and Preprocessing
MR images were collected on a 1.5T scanner (Discovery MR450, GE Healthcare). 2D T2-weighted images were acquired in coronal, sagittal, and axial planes with 3 repetitions using the following parameters: FOV = 32 cm, matrix size = 256 × 256, section thickness = 2 mm, TE = 160 ms, TR = 1100 ms. All pregnant women were scanned without sedation.
Images were reconstructed to a high-resolution 3D volume (resolution = 0.875 × 0.875 × 0.875 mm) using a validated section-to-volume method with motion correction.31 3D images were re-oriented manually. Skull stripping was performed using the FSL Brain Extraction Tool (http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/BET),32 and whole-brain masks were manually corrected as needed. Intensity inhomogeneities were corrected using the N4ITK algorithm.33
Deep Learning Segmentation with 3D U-Net
Fetal brain images were cropped at the edges and rescaled to a matrix size of 80 × 110 × 90. Image patches were randomly extracted with a size of 64 × 64 × 64. Patches were normalized by subtracting the mean and scaling by the SD so that values within the patch were between 0 and 1. A stride of 1 × 1 × 1 was used in the training patches, and 4 × 4 × 4 was used in the prediction patches. Furthermore, images were flipped along the left-right direction to generate additional data, and the labels of overlapped patched regions were decided by a majority voting approach in the prediction (Online Supplemental Data).
Compared with the standard U-Net, a parametric Rectified Linear Unit activation function was used. There were 96 initial features used. The Adam optimizer was used with a learning rate of 1e-4. Cross-entropy was used as the loss function. The model was trained for 20 epochs and was validated every 128 steps; the batch size was set at 4.
To optimize model performance, we tested image normalization and 3 augmentation methods, including no augmentation, left-right flip, and 3-direction flip. The tests were repeated 5 times to assess stability.
Performance Evaluation
The healthy fetal brain was segmented by registering a GA-matched T2 template from a 4D fetal brain atlas15 to the subject’s brain using Advanced Normalization Tools (ANTS; http://stnava.github.io/ANTs/).34 After transforming template tissue labels to the subject’s brain, segmentations were corrected manually by a senior physician-neuroscientist (J.D.A.-C.) with expertise in MR imaging–based fetal-neonatal brain segmentation. These manually refined images served as ground truth data. The 6 tissue classes of interest were the cortical gray matter (CGM), WM, CSF, deep gray matter (DGM), cerebellum, and brain stem (BS). The proposed 3D U-Net method was compared with segmentations generated by the Developing Brain Region Annotation With Expectation-Maximization (DRAW-EM) package (BioMedIa),35 a widely used and previously validated atlas-based method. The MR images of fetuses with CHD were segmented using the DRAW-EM method and were manually corrected by an MR imaging engineer (K.K.), highly trained in perinatal segmentation. Using a second atlas as the basis for the ground truth data for the CHD fetal brain segmentation allowed us to examine the performance of the proposed model with minimal bias.
The proposed method was evaluated on the healthy fetal data using 10-fold cross-validation. Performance of the 3D U-Net was compared with the atlas-based method. Outputs from both approaches were compared with ground truth data (ie, manually-corrected labels). Segmentation performance metrics, Dice score, 95% Hausdorff distance, sensitivity, and specificity for each brain tissue class were calculated and compared using the Wilcoxon signed-rank test. The trained 3D U-Net model was then used to segment brain MR imaging of fetuses with CHD to assess the generalizability of the model to the clinical milieu.
RESULTS
Study Population
The first data set included fetal brain MR images from healthy pregnancies. After we excluded images that contained severe motion artifacts, 65 fetal MR images from 54 fetuses (ie, 11 study participants underwent a second MR imaging 5–8 weeks later) between 24.4 and 39.4 weeks GA (mean, 32.5 [SD, 4.5] weeks) were evaluated. The second data set included brain MR images from 41 fetuses with CHD between 22.9 and 38.6 weeks GA (mean, 32.5 [SD, 3.8] weeks).
Performance with Augmentation and Normalization
The proposed method was more time-efficient than the atlas-based method. 3D U-Net segmentation took 2 minutes and 30 seconds to complete compared with 22 minutes for the atlas-based approach using 28 CPUs.
The proposed method had the best performance with image normalization and data augmentation using a left-right flip (Table 1). The training process using no augmentation resulted in a lower cross-entropy and a higher Dice score compared with the one using left-right flip augmentation. However, the validation performance was the opposite. This finding indicated that training without augmentation tended to overfit the data. With augmentation in 3 directions, the performance of training and validation was reduced, likely because of the unrealistic brain orientations produced. Furthermore, high Dice scores were achieved with normalized images, likely because of improved consistency among subjects and improved data balance from the reduced background.
Table 1:
Average performance of 3D U-Net augmentation methods and normalization across 5 repetitions
| Flip Augmentation |
No Normalization |
|||
|---|---|---|---|---|
| None | 3 Direction | Left-Right | Left-Right | |
| Training | ||||
| Cross-entropy | 0.0461 (SD, 0.0017) | 0.0596 (SD, 0.0006) | 0.0502 (SD, 0.0008) | 0.03504 (SD, 0.0010) |
| Dice score | 0.9380 (SD, 0.0016) | 0.9140 (SD, 0.0004) | 0.9315 (SD, 0.0012) | 0.9284 (SD, 0.0008) |
| Validation | ||||
| Cross-entropy | 0.0726 (SD, 0.0041) | 0.07472 (SD, 0.0050) | 0.06771 (SD, 0.0039) | 0.04062 (SD, 0.0023) |
| Dice score | 0.8698 (SD, 0.0136) | 0.8518 (SD, 0.0163) | 0.8819 (SD, 0.0122) | 0.8419 (SD, 0.0279) |
Accuracy of the 3D U-Net
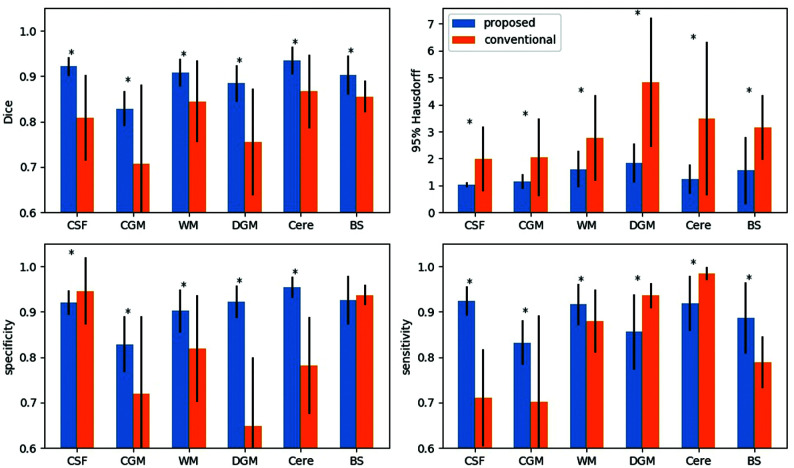
The proposed method showed high segmentation accuracy on our normative fetal sample. On the cross-validation of healthy fetuses, the proposed method yielded an average Dice score of 0.897 across the 6 brain regions compared with 0.806 for the atlas-based method. The Dice score per region was also significantly higher (P < .001) for the proposed method (Table 2).
Table 2:
Dice scores per region
| CSF | CGM | WM | DGM | Cere | BS | |
|---|---|---|---|---|---|---|
| 3D U-Net | 0.922 | 0.828 | 0.908 | 0.884 | 0.935 | 0.902 |
| Altas-based | 0.808 | 0.707 | 0.845 | 0.755 | 0.867 | 0.855 |
Note:—Cere indicates cerebellum
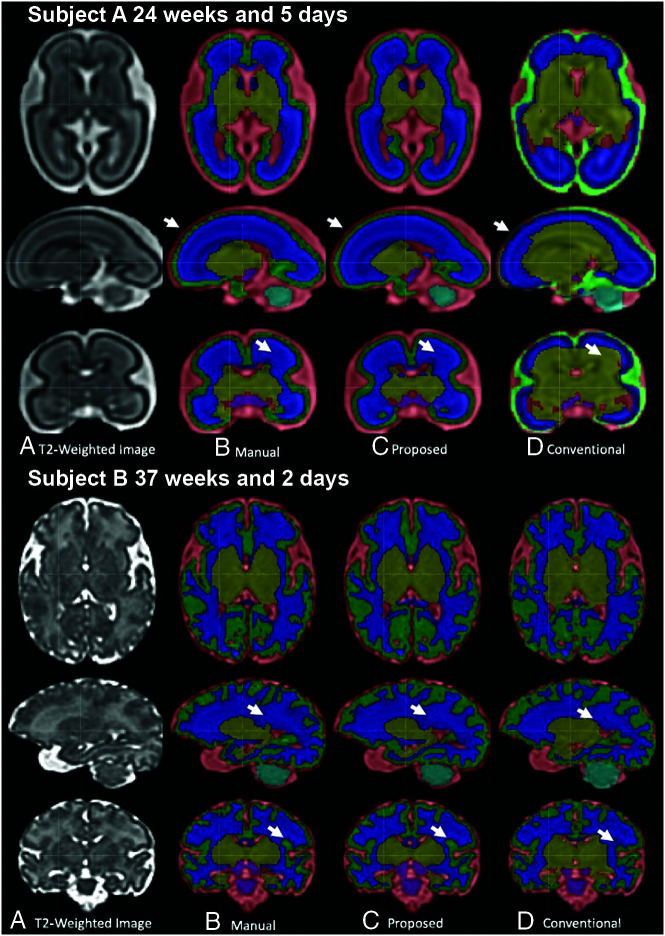
Figure 1A shows the segmentation results for a fetal brain at an early GA of 24 weeks and 5 days. The atlas-based method mislabeled the CSF as cortical gray matter, shown as light green when overlaid on the high-intensity signal of CSF in Fig 1 (upper row). The arrows on the sagittal/coronal images point to incorrectly labeled DGM, CSF, and CGM using the atlas-based approach. In contrast, the proposed method provided high consistency with the ground truth. Similarly, Fig 1B shows high consistency between the 3D U-Net and ground truth segmentation in a fetus at a late GA of 37 weeks and 2 days. In general, the proposed method resulted in smoother and continuous segmentation in the CGM compared with the atlas-based method.
FIG 1.
Comparison of segmentation methods on healthy fetuses of early and late GAs.
In all brain regions, the segmentation performance, measured with the Dice score and 95% Hausdorff distance, was significantly better (P < .001) for the proposed method compared with the atlas-based technique (Fig 2). Improved specificity and sensitivity scores were also noted in the CGM and WM regions for the 3D U-Net method.
FIG 2.
Regional comparisons between the proposed and conventional methods. The asterisk indicates P < .001. Cere indicates cerebellum.
Performance across GA
The proposed method showed consistent performance across GAs. As shown in Fig 3, the Dice score at each ROI was generally higher compared with the atlas-based method at each GA. In the CGM, the proposed method showed consistent performance from 24 to 39 weeks. In contrast, the atlas-based method resulted in reduced accuracy in the CSF and CGM at around 35 weeks, during which the secondary sulci develop. Furthermore, the conventional method resulted in reduced accuracy in the GM and WM regions around 26 weeks, during which early myelination occurs in the thalamus.
FIG 3.
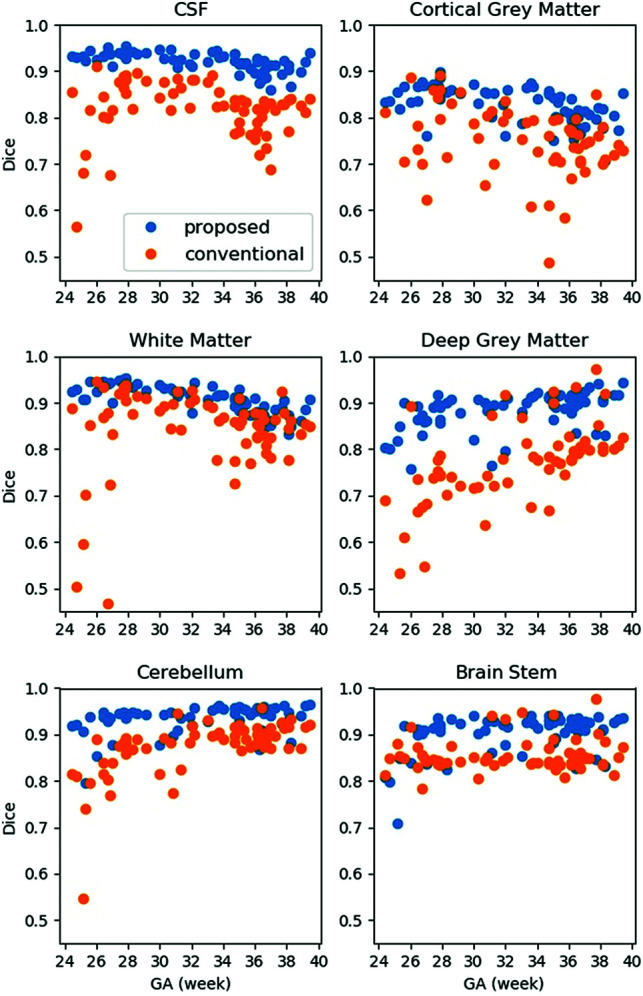
Regional performance across GAs.
Performance in the Fetus with CHD
The proposed model trained on the healthy fetal brain provided high accuracy in fetuses with CHD, as shown in Fig 4. The proposed method provided an average Dice score of 0.831 (0.802 in CSF, 0.744 in CGM, 0.871 in WM, 0.815 in DGM, 0.887 in the cerebellum, and 0.869 in the BS. This result was 7% lower than that of healthy fetuses.
FIG 4.
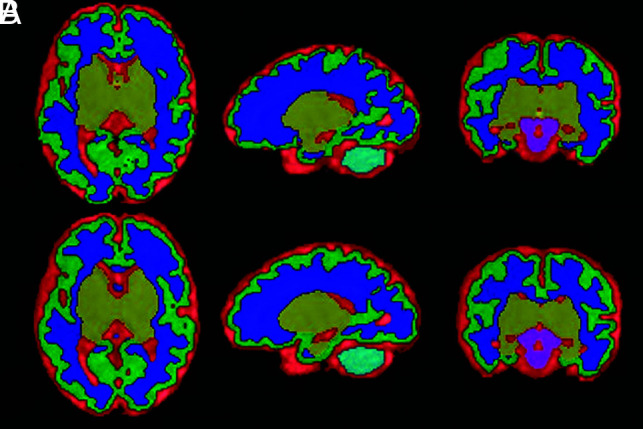
Brain segmentation in a fetus with CHD. A, Manually corrected segmentation. B, The proposed method.
DISCUSSION
In this work, we implemented a 3D U-Net model for fetal brain MR imaging segmentation and demonstrated superior performance compared with the atlas-based technique. The tissue labels generated by the proposed method were highly consistent with manual segmentations and were more accurate compared with segmentations produced using a spatiotemporal atlas. The superiority of the proposed method likely stems from the learning model, which enabled the identification of high-dimensional and intrinsic features of the fetal brains. Notably, the proposed approach provided more consistent performance across the evaluated GA range (ie, 24–39 weeks) compared with the atlas-based method. We speculate that this will provide more reliable fetal segmentations for future large-scale studies. This method has since been implemented in an automatic image-processing pipeline that provides regional segmentation for quantitative fetal MR imaging measures in our clinical and research studies.
The proposed method demonstrated superior segmentation performance for all regions compared with conventional segmentation based on the Dice scores and the 95% Hausdorff distance. Similarly, specificity and sensitivity scores for CGM and WM regions were higher using our proposed method. The atlas-based method tended to overestimate the segmentation of the cerebellum and DGM so that the labels for these tissues extended beyond the boundaries defined in the ground truth segmentation, as shown in Fig 2. This feature resulted in more accurate label overlap with the ground truth and higher sensitivity scores but much lower specificity scores than the proposed method. In contrast, the atlas-based technique tended to cover smaller CSF regions than the ground truth. Therefore, the segmented region was always inside the ground truth, leading to a higher specificity score. However, this segmentation approach also missed some true CSF regions, which resulted in lower sensitivity. Thus, the differences between our sensitivity and specificity scores appear to demonstrate inaccuracies of the conventional atlas-based method.
Data quality and preprocessing highly influence the quality of the image segmentation. In this work, the same data sets and preprocessing pipelines were used.15 Thus, the difference in segmentation performance was likely due to the segmentation method, but not the data quality and preprocessing. We expect that the superior performance of the proposed method will be preserved, given alternative data sets and processing steps; this expectation, however, needs to be empirically evaluated in future studies.
This study has several limitations. First, we used fewer data sets for training compared with adult brain segmentation studies. However, with 65 scans from healthy fetuses, the size of our data set is larger compared with previous fetal brain MR imaging studies (12–50 fetal scans). Second, the data in this study were acquired from the same scanner using an identical protocol. Thus, the reproducibility of the proposed method on other scanners requires further evaluation. Third, there are minor differences in the atlases used in the manual and conventional segmentations. However, because the ground truth was manually corrected, such mismatches were assumed to be removed. The definitions of the CGM and WM were similar in both atlases. Therefore, the performance of the proposed method can be confirmed reliably in these regions. Fourth, the proposed model was trained using healthy fetal data and was tested on fetuses with CHD. Inherent differences between the 2 data sets likely account for the reduced performance of the proposed method on the clinical CHD cohort. Nevertheless, an improved model based on transfer learning should be investigated further.
CONCLUSIONS
Our work demonstrated the feasibility and superior performance of the 3D U-Net method for fetal brain segmentation. The proposed method provided faster, higher accuracy, and more consistent segmentation across GAs compared with the conventional method based on atlases. Such advantages can provide reliable information for morphologic analysis and accurate quantitative criteria to support radiologists’ clinical diagnoses. Furthermore, the proposed pipeline will promote a standardized procedure and significantly facilitates the fetal brain image processing for large-cohort studies.
ABBREVIATIONS:
- BS
brain stem
- CGM
cortical GM
- CNN
convolutional neural network
- CHD
congenital heart disease
- DGM
deep GM
- GA
gestational age
Footnotes
This work was supported, in part, by R01HL116585 from the National Institutes of Health National Heart, Lung, and Blood Institute, NIH R21EB022309 from the National Institute of Biomedical Imaging and Bioengineering, and UL1TR001876 National Institutes of Health National Center for Advancing Translation Sciences.
Disclosure forms provided by the authors are available with the full text and PDF of this article at www.ajnr.org.
References
- 1.Clouchoux C, Guizard N, Evans AC, et al. Normative fetal brain growth by quantitative in vivo magnetic resonance imaging. Am J Obstet Gynecol 2012;206:173.e1–8 10.1016/j.ajog.2011.10.002 [DOI] [PubMed] [Google Scholar]
- 2.Limperopoulos C. Disorders of the fetal circulation and the fetal brain. Clin Perinatol 2009;36:561–77 10.1016/j.clp.2009.07.005 [DOI] [PubMed] [Google Scholar]
- 3.Despotović I, Goossens B, Philips W. MRI segmentation of the human brain: challenges, methods, and applications. Comput Math Methods Med 2015;2015:450341 10.1155/2015/450341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xue H, Srinivasan L, Jiang S, et al. Automatic segmentation and reconstruction of the cortex from neonatal MRI. Neuroimage 2007;38:461–77 10.1016/j.neuroimage.2007.07.030 [DOI] [PubMed] [Google Scholar]
- 5.Sui Y, Afacan O, Gholipour A, et al. Fast and high-resolution neonatal brain MRI through super-resolution reconstruction from acquisitions with variable slice selection direction. Front Neurosci 2021;15:636268 10.3389/fnins.2021.636268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Serag A, Aljabar P, Ball G, et al. Construction of a consistent high-definition spatio-temporal atlas of the developing brain using adaptive kernel regression. Neuroimage 2012;59:2255–65 10.1016/j.neuroimage.2011.09.062 [DOI] [PubMed] [Google Scholar]
- 7.Altaye M, Holland SK, Wilke M, et al. Infant brain probability templates for MRI segmentation and normalization. Neuroimage 2008;43:721–30 10.1016/j.neuroimage.2008.07.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi F, Yap PT, Wu G, et al. Infant brain atlases from neonates to 1- and 2-year-olds. PLoS One 2011;6:e18746 10.1371/journal.pone.0018746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi F, Shen D, Yap PT, et al. CENTS: cortical enhanced neonatal tissue segmentation. Hum Brain Mapp 2011;32:382–96 10.1002/hbm.21023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang L, Shi F, Li G, et al. Segmentation of neonatal brain MR images using patch-driven level sets. Neuroimage 2014;84:141–58 10.1016/j.neuroimage.2013.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devi CN, Chandrasekharan A, Sundararaman VK, et al. Neonatal brain MRI segmentation: a review. Comput Biol Med 2015;64:163–78 10.1016/j.compbiomed.2015.06.016 [DOI] [PubMed] [Google Scholar]
- 12.Bouyssi-Kobar M, Du Plessis AJ, McCarter R, et al. Third trimester brain growth in preterm infants compared with in utero healthy fetuses. Pediatrics 2016;138:e20161640 10.1542/peds.2016-1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomason ME, Scheinost D, Manning JH, et al. Weak functional connectivity in the human fetal brain prior to preterm birth. Sci Rep 2017;7:39286 10.1038/srep39286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Makropoulos A, Counsell SJ, Rueckert D. A review on automatic fetal and neonatal brain MRI segmentation. Neuroimage 2018;170:231–48 10.1016/j.neuroimage.2017.06.074 [DOI] [PubMed] [Google Scholar]
- 15.Habas PA, Kim K, Rousseau F, et al. A spatio-temporal atlas of the human fetal brain with application to tissue segmentation. Med Image Comput Comput Assist Interv 2009;12(Pt 1):289–96 10.1007/978-3-642-04268-3_36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gholipour A, Rollins CK, Velasco-Annis C, et al. A normative spatiotemporal MRI atlas of the fetal brain for automatic segmentation and analysis of early brain growth. Sci Rep 2017;7:476 10.1038/s41598-017-00525-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vasung L, Rollins CK, Velasco-Annis C, et al. Spatiotemporal differences in the regional cortical plate and subplate volume growth during fetal development. Cereb Cortex 2020;30:4438–53 10.1093/cercor/bhaa033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baumgartner CF, Kamnitsas K, Matthew J, et al. SonoNet: real-time detection and localisation of fetal standard scan planes in freehand ultrasound. IEEE Trans Med Imaging 2017;36:2204–15 10.1109/TMI.2017.2712367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu L, Xin Y, Li S, et al. Cascaded fully convolutional networks for automatic prenatal ultrasound image segmentation. In: Proceedings of the 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017). Melbourne, Australia, April 18–21, 2007 [Google Scholar]
- 20.Joskowicz L, Event A, Leo AJ. Interactive fetal envelope segmentation in MRI scans by real-time fine-tuning of a fully convolutional network. In: Proceedings of the Israel Radiological Association Annual Meeting. Eliat, Israel. October 23–25, 2019 [Google Scholar]
- 21.Li Y, Xu R, Ohya J, et al. Automatic fetal body and amniotic fluid segmentation from fetal ultrasound images by encoder-decoder network with inner layers. Annu Int Conf IEEE Eng Med Biol Soc 2017;2017:1485–88 10.1109/EMBC.2017.8037116 [DOI] [PubMed] [Google Scholar]
- 22.Yu L, Guo Y, Wang Y, et al. Segmentation of fetal left ventricle in echocardiographic sequences based on dynamic convolutional neural networks. IEEE Trans Biomed Eng 2017;64:1886–95 10.1109/TBME.2016.2628401 [DOI] [PubMed] [Google Scholar]
- 23.Rajchl M, Lee MCH, Oktay O, et al. Deepcut: object segmentation from bounding box annotations using convolutional neural networks. IEEE Trans Med Imaging 2017;36:674–83 10.1109/TMI.2016.2621185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mori K, Sakuma I, Sato Y, et al. eds. Medical image computing and computer-assisted intervention. In: Proceedings Part 1 of MICCAI 2013: 16th International Conference. Nagoya, Japan. September 22–26, 2013 [PubMed] [Google Scholar]
- 25.Rong Y, Xiang D, Zhu W, et al. Deriving external forces via convolutional neural networks for biomedical image segmentation. Biomed Opt Express 2019;10:3800–14 10.1364/BOE.10.003800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salehi SS, Hashemi SR, Velasco-Annis C, et al. Real-time automatic fetal brain extraction in fetal MRI by deep learning. In: Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018). Washington, D.C. April 4–7, 2018 10.1109/ISBI39256.2018 [DOI] [Google Scholar]
- 27.Lou J, Li D, Bui TD, et al. Automatic fetal brain extraction using multi-stage U-Net with deep supervision. In: Suk HI, Liu M, Yan P, et al. eds. Machine Learning in Medical Imaging: 10th International Workshop; MLMI 2019, Held in Conjunction with MICCAI 2019, Shenzhen, China, October 13, 2019, Proceedings. Vol 11861. Springer; 2019:592–600 [Google Scholar]
- 28.Cerrolaza JJ, Sinclair M, Li Y, et al. Deep learning with ultrasound physics for fetal skull segmentation. In: Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018). Washington, D.C. April 4–7, 2018 10.1109/ISBI39256.2018 [DOI] [Google Scholar]
- 29.Khalili N, Lessmann N, Turk E, et al. Automatic brain tissue segmentation in fetal MRI using convolutional neural networks. Magn Reson Imaging 2019;64:77–89 10.1016/j.mri.2019.05.020 [DOI] [PubMed] [Google Scholar]
- 30.Payette K, de Dumast P, Kebiri H, et al. An automatic multi-tissue human fetal brain segmentation benchmark using the Fetal Tissue Annotation Dataset. Sci Data 2021;8:167 10.1038/s41597-021-00946-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kainz B, Steinberger M, Wein W, et al. Fast volume reconstruction from motion corrupted stacks of 2D slices. IEEE Trans Med Imaging 2015;34:1901–13 10.1109/TMI.2015.2415453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith SM. Fast robust automated brain extraction. Hum Brain Mapp 2002;17:143–55 10.1002/hbm.10062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tustison NJ, Avants BB, Cook PA, et al. N4ITK: Improved N3 bias correction with robust B-spline approximation. In: Proceedings of the 2010 IEEE International Symposium on Biomedical Imaging: From Nano to Macro. Rotterdam, The Netherlands. April 14–17, 2010 10.1109/ISBI.2010.5490078 [DOI] [Google Scholar]
- 34.Avants BB, Tustison NJ, Song G, et al. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage 2011;54:2033–44 10.1016/j.neuroimage.2010.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Makropoulos A, Gousias IS, Ledig C, et al. Automatic whole brain MRI segmentation of the developing neonatal brain. IEEE Trans Med Imaging 2014;33:1818–31 10.1109/TMI.2014.2322280 [DOI] [PubMed] [Google Scholar]