Abstract
Background:
DNA methylation alterations may underlie associations between gestational perfluoroalkyl substances (PFAS) exposure and later-life health outcomes. To the best of our knowledge, no longitudinal studies have examined the associations between gestational PFAS and DNA methylation.
Objectives:
We examined associations of gestational PFAS exposure with longitudinal DNA methylation measures at birth and in adolescence using the Health Outcomes and Measures of the Environment (HOME) Study (2003–2006; Cincinnati, Ohio).
Methods:
We quantified serum concentrations of perfluorooctanoate (PFOA), perfluorooctane sulfonate (PFOS), perfluorononanoate (PFNA), and perfluorohexane sulfonate (PFHxS) in mothers during pregnancy. We measured DNA methylation in cord blood () and peripheral leukocytes at 12 years of age () using the Illumina HumanMethylation EPIC BeadChip. We analyzed associations between PFAS concentrations and repeated DNA methylation measures using linear regression with generalized estimating equations. We included interaction terms between children’s age and gestational PFAS. We performed Gene Ontology enrichment analysis to identify molecular pathways. We used Project Viva (1999–2002; Boston, Massachusetts) to replicate significant associations.
Results:
After adjusting for covariates, 435 cytosine–guanine dinucleotide (CpG) sites were associated with PFAS (false discovery rate, ). Specifically, we identified 2 CpGs for PFOS, 12 for PFOA, 8 for PFHxS, and 413 for PFNA; none overlapped. Among these, 2 CpGs for PFOA and 4 for PFNA were replicated in Project Viva. Some of the PFAS-associated CpG sites annotated to gene regions related to cancers, cognitive health, cardiovascular disease, and kidney function. We found little evidence that the associations between PFAS and DNA methylation differed by children’s age.
Discussion:
In these longitudinal data, PFAS biomarkers were associated with differences in several CpGs at birth and at 12 years of age in or near genes linked to some PFAS-associated health outcomes. Future studies should examine whether DNA methylation mediates associations between gestational PFAS exposure and health. https://doi.org/10.1289/EHP10118
Introduction
Perfluoroalkyl substances (PFAS) are a family of persistent synthetic chemicals with unique properties that allow them to resist water, oil, heat, and chemical reactions. PFAS have been used in industrial processes and commercial products including fire-fighting foams, paints, waxes, cleaning products, and textiles (Glüge et al. 2020). The major exposure pathway for PFAS is through the ingestion of contaminated food and drinking water, with inhalation of indoor air and house dust being minor pathways (ATSDR 2018). PFAS-contaminated drinking water is a growing concern; at least Americans have PFAS in their drinking water (Andrews and Naidenki 2020). A growing number of studies have linked prenatal exposure to PFAS to a range of health outcomes in children, such as low birth weight and preterm birth, decreased immune responses, cognitive function, later pubertal onset, obesity, and unfavorable cardiometabolic profiles (Braun 2017; Li et al. 2021; Liu and Peterson 2015; Liu et al. 2020; Rappazzo et al. 2017). However, the biological mechanisms by which gestational PFAS exposure may affect child health are not well understood.
Gestational exposure to PFAS may alter epigenetic modifications, including DNA methylation in fetuses, and these changes may in turn affect gene expression and related health outcomes in later life (Baccarelli and Bollati 2009; Kim et al. 2021a). DNA methylation, the most-studied epigenetic mark, is the attachment of a methyl group () to the C-5 position of a cytosine ring, which generally occurs at a cytosine–guanine dinucleotide (CpG) site. Alterations in DNA methylation play an important role in embryonic development and cell differentiation and are relatively stable and mitotically inheritable (Barrett 2017; Breton et al. 2017; Tang et al. 2015). In utero and neonatal exposure to PFAS have been linked to reduced global DNA and insulin like growth factor 2 (IGF2) methylation in cord blood (Guerrero-Preston et al. 2010; Kobayashi et al. 2017; Liu et al. 2018). IGF2, expressed in early embryonic development, is an important determinant of fetal growth (Randhawa and Cohen 2005). Thus, PFAS-induced DNA methylation changes in early development are a potential molecular pathway linking PFAS to human health.
Few studies have examined the associations of gestational PFAS exposure with changes in epigenome-wide DNA methylation in cord blood or placenta (Miura et al. 2018; Ouidir et al. 2020; Starling et al. 2020). Two studies found that higher serum perfluorooctanoate (PFOA) was significantly associated with lower cord blood DNA methylation at specific CpG sites (Miura et al. 2018; Starling et al. 2020), whereas one study found no significant associations (Ouidir et al. 2020). Some of these studies have reported significant associations of DNA methylation with serum perfluorooctane sulfonate (PFOS), perfluorohexane sulfonate (PFHxS) and perfluorononanoic acid (PFNA) concentrations (Miura et al. 2018; Ouidir et al. 2020). In addition, cross-sectional epigenome-wide association studies (EWAS) of PFAS with DNA methylation in adults have reported inconsistent findings (van den Dungen et al. 2017; Xu et al. 2020). Current studies are limited by their cross-sectional design, and we are unaware of studies examining longitudinal changes in DNA methylation. Identifying CpGs that are persistently associated with gestational PFAS exposure is critical because these CpGs may provide insights about biological pathways underlying the health effects of PFAS exposure (Baccarelli and Bollati 2009).
In this study, we used data from the Health Outcomes and Measures of the Environment (HOME) Study to characterize the association of maternal serum concentrations of PFOA, PFOS, PFNA and PFHxS with repeated measures of DNA methylation at birth and at 12 years of age. We used data from a different cohort, Project Viva, to replicate our findings.
Methods
Study Participants
For this analysis, we collected data from mother–child pairs who were enrolled in the HOME Study (discovery cohort). From March 2003 to January 2006, pregnant women living in the Cincinnati, Ohio, metropolitan area were recruited from nine prenatal clinics affiliated with three hospitals. Detailed information regarding the eligibility and recruitment methods can be found elsewhere (Braun et al. 2017). Briefly, pregnant women were enrolled at of gestation and their children were followed at 4 wk and at 1, 2, 3, 4, 5, 8, and 12 years of age. Originally, 468 pregnant women were recruited in the HOME Study and 401 completed the baseline visit. We excluded children who were twins. For follow-up visits at of age, 390 singleton children were eligible and 242 completed the follow-up assessments. Of these, 329 participants at baseline and 199 participants at follow-up had a sufficient blood sample volume available for DNA methylation measurements. We excluded 4 participants whose samples did not pass the quality control for DNA methylation measurements (see the section “DNA Methylation Analysis”). Of these, we included 291 participants (266 at baseline and 160 at follow-up) who had gestational PFAS measurements and complete covariate information. There were 135 participants who had data on both time points, and 156 participants who had data on a single time point. We did not find substantial differences in baseline sociodemographic features or PFAS concentrations between mother–child pairs with one or both time points (Table S1). Additional information including participant characteristics and collection of child information for the follow-up visit at 12 years of age have been published previously (Braun et al. 2020).
The institutional review boards at the Cincinnati Children’s Hospital Medical Center and the participating hospitals approved this study prior to enrollment. Pregnant women provided written informed consent upon enrollment at each study visit; their children provided written informed assent at 12 years of age.
Gestational PFAS Measurements
To estimate gestational exposure to PFAS, we quantified the concentrations of PFOA, PFOS, PFNA, and PFHxS in serum collected from pregnant women at of gestation (10.4–30.3 wk). Serum PFAS concentrations were measured using online solid-phase extraction coupled to high-performance liquid chromatography–isotope dilution tandem mass spectrometry (HPLC-MS/MS) at the Centers for Disease Control and Prevention (CDC) laboratory following a modified analytical method (CDC 2015; Kato et al. 2011, 2018). The limits of detection (LODs) were for PFNA, for PFOA and PFHxS, and for PFOS; each of the four PFAS were detectable in at least 98% of serum samples. PFAS concentrations below the LOD were replaced with the LOD divided by the square root of 2.
DNA Methylation Analysis
Blood samples were collected at delivery (cord blood) and at 12 years of age and then stored at before performing DNA methylation analysis. Venous cord blood samples were collected immediately after birth via venipuncture. Trained phlebotomists obtained fasting blood samples from the children at 12 years of age. We extracted genomic DNA (gDNA) from whole blood using the DNeasy Blood & Tissue Kit (Qiagen) with RNase A (; cat. no. 19101), following the protocols provided by the manufacturer. We quantified DNA methylation at 866,836 CpG sites per sample using the Infinium HumanMethylationEPIC BeadChip (EPIC) array (Illumina). The EPIC array covers of the CpGs quantified by the Infinium HumanMethylation450 BeadChip (450K) array (Solomon et al. 2018). Briefly, gDNA () was subject to sodium bisulfite conversion using the EZ DNA Methylation Kit (Zymo Research). Bisulfite-modified DNA was then whole-genome amplified, enzymatically fragmented, and hybridized to the Infinium MethylationEPIC BeadChip (Illumina), which was analyzed using an Illumina Hi-Scan System. All the laboratory work was performed according to Illumina’s protocols at the Genomics, Epigenomics and Sequencing Core at the University of Cincinnati College of Medicine. Paired samples (at delivery and at 12 years of age) were randomly assigned to positions on the same BeadChip, balanced by child sex.
Standard protocols were followed for the normalization and quality control of raw DNA methylation data (Kelsey et al. 2019; Wilhelm-Benartzi et al. 2013). Briefly, we removed samples with detection -values (; ) and probes with at least one high-detection -value using the same cutoff. We also removed sex chromosomes, loci associated with single-nucleotide polymorphisms and cross-hybridizing probes, as previously described (Zhou et al. 2017). This resulted in 669,622 autosomal probes being included in the analysis. We used Noob (preprocessNoob: minfi Bioconductor package) for background correction and dye bias normalization. In addition, we used Beta MIxture Quantile dilation (BMIQ) (wateRmelon) to correct the probe design bias (Teschendorff et al. 2013), and we used ComBat to adjust for potential batch effects (Wilhelm-Benartzi et al. 2013). The ComBat() function of the SVA package (Leek et al. 2012) was used to correct for batch effects at the BeadChip level using a parametric empirical Bayes procedure (Johnson et al. 2007). We performed all statistical analyses using logit-transformed , where beta is the ratio of intensities measured by methylated probes and the sum of intensities measured by both methylated and unmethylated probes (Du et al. 2010).
To account for heterogeneity in leukocyte and nucleated red blood cells across participants, we estimated the relative proportions of cell types in blood samples using the estimateCellCounts2 function in the R package FlowSorted.Blood.EPIC (Salas et al. 2018). We calculated the proportions of seven cell types (B lymphocytes, and T cells, neutrophils, monocytes, natural killer (NK) cells, and nucleated red blood cells) in cord blood, using an umbilical cord blood reference data set (Gervin et al. 2019). For child blood samples at 12 years of age, we estimated the proportions of six cell types (B lymphocytes, and T cells, neutrophils, monocytes, and NK cells) using an adult reference data set (Salas et al. 2018).
Covariates
We adjusted for covariates that were associated with gestational PFAS concentrations and DNA methylation based on a priori knowledge (Braid et al. 2017; Kingsley et al. 2018) (Figure S1). In addition to cell type composition, we adjusted for child age (continuous) and sex, annual household income (continuous), maternal race/ethnicity (non-Hispanic white vs. non-Hispanic black, and other) and maternal smoking during pregnancy (active smoking defined as serum cotinine vs. nonsmoking) in the models. We measured serum cotinine during pregnancy or at delivery using HPLC-MS/MS (Braun et al. 2010). Maternal race/ethnicity may be a confounder (Adkins et al. 2011; Boronow et al. 2019; Park et al. 2019) for the associations between gestational PFAS and DNA methylation. Other covariates were obtained using standardized computer-assisted interviews conducted by trained staff. Race/ethnicity (non-Hispanic white; non-Hispanic black; American Indian; Asian/Pacific; Hispanic) was self-reported by participants using standardized questionnaires, and only one category could be selected. In the analysis, race was collapsed into non-Hispanic white, non-Hispanic black, and other. We did this to reduce the degrees on freedom in the regression models and maximize precision. We did not adjust for premature birth, gestational age, mode of delivery, or pubarche in our analysis because these variables could be mediators of the associations between gestational PFAS and DNA methylation.
Statistical Analyses
We performed univariate analysis on the pregnancy concentrations of PFOA, PFOS, PFNA, and PFHxS in serum [median (25th, 75th percentiles)] and the levels of covariates [ (SD) or proportion (%)]. Prior to analysis, we PFAS concentrations to reduce the potential impact of outliers.
We analyzed longitudinal associations between PFAS concentrations and repeated DNA methylation measures using a multivariable linear regression model with generalized estimating equations (GEE) approach (Zeger and Liang 1986) with an identity link function, a working independence correlation matrix, and normal errors. The R package gee (available at https://cran.r-project.org/web/packages/gee) was used for estimation purposes.
For each of 669,662 autosomal CpG markers that survived quality control, we first estimated the main effect model: . Here represents the logit of the DNA methylation proportions for subject at each visit ( at baseline, at follow-up) and represents the proportion of cell types estimated at each visit. In this model, we were interested in , which can be interpreted as the differences in the logits of DNA methylation proportions across the two visits per 2-fold increase in gestational PFAS concentrations. Second, we examined the stability of PFAS-associated differences in DNA methylation across visits by adding the interaction of (PFAS) concentrations to the model above. In addition, we performed analyses examining associations of PFAS concentrations with DNA methylation in cord blood and at 12 years of age individually for those CpG sites that were significant in the main effect model to evaluate the stability of these differences over time.
To adjust for multiple comparisons across the epigenome, we used a modification of the Benjamini-Hochberg procedure (Benjamini et al. 2001) to control for false discovery rate (FDR) at the 5% level across CpG markers for each PFAS exposure. Storey’s -values (Storey and Tibshirani 2003) were obtained by shrinking the Benjamini-Hochberg adjusted -values by an estimate of the proportion of truly null markers (), and any was considered significant for both main and interaction effects. We evaluated genomic inflation by calculating lambda and created quantile–quantile plots for epigenome-wide analyses for each PFAS. We used the University of California, Santa Cruz (UCSC) Genome Browser database to annotate CpGs by assigning genomic features using the human reference genome GRCh37 (Haeussler et al. 2019), and CpGs assigned to TSS200 or TSS1500 (200 or 1,500 bases from the transcription start site) were annotated as being located in a promoter region.
To improve the biological interpretation of the microarray data, we performed gene ontology enrichment analysis on the top 500 CpGs with the lowest FDR -values for each PFAS from the main effect models to identify putative biological pathways. We decided to include 500 CpGs for this analysis because it is difficult to detect enriched pathways using the small number of CpGs associated with PFAS based on a stringent FDR . We used the gometh function in the R package version 1.28.0, missMethyl to perform the enrichment test for Gene Ontology (GO) terms using Wallenius’ noncentral hypergeometric distribution, taking into account the number of probes per gene on EPIC array and CpGs annotated to multiple genes (Phipson and Maksimovic 2020). GO annotates genes to three biological domains including biological processes, molecular functions, and cellular components. GO terms with FDR were considered significant.
In the secondary analysis, we examined the associations of gestational PFAS concentrations with cell type composition in cord blood and adolescent blood at 12 years of age to improve our understanding of the potential immunotoxicity of PFAS. We assessed these associations using separate linear regression models for each cell type, adjusting for child sex, household income, maternal race/ethnicity, serum cotinine in pregnancy, and child age (for adolescence models).
Replication Analysis
We used data collected from Project Viva to replicate EWAS findings from the HOME Study. Project Viva is a longitudinal prebirth cohort established to examine the effects of events during early development on lifetime health outcome. Between April 1999 and November 2002, the study recruited women in early pregnancy from eight obstetric offices of Atrius Harvard Vanguard Medical Associates, a multispecialty group practice in eastern Massachusetts. Exclusion criteria included multiple gestation, inability to answer questions in English, gestational age at recruitment, and plans to move away from the study area before delivery. Of 2,670 enrolled participants, 2,128 were still enrolled at delivery and had a live birth. Recruitment and characteristics of the cohort have been described in detail elsewhere (Oken et al. 2015). Gestational PFAS concentrations, DNA methylation, and covariate data were available for 371 mother–child pairs at delivery and 342 children at 7 years of age (Table S2).
The concentrations of PFOS, PFOA, PFNA, and PFHxS in plasma collected from pregnant women at 30.9–42.6 wk of gestation were quantified at the CDC laboratory using the same analytical method used for the HOME Study. Venous cord blood was collected. At the mid-childhood visit ( of age; range: 6.7–10.5), trained research assistants obtained whole blood from the antecubital vein. Genomic DNA was extracted from nucleated cells using commercially available Qiagen PureGene Kits and frozen at . DNA underwent sodium bisulfite conversion using the EZ DNA Methylation-Gold Kit (Zymo Research) was analyzed for DNA methylation using the Infinium Human Methylation450 BeadChip (Illumina). DNA methylation in cord blood and at of age was measured using the Illumina Infinium HumanMethylation 450 BeadChip. Following the same normalization and quality control pipeline for DNA methylation data in the HOME Study (see the section “DNA Methylation Analysis”), we excluded samples that failed on the array (cord blood , mid-childhood blood ), that were low-quality (cord blood , mid-childhood blood ), that had a genotype mismatch when compared with a blood sample from the same individual at a different time point (cord blood , mid-childhood blood ), and that had a predicted sex mismatch with recorded sex in our phenotypic data set (cord blood , mid-childhood ). We excluded probes that were on the sex chromosomes or that had a detection in of samples, contributing a total of 467,471 probes to this replication analysis. The associations between PFAS concentrations and repeated blood DNA methylation measures were evaluated using the same GEEs that were used for the HOME Study data with adjustment for the same covariates except for smoking, which was collected using self-reported questionnaire. CpGs with a and the same direction of associations observed in the HOME Study were considered successful replications. We performed all the statistical analyses using R (version 4.0.5; R Development Core Team) and Bioconductor (version 3.12; Bioconductor).
Results
Among the 266 mother–offspring pairs in the HOME Study at baseline, 53% of the children were girls, 8% of the mothers were active smokers during pregnancy (serum cotinine concentrations ), 66% were non-Hispanic white, and 17% had annual household income of (Table 1). Median serum concentrations of PFOA, PFOS, PFHxS, and PFNA during pregnancy were 5.4, 13.7, 1.5, and , respectively (Figure S2, Table S3). We observed moderate correlations among serum PFAS concentrations during pregnancy () (Table S4). The mean age of the 160 children at follow-up was 12.3 y (range: 11.1–14.0).
Table 1.
Sociodemographic and perinatal characteristics and maternal serum PFAS concentrations in pregnancy for participants at delivery and at 12 years of age in the HOME Study (Cincinnati, Ohio; enrolled 2003–2006).
| Characteristics | Delivery () | Follow-up () |
|---|---|---|
| Child sex [ (%)] | ||
| Girls | 140 (53) | 85 (53) |
| Boys | 126 (47) | 75 (47) |
| Child age [mean (SD)] | — | 12.3 (0.6) |
| Maternal serum cotinine (ng/mL) [ (%)] | ||
| (Unexposed)a | 89 (34) | 44 (28) |
| (secondhand tobacco smoke exposure) | 155 (58) | 103 (64) |
| (active smoking) | 22 (8) | 13 (8) |
| Maternal race/ethnicity [ (%)] | ||
| Non-Hispanic white | 175 (66) | 92 (57) |
| Non-Hispanic black | 74 (28) | 59 (37) |
| All others | 17 (6) | 9 (6) |
| Annual household income [ (%)] | ||
| 81 (31) | 44 (28) | |
| 94 (35) | 50 (31) | |
| 46 (17) | 28 (17) | |
| 45 (17) | 38 (24) | |
| Prenatal PFAS concentrations (ng/mL) [median (25th, 75th percentiles)] | ||
| PFOA | 5.5 (3.9, 7.9) | 5.1 (3.6, 7.6) |
| PFOS | 14.0 (9.9, 17.8) | 13.4 (8.9, 18.5) |
| PFHxS | 1.5 (0.9, 2.4) | 1.4 (0.8, 2.3) |
| PFNA | 0.9 (0.7, 1.2) | 0.9 (0.7, 1.2) |
Note: Data were complete for all variables. PFAS concentrations below LOD were replaced with the LOD divided by the square root of 2. —, no data; HOME, Health Outcomes and Measures of the Environment; LOD, limit of detection; PFAS, perfluoroalkyl substances; PFHxS, perfluorohexane sulfonate; PFNA, perfluorononanoate; PFOA, perfluorooctanoate; PFOS, perfluorooctane sulfonate; SD, standard deviation.
Below detection limit. PFAS concentrations were measured in maternal serum at 10.4–30.3 wk of gestation.
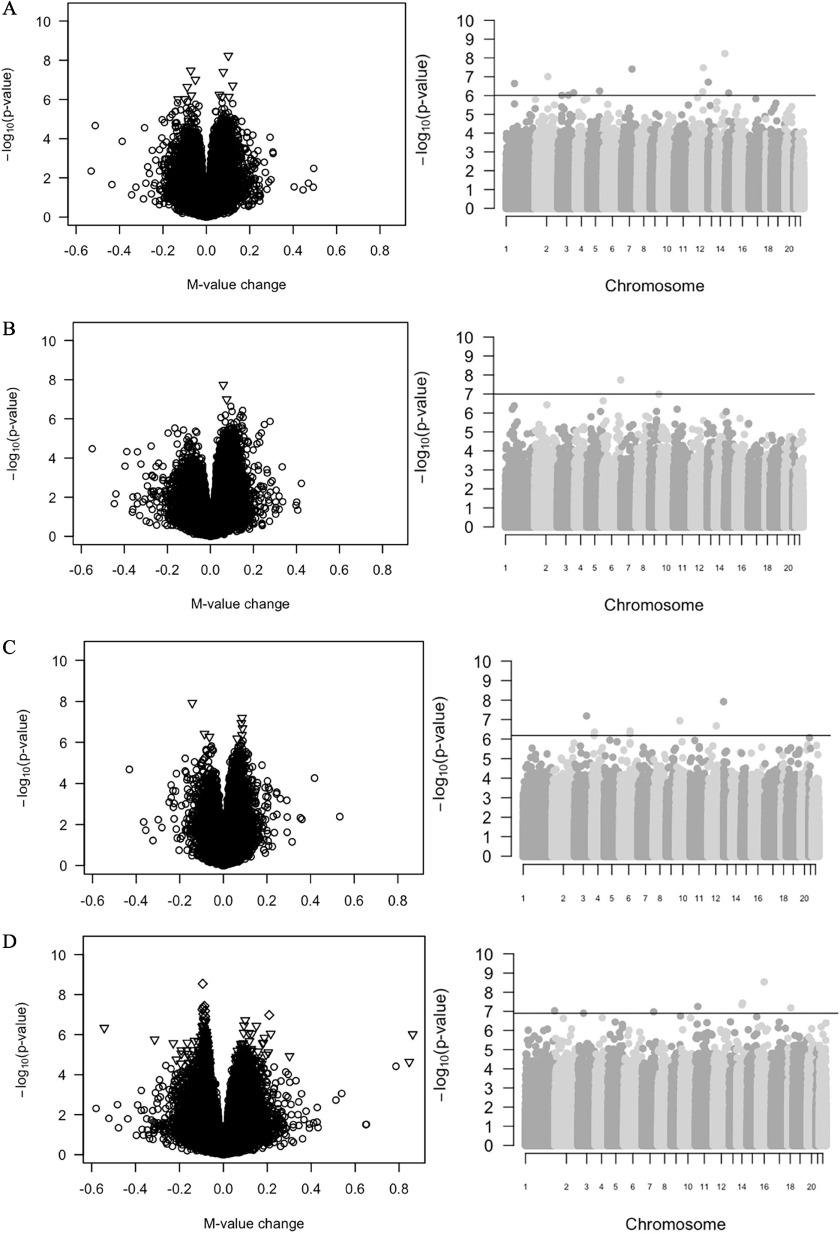
After adjusting for potential confounders and multiple comparisons, 435 CpGs were significantly associated with PFAS concentrations at FDR (Excel Tables S1–S4). We identified 12 significant CpGs for PFOA, 2 for PFOS, 8 for PFHxS, and 413 for PFNA; there were no overlapping CpGs across different PFAS. Because of the large number of CpG loci with significant associations with PFNA, only the top 8 loci with FDR are reported in the text (total CpGs) (Table 2, Figure 1) and the 413 PFNA-associated CpGs at FDR are included in Excel Table S4. The quantile–quantile plots for each PFAS did not show noteworthy inflation in the distribution of observed -values (Figure S3; genomic inflation factor: PFOA, ; PFOS, ; PFHxS, ; PFNA, ). The list of CpGs associated with gestational PFAS concentrations (HOME Study) with along with coefficients, -values, and FDR -values can be found in the Excel Tables S1–S4.
Table 2.
Top CpG sites from adjusted associations of gestational PFAS concentrations with repeated measures of DNA methylation in the HOME Study (at delivery and at 12 years of age) and Project Viva (at delivery and at 7 years of age).
| CpG | Chr | Position | CpG location | Gene | Gene region | FANTOM5 enhancers | ENCODE DNAse hypersensitivity sites | Discovery cohort [ (-value)] | FDR -value | Replication cohort [ (-value)] | Replicateda |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PFOA | |||||||||||
| cg25869005 | 14 | 95048198 | Open sea | SERPINA5 | 5′UTR | — | chr14:95047965-95048250 | 0.101 () | 0.003 | NA | — |
| cg03707973 | 12 | 54826237 | Open sea | LOC102724050 | Body | — | chr12:54826045-54826655 | () | 0.008 | NA | — |
| cg04042784 | 7 | 42884749 | Open sea | — | — | — | — | 0.078 () | 0.008 | NA | — |
| cg09309282 | 2 | 236523347 | Open sea | AGAP1 | Body | — | chr2:236523145-236523410 | () | 0.015 | NA | — |
| cg22971191 | 13 | 103719706 | Open sea | SLC10A2 | Promoter | — | — | 0.121 () | 0.023 | (0.435) | |
| cg01787798 | 1 | 175474437 | Open sea | TNR | 5′UTR | — | chr1:175474340-175474655 | () | 0.023 | (0.001) | |
| cg04977784 | 5 | 53972541 | Open sea | LOC102467080 | Body | chr5:53972314-53972314 | chr5:53972220-53972635 | 0.060 () | 0.043 | NA | — |
| cg14212748 | 12 | 53343703 | Shore | KRT18 | Body | — | chr12:53343445-53343855 | () | 0.043 | (0.492) | — |
| cg05195288 | 3 | 65582834 | Open sea | MAGI1 | Body | — | — | 0.073 () | 0.043 | NA | — |
| cg21821684 | 15 | 47686828 | Open sea | — | — | — | chr15:47686685-47686890 | 0.104 () | 0.043 | (0.379) | — |
| cg16415457 | 3 | 23713441 | Open sea | — | — | — | chr3:23713340-23713815 | () | 0.048 | (0.007) | |
| cg03760072 | 3 | 133502685 | Island | SRPRB | Promoter | — | chr3:133502565-133503275 | () | 0.048 | NA | — |
| PFOS | |||||||||||
| cg16381104 | 6 | 99578913 | Open sea | — | — | — | chr6:99578800-99579035 | 0.060 () | 0.011 | NA | — |
| cg21421331 | 10 | 100618466 | Open sea | HPSE2 | Body | — | chr10:100618360-100618610 | 0.076 () | 0.033 | NA | — |
| PFHxS | |||||||||||
| cg02794779 | 13 | 100150711 | Island | — | — | — | — | () | 0.007 | (0.106) | — |
| cg11331322 | 3 | 42530661 | Open sea | VIPR1 | Promoter | — | — | 0.086 () | 0.018 | NA | — |
| cg00983482 | 10 | 130083707 | Open sea | — | — | — | chr10:130082925-130083755 | 0.082 () | 0.022 | (0.440) | — |
| cg26477711 | 12 | 39518309 | Open sea | — | — | — | — | 0.088 () | 0.030 | NA | — |
| cg24735489 | 6 | 31088352 | Open sea | CDSN;PSORS1C1 | Promoter;5′UTR | — | chr6:31088120-31088430 | () | 0.042 | 0.019 (0.227) | — |
| cg03639671 | 4 | 145430689 | Open sea | — | — | — | — | 0.085 () | 0.042 | (0.087) | — |
| cg00457165 | 6 | 31088366 | Open sea | CDSN;PSORS1C1 | Promoter;5′UTR | — | chr6:31088120-31088430 | () | 0.043 | NA | — |
| cg00647458 | 4 | 139760831 | Open sea | — | — | — | — | 0.063 () | 0.046 | NA | — |
| PFNA | |||||||||||
| cg25601695 | 16 | 55690178 | Island | SLC6A2 | 5′UTR;Promoter | — | chr16:55689225-55691015 | () | 0.002 | NA | — |
| cg04509825 | 14 | 62162178 | Island | HIF1A | 1stExon;5′UTR | — | chr14:62161865-62162315 | () | 0.008 | 0.002 (0.807) | — |
| cg01713535 | 14 | 57046409 | Shore | C14orf101 | Promoter | — | chr14:57046105-57047510 | () | 0.008 | 0.019 (0.525) | — |
| cg11506907 | 11 | 118972568 | Shore | DPAGT1 | 5′UTR;1stExon | — | chr11:118972040-118973615 | () | 0.008 | 0.005 (0.672) | — |
| cg12967001 | 18 | 5544089 | Island | EPB41L3 | Promoter | — | chr18:5543880-5544455 | () | 0.008 | (0.681) | — |
| cg05792003 | 1 | 95582492 | Shore | TMEM56 | Promoter | — | chr1:95582305-95582570 | () | 0.009 | 0.002 (0.874) | — |
| cg24620436 | 7 | 4918906 | Shelf | RADIL | 5′UTR | — | chr7:4918205-4919455 | 0.209 () | 0.009 | 0.118 (0.055) | — |
| cg09861318 | 3 | 149530987 | Island | RNF13 | 1stExon;5′UTR | — | chr3:149530185-149531075 | () | 0.009 | 0.011 (0.544) | — |
| cg26975787 | 12 | 2613778 | Open sea | CACNA1C | Body | — | — | () | 0.029 | (0.010) | |
| cg24822529 | 5 | 134582880 | Open sea | — | — | — | chr5:134582625-134582950 | () | 0.033 | (0.005) | |
| cg19497517 | 8 | 145028257 | Island | PLEC1 | Body;TSS200 | — | chr8:145028185-145028535 | 0.194 () | 0.035 | 0.073 (0.027) | |
| cg02842197 | 17 | 5389625 | Island | DERL2;MIS12 | TSS200;TSS1500 | — | chr17:5389485-5389890 | 0.136 () | 0.042 | 0.048 (0.026) | |
Note: All models were adjusted for child age and sex, annual household income (median income of each category), maternal race/ethnicity (non-Hispanic white, other) and smoking during pregnancy (yes, no), and cell type composition. CpG sites were annotated to a promoter region if they were assigned toTSS200 or TSS1500. Data were complete for all variables. PFAS concentrations below the LOD were replaced with the LOD divided by the square root of 2. We analyzed longitudinal associations between PFAS concentrations and repeated DNA methylation measures using a generalized estimating equations (GEE) approach with an identity link function, a working independence correlation matrix and normal errors. can be interpreted as the differences in the logits of DNA methylation proportions across the two visits per 2-fold increase in gestational PFAS concentrations. Statistically significant CpG sites were defined as having an FDR (PFOA, PFOS, and PFHxS) or (PFNA). —, not annotated to any known gene; 5′UTR, between the TSS and the ATG start site; 1stExon, the first exon of the gene; Chr, chromosome; CpG, cytosine–guanine dinucleotide; Body, between the ATG and stop codon; FDR, false discovery rate; HOME, Health Outcomes and Measures of the Environment; LOD, limit of detection; NA, not available in 450K microarray; PFAS, perfluoroalkyl substances; PFHxS, perfluorohexane sulfonate; PFNA, perfluorononanoate; PFOA, perfluorooctanoate; PFOS, perfluorooctane sulfonate; TSS, transcription start site; TSS200, 200 bases from TSS; TSS1500, 1,500 bases from TSS, UTR, untranslated region.
CpGs with a and the same direction of associations observed in the HOME Study were considered as successful replications.
Figure 1.
Volcano and Manhattan plots for the epigenome-wide adjusted associations of (A) gestational serum concentrations of PFOA or (B) PFOS or (C) PFHxS or (D) PFNA with repeated measures of DNA methylation with in the HOME Study (Cincinnati, Ohio; enrolled 2003–2006). These models were adjusted for child age and sex, annual household income (median income of each category), maternal race/ethnicity (non-Hispanic white vs. non-Hispanic black and other) and smoking during pregnancy (active smoking for serum cotinine, not active smoking), and cell type composition. Left panels are volcano plots showing the difference in leukocyte DNA methylation (magnitude of effect on M-value: x-axis) associated with gestational PFAS concentrations for each CpG site plotted against its negative -value (y-axis). Triangles represent the CpG sites with FDR . For PFNA, diamonds represent the CpG sites with FDR . Right panels are Manhattan plots showing negative -values for the associations between gestational PFAS concentrations and DNA methylation across chromosomes. Statistically significant CpG sites were defined as having an FDR (PFOA, PFOS, and PFHxS) or 0.01 (PFNA). Horizontal lines denote FDR or 0.01. Note: CpG, cytosine–guanine dinucleotide; FDR, false discovery rate; HOME, Health Outcomes and Measures of the Environment; PFHxS, perfluorohexane sulfonate; PFNA, perfluorononanoate; PFOA, perfluorooctanoate; PFOS, perfluorooctane sulfonate.
Of the 435 significant associations discovered in the HOME Study using the Infinium Methylation EPIC array, 315 CpGs were present on the 450K array used by Project Viva, and we successfully replicated 6 CpG sites with the same direction of associations and a in Project Viva (2 CpGs for PFOA, 4 CpGs for PFNA) (Table 2, Figure 1).
In the HOME Study, few interactions reached statistical significance (FDR ) (Table 3). Only four CpGs for PFHxS (cg12507840, cg12119988, cg11035296, and cg21869609) and one CpG for PFNA (cg24155143) had significant interaction terms. Among the five CpGs with significant interaction terms in the HOME Study, two CpGs (cg12119988 and cg21869609) were available using 450K array in Project Viva, and neither one was successfully replicated ( interaction ) (Table 3). Among CpGs significantly associated with PFAS in our main effects model (no age interaction), these associations were consistent at birth and at 12 years of age, having the same direction and comparable effect size for both time points (Excel Table S5).
Table 3.
Statistically significant CpG sites whose PFAS-associated methylation differences differed over time ( FDR ) in the HOME Study (Cincinnati, Ohio; enrolled 2003–2006) and associations in Project Viva (eastern Massachusetts; enrolled 1999–2002).
| CpGa | Gene | Chr | HOME Study ( value) |
FDR -value | HOME Study ( value) |
HOME Study ( value) |
Project Viva ( value) |
Replicated |
|---|---|---|---|---|---|---|---|---|
| PFHxS | ||||||||
| cg12507840 | UBAC2; MIR548AN | 13 | () | 0.011 | 0.041 (0.009) | (0.021) | NA | No |
| cg12119988 | ANKDD1A | 15 | 0.012 () | 0.011 | (0.290) | 0.093 (0.0004) | (0.766) | No |
| cg11035296 | DLGAP2-AS1 | 8 | () | 0.030 | 0.044 (0.028) | (0.020) | NA | No |
| cg21869609 | LINGO3 | 19 | () | 0.030 | 0.028 (0.093) | (0.160) | (0.343) | No |
| PFNA | ||||||||
| cg24155143 | — | 4 | () | 0.021 | 0.14 (0.004) | (0.0001) | NA | No |
Note: Adjusted for child age and sex, annual household income, maternal smoking during pregnancy and race, and cell type composition. —, not annotated to any known gene; , shows the association between gestational PFAS and DNA methylation at birth; , shows the association between gestational PFAS and DNA methylation at 12 years of age; CpG, cytosine–guanine dinucleotide; Chr, chromosome; FDR, false discovery rate; HOME, Health Outcomes and Measures of the Environment; NA, not available in 450K microarray; PFAS, perfluoroalkyl substances; PFHxS, perfluorohexane sulfonate; PFNA, perfluorononanoate; PFOA, perfluorooctanoate; PFOS, perfluorooctane sulfonate.
Statistically significant CpG sites were defined as having an FDR for .
Among the top 30 CpGs associated with PFAS in the HOME Study, we annotated these CpGs to 20 unique genes, 2 CpGs associated with PFHxS were mapped to the same gene [corneodesmosin (CDSN); psoriasis susceptibility 1 candidate 1 (PSORS1C1)] (Table 2). All CpGs associated with PFOS were hypermethylated, whereas the majority of the CpGs associated with PFNA concentrations were hypomethylated. A mix of hypermethylated and hypomethylated CpGs were observed for PFOA and PFHxS. Most CpGs for PFOA, PFOS, and PFHxS were in an open sea region, whereas the majority of CpGs were in a CpG island for PFNA. In addition, we found 1 CpG (cg04977784) is in an enhancer region, and the majority of these CpGs reside in a DNase I hypersensitivity region (a mark for transcriptionally competent chromatin). Finally, these CpGs annotated to the nearest genes, which have been linked to several PFAS-associated health outcomes including cancers, cognitive health, cardiovascular disease, inflammatory bowel disease, and kidney function (Table S5).
In the enrichment analysis using the top 500 CpGs for each PFAS (mapped to 239–432 unique genes), we observed significant enrichment of gene sets annotating to a total number of 39 unique GO terms with an FDR (Table 4). Among them, we identified 14 significant GO terms for PFOA, 13 for PFOS, 22 for PFNA, and none for PFHxS; there were 10 overlapping terms between PFOA and PFOS. The most significant GO term for both PFOA and PFOS was homophilic cell adhesion via plasma membrane adhesion molecules in the category of biological process (BP) (FDR -value: ; ). GO cellular component (CC) terms, such as cytoplasm and intracellular organelle, were the most significant for PFNA (FDR ).
Table 4.
GO Terms significantly enriched (FDR ) among top 500 genes associated with each gestational PFAS concentration ordered by FDR -value in the HOME Study (Cincinnati, Ohio; enrolled 2003–2006).
| ID | Ontology | Description | PFAS | -Value | FDR -value |
|---|---|---|---|---|---|
| GO:0007156 | BP | Homophilic cell adhesion via plasma membrane adhesion molecules | PFOA | ||
| GO:0098742 | BP | Cell–cell adhesion via plasma membrane adhesion molecules | PFOA | ||
| GO:0005509 | MF | Calcium ion binding | PFOA | ||
| GO:0022610 | BP | Biological adhesion | PFOA | ||
| GO:0098609 | BP | Cell–cell adhesion | PFOA | ||
| GO:0007155 | BP | Cell adhesion | PFOA | ||
| GO:0005887 | CC | Integral component of plasma membrane | PFOA | ||
| GO:0031226 | CC | Intrinsic component of plasma membrane | PFOA | ||
| GO:0071944 | CC | Cell periphery | PFOA | 0.00124 | |
| GO:0005886 | CC | Plasma membrane | PFOA | 0.00488 | |
| GO:0043169 | MF | Cation binding | PFOA | 0.01497 | |
| GO:0046872 | MF | Metal ion binding | PFOA | 0.01497 | |
| GO:1903367 | BP | Positive regulation of fear response | PFOA | 0.03098 | |
| GO:2000987 | BP | Positive regulation of behavioral fear response | PFOA | 0.03098 | |
| GO:0007156 | BP | Homophilic cell adhesion via plasma membrane adhesion molecules | PFOS | ||
| GO:0098742 | BP | Cell–cell adhesion via plasma membrane adhesion molecules | PFOS | ||
| GO:0098609 | BP | Cell–cell adhesion | PFOS | ||
| GO:0005887 | CC | Integral component of plasma membrane | PFOS | ||
| GO:0031226 | CC | Intrinsic component of plasma membrane | PFOS | ||
| GO:0005509 | MF | Calcium ion binding | PFOS | ||
| GO:0007155 | BP | Cell adhesion | PFOS | ||
| GO:0022610 | BP | Biological adhesion | PFOS | ||
| GO:0005886 | CC | Plasma membrane | PFOS | ||
| GO:0071944 | CC | Cell periphery | PFOS | ||
| GO:0016021 | CC | Integral component of membrane | PFOS | 0.000547 | |
| GO:0031224 | CC | Intrinsic component of membrane | PFOS | 0.000563 | |
| GO:0016020 | CC | Membrane | PFOS | 0.039281 | |
| GO:0005737 | CC | Cytoplasm | PFNA | 0.001140 | |
| GO:0043229 | CC | Intracellular organelle | PFNA | 0.001140 | |
| GO:0043226 | CC | Organelle | PFNA | 0.001140 | |
| GO:0043227 | CC | Membrane-bounded organelle | PFNA | 0.001309 | |
| GO:0005622 | CC | Intracellular | PFNA | 0.001309 | |
| GO:0043231 | CC | Intracellular membrane-bounded organelle | PFNA | 0.002074 | |
| GO:0044237 | BP | Cellular metabolic process | PFNA | 0.002542 | |
| GO:0005654 | CC | Nucleoplasm | PFNA | 0.005690 | |
| GO:0031974 | CC | Membrane-enclosed lumen | PFNA | 0.005690 | |
| GO:0043233 | CC | Organelle lumen | PFNA | 0.005690 | |
| GO:0070013 | CC | Intracellular organelle lumen | PFNA | 0.005690 | |
| GO:0044249 | BP | Cellular biosynthetic process | PFNA | 0.005690 | |
| GO:0044238 | BP | Primary metabolic process | PFNA | 0.009058 | |
| GO:0031981 | CC | Nuclear lumen | PFNA | 0.009495 | |
| GO:0009058 | BP | Biosynthetic process | PFNA | 0.014653 | |
| GO:0034641 | BP | Cellular nitrogen compound metabolic process | PFNA | 0.014653 | |
| GO:0008152 | BP | Metabolic process | PFNA | 0.014653 | |
| GO:1901576 | BP | Organic substance biosynthetic process | PFNA | 0.014653 | |
| GO:0071704 | BP | Organic substance metabolic process | PFNA | 0.01594566 | |
| GO:1901360 | BP | Organic cyclic compound metabolic process | PFNA | 0.02246282 | |
| GO:0006807 | BP | Nitrogen compound metabolic process | PFNA | 0.03161224 | |
| GO:0031076 | BP | Embryonic camera-type eye development | PFNA | 0.04725753 |
Note: Top 500 CpG sites for each PFAS ranked by FDR -value with corresponding genes were included in the pathway analysis. At FDR , we found 14 significant GO terms for PFOA, 13 for PFOS, 22 for PFNA, and none for PFHxS, and 10 overlapped between PFOA and PFOS. BP, biological process; CC, cellular component; FDR, false discovery rate; GO, Gene Ontology; HOME, Health Outcomes and Measures of the Environment; ID, identifier; MF, molecular function; PFAS, perfluoroalkyl substances; PFHxS, perfluorohexane sulfonate; PFNA, perfluorononanoate; PFOA, perfluorooctanoate; PFOS, perfluorooctane sulfonate.
In general, gestational PFAS concentrations were not associated with estimated cell type composition in cord blood (Table 5) or child blood collected at 12 years of age (Table 6). However, we observed a monotonic decrease in the percentage of B lymphocytes in cord blood () across tertiles of PFOS and a monotonic decrease in the percentage of nucleated red blood cells at delivery () across tertiles of PFNA.
Table 5.
Adjusted mean cell type composition in cord blood by gestational PFAS concentration (ng/mL) tercile in the HOME Study (Cincinnati, Ohio; enrolled 2003–2006).
| PFAS [tercile (range)] | Cell type | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| B cell (%) | CD4 (%) | CD8 (%) | Monocyte (%) | Neutrophil (%) | NK (%) | nRBC (%) | CD4/CD8 | NLR | LMR | |
| PFOA | ||||||||||
| T1 (1.1–4.4) | 4.0 | 19.0 | 3.5 | 9.0 | 56.0 | 1.8 | 5.0 | 33.6 | 2.41 | 3.74 |
| T2 (4.4–6.9) | 5.0 | 20.0 | 3.0 | 9.0 | 57.0 | 1.7 | 5.0 | 38.1 | 2.30 | 6.35 |
| T3 (6.9–26.4) | 4.0 | 19.0 | 2.5 | 9.0 | 58.0 | 2.3 | 5.0 | 31.6 | 2.44 | 4.11 |
| a | 0.083 | 0.31 | 0.2 | 0.98 | 0.23 | 0.59 | 0.56 | 0.8 | 0.8 | 0.83 |
| PFOS | ||||||||||
| T1 (1.4–11.7) | 4.6 | 19.0 | 3.7 | 9.0 | 55.0 | 2.0 | 6.0 | 34.4 | 2.15 | 6.33 |
| T2 (11.7–16.5) | 4.3 | 19.0 | 2.5 | 9.0 | 59.0 | 2.0 | 5.0 | 31.3 | 2.67 | 3.95 |
| T3 (16.5–57.2) | 4.0 | 20.0 | 2.8 | 9.0 | 58.0 | 2.0 | 4.0 | 37.7 | 2.35 | 3.86 |
| a | 0.031 | 0.63 | 0.53 | 0.67 | 0.23 | 0.21 | 0.17 | 0.56 | 0.56 | 0.39 |
| PFHxS | ||||||||||
| T1 (0.1–1.1) | 4.4 | 19.0 | 4.0 | 9.0 | 44.0 | 1.8 | 6.0 | 25.7 | 2.06 | 5.90 |
| T2 (1.1–2.0) | 3.9 | 19.0 | 2.0 | 9.0 | 61.0 | 1.6 | 3.0 | 39.9 | 2.91 | 4.09 |
| T3 (2.0–30.9) | 4.7 | 20.0 | 3.0 | 9.0 | 56.0 | 2.4 | 5.0 | 37.6 | 2.22 | 4.03 |
| a | 0.29 | 0.94 | 0.13 | 0.31 | 0.23 | 0.69 | 0.79 | 0.35 | 0.35 | 0.81 |
| PFNA | ||||||||||
| T1 (0.3–-0.8) | 4.7 | 20.0 | 3.0 | 9.0 | 56.0 | 2.0 | 6.0 | 42.3 | 2.22 | 6.03 |
| T2 (0.8–1.1) | 4.1 | 18.0 | 3.0 | 9.0 | 59.0 | 2.0 | 5.0 | 30.7 | 2.67 | 3.86 |
| T3 (1.1–2.9) | 4.2 | 21.0 | 3.0 | 10.0 | 57.0 | 2.0 | 3.0 | 28.2 | 2.29 | 3.94 |
| a | 0.06 | 0.54 | 0.89 | 0.41 | 0.23 | 0.68 | 0.019 | 0.49 | 0.49 | 0.84 |
Note: All models were adjusted for child sex, maternal pregnancy cotinine concentrations, race, and annual household income. CD4, T helper cells; CD8, cytotoxic T cells; CD4/CD8, the ratio of T helper cells to cytotoxic T cells; HOME, Health Outcomes and Measures of the Environment; NK, natural killer cells; NLR, the ratio of neutrophils to lymphocytes; LMR, the ratio of lymphocytes to monocytes; nRBC, nucleated red blood cells; PFAS, perfluoroalkyl substances; PFHxS, perfluorohexane sulfonate; PFNA, perfluorononanoate; PFOA, perfluorooctanoate; PFOS, perfluorooctane sulfonate.
The for cell type composition differences across median PFAS concentrations in each tercile.
Table 6.
Adjusted mean cell type composition based on methylation at 12 years of age by gestational PFAS concentration (ng/mL) tercile in the HOME Study (Cincinnati, Ohio; enrolled 2003–2006).
| PFAS [tercile (range)] | Cell type | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| B cell (%) | CD4 (%) | CD8 (%) | Monocyte (%) | Neutrophil (%) | NK (%) | CD4/CD8 | NLR | LMR | |
| PFOA | |||||||||
| T1 (1.1–4.2) | 8.0 | 17.0 | 13.0 | 7.5 | 49.0 | 5.4 | 1.37 | 1.19 | 6.20 |
| T2 (4.2–6.3) | 9.0 | 18.0 | 13.0 | 7.7 | 48.0 | 5.0 | 1.52 | 1.13 | 6.41 |
| T3 (6.3–17.4) | 8.0 | 17.0 | 13.0 | 7.1 | 51.0 | 4.7 | 1.39 | 1.29 | 6.26 |
| a | 0.2 | 0.61 | 0.64 | 0.56 | 0.14 | 0.089 | 0.17 | 0.17 | 0.84 |
| PFOS | |||||||||
| T1 (1.4–10.0) | 9.0 | 18.0 | 13.0 | 7.0 | 47.0 | 5.3 | 1.38 | 1.12 | 6.63 |
| T2 (10.0–16.0) | 8.0 | 17.0 | 13.0 | 8.0 | 50.0 | 4.6 | 1.40 | 1.28 | 5.74 |
| T3 (16.0–57.2) | 8.0 | 18.0 | 13.0 | 7.0 | 50.0 | 5.2 | 1.50 | 1.22 | 6.49 |
| a | 0.14 | 0.85 | 0.38 | 0.56 | 0.23 | 0.61 | 0.22 | 0.22 | 0.70 |
| PFHxS | |||||||||
| T1 (0.1–1.0) | 9.0 | 18.0 | 13.0 | 8.0 | 48.0 | 5.0 | 1.40 | 1.14 | 6.23 |
| T2 (1.0–2.0) | 8.0 | 17.0 | 13.0 | 7.0 | 50.0 | 5.0 | 1.48 | 1.26 | 6.28 |
| T3 (2.0–32.5) | 8.0 | 17.0 | 13.0 | 7.0 | 49.0 | 5.0 | 1.40 | 1.22 | 6.37 |
| a | 0.23 | 0.28 | 0.39 | 0.41 | 0.23 | 0.29 | 0.19 | 0.19 | 0.58 |
| PFNA | |||||||||
| T1 (0.3–0.7) | 8.0 | 17.0 | 13.0 | 7.0 | 49.0 | 5.0 | 1.41 | 1.20 | 6.28 |
| T2 (0.7–1.1) | 8.0 | 17.0 | 13.0 | 8.0 | 50.0 | 5.0 | 1.37 | 1.25 | 6.10 |
| T3 (1.1–2.9) | 8.0 | 19.0 | 13.0 | 7.0 | 48.0 | 5.0 | 1.54 | 1.14 | 6.59 |
| a | 0.62 | 0.26 | 0.84 | 0.99 | 0.72 | 0.9 | 0.74 | 0.74 | 0.42 |
Note: All models were adjusted for child sex, child age, maternal pregnancy cotinine concentrations, race, and annual household income. CD4, T helper cells; CD8, cytotoxic T cells; CD4/CD8, the ratio of T helper cells to cytotoxic T cells; HOME, Health Outcomes and Measures of the Environment; NK, natural killer cells; NLR, ratio of neutrophils to lymphocytes; LMR, the ratio of lymphocytes to monocytes; PFAS, perfluoroalkyl substances; PFHxS, perfluorohexane sulfonate; PFNA, perfluorononanoate; PFOA, perfluorooctanoate; PFOS, perfluorooctane sulfonate.
The for cell type composition differences across median PFAS concentrations in each tercile.
Discussion
In this longitudinal EWAS analysis using data from the HOME Study, we found evidence that gestational PFAS exposure was associated with differences in offspring peripheral leukocyte DNA methylation at several CpGs at birth and at 12 years of age. These associations were consistent in direction and magnitude at both time points, indicating that these associations are persistent over time. To the best of our knowledge, these CpGs have not been reported in previous studies and were annotated to gene regions linked to various PFAS-associated health outcomes, including cancers, cognitive health, cardiovascular disease, inflammatory bowel disease, and kidney function. In addition, we successfully replicated associations for six CpG sites in Project Viva.
Previous EWAS studies have found statistically significant associations between gestational PFAS concentrations and DNA methylation at birth or in newborns (Miura et al. 2018; Ouidir et al. 2020; Starling et al. 2020; van den Dungen et al. 2017). Specifically, two studies reported significant associations between higher pregnancy PFOA concentrations and lower cord blood DNA methylation at two CpGs (cg11260715: AC002480.3 and cg04461802: GPR126) in 190 Japanese mother–offspring pairs (Miura et al. 2018), and at one CpG (cg18587484: TJAP1) in 583 mother–offspring pairs from Colorado. (Starling et al. 2020). A cross-sectional study reported that higher neonatal PFOA concentrations were significantly associated with lower DNA methylation at one CpG (cg15557840: SCRT2; SRXN1) in 597 newborns from New York. (Robinson et al. 2021). In contrast, no significant CpGs were observed for the associations of pregnancy PFOA concentrations with placental DNA methylation in a U.S. study () (Ouidir et al. 2020) or the cross-sectional associations of PFOA concentrations with DNA methylation in 80 adult Dutch men (van den Dungen et al. 2017). Although we were not able to identify the same CpGs in the previous studies, these CpGs were linked to genes that are associated with similar health outcomes.
We found that PFOS concentrations in pregnancy were significantly associated with DNA methylation at two CpGs (cg16381104 and cg21421331: HPSE2), and both were hypermethylated. In other studies, pregnancy PFOS concentrations were positively associated with cord blood DNA methylation at two CpGs (cg02044327: CXADRP3 and cg25705526: SNAPIN) (Miura et al. 2018), placental DNA methylation at two CpGs (cg17921248: PRKCA and cg11891579: EBF1) (Ouidir et al. 2020), and newborn blood DNA methylation at two CpGs (cg19039925: GVIN1 in boys and cg05754408: ZNF26 in girls) (Robinson et al. 2021). In adults, PFOS concentrations were significantly associated with DNA methylation at 117 CpGs in Swedish women (Xu et al. 2020). However, PFOS was not significantly associated with DNA methylation in Dutch men (van den Dungen et al. 2017).
In the HOME Study, both PFNA and PFHxS concentrations were significantly associated with DNA methylation at specific CpG sites. Ouidir et al. (2020) found two CpGs significantly related to PFNA (cg26808417: ARL8A and cg21502999: ELK3) and three CpGs for PFHxS (cg21058927: CISD2, cg11428546: AFF3, and cg10530492: NOL7). However, Starling et al. (2020) observed no significant associations for PFOS, PFNA, or PFHxS. In the present study, gestational PFNA concentrations were associated with a greater number of CpGs than other PFAS. It is possible that the distribution of PFNA concentrations may have impacted these findings. PFNA has a relatively narrow exposure range (), and each participant can take on only one of 24 values (machine rounds levels to 0.1). It is also possible that blood DNA methylation is more sensitive to the effect of PFNA exposure; however, the potential biological mechanism is not clear based on the current literature.
We compared our results with those from prior studies that listed CpGs with a for their associations (Table S6). Among these CpG sites, 3–4% for PFOS and PFOA in the study by Miura et al. (2018), 3–6% for PFOS, PFOA, PFNA, and PFHxS in the study by Starling et al. (2020), and 7% in the study by Xu et al. (2020) had the same direction of the significant associations for the respective PFAS that we observed in the HOME Study.
The inconsistent findings across existing EWAS studies of PFAS and DNA methylation may be explained by differences in the study source population and design, sample size, data analysis methods, concentration ranges and timing of PFAS measurements, and the timing and methods of measuring DNA methylation. The existing studies were generally cross-sectional or had a single measurement of DNA methylation, whereas we were able to collect longitudinal DNA methylation data at two time points. The longitudinal design enabled us to evaluate whether these changes are persistent over time. Most prior studies used a less comprehensive microarray assay (i.e., the 450K array) to evaluate DNA methylation covering probes, which may contribute to variations in observed DNA methylation differences. In addition, it is known that DNA methylation profiles differ between newborns and adults (Bjornsson et al. 2008). Thus, studies examining different exposure periods may identify inconsistent findings (van den Dungen et al. 2017; Xu et al. 2020). Although serum concentrations of most PFAS during pregnancy (, , ) in the HOME Study were similar to those of pregnant women measured in the National Health and Nutrition Examination Survey 2003–2008, PFOA concentrations were higher () (Braun et al. 2016). Moreover, PFOA and PFOS concentrations among HOME Study women were higher than levels in the other EWAS studies of DNA methylation at delivery or in newborns in the United States (, ) (Ouidir et al. 2020; Robinson et al. 2021; Starling et al. 2020) and Japan (, ) (Miura et al. 2018), but they were lower than the PFOS concentrations in Swedish women living in communities with contaminated drinking water (median PFOS by exposure groups: low, ; medium, ; high, ) (Xu et al. 2020).
In our analysis, serum PFOA and PFOS concentrations were significantly associated with DNA methylation at several CpGs annotated to gene regions that have been linked to breast, prostate, and pancreatic cancers (Table S5), which is consistent with prior studies showing higher PFOA/PFOS concentrations were associated with increased risk of breast cancer (Bonefeld-Jorgensen et al. 2011; Tsai et al. 2020; Wielsøe et al. 2017), prostate cancer (Lundin et al. 2009; Vieira et al. 2013), and pancreatic cancer (Raleigh et al. 2014). For instance, in our study, we found that PFOA was associated with MAGI1, which is a potential tumor suppressor gene in breast cancer (Alday-Parejo et al. 2020); PFOS was associated with HPSE2 that encodes a heparanase enzyme and plays a role in breast cancer (Wu et al. 2020). In addition, PFOA was associated with KRT18, an intermediate filament gene related to both prostate cancer and breast cancer-associated fibroblasts (Davalieva et al. 2015; Vastrad et al. 2018). Finally, PFOA was associated with SRPRB, a gene that regulates cell apoptosis and expression and may be associated with pancreatic ductal adenocarcinoma (Ma et al. 2017; Mez et al. 2017).
Several novel CpGs identified in our analysis were located in gene regions that have been linked to cognitive abilities, which is consistent with some prior epidemiological studies showing associations of higher prenatal or childhood PFAS concentrations with poor cognition performance, increased odds of attention deficit hyperactivity disorder (ADHD), or ADHD-related outcomes in children (Harris et al. 2018; Hoffman et al. 2010; Vuong et al. 2021). Our results suggested that PFOA may be associated with tenascin R (TNR) and solute carrier family 10 member 2 (SLC10A2), which have been shown to affect neural development and Alzheimer’s disease (Chiovaro et al. 2015; Mez et al. 2017); PFNA was associated with dolichyl-phosphate N-acetylglucosaminephosphotransferase 1 (DPAGT1), solute carrier family 6 member 2 (SLC6A2), transmembrane protein 56 (TMEM56), and these genes were associated with neuromuscular transmission disorders, ADHD, depression, and bipolar disorder (Belaya et al. 2012; Bi et al. 2017; Kim et al. 2021b; Song et al. 2011; Yao et al. 2021). In addition to cancers and cognitive outcomes, we found a few genes that have been linked to the development of obesity [ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 (AGAP1)], coronary artery disease [ring finger protein 13 (RNF13)], inflammatory bowel disease [vasoactive intestinal peptide receptor 1 (VIPR1) and CDSN; PSORS1C1], and kidney function [erythrocyte membrane protein band 4.1 like 3 (EPB41L3) and serpin family A member 5 (SERPINA5)] (D’Angelo et al. 2018; Gorski et al. 2017; Vilander et al. 2017; Wakil et al. 2016; Yukawa et al. 2007). These findings are consistent with previous epidemiological studies reporting similar child and adult health conditions in relation to PFAS exposure (Blake and Fenton 2020; Rappazzo et al. 2017).
We found that PFAS were associated with a lower percentage of B lymphocytes in cord blood. Some studies have shown that PFAS may decrease B cell populations (Rockwell et al. 2017) and suppress T-cell antigen response in adult mice (DeWitt et al. 2016), decrease the production of vaccine antibodies against tetanus and diphtheria in children (Abraham et al. 2020), and increase the severity of COVID-19 in adults (Grandjean et al. 2020). Future studies are needed to determine the role of PFAS exposure in vaccine response against infectious diseases, particularly COVID-19.
Strengths of the present study include the longitudinal design with DNA methylation assessed at two time points in children, using the most recent microarray for genome-wide DNA methylation assessment, the EPIC array, which interrogates almost twice the measured CpGs as the 450K array, with higher coverage within enhancer regions and distal regulatory elements (Pidsley et al. 2016). Another strength is our replication study using Project Viva, with repeated measures of DNA methylation across childhood. This allowed us to validate the novel PFAS-associated CpGs identified in our study.
Our study has some limitations. We assessed DNA methylation in leukocytes, which may not accurately represent the levels of DNA methylation in other tissues. We also did not measure mRNA levels to evaluate gene expression related to DNA methylation differences observed in our analysis. In addition, given that the DNA methylation varies by genotype, our results may not be generalized to populations with different genetic backgrounds. In addition, this study was underpowered to identify sex-specific associations between PFAS and repeated measures of DNA methylation. Robinson et al. (2021) found evidence that sex may modify the associations of DNA methylation with PFOS concentrations in newborns, but the interaction terms between sex and PFAS concentrations were not significant in the analysis by Starling et al. (2020). Therefore, future longitudinal studies with larger samples are needed to confirm the sex-specific associations. Although we did control for multiple comparisons in our analysis, we did not control for multiplicity of exposures, but this may not be appropriate given that PFAS concentrations were correlated with each other.
Another limitation is the timing of measurement for gestational PFAS concentrations. Concentrations were measured across a range of gestational ages (10.4–30.3 wk) across the HOME Study and Project Viva, and in different trimesters. This may have resulted in different patterns of associations across the two studies given that PFAS concentrations may decline over pregnancy owing to hemodynamic changes (e.g., plasma volume expansion). Although concentrations decline over pregnancy, we previously showed that PFAS concentrations across pregnancy are highly correlated in the HOME Study (Kato et al. 2014), and the preservation of rank order in exposure is likely to maintain the associations of PFAS levels with DNA methylation regardless of the timing of exposure assessment. Relatedly, it is unlikely that maternal plasma volume changes during pregnancy are related to child leukocyte DNA methylation given that they will not alter the relative blood cell profile collected in offspring. In a prior study from Project Viva, adjusting for pregnancy hemodynamics did not meaningfully change the associations of birth outcomes with PFAS concentrations in pregnancy (Sagiv et al. 2018).
Finally, given that two different chips were used for the HOME Study and Project Viva, 28% of the 435 significant CpG sites detected on the EPIC array were missing from the 450K array. We were not able to attempt to replicate these CpG sites. Last, we cannot rule out the possibility that residual confounding may bias our results.
Conclusions
Using longitudinal data, serum PFAS concentrations during pregnancy were associated with stable differences in several CpGs at delivery and in adolescence. These CpGs were in or near genes linked to PFAS-associated health outcomes. To the best of our knowledge, this is the first study to examine the association between gestational PFAS exposure and longitudinal epigenome-wide DNA methylation. Future studies are needed to replicate our findings and verify whether DNA methylation is a mechanism linking early life PFAS exposure to human health.
Supplementary Material
Acknowledgments
We are grateful to the study participants for the time they have given to our study. We thank A. Calafat and the laboratory staff at the Centers for Disease Control and Prevention for performing the perfluoroalkyl substances measurements. This work was supported by the National Institute of Environmental Health Sciences [P01 ES011261 (to BPL), R01 ES020349 (to K.Y.), R01 ES027224 (to K.M.C.), R01 ES025214 (to J.M.B.), and R01 ES030078 (to J.B.)]. Project Viva is supported by the National Institute of Child Health and Human Development [R01 HD034568 (to E.O.)] and National Institutes of Health [UH3 OD023286 (to E.O.)].
References
- Abraham K, Mielke H, Fromme H, Volkel W, Menzel J, Peiser M, et al. 2020. Internal exposure to perfluoroalkyl substances (PFASs) and biological markers in 101 healthy 1-year-old children: associations between levels of perfluorooctanoic acid (PFOA) and vaccine response. Arch Toxicol 94:2131–2147, PMID: , 10.1007/s00204-020-02715-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adkins RM, Krushkal J, Tylavsky FA, Thomas F. 2011. Racial differences in gene-specific DNA methylation levels are present at birth. Birth Defects Res A Clin Mol Teratol 91(8):728–736, PMID: , 10.1002/bdra.20770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alday-Parejo B, Richard F, Wörthmüller J, Rau T, Galvan JA, Desmedt C, et al. 2020. MAGI1, a new potential tumor suppressor gene in estrogen receptor positive breast cancer. Cancers (Basel) 12(1):223, PMID: , 10.3390/cancers12010223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews DQ, Naidenki OV. 2020. Population-wide exposure to per- and polyfluoroalkyl substances from drinking water in the United States. Environ Sci Technol Lett 7(12):931–936, 10.1021/acs.estlett.0c00713. [DOI] [Google Scholar]
- ATSDR (Agency for Toxic Substances and Disease Registry). 2018. Toxicological Profile for Perfluoroalkyls. https://wwwn.cdc.gov/TSP/ToxProfiles/ToxProfiles.aspx?id=1117&tid=237 [accessed 13 December 2021]. [PubMed]
- Baccarelli A, Bollati V. 2009. Epigenetics and environmental chemicals. Curr Opin Pediatr 21(2):243–251, PMID: , 10.1097/MOP.0b013e32832925cc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JR. 2017. Programming the future: epigenetics in the context of DOHad. Environ Health Perspect 125(4):A72, PMID: , 10.1289/ehp.125-A72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belaya K, Finlayson S, Slater CR, Cossins J, Liu WW, Maxwell S, et al. 2012. Mutations in DPAGT1 cause a limb-girdle congenital myasthenic syndrome with tubular aggregates. Am J Hum Genet 91(1):193–201, PMID: , 10.1016/j.ajhg.201ok2.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. 2001. Controlling the false discovery rate in behavior genetics research. Behav Brain Res 125(1–2):279–284, PMID: , 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- Bi Y, Huang X, Niu W, Chen S, Wu X, Cao Y, et al. 2017. Common variants in SLC6A2, SLC6A3, DRD2, and major depressive disorder: an association study in the Chinese Han population. Psychiatr Genet 27(3):103–104, PMID: , 10.1097/YPG.0000000000000163. [DOI] [PubMed] [Google Scholar]
- Bjornsson HT, Sigurdsson MI, Fallin MD, Irizarry RA, Aspelund T, Cui H, et al. 2008. Intra-individual change over time in DNA methylation with familial clustering. JAMA 299(24):2877–2883, PMID: , 10.1001/jama.299.24.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake BE, Fenton SE. 2020. Early life exposure to per- and polyfluoroalkyl substances (PFAS) and latent health outcomes: a review including the placenta as a target tissue and possible driver of peri- and postnatal effects. Toxicology 443:152565, PMID: , 10.1016/j.tox.2020.152565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonefeld-Jorgensen EC, Long M, Bossi R, Ayotte P, Asmund G, Kruger T, et al. 2011. Perfluorinated compounds are related to breast cancer risk in Greenlandic Inuit: a case control study. Environ Health 10:88, PMID: , 10.1186/1476-069X-10-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boronow KE, Brody JG, Schaider LA, Peaslee GF, Havas L, Cohn BA. 2019. Serum concentrations of PFASs and exposure-related behaviors in African American and non-Hispanic white women. J Expo Sci Environ Epidemiol 29(2):206–217, PMID: , 10.1038/s41370-018-0109-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braid SM, Okrah K, Shetty A, Corrada Bravo H. 2017. DNA methylation patterns in cord blood of neonates across gestational age: association with cell-type proportions. Nurs Res 66(2):115–122, PMID: , 10.1097/NNR.0000000000000210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun JM. 2017. Early-life exposure to EDCs: role in childhood obesity and neurodevelopment. Nat Rev Endocrinol 13(3):161–173, PMID: , 10.1038/nrendo.2016.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun JM, Buckley JP, Cecil KM, Chen A, Kalkwarf HJ, Lanphear BP, et al. 2020. Adolescent follow-up in the Health Outcomes and Measures of the Environment (HOME) study: cohort profile. BMJ Open 10(5):e034838, PMID: , 10.1136/bmjopen-2019-034838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun JM, Chen A, Romano ME, Calafat AM, Webster GM, Yolton K, et al. 2016. Prenatal perfluoroalkyl substance exposure and child adiposity at 8 years of age: the HOME study. Obesity (Silver Spring) 24(1):231–237, PMID: , 10.1002/oby.21258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun JM, Daniels JL, Poole C, Olshan AF, Hornung R, Bernert JT, et al. 2010. A prospective cohort study of biomarkers of prenatal tobacco smoke exposure: the correlation between serum and meconium and their association with infant birth weight. Environ Health 9:53, PMID: , 10.1186/1476-069X-9-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun JM, Kalloo G, Chen A, Dietrich KN, Liddy-Hicks S, Morgan S, et al. 2017. Cohort profile: the Health Outcomes and Measures of the Environment (HOME) study. Int J Epidemiol 46(1):24, PMID: , 10.1093/ije/dyw006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton CV, Marsit CJ, Faustman E, Nadeau K, Goodrich JM, Dolinoy DC, et al. 2017. Small-magnitude effect sizes in epigenetic end points are important in children’s environmental health studies: the Children’s Environmental Health and Disease Prevention Research Center’s Epigenetics Working Group. Environ Health Perspect 125(4):511–526, PMID: , 10.1289/EHP595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC (Centers for Disease Control and Prevention). 2015. Laboratory Procedure Manual. https://wwwn.cdc.gov/nchs/data/nhanes/2015-2016/labmethods/PFAS_I_MET.pdf [accessed 13 December 2021].
- Chiovaro F, Chiquet-Ehrismann R, Chiquet M. 2015. Transcriptional regulation of tenascin genes. Cell Adh Migr 9(1–2):34–47, PMID: , 10.1080/19336918.2015.1008333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo CS, Varela MC, de Castro CIE, Otto PA, Perez ABA, Lourenço CM, et al. 2018. Chromosomal microarray analysis in the genetic evaluation of 279 patients with syndromic obesity. Mol Cytogenet 11:14, PMID: , 10.1186/s13039-018-0363-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davalieva K, Kostovska IM, Kiprijanovska S, Markoska K, Kubelka-Sabit K, Filipovski V, et al. 2015. Proteomics analysis of malignant and benign prostate tissue by 2d DIGE/MS reveals new insights into proteins involved in prostate cancer. Prostate 75(14):1586–1600, PMID: , 10.1002/pros.23034. [DOI] [PubMed] [Google Scholar]
- DeWitt JC, Williams WC, Creech NJ, Luebke RW. 2016. Suppression of antigen-specific antibody responses in mice exposed to perfluorooctanoic acid: role of PPARα and T- and B-cell targeting. J Immunotoxicol 13(1):38–45, PMID: , 10.3109/1547691X.2014.996682. [DOI] [PubMed] [Google Scholar]
- Du P, Zhang X, Huang CC, Jafari N, Kibbe WA, Hou L, et al. 2010. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics 11:587, PMID: , 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gervin K, Salas LA, Bakulski KM, van Zelm MC, Koestler DC, Wiencke JK, et al. 2019. Systematic evaluation and validation of reference and library selection methods for deconvolution of cord blood DNA methylation data. Clin Epigenetics 11(1):125, PMID: , 10.1186/s13148-019-0717-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glüge J, Scheringer M, Cousins IT, DeWitt JC, Goldenman G, Herzke D, et al. 2020. An overview of the uses of per- and polyfluoroalkyl substances (PFAS). Environ Sci Process Impacts 22(12):2345–2373, PMID: , 10.1039/d0em00291g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski M, van der Most PJ, Teumer A, Chu AY, Li M, Mijatovic V, et al. 2017. 1000 Genomes-based meta-analysis identifies 10 novel loci for kidney function. Sci Rep 7:45040, PMID: , 10.1038/srep45040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandjean P, Timmermann CAG, Kruse M, Nielsen F, Vinholt PJ, Boding L, et al. 2020. Severity of COVID-19 at elevated exposure to perfluorinated alkylates. PLoS One 15:e0244815, PMID: , 10.1371/journal.pone.0244815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Preston R, Goldman LR, Brebi-Mieville P, Ili-Gangas C, Lebron C, Witter FR, et al. 2010. Global DNA hypomethylation is associated with in utero exposure to cotinine and perfluorinated alkyl compounds. Epigenetics 5(6):539–546, PMID: , 10.4161/epi.5.6.12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeussler M, Zweig AS, Tyner C, Speir ML, Rosenbloom KR, Raney BJ, et al. 2019. The UCSC Genome Browser database: 2019 update. Nucleic Acids Res 47(D1):D853–D858, PMID: , 10.1093/nar/gky1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MH, Oken E, Rifas-Shiman SL, Calafat AM, Ye X, Bellinger DC, et al. 2018. Prenatal and childhood exposure to per- and polyfluoroalkyl substances (PFASs) and child cognition. Environ Int 115:358–369, PMID: , 10.1016/j.envint.2018.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman K, Webster TF, Weisskopf MG, Weinberg J, Vieira VM. 2010. Exposure to polyfluoroalkyl chemicals and attention deficit/hyperactivity disorder in U.S. children 12–15 years of age. Environ Health Perspect 118(12):1762–1767, PMID: , 10.1289/ehp.1001898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A. 2007. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8(1):118–127, PMID: , 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Kato K, Basden BJ, Needham LL, Calafat AM. 2011. Improved selectivity for the analysis of maternal serum and cord serum for polyfluoroalkyl chemicals. J Chromatogr A 1218(15):2133–2137, PMID: , 10.1016/j.chroma.2010.10.051. [DOI] [PubMed] [Google Scholar]
- Kato K, Kalathil AA, Patel AM, Ye X, Calafat AM. 2018. Per- and polyfluoroalkyl substances and fluorinated alternatives in urine and serum by on-line solid phase extraction–liquid chromatography–tandem mass spectrometry. Chemosphere 209:338–345, PMID: , 10.1016/j.chemosphere.2018.06.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Wong LY, Chen A, Dunbar C, Webster GM, Lanphear BP, et al. 2014. Changes in serum concentrations of maternal poly- and perfluoroalkyl substances over the course of pregnancy and predictors of exposure in a multiethnic cohort of Cincinnati, Ohio pregnant women during 2003–2006. Environ Sci Technol 48(16):9600–9608, PMID: , 10.1021/es501811k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsey KT, Rytel M, Dere E, Butler R, Eliot M, Huse SM, et al. 2019. Serum dioxin and DNA methylation in the sperm of Operation Ranch Hand veterans exposed to Agent Orange. Environ Health 18(1):91, PMID: , 10.1186/s12940-019-0533-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Thapar I, Brooks BW. 2021a. Epigenetic changes by per- and polyfluoroalkyl substances (PFAS). Environ Pollut 279:116929, PMID: , 10.1016/j.envpol.2021.116929. [DOI] [PubMed] [Google Scholar]
- Kim SY, Kim HN, Jeon SW, Lim WJ, Kim SI, Lee YJ, et al. 2021b. Association between genetic variants of the norepinephrine transporter gene (SLC6A2) and bipolar I disorder. Prog Neuropsychopharmacol Biol Psychiatry 107:110227, PMID: , 10.1016/j.pnpbp.2020.110227. [DOI] [PubMed] [Google Scholar]
- Kingsley SL, Eliot MN, Kelsey KT, Calafat AM, Ehrlich S, Lanphear BP, et al. 2018. Variability and predictors of serum perfluoroalkyl substance concentrations during pregnancy and early childhood. Environ Res 165:247–257, PMID: , 10.1016/j.envres.2018.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S, Azumi K, Goudarzi H, Araki A, Miyashita C, Kobayashi S, et al. 2017. Effects of prenatal perfluoroalkyl acid exposure on cord blood IGF2/H19 methylation and ponderal index: the Hokkaido Study. J Expo Sci Environ Epidemiol 27(3):251–259, PMID: , 10.1038/jes.2016.50. [DOI] [PubMed] [Google Scholar]
- Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. 2012. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28(6):882–883, PMID: , 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Liu Y, Papandonatos GD, Calafat AM, Eaton CB, Kelsey KT, et al. 2021. Gestational and childhood exposure to per- and polyfluoroalkyl substances and cardiometabolic risk at age 12 years. Environ Int 147:106344, PMID: , 10.1016/j.envint.2020.106344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CY, Chen PC, Lien PC, Liao YP. 2018. Prenatal perfluorooctyl sulfonate exposure and Alu DNA hypomethylation in cord blood. Int J Environ Res Public Health 15(6):1066, PMID: , 10.3390/ijerph15061066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Li N, Papandonatos GD, Calafat AM, Eaton CB, Kelsey KT, et al. 2020. Exposure to per- and polyfluoroalkyl substances and adiposity at age 12 years: evaluating periods of susceptibility. Environ Sci Technol 54(24):16039–16049, PMID: , 10.1021/acs.est.0c06088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Peterson KE. 2015. Maternal exposure to synthetic chemicals and obesity in the offspring: recent findings. Curr Environ Health Rep 2(4):339–347, PMID: , 10.1007/s40572-015-0068-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin JI, Alexander BH, Olsen GW, Church TR. 2009. Ammonium perfluorooctanoate production and occupational mortality. Epidemiology 20(6):921–928, PMID: , 10.1097/EDE.0b013e3181b5f395. [DOI] [PubMed] [Google Scholar]
- Ma Q, Wu X, Wu J, Liang Z, Liu T. 2017. SERP1 is a novel marker of poor prognosis in pancreatic ductal adenocarcinoma patients via anti-apoptosis and regulating SRPRB/NF-κB axis. Int J Oncol 51(4):1104–1114, PMID: , 10.3892/ijo.2017.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mez J, Chung J, Jun G, Kriegel J, Bourlas AP, Sherva R, et al. 2017. Two novel loci, COBL and SLC10A2, for Alzheimer’s disease in African Americans. Alzheimers Dement 13(2):119–129, PMID: , 10.1016/j.jalz.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura R, Araki A, Miyashita C, Kobayashi S, Kobayashi S, Wang SL, et al. 2018. An epigenome-wide study of cord blood DNA methylations in relation to prenatal perfluoroalkyl substance exposure: the Hokkaido study. Environ Int 115:21–28, PMID: , 10.1016/j.envint.2018.03.004. [DOI] [PubMed] [Google Scholar]
- Oken E, Baccarelli AA, Gold DR, Kleinman KP, Litonjua AA, De Meo D, et al. 2015. Cohort profile: Project Viva. Int J Epidemiol 44(1):37–48, PMID: , 10.1093/ije/dyu008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouidir M, Mendola P, Buck Louis GM, Kannan K, Zhang C, Tekola-Ayele F. 2020. Concentrations of persistent organic pollutants in maternal plasma and epigenome-wide placental DNA methylation. Clin Epigenet 12(1):103, PMID: , 10.1186/s13148-020-00894-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SK, Peng Q, Ding N, Mukherjee B, Harlow SD. 2019. Determinants of per- and polyfluoroalkyl substances (PFAS) in midlife women: evidence of racial/ethnic and geographic differences in PFAS exposure. Environ Res 175:186–199, PMID: , 10.1016/j.envres.2019.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipson B, Maksimovic J. 2020. Package ‘missMethyl’: analysing Illumina HumanMethylation BeadChip data. Version 1.28.0. https://bioconductor.org/packages/release/bioc/manuals/missMethyl/man/missMethyl.pdf [accessed 13 December 2021].
- Pidsley R, Zotenko E, Peters TJ, Lawrence MG, Risbridger GP, Molloy P, et al. 2016. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol 17(1):208, PMID: , 10.1186/s13059-016-1066-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raleigh KK, Alexander BH, Olsen GW, Ramachandran G, Morey SZ, Church TR, et al. 2014. Mortality and cancer incidence in ammonium perfluorooctanoate production workers. Occup Environ Med 71(7):500–506, PMID: , 10.1136/oemed-2014-102109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randhawa R, Cohen P. 2005. The role of the insulin-like growth factor system in prenatal growth. Mol Genet Metab 86(1–2):84–90, PMID: , 10.1016/j.ymgme.2005.07.028. [DOI] [PubMed] [Google Scholar]
- Rappazzo KM, Coffman E, Hines EP. 2017. Exposure to perfluorinated alkyl substances and health outcomes in children: a systematic review of the epidemiologic literature. Int J Environ Res Public Health 14(7):691, PMID: , 10.3390/ijerph14070691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson SL, Zeng X, Guan W, Sundaram R, Mendola P, Putnick DL, et al. 2021. Perfluorooctanoic acid (PFOA) or perfluorooctane sulfonate (PFOS) and DNA methylation in newborn dried blood spots in the Upstate KIDS cohort. Environ Res 194:110668, PMID: , 10.1016/j.envres.2020.110668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell CE, Turley AE, Cheng X, Fields PE, Klaassen CD. 2017. Persistent alterations in immune cell populations and function from a single dose of perfluorononanoic acid (PFNA) in C57Bl/6 mice. Food Chem Toxicol 100:24–33, PMID: , 10.1016/j.fct.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagiv SK, Rifas-Shiman SL, Fleisch AF, Webster TF, Calafat AM, Ye X, et al. 2018. Early-pregnancy plasma concentrations of perfluoroalkyl substances and birth outcomes in Project Viva: confounded by pregnancy hemodynamics? Am J Epidemiol 187(4):793–802, PMID: , 10.1093/aje/kwx332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas LA, Koestler DC, Butler RA, Hansen HM, Wiencke JK, Kelsey KT, et al. 2018. An optimized library for reference-based deconvolution of whole-blood biospecimens assayed using the Illumina HumanMethylationEPIC BeadArray. Genome Biol 19(1):64, PMID: , 10.1186/s13059-018-1448-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon O, MacIsaac J, Quach H, Tindula G, Kobor MS, Huen K, et al. 2018. Comparison of DNA methylation measured by Illumina 450K and EPIC BeadChips in blood of newborns and 14-year-old children. Epigenetics 13(6):655–664, PMID: , 10.1080/15592294.2018.1497386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Song DH, Jhung K, Cheon KA. 2011. Norepinephrine transporter gene (SLC6A2) is involved with methylphenidate response in Korean children with attention deficit hyperactivity disorder. Int Clin Psychopharmacol 26(2):107–113, PMID: , 10.1097/YIC.0b013e32834152d1. [DOI] [PubMed] [Google Scholar]
- Starling AP, Liu C, Shen G, Yang IV, Kechris K, Borengasser SJ, et al. 2020. Prenatal exposure to per- and polyfluoroalkyl substances, umbilical cord blood DNA methylation, and cardio-metabolic indicators in newborns: the Healthy Start Study. Environ Health Perspect 128(12):127014, PMID: , 10.1289/EHP6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. 2003. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A 100(16):9440–9445, PMID: , 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WWC, Dietmann S, Irie N, Leitch HG, Floros VI, Bradshaw CR, et al. 2015. A unique gene regulatory network resets the human germline epigenome for development. Cell 161(6):1453–1467, PMID: , 10.1016/j.cell.2015.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff AE, Marabita F, Lechner M, Bartlett T, Tegner J, Gomez-Cabrero D, et al. 2013. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics 29(2):189–196, PMID: , 10.1093/bioinformatics/bts680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MS, Chang SH, Kuo WH, Kuo CH, Li SY, Wang MY, et al. 2020. A case-control study of perfluoroalkyl substances and the risk of breast cancer in Taiwanese women. Environ Int 142:105850, PMID: , 10.1016/j.envint.2020.105850. [DOI] [PubMed] [Google Scholar]
- van den Dungen MW, Murk AJ, Kampman E, Steegenga WT, Kok DE. 2017. Association between DNA methylation profiles in leukocytes and serum levels of persistent organic pollutants in Dutch men. Environ Epigenet 3(1):dvx001, PMID: , 10.1093/eep/dvx001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastrad B, Vastrad C, Tengli A, Iliger S. 2018. Identification of differentially expressed genes regulated by molecular signature in breast cancer-associated fibroblasts by bioinformatics analysis. Arch Gynecol Obstet 297(1):161–183, PMID: , 10.1007/s00404-017-4562-y. [DOI] [PubMed] [Google Scholar]
- Vieira VM, Hoffman K, Shin HM, Weinberg JM, Webster TF, Fletcher T. 2013. Perfluorooctanoic acid exposure and cancer outcomes in a contaminated community: a geographic analysis. Environ Health Perspect 121(3):318–323, PMID: , 10.1289/ehp.1205829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilander LM, Kaunisto MA, Vaara ST, Pettilä V; FINNAKI study group. 2017. Genetic variants in SERPINA4 and SERPINA5, but not BCL2 and SIK3 are associated with acute kidney injury in critically ill patients with septic shock. Crit Care 21(1):47, PMID: , 10.1186/s13054-017-1631-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong AM, Webster GM, Yolton K, Calafat AM, Muckle G, Lanphear BP, et al. 2021. Prenatal exposure to per- and polyfluoroalkyl substances (PFAS) and neurobehavior in US children through 8 years of age: the HOME study. Environ Res 195:110825, PMID: , 10.1016/j.envres.2021.110825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakil SM, Ram R, Muiya NP, Mehta M, Andres E, Mazhar N, et al. 2016. A genome-wide association study reveals susceptibility loci for myocardial infarction/coronary artery disease in Saudi Arabs. Atherosclerosis 245:62–70, PMID: , 10.1016/j.atherosclerosis.2015.11.019. [DOI] [PubMed] [Google Scholar]
- Wielsøe M, Kern P, Bonefeld-Jørgensen EC. 2017. Serum levels of environmental pollutants is a risk factor for breast cancer in Inuit: a case control study. Environ Health 16(1):56, PMID: , 10.1186/s12940-017-0269-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm-Benartzi CS, Koestler DC, Karagas MR, Flanagan JM, Christensen BC, Kelsey KT, et al. 2013. Review of processing and analysis methods for DNA methylation array data. Br J Cancer 109(6):1394–1402, PMID: , 10.1038/bjc.2013.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Liu G, Jin Y, Yang T, Zhang D, Ding L, et al. 2020. miR-15b-5p promotes growth and metastasis in breast cancer by targeting HPSE2. Front Oncol 10:108, PMID: , 10.3389/fonc.2020.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Jurkovic-Mlakar S, Lindh CH, Scott K, Fletcher T, Jakobsson K, et al. 2020. Associations between serum concentrations of perfluoroalkyl substances and DNA methylation in women exposed through drinking water: a pilot study in Ronneby, Sweden. Environ Int 145:106148, PMID: , 10.1016/j.envint.2020.106148. [DOI] [PubMed] [Google Scholar]
- Yao X, Glessner JT, Li J, Qi X, Hou X, Zhu C, et al. 2021. Integrative analysis of genome-wide association studies identifies novel loci associated with neuropsychiatric disorders. Transl Psychiatry 11(1):69, PMID: , 10.1038/s41398-020-01195-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yukawa T, Oshitani N, Yamagami H, Watanabe K, Higuchi K, Arakawa T. 2007. Differential expression of vasoactive intestinal peptide receptor 1 expression in inflammatory bowel disease. Int J Mol Med 20(2):161–167, PMID: , 10.3892/ijmm.20.2.161. [DOI] [PubMed] [Google Scholar]
- Zeger SL, Liang KY. 1986. Longitudinal data analysis for discrete and continuous outcomes. Biometrics 42(1):121–130, PMID: , 10.2307/2531248. [DOI] [PubMed] [Google Scholar]
- Zhou W, Laird PW, Shen H. 2017. Comprehensive characterization, annotation and innovative use of Infinium DNA methylation BeadChip probes. Nucleic Acids Res 45(4):e22, PMID: , 10.1093/nar/gkw967. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.