Abstract
The new rifamycin derivatives KRM-1657 and KRM-1648 were evaluated for their in vitro antimicrobial activities against 44 strains of Helicobacter pylori. Although the drugs were not very active against other gram-negative bacteria, the MICs at which 90% of isolates are inhibited for these drugs were lower (0.002 and 0.008 μg/ml, respectively) than those of amoxicillin and rifampin for H. pylori. Time-kill studies revealed that the bactericidal activities of these agents were due to cell lysis. The results presented here indicate that these new rifamycin derivatives may be useful for the eradication of H. pylori infections.
Helicobacter pylori is associated with chronic gastritis and peptic ulcer disease (3, 12), as well as gastric carcinoma (20, 21). In patients with mucosal H. pylori infection, eradication of this microorganism seems to cure both infection and ulcer disease (18, 23). Although H. pylori is susceptible to most antimicrobial agents in vitro, in vivo eradication of this pathogen has been difficult (11). The highest cure rates have required multidrug antimicrobial therapies, including those with combinations of clarithromycin, metronidazole, or amoxicillin in association with a proton pump inhibitor, e.g., omeprazole (8, 24). It has been reported, however, that the presence of strains resistant to clarithromycin and metronidazole pre- and posttreatment resulted in treatment failure (11, 13, 22), and thus, new drugs with activity against H. pylori are crucial.
The recently synthesized rifamycin derivatives 3′-hydroxy-5′-(4-isobutyl-1-piperazinyl)benzoxazinorifamycin (KRM-1648) and 3′-hydroxy-5′-(4-propyl-1-piperazinyl)benzoxazinorifamycin (KRM-1657) (25, 28) exhibit potent activities against a variety of gram-positive bacteria such as Mycobacterium tuberculosis, Mycobacterium avium complex, Staphylococcus aureus, Streptococcus pneumoniae, and Streptococcus pyogenes (10). On the other hand, these drugs have little activity against gram-negative bacteria such as Escherichia coli, Klebsiella pneumoniae, Haemophilus influenzae, and Neisseria gonorrhoeae (9, 10).
In the present study, we determined the antibacterial activities of the KRM compounds against H. pylori. Both KRM-1648 and KRM-1657 exhibited potent antimicrobial activities against clinical isolates of H. pylori, a gram-negative bacterium.
MATERIALS AND METHODS
Antimicrobial agents.
KRM-1648, KRM-1657, and 14C-KRM-1648 (0.51 MBq/mg) were obtained from KANEKA Corp. (Hyogo, Japan). Rifampin, amoxicillin, and clarithromycin were obtained from Calbiochem (La Jolla, Calif.), Wako (Tokyo, Japan), and Taisho Pharmaceutical Co., Ltd. (Tokyo, Japan), respectively. The antibiotics were dissolved in dimethyl sulfoxide at 5.0 mg/ml (stock solution) and were stored at −20°C until use.
Bacterial strains and growth conditions.
H. pylori NCTC11637 (the type strain) and 34 strains including strain HPK5 and numbered CPY strains isolated from biopsy specimens from patients undergoing upper gastrointestinal endoscopy at the University Hospital, Yamaguchi University School of Medicine, Ube, Japan, were studied. Four clarithromycin-resistant strains, three metronidazole-resistant strains, and two strains resistant to both clarithromycin and metronidazole were isolated in Oita Medical University, Oita, Japan, and were also studied. The sources of these strains were patients with gastric ulcers (20 strains), duodenal ulcers (6 strains), gastroduodenal ulcers (2 strains), gastric cancer (8 strains), chronic gastritis (6 strains), and duodenitis (1 strain). We used Mueller-Hinton broth (Difco Laboratories, Detroit, Mich.) containing 5% (wt/vol) horse serum (Gibco BRL, Rockville, Md.) and solidified it with 1.3% agar (Nakarai, Kyoto, Japan) to make an agar medium. The bacteria were inoculated into 18-mm-diameter tubes containing 5 ml of broth medium or into 25-ml flasks containing 10 ml of broth medium, and the tubes and flasks were incubated at 37°C with shaking on a rotating shaker (150 rpm) in a chamber filled with 15% CO2. The agar plates were incubated in a gas mixture (10% CO2, 5% O2, 85% N2) at 37°C for 4 days under high humidity.
MICs.
The MICs of KRM-1648, KRM-1657, rifampin, and amoxicillin were determined in duplicate by the twofold serial agar dilution method. Stock solutions were diluted in 0.1 M sodium-potassium phosphate buffer (pH 7.0) and were serially diluted with the buffer. Drug solutions with volumes equal to 1/10 the volume of the agar medium were added to make drug-supplemented agar plates. Agar plates with drug concentrations greater than 1 μg/ml were prepared by adding a stock solution directly to the agar medium. Overnight cultures in 5 ml of broth medium were adjusted to an optical density at 590 nm of 0.2, and inocula were spotted onto drug-supplemented agar plates by using a 96-well inoculator. The bacterial counts in each spot (approximately 1 μl) were between 5 × 103 and 1 × 104 CFU (corresponding to 5 × 106 and 1 × 107 CFU/ml, respectively).
To determine the MICs at a lower pH, Muller-Hinton broth was adjusted to pH 7.0 or pH 5.0 with HCl, solidified, autoclaved, and supplemented with 1/20 the final volume of 5 horse serum and 1/10 the volume of the drug solutions serially diluted with 0.1 M sodium-potassium phosphate buffer (pH 7.0 or pH 5.0) to make the agar plates. The final pH values were 7.4 and 5.4, as determined with broth medium without agar prepared by the same procedure described above. Two of the several strains tested showed satisfactory growth on the agar medium at pH 5.4.
Stability of KRM-1648 in an acidic environment.
To determine the stability of KRM-1648 in medium with an acidic pH, 20 mg of KRM-1648 dissolved in 200 ml of 0.01 N HCl (0.1 mg/ml; pH 2) in duplicate was incubated at 37°C. After 1, 2, 5, 8, and 24 h, the amounts of KRM-1648 in a 20-μl solution were determined by high-pressure liquid chromatography.
Time-kill bactericidal activity studies.
Time-kill studies were performed by the method of Coudron and Stratton (6, 7). H. pylori HPK5 bacteria cultured overnight in 10 ml of broth medium were inoculated at a density of 3 × 106 CFU/ml into a 25-ml flask containing 10 ml of drug-supplemented broth medium. At 0, 3, and 24 h, the samples were removed and serially diluted with saline, 20-μl aliquots were spotted in duplicate onto agar plates, the agar plates were incubated, and the numbers of CFU were determined. The lowest level of accurate cell detection was 100 CFU/ml. In addition, undiluted samples were spread onto glass slides and were Gram stained to determine the cell morphology.
Antibiotic-resistant mutants.
The occurrence of spontaneous antibiotic-resistant mutants was determined with six strains of H. pylori. The bacteria were grown in 10 ml of broth medium overnight; 1-ml aliquots of each culture containing approximately 108 bacteria were centrifuged, suspended in 0.2 ml of broth medium, and plated onto drug-supplemented agar medium; and the plates were incubated for 4 days. In addition, the inoculum of each strain was determined after appropriate dilution and plating on drug-free agar plates. The number of bacterial colonies on agar plates was counted, and the frequency of occurrence of spontaneous mutants was determined. Resistant mutants were isolated by a single-colony isolation procedure by streaking a colony once on the same drug-supplemented agar medium followed by streaking of the colony on drug-free agar medium.
Incorporation of 14C-labeled KRM-1648 into H. pylori cells.
Measurement of the incorporation of KRM-1648 was performed as described previously (9). In brief, H. pylori NCTC11637 and its KRM-1648-resistant mutant, NCTC11637-K48r, which were grown in broth medium, were inoculated into 10 ml of broth medium to an optical density at 590 nm of 0.1. [14C]KRM-1648 (0.51 kBq/μmol) was added to a final concentration of 1 μg/ml, and the reaction mixture was incubated at 37°C. At indicated times, 1 ml of each of the incubated suspensions was applied to a cellulose-nitrate filter (pore size, 0.45 μm; ADVANTEC, Tokyo, Japan); filtered; washed successively with distilled water (30 ml), 0.5% trichloroacetic acid (30 ml), and 95% ethanol (2 ml); and air dried. The radioactivity was then determined with a Packard 3255 liquid scintillation counter. The incorporation experiment was performed in triplicate.
RESULTS
MICs of KRM-1648 and KRM-1657.
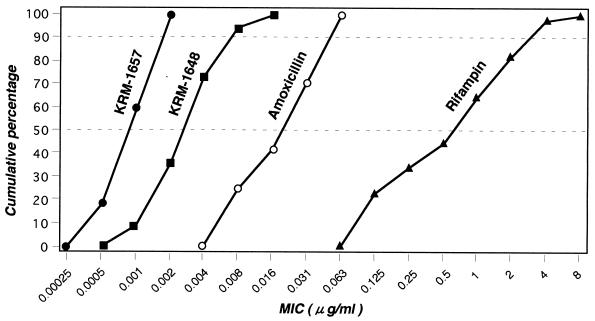
The MICs of KRM-1648, KRM-1657, and rifampin for 44 H. pylori strains and those of amoxicillin for 24 strains are presented in Fig. 1. KRM-1657 and KRM-1648 exhibited more potent antimicrobial activities (MICs at which 50 and 90% of isolates are inhibited [MIC50 and MIC90, respectively] for KRM-1657, 0.001 and 0.002 μg/ml, respectively; MIC50 and MIC90 of KRM-1648, 0.004 and 0.008 μg/ml, respectively) than amoxicillin (MIC50 and MIC90, 0.031 and 0.063 μg/ml, respectively) and rifampin (MIC50 and MIC90, 1 and 4 μg/ml, respectively). When the MICs and the type of disease in the patient from whom the strain was isolated were compared, there was no correlation between the MICs and disease type for any drug (data not shown). Clinically isolated strains that are resistant to clarithromycin and/or metronidazole showed levels of susceptibility to rifampin, KRM-1648, and KRM-1657 similar to those of the drug-sensitive strains. An antimicrobial agent that is commonly used for the treatment of H. pylori infection (clarithromycin) is known to be less active at lower pH (5). In fact, as shown in Table 1, lowering of the pH resulted in a more than eightfold increase in the MICs of clarithromycin. On the other hand, the MICs of KRM-1648 and KRM-1657 did not change by lowering the pH. We could not determine the MICs at pH values lower than 5.4 because of the inability of most H. pylori strains to grow at those pHs.
FIG. 1.
Distribution of susceptibilities of 44 H. pylori strains to KRM-1657 (●), KRM-1648 (■), and rifampin (▴), and 24 strains to amoxicillin (○).
TABLE 1.
MICs of clarithromycin, KRM-1648, and KRM-1657 for H. pylori NCTC11637 and CPY3281 at pH 7.4 or 5.4
| Drug | MIC (μg/ml)
|
|||
|---|---|---|---|---|
| NCTC11637
|
CPY3281
|
|||
| pH 7.4 | pH 5.4 | pH 7.4 | pH 5.4 | |
| Clarithromycin | 0.063 | 0.5 | 0.031 | 0.5 |
| KRM-1648 | 0.004 | 0.002 | 0.008 | 0.004 |
| KRM-1657 | 0.001 | 0.001 | 0.001 | 0.002 |
Stability of KRM-1648 in an acidic environment.
To determine the stability of KRM-1648 in the acidic stomach, we incubated KRM-1648 (0.1 mg/ml) at pH 2.0 and 37°C. After incubation for 8 and 24 h, 95.8 and 87% of the KRM-1648, respectively, remained, suggesting that it is stable in an acidic environment.
Time-kill studies.
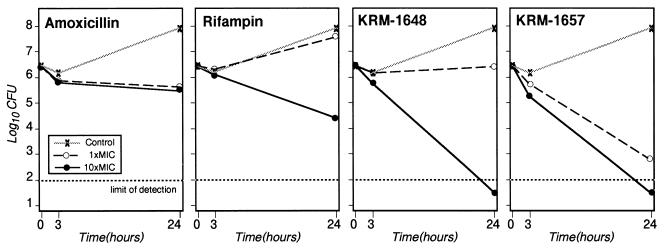
H. pylori HPK5 was inoculated in broth medium containing various drugs at the MIC or at 10× the MIC, and the CFUs were determined at 0, 3, and 24 h (Fig. 2). KRM-1657 at 0.001 μg/ml (1× the MIC) produced a 1-log reduction in the numbers of CFU per milliliter relative to the numbers in the inoculum at 3 h and produced more than a 3-log decrease at 24 h. The numbers of CFU at 24 h in the broth medium containing KRM-1657 at 0.01 μg/ml (10× the MIC) or KRM-1648 at 0.04 μg/ml (10× the MIC) were more than 4.5 logs lower than those in the control at 0 h, indicating the potent bactericidal activities of the drugs. Rifampin at 2.5 μg/ml (10× the MIC) also produced a 2-log reduction at 24 h, whereas amoxicillin at 0.031 μg/ml (1× the MIC) and 0.31 μg/ml (10× the MIC) produced only a 1-log decrease in the numbers of CFU per milliliter after 24 h.
FIG. 2.
Time-kill bactericidal activities of amoxicillin, rifampin, KRM-1648, and KRM-1657 at the MICs (0.031, 0.25, 0.004, and 0.001 μg/ml, respectively) and at 10× the MICs (0.31, 2.5, 0.04, and 0.01 μg/ml, respectively) against H. pylori HPK5.
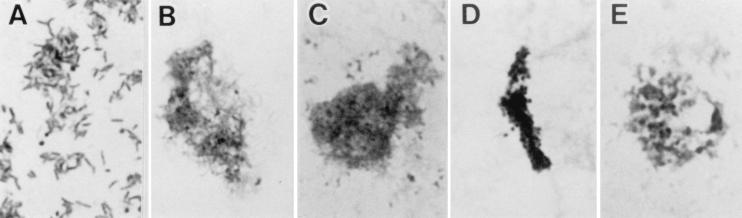
Morphological studies performed by Gram staining revealed that cell lysis occurred after incubation with 0.001 μg of KRM-1657 per ml for 24 h (Fig. 3B), whereas the control culture did not show cell lysis (Fig. 3A). Similar morphological changes were observed when the cells were incubated with 0.04 μg of KRM-1648 per ml for 24 h (Fig. 3C). In contrast, amoxicillin treatment produced coccoid forms. When the cells were incubated with 0.31 μg of amoxicillin per ml for 3 h, cell aggregates consisting mostly of small coccoid forms formed (Fig. 3D). When the cells were incubated with 0.031 μg of amoxicillin per ml for 24 h, large coccoid forms were predominant (Fig. 3E). Only a few coccoid forms with irregular morphologies appeared among the KRM-treated cells.
FIG. 3.
Gram staining of H. pylori HPK5 sampled from the time-kill bactericidal activity test (Fig. 2). (A) No-drug control at 24 h. A stationary-phase culture shows many spiral cells and a few coccoid cells. (B) KRM-1657 (0.001 μg/ml; 1× the MIC) at 24 h. (C) KRM-1648 (0.04 μg/ml; 10× the MIC) at 24 h. (D) Amoxicillin (0.31 μg/ml; 10× the MIC) at 3 h. (E) Amoxicillin (0.031 μg/ml; 1× the MIC) at 24 h. Magnifications, ×1,000.
Antibiotic-resistant mutants.
The frequency of occurrence of spontaneous antibiotic-resistant mutants from six strains of H. pylori is presented in Table 2. The concentrations of each drug used for selection were 50 μg/ml for rifampin, 3.1 μg/ml for KRM-1648, 0.1 μg/ml for KRM-1657, and 3.1 μg/ml for amoxicillin. Colonies that grew on selective agar plates were considered resistant to the respective drug. The frequencies of occurrence of mutants resistant to KRM-1657, KRM-1648, and rifampin were similar for a given strain but varied from strain to strain, ranging from 2.0 × 10−6 to 1.9 × 10−8. No amoxicillin-resistant mutant appeared. We then isolated mutant clones resistant to KRM-1657, KRM-1648, or rifampin, and the MICs of these drugs were evaluated (Table 2). All mutant clones selected on plates containing one of the rifamycin drugs was resistant to the other agents as well, indicating cross-resistance. On the contrary, no cross-resistance with rifamycin agents was seen for clarithromycin or amoxicillin (data not shown).
TABLE 2.
Frequency of occurrence of spontaneous mutants in H. pylori strains resistant to rifampin, KRM-1648, KRM-1657, and amoxicillin and MICs for parental strain and mutant clone
| Strain | Druga | Frequency of mutation | Mutant cloneb | MIC (μg/ml)
|
||
|---|---|---|---|---|---|---|
| Rifampin | KRM-1648 | KRM-1657 | ||||
| NCTC11637 | 0.25 | 0.004 | 0.001 | |||
| Rifampin | 2.1 × 10−8 | NCTC11637-RFPr | >50 | >10 | 10 | |
| KRM-1648 | 7.1 × 10−8 | NCTC11637-K48r | >50 | >10 | 1.6 | |
| KRM-1657 | 7.1 × 10−8 | NCTC11637-K57r | >50 | >10 | 3.2 | |
| Amoxicillin | <7.1 × 10−9 | |||||
| HPK5 | 0.25 | 0.004 | 0.001 | |||
| Rifampin | 2.9 × 10−7 | HPK5-RFPr | >50 | >10 | >10 | |
| KRM-1648 | 2.1 × 10−7 | HPK5-K48r | >50 | >10 | >10 | |
| KRM-1657 | 5.3 × 10−7 | HPK5-K57r | >50 | >10 | >10 | |
| Amoxicillin | <5.1 × 10−9 | |||||
| CPY1124 | 8 | 0.004 | 0.001 | |||
| Rifampin | 2.0 × 10−6 | CPY1124-RFPr | >50 | >10 | >10 | |
| KRM-1648 | 1.5 × 10−6 | CPY1124-K48r | >50 | >10 | >10 | |
| KRM-1657 | 8.7 × 10−7 | CPY1124-K57r | >50 | >10 | >10 | |
| Amoxicillin | <5.0 × 10−9 | |||||
| CPY3281 | 4 | 0.016 | 0.001 | |||
| Rifampin | 1.2 × 10−6 | CPY3281-RFPr | >50 | >10 | 10 | |
| KRM-1648 | 6.4 × 10−7 | CPY3281-K48r | >50 | >10 | 10 | |
| KRM-1657 | 7.9 × 10−7 | CPY3281-K57r | >50 | >10 | 10 | |
| Amoxicillin | <3.0 × 10−9 | |||||
| CPY3401 | 0.125 | 0.004 | 0.0005 | |||
| Rifampin | 6.6 × 10−7 | CPY3401-RFPr | >50 | >10 | 1 | |
| KRM-1648 | 4.6 × 10−8 | CPY3401-K48r | >50 | >10 | >10 | |
| KRM-1657 | 2.3 × 10−7 | CPY3401-K57r | >50 | >10 | >10 | |
| Amoxicillin | <1.5 × 10−9 | |||||
| CPY6071 | 4 | 0.008 | 0.001 | |||
| Rifampin | 4.5 × 10−7 | CPY6071-RFPr | >50 | >10 | >10 | |
| KRM-1648 | 8.2 × 10−8 | CPY6071-K48r | >50 | >10 | >10 | |
| KRM-1657 | 1.9 × 10−8 | CPY6071-K57r | >50 | >10 | 3.2 | |
| Amoxicillin | <4.5 × 10−9 | |||||
The concentrations selected for isolation were 50, 3.1, 0.1, and 3.1 μg/ml for rifampin, KRM-1648, KRM-1657, and amoxicillin, respectively.
RFPr, K48r, and K57r represent clones of each strain resistant to rifampin, KRM-1648, and KRM-1657, respectively.
We could see some correlation between the MICs of rifampin and selection frequency; selection frequencies for strains for which rifampin MICs were lower, such as NCTC11637 and CPY3401, were lower, while selection frequencies for strains for which MICs were higher, such as CPY1124 and CPY3281, were higher. Such a correlation was not observed with KRM-1648 or KRM-1657, which have potent antimicrobial activities.
Incorporation of [14C]KRM-1648 into H. pylori.
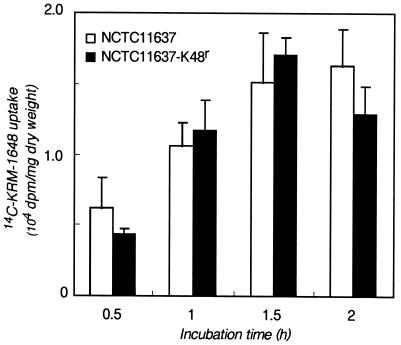
The cross-resistance among rifamycin agents suggested that drug resistance is caused by point mutations in RNA polymerase, as for other bacteria (16). However, previous results suggested the involvement in gram-negative bacteria of a multidrug efflux system that causes an apparent decrease in the level of drug incorporation (9). To see whether H. pylori NCTC11637 and a KRM-1648-resistant clone (NCTC11637-K48r) have such an efflux system, we determined the rate of incorporation of [14C]KRM-1648 (Fig. 4). The radioactivity of KRM-1648 in both the parental and the mutant cells increased in a time-dependent manner, reaching ca. 1,800 to 2,000 dpm/mg (dry weight) during the first 1.5 h. Thus, the rifamycin agent is incorporated irrespective of the resistance phenotype.
FIG. 4.
Uptake of [14C]KRM-1648 by parental type strain H. pylori NCTC11637 (open bars) and a KRM-1648-resistant mutant (solid bars). Each bar indicates the mean ± standard deviation of three determinations.
DISCUSSION
Following the recognition of the important pathogenic role of H. pylori infection in the development of gastroduodenal diseases, there has been a continuous search for improved eradication therapy. In the present study we tested the antimicrobial activities of newly synthesized rifamycin derivatives against 44 strains of H. pylori. The MIC90s of KRM-1657 and KRM-1648 were 0.002 and 0.008 μg/ml, respectively, which were lower than those of amoxicillin (0.063 μg/ml; this study) (14), clarithromycin (14), ciprofloxacin (14), tetracycline (14), tobramycin (14), metronidazole (14), rifampin (4 μg/ml; this study), and rifaximin (15). Time-kill studies revealed the antimicrobial activities of the KRM compounds to be bactericidal, resulting in cell lysis after incubation for 24 h with KRM-1657 at a concentration 0.001 μg/ml.
Several studies demonstrated that amoxicillin treatment of H. pylori can result in the development of coccoid forms (2, 4), as was also found in the present study. It has been postulated that the coccoid forms are a nonculturable, dormant stage of H. pylori and that they play a role in the survival of the bacterium in hostile environments (1, 4, 26). Although the coccoid forms induced by aerobiosis in the presence of a bactericidal antibiotic have been postulated to be a manifestation of cell death (17), under microaerobic conditions the amoxicillin-induced coccoid form might not be such a manifestation. H. pylori is highly susceptible to amoxicillin in vitro, but amoxicillin alone has little curative effect in clinical applications. The possibility remained, therefore, that the coccoid form, if induced in the stomach by amoxicillin, might be viable and even infectious. In contrast to amoxicillin, KRM-1657 and KRM-1648 do not induce a transition from the spiral to the coccoid form. In addition, these agents appear to be stable under acidic conditions, which may help to preserve their antimicrobial activities in the stomach.
KRM-1648 is known to exhibit potent activity in vitro and in vivo against gram-positive bacteria, but it is not generally effective against gram-negative bacteria (10, 25). It is well established that the antimicrobial activity of rifamycin is due to inhibition of microbial DNA-dependent RNA polymerases (9, 16). The mechanisms of action of the antimicrobial activity of KRM-1648 against M. avium and E. coli have been studied previously (9). RNA polymerases from M. avium and E. coli are similarly inhibited, but the level of uptake of [14C]KRM-1648 by M. avium is much greater than that by E. coli, possibly because E. coli has AcrAB, a multidrug efflux system involved in resistance to antibiotics such as β-lactams, tetracycline, and rifampin (19). We isolated spontaneous mutants resistant to rifampin and showed that cross-resistance to KRM-1648 or KRM-1657 but not to clarithromycin or amoxicillin occurred. In addition, radioactive KRM-1648 was taken up by H. pylori NCTC11637 and by a KRM-1648-resistant mutant at similar rates. These results are consistent with the assumption that the antimicrobial activities of these KRM agents are due to their inhibitory actions on RNA polymerase. Genomic sequencing of H. pylori 26695 revealed that the genes encoding the β and β′ subunits of RNA polymerase are fused and that an AcrB orthologue that is involved in the efflux of rifampin in other bacteria is present (27). The extremely high degree of susceptibility of H. pylori to KRM-1657 and KRM-1648 might be related to some unique features of the RNA polymerase and/or to the specificity of the multidrug efflux system.
Taken together, the results presented here indicate that the new rifamycin derivatives KRM-1657 and KRM-1648 are active against H. pylori at very low concentrations. Further studies are needed to determine if these compounds can be used to eradicate H. pylori in vivo.
ACKNOWLEDGMENTS
This work was supported by a Grant-in-Aid for Scientific Research (grant B08457089) from the Ministry of Education, Science, Culture, and Sports of Japan.
We thank T. Fujioka, Oita Medical University, Oita, Japan, for the kind gift of clarithromycin- and/or metronidazole-resistant strains of H. pylori.
REFERENCES
- 1.Benaissa M, Babin P, Quellard N, Pezennec L, Cenatiempo Y, Fauchere J L. Changes in Helicobacter pylori ultrastructure and antigens during conversion from the bacillary to the coccoid form. Infect Immun. 1996;64:2331–2335. doi: 10.1128/iai.64.6.2331-2335.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berry V, Jennings K, Woodnutt G. Bactericidal and morphological effects of amoxicillin on Helicobacter pylori. Antimicrob Agents Chemother. 1995;39:1859–1861. doi: 10.1128/aac.39.8.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blaser M J. Helicobacter pylori: its role in disease. Clin Infect Dis. 1992;15:386–391. doi: 10.1093/clind/15.3.386. [DOI] [PubMed] [Google Scholar]
- 4.Bode G, Mauch F, Malfertheiner P. The coccoid forms of Helicobacter pylori. Criteria for their viability. Epidemiol Infect. 1993;111:483–490. doi: 10.1017/s0950268800057216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carbone M, Fera M T, Cecchetti V, Tabarrini O, Losi E, Cusumano V, Teti G. In vitro activities of new quinolones against Helicobacter pylori. Antimicrob Agents Chemother. 1997;41:2790–2792. doi: 10.1128/aac.41.12.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coudron P E, Stratton C W. Utilization of time-kill kinetics methodologies for assessing the bactericidal activities of ampicillin and bismuth, alone and in combination, against Helicobacter pylori in stationary and logarithmic growth phases. Antimicrob Agents Chemother. 1995;39:66–69. doi: 10.1128/aac.39.1.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coudron P E, Stratton C W. Use of time-kill methodology to assess antimicrobial combinations against metronidazole-susceptible and metronidazole-resistant strains of Helicobacter pylori. Antimicrob Agents Chemother. 1995;39:2641–2644. doi: 10.1128/aac.39.12.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.European Helicobacter pylori Study Group. Current European concepts in the management of Helicobacter pylori infection. Gut. 1997;41:8–13. doi: 10.1136/gut.41.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujii K, Saito H, Tomioka H, Mae T, Hosoe K. Mechanism of action of antimycobacterial activity of the new benzoxazinorifamycin KRM-1648. Antimicrob Agents Chemother. 1995;39:1489–1492. doi: 10.1128/aac.39.7.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujii K, Tsuji A, Miyazaki S, Yamaguchi K, Goto S. In vitro and in vivo antimicrobial activities of KRM-1648 and KRM-1657, new rifamycin derivatives. Antimicrob Agents Chemother. 1994;38:1118–1122. doi: 10.1128/aac.38.5.1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goddard A F, Logan R P H. Antimicrobial resistance and Helicobacter pylori. J Antimicrob Chemother. 1996;37:639–643. doi: 10.1093/jac/37.4.639. [DOI] [PubMed] [Google Scholar]
- 12.Graham D Y. Campylobacter pylori and peptic ulcer disease. Gastroenterology. 1989;96:615–625. doi: 10.1016/s0016-5085(89)80057-5. [DOI] [PubMed] [Google Scholar]
- 13.Graham D Y, de Boer W A, Tytgat G N J. Choosing the best anti-Helicobacter pylori therapy: effect of antimicrobial resistance. Am J Gastroenterol. 1996;91:1072–1076. [PubMed] [Google Scholar]
- 14.Hartzen S H, Andersen L P, Bremmelgaard A, Golding H, Arpi M, Kristiansen J, Justesen T, Espersen F, Frimodt-Møller N, Bonnevie O. Antimicrobial susceptibility testing of 230 Helicobacter pylori strains: importance of medium, inoculum, and incubation time. Antimicrob Agents Chemother. 1997;41:2634–2639. doi: 10.1128/aac.41.12.2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holton J, Vbaira D, Menegatti M, Barbara L. The susceptibility of Helicobacter pylori to the rifamycin, rifaximin. J Antimicrob Chemother. 1995;35:545–549. doi: 10.1093/jac/35.4.545. [DOI] [PubMed] [Google Scholar]
- 16.Jin D J, Gross C A. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampin resistance. J Mol Biol. 1988;202:45–58. doi: 10.1016/0022-2836(88)90517-7. [DOI] [PubMed] [Google Scholar]
- 17.Kusters J G, Gerrits M M, Van Strijp J A G, Vandenbroucke-Grauls C M J E. Coccoid forms of Helicobacter pylori are the morphologic manifestation of cell death. Infect Immun. 1997;65:3672–3679. doi: 10.1128/iai.65.9.3672-3679.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marshall B J, Goodwin C S, Waren J R, Murray R, Blincow E D, Blackbourn S J, Phillips M, Waters T E, Sanderson C R. Prospective double-blind trial of duodenal ulcer relapse after eradication of Campylobacter pylori. Lancet. 1988;ii:1437–1441. doi: 10.1016/s0140-6736(88)90929-4. [DOI] [PubMed] [Google Scholar]
- 19.Nikaido H. Multidrug efflux pumps of gram-negative bacteria. J Bacteriol. 1996;178:5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nomura A, Stemmermann G N, Chyou P H, Kato I, Perez-Perez G I, Blaser M J. Helicobacter pylori infection and gastric carcinoma among Japanese Americans in Hawaii. N Engl J Med. 1991;325:1132–1136. doi: 10.1056/NEJM199110173251604. [DOI] [PubMed] [Google Scholar]
- 21.Parsonnet J, Friedman G D, Vandersteen D P, Chang Y, Vogelman J H, Orentreich N, Sibley R K. Helicobacter pylori infection and the risk of gastric carcinoma. N Engl J Med. 1991;325:1127–1131. doi: 10.1056/NEJM199110173251603. [DOI] [PubMed] [Google Scholar]
- 22.Rautelin H, Seppala K, Renkonen O V, Vainio U, Kosunen T U. Role of metronidazole resistance in therapy of Helicobacter pylori infections. Antimicrob Agents Chemother. 1992;36:163–166. doi: 10.1128/aac.36.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rauws E A J, Tytgat G N J. Cure of duodenal ulcer associated with eradication of Helicobacter pylori. Lancet. 1990;335:1233–1235. doi: 10.1016/0140-6736(90)91301-p. [DOI] [PubMed] [Google Scholar]
- 24.Rene W M, Hulst V D, Keller J J, Rauws E A J, Tytgat G N J. Treatment of Helicobacter pylori infection: a review of the world literature. Helicobacter. 1996;1:6–19. doi: 10.1111/j.1523-5378.1996.tb00003.x. [DOI] [PubMed] [Google Scholar]
- 25.Saito H, Tomioka H, Sato K, Emori M, Yamane T, Yamashita K, Hosoe K, Hidaka T. In vitro antimycrobacterial activities of newly synthesized benzoxazinorifamycins. Antimicrob Agents Chemother. 1991;35:542–547. doi: 10.1128/aac.35.3.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shahamat M, Mai U, Paszko-Kova C, Kessel M, Colwell R R. Use of autoradiography to assess viability of Helicobacter pylori in water. Appl Environ Microbiol. 1993;59:1231–1235. doi: 10.1128/aem.59.4.1231-1235.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomb J-F, White O, Kerlavage A R, Clayton A, Sutton G G, Fleischmann R D, Keychum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenney K, Fitzegerald L M, Lee N, Adams M D, Hickey E K, Berg D E, Gocayne J D, Utterback T R, Peterson J D, Kelley J M, Cotton M D, Weidman J M, Fujii C, Bowman C, Watthey L, Wallin E, Hayes W S, Borodovsky M, Karp P D, Smith H O, Fraser C M, Venter J C. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 28.Yamane T, Hashizume T, Yamashita K, Konishi E, Hosoe K, Hidaka T, Watanabe K, Kawaharada H, Yamamoto T, Kuze F. Synthesis and biological activity of 3′-hydroxy-5′-aminobenzoxanorifamycin derivatives. Chem Pharm Bull. 1993;41:148–155. doi: 10.1248/cpb.41.148. [DOI] [PubMed] [Google Scholar]