Abstract
CCN1 is well studied in terms of its functions in injury repair, cell adhesion survival and apoptosis, bacterial clearance and mediation of inflammation-related pathways, such as the TLR2/4 pathways. However, the role of CCN1 protein and its interaction with TLR2/4 pathways in intestinal epithelial cells was not elucidated after Listeria monocytogenes infection. The results of this study confirm that L. monocytogenes infection induced intestinal inflammation and increased the protein expression of CCN1, TLR2, TLR4 and p38, which followed a similar tendency in the expression of genes related to the TLR2/4 pathways. In addition, organoids infected by L. monocytogenes showed a significant increase in the expression of CCN1 and the activation of TLR2/4 pathways. Furthermore, pre-treatment with CCN1 protein to organoids infected by L. monocytogenes could increase the related genes of TLR2/4 pathways and up-regulate the expression of TNF, and increase the count of pathogens in organoids, which indicates that the interaction between the CCN1 protein and TLR2/4 signaling pathways in intestinal epithelial cells occurred after L. monocytogenes infection. This study will provide a novel insight of the role of CCN1 protein after L. monocytogenes infection in the intestine.
Keywords: Listeria monocytogenes, intestine epithelial cells, CCN1, TLR2/4 pathways
1. Introduction
Listeria monocytogenes is a short Gram-positive flagellar and ubiquitous intracellular bacterium that causes listeriosis in immunocompromised individuals, with a case fatality rate of up to 30% [1]. While L. monocytogenes outbreaks are common and generally attributable to highly contaminated foods, most cases of listeriosis are sporadic.
Upon infection, intestinal epithelial cells, which respond to intruders and their cellular molecules by displaying an inflammatory state, play a crucial role in the immune response to pathogens [2]. Toll-like receptors (TLRs), expressed in the intestinal epithelial cells, are the phylogenetically conserved mediators of innate immunity that can discriminate intestinal microbiota and respond to pathogenic microbes by recognizing pathogen-associated molecular patterns (PAMPs) [3,4]. Intestinal epithelial cells, which constitute a single monolayer of cells found at the mucosal surface, express several TLRs, including TLR2, TLR4, TLR5 and TLR9, with the location of these being restricted to either the apical or the basolateral surface, or both [5]. During L. monocytogenes infection, TLR2 could recognize peptide glycan and lipoproteins [6], and TLR5 and TLR9 could recognize flagellin and bacterial DNA, respectively [7]. Although TLR4 was commonly reported to act as a signaling receptor of lipopolysaccharide (LPS), a Gram-negative bacterial component, in a recent study, TLR4 of macrophages were activated through cellular communication network factor 1 (CCN1), which could recognize peptidoglycan (PGN) of S. aureus [8]. Upon recognizing PAMPs, TLR2 or TLR4 could lead signaling to produce various inflammatory cytokines through the activation of myeloid differentiation factor 88 (MyD88)-dependent and -independent pathways [9]. In the MyD88-dependent pathway, TLR2 or TLR4 could activate IL-1 receptor-associated kinase (IRAK), which then proceeded to activate a complex signaling of TGF-β-activated kinase (TAK1). Lastly, the gene expression of regulatory factors of p38 (the MAPKs family) was activated, so as to induce the production of proinflammatory cytokines and chemokines [10].
CCN1 encodes a 42 kDa matricellular protein that could exhibit diverse and different functions, thereby involving it in distinct and complex processes, such as inflammation and injury repair in the skin, liver and gut [11,12,13]. For instance, a study showed that the CCN1 protein was up-regulated in the intestinal tissue that was subjected to ischemia [14]. In addition, Jun et al. (2020) found that CCN1 protein could activate TLR2 or TLR4 by direct binding to those receptors, leading to MyD88-dependent expression of inflammatory cytokines and chemokines [8], whereas, the role of CCN1 protein and the interaction between the CCN1 protein and TLR2/4 pathways in regulating innate immune responses of intestinal epithelial cells during L. monocytogenes infection is largely unknown.
Investigations of pattern recognition receptors (PRRs) were mostly focused on the immune cell’s response to enteric pathogens [15,16,17]; however, PRRs of the intestinal epithelium also played an important role in innate immunity after bacterial infection [18]. As for the cell models in vitro, compared with other traditional cell models, intestinal organoids were cell spheroids that could differentiate into the various intestinal epithelial cell types, which could express several TLRs, including TLR2, TLR4, TLR5 and TLR9 [5]. The model allowed the investigation of intestinal function and direct interactions with microbes. In addition, researchers had used intestinal organoids to model the natural infection of L. monocytogenes [19], Salmonella [20] and pathogenic Escherichia coli strains [21,22], which provided important insights into pathogenesis in the intestine.
Therefore, this study, based on mice and intestinal organoids as in vivo and in vitro models, respectively, aimed to explore the changes in the CCN1 protein and TLR2/4 signaling pathways of intestinal epithelial cells, as well as their interaction after L. monocytogenes infection, which were expected to provide a novel insight on the role of CCN1 protein after L. monocytogenes infection in the intestine.
2. Results
2.1. The Pathological Changes in Mice after L. monocytogenes Infection
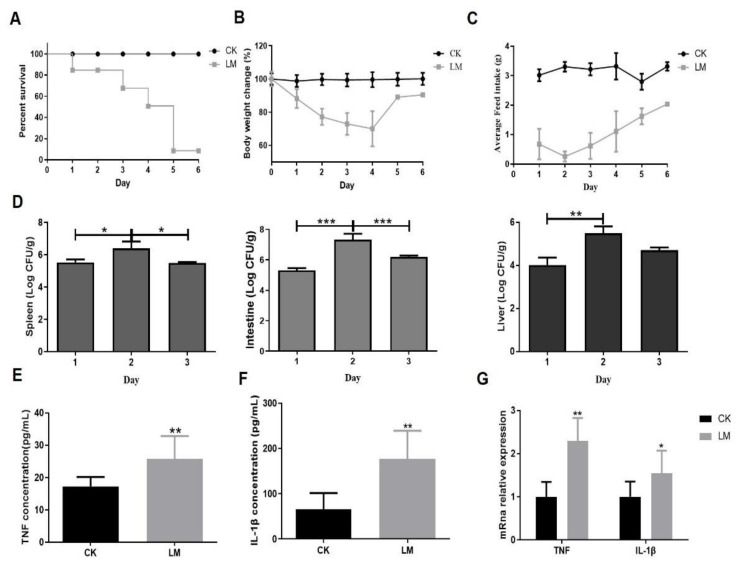
The pathological changes were analyzed in mice gavaged with either PBS (CK) or L. monocytogenes (LM). Compared with the CK group (mice or organoids without infection, CK group), mice infected with L. monocytogenes by oral gavage died starting from day one, and the survival rate dropped sharply at 5 days (Figure 1A). Moreover, the body weight and feed intake of mice were much lower than the control group after infection (Figure 1B,C). The colonization of L. monocytogenes in the spleen, intestine and liver reached a peak on day two (Figure 1D). Figure 1E,F show that the concentrations of IL-1β and TNF in jejunum homogenate were significantly higher than that in the CK group, which was in agreement with the results of mRNA expression levels of IL-1β and TNF in the jejunum (Figure 1G). These results confirm that infection of this L. monocytogenes strain could lead to serious pathological changes in mice.
Figure 1.
The pathological changes in mice after L. monocytogenes infection. (A) Time course of survival rate of mice. (B) Time course of body weight change in mice. (C) Time course of average feed intake of mice. (D) L. monocytogenes burden in the spleen, intestine and liver. (E,F) The concentration of TNF and IL-1β in jejunum homogenate of mice. (G) mRNA levels of TNF and IL-1β from homogenized jejunum samples. * p < 0.05, ** p < 0.01, *** p < 0.001.
2.2. The Changes in CCN1 Protein and TLR2/4 Signaling Pathways Response to L. monocytogenes Infection in Mice
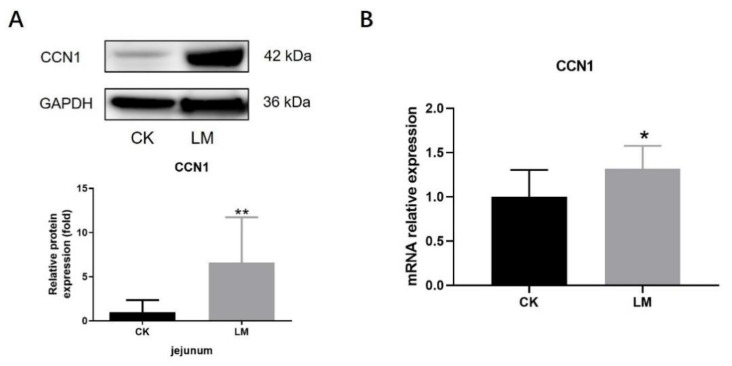
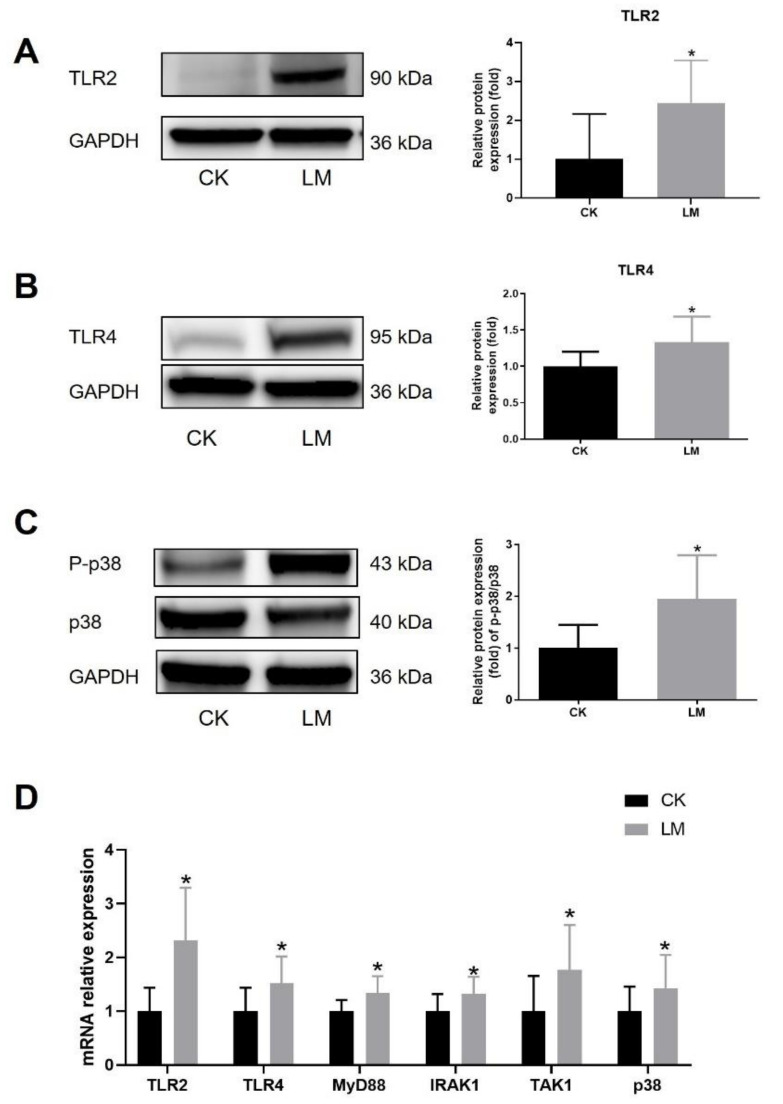
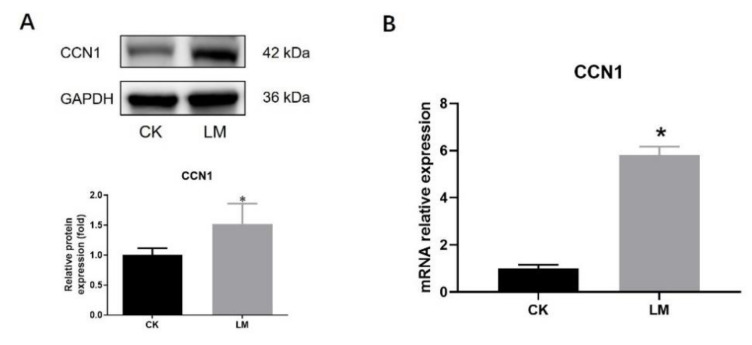
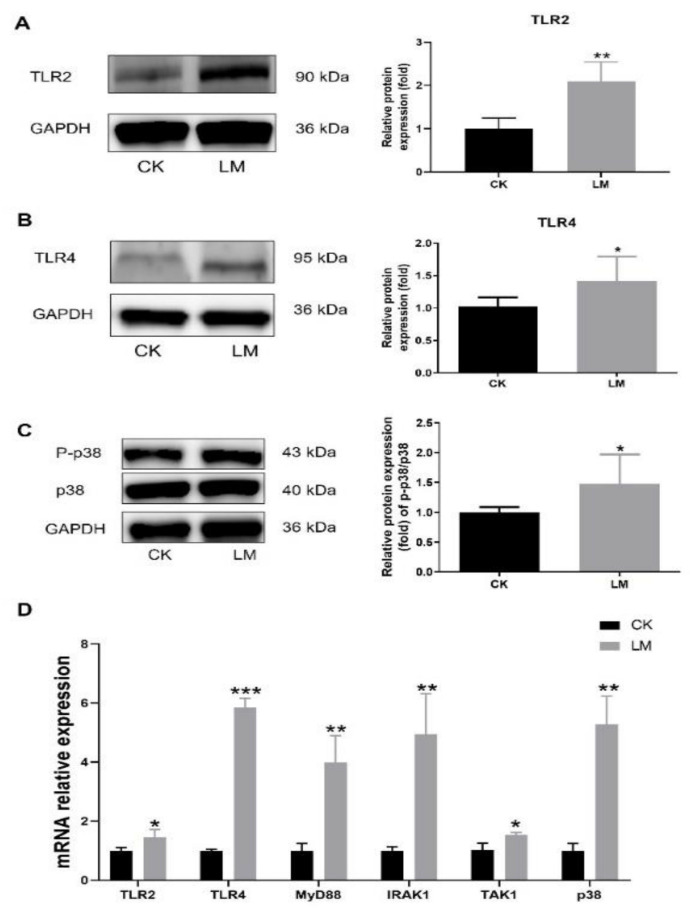
In order to determine the changes in CCN1 protein and TLR2/4 signaling pathways, the jejuna of mice, gavaged with either PBS (CK) or L. monocytogenes (LM), were collected. After L. monocytogenes infection, the protein expression of CCN1 had a 6.63-fold increase (p < 0.05) (Figure 2A), and the mRNA relative expression of CCN1 also increased significantly (Figure 2B), which indicated that L. monocytogenes infection stimulated the expression increase in CCN1 in the jejuna of mice. Under this circumstance, Western blot analysis showed that the protein expression of TLR2 and TLR4, which could be activated through CCN1 protein in the macrophages, increased significantly in the LM group (Figure 3A,B), and the protein expression of P-p38/p38 that was downstream of the TLR2/4 signaling pathways increased 1.94-fold in the LM group (Figure 3C). In the meantime, the mRNA expression levels of genes related to the TLR2/4 signaling pathway (TLR2, TLR4, MyD88, IRAK1, TAK1 and p38) were significantly increased after L. monocytogenes infection (Figure 3D). These results indicate that L. monocytogenes infection increased the expression of CCN1, which followed a similar tendency in the expression of the TLR2/4 signaling pathways.
Figure 2.
The changes in CCN1 response to L. monocytogenes infection in mice. (A) Western blot of CCN1 in the jejunum. (B) mRNA levels of CCN1 in the jejunum. * p < 0.05, ** p < 0.01.
Figure 3.
The increase in components in TLR2/4 signaling pathways upon L. monocytogenes infection in mice. (A–C) Western blot of TLR2, TLR4 and P-p38/p38 in the jejunum. (D) mRNA levels of TLR2, TLR4, MyD88, IRAK1, TAK1 and p38 from homogenized jejunum samples. * p < 0.05.
2.3. The Damage of Small Intestinal Organoids after L. monocytogenes Infection
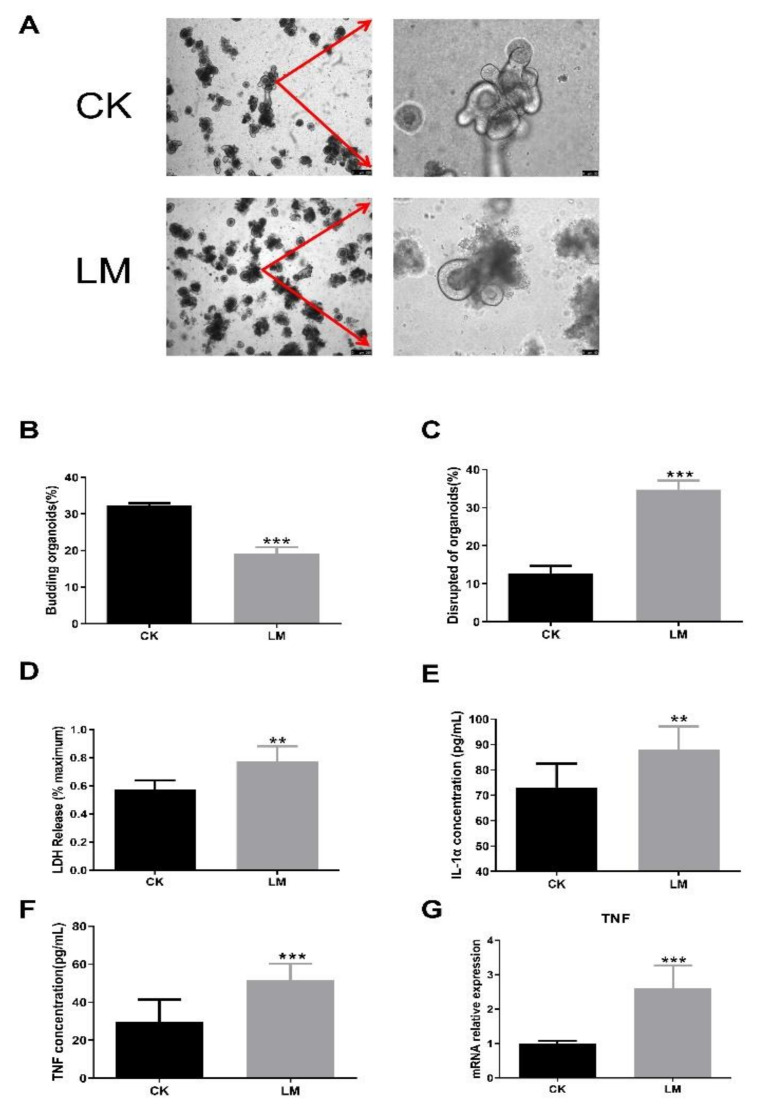
Intestinal organoids, treated with culture medium (CK) or L. monocytogenes (LM), were used to verify the effect of L. monocytogenes infection on the intestinal epithelial cells, which was also an important line against infection except for immune cells in the intestine. Figure 4A shows that organoids infected by L. monocytogenes were partially lysed at 18 h compared with the normal budding in the control group, and the budding rate of organoids was significantly decreased, and the mortality of organoids was significantly increased, in the LM group (Figure 4B,C). Moreover, LDH (lactate dehydrogenase) release in the supernatant was increased significantly in the LM group, which indicated the damage to the cell membrane of small intestinal organoids (Figure 4D). As shown in Figure 4E,F, L. monocytogenes infection induced a significant increase in the protein expression levels of TNF and IL-1β at 18 h of co-culture, and the mRNA expression levels of TNF followed a similar significant increase to the protein expression (Figure 4G). These results reflect that L. monocytogenes infection had significant damage to the organoids.
Figure 4.
The damage of small intestinal organoids after L. monocytogenes infection. (A) The morphological changes in organoids after L. monocytogenes infection. (B,C) The budding percentage, and mortality, of organoids after L. monocytogenes infection. (D) LDH release in the supernatant. (E,F) The concentration of TNF and IL-1β in the supernatant. (G) mRNA levels of TNF from organoids samples. ** p < 0.01, *** p < 0.001.
2.4. The Changes in CCN1 Protein and TLR2/4 Signaling Pathways Response to L. monocytogenes Infection in Organoids
Intestinal organoids were used to further confirm the changes in the CCN1 protein and TLR2/4 signaling pathways of intestinal epithelial cells in response to L. monocytogenes infection. Obviously, the protein and mRNA expression levels of CCN1 were increased significantly in the LM group (Figure 5A,B). Furthermore, compared with the control group, the protein expression of TLR2, TLR4 and P-p38/p38 also showed a significant increase (Figure 6A–C), and the mRNA expression levels of TLR2/4-pathway-related genes (TLR2, TLR4, MyD88, IRAK1, TAK1, and p38) increased significantly (Figure 6D). Overall, the results of organoids confirmed those of the mice findings, which indicated that the expression of CCN1 was increased and the TLR2/4 signaling pathway was activated after L. monocytogenes infection in the intestinal epithelial cells.
Figure 5.
The changes in CCN1 protein response to L. monocytogenes infection in organoids. (A) Western blot of CCN1 in organoids. (B) mRNA levels of CCN1 from organoids samples. * p < 0.05.
Figure 6.
The increase in components in TLR2/4 signaling pathways upon L. monocytogenes infection in organoids. (A–C) Western blot of TLR2, TLR4 and P-p38/p38 in organoids. (D) mRNA levels of TLR2, TLR4, MyD88, IRAK1, TAK1 and p38 from organoids samples. * p < 0.05, ** p < 0.01, *** p < 0.001.
2.5. The Changes in TLR2/4 Signaling Pathways Response to L. monocytogenes Infection in Organoids Pre-Treated with CCN1 Protein
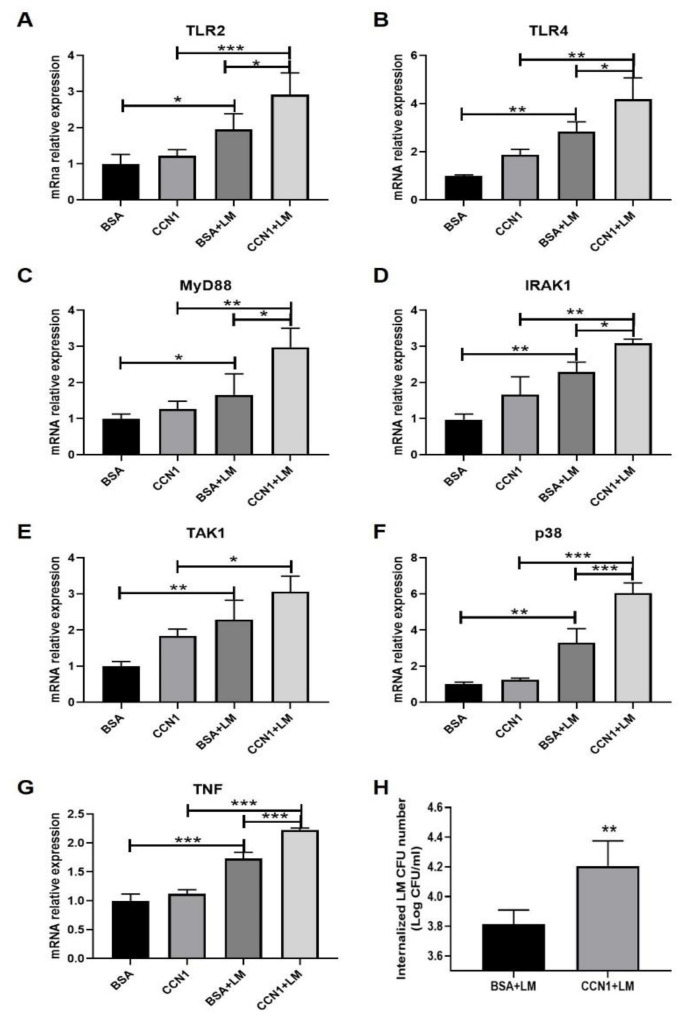
The results show that the pre-treatment of CCN1 protein significantly increased the mRNA expression levels of TLR2/4-pathway-related genes (TLR2, TLR4, MyD88, IRAK1, TAK1 and p38) (Figure 7A–F), which could lead to the increase in the mRNA expression level of TNF after L. monocytogenes infection (Figure 7G). Furthermore, it was found that the pre-treatment of CCN1 protein could increase the counts of L. monocytogenes in intestinal organoids (Figure 7H). These results illustrate that pre-treatment of CCN1 protein could increase the expression of the TLR2/4 signaling pathways of intestinal epithelial cells and the susceptibility of intestinal organoids to L. monocytogenes infection.
Figure 7.
The changes in TLR2/4 signaling pathways in response to L. monocytogenes infection in organoids pre-treated with CCN1 protein. (A–G) mRNA levels of TLR2, TLR4, MyD88, IRAK1, TAK1 and p38 from organoids samples. (H) The number of L. monocytogenes in the organoids. * p < 0.05, ** p < 0.01, *** p < 0.001.
3. Discussion
The innate immunity of intestinal epithelial cells plays an important part in defending against many enteric bacterial pathogens [23]. The results of this study show a significant up-regulated response in TNF and IL-1β expression levels caused by L. monocytogenes infection in mice and organoids, which indicates that L. monocytogenes caused the inflammation of intestinal epithelial cells. This corroborated the studies of He et al., suggesting that L. monocytogenes infection could cause the increased production of TNF and IL-1β in intestinal cells [24].
CCN1 protein has been found to exhibit diverse functions, including cell adhesion survival and apoptosis, and plays an important role in inflammation and tissue repair [8,25,26,27]. In this study, it was first found that L. monocytogenes infection could increase the mRNA and protein expression of CCN1 in mice and intestinal organoids. However, as for other pathogen infections, it was found that S. aureus and its supernatants could induce the expression of CCN1 in epithelial cells [28], while the expression of CCN1 was inhibited by Salmonella enterica [29]. Furthermore, a recent reference reported that CCN1 of macrophages, functioning as a pattern recognition receptor (PRR), opsonized Gram-positive and -negative bacteria through binding peptide glycan (PGN) and lipopolysaccharide (LPS), respectively [8]. Therefore, after L. monocytogenes infection in vivo and in vitro, the PGN of bacteria and the damage of intestines may be the reasons for the increase in CCN1 expression in intestinal epithelial cells.
The stimulation of TLRs activates at least two major downstream signaling pathways, nuclear factor (NF)-κB and mitogen-activated protein kinases (MAPKs), to induce inflammation after pathogen infection [30]. This study showed that L. monocytogenes infection caused the increase in TLR2 expression in mice and organoids, which was consistent with the previous studies [15,31,32]. In addition, to our surprise, it was found that the mRNA and protein expression levels of TLR4, in vivo and in vitro, were increased after L. monocytogenes infection. In the literature, it was generally acknowledged that TLR4 did not recognize any components of L. monocytogenes based on the data generated from TLR4-deficient mice and in vitro activation assays using TLR4 transfected human embryonic kidney (HEK) 293 cells [33,34]. However, a study demonstrated that the TLR4 signaling pathway was activated in Pregnane X Receptor (PXR) deficient mice after L. monocytogenes infection, and the inflammation induced by TLR4 signaling was detrimental to host defense against the infection [35]. Our study found that the related genes of the MyD88-dependent signaling pathway (MyD88, IRKA1 and TAK1), which could be activated by TLR2 or TLR4, were increased significantly in intestinal epithelial cells infected by L. monocytogenes. This phenomenon was consistent with the previous report of L. monocytogenes infection [36,37]. In addition, the ratio of P-p38/p38 in protein level and the p38 in mRNA level was also up-regulated in mice and intestinal organoids, which indicates that the downstream MAPK pathway had been activated to induce inflammation after L. monocytogenes infection [38].
CCN1 can, by itself, induce inflammation through physical interaction with TLR2 or TLR4 to activate MyD88-dependent signaling in mice and macrophages [8]. Based on the results of the increase in the expression of CCN1 and the activation of TLR2/4 signaling pathways in vivo and in vitro, we speculated that the interaction between CCN1 protein and TLR2/4 signaling pathways occurred after L. monocytogenes infection. To verify this hypothesis, intestinal organoids were pre-treated with or without CCN1 protein prior to infection, and the results show that pre-treatment with CCN1 protein increased the expression of the related genes in the TLR2/4 signaling pathways, which followed a similar pattern to the mRNA expression level of TNF after infection. Moreover, the pre-treatment of CCN1 protein could increase the counts of L. monocytogenes in intestinal organoids.
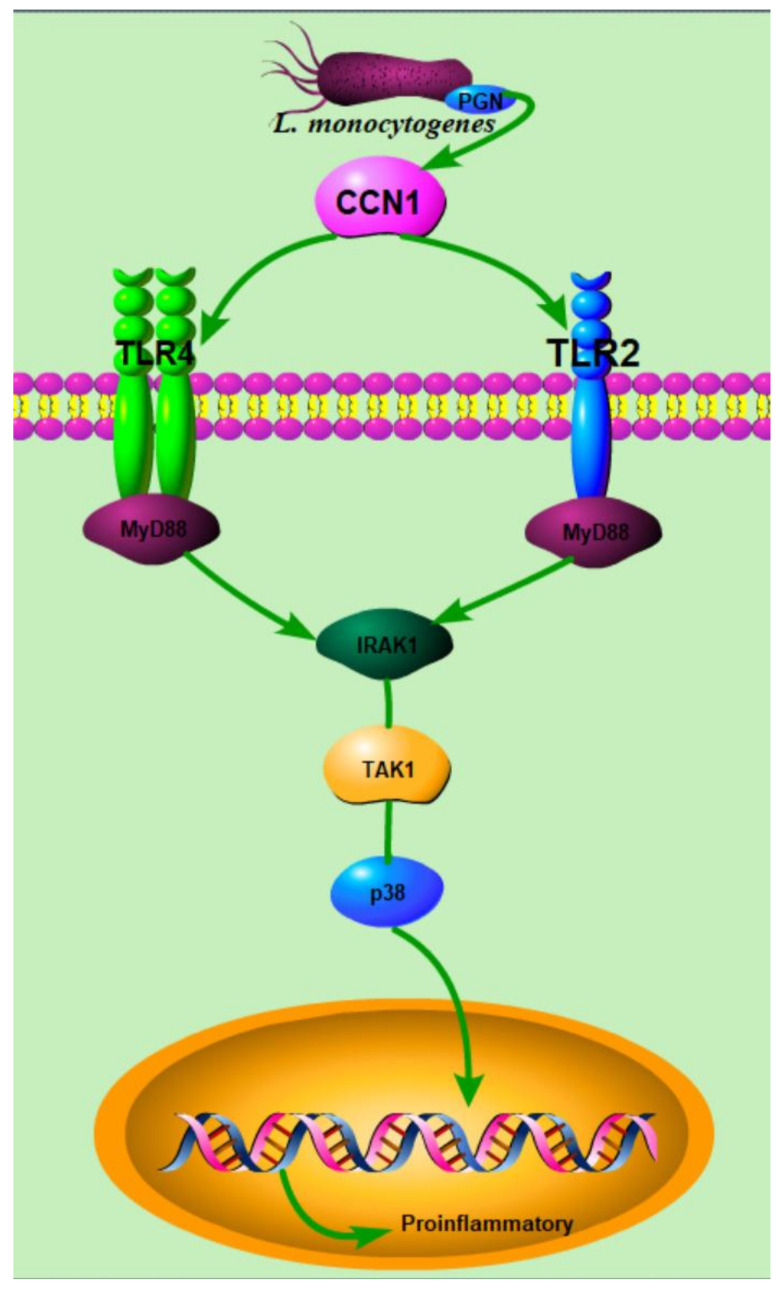
In summary, this study, using mice and intestinal organoids, indicates that L. monocytogenes infection caused the pathological changes in mice and the disruption of organoids, and induced the inflammation of the intestinal epithelial cells. Furthermore, L. monocytogenes infection caused the increased expression of CCN1 and the activation of TLR2/4 signaling pathways, and the interaction between these occurred after infection, which may be the reason for the increase in pro-inflammatory cytokines secretion (Figure 8). Moreover, the pre-treatment of CCN1 protein could increase the susceptibility of intestinal organoids to L. monocytogenes. Further studies can be focused on CCN1-deficient mice to confirm the role of CCN1 and the specific mode of interaction between CCN1 and TLR2/4 signaling pathways after infection, which may prompt novel therapies for pathogen infections.
Figure 8.
The conjectured mechanism of the intestinal epithelial cell’s response to L. monocytogenes infection.
4. Materials and Methods
4.1. Bacterial Strain Culture
The challenge organism for this experiment was L. monocytogenes 10403s. It was grown with agitation overnight at 37 °C in Brain Heart Infusion (BHI) broth, and supplemented with 5 μg mL−1 erythromycin.
4.2. Animals and Intestinal Organoids
4.2.1. Ethics Statement
This study was carried out in accordance with the National Institutes of Health guidelines for the performance of animal experiments. All operations related to animal experiments were examined and approved by the Nanjing Agriculture University Committee on Animal Resources Committee (Approval ID: NJAU.No20210305009).
4.2.2. Animals
Four-week-old C57BL/6 mice (specific-pathogen-free (SPF) female) were purchased from the Animal Research Centre of Yang Zhou University (production license number: SCXK(SU)2017-0007). Three independent animal experiments were carried out in this study. For the first animal experiment, 22 mice were randomly divided into two groups, which were orally administered sterile PBS (200 μL, control group, CK, n = 10) and L. monocytogenes 10403s (200 μL, 109 CFU/mL, LM, n = 12). Then, the survival, average feed intake and average body weight of mice were recorded every day. For the second animal experiment, 15 mice were orally administered L. monocytogenes 10403s (200 μL, 109 CFU/mL) for determining the number of bacteria in the spleen, liver and jejunum after 24, 48, and 72 h of infection (3 survivors were used in each time point). For the third animal experiment, mice were randomly divided into two groups, which were orally administrated sterile PBS (200 μL, control group, CK, n = 10) and L. monocytogenes 10403s (200 μL, 109 CFU/mL, LM, n = 20). They were sacrificed on day 4, and jejunum tissue samples and contents were collected for further analysis.
4.2.3. Intestinal Organoids
Isolation and Culture of Intestinal Organoids
The small intestine of 4-week-old C57BL/6 wild-type mice was cut into small pieces and washed several times with cold PBS. The pieces were then incubated with Gentle Cell Dissociation Reagent (Stem Cell, Vancouver, Canada) for 15 min at 20 °C, and crypts were detached from basal membrane by vigorous shaking. After incubation, crypts enriched in the supernatant were passed through a 70 μm strainer and centrifuged at 300× g for 5 min at 4 °C. The cells were counted, and then, about 200 crypts in each well were resuspended by 25 μL Matrigel (Corning, NY, USA) and 25 μL IntestiCultTM OGM Mouse Basal Medium (Stem cell, Vancouver, Canada), and then plated in 24-well plates. After polymerizing at 37 °C for 20 min, the culture medium supplemented with penicillin–streptomycin (100 U/mL) was added into the gel in each well. The medium was changed every 2–3 days.
L. monocytogenes Infection of Organoid Cells
The methods of organoid infection were referenced by Huang et al. [19], with some modifications. Briefly, after removing the Matrigel with cold PBS, organoids of each well were exposed to the culture medium with L. monocytogenes (100 μL, 108 CFU/mL) for 1 h. Subsequently, the organoids were reseeded with Matrigel and cultured with medium containing gentamicin (100 μg/mL, Gbico) for 18 h. Finally, in order to evaluate infection damage, total organoid numbers, budding organoids and dead organoid numbers per well were counted under a light microscope (Leica, Wetzlar, Germany).
To verify the interaction between CCN1 protein and TLR2/4 signaling pathways, organoids of each well were pre-treated with 100 μL CCN1 protein (4 μg/mL, CUSABIO) for 1 h (marked as CCN1), and organoids of each well were pre-treated with 100 μL BSA (4 μg/mL) as the control group (marked as BSA). Then, the intestinal organoids of each well were infected by L. monocytogenes (100 μL, 108 CFU/mL) for 1 h (BSA + LM: organoids pre-treated with BSA for 1 h prior to infection; CCN1 + LM: organoids pre-treated with CCN1 protein for 1 h prior to infection). Subsequently, the organoids were reseeded with Matrigel and cultured with medium containing gentamicin (100 μg/mL, Gbico) for 18 h.
4.3. The Number of L. monocytogenes in Target Organs and Organoids
After 24, 48 and 72 h of infection, the jejuna, spleens and livers were weighed and grounded with a 200-mesh sieve, respectively. Then, the tissue homogenate was resuspended with 1 mL sterile water for 30 min at room temperature. After performing 1:10 serial dilutions, 1 mL suspension of each dilution was inoculated onto BHI agar plates supplemented with 5 μg/mL erythromycin and incubated for 48 h at 37 °C to determine the count of the L. monocytogenes (Log CFU/g).
To assess the total number of L. monocytogenes infecting organoids with or without CCN1 pre-treatment, the organoids were resuspended with 1 mL sterile water for 30 min at room temperature, and the serial dilution and incubation of L. monocytogenes was described above. After incubation, the CFU/mL was counted.
4.4. ELISA
After L. monocytogenes infection, the production of TNF and IL-1β in jejunum contents and organoid culture supernatants were analyzed using Mouse TNF ELISA kits (NEBIOSCIENC, China) and Mouse IL-1β ELISA kits (NEBIOSCIENC, China) according to the manufacturer’s protocols, respectively.
4.5. LDH Release Assay
After the organoids were co-cultured with L. monocytogenes for 18 h, the LDH in cell culture supernatants was collected and detected by LDH Cytotoxicity Assay Kit according to the manufacturer’s protocols.
4.6. Real-Time Quantitative PCR
Total RNA from the jejunum and organoid samples was extracted with TRIzol (Ambion, Austin, TX, USA) and quantified by spectrophotometry (NanoDrop ND1000, USA), after which reverse transcription PCR was performed. Then, 1 μL of template cDNA was reacted with a master mix in a final volume of 10 μL. The qPCR thermal cycling conditions were as follows: 30 s at 95 °C, followed by 40 cycles of 10 s at 95 °C and 30 s at 60 °C. Primers for specific genes are listed in Table 1.
Table 1.
Primer sequences used for RT-qPCR.
| Target Genes | Primer Sense (5′-3′) | Primer Antisense (5′-3′) |
|---|---|---|
| CCN1 | CTGCGCTAAACAACTCAACGA | GCAGATCCCTTTCAGAGCGG |
| TLR2 | GGACATCCCCTTCCCTCACTTC | ACGGGCAGTGGTGAAAACT |
| TLR4 | TTCAGAGCCGTTGGTGTATC | CCCATTCCAGGTAGGTGTTT |
| MyD88 | CCTGCGGTTCATCACTAT | GGCTCCGCATCAGTCT |
| IRAK1 | CCACCCTGGGTTATGTGCC | GAGGATGTGAACGAGGTCAGC |
| TAK1 | ATGTTTGTCGTGCCTTTCTCT | AAGGGTTTCCGGCGTGTTAT |
| p38 | CAGAAACTGACGGACGACCA | CAGCTCGGCCATAATGCAAC |
| TNF | TACTGAACTTCGGGGTGATTGGTCC | CAGCCTTGTCCCTTGAAGAGAACC |
| IL-1β | GCTTGGTGATGTCTGGTCCA | AACACGCAGGACAGGTACAG |
| GAPDH | ATGGTGAAGGTCGGTGTGAA | TGGAAGATGGTGATGGGCTT |
4.7. Western Blot
Tissue samples and organoids were lysed in RIPA buffer containing a protease and phosphatase inhibitor cocktail. Protein concentrations of the lysed samples were detected using a bicinchoninic acid (BCA) assay kit (Thermo Scientific, Waltham, MA, USA), after which samples containing 5× load buffer were heated at 95 °C for 5 min. Equal amounts of protein in various samples were separated by 4–20% SDS-PAGE and transferred to PVDF membranes (BIO-RAD, Hercules, CA, USA). After that, the membranes were blocked with 5% non-fat milk in TBS with 0.1% Tween-20 for 1 h and then incubated with rabbit anti-GAPDH (abcam, Cambridge, UK, RRID: AB_2107448), rabbit anti-CCN1(1:1000, Affinity, Cincinnati, OH, USA, RRID: AB_2838216), rabbit anti-TLR2 (1:1000, Affinity, Cincinnati, OH, USA, RRID: AB_2838958), rabbit anti-TLR4 (1:1000, abcam, Cambridge, UK, RRID: AB_300696), rabbit anti-p38 (1:1000, CST, Beverly, MA, USA, RRID: AB_10999090) and rabbit p-p38 (1:1000, CST, Beverly, MA, USA, RRID: AB_331641) overnight. After washing, goat anti-rabbit secondary antibodies (Bioss, 1:1000) were used to incubate the membranes for 2 h. Finally, the optical protein bands were developed using the efficient chemiluminescence (ECL) kit, and light emission was captured using the Versa DOC 4000 imaging system.
4.8. Statistical Analysis
The results are expressed as the means ± SD. A t-test was employed to determine the significant differences between two groups, and one-way ANOVA was employed to determine the significant differences among multiple groups. The significance levels are shown as * p < 0.05, ** p < 0.01 and *** p < 0.001. Data were combined from at least three independent experiments unless otherwise stated.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (32172267) and Program for Student Innovation through Research and Training (202110307047).
Author Contributions
Conceptualization, C.Z. and K.Y.; Data curation, C.Z., Y.Z. (Yafang Zou), Y.Z. (Yuanyuan Zhang), S.T. and K.Y.; Formal analysis, C.Z. and K.Y.; Investigation, C.Z. and K.Y.; Methodology, C.Z., Y.Z. (Yafang Zou) and S.T.; Project administration, K.Y.; Software, C.Z.; Supervision, K.Y.; Validation, C.Z.; Writing—original draft, C.Z., Y.Z. (Yafang Zou), Y.Z. (Yuanyuan Zhang), S.T. and K.Y.; Writing—review and editing, K.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China, grant number 32172267 and Program for Student Innovation through Research and Training, grant number 202110307047.
Institutional Review Board Statement
The animal study protocol was approved by the Institutional Review Board of Nanjing Agriculture University Committee on Animal Resources Committee, and “National Institutes of Health guidelines for the performance of animal experiments” for studies involving animals.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.De Noordhout C.M., Devleesschauwer B., Angulo F.J., Verbeke G., Haagsma J., Kirk M., Havelaar A., Speybroeck N. The global burden of listeriosis: A systematic review and meta-analysis. Lancet Infect. Dis. 2014;14:1073–1082. doi: 10.1016/S1473-3099(14)70870-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shimazu T., Villena J., Tohno M., Fujie H., Hosoya S., Shimosato T., Aso H., Suda Y., Kawai Y., Saito T., et al. Immunobiotic Lactobacillus jensenii Elicits Anti-Inflammatory Activity in Porcine Intestinal Epithelial Cells by Modulating Negative Regulators of the Toll-Like Receptor Signaling Pathway. Infect. Immun. 2012;80:276–288. doi: 10.1128/IAI.05729-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hosoya S., Villena J., Shimazu T., Tohno M., Fujie H., Chiba E., Shimosato T., Aso H., Suda Y., Kawai Y., et al. Immunobiotic lactic acid bacteria beneficially regulate immune response triggered by poly(I:C) in porcine intestinal epithelial cells. Vet. Res. 2011;42:111. doi: 10.1186/1297-9716-42-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tomosada Y., Villena J., Murata K., Chiba E., Shimazu T., Aso H., Iwabuchi N., Xiao J., Saito T., Kitazawa H. Immunoregulatory Effect of Bifidobacteria Strains in Porcine Intestinal Epithelial Cells through Modulation of Ubiquitin-Editing Enzyme A20 Expression. PLoS ONE. 2013;8:e59259. doi: 10.1371/journal.pone.0059259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harris G., KuoLee R., Chen W.X. Role of Toll-like receptors in health and diseases of gastrointestinal tract. World J. Gastroenterol. 2006;12:2149–2160. doi: 10.3748/wjg.v12.i14.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seki E., Tsutsui H., Tsuji N.M., Hayashi N., Adachi K., Nakano H., Futatsugi-Yumikura S., Takeuchi O., Hoshino K., Akira S., et al. Critical roles of myeloid differentiation factor 88-dependent proinflammatory cytokine release in early phase clearance of Listeria monocytogenes in mice. J. Immunol. 2002;169:3863–3868. doi: 10.4049/jimmunol.169.7.3863. [DOI] [PubMed] [Google Scholar]
- 7.D’Orazio S.E.F. Innate and Adaptive Immune Responses during Listeria monocytogenes Infection. Microbiol. Spectr. 2019;7:3–7. doi: 10.1128/microbiolspec.GPP3-0065-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jun J.I., Lau L.F. CCN1 is an opsonin for bacterial clearance and a direct activator of Toll-like receptor signaling. Nat. Commun. 2020;11:1242. doi: 10.1038/s41467-020-15075-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zughaler S.M., Zimmer S.M., Datta A., Carlson R.W., Stephens D.S. Differential induction of the toll-like receptor 4-MyD88-dependent and -independent signaling pathways by endotoxins (vol 73, pg 2940, 2005) Infect. Immun. 2006;74:3077. doi: 10.1128/IAI.74.5.3077.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McClure R., Massari P. TLR-dependent human mucosal epithelial cell responses to microbial pathogens. Front. Immunol. 2014;5:386. doi: 10.3389/fimmu.2014.00386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi J.S., Kim K.H., Lau L.F. The matricellular protein CCN1 promotes mucosal healing in murine colitis through IL-6. Mucosal. Immunol. 2015;8:1285–1296. doi: 10.1038/mi.2015.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jun J.I., Kim K.H., Lau L.F. The matricellular protein CCN1 mediates neutrophil efferocytosis in cutaneous wound healing. Nat. Commun. 2015;6:7386. doi: 10.1038/ncomms8386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim K.H., Chen C.C., Alpini G., Lau L.F. CCN1 induces hepatic ductular reaction through integrin alpha(v)beta(5)-mediated activation of NF-kappa B. J. Clin. Investig. 2015;125:1886–1900. doi: 10.1172/JCI79327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shegarfi H., Krohn C.D., Gundersen Y., Kjeldsen S.F., Hviid C.V.B., Ruud T.E., Aasen A.O. Regulation of CCN1 (Cyr61) in a porcine model of intestinal ischemia/reperfusion. Innate Immun. 2015;21:453–462. doi: 10.1177/1753425915569089. [DOI] [PubMed] [Google Scholar]
- 15.Shihab P.K., Al-Roub A., Al-Ghanim M., Al-Mass A., Behbehani K., Ahmad R. TLR2 and AP-1/NF-kappaB are involved in the regulation of MMP-9 elicited by heat killed Listeria monocytogenes in human monocytic THP-1 cells. J. Inflamm. 2015;12:32. doi: 10.1186/s12950-015-0077-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilharm A., Brigas H.C., Sandrock I., Ribeiro M., Amado T., Reinhardt A., Demera A., Hoenicke L., Strowig T., Carvalho T., et al. Microbiota-dependent expansion of testicular IL-17-producing Vγ6+ γδ T cells upon puberty promotes local tissue immune surveillance. Mucosal Immunol. 2020;14:242–252. doi: 10.1038/s41385-020-0330-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fernandes-Alnemri T., Kang S., Anderson C., Sagara J., Fitzgerald† K., Alnemri E.S. Toll-Like Receptor Signaling Licenses IRAK1 for Rapid Activation of The NLRP3 Inflammasome1. J. Immunol. 2013;191:3995–3999. doi: 10.4049/jimmunol.1301681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Co J.Y., Margalef-Catala M., Li X., Mah A.T., Kuo C.J., Monack D.M., Amieva M.R. Controlling Epithelial Polarity: A Human Enteroid Model for Host-Pathogen Interactions. Cell Rep. 2019;26:2509–2520.e4. doi: 10.1016/j.celrep.2019.01.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang J., Zhou C., Zhou G., Li H., Ye K. Effect of Listeria monocytogenes on intestinal stem cells in the co-culture model of small intestinal organoids. Microb. Pathog. 2021;153:104776. doi: 10.1016/j.micpath.2021.104776. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y.G., Wu S., Xia Y., Sun J. Salmonella-infected crypt-derived intestinal organoid culture system for host–bacterial interactions. Physiol. Rep. 2014;2:e12147. doi: 10.14814/phy2.12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vandussen K.L., Marinshaw J.M., Shaikh N., Miyoshi H., Stappenbeck T.S. Development of an enhanced human gastrointestinal epithelial culture system to facilitate patient-based assays. Gut. 2014;64:911–920. doi: 10.1136/gutjnl-2013-306651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rajan A., Vela L., Zeng X.L., Yu X., Shroyer N., Blutt S.E., Poole N.M., Carlin L.G., Nataro J.P., Estes M.K., et al. Novel Segment- and Host-Specific Patterns of Enteroaggregative Escherichia coli Adherence to Human Intestinal Enteroids. MBio. 2018;9:e02419-17. doi: 10.1128/mBio.02419-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garner C.D., Antonopoulos D.A., Wagner B., Duhamel G.E., Keresztes I., Ross D.A., Young V.B., Altier C. Perturbation of the Small Intestine Microbial Ecology by Streptomycin Alters Pathology in a Salmonella enterica Serovar Typhimurium Murine Model of Infection. Infect. Immun. 2009;77:2691–2702. doi: 10.1128/IAI.01570-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He Y., Yang Q., Tian L.L., Zhang Z.H., Qiu L., Tao X.Y., Wei H. Protection of surface layer protein from Enterococcus faecium WEFA23 against Listeria monocytogenes CMCC54007 infection by modulating intestinal permeability and immunity. Appl. Microbiol. Biotechnol. 2021;105:4269–4284. doi: 10.1007/s00253-021-11240-y. [DOI] [PubMed] [Google Scholar]
- 25.Lau L.F. CCN1/CYR61: The very model of a modern matricellular protein. Cell. Mol. Life Sci. 2011;68:3149–3163. doi: 10.1007/s00018-011-0778-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moon H.G., Qin Z., Quan T., Xie L., Dela Cruz C.S., Jin Y. Matrix protein CCN1 induced by bacterial DNA and CpG ODN limits lung inflammation and contributes to innate immune homeostasis. Mucosal Immunol. 2015;8:243–253. doi: 10.1038/mi.2014.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao X., Ding E.Y., Yu O.M., Xiang S.Y., Tan-Sah V.P., Yung B.S., Hedgpeth J., Neubig R.R., Lau L.F., Brown J.H., et al. Induction of the matricellular protein CCN1 through RhoA and MRTF-A contributes to ischemic cardioprotection. J. Mol. Cell. Cardiol. 2014;75:152–161. doi: 10.1016/j.yjmcc.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiedmaier N., Wuller S., Koberle M., Manncke B., Krejci J., Autenrieth I.B., Bohn E. Bacteria induce CTGF and CYR61 expression in epithelial cells in a lysophosphatidic acid receptor-dependent manner. Int. J. Med. Microbiol. 2008;298:231–243. doi: 10.1016/j.ijmm.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 29.Higgins S.E., Wolfenden A.D., Tellez G., Hargis B.M., Porter T.E. Transcriptional profiling of cecal gene expression in probiotic- and Salmonella-challenged neonatal chicks. Poult. Sci. 2011;90:901–913. doi: 10.3382/ps.2010-00907. [DOI] [PubMed] [Google Scholar]
- 30.Smale S.T. Selective Transcription in Response to an Inflammatory Stimulus. Cell. 2010;140:833–844. doi: 10.1016/j.cell.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang B., Zhao J., Shen S.Q., Li H.X., He K.L., Shen G.X., Mayer L., UnkelesS J., Li D., Yuan Y., et al. Listeria monocytogenes promotes tumor growth via tumor cell toll-like receptor 2 signaling. Cancer Res. 2007;67:4346–4352. doi: 10.1158/0008-5472.CAN-06-4067. [DOI] [PubMed] [Google Scholar]
- 32.Chen S.T., Li F.J., Hsu T.Y., Liang S.M., Yeh Y.C., Liao W.Y., Chou T.Y., Chen N.J., Hsiao M., Yang W.B., et al. CLEC5A is a critical receptor in innate immunity against Listeria infection. Nat. Commun. 2017;8:299. doi: 10.1038/s41467-017-00356-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Janot L., Secher T., Torres D., Maillet I., Pfeilschifter J., Quesniaux V.F.J., Landmann R., Ryffel B., Erard F. CD14 works with toll-like receptor 2 to contribute to recognition and control of Listeria monocytogenes infection. J. Infect. Dis. 2008;198:115–124. doi: 10.1086/588815. [DOI] [PubMed] [Google Scholar]
- 34.Van Riet E., Everts B., Retra K., Phylipsen M., van Hellemond J.J., Tielens A.G.M., van der Kleij D., Hartgers F.C., Yazdanbakhsh M. Combined TLR2 and TLR4 ligation in the context of bacterial or helminth extracts in human monocyte derived dendritic cells: Molecular correlates for Th1/Th2 polarization. BMC Immunol. 2009;10:9. doi: 10.1186/1471-2172-10-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qiu Z.J., Cervantes J.L., Cicek B.B., Mukherjee S., Venkatesh M., Maher L.A., Salazar J.C., Mani S., Khanna K.M. Pregnane X Receptor Regulates Pathogen-Induced Inflammation and Host Defense against an Intracellular Bacterial Infection through Toll-like Receptor 4 (vol 6, 2016) Sci. Rep. 2017;7:31936. doi: 10.1038/srep31936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Popovic N., Djokic J., Brdaric E., Dinic M., Terzic-Vidojevic A., Golic N., Veljovic K. The Influence of Heat-Killed Enterococcus faecium BGPAS1-3 on the Tight Junction Protein Expression and Immune Function in Differentiated Caco-2 Cells Infected With Listeria monocytogenes ATCC 19111. Front. Microbiol. 2019;10:412. doi: 10.3389/fmicb.2019.00412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Theisen E., Sauer J.D. Listeria monocytogenes and the Inflammasome: From Cytosolic Bacteriolysis to Tumor Immunotherapy. Curr. Top. Microbiol. 2016;397:133–160. doi: 10.1007/978-3-319-41171-2_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soria-Castro R., Alfaro-Doblado A.R., Rodriguez-Lopez G., Campillo-Navarro M., Meneses-Preza Y.G., Galan-Salinas A., Alvarez-Jimenez V., Yam-Puc J.C., Munguia-Fuentes R., Dominguez-Flores A., et al. TLR2 Regulates Mast Cell IL-6 and IL-13 Production During Listeria monocytogenes Infection. Front. Immunol. 2021;12:2231. doi: 10.3389/fimmu.2021.650779. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.