Abstract
Pseudomonas aeruginosa OprD is a specific porin which facilitates the uptake of basic amino acids and imipenem, a carbapenem antibiotic. Resistance to imipenem due to the loss of OprD is an important mechanism for the loss of clinical effectiveness. To investigate the negative regulatory mechanisms influencing oprD expression, a gene upstream of the coregulated mexEF-oprN efflux operon, designated mexT, was cloned. The predicted 304-amino-acid mature MexT protein showed strong homology to LysR-type regulators. When overexpressed it induced the expression of the mexEF-oprN efflux operon while decreasing the level of expression of OprD. The use of an oprD::xylE transcriptional fusion indicated that it acted by repressing the transcription of oprD. Salicylate, a weak aromatic acid known to reduce porin expression and induce low levels of multiple antibiotic resistance in Escherichia coli, was able to induce imipenem resistance and reduce the expression of OprD but not multiple antibiotic resistance or OprN expression in P. aeruginosa. This was also demonstrated to occur at the level of transcription. Acetyl salicylate and benzoate, but not catechol, were also able to reduce the levels of OprD in the P. aeruginosa outer membranes. These OprD-suppressing compounds increased imipenem resistance even in a mexT-overexpressing and nfxC mutant backgrounds, suggesting that such resistance is independent of the MexT repressor and that oprD is influenced by more than a single mechanism of repression.
Pseudomonas aeruginosa infections are a major clinical problem and can be one of the more difficult infections to treat, due to the organisms’s high intrinsic resistance to many of the most commonly used antibiotics (12). This phenomenon is partly due to its relatively low outer membrane permeability, in conjunction with secondary resistance mechanisms like β-lactamase and active efflux (11, 22). Most small hydrophilic molecules, including antibiotics, enter the periplasm by diffusion through the channels of nonspecific porins that are present in the outer membrane, and it has been shown that this pathway in P. aeruginosa is 10- to 100-fold less efficient than, e.g., that in Escherichia coli. In contrast, it has been shown that the basic amino acid-specific OprD porin of P. aeruginosa mediates the uptake of the β-lactam antibiotic imipenem (13, 35).
Imipenem, a carbapenem β-lactam, is of particular interest because of its high potency, broad spectrum, and general lack of microbial cross-resistance to other β-lactam antibiotics (26). At least two types of imipenem-resistant mutants are observed in clinics. The major type involves the loss by mutation of OprD, and such mutants are resistant only to zwitterionic carbapenem antibiotics (17). Genetic analysis of laboratory-derived mutants has shown that the loss of OprD expression is due to deletions in the oprD coding region and the upstream promoter region (36). A second common type of resistance observed involves multiple antibiotic resistance to both imipenem and other unrelated classes of antibiotics (4, 9). Such mutants are of the nfxC class (8), which show an increased resistance to chloramphenicol, quinolones, and imipenem. They tend to show increased susceptibility towards most β-lactam antibiotics (except imipenem) and aminoglycoside antibiotics. There are two changes in the outer membrane profile of nfxC mutants, including a strong reduction in the expression of OprD and new expression of a 51-kDa protein designated OprN (20). Kohler et al. (15) recently cloned the operon responsible for the nfxC-type phenotype. It consisted of three open reading frames (ORFs), which encoded the cytoplasmic membrane protein MexF (115 kDa), the membrane fusion protein MexE (45 kDa), and the outer membrane protein OprN (51 kDa). MexE, MexF, and OprN showed high similarity to components of the major efflux operon involved in intrinsic antibiotic resistance in Pseudomonas, namely, MexA, MexB, and OprM, respectively. Kohler et al. (15) reported the existence (but not the sequence) of an ORF upstream of this operon, which encoded a putative LysR-type transcriptional activator. The overexpression of this fragment resulted in a multiple-drug-resistant phenotype including resistance to imipenem. Recently we have found that OprD expression is positively regulated in a variety of different ways, including arginine-responsive activation mediated by the AraC-type regulator ArgR (24). A prior study in our laboratory had indicated that a protein termed OpdE was also involved in the regulation of OprD (14). Therefore, we set out to determine the roles of OpdE and the putative transcriptional regulator defined by Kohler et al. (15) and in particular to determine whether they regulate oprD at the transcriptional level. It had been previously shown that salicylate caused reduced porin expression, induced low levels of multiple antibiotic resistance in E. coli (7, 27), and was also able to reduce the expression of a 45-kDa outer membrane protein in P. aeruginosa (34). Therefore, we examined if salicylate exerted its effect through MexT. We have demonstrated here that our previous conclusions as to the function of OpdE were incorrect and that MexT and salicylate independently repress the synthesis of OprD at the level of transcription.
MATERIALS AND METHODS
Strains and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. All medium components were purchased from Difco Laboratories (Detroit, Mich.). E. coli strains were routinely grown in Luria-Bertani broth (1.0% tryptone, 0.5% yeast extract, 0.5% NaCl) containing 0.5% glucose; to obtain a solid medium, agar was added to 1.5% (wt/vol). P. aeruginosa strains were routinely grown in Mueller-Hinton broth; to obtain a solid medium, agar was added to 1.5% (wt/vol). BM2 minimal glucose medium was made as previously described (10). Solutions of acetyl salicylate, salicylate, benzoate, and catechol were prepared fresh daily, and the pH was adjusted to 7 with NaOH as required. For selection purposes, 100 μg of ampicillin per ml and 10 μg of gentamicin per ml were used for E. coli, whereas for P. aeruginosa, carbenicillin at 400 μg/ml, norfloxacin at 1 μg/ml, and gentamicin at 10 μg/ml were used.
TABLE 1.
Bacterial strains and plasmids relevant to this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| P. aeruginosa | ||
| H103 | Wild-type PAO1 | 10 |
| H673 | H103 opdE::Tn501 | 14 |
| H847 | H103 opdE::xylE-Gmr | This study |
| H846 | H103 oprD::xylE-Gmr | This study |
| H729 | H103 oprD::Kmr | 13 |
| 1008 | Clinical isolate | 20 |
| 1008OIR01 | 1008 nfxC phenotype | 20 |
| E. coli S17-1 | thi pro hsdR recA Tra+ | 33 |
| Plasmids | ||
| pBK19R | ColE1-derived plasmid, Apr plac oprD | 14 |
| pX1918GT | pUC-based plasmid containing xylE-Gmr cassette flanked by MCS of pUC19, Apr | 32 |
| pEX100T | AproriV sacB; gene replacement vector | 32 |
| pD2-45 | oriV mob+ORF2 opdE opdR Tcr | 14 |
| pUCP19 | Escherichia-Pseudomonas shuttle vector; Apr plac | 31 |
| pMC2 | pUCP19 mexT | This study |
| pET28a | Expression vector, Kmr, pT7, lac operator, lacI | Novagen, Inc. |
| pMC5 | pET28a mexT | This study |
Reagents.
Gentamicin and carbenicillin were purchased from ICN Biomedicals Inc. (Aurora, Ohio), imipenem was obtained from Merck, Sharp and Dohme (West Point, Pa.), meropenem was obtained from ICI Pharmaceuticals (Macclesfield, U.K.), cefpirome was purchased from Roussel (Paris, France), and ciprofloxacin was supplied by Bayer (Elberfeld, Germany). All other chemical reagents used in this study were purchased from Sigma Chemical (St. Louis, Mo.). The meganuclease I-SceI was purchased from Boehringer Mannheim (Laval, Quebec, Canada). Restriction endonucleases, DNA modifying enzymes, and other molecular biology reagents were purchased from Gibco BRL (Burlington, Ontario, Canada) and New England Biolabs (Mississauga, Ontario, Canada). Mouse anti-OprD (YD11), mouse anti-OprM (TM01), mouse anti-OprJ (HJ004), and mouse anti-OprN (TN009) monoclonal antibodies (MAbs) were a kind gift from Naomasa Gotoh (Kyoto Pharmaceutical University, Kyoto, Japan). Alkaline phosphatase-conjugated goat anti-mouse antibodies were purchased from Bio-Rad (Richmond, Calif.).
Genetic manipulations.
General molecular biology protocols were performed according to the method of Sambrook et al. (28). DNA fragments were isolated by using the Gene Clean kit (Bio 101 Inc., Vista, Calif.). Sticky-end ligations were performed at 23°C for 30 to 60 min; blunt-end ligations were done overnight in a thermocycler programmed to continuously cycle between 10 and 30°C for 30 s at each temperature. DNA sequencing was done with an ABI 373 automated DNA sequencer (Applied Biosystems, Foster City, Calif.) and the ABI Prism dye terminator sequencing kit according to the manufacturer’s directions. Plasmid DNA was prepared for sequencing with Qiagen columns (Qiagen, Inc., Chatsworth, Calif.), and plasmid DNA was quantified with a Hoefer TKO 100 minispectrofluorimeter. Oligonucleotide sequencing primers were synthesized as needed on an ABI 392 oligonucleotide synthesizer according to the manufacturer’s directions. Nucleotide and deduced amino acid sequences were analyzed with the PC Gene software package (Intelligenetics Inc., Mountain View, Calif.). The DNA sequence was used for homology searches of the National Center for Biotechnology Information nonredundant database with the BLASTX program (21a) by using the default settings for all searches. The DNA sequence was also used for homology searches of the P. aeruginosa genome sequencing project with the BLASTN program (21a). Plasmids were transferred into P. aeruginosa by transformation (25).
Cloning of the mexT gene.
According to Kohler et al. (15), a gene encoding a putative regulator of mexEF-oprN, named here the mexT gene consistent with the GenBank entry accession no. AJ007825, was directly upstream and in the same orientation as the mexE gene. The sequence of mexE was retrieved from the National Center for Biotechnology Information data bank (accession no. X99514) and used to search the contents of the P. aeruginosa genome sequencing project (21a), and contig 1523 was retrieved and found to contain the 5′ end of mexE and one other upstream ORF. The sequence of this contig was used to design primers for PCR amplification of the mexT gene from strain H103 by using Vent polymerase and primers with overhangs containing a BamHI site and an EcoRI site to permit easy cloning. To ensure the expression of mexT, the 1.2-kb BamHI/EcoRI fragment containing the mexT gene was cloned into pUCP19 to form the plasmid pMC2, so that the direction of expression of the mexT gene was in the same orientation as the lac promoter. It was then transformed into P. aeruginosa H103 and H846 (oprD::xylE-Gmr). The mexT gene was also amplified by PCR by using primers with overhangs containing a BamHI site and an NdeI site, to permit cloning into the BamHI/NdeI sites of expression vector pET28a (Novagen, Madison, Wis.), creating pMC5.
Construction of oprD::xylE-Gmr and opdE::xylE-Gmr mutants.
A 2.1-kb SstI/HindIII fragment containing the oprD gene was excised from plasmid pBK19R, treated with Klenow polymerase, and cloned into the SmaI site of the gene replacement vector pEX100T. The oprD gene was disrupted by inserting a 2.5-kb EcoRI fragment containing the xylE-Gmr cassette of plasmid pX1918GT into the EcoRI site of oprD. The orientation of the xylE-Gmr cassette was determined by restriction analysis, and a plasmid carrying an oprD::xylE transcriptional fusion was transformed into the mobilizing E. coli strain S17-1 and subsequently transferred into P. aeruginosa H103 by biparental mating. Gentamicin- and carbenicillin-resistant exconjugants were isolated, which had integrated the entire plasmid into the chromosome. Vector pEX100T carries also the sacB gene from Bacillus subtilis, allowing sucrose-inducible expression, which in turn is lethal to P. aeruginosa. Consequently, a positive selection of true mutants was permitted by streaking the exconjugants on a medium containing 10% sucrose and gentamicin. Single colonies were isolated, and the loss of vector sequences was confirmed by testing for carbenicillin sensitivity. The isolated oprD::xylE-Gmr mutant was resistant to imipenem as expected, and the insertion of the xylE-Gmr cassette in the oprD gene was confirmed by Southern blot analysis of chromosomal DNA. This strain was named H846. For the inactivation of the opdE gene the following cloning steps were performed: a 1,680-bp PstI/SstI fragment containing the opdE gene was excised from plasmid pD2-45, treated with Klenow polymerase, and cloned into the SmaI site of vector pUCSce (32). The 2.5-kb xylE-Gmr cassette from plasmid pX1918GT was excised with SphI and cloned into the SphI site of the opdE gene, disrupting the gene. The SmaI site of pUCSce was flanked by restriction sites for the meganuclease I-SceI, allowing the excision of the 4.2-kb DNA fragment containing the opdE::xylE-Gmr DNA and cloning into the I-SceI sites of the gene replacement vector pEX100T. The opdE::xylE-Gmr mutant strain H847 was constructed from H103 as described above.
Determination of catechol 2,3-dioxygenase activity.
P. aeruginosa strains containing the chromosomal oprD::xylE transcriptional fusion and either no plasmid, pUCP19, or pMC2 were harvested in mid-logarithmic growth phase (optical density at 600 nm of 0.5), and cell extracts were assayed for activity of catechol 2,3-dioxygenase, the xylE gene product, as described previously (16). To ensure that salicylate or acetyl salicylate did not influence the dioxygenase activity, cell extracts from cultures grown in Mueller-Hinton broth only were incubated with various concentrations of salicylate or acetyl salicylate prior to the addition of catechol. We did not observe any substantial influence of acetyl salicylate or salicylate on the dioxygenase activities.
Outer membrane isolation and immunodetection.
Outer membranes were prepared from cultures grown to an optical density at 600 nm of 1.0 by sucrose gradient density centrifugation (10). Analysis of protein profiles was done by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10). Immunoblotting procedures were performed as described previously (21).
MIC determination.
The MIC was determined in Mueller-Hinton broth by the broth microdilution method (1). Microtiter plates were inoculated with a final concentration of 2 × 104 to 2 × 105 CFU/ml and incubated for 18 h at 37°C.
RESULTS
Characterization of OpdE.
Previously we isolated a Tn501 mutant of P. aeruginosa H103, named H673, which was resistant to imipenem and deficient in OprD (14). The DNA surrounding the Tn501 insertion site was cloned and sequenced, and the gene carrying Tn501 was named opdE for the putative OprD expression regulator (14). A recent comparison to protein sequence databases revealed that OpdE was most homologous to two proteins, f451 and AraJ, from E. coli and several cytoplasmic membrane proteins from gram-positive bacteria that were linked to chloramphenicol resistance, and analysis of the OpdE protein sequence indicated that it was a cytoplasmic membrane protein. Subsequent experiments showed that H673 was resistant not only to imipenem but also to chloramphenicol and ciprofloxacin, as well as hypersusceptible to carbenicillin. This led us to reexamine here the role of OpdE in the regulation of oprD. An opdE interposon mutant was constructed, which showed wild-type susceptibility to all antibiotics tested (imipenem, chloramphenicol, ciprofloxacin, carbenicillin, and tetracycline), and there was no change in OprD expression. Similarly, opdE provided on a plasmid failed to alter the antibiotic susceptibility of the wild type, H103. The original complementation experiment with strain H673 (14) could not be repeated and probably was due to a reversion mutation. Therefore, we conclude that the opdE::Tn501 mutation was not the cause of the imipenem resistance and OprD deficiency phenotypes and furthermore that this protein is not involved in chloramphenicol resistance as in its gram-positive counterparts. Preliminary data indicated overexpression of OprN in strain H673, indicating along with the characteristic pattern of antibiotic susceptibilities and OprD deficiency that strain H673 carried a separate nfxC mutation.
Characterization of the mexT gene.
The putative regulatory gene located upstream of the mexE gene (15) was amplified by PCR in separate reactions from P. aeruginosa H103. The PCR products were cloned into pUCP19 and pET28a and sequenced. Compared to the recent GenBank entry by T. Kohler and coworkers (accession no. AJ007825), which designated this gene mexT, the sequences of our PCR products contained four base pair changes, of which only one resulted in a conservative amino acid change, L26V. The overexpression of the mexT gene, with the pET28a-based expression system, yielded a major protein band with an Mr of approximately 33 kDa (data not shown). There were two potential ATG start codons at the predicted 5′ end of the mexT gene, although the largest ORF was likely to be correct, based on the alignments of the largest deduced MexT product with other bacterial proteins (see below).
The deduced MexT protein sequence was compared with sequences in the GenBank database by using the BLAST algorithm. The search for amino acid sequences similar to MexT identified 14 proteins with similarity scores higher than 115. Thirteen of the first 14 were NodD proteins from various species of Rhizobium. The protein with the highest similarity was NahR, a LysR-type transcriptional regulator that regulates the production of enzymes for the metabolism of naphthalene and salicylate as sole carbon sources in Pseudomonas putida (37). An alignment of the two sequences (MexT, 304 amino acids, and NahR, 300 amino acids) revealed a total of 33% identical residues and 14.5% conserved residue changes. The regions of similarity were concentrated somewhat at the N terminus. This region of NahR contains the DNA binding helix-turn-helix domain of the protein. A number of very highly conserved residues in NahR and other LysR-type regulators essential for DNA binding (Ala 27, Pro 35, and Arg 43) (30) were also conserved in MexT. Analysis of the region of NahR which binds salicylate as a coinducer (5) and the residues important for binding showed only 18% sequence identity and no conservation of key residues in the same region of MexT, consistent with results observed for other LysR-type transcriptional regulators which show little similarity in this region.
Expression of the mexT gene in P. aeruginosa.
To determine the effect of overexpressing mexT in P. aeruginosa, plasmid pMC2, in which mexT was cloned in the same orientation as the lac promoter, was transformed into a P. aeruginosa wild-type strain, H103. The antimicrobial susceptibility and outer membrane profiles of this strain were examined. In comparison to the wild-type strain with and without the vector, the overexpression of mexT resulted in a two- to fourfold increase in resistance to imipenem, a 32-fold increase in resistance to chloramphenicol, and an 8- to 16-fold increase in resistance to norfloxacin (Table 2). There were no significant changes in the susceptibilities to gentamicin, cefpirome, and tetracycline. The antibiotic susceptibility profile when mexT was overexpressed was consistent with that of the nfxC-type mutant 1008OIR01 compared to its parent 1008 (Table 2) (20). The characteristic increase in susceptibility to carbenicillin seen in nfxC-type mutants could not be confirmed, due to the plasmid-encoded carbenicillin resistance marker in the strains containing pUCP19 and pMC2.
TABLE 2.
Influence of various additives to the growth medium on the MIC of different antibiotics
| P. aeruginosa strain | Additiveb | MIC (μg/ml)a
|
||||
|---|---|---|---|---|---|---|
| IPM | CHL | NOR | TET | CFP | ||
| H103 | None | 0.5 | 25 | 0.1 | 3.1 | 1 |
| Salicylate | 8 | 25 | 0.1 | 1.6 | 1 | |
| Acetyl salicylate | 4 | 25 | 0.2 | 1.6 | 1 | |
| Benzoate | 4 | 25 | 0.1 | 3.1 | 1 | |
| H103/pUCP19 | None | 0.5 | 25 | 0.2 | 1.6 | 1 |
| Salicylate | 8 | 25 | 0.1 | 3.1 | 2 | |
| H103/pMC2::mexT | None | 2 | >400 | 1.6 | 6.3 | 1 |
| Salicylate | 8 | >400 | 1.6 | 3.1 | 2 | |
| H729 oprD::Kmr | None | 8 | 25 | 0.1 | 1.6 | 1 |
| Salicylate | 8 | 25 | 0.2 | 1.6 | 1 | |
| 1008 | None | 1 | 50 | 0.1 | 3.1 | 1 |
| Salicylate | 8 | 50 | 0.2 | 1.6 | 2 | |
| 1008OIR01 nfxC | None | 4 | >400 | 3.1 | 6.3 | 1 |
| Salicylate | 8 | >400 | 3.1 | 3.1 | 1 | |
IPM, imipenem; CHL, chloramphenicol; NOR, norfloxacin; TET, tetracycline; CFP, cefpirome. Results are the mean MICs from four to eight determinations.
All additives were used at a concentration of 16 mM.
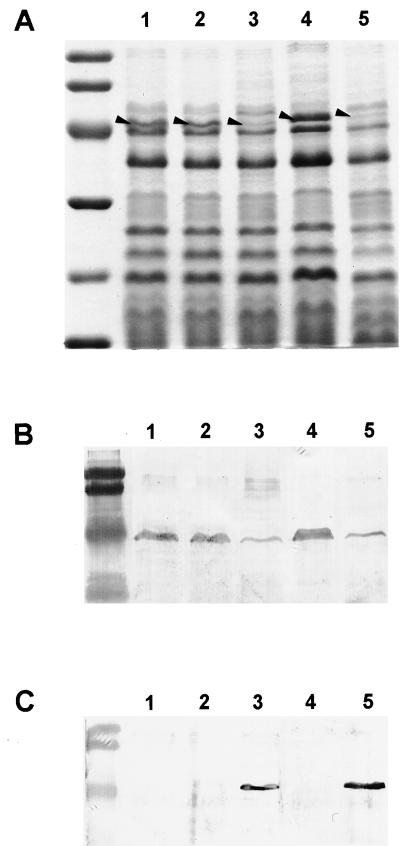
Consistent with the findings of Kohler et al. (15), analysis of the outer membrane proteins showed substantially reduced expression of a 45-kDa protein (Fig. 1A, lane 3) in the mexT-overexpressing strain compared to that of the control strains (Fig. 1A, lanes 1 and 2). This underexpressed protein was confirmed to be OprD by Western blot analysis with an OprD-specific MAb (Fig. 1B). This reduction of OprD in the outer membrane was consistent with the results for the nfxC mutant (Fig. 1A, lane 5). Outer membrane preparations were further analyzed by Western blotting with anti-OprM, -OprJ, and -OprN specific MAbs. OprN was immunodetected only in the mexT-overexpressing strain H103/pMC2 and the nfxC mutant (Fig. 1C, lanes 3 and 5). No cross-reactive bands were seen with the OprJ antibody, while the OprM MAb revealed only wild-type levels of OprM in all strains (data not shown). No other changes in the outer membrane profiles were observed. These results confirm that the overexpression of mexT is itself sufficient to reproduce an nfxC-type phenotype.
FIG. 1.
Expression of the cloned mexT gene in P. aeruginosa. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of outer membrane proteins (A) and Western immunoblots of outer membrane proteins with an anti-OprD MAb (B) and an anti-OprN MAb (C). Lanes 1, H103; lanes 2, H103/pUCP19; lanes 3, H103/pMC2::mexT; lanes 4, 1008; lanes 5, 1008OIR01 (nfxC). The positions of OprD in panel A are labelled with arrowheads. Molecular weight standards in the left-hand lane of panel A are, from top to bottom, 94,000, 67,000, 43,000, 30,000, 20,100, and 14,400. Molecular weight standards in the left-hand lanes of panels B and C are 104,000, 81,000, 47,700, and 34,600.
Reduction of transcription of oprD by MexT.
To directly test whether MexT exerted its effect at the transcriptional level, plasmid pMC2 was transformed into P. aeruginosa H846, which had an oprD::xylE transcriptional fusion in the chromosome. The overexpression of mexT in this strain resulted in a 2.5-fold reduction in activity of catechol 2,3-dioxygenase, the xylE gene product (Table 3), whereas the vector control plasmid had no significant effect.
TABLE 3.
Effects of mexT, salicylate, and acetyl salicylate on the catechol 2,3-dioxygenase activity of the chromosomal oprD::xylE transcriptional fusion in P. aeruginosa H846
| Plasmid | Growth additive | Catechol 2,3-dioxygenase activity (pmol/min/μg of extract) (%)a |
|---|---|---|
| None | None | 61 ± 5 (115) |
| pUCP19 | None | 55 ± 12 (104) |
| pMC2::mexT | None | 22 ± 5 (42) |
| None | None | 53 ± 14 (100) |
| None | 16 mM salicylate | 16 ± 6 (30) |
| None | 16 mM acetyl salicylate | 29 ± 10 (55) |
Results are means ± standard deviations of at least three independent experiments. The second and fourth rows present data for independent control experiments performed at separate times for the two experiments below each control. The value 53 was used as the 100% activity to calculate the percentages in the third column.
Influence of organic acids on oprD expression.
In P. aeruginosa, it has been shown that salicylate reduces the expression of an outer membrane protein with a molecular mass of 45 kDa (34). Salicylate has also been reported to induce the expression of a 50-kDa protein thought to be OprN (20). For this reason we were interested in investigating the possibility that salicylate or other structurally related compounds may be inducers of the mexEF-oprN operon and a repressor of oprD, by acting as a cofactor of the MexT protein.
The antibiotic susceptibilities of P. aeruginosa H103 were tested in the presence of salicylate and a number of related compounds known to induce multiple antibiotic resistance in E. coli (Table 2). Salicylate induced a 16-fold increase in resistance to imipenem but did not lead to significant changes in resistance to chloramphenicol, norfloxacin, tetracycline, or the control β-lactam antibiotic cefpirome (Table 2). Similarly, the only significant change in MIC with benzoate or acetyl salicylate was an eightfold increase in resistance to imipenem (Table 2). Catechol, dinitrophenol, naphthalene, or naphthaquinone, as well as acetate, induced no significant changes in susceptibility to any of the antibiotics tested.
All compounds were tested for their abilities to influence the antibiotic susceptibilities of strains H103/pUCP19, the mexT-overexpressing clone H103/pMC2, the OprD-deficient mutant H729, the nfxC mutant 1008OIR01, and its parent strain 1008. For clarity, only the data for salicylate are presented in Table 2. In the oprD::Kmr mutant H729, there were no significant changes in MIC of any antibiotic in the presence of salicylate (Table 2), benzoate, acetyl salicylate, or catechol, indicating that the imipenem resistance induced in the isogenic parent strain H103 was entirely due to oprD repression. In contrast, all agents able to influence imipenem resistance in H103 had similar effects on the other strains. Maximal levels of imipenem resistance induced in the mexT-overexpressing clone and the nfxC mutant were to a MIC of 8 μg/ml, which was the same MIC observed for the OprD-deficient mutant H729.
To determine whether the increased imipenem resistance was due to decreased expression of OprD and/or one of the efflux operons, we examined the outer membrane profiles of P. aeruginosa when grown in the presence of 32 mM salicylate or 16 mM benzoate. The presence of salicylate or benzoate led to reduced levels of OprD, as confirmed by Western immunoblotting with an anti-OprD MAb (Fig. 2A). Wild-type levels of OprM were detected in all samples by using an anti-OprM MAb (Fig. 2C), and no cross-reacting bands were revealed with the OprN antibody (Fig. 2B) or the OprJ antibody (data not shown). The same results were obtained in the presence of 16 mM acetyl salicylate, and we also detected no salicylate-mediated changes in the levels of OprN when mexT was expressed from plasmid pMC2 (data not shown).
FIG. 2.
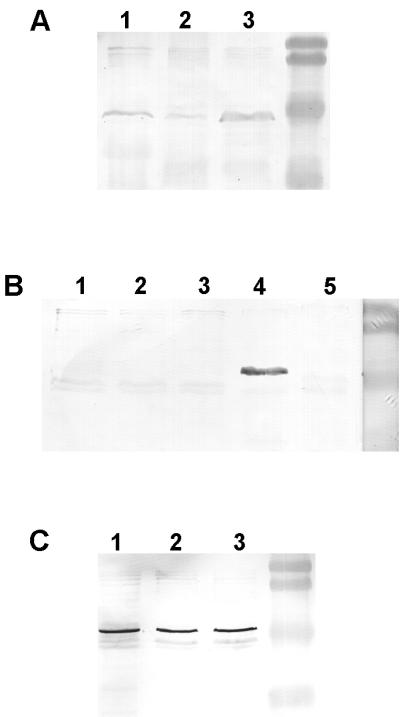
The effects of addition of salicylate and benzoate to the growth medium on the expression of OprD, OprM, and OprN. Western immunoblots of outer membrane proteins of strain H103 grown in Mueller-Hinton medium with 16 mM benzoate (lane 1) or 32 mM salicylate (lane 2) or without any additives (lane 3). In lanes 4 and 5 of panel B, strains 1008OIR01 (nfxC) and 1008, respectively, were grown without any additives. Proteins were detected with anti-OprD (A), anti-OprN (B), and anti-OprM (C) MAbs, respectively. Molecular weight standards in the right-hand lanes are, from top to bottom, 104,000, 81,000, 47,700, and 34,600.
Influence of organic acids on oprD gene transcription.
To further examine the mechanism through which salicylate and acetyl salicylate reduced OprD levels in the outer membrane, we assessed the expression of the oprD::xylE transcriptional fusion in strain H846 (Table 3). The presence of 16 or 32 mM salicylate led to a 3.3-fold reduction of catechol 2,3-dioxygenase activity, and a similar but somewhat lesser reduction was observed with acetyl salicylate. This indicated that salicylate and acetyl salicylate led to reduced levels of OprD by causing reduction of oprD transcription. We could not do these same studies in the presence of benzoate, because these cultures had a strong increase in yellow coloration due presumably to the bioconversion of benzoate to catechol. We also tested acetate and observed a slight increase of catechol 2,3-dioxygenase activity (data not shown).
DISCUSSION
The existence of the LysR-type regulator MexT was predicted by Kohler et al. (15), and the P. aeruginosa genome sequencing project proved to be very useful in designing primers that allowed PCR amplification of the mexT structural gene from the genome of P. aeruginosa PAO1. The overexpression of the cloned mexT gene in P. aeruginosa confirmed the previous notion (15) that it is sufficient to reproduce the multiple-drug-resistant nfxC phenotype. Outer membrane profiles were also changed by mexT overexpression, with a reduction of OprD and expression of the outer membrane component of the mexEF-oprN efflux operon, OprN. Significantly, by using a chromosomal oprD::xylE transcriptional fusion, MexT was shown to cause reduced levels of transcription of the oprD gene. Thus, it appears that MexT is a regulator that can presumably activate the efflux operon while repressing OprD. While unusual, this is not the first example of a LysR-type regulator acting both as a repressor and an activator. The AmpR proteins from Citrobacter freundii and Enterobacter cloacae repress or activate the expression of ampC depending on the conditions. The level of expression of ampC is two- to threefold higher in an ampR-negative situation than in the presence of AmpR under noninducing conditions (2). However, although it seems unlikely, we cannot exclude the possibility that the repression of oprD by MexT is mediated by an unknown regulatory mechanism, and there was no obvious concensus region in the upstream regions of these two genes that would be a candidate for the MexT binding region.
In E. coli, the plant-derived compound salicylate has been shown to reduce the expression of outer membrane porins as well as to induce low-level resistance to some antibiotics (7, 27). Salicylate does this in part by antagonizing the repressor activity of the MarR protein, which negatively regulates the marRAB locus. By reducing the binding of MarR to the promoter region of the marRAB genes, salicylate relieves the repression of MarA, which in turn results in changes in the expression of several unlinked target genes (19). Interestingly, the P. aeruginosa genome has no obvious homologs to the marA or marB gene (data not shown), and it was reported that salicylate can also act in E. coli by a mar-independent pathway (7). Salicylate is a membrane-permeative weak acid and is known to also suppress the synthesis of certain outer membrane proteins in a number of gram-negative bacteria (3, 29), although the mechanism by which this occurs has not been elucidated to date. Given the observed apparent homology between the transcriptional regulators MexT and NahR and the previous reports that salicylate, one of the natural cofactors of NahR, reduced the expression of a 45-kDa outer membrane protein in P. aeruginosa (20, 34), we were intrigued with the possibility that salicylate reduced oprD expression via the MexT protein. In the presence of 16 or 32 mM salicylate, the levels of OprD protein in the outer membrane and the susceptibility of P. aeruginosa to the carbapenem imipenem were reduced. This was consistent with previous reports, but we demonstrated by Western immunoblotting that the 45-kDa protein with reduced expression was indeed OprD (an important point given the 17 OprD homologs of similar molecular weight in the genome). By using a chromosomally encoded oprD::xylE fusion, it was determined that the presence of salicylate caused decreased oprD expression at the level of transcription by 3.3-fold, a level similar to that seen when mexT was overexpressed.
Masuda et al. (20) reported that increased resistance to ofloxacin and the induction of an outer membrane protein of approximately 50 kDa occurred in the presence of 32 mM salicylate. This 50-kDa protein was not heat modifiable and was therefore thought to be OprN rather than OprM. In the publication by Sumita and Fukasawa (34), however, no change in the MIC of quinolones was seen and no expression of a non-heat-modifiable protein was noted in the presence of 32 mM salicylate. Our results showed some minor variations of MICs, which were not considered significant enough to suspect a meaningful induction of an efflux operon. An examination of outer membrane profiles showed that only OprD expression was changed and that there were wild-type levels of expression of OprM and the usual lack of expression of either OprJ or OprN. These data indicate that salicylate alone does not increase the expression of the mexEF-oprN operon. Similarly, acetyl salicylate and benzoate reduced the levels of OprD, and we could demonstrate in the former case that this was mediated at the level of transcription. In contrast, the structurally related compounds naphthalene, naphthaquinone, catechol, and dinitrophenol, the latter of which strongly induced the marRAB operon in E. coli (7), resulted in no significant increase in resistance to any of the antibiotics tested. We also observed that the weak membrane-solubile acid acetate had no effect on the transcription of OprD, indicating that salicylate and related compounds are not acting as weak membrane-solubile acids perturbing the pH gradient across the cytoplasmic membrane. Furthermore, it seems that only molecules with an ionizable carboxyl group mediated the repression of oprD. We propose that there is at least one more negative regulatory mechanism acting on oprD, independently of MexT. Recent data suggested that the transcription of oprD decreased in stationary phase (23). In addition we have demonstrated that ArgR can act as an activator of oprD expression, that independent glutamate-mediated activation occurs, and that catabolite repression by succinate and probably positive regulation mediated by nitrogen sources are operative (24). Thus, OprD appears to be a highly regulated protein. While this is probably related in part to the function of OprD in the uptake of basic amino acids and peptides and to the ability to grow on these as the sole carbon and nitrogen source, the physiological purpose of coregulation with a broad specificity antibiotic efflux operon through MexT is not as easy to understand. Possibly MexT serves a function in P. aeruginosa analogous to that of the E. coli mar operon which also upregulates the efflux of antibiotics (18) and downregulates through micF the major porin OmpF (6). In this regard, it should be noted that OprD, while demonstrating specificity for imipenem and basic amino acids, also has a nonspecific role in the uptake of small compounds like gluconate (13).
ACKNOWLEDGMENTS
This work was supported by the Medical Research Council of Canada. R.E.W.H. was an MRC Distinguished Scientist and M.M. was sponsored by an MRC studentship. M.M.O. was a Canadian Cystic Fibrosis Foundation Postdoctoral Fellow.
REFERENCES
- 1.Amsterdam D. Susceptibility testing of antimicrobials in liquid media. In: Lorian V, editor. Antibiotics in laboratory medicine. Baltimore, Md: Williams & Wilkins; 1991. pp. 72–78. [Google Scholar]
- 2.Bennett P M, Chopra I. Molecular basis of β-lactamase induction in bacteria. Antimicrob Agents Chemother. 1993;37:153–158. doi: 10.1128/aac.37.2.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burns J L, Clark D K. Salicylate-inducible antibiotic resistance in Pseudomonas cepacia associated with absence of a pore-forming outer membrane protein. Antimicrob Agents Chemother. 1992;36:2280–2285. doi: 10.1128/aac.36.10.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cambau E, Perani E, Dib C, Petinon C, Trias J, Jarlier V. Role of mutations in DNA gyrase genes in ciprofloxacin resistance of Pseudomonas aeruginosa susceptible or resistant to imipenem. Antimicrob Agents Chemother. 1995;39:2248–2252. doi: 10.1128/aac.39.10.2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cebolla A, Sousa C, de Lorenzo V. Effector specificity mutants of the transcriptional activator NahR of naphthalene degrading Pseudomonas define protein sites involved in binding of aromatic inducers. J Biol Chem. 1997;272:3986–3992. doi: 10.1074/jbc.272.7.3986. [DOI] [PubMed] [Google Scholar]
- 6.Cohen S P, McMurry L M, Levy S B. marA locus causes decreased expression of OmpF porin in multiple-antibiotic-resistant (Mar) mutants of Escherichia coli. J Bacteriol. 1988;170:5416–5422. doi: 10.1128/jb.170.12.5416-5422.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen S P, Levy S B, Foulds J, Rosner J L. Salicylate induction of antibiotic resistance in Escherichia coli: activation of the mar operon and a mar-independent pathway. J Bacteriol. 1993;175:7856–7862. doi: 10.1128/jb.175.24.7856-7862.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukuda, H., M. Hosaka, K. Hirai, and S. Iyobe. 1990. New norfloxacin resistance gene in Pseudomonas aeruginosa PAO. 34:1757–1761. [DOI] [PMC free article] [PubMed]
- 9.Fukuda H, Hosaka M, Iyobe S, Gotoh N, Nishino T, Hirai K. nfxC-type quinolone resistance in a clinical isolate of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:790–792. doi: 10.1128/AAC.39.3.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hancock R E W, Carey A M. Outer membrane of Pseudomonas aeruginosa: heat- and 2-mercaptoethanol-modifiable proteins. J Bacteriol. 1979;140:902–910. doi: 10.1128/jb.140.3.902-910.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hancock R E W. The bacterial outer membrane as a drug barrier. Trends Microbiol. 1997;5:37–42. doi: 10.1016/S0966-842X(97)81773-8. [DOI] [PubMed] [Google Scholar]
- 12.Hancock R E W, Speert D P. Antibiotics for Pseudomonas and related infections. In: Dodge J A, Brock D J H, Widdicombe J H, editors. Cystic fibrosis—current topics. Vol. 3. New York, N.Y: John Wiley and Sons Ltd.; 1996. pp. 245–266. [Google Scholar]
- 13.Huang H, Hancock R E W. Genetic definition of the substrate selectivity of outer membrane protein OprD of Pseudomonas aeruginosa. J Bacteriol. 1993;175:7793–7800. doi: 10.1128/jb.175.24.7793-7800.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang H, Siehnel R, Bellido F, Rawling E, Hancock R E W. Analysis of two gene regions involved in the expression of the imipenem-specific outer membrane porin protein OprD of Pseudomonas aeruginosa. FEMS Microbiol Lett. 1992;97:267–274. doi: 10.1016/0378-1097(92)90347-q. [DOI] [PubMed] [Google Scholar]
- 15.Kohler T, Michea-Hamzehpour M, Henze U, Gotoh N, Curty L K, Pechere J C. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol Microbiol. 1997;23:345–354. doi: 10.1046/j.1365-2958.1997.2281594.x. [DOI] [PubMed] [Google Scholar]
- 16.Konyecsni W M, Deretic V. Broad-host-range plasmid and M13 bacteriophage-derived vectors for promoter analysis in Escherichia coli and Pseudomonas aeruginosa. Gene. 1988;74:375–386. doi: 10.1016/0378-1119(88)90171-0. [DOI] [PubMed] [Google Scholar]
- 17.Lynch M J, Drusano G L, Mobley H L T. Emergence of resistance to imipenem in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1987;31:1892–1896. doi: 10.1128/aac.31.12.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma D, Cook D N, Alberti M, Pon N G, Nikaido H, Hearst J E. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol Microbiol. 1995;16:45–55. doi: 10.1111/j.1365-2958.1995.tb02390.x. [DOI] [PubMed] [Google Scholar]
- 19.Martin R G, Jair K-W, Wolf R E, Jr, Rosner J L. Autoactivation of the marRAB multiple antibiotic resistance operon by the MarA transcriptional activator in Escherichia coli. J Bacteriol. 1996;178:2216–2223. doi: 10.1128/jb.178.8.2216-2223.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masuda N, Sakagawa E, Ohya S. Outer membrane proteins responsible for multiple drug resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:645–649. doi: 10.1128/AAC.39.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mutharia L M, Hancock R E W. Surface localization of Pseudomonas aeruginosa outer membrane porin protein F by using monoclonal antibodies. Infect Immun. 1983;42:1027–1033. doi: 10.1128/iai.42.3.1027-1033.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21a.National Center for Biotechnology Investigation. BLASTX and BLASTN. [Online.] http://www.ncbi.nlm.nih.gov/BLAST/. [October 1998, last date accessed.]
- 22.Nikaido H, Okusu H, Ma D, Li X-Z. Multidrug efflux pumps make a major contribution to drug resistance in pseudomonads. In: Nakazowa T, Furukawa K, Haas D, Silver S, editors. Molecular biology of pseudomonads. Washington, D.C: ASM Press; 1996. pp. 353–362. [Google Scholar]
- 23.Ochs, M. M. Unpublished data.
- 24.Ochs, M. M., C.-D. Lu, A. T. Abdelal, and R. E. W. Hancock. Submitted for publication.
- 25.Olsen R H, DeBusscher G, McCombie W R. Development of broad-host-range vectors and gene banks: self-cloning of the Pseudomonas aeruginosa PAO chromosome. J Bacteriol. 1982;150:60–69. doi: 10.1128/jb.150.1.60-69.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rohnson G N. β-Lactam antibiotics. J Antimicrob Chemother. 1986;17:5–36. doi: 10.1093/jac/17.1.5. [DOI] [PubMed] [Google Scholar]
- 27.Rosner J L. Nonheritable resistance to chloramphenicol and other antibiotics induced by salicylates and other chemotactic repellents in Escherichia coli K-12. Proc Natl Acad Sci USA. 1985;82:8771–8774. doi: 10.1073/pnas.82.24.8771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Sawai T, Hirano S, Yamaguchi A. Repression of porin synthesis by salicylate in Escherichia coli, Klebsiella pneumoniae and Serratia marcescens. FEMS Microbiol Lett. 1987;40:233–237. [Google Scholar]
- 30.Schell M A, Brown P H, Raju S. Use of saturation mutagenesis to localize probable functional domains in the NahR protein, a LysR-type transcriptional activator. J Biol Chem. 1990;265:3844–3850. [PubMed] [Google Scholar]
- 31.Schweizer H P. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene. 1991;97:109–112. doi: 10.1016/0378-1119(91)90016-5. [DOI] [PubMed] [Google Scholar]
- 32.Schweizer H P, Hoang T T. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene. 1995;158:15–22. doi: 10.1016/0378-1119(95)00055-b. [DOI] [PubMed] [Google Scholar]
- 33.Simon R, Preifer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology. 1983;1:784–790. [Google Scholar]
- 34.Sumita Y, Fukasawa M. Transient carbapenem resistance induced by salicylate in Pseudomonas aeruginosa associated with suppression of outer membrane protein D2 synthesis. Antimicrob Agents Chemother. 1993;37:2743–2746. doi: 10.1128/aac.37.12.2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trias J, Nikaido H. Protein D2 channel of the Pseudomonas aeruginosa outer membrane has a binding site for basic amino acids and peptides. J Biol Chem. 1990;265:15680–15684. [PubMed] [Google Scholar]
- 36.Yoneyama H, Nakae T. Mechanism of efficient elimination of protein D2 in outer membrane of imipenem-resistant Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1993;37:2385–2390. doi: 10.1128/aac.37.11.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.You I S, Ghosal D, Gunsalus I C. Nucleotide sequence of plasmid NAH7 gene nahR and DNA binding of the nahR product. J Bacteriol. 1988;170:5409–5415. doi: 10.1128/jb.170.12.5409-5415.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]