Abstract
A new flavonoid, Jusanin, (1) has been isolated from the aerial parts of Artemisia commutata. The chemical structure of Jusanin has been elucidated using 1D, 2D NMR, and HR-Ms spectroscopic methods to be 5,2′,4′-trihydroxy-6,7,5′-trimethoxyflavone. Being new in nature, the inhibition potential of 1 has been estimated against SARS-CoV-2 using different in silico techniques. Firstly, molecular similarity and fingerprint studies have been conducted for Jusanin against co-crystallized ligands of eight different SARS-CoV-2 essential proteins. The studies indicated the similarity between 1 and X77, the co-crystallized ligand SARS-CoV-2 main protease (PDB ID: 6W63). To confirm the obtained results, a DFT study was carried out and indicated the similarity of (total energy, HOMO, LUMO, gap energy, and dipole moment) between 1 and X77. Accordingly, molecular docking studies of 1 against the target enzyme have been achieved and showed that 1 bonded correctly in the protein’s active site with a binding energy of −19.54 Kcal/mol. Additionally, in silico ADMET in addition to the toxicity evaluation of Jusanin against seven models have been preceded and indicated the general safety and the likeness of Jusanin to be a drug. Finally, molecular dynamics simulation studies were applied to investigate the dynamic behavior of the Mpro-Jusanin complex and confirmed the correct binding at 100 ns. In addition to 1, three other metabolites have been isolated and identified to be сapillartemisin A (2), methyl-3-[S-hydroxyprenyl]-cumarate (3), and β-sitosterol (4).
Keywords: Artemisia commutata, new flavonoid, Jusanin, COVID-19 main protease, molecular similarity, DFT, molecular docking, ADMET, toxicity, molecular dynamic simulations
1. Introduction
Natural products served for thousands of years as the major source for the basic needs of treatment and food in human life [1,2]. Scientists investigated plants [3,4] and microbes [5,6] to deeply understand the causatives of these powers. Various metabolites have been isolated and exhibited promising activities as alkaloids [7], flavonoids [8,9], isochromenes [10], α-pyrones [11], diterpenes [12], sesquiterpenes [13,14,15], steroids [16], and saponins [17,18].
Artemisia commutata Besser is a perennial herb found in northern and eastern Kazakhstan. The plant is also found in Europe, Mongolia, Altai, western and eastern Siberia, Primorsky Krai, Amur Region, and Sakhalin in the Far East, as well as in the Volga-Kama region of the European part of Russia [19].
The genus Artemisia was and still is an interesting target for the scientist to study. The isolation and identification of sesquiterpene lactones have been recorded severally in a lot of Artemisia species such as А. tschernieviana and A. sublessingiana [20]. Furthermore, the presence of flavonoids was also reported in A. albida [21] and A. santolinifolia [22]. The isolation of promising secondary metabolites was reported before as epi-аshantin from A. sieversiana [23], anhydroaustricin from A. albida [24], matricarin from A. austriaca [25], cirsineol and cubreva lactone in A. umbrosa [26]. Recently, the in silico anti-SARS-CoV-2 papain-like protease activities of two derivatives of 2-Phenoxychromone of Artemisia spp. has been reported [27]. Additionally, chemical and biological properties of essential oils of diverse Artemisia species were discussed in various records as A. kasakorum [28] A. lercheana, A. sieversiana [29], A. species [30], A. umbrosa [31], five Artemisia species [32], A. gurganica [33], A. proceriformis [34], A. terrae-albae [35], A. keiskeana [36], A. littoricola, A. mandshurica [37], and A. commutata [38].
The WHO stated on the COVID-19 dashboard on 27 December 2021, that the global confirmed infections had reached 279,114,972 and the number of deaths passed 5,397,580 [39]. These alarming numbers require massive and continuous work in the field of drug discovery to face the COVID-19 pandemic.
The computer-aided (in silico, computational or cheminformatics) drug design and discovery is a widely employed approach. The ability to determine both the physical and chemical properties of a molecule enabled the identification of molecular similarities and hence the prediction of activity. Additionally, molecular docking and dynamic simulations studies can determine the interaction of a specific molecule with a certain protein successfully [40]. Our team presented several reports in which they utilized computational chemistry to find a cure against COVID-19 [41,42,43,44,45].
Interestingly, the inhibitory effect of flavonoids has been reported before. For instance, rutin was predicted to active against COVID-19 3CL proteinase through exhibiting the best binding affinity among 2030 other natural compounds [46].
We herein report the isolation and structure elucidation of the new flavonoid, Jusanin, (jusan is the original kazak name of wormwood) from the aerial parts of A. commutata. Because Jusanin is a new compound, its potential anti-COVID-19 effect was investigated. Additionally, ADMET, toxicity, and DFT properties of Jusanin have been studied. Finally, molecular dynamics simulation studies confirmed the expected affinity of Jusanin. Besides Jusanin, three known metabolites have been isolated and identified as сapillartemisin A (2), methyl-3-[S-hydroxyprenyl]-cumarate (3), and β-sitosterol (4).
2. Results and Discussion
2.1. Isolation of Jusanin
To study the composition of A. commutata [47], 0.94 kg of the aboveground parts were collected in the East Kazakhstan region (Western Altai Mountains). Raw materials afforded 20 g of chloroform extract.
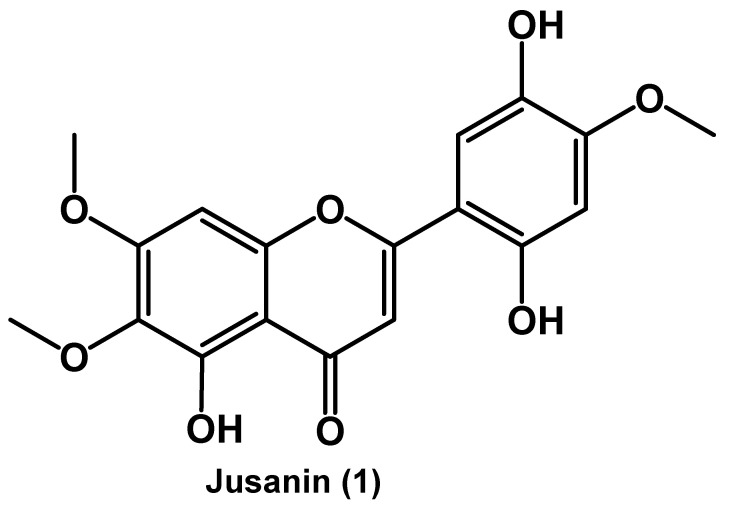
The chloroform extract was subjected to a further isolation by silica gel column using heptane-ethyl acetate-methanol as a mobile phase in a manner of increasing polarity. Fraction 5 was further purified utilizing Sephadex LH-20 to afford the crystalline substance (1) a yellow color with a m.p. more than 350 °C. Based on the spectral data of UV, 1H, and 13C NMR, with two-dimensional experiments, HSQC, Dept, COSY, and HMBC (Table 1), compound 1 was identified to be the new compound 5,2′,4′-trihydroxy-6,7,5′-trimethoxyflavone, Jusanin (Figure 1). The molecular formula C18H16O8 was decided by the HR-ESI-MS experiment, (+ve mode), that displayed a pseudo molecular ion peak [M + H]+ at m/z 361.0934 (calcd. for C18H17O8, 361.0923).
Table 1.
1H and 13C spectral data of Jusanin (DMSO).
| Position | 1H (J = Hz) | 13С | Position | 1H (J = Hz) | 13С |
|---|---|---|---|---|---|
| 2 | 161.97 | 2′ | 152.4 | ||
| 3 | 7.11 s | 106.71 | 3′ | 6.58 s | 104.39 |
| 4 | 182.22 | 4′ | 142.0 | ||
| 5 | 152.6 | 5′ | 139.7 | ||
| 6 | 131.7 | 6′ | 7.45 s | 111.94 | |
| 7 | 158.45 | 6-OCH3 | 3.74 s | 60.0 | |
| 8 | 6.97 s | 91.57 | 7-OCH3 | 3.82 s | 56.7 |
| 9 | 151.95 | 4′-OCH3 | 3.94 s | 56.5 | |
| 10 | 105.3 | 5-OH | 13.06 s | ||
| 1′ | 107.24 |
Figure 1.
The chemical structure of Jusanin (1).
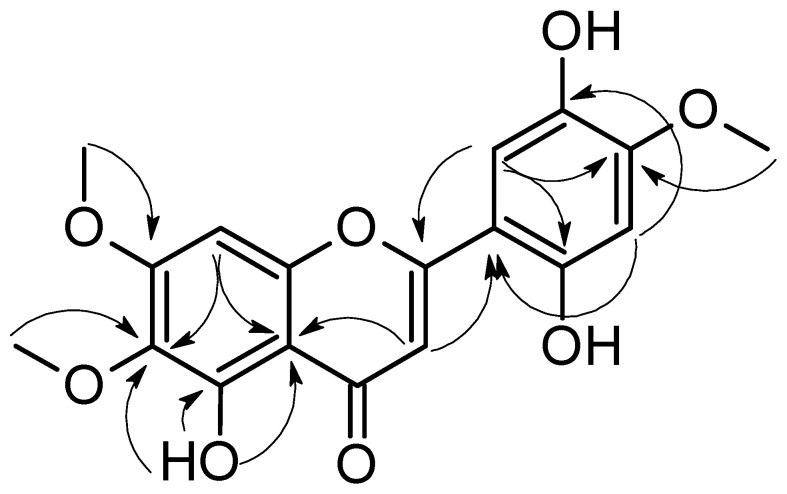
Compound 1 was isolated as a yellow powder with a m.p. more than 350 °C. The 1H NMR spectrum of 1 showed four singlet signals of aromatic protons at δH 7.11 (s, H-3), δH 6.97 (s, H-8), δH 6.58 (s, H-3′), and δH 7.45 (s, H-6′). Furthermore, three signals of methoxy groups were detected at δH 3.74 (s), δH 3.82 (s), and δH 3.94 (s) ppm. The characteristic chelated signal of the OH at C-5 appeared as a sharp singlet at δH 13.06 (due to intramolecular hydrogen bonding) (see Table 1). The 13С, HSQC, and HMBC spectra of 1 indicated the presence of 15 carbon atoms in addition to 3 methoxy groups. The chemical structure of 1 was further confirmed by the heteronuclear multiple bond correlation (HMBC) experiment (Figure 2). Two HMBC correlations were detected between the proton at δH 7.11 (H, s, H-3) and C-10 (δC 105.3) and C-1′ (δC 107.2). Moreover, the OH at C-5 was engaged in three HMBC correlations with C-5 (δC 152.6), C-6 (δC 131.7), and C-10 (δC 105.3). The ring B was confirmed through the HMBC correlations between the proton at δH 6.58 (H, s, H-3′) and C-1′ (δC 107.2) as well as C-5′ (δC 139.7) in addition to the correlations between the proton at δH 7.45 (H, s, H-6′) C-2′ (δC 152.4), and C-4′ (δC 142.0). The HMBC spectrum indicated the attachment of the methoxy groups at δH 3.74 (s), δH 3.82 (s), and δH 3.94 (s) ppm to the carbon atoms C-6 (δC 60.0), C-7 (δC 56.7), and C-4′ (δC 56.5), respectively.
Figure 2.
Key HMBC correlations in Jusanin.
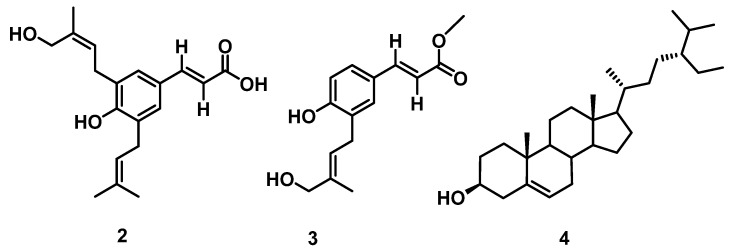
Further chromatographic techniques led to the isolation of 2 (200 mg), 3 (20 mg), and 4 (50 mg). The spectral data of 2–4 were found to correspond to capillartemisin A [48], methyl-3-[S-hydroxyprenyl]-cumarate [49], and β-sitosterol [50], respectively (Figure 3).
Figure 3.
Chemical structures of compounds 2–4.
2.2. Molecular Similarity Study
The basic principle to understand the molecular similarity technique is the well-known link between the bioactivity of a ligand and interactions of that ligand with a protein target. These interactions are based on physical and chemical properties as the ability to donate or accept hydrogen bond interactions as well as the hydrophobic interactions. In consequence, if two molecules have similar chemical structures, they are expected to have similar steric configuration in addition to similar atoms that act as H-bond donors, acceptors, or hydrophobic centers. Accordingly, these two compounds are predicted to exhibit similar bioactivity too [51].
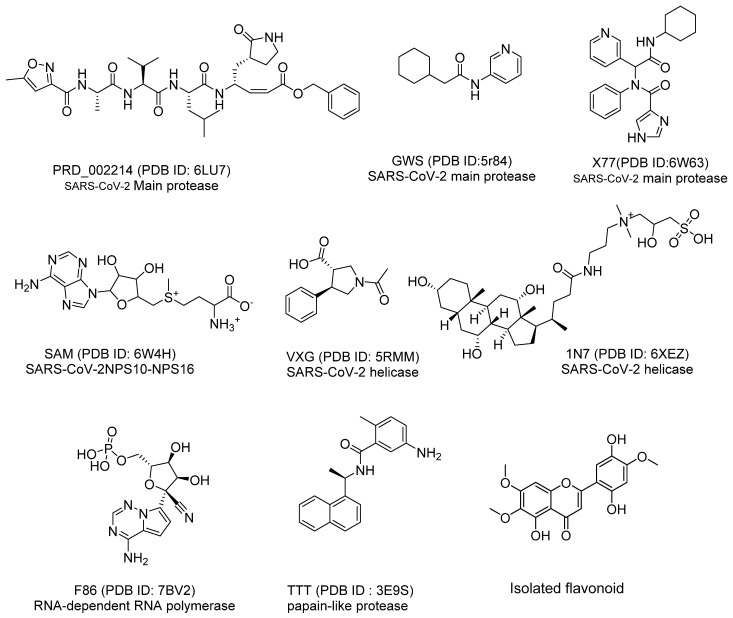
The chemical structure of Jusanin was compared to the chemical structures of eight co-crystallized ligands of eight essential SARS-CoV-2 proteins (Figure 4). This experiment was designed to check if Jusanin has a similarity with one of these ligands and hence it may show a potential inhibitory impact anti-COVID-19 effect.
Figure 4.
The chemical structures of co-crystallized ligands of SARS-Cov-2 proteins and Jusanin (the isolated flavonoid).
Discovery studio software was utilized to explore the following molecular properties in Jusanin and the nine ligands; partition coefficient (ALog p) [52], molecular weight (M. Wt) [53], hydrogen bond (H-bond) donors (HBA) [54], H-bond acceptors (HBD) [55], number of rotatable bonds [56], number of rings, and also aromatic rings [57], as well as molecular fractional polar surface area (MFPSA) [58].
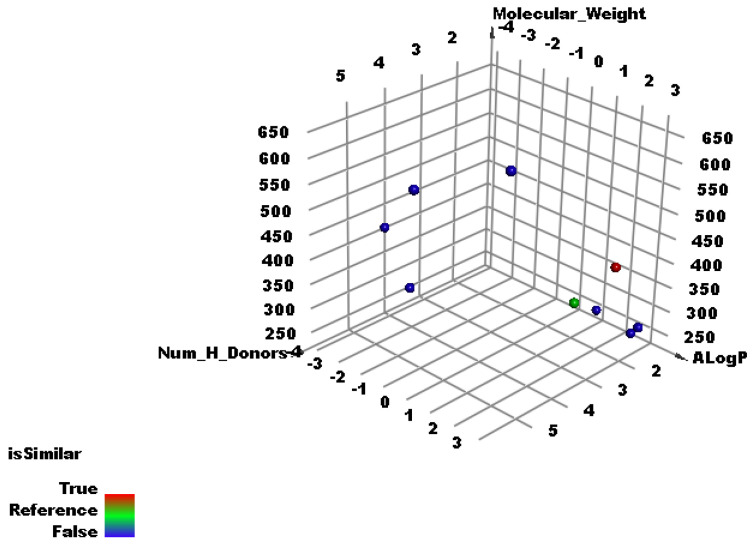
As shown in Figure 5 which clarifies the correlation between Alog p, molecular weight, and number of hydrogen bond donors, the co-crystalized ligand (X77) of the SARS-Cov-2 main protease (PDB ID: 6W63) (red ball) has good similarity with Jusanin (green ball). The minimum distance between the red and green ball is 0.92. and on the other hand, the co-crystallized ligands of the other SARSCoV-2 proteins (blue balls) did not show similarity with Jusanin (green ball).
Figure 5.
The results of the similarity analysis of Jusanin.
The results revealed a high degree of similarity between the Jusanin and X77 (Table 2 and Figure 5).
Table 2.
Structural properties of Jusanin and the different co-crystallized ligands of SARSCoV-2 proteins.
| Compound | ALog p | M. Wt | HBA | HBD | Rotatable Bonds | Rings | Aromatic Rings | MFPSA | Minimum Distance |
|---|---|---|---|---|---|---|---|---|---|
| Jusanin | 2.361 | 360.315 | 8 | 3 | 4 | 3 | 2 | 0.325 | 0 |
| X77 | 2.622 | 403.477 | 4 | 2 | 6 | 4 | 3 | 0.22 | 0.92 |
| TTT | 3.647 | 304.386 | 2 | 2 | 3 | 3 | 3 | 0.171 | 0.95 |
| VXG | 0.711 | 233.263 | 3 | 1 | 2 | 2 | 1 | 0.237 | 1.10 |
| F86 | −1.502 | 371.243 | 11 | 5 | 4 | 3 | 2 | 0.612 | 1.12 |
| SAM | −4.254 | 399.445 | 9 | 4 | 7 | 3 | 2 | 0.483 | 1.17 |
| GWS | 2.171 | 218.295 | 2 | 1 | 3 | 2 | 1 | 0.179 | 1.20 |
| PRD_002214 | 2.453 | 680.791 | 8 | 5 | 18 | 3 | 2 | 0.273 | 1.38 |
| 1N7 | 0.231 | 631.884 | 8 | 6 | 12 | 4 | 0 | 0.256 | 1.52 |
2.3. Structural Fingerprints Study
In order to verify the obtained molecular similarity results, a fingerprint study was carried out using Discovery Studio. The fingerprint technique calculates the 2D molecular features of two different compounds or more in a binary format. The fingerprint study examined the presence or absence of the coming parameters: charge [59], hybridization [60], H-bond acceptor and donor [61], positive and negative ionizable [62], halogen [63], aromatic [64], and ALogP [65].
In structural keys, the chemical structure of a molecule is encoded into a binary bit string (0′s and 1′s), each bit of which corresponds to a “pre-defined” structural feature (e.g., substructure or fragment). If the molecule has a pre-defined feature, the bit position corresponding to this feature is set to 1 (ON). Otherwise, it is set to 0 (OFF) [66].
Fingerprint approach depends on the Tanimoto coefficient. In this work, there are three parameters controlling the binary bit string. SA refers to the bits number that computed in the co-crystallized ligands of SARSCoV-2 proteins and Jusanin. SB refers to the bits number that computed in the co-crystallized ligands of SARSCoV-2 proteins but not in Jusanin. SC refers to the bits number that computed in Jusanin but not in the co-crystallized ligands of SARSCoV-2 proteins.
The results of the fingerprint study revealed that X77 has the highest similarity value (0.512) with Jusanin. In addition, X77 showed high SA value with acceptable rage of SB and SC values indicating the presence of a high number of bits in both X77 and Jusanin. These findings verified the significant fingerprint similarity of Jusanin with X77 (Table 3).
Table 3.
Fingerprint similarity between Jusanin and the different co-crystallized ligands of SARSCoV-2 proteins.
| Comp. | Similarity | SA | SB | SC |
|---|---|---|---|---|
| Jusanin | 1 | 331 | 0 | 0 |
| X77 | 0.512 | 266 | 188 | 65 |
| TTT | 0.489 | 258 | 196 | 73 |
| VXG | 0.436 | 151 | 15 | 180 |
| F86 | 0.339 | 136 | 70 | 195 |
| GWS | 0.310 | 120 | 56 | 211 |
| SAM | 0.299 | 131 | 106 | 200 |
| PRD_002214 | 0.291 | 327 | 789 | 4 |
| 1N7 | 0.138 | 216 | 1231 | 115 |
2.4. Flexible Alignment Studies
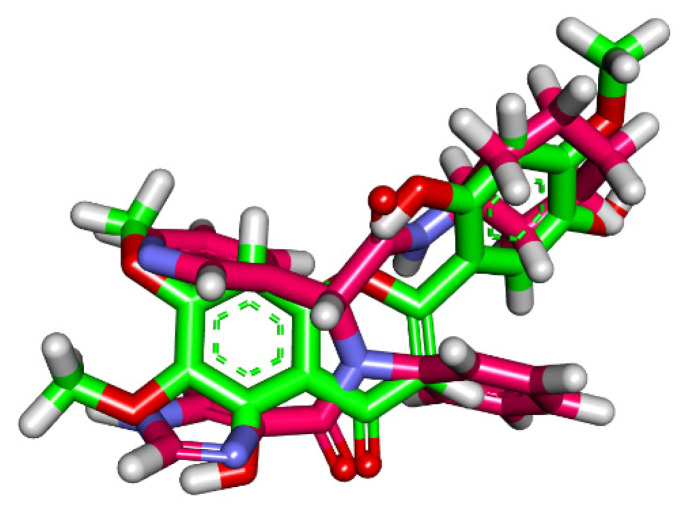
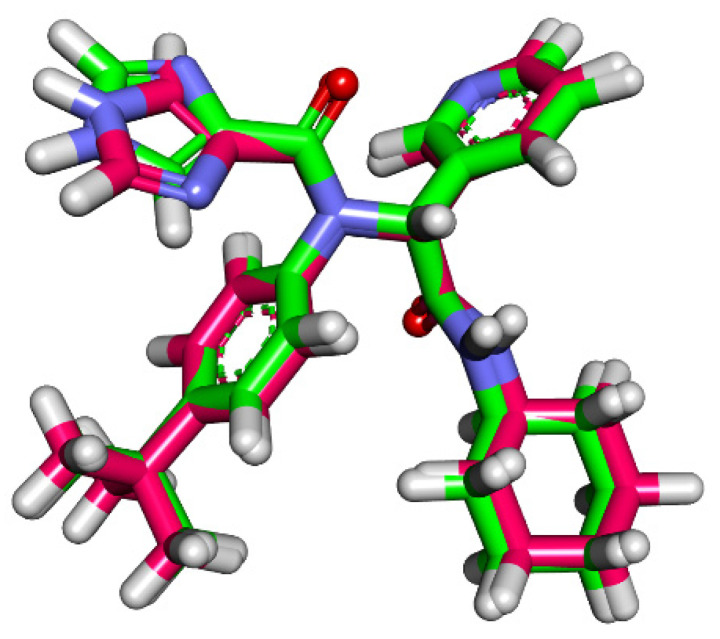
Three-dimensional flexible alignment of Jusanin with the co-crystallized ligands (X77) was studied. The result of flexible alignment revealed the generally good overlap of Jusanin with the reference molecule (X77). In addition, Jusanin showed the same spatial orientation of X77. In detail, the 2,3-dimethoxyphenol moiety showed the same orientation of both pyridine and 1H-imidazole moieties of X77. In addition, the 4H-pyran-4-one moiety of Jusanin exhibited close orientation to the phenyl ring of X77. Furthermore, the 2-methoxybenzene-1,4-diol moiety of Jusanin showed the same orientation of the cyclohexyl moiety of X77 (Figure 6).
Figure 6.
Flexible alignment of Jusanin (green) with X77 (pink).
2.5. DFT Studies
The DFT study is an in silico method that calculates the molecular orbital analysis in addition to the molecular electrostatic potential maps of a certain compound based on the examined parameters (Table 4) [67,68]. The DFT study determines the degree of reactivity of a compound. Accordingly, the DFT properties of Jusanin and X77 were studied by Discovery Studio software to disclose the similarity between them from this point. The results were summarized in Table 4, Figure 7 and Figure 8. The functional used in this test was PWC of local density approximation (LDA). In addition, the quality was adjusted to be Coarse which uses DN basis set with SCF density convergence of 1.0e-4 as utilized from Accelrys in the DMol3 module of Materials Studio package.
Table 4.
MO spatial distribution of Jusanin and X77.
| Name | Binding Energy (Ha) | HOMO Energy (Ha) | LUMO Energy (Ha) | Dipole Mag | Band Gap Energy (Ha) |
|---|---|---|---|---|---|
| Jusanin | −8.558 | −0.180 | −0.098 | 2.395 | 0.083 |
| X77 | −10.830 | −0.158 | −0.062 | 2.906 | 0.096 |
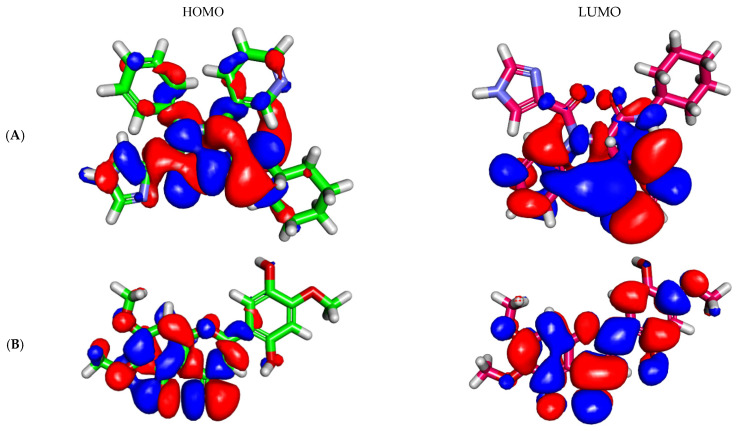
Figure 7.
MO spatial distribution of (A) Jusanin and (B) X77.
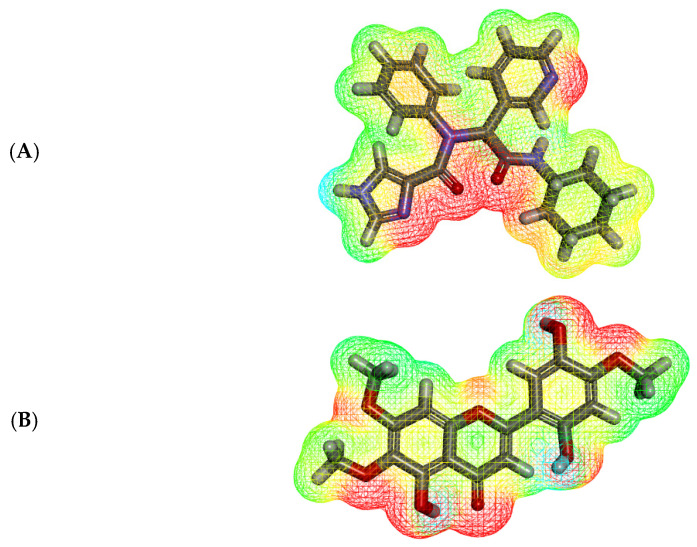
Figure 8.
MEP map of (A) X77 and (B) Jusanin.
2.5.1. Molecular Orbital (MO) Analysis
The compounds also exhibited almost equal dipole moment values of 2.395 and 2.906. Additionally, the gap energy of Jusanin (0.083 Ha) was less than that of X77 (0.096 Ha) which denotes a higher degree of reactivity. Consequently, Jusanin may serve as a promising candidate for further studies.
As shown in Figure 7, the analysis of EHOMO and ELUMO, which are corresponding to the highest occupied and lowest unoccupied molecular orbital energies, respectively. EHOMO and ELUMO were related to the chemical reactivity of the investigated compound and its stability. The HOMO spatial (the electron transfer zones) distributions of X77 are mainly on the 1H-imidazole and the two amide moieties, while its LUMO spatial (the electron acceptor zones) distributions are on the pyridine and phenyl moieties. For compound Jusanin, the HOMO spatial distributions are mainly distributed on 5-hydroxy-6,7-dimethoxy-4H-chromen-4-one moiety, while its LUMO spatial distributions are located on the 2-methoxybenzene-1,4-diol.
2.5.2. Molecular Electrostatic Potential Maps (MEP)
MEP is an in silico method that calculates partial charges [69], electronegativity [70], intermolecular interaction [71], and chemical reactivity [71] to determine the electrostatic potential of a compound in a 3D form [72]. In MEP study, the electronegative atoms (H-bond acceptors) appear in red. On the other side, the electron-poor atoms (H-bond donors) appear in blue. Additionally, the neutral atoms (hydrophobic interactors) appear in a green to yellow color [73].
The MEP of Jusanin and X77 was illustrated in Figure 8A,B, respectively. The ligand, X77, showed four red patches indicating H-bond acceptors and one blue patch (an H-bond donor). Additionally, the high possibility for hydrophobic interactions was indicated by the presence of yellow patches on the aromatic and aliphatic moieties.
For Jusanin, it showed eight red patches (H-bond acceptors) and two blue patches (H-bond donors). Additionally, there is a yellow patch on the aromatic system indicating the possibility of hydrophobic interaction. These findings validate the ability of Jusanin to interact with Mpro like X77.
2.6. Docking Studies
To validate the results of the previous experiments, the binding interaction of Jusanin was investigated against the SARS-CoV-2 main protease (PDB ID: 6W63) through docking studies. Compound X77, was used as a reference. The binding free energy (∆G) and the correct binding mode were employed as the base to evaluate the biding. First of all, a validation procedure of the docking process was applied via re-docking of X77 against Mpro. The resulting RMSD value was 1.7 °A to indicate the validity of the docking process (Figure 9).
Figure 9.
Superimposition of the docking pose (dark green) and the co-crystallized (pink) of the same molecule.
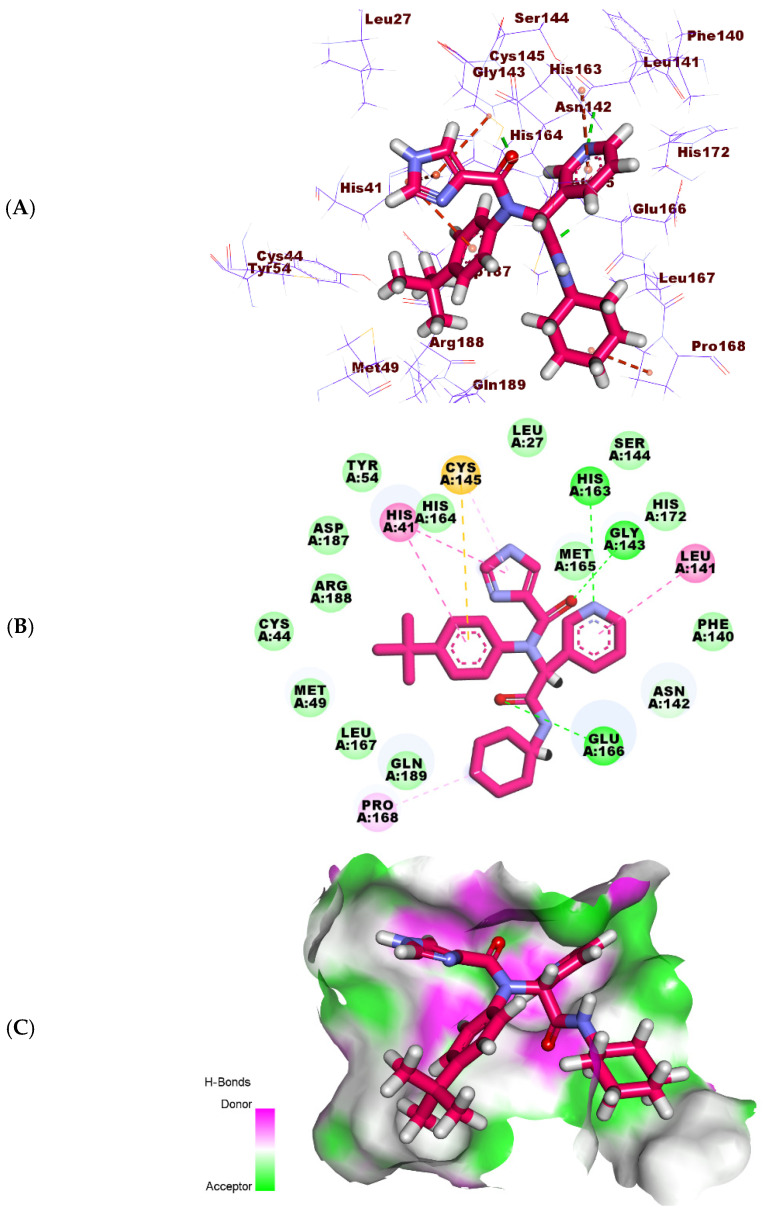
The binding of X77 exhibited a binding free energy of −22.81 kcal/mol. The pyridine moiety was buried in the first pocket of the receptor to form one H-bond with His163 and one hydrophobic interaction with Leu141. Furthermore, the 1H-imidazole was oriented into the second pocket and interacted hydrophobically with Cys145 and His41. The tert-butylbenzene moiety occupied the third pocket closely to the amino acids Arg188, Met49, and Leu167. The same moiety reacted with two hydrophobic interactions with Cys145 and His41. The cyclohexyl moiety was buried in the fourth pocket forming a hydrophobic interaction with the amino acid Pro168. The amide moiety formed one H-bond interaction with Glu166 (Figure 10).
Figure 10.
(A) Three-dimensional, (B) two-dimensional, and (C) surface mapping of X77 docked into Mpro.
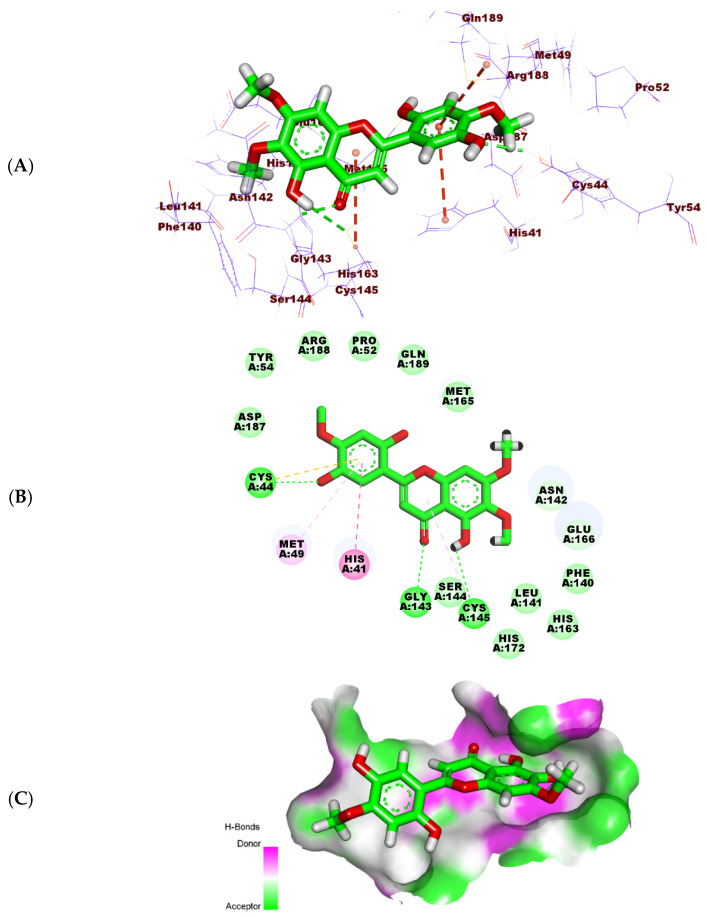
The binding mode of the Jusanin showed a binding free energy of -19.54 kcal/mol (Table 5). The 2,3-dimethoxyphenol moiety was oriented into the first pocket of the receptor to form one H-bond with Cys145. It showed close contact with Asn142, Glu166, Phe140, His163, and Leu141. In addition, the 4H-pyran-4-one was buried in the second pocket forming one hydrophobic interaction with Cys145. It formed one H-bond with Gly143. The 2-methoxybenzene-1,4-diol moiety occupied the third pocket in close contact with Arg188 and Met49. It formed three hydrophobic bonds with Cys44, Met49, and His41. Additionally, it formed one H-bond with Cys44 (Figure 11).
Table 5.
Predicted ADMET for Jusanin and reference.
| Comp. | BBB Level a | Solubility Level | Absorption Level | CYP2D6 Prediction b | PPB Prediction c |
|---|---|---|---|---|---|
| Jusanin | V. low | good | good | non inhibitor | >90% |
| Simeprevir | V. low | low | V. poor | non inhibitor | >90% |
a BBB level, blood–brain barrier penetration level; b CYP2D6, cytochrome P2D6 inhibition; c PBB, plasma protein binding level.
Figure 11.
(A) Three-dimensional, (B) two-dimensional, and (C) surface mapping of Jusanin docked into Mpro.
2.7. In Silico ADMET Analysis
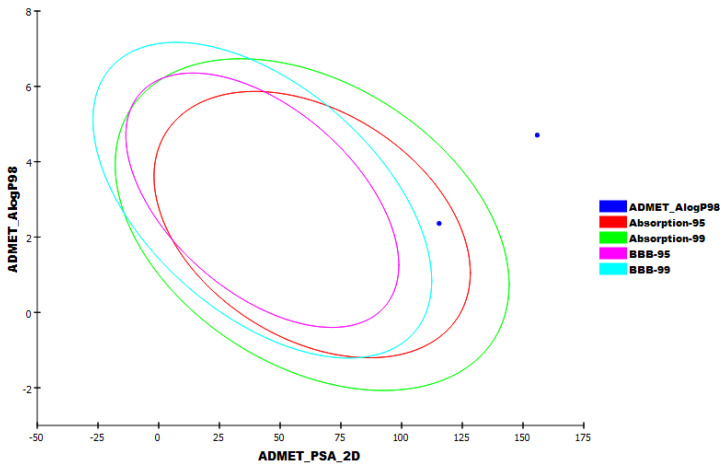
The possibility of a molecule being a drug depends on its activity as well as its bioavailability [74]. In consequence, the in silico ADMET parameters of Jusanin were determined by Discovery Studio software. Simeprevir was used as a reference drug. The results were illustrated in Table 5 and Figure 12.
Figure 12.
The expected ADMET study. ADMET_AlogP98: lipid-water partition coefficient; ADMET_PSA_2D: polar molecular surface area. Two-dimensional polar surface area (PSA_2D) each drug is plotted against their computed atom-type partition coefficient (ALogP98). The area encompassed by the ellipse represents good absorption without any violation of the ADMET properties. Based on Egan et al. [75] absorption model the 95% and 99% confidence limit ellipses corresponding to the blood–brain barrier (BBB) and intestinal absorption models are indicated.
The results revealed that Jusanin has a very low chance to penetrate BBB indicating its high safety margin against CNS. Moreover, Jusanin exhibited good aqueous solubility and intestinal absorption. Finally, Jusanin was predicted to be CYP2D6 non-inhibitor and can bind plasma protein by more than 90%.
2.8. In Silico Toxicity Studies
The proposed toxicity of Jusanin was determined in silico utilizing Discovery Studio software. Seven different toxicity models were applied using Simeprevir as a reference drug. The results were summarized in Table 6.
Table 6.
Toxicity properties of Jusanin and reference.
| Comp. | FDA Rodent Carcinogenicity (Mouse-Male) |
Carcinogenic Potency TD50 (Rat) a |
Rat Maximum Tolerated Dose (Feed) b |
Rat Oral LD50 b | Rat Chronic LOAEL b | Ocular Irritancy | Skin Irritancy |
|---|---|---|---|---|---|---|---|
| Jusanin | Non-Carcinogen | 13.5782 | 0.3502 | 0.4621 | 0.0624 | Mild | Non-Irritant |
| Simeprevir | Non-Carcinogen | 0.2803 | 0.0030 | 0.2088 | 0.0021 | Mild | Non-Irritant |
a Unit: mg/kg body weight/day b Unit: g/kg body weight.
FDA rodent carcinogenicity in male mice expected that Jusanin is non-carcinogenic. Additionally, Jusanin showed high carcinogenic potency TD50 value of 13.5782 mg/kg body weight/day, comparing to Simeprevir (0.2803 mg/kg/day). Additionally, Jusanin showed a high rat maximum tolerated dose value of 0.3502 g/kg compared to Simeprevir (0.0030 g/kg body weight). Jusanin showed a rat oral LD50 value of 0.4621 g/kg which was higher than the reference drug LD50 = 0.2088 g/kg. Regarding the rat chronic LOAEL model, Jusanin exhibited a high value of 0.0624 g/kg, higher than Simeprevir (0.0021 g/kg body weight). Finally, Jusanin was predicted to have mild irritancy against the ocular irritancy model and non-irritant against the skin irritancy model.
2.9. Molecular Dynamics (MD) Simulations Studies
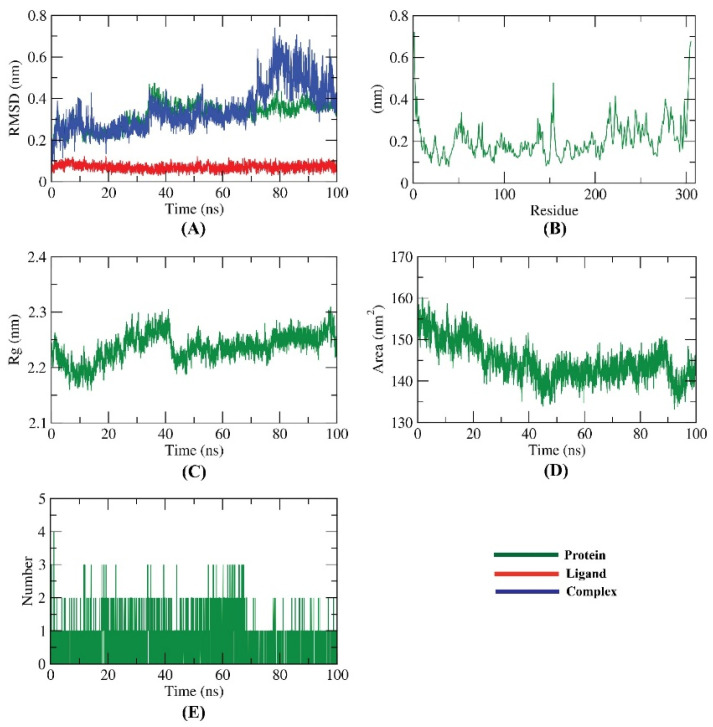
MD simulation is a powerful accurate in silico method that can explore the exact binding mode, flexibility, and stability of a specific receptor–ligand complex for a certain time [76]. Molecular dynamics (MD) simulations studies were employed to mimic the dynamic nature of Mpro–Jusanin interaction investigating the simulation for binding complex stability at 100 ns.
The dynamic changes of atoms in addition to the conformational variations of atoms backbone in the Mpro-Jusanin complex were estimated by RMSD to investigate their stability in both apo- and ligand-bonded states. The Mpro-Jusanin exhibited low RMSD values (less than 0.6 nm) with no major fluctuations declaring the great stability of the complex. The Mpro-Jusanin complex was stable till 70 ns~. Although the Mpro-Jusanin complex showed a minor fluctuation later, it started to be more stable again at 85 ns~ (Figure 13A).
Figure 13.
Molecular dynamics simulations results; (A) RMSD values of Jusanin, Mpro, and Jusanin-Mpro complex during MD runs. (B) RMSF for Mpro in the MD run. (C) Radius of gyration of Mpro in the MD run. (D) SASA of Mpro in the MD run. (E) H-bonding between Jusanin-Mpro complex in the MD run.
The flexibility of Mpro was estimated in terms of RMSF to identify the regions of Mpro that are being fluctuated through the simulation. Figure 13B shows that the binding of Mpro with Jusanin did not change the flexibility of Mpro. The compactness of the Mpro-Jusanin was represented by the radius of gyration (Rg). The decrease in fluctuation throughout the simulation period indicates the higher compactness of the examined system. The Rg of the Mpro-Jusanin complex reached a stable conformation with the radius of gyration fluctuating around 2.2 nm (Figure 13C). The interaction between Mpro-Jusanin complex and solvents was investigated by solvent accessible surface area (SASA) over the simulation time. Consequently, SASA of the Mpro-Jusanin complex was calculated to investigate the degree of the conformational changes that happened after the interaction. Interestingly, as shown in Figure 13D, Mpro exhibeted a decrease in the surface area showing a comparatively lower value of SASA than the starting period. H-bonding between the Mpro-Jusanin complex as anessential factor to stabilize the complex observed (Figure 13E). The highest number of conformations of the Mpro formed up to three H-bonds with Jusanin.
3. Experimental
3.1. General Experimental Section
NMR spectra were carried out on a commercial instrument (Bruker Avance 300 and 600 MHz), chemical shifts (δ) are presented in parts per million (ppm) and re-calculated with respect to tetramethylsilane (TMS) (1H) or carbon signals of deuterium solvents (13C). Spin–spin coupling constants (J) are given in hertz (Hz). The refinement of 13C NMR spectra signals was carried out using Dept, HSQC, HMBC NMR spectra. Mass spectra were recorded on an HP5989A instrument (CI and EI, ionization energy 70 eV) with Apollo 300 data, and on a Kratos MS50TC instrument for accurate calculations (reaching by electric shock (ESI), common solvent mixture: CH2Cl2−MeOH+ NH4OAc) with MASSLYNX system data. UV spectra were obtained on a Perkin-Elmer Lambda 20 Spectrometer instrument. Melting points were determined on Reichert Thermovar. For column chromatography, silica gel 0.06–0.2 mm (Acros) was used as the stationary phase. Silica gel 32–63 mesh was used for flash column chromatography.
3.2. Plant Material
To study the component composition of Artemisia commutata Besser, or wormwood substitute (family Asteraceae), the aboveground part of the plant is collected in the eastern Kazakhstan region (Western Altai Mountains) on July, phase of blooming—beginning of flowering.
Species is identified by botanists of the Altai Botanical Garden (Rider city, eastern Kazakhstan). The herbarium sample is stored in the International Scientific Research Holding «Phytochemistry» Fund. The herbarium sample code is 2007.10.02.02.03.
3.3. Extraction and Isolation
In total, 1.04 kg of raw material was placed in a round-bottomed flask and filled with chloroform and heated to the boiling point of solvents. This operation was repeated three times. The solvent was evaporated on a rotary evaporator under the vacuum of a water-jet pump to obtain an extract weighing 20 g, which was used for preparative chromatographic separation by column chromatography on silica gel using Heptane-Ethylacetate-Methanol followed by Sephadex LH-20 (Pharmacia Fine Chemicals), Uppsala, Sweden.
3.4. Molecular Similarity
The molecular similarity of Jusanin against the co-crystallized ligands of SARS-Cov-2 was carried out and calculated using Discovery Studio 4.0 [77] (See Supplementary Materials).
3.5. Fingerprint Study
A fingerprint study of Jusanin against the nine co-crystallized ligands of SARS-Cov-2 was carried out and calculated using Discovery Studio 4.0 (See Supplementary Materials).
3.6. DFT
The DFT parameters of Jusanin were calculated using Discovery Studio software [78] (See Supplementary Materials).
3.7. Docking Studies
The docking investigation was carried out for Jusanin using MOE2014 software. The results of the docking process were then visualized using Discovery Studio 4.0 software [79,80,81] (See Supplementary Materials).
3.8. ADMET
ADMET descriptors of Jusanin were determined using Discovery Studio 4.0 [82,83] (See Supplementary Materials).
3.9. Toxicity Studies
The toxicity parameters of Jusanin were calculated using Discovery Studio 4.0 [84,85,86] (See Supplementary Materials).
3.10. Molecular Dynamics Simulations
The system was prepared using the web-based CHARMM-GUI [87,88,89] interface with the CHARMM36 force field [90]. All the simulations were carried out using the NAMD 2.13 [91] package. The TIP3P explicit solvation model and CHARMM general force field were used [92,93] (See Supplementary Materials).
4. Conclusions
A new flavonoid, Jusanin, (1) has been isolated from the aerial parts of Artemisia commutata. Jusanin showed a high structural similarity degree with X77, the co-crystallized ligand of the COVID-19 main protease (PDB ID: 6W63), Mpro. This result was indicated by molecular similarity, fingerprint, and DFT studies. The molecular docking of 1 against Mpro confirmed the correct binding of 1 inside Mpro exhibiting a binding energy of −19.54 Kcal/mol. ADMET and toxicity properties of 1 indicated its likeness to be a drug as well as its general safety. The MD simulation studies at 100 ns confirmed the correct binding of the Mpro-Jusanin complex. These interesting results open the door to finding a treatment against COVID-19 after in vitro and in vivo studies.
Acknowledgments
The authors are grateful to W. De Borggraeve and B. Demarsin (KULeuven, Belgium) for help in recording and interpreting mass- and NMR spectra.
Supplementary Materials
The following data are available online, 1D, 2D NMR and HR-Ms spectra data of Jusanin, detailed toxicity report of Jusanin, in addition to the utilized in silico methods.
Author Contributions
Conceptualization, Y.M.S., R.N.S. and A.M.M.; Funding acquisition, B.A.A.; Investigation, Y.M.S., R.A.J., R.N.S., C.A., M.Y.I., S.T., W.D., A.M.M. and I.H.E.; Software, E.B.E., I.H.E. and A.M.M.; Writing, review & editing, B.A.A., E.B.E., I.H.E. and A.M.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research is funded by the Science Committee of the Ministry of Education and Science of the Republic of Kazakhstan (Grant OR11465530 «Creation and replenishment of the collection of industrially valuable microorganisms, study and preservation of their biological diversity for the needs of biotechnology, medicine and agriculture», Grant AP13067578 «Comprehensive phytochemical study of a number of endemic plants of Kazakhstan» and Grant AP08051842 «Study of the composition and biological activity of the essential oils of plants of Central and Southeast Asia»).This research was funded by Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2022R142), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data is contained in the published article.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Samples of the compounds are available from the authors.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Metwaly A.M., Ghoneim M.M., Eissa I.H., Elsehemy I.A., Mostafa A.E., Hegazy M.M., Afifi W.M., Dou D. Traditional ancient Egyptian medicine: A review. Saudi J. Biol. Sci. 2021;28:5823–5832. doi: 10.1016/j.sjbs.2021.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Han X., Yang Y., Metwaly A.M., Xue Y., Shi Y., Dou D. The Chinese herbal formulae (Yitangkang) exerts an antidiabetic effect through the regulation of substance metabolism and energy metabolism in type 2 diabetic rats. J. Ethnopharmacol. 2019;239:111942. doi: 10.1016/j.jep.2019.111942. [DOI] [PubMed] [Google Scholar]
- 3.Metwaly A.M., Lianlian Z., Luqi H., Deqiang D. Black ginseng and its saponins: Preparation, phytochemistry and pharmacological effects. Molecules. 2019;24:1856. doi: 10.3390/molecules24101856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y.-M., Ran X.-K., Riaz M., Yu M., Cai Q., Dou D.-Q., Metwaly A.M., Kang T.-G., Cai D.-C. Chemical constituents of stems and leaves of Tagetespatula L. and its fingerprint. Molecules. 2019;24:3911. doi: 10.3390/molecules24213911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Metwaly A. Comparative biological evaluation of four endophytic fungi isolated from nigella sativa seeds. Al-Azhar J. Pharm. Sci. 2019;59:123–136. doi: 10.21608/ajps.2019.64111. [DOI] [Google Scholar]
- 6.Metwaly A.M., Wanas A.S., Radwan M.M., Ross S.A., ElSohly M.A. New α-Pyrone derivatives from the endophytic fungus Embellisia sp. Med. Chem. Res. 2017;26:1796–1800. doi: 10.1007/s00044-017-1889-7. [DOI] [Google Scholar]
- 7.Metwaly A.M., Ghoneim M.M., Musa A. Two new antileishmanial diketopiperazine alkaloids from the endophytic fungus Trichosporum sp. Derpharmachemica. 2015;7:322–327. [Google Scholar]
- 8.Ghoneim M.M., Afifi W.M., Ibrahim M., Elagawany M., Khayat M.T., Aboutaleb M.H., Metwaly A.M. Biological evaluation and molecular docking study of metabolites from Salvadora Persica L. Growing in Egypt. Pharmacogn. Mag. 2019;15:232. [Google Scholar]
- 9.Liu L., Luo S., Yu M., Metwaly A.M., Ran X., Ma C., Dou D., Cai D. Chemical Constituents of Tagetes patula and Their Neuroprotecting Action. Nat. Prod. Commun. 2020;15:1934578X20974507. [Google Scholar]
- 10.Metwaly A.M., Kadry H.A., Atef A., Mohammad A.-E.I., Ma G., Cutler S.J., Ross S.A. Nigrosphaerin A a new isochromene derivative from the endophytic fungus Nigrospora sphaerica. Phytochem. Lett. 2014;7:1–5. doi: 10.1016/j.phytol.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Metwaly A.M., Fronczek F.R., Ma G., Kadry H.A., Atef A., Mohammad A.-E.I., Cutler S.J., Ross S.A. Antileukemic α-pyrone derivatives from the endophytic fungus Alternaria phragmospora. Tetrahedron Lett. 2014;55:3478–3481. doi: 10.1016/j.tetlet.2014.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhanzhaxina A., Suleimen Y., Metwaly A.M., Eissa I.H., Elkaeed E.B., Suleimen R., Ishmuratova M., Akatan K., Luyten W. In Vitro and In Silico Cytotoxic and Antibacterial Activities of a Diterpene from Cousinia alata Schrenk. J. Chem. 2021;2021:5542455. doi: 10.1155/2021/5542455. [DOI] [Google Scholar]
- 13.Imieje V.O., Zaki A.A., Metwaly A.M., Mostafa A.E., Elkaeed E.B., Falodun A. Comprehensive In Silico Screening of the Antiviral Potentialities of a New Humulene Glucoside from Asteriscus hierochunticus against SARS-CoV-2. J. Chem. 2021;2021:5541876. doi: 10.1155/2021/5541876. [DOI] [Google Scholar]
- 14.Imieje V.O., Zaki A.A., Metwaly A.M., Eissa I.H., Elkaeed E.B., Ali Z., Khan I.A., Falodun A. Antileishmanial Derivatives of Humulene from Asteriscus hierochunticus with in silico Tubulin Inhibition Potential. Rec. Nat. Prod. 2021;16:150–171. [Google Scholar]
- 15.Jalmakhanbetova R., Elkaeed E.B., Eissa I.H., Metwaly A.M., Suleimen Y.M. Synthesis and Molecular Docking of Some Grossgemin Amino Derivatives as Tubulin Inhibitors Targeting Colchicine Binding Site. J. Chem. 2021;2021:5586515. doi: 10.1155/2021/5586515. [DOI] [Google Scholar]
- 16.Suleimen Y.M., Metwaly A.M., Mostafa A.E., Elkaeed E.B., Liu H.-W., Basnet B.B., Suleimen R.N., Ishmuratova M.Y., Turdybekov K.M., Van Heсke K. Isolation, Crystal Structure, and In Silico Aromatase Inhibition Activity of Ergosta-5, 22-dien-3β-ol from the Fungus Gyromitra esculenta. J. Chem. 2021;2021:5529786. doi: 10.1155/2021/5529786. [DOI] [Google Scholar]
- 17.Yassin A.M., El-Deeb N.M., Metwaly A.M., El Fawal G.F., Radwan M.M., Hafez E.E. Induction of apoptosis in human cancer cells through extrinsic and intrinsic pathways by Balanites aegyptiaca furostanol saponins and saponin-coated silvernanoparticles. Appl. Biochem. Biotechnol. 2017;182:1675–1693. doi: 10.1007/s12010-017-2426-3. [DOI] [PubMed] [Google Scholar]
- 18.Sharaf M.H., El-Sherbiny G.M., Moghannem S.A., Abdelmonem M., Elsehemy I.A., Metwaly A.M., Kalaba M.H. New combination approaches to combat methicillin-resistant Staphylococcus aureus (MRSA) Sci. Rep. 2021;11:4240. doi: 10.1038/s41598-021-82550-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vasilyeva A.P.G., Goloskokov V.P., Zaitseva L.G. Flora of Kazakhstan. Volume 9. Nauka; Alma-Ata, Kazakhstan: 1966. pp. 106–107. [Google Scholar]
- 20.Sisengalieva G., Suleimen E., Ishmuratova M.Y., Iskakova Z.B., Van Hecke K. Constituents of Artemisia tschernieviana and their biological activity. Chem. Nat. Compd. 2015;51:544–547. doi: 10.1007/s10600-015-1337-1. [DOI] [Google Scholar]
- 21.Suleimenov E., Smagulova F., Morozova O., Raldugin V., Bagryanskaya I.Y., Gatilov Y.V., Yamovoi V., Adekenov S. Sesquiterpene lactones and flavonoids from Artemisia albida. Chem. Nat. Compd. 2005;41:689–691. doi: 10.1007/s10600-006-0013-x. [DOI] [Google Scholar]
- 22.Suleimen E., Dzhalmakhanbetova R., Ishmuratova M.Y. Flavonoids from Artemisia santolinifolia. Chem. Nat. Compd. 2014;50:918–919. doi: 10.1007/s10600-014-1117-3. [DOI] [Google Scholar]
- 23.Suleimenov E., Smagulova F., Seidakhmetova R., Aksartov R., Raldugin V., Adekenov S. 4-Epiashantin from Artemisia sieversiana. Chem. Nat. Compd. 2007;43:232–233. doi: 10.1007/s10600-007-0090-5. [DOI] [Google Scholar]
- 24.Suleimenov E., Raldugin V., Adekenov S. Anhydroaustricin from Artemisia albida. Chem. Nat. Compd. 2008;44:541–542. doi: 10.1007/s10600-008-9118-8. [DOI] [Google Scholar]
- 25.Kikhanova Z.S., Iskakova Z.B., Dzhalmakhanbetova R., Seilkhanov T., Ross S., Suleimen E. Constituents of Artemisia austriaca and their biological activity. Chem. Nat. Compd. 2013;49:967–968. doi: 10.1007/s10600-013-0796-5. [DOI] [Google Scholar]
- 26.Tashenov E., Dzhalmakhanbetova R., Smagulova F., Dudkin R., Gorovoi P., Suleiman E., Ross S. Cirsilineol and cubreuva lactone from Artemisia umbrosa and their biological activity. Chem. Nat. Compd. 2013;49:97–98. doi: 10.1007/s10600-013-0517-0. [DOI] [Google Scholar]
- 27.Suleimen Y.M., Jose R.A., Suleimen R.N., Arenz C., Ishmuratova M., Toppet S., Dehaen W., Alsfouk A.A., Elkaeed E.B., Eissa I.H., et al. Isolation and In Silico Anti-SARS-CoV-2 Papain-like Protease Potentialities of Two Rare 2-Phenoxychromone Derivatives from Artemisia spp. Molecules. 2022;27:1216. doi: 10.3390/molecules27041216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suleimenov E., Ozek T., Demirci F., Demirci B., Baser K., Adekenov S. Component composition and antimicrobial activity of essential oil from Artemisia kasakorum. Chem. Nat. Compd. 2008;44:263–265. doi: 10.1007/s10600-008-9034-y. [DOI] [Google Scholar]
- 29.Suleimenov E., Ozek T., Demirci F., Demirci B., Baser K., Adekenov S. Component composition of essential oils of Artemisia lercheana and A. sieversiana of the flora of Kazakhstan. Antimicrobial activity of A. sieversiana essential oil. Chem. Nat. Compd. 2009;45:120–123. doi: 10.1007/s10600-009-9232-2. [DOI] [Google Scholar]
- 30.Suleimenov E.M., Tkachev A.V., Adekenov S.M. Essential oil from Kazakhstan Artemisia species. Chem. Nat. Compd. 2010;46:135–139. doi: 10.1007/s10600-010-9548-y. [DOI] [Google Scholar]
- 31.Suleimen E.M., Dudkin R.V., Gorovoi P.G., Wang M., Khan I., Ross S.A. Composition and Bioactivity of Artemisia umbrosa Essential Oil. Chem. Nat. Compd. 2014;50:545–546. doi: 10.1007/s10600-014-1011-z. [DOI] [Google Scholar]
- 32.Ozek G., Suleimen Y., Tabanca N., Doudkin R., Gorovoy P.G., Goger F., Wedge D.E., Ali A., Khan I.A., Baser K.H. Chemical diversity and biological activity of the volatiles of five Artemisia species from Far East Russia. Rec. Nat. Prod. 2014;8:242–261. [Google Scholar]
- 33.Suleimen E.M., Ibataev Z.A., Iskakova Z.B., Ishmuratova M.Y. Constituent Composition and Biological Activity of Essential Oil from Artemisia gurganica. Chem. Nat. Compd. 2015;51:1184–1185. doi: 10.1007/s10600-015-1526-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sampietro D.A., Lizarraga E.F., Ibatayev Z.A., Omarova A.B., Suleimen Y.M., Catalán C.A.N. Chemical composition and antimicrobial activity of essential oils from Acantholippia deserticola, Artemisia proceriformis, Achillea micrantha and Libanotis buchtormensis against phytopathogenic bacteria and fungi. Nat. Prod. Res. 2016;30:1950–1955. doi: 10.1080/14786419.2015.1091453. [DOI] [PubMed] [Google Scholar]
- 35.Suleimen E.M., Ibataev Z.A., Iskakova Z.B., Ishmuratova M.Y., Ross S.A., Martins C.H.G. Constituent Composition and Biological Activity of Essential Oil from Artemisia terrae-albae. Chem. Nat. Compd. 2016;52:173–175. doi: 10.1007/s10600-016-1584-9. [DOI] [Google Scholar]
- 36.Suleimen E.M., Sisengalieva G.G., Adilkhanova A.A., Dudkin R.V., Gorovoi P.G., Iskakova Z.B. Composition and Biological Activity of Essential Oil from Artemisia keiskeana. Chem. Nat. Compd. 2019;55:154–156. doi: 10.1007/s10600-019-02641-7. [DOI] [Google Scholar]
- 37.Suleimen E.M., Dudkin R.V., Gorovoi P.G., Wang M., Khan I., Ross S.A. Constituent Compositions of Essential Oils from Artemisia littoricola and A. mandshurica. Chem. Nat. Compd. 2015;51:790–792. doi: 10.1007/s10600-015-1414-5. [DOI] [Google Scholar]
- 38.Shatar S., Altantsetseg S., Darijimaa S. The essential oil composition of six Artemisia species from Mongolia. J. Essent. Oil Bear. Plants. 1999;2:56–67. [Google Scholar]
- 39.WHO WHO Coronavirus (COVID-19) Dashboard. [(accessed on 25 December 2021)]. Available online: https://covid19.who.int/
- 40.Sliwoski G., Kothiwale S., Meiler J., Lowe E.W. Computational methods in drug discovery. Pharmacol. Rev. 2014;66:334–395. doi: 10.1124/pr.112.007336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jalmakhanbetova R.I., Suleimen Y.M., Oyama M., Elkaeed E.B., Eissa I., Suleimen R.N., Metwaly A.M., Ishmuratova M.Y. Isolation and In Silico Anti-COVID-19 Main Protease (Mpro) Activities of Flavonoids and a Sesquiterpene Lactone from Artemisia sublessingiana. J. Chem. 2021;2021:5547013. doi: 10.1155/2021/5547013. [DOI] [Google Scholar]
- 42.Al-Karmalawy A.A., Dahab M.A., Metwaly A.M., Elhady S.S., Elkaeed E.B., Eissa I.H., Darwish K.M. Molecular Docking and Dynamics Simulation Revealed the Potential Inhibitory Activity of ACEIs Against SARS-CoV-2 Targeting the hACE2 Receptor. Front. Chem. 2021;9:661230. doi: 10.3389/fchem.2021.661230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alesawy M.S., Abdallah A.E., Taghour M.S., Elkaeed E.B., Eissa I.H., Metwaly A.M. In Silico Studies of Some Isoflavonoids as Potential Candidates against COVID-19 Targeting Human ACE2 (hACE2) and Viral Main Protease (Mpro) Molecules. 2021;26:2806. doi: 10.3390/molecules26092806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.El-Demerdash A., Metwaly A.M., Hassan A., El-Aziz A., Mohamed T., Elkaeed E.B., Eissa I.H., Arafa R.K., Stockand J.D. Comprehensive virtual screening of the antiviral potentialities of marine polycyclic guanidine alkaloids against SARS-CoV-2 (COVID-19) Biomolecules. 2021;11:460. doi: 10.3390/biom11030460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eissa I.H., Khalifa M.M., Elkaeed E.B., Hafez E.E., Alsfouk A.A., Metwaly A.M. In Silico Exploration of Potential Natural Inhibitors against SARS-Cov-2 nsp10. Molecules. 2021;26:6151. doi: 10.3390/molecules26206151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu Z., Yang L., Zhang X., Zhang Q., Yang Z., Liu Y., Wei S., Liu W. Discovery of potential flavonoid inhibitors against COVID-19 3CL proteinase based on virtual screening strategy. Front. Mol. Biosci. 2020;7:556481. doi: 10.3389/fmolb.2020.556481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Plantarium Artemisia Commutata Besser//Plantarium. Plants and Lichens of Russia and Neighboring Countries: Open Online Galleries and Plant Identification Guide. [(accessed on 24 September 2021)]. Available online: https://www.plantarium.ru/page/view/item/3849.html.
- 48.Kitagawa I., Fukuda Y., Yoshihara M., Yamahara J., Yoshikawa M. Capillartemisin A and B, two new choleretic principles from Artemisiae capillaris Herba. Chem. Pharm. Bull. 1983;31:352–355. doi: 10.1248/cpb.31.352. [DOI] [PubMed] [Google Scholar]
- 49.Jakupovic J., Tan R., Bohlmann F., Jia Z., Huneck S. Prenylated coumarates from Artemisia xanthochroa. Phytochemistry. 1990;29:3683–3685. doi: 10.1016/0031-9422(90)85307-2. [DOI] [Google Scholar]
- 50.De-Eknamkul W., Potduang B. Biosynthesis of β-sitosterol and stigmasterol in Croton sublyratus proceeds via a mixed origin of isoprene units. Phytochemistry. 2003;62:389–398. doi: 10.1016/S0031-9422(02)00555-1. [DOI] [PubMed] [Google Scholar]
- 51.Nasser M., Salim N., Hamza H., Saeed F., Rabiu I. Improved deep learning based method for molecular similarity searching using stack of deep belief networks. Molecules. 2021;26:128. doi: 10.3390/molecules26010128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Turchi M., Cai Q., Lian G. An evaluation of in-silico methods for predicting solute partition in multiphase complex fluids—A case study of octanol/water partition coefficient. Chem. Eng. Sci. 2019;197:150–158. doi: 10.1016/j.ces.2018.12.003. [DOI] [Google Scholar]
- 53.Sullivan K.M., Enoch S.J., Ezendam J., Sewald K., Roggen E.L., Cochrane S. An adverse outcome pathway for sensitization of the respiratory tract by low-molecular-weight chemicals: Building evidence to support the utility of in vitro and in silico methods in a regulatory context. Appl. Vitr. Toxicol. 2017;3:213–226. doi: 10.1089/aivt.2017.0010. [DOI] [Google Scholar]
- 54.Altamash T., Amhamed A., Aparicio S., Atilhan M. Effect of hydrogen bond donors and acceptors on CO2 absorption by deep eutectic solvents. Processes. 2020;8:1533. doi: 10.3390/pr8121533. [DOI] [Google Scholar]
- 55.Wan Y., Tian Y., Wang W., Gu S., Ju X., Liu G. In silico studies of diarylpyridine derivatives as novel HIV-1 NNRTIs using docking-based 3D-QSAR, molecular dynamics, and pharmacophore modeling approaches. RSC Adv. 2018;8:40529–40543. doi: 10.1039/C8RA06475J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Escamilla-Gutiérrez A., Ribas-Aparicio R.M., Córdova-Espinoza M.G., Castelán-Vega J.A. In silico strategies for modeling RNA aptamers and predicting binding sites of their molecular targets. Nucleosides Nucleotides Nucleic Acids. 2021;40:798–807. doi: 10.1080/15257770.2021.1951754. [DOI] [PubMed] [Google Scholar]
- 57.Kaushik A.C., Kumar A., Bharadwaj S., Chaudhary R., Sahi S. Bioinformatics Techniques for Drug Discovery. Springer; Berlin/Heidelberg, Germany: 2018. Ligand-Based Approach for In-silico Drug Designing; pp. 11–19. [Google Scholar]
- 58.Zhang H., Ren J.-X., Ma J.-X., Ding L. Development of an in silico prediction model for chemical-induced urinary tract toxicity by using naïve Bayes classifier. Mol. Divers. 2019;23:381–392. doi: 10.1007/s11030-018-9882-8. [DOI] [PubMed] [Google Scholar]
- 59.Ieritano C., Campbell J.L., Hopkins W.S. Predicting differential ion mobility behaviour in silico using machine learning. Analyst. 2021;146:4737–4743. doi: 10.1039/D1AN00557J. [DOI] [PubMed] [Google Scholar]
- 60.Taha M., Ismail N.H., Ali M., Rashid U., Imran S., Uddin N., Khan K.M. Molecular hybridization conceded exceptionally potent quinolinyl-oxadiazole hybrids through phenyl linked thiosemicarbazide antileishmanial scaffolds: In silico validation and SAR studies. Bioorg. Chem. 2017;71:192–200. doi: 10.1016/j.bioorg.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 61.Chu H., He Q.-X., Wang J., Hu Y., Wang Y.-Q., Lin Z.-H. In silico design of novel benzohydroxamate-based compounds as inhibitors of histone deacetylase 6 based on 3D-QSAR, molecular docking, and molecular dynamics simulations. New J. Chem. 2020;44:21201–21210. doi: 10.1039/D0NJ04704J. [DOI] [Google Scholar]
- 62.Opo F.A.D.M., Rahman M.M., Ahammad F., Ahmed I., Bhuiyan M.A., Asiri A.M. Structure based pharmacophore modeling, virtual screening, molecular docking and ADMET approaches for identification of natural anti-cancer agents targeting XIAP protein. Sci. Rep. 2021;11:4049. doi: 10.1038/s41598-021-83626-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Heidrich J., Sperl L.E., Boeckler F.M. Embracing the diversity of halogen bonding motifs in fragment-based drug discovery—construction of a diversity-optimized halogen-enriched fragment library. Front. Chem. 2019;7:9. doi: 10.3389/fchem.2019.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Szatylowicz H., Stasyuk O.A., Krygowski T.M. Calculating the aromaticity of heterocycles. Adv. Heterocycl. Chem. 2016;120:301–327. [Google Scholar]
- 65.Andreeva E., Raevsky O. Lipophilicity of organic compounds calculated using structural similarity and molecular physicochemical descriptors. Pharm. Chem. J. 2009;43:258. doi: 10.1007/s11094-009-0280-5. [DOI] [Google Scholar]
- 66.Durant J.L., Leland B.A., Henry D.R., Nourse J.G. Reoptimization of MDL keys for use in drug discovery. J. Chem. Inf. Comput. Sci. 2002;42:1273–1280. doi: 10.1021/ci010132r. [DOI] [PubMed] [Google Scholar]
- 67.Fleming I. Frontier Orbitals and Organic Chemical Reactions. Wiley; Hoboken, NJ, USA: 1977. [Google Scholar]
- 68.El-Nahass M., Kamel M., El-Deeb A., Atta A., Huthaily S. Ab initio HF, DFT and experimental (FT-IR) investigation of vibrational spectroscopy of PN, N-dimethylaminobenzylidenemalononitrile (DBM) Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2011;79:443–450. doi: 10.1016/j.saa.2011.02.055. [DOI] [PubMed] [Google Scholar]
- 69.Wang J. On contribution of known atomic partial charges of protein backbone in electrostatic potential density maps. Protein Sci. 2017;26:1098–1104. doi: 10.1002/pro.3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Berente I., Czinki E., Náray-szabó G. A combined electronegativity equalization and electrostatic potential fit method for the determination of atomic point charges. J. Comput. Chem. 2007;28:1936–1942. doi: 10.1002/jcc.20676. [DOI] [PubMed] [Google Scholar]
- 71.Alkorta I., Perez J.J., Villar H.O. Molecular polarization maps as a tool for studies of intermolecular interactions and chemical reactivity. J. Mol. Graph. 1994;12:3–13. doi: 10.1016/0263-7855(94)80002-2. [DOI] [PubMed] [Google Scholar]
- 72.Suhasini M., Sailatha E., Gunasekaran S., Ramkumaar G. Vibrational and electronic investigations, thermodynamic parameters, HOMO and LUMO analysis on Lornoxicam by density functional theory. J. Mol. Struct. 2015;1100:116–128. doi: 10.1016/j.molstruc.2015.07.003. [DOI] [Google Scholar]
- 73.Matin M.M., Hasan M.S., Uzzaman M., Bhuiyan M.M.H., Kibria S.M., Hossain M.E., Roshid M.H. Synthesis, spectroscopic characterization, molecular docking, and ADMET studies of mannopyranoside esters as antimicrobial agents. J. Mol. Struct. 2020;1222:128821. doi: 10.1016/j.molstruc.2020.128821. [DOI] [Google Scholar]
- 74.Ferreira L.L., Andricopulo A.D. ADMET modeling approaches in drug discovery. Drug Discov. Today. 2019;24:1157–1165. doi: 10.1016/j.drudis.2019.03.015. [DOI] [PubMed] [Google Scholar]
- 75.Egan W.J., Merz K.M., Baldwin J.J. Prediction of drug absorption using multivariate statistics. J. Med. Chem. 2000;43:3867–3877. doi: 10.1021/jm000292e. [DOI] [PubMed] [Google Scholar]
- 76.Hansson T., Oostenbrink C., van Gunsteren W. Molecular dynamics simulations. Curr. Opin. Struct. Biol. 2002;12:190–196. doi: 10.1016/S0959-440X(02)00308-1. [DOI] [PubMed] [Google Scholar]
- 77.Mohammed S.O., El Ashry E.S.H., Khalid A., Amer M.R., Metwaly A.M., Eissa I.H., Elkaeed E.B., Elshobaky A., Hafez E.E. Expression, Purification, and Comparative Inhibition of Helicobacter pylori Urease by Regio-Selectively Alkylated Benzimidazole 2-Thione Derivatives. Molecules. 2022;27:865. doi: 10.3390/molecules27030865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Parmar D.R., Soni J.Y., Guduru R., Rayani R.H., Kusurkar R.V., Vala A.G., Talukdar S.N., Eissa I.H., Metwaly A.M., Khalil A. Discovery of new anticancer thiourea-azetidine hybrids: Design, synthesis, in vitro antiproliferative, SAR, in silico molecular docking against VEGFR-2, ADMET, toxicity, and DFT studies. Bioorg. Chem. 2021;115:105206. doi: 10.1016/j.bioorg.2021.105206. [DOI] [PubMed] [Google Scholar]
- 79.Amer H.H., Alotaibi S.H., Trawneh A.H., Metwaly A.M., Eissa I.H. Anticancer activity, spectroscopic and molecular docking of some new synthesized sugar hydrazones, Arylidene and α-Aminophosphonate derivatives. Arabian J. Chem. 2021;14:103348. doi: 10.1016/j.arabjc.2021.103348. [DOI] [Google Scholar]
- 80.El-Adl K., Sakr H.M., Yousef R.G., Mehany A.B., Metwaly A.M., Elhendawy M.A., Radwan M.M., ElSohly M.A., Abulkhair H.S., Eissa I.H. Discovery of new quinoxaline-2 (1H)-one-based anticancer agents targeting VEGFR-2 as inhibitors: Design, synthesis, and anti-proliferative evaluation. Bioorg. Chem. 2021;114:105105. doi: 10.1016/j.bioorg.2021.105105. [DOI] [PubMed] [Google Scholar]
- 81.Eissa I.H., Ibrahim M.K., Metwaly A.M., Belal A., Mehany A.B., Abdelhady A.A., Elhendawy M.A., Radwan M.M., ElSohly M.A., Mahdy H.A. Design, molecular docking, in vitro, and in vivo studies of new quinazolin-4 (3H)-ones as VEGFR-2 inhibitors with potential activity against hepatocellular carcinoma. Bioorg. Chem. 2021;107:104532. doi: 10.1016/j.bioorg.2020.104532. [DOI] [PubMed] [Google Scholar]
- 82.Yousef R.G., Sakr H.M., Eissa I.H., Mehany A.B., Metwaly A.M., Elhendawy M.A., Radwan M.M., ElSohly M.A., Abulkhair H.S., El-Adl K. New quinoxaline-2 (1 H)-ones as potential VEGFR-2 inhibitors: Design, synthesis, molecular docking, ADMET profile and anti-proliferative evaluations. New J. Chem. 2021;45:16949–16964. doi: 10.1039/D1NJ02509K. [DOI] [PubMed] [Google Scholar]
- 83.Eissa I.H., El-Helby A.-G.A., Mahdy H.A., Khalifa M.M., Elnagar H.A., Mehany A.B., Metwaly A.M., Elhendawy M.A., Radwan M.M., ElSohly M.A. Discovery of new quinazolin-4 (3H)-ones as VEGFR-2 inhibitors: Design, synthesis, and anti-proliferative evaluation. Bioorg. Chem. 2020;105:104380. doi: 10.1016/j.bioorg.2020.104380. [DOI] [PubMed] [Google Scholar]
- 84.El-Adl K., El-Helby A.-G.A., Ayyad R.R., Mahdy H.A., Khalifa M.M., Elnagar H.A., Mehany A.B., Metwaly A.M., Elhendawy M.A., Radwan M.M. Design, synthesis, and anti-proliferative evaluation of new quinazolin-4 (3H)-ones as potential VEGFR-2 inhibitors. Bioorg. Med. Chem. 2021;29:115872. doi: 10.1016/j.bmc.2020.115872. [DOI] [PubMed] [Google Scholar]
- 85.El-Helby A.-G.A., Sakr H., Ayyad R.R., Mahdy H.A., Khalifa M.M., Belal A., Rashed M., El-Sharkawy A., Metwaly A.M., Elhendawy M.A. Design, synthesis, molecular modeling, in vivo studies and anticancer activity evaluation of new phthalazine derivatives as potential DNA intercalators and topoisomerase II inhibitors. Bioorg. Chem. 2020;103:104233. doi: 10.1016/j.bioorg.2020.104233. [DOI] [PubMed] [Google Scholar]
- 86.Eissa I.H., Metwaly A.M., Belal A., Mehany A.B., Ayyad R.R., El-Adl K., Mahdy H.A., Taghour M.S., El-Gamal K.M., El-Sawah M.E. Discovery and antiproliferative evaluation of new quinoxalines as potential DNA intercalators and topoisomerase II inhibitors. Arch. Pharm. 2019;352:1900123. doi: 10.1002/ardp.201900123. [DOI] [PubMed] [Google Scholar]
- 87.Jo S., Kim T., Iyer V.G., Im W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 88.Brooks B.R., Brooks C.L., III, Mackerell A.D., Jr., Nilsson L., Petrella R.J., Roux B., Won Y., Archontis G., Bartels C., Boresch S., et al. CHARMM: The biomolecular simulation program. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lee J., Cheng X., Swails J.M., Yeom M.S., Eastman P.K., Lemkul J.A., Wei S., Buckner J., Jeong J.C., Qi Y., et al. CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. J. Chem. Theory Comput. 2016;12:405–413. doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Best R.B., Zhu X., Shim J., Lopes P.E., Mittal J., Feig M., Mackerell A.D., Jr. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone phi, psi and side-chain chi(1) and chi(2) dihedral angles. J. Chem. Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Phillips J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R.D., Kale L., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- 93.Yu W., He X., Vanommeslaeghe K., MacKerell A.D., Jr. Extension of the CHARMM General Force Field to sulfonyl-containing compounds and its utility in biomolecular simulations. J. Comput. Chem. 2012;33:2451–2468. doi: 10.1002/jcc.23067. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data is contained in the published article.