Abstract
We describe a new, sensitive, rapid, and nonradioactive method involving the use of the commercially available BOCILLIN FL, a fluorescent penicillin, as a labeling reagent for the detection and study of penicillin-binding proteins (PBPs). This method allowed rapid detection of 30 ng of a purified PBP protein under UV light and of 2 to 4 ng of the protein with the aid of a FluorImager. This method also allowed rapid determination of the PBP profiles of Escherichia coli, Pseudomonas aeruginosa, and Streptococcus pneumoniae. The PBP profiles obtained are virtually identical to those reported previously with 3H-, 14C-, or 125I-labeled penicillin. Using this method enabled us to determine the 50% inhibitory concentrations of the penicillin-sensitive and -resistant PBP2x proteins of S. pneumoniae for penicillin G, thereby allowing a direct evaluation of their relative affinities for penicillin G. Finally, this method also allowed us to compare relative affinities of a PBP2x protein for different β-lactam antibiotics with the aid of fluorescence polarization technology and to monitor a PBP2x protein during purification.
Penicillin-binding proteins (PBPs) are the enzymes that are required for the biosynthesis of the bacterial cell wall (10, 11, 23, 31). PBPs catalyze the final steps of the polymerization (transglycosylation) and cross-linking (transpeptidation) of peptidoglycan, an essential component of the bacterial cell wall (10, 11, 23, 31). PBPs are membrane-bound enzymes and targets of β-lactam antibiotics (6, 7, 10, 11, 14, 24, 28–31). The emerging resistance of pathogenic gram-positive bacteria to β-lactam antibiotics is a serious clinical problem (6, 25, 29, 30). Resistance to β-lactam antibiotics in some species such as Streptococcus pneumoniae has occurred by development of altered high-molecular-mass PBPs which have reduced affinity for the antibiotics (6, 7, 10, 11, 14, 24, 25, 29, 30). Extensive molecular and genetic studies have been devoted to the understanding of the mechanisms of bacterial resistance to β-lactam antibiotics (6, 7, 10, 11, 25, 29, 30).
PBPs have often been detected by labeling bacterial membrane preparations with 3H-, 14C-, or 125I-labeled penicillin, separating the labeled proteins by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and exposing the gels to X-ray films (22, 27, 28). The major limitations of this type of methodology are time intensiveness (days to weeks), accumulation of hazardous materials, and lack of commercial availability.
In this study, we describe a new, rapid, sensitive, and nonradioactive method for the detection and study of bacterial PBPs. This method involves the use of BOCILLIN FL (Fig. 1), a newly synthesized and commercially available fluorescent penicillin, as a labeling reagent (Molecular Probes, Inc., Eugene, Oreg.). The BOCILLIN FL-labeled PBPs were separated by SDS-PAGE and detected with a FluorImager or with the naked eye under UV light.
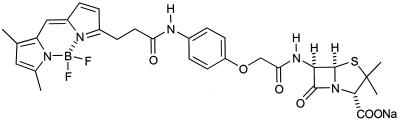
FIG. 1.
Structure of BOCILLIN FL.
MATERIALS AND METHODS
Materials.
BOCILLIN FL (Fig. 1), a derivative of penicillin V, is an orange solid with an extinction coefficient of 68,000 and a maximal absorption at 504 nm (Molecular Probes, Inc.). It fluoresces at 511 nm upon excitation at 504 nm (Molecular Probes, Inc.). BOCILLIN FL was stable for months with no apparent loss of its activity when it was stored at −20°C, even after repeated freezing and thawing (this study).
Cefaclor, cephalexin, penicillin V, and penicillin G were from Eli Lilly. Methicillin and cefotaxime were obtained from Sigma Chemical Company (St. Louis, Mo.). Imipenem was obtained from Merck, Sharp, and Dohme (Westpoint, Pa.). Piperacillin was obtained from Cyanamid (Wayne, N.J.).
Detection of PBPs from bacterial membrane preparations.
For preparation of membranes for the detection of PBPs with BOCILLIN FL as a labeling reagent, the following bacterial strains were used: Escherichia coli MC6RP1 (F− thrA leuA proA dra drm lysA) (9), Pseudomonas aeruginosa PAO1 (16), and S. pneumoniae (hex) R6, a penicillin-sensitive and uncapsulated derivative of Rockefeller University strain R36A that was kindly provided by A. Tomasz (Rockefeller University). E. coli and P. aeruginosa were first grown in Luria-Bertani (LB) medium (Difco Laboratories, Detroit, Mich.), with vigorous shaking, at 33°C overnight. The overnight cultures (10 ml each) were inoculated into 350 ml of fresh LB medium, allowed to grow to an optical density at 600 nm (OD600) of 1.0, and harvested by centrifugation at 4,400 × g for 8 min. Cells were washed once with 20 mM potassium phosphate (pH 7.5) and 140 mM NaCl, resuspended in the same buffer, and disrupted by passing through a French press cell (Aminco Laboratories, Inc., Rochester, N.Y.) twice at 20,000 lb/in2. The resulting cell lysates were centrifuged at 12,000 × g for 10 min. The supernatant fractions were collected and centrifuged at 150,000 × g for 35 min. The pellets were collected, washed once, and resuspended in the same phosphate buffer (1 ml each). The resulting suspensions were designated as membrane preparations and used for fluorescent BOCILLIN FL binding assays. The protein concentrations of the membrane preparations were determined by using the Bradford protein assay kit (1), with bovine serum albumin as a standard (Bio-Rad Laboratories, Hercules, Calif.).
S. pneumoniae (hex) R6 was grown in 500 ml of brain heart infusion medium (Difco), without shaking, at 37°C. Cells were collected at an OD600 of 0.5 to 0.8, and membranes were prepared as described above.
For detection of the PBPs of E. coli, P. aeruginosa, and S. pneumoniae (hex) R6, reaction mixtures (100 μl each) contained 75 μl of each membrane preparation (≈300 μg of protein) and 25 μl of various amounts of BOCILLIN FL ranging from 0.4 to 50 μM (final concentration). The reaction mixtures were incubated at 35°C for 30 min and denatured with 100 μl each of SDS-denaturing solution (19) at 100°C for 3 min. Then, 5 to 10 μl of each reaction mixture (≈7.5 to 15 μg of protein) was subjected to SDS-PAGE analysis (10% polyacrylamide gel; Bio-Rad Laboratories) (19). The protein gels were rinsed with water immediately after electrophoresis. To visualize the labeled PBPs, the gels were directly scanned with a FluorImager 575 (excitation at 488 nm and emission at 530 nm) (Molecular Dynamics, Inc., Sunnyvale, Calif.).
Sensitivity of BOCILLIN FL for detection of PBPs and IC50 (50% inhibitory concentration) determinations.
The penicillin-sensitive and -resistant PBP2x proteins from S. pneumoniae (hex) R6 (a penicillin-sensitive isolate) and 328 (a penicillin-resistant clinical isolate), respectively, were purified as described previously (33). Protein concentrations were determined as described above (1).
For assessment of the sensitivity of BOCILLIN FL for the detection of PBPs, reaction mixtures (10 μl each) contained 2 to 48 ng of the purified penicillin-sensitive PBP2x protein of S. pneumoniae, 10 μM BOCILLIN FL, 20 mM potassium phosphate (pH 7.5), and 140 mM NaCl. The reaction mixtures were incubated at 35°C for 30 min, denatured with 10 μl of SDS solution, and subjected to SDS-PAGE as described above. The labeled protein was visualized with a FluorImager as described above or with the naked eye under UV light (290 nm). For quantitation of the protein, the labeled protein was scanned with a FluorImager, and fluorescence intensities of each band were quantified with an Ultrascan XL laser densitometer (Pharmacia LKB Biotechnology, Alameda, Calif.).
For comparison with the 125I-labeled penicillin V method, both penicillin-sensitive and -resistant PBP2x proteins of S. pneumoniae were labeled with 125I-labeled penicillin V and separated by SDS-PAGE as described previously (27, 33). The resulting protein gels were dried and exposed to X-ray films as described previously (27, 33).
For determination of IC50s of the two PBP2x proteins for penicillin G, reaction mixtures (20 μl each) contained 25 mM potassium phosphate (pH 7.5), 3 μg of each PBP2x protein, 10 μM BOCILLIN FL, and various amounts of penicillin G. The reaction mixtures were incubated at 35°C for 15 min, denatured with 20 μl of SDS solution, and subjected to SDS-PAGE as described above. Both PBP2x proteins were detected by using the FluorImager. Fluorescence intensities of each band were quantified by using an Ultrascan XL laser densitometer (Pharmacia LKB Biotechnology).
Fluorescence polarization.
To determine IC50s of the penicillin-sensitive PBP2x protein for different β-lactam antibiotics, fluorescence polarization binding assays were performed in quadruplicate by using 96-well black opaque microplates (Costar, Cambridge, Mass.). All reagents were diluted in BGG buffer (100 mM potassium phosphate [pH 7.4], 100 μg of bovine gamma globulin/ml; PanVera, Inc.) prior to use. In the standard assay, 50 μl of BOCILLIN FL (final assay concentration, 2 nM) was added to the well, followed by 50 μl of each β-lactam compound (final concentrations ranging from 0.01 to 10,000 nM) and 100 μl of penicillin-sensitive PBP2x enzyme (final concentration, 1.3 nM). After shaking for 1 min, the plate was incubated at room temperature for 2 h. To determine maximum binding, buffer was used instead of added compound, and to determine the basal polarization associated with unbound BOCILLIN FL probe, buffer was substituted for the enzyme. Polarization measurements were made on a multimode Analyst instrument (LJL Biosystems, Inc., Sunnyvale, Calif.) equipped with excitation and emission filters of 485 and 520 nm, respectively, and a fluorescein-specific beamsplitter. The focusing optics were directed at a point in the well 4 mm above the bottom (z height) which minimizes the polarization effects from fluorophores that may be bound to the surfaces of the well. A dynamic polarization filter switches automatically to record intensity signals both vertically and horizontally. Intensities measured were corrected for background from the buffer-only condition.
Polarization (P) was calculated by using the following equation:
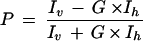 |
where P is polarization, a unit-dimensionless number representing the ratio of the light intensities, expressed in millipolarization (mP), Iv is the fluorescence intensity measured when the excitation and emission polarizers are parallel, Ih is the fluorescence intensity measured when the excitation and emission polarizers are perpendicular, and G is the grating factor that corrects for instrument bias which may be contributed by excitation and emission filters, beamsplitters, and polarizers. The G factor was calculated for each experiment by using the basal polarization value determined with the BOCILLIN FL-only wells.
RESULTS AND DISCUSSION
Sensitivity of BOCILLIN FL for the detection of purified PBP2x proteins of S. pneumoniae.
To assess the sensitivity of BOCILLIN FL, a derivative of penicillin V (Fig. 1), for the detection of PBPs, we performed fluorescent BOCILLIN FL and 125I-labeled penicillin V binding experiments with purified penicillin-sensitive and -resistant PBP2x proteins. When the PBP2x proteins were labeled with 125I-labeled penicillin V, 7.5 to 15 ng of the purified PBP2x proteins was detectable after overnight exposure of the gels (data not shown). When both proteins were labeled with BOCILLIN FL, 30 to 60 ng of the proteins was detectable with the naked eye under UV light (data not shown). The sensitivity of this reagent for detection of PBPs is similar to that observed with fluorescein-labeled compounds (8). With the aid of a FluorImager, 2 to 4 ng of the penicillin-sensitive PBP2x protein was clearly detected when labeled with BOCILLIN FL (Fig. 2A). A linear increase in the fluorescence intensity of the enzyme was observed when the amount of the enzyme increased (Fig. 2B). Thus, these results suggest that the BOCILLIN FL method is at least as sensitive as the 125I-labeled penicillin V method.
FIG. 2.
Detection and quantitation of the penicillin-sensitive PBP2x protein of S. pneumoniae by BOCILLIN FL-binding assays. The penicillin-sensitive PBP2x protein of S. pneumoniae was purified (33), labeled with BOCILLIN FL (10 μM) for 30 min, separated by SDS-PAGE, and visualized by using a FluorImager. (A) Detection of the PBP2x protein; lanes 1 to 8: 2, 4, 8, 12, 16, 24, 36, and 48 ng of the purified PBP2x protein, respectively. (B) Quantitation of the PBP2x protein; fluorescence intensities of each band from panel A were quantified and plotted.
Detection of PBPs in bacterial cell membrane preparations.
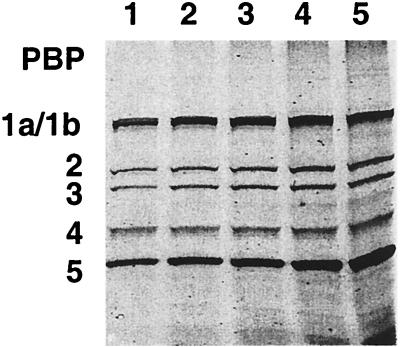
To further evaluate the utility of BOCILLIN FL for the detection of PBPs, we carried out fluorescent BOCILLIN FL binding assays with the membrane preparations of E. coli, P. aeruginosa, and S. pneumoniae. As shown in Fig. 3, PBPs from these organisms were clearly detected when labeled with BOCILLIN FL, and their profiles are very similar to those reported previously with 3H-, 14C-, or 125I-labeled penicillin used as a labeling reagent (14, 21, 22, 26–28, 32). Using a mini-gel (SDS-PAGE) system, we were unable to separate the S. pneumoniae PBP1a protein from its PBP1b protein (Fig. 3, lane 1).
FIG. 3.
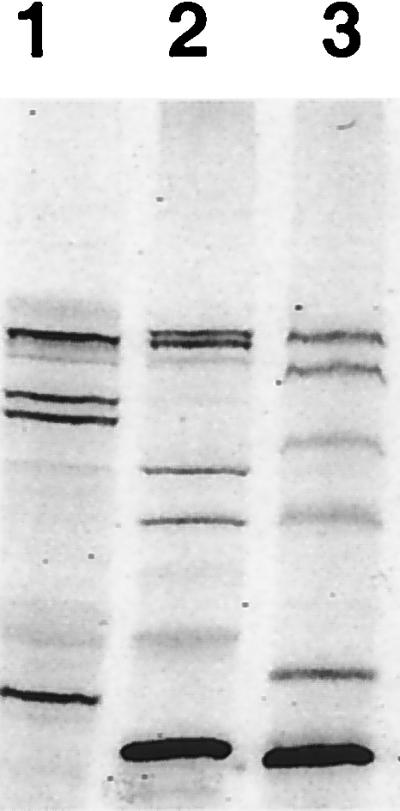
Detection of PBPs of S. pneumoniae, E. coli, and P. aeruginosa. The membrane fractions of S. pneumoniae, E. coli, and P. aeruginosa were prepared and labeled with BOCILLIN FL (25 μM). The labeled membrane preparations (≈7.5 μg of protein each) were separated by SDS-PAGE and visualized by using a FluorImager. Lanes 1 to 3: BOCILLIN FL-labeled membrane preparations of S. pneumoniae, E. coli, and P. aeruginosa, respectively.
As shown in Fig. 4, the intensities of most PBP bands, except PBP4, increased when the E. coli membrane preparation was labeled with BOCILLIN FL concentrations ranging from 1.6 to 25 μM. When concentrations of BOCILLIN FL were lower than 0.8 μM, BOCILLIN FL failed to clearly detect PBPs (data not shown). At concentrations of BOCILLIN FL of 25 μM or higher, the E. coli PBPs appeared to be saturated (data not shown). Thus, we conclude that 1.6 μM BOCILLIN FL is minimal for routine labeling experiments for E. coli.
FIG. 4.
Minimal amount of BOCILLIN FL required for the detection of E. coli PBPs. The E. coli membrane preparation (≈15 μg of protein each) was labeled with BOCILLIN FL (final concentration, 1.6 to 25 μM). The labeled PBPs are separated by SDS-PAGE and detected by using a FluorImager. Lanes 1 to 5: membrane preparation labeled with 1.6, 3.2, 6.4, 12.5, and 25 μM BOCILLIN FL, respectively.
Comparison of the penicillin-sensitive and -resistant PBP2x proteins of S. pneumoniae (hex) R6 and 328, respectively, by fluorescent BOCILLIN FL binding assays.
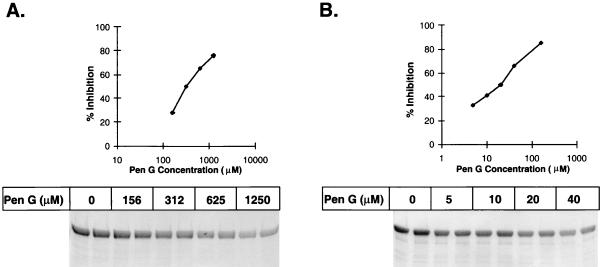
To further examine the utility of BOCILLIN FL for routine comparisons of PBPs for their affinities for β-lactams, we labeled both purified PBP2x proteins with BOCILLIN FL and determined their IC50s for penicillin G. As shown in Fig. 5, the IC50s of the penicillin-sensitive and -resistant PBP2x proteins for penicillin G were determined to be 22 and 312 μM, respectively. Previously, the Kd values of the penicillin-sensitive and -resistant PBP2x proteins for penicillin V were determined to be 0.11 and 1.28 μM, respectively (33). The affinities of the two PBP2x proteins for BOCILLIN FL have not been determined and may be different. Nevertheless, these results are consistent with our previous finding that the affinity of the penicillin-sensitive PBP2x protein for 125I-labeled penicillin V is about 12-fold higher than that of the penicillin-resistant PBP2x protein (33).
FIG. 5.
Determination of IC50s of the penicillin-sensitive and -resistant PBP2x proteins of S. pneumoniae (hex) R6 and 328, respectively, for penicillin G (Pen G). Both PBP2x proteins were purified (33), labeled with BOCILLIN FL in the presence of various amounts of penicillin G, separated by SDS-PAGE, and visualized by using a FluorImager. (A) Penicillin-resistant PBP2x protein of S. pneumoniae 328. (B) Penicillin-sensitive PBP2x protein of S. pneumoniae (hex) R6.
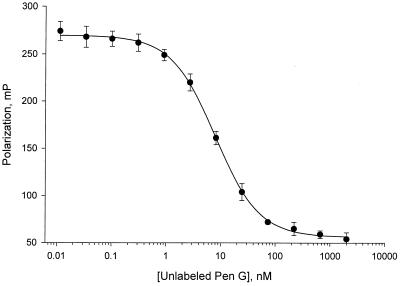
We also employed fluorescence polarization technology to compare the affinities of the penicillin-sensitive PBP2x protein for penicillin G and other β-lactam antibiotics. As shown in Fig. 6, the IC50 of the penicillin-sensitive PBP2x protein for penicillin G was determined to be 7.9 nM. The IC50s of the PBP2x protein for cefotaxime, imipenem, piperacillin, methicillin, cefaclor, and cephalexin were determined to be 4.4, 5.1, 17, 59, 294, and 1,627 nM, respectively. The k2/K value or acylation efficiency of BOCILLIN FL for PBP2x can be derived (7a, 13) from the k2/K value of penicillin G for the penicillin-sensitive PBP2x protein of S. pneumoniae, which is known to be 58,000 M−1s−1 (17, 18). Assuming the IC50 of the penicillin-sensitive PBP2x enzyme for penicillin G is 22 μM, as determined by the gel-based assay (Fig. 5B), the k2/K value of BOCILLIN FL for this PBP2x is estimated to be 128,000 M−1s−1. If the IC50 of the PBP2x enzyme for penicillin G is 7.9 nM as determined by fluorescence polarization assay (Fig. 6), the k2/K value for the enzyme is approximately 232,000 M−1s−1. Thus, with the IC50 determined either by gel-based assay or fluorescence polarization method, a similar k2/K value for BOCILLIN FL was derived. On the basis of these two derived k2/K values of BOCILLIN FL for the PBP2x enzyme, the following k2/K values can be derived for these antibiotics from their IC50s as follows (12, 13): 58,000 to 128,000, 51,000 to 102,000, 15,000 to 27,000, 4,000 to 8,000, and 160 to 300 M−1s−1 for cefotaxime, imipenem, piperacillin, methicillin, and cephalexin, respectively. The k2/K values of these antibiotics for the PBP2x enzyme were previously reported to be 162,000, 107,000, 53,000, 4,900, and 1,600 M−1s−1, respectively (17, 18). Thus, our k2/K values are in agreement with those reported (17, 18), except that our k2/K value for cephalexin was significantly lower. Nevertheless, cephalexin had the lowest k2/K value in both cases (17, 18; this study).
FIG. 6.
Determination of the IC50 of the penicillin-sensitive PBP2x protein of S. pneumoniae (hex) R6 for penicillin G (Pen G). The fluorescence polarization for the penicillin-sensitive PBP2x protein (1.3 nM) in a competitive interaction with BOCILLIN FL (2 nM) and increasing concentrations of unlabeled penicillin G (0.01 to 10,000 nM) were measured as described previously. Data points represent the average of four replicates (± standard deviations), and the curve is the predicted nonlinear regression result.
The IC50 of the penicillin-sensitive PBP2x protein for penicillin G determined by the gel-based assay (22 μM) was about 3,000 times higher than that determined by the fluorescence polarization assay (7.9 nM). This difference is not surprising, since the concentration of BOCILLIN FL (labeling reagent) used in the gel-based assay (10 μM) was 5,000 times higher than the concentration used in the fluorescence polarization assay (2 nM). When IC50s are determined by a true competition assay, the use of higher substrate concentrations generates higher IC50s with a fixed amount of enzyme (2, 4, 12). In addition, the molar ratio of BOCILLIN FL to the enzyme for the gel-based assay (5 ≈ 10 μM BOCILLIN/FL/2 μM PBP2x) was approximately three times higher than that for the fluorescence polarization assay (1.5 ≈ 2 nM BOCILLIN FL/1.3 nM PBP2x). The concentration of the enzyme used in the gel-based assay (2 μM ≈ 3 μg in 20 μl) was approximately 1,500 times higher than that used in the fluorescence polarization assays (1.3 nM). Despite the very different conditions employed in the two types of assays (gel-based and fluorescence polarization assays), especially the different concentrations of BOCILLIN FL used in the two assays, the derived k2/K values for BOCILLIN FL were in excellent agreement (see above). Together, these results suggest that BOCILLIN FL can be used to evaluate relative affinities of PBPs for β-lactam antibiotics.
Finally, the standard assay conditions for the determination of IC50s for β-lactam antibiotics include prelabeling enzymes with β-lactam compounds to be tested, followed by incubation with a labeled β-lactam compound (3, 28). In this study, the IC50s were determined by a true competition assay and were thus strictly dependent on the BOCILLIN FL concentrations utilized in the assay (12).
Implications and conclusions.
In this study, we describe a new, rapid, and sensitive method for the detection and study of PBPs. We used this method to detect PBPs from membrane preparations of three different organisms and to compare two penicillin-sensitive and -resistant PBP2x proteins for their relative affinities for penicillin G. The PBP profiles generated by this method are very similar to those reported before by the method of 3H-, 14C-, or 125I-labeled penicillin (14, 21, 22, 26–28, 32). Using this method, we were also able to demonstrate that a PBP1a mutant of S. pneumoniae was indeed missing the PBP1a protein compared with its parent strain (data not shown). The IC50s determined for the penicillin-sensitive and -resistant PBP2x proteins allowed us to directly evaluate their relative affinities for penicillin G and other β-lactam antibiotics. The results of this comparative study are consistent with those of the previous studies with 125I- and 14C-labeled penicillin and also a thioester substrate (17, 18, 33). Finally, this methodology was also successfully applied to the detection of a PBP2x protein during its purification (data not shown). The results of our study, therefore, have validated BOCILLIN FL as a labeling reagent for the detection of PBPs and the evaluation of relative affinities of PBPs for different β-lactam antibiotics.
The use of BOCILLIN FL as a labeling reagent for the detection of PBPs offers several advantages over the radioisotope methods (22, 27, 28). By this method, results can be obtained immediately after the completion of SDS-PAGE, since no gel manipulations are required for detection. Routinely, the 125I-labeled penicillin V and 3H- or 14C-labeled penicillin methods require days to weeks. This BOCILLIN FL method is sensitive, allowing rapid detection of quantities of proteins in nanograms with the naked eye under UV light or with the aid of a FluorImager. The sensitivity of this method is similar to that of the 125I-labeled penicillin V method, which also detected quantities of the proteins as small as nanograms but required overnight exposure. The BOCILLIN FL method does not involve the use of radioactive materials, in contrast to the 3H-, 14C-, and 125I-labeled penicillin methods. Thus, no hazardous materials are produced. Only small amounts of BOCILLIN FL are needed for routine labeling studies. We established that a concentration as low as 1.6 μM the reagent could be used for labeling bacterial membrane preparations.
BOCILLIN FL and 3H-labeled penicillin are both commercially available (28), but the latter methodology typically requires weeks for the development of PBP bands. 125I-labeled penicillin V has considerably shortened the amount of time that is required, but it is not commercially available (27). Fluorescein-labeled and biotinylated penicillins offer similar advantages (5, 8, 15, 20), yet they are not commercially available (5, 8, 15, 20). Also, the use of biotinylated penicillins requires transferring proteins to membranes and blocking and developing the membranes, processes which usually require at least a full day. BOCILLIN FL is a safe and sensitive reagent for the detection and study of bacterial PBPs.
Finally, a comparison of the PBP binding properties of BOCILLIN FL and those of its parent molecule, penicillin V, has not yet been carried out. Therefore, the effects of the addition of the fluorophore on the PBP binding properties of BOCILLIN FL are unknown. Further study is also needed to address the suitability of BOCILLIN FL for kinetic studies of PBPs.
ACKNOWLEDGMENTS
We thank A. Tomasz of Rockefeller University, R. A. Jensen of the University of Florida, and M. A. de Pedro of the Universidad of Autonoma de Madrid, Spain, for providing bacterial strains used in this study. We also thank J. Flokowitsch and P. Matsushima for their excellent technical assistance and J. Colacino, T. Nicas, and M. Smith for their critical review of the manuscript.
REFERENCES
- 1.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 2.Bush K. Screening and characterization of enzyme inhibitors as drug candidates. Drug Metab Rev. 1983;14:689–708. doi: 10.3109/03602538308991405. [DOI] [PubMed] [Google Scholar]
- 3.Bush K, Smith S A, Ohringer S, Takana S K, Bonner D P. Improved sensitivity in assays for binding of novel β-lactam antibiotics to penicillin-binding proteins of Escherichia coli. Antimicrob Agents Chemother. 1987;31:1271–1273. doi: 10.1128/aac.31.8.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Copeland R A. Enzymes: a practical introduction to structure, mechanisms, and data analysis. New York, N.Y: Wiley-VCH; 1996. [Google Scholar]
- 5.Dargis M, Malouin F. Use of biotinylated β-lactams and chemiluminescence for study and purification of penicillin-binding proteins in bacteria. Antimicrob Agents Chemother. 1994;38:973–980. doi: 10.1128/aac.38.5.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dowson C G, Coffey T J, Spratt B G. Origin and molecular epidemiology of penicillin-binding protein-mediated resistance to β-lactam antibiotics. Trends Microbiol. 1994;2:361–366. doi: 10.1016/0966-842x(94)90612-2. [DOI] [PubMed] [Google Scholar]
- 7.Dowson C G, Hutchison A, Spratt B G. Extensive remodeling of the transpeptidase domain of penicillin-binding protein 2B of a penicillin-resistant South African isolate of Streptococcus pneumoniae. Mol Microbiol. 1989;3:95–102. doi: 10.1111/j.1365-2958.1989.tb00108.x. [DOI] [PubMed] [Google Scholar]
- 7a.Frere J-M, Nguyen-Disteche M, Coyette J, Joris B. Mode of action: interaction with the penicillin-binding proteins. In: Page M I, editor. The chemistry of β-lactams. Glasgow, Scotland: Chapman and Hall; 1992. pp. 148–197. [Google Scholar]
- 8.Galleni M, Lakaye B, Lepage S, Jamin M, Thamn I, Joris B, Frere J-M. A new, highly sensitive method for the detection and quantification of penicillin-binding proteins. Biochem J. 1993;291:19–21. doi: 10.1042/bj2910019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia Del Portillo F, De Pedro M A. Differential effects of mutational impairment of penicillin-binding proteins 1A and 1B on Escherichia coli strains harboring thermosensitive mutations in the cell division genes ftsA, ftsQ, ftsZ, and pbpB. J Bacteriol. 1990;172:5863–5870. doi: 10.1128/jb.172.10.5863-5870.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghuysen J-M. Serine β-lactamases and penicillin-binding proteins. Annu Rev Microbiol. 1991;45:37–67. doi: 10.1146/annurev.mi.45.100191.000345. [DOI] [PubMed] [Google Scholar]
- 11.Ghuysen J-M, Dive G. Biochemistry of the penicilloyl serine transferases. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier Science B.V.; 1994. pp. 103–129. [Google Scholar]
- 12.Ghuysen J-M, Frere J-M, Leyh-Bouille M, Nguyen-Disteche M, Coyette J. Active-site-serine D-alanyl-D-alanine-cleaving-peptidase-catalyzed acyl-transfer reactions. Biochem J. 1986;235:159–165. doi: 10.1042/bj2350159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Granier B, Jamin M, Adam M, Galleni M, Lakaye B, Zorzi W, Grandchamps J, Wilkin J-M, Fraipont C, Joris B, Duez C, Nguyen-Disteche M, Coyette J, Leyh-Bouille M, Dusart J, Christiaens L, Frere J-M, Ghuysen J-M. Serine-type D-ala-D-ala peptidases and penicillin-binding proteins. Methods Enzymol. 1994;244:249–265. doi: 10.1016/0076-6879(94)44021-2. [DOI] [PubMed] [Google Scholar]
- 14.Hakenbeck R, Konig A, Kern I, Van Der Linden M, Keck W, Billot-Klein D, Legrand R, Schoot B, Gutmann L. Acquisition of five high-Mr penicillin-binding protein variants during transfer of high-level β-lactam resistance from Streptococcus mitis to Streptococcus pneumoniae. J Bacteriol. 1998;180:1831–1840. doi: 10.1128/jb.180.7.1831-1840.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hao J, Kendrick K E. Visualization of penicillin-binding proteins during sporulation of Streptomyces griseus. J Bacteriol. 1998;180:2125–2132. doi: 10.1128/jb.180.8.2125-2132.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holloway B W. Genetic recombination in Pseudomonas aeruginosa. J Gen Microbiol. 1955;13:572–581. doi: 10.1099/00221287-13-3-572. [DOI] [PubMed] [Google Scholar]
- 17.Jamin M, Damblon C, Miller S, Hakenbeck R, Frere J-M. Penicillin-binding protein 2x of Streptococcus pneumoniae: enzymic activities and interactions with β-lactams. Biochem J. 1993;292:735–741. doi: 10.1042/bj2920735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jamin M, Hakenbeck R, Frere J-M. Penicillin binding protein 2x protein as a major contributor to intrinsic β-lactam resistance of Streptococcus pneumoniae. FEBS Lett. 1993;331:101–104. doi: 10.1016/0014-5793(93)80305-e. [DOI] [PubMed] [Google Scholar]
- 19.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Lakaye B, Damblon C, Jamin M, Galleni M, Lepage S, Joris B, Marchand-Brynaert J, Frydrych C, Frere J-M. Synthesis, purification, and kinetic properties of fluorescein-labelled penicillins. Biochem J. 1994;300:141–145. doi: 10.1042/bj3000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liao X, Hancock R W. Identification of a penicillin-binding protein 3 homolog, PBP3x, in Pseudomonas aeruginosa: gene cloning and growth phase-dependent expression. J Bacteriol. 1997;179:1490–1496. doi: 10.1128/jb.179.5.1490-1496.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masson J M, Baron P, Michelot D, Labia R. Evaluation of 125I-radiolabelled penicillin-X in penicillin-binding protein studies. Drugs Exp Clin Res. 1983;9:821–825. [Google Scholar]
- 23.Matsuhashi M. Utilization of lipid-linked precursors and the formation of peptidoglycan in the process of cell growth and division: membrane enzymes involved in the final steps of peptidoglycan synthesis and the mechanism of their regulation. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier Science B.V.; 1994. pp. 55–72. [Google Scholar]
- 24.Munoz R T, Dowson C G, Daniels M, Coffey T J, Martin C, Hakenbeck R, Spratt B G. Genetics of resistance to third-generation cephalosporins in clinical isolates of Streptococcus pneumoniae. Mol Microbiol. 1992;6:2461–2465. doi: 10.1111/j.1365-2958.1992.tb01422.x. [DOI] [PubMed] [Google Scholar]
- 25.Neu H C. The crisis in antibiotic resistance. Science. 1992;257:1064–1073. doi: 10.1126/science.257.5073.1064. [DOI] [PubMed] [Google Scholar]
- 26.Noguchi H, Matsuhashi M, Mitsuhasi S. Comparative studies of penicillin-binding proteins in Pseudomonas aeruginosa and Escherichia coli. Eur J Biochem. 1979;100:41–49. doi: 10.1111/j.1432-1033.1979.tb02031.x. [DOI] [PubMed] [Google Scholar]
- 27.Preston D A, Wu C Y E, Blaszczak L C, Seitz D E, Halligan N G. Biological characterization of a new radioactive labeling reagent for bacterial penicillin-binding proteins. Antimicrob Agents Chemother. 1990;34:718–721. doi: 10.1128/aac.34.5.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spratt B G. Properties of the penicillin binding proteins of Escherichia coli K12. Eur J Biochem. 1977;72:341–352. doi: 10.1111/j.1432-1033.1977.tb11258.x. [DOI] [PubMed] [Google Scholar]
- 29.Spratt B G. Resistance to β-lactam antibiotics. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier Science B.V.; 1994. pp. 517–534. [Google Scholar]
- 30.Tomasz A, Munoz R. β-Lactam antibiotic resistance in Gram-positive bacterial pathogens of the upper respiratory tract: a brief overview of mechanisms. Microb Drug Resist. 1995;1:103–109. doi: 10.1089/mdr.1995.1.103. [DOI] [PubMed] [Google Scholar]
- 31.Van Heijenoort J. Murein synthesis. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W C, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 1025–1034. [Google Scholar]
- 32.Yousif S Y, Broome-Smith J K, Spratt B G. Lysis of Escherichia coli by β-lactam antibiotics: deletion analysis of the role of penicillin-binding proteins 1A and 1B. J Gen Microbiol. 1985;131:2839–2845. doi: 10.1099/00221287-131-10-2839. [DOI] [PubMed] [Google Scholar]
- 33.Zhao G, Yeh W-K, Carnahan R H, Flokowitsch J, Meier T I, Alborn W E, Jr, Becker G W, Jaskunas S R. Biochemical characterization of penicillin-resistant and -sensitive penicillin-binding protein 2x transpeptidase activities of Streptococcus pneumoniae and mechanistic implications in bacterial resistance to β-lactam antibiotics. J Bacteriol. 1997;179:4901–4908. doi: 10.1128/jb.179.15.4901-4908.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]