Abstract
Apocynum venetum L. (Apocynaceae) is valuable for its medicinal compounds and fiber content. Native A. venetum populations are threatened and require protection. Wild A. venetum resources are limited relative to market demand and a poor understanding of the composition of A. venetum at the molecular level. The chloroplast genome contains genetic markers for phylogenetic analysis, genetic diversity evaluation, and molecular identification. In this study, the entire genome of the A. venetum chloroplast was sequenced and analyzed. The A. venetum cp genome is 150,878 bp, with a pair of inverted repeat regions (IRA and IRB). Each inverted repeat region is 25,810 bp, which consist of large (LSC, 81,951 bp) and small (SSC, 17,307 bp) single copy areas. The genome-wide GC content was 38.35%, LSC made up 36.49%, SSC made up 32.41%, and IR made up 43.3%. The A. venetum chloroplast genome encodes 131 genes, including 86 protein-coding genes, eight ribosomal RNA genes, and 37 transfer RNA genes. This study identified the unique characteristics of the A. venetum chloroplast genome, which will help formulate effective conservation and management strategies as well as molecular identification approaches for this important medicinal plant.
Introduction
Apocynum venetum L. (Apocynaceae) (Luobuma in Chinese) is a perennial herb distributed in Eurasia from Southeast Europe to Northern China. It occurs in floodplains and valleys along rivers such as the Tarim River [1, 2]. The roots, stems, leaves, and flowers of A. venetum have medicinal uses [3, 4] and these uses were documented in the “Compendium of Materia Medica.” In 1977, A. venetum was listed in the Pharmacopoeia of the People’s Republic of China as a primary treatment for hypertension and hyperlipidemia [5–8], and pharmacological studies have demonstrated that A. venetum possesses many pharmacological activities including cardiotonic [9], hepatoprotective [10, 11], antioxidant [12–14], antidepressant and anxiolytic effects [15–18]. A. venetum maybe useful for the prevention and treatment of cardiovascular and neurological diseases such as high blood pressure, high cholesterol, neurasthenia, depression, and anxiety [19–23].
A. venetum has relatively high salt tolerance, cold tolerance, drought tolerance, high temperature tolerance, and wind resistance [24, 25]. It is an important plant for the wind proofing and sand-stabilization of desert grasslands in Central Asia. A. venetum therefore combines ecological benefits and economic benefits [24, 26]. Overharvesting of wild A. venetum and environmental degradation have reduced Apocynum populations and protection of Apocynum germplasm resources is needed. Studies of A. venetum have mainly focused on its medicinal effects and physiological characteristics such as photosynthesis and water absorption [27, 28]. However, there are few studies on the genetic diversity and genetic structure of wild A. venetum populations [29, 30].
Chloroplasts (cps) are the descendants of ancient bacteria endosymbionts. They are important organelles in plant cells that are responsible for photosynthesis and other aspects of metabolism [31]. Cp DNA is independent of the nuclear genome and exhibits semi-autonomous genetic characteristics. The characteristics of maternal and highly conserved genes in the cp genome are favorable for studying plant phylogeny [32, 33]. Molecular barcodes based on the cp genome have potential for species identification, especially among closely related taxa [34, 35]. The complete cp genome sequence may provide reliable barcodes for accurate plant identification at species and population levels [36, 37]. In higher plants, photosynthesis occurs in cp, which provides the necessary energy for plant growth and survival.
There are many counterfeit A. venetum products on the market, and they are difficult to detect based on appearance. There is a need for a molecular method to distinguish counterfeit products. DNA barcode sequence analysis is a molecular identification technology that uses standardized DNA sequence fragments to provide a fast, accurate, and automated species identification method [38–41]. The non-coding region of the cp has been successfully used in research on the DNA barcode. A. venetum cp genome information can provide candidate DNA barcodes for the identification of A. venetum and counterfeit products.
In this study, we assembled and analyzed the A. venetum cp genome sequence based on Illumina paired-end (PE) sequencing data. Through bioinformatics analysis, the sequence was compared with other known cp genome sequences. The information helped us determine the phylogeny of this species.
Materials and methods
Sampling, DNA extraction, sequencing, and assembly
A. venetum seeds were collected from wild plants in Shaya County in the Xinjiang Uygur Autonomous Region, China (40°92´N, 82°21´E; 957 m). After removal of the bracts, seeds were surface sterilized for 1 min in 75% ethanol (v/v), rinsed three times with distilled water, and then germinated at 25°C in the dark on filter paper dampened with distilled water. When the plumule emerged, uniform seedlings were transplanted into plugged holes in plastic containers (5 cm × 5 cm × 5 cm, 1 seedling/container) filled with vermiculite and watered with modified Hoagland nutrient solution containing 2 mM KNO3, 0.5 mM NH4H2PO4, 0.25 mM MgSO4·7H2O, 0.1 mM Ca(NO3)2·4H2O, 50 μM Fe-citrate, 92 μM H3BO3, 18 μM MnCl2·4H2O, 1.6 μM ZnSO4·7H2O, 0.6 μM CuSO4·5H2O and 0.7 μM (NH4)6Mo7O24·4H2O. Solutions were renewed every 3 d. Seedlings were grown in a greenhouse at a temperature of 28°C/23°C (day/night) and photoperiod of 16:8 h (light:dark). The flux density was approximately 800 μmol m−2 s−1) and the relative humidity was 65%. Fresh leaves were collected on October 18, 2019, frozen in liquid nitrogen and then stored at −80°C until analysis [42].
Genomic DNA was isolated by the modified CTAB method. Agarose gel electrophoresis and a one drop spectrophotometer (OD-1000, Shanghai, China) were used to detect DNA integrity and quality. One library (250 bp) was constructed using pure DNA according to the manufacturer’s instructions (NEBNext® UltraTM DNA Library Prep Kit for Illumina®). The library was constructed with an Illumina NovaSeq platform (Benagen Tech Solution Co. Ltd., Wuhan, China) and 150-bp paired-end reads were generated. The Illumina PCR adapter reads, low-quality reads and reads containing more than 5% unknown nucleotides “Ns” were filtered from the paired-end raw reads in the quality control step. All good-quality paired clean reads were obtained using SOAPnuke software (version: 1.3.0). The assembled reads were joined into a bidirectional iterative derivation using NOVOPlasty (version:3.13.1, parameter:k-mer = 127) to obtain the whole-genome sequence. The cp-like reads were used to assemble sequences using NOVOPlasty. NOVOPlasty assembled the partial reads and stretched as far as possible until a circular genome was formed. All circled sequences were searched by BLASTN (version: BLAST 2.2.30+, E-value ≤ 1e-5) against the reference database. Sequences with alignment greater than 1,000 bp and coverage greater than 90% were retained. Based on the depth of sequencing, PE reads alignment, and alignment with closely species to A. venetum, the candidate sequences were connected in order to determine whether they formed a loop. When a gap (including N sequence) appeared, Gapcloser (Version: 1.12) was used to fill in the hole to obtain the final splicing result [43]. After filtering the repeated sequences and the sequences with lengths less than 300 bp, 48 sequences with start codons of ATG, TTG, CTG, ATT, ATC, GTG, and ATA and end codons of TGA, TAG, and TAA, were retained to conduct subsequent analysis.
Annotation and analysis of the cpDNA sequences
The cp genome sequence was annotated using the DOGMA program (http://dogma.ccbb.utexas.edu/) [44], and the tRNAscan-SE program was used to predict tRNAs in the genome [45]. The circular maps were drawn by the OGDRAWv1.2 program [46] (http://ogdraw.mpimp-golm.mpg.de/). In order to eliminate the influence of amino acid composition on codon usage, the characteristics of the variations in synonymous codon usage, the relative synonymous codon usage values (RSCU), base composition and codon content were analyzed using MEGA 7.0. Simple sequence repeats (SSRs) in the cp genome were identified using SSRHunter software (http://www.biosoft.net) [47, 48]. The parameters were set to five repeat units for mononucleotide SSRs, five repeat units for dinucleotide SSRs, three repeat units for trinucleotide SSRs, and three repeat units each for tetranucleotides, and pentanucleotide SSRs.
Genome comparison
The pairwise alignments of cp genomes was conducted by MUMmer [42]. The mVISTA software was used to compere the A. venetum cp genome with three other cp genomes. Nicotiana attenuata, Gossypium hirsutum, and Arabidopsis thaliana (NC_035952.1, DQ345959, and NC_000932.1, respectively) using the annotation of Sophora japonica L. as reference [44, 45]. We determined the repeat structure, including forward and reverse repeats, using the REPuter software [46–49].
Phylogenetic analysis
We downloaded 21 cp genome sequences from the NCBI organelle genome and nucleotide resource database, and used all genomes for phylogenetic analysis. Clustalw2 software (Conway Institute of Biomolecular and Biomedicine, Dublin, Ireland) was used to sequence the genome [50–53]. We used MEGA7.0 to analyze and draw a phylogenetic tree with ML (maximum likelihood). Bootstrap analysis was performed using 1,000 repetitions and TBR branch exchanges [54–56]. We used 1,000 replicates and TBR branch exchanges to complete the bootstrap analysis.
Results
Features of A. venetum cpDNA
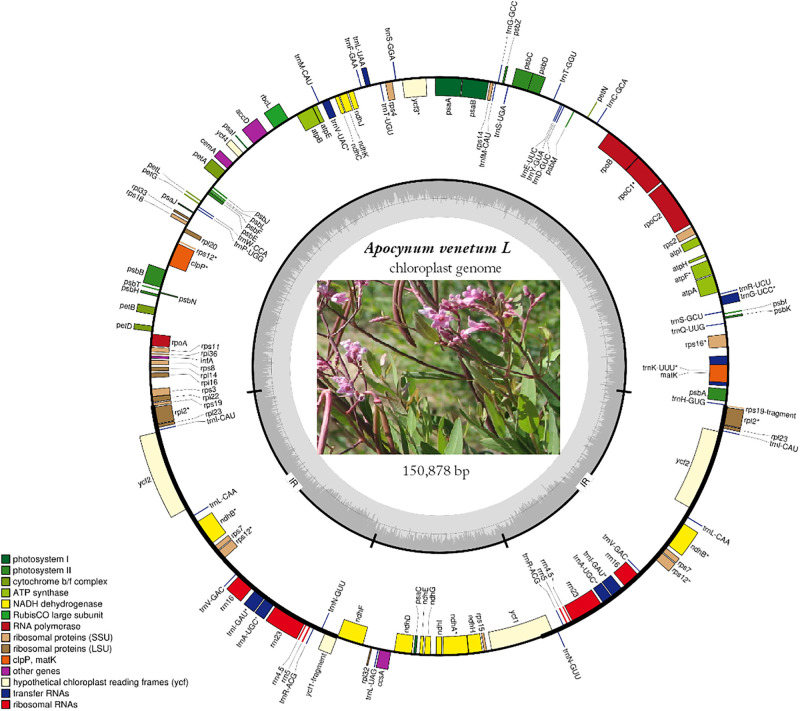
The complete cp genome of A. venetum is 150,878 bp in length (GenBank accession number: MT568765) (Fig 1), and includes a pair of inverted repeats (IR) 25,810 bp long, separated by a large single region (LSC) and a small copy region (SSC) of 81,951 bp and 17,307 bp, respectively (Table 1). It is similar to the cp genome of other Apocynaceae species [57]
Fig 1. Map of A. venetum cpgenome.
Thick lines indicate the extent of the inverted repeat regions (Ira and Irb), which separate the genome into small (SSC) and large (LSC) single copy regions. Genes drawn inside the circle are transcribed clockwise, and those outside are transcribed counterclockwise. Different colors represent different functional groups of genes.
Table 1. Base composition in the A. venetum chloroplast genome.
| Region | Length | A (%) | T (%) | C (%) | G (%) | AT (%) | GC (%) |
|---|---|---|---|---|---|---|---|
| Total_genome | 150878 | 30.43 | 31.21 | 19.52 | 18.83 | 61.64 | 38.35 |
| LSC | 81951 | 31.02 | 32.49 | 18.69 | 17.8 | 63.51 | 36.49 |
| IRA | 25810 | 28.59 | 28.11 | 20.84 | 22.46 | 56.7 | 43.3 |
| SSC | 17307 | 33.85 | 33.65 | 17.09 | 15.32 | 67.49 | 32.41 |
| IRB | 25810 | 28.11 | 28.59 | 22.46 | 20.84 | 56.7 | 43.3 |
In the A. venetum cp genome, 131 functional genes were predicted, including eight rRNA genes, 37 tRNA genes, and 86 protein-coding genes (Table 2) Cp genomes in the IR regions include 33 duplicated genes, with approximately 15 tRNA genes (tRNAs), eight rRNA genes (rRNAs), and nine protein-coding genes (PCGs) (Fig 1). The LSC region includes 58 protein-coding and 22 tRNA genes, while the SSC region includes one tRNA gene and 11 protein-coding genes.
Table 2. Genes present in the A. venetum chloroplast genome.
| Category for genes | Group of genes | Name of genes |
|---|---|---|
| Transcription and translation-related genes | transfer RNAs | trnM-CAU, trnR-ACG, trnY-GUA, trnG-UCC, trnL-UAG, trnI-GAU, trnW-CCA, trnR-UCU, trnQ-UUG, trnL-UAA, trnS-GGA, trnH-GUG, trnT-GGU, trnT-UGU, trnP-UGG, trnK-UUU, trnN-GUU, trnG-GCC, trnI-CAU, trnD-GUC, trnF-GAA, trnS-GCU, trnS-UGA, trnfM-CAU, trnE-UUC, trnV-GAC, trnA-UGC, trnV-UAC, trnL-CAA, trnC-GCA |
| RNA polymerase | rpoB, rpoA, rpoC1, rpoC2 | |
| ribosomal proteins(SSU) | rps8, rps4, rps16, rps14, rps7, rps12, rps2, rps11, rps19-fragment, rps19, rps18, rps3, rps15 | |
| ribosomal proteins(LSU) | rpl2, rpl23, rpl32, rpl33, rpl36, rpl14, rpl16, rpl22, rpl20 | |
| Translational initiation factor | infA | |
| ribosomal RNAs | rrn4.5, rrn5, rrn23, rrn16 | |
| Photosynthesis-related genes | NADH dehydrogenase | ndhA, ndhH, ndhF, ndhJ, ndhE, ndhI, ndhG, ndhK, ndhC, ndhD, ndhB |
| photosystem I | psaI, psaJ, psaC, psaB, psaA | |
| photosystem II | psbA, psbL, psbF, psbB, psbK, psbJ, psbM, psbT, psbE, psbD, psbC, psbH, psbI, psbN, psbZ | |
| cytochrome b/f complex | petL, petN, petB, petG, petA, petD | |
| RubisCO | rbcL | |
| ATP synthase | atpA, atpE, atpH, atpI, atpB, atpF | |
| hypothetical chloroplast reading frames(ycf) | ycf2, ycf4, ycf1, ycf3, ycf1-fragment | |
| Other genes | Maturase | matK |
| Protease | clpP | |
| Envelope membrane protein | cemA | |
| Subunit of Acetyl-CoA carboxylase | accD | |
| C-type cytochrome synthesis gene | ccsA |
The tRNA and protein-encoding gene sequences of the A. venetum cp were analyzed, and the codon usage frequency of the cp genome of A. venetum was inferred and summarized. A total of 17,318 codons represent the coding ability of 86 protein-coding genes and tRNA genes of A. venetum (Table 4), of which 1,814 codons code for leucine (10.47%), and 319 codons code for tryptophan (1.84%), which are the most common and least common amino acids in the cp genome of A. venetum, respectively. Codons ending in A and U are very common. Except for trtl-caa, all preferred synonymous codons (RSCU > 1) end in A or U. There are 14 intron-containing genes, including nine protein-coding genes and five tRNA genes (Table 3). Twelve genes (seven protein-coding and five tRNA genes) contain an intron, and two genes (ycf3 and clpP) contain two introns of the intragene region (Table 3). The size of the intron-containing matK gene in the trnK-UUU gene was 2,474 bp. The Rps12 gene is a trans-splicing gene with the 5’ end in the LSC region and the 3’ end in the IR region.
Table 4. Codon-anticodon recognition patterns and codon usage of the A. venetum chloroplast genome.
| Amino Acid | Codon | Number | RSCU* | tRNA | Amino Acid | Codon | Number | RSCU* | tRNA |
|---|---|---|---|---|---|---|---|---|---|
| Stop | UAA | 23 | 1.53 | Met | AUG | 401 | 1 | trnM-CAU | |
| Stop | UAG | 9 | 0.6 | Asn | AAU | 627 | 1.56 | ||
| Stop | UGA | 13 | 0.87 | Asn | AAC | 178 | 0.44 | ||
| Ala | GCU | 459 | 1.78 | Pro | CCU | 270 | 1.49 | ||
| Ala | GCC | 174 | 0.67 | Pro | CCC | 153 | 0.84 | ||
| Ala | GCA | 281 | 1.09 | Pro | CCA | 191 | 1.05 | trnP-UGG | |
| Ala | GCG | 118 | 0.46 | Pro | CCG | 111 | 0.61 | ||
| Cys | UGU | 128 | 1.44 | Gln | CAA | 495 | 1.54 | trnQ-UUG | |
| Cys | UGC | 50 | 0.56 | trnC-GCA | Gln | CAG | 148 | 0.46 | |
| Asp | GAU | 578 | 1.59 | Arg | CGU | 227 | 1.32 | trnR-ACG | |
| Asp | GAC | 147 | 0.41 | trnD-GUC | Arg | CGC | 78 | 0.45 | |
| Glu | GAA | 682 | 1.51 | trnE-UUC | Arg | CGA | 240 | 1.39 | |
| Glu | GAG | 220 | 0.49 | Arg | CGG | 90 | 0.52 | ||
| Phe | UUU | 611 | 1.28 | Arg | AGA | 285 | 1.65 | trnR-UCU | |
| Phe | UUC | 343 | 0.72 | trnF-GAA | Arg | AGG | 115 | 0.67 | |
| Gly | GGU | 412 | 1.28 | Ser | UCU | 366 | 1.67 | ||
| Gly | GGC | 144 | 0.45 | trnG-GCC | Ser | UCC | 223 | 1.02 | trnS-GGA |
| Gly | GGA | 474 | 1.48 | Ser | UCA | 246 | 1.12 | trnS-UGA | |
| Gly | GGG | 253 | 0.79 | Ser | UCG | 131 | 0.6 | ||
| His | CAU | 315 | 1.45 | Ser | AGU | 269 | 1.23 | ||
| His | CAC | 120 | 0.55 | trnH-GUG | Ser | AGC | 81 | 0.37 | trnS-GCU |
| Ile | AUU | 722 | 1.5 | Thr | ACU | 353 | 1.63 | ||
| Ile | AUC | 294 | 0.61 | trnI-CAU | Thr | ACC | 176 | 0.81 | trnT-GGU |
| Ile | AUA | 431 | 0.89 | Thr | ACA | 238 | 1.1 | trnT-UGU | |
| Lys | AAA | 594 | 1.49 | Thr | ACG | 98 | 0.45 | ||
| Lys | AAG | 204 | 0.51 | Val | GUU | 361 | 1.49 | ||
| Leu | UUA | 597 | 1.97 | Val | GUC | 114 | 0.47 | trnV-GAC | |
| Leu | UUG | 365 | 1.21 | trnL-CAA | Val | GUA | 360 | 1.48 | |
| Leu | CUU | 388 | 1.28 | Val | GUG | 135 | 0.56 | ||
| Leu | CUC | 107 | 0.35 | Trp | UGG | 319 | 1 | trnW-CCA | |
| Leu | CUA | 225 | 0.74 | Tyr | UAU | 512 | 1.64 | ||
| Leu | CUG | 132 | 0.44 | Tyr | UAC | 114 | 0.36 | trnY-GUA |
RSCU *: relative synonymous codon usage.
Table 3. Length of exons and introns in genes with introns in the A. venetum chloroplast genome.
| Gene | Location | Exon I (bp) | Intron I (bp) | Exon II (bp) | Intron II (bp) | Exon III (bp) |
|---|---|---|---|---|---|---|
| trnK-UUU | LSC | 35 | 2474 | 37 | ||
| rps16 | LSC | 226 | 837 | 41 | ||
| trnG-UCC | LSC | 23 | 672 | 48 | ||
| atpF | LSC | 411 | 706 | 144 | ||
| rpoC1 | LSC | 1599 | 748 | 451 | ||
| ycf3 | SSC | 155 | 794 | 226 | 717 | 126 |
| trnV-UAC | LSC | 37 | 588 | 36 | ||
| rps12 | LSC | 114 | 536 | 234 | ||
| clpP | LSC | 228 | 642 | 291 | 763 | 69 |
| rpl2 | IR | 434 | 649 | 391 | ||
| ndhB | IR | 756 | 685 | 777 | ||
| trnI-GAU | IR | 37 | 952 | 35 | ||
| trnA-UGC | IR | 38 | 817 | 35 | ||
| ndhA | SSC | 545 | 1039 | 553 |
Comparative analysis of genomic structure
Comparative genome analysis permits the examination of how DNA sequences diverge among related species. The whole cp genome sequence of A. venetum was compared to the sequences of N. attenuata, G. hirsutum, and A. thaliana. The identities of the entire sequence of the four cp genomes were drawn using the annotation mVISTA N. attenuata as a reference (Fig 2). The variation of the LSC and SSC regions were significantly greater than that of the IR regions. Moreover, the coding regions were more conserved than the non-coding regions. The most divergent coding regions of the four cp genomes were rnH-psbA, psbM-petN, trnC-GCA-petN, trnE-UUC-rpoB, trnY-GUA-trnE-UUC, trnV-UAC-ndhC, rbcL-accD, accD- psaI, LSC rpl32-trnL-UAG, and ndhI-ndhG ycf1-rps15 SSC, and the distribution of plastid rRNAs (rrn4.5, rrn5, rrn16, and rrn23) was the most conserved.
Fig 2. Comparison of the cpgenome sequences of four plants.
Comparison of the cp genome sequences of N attenuata, G. hirsutum, A, thaliana, and A. venetum generated with mVISTA. Gray arrows indicate the position and direction of each gene. Red and blue areas indicate the intergenic and genic regions, respectively. The vertical scale indicates the percentage of identity, ranging from 50% to 100%.
Repeat sequence analysis
We studied the type, existence, and distribution of SSR in the cp genome of A. venetum. A total of 273 SSRs were found in A. venetum, most of which were distributed in LSC and SSC, and some in IR. These included 105 single nucleotide SSRs (38.46%), 142 dinucleotide SSRs (50.01%), 10 trinucleotides, 14 tetranucleotides, and two pentanucleotide repeats. The mononucleotide A and T repeat units accounted for the largest portion.
Phylogenetic analysis
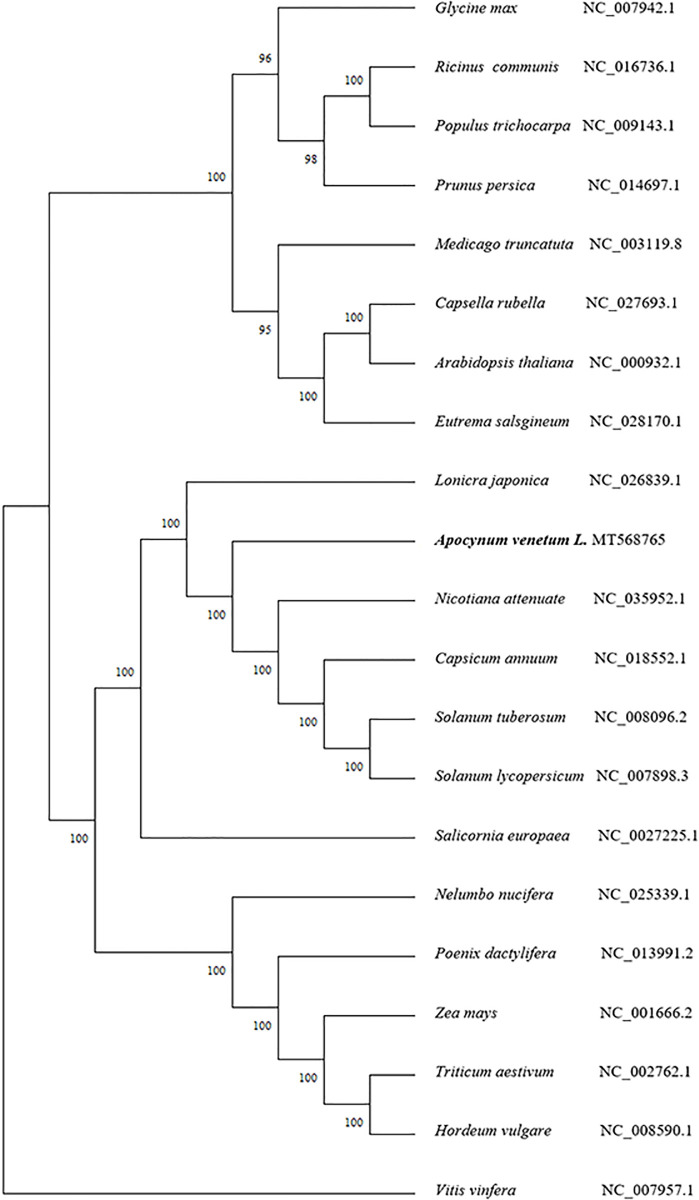
The cpDNA gene content is highly conserved in most land plants. We downloaded 21 complete cp genome sequences from the NCBI Organelle Genome Resources database to reveal the phylogenetic location of A. venetum (Fig 3). In this study, we constructed a phylogenetic tree to infer the phylogenetic positions of A. venetum cp genomes. The evolutionary tree was separated into four clusters. The phylogenetic tree showed that Vitis vinfera were clustered on a single terminal branch. Phylogeny analysis showed that Glycine max, Ricinus communis, Populus trichocarpa, Prunus persica, Medicago truncatuta, Capsella rubella, A. thaliana, and Eutrema salsgineum formed an independent branch. We found that A. venetum L. was grouped into a terminal branch with Lonicra japonica and N. atienuate, Capsicum annuum, Solanum tuberosum, Solanum lycopersicum and Salicornia europaea. Meanwhile, Nelumbo nucifera, Poenix dactylifera, Zea mays, Triticum aestivum, and Hordeum vulgare were clustered on a branch.
Fig 3. Phylogenetic tree analysis of whole chloroplast genome.
Maximum likelihood (ML) phylogenetic tree reconstruction including 21 species based on all chloroplast genomes. The bootstrap value, based on 1,000 replicates, is shown on each node. V. vinfera was used as the outgroup. The GenBank accession numbers are listed following the species name.
Discussion
In this study, we assembled, annotated and analyzed the complete cp sequence of A. venetum. We then analyzed its features, GC content, gene structure, and repeat sequences. The complete cp genome of A. venetum has a total length of 150,878 bp, with a pair of IRs of 25,810 bp that separate an LSC region of 81,951 bp and an SSC region of 17,307 bp. The DNA GC content of LSC, SSC, IR, and the whole genome were 36.49%, 32.41%, 43.3%, and 38.36%, respectively, which were similar to those of other species in Nerium. DNA GC content is an important index to evaluate the genetic relationship of Nerium oleracea, and the cpDNA GC content of Nerium indicum is similar to that in other species of Apocynaceae [58–63]. The content of DNA GC in the IR region is higher than that in other regions (LSC, SSC); this phenomenon is common in other plants [64–66]. The relatively high DNA GC content in the IR region was mainly attributed to the rRNA gene and the tRNA gene [67, 68].
Cp sequences have been used to compare the genetics of plant species, gene flow between species, and the size of ancestral populations of sister species [69]. Therefore, it is necessary to understand cp differences among species. We observed the order of approximately the same genes and the coding regions in the organization of the cp genome (Fig 2). The cp genome is considered to be highly conservative compared to the non-coding region, and the two infrared regions are less divergent than the LSC and SSC regions. The four cp genomes with the most different coding regions (rnH-psbA, psbM-petN, trnC-GCA-petN, trnE-UUC-rpoB, trnY-GUA-trnE-UUC, trnV-UAC-ndhC, rbcL-accD, accD-psaI, LSC rpl32-trnL-UAG, and ndhI-ndhG ycf1-rps15 SSC) and the four ribosomal RNA genes (rrn4.5, rrn5, rrn16, and rrn23) were the most conserved. Similar results have been observed in other plant cp genomes.
Cp genomes are highly conserved and contain a large amount of genetic information. The noncoding regions are less conserved than the coding regions [70, 71]. The genes trnK-UUU, rps16, trnG-UCC, atpF, rpoC1, trnV-UAC, rps12, rpl2, ndhB, trnI-GAU, trnA-UGC, and ndhA have one intron each, while clpP and ycf3 contain two introns. A trans-splicing event was also observed in the rps12 gene (Table 4). Previous studies have reported that ycf3 is necessary for the stable accumulation of photosystem I complexes [42, 72]. Therefore, we believe that the intron gain in ycf3 of A. venetum provides insight into the evolution of photosynthesis. As cp-specific SSRs are inherited from one parent and are mainly formed by the chain mismatch caused by the sliding of polymerase during DNA replication, they are often used in population genetics, species identification, and evolutionary process research on wild plants. In addition, the cp genome sequence is highly conserved, and SSR primers of cp genome can be transferred across species and genera. There were 273 SSRs detected in in the CP genome of A. venetum. Among these SSRs, mono-, di-, tri-, tetra-, and pentanucleotide were detected. The average density of SSRs was 1.809 SSR/kb in A. venetum (A/T as the main component). These cpSSR markers could be used for future studies of the genetic structure, diversity, and differentiation of A. venetum and its related species.
The phylogenetic positions of 21 cp genomes were successfully analyzed with the support of full bootstrap at almost all nodes. A phylogenetic tree was constructed for the data by ML, and V. vinfera was used as an outgroup. In this method, an initial tree is first built using a fast but suboptimal method such as the neighbor-joining method, and its branch lengths are adjusted to maximize the likelihood of the data set for that tree topology under the desired model of evolution. The results show that A. venetum has the closest relationship with L. japonica, N. attenuata, C. annuum, S. tuberosum, and S. lycopersicum.
Conclusion
We analyzed and illustrated the complete cp genome of A. venetum. The cp genome is conservative and similar to other species of Apocynum. These results provide a reference for the complete assembly of the cp genome of Apocynaceae, which may aid future breeding and research efforts. It may also assist in the development of unique Apocynaceae DNA barcodes of Apocynaceae and in determining the evolutionary history of Apocynaceae.
Supporting information
(ZIP)
(RAR)
(ZIP)
Acknowledgments
We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of manuscript.
Data Availability
All relevant data are within the paper and its Supporting information files.
Funding Statement
The National Natural Science Foundation of China (Grant No. 31760242), Gansu Provincial Natural Science Foundation (Grant No. 20JR10RA120), Gansu Provincial Natural Science Foundation the Ministry of Education of China for an Innovative Research Team in University (IRT 17R88), and the Fundamental Research Funds for the Central Universities (Grant No. 31920190021) The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Chen M, Zhao X-y, Zuo X-a. Comparative reproductive biology of Apocynum venetum L. in wild and managed populations in the arid region of NW China. Plant Syst Evol. 2015; 301(6): 1735–45. doi: 10.1007/S00606-014-1192-8 [DOI] [Google Scholar]
- 2.Dong Z-J. A new advanced textile fiber plant in China-Apocynum. Chin. Sci. Bull. 1957; 19: 607–8. [Google Scholar]
- 3.Peng X-M, Zhang W-M, Wang M-L, Lu C-M, Gu G-P. Molecular Identification of Apoacynum venetum and its Confusable Species. Bull Bot Res. 2007; (03): 302–307. [Google Scholar]
- 4.Su Q, Qiu L-Y. Study on the genetic diversity of Apocynum in Xinjiang based on RAPD technique. J Agric Catastrophol. 2015; 5(07): 1–17. [Google Scholar]
- 5.Xie W, Zhang X, Wang T, Hu J. Botany, traditional uses, phytochemistry and pharmacology of Apocynum venetum L. (Luobuma): A review. J Ethnopharmacol. 2012; 141(1): 1–8. doi: 10.1016/j.jep.2012.02.003 [DOI] [PubMed] [Google Scholar]
- 6.Kim DW, Yokozawa T, Hattori M, Kadota S, Namba T. Inhibitory effects of an aqueous extract of Apocynum venetum leaves and its constituents on Cu2+-induced oxidative modification of low density lipoprotein. Phtother Res. 2000; 14(7): 501–504. [DOI] [PubMed] [Google Scholar]
- 7.Yokozawa T, Nakagawa T. Inhibitory effects of Luobuma tea and its components against glucose-mediated protein damage. Food Chem. Toxicol. 2004; 42(6): 975–81. doi: 10.1016/j.fct.2004.02.010 [DOI] [PubMed] [Google Scholar]
- 8.Kim D-W, Yokozawa T, Hattori M, Kadota S, Namba T. Effects of aqueous extracts of Apocynum venetum leaves on spontaneously hypertensive, renal hypertensive and NaCl-fed-hypertensive rats. J Ethnopharmacol. 2000; 72(1): 53–59. doi: 10.1016/s0378-8741(00)00197-5 [DOI] [PubMed] [Google Scholar]
- 9.Irie K, Sato T, Tanaka I, Nakajima J-i, Kawaguchi M, Himi T. Cardiotonic effect of Apocynum venetum L. extracts on isolated guinea pig atrium. J Nat Med. 2009; 63(2): 111–116. doi: 10.1007/s11418-008-0296-2 [DOI] [PubMed] [Google Scholar]
- 10.Xiong Q, Fan W, Tezuka Y, Adnyana IK, Stampoulis P, Hattori M, et al. Hepatoprotective effect of Apocynum venetum and its active constituents. Planta Med. 2000; 66(2): 127–133. doi: 10.1055/s-2000-11135 [DOI] [PubMed] [Google Scholar]
- 11.Yang X-B, Wu X-Q, Yang J-R, Huang Z-M, Chen H-Y, Cao W-B, et al. Hepatoprotective effect of Apocynum venetum L extract on fatty liver disease of 2K1C rats with high-fat and refined-carbohydrate diet. World Chin. J.Digestol. 2009; 17(2). doi: 10.11569/WCJD.V17.I2.135 [DOI] [Google Scholar]
- 12.Chan C-O, Lau C-C, Ng Y-F, Xu L-J, Chen S-B, Chan S-W, et al. Discrimination between Leave of Apocynum venetum and Its Adulterant, A. pictum Based on Antioxidant Assay and Chemical Profiles Combined with Multivariate Statistical Analysis. Antioxidants. 2015; 4(2): 359–372. doi: 10.3390/antiox4020359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yokozawa T, Kashiwada Y, Hattori M, Chung HY. Study on the components of luobuma with peroxynitrite-scavenging activity. Biol Pharm Bull. 2002; 25(6): 748–52. doi: 10.1248/bpb.25.748 [DOI] [PubMed] [Google Scholar]
- 14.Liang T, Yue W, Li Q. Comparison of the Phenolic Content and Antioxidant Activities of Apocynum venetum L. (Luo-Bu-Ma) and Two of Its Alternative Species. IJMS. 2010; 11(11): 4452–4464. doi: 10.3390/ijms11114452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Butterweck V, Nishibe S, Sasaki T, Uchida M. Antidepressant Effects of Apocynum venetum Leaves in a Forced Swimming Test. Biol Pharm Bull. 2001; 24(7): 848–851. doi: 10.1248/bpb.24.848 [DOI] [PubMed] [Google Scholar]
- 16.Grundmann O, Nakajima J-I, Kamata K, Seo S, Butterweck V. Kaempferol from the leaves of Apocynum venetum possesses anxiolytic activities in the elevated plus maze test in mice. Phytomedicine. 2009; 16(4): 295–302. doi: 10.1016/j.phymed.2008.12.020 [DOI] [PubMed] [Google Scholar]
- 17.Butterweck V, Simbrey K, Seo S, Sasaki T, Nishibe S. Long-term effects of an Apocynum venetum extract on brain monoamine levels and β-AR density in rats. Pharmacol Biochem Behav. 2003; 75(3): 557–64. doi: 10.1016/s0091-3057(03)00118-7 [DOI] [PubMed] [Google Scholar]
- 18.Grundmann O, Nakajima J-I, Seo S, Butterweck V. Anti-anxiety effects of Apocynum venetum L. in the elevated plus maze test. J Ethnopharmacol. 2007; 110(3): 406–11. doi: 10.1016/j.jep.2006.09.035 [DOI] [PubMed] [Google Scholar]
- 19.Fan W, Tezuka Y, Xiong Q, Hattori M, Namba T, Kadota S. Apocynins A-D: New Phenylpropanoid-substituted Flavan-3-ols Isolated from Leaves of Apocynum venetum (Luobuma-Ye). Chem Pharm Bull. 1999; 47(7): 1049–50. doi: 10.1248/CPB.47.1049 [DOI] [Google Scholar]
- 20.Murakami T, Kishi A, Matsuda H, Hattori M, Yoshikawa M. Medicinal foodstuffs. XXIV. Chemical constituents of the processed leaves of Apocynum venetum L.: absolute stereostructures of apocynosides I and II. Chem Pharm Bull. 2001; 49(7): 845–8. doi: 10.1248/cpb.49.845 [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi M, Saitoh H, Seo S, Butterweck V, Nishibe S. Apocynum venetum extract does not induce CYP3A and P-glycoprotein in rats. Biol Pharm Bull. 2004;27(10):1649–52. doi: 10.1248/bpb.27.1649 [DOI] [PubMed] [Google Scholar]
- 22.Kwan C-Y, Zhang W-B, Nishibe S, Seo S. A novel in vitro endothelium-dependent vascular relaxant effect of Apocynum venetum leaf extract. Clin Exp Pharmacol Physiol. 2005; 32(9): 789–795. doi: 10.1111/j.1440-1681.2005.04255.x [DOI] [PubMed] [Google Scholar]
- 23.Kuo C-S, Kwan C-Y, Gong C-L, Tsai M-F, Nishibe S, Tatsuzaki J, et al. Apocynum venetum leaf aqueous extract inhibits voltage-gated sodium channels of mouse neuroblastoma N2A cells. J Ethnopharmacol. 2011; 136(1): 149–55. doi: 10.1016/j.jep.2011.04.035 [DOI] [PubMed] [Google Scholar]
- 24.Xu Z-C, Zhou J-H, Zhang C-S, Li Y-Q. Review of Current Research and Utilization Status of Apocynum venetum Germplasm in China. 2018, 53 (3): 382–390. [Google Scholar]
- 25.Xu Z, Zhou J, Ren T, Du H, Liu H, Li Y, et al. Salt stress decreases seedling growth and development but increases quercetin and kaempferol content in Apocynum venetum. Plant Biology. 2020; 22(5): 813–821. doi: 10.1111/plb.13128 [DOI] [PubMed] [Google Scholar]
- 26.Bai L, Luo MB, Chuan LC, Wang SL, Amina. Short report on planting techniques of Apocynum venetum. Chin Wild Plant Resour. 2005; 24: 65–68. [Google Scholar]
- 27.Chen M, Zhao X-Y, Zuo X-A, Lian J. Comparative pollination biology of Apocynum venetum at different desert landscapes. J Desert Res. 2016; (1): 124–30. [Google Scholar]
- 28.Wang DQ, Li GQ, Wang L. Daily dynamics of photosynthesis and water physiological characteristics of Apocynum venetum and A.cannabinum under drought stress. Acta Botan. Bor. Sin. 2012; 32: 1198–1205. [Google Scholar]
- 29.Liu ZH, Yu Z, Dong P, Zhou YX, Li XL. Genetic Diversity of Apocynum venetum Based on ISSR. Chin J Grassland. 2009; 31: 96–101. [Google Scholar]
- 30.Yuan N, Li M, Jia C. De novo transcriptome assembly and population genetic analyses of an important coastal shrub, Apocynum venetum L. BMC Plant Biology. 2020; 20(1): 1–15. doi: 10.1186/s12870-019-2170-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brunkard JO, Runkel AM, Zambryski PC. Chloroplasts extend stromules independently and in response to internal redox signals. Proc Natl Acad Sci. USA. 2015; 112(32): 10044–9. doi: 10.1073/pnas.1511570112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lilly JW, Havey MJ, Jackson SA, Jiang J. Cytogenomic Analyses Reveal the Structural Plasticity of the Chloroplast Genome in Higher Plants. Plant Cell. 2001; 13(2): 245–254. doi: 10.1105/tpc.13.2.245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang W, Lanfear R. Long-Reads Reveal That the Chloroplast Genome Exists in Two Distinct Versions in Most Plants. Genome Biol Evol. 2019; 11(12): 3372–3381. doi: 10.1093/gbe/evz256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amiryousefi A, Hyvönen JT, Poczai P. The chloroplast genome sequence of bittersweet (Solanum dulcamara): Plastid genome structure evolution in Solanaceae. PLoS ONE. 2018; 13(4). 1–23. doi: 10.1371/journal.pone.0196069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dong W, Xu C, Li C, Sun J, Zuo Y, Shi S, et al. ycf1, the most promising plastid DNA barcode of land plants. Sci Rep. 2015; 5(1): 8348-. doi: 10.1038/srep08348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neubig KM, Whitten WM, Carlsward BS, Blanco MA, Endara L, Williams NH, et al. Phylogenetic utility of ycf1 in orchids: a plastid gene more variable than matK. Plant Syst Evol. 2009; 277(1): 75–84. doi: 10.1007/S00606-008-0105-0 [DOI] [Google Scholar]
- 37.Hernández-León S, Gernandt DS, Rosa JAPdl, Jardón-Barbolla L. Phylogenetic relationships and species delimitation in pinus section trifoliae inferrred from plastid DNA. PLoS ONE. 2013; 8(7). doi: 10.1371/journal.pone.0070501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu C, Zhang W, Peng X, Gu G, Chen M, Tang Z. Development of randomly amplified polymorphic DNA-sequence characterized amplified region marker for identification of Apocynum venetum LINN. from A. pictum SCHRENK. Biol Pharm Bull. 2010; 33(3): 522–6. doi: 10.1248/bpb.33.522 [DOI] [PubMed] [Google Scholar]
- 39.Jh Yuan, Cheng FY, Zhou SL. Hybrid Origin of Paeonia × yananensis Revealed by Microsatellite Markers, Chloroplast Gene Sequences, and Morphological Characteristics. Int J Plant Sci. 2010; 171(4): 409–20. doi: 10.1086/651228 [DOI] [Google Scholar]
- 40.Zhao X, Zhou Z-Q, Lin Q-B, Pan K-Y, Li M-Y. Phylogenetic analysis of Paeonia sect. Moutan (Paeoniaceae) based on multiple DNA fragments and morphological data. J Syst Evol. 2007; 46(4): 563–572. doi: 10.3724/SP.J.1002.2008.06197 [DOI] [Google Scholar]
- 41.Zhang J-M, Liu J, Sun H-L, Yu J, Wang J-X, Zhou S-L. Nuclear and chloroplast SSR markers in Paeonia delavayi (Paeoniaceae) and cross-species amplification in P. ludlowii. Am J Bot. 2011; 98(12): 346–348. doi: 10.3732/AJB.1100240 [DOI] [PubMed] [Google Scholar]
- 42.Liu X, Chang E-M, Liu J-F, Huang Y-N, Wang Y, Yao N, et al. Complete Chloroplast Genome Sequence and Phylogenetic Analysis of Quercus bawanglingensis Huang, Li et Xing, a Vulnerable Oak Tree in China. Forests. 2019, 10, 587. doi: 10.3390/f10070587 [DOI] [Google Scholar]
- 43.Dierckxsens N., Mardulyn P. and Smits G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Research. 2016; doi: 10.1093/nar/gkw955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA, et al. VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics. 2000; 16(11): 1046–1047. doi: 10.1093/bioinformatics/16.11.1046 [DOI] [PubMed] [Google Scholar]
- 45.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004; 32: 273–279. doi: 10.1093/nar/gkh458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001; 29(22): 4633–4642. doi: 10.1093/nar/29.22.4633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen P, Gao G, Yu C, Chen J, Chen K, Zhu A. Data set for transcriptome analysis of Apocynum venetum L. Data Brief. 2018; 20: 1739–1744. doi: 10.1016/j.dib.2018.08.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Q, Wan JM. SSR Hunter: development of a local searching software for SSR sites. Yi Chuan. 2005; 27: 808–810. [PubMed] [Google Scholar]
- 49.Gao G, Chen P, Chen J, Chen K, Wang X, Abubakar AS, et al. Genomic Survey, Transcriptome, and Metabolome Analysis of Apocynum venetum and Apocynum hendersonii to Reveal Major Flavonoid Biosynthesis Pathways. Metabolites. 2019; 9(12). doi: 10.3390/METABO9120296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu J, Shen X, Liao B, Xu J, Hou D. Comparing and phylogenetic analysis chloroplast genome of three Achyranthes species. Sci Rep. 2020; 10(1): 10818. doi: 10.1038/s41598-020-67679-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007; 23(21): 2947–8. doi: 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 52.Plovanich M, Bogorad RL, Sancak Y, Kamer KJ, Strittmatter L, Li AA, et al. MICU2, a paralog of MICU1, resides within the mitochondrial uniporter complex to regulate calcium handling. PLoS ONE. 2013; 8(2): e55785. doi: 10.1371/journal.pone.0055785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu H-Y, Yu Y, Deng Y-Q, Li J, Huang Z-X, Zhou S-D. The Chloroplast Genome of Lilium henrici: Genome Structure and Comparative Analysis. Molecules. 2018; 23(6): 1276. doi: 10.3390/molecules23061276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen X, Guo S, Yin Y, Zhang J, Yin X, Liang C, et al. Complete Chloroplast Genome Sequence and Phylogenetic Analysis of Aster tataricus. Molecules. 2018; 23(10): 2426. doi: 10.3390/molecules23102426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li W, Liu Y, Yang Y, Xie X, Lu Y, Yang Z, et al. Interspecific chloroplast genome sequence diversity and genomic resources in Diospyros. BMC Plant Biol. 2018; 18(1): 210-. doi: 10.1186/s12870-018-1421-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016; 33(7): 1870. doi: 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chang L, Yu X, Wang W, Tian X. The complete chloroplast genome of Apocynum venetum (Apocynaceae). Mitochondrial DNA Part B. 2020; 5: 2601–2602. doi: 10.1080/23802359.2020.1781567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang D-L, Liu Y-Y, Tian D, Yu L-Y, Gui L-J. Characterization of the complete chloroplast genome of Plumeria rubra cv. Acutifolia (Apocynaceae). Mitochondrial DNA Part B. 2020; 5(1): 927–928. doi: 10.1080/23802359.2020.1721023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ali MA. Complete chloroplast genome of medicinally important poisonous shrub Adenium obesum (Forssk.) Roem. & Schult. (Apocynaceae). Mitochondrial DNA Part B. 2020; 5(1): 568–569. doi: 10.1080/23802359.2019.1710292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Park S, Ruhlman TA, Sabir JSM, Mutwakil MHZ, Baeshen MN, Sabir MJ, et al. Complete sequences of organelle genomes from the medicinal plant Rhazya stricta (Apocynaceae) and contrasting patterns of mitochondrial genome evolution across asterids. BMC Genomics. 2014; 15(1): 405-. doi: 10.1186/1471-2164-15-405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oliveira EJdS, Marques A, Almeida C. The chloroplast genome of Hancornia speciosa Gomes: structural organization and phylogenomic studies in Rauvolfioideae (Apocynaceae). BRAZ J BOT. 2019; 42(3): 449–455. doi: 10.1007/S40415-019-00549-8 [DOI] [Google Scholar]
- 62.Guan M, Zhang R. The complete chloroplast genome of Biondia insignis Tsiang (Apocynaceae). Mitochondrial DNA Part B. 2019; 4(1): 280–281. doi: 10.1080/23802359.2018.1541722 [DOI] [Google Scholar]
- 63.Jansen RK, Raubeson LA, Boore JL, dePamphilis CW, Chumley TW, Haberle RC, et al. Methods for Obtaining and Analyzing Whole Chloroplast Genome Sequences. Methods Enzymol. 2005; 395: 348–384. doi: 10.1016/S0076-6879(05)95020-9 [DOI] [PubMed] [Google Scholar]
- 64.Shao X. Progress in Chloroplast Genome Analysis. progress in biochemistry and biophysics. 2008. [Google Scholar]
- 65.Zhou SL, Zou XH, Zhou ZQ, Liu J, Xu C, Yu J, et al. Multiple species of wild tree peonies gave rise to the ‘king of flowers’, Paeonia suffruticosa Andrews. Proc. Biol. Sci. 2014; 281(1797). doi: 10.1098/RSPB.2014.1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shen X, Wu M, Liao B, Liu Z, Bai R, Xiao S, et al. Complete Chloroplast Genome Sequence and Phylogenetic Analysis of the Medicinal Plant Artemisia annua. Molecules. 2017; 22(8): 1330. doi: 10.3390/molecules22081330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.He Y, Xiao H, Deng C, Xiong L, Yang J, Peng C. The Complete Chloroplast Genome Sequences of the Medicinal Plant Pogostemon cablin. Int J Mol Sci. 2016;17(6): 820. doi: 10.3390/ijms17060820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Boudreau E, Takahashi Y, Lemieux C, Turmel M, Rochaix JD. The chloroplast ycf3 and ycf4 open reading frames of Chlamydomonas reinhardtii are required for the accumulation of the photosystem I complex. EMBO J 1997; 16(20): 6095–6104. doi: 10.1093/emboj/16.20.6095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li X, Li Y, Zang M, Li M, Fang Y. Complete Chloroplast Genome Sequence and Phylogenetic Analysis of Quercus acutissima. Int J Mol Sci. 2018; 19(8): 2443–2459. doi: 10.3390/ijms19082443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Souza UJBd, Nunes R, Targueta CP, Diniz-Filho JAF, Telles MPdC. The complete chloroplast genome of Stryphnodendron adstringens (Leguminosae—Caesalpinioideae): comparative analysis with related Mimosoid species. Sci Rep. 2019; 9(1): 14206–14206. doi: 10.1038/s41598-019-50620-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Naver H, Boudreau E, Rochaix J-D. Functional Studies of Ycf3: Its Role in Assembly of Photosystem I and Interactions with Some of Its Subunits. the Plant Cell. 2001; 13(12): 2731–2745. doi: 10.1105/tpc.010253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guo XN, Wang YL, Wang SM. Complete chloroplast genome sequences from yellowhorn (Xanthoceras sorbifolia) and evolution analysis based on codon usage bias. Intl J Agric Biol. 2020; 24: 676–684. [Google Scholar]