Abstract
Vibrio parahaemolyticus causes seafood-borne gastroenteritis in humans. It is particularly important in Japan, where raw seafood is frequently consumed. Fluoroquinolone is one of the current drugs of choice for treating patients infected by V. parahaemolyticus because resistant strains are rarely found. To study a possible fluoroquinolone resistance mechanism in this organism, nucleotide sequences that are homologous to known gyrA and parC genes have been cloned from V. parahaemolyticus AQ3815 and sequenced by amplification with degenerate primers of the quinolone resistance-determining region (QRDR), followed by cassette ligation-mediated PCR. Open reading frames encoding polypeptides of 878 and 761 amino acid residues were detected in the gyrA and parC homologues, respectively. The V. parahaemolyticus GyrA and ParC sequences were most closely related to Erwinia carotovora GyrA (76% identity) and Escherichia coli ParC (69% identity) sequences, respectively. Ciprofloxacin-resistant mutants of AQ3815 were obtained on an agar medium by multistep selection with increasing levels of the quinolone. One point mutation only in the gyrA QRDR was detected among mutants with low- to intermediate-level resistance, while point mutations in both the gyrA and parC QRDRs were detected only in strains with high-level resistance. These results strongly suggest that, as in other gram-negative bacteria, GyrA and ParC are the primary and secondary targets, respectively, of ciprofloxacin in V. parahaemolyticus.
Bacterial species that have developed clinical resistance to quinolones are increasing in numbers, and the mechanisms of their resistance have been studied. Most of the acquired resistance can be attributed to mutations in the genes encoding DNA gyrase or topoisomerase IV (Topo IV) (30).
DNA gyrase, a type II DNA topoisomerase, catalyzes ATP-dependent negative supercoiling of DNA and is involved in DNA replication, recombination, and transcription (43). The enzyme consists of GyrA and GyrB subunits, encoded by the gyrA and gyrB genes, respectively (1, 39, 46). The two genes are unlinked in Escherichia coli (39, 46) and map to 48 (gyrA) and 83 (gyrB) min on the chromosomal map (3) but are contiguous in Staphylococcus aureus (16). GyrA is considered to be responsible for DNA strand cleavage and rejoining (14, 43), and GyrB contains ATPase activity (41). Topo IV is a type II topoisomerase composed of ParC and ParE subunits, the amino acid sequences of which are homologous to some degree with those of GyrA and GyrB, respectively (20, 35). Topo IV has DNA decatenating and relaxing activities and plays an essential role in partitioning chromosomes at the terminal stage of chromosome replication (2). The majority of the quinolone resistance mutations in 17 bacterial species have been shown to map to a relatively small region at the N terminus of GyrA, corresponding to the 67th through the 106th amino acid residues in E. coli K-12; this region is called the quinolone resistance-determining region (QRDR) (30, 47). Quinolone-resistance mutations in the parC genes of E. coli, Klebsiella pneumoniae, Neisseria gonorrhoeae, S. aureus, and Streptococcus pneumoniae have been analyzed; the mutations were detected in the region corresponding to the QRDR of GyrA (30).
High-level resistance to quinolones appears to emerge in a stepwise fashion. E. coli and N. gonorrhoeae strains with low-level resistance to quinolones have a gyrA mutation alone, and strains with high-level resistance possess both gyrA and parC mutations (5, 15). Therefore, GyrA and ParC are considered to be the primary and secondary targets, respectively, of quinolones in these organisms. On the other hand, ParC (GrlA) and GyrA seem to be the primary and secondary targets, respectively, of ciprofloxacin in S. aureus (9, 10, 30).
Vibrio parahaemolyticus is a gram-negative marine bacterium. Strains producing a thermostable direct hemolysin or related hemolysins can cause gastroenteritis in humans who eat contaminated seafood (31, 32). This is of particular importance in Japan, where raw seafood is frequently consumed. V. parahaemolyticus is usually susceptible to tetracycline, chloramphenicol, gentamicin, and nalidixic acid but resistant to ampicillin, carbenicillin, and cephalothin (18, 19, 25, 28). New quinolones such as norfloxacin and enoxacin have potent inhibitory activity against V. parahaemolyticus (7, 29, 38). Quinolone-resistant strains have rarely been found among strains of V. parahaemolyticus isolated from the environment and clinical sources. Fluoroquinolone is one of the current drugs of choice for treating patients infected by this organism, because it is considered that this marine bacterium is not exposed to quinolones in its natural habitat. However, quinolone-resistant V. parahaemolyticus strains might emerge in the future if spontaneous mutation of the gyrA or parC gene, followed by selection of the mutant in the presence of quinolones, is the general mechanism of quinolone resistance. To examine this hypothesis, we first identified the gyrA and parC homologues in V. parahaemolyticus by cassette ligation-mediated PCR, in which well-conserved amino acid sequences of the QRDR of GyrA and the corresponding ParC region (hereinafter called the QRDR of ParC) are utilized. We then obtained quinolone-resistant mutants in vitro and examined whether the mutation can be detected in the gyrA or parC gene.
MATERIALS AND METHODS
Bacterial strains and growth medium.
A clinical strain of V. parahaemolyticus, AQ3815, and its ciprofloxacin (CIP)-resistant mutants (described below) were grown at 37°C with Luria-Bertani (LB) broth or agar (26) unless otherwise specified. AQ3815 was isolated at Osaka Quarantine Station, Osaka, Japan, in 1983 from a traveler with diarrhea arriving from the Philippines. This strain carried the tdh genes, encoding thermostable direct hemolysin, a major virulence factor of V. parahaemolyticus (31).
Nucleotide sequence determination of the gyrA and parC genes.
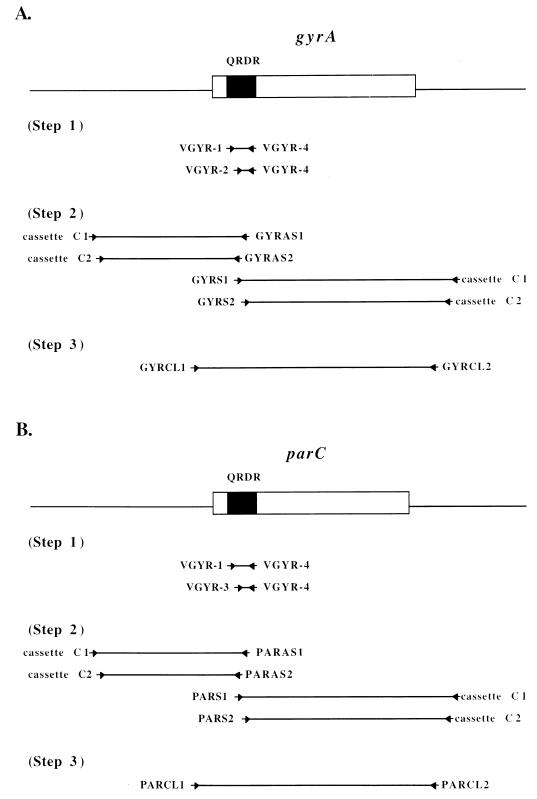
The nucleotide sequences of the gyrA and parC genes of V. parahaemolyticus AQ3815 were determined in three steps, as shown schematically in Fig. 1. The sequences of the QRDRs of the genes were determined first, and then their flanking sequences were determined by a walking method. Finally, the entire gene sequences were determined. In each step, the target sequence was first amplified by a PCR method, as described below. PCR products were then cloned into vector pGEM-T (Promega, Madison, Wis.) and sequenced with BcaBEST dideoxy sequencing kit (Takara) and [α-32P]dCTP or with the ABI-Prism Big Dye determinator cycle sequencing ready reaction kit and an ABI-Prism 377 sequencer (Perkin-Elmer, Applied Biosystems Division, Foster City, Calif.). The nucleotide sequences of both strands of the cloned sequence were determined. The sequences of the PCR primers are listed in Table 1.
FIG. 1.
PCR strategy used to determine the nucleotide sequences of the gyrA and parC genes. (A) gyrA gene (boxed) and flanking sequences (straight lines). (B) parC gene (boxed) and flanking sequences (straight lines). The distances between the primers are not drawn to scale. Arrows indicate locations and directions of PCR primers.
TABLE 1.
Oligonucleotide primers used in this study
| Primer | Orientation when used as primer | Positionsa | Base sequence |
|---|---|---|---|
| VGYR-1 | Sense | GyrA, 41–46; ParC, 43–48 | 5′-T(A/C/G/T)AA(A/G)CC(A/C/G/T)GT(A/C/G/T)CA(C/T)(A/C)G-3′ |
| VGYR-2 | Sense | GyrA, 57–62 | 5′-AA(C/T)GA(C/T)TGGAA(C/T)AA(A/G)(G/C)C-3′ |
| VGYR-3 | Sense | ParC, 78–83 | 5′-AA(A/G)TA(C/T)CA(C/T)CC(A/C/G/T)CA(C/T)G-3′ |
| VGYR-4 | Antisense | GyrA, 124–119; ParC, 127–122 | 5′-TC(A/C/G/T)GT(A/G)TA(A/C/G/T)C(G/T)CAT(A/C/G/T)GC-3′ |
| GYRAS1 | Antisense | 564–534 | 5′-CGGTGTAACGCATTGCCGCAGCAGAGTCGCC-3′ |
| GYRAS2 | Antisense | 395–365 | 5′-GGCTGATTTTTTATATGGTTTGTTCCAATCG-3′ |
| GYRS1 | Sense | 365–395 | 5′-CGATTGGAACAAACCATATAAAAAATCAGCC-3′ |
| GYRS2 | Sense | 534–564 | 5′-GGCGACTCTGCTGCGGCAATGCGTTACACCG-3′ |
| PARAS1 | Antisense | 465–434 | 5′-GGAGCACCCCAGTTTCCCTGACCATCAACCAG-3′ |
| PARAS2 | Antisense | 441–408 | 5′-TCAACCAGTGGGTAACGGTAAGAGAATGGTTGCG-3′ |
| PARS1 | Sense | 403–436 | 5′-AATGGCGCAACCATTCTCTTACCGTTACCCACTG-3′ |
| PARS2 | Sense | 434–465 | 5′-CTGGTTGATGGTCAGGGAAACTGGGGTGCTCC-3′ |
| GYRCL1 | Sense | 11–42 | 5′-AAATATAGTCAGTTTCAGAAAGATATGGGATG-3′ |
| GYRCL2 | Antisense | 2895–2864 | 5′-TGATGAAGATACAAAAAAGACGAGAATTTTGC-3′ |
| PARCL1 | Sense | 11–40 | 5′-ATCGACGACTCTGAAGCCACTATGGAAATG-3′ |
| PARCL2 | Antisense | 2523–2495 | 5′-GTTGGTATCATTCGTAAAGGCAAGGAATG-3′ |
| GYRM1 | Sense | 365–386 | 5′-CGATTGGAACAAACCATATAAA-3′ |
| GYRM2 | Antisense | 564–545 | 5′-CGGTGTAACGCATTGCCGCA-3′ |
| PARM1 | Sense | 293–312 | 5′-CTTGGTCTTTCGGCATCAGC-3′ |
| PARM2 | Antisense | 506–487 | 5′-CTTCGGTATAACGCATTGCC-3′ |
| Cassette C1 | 5′-GTACATATTGTCGTTAGAACGCGTAATACGACTCA-3′ | ||
| Cassette C2 | 5′-CGTTAGAACGCGTAATACGACTCACTATAGGGAGA-3′ |
The positions of VGYR-1, VGYR-2, VGYR-3, and VGYR-4 degenerate primers correspond to amino acid residues of E. coli GyrA and ParC (Fig. 2 and Fig. 3). The positions of other primers correspond to base positions of the V. parahaemolyticus gyrA and parC genes deposited in GenBank (accession no. AB023569 and AB023570, respectively); primer designations starting with GYR and PAR indicate primers for the gyrA and parC genes, respectively.
To determine the QRDR sequence of the gyrA gene, the sequence was amplified by a semi-nested PCR method (Fig. 1A, step 1). The nucleotide sequences of the degenerate primers used in PCR were deduced from the amino acid sequences of highly conserved regions of GyrA of Pseudomonas aeruginosa, S. aureus, Aeromonas salmonicida, E. coli, and Campylobacter jejuni (22, 24, 33, 39, 44). The first PCR was carried out in a 50-μl reaction mix containing 200 μM (each) deoxynucleoside triphosphates, 10 ng of total DNA of AQ3815, 2 μl of each primer (VGYR-1 and -4), 5 μl of GeneTaq buffer (Wako, Osaka, Japan), and 3 U of GeneTaq polymerase (Wako). Thermocycling was one cycle of 95°C for 5 min, followed by three cycles of 95°C for 30 s, 37°C for 30 s, and 72°C for 2 min, and then 30 cycles of 95°C for 30 s, 45°C for 30 s, and 72°C for 2 min. The second PCR was carried out in a 50-μl reaction mix containing 200 μM (each) deoxynucleoside triphosphates, 2 μl of the first PCR product, 2 μl of each primer (VGYR-2 and -4), 5 μl of GeneTaq buffer, and 3 U of GeneTaq polymerase. Thermocycling was one cycle of 95°C for 5 min, followed by three cycles of 95°C for 30 s, 37°C for 30 s, and 72°C for 2 min, and then 30 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 2 min. A DNA fragment of ca. 0.2 kb was produced. This fragment was cloned and the nucleotide sequence was determined.
The upstream and downstream sequences of the gyrA QRDR were amplified by cassette ligation-mediated PCR (step 2 of Fig. 1A). This PCR method allows amplification of nucleotide sequences from the DNA region for which the nucleotide sequence is known (17). AQ3815 total DNA was digested separately with SalI, PstI, HindIII, XbaI, and EcoRI. Each enzyme digest was ligated with the double-stranded ligation cassette containing the respective sticky end for ligation and the nucleotide sequence complementary to two PCR primers (cassette C1 and cassette C2). The ligation products were used as templates for PCR. One PCR primer pair was cassette C1 or cassette C2, the 5′ end of which was not phosphorylated. The ligation cassettes, cassette C1, and cassette C2 were included in an LA PCR in vitro cloning kit purchased from Takara, Shiga, Japan. The other PCR primer was designed so that it was complementary to a part of the gyrA QRDR sequence. Specific amplification is expected to result between the cassette primer and the gyrA QRDR-complementary primer because of the nonphosphorylation of the 5′ end of the cassette primer (17). Cassette ligation-mediated PCR was carried out in a nested fashion (Fig. 1A, step 2). The first PCR was performed in a 50-μl reaction mix containing 400 μM (each) deoxynucleoside triphosphates, 500 ng of template DNA (each of the ligation products), 0.2 μM (each) primers (cassette C1 and GYRAS1 for the QRDR upstream, cassette C1 and GYRS1 for the QRDR downstream), 5 μl of Takara LA PCR buffer II (Mg2+) (Takara), and 2.5 U of Takara LA Taq polymerase (Takara). Takara LA Taq polymerase was employed to achieve long-range and high-proofreading PCR. Thermocycling was 30 cycles of 94°C for 30 s, 55°C for 2 min, and 72°C for 1 min. The second PCR was performed in a 50-μl reaction mix containing 400 μM (each) deoxynucleoside triphosphates, 2 μl of the first PCR product, 0.2 μM (each) primers (cassette C2 and GYRAS2 for the QRDR upstream, cassette C2 and GYRS2 for the QRDR downstream), 5 μl of Takara LA PCR buffer II (Mg2+), and 2.5 U of Takara LA Taq polymerase. Thermocycling was 30 cycles of 94°C for 30 s, 63°C for 2 min, and 72°C for 1 min. Each restriction enzyme digest of AQ3815 DNA yielded a PCR product of a certain size. PstI digest resulted in a 2-kb PCR product for the gyrA QRDR upstream sequence. HindIII digest resulted in a 2.4-kb PCR product for the gyrA QRDR downstream sequence. These PCR products were judged to be of reasonable sizes and thus were cloned and the nucleotide sequences were determined.
The determined upstream and downstream sequences of the gyrA gene allowed design of the PCR primers, GYRCL1 and GYRCL2, for amplification of the sequence containing the complete gyrA gene (Fig. 1A, step 3). PCR was carried out in a 50-μl reaction mix containing 400 μM (each) deoxynucleoside triphosphates, 50 ng of AQ3815 total DNA, 0.2 μM (each) primers, 5 μl of Takara LA PCR buffer II (Mg2+), and 2.5 U of Takara LA Taq polymerase. Thermocycling was 30 cycles of 94°C for 30 s, 55°C for 2 min, and 72°C for 1 min. An amplicon of 3 kb was cloned and the nucleotide sequence was determined.
The nucleotide sequence of the parC gene was determined by the same method as for the gyrA sequence determination, with minor modifications (Fig. 1B). The modifications were as follows. To determine the QRDR sequence of the parC gene, the nucleotide sequences of the degenerate primers used in PCR were deduced from the amino acid sequences of highly conserved regions of ParC of E. coli, S. aureus, and N. gonorrhoeae (5, 11, 20). To amplify the QRDR sequence, PCR primer VGYR-3 was used in place of VGYR-2 in the second PCR (Fig. 1B, step 1). This resulted in an amplicon of ca. 0.15 kb. To amplify the upstream and downstream sequences of the QRDR, PCR primers PARAS1, PARAS2, PARS1, and PARS2 were used in place of GYRAS1, GYRAS2, GYRS1, and GYRS2, respectively (Fig. 1B, step 2). An annealing temperature of 58°C was employed in place of 63°C in the second of the nested PCRs. As a result, PstI digest of AQ3815 DNA yielded a 1.4-kb PCR product for the parC QRDR upstream sequence and SalI digest yielded a 2.1-kb PCR product for the parC QRDR downstream sequence. These fragments were judged to be of reasonable sizes and thus were cloned, and the nucleotide sequences were determined. To amplify the sequence containing the complete parC gene, PCR primers PARCL1 and PARCL2 were used in place of GYRCL1 and GYRCL2 (Fig. 1B, step 3). This resulted in an amplicon of 2.5 kb, and this amplicon was cloned and the nucleotide sequence was determined.
To compare the QRDRs of AQ3815 and its CIP-resistant mutants, the QRDR sequences of the gyrA and parC genes of these strains were amplified as follows. PCR was carried out in a 50-μl reaction mix containing 200 μM (each) deoxynucleoside triphosphates, 30 μl of 10-fold-diluted boiled overnight culture, 0.2 μM (each) primers (GYRM1 and GYRM2 for the gyrA QRDR, PARM1 and PARM2 for the parC QRDR), 5 μl of GeneTaq buffer, and 2.5 U of GeneTaq polymerase. Thermocycling was 30 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 2 min. PCR was expected to yield amplicons of 200 and 214 bp for the gyrA QRDR and the parC QRDR, respectively. These amplicons were cloned and the nucleotide sequences were determined.
MIC.
MICs were determined by the twofold agar dilution method recommended by the Japan Society of Chemotherapy. The test strain was grown in LB broth to mid-logarithmic phase at 37°C. The concentration of the organism was then adjusted to 106 CFU/ml, and 5 μl of the culture was inoculated onto LB agar. The MIC was determined after an 18-h incubation at 37°C.
Antibiotics.
The following antibiotics were purchased from the indicated manufacturers: CIP, Bayer (Osaka, Japan); enoxacin, Dainippon Pharmaceutical Co. (Osaka, Japan); nadifloxacin, Otsuka Pharmaceutical Co. (Tokyo, Japan); nalidixic acid, Dainippon Pharmaceutical Co.; norfloxacin, Kyorin Pharmaceutical Co. (Tokyo, Japan); ofloxacin, Daiichi Pharmaceutical Co., Ltd. (Tokyo, Japan); and sparfloxacin, Dainippon Pharmaceutical Co.
In vitro selection of CIP-resistant mutants.
CIP-resistant mutants of AQ3815 were obtained by plating the test strain onto LB agar containing CIP. A mutant resistant to CIP at 0.78 μg/ml was selected first, and mutants resistant to higher concentrations of CIP (3.13, 25, and 50 μg/ml) were obtained from the mutant step by step (explained below).
Nucleotide sequence accession numbers.
The nucleotide sequences of the gyrA and parC genes will appear in the GenBank nucleotide sequence database under accession no. AB023569 and AB023570, respectively.
RESULTS
Nucleotide sequences of the gyrA and parC genes.
The nucleotide sequences of the gyrA and parC genes of V. parahaemolyticus AQ3815 were determined by PCR amplification, cloning, and determination of their sequences in three steps, as illustrated in Fig. 1 and described in Materials and Methods.
A 203-bp PCR product, for the gyrA QRDR, and a 146-bp PCR product, for the parC QRDR, were amplified (Fig. 1, steps 1). To confirm that the two fragments resulted from amplification of the gyrA gene and the parC gene, respectively, the amplified fragments were sequenced and the deduced amino acid sequences were compared with GyrA and ParC of other bacterial species so far reported. The deduced amino acid sequence of the 203-bp fragment had high identity with the corresponding sequences of GyrA of E. coli (100%), Haemophilus influenzae (100%), K. pneumoniae (97.5%), and Erwinia carotovora (97.5%) (9, 12, 36, 39). The deduced amino acid sequence of the 146-bp fragment showed high identity with the corresponding sequences of ParC of E. coli (100%), Salmonella typhimurium (100%), H. influenzae (97.5%), and N. gonorrhoeae (75%) (5, 12, 20, 23).
The upstream and downstream sequences of the QRDR were determined in the second step, and a stretch of the sequence containing the complete gene was amplified and sequenced in the final step. There were no discrepancies among the sequences determined in the three steps. The gyrA gene had a predicted open reading frame (ORF) encoding 878 amino acids, preceded by putative promoter and ribosome binding sequences (data not shown). The amino acid sequence encoded by this ORF was homologous with that of E. coli GyrA not only in the QRDR but also in the other regions (Fig. 2). The amino acid sequence identities of this ORF with GyrA and ParC proteins of other bacterial species were compared. So far, complete sequences of both GyrA and ParC have been reported for seven species (4–6, 8, 9, 11–13, 20–24, 27, 33, 34, 36, 37, 40, 42, 45). When the identities of the ORF with the sequences of the seven species were compared, the ORF had significantly higher sequence identity with GyrA proteins (56.3% ± 12.4%) than with ParC proteins (38.3% ± 2.1%) (P < 0.001). Gram-negative bacteria exhibited higher identities for GyrA than did gram-positive bacteria (data not shown); E. carotovora GyrA was most closely related (76% identity). Based on these results and the results of mutational analysis (described below), we identified this ORF as the gyrA coding region of V. parahaemolyticus.
FIG. 2.
Comparison by alignment of V. parahaemolyticus (upper) and E. coli (lower) GyrA amino acid sequences. Residue numbers are given for the sequence of V. parahaemolyticus. An asterisk indicates absence of a residue. Identical residues are indicated by dashes. Arrows above the amino acid sequences correspond to the orientations and positions of VGYR-1, VGYR-2, and VGYR-4 degenerated primers (Table 1) used to amplify the QRDR of V. parahaemolyticus GyrA.
The parC gene sequence contained a predicted ORF encoding 761 amino acids, the sequence of which was homologous with that of E. coli ParC (Fig. 3). The ORF sequence was preceded by putative promoter and ribosome binding sequences (data not shown). The amino acid sequence identities of this ORF with ParC and GyrA proteins of other bacterial species were compared. This ORF sequence was significantly more homologous with the ParC sequences (49.4% ± 15.7% identity) than with the GyrA sequences (37.2% ± 1.7% identity) of the seven species for which both GyrA and ParC sequences are known (P < 0.005). Gram-negative bacteria exhibited higher identities for ParC than did gram-positive bacteria (data not shown); E. coli ParC was most closely related (69% identity). These results and the results of mutational analysis (described below) permitted us to identify this ORF as the parC coding region of V. parahaemolyticus.
FIG. 3.
Comparison by alignment of V. parahaemolyticus (upper) and E. coli (lower) ParC amino acid sequences. Residue numbers are given for the sequence of V. parahaemolyticus. An asterisk indicates absence of a residue. Identical residues are indicated by dashes. Arrows above the amino acid sequences correspond to the orientations and positions of VGYR-1, VGYR-3, and VGYR-4 degenerated primers (Table 1) used to amplify the QRDR of V. parahaemolyticus ParC.
Isolation and characterization of CIP-resistant mutants.
Mutants of AQ3815 that are resistant to CIP at various concentrations were isolated in a stepwise manner, as shown in Table 2. These mutants were also resistant to various other quinolone antibacterials (Table 2).
TABLE 2.
Isolation of CIP-resistant mutants of V. parahaemolyticus AQ3815 and their susceptibilities to various quinolone antibacterials
| Strain | Selection step | Concn (μg/ml) of CIP used for isolation | Isolation frequency | MIC (μg/ml)a
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CIP | ENX | NAD | NOR | OFX | SPX | NAL | ||||
| AQ3815b | nac | na | na | 0.20 | 0.39 | 0.39 | 0.20 | 0.39 | 0.39 | 0.39 |
| VP-M1 | 1st | 0.78 | 6 × 10−8 | 0.78 | 3.13 | 3.13 | 0.78 | 0.78 | 0.39 | 12.5 |
| VP-M2 | 2nd | 3.13 | 4 × 10−7 | 6.25 | 25 | 6.25 | 25 | 6.25 | 3.13 | 50 |
| VP-M3 | 3rd | 25 | 6 × 10−8 | 50 | 100 | 25 | 100 | 50 | 12.5 | 100 |
| VP-M4 | 4th | 50 | 1 × 10−7 | 200 | 100 | 100 | >200 | 200 | 50 | 100 |
ENX, enoxacin; NAD, nadifloxacin; NOR, norfloxacin; OFX, ofloxacin; SPX, sparfloxacin; NAL, nalidixic acid.
Wild-type strain.
Not applicable.
The QRDRs of the gyrA and parC genes of AQ3815 and the CIP-resistant mutants were amplified by single-step PCR with specific primers (Table 1). The amplified sequences were cloned, and the nucleotide sequences were determined and compared. Mutations detected within the amplified regions of the mutant strains are summarized in Table 3. One base change in the gyrA sequence responsible for a Ser-to-Ile change at residue position 83 was detected in all four mutant strains. In addition, one base change in the parC gene causing a Ser-to-Phe change at residue position 85 was detected in two of the mutant strains that were resistant to high concentrations of quinolones (for VP-M3 and -M4, MIC of CIP was ≧50 μg/ml [Tables 2 and 3]).
TABLE 3.
Mutations detected in the gyrA and parC sequences of CIP-resistant mutants of V. parahaemolyticus AQ3815
| Strain | MIC of CIP (μg/ml) | Mutation in:
|
|||||
|---|---|---|---|---|---|---|---|
|
gyrA
|
parC
|
||||||
| Amino acid residue
|
Base change | Amino acid residue
|
Base change | ||||
| Positiona | Mutation | Positiona | Mutation | ||||
| VP-M1 | 0.78 | 83 | Ser→Ile | AGG→ATT | —b | — | — |
| VP-M2 | 6.25 | 83 | Ser→Ile | AGG→ATT | — | — | — |
| VP-M3 | 50 | 83 | Ser→Ile | AGG→ATT | 85 | Ser→Phe | TCT→TTT |
| VP-M4 | 200 | 83 | Ser→Ile | AGG→ATT | 85 | Ser→Phe | TCT→TTT |
Residue numbers counted from the N-terminal residue.
—, no change.
DISCUSSION
This study demonstrated that V. parahaemolyticus carries nucleotide sequences highly homologous to the gyrA and parC genes of other bacterial species and that these sequences are mainly involved in experimentally induced resistance to quinolone antibacterials. Based on these results, we designated these sequences the gyrA and parC genes of V. parahaemolyticus. The approach that we used to determine the nucleotide sequences of these genes is based on conservation of the QRDR sequences and thus would be applicable to sequence determinations of the gyrA and parC homologues in many bacterial species.
Mutant strains of AQ3815 that became resistant to CIP had a base substitution at residue position 83 of GyrA or substitutions at both position 83 of GyrA and position 85 of ParC (Table 3). Mutations in the corresponding residues of GyrA and ParC in other bacterial species were shown to be associated with quinolone resistance (30). The stepwise appearance of a gyrA mutation followed by a parC mutation in AQ3815 derivatives with increasing CIP concentrations suggests that DNA gyrase and Topo IV may be the primary and secondary targets of CIP, respectively, in V. parahaemolyticus. Similar observations were made for other gram-negative bacteria, such as E. coli and N. gonorrhoeae (5, 15). Since no mutations other than the above were found in the gyrA and parC QRDRs of the AQ3815-derived mutants, increases in the level of resistance from VP-M1 to -M2 and from VP-M3 to -M4 must be explained by a mutation(s) in non-QRDRs. Mutations in the gyrB gene or those that can cause decreased drug permeability or increased drug efflux are possible explanations (30). These possibilities need to be examined in a future study. In any event, the use of quinolones should be avoided as much as possible to prevent the emergence and spread of quinolone-resistant strains of V. parahaemolyticus in the environment.
ACKNOWLEDGMENTS
This research was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture, Japan.
REFERENCES
- 1.Adachi T, Mizuuchi M, Robinson E, Appella E, O’Dea M, Gellert M, Mizuuchi K. DNA sequence of the E. coli gyrB gene: application of a new sequencing strategy. Nucleic Acids Res. 1987;15:771–783. doi: 10.1093/nar/15.2.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams D E, Shekhtman E M, Zechiedrich E L, Schmid M B, Cozzarelli N R. The role of topoisomerase IV in partitioning bacterial replicons and the structure of catenated intermediates in DNA replication. Cell. 1992;71:277–288. doi: 10.1016/0092-8674(92)90356-h. [DOI] [PubMed] [Google Scholar]
- 3.Bachmann B J. Linkage map of Escherichia coli K-12, edition 8. Microbiol Rev. 1990;54:130–197. doi: 10.1128/mr.54.2.130-197.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balas D, Fernandez-Moreira E, De La Campa A G. Molecular characterization of the gene encoding the DNA gyrase A subunit of Streptococcus pneumoniae. J Bacteriol. 1998;180:2854–2861. doi: 10.1128/jb.180.11.2854-2861.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belland R, Morrison S G, Ison C, Huang W M. Neisseria gonorrhoeae acquires mutations in analogous regions of gyrA and parC in fluoroquinolone-resistant isolates. Mol Microbiol. 1994;14:371–380. doi: 10.1111/j.1365-2958.1994.tb01297.x. [DOI] [PubMed] [Google Scholar]
- 6.Calcutt M J. Gene organization in the dnaA-gyrA region of the Streptomyces coelicolor chromosome. Gene. 1994;151:23–28. doi: 10.1016/0378-1119(94)90628-9. [DOI] [PubMed] [Google Scholar]
- 7.Carlson J R, Thornton S A, DuPont H L, West A H, Mathewson J J. Comparative in vitro activities of ten antimicrobial agents against bacterial enteropathogens. Antimicrob Agents Chemother. 1983;24:509–513. doi: 10.1128/aac.24.4.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deguchi T, Fukuoka A, Yasuda M, Nakano M, Ozeki S, Kanematsu E, Nishino Y, Ishihara S, Ban Y, Kawada Y. Alterations in the GyrA subunit of DNA gyrase and the ParC subunit of topoisomerase IV in quinolone-resistant clinical isolates of Klebsiella pneumoniae. Antimicrob Agents Chemother. 1997;41:699–701. doi: 10.1128/aac.41.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dimri G P, Das H K. Cloning and sequence analysis of the gyrA gene of Klebsiella pneumoniae. Nucleic Acids Res. 1990;18:151–156. doi: 10.1093/nar/18.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrero L, Cameron B, Crouzet J. Analysis of gyrA and grlA mutations in step-wise-selected ciprofloxacin-resistant mutants of Staphylococcus aureus. Antimicrob Agents Chemother. 1995;39:1554–1558. doi: 10.1128/aac.39.7.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferrero L, Cameron B, Manse B, Lagneaux D, Crouzet J, Famechon A, Blanche F. Cloning and primary structure of Staphylococcus aureus DNA topoisomerase IV: a primary target of fluoroquinolones. Mol Microbiol. 1994;13:641–653. doi: 10.1111/j.1365-2958.1994.tb00458.x. [DOI] [PubMed] [Google Scholar]
- 12.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J, Dougherty B A, Merrick J M, McKenney K, Sutton G G, FitzHugh W, Fields C A, Gocayne J D, Scott J D, Shirley R, Liu L I, Glodek A, Kelley J M, Weidman J F, Phillips C A, Spriggs T, Hedblom E, Brandon R C, Fine L D, Fritchman J L, Fuhrmann J L, Geoghagen N S, Gnehm C L, McDonald L A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 13.Fraser C M, Gocayne J D, White O, Adams M D, Clayton R A, Fleischmann R D, Bult C J, Kerlavage A R, Sutton G, Kelly J M, Fritchman J L, Weidman J F, Small K V, Sandusky M, Fuhrmann J L, Nguyen D T, Utterback T R, Saudek D M, Phillips C A, Merrick J M, Tomb J-F, Dougherty B A, Bott K F, Hu P-C, Lucier T S, Peterson S N, Smith H O, Hutchison C A, Venter J C. The minimal gene complement of Mycoplasma genitalium. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 14.Gellert M. DNA topoisomerases. Annu Rev Biochem. 1981;50:879–910. doi: 10.1146/annurev.bi.50.070181.004311. [DOI] [PubMed] [Google Scholar]
- 15.Heisig P. Genetic evidence for a role of parC mutations in development of high-level fluoroquinolone resistance in Escherichia coli. Antimicrob Agents Chemother. 1996;37:696–701. doi: 10.1128/aac.40.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hopewell R, Oram M, Briesewitz R, Fisher L M. DNA cloning and organization of the Staphylococcus aureus gyrA and gyrB genes: close homology among gyrase proteins and implications for 4-quinolone action and resistance. J Bacteriol. 1990;172:3481–3484. doi: 10.1128/jb.172.6.3481-3484.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Isegawa Y, Sheng J, Sokawa Y, Yamanishi K, Nakagomi O, Ueda S. Selective amplification of cDNA sequence from total RNA by cassette-ligation mediated polymerase chain reaction (PCR): application to sequencing 6.5kb genome segment of hantavirus strain B-1. Mol Cell Probes. 1992;6:467–475. doi: 10.1016/0890-8508(92)90043-w. [DOI] [PubMed] [Google Scholar]
- 18.Janda J M, Powers C, Bryant R G, Abbott S L. Current perspectives on the epidemiology and pathogenesis of clinically significant Vibrio spp. Clin Microbiol Rev. 1988;1:245–267. doi: 10.1128/cmr.1.3.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joseph S W, DeBell R M, Brown W P. In vitro response to chloramphenicol, tetracycline, ampicillin, gentamicin, and beta-lactamase production by halophilic vibrios from human and environmental sources. Antimicrob Agents Chemother. 1978;13:244–248. doi: 10.1128/aac.13.2.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato J, Nishimura Y, Imamura R, Niki H, Hiraga S, Suzuki H. New topoisomerase essential for chromosome segregation in E. coli. Cell. 1990;63:393–404. doi: 10.1016/0092-8674(90)90172-b. [DOI] [PubMed] [Google Scholar]
- 21.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S K, Codani J J, Connerton I F, Cummings N J, Daniel R A, Denizot F, Devine K M, Dusterhoft A, Ehrlich S D, Emmerson P T, Entian K D, Errington J, Fabret C, Ferrari E, Foulger D, Fritz C, Fujita M, Fujita Y, Fuma S, Galizzi A, Galleron N, Ghim S Y, Glaser P, Goffeau A, Golightly E J, Grandi G, Guiseppi G, Guy B J, Haga K, Haiech J, Harwood C R, Henaut A, Hilbert H, Holsappel S, Hosono S, Hullo M F, Itaya M, Jones L, Joris B, Karamata D, Kasahara Y, Klaerr-Blanchard M, Klein C, Kobayashi Y, Koetter P, Koningstein G, Krogh S, Kumano M, Kurita K, Lapidus A, Lardinois S, Lauber J, Lazarevic V, Lee S M, Levine A, Liu H, Masuda S, Mauel C, Medigue C, Medina N, Mellado R P, Mizuno M, Moestl D, Nakai S, Noback M, Noone D, O’Reilly M, Ogawa K, Ogiwara A, Oudega B, Park S H, Parro V, Pohl T M, Portetelle D, Porwollik S, Prescott A M, Presecan E, Pujic P, Purnelle B, Rapoport G, Rey M, Reynolds S, Rieger M, Rivolta C, Rocha E, Roche B, Rose M, Sadaie Y, Sato T, Scanlan E, Schleich S, Schroeter R, Scoffone F, Sekiguchi J, Sekowska A, Seror S J, Serror P, Shin B S, Soldo B, Sorokin A, Tacconi E, Takagi T, Takahashi H, Takemaru K, Takeuchi M, Tamakoshi A, Tanaka T, Terpstra P, Tognoni A, Tosato V, Uchiyama S, Vandenbol M, Vannier F, Vassarotti A, Viari A, Wambutt R, Wedler E, Wedler H, Weitzenegger T, Winters P, Wipat A, Yamamoto H, Yamane K, Yasumoto K, Yata K, Yoshida K, Yoshikawa H F, Zumstein E, Yoshikawa H, Danchin A. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 22.Kureishi A, Diver J M, Beckthold B, Schollaardt T, Bryan L E. Cloning and nucleotide sequence of Pseudomonas aeruginosa DNA gyrase gyrA gene from strain PAO1 and quinolone-resistant clinical isolates. Antimicrob Agents Chemother. 1994;38:1944–1952. doi: 10.1128/aac.38.9.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luttinger A L, Springer A L, Schmid M B. A cluster of genes that affects nucleoid segregation in Salmonella typhimurium. New Biol. 1991;3:687–697. [PubMed] [Google Scholar]
- 24.Margerrison E E C, Hopewell R, Fisher L M. Nucleotide sequence of the Staphylococcus aureus gyrB-gyrA locus encoding the DNA gyrase A and B proteins. J Bacteriol. 1992;174:1596–1603. doi: 10.1128/jb.174.5.1596-1603.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsushita S, Yamada S, Kudoh Y, Ohashi M. Drug-resistance and transferable R plasmids in Vibrio cholerae O-1 and non O-1, V. fluvialis and V. parahaemolyticus recently isolated from human sources. Kansenshogaku Zasshi. 1987;61:109–117. doi: 10.11150/kansenshogakuzasshi1970.61.109. . (In Japanese.) [DOI] [PubMed] [Google Scholar]
- 26.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 27.Moore R A, Backthold B, Wong S, Kureishi A, Bryan L E. Nucleotide sequence of the gyrA gene and characterization of ciprofloxacin-resistant mutants of Helicobacter pylori. Antimicrob Agents Chemother. 1995;39:107–111. doi: 10.1128/aac.39.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morris J G, Jr, Black R E. Cholera and other vibrioses in the United States. N Engl J Med. 1985;312:343–350. doi: 10.1056/NEJM198502073120604. [DOI] [PubMed] [Google Scholar]
- 29.Morris J G, Jr, Tenney J H, Drusano G L. In vitro susceptibility of pathogenic Vibrio species to norfloxacin and six other antimicrobial agents. Antimicrob Agents Chemother. 1985;28:442–445. doi: 10.1128/aac.28.3.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakamura S. Mechanisms of quinolone resistance. J Infect Chemother. 1997;3:128–138. [Google Scholar]
- 31.Nishibuchi M, Fasano A, Russell R G, Kaper J B. Enterotoxigenicity of Vibrio parahaemolyticus with and without genes encoding thermostable direct hemolysin. Infect Immun. 1992;60:3539–3545. doi: 10.1128/iai.60.9.3539-3545.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishibuchi M, Kaper J B. Thermostable direct hemolysin gene of Vibrio parahaemolyticus: a virulence gene acquired by a marine bacterium. Infect Immun. 1995;63:2093–2099. doi: 10.1128/iai.63.6.2093-2099.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oppegaard H, Sorum H. gyrA mutations in quinolone-resistant isolates of the fish pathogen Aeromonas salmonicida. Antimicrob Agents Chemother. 1994;38:2460–2464. doi: 10.1128/aac.38.10.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pan X-S, Fisher L M. Cloning and characterization of the parC and parE genes of Streptococcus pneumoniae encoding DNA topoisomerase IV: role in fluoroquinolone resistance. J Bacteriol. 1996;178:4060–4069. doi: 10.1128/jb.178.14.4060-4069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng H, Marians K V. Escherichia coli topoisomerase IV. Purification, characterization, subunit structure, and subunit interactions. J Biol Chem. 1993;268:24481–24490. [PubMed] [Google Scholar]
- 36.Rosanas A, Barbe J, Gilbert I. Cloning and sequencing of the gyrA gene from the plant pathogen Erwinia carotovora. Gene. 1995;161:11–14. doi: 10.1016/0378-1119(95)00267-a. [DOI] [PubMed] [Google Scholar]
- 37.Salazar L, Fsihi H, DeRossi E, Riecardi G, Rios C, Cole S T, Takiff H E. Organization of the origins of replication of the chromosomes of Mycobacterium smegmatis, Mycobacterium leprae and Mycobacterium tuberculosis and isolation of a functional origin from M. smegmatis. Mol Microbiol. 1996;20:283–293. doi: 10.1111/j.1365-2958.1996.tb02617.x. [DOI] [PubMed] [Google Scholar]
- 38.Shungu D L, Weinberg E, Gadebusch H H. In vitro antibacterial activity of norfloxacin (MK-0366, AM-715) and other agents against gastrointestinal tract pathogens. Antimicrob Agents Chemother. 1983;23:86–90. doi: 10.1128/aac.23.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swanberg S, Wang J. Cloning and sequencing of the Escherichia coli gyrA gene coding for the A subunit of DNA gyrase. J Mol Biol. 1987;197:729–736. doi: 10.1016/0022-2836(87)90479-7. [DOI] [PubMed] [Google Scholar]
- 40.Takiff H E, Salazar I, Guerrero C, Philipp W, Huang W M, Kreiswirth B, Cole S T, Jacobs W R, Jr, Telenti A. Cloning and nucleotide sequence of Mycobacterium tuberculosis gyrA and gyrB genes and detection of quinolone resistance mutations. Antimicrob Agents Chemother. 1994;38:773–780. doi: 10.1128/aac.38.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tamura J, Gellert M. Characterization of the ATP binding site on Escherichia coli DNA gyrase. Affinity labelling of lys-103 and lys-110 of the B subunit by pyridoxal 5′-diphospho-5′-adenosine. J Biol Chem. 1990;265:21342–21349. [PubMed] [Google Scholar]
- 42.Taylor D E, Chau A S-S. Cloning and nucleotide sequence of the gyrA gene from Campylobacter fetus subsp. fetus ATCC 27374 and characterization of ciprofloxacin-resistant laboratory and clinical isolates. Antimicrob Agents Chemother. 1997;41:665–671. doi: 10.1128/aac.41.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang J C. DNA topoisomerases. Annu Rev Biochem. 1985;54:665–697. doi: 10.1146/annurev.bi.54.070185.003313. [DOI] [PubMed] [Google Scholar]
- 44.Wang Y, Huang W M, Taylor D E. Cloning and nucleotide sequence of the Campylobacter jejuni gyrA gene and characterization of quinolone resistance mutations. Antimicrob Agents Chemother. 1993;37:457–463. doi: 10.1128/aac.37.3.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wood D O, Waite R T. Sequence analysis of the Rickettsia prowazekii gyrA gene. Gene. 1994;151:191–196. doi: 10.1016/0378-1119(94)90655-6. [DOI] [PubMed] [Google Scholar]
- 46.Yamagishi J, Yoshida H, Yamayoshi M, Nakamura S. Nalidixic acid-resistant mutations of the gyrB gene of Escherichia coli. Mol Gen Genet. 1986;204:367–373. doi: 10.1007/BF00331012. [DOI] [PubMed] [Google Scholar]
- 47.Yoshida H, Bogaki M, Nakamura M, Nakamura S. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob Agents Chemother. 1990;34:1271–1272. doi: 10.1128/aac.34.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]