Abstract
The sterol 14-demethylase P450 (CYP51) of a fluconazole-resistant isolate of Candida albicans, DUMC136, showed reduced susceptibility to this azole but with little change in its catalytic activity. Twelve nucleotide substitutions, resulting in four amino acid changes, were identified in the DUMC136 CYP51 gene in comparison with a reported CYP51 sequence from a wild-type, fluconazole-susceptible C. albicans strain. Seven of these substitutions, including all of those causing amino acid changes, were located within a region covering one of the putative substrate recognition sites of the enzyme (SRS-1). Polymorphisms within this region were observed in several C. albicans isolates, and some were found to be CYP51 heterozygotes. Among the amino acid changes occurring in this region, only an alteration of Y132 was common among these fluconazole-resistant isolates, which suggests the importance of this residue to the fluconazole resistance of the target enzyme. DUMC136 and another fluconazole-resistant isolate were homozygotes with respect to CYP51, although the typical wild-type, fluconazole-susceptible C. albicans was a CYP51 heterozygote. These findings suggest that part of the fluconazole-resistant phenotype of C. albicans DUMC136 was acquired through a mutation-prone area of CYP51, an area which might promote the formation of fluconazole-resistant CYP51, along with a mechanism(s) which allows the formation of a homozygote of this altered CYP51 in this diploid pathogenic yeast.
Candida albicans is one of the major pathogenic fungi causing systemic infection among immunocompromised hosts. Azole antifungal agents, such as fluconazole and itraconazole, are commonly prescribed to treat systemic and mucocutaneous candidiasis. However, emergence of fluconazole-resistant C. albicans strains in patients receiving triazole treatment has become a serious problem, particularly in the treatment of oral candidiasis in AIDS patients (15, 23, 24).
Fluconazole resistance has been proposed to occur through several mechanisms: (i) failure of cells to accumulate the agent (1, 4, 27); (ii) alteration of the ergosterol biosynthetic pathway through a defect in sterol Δ5,6-desaturase (16); (iii) an increase in the cellular content of sterol 14-demethylase P450 (CYP51), the primary target of the azole (30); and (iv) a decrease in the affinity of CYP51 for fluconazole (19, 26, 31). It has now been shown that these distinct mechanisms can develop in single Candida isolates in a stepwise process over the period of antifungal treatment (30). The most common mechanism of azole resistance appears to involve the enhancement of efflux pumps that remove azole compounds from the cytoplasm (1, 4, 27). Expression of the multidrug efflux transporters of the ATP-binding cassette superfamily, such as CDR1, and of the major facilitator class, such as MDR1, has been frequently reported in fluconazole-resistant strains. Also, alterations in the cell membrane through changes in sterol and/or lipid content (12) or formation of a biofilm (11) may confer azole resistance.
The importance of alteration of the affinity of CYP51 for azoles due to some mutations as a mechanism of drug resistance has been deduced from the fact that mutations in penicillin-binding proteins and DNA gyrase have largely diminished the efficacies of β-lactam antibiotics and new quinolones, respectively, in antibacterial chemotherapy (13, 25). Alterations of CYP51 can have further implications with regard to the development of newer azoles. For instance, it was shown that an artificial mutation in CYP51 reduced both its azole susceptibility and catalytic activity (18). Genetic alterations of CYP51 were also reported in fluconazole-resistant clinical isolates of C. albicans (19, 26, 31). However, there have been few systematic studies of both the biochemistry and molecular biology of CYP51 in clinical isolates of azole-resistant C. albicans. This paper examines biochemical and molecular biological studies of CYP51 from fluconazole-resistant strains of C. albicans isolated from AIDS patients receiving long-term fluconazole therapy. The strains were found to be homozygous for an altered CYP51 gene, and the encoded enzyme exhibited a low affinity for fluconazole but normal sterol 14-demethylase activity.
MATERIALS AND METHODS
Azole-resistant isolates of C. albicans.
The fluconazole-resistant C. albicans DUMC136 and S78941 were isolated at Duke University Medical Center from two AIDS patients who had received either 3 months of continuous fluconazole therapy or approximately 3 years of intermittent azole therapy, respectively, for mucosal candidiasis. C. albicans ATCC 90028, which is recommended for use as the quality control strain for determining MICs of azole compounds (20), was obtained from the American Type Culture Collection.
Chemicals.
Fluconazole was extracted from a commercially available intravenous-injection material (a product of Pfizer, Tokyo, Japan) and purified. Other chemicals used in this study were obtained commercially.
Fluconazole susceptibility testing of C. albicans strains.
MICs of fluconazole for C. albicans strains were determined by the agar dilution method, using RPMI 1640 medium (Gibco) with 0.7% agar as reported previously (32). Results obtained by this assay were comparable to those of studies performed according to National Committee for Clinical Laboratory Standards criteria (32).
Preparation of C. albicans microsomes.
C. albicans was grown at 30°C for 15 h in Sabouraud dextrose broth (Difco). Collected and washed cells were suspended in 0.65 M sorbitol and disrupted with a French press. The cell homogenate was centrifuged successively at 5,000 × g for 20 min and 120,000 × g for 90 min. The precipitate obtained after the second centrifugation was suspended in 0.1 M potassium phosphate buffer (pH 7.4) containing 0.1 mM EDTA to give a protein concentration of 50 mg/ml of microsomes.
Lanosterol 14-demethylase assay.
Lanosterol 14-demethylase activity was assayed by the method of Aoyama et al. (3). Briefly, 2 ml of a reaction mixture containing 3 to 5 mg of microsomal protein/ml, 23.5 nmol of lanosterol/ml, and an NADPH generator in 0.1 M potassium phosphate buffer (pH 7.4) was incubated aerobically at 30°C. After saponification with 5 ml of 10% (wt/vol) KOH in methanol at 80°C for 60 min, nonsaponifiable lipids were extracted with petroleum ether and the lanosterol fraction was separated by thin-layer chromatography. This lanosterol fraction was extracted, trimethylsilylated, and analyzed by gas-liquid chromatography. The 14-demethylase activity was calculated from the chromatographically determined conversion ratio and the initial amount of the substrate.
Measurement of ergosterol biosynthesis by cell homogenate.
Incorporation of [14C]mevalonic acid into ergosterol by the cell homogenates was determined by a previously described method (5). Briefly, 4 mg of protein from a cell homogenate was incubated aerobically at 37°C for 60 min with 0.25 μCi of [14C]mevalonic acid (40 mCi/mmol; Amersham) and an NADPH generator in 0.5 ml of 0.1 M potassium phosphate buffer (pH 7.4). After saponification with 1 ml of 2.7 M KOH in 90% (vol/vol) methanol at 80°C for 60 min, the nonsaponifiable lipids were extracted and analyzed by thin-layer chromatography. The radioactivity incorporated into sterols was measured by autoradiography.
Determination of P450 and sterol contents.
The P450 content of the microsomes was determined spectrophotometrically by the method of Omura and Sato (21). The sterol content of the yeast cells was determined as follows. Cells corresponding to 140 mg of the dry weight were suspended in 2 ml of 0.1 M potassium phosphate buffer (pH 7.4) and saponified with 5 ml of 10% KOH in methanol at 80°C for 2.5 h. Sterols were extracted from the saponified mixture, trimethylsilylated, and analyzed by gas-liquid chromatography with cholesterol as the internal standard.
Cloning and nucleotide sequencing of CYP51.
DNA fragments covering the coding region for CYP51 (17) were cloned by PCR. A sense primer, Um87 (5′-AGGGAATTCAATCGTTATTC-3′), and an antisense primer, D1625 (5′-CAATCAGAACACTGAATCGA-3′), were used for amplification of the CYP51 genes of C. albicans ATCC 90028 and S78941. The CYP51 gene of C. albicans DUMC136 was amplified as two partially overlapping fragments. A sense (Um15; 5′-AGATCATAACTCAATATGGC-3′) and an antisense (D938; 5′-GCAGAAGTATGTTGACCACC-3′) primer pair was used for amplifying the upstream fragment, and another sense (U639; 5′-TGACCGTTCATTTGCTCAAC-3′) and antisense (D1625) primer pair was used for amplifying the downstream fragment. The PCR products were ligated into SmaI-cut pBluescript II SK(+) vector (Stratagene) and transformed into competent Escherichia coli XL1-Blue cells (Stratagene). Sequencing was carried out with an ABI Prism Dye Terminator Cycle Sequencing Ready Reaction Kit (Perkin-Elmer). Sequencing was performed in triplicate with three independent clones.
Restriction fragment length polymorphism (RFLP) analysis.
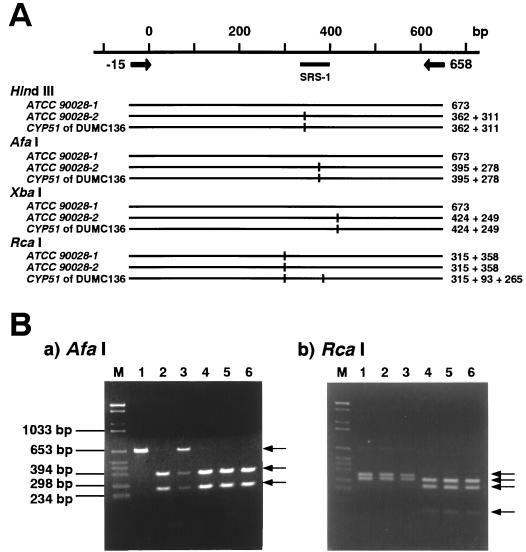
A 673-bp fragment covering the potential polymorphic region (see Fig. 3) of CYP51 was prepared from genomic DNAs of various C. albicans isolates (see Table 4) by PCR with primers Um15 and D658 (5′-GTTGAGCAAATGAACGGTCA-3′). The PCR products were digested overnight at 37°C with AfaI, HindIII, XbaI, or RcaI. The digested fragments were then subjected to electrophoresis in a 1.5% agarose gel.
FIG. 3.
RFLPs observed on the 673-bp PCR products from the CYP51 genes of ATCC 90028 and DUMC136. (A) The restriction maps of the 673-bp PCR products (nucleotide positions −15 to 658) from the alleles encoding ATCC 90028-1 and ATCC 90028-2 and from the DUMC136 CYP51 gene. (B) DNA fragments obtained by digestion of the 673-bp PCR products with AfaI (a) or RcaI (b) are shown. The templates used for the PCR amplifications were as follows: lane 1, the cloned ATCC 90028-1 DNA; lane 2, the cloned ATCC 90028-2 DNA; lane 3, whole genomic DNA of C. albicans ATCC 90028; lane 4 to 6, whole genomic DNA of three independent colonies of C. albicans DUMC136. Lane M is the DNA size marker (BglII- and HinfI-digested pBR328).
TABLE 4.
Polymorphisms observed around SRS-1 of C. albicans CYP51
| Template DNA | Restriction sitea
|
Fluconazole MIC (μM) for the strain | ||
|---|---|---|---|---|
| HindIII | AfaI | XbaI | ||
| Cloned ATCC 90028-1 | A | A | A | |
| Cloned ATCC 90028-2 | P | P | P | |
| DUMC136 genomic | P | P | P | >200 |
| S78941 genomic | P | P | P | >200 |
| ATCC 90028 genomic | P/A | P/A | P/A | 1.6 |
| CA1 genomic | P/A | P/A | P/A | 0.8 |
| CA2 genomic | A | A | P/A | 0.4 |
| CA3 genomic | A | A | P/A | 0.4 |
| CA4 genomic | P | A | P | 0.8 |
| CA5 genomic | P | A | P | 1.6 |
| CA6 genomic | A | A | P | 26 |
| CA7 genomic | A | A | P | 6.6 |
| CA8 genomic | A | A | P | 3.3 |
| CA9 genomic | A | A | P | 0.8 |
| CA10 genomic | A | A | A | 1.6 |
The restriction sites of the 673-bp PCR products obtained from the indicated templates were analyzed as described in the legend to Fig. 3. P, present; A, absent; P/A, present on one allele and absent on another allele.
Nucleotide sequence accession numbers.
The nucleotide sequences of the C. albicans DUMC136 and S78941 CYP51 genes and the ATCC 90028-2 CYP51 allele have been deposited in the GenBank, EMBL, and DDBJ databases under accession no. AB006855, AB006856, and AB006854, respectively.
RESULTS
Biochemical characterization of novel fluconazole-resistant isolates of C. albicans strains.
The MICs of fluconazole for two novel clinical isolates of C. albicans, DUMC136 and S78941, were more than 125 times higher than that for a fluconazole-susceptible standard strain of C. albicans, ATCC 90028 (Table 1). The 50% inhibitory concentrations (IC50s) of fluconazole for in vitro ergosterol synthesis by the cell-free preparations of DUMC136 and S78941 were more than 20 times higher than that for ATCC 90028 (Table 1). Since fluconazole inhibits ergosterol synthesis at the CYP51-catalyzed 14-demethylation step, these observations suggested that the reduced fluconazole susceptibility of the CYP51s of DUMC136 and S78941 was in part due to the inability of fluconazole to efficiently interact with CYP51.
TABLE 1.
MICs and IC50s of fluconazole for ergosterol biosynthesis in cell lysates of C. albicans ATCC 90028, DUMC136, and S78941
| Strain | MIC (μM) | IC50 on incorporation of [14C]mevalonic acida (nM) |
|---|---|---|
| ATCC 90028 | 1.6 | 16 |
| DUMC136 | >200 | 520 |
| S78941 | >200 | 370 |
The concentration of fluconazole causing 50% inhibition of incorporation of [14C]mevalonic acid into ergosterol in the cell lysate.
To obtain further information, the lanosterol demethylase activities of DUMC136 and ATCC 90028 microsomes were compared. The IC50 of fluconazole for the demethylase activity of DUMC136 microsomes was about 10 times higher than that for the ATCC 90028 microsomes. The specific activity of the lanosterol 14-demethylase of DUMC136 microsomes was found to be lower than that of the ATCC 90028 microsomes. However, when these activities were calibrated with the P450 contents, no significant difference was observed because of the low P450 content of the DUMC136 microsomes (Table 2). These observations indicated that the catalytic activity retained by CYP51 of DUMC136 was comparable to that of the wild-type enzyme in spite of the low overall activity. The reason for the low overall activity of CYP51 in DUMC136 may be either a low expression level or the instability of this enzyme.
TABLE 2.
IC50s of fluconazole for the lanosterol 14-demethylase activity, lanosterol 14-demethylase activity, and P450 contents of the C. albicans ATCC 90028 and DUMC136 microsomes
| Strain | IC50 of fluconazole on lanosterol 14-demethylationa (nM) | Demethylase activityb
|
P450 contentc (pmol/mg of protein) | |
|---|---|---|---|---|
| pmol/min/mg of protein | pmol/min/pmol of P450 | |||
| ATCC 90028 | 50 | 51 | 4.0 | 13 |
| DUMC136 | 490 | 8.7 | 3.1 | 2.9 |
The concentration of fluconazole resulting in 50% inhibition of lanosterol 14-demethylase activity.
Lanosterol 14-demethylase activity was assayed according to the method of Aoyama et al. (3).
P450 content was determined from the carbon monoxide difference spectrum of sodium dithionite-reduced microsomes by using Δɛ450–490 = 91 mM−1 cm−1 (21).
DUMC136 grew normally in the absence of fluconazole (data not shown), and the ergosterol content of DUMC136 cells was comparable to that of ATCC 90028 cells (Table 3). Therefore, the level of CYP51 in DUMC136 microsomes appeared to be sufficient for the production of normal amounts of ergosterol by this strain.
TABLE 3.
Sterol contents of C. albicans ATCC 90028 and DUMC136
| Strain | Sterol content (pmol/mg of dry cells)a
|
||
|---|---|---|---|
| Lanosterol | 24-Methylene DHLb | Ergosterol | |
| ATCC 90028 | 91 | NDc | 1,200 |
| DUMC136 | 80 | 6.2 | 1,300 |
The amount of sterol was determined by gas chromatography, using cholesterol as the internal standard.
24-Methylene DHL, 24-methylene-24,25-dihydrolanosterol.
ND, not detectable.
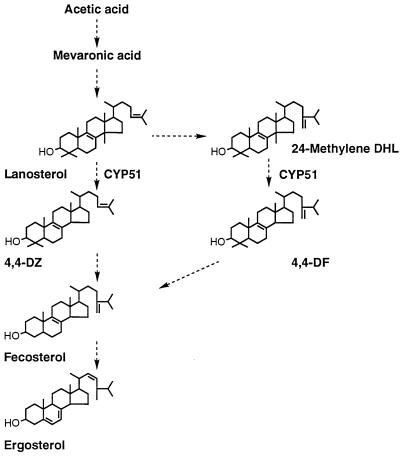
A significant amount of 24-methylene-24,25-dihydrolanosterol was also detected in DUMC136 cells, though it was not detected in ATCC 90028 cells (Table 3). This observation suggests that in DUMC136 a part of the lanosterol is metabolized through 24-transmethylation prior to the 14-demethylation, whereas in the wild-type strain most of the lanosterol first undergoes the 14-demethylation (Fig. 1). Although neither the exact reason for nor the specific consequence of this shunt is yet known, the decreased 14-demethylation rate due to the reduced level of P450 in DUMC136 or the slight modification of the substrate specificity of the DUMC136 CYP51 due to some structural alteration is a possible explanation.
FIG. 1.
Biosynthetic pathway for ergosterol in C. albicans. Note that two alternative pathways for conversion of lanosterol to fecosterol exist. Abbreviations: 4,4-DZ, 4,4-dimethylzymosterol; 24-Methylene DHL, 24-methylene-24,25-dihydrolanosterol; 4,4-DF, 4,4-dimethylfecosterol.
Nucleotide substitutions occurring in the CYP51 gene of fluconazole-resistant C. albicans, and their resulting amino acid substitutions.
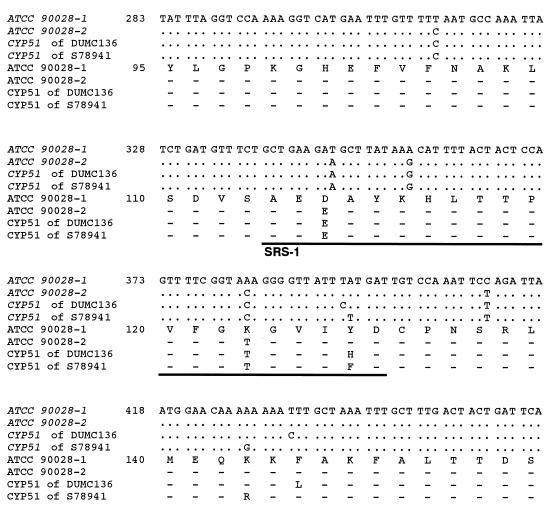
The nucleotide sequence of the cloned DUMC136 CYP51 gene was determined (GenBank/EMBL/DDBJ accession no. AB006855) and compared with a previously reported standard (17) (GenBank/EMBL accession no. X13296). Twelve nucleotide substitutions, resulting in four amino acid changes (shown in parentheses)—T315C (replacement of the thymine at position 315 by cytosine), T348A (D116E), A357G, A383C (K128T), T394C (Y132H), C411T, T433C (F145L), C658T, A1020G, C1110T, A1440G, and T1470C—were identified. (The first nucleotide of the initiation codon, which corresponds to nucleotide 148 of the reported sequence [17], has been denoted nucleotide 1.) Two different CYP51 sequences were identified in the ATCC 90028 genome; one of them, called the ATCC 90028-1 gene in this paper, was identical to the originally reported sequence (17), and the other, called the ATCC 90028-2 gene was a novel sequence (GenBank/EMBL/DDBJ accession no. AB006854). The ATCC 90028-2 gene sequence had 10 nucleotide differences from the reported sequence (17) and that of the ATCC 90028-1 gene, and all of these substitutions were included in the above-mentioned 12 nucleotide substitutions found in DUMC136 CYP51. CYP51 of DUMC136 had two unique nucleotide substitutions resulting in amino acid changes, T394C (Y132H) and T433C (F145L), compared to the ATCC 90028-2 gene sequence.
Seven of the 12 nucleotide substitutions, including all of those causing amino acid changes, occurred in the 119-nucleotide-long span (T315C to T433C) within the 1,584-nucleotide-long encoding sequence. Figure 2 represents the alignment of nucleotide (and resulting amino acid) sequences including the variable region of CYP51 from ATCC 90028 (genes ATCC 90028-1 and ATCC 90028-2), DUMC136, and S78941. As shown in Fig. 2, this region includes one of the putative substrate recognition sites of CYP51, named SRS-1 (2), and a unique amino acid residue, Y132, which was commonly changed in both the DUMC136 and S78941 CYP51s. Y132 exists within SRS-1. Since Y132 is one of the highly conserved residues of CYP51 (2), its replacement by another amino acid may affect its recognition by compounds that interact with its active site. Replacement of Y132 has been reported for another fluconazole-resistant isolate of C. albicans (26). Therefore, replacement of Y132 may make an important contribution to the reduction of fluconazole susceptibility at the CYP51 locus.
FIG. 2.
The nucleotide (and deduced amino acid) sequence of the highly substituted region observed in CYP51 clones from wild-type C. albicans ATCC 90028 and fluconazole-resistant C. albicans isolates DUMC136 and S78941. The nucleotides and corresponding amino acids of the ATCC 90028-1 allele which are identical to those of the reported C. albicans CYP51 sequences (17) are fully indicated, and only substituted nucleotides and amino acids are shown for ATCC 90028, DUMC136, and S78941. The underlined region is one of the putative substrate recognition sites of CYP51, named SRS-1 (2). Dashes indicate amino acids identical to those of the ATCC 90028-1, and dots indicate nucleotides identical to those of the ATCC 90028-1 gene.
Polymorphisms of C. albicans CYP51 around SRS-1.
Sequence alignments indicate that the nucleotide substitution occurring in the ATCC 90028-2 gene and CYP51 genes of DUMC136 and S78941, respectively, introduced a few restriction sites, resulting in a characteristic RFLP pattern for this region (Fig. 3A).
PCR products (673 bp; nucleotide positions −15 to 658) covering this polymorphic region were prepared from genomic DNAs of ATCC 90028 and DUMC136, and RFLP analysis was performed with AfaI and RcaI. The 673-bp PCR product from the genomic DNA of ATCC 90028 gave three fragments, including the originally sized PCR product, after digestion with AfaI (Fig. 3B, panel a, lane 3). AfaI cleaves only the ATCC 90028-2 gene sequence (Fig. 3A), and this indicates that ATCC 90028 is a heterozygote having ATCC 90028-1 and -2 genes. In contrast, AfaI digestion of the comparable PCR product from the DUMC136 genomic DNA gave only two fragments, and no 673-bp band was observed (Fig. 3B, panel a, lane 4 to 6), indicating the absence of the ATCC 90028-1 gene sequence in the DUMC136 genome. DUMC136 CYP51 has two RcaI sites, but the ATCC 90028-1 and -2 genes each have only one RcaI site (Fig. 3A). The PCR product from DUMC136 genomic DNA was completely cleaved into three fragments by RcaI, and no 358-bp fragment, which was found in the ATCC 90028-1 or -2 gene sequence, was observed (Fig. 3B, panel b). These results clearly indicate that the complete sequence of neither the ATCC 90028-1 nor the ATCC 90028-2 gene is present in the DUMC136 genome. It is concluded that DUMC136 is a homozygote with the altered CYP51.
Table 4 is a summary of results of RFLP analysis of the 673-bp PCR products from 13 independent clinical isolates of C. albicans, obtained by HindIII, AfaI, and XbaI digestion. These results show that genetic polymorphism at this site is generally observed among natural isolates of C. albicans. Among the isolates tested, DUMC136 and S78941 were highly fluconazole resistant (MICs, >200 μM) and CA6 and CA7 were moderately fluconazole resistant (MICs, 26 and 6.6 μM, respectively), and they are all homozygotes with this allele. In contrast, four heterozygous strains, including ATCC 90028, had considerably lower fluconazole MICs (<2 μM).
DISCUSSION
A fluconazole-resistant strain of C. albicans, DUMC136, contained an altered CYP51 protein that showed a reduced affinity for fluconazole but demonstrated normal catalytic activity (Table 2). Although the CYP51 level in DUMC136 cells was lower than that of a fluconazole-susceptible strain, ATCC 90028, DUMC136 grew normally and produced ergosterol in amounts similar to those produced by strain ATCC 90028 (Table 3). Reduced activity of CYP51 would actually be considered a disadvantage for surviving in the presence of azoles, but DUMC136 was highly resistant to fluconazole. A shunt via 24-methylene-24,25-dihydrolanosterol, which is found in DUMC136, can increase ergosterol synthesis and may be relevant to the observed fluconazole resistance. Thus, our lines of evidence suggest that the reduced affinity of DUMC136 CYP51 for fluconazole effectively contributed to the fluconazole-resistant phenotype of this strain. However, this may not be the sole mechanism of its fluconazole resistance, because the difference in the fluconazole MICs for ATCC 90028 and DUMC136 was even greater than the difference in their IC50s for CYP51 activity. The most probable additional explanation for this observation is the enhancement of efflux pumps in this strain, such as the ATP-binding cassette transporter, which have been accepted as the most general mechanism for azole resistance among the pathogenic fungi (1, 4, 27).
Nucleotide sequence analysis of the CYP51 gene of DUMC136 revealed 12 nucleotide substitutions compared to the standard C. albicans CYP51 gene sequence (17). However, it was found that DUMC136 CYP51 had a much higher degree of similarity to the newly identified allele of CYP51 called the ATCC 90028-2 gene. Only two nucleotide differences resulting in amino acid changes (Y132H and F145L) were identified between DUMC136 CYP51 and the ATCC 90028-2 gene (Fig. 2). These findings strongly suggest that a CYP51 gene having a sequence that is either the same as or very similar to that of the ATCC 90028-2 gene is a close ancestor of DUMC136 CYP51 and that the amino acid changes may contribute to the reduced susceptibility of DUMC136 CYP51 to fluconazole. One of the residues, Y132, which is highly conserved among fungal and animal CYP51s and is included in a putative substrate recognition site named SRS-1 (2), was replaced in DUMC136 CYP51. Substitution at Y132 was also found in the CYP51 proteins of another fluconazole-resistant isolate, S78941 (Fig. 2), and a strain reported by Sanglard et al. (26), suggesting a critical role for this residue in the fluconazole susceptibility of CYP51. Since azole antifungal agents interact with the substrate-binding site of CYP51 (33, 34), mutations occurring in such substrate recognition sites will probably affect the azole susceptibility of the enzyme. Lamb et al. (18) and White (31) reported that the T315A mutation in SRS-4 and the R467K mutation in SRS-6, respectively, were involved in azole resistance of C. albicans, and Délye et al. (6) inferred that the F136Y mutation in SRS-1 caused the triadimenol resistance of Uncinula necator, a plant pathogen. A considerable number of nucleotide substitutions and polymorphisms were observed around SRS-1 of C. albicans CYP51 (Fig. 2 and 3 and Table 4). This fact suggests that the substrate recognition site is mutation prone, as is the case in the CYP2 family (9). Amino acid changes in the substrate recognition site could impair the catalytic activity of the enzyme. A mutation that impairs the catalytic activity of the enzyme will be unfavorable for cellular growth and thus may be eliminated from the infectious population. In fact, the above-mentioned T315A mutation was reported to reduce both catalytic activity and fluconazole susceptibility (18). In contrast, the mutation occurring within SRS-1 of DUMC136 CYP51 reduced fluconazole susceptibility without significantly changing the enzyme activity, and such alterations appear to be selected in fluconazole-containing environments.
In diploid C. albicans, a mutation occurring on one allele may not appear as a phenotype unless it becomes dominant. The CYP51 level in DUMC136 was one-fifth the level in the wild-type strain ATCC 90028 (Table 2). The DUMC136 CYP51 gene was highly similar to the ATCC 90028-2 allele. These results pose the possibility that the ATCC 90028-2 protein level is also low. If this is a case, a fluconazole resistance-inducing mutation of CYP51 occurring on only one allele, such as ATCC 90028-2 allele, may not significantly contribute to the survival of this yeast in an environment containing fluconazole, and only formation of the homozygote of the gene encoding fluconazole-resistant CYP51 would be essential for survival in a fluconazole-rich environment. For instance, DUMC136 and another fluconazole-resistant isolate, S78941, were homozygote with the altered CYP51 gene encoding fluconazole-resistant CYP51. Fluconazole-resistant C. albicans strains reported by White (31) and Franz et al. (8) were also CYP51 homozygotes. These facts suggest that homozygosity for the gene encoding azole-resistant CYP51 is observed generally in azole-resistant C. albicans.
In diploid cells, mutations usually occur randomly on each allele and result in heterozygocity. It is generally considered that meiosis and mating are included in the formation of a homozygote for an altered gene. Frequent mitotic recombination and gene conversion are also possible mechanisms for forming a homozygote. Since C. albicans is known as an incomplete fungus, the occurrence of a sexual generation is unlikely (22, 29), and mitotic recombination or gene conversion is a more likely event in the formation of a CYP51 homozygote in fluconazole-resistant C. albicans strains such as DUMC136. Sexual processes, however, may not completely be excluded, since sequences homologous to meiosis-related genes of Saccharomyces cerevisiae have been identified in C. albicans (7, 14), and several papers (7, 10, 28) have suggested the possibility of sexual reproduction of C. albicans.
In conclusion, we propose the following scenario for a possible mechanism for emergence of azole-resistant C. albicans in azole-treated patients. The mutation-prone nature of a substrate recognition site of CYP51 leads to the introduction of mutations in one allele of CYP51, and a yeast that is a homozygote with the gene encoding azole-resistant CYP51 is formed and selected for survival under azole-rich environmental conditions. Screening of azole-resistant yeast strains by RFLP analysis of SRS-1 can be done to determine the magnitude of emergence of azole-resistant C. albicans by this mechanism.
ACKNOWLEDGMENTS
We thank M. Fukuda and C. Yamashita of Mukogawa Women’s University for technical assistance.
REFERENCES
- 1.Albertson G D, Niimi M, Cannon R D, Jenkinson H F. Multiple efflux mechanisms are involved in Candida albicans fluconazole resistance. J Antimicrob Chemother. 1996;40:2835–2841. doi: 10.1128/aac.40.12.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aoyama Y, Noshiro M, Gotoh O, Imaoka S, Funae Y, Kurosawa N, Horiuchi T, Yoshida Y. Sterol 14-demethylase P450 (P45014DM) is one of the most ancient and conserved P450 species. J Biochem. 1996;119:926–933. doi: 10.1093/oxfordjournals.jbchem.a021331. [DOI] [PubMed] [Google Scholar]
- 3.Aoyama Y, Yoshida Y, Sato R. Yeast cytochrome P-450 catalyzing lanosterol 14α-demethylation. J Biol Chem. 1984;259:1661–1666. [PubMed] [Google Scholar]
- 4.Balan I, Alarco A-M, Raymond M. The Candida albicans CDR3 gene codes for an opaque-phase ABC transporter. J Bacteriol. 1997;179:7210–7218. doi: 10.1128/jb.179.23.7210-7218.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrett-Bee K J, Lane A C, Turner R W. The mode of antifungal action of tolnaftate. J Med Vet Mycol. 1986;24:155–160. doi: 10.1080/02681218680000221. [DOI] [PubMed] [Google Scholar]
- 6.Délye C, Laigret F, Corio-Costet M-F. A mutation in the 14α-demethylase gene of Uncinula necator that correlates with resistance to a sterol biosynthesis inhibitor. Appl Environ Microbiol. 1997;63:2966–2970. doi: 10.1128/aem.63.8.2966-2970.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diener A C, Fink G R. DLH1 is a functional Candida albicans homologue of the meiosis-specific gene DMC1. Genetics. 1996;143:769–776. doi: 10.1093/genetics/143.2.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franz R, Kelly S L, Lamb D C, Kelly D E, Ruhnke M, Morschhäuser J. Multiple molecular mechanisms contribute to a stepwise development of fluconazole resistance in clinical Candida albicans strains. Antimicrob Agents Chemother. 1998;42:3065–3072. doi: 10.1128/aac.42.12.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gotoh O. Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J Biol Chem. 1992;267:83–90. [PubMed] [Google Scholar]
- 10.Gräser Y, Volovsek M, Arrington J, Schönian G, Presber W, Mitchell T G, Vilgalys R. Molecular markers reveal that population structure of the human pathogen Candida albicans exhibits both clonality and recombination. Proc Natl Acad Sci USA. 1996;93:12473–12477. doi: 10.1073/pnas.93.22.12473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hawser S P, Douglas L J. Resistance of Candida albicans biofilms to antifungal agents in vitro. Antimicrob Agents Chemother. 1995;39:2128–2131. doi: 10.1128/aac.39.9.2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hitchcock C A, Barrett-Bee K J, Russell N J. The lipid composition of azole-sensitive and azole-resistant strains of Candida albicans. J Gen Microbiol. 1986;132:2421–2431. doi: 10.1099/00221287-132-9-2421. [DOI] [PubMed] [Google Scholar]
- 13.Hooper D C, Wolfson J S. Mode of action of the new quinolones: new data. Eur J Clin Microbiol Infect Dis. 1991;10:223–231. doi: 10.1007/BF01966994. [DOI] [PubMed] [Google Scholar]
- 14.Hoyer L L, Scherer S, Shatzman A R, Livi G P. Candida albicans ALS1: domains related to a Saccharomyces cerevisiae sexual agglutinin separated by a repeating motif. Mol Microbiol. 1995;15:39–54. doi: 10.1111/j.1365-2958.1995.tb02219.x. [DOI] [PubMed] [Google Scholar]
- 15.Johnson E M, Warnock D W, Luker J, Porter S R, Scully C. Emergence of azole drug resistance in Candida species from HIV-infected patients receiving prolonged fluconazole therapy for oral candidosis. J Antimicrob Chemother. 1995;35:103–114. doi: 10.1093/jac/35.1.103. [DOI] [PubMed] [Google Scholar]
- 16.Kelly S L, Lamb D C, Kelly D E, Manning N J, Loeffler J, Hebart H, Schumacher U, Einsele H. Resistance to fluconazole and cross-resistance to amphotericin B in Candida albicans from AIDS patients caused by defective sterol Δ5,6-desaturation. FEBS Lett. 1997;400:80–82. doi: 10.1016/s0014-5793(96)01360-9. [DOI] [PubMed] [Google Scholar]
- 17.Lai M H, Kirsch D R. Nucleotide sequence of cytochrome P450 L1A1 (lanosterol 14α-demethylase) from Candida albicans. Nucleic Acids Res. 1989;17:804. doi: 10.1093/nar/17.2.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamb D C, Kelly D E, Schunck W-H, Shyadehi A Z, Akhtar M, Lowe D J, Baldwin B C, Kelly S L. The mutation T315K in Candida albicans sterol 14α-demethylase causes reduced enzyme activity and fluconazole resistance through reduced affinity. J Biol Chem. 1997;272:5682–5688. doi: 10.1074/jbc.272.9.5682. [DOI] [PubMed] [Google Scholar]
- 19.Löffler J, Kelly S L, Hebart H, Schumacher U, Lass-Flörl C, Einsele H. Molecular analysis of cyp51 from fluconazole-resistant Candida albicans strains. FEMS Microbiol Lett. 1997;151:263–268. doi: 10.1111/j.1574-6968.1997.tb12580.x. [DOI] [PubMed] [Google Scholar]
- 20.National Committee for Clinical Laboratory Standards. Reference method for broth dilution antifungal susceptibility testing of yeasts. Proposed standard document M27-P. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1992. [Google Scholar]
- 21.Omura T, Sato R. The carbon monoxide-binding pigment of liver microsomes. J Biol Chem. 1964;239:2370–2378. [PubMed] [Google Scholar]
- 22.Pujol C, Reynes J, Renaud F, Raymond M, Tibayrenc M, Ayala F J, Janbon F, Mallie M, Bastide J-M. The yeast Candida albicans has a clonal mode of reproduction in a population of infected human immunodeficiency virus-positive patients. Proc Natl Acad Sci USA. 1993;90:9456–9459. doi: 10.1073/pnas.90.20.9456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Revankar S G, Kirkpatrick W R, McAtee R K, Dib O P, Fothergill A W, Redding S W, Rinaldi M G, Patterson T F. Detection and significance of fluconazole resistance in oropharyngeal candidiasis in human immunodeficiency virus-infected patients. J Infect Dis. 1996;174:821–827. doi: 10.1093/infdis/174.4.821. [DOI] [PubMed] [Google Scholar]
- 24.Rex J H, Rinaldi M G, Pfaller M A. Resistance of Candida species to fluconazole. Antimicrob Agent Chemother. 1995;39:1–8. doi: 10.1128/aac.39.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rybkine T, Mainardi J-L, Sougakoff W, Collatz E, Gutmann L. Penicillin-binding protein 5 sequence alterations in clinical isolates of Enterococcus faecium with different levels of β-lactam resistance. J Infect Dis. 1998;178:159–163. doi: 10.1086/515605. [DOI] [PubMed] [Google Scholar]
- 26.Sanglard D, Ischer F, Koymans L, Bille J. Amino acid substitutions in the cytochrome P-450 lanosterol 14α-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob Agents Chemother. 1998;42:241–253. doi: 10.1128/aac.42.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanglard D, Kuchler K, Ischer F, Pagani J-L, Monod M, Bille J. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob Agents Chemother. 1995;39:2378–2386. doi: 10.1128/aac.39.11.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tibayrenc M. Are Candida albicans natural populations subdivided? Trends Microbiol. 1997;5:253–257. doi: 10.1016/S0966-842X(97)01068-8. [DOI] [PubMed] [Google Scholar]
- 29.Tibayrenc M, Kjellberg F, Arnaud J, Oury B, Breniere S F, Darde M-L, Ayala F J. Are eukaryotic microorganisms clonal or sexual? A population genetics vantage. Proc Natl Acad Sci USA. 1991;88:5129–5133. doi: 10.1073/pnas.88.12.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White T C. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob Agents Chemother. 1997;41:1482–1487. doi: 10.1128/aac.41.7.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.White T C. The presence of an R467K amino acid substitution and loss of allelic variation correlate with an azole-resistant lanosterol 14α demethylase in Candida albicans. Antimicrob Agents Chemother. 1997;41:1488–1494. doi: 10.1128/aac.41.7.1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshida T, Jono K, Okonogi K. Modified agar dilution susceptibility testing method for determining in vitro activities of antifungal agents, including azole compounds. Antimicrob Agents Chemother. 1997;41:1349–1351. doi: 10.1128/aac.41.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshida Y. Cytochrome P450 of fungi: primary target for azole antifungal agents. In: McGinnis M R, editor. Current topics in medical mycology. Vol. 2. New York, N.Y: Springer-Verlag; 1988. pp. 388–418. [DOI] [PubMed] [Google Scholar]
- 34.Yoshida Y, Aoyama Y. Stereoselective interaction of an azole antifungal agent with its target, lanosterol 14α-demethylase (cytochrome P-45014DM): a model study with stereoisomers of triadimenol and purified cytochrome P-45014DM from yeast. Chirality. 1990;2:10–15. doi: 10.1002/chir.530020103. [DOI] [PubMed] [Google Scholar]