Abstract
The outbreak of COVID-19 has caused a severe shortage of healthcare resources. Ground Glass Opacity (GGO) and consolidation of chest CT scans have been an essential basis for imaging diagnosis since 2020. The similarity of imaging features between COVID-19 and other pneumonia makes it challenging to distinguish between them and affects radiologists' diagnosis. Recently, deep learning in COVID-19 has been mainly divided into disease classification and lesion segmentation, yet little work has focused on the feature correlation between the two tasks. To address these issues, in this study, we propose MultiR-Net, a 3D deep learning model for combined COVID-19 classification and lesion segmentation, to achieve real-time and interpretable COVID-19 chest CT diagnosis. Precisely, the proposed network consists of two subnets: a multi-scale feature fusion UNet-like subnet for lesion segmentation and a classification subnet for disease diagnosis. The features between the two subnets are fused by the reverse attention mechanism and the iterable training strategy. Meanwhile, we proposed a loss function to enhance the interaction between the two subnets. Individual metrics can not wholly reflect network effectiveness. Thus we quantify the segmentation results with various evaluation metrics such as average surface distance, volume Dice, and test on the dataset. We employ a dataset containing 275 3D CT scans for classifying COVID-19, Community-acquired Pneumonia (CAP), and healthy people and segmented lesions in pneumonia patients. We split the dataset into 70% and 30% for training and testing. Extensive experiments showed that our multi-task model framework obtained an average recall of 93.323%, an average precision of 94.005% on the classification test set, and a 69.95% Volume Dice score on the segmentation test set of our dataset.
Keywords: COVID-19, Multi-task learning, Multi-scale, Interpretability, V-Net
1. Introduction
COVID-19 has spread rapidly around the world. Reverse Transcription-Polymerase Chain Reaction (RT-PCR) is considered to be the primary diagnostic method for COVID-19, but it is time-consuming and may produce false-positive cases [1]. Thus, the false-negative cases of RT-PCR tests are a potential threat to public wellness, and the missing of any COVID-19 cases will probably cause secondary infections of large areas.
Meanwhile, the works of [2,3] show that chest Computed Tomography (CT) scans have higher recall in diagnosing COVID-19 which is particularly important in epidemic stricken regions [4,5]. In addition, CT scans are necessary to monitor the severity of the disease [6] due to the high density and spatial resolution. CT scans of patients with COVID-19 mainly show the GGO, consolidation, and other symptoms of the lung, which are also one of the important diagnostic indicators for diagnosis [7]. Therefore, chest CT scans are considered the primary diagnostic modality for COVID-19. There are hundreds of CT slices in each case, and it can be very time-consuming for radiologists to make a diagnosis based on the slices. Even an experienced radiologist can only diagnose an average of seven chest CT scans per hour [8,9].
Deep learning is widely used in medical image processing due to its high accuracy and fast response time. Convolutional Neural Networks (CNNs) significantly impact medical image segmentation and classification tasks. Several CT scan diagnostic systems have been established to assist with COVID-19 diagnosis [[10], [11], [12]], but most suffer from four problems:
-
(1)
For the segmentation task, the infected area of COVID-19 includes nodules, GGO, and other manifestations, as shown in Fig. 1 . They have shortcomings of low contrast between GGO and background, boundary-blurring, making it hard to segment lesions [13] accurately.
-
(2)
For the classification task, Viral pneumonia has a wide range of imaging manifestations. It overlaps with other non-viral pneumonia, with COVID-19 having many similarities to other pneumonia, so how to differentiate is a major clinical challenge [14].
-
(3)
Most existing systems are single-task, which ignores the correlation between segmentation and classification tasks [15].
-
(4)
CNNs are black-box models without interpretability, and physicians do not have sufficient confidence in the model's diagnostic results [16].
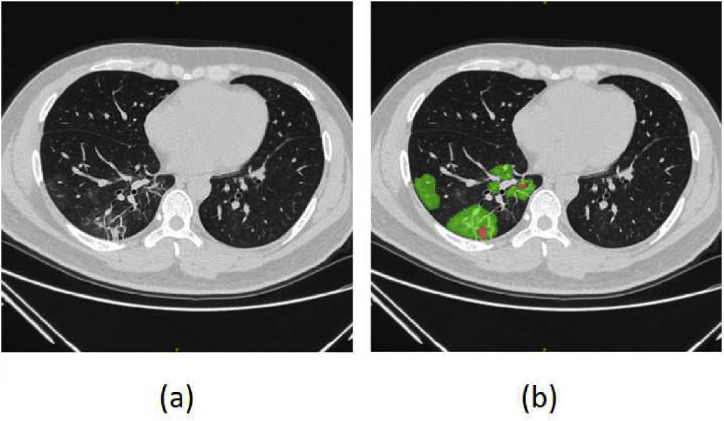
Fig. 1.
The example of COVID-19 lesions, where the red and green areas denote the consolidation and GGO respectively.
To solve the above problems and assist radiologists with COVID-19 diagnosis, in this study, we proposed MultiR-Net, a joint classification and segmentation deep learning network for segmenting COVID-19 lesions and performing diagnosis. MultiR-Net consists of two subnets: a UNet-like subnet for segmentation of 3D lesions with multi-scale feature fusion and a classification subnet for disease diagnosis. Precisely, we weight summed the feature maps at each level of the segmentation subnet, using multi-scale fusion to obtain the final segmentation mask while including the enhanced reversed attention mechanism in the skip connection to improve the network for boundary identification. The segmentation mask obtained from the segmentation subnet is used to strengthen the input volume in the classification subnet. With the assistance of a segmentation mask, the classification subnet can enhance focus on lesions and has a better classification effect. Furthermore, in the training phase, we improve the loss function of the segmentation network by introducing the Focal Tversky loss to enhance the focus of the segmentation network on complex regions. Meanwhile, the iterative training strategy is employed to refine the features by using the predicted segmentation map as part of the input to guide feature extraction.
In short, our contributions are as follows:
-
(1)
We present an interpretable multi-task model based on deep learning for 3D COVID-19 lesion segmentation and disease classification tasks that offers significant advantages over previous systems.
-
(2)
We adopt an iterative training strategy to improve the effectiveness of the network by refining the features.
-
(3)
To comprehensively judge our model's effectiveness, we introduce a variety of surface and volume evaluation metrics to prove the vast superiority of the model segmentation results in terms of surface and volume.
2. Related works
2.1. Automatic COVID-19 diagnosis systems
CT scan is an essential medical imaging diagnostic method, which plays a vital role in the diagnosis of lung diseases [17,18]. Since the outbreak of COIVD-19, a variety of deep learning-based methods have been developed to diagnose COVID-19. Two major categories of tasks are disease classification and lesion segmentation.
The aim of the disease classification task is automatic COVID-19 diagnosis. Aayush Jaiswal et al. [19] proposed a DenseNet201 based network to detect and diagnose COVID-19 in chest CT scans. The network was pre-trained on ImageNet dataset to extract features, then transferred to COVID-19 diagnosis with a transfer learning strategy. Li et al. [20] proposed COVNet, a COVID-19 detection network using ResNet-50 as the backbone, which identified 400 COVID-19 patients with a sensitivity of 90% and a specificity of 96% by generating a classification result from a series of CT slices. Wang et al. [21] developed a weakly-supervised deep learning framework. Specifically, they proposed an unsupervised training strategy for localization by combining active regions and unsupervised connected components in the classification network.
In practice, the segmentation of lesions plays a positive role in improving diagnostic accuracy, reducing misdiagnosis, and assisting doctors in diagnosing lung diseases [22]. Several 3D U-Net [23] variants have been proposed to obtain more accurate results. Attention mechanisms can learn some of the most discriminative features in the network. Oktay et al. [24] proposed attention U-Net capture fine structures in medical images and be suitable for the COVID-19 applications to segment lesions and pulmonary nodules. Fan et al. [25] proposed Inf-Net, a semi-supervised segmentation framework to reduce the need for annotated data. Wang et al. [26] introduced a noise-robust Dice loss to the network to better deal with the noisy training labels for COVID-19. UNet++ [27]is also introduced in COVID-19 lesion segmentation, Jin et al. [28] used UNet++ as the backbone segmentation network to build a system for detecting COVID-19 infection regions. At the same time, UNet++ is hard to train due to its high calculation cost. Zhao et al. [29] proposed a lightweight 3D CNN for COVID-19 segmentation by replacing conventional 3D convolution layers with an attention-based convolutional block. Yan et al. [30] introduced a feature variation block that can adaptively adjust the global features into VNet to enhance the capability of feature representation for different cases.
In recent years, transformer structure has been applied to medical image segmentation tasks and has achieved good performance. Chen et al. [31] combined transformer and U-Net, the transformer encoded image patches from a CNN feature map as input sequences for extracting global features, and the decoder upsampled the encoded features and combined them with high-resolution CNN features for precise localization. Valanarasu et al. [32] proposed Medical Transformer (MedT), a Gated Axial-Attention model, by introducing an additional control mechanism in the self-attention module. Transformer has also been applied in the COVID-19 segmentation task. Roy et al. [33] presented a network derived from Spatial Transformer Networks, which simultaneously predicted the disease severity score associated with the input frame and provides localization of pathological artifacts in a weakly supervised manner.
However, all of the above networks ignore the connectivity between disease classification and lesion segmentation tasks. The classification of pneumonia is highly correlated with the characteristics of its lesions. Interpretable classification results allow the network to focus on the features of the lesion part and can further improve the accuracy of the lesion segmentation results. In the meantime, the segmentation results can be used as an essential diagnostic basis for interpretable disease classification.
2.2. Multi-task learning for COVID-19
In contrast to the single-task classification and segmentation tasks described above, several deep learning methods have also been developed to implement multi-task segmentation and classification of COVID-19. Mahmud [34] proposed a tri-level attention mechanism-based network for disease classification, lesion segmentation, and disease severity prediction. Specifically, they pre-trained a subnet for lesion segmentation and integrated the weights obtained into two other tasks. Similarly, Wu et al. [16] proposed a COVID-19 automatic diagnosis system. They first trained a separate classification network withRes2Net as the backbone. They then used the weights of the classification network for feature fusion with the segmentation network to obtain lesion segmentation results. Similarly, Jin et al. [28] proposed a linear optimization procedure, in contrast to Wu, who used the results of the segmentation network to guide the classification.
However, the above approaches split the learning steps of segmentation and classification tasks in the training strategy, ignoring the feature sharing between two tasks, and thus cannot be regarded as multi-task learning in the true sense. Currently, only a few studies have focused on feature correlation between tasks and jointly learned both 3D lesion segmentation and classification of covid-19. Wang et al. [15] proposed a joint deep learning model of 3D lesion segmentation and classification for diagnosing COVID-19. Specifically, the network contains three subnetworks, extracts shared features through a cross-task feature subnet and proposes task-aware loss-enhanced task interaction between the classification and segmentation subnetworks.
3. Methodology
Inspired by V-Net [35] and Liu's network [36], we propose MultiR-Net. Suppose a given 3D CT scan , where , , denote the length, width, and depth of the 3D CT scan respectively. Manually labeled ground truth areas and prediction results have the same size as . Firstly, will be divided into a series of -slice sequence , which is the basic unit of network input. Then MultiR-Net will derive the predicted volumes and classification results . The network consists of two subnets, the segmentation subnet and the classification subnet. The UNet-like network can integrate high semantic-level and low semantic-level features in the segmentation subnet; in the classification subnet, the input volume will be added to the segmentation map from the segmentation subnetwork, facilitating the focus of the subnet on the lesion. Meanwhile, the Iterative learning strategy is applied to guide the network training with the segmentation results.
The following sections will introduce: 1) network architecture; 2) Reverse attention module; 3) Loss Function; 4) Iterative learning strategy; 5) Evaluate metrics.
3.1. Network architecture
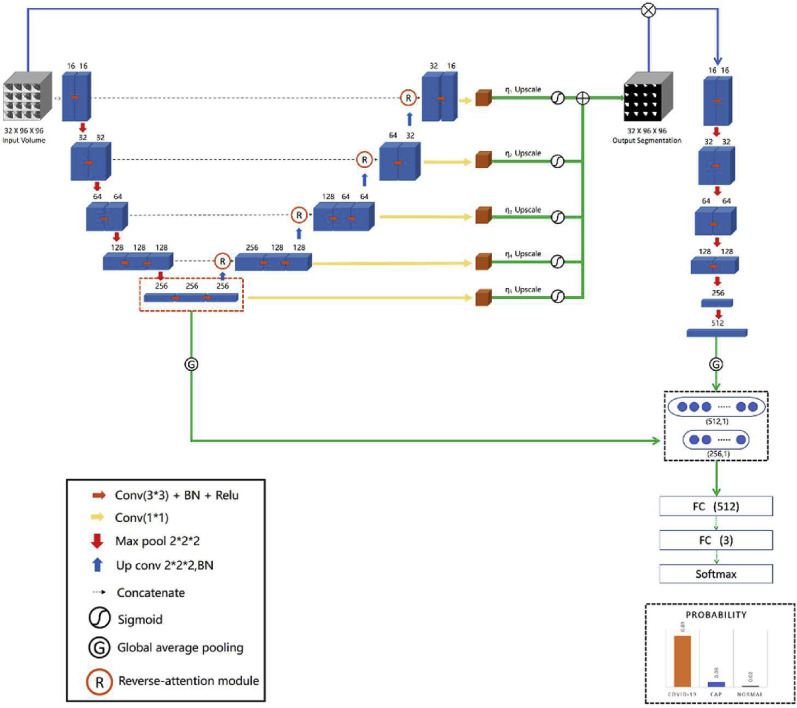
Our proposed network is illustrated by Fig. 2 , where we use the V-Net as the backbone of the segmentation subnet due to the outstanding performance in various tasks [37]. Our segmentation subnet consists of encoding and decoding paths. Encoding path extracts features of the input volume and reduces to 16 times through down-sampling while preserving the information of each level; the symmetric decoding path restores the high-level semantic features obtained by an encoding path to the original dimension through up-sampling. The reverse attention module in skip connection can improve the segmentation of the edge of the lesion. In order to make full use of the features extracted by the classification subnet at different scales, we use 1*1 convolution to process further the feature maps obtained from each layer of the decoding layer to generate a multi-scale segmentation map. Assuming that is the segmentation map at the i-th level, the final segmentation mask can be formalized as:
| (1) |
where is the hyperparameter used to balance the scale of features at each scale, σ is sigmoid operation, and UP(·) is the t-fold upsampling operation using deconvolution layer.
Fig. 2.
The overall architecture of MultiR-Net.
The classification subnet consists of six layers of convolutional modules, and we add a residual structure between each convolutional module to alleviate the problem of difficult gradient back-propagation. The segmentation results are element-wise multiplied with the original input and then fed to the classification subnet to enhance the interaction between the tasks. The feature maps obtained will have higher activation values in the lesion region and make the classification subnet focus on the lesion.
3.2. Reverse attention module
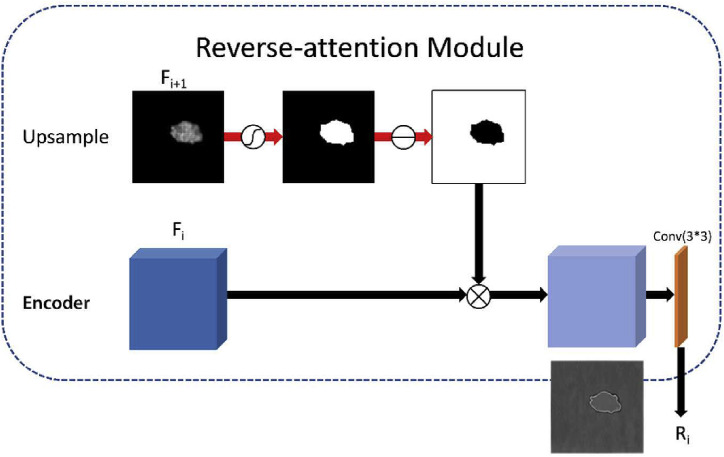
The reverse attention mechanism is used for the salient object detection edge refinement [38], a different network structure from the U-Net. Our segmentation subnet was able to localize the lesions in the input volume; however, due to the blurred boundaries and low contrast of the lesions in COVID-19. They have shortcomings of low contrast between GGO and background, blurring boundary, making it hard to segment accurately. We modified the number of channels in the reverse attention module to segment these lesions accurately. We added the reverse attention module to the skip connection as a fine marker to identify infected regions in an erasing-strategy. By erasing the current prediction result from skip-connection features obtained by the encoder, where the current prediction result is upsampled from the high-level features, the network can learn the details of the complementary regions to focus on the segmentation of the boundary regions. The top-down erasing strategy can eventually refine the coarse prediction into a complete and high-resolution prediction result with these explored regions and details. The architecture of the RA module is illustrated in Fig. 3 . Notably, we obtain the output reverse attention features R by multiplying(element-wise ) the high-level features by the reverse attention weights , it is formalized as:
| (2) |
where can be obtained from the following formula:
| (3) |
where is a reverse operation subtracting the input feature from the matrix where all the elements are 1, as shown in Fig. 3, the reverse attention features enable increased attention to boundaries.
Fig. 3.
The architecture reverse attention module.
In order to make the reverse attention module work better in our network, we improve the numbers of output channels from 64 to be the same as each level of the segmentation subnet. The variable number of channels ensures that the reverse attention mechanism can refine all feature maps connected by skip connection, improving segmentation by adjusting imprecise and rough estimates to accurate and complete prediction maps.
3.3. Loss function
The loss functions are introduced for training the MultiR-Net, including segmentation and classification. We will then introduce the two parts of the loss function separately.
-
(1)
Segmentation loss
Dice Score Coefficient (DSC) is widely used in medical image segmentation to assess the overlapping rate between the segmented region and ground truth. It is a significant metric to evaluate the segmentation performance. The two-class DSC and DSC Loss are defined in (4), (5).
| (4) |
| (5) |
where N denotes the number of the voxels in a CT scan, denotes the predicted classes, denotes the probability that voxel is of the class , denotes the ground truth that voxel is of the class , and is a constant to prevent division by zero. Using DSC Loss may encounter the problem that it equally weighs False-Positive (FP) and False-Negative (FN), which leads to the segment results with high precision but low recall. Experimental results prove that FN needs to be higher than FP in a highly imbalanced dataset such as COVID-19 lesions to improve recall rate [39]. Tversky similarity index [40] is a generalization of the DSC, which allows balancing FP and FN flexibly. Tversky similarity index is defined in (6).
| (6) |
where denotes the probability that voxel is of the class , denotes the probability that voxel is of the non-class , denotes the ground truth that voxel is of the class c, denotes the ground truth that voxel is of the non-class , and are hyperparameters that can shift the emphasis between FP and FN. Another problem in the COVID-19 dataset is the small Regions Of Interest (ROI), resulting in a low contribution to the loss. The Focal Tversky Loss function (FTL) [41] adds a hyperparameter to reduce the loss of easily classified samples. FTL is defined in (7).
| (7) |
where is a hyperparameter that varies in the range [1,3]. If the Tversky index is small and the misclassified pixel, the FTL will decrease significantly. We use FTL to train the network for segmenting small ROIs in the COVID-19 dataset.
-
(2)
Classification loss
For classification task, we use Cross Entropy loss as the classification loss function:
| (8) |
where is the number of class, is the true classification of the data, and is the predicted probability.
-
(3)
Total loss
Finally, to balance the losses of segmentation and classification tasks to the same order of magnitude and thus improve the effectiveness of multi-task learning, the classification loss and the segmentation loss are combined linearly by the hyperparameter as a multitasking loss. The multi-task loss is defined as:
| (9) |
where ∈ [0, 1], is the weight of the classification loss.
3.4. Iterative learning strategy
COVID-19 lesions show blurred boundaries and variable shapes in CT scans, complicating lesion segmentation. In order to improve the performance of the edge and improve the segmentation performance while further enhancing the interaction between tasks, we applied an iterative training strategy. We use the segmentation feature map from the previous iteration to update the features, as shown in Algorithm 1. The input volume is the original 3D chest CT scan in the first iteration. The output feature map contains contextual information that can direct the network to focus on the lesion region. In practice, it is performed on a feature map obtained after multi-scale feature fusion. In subsequent iterations, the input changes to the original chest CT scan with the segmentation feature map obtained in the previous iteration.
Algorithm 1
Iterative learning strategy.
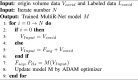
3.5. Evaluate metrics
Segmentation accuracy determines the eventual performance of segmentation procedures. To measure the segmentation performance of the proposed methods, four evaluation metrics: Average Surface Distance (), Average Surface Overlap (), Surface Dice (), and Volumetric Dice () are used to obtain quantitative measurements of the segmentation accuracy.
ASD determines the average difference between the surface of the segmented area and the ground truth in 3D. This metric is also used in the medical image segmentation challenges, such as CHAOS [42] and RT-MAC [43]. ASD can be defined as follows:
| (10) |
where denotes the set of surface voxels of the predicted mask, denotes the set of ground truth surface voxels. ||·|| denotes the Euclidean distance.
Average determines the overlapping parts of segmented and reference volumes. Standard are defined in (4).
COVID-19 lesions are deformable without a uniform approximate shape. Thus, assessment of lesion segmentation quality may not be optimal in a volume-based comparison. Therefore, we also calculated and for our dataset, which are also used in neuroimaging [44].
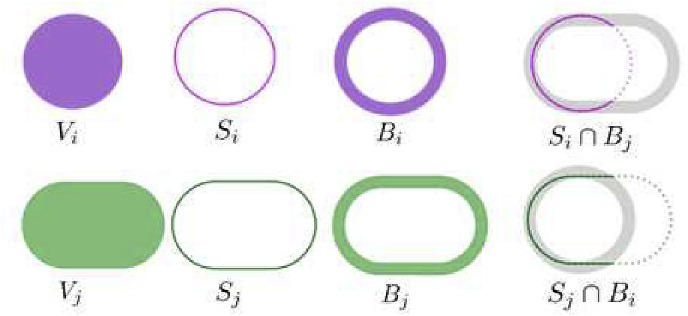
As shown in Fig. 4 , calculates the surface overlap ratio of the segmented area and the reference within a given tolerance range and while calculates the surface dice as the same way. Here, we set the tolerance range to be 1.0 mm. The surface of a volumetric object can be defined as (11):
| (11) |
where denotes the point on the surface . Therefore, can be defined as:
| (12) |
Fig. 4.
The diagram for calculating .
And can be defined as:
| (13) |
For classification task, we employ Recall , Precision , Accuracy (), F1-score for quantitative evaluation of:
| (14) |
| (15) |
| (16) |
where TP, FP, TN, FN are the number of true positives, false positives, true negatives, and false negatives, respectively.
4. Experiments
To evaluate the effectiveness of the proposed network MultiR-Net, we conduct several ablation experiments. Furthermore, we systematically compare the MultiR-Net with existing multi-task learning methods, including Joint Classification and Segmentation System(JCS) [16]and Zhou's network [45]. Also, we compare the method with existing single-task segmentation and classification networks to give a comprehensive picture of the performance of the proposed network. The dataset, implementation details, and experimental results are shown as follows.
4.1. Dataset and preprocessing
The dataset used in this paper includes CT scans from 275 patients with COVID-19, CAP, and ordinary people, over 20000 CT images. The spacing of slices between CT scans varies considerably depending on the machine used for acquisition (from 1.0 mm to 5.0 mm). CT data is collected from a Class A tertiary hospital in Shanghai, 96 of which are positive cases of COVID-19, 107 cases are CAP patients, and the other 72 cases are ordinary people. RT-PCR tests confirm all cases. Two radiologists labeled the images using two labels: GGO and consolidation. While due to the severe data imbalance, the number of slices with consolidation is much smaller than that with GGO, we take all the labels as unified COVID-19 lesion labels. The difficulty level of the dataset was proved to be balanced by radiologists. We randomly split the datasets into 70% and 30% for training and testing. The division of the dataset is shown in Table 1 .
Table 1.
Division of the dataset.
| COVID-19 | CAP | Ordinary People | |
|---|---|---|---|
| Training | 67 | 75 | 50 |
| Testing | 29 | 32 | 22 |
| Overall | 96 | 107 | 72 |
Excessive spacing gaps can make it hard for the network to effectively learn the space contextual information. Thus, before sending CT scans data to network training, we need to use Algorithm 2 for resampling and normalization operation to enhance the region to be segmented. Then, COVID-19 lesion ROIs with the size of 32 × 96 × 96 voxels were extracted from a scan at the approximate lesion center.
Algorithm 2
COVID-19 Normalization.

4.2. Implementation details
Our network is implemented in Tensorflow. The experiments are run on a computer with a single Nvidia GPU Tesla V100 and an Intel(R) Xeon(R) Gold 5115 CPU @ 2.40 GHz. The network is optimized using the Adam optimizer and set , as 0.9 and 0.999. The initial learning rate is set to 3e-4 with the patience of 100 epochs. We set the batch size to 4, and early stopping is employed to avoid overfitting. CT slices and the masks are resized into 32 × 96 × 96. For the Focal Tversky loss, the network is trained with = 0.7, = 0.3 and = 0.9 to get the best performance. To evaluate the effectiveness of the proposed method, we employed four-fold cross-validation for all experiments.
4.3. Lesion segmentation results
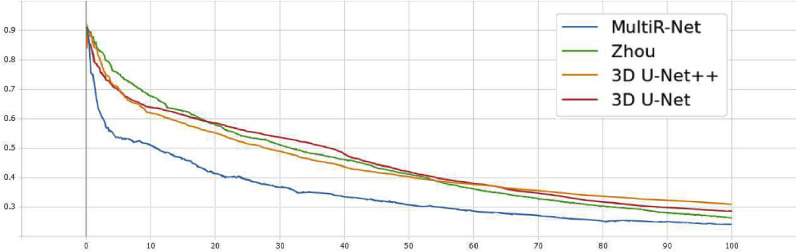
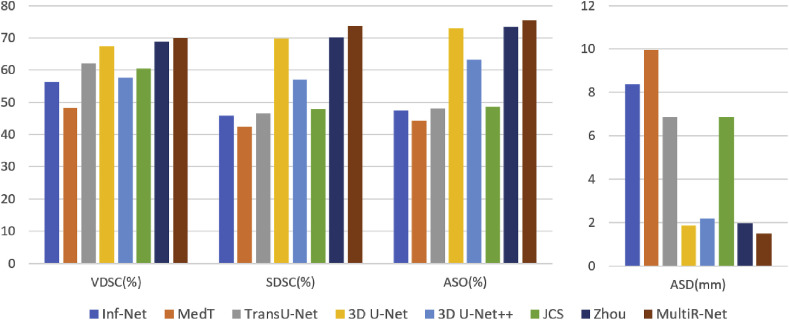
To comprehensively evaluate the segmentation performance of the proposed method, we compare MultiR-Net with generic methods such as 3D U-Net, 3D U-Net++, MedT, TransU-Net, and the specialized segmentation method Inf-Net of COVID-19 segmentation. Also, to compare the effectiveness of our network on multi-task classification segmentation networks, we compare it with Zhou's method and JCS, a dedicated COVID-19 multi-task classification segmentation network. All methods use their original implementation, except for Zhou's method, and 3D U-Net++ are reimplemented. The training loss curves of the networks are shown in Fig. 5 . The final results on the testing set are shown in Table 2 and Fig. 6 .
Fig. 5.
The training loss curves of the networks.
Table 2.
Segmentation performance of nine networks tested on dataset.
| Inf-Net(2D) [25] | 8.3673* | 56.21* | 45.83* | 47.43* |
| MedT(2D) [32] | 9.9634* | 48.25* | 42.40* | 44.33 |
| TransU-Net(2D) [31] | 6.8709* | 62.15* | 46.63* | 48.02 |
| 3D U-Net | 1.8579* | 67.41* | 69.69 | 72.96 |
| 3D U-Net++ | 2.2089** | 57.53** | 57.10** | 63.19** |
| JCS [16] | 6.8589* | 60.38* | 47.94* | 48.59 |
| Zhou's [45] | 1.9771* | 68.87* | 70.24* | 72.09* |
| MultiR-Net w/o cls | 1.6334** | 68.73** | 71.63** | 73.38** |
| MultiR-Net with cls | 1.4897 | 69.95 | 73.68 | 75.32 |
Correction for comparisons is performed with the T test. ∗ Highest average metric values are <0.05 for all comparisons. ∗∗ Highest average metric values are <0.0001 for all comparisons.
Fig. 6.
The segmentation performance of the networks.
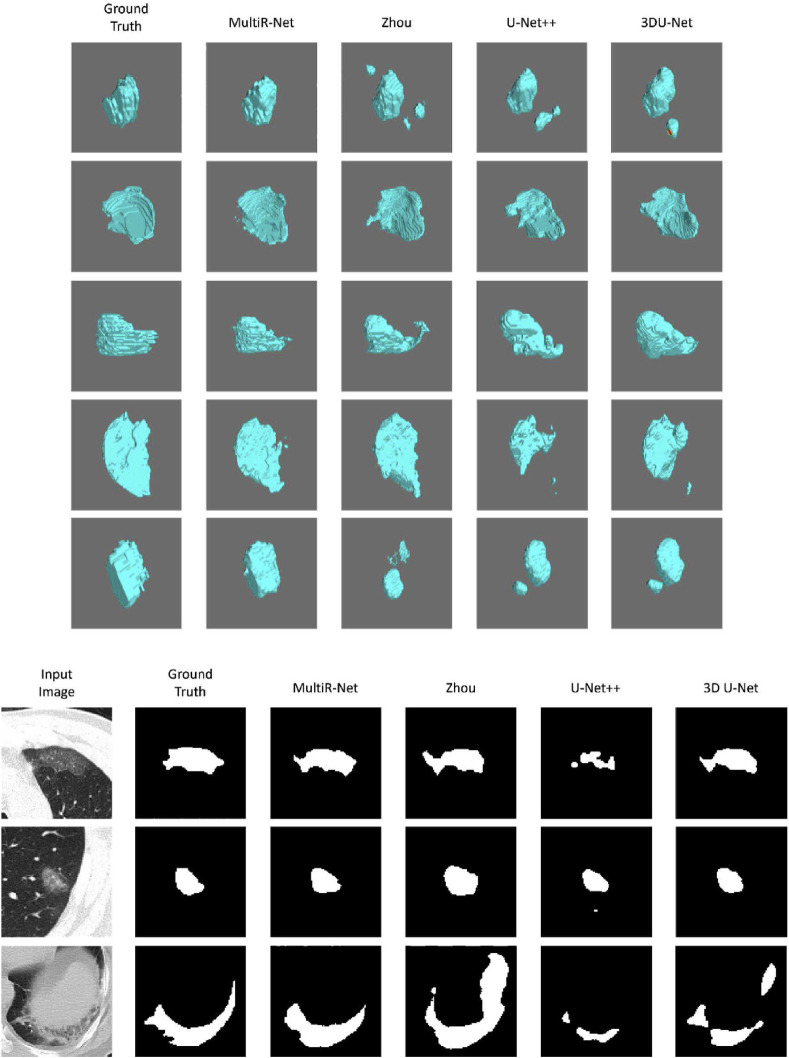
As shown in Table 2, it can be seen that 2D networks are all lower in overall performance than networks with 3D input. This is mainly due to the characteristics of the COVID-19 dataset. COVID-19 lesions have a strong 3D consecutiveness and correlation between slices. 3D convolution can better extract the related information in three dimensions. At the same time, the 2D network extracts global features, while the 3D network with a patch extracts local features so that the 3D network performs better in the segmentation of edges. Our method achieved the best accuracy in all metrics with 1.48 mm,70.0%,73.7% and 75.3% in , , , respectively. Compared with our model, the performance of other single-task models is lower. It is worth noting that the results of 3D U-Net++ are worse compared to U-Net, presumably because the U-Net++ decoder part is too complex and the number of parameters is too large to learn the volume features. At the same time, it can be seen that our method does not improve significantly in , with a difference of 1.2% over the second-best model. However, our method plays a better role in the other three metrics evaluating the degree of surface overlap, improving by 0.49 mm, 3.4%, and 3.2%, respectively, indicating that our network fits better in the volume surface, which is also shown in Fig. 7 . Fig. 7 shows the visual segmentation model's qualitative results. We use the binary segmentation result for 2D visualization, and we convert the segmentation result to a 3D NumPy array and use the Mayavi package in Python for 3D scientific data visualization. Our method outperforms the other models in COVID-19 and CAP segmentation.
Fig. 7.
The visualization images of 3D and 2D lesion segmentation results.
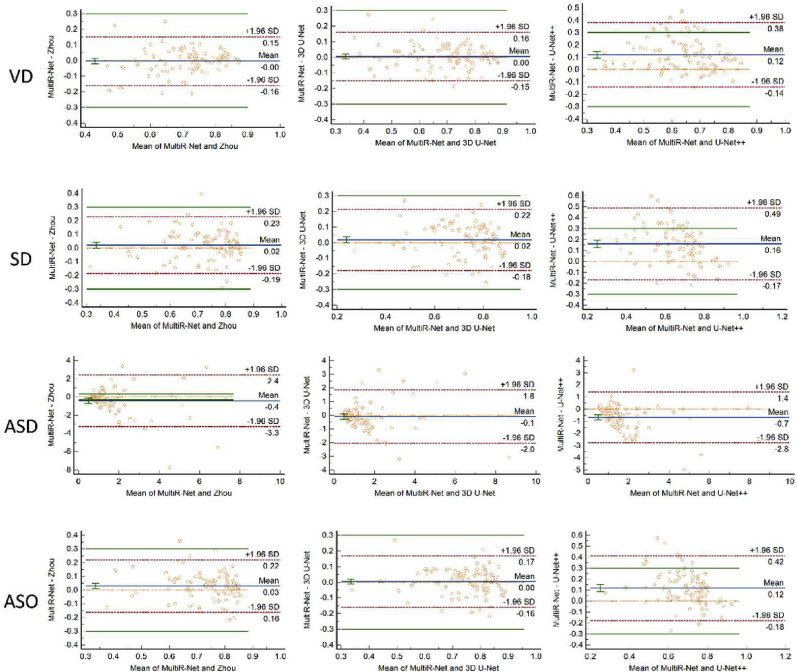
Meanwhile, we add Bland-Altman [46] plots to show the exact type of potential misbehaviors in the segmentation networks, as shown in Fig. 8 . Bland-Altman is used to tell how far apart measurements by two methods are more likely to be for most individuals. It can be seen that most of the differences are within ±1.96 standard deviation, indicating that the two methods are in good agreement and may be used interchangeably.
Fig. 8.
The Bland-Altman plots of the segmentation networks.
Also, to evaluate the segmentation effect of our multi-task model, we kept only the segmentation subnet of the network. We tested its effect on the dataset, as shown in Table 2. It can be seen that in terms of segmentation, the overall accuracy of the single-task network is lower than that of the multi-task learning network, and the multi-task gains in terms of , , and are 0.14 mm, 1.2%, 2.1%, and 1.9% respectively.
4.4. Explainable disease classification results
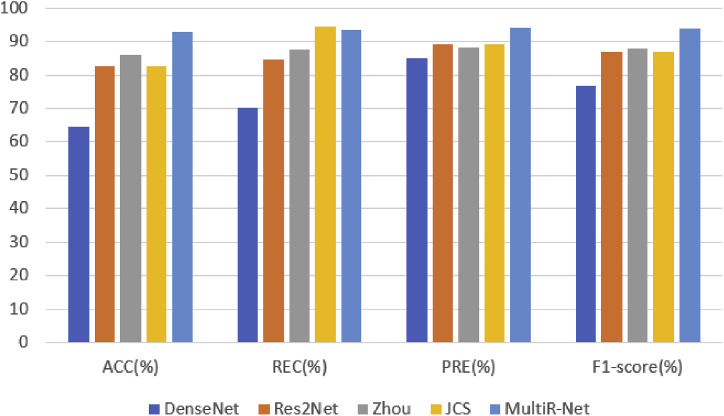
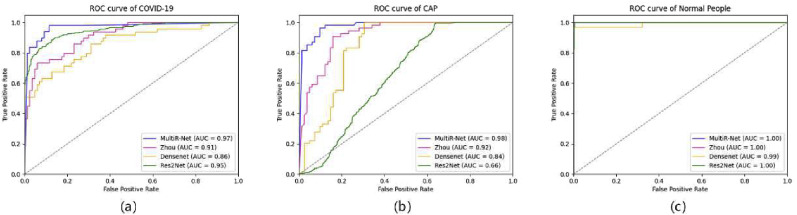
To comprehensively evaluate the classification performance of the proposed method, we compare MultiR-Net with DenseNet, Res2Net [47], and other state-of-the-art multi-task models. JCS has consistent metrics for both in the classification task due to the Res2Net classification model being trained separately first and then the fusion of features for multiple tasks. The quantitative results on the testing set are shown in Table 3 and Fig. 9 . The Roc curves of each classification are shown in Fig. 10 .
Table 3.
Classification performance of five networks tested on dataset.
| DenseNet | 64.706* | 70.068* | 85.034* | 76.829* |
| Res2Net | 82.482* | 84.495* | 89.322* | 86.841* |
| Zhou's | 86.029* | 87.453** | 88.134* | 87.792* |
| JCS | 82.482* | 84.495* | 89.322 | 86.841* |
| MultiR-Net | 92.647 | 93.323 | 94.005 | 93.663 |
Correction for comparisons is performed with the T test. ∗ Highest average metric values are <0.05 for all comparisons. ∗∗ Highest average metric values are <0.0001 for all comparisons.
Fig. 9.
The classification performance of the networks.
Fig. 10.
The ROC curves of our MultiR-Net and other models of (a) COVID-19, (b) CAP and (c) Normal people.
As shown in Table 3, the classification performance of our method was significantly higher than other methods, achieving results of 92.6%, 93.3%, and 94.0% in ACC, REC, and PRE, with improvements of 6.6%, 5.9%, and 5.9% in each metric, compared to the suboptimal performing model.
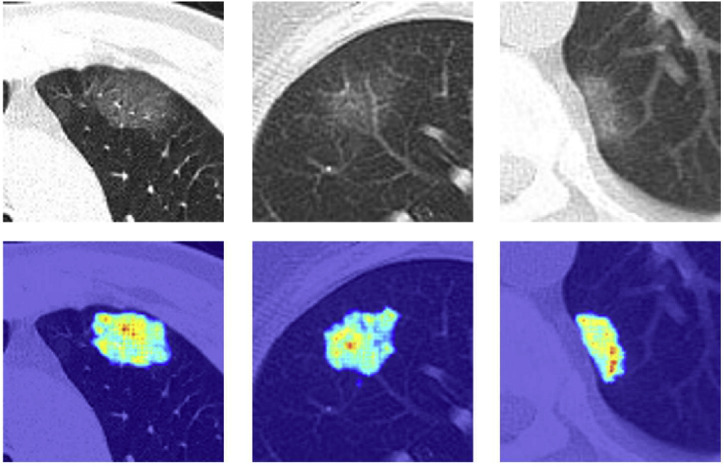
Meanwhile, to verify the interpretability of the method for classification, we output the class activation map of MultiR-Net for COVID-19 using the Grad-CAM [48] method, as shown in Fig. 11 . It can be seen that as the network performs a Global Average Pooling operation on the feature map of the segmentation subnet together with the classification subnet to obtain the final classification results, the heatmap is highly consistent with the segmentation results, proving that our model does focus on the focal regions of the image and is interpretable.
Fig. 11.
The interpretable visualizations of Grad-CAM.
4.5. Ablation study of MultiR-Net
This section conducts an ablation study to show that each module in the proposed method contributes to the segmentation and classification results. Specifically, we divide the modules in the proposed method into three parts: the reverse attention module, the Focal Tversky loss function, and the iterative learning strategy. We remove each part from the network while retaining the rest and analyzing the impact of different modules on network training.
-
1)
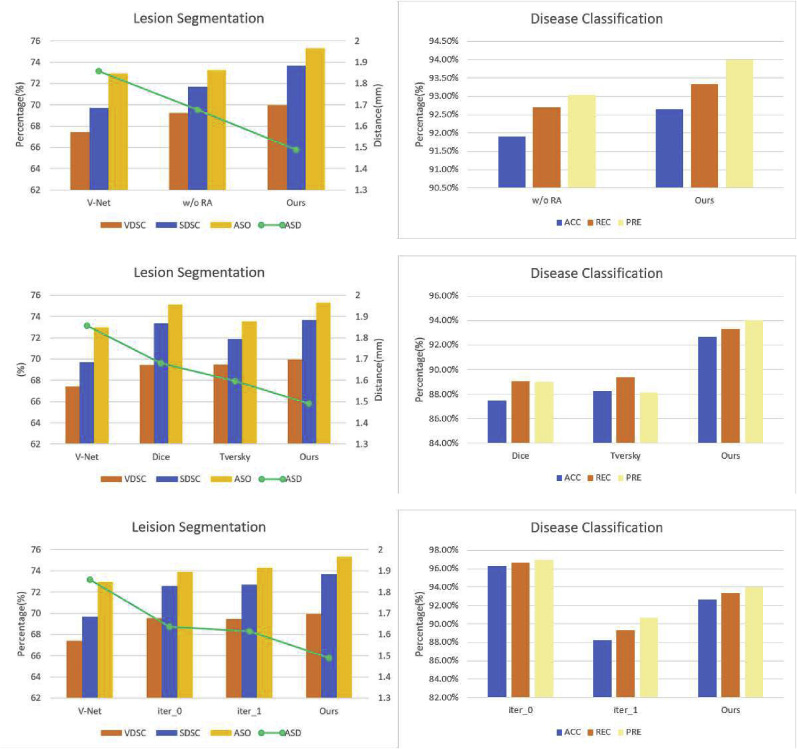
Effectiveness of Reverse attention module: We first analyze the impact of the reverse attention module on 3D lesion segmentation and disease classification. Specifically, we remove the reverse attention module from the skip connection. The specific segmentation and classification results are shown in Fig. 12 . It can be seen that after removing the reverse attention module, the performance of the segmentation task was significantly reduced, especially for the three metrics , and , by 0.19 mm, 2% and 2.1%, indicating the effectiveness of the reverse attention module for segmentation.
-
2)
Effectiveness of Reverse attention module: Secondly, we carry out further ablation experiments to analyze the impact of our improvements on the loss function. To prove the effectiveness of the Focal Tversky loss in balancing FP and FN, we compare three different loss functions: (1) Dice Loss, (2) Tversky Loss, and (3) Focal Tversky Loss.
Fig. 12.
The effect of Reserve-attention module, Focal Tversky Loss and Iterative Learning Strategy on the performance of segmentation and classification.
Dice loss is a commonly used loss function in medical image segmentation and can be used as a baseline to compare the effects of different loss functions. The results of different loss functions are shown in Fig. 12. It can be observed that the introduced Focal Tversky Loss can deliver the best performance.
-
3)
Effectiveness of Iterative Learning Strategy: Finally, to verify the effectiveness of the iterative learning strategy, the following experiments are conducted. Expressly, we set different numbers of iterations n, n = 0, n = 1, and n = 2, respectively. Note that n = 0 indicates that no iterative strategy is applied. Fig. 12 shows the segmentation and classification results. It can be seen that the segmentation performance of the network gradually improves as the number of iterations increases, illustrating the effectiveness of the iterative training strategy. It is worth noting that the classification accuracy of the network reaches its maximum when n = 0, i.e., no iterative training strategy is used.
4.6. Discussion
-
1)
Effectiveness and application
We demonstrate the effectiveness of our method in COVID-19 segmentation and classification. Considering that the commonly used Dice coefficients do not give a comprehensive representation of the segmentation effect, we use several metrics to measure the effectiveness of the network at the volume level and the surface level. We obtain superior performance on all the metrics evaluated, meaning that the proposed method correctly segments the COVID-19 foci in our test data. The introduction of reverse attention into the network proves to be effective.
Our main goal is to assist physicians in the diagnosis of COVID-19:
-
1.
The model can segment and display portions of COVID-19 lesion at the volumetric level.
-
2.
The classification results can provide a quick prediagnosis for physicians.
-
3.
Based on the location and size of the segmentation results, our network can help physicians determine the condition's severity.
-
2)
Limitations and future work:
Dataset Limitations: The lesions in the COVID-19 dataset are much smaller than the background, presenting a class imbalance. We try to attenuate the effect of class imbalance by improving the loss function, but it still affects the network's training to some extent.
Meanwhile, since COVID-19 multi-task learning is an emerging field with no available public datasets and corresponding methods for comparison, the performance improvement of our proposed network cannot be fully demonstrated when compared with the standard algorithms for medical image segmentation.
Method Limitations: Due to the high computational cost of a 3D input, a CT scan needs to be cropped to several small volumes. Thus the network is not an end-to-end framework. Meanwhile, our COVID-19 dataset label is divided into consolidation and GGO. At the same time, the training uniformly views both as the lesion part for single-class segmentation, which may lead to unsatisfactory learning performance.
Based on the above limitations, we plan to make the following improvements to the network:
Dataset improvement: we will add more labeled COVID-19 CT scans to the existing dataset for training while collecting more data from different pneumonia cases to enhance the robustness of the network and reduce the impact of class imbalance.
End-to-end framework: the preprocessing process will be improved with a multi-stage strategy that firstly detects the location of the focal part of the network and crops it to a small volume for segmentation and classification. In future work, we will investigate how to build an end-to-end framework for the segmentation task.
Multi-classification segmentation: the existing single-class segmentation network will be enhanced to a multi-class segmentation network to accommodate the different characterization of the COVID-19 focal part and improve the network's performance.
5. Conclusion
This study proposes a multi-task learning COVID-19 segmentation and classification network that utilizes the reverse attention mechanism to identify infected regions in an erasure fashion. The network can learn details of complementary regions to focus on the segmentation of boundary regions. In addition, we modify the loss function and propose an iterative learning strategy to enhance the interaction between tasks. To fully demonstrate the network's performance, experimental and visualization results on the dataset show the existing frontier segmentation model of the proposed network as a whole. Meanwhile, various evaluation metrics show the combined advantages of the proposed network in terms of volume and surface. Our system holds great promise in assisting physicians with COVID-19 diagnostics, such as rapid localization of lesions and quantification of infected areas and exemplary performance in low-contrast, border-blurring lesion segmentation. In the future, we will add more labeled COVID-19 CT scans to the existing dataset for training and develop an end-to-end framework for segmentation and classification.
APPROVAL
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Ethics Committee of Shanghai Jiao Tong University School of Medicine, Shanghai Ninth People's Hospital (Approval code: SH9H-2020-TK9-1).
Declaration of competing interest
We declare that we have no financial and personal relationships with other people or organizations that can inappropriately influence our work, there is no professional or other personal interest of any nature or kind in any product, service and/or company that could be construed as influencing the position presented in, or the review of, the manuscript entitled, “MultiR-Net: A Novel Joint Learning Network for COVID-19 Segmentation and classification”.
References
- 1.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of chest CT and RT-PCR testing for coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020;296 doi: 10.1148/radiol.2020200642. 02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu B., Xing Y., Peng J., Zheng Z., Tang W., Sun Y., Xu C., Peng F. Chest CT for detecting COVID-19: a systematic review and meta-analysis of diagnostic accuracy. Eur. Radiol. 2020;30:5720–5727. doi: 10.1007/s00330-020-06934-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang L., Han R., Ai T., Yu P., Kang H., Tao Q., Xia L. Serial quantitative chest CT assessment of COVID-19: a deep learning approach. Radiology: Cardiothor. Imaging. 2020;2(2) doi: 10.1148/ryct.2020200075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lei J., Li J., Li X., Qi X. CT imaging of the 2019 novel coronavirus (2019-nCoV) pneumonia. Radiology. 2020;295(1) doi: 10.1148/radiol.2020200236. 18–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang W., Sirajuddin A., Zhang X., Liu G., Teng Z., Zhao S., Lu M. The role of imaging in 2019 novel coronavirus pneumonia (COVID-19) Eur. Radiol. 2020:1–9. doi: 10.1007/s00330-020-06827-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi F., Wang J., Shi J., Wu Z., Wang Q., Tang Z., He K., Shi Y., Shen D. Review of artificial intelligence techniques in imaging data acquisition, segmentation, and diagnosis for COVID-19. IEEE Rev. Biomed. Eng. 2020;14:4–15. doi: 10.1109/RBME.2020.2987975. [DOI] [PubMed] [Google Scholar]
- 7.Wang Tianming, Zeng Xiong, Zhou Hui, Luo Weijun, Tang Haixiong, Liu Jinkang. Design, validation, and clinical practice of standardized imaging diagnostic report for covid-19. Zhong nan da xue xue bao. Yi xue ban = J. Centr. South Univ. Med. Sci. 2020;45(3):229–235. doi: 10.11817/j.issn.1672-7347.2020.200152. [DOI] [PubMed] [Google Scholar]
- 8.Cowan I.A., MacDonald S.L.S., Floyd R.A. Measuring and managing radiologist workload: measuring radiologist reporting times using data from a Radiology Information System. J. Med. Imaging Radiat. Oncol. 2013;57(5):558–566. doi: 10.1111/1754-9485.12092. [DOI] [PubMed] [Google Scholar]
- 9.An P., Ye Y., Chen M., Chen Y., Fan W., Wang Y. Management strategy of novel coronavirus (COVID-19) pneumonia in the radiology department: a Chinese experience. Diagn. Interventional Radiol. 2020;26(3):200. doi: 10.5152/dir.2020.20167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mei X., Lee H.C., Diao K., Huang M., Lin B., Liu C., Xie Z., Ma Y., Robson P.M., Chung M., et al. Artificial intelligence–enabled rapid diagnosis of patients with COVID-19. Nat. Med. 2020;26(8):1224–1228. doi: 10.1038/s41591-020-0931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang K., Liu X., Shen J., Li Z., Sang Y., Wu X., Zha Y., Liang W., Wang C., Wang K., et al. Clinically applicable AI system for accurate diagnosis, quantitative measurements, and prognosis of COVID-19 pneumonia using computed tomography. Cell. 2020;181(6):1423–1433. doi: 10.1016/j.cell.2020.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shoeibi A., Khodatars M., Alizadehsani R., Ghassemi N., Jafari M., Moridian P., Khadem A., Sadeghi D., Hussain S., Zare A., et al. 2020. Automated Detection and Forecasting of Covid-19 Using Deep Learning Techniques: A Review. arXiv preprint arXiv:2007.10785. [Google Scholar]
- 13.Wu J., Wu X., Zeng W., Guo D., Fang Z., Chen L., Huang H., Li C. Chest CT findings in patients with coronavirus disease 2019 and its relationship with clinical features. Invest. Radiol. 2020;55(5):257. doi: 10.1097/RLI.0000000000000670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanne J.P. 2020. Chest CT Findings in 2019 Novel Coronavirus (2019-nCoV) Infections from Wuhan, China: Key Points for the Radiologist. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X., Jiang L., Li L., Xu M., Deng X., Dai L., Xu X., Li T., Guo Y., Wang Z., et al. Joint learning of 3D lesion segmentation and classification for explainable COVID-19 diagnosis. IEEE Trans. Med. Imag. 2021 doi: 10.1109/TMI.2021.3079709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Y.H., Gao S.H., Mei J., Xu J., Fan D.P., Zhang R.G., Cheng M.M. Jcs: An explainable covid-19 diagnosis system by joint classification and segmentation. IEEE Trans. Image Process. 2021;30:3113–3126. doi: 10.1109/TIP.2021.3058783. [DOI] [PubMed] [Google Scholar]
- 17.Sluimer I., Schilham A., Prokop M., Van Ginneken B. Computer analysis of computed tomography scans of the lung: a survey. IEEE Trans. Med. Imag. 2006;25(4):385–405. doi: 10.1109/TMI.2005.862753. [DOI] [PubMed] [Google Scholar]
- 18.Kamble B., Sahu S.P., Doriya R. Advances in Data and Information Sciences. Springer; 2020. A review on lung and nodule segmentation techniques; pp. 555–565. [Google Scholar]
- 19.Jaiswal A., Gianchandani N., Singh D., Kumar V., Kaur M. Classification of the COVID-19 infected patients using DenseNet201 based deep transfer learning. J. Biomol. Struct. Dyn. 2020:1–8. doi: 10.1080/07391102.2020.1788642. [DOI] [PubMed] [Google Scholar]
- 20.Li L., Qin L., Xu Z., Yin Y., Wang X., Kong B., Bai J., Lu Y., Fang Z., Song Q., et al. Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology. 2020 doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X., Deng X., Fu Q., Zhou Q., Feng J., Ma H., Liu W., Zheng C. A weakly-supervised framework for COVID-19 classification and lesion localization from chest CT. IEEE Trans. Med. Imag. 2020;39(8):2615–2625. doi: 10.1109/TMI.2020.2995965. [DOI] [PubMed] [Google Scholar]
- 22.Gordaliza P.M., Muñoz-Barrutia A., Abella Mónica, Desco M., Sharpe S., Vaquero J.J. Unsupervised CT lung image segmentation of a mycobacterium tuberculosis infection model. Sci. Rep. 2018;8(1):1–10. doi: 10.1038/s41598-018-28100-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Çiçek Ö., Abdulkadir A., Lienkamp S.S., Brox T., Ronneberger O. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2016. 3D U-Net: learning dense volumetric segmentation from sparse annotation; pp. 424–432. [Google Scholar]
- 24.Oktay O., Schlemper J., Folgoc L.L., Lee M., Heinrich M., Misawa K., Mori K., McDonagh S., Hammerla N.Y., Kainz B., et al. 2018. Attention U-Net: Learning where to Look for the Pancreas. arXiv preprint arXiv:1804.03999. [Google Scholar]
- 25.Fan D.P., Zhou T., Ji G.P., Zhou Y., Chen G., Fu H., Shen J., Shao L. Inf-net: Automatic covid-19 lung infection segmentation from ct images. IEEE Trans. Med. Imag. 2020;39(8):2626–2637. doi: 10.1109/TMI.2020.2996645. [DOI] [PubMed] [Google Scholar]
- 26.Wang G., Liu X., Li C., Xu Z., Ruan J., Zhu H., Meng T., Li K., Huang N., Zhang S. A noise-robust framework for automatic segmentation of COVID-19 pneumonia lesions from CT images. IEEE Trans. Med. Imag. 2020;39(8):2653–2663. doi: 10.1109/TMI.2020.3000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Z., Siddiquee M.M.R., Tajbakhsh N., Liang J. Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support. Springer; 2018. Unet++: A nested u-net architecture for medical image segmentation; pp. 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin S., Wang B., Xu H., Luo C., Wei L., Zhao W., Hou X., Ma W., Xu Z., Zheng Z., et al. AI-assisted CT imaging analysis for COVID-19 screening: building and deploying a medical AI system in four weeks. medRxiv. 2020 doi: 10.1016/j.asoc.2020.106897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao Q., Wang H., Wang G. 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI) IEEE; 2021. LCOV-NET: A lightweight neural network for COVID-19 pneumonia lesion segmentation from 3D CT images; pp. 42–45. [Google Scholar]
- 30.Yan Q., Wang B., Gong D., Luo C., Zhao W., Shen J., Ai J., Shi Q., Zhang Y., Jin S., et al. COVID-19 chest CT image segmentation network by multi-scale fusion and enhancement operations. IEEE Trans. Big Data. 2021;7(1):13–24. doi: 10.1109/TBDATA.2021.3056564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Jieneng, Lu Yongyi, Yu Qihang, Luo Xiangde, Adeli Ehsan, Wang Yan, Lu Le, Yuille Alan L., Zhou Yuyin. 2021. Transunet: Transformers Make Strong Encoders for Medical Image Segmentation. arXiv preprint arXiv:2102.04306. [Google Scholar]
- 32.Jose Valanarasu Jeya Maria, Oza Poojan, Hacihaliloglu Ilker, Patel Vishal M. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2021. Medical transformer: Gated axial-attention for medical image segmentation; pp. 36–46. [Google Scholar]
- 33.Roy Subhankar, Menapace Willi, Oei Sebastiaan, Ben Luijten, Fini Enrico, Saltori Cristiano, Huijben Iris, Chennakeshava Nishith, Mento Federico, Sentelli Alessandro, et al. Deep learning for classification and localization of covid-19 markers in point-of-care lung ultrasound. IEEE Trans. Med. Imag. 2020;39(8):2676–2687. doi: 10.1109/TMI.2020.2994459. [DOI] [PubMed] [Google Scholar]
- 34.Mahmud T., Alam M.J., Chowdhury S., Ali S.N., Rahman M.M., Fattah S.A., Saquib M. CovTANet: a hybrid tri-level attention-based network for lesion segmentation, diagnosis, and severity prediction of COVID-19 chest CT scans. IEEE Trans. Ind. Inf. 2020;17(9):6489–6498. doi: 10.1109/TII.2020.3048391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Milletari F., Navab N., Ahmadi S.A., V-net . 2016 Fourth International Conference on 3D Vision (3DV) IEEE; 2016. Fully convolutional neural networks for volumetric medical image segmentation; pp. 565–571. [Google Scholar]
- 36.Liu M., Li F., Yan H., Wang K., Ma Y., Shen L., Xu M. Alzheimer's Disease Neuroimaging Initiative, et al. A multi-model deep convolutional neural network for automatic hippocampus segmentation and classification in Alzheimer's disease. Neuroimage. 2020;208 doi: 10.1016/j.neuroimage.2019.116459. [DOI] [PubMed] [Google Scholar]
- 37.Litjens Geert, Kooi Thijs, Bejnordi Babak Ehteshami, Adiyoso Setio Arnaud Arindra, Ciompi Francesco, Ghafoorian Mohsen, Van Der Laak Jeroen Awm, Van Ginneken Bram, I Sánchez Clara. A survey on deep learning in medical image analysis. Med. Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 38.Chen Shuhan, Tan Xiuli, Wang Ben, Hu Xuelong. Proceedings of the European Conference on Computer Vision. ECCV); 2018. Reverse attention for salient object detection; pp. 234–250. [Google Scholar]
- 39.Zhou Tongxue, Canu Stéphane, Su Ruan. 2020. An Automatic Covid-19 Ct Segmentation Based on U-Net with Attention Mechanism. arXiv preprint arXiv:2004.06673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hashemi S.R., Salehi S.S.M., Erdogmus D., Prabhu S.P., Warfield S.K., Gholipour A. 2018. Tversky as a Loss Function for Highly Unbalanced Image Segmentation Using 3d Fully Convolutional Deep Networks. arXiv preprint arXiv:1803.11078. [Google Scholar]
- 41.Abraham N., Khan N.M. 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019) IEEE; 2019. A novel focal tversky loss function with improved attention u-net for lesion segmentation; pp. 683–687. [Google Scholar]
- 42.Emre Kavur A., Sinem Gezer N., Barıs Mustafa, Aslan Sinem, Conze PierreHenri, Groza Vladimir, Pham Duc Duy, Chatterjee Soumick, Ernst Philipp, Özkan Savas, Baydar Bora, Lachinov Dmitry, Han Shuo, Pauli Josef, Isensee Fabian, Perkonigg Matthias, Sathish Rachana, Rajan Ronnie, Sheet Debdoot, Dovletov Gurbandurdy, Speck Oliver, Nürnberger Andreas, Maier-Hein Klaus H., Bozdăgı Akar Gözde, Ünal Gözde, Dicle Ŏguz, Alper Selver M. CHAOS Challenge – combined (CT-MR) healthy abdominal organ segmentation. Med. Image Anal. April 2021;69 doi: 10.1016/j.media.2020.101950. [DOI] [PubMed] [Google Scholar]
- 43.Sharp G.C., Yang J., Gooding M.J., Cardenas C.E., Mohamed A.S.R., Veeraraghavan H. 2019. Aapm Rt-Mac Challenge.https://www.aapm.org/GrandChallenge/RT-MAC/ [Google Scholar]
- 44.Henschel Leonie, Conjeti Sailesh, Santiago Estrada, Diers Kersten, Fischl Bruce, Reuter Martin. Fastsurfer-a fast and accurate deep learning based neuroimaging pipeline. Neuroimage. 2020;219 doi: 10.1016/j.neuroimage.2020.117012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou Y., Chen H., Li Y., Liu Q., Xu X., Wang S., Yap P.T., Shen D. Multi-task learning for segmentation and classification of tumors in 3D automated breast ultrasound images. Med. Image Anal. 2021;70 doi: 10.1016/j.media.2020.101918. [DOI] [PubMed] [Google Scholar]
- 46.Martin Bland J., Altman DouglasG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;327(8476):307–310. [PubMed] [Google Scholar]
- 47.Gao S., Cheng M.M., Zhao K., Zhang X.Y., Yang M.H., Torr P.H.S. IEEE transactions on pattern analysis and machine intelligence; 2019. Res2net: A New Multi-Scale Backbone Architecture. [DOI] [PubMed] [Google Scholar]
- 48.Selvaraju R.R., Cogswell M., Das A., Vedantam R., Parikh D., Batra D. Proceedings of the IEEE International Conference on Computer Vision. 2017. Grad-cam: visual explanations from deep networks via gradient-based localization; pp. 618–626. [Google Scholar]