Abstract
Because of COVID-19's effect on pulmonary tissues, Chest X-ray(CXR) and Computed Tomography (CT) images have become the preferred imaging modality for detecting COVID-19 infections at the early diagnosis stages, particularly when the symptoms are not specific. A significant fraction of individuals with COVID-19 have negative polymerase chain reaction (PCR) test results; therefore, imaging studies coupled with epidemiological, clinical, and laboratory data assist in the decision making. With the newer variants of COVID-19 emerging, the burden on diagnostic laboratories has increased manifold. Therefore, it is important to employ beyond laboratory measures to solve complex CXR image classification problems. One such tool is Convolutional Neural Network (CNN), one of the most dominant Deep Learning (DL) architectures. DL entails training a CNN for a task such as classification using extensive datasets. However, the labelled data for COVID-19 is scarce, proving to be a prime impediment to applying DL-assisted analysis. The available datasets are either scarce or too diversified to learn effective feature representations; therefore Transfer Learning (TL) approach is utilized. TL-based ResNet architecture has a powerful representational ability, making it popular in Computer Vision. The aim of this study is two-fold- firstly, to assess the performance of ResNet models for classifying Pneumonia cases from CXR images and secondly, to build a customized ResNet model and evaluate its contribution to the performance improvement. The global accuracies achieved by the five models i.e., ResNet18_v1, ResNet34_v1, ResNet50_v1, ResNet101_v1, ResNet152_v1 are 91.35%, 90.87%, 92.63%, 92.95%, and 92.95% respectively. ResNet50_v1 displayed the highest sensitivity of 97.18%, ResNet101_v1 showed the specificity of 94.02%, and ResNet18_v1 had the highest precision of 93.53%. The findings are encouraging, demonstrating the effectiveness of ResNet in the automatic detection of Pneumonia for COVID-19 diagnosis. The customized ResNet model presented in this study achieved 95% global accuracy, 95.65% precision, 92.74% specificity, and 95.9% sensitivity, thereby allowing a reliable analysis of CXR images to facilitate the clinical decision-making process. All simulations were carried in PyTorch utilizing Quadro 4000 GPU with Intel(R) Xeon(R) CPU E5-1650 v4 @ 3.60 GHz processor and 63.9 GB useable RAM.
Keywords: Transfer learning, Deep learning, CNN, COVID-19, ResNet
1. Section I
1.1. Introduction
SARS-CoV-2(Severe Acute Respiratory Syndrome Corona Virus-2), a novel coronavirus, was discovered in Wuhan, China, in December 2019 [1]. Commonly referred to as COVID-19, the World Health Organisation(WHO) declared it a global pandemic in March 2020. After more than 1.5 years, newer variants of COVID-19 are emerging, burdening the healthcare systems globally [2]. The usual clinical presentation for COVID-19 includes cough, dyspnoea, fever, and radiological abnormalities [3]. The measures of COVID-19 testing globally are swab tests from the nose and throat for reverse-transcribed polymerase chain reaction (RT-PCR) and rapid antigen test (RAT) [4]. However, RT-PCR tests have low sensitivity (69%), resulting in an aberration in various inspections; therefore, numerous studies have given a pivotal role to imaging techniques in the initial diagnosis and management of the disease with CXR imaging as a front-line technique to confirm the presence of infection [5,6].
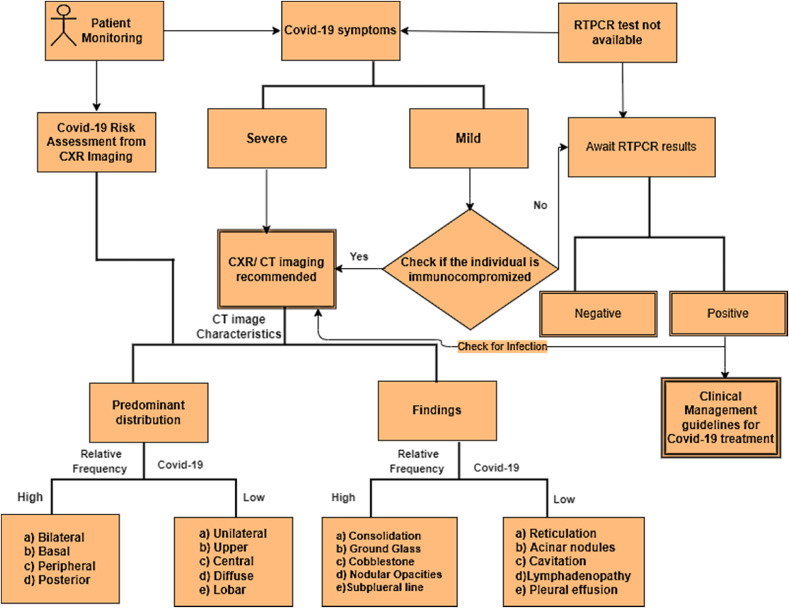
Community-acquired coronavirus linked with SARS causes atypical Pneumonia [7]. The “American Thoracic Society” (ATS) guidelines recommend a chest radiograph to confirm the diagnosis in all cases with suspected Community-acquired Pneumonia [7]. The “Canadian Society of Thoracic Radiology and the Canadian Association of Radiologists” has provided a detailed account of the role of CXR and CT imaging in complementing COVID-19 management in their article [8]. The study reveals that CT/CXR is ideal for determining COVID-19 infection in patients with worsening symptoms, immunocompromised individuals and if RT-PCR tests are unavailable. CT/CXR imaging is also essential for monitoring the disease progression, especially in high-risk patients. However, although standard CXR scans can aid in the early detection of suspected instances, the imaging of diverse viral pneumonia cases is comparable and overlaps with other infectious and inflammatory lung disorders. Fig. 1 represents the potential role of CT/CXR imaging in COVID-19 diagnosis based on studies in Refs. [6,8].
Fig. 1.
Role of CT/CXR imaging in COVID-19 diagnosis.
Two critical parameters of COVID-19 diagnosis are accuracy and timeliness. In the current situation, when COVID-19 has had a significant impact on clinical diagnostic laboratories [9] and the side symptoms of COVID-19 are similar to viral Pneumonia, the possibility of late or incorrect diagnosis increases. Experts are harnessing beyond laboratory measures such as DL, especially CNNs, for CXR/CT image classification. DL techniques contribute to automatic mass feature extraction using convolution operation on the input images at the lower layers and amplify the distinguishing characteristics at the higher level [10]. Traditional DL technology has found a place in numerous practical applications [11], but it has certain limitations in real-world domains. Sophisticated models such as CNNs require large-scale datasets to accomplish effective feature extraction and classification. Furthermore, DL ideally requires labelled training examples with the same distribution as the test data. However, gathering enough training data is sometimes prohibitively expensive, time-consuming, or impossible.
The application of DL for analysis of COVID-19 data also faces significant hindrances, including lack of a standard global dataset, mislabelled data, class imbalance, ambiguity, noise, sparsity, and redundancy in the available data [12,13]. A preferred method to counter the problem of the limited size of the samples connected to COVID-19 is utilizing pre-trained models using TL. TL is an approach in which a CNN's knowledge extracted from one dataset solves a second but similar task needing new data, usually due to the under-availability of data and the high computational cost of training a new CNN from scratch [14,15]. TL aims to improve performance on the target domains by reutilizing and transferring learned information from correlated source domains [16].
The success of TL is attributed to feature reuse, and the authors of [17] confirm in their research that the performance gains occur even for the most remote target domains. The authors of [18] demonstrate that a fine-tuned pre-trained CNN can outperform or perform equally well as a fully trained CNN in medical imaging applications. TL has found space in various applications such as drug discovery [19], plant disease identification [20], plant species classification [21], medical imaging [22], fault diagnosis [23], natural language processing [24], and many domains with underrepresented data. Various Deep Transfer Learning (DTL) based pre-trained models are utilized for image classification, key point detection, segmentation, and object detection.
TL is commonly expressed in computer vision through the usage of pre-trained models. Popular pre-trained models for feature extraction in images include AlexNet [25], VGG [26], SqueezeNet [27], DenseNet [28], MobileNet [29], Inception [30], GoogLeNet [31], Xception [32], ResNet [33], and EfficientNet [34] with architectural variations existing in model versions. ImageNet is predominantly used as a transferred source [35]. Researchers have widely used these models in CXR/CT imaging on different datasets to detect COVID-19 Pneumonia. The authors of [36] employ ResNet32 as their DTL model with a top-2 smooth loss function with cost-sensitive attributes to deal with noisy and imbalanced data. The dataset taken is small with 852 images (413 Covid positives and 439 of Normal Pneumonia) and obtained a precision of 0.95 with 0.91 sensitivity, 0.95 specificity, and 0.93 validation accuracy. The authors of [37] have gathered 1065 CT images of pathogen-confirmed COVID-19 cases and modified the Inception TL for their model. The external testing dataset demonstrates total sensitivity of 0.67, specificity of 0.83, and accuracy of 0.79.
The authors of [38] have worked on a collection of two datasets, one with 1428 X-ray photos(224 with COVID-19 illness, 700 with typical bacterial Pneumonia, and 504 of healthy images) and the other with 1422 images(224 with COVID-19 condition, 714 with bacterial and viral Pneumonia with typical bacterial Pneumonia, and 504 Healthy images) and show a comparative analysis on CNN models using TL. The authors of [39] propose a Deep CNN-based Inception V3 model to detect Covid Pneumonia infected patients using CXR imaging. The dataset contains 3550 images (864 Covid positive, 1345 viral pneumonia, and 1341 Healthy images), and the authors have attained a validation accuracy of 0.93. The authors of [40] have divided the dataset with 1616 CXR images into three classes(728 Normal, 648 Pathological but non-Covid, and 240 Covid positives). In their experiment, the authors have adopted a DenseNet based architecture to distinguish between Healthy and Pathological/Covid positives through binary classification and attained a global accuracy of 0.7962.
Our research aims to assess the effectiveness of ResNet architectures for Pneumonia classification on CXR images in the early stages of Covid diagnosis for determining the efficacy of raise in pre-test probability. Our findings reveal that TL with ResNet variants for Pneumonia detection to assist COVID-19 diagnosis is effective, has stable performance, and is simple to implement. The rest of the paper is organized as follows.
-
•
Section II presents the description of the dataset, pre-processing methods, models, and statistical analysis done in this study. A detailed background of architectural tweaks on ResNet variants carried before simulation, training details, hyperparameter tuning, and architecture of our customized ResNet model are presented.
-
•
Section III presents two sets of experimental results. Firstly a comprehensive comparative analysis of the performance of ResNet architectures for classifying Pneumonia from CXR images and secondly, the evaluation of customized ResNet18 model.
-
•
Lastly, in Section IV, we present the limitations of the study and the conclusion.
2. Section II
2.1. Materials and methods
-
A)
Dataset Preparation and training
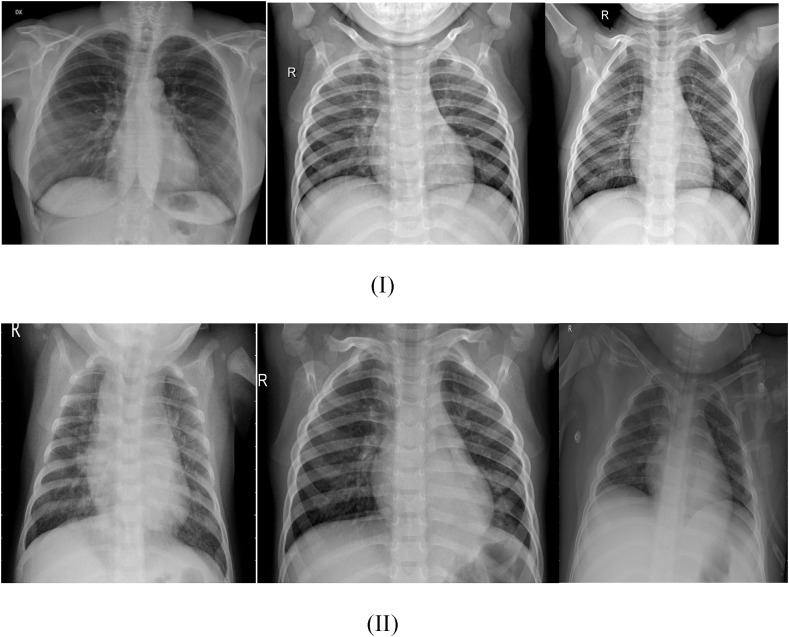
Due to geographical bias, non-uniformity, construction methods, and data imbalance, there is currently no dataset that can serve as a benchmark for COVID-19. We have compiled the data used in our study from the public database ‘CoronaHack-Chest X-Ray-Dataset’ on Kaggle, consisting of 5910 unique CXR images corresponding to 2 clinical categories- Pneumonia (4334 images)and ‘Not Pneumonia’ (1576 images). Fig. 2 displays representative samples of CXR images corresponding to the two classes. The dataset was split into two mutually exclusive sets- 89% for training/validation and 11% for testing. The 5286 images constituting the training set contain 1342 images belonging to the ‘Not Pneumonia’ class and 3944 to ‘Pneumonia’ class, while in the test set containing 624 images, 234 correspond to the ‘Not Pneumonia’ class and 390 to the ‘Pneumonia’ class. All images were rescaled to a size of 224 × 224, and data augmentation was performed before every simulation to prevent overfitting and improve the network's generalization. Generative adversarial networks (GANs) have recently gained popularity as a new method of data augmentation. Through GANs, it is possible to synthesize images from scratch. However, GANs achieve adequate results when combined with other methods, computation time is very high, face problems with counting, lack comprehension of the perspective, and difficulty coordinating global structures. For faster results, we have used the traditional data augmentation method based on a combination of affine changes and color modification. This method is relatively fast and straightforward to implement and has effectively expanded the training dataset.
Fig. 2.
CXR images corresponding to (I)-‘Not Pneumonia’ and (II)-‘Pneumonia’ clinical category.
In our model, TL was used for the training procedure, and the model weights from the ImageNet dataset were used as the initialization weights for the ResNet architecture. The utilization of a pre-trained model reduces the number of labelled examples needed for an acceptable training model, as is the case in our study. During training, model checkpoints were employed to save the best model weights for further analysis. Since a higher number of epochs does not necessarily lead to a substantial increase in classification accuracy, we have performed each training process with the ‘Early stopping’ concept for regularization and preventing overfitting. In early stopping, a large number of training epochs are specified, and training is stopped once the model's performance no longer improves on a specific metric in the validation dataset.
-
B)
ResNet Network Architecture
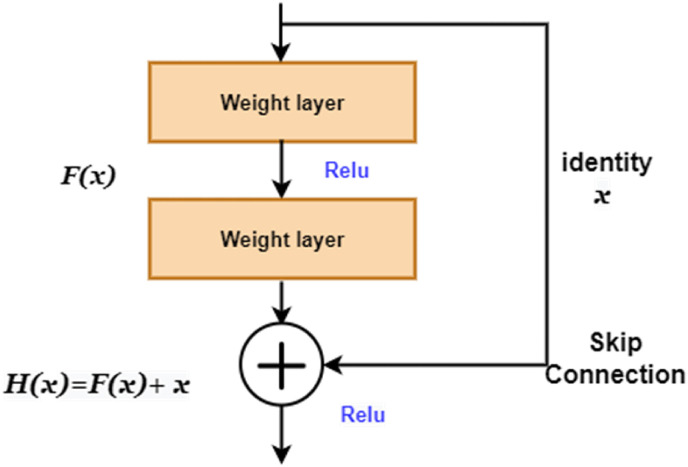
ResNet was introduced to address the problem of vanishing gradients [41,42]. ResNets alleviate this issue through shortcut connections, enabling the gradient to flow along an additional shortcut channel. The authors of [33] suggest optimizing residual mapping is simpler than optimizing and can be achieved in feed-forward neural networks through shortcut connections. The primary working principle is that if the additional layers are built as identity mappings, a deeper model should have no more training error than the corresponding shallower equivalent [33]. Instead of directly stacking layers to meet a particular underlying mapping, the layers are stacked to fit a residual mapping. Let represent the required underlying mapping, the nonlinear layers are designed to match a different mapping such that
| (1) |
| (2) |
Through shortcut connections, layer(s) are skipped. In Residual learning, backpropagation is done through the identity function using only vector addition, as depicted in Fig. 3 . Identity mappings do not contribute any extra parameters [43].
Fig. 3.
Residual learning.
The different variations of ResNet architecture work on the same principle but differ in the number of layers. The architecture of 18-layer and 34-layer, 50-layer, 101-layer, and 152-layer ResNet is depicted in Table 1 .
Table 1.
Architectural variation in different ResNet models [33].
| Layer name | Output Size | 18 layer | 34 layer | 50 layer | 101 layer | 152 layer | ||
|---|---|---|---|---|---|---|---|---|
| Convol_1 | 112 × 112 | 7 × 7,64,stride = 2 | ||||||
| Convol_2 | 56 × 56 | 3 × 3, Max pool, stride = 2 | ||||||
| Convol_3 | 28 × 28 | |||||||
| Convol_4 | 14 × 14 | |||||||
| Convol_5 | 7 × 7 | |||||||
| 1 × 1 | Average Pooling 1000, Softmax function | |||||||
-
C)
Layer Freezing
Two popular approaches exist for utilizing a pre-trained CNN's abilities. In the first approach, the pre-trained CNN model retains its basic architecture and learned weights and is only used for feature extraction, which is then introduced into the network performing the classification task. The second approach involves specific modifications to the pre-trained model to get optimal results, such as architectural tuning and parameter optimization. Only specific information from the previous task is kept while new trainable parameters are added to the network. The second approach is complex, while the first avoids the computational overheads of fully training a deep network.
Deep networks typically perform poorly on test data when trained on a small dataset, causing the model to overfit and can be alleviated by eliminating some feature detectors during each epoch [44]. The top layers of a deep network may have the fewest parameters but are the most computationally intensive [45]. All CNNs share some hyper-parameters, and saving the relevant feature extractors learned during the initial step avoids additional computational complexity. The top layers in the convolutional base that perform feature extraction are frozen and made untrainable, while the late convolutional layers, closer to the output features, are made trainable to allow for better extraction of information. Based on the large variety of contexts in which the network must operate, the goal is to detect the features that are generally helpful for producing the correct classification result.
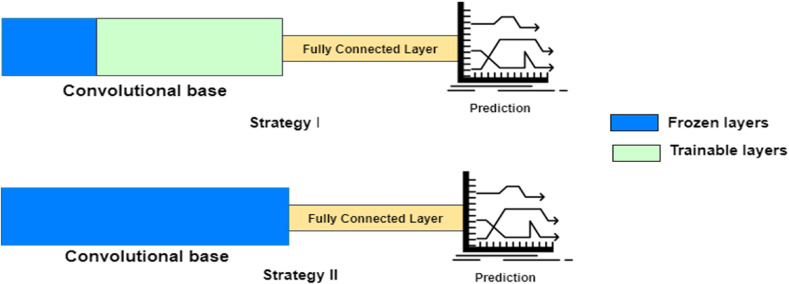
The goal of layer freezing is to control how weights are updated; the weights cannot be changed further by freezing a layer. This accelerates the training process, prevents complex co-adaptations, and reduces the computational time required for training without compromising accuracy. Two prime strategies are available for TL with smaller datasets; the choice is dictated by the similarity to the dataset in which the pre-trained model was trained, as depicted in Fig. 4 . In strategy I, only the first few layers are frozen while the end layers are made trainable. Additional trainable layers can be added to the model if the dataset is comparatively larger. In strategy II, the previously trained model is run as a fixed feature extractor, and then the features are then used to train a new classifier.
Fig. 4.
Concept of Layer Freezing.
Strategy I is followed when the dataset is dissimilar to the dataset in the pre-trained model.
Strategy II is followed when the dataset is similar to the dataset in the pre-trained model.
-
D)
Model Configuration
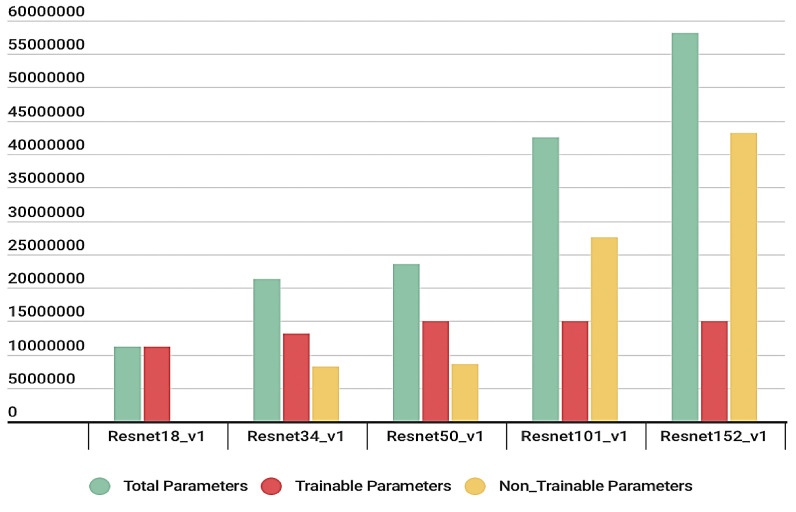
We have tested a range of ImageNet pre-trained ResNet models, including a) ResNet18, b) ResNet34, c) ResNet50, d) ResNet101, e) ResNet152, and a customized ResNet model. We have followed Strategy I during training; the first few layers were frozen, and only the last layers were trained. The modified trainable parameters are displayed in Fig. 5 : the models with parameter reduction are named ResNet18_v1, ResNet34_v1, ResNet50_v1, ResNet101_v1, and ResNet152_v1. The hyperparameters were determined after multiple experiments in each model. ResNet18 utilized lesser parameters and showed at par performance with the others; therefore, we chose it as the base for the customized model.
Fig. 5.
Comparison of the number of parameters in different ResNet models with Layer Freezing.
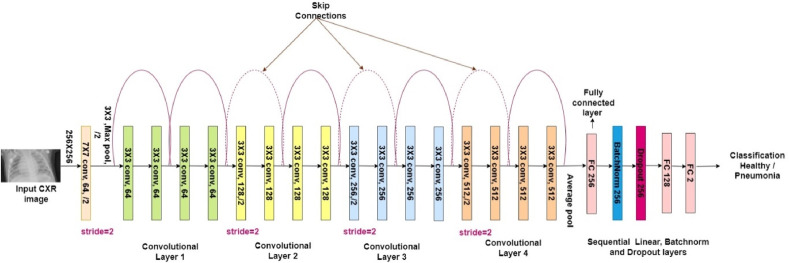
Customized ResNet model: The architecture of the customized ResNet employed in this investigation is shown in Fig. 6 . ResNet18 has been taken as the base for our customized model. The dropout layer was appended to the fully connected (FC) layer to alleviate model overfitting difficulties by lowering model sensitivity to the peculiarities of the training input. Dropout forces neurons to rely on population behavior rather than an individual activity to prevent overfitting and co-adaptation of feature detectors, increasing generalization accuracy [46]. Dropouts in CNNs regularize the networks through the addition of noise to each layer's output feature maps, resulting in robustness for varying images [47]. Taking a cue from the authors of [48], who proposed that adding batch normalization(BN) results in robust training and is a prerequisite for achieving convergence in many cases, we have added a BN to the FC layer. With BN, gradients behave more predictably and are more stable, resulting in faster training [49].
Fig. 6.
Architecture of the customized ResNet model.
-
E)
Metrics
Specific metrics were documented to evaluate the classification task performance: These include (a) correctly characterized Pneumonia images (True Positives, TP), (b) correctly characterized ‘Not-Pneumonia’ images (True Negatives, TN), c) incorrectly characterized Pneumonia images (False Negatives, FN), and (d) incorrectly characterized ‘Not-Pneumonia’ images (False Positives, FP). The statistical measures calculated for the classification report include [50].
| (3) |
| (4) |
| (5) |
| (6) |
| (7) |
| (8) |
| (9) |
| (10) |
| (11) |
| (12) |
3. Section III
3.1. Experimental results
We have carried out multiple sets of experiments in this investigation; the results of each are detailed in this section. We have carried out individual experiments on the fine-tuned ResNet architectures and the customized ResNet18 model, tested them on the same metrics and recorded the detailed comparisons of their performance.
-
A)
Performance comparison of ResNet Architectures
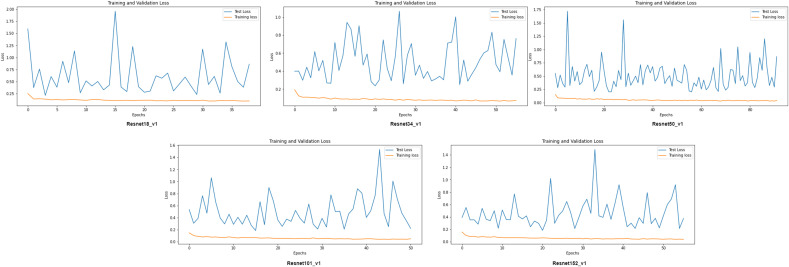
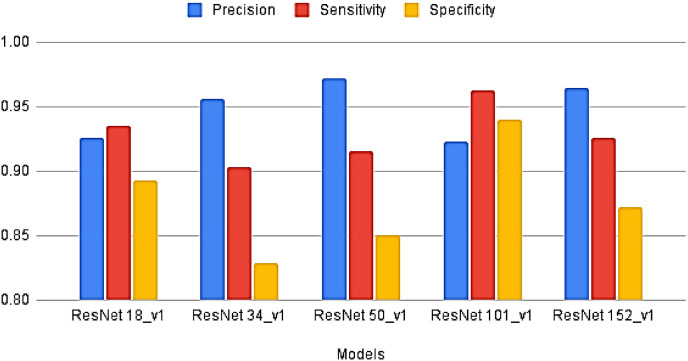
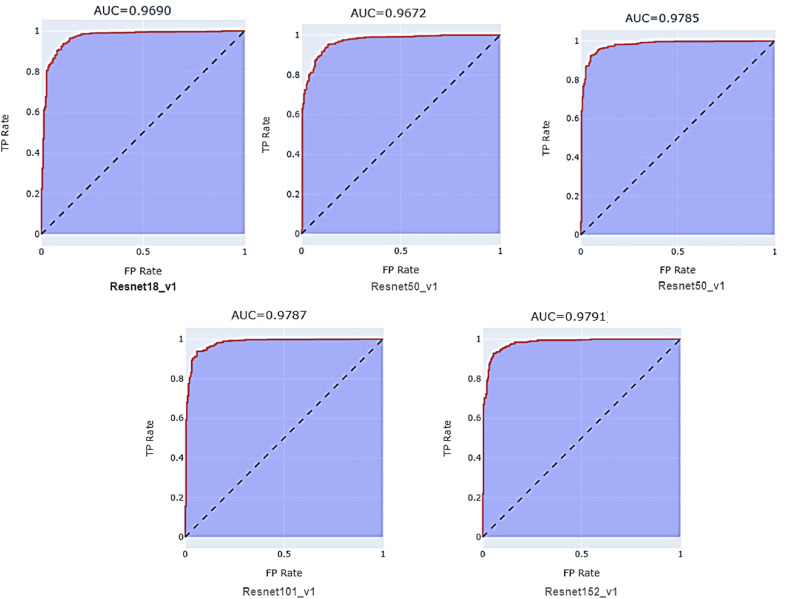
We present a comparative analysis of the performances of ResNet configurations on different metrics in this section. We have trained all finetuned pre-trained ResNet models- ResNet18_v1, ResNet34_v1, ResNet50_v1, ResNet101_v1, and ResNet152_v1 separately on the same dataset. Each model was end-end trained with binary cross-entropy (BCE)loss function, and the optimization during the training stage was performed through Adam. The training was carried with a holdout factor of 35 epochs for each model depicted in Fig. 7 . The models were trained with different learning rates and batch sizes to determine the optimal hyperparameters. The final results have been recorded after multiple simulations. Fig. 8 depicts the comparison of the models on sensitivity, specificity, and precision performance metrics. The performance comparison on the remaining metrics has been recorded in Table 2 . The receiver operating characteristic (ROC) curve analysis graph displaying the performance of the models at different classification thresholds is presented in Fig. 9 .
Fig. 7.
Training process of ResNet variants; Training Vs Validation loss. ResNet18_v1, ResNet34_v1, ResNet50_v1, ResNet101_v1, ResNet152_v1 converged after 40, 56, 92, 50, 56 epochs respectively.
Fig. 8.
Comparison of Precision, Specificity, and Sensitivity achieved by models.
Table 2.
Comparative Analysis of the performance of ResNet variants for classifying Pneumonia from CXR images.
| Metric | 18_v1 | 34_v1 | 50_v1 | 101_v1 | 152_v1 |
|---|---|---|---|---|---|
| NPV | 0.8782 | 0.9194 | 0.9476 | 0.88 | 0.9358 |
| FPR | 0.1068 | 0.1709 | 0.1496 | 0.0598 | 0.1282 |
| FDR | 0.0648 | 0.0969 | 0.0845 | 0.0374 | 0.0739 |
| FNR | 0.0744 | 0.0436 | 0.0282 | 0.0769 | 0.0359 |
| Accuracy | 0.9135 | 0.9087 | 0.9263 | 0.9295 | 0.9295 |
| F1 Score | 0.9304 | 0.929 | 0.9428 | 0.9424 | 0.9447 |
| MCC | 0.8161 | 0.8038 | 0.8424 | 0.8528 | 0.8488 |
ROC curves: The ROC curve graphs (1 – Specificity) Vs. Sensitivity.
Fig. 9.
ROC Curve Analysis of ResNet models.
-
B)
Customized DTL ResNet18
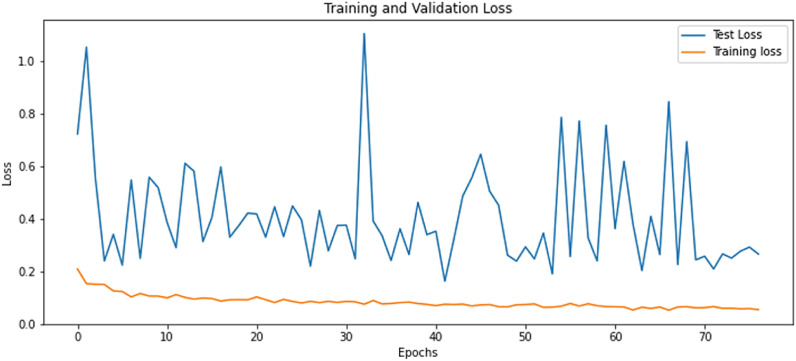
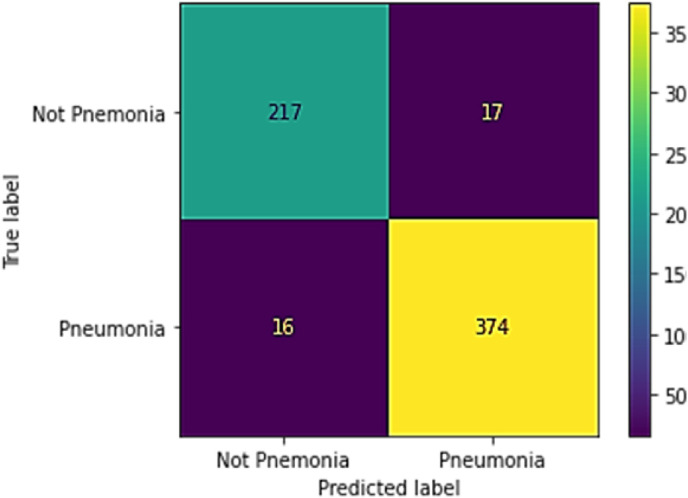
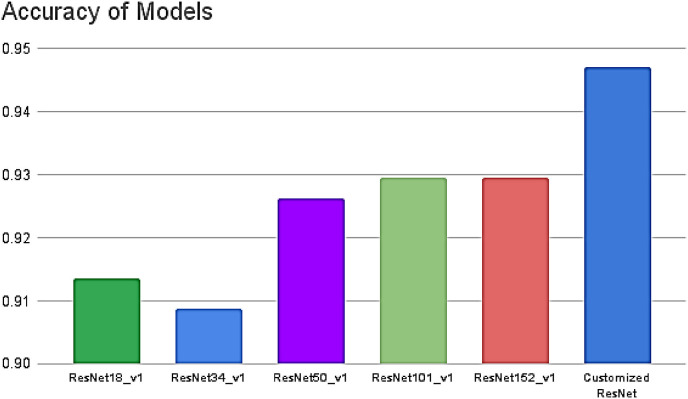
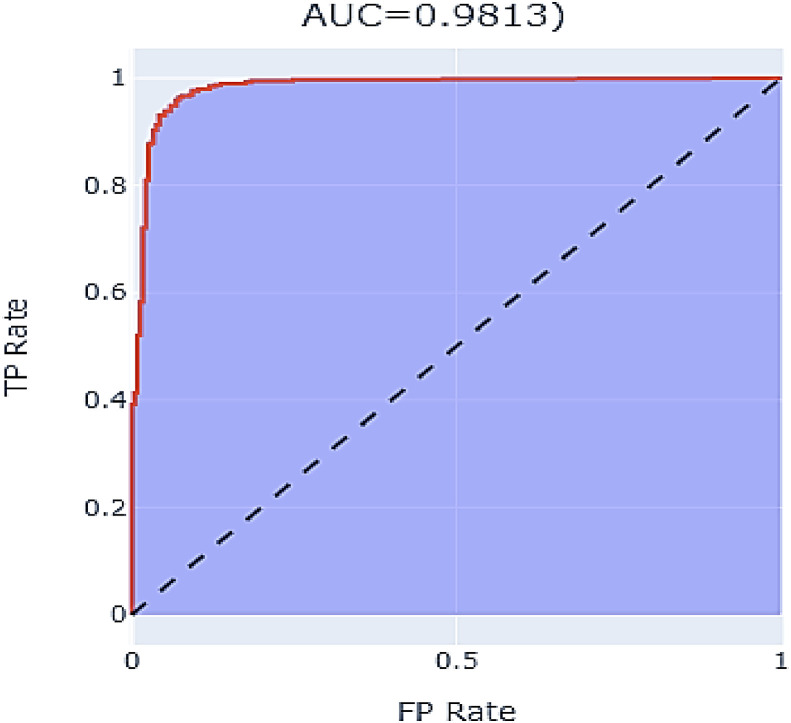
The customized ResNet model was trained with BCE as the loss function, a learning rate of 0.0001, Adam optimizer, and batch size of 64. The final parameters were defined after multiple experiments, but many other options can be studied in the future to see if they contribute to performance improvement. Data augmentation was carried out to obtain a stable model. The model was trained with a holdout factor of 35 and trained for 77 epochs, displayed in Fig. 10 . The model achieved 0.959 sensitivity, 0.9274 specificity, 0.9565 precision, 0.9577 F1 score, 0.9313 NPV, 0.0726 FPR, 0.0435 FDR and 0.041 FNR. The confusion matrix and classification report are recorded in Table 3 and Fig. 11 . The custom ResNet model displays significant improvement in the global accuracy compared to base ResNet models; a contrast of the same is shown in Fig. 13. The AUC-ROC value recorded with the finalized parameters is presented in Fig. 12 .
Fig. 10.
Training of Customized ResNet Model: Training and Validation loss Vs. Epochs.
Table 3.
Classification report and Confusion matrix recorded on customized ResNet.
| Recall | Precision | F1-score | Support | |
|---|---|---|---|---|
| Not Pneumonia | 0.93 | 0.93 | 0.93 | 234 |
| Pneumonia | 0.96 | 0.96 | 0.96 | 390 |
| Accuracy | 0.95 | 624 | ||
| Macro Average | 0.94 | 0.94 | 0.94 | 624 |
| Weighted Average | 0.95 | 0.95 | 0.95 | 624 |
Fig. 11.
Classification report and confusion matrix recorded on customized ResNet model.
Fig. 13.
Comparison of Accuracy achieved by our customized model Vs. ResNet variants.
Fig. 12.
ROC curve of customized ResNet model.
4. Section IV
4.1. Discussions, limitations and conclusion
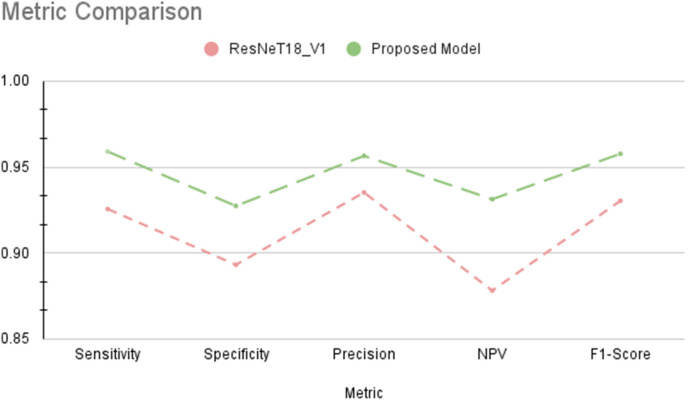
CNNs learn to extract essential features of an image; therefore, data availability for initial training is the most important aspect of practical training. Most of the datasets are small and suffer from the issue of class imbalance. The uneven distribution of classes and lack of sufficient data affects the robustness of the DL models. Some data samples may overlap, resulting in the use of the same images multiple times during training. Our study has countered these limitations through a combination of pre-trained models with TL, parameter reduction and feature reuse through layer freezing and data augmentation, providing faster training with smaller datasets. Instead of training a more in-depth model from scratch on a small database, our proposed model works in a transfer setting, where a pre-trained ResNet 18 model is employed, and utilizing the available dataset to refine the base model. This allows for parameter reduction with data-specific customization-the top layers near the fully connected layers finetune the model through data-specific training. A comparison of customized ResNet18 and base ResNet18 is displayed in Fig 14 ; the proposed customized model achieved better values on all metrics.
Fig. 14.
Performance comparison of proposed customized ResNet18 model with ResNet18_v1.
Limitations: Although standard CXR scans can aid in the early detection of suspected instances, the imaging of diverse viral pneumonia cases is comparable and overlaps with other infectious and inflammatory lung disorders. Thus, the classified Pneumonia cases may include subjects with pathological conditions unrelated to the traits of COVID-19. To address this problem, medical experts must be more involved in all stages of DL model creation, assessment to integrate medical domain knowledge into models [51]. The prediction accuracy and model transparency would improve if indicators’ detection were included with the categorization output [52].
Conclusion: The increase in the COVID-19 cases worldwide and the limitations of the available diagnostic tools have enforced a big challenge in handling the pandemic effectively. Radiologists have recorded a variety of abnormalities found in the CXR/CT scans of COVID-19 affected individuals, and many studies have confirmed that these images are decisive for identifying high-risk patients who need immediate attention and support. DL techniques, particularly CNNs, have significantly improved image analysis performed in the medical imaging domain, but face multiple challenges, with COVID-19 data not being an exception. Thus, instead, we have applied the TL approach, which counters the limitations of the dataset through feature reuse by transferring the knowledge between domains. The initial layers are computationally expensive to train in a deep network such as CNN. Furthermore, all CNNs share some hyper-parameters in the initial feature extraction stages; therefore, we have applied the layer freezing concept on the initial layers in the convolutional base to avoid additional computational complexity. This study has examined the effects of five versions of TL- based pre-trained ResNet models for automated Pneumonia detection from CXR images. We have performed a detailed experimental analysis evaluating the performance of each of these five models in terms of sensitivity, specificity, accuracy and AUC-ROC. The results showed that the pre-trained ResNet models provide significant gains and display good classification reports even when trained on a limited-sized dataset. For faster training, avoiding computational costs and timeliness, we have utilized ResNet 18 as the base for the customized model and finetuned it through additional layers such as BN, enabling faster and more stable training and Dropout that regularizes the network. The customized model is lightweight, with much lesser parameters than a Deep model, and attains higher classification accuracy even when trained on a small dataset. The performance achieved is encouraging, but there is still much room for improvement. As a future study, we intend to test the potential of our finetuned ResNet model on more diversified datasets to achieve a complete validation of their potential for assisting COVID-19 diagnosis.
Authorship contributions
Category 1.
Conception and design of study: Sadia Showkat and Dr. Shaima Qureshi.
Acquisition of data: Sadia Showkat.
Analysis and/or interpretation of data: Sadia Showkat.
Category 2.
Drafting the manuscript: Sadia Showkat.
Revising the manuscript critically for important intellectual content: Sadia Showkat and Dr. Shaima Qureshi.
Category 3.
Approval of the version of the manuscript to be published (the names of all authors must be listed):
Sadia Showkat and Dr. Shaima Qureshi.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
All persons who have made substantial contributions to the work reported in the manuscript (e.g., technical help, writing and editing assistance, general support), but who do not meet the criteria for authorship, are named in the Acknowledgements and have given us their written permission to be named. If we have not included an Acknowledgements, then that indicates that we have not received substantial contributions from non-authors.
References
- 1.Leung K.S.-S., et al. Territorywide study of early coronavirus disease outbreak, Hong Kong, China. Emerging Infectious Diseases journal - CDC. January 2021;27(Number 1) doi: 10.3201/eid2701.201543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parwanto M.L.E. Response to mutation and variants of the SARS-CoV-2 gene. Universa Medicina. May 2021;40(2) doi: 10.18051/UnivMed.2021.v40.77-78. Art. no. 2. [DOI] [Google Scholar]
- 3.Singh R., et al. Corona virus (COVID-19) symptoms prevention and treatment: a short review. J. Drug Deliv. Therapeut. Apr. 2021;11(2-S) doi: 10.22270/jddt.v11i2-S.4644. Art. no. 2-S. [DOI] [Google Scholar]
- 4.Tsang N.N.Y., So H.C., Ng K.Y., Cowling B.J., Leung G.M., Ip D.K.M. Diagnostic performance of different sampling approaches for SARS-CoV-2 RT-PCR testing: a systematic review and meta-analysis. Lancet Infect. Dis. Sep. 2021;21(9):1233–1245. doi: 10.1016/S1473-3099(21)00146-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen J.P., Morrison P., Dao L., Roth K., Duong T.Q., Ghassemi M. Dec. 2020. COVID-19 Image Data Collection: Prospective Predictions Are the Future. arXiv:2006.11988 [cs, eess, q-bio] [Google Scholar]
- 6.Arenas-Jiménez J.J., Plasencia-Martínez J.M., García-Garrigós E. When pneumonia is not COVID-19. Radiol. 2021;63(2):180–192. doi: 10.1016/j.rxeng.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Franquet T. Imaging of community-acquired pneumonia. J. Thorac. Imag. Sep. 2018;33(5):282–294. doi: 10.1097/RTI.0000000000000347. [DOI] [PubMed] [Google Scholar]
- 8.Dennie C., et al. Canadian society of thoracic Radiology/Canadian association of Radiologists consensus statement regarding chest imaging in suspected and confirmed COVID-19. Can. Assoc. Radiol. J. Nov. 2020;71(4):470–481. doi: 10.1177/0846537120924606. [DOI] [PubMed] [Google Scholar]
- 9.Sidiq Z., Hanif M., Dwivedi K.K., Chopra K.K. Laboratory diagnosis of novel corona virus (2019-nCoV)-present and the future. Indian J. Tubercul. Dec. 2020;67(4):S128–S131. doi: 10.1016/j.ijtb.2020.09.023. Supplement. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Albawi S., Mohammed T.A., Al-Zawi S. 2017 International Conference on Engineering and Technology. ICET); Aug. 2017. Understanding of a convolutional neural network; pp. 1–6. [DOI] [Google Scholar]
- 11.Singha A., Thakur R.S., Patel T. Biomedical Data Mining for Information Retrieval. John Wiley & Sons, Ltd; 2021. Deep learning applications in medical image analysis; pp. 293–350. [DOI] [Google Scholar]
- 12.Islam Md M., Karray F., Alhajj R., Zeng J. A review on deep learning techniques for the diagnosis of novel coronavirus (COVID-19) IEEE Access. 2021;9:30551–30572. doi: 10.1109/ACCESS.2021.3058537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rajaraman S., Antani S. medRxiv; May 2020. Training Deep Learning Algorithms with Weakly Labeled Pneumonia Chest X-Ray Data for COVID-19 Detection. 2020.05.04.20090803. [DOI] [Google Scholar]
- 14.Torrey L., Shavlik J. 2010. Transfer Learning,” Handbook of Research on Machine Learning Applications and Trends: Algorithms, Methods, and Techniques. [Google Scholar]
- 15.Weiss K., Khoshgoftaar T.M., Wang D. A survey of transfer learning. Journal of Big Data. May 2016;3(1):9. doi: 10.1186/s40537-016-0043-6. [DOI] [Google Scholar]
- 16.Zhuang F., et al. A comprehensive survey on transfer learning. Proc. IEEE. Jan. 2021;109(1):43–76. doi: 10.1109/JPROC.2020.3004555. [DOI] [Google Scholar]
- 17.Neyshabur B., Sedghi H., Zhang C. Jan. 2021. What Is Being Transferred in Transfer Learning? arXiv:2008.11687 [cs, stat] [Google Scholar]
- 18.Tajbakhsh N., et al. Convolutional neural networks for medical image analysis: full training or fine tuning? IEEE Trans. Med. Imag. May 2016;35(5):1299–1312. doi: 10.1109/TMI.2016.2535302. [DOI] [PubMed] [Google Scholar]
- 19.Cai C., et al. Transfer learning for drug discovery. J. Med. Chem. Aug. 2020;63(16):8683–8694. doi: 10.1021/acs.jmedchem.9b02147. [DOI] [PubMed] [Google Scholar]
- 20.Chen J., Chen J., Zhang D., Sun Y., Nanehkaran Y.A. Using deep transfer learning for image-based plant disease identification. Comput. Electron. Agric. Jun. 2020;173:105393. doi: 10.1016/j.compag.2020.105393. [DOI] [Google Scholar]
- 21.Kaya A., Keceli A.S., Catal C., Yalic H.Y., Temucin H., Tekinerdogan B. Analysis of transfer learning for deep neural network based plant classification models. Comput. Electron. Agric. Mar. 2019;158:20–29. doi: 10.1016/j.compag.2019.01.041. [DOI] [Google Scholar]
- 22.Raghu M., Zhang C., Kleinberg J., Bengio S. Oct. 2019. Transfusion: Understanding Transfer Learning for Medical Imaging. arXiv:1902.07208 [cs, stat] [Google Scholar]
- 23.Shao S., McAleer S., Yan R., Baldi P. Highly accurate machine fault diagnosis using deep transfer learning. IEEE Trans. Ind. Inf. Apr. 2019;15(4):2446–2455. doi: 10.1109/TII.2018.2864759. [DOI] [Google Scholar]
- 24.Ruder S., Peters M.E., Swayamdipta S., Wolf T. Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Tutorials. Jun. 2019. Transfer learning in natural language processing; pp. 15–18. Minneapolis, Minnesota. [DOI] [Google Scholar]
- 25.Yuan Z.-W., Zhang J. Eighth International Conference on Digital Image Processing. vol. 10033. ICDIP 2016; Aug. 2016. Feature extraction and image retrieval based on AlexNet; pp. 65–69. [DOI] [Google Scholar]
- 26.W. Yu, K. Yang, Y. Bai, T. Xiao, H. Yao, and Y. Rui, “Visualizing and Comparing AlexNet and VGG Using Deconvolutional Layers,” p. 7.
- 27.Iandola F.N., Han S., Moskewicz M.W., Ashraf K., Dally W.J., Keutzer K. Nov. 2016. SqueezeNet: AlexNet-Level Accuracy with 50x Fewer Parameters and <0.5MB Model Size. arXiv:1602.07360 [cs] [Google Scholar]
- 28.Iandola F., Moskewicz M., Karayev S., Girshick R., Darrell T., Keutzer K. Apr. 2014. DenseNet: Implementing Efficient ConvNet Descriptor Pyramids. arXiv:1404.1869 [cs] [Google Scholar]
- 29.Howard A.G., et al. Apr. 2017. MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications. arXiv:1704.04861 [cs] [Google Scholar]
- 30.Soria X., Riba E., Sappa A. 2020 IEEE Winter Conference on Applications of Computer Vision (WACV) Snowmass Village; CO, USA: Mar. 2020. Dense extreme inception network: towards a robust CNN model for edge detection; pp. 1912–1921. [DOI] [Google Scholar]
- 31.Szegedy C., et al. 2015 IEEE Conference on Computer Vision and Pattern Recognition. CVPR); Boston, MA, USA: Jun. 2015. Going deeper with convolutions; pp. 1–9. [DOI] [Google Scholar]
- 32.Chollet F. 2017 IEEE Conference on Computer Vision and Pattern Recognition. CVPR; Honolulu, HI: Jul. 2017. Xception: deep learning with depthwise separable convolutions; pp. 1800–1807. [DOI] [Google Scholar]
- 33.He K., Zhang X., Ren S., Sun J. Dec. 2015. Deep Residual Learning for Image Recognition. arXiv:1512.03385 [cs] [Google Scholar]
- 34.Tan M., Le Q. Proceedings of the 36th International Conference on Machine Learning. May 2019. EfficientNet: rethinking model scaling for convolutional neural networks; pp. 6105–6114. [Google Scholar]
- 35.Krizhevsky A., Sutskever I., Hinton G.E. ImageNet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012;25 [Google Scholar]
- 36.Pathak Y., Shukla P.K., Tiwari A., Stalin S., Singh S., Shukla P.K. IRBM; May 2020. Deep Transfer Learning Based Classification Model for COVID-19 Disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang S., et al. A deep learning algorithm using CT images to screen for Corona virus disease (COVID-19) Eur. Radiol. Aug. 2021;31(8):6096–6104. doi: 10.1007/s00330-021-07715-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Apostolopoulos I.D., Mpesiana T.A. Covid-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med. Jun. 2020;43(2):635–640. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Asif S., Wenhui Y., Jin H., Tao Y., Jinhai S. preprint; May 2020. Classification of COVID-19 from Chest X-Ray Images Using Deep Convolutional Neural Networks,” Radiology and Imaging. [DOI] [Google Scholar]
- 40.De Moura J., et al. Deep convolutional approaches for the analysis of COVID-19 using chest X-ray images from portable devices. IEEE Access. 2020;8:195594–195607. doi: 10.1109/ACCESS.2020.3033762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bengio Y., Simard P., Frasconi P. Learning long-term dependencies with gradient descent is difficult. IEEE Trans. Neural Network. Mar. 1994;5(2):157–166. doi: 10.1109/72.279181. [DOI] [PubMed] [Google Scholar]
- 42.Hochreiter S. The vanishing gradient problem during learning recurrent neural nets and problem solutions. Int. J. Unc. Fuzz. Knowl. Based Syst. Apr. 1998;6(2):107–116. doi: 10.1142/S0218488598000094. [DOI] [Google Scholar]
- 43.He K., Zhang X., Ren S., Sun J. Computer Vision. ECCV 2016; Cham: 2016. Identity mappings in deep residual networks; pp. 630–645. [DOI] [Google Scholar]
- 44.Hinton G.E., Srivastava N., Krizhevsky A., Sutskever I., Salakhutdinov R.R. Jul. 2012. Improving Neural Networks by Preventing Co-adaptation of Feature Detectors. arXiv:1207.0580 [cs] [Google Scholar]
- 45.Brock A., Lim T., Ritchie J.M., Weston N. Jun. 2017. FreezeOut: Accelerate Training by Progressively Freezing Layers. arXiv:1706.04983 [cs, stat] [Google Scholar]
- 46.Wang T., Huan J., Li B. 2018 IEEE 30th International Conference on Tools with Artificial Intelligence. ICTAI; Nov. 2018. Data dropout: optimizing training data for convolutional neural networks; pp. 39–46. [DOI] [Google Scholar]
- 47.Park S., Kwak N. Computer Vision. ACCV 2016; Cham: 2017. Analysis on the dropout effect in convolutional neural networks; pp. 189–204. [DOI] [Google Scholar]
- 48.Amthor M., Rodner E., Denzler J. Oct. 2016. Impatient DNNs - Deep Neural Networks with Dynamic Time Budgets. arXiv:1610.02850 [cs] [Google Scholar]
- 49.Santurkar S., Tsipras D., Ilyas A., Madry A. How does batch normalization help optimization? Adv. Neural Inf. Process. Syst. 2018;31 [Google Scholar]
- 50.Martin Sagayam K., Felix Jacob Edwin J., Sujith Christopher J., Reddy G.V., Bestak R., Hun L.C. In: Hemanth J., Balas V., editors. vol. 25. Springer; Cham: 2018. Survey on the classification of intelligence-based biometric techniques. (Biologically Rationalized Computing Techniques for Image Processing Applications. Lecture Notes in Computational Vision and Biomechanics). [DOI] [Google Scholar]
- 51.Xie X., Niu J., Liu X., Chen Z., Tang S., Yu S. A survey on incorporating domain knowledge into deep learning for medical image analysis. Med. Image Anal. Apr. 2021;69:101985. doi: 10.1016/j.media.2021.101985. [DOI] [PubMed] [Google Scholar]
- 52.Alghamdi H.S., Amoudi G., Elhag S., Saeedi K., Nasser J. Deep learning approaches for detecting COVID-19 from chest X-ray images: a survey. IEEE Access. 2021;9:20235–20254. doi: 10.1109/ACCESS.2021.3054484. [DOI] [PMC free article] [PubMed] [Google Scholar]