Abstract
The glycolytic enzyme phosphoglycerate mutase (PGM) is of utmost importance for overall cellular metabolism and has emerged as a novel therapeutic target in cancer cells. This enzyme is also conserved in the rapidly proliferating malarial parasite Plasmodium falciparum, which have a similar metabolic framework as cancer cells and rely on glycolysis as the sole energy-yielding process during intraerythrocytic development. There is no redundancy among the annotated PGM enzymes in Plasmodium, and PfPGM1 is absolutely required for the parasite survival as evidenced by conditional knockdown in our study. A detailed comparison of PfPGM1 with its counterparts followed by in-depth structure-function analysis revealed unique attributes of this parasitic protein. Here, we report for the first time the importance of oligomerization for the optimal functioning of the enzyme in vivo, as earlier studies in eukaryotes only focused on the effects in vitro. We show that single point mutation of the amino acid residue W68 led to complete loss of tetramerization and diminished catalytic activity in vitro. Additionally, ectopic expression of the WT PfPGM1 protein enhanced parasite growth, whereas the monomeric form of PfPGM1 failed to provide growth advantage. Furthermore, mutation of the evolutionarily conserved residue K100 led to a drastic reduction in enzymatic activity. The indispensable nature of this parasite enzyme highlights the potential of PfPGM1 as a therapeutic target against malaria, and targeting the interfacial residues critical for oligomerization can serve as a focal point for promising drug development strategies that may not be restricted to malaria only.
Keywords: Plasmodium falciparum, phosphoglycerate mutase, tetramer, enzyme kinetics, knockdown, glmS
Abbreviations: GlcN, glucosamine; IDC, intra-erythrocytic developmental cycle; Pf, Plasmodium falciparum; PG, phosphoglycerate; PGM, phosphoglycerate mutase
Malaria, a highly prevalent infection caused by the protozoan parasite Plasmodium, affects mankind and global economy till date. The World Health Organization’s World Malaria Report 2020 revealed an estimated 229 million malaria cases and 409,000 related deaths in 2019 globally (1). Despite recent interventions, this devastating disease is still far from elimination and eradication. A profound knowledge of the parasite biology can help annihilate this malady. Infections with Plasmodium falciparum are most severe and can be fatal (2). Erythrocytes infected by the malarial parasites rely mainly on glycolysis for energy and utilize glucose at higher rates, 20 to 100 times, than the normal uninfected red blood cells (3, 4, 5). The eighth step of this energy-yielding machinery, the conversion of 3-phosphoglycerate to 2-phosphoglycerate, is catalyzed by phosphoglycerate mutase (PGM) enzyme (6). The reaction is reversible in nature and hence the enzyme takes part in both glycolysis and gluconeogenesis.
Phosphoglycerate mutase acts as a rate-limiting enzyme in heart tissue, leukocytes, and tumor cells (7). The enzyme is involved in multiple metabolic reactions of the cell directly or indirectly. It regulates the pentose phosphate pathway and amino acid biosynthesis, thereby modulating cell development and proliferation (8, 9). Moreover, PGM has the potential to immortalize mouse fibroblasts (10). In yeast, PGM helps in fungal adhesion and invasion in the human host through plasminogen binding (11). Therefore, PGM has many vital functions to play across species. Strikingly, cancer cells that undergo uncontrolled growth and rely on enhanced glycolysis show overexpression of mammalian counterpart PGAM1 citing the analogy between cancer cells and Plasmodium parasites for their dependence on this unique enzyme to support their unperturbed growth (12).
Different oligomeric forms of the enzyme have been reported in different organisms. In yeast, tetrameric form of PGM is seen, whereas the human counterpart exists as a dimer (13, 14). In another parasite, Leishmania Mexicana, PGM exists as monomer and dimer (15). Monomeric forms are mainly seen in bacteria (16, 17). The oligomeric properties of an enzyme play key role in affecting kinetic properties, cellular localization, and protein–protein interactions (18, 19, 20, 21). Most of the findings related to oligomerization of the enzyme and activity rely on in vitro data. Therefore, in vivo validation of oligomerization property of the enzyme and its effects still remain elusive.
The homolog of the PGM enzyme is present in the P. falciparum genome. Of the two P. falciparum pgm annotated genes, Pfpgm1 and Pfpgm2, the protein encoded by Pfpgm2 has been characterized as a phosphatase with weak mutase activity (22). However, the functionality of PfPGM1 and structure-function relationship have not been elucidated earlier. Further, the oligomeric status of PfPGM1 can be investigated to discover whether the parasitic enzyme resembles the eukaryotic homolog in yeast or the host enzyme, which would be useful in generating parasite-specific inhibitors.
PfPGM1 is indispensable for the parasite survival as studied by saturation mutagenesis and deletion analysis in P. falciparum and Plasmodium berghei respectively (23, 24). However, both the above studies focused on the overall growth phenotype as a read out for essentiality. The information is limiting, and the corresponding protein level to ascertain the functional correlation and specific effects on the parasite upon PfPGM1 deletion are lacking. PfPGM1 is one of the highly expressed genes as determined through mRNA abundance studies (25). Moreover, the enzyme is among the 35 proteins that are most abundant in the parasite (26). The gene is conserved throughout the Plasmodia species and is expressed in the different parasitic life stages (http://www.plasmoDB.org) (27). Interestingly, PfPGM1 also has the potential to be exploited as a biomarker for P. falciparum infection (28). In patients with severe malaria, high levels of PfPGM1 were detected in the plasma via proteome studies. Additionally, the recombinant PGM protein from another protozoan parasite Toxoplasma gondii (rTgPGAM2), when administered in BALB/c mice, provided immunity to the mice against T. gondii infection (29). The enzyme has clinical relevance as well, consequently notching up the inquisitiveness to explore it further. Thus, a targeted approach toward designing inhibitors against PfPGM1 could be explored for developing antimalarial medications. Even though the glycolytic enzymes are conserved across species, but they still display certain characteristic structural differences from the host enzymes that can be utilized for developing target-specific inhibitors against P. falciparum (5).
In the present study, we have characterized PfPGM1 for the first time and report that the enzyme is indispensable for parasite growth. Biochemical studies of the enzyme revealed that it exists as a tetramer as compared to the dimeric human counterpart and exhibits remarkable mutase activity in comparison with the yeast enzyme. Further, in-depth oligomerization analysis along with experimental validation highlighted key role of a single residue in maintaining the tetrameric structure. The tetramerization of this enzyme contributes to optimal functioning both in vitro and in vivo. Collectively, our data highlight the fundamental role of PGM in the malarial parasite, and we propose this enzyme as a potential target for antimalarial drug development. Moreover, our findings point toward a new strategy for drug development keeping oligomerization at the center point that may be of general interest and not limited to the parasites.
Results
P. falciparum PGM is indispensable for parasite development
Pfpgm1 gene (PF3D7_1120100) consists of two introns and codes for a 250 amino acid long protein. Motif search using ScanProsite tool revealed that the phosphohistidine signature motif of PGM family is located at the extreme N-terminus of the protein. The schematic of the protein is represented in Fig. S1A.
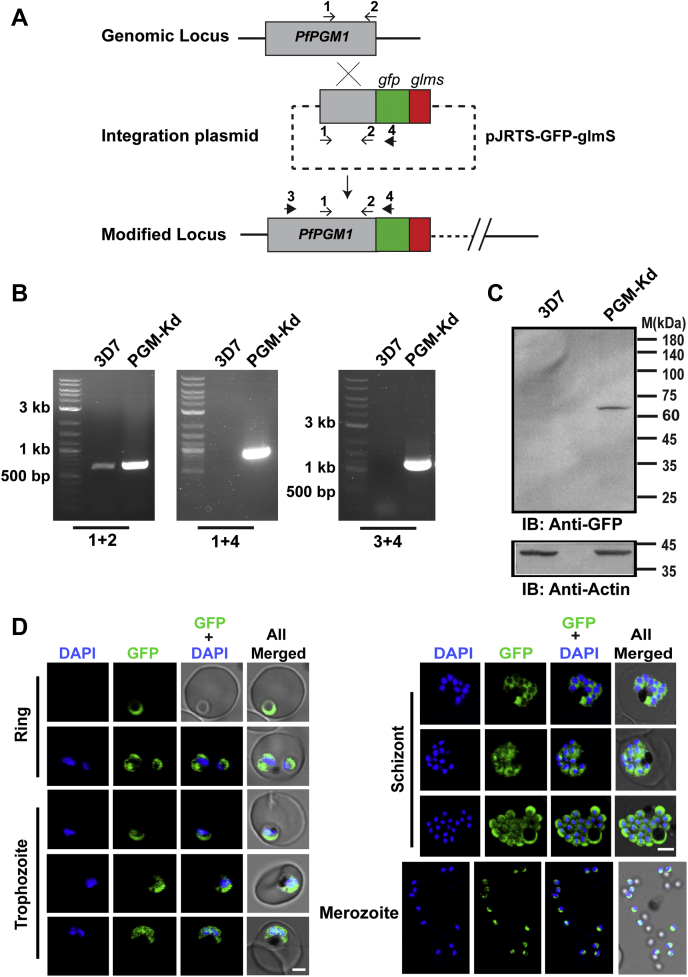
The essential role of PfPGM1 and its absolute requirement for the parasite survival was studied by conditional knockdown of the gene in order to have a quantitative and thorough analysis to validate the genotype–phenotype correlation. Glucosamine (GlcN)-induced ribozyme based glmS strategy (Fig. 1A) was exploited to deplete PfPGM1 levels within the parasites (30). The successfully appended GFP-glmS tag at the 3' region of the genomic locus of Pfpgm1 was confirmed via diagnostic PCR (Fig. 1B) using primers outside the homology region as shown in Figure 1A and mentioned in the methodology section. Western blot analysis using antibodies against GFP revealed the expression of GFP-fusion protein in the integrant parasites (Fig. 1C). The protein is constitutively expressed throughout the three developmental intraerythrocytic stages namely, ring, trophozoites, and schizonts in consonance with the glycolytic needs of the parasite (Fig. 1D). The live cell microscopy also demonstrated that PfPGM1 is predominantly localized in the cytosol of the parasite where the glycolytic process takes place. A significant fraction of the protein was also detected in the nucleus which is suggestive of moonlighting function. Western blot analysis and immunofluorescence assay using the polyclonal antisera directed against PfPGM1 represented the protein abundance in the different stages of the parasite (Fig. S1, B–E).
Figure 1.
PfPGM1 is expressed throughout the parasite developmental stages.A, the integration strategy schematic for endogenous tagging of PfPGM1 at the genomic locus by single crossover homologous recombination. The primers used in cloning (1, 2) and the diagnostic PCR (3, 4) are indicated by arrows and numbered accordingly. GFP and glmS tag are represented by green and red boxes, respectively. B, diagnostic PCR with the indicated primer sets to confirm integration at the PfPGM1 genomic locus in the obtained transgenic parasites (denoted PGM-Kd). 3D7 parasites, having the unmodified genomic locus, were used as control. C, immunoblotting using antibodies against GFP confirms the expression of GFP-fusion protein in the PGM-Kd parasite line. PfActin served as a loading control. D, live cell microscopy images demonstrating the abundance and subcellular localization of the GFP-tagged PfPGM1 protein (green) in the PGM-Kd parasites. 4′,6-diamidino-2-phenylindole (DAPI) was used to stain the nuclei. The scale bar represents 2 μm. The sequences of the primers (1–4) are listed in the Table S3 (S.No. 11–14). Kd, knockdown; PGM, phosphoglycerate mutase.
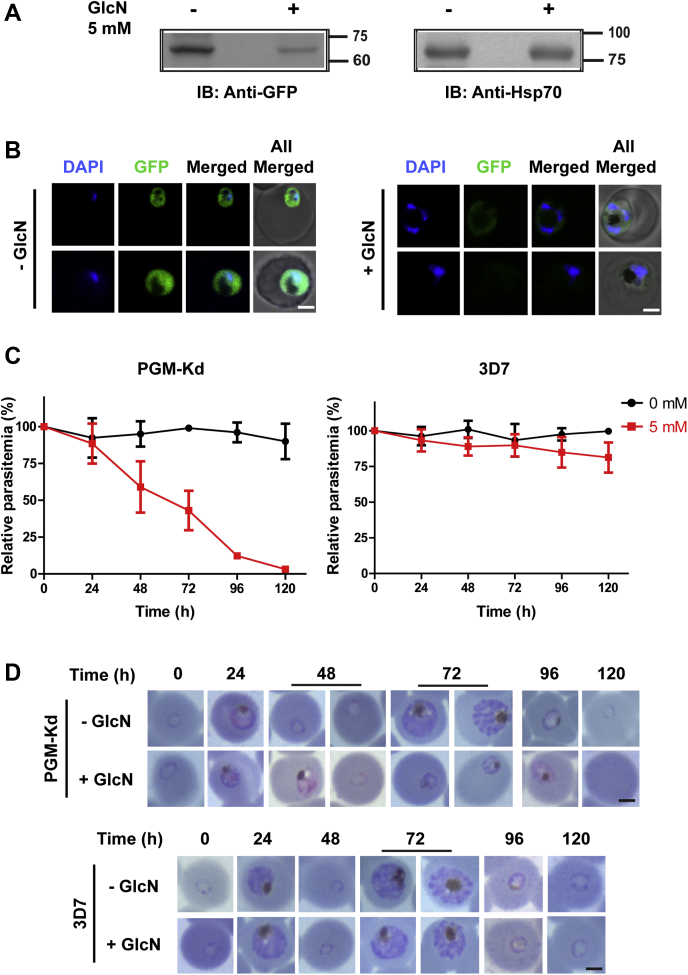
Subsequently, the regulation of protein expression by the glmS riboswitch and its influence on the parasite was assessed by the addition of 5 mM GlcN (31, 32, 33). Interestingly, PfPGM1 protein levels went down, 24 h post-GlcN addition, as detected by Western blot analysis and live cell microscopy, in comparison to the untreated parasites (Fig. 2, A and B). The residual expression in the treated parasites could be due to leaky expression or the already synthesized protein in the early ring stage. The depletion of PfPGM1 resulted in pronounced growth defect as evident from the parasite growth assay in the presence and absence of GlcN (Fig. 2C). Absence of the parasites after two cycles following GlcN addition in the case of PGM-knockdown parasite line indicates the critical role of PfPGM1 for parasite growth and proliferation. Giemsa-stained blood smears revealed that the parasite development was severely affected upon PfPGM1 depletion (Fig. 2D). Further, the parasite maturation to later stages was impeded in the absence of this key glycolytic enzyme (Fig. 2, B and D).
Figure 2.
PfPGM1 is indispensable for the parasite growth and development.A, Western blot analysis using antibodies against GFP illustrating the regulation of PfPGM1 expression via glmS riboswitch. The PfPGM1 expression levels were downregulated following 24 h of glucosamine treatment (5 mM) in synchronized ring parasites. PfHsp70 was used as a loading control. B, confocal microscopy live cell images illustrating the depletion of PfPGM1 and its effect on parasite phenotype. The scale bar denotes 2 μm. C, parasite survival curves demonstrating relative parasitemia in the absence and presence of 5 mM glucosamine in PGM-Kd (left) and 3D7 parasites taken as control (right). The graphs represent mean ± SD of at least three independent experiments. D, Giemsa-stained erythrocyte smears depicting the progression of the parasite in the presence and absence of PfPGM1 and the effect of PfPGM1 knockdown on parasite phenotype (top panel). Effect of 5 mM glucosamine on the phenotype of 3D7 parasites, taken as control (bottom panel). The scale bar denotes 2 μm. Kd, knockdown; PGM, phosphoglycerate mutase.
Overall, the presence or absence of GlcN did not show drastic effect on the survival and phenotype of the parental 3D7 parasites compared to the PGM-knockdown parasite line (Fig. 2, C and D). Thus, PfPGM1 is crucial for the parasite survival, and its depletion is deleterious to the parasite development.
PfPGM1 enzyme exists as a tetramer
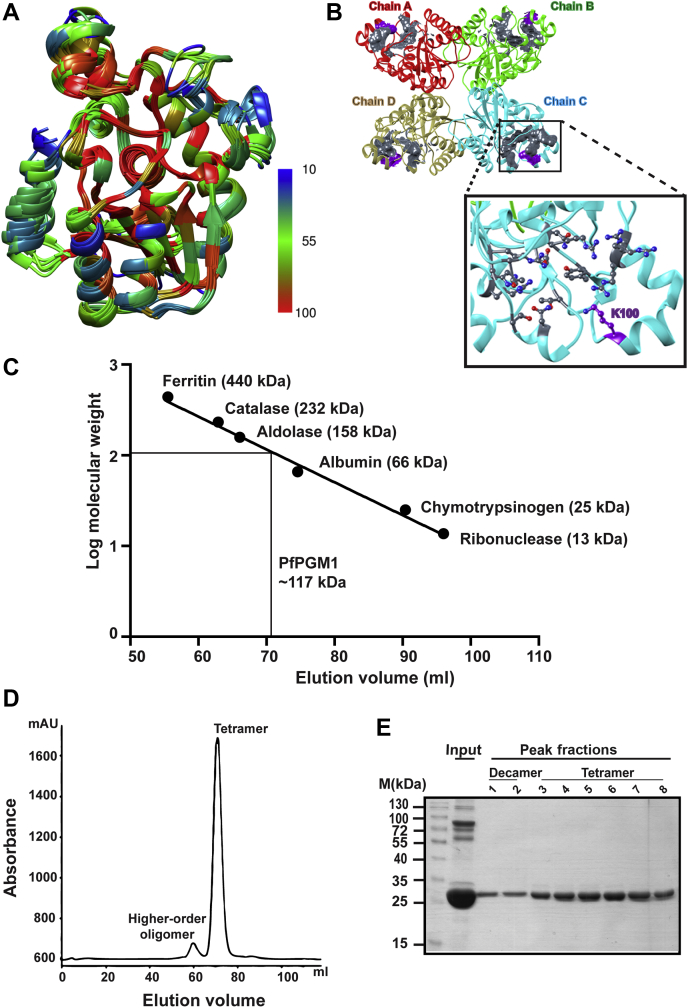
Having established the absolute requirement of PfPGM1 by the parasites, we next focused on the structure-function studies of the enzyme. A comparative structure and sequence analysis of PfPGM1 with its homologs from diverse species (Table S1) revealed that the PGM structures are highly conserved. The RMSD values of the PfPGM1 structure with respect to the homologs were found to be below 1 Å over the aligned residues. However, the sequences of these homologs are more diverse. PfPGM1 sequence identity was found to vary from 47.72 percent with its counterpart from Mycobacterium tuberculosis to 73.44 percent with T. gondii enzyme (Fig. S2). Structure-sequence mapping suggests that the maximum sequence variations (least conserved) are displayed in the residues forming the PfPGM1 surface (Fig. 3A).
Figure 3.
PfPGM1 structure and its oligomeric status.A, structural comparison of PfPGM1 and its homologs. Monomeric subunits of each homolog were structurally aligned to PfPGM1 structure, and the corresponding RMSD values are provided in Table S1. Amino acid residues are colored as per the color code. Blue and red colors denote the least and maximum conservation, respectively at the corresponding amino acid position among the homologs. B, the tetrameric structure of PfPGM1 illustrating the active site in gray surface view and labeled. The position of K100 is shown in the inset in purple. C, standard curve of known molecular mass proteins separated with size-exclusion chromatography. The solid line interpolation corresponds to the tetrameric form of PfPGM1. D, size-exclusion chromatogram of PfPGM1 displaying elution peaks of the two oligomeric forms. E, SDS-PAGE of the peak fractions in comparison with input highlighting the purity of the eluted fractions. PGM, phosphoglycerate mutase.
The oligomeric structure of an enzyme is crucial, and its disruption affects enzymatic activity and function in the cell (34, 35, 36, 37). The yeast PGM enzyme is a tetramer (38) whereas the human enzyme exists in a dimeric form (39). To decipher whether the P. falciparum PGM enzyme resembles the eukaryotic model organism yeast or the secondary host of the parasite, human enzyme, in the oligomerization status, we performed in silico studies followed by size-exclusion chromatography. The available crystal structure of PfPGM1 (PDB ID: 1XQ9) (40, 41) shows tetrameric organization of the protein (Fig. 3B). In this oligomeric state, the four catalytic sites are located away from each other and can be accessed freely from outside (Fig. 3B).
Subsequent to in silico analysis, experimental validation of the oligomeric status of PfPGM1 was performed. Pfpgm1 gene was successfully cloned, and the expression of recombinant His6-PGM1 protein was validated using anti-6X His-tag antibodies (Fig. S3).
Further, the recombinant protein was nickel-nitrilotriacetic acid purified and size-exclusion chromatography was performed to ascertain the oligomeric structure of the protein. Prior to this, proteins with known molecular mass were run to plot a standard curve of molecular mass versus elution volume (Fig. 3C). The chromatogram of PfPGM1 revealed that the protein exists in multimeric forms in solution, mainly as a tetramer, and a minute fraction is also present as a higher-order oligomer (Fig. 3D). The peak fractions were subjected to SDS-PAGE analysis reflecting the purity of the eluted protein as compared to the input (Fig. 3E). Different multimeric structures exist for PGM across organisms. Here, we comprehend that the oligomeric status of the parasitic enzyme differs from the human enzyme.
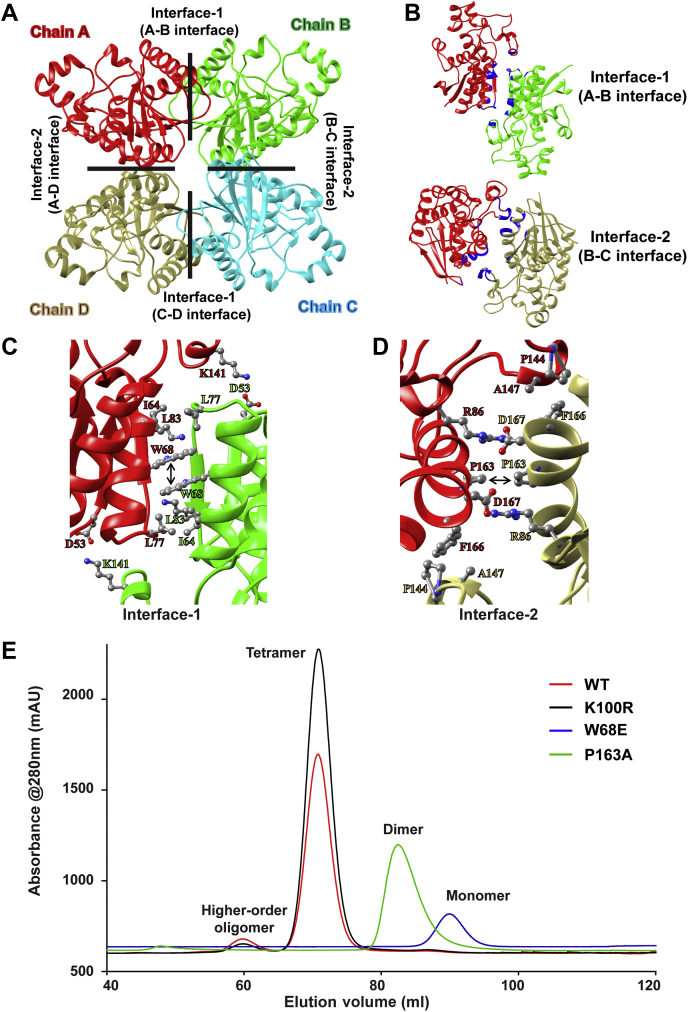
PfPGM1 oligomerization is mediated by the hydrophobic patches
The tetrameric form of PfPGM1 is achieved via interaction of each monomer with two other monomeric units. Therefore, the monomeric subunit has two interaction interfaces and altogether, the tetrameric unit has four interaction interfaces (Fig. 4A). The amino acid composition at the interface between chain A-B and chain C-D are identical, hereafter referred to as interface-1. Similarly, the composition at interfaces between chain B-C and chain A-D are alike, hereafter referred to as interface-2 (Fig. 4B). The oligomeric structures of PGM have been reported across many species. To gain further insight into the possibility of sequentially conserved oligomeric interfaces in PfPGM1, we closely inspected the interfacial residues at both interface-1 and interface-2.
Figure 4.
Key amino acid residues involved in the oligomerization of PfPGM1.A, the four subunits of PfPGM1 are represented as Chains A to D. The interface-1 formed between chains A-B and C-D and the interface-2 formed between chains B-C and A-D are marked by black lines and labeled accordingly. B, depicts interface-1 (chain A-B interface) on top and interface-2 (chain A-D interface) at the bottom. The interface residues are highlighted in blue color. Amino acid residues at PfPGM1 oligomerization interface-1 and interface-2 are shown in (C) and (D) respectively. The residues forming the hydrophobic interactions (W68-W68 at interface-1 and P163-P163 at interface-2) in the center of both the interfaces is marked by the black arrow. The interaction profile at the respective interfaces is shown in Fig. S4 in more detail. E, size-exclusion chromatogram demonstrating the elution peaks and oligomeric forms of WT protein and mutants. PGM, phosphoglycerate mutase.
We observed that the interface-1 of PfPGM1 is formed by 15 residues, out of which only D53 is 100% conserved across all the homologs (Table 1 and Fig. S2). This residue is responsible for hydrogen bond interaction with K141, which is 50% conserved (Figs. 4C and S4). Another residue, I64 (80% conserved) is involved in hydrophobic interactions with the neighboring residues. Importantly, the residue W68 (70% conserved) of one monomer establishes hydrophobic interaction (π-π stacking) with W68 of another monomer, at the center of interface-1. Additionally, W68 is sequentially surrounded by other hydrophobic residues (A67, V70, and L71) and a polar but uncharged one (N69), altogether forming a hydrophobic patch at the center of the interface. This hydrophobic patch is conserved in PGM across the diverse species (Fig. S2). Hence, the hydrophobic patch formed by A67, W68, N69, V70, and L71 might be very crucial for oligomerization at interface-1, especially W68 which forms π–π stacking interaction with W68 from the interacting partner. Only 6 PfPGM1 interface-1 residues are positionally identical (D53, V59, K61, I64, W68, and D75) to the human PGAM enzyme interface-1 residues (Table 1). The hydrophobic patch at the center of interface 1 in human PGAM constitutes L67, W68, T69, V70, and L71 residues. However, W68, V70, and L71 are identical in human and Plasmodium, the presence of L67 and T69 in human instead of A67 and N69 respectively makes it slightly more hydrophobic in human (Fig. S2).
Table 1.
Amino acid composition of the dimer interfaces formed in PfPGM1 tetrameric state and their sequential conservation
| Interface-1 | Sequence conservation (%) | Interface-2 | Sequence conservation (%) |
|---|---|---|---|
| E35 (SB) [H35] | 20 | R86 (HB) [R86]a | 90 |
| D53 (HB) [D53]a | 100 | K141 (HPB) [A143] | 50 |
| V59 (HPB) [V59]a | 60 | N142 (HPB) [D144] | 10 |
| K61 (HB) [K61]a | 60 | P144 (HPB) [T146] | 40 |
| I64 (HPB) [I64]a | 80 | K145 (HPB) [E147] | 10 |
| C65 (HPB)/(HB) [R65] | 10 | D146 (HB) [D148]a | 20 |
| W68 (HPB) [W68]a | 70 | A147 (HPB) [Q149] | 20 |
| K72 (HPB)/SB [D72] | 30 | P163 (HPB) [P165]a | 100 |
| D75 (HB) [D75]a | 80 | F166 (HPB) [N168] | 20 |
| L77 (HB) [M77] | 30 | D167 (HB) [E169] | 70 |
| H78 (HPB) [W78] | 20 | A170 (HPB) [V172] | 70 |
| V79 (HB) [L79] | 20 | P171 (HPB) [P173]a | 70 |
| K83 (HB) [R83] | 30 | L174 (HPB) [K176] | 30 |
| V139 (HPB) [R141] | 20 | H194 (HPB)/(HB)/SB [H196]a | 50 |
| K141 (HB) [A143] | 50 | N221 (HPB) [N223]a | 50 |
The interface residues were identified using LigPlot software. Sequence conservation (%) represents the positional conservation of respective amino acid in the PfPGM1 protein across the 10 organisms taken into account in this study (Table S1). HB, hydrogen bonding interaction; HPB, hydrophobic interaction SB, salt-bridges interactions are labeled for PfPGM1 protein, as illustrated in Fig. S4, C and D. The corresponding residues of human PGAM located at the same position on the structure are mentioned in square bracket.
Shows that the residues are positionally conserved between Plasmodium and human.
Similar to interface-1, interface-2 of PfPGM1 is also formed by the participation of 15 residues establishing hydrophobic and hydrogen bonding interactions (Table 1). The interface 2 residues also hold significant differences from the corresponding residues in human PGAM. Only residues R86, D146, P163, P171, H194, and N221 of PfPGM1 interface-2 are positionally identical to R8, D148, P165, P173, H196, and N223, respectively in human PGAM (Table 1). Compared to interface-2, interface-1 has more hydrogen bonds suggesting that oligomerization at interface-1 might be stronger than oligomerization at interface-2. Interface-2 also exhibits a hydrophobic patch formed by P163, F164, W165, and F166 at the center of the interface and lined by hydrogen bond interactions between R86-D167 at the ends (Figs. 4D and S4). In human PGAM, the amino acids located at identical position to the residues forming PfPGM1 interface-2 hydrophobic patch are P165, F166, W167, and N168. The residues corresponding to R86 and D167 of PfPGM1 are R86 and E169, respectively (Fig. S2). Due to the presence of N168 in human PGAM instead of F166 as in PfPGM1, the interface-2 hydrophobic patch in human is slightly less hydrophobic than in Plasmodium. The residue P163 is completely conserved across diverse species. Due to the constrained phi torsional angles, proline is considered to be very important for local conformations in the protein structures (42). In this scenario, the hydrophobic patch formed by P163, F164, W165, and F166 might also be important for oligomerization at interface-2.
Based on the above observations, two residues, W68 at interface-1 and P163 at interface-2 were chosen for experimental validation and characterization along with an active site residue K100 as a control. The mutants and WT protein were then purified (Fig. S5) and subjected to size-exclusion chromatography to determine the effect of single point mutations on the oligomeric status of the protein (Fig. 4E). Interestingly, the mutation of single amino acid tryptophan (W) to glutamate (E) at the 68th position completely disrupted the tetrameric structure of PfPGM1. The mutation of this residue to the polar amino acid glutamate hinders the interaction between the interface residues and slightly changes the pI of the protein thus preventing tetramerization. The chromatogram of W68E revealed a characteristic single elution peak at monomeric size highlighting the essentiality of tryptophan at that position for the correct oligomerization of the protein. The role of this single residue in maintaining the oligomerization of protein is quite astonishing and note-worthy.
Another mutant, P163A, eluted as dimeric form of the protein highlighting the significance of P163 at interface-2 for oligomerization. However, the active site mutant, K100R maintained the oligomerization status of the protein and eluted as tetramer and a higher-order oligomer similar to the WT protein.
The tetrameric structure of PfPGM1 contributes to optimal enzyme activity
Enzyme-kinetics define the catalytic activity and efficiency of an enzyme. The kinetic parameters of P. falciparum PGM enzyme were determined using PGM assay (Fig. S6A). The formation of the product 2-phosphoglycerate (PG) linked with NADH consumption via coupled-enzyme assay revealed that the enzyme follows Michaelis–Menten kinetics (Fig. S6B). The kinetic constants were determined by nonlinear regression using GraphPad Prism. The analysis yielded a KM value of 182 ± 56 μM and kcat value of 714 ± 68 s−1 for the WT enzyme. Interestingly, the substitution of Lys-100 to Arg resulted in dramatic loss in catalytic activity, thus evincing the essentiality of K100 for the catalytic activity (Fig. S6B).
A comparison with the yeast enzyme revealed that PfPGM1 has more affinity towards the substrate and has better turnover number highlighting its efficiency (Table 2). The higher catalytic activity and substrate affinity of the P. falciparum enzyme is beneficial for the rapidly proliferating parasites dependent mainly on glycolysis for energy.
Table 2.
Comparison of the kinetic constants of ScPGM, PfPGM1 WT, and mutant proteins
| Kinetic constants | ScPGMa | PfPGM1 | PfPGM1 W68E | PfPGM1 P163A |
|---|---|---|---|---|
| KM 3-PG (μM) | 510 ± 60 | 182 ± 56 | 175 ± 64 | 197 ± 68 |
| kcat (s−1) | 380 ± 10 | 714 ± 68 | 215 ± 23 | 335 ± 35 |
| kcat/KM (M−1 s−1) | 7.4 × 105 | 3.9 × 106 | 1.2 × 106 | 1.6 × 106 |
The data for ScPGM was adapted from reference (17). The results represent mean ± SD of three independent experiments for PfPGM1 WT and mutant proteins.
Next, structure-function relationship of the oligomerization defective mutants was analyzed. Subsequently, the catalytic activity of the mutants at positions 68 and 163 was determined and compared with the WT protein (Table 2). The dimer (P163A) and monomer (W68E) mutants exhibited reduced enzymatic activity in comparison with the WT protein. The catalytic efficiency (kcat/KM) of P163A and W68E mutant proteins reduced by ∼56.8% and 68.6% of the WT PfPGM1 protein, respectively (Table 2). The KM values of the mutant enzymes were comparable to the WT enzyme. These results strongly suggest that the tetrameric structure of the protein contributes to optimal enzymatic activity.
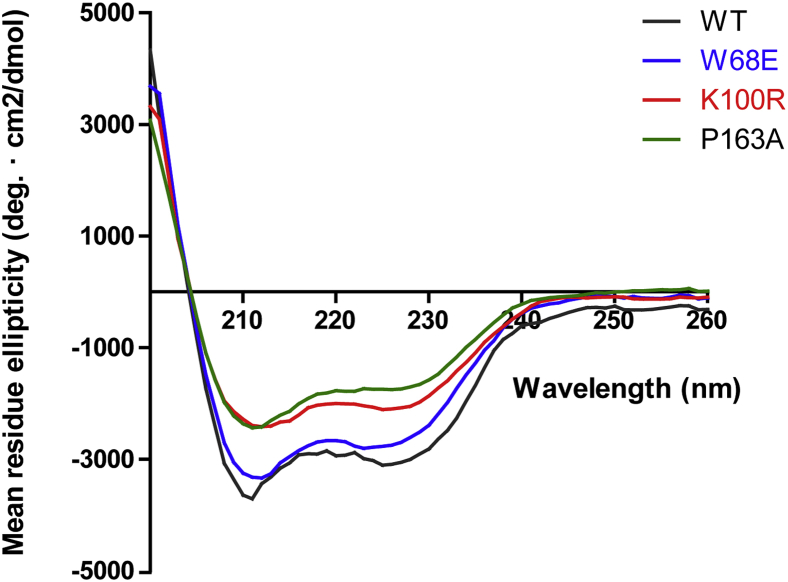
Subsequently, far-UV CD spectra depicting the secondary structure characteristics of the WT and mutant proteins were recorded (Fig. 5). Analysis of the spectra using BeStSel (Beta Structure Selection) tool revealed the percentage of different secondary structures in the respective proteins (Table S2) (43). The MRE values are slightly different for mutant proteins, K100R and P163A, however, the analysis indicate that the overall secondary structures of the mutant proteins do not change drastically when compared with the WT protein.
Figure 5.
Comparison of Far-UV CD spectra of PfPGM1 mutant proteins with the WT protein. The graph is representative of data of one experiment from two independent repeats. PGM, phosphoglycerate mutase.
Ectopic expression of WT PfPGM1 enhances parasite proliferation rate
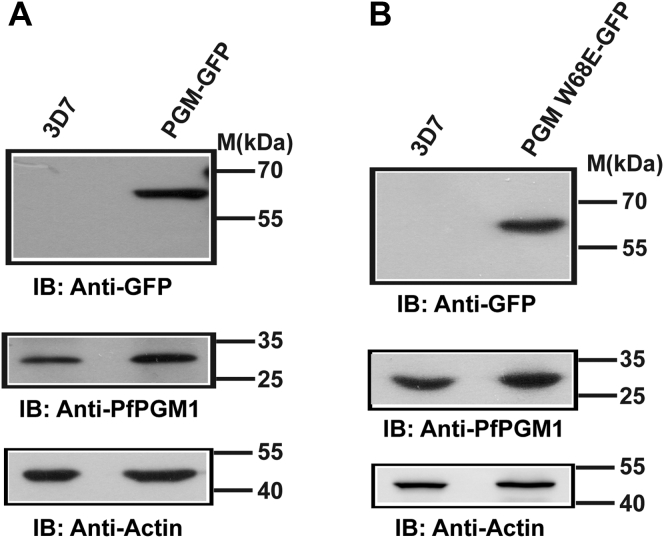
The increased PGM expression in cancer cells helps to augment proliferation and tumor growth (9). The metabolic framework of the parasite is similar to the cancerous cells and is required to support the rapid multiplication rates of the parasite (44). Consequently, we determined whether the ectopic expression of the glycolytic protein PfPGM1 episomally in the parasites will lead to better parasite growth and proliferation. Additionally, to better understand the in vivo structure-function relationship of PfPGM1, GFP expressing WT and monomer mutant (W68E) transgenic parasite lines were generated. Western blot analysis using antibodies against GFP confirmed the expression of PfPGM1-GFP and PfPGM1 W68E-GFP (∼60 kDa) in the respective transgenic parasites (Fig. 6, A and B, top panels). The status of the endogenous PfPGM1 protein in the transgenic parasites and 3D7 parasites is shown using antibodies against PfPGM1 (Fig. 6, A and B, middle panel). Live cell imaging revealed the expression and localization of the GFP-fusion protein (WT and mutant forms of PfPGM1 protein respectively) in the transgenic parasite lines (Fig. S7, A and B).
Figure 6.
Ectopic expression of PfPGM1-GFP and PfPGM1 W68E-GFP. Immunoblotting with antibodies against GFP and PfPGM1 in the transgenic parasites expressing, (A) PfPGM1-GFP and (B) PfPGM1 W68E-GFP, illustrating the expression profile of GFP-fusion protein (top panel) and the endogenous protein (middle panel). PfActin served as a loading control (bottom panel). 3D7 parasites were taken as control. PGM, phosphoglycerate mutase.
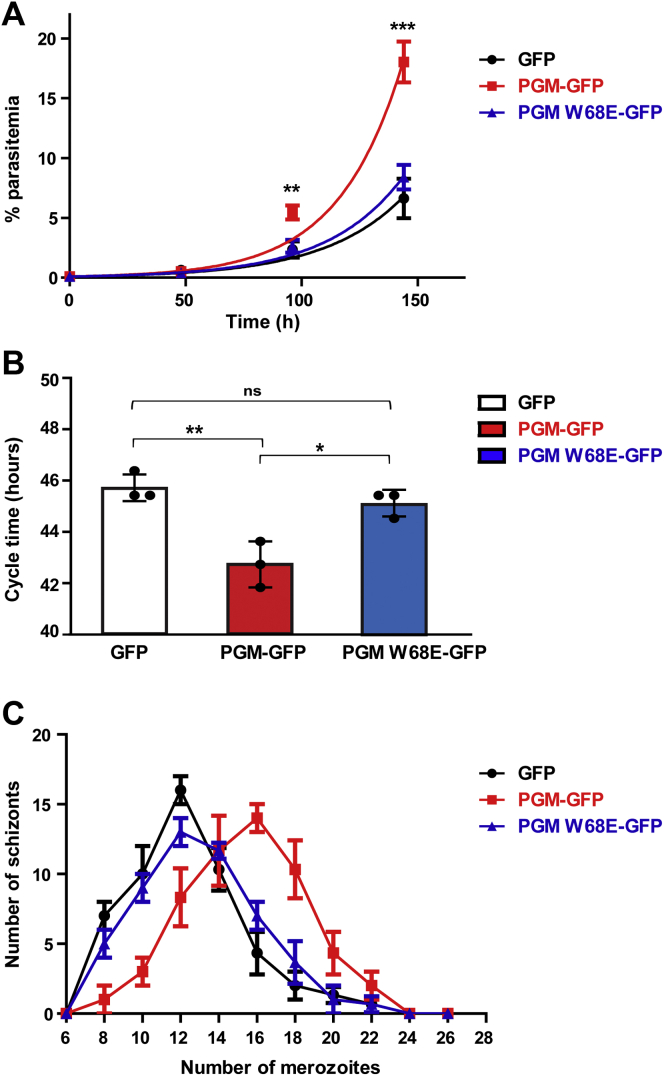
Further, the growth of the transgenic parasite lines expressing PGM-GFP and PGM W68E-GFP was monitored for three cycles (Fig. 7A). Interestingly, there was a robust increase in the parasitemia of PGM-GFP transgenic parasites as compared to the PfPGM1 W68E-GFP expressing parasites, and the difference was evident after the first cycle (Fig. 7A). GFP-only expressing parasites were taken as control. The doubling time of the respective parasite lines was calculated by fitting the data to an exponential growth curve. PfPGM1-GFP exhibited a shorter doubling time of 19.16 ± 0.36 h than PfPGM1 W68E-GFP (22.46 ± 0.46 h) and GFP (23.65 ± 0.98 h). The ectopic expression of PfPGM1-GFP protein accelerated the parasite proliferation rate indicating the crucial role of PfPGM1 in multiple pathways of the parasite including glycolytic flux.
Figure 7.
PfPGM1 enhances parasite growth and proliferation rate.A, the growth of transgenic parasite lines expressing the WT tetrameric protein, monomer mutant W68E, and GFP only were monitored for three cycles. B, duration of IDC for the three parasite lines. C, number of merozoites produced in different schizonts in the three transgenic parasite lines. The graphs represent mean ± SD of three independent experiments. Statistical significance of the data is represented as: ns p > 0.05, ∗ p ≤ 0.05, ∗∗ p ≤ 0.01 and ∗∗∗ p ≤ 0.001. IDC, intra-erythrocytic developmental cycle; PGM, phosphoglycerate mutase.
For a detailed analysis of alterations in cell proliferation, the length of intraerythrocytic cycle of the parasites was compared for the transgenic parasite lines expressing WT and mutant form of PfPGM1 (Fig. 7B). Interestingly, the PGM-GFP parasite line exhibited a faster cycle of 43 ± 1 h, while the parasites expressing mutant PGM W68E-GFP completed intra-erythrocytic developmental cycle (IDC) in 45.67 ± 0.57 h. Giemsa-stained smears made every 2 h demonstrated faster progression of the transgenic parasites expressing PGM-GFP in comparison with the parasites expressing PGM W68E-GFP or GFP only, through the three asexual stages of the parasite life cycle namely, rings, trophozoites, and schizonts (Fig. S8). Further, the progeny merozoites produced per schizont were counted from Giemsa-stained smears (Fig. 7C) and found to be more in PGM-GFP parasite line (15.39 ± 0.36) than the parasite line expressing the mutant PGM W68E-GFP (12.98 ± 0.47). The control GFP expressing parasites completed IDC in 46.33 ± 0.57 h and produced 12.36 ± 0.49 merozoites per parasite.
These results suggest that the monomeric structure of PfPGM1 fails to provide growth advantage when expressed ectopically in the parasites. Therefore, the tetrameric structure of PfPGM1 is imperative for efficient functioning of the protein and regulation of parasite growth rate in vivo.
Discussion
Our study reports the first functional characterization of P. falciparum glycolytic enzyme, PfPGM1 with emphasis on the biological significance of the oligomerization form of the enzyme in vivo. The human counterpart, PGAM, is highly overexpressed during cancer and the inhibition of this enzyme is lethal (45, 46, 47, 48, 49). The increased PGAM expression favors cancer cell proliferation and tumor growth by regulating glycolysis, anabolic biosynthesis, and other nonmetabolic functions (50, 51, 52, 53, 54, 55). Similar to the highly proliferating cancer cells, intraerythrocytic Plasmodium parasite metabolism is not only required to generate ATP but also to support the rapid rates of proliferation and replication (44, 56). Glycolysis is the major energy-yielding source for P. falciparum. However, the role of the essential glycolytic protein, PGM, was not characterized earlier and by doing so, we have tried to elaborate its functionality and structural aspect in P. falciparum.
In vivo characterization of PfPGM1 revealed that the protein is expressed throughout the asexual developmental stages; rings, trophozoites, and schizonts highlighting the metabolic needs of the parasite. Majority of the protein localizes in the parasite cytoplasm, in agreement with the subcellular occurrence of the glycolytic process. Intriguingly, the protein was also present in the nucleus, suggestive of moonlighting functions. Previous proteomics analysis of the nuclear fraction also identified PfPGM1 peptides in the different asexual stages of the parasite (57, 58). The human counterpart has been reported to be present in the nucleolus and regulate ribosome biogenesis (59). Whether PfPGM1 also localizes in the parasite nucleolus or performs noncanonical functions in the nucleus as detected for different metabolic enzymes (60) requires further detailed mechanistic studies.
The aftermath of PfPGM1 knockdown was investigated using the glmS riboswitch. This knockdown study of a glycolytic enzyme in P. falciparum is novel and highlights the indispensable role of a single enzyme PfPGM1 in parasite development. Further, the conditional knockdown allowed us to monitor parasite growth in real time while depleting PfPGM1. The depletion of PfPGM1 had deleterious effects on parasite progression and survival. Absence of PfPGM1 resulted in several defects including poor schizont maturation, prolonged cycle, delayed proliferation, and limited occupancy of the RBC by schizonts, suggesting either decreased digestive ability or lower nutrient uptake by the parasites.
It was reported earlier that the knockout of aldolase (another glycolytic enzyme) did not affect survival of the closely related parasite T. gondii and therefore is dispensable (61). Thus, the absolute requirement of the glycolytic enzyme PfPGM1 in P. falciparum highlights its crucial role and reflects that there is no redundancy between PfPGM1 and PfPGM2. Furthermore, the effects of deletion of PfPGM1 on global gene expression, ribosome biogenesis, total proteome, and metabolome of the parasite can be explored in future. Additionally, the parasite susceptibility to different genotoxic and oxidative stress, in the absence of PfPGM1, can be investigated.
The structure-function properties of an enzyme act as “footprints” and thus reflect a comprehensive evaluation of a particular enzyme which differentiates the enzyme from its homologs. The kinetic constants and oligomerization status of an enzyme are key determinants which can be exploited for pharmaceutical aspects (62). Overall, the biochemical studies of an enzyme are fundamental in understanding the mode of action, evolutionary process, differences from homologs, and the rate of catalysis of an enzyme which can be beneficial for targeted-drug development approaches.
In our present study, we have established that unlike the dimeric enzyme seen in Homo sapiens, PfPGM1 exists as a tetramer. This could be attributed to the differences observed in amino acid constituent of the dimeric interfaces 1 and 2. The detailed in silico analysis and size-exclusion chromatography revealed that the parasite enzyme has multimeric forms and resembles the eukaryotic model organism, yeast, that also shows the tetrameric oligomerization status of the protein. A higher-order oligomer is also formed by PfPGM1, a multimeric form peculiar to the parasitic enzyme although its functional significance remains elusive at this point.
Subsequently, we investigated the functional characteristics of the parasite protein. The biochemical characterization revealed that the kinetic parameters of P. falciparum PGM enzyme are better than its tetrameric counterpart in Saccharomyces cerevisiae. Further, the kinetic constants of PfPGM1 are also better than the kinetic parameters of the human enzyme that exists as a dimer (KM is 0.4 mM and kcat is 350 s−1) (63, 64). As the malarial parasite is very much dependent on the fundamental glucose metabolism for energy purposes, a highly efficient glycolytic enzyme is advantageous to support the rapid multiplication and proliferation within the host erythrocytes.
Further, we identified that W68 is a crucial interfacial residue and regulates the oligomerization status of the protein. Intriguingly, the mutation of this residue completely disrupted the tetrameric and higher-order oligomeric structures of PfPGM1, giving rise to monomeric form. This is the first report depicting the role of tryptophan in maintaining the oligomeric structure of PGM. Additionally, the mutation of another critical residue P163, resulted in dimeric form of the protein. Subsequently, the functional analysis of the oligomeric mutants W68E and P163A revealed considerable reduction in the in vitro enzyme activity (kcat/KM) compared to WT enzyme (∼68.6% and 56.8% respectively), implying that the tetrameric form has the maximum activity. The mutant enzymes exhibited KM values comparable to the WT enzyme suggesting that the mutations do not influence the substrate binding, but affect the catalytic process. The active site residue mutation (K100R) led to a dramatic reduction in mutase activity. The data highlight the critical role of the conserved residue, K100, for enzymatic activity.
Interestingly, PGM is upregulated in different cancer types and is a potential drug target in cancer treatment (51, 65, 66, 67). It is reported that the enzyme benefits tumor growth, increases the cellular life span, and has the ability to immortalize the cells. Quite intriguingly, the ectopic expression of PfPGM1 in the parasites, akin to cancer cells, demonstrated accelerated growth rate. The transgenic parasites expressing monomer mutant, W68E, were inefficient in comparison to the WT and did not exhibit increased proliferation. Additionally, the parasites expressing the WT protein completed the intraerythrocytic cycle faster and produced more number of merozoites than the mutant parasite line. Thus, PfPGM1 is critical for the overall growth and development of the parasite. Faster IDC and more number of progeny merozoites collectively may lead to faster growth rate over the cycles following ectopic expression of WT PfPGM1. Thus, the tetrameric form indeed contributes to enhanced proliferation and optimal activity of the enzyme in vivo. Nevertheless, the importance of oligomerization in vivo for optimal functioning of PGM is highlighted for the first time as earlier reports focused only on in vitro studies (15, 38, 68).
Overall, our findings on indispensable nature of PfPGM1, growth defect in parasites upon protein depletion, growth advantage to parasites following ectopic expression of the protein episomally, structure-function relation of PfPGM1, and the critical role of W68 and K100 residue for oligomerization and catalytic activity, respectively, highlight the fundamental aspects of a key glycolytic enzyme in general as well as unique aspects of parasite biology. Therefore, we propose PfPGM1 as a potential and promising drug target for antimalarial development.
Additionally, as oligomerization is a key parameter of the enzyme for its optimal function in vivo, screening small molecule inhibitors that can affect oligomerization will be a potential future strategy to specifically inhibit the parasite enzyme without affecting the host enzyme. Precluding protein oligomerization is emerging as a novel drug design approach and the inhibition of Hif1 dimerization using small molecule has been proven useful as potential cancer therapy (35, 69, 70, 71). Therefore, our study highlights the importance of oligomerization of an essential metabolic enzyme, the implication and usefulness of the same may not be restricted to the parasites only.
Experimental procedures
Prediction of PfPGM1 residues critical for oligomerization
The available PGM structures from P. falciparum and nine other organisms including human were obtained from Protein Data Bank and compared using UCSF Chimera (72, 73). The structures were aligned, and sequence conservation at each position over the structurally aligned region was calculated. Additionally, the critical residues at PfPGM-1 oligomerization interfaces were identified using LigPlot+ v.2.2.4 (74). These residues were mapped to their respective sequence conservation and visually inspected for their solvent accessibility, hydrophobic, and hydrophilic properties. This led to the identification of conserved hydrophobic patches at each interface of PfPGM1. Further, the potential interfacial residues critical for PfPGM1 oligomerization were identified and taken for experimental validation.
Generation and expression of His6-PfPGM1 and mutants
PfPGM1 (PF3D7_1120100) gene was PCR amplified and cloned in pET21c plasmid (Novagen) using primers 1 and 2 (Table S3) to obtain 6X-His tagged WT protein. Generation of the mutant proteins, K100R, W68E, and P163A, was achieved by site-directed mutagenesis (75) using respective primer set mentioned in Table S3. The WT clone and mutants were verified using Sanger sequencing (Eurofins Genomics India). Chemically competent Escherichia coli BL21 CodonPlus cells were transformed with the manipulated plasmids for the expression of WT and mutant proteins.
Protein purification and size-exclusion chromatography
The expression of the proteins was induced by 0.5 mM IPTG at 22 °C for 14 to 16 h. Recombinant His-tag WT and mutant PfPGM1 proteins were purified using nickel-nitrilotriacetic acid affinity protocol as described elsewhere with slight modifications (76). The protein-bound beads were washed with increasing NaCl (300–700 mM) and imidazole (10–40 mM) concentrations in lysis buffer containing 50 mM Tris–Cl (pH 7.5), 300 mM NaCl, 10 mM imidazole, 5 mM βME, 100 μM PMSF, and protease inhibitor cocktail. Proteins were finally eluted using increasing imidazole concentrations (100–500 mM) in lysis buffer.
The eluted protein was subjected to size-exclusion chromatography using HiLoad 16/600 Superdex 200 pg (GE Healthcare) in the presence of 50 mM Tris–Cl (pH 7.5), 150 mM NaCl, 5 mM βME, 100 μM PMSF, and protease inhibitor cocktail for second-column purification to obtain highly purified protein and deciphering oligomerization status as previously described (77).
Phosphoglycerate mutase assay
Phosphoglycerate mutase assay was performed as described elsewhere (7). Briefly, the formation of the product, 2-PG, was monitored by NADH-coupled assay. The linear portion of the curve (till 3 min) was used for the calculation of kinetic parameters. The reaction was done at 37 °C and the absorbance measured at 340 nm using Beckmann Coulter DU 800 UV-Vis Spectrophotometer. The reaction mixture comprised purified enzyme and increasing concentrations of 3-PG substrate. The buffer contained 100 mM Tris–HCl, pH 7.5, 100 mM KCl, 2 mM MgCl2, 0.5 mM EDTA, 10 μM 2,3-bisphosphoglycerate, 1.5 mM ADP, enolase (0.3 unit/ml), pyruvate kinase (0.5 unit/ml), lactate dehydrogenase (0.6 unit/ml), and 0.5 mM NADH. Five hundred nanograms of WT enzyme, 1 μg of P163A mutant, and 1.5 μg of W68E mutant protein were used during the assay. The experiments were performed thrice, and the graphs were plotted using GraphPad Prism followed by the calculation of kinetics parameters using nonlinear regression in GraphPad from at least two values. All the chemicals were purchased from Sigma-Aldrich.
Circular dichroism
The changes in secondary structure after mutations were studied using CD in far-UV range (78). The highly purified proteins were dialyzed against 10 mM phosphate buffer pH 7.4. The readings were taken in Chirascan Applied photophysics U.K CD spectrometer in at AIRF, JNU at 25 °C (wavelength range of 190 nm–260 nm). Mean residue Ellipticity values, [θ], in deg.cm2/dmol, were calculated and the graph was plotted. The data was used to calculate the secondary structure content using BeStSel online tool.
P. falciparum culture and transfection
P. falciparum 3D7 parasites were cultured using standard procedures (79, 80). Briefly, the parasites were nurtured in O+ human erythrocytes (obtained from Rotary Blood Bank) in RPMI 1640 medium supplemented with 0.5% Albumax II (Invitrogen), 27.2 mg/L hypoxanthine (Sigma), 0.2% sodium bicarbonate (Sigma), and 50 mg/L gentamicin (Himedia). The parasites were grown in an atmosphere of 5% CO2, 5% O2, and 90% N2 and maintained at 37 °C. Synchronization of parasites was performed with 5% sorbitol treatment, as described previously (81).
For generation of transgenic parasite lines with GFP-tagged proteins (82), the respective gene of interest were PCR amplified and cloned in pARL-GFP vector using primer pair 9 and 10 (Table S3). Transfection of P. falciparum 3D7 parasites with GFP-fusion constructs was performed using standard protocol (83). The transfectants were obtained after 21 to 30 days of selection using WR99210 (Jacobus Pharmaceuticals).
Generation and characterization of PfPGM1-GFPglmS transgenic parasite line
The 3′ end of PfPGM1 genomic locus was tagged using GFP-glmS. The region from 482 to 1124 bp of the gene was cloned via SpeI and KpnI restriction sites in the pJRTS_GFP_glmS plasmid (30). 3D7 parasites were then transfected with the modified plasmid using the method described above. Blasticidin (Sigma-Aldrich) was used for selection at a concentration of 2 μg/ml. The parasites obtained after 21 to 30 days of transfection were further selected using 2 on/off cycles of blasticidin of 2 weeks duration each. The integrants obtained were further purified with the help of limiting dilution.
The transgenic parasite line obtained was characterized by diagnostic PCR using DreamTaq DNA polymerase (Thermo Fisher Scientific) and primers from outside the homology region and GFP region (Primer no. 13 and 14 in Table S3). The genomic DNA from the transgenic parasites was purified using Genomic DNA isolation kit (mdi Genomic DNA Miniprep Kit). The conditions of diagnostic PCR were as follows: initial denaturation at 95 °C for 5 min, 35 cycles of denaturation at 95 °C for 40 s, annealing at 50 °C for 35 s, and extension at 68 °C for 1 min 15 s and followed by final extension at 68 °C for 10 min.
PfPGM1 knockdown was induced in tightly synchronized ring stage parasites using 5 mM GlcN (Sigma-Aldrich) for 24 h (30). Subsequently, PfPGM1 expression in GlcN-treated parasites and untreated parasites was analyzed using Western blot and confocal microscopy.
Growth curve analysis of untreated and GlcN-treated parasites was followed for three cycles. In 6-well plate, 3% hematocrit and initial parasitemia of 0.5% was set. Subsequently, every alternate day, culture media with or without sugar (GlcN) were added accordingly. Parasites were diluted depending on parasitemia and while calculating final parasitemia, the dilution factor was incorporated. Approximately, 1000 RBCs (Giemsa-stained thin blood smears) were counted to determine the parasitemia. Relative parasitemia values were calculated by dividing the parasitemia of treated culture by the corresponding parasitemia of untreated parasite culture taken as 100% at each time point (84). Each time point represents mean and SD obtained from at least three independent experiments.
Generation of polyclonal antibodies in mice
Immune sera directed against PfPGM1 was generated as per CPCSEA guidelines and following standard protocol (85) in 4 to 6 weeks old swiss albino mice. The generated polyclonal antibodies were checked for specificity against PfPGM1 using preimmune sera and immune sera in 3D7 parasite and RBC lysate.
Western blot analysis, immunofluorescence, and live cell imaging
Immunoblotting and immunofluorescence assay were performed, as described previously (86). Briefly, P. falciparum-infected erythrocytes or uninfected RBC were lysed with 0.15% saponin. The samples were prepared using 2X Laemmli SDS loading buffer and separated by SDS-PAGE followed by transfer on polyvinylidene difluoride membranes. Later, it was incubated with desired primary antibodies. The dilutions of the primary antibodies used in this study were as follows: anti-PfPGM1 1:2500, anti-GFP (Sigma) 1:5000. Subsequently, the blots were incubated with HRP-conjugated secondary antibodies (1:5000) as applicable and developed using enhanced chemiluminescence method. ImageJ software (NIH) was used for densitometric analysis. The slides prepared for immunofluorescence assay were visualized using Olympus confocal microscope. 3D reconstruction of Z-sections of selected images was made using Olympus FV31S-SW software.
Live cell imaging of GFP-expressing parasites was done using the method described elsewhere (87). The cells were immediately visualized under Olympus Confocal Laser Scanning Microscope to prevent the echinocytosis of the erythrocytes or drying out.
Proliferation and phenotype analysis
Proliferation rate analysis, duration of asexual growth cycle, and the number of merozoites produced per schizont were performed as described elsewhere (88, 89). The parasites in the three transgenic parasite lines expressing GFP, PGM-GFP, and PGM W68E-GFP proteins episomally were tightly synchronized using sorbitol treatment thrice prior to analysis. For proliferation analysis, parasites at 0.1% starting parasitemia were taken, and their proliferation was monitored for 7 days. Parasitemia was calculated from Giemsa-stained thin blood smears by counting approximately 1000 RBCs. The cycle duration was determined from the time taken between peak schizonts of two cycles monitored through Giemsa-stained blood smears made every 2 h. The number of merozoites produced per schizont was counted from mature schizonts in Giemsa-stained blood smears. Fifty mature schizonts per replicate were counted approximately.
Statistical analysis
p values were calculated using t test function in GraphPad Prism to determine the statistical significance of the data. All the experiments were at least performed thrice.
Data availability
All data are contained within the article or supporting information.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflict of interest with the contents of this article.
Acknowledgments
The authors acknowledge Dr Meetu Agarwal for the help with the generation of transgenic parasite line expressing GFP, Dr Abhik Saha for the help with size-exclusion chromatography, and Dr Philip J. Shaw from National Center for Genetic Engineering and Biotechnology (BIOTEC), National Science and Technology Development Agency (NSTDA), Pathum Thani, Thailand, for providing the pJRTS_GFP_glmS plasmid.
Author contributions
A. T., K. B., and S. K. D. conceptualization; A. T., K. B., and A. K. methodology; A. T., K. B., and A. K. validation; A. T., K. B., A. K., N. S., and S. K. D. formal analysis; A. T., K. B., and A. K. investigation; N. S. and S. K. D. resources; A. T., K. B., and A. K. data curation; A. T. writing–original draft; A. T., K. B., A. K., N. S., and S. K. D. writing–review & editing; A. T., K. B., A. K., N. S., and S. K. D. visualization; N. S. and S. K. D. supervision; S. K. D. funding acquisition.
Funding and additional information
S. K. D. acknowledges the Department of Biotechnology (BT/PR15639/Med/29/1145/2016) and Department of Science and Technology, SERB, Govt. of India (DPRP) for financial assistance. A. T. acknowledges CSIR, India for fellowship.
Edited by Ruma Banerjee
Supporting information
References
- 1.World Health Organization . World Health Organization; Geneva: 2020. World Malaria Report 2020: 20 Years of Global Progress and Challenges. [Google Scholar]
- 2.Maier A.G., Matuschewski K., Zhang M., Rug M. Plasmodium falciparum. Trends Parasitol. 2019;35:481–482. doi: 10.1016/j.pt.2018.11.010. [DOI] [PubMed] [Google Scholar]
- 3.Roth E., Jr. Plasmodium falciparum carbohydrate metabolism: A connection between host cell and parasite. Blood Cells. 1990;16:453–460. [PubMed] [Google Scholar]
- 4.Mehta M., Sonawat H.M., Sharma S. Glycolysis in Plasmodium falciparum results in modulation of host enzyme activities. J. Vector Borne Dis. 2006;43:95–103. [PubMed] [Google Scholar]
- 5.Alam A., Neyaz M.K., Ikramul Hasan S. Exploiting unique structural and functional properties of malarial glycolytic enzymes for antimalarial drug development. Malar. Res. Treat. 2014;2014:451065. doi: 10.1155/2014/451065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Britton H.G., Clarke J.B. Mechanism of the 2,3-diphosphoglycerate-dependent phosphoglycerate mutase from rabbit muscle. Biochem. J. 1972;130:397–410. doi: 10.1042/bj1300397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hallows W.C., Yu W., Denu J.M. Regulation of glycolytic enzyme phosphoglycerate mutase-1 by Sirt1 protein-mediated deacetylation. J. Biol. Chem. 2012;287:3850–3858. doi: 10.1074/jbc.M111.317404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oslund R.C., Su X., Haugbro M., Kee J.M., Esposito M., David Y., Wang B., Ge E., Perlman D.H., Kang Y., Muir T.W., Rabinowitz J.D. Bisphosphoglycerate mutase controls serine pathway flux via 3-phosphoglycerate. Nat. Chem. Biol. 2017;13:1081–1087. doi: 10.1038/nchembio.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hitosugi T., Zhou L., Elf S., Fan J., Kang H.B., Seo J.H., Shan C., Dai Q., Zhang L., Xie J., Gu T.L., Jin P., Aleckovic M., LeRoy G., Kang Y., et al. Phosphoglycerate mutase 1 coordinates glycolysis and biosynthesis to promote tumor growth. Cancer Cell. 2012;22:585–600. doi: 10.1016/j.ccr.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kondoh H., Lleonart M.E., Gil J., Wang J., Degan P., Peters G., Martinez D., Carnero A., Beach D. Glycolytic enzymes can modulate cellular life span. Cancer Res. 2005;65:177–185. [PubMed] [Google Scholar]
- 11.Lopez C.M., Wallich R., Riesbeck K., Skerka C., Zipfel P.F. Candida albicans uses the surface protein Gpm1 to attach to human endothelial cells and to keratinocytes via the adhesive protein vitronectin. PLoS One. 2014;9 doi: 10.1371/journal.pone.0090796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharif F., Rasul A., Ashraf A., Hussain G., Younis T., Sarfraz I., Chaudhry M.A., Bukhari S.A., Ji X.Y., Selamoglu Z., Ali M. Phosphoglycerate mutase 1 in cancer: A promising target for diagnosis and therapy. IUBMB Life. 2019;71:1418–1427. doi: 10.1002/iub.2100. [DOI] [PubMed] [Google Scholar]
- 13.Rigden D.J., Alexeev D., Phillips S.E., Fothergill-Gilmore L.A. The 2.3 A X-ray crystal structure of S. cerevisiae phosphoglycerate mutase. J. Mol. Biol. 1998;276:449–459. doi: 10.1006/jmbi.1997.1554. [DOI] [PubMed] [Google Scholar]
- 14.Sakoda S., Shanske S., DiMauro S., Schon E.A. Isolation of a cDNA encoding the B isozyme of human phosphoglycerate mutase (PGAM) and characterization of the PGAM gene family. J. Biol. Chem. 1988;263:16899–16905. [PubMed] [Google Scholar]
- 15.Blackburn E.A., Fuad F.A., Morgan H.P., Nowicki M.W., Wear M.A., Michels P.A., Fothergill-Gilmore L.A., Walkinshaw M.D. Trypanosomatid phosphoglycerate mutases have multiple conformational and oligomeric states. Biochem. Biophys. Res. Commun. 2014;450:936–941. doi: 10.1016/j.bbrc.2014.06.113. [DOI] [PubMed] [Google Scholar]
- 16.Chander M., Setlow P., Lamani E., Jedrzejas M.J. Structural studies on a 2,3-diphosphoglycerate independent phosphoglycerate mutase from Bacillus stearothermophilus. J. Struct. Biol. 1999;126:156–165. doi: 10.1006/jsbi.1999.4112. [DOI] [PubMed] [Google Scholar]
- 17.Fraser H.I., Kvaratskhelia M., White M.F. The two analogous phosphoglycerate mutases of Escherichia coli. FEBS Lett. 1999;455:344–348. doi: 10.1016/s0014-5793(99)00910-2. [DOI] [PubMed] [Google Scholar]
- 18.Baisamy L., Jurisch N., Diviani D. Leucine zipper-mediated homo-oligomerization regulates the Rho-GEF activity of AKAP-Lbc. J. Biol. Chem. 2005;280:15405–15412. doi: 10.1074/jbc.M414440200. [DOI] [PubMed] [Google Scholar]
- 19.Hashimoto K., Panchenko A.R. Mechanisms of protein oligomerization, the critical role of insertions and deletions in maintaining different oligomeric states. Proc. Natl. Acad. Sci. U. S. A. 2010;107:20352–20357. doi: 10.1073/pnas.1012999107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodsell D.S., Olson A.J. Structural symmetry and protein function. Annu. Rev. Biophys. Biomol. Struct. 2000;29:105–153. doi: 10.1146/annurev.biophys.29.1.105. [DOI] [PubMed] [Google Scholar]
- 21.Koike R., Kidera A., Ota M. Alteration of oligomeric state and domain architecture is essential for functional transformation between transferase and hydrolase with the same scaffold. Protein Sci. 2009;18:2060–2066. doi: 10.1002/pro.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hills T., Srivastava A., Ayi K., Wernimont A.K., Kain K., Waters A.P., Hui R., Pizarro J.C. Characterization of a new phosphatase from Plasmodium. Mol. Biochem. Parasitol. 2011;179:69–79. doi: 10.1016/j.molbiopara.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 23.Bushell E., Gomes A.R., Sanderson T., Anar B., Girling G., Herd C., Metcalf T., Modrzynska K., Schwach F., Martin R.E., Mather M.W., McFadden G.I., Parts L., Rutledge G.G., Vaidya A.B., et al. Functional profiling of a Plasmodium genome reveals an abundance of essential genes. Cell. 2017;170:260–272.e8. doi: 10.1016/j.cell.2017.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang M., Wang C., Otto T.D., Oberstaller J., Liao X., Adapa S.R., Udenze K., Bronner I.F., Casandra D., Mayho M., Brown J., Li S., Swanson J., Rayner J.C., Jiang R.H.Y., et al. Uncovering the essential genes of the human malaria parasite Plasmodium falciparum by saturation mutagenesis. Science. 2018;360 doi: 10.1126/science.aap7847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le Roch K.G., Zhou Y., Blair P.L., Grainger M., Moch J.K., Haynes J.D., De La Vega P., Holder A.A., Batalov S., Carucci D.J., Winzeler E.A. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003;301:1503–1508. doi: 10.1126/science.1087025. [DOI] [PubMed] [Google Scholar]
- 26.Foth B.J., Zhang N., Chaal B.K., Sze S.K., Preiser P.R., Bozdech Z. Quantitative time-course profiling of parasite and host cell proteins in the human malaria parasite Plasmodium falciparum. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M110.006411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.The Plasmodium Genome Database Collaborative PlasmoDB: An integrative database of the Plasmodium falciparum genome. Tools for accessing and analyzing finished and unfinished sequence data. The Plasmodium Genome Database Collaborative. Nucleic Acids Res. 2001;29:66–69. doi: 10.1093/nar/29.1.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thezenas M.L., Huang H., Njie M., Ramaprasad A., Nwakanma D.C., Fischer R., Digleria K., Walther M., Conway D.J., Kessler B.M., Casals-Pascual C. PfHPRT: A new biomarker candidate of acute Plasmodium falciparum infection. J. Proteome Res. 2013;12:1211–1222. doi: 10.1021/pr300858g. [DOI] [PubMed] [Google Scholar]
- 29.Wang H.L., Wen L.M., Pei Y.J., Wang F., Yin L.T., Bai J.Z., Guo R., Wang C.F., Yin G.R. Recombinant Toxoplasma gondii phosphoglycerate mutase 2 confers protective immunity against toxoplasmosis in BALB/c mice. Parasite. 2016;23:12. doi: 10.1051/parasite/2016012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prommana P., Uthaipibull C., Wongsombat C., Kamchonwongpaisan S., Yuthavong Y., Knuepfer E., Holder A.A., Shaw P.J. Inducible knockdown of Plasmodium gene expression using the glmS ribozyme. PLoS One. 2013;8 doi: 10.1371/journal.pone.0073783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jankowska-Dollken M., Sanchez C.P., Cyrklaff M., Lanzer M. Overexpression of the HECT ubiquitin ligase PfUT prolongs the intraerythrocytic cycle and reduces invasion efficiency of Plasmodium falciparum. Sci. Rep. 2019;9:18333. doi: 10.1038/s41598-019-54854-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parkyn Schneider M., Liu B., Glock P., Suttie A., McHugh E., Andrew D., Batinovic S., Williamson N., Hanssen E., McMillan P., Hliscs M., Tilley L., Dixon M.W.A. Disrupting assembly of the inner membrane complex blocks Plasmodium falciparum sexual stage development. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Florentin A., Cobb D.W., Fishburn J.D., Cipriano M.J., Kim P.S., Fierro M.A., Striepen B., Muralidharan V. PfClpC is an essential Clp chaperone required for plastid integrity and Clp protease stability in Plasmodium falciparum. Cell Rep. 2017;21:1746–1756. doi: 10.1016/j.celrep.2017.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delto C.F., Heisler F.F., Kuper J., Sander B., Kneussel M., Schindelin H. The LisH motif of muskelin is crucial for oligomerization and governs intracellular localization. Structure. 2015;23:364–373. doi: 10.1016/j.str.2014.11.016. [DOI] [PubMed] [Google Scholar]
- 35.Jastrzebska B., Chen Y., Orban T., Jin H., Hofmann L., Palczewski K. Disruption of rhodopsin dimerization with synthetic peptides targeting an interaction interface. J. Biol. Chem. 2015;290:25728–25744. doi: 10.1074/jbc.M115.662684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rofougaran R., Crona M., Vodnala M., Sjoberg B.M., Hofer A. Oligomerization status directs overall activity regulation of the Escherichia coli class Ia ribonucleotide reductase. J. Biol. Chem. 2008;283:35310–35318. doi: 10.1074/jbc.M806738200. [DOI] [PubMed] [Google Scholar]
- 37.Xu H., Qing X., Wang Q., Li C., Lai L. Dimerization of PHGDH via the catalytic unit is essential for its enzymatic function. J. Biol. Chem. 2021;296:100572. doi: 10.1016/j.jbc.2021.100572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.White M.F., Fothergill-Gilmore L.A., Kelly S.M., Price N.C. Substitution of His-181 by alanine in yeast phosphoglycerate mutase leads to cofactor-induced dissociation of the tetrameric structure. Biochem. J. 1993;291:479–483. doi: 10.1042/bj2910479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsujino S., Shanske S., Sakoda S., Fenichel G., DiMauro S. The molecular genetic basis of muscle phosphoglycerate mutase (PGAM) deficiency. Am. J. Hum. Genet. 1993;52:472–477. [PMC free article] [PubMed] [Google Scholar]
- 40.Burley S.K., Bhikadiya C., Bi C., Bittrich S., Chen L., Crichlow G.V., Christie C.H., Dalenberg K., Di Costanzo L., Duarte J.M., Dutta S., Feng Z., Ganesan S., Goodsell D.S., Ghosh S., et al. RCSB Protein Data Bank: Powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences. Nucleic Acids Res. 2021;49:D437–D451. doi: 10.1093/nar/gkaa1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caruthers J.M., Hol W.G.J. Structural Genomics of Pathogenic Protozoa Consortium (SGPP) 2004. Structure of phosphoglycerate mutase from Plasmodium falciparum at 2.6 resolution. [DOI] [Google Scholar]
- 42.MacArthur M.W., Thornton J.M. Influence of proline residues on protein conformation. J. Mol. Biol. 1991;218:397–412. doi: 10.1016/0022-2836(91)90721-h. [DOI] [PubMed] [Google Scholar]
- 43.Micsonai A., Wien F., Bulyaki E., Kun J., Moussong E., Lee Y.H., Goto Y., Refregiers M., Kardos J. BeStSel: A web server for accurate protein secondary structure prediction and fold recognition from the circular dichroism spectra. Nucleic Acids Res. 2018;46:W315–W322. doi: 10.1093/nar/gky497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salcedo-Sora J.E., Caamano-Gutierrez E., Ward S.A., Biagini G.A. The proliferating cell hypothesis: A metabolic framework for plasmodium growth and development. Trends Parasitol. 2014;30:170–175. doi: 10.1016/j.pt.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li C., Xiao Z., Chen Z., Zhang X., Li J., Wu X., Li X., Yi H., Li M., Zhu G., Liang S. Proteome analysis of human lung squamous carcinoma. Proteomics. 2006;6:547–558. doi: 10.1002/pmic.200500256. [DOI] [PubMed] [Google Scholar]
- 46.Ren F., Wu H., Lei Y., Zhang H., Liu R., Zhao Y., Chen X., Zeng D., Tong A., Chen L., Wei Y., Huang C. Quantitative proteomics identification of phosphoglycerate mutase 1 as a novel therapeutic target in hepatocellular carcinoma. Mol. Cancer. 2010;9:81. doi: 10.1186/1476-4598-9-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takahashi Y., Takahashi S., Yoshimi T., Miura T. Hypoxia-induced expression of phosphoglycerate mutase B in fibroblasts. Eur. J. Biochem. 1998;254:497–504. doi: 10.1046/j.1432-1327.1998.2540497.x. [DOI] [PubMed] [Google Scholar]
- 48.Evans M.J., Morris G.M., Wu J., Olson A.J., Sorensen E.J., Cravatt B.F. Mechanistic and structural requirements for active site labeling of phosphoglycerate mutase by spiroepoxides. Mol. Biosyst. 2007;3:495–506. doi: 10.1039/b705113a. [DOI] [PubMed] [Google Scholar]
- 49.Liu Z.G., Ding J., Du C., Xu N., Wang E.L., Li J.Y., Wang Y.Y., Yu J.M. Phosphoglycerate mutase 1 is highly expressed in C6 glioma cells and human astrocytoma. Oncol. Lett. 2018;15:8935–8940. doi: 10.3892/ol.2018.8477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lincet H., Icard P. How do glycolytic enzymes favour cancer cell proliferation by nonmetabolic functions? Oncogene. 2015;34:3751–3759. doi: 10.1038/onc.2014.320. [DOI] [PubMed] [Google Scholar]
- 51.Jiang X., Sun Q., Li H., Li K., Ren X. The role of phosphoglycerate mutase 1 in tumor aerobic glycolysis and its potential therapeutic implications. Int. J. Cancer. 2014;135:1991–1996. doi: 10.1002/ijc.28637. [DOI] [PubMed] [Google Scholar]
- 52.Chaneton B., Gottlieb E. PGAMgnam style: A glycolytic switch controls biosynthesis. Cancer Cell. 2012;22:565–566. doi: 10.1016/j.ccr.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 53.Zhang D., Jin N., Sun W., Li X., Liu B., Xie Z., Qu J., Xu J., Yang X., Su Y., Tang S., Han H., Chen D., Ding J., Tan M., et al. Phosphoglycerate mutase 1 promotes cancer cell migration independent of its metabolic activity. Oncogene. 2017;36:2900–2909. doi: 10.1038/onc.2016.446. [DOI] [PubMed] [Google Scholar]
- 54.Liu X., Tan X., Liu P., Wu Y., Qian S., Zhang X. Phosphoglycerate mutase 1 (PGAM1) promotes pancreatic ductal adenocarcinoma (PDAC) metastasis by acting as a novel downstream target of the PI3K/Akt/mTOR pathway. Oncol. Res. 2018;26:1123–1131. doi: 10.3727/096504018X15166223632406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mikawa T., Shibata E., Shimada M., Ito K., Ito T., Kanda H., Takubo K., Lleonart M.E., Inagaki N., Yokode M., Kondoh H. Phosphoglycerate mutase cooperates with Chk1 kinase to regulate glycolysis. iScience. 2020;23:101306. doi: 10.1016/j.isci.2020.101306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ghashghaeinia M., Koberle M., Mrowietz U., Bernhardt I. Proliferating tumor cells mimick glucose metabolism of mature human erythrocytes. Cell Cycle. 2019;18:1316–1334. doi: 10.1080/15384101.2019.1618125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Batugedara G., Lu X.M., Saraf A., Sardiu M.E., Cort A., Abel S., Prudhomme J., Washburn M.P., Florens L., Bunnik E.M., Le Roch K.G. The chromatin bound proteome of the human malaria parasite. Microb. Genom. 2020;6 doi: 10.1099/mgen.0.000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oehring S.C., Woodcroft B.J., Moes S., Wetzel J., Dietz O., Pulfer A., Dekiwadia C., Maeser P., Flueck C., Witmer K., Brancucci N.M., Niederwieser I., Jenoe P., Ralph S.A., Voss T.S. Organellar proteomics reveals hundreds of novel nuclear proteins in the malaria parasite Plasmodium falciparum. Genome Biol. 2012;13 doi: 10.1186/gb-2012-13-11-r108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gizak A., Grenda M., Mamczur P., Wisniewski J., Sucharski F., Silberring J., McCubrey J.A., Wisniewski J.R., Rakus D. Insulin/IGF1-PI3K-dependent nucleolar localization of a glycolytic enzyme--phosphoglycerate mutase 2, is necessary for proper structure of nucleolus and RNA synthesis. Oncotarget. 2015;6:17237–17250. doi: 10.18632/oncotarget.4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boukouris A.E., Zervopoulos S.D., Michelakis E.D. Metabolic enzymes moonlighting in the nucleus: Metabolic regulation of gene transcription. Trends Biochem. Sci. 2016;41:712–730. doi: 10.1016/j.tibs.2016.05.013. [DOI] [PubMed] [Google Scholar]
- 61.Shen B., Sibley L.D. Toxoplasma aldolase is required for metabolism but dispensable for host-cell invasion. Proc. Natl. Acad. Sci. U. S. A. 2014;111:3567–3572. doi: 10.1073/pnas.1315156111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nagar S., Argikar U.A., Tweedie D.J. Enzyme kinetics in drug metabolism: Fundamentals and applications. Methods Mol. Biol. 2014;1113:1–6. doi: 10.1007/978-1-62703-758-7_1. [DOI] [PubMed] [Google Scholar]
- 63.de Atauri P., Repiso A., Oliva B., Vives-Corrons J.L., Climent F., Carreras J. Characterization of the first described mutation of human red blood cell phosphoglycerate mutase. Biochim. Biophys. Acta. 2005;1740:403–410. doi: 10.1016/j.bbadis.2004.11.023. [DOI] [PubMed] [Google Scholar]
- 64.Wisniewski J.R., Gizak A., Rakus D. Integrating proteomics and enzyme kinetics reveals tissue-specific types of the glycolytic and gluconeogenic pathways. J. Proteome Res. 2015;14:3263–3273. doi: 10.1021/acs.jproteome.5b00276. [DOI] [PubMed] [Google Scholar]
- 65.Huang K., Liang Q., Zhou Y., Jiang L.L., Gu W.M., Luo M.Y., Tang Y.B., Wang Y., Lu W., Huang M., Zhang S.Z., Zhuang G.L., Dai Q., Shen Q.C., Zhang J., et al. A novel allosteric inhibitor of phosphoglycerate mutase 1 suppresses growth and metastasis of non-small-cell lung cancer. Cell Metab. 2019;30:1107–1119.e8. doi: 10.1016/j.cmet.2019.09.014. [DOI] [PubMed] [Google Scholar]
- 66.Huang K., Jiang L., Liang R., Li H., Ruan X., Shan C., Ye D., Zhou L. Synthesis and biological evaluation of anthraquinone derivatives as allosteric phosphoglycerate mutase 1 inhibitors for cancer treatment. Eur. J. Med. Chem. 2019;168:45–57. doi: 10.1016/j.ejmech.2019.01.085. [DOI] [PubMed] [Google Scholar]
- 67.Liang Q., Gu W.M., Huang K., Luo M.Y., Zou J.H., Zhuang G.L., Lei H.M., Chen H.Z., Zhu L., Zhou L., Shen Y. HKB99, an allosteric inhibitor of phosphoglycerate mutase 1, suppresses invasive pseudopodia formation and upregulates plasminogen activator inhibitor-2 in erlotinib-resistant non-small cell lung cancer cells. Acta Pharmacol. Sin. 2021;42:115–119. doi: 10.1038/s41401-020-0399-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.White M.F., Fothergill-Gilmore L.A., Kelly S.M., Price N.C. Dissociation of the tetrameric phosphoglycerate mutase from yeast by a mutation in the subunit contact region. Biochem. J. 1993;295:743–748. doi: 10.1042/bj2950743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gabizon R., Friedler A. Allosteric modulation of protein oligomerization: An emerging approach to drug design. Front. Chem. 2014;2:9. doi: 10.3389/fchem.2014.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee K., Zhang H., Qian D.Z., Rey S., Liu J.O., Semenza G.L. Acriflavine inhibits HIF-1 dimerization, tumor growth, and vascularization. Proc. Natl. Acad. Sci. U. S. A. 2009;106:17910–17915. doi: 10.1073/pnas.0909353106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 71.Miranda E., Nordgren I.K., Male A.L., Lawrence C.E., Hoakwie F., Cuda F., Court W., Fox K.R., Townsend P.A., Packham G.K., Eccles S.A., Tavassoli A. A cyclic peptide inhibitor of HIF-1 heterodimerization that inhibits hypoxia signaling in cancer cells. J. Am. Chem. Soc. 2013;135:10418–10425. doi: 10.1021/ja402993u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 74.Laskowski R.A., Swindells M.B. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011;51:2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 75.Bachman J. Site-directed mutagenesis. Methods Enzymol. 2013;529:241–248. doi: 10.1016/B978-0-12-418687-3.00019-7. [DOI] [PubMed] [Google Scholar]
- 76.Dar A., Prusty D., Mondal N., Dhar S.K. A unique 45-amino-acid region in the toprim domain of Plasmodium falciparum gyrase B is essential for its activity. Eukaryot. Cell. 2009;8:1759–1769. doi: 10.1128/EC.00149-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Verma V., Kumar A., Nitharwal R.G., Alam J., Mukhopadhyay A.K., Dasgupta S., Dhar S.K. 'Modulation of the enzymatic activities of replicative helicase (DnaB) by interaction with Hp0897: A possible mechanism for helicase loading in Helicobacter pylori'. Nucleic Acids Res. 2016;44:3288–3303. doi: 10.1093/nar/gkw148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Banu K., Mitra P., Subbarao N., Dhar S.K. Role of tyrosine residue (Y213) in nuclear retention of PCNA1 in human malaria parasite Plasmodium falciparum. FEMS Microbiol. Lett. 2018;365 doi: 10.1093/femsle/fny182. [DOI] [PubMed] [Google Scholar]
- 79.Moll K., Kaneko A., Scherf A., Wahlgren M. EVIMalaR; Glasgow, UK: 2013. Methods in Malaria Research. [Google Scholar]
- 80.Mehra P., Biswas A.K., Gupta A., Gourinath S., Chitnis C.E., Dhar S.K. Expression and characterization of human malaria parasite Plasmodium falciparum origin recognition complex subunit 1. Biochem. Biophys. Res. Commun. 2005;337:955–966. doi: 10.1016/j.bbrc.2005.09.131. [DOI] [PubMed] [Google Scholar]
- 81.Bhowmick K., Dhar S.K. Plasmodium falciparum single-stranded DNA-binding protein (PfSSB) interacts with PfPrex helicase and modulates its activity. FEMS Microbiol. Lett. 2014;351:78–87. doi: 10.1111/1574-6968.12343. [DOI] [PubMed] [Google Scholar]
- 82.Deshmukh A.S., Srivastava S., Herrmann S., Gupta A., Mitra P., Gilberger T.W., Dhar S.K. The role of N-terminus of Plasmodium falciparum ORC1 in telomeric localization and var gene silencing. Nucleic Acids Res. 2012;40:5313–5331. doi: 10.1093/nar/gks202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fidock D.A., Wellems T.E. Transformation with human dihydrofolate reductase renders malaria parasites insensitive to WR99210 but does not affect the intrinsic activity of proguanil. Proc. Natl. Acad. Sci. U. S. A. 1997;94:10931–10936. doi: 10.1073/pnas.94.20.10931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Burda P.C., Crosskey T., Lauk K., Zurborg A., Sohnchen C., Liffner B., Wilcke L., Pietsch E., Strauss J., Jeffries C.M., Svergun D.I., Wilson D.W., Wilmanns M., Gilberger T.W. Structure-based identification and functional characterization of a lipocalin in the malaria parasite Plasmodium falciparum. Cell Rep. 2020;31:107817. doi: 10.1016/j.celrep.2020.107817. [DOI] [PubMed] [Google Scholar]
- 85.Harlow E., Lane D. CSHL Press; Long Island, NY: 1988. Antibodies: A Laboratory Manual. [Google Scholar]
- 86.Bhowmick K., Tehlan A., Sunita, Sudhakar R., Kaur I., Sijwali P.S., Krishnamachari A., Dhar S.K. Plasmodium falciparum GCN5 acetyltransferase follows a novel proteolytic processing pathway that is essential for its function. J. Cell Sci. 2020;133 doi: 10.1242/jcs.236489. [DOI] [PubMed] [Google Scholar]
- 87.Gruring C., Spielmann T. Imaging of live malaria blood stage parasites. Methods Enzymol. 2012;506:81–92. doi: 10.1016/B978-0-12-391856-7.00029-9. [DOI] [PubMed] [Google Scholar]
- 88.Miao J., Fan Q., Cui L., Li X., Wang H., Ning G., Reese J.C. The MYST family histone acetyltransferase regulates gene expression and cell cycle in malaria parasite Plasmodium falciparum. Mol. Microbiol. 2010;78:883–902. doi: 10.1111/j.1365-2958.2010.07371.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Reilly H.B., Wang H., Steuter J.A., Marx A.M., Ferdig M.T. Quantitative dissection of clone-specific growth rates in cultured malaria parasites. Int. J. Parasitol. 2007;37:1599–1607. doi: 10.1016/j.ijpara.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article or supporting information.