Background
Mucopolysaccharidosis type II (MPS II), also known as Hunter syndrome, is a rare X-linked recessive disease caused by a deficiency of the lysosomal enzyme iduronate-2-sulfatase (IDS), which activates intracellular accumulation of nonmetabolized glycosaminoglycans such as heparan sulfate and dermatan sulfate. This accumulation causes severe damage to several tissues, principally the central nervous system. Previously, we identified 187 IDS-protein interactions in the mouse brain. To validate a subset of these interactions, we selected and cloned the coding regions of 10 candidate genes to perform a targeted yeast two-hybrid assay. The results allowed the identification of the physical interaction of IDS with LSAMP and SYT1. Although the physiological relevance of these complexes is unknown, recent advances allow us to point out that these interactions could be involved in vesicular trafficking of IDS through the interaction with SYT1, as well as to the ability to form a transcytosis module between the cellular components of the blood-brain-barrier (BBB) through its interaction with LSAMP. These results may shed light on the role of IDS on cellular homeostasis and may also contribute to the understanding of MPS II physiopathology and the development of novel therapeutic strategies to transport recombinant IDS through the brain endothelial cells toward the brain parenchyma.
Keywords: Lysosome, Hunter syndrome, Yeast two-hybrid, Proteomics, Interactome
Lysosome; Hunter syndrome; Yeast two-hybrid; Proteomics; Interactome.
1. Introduction
MPS II (OMIM 309900) is a metabolic disorder caused by mutations affecting the gene IDS (3.1.6.13), encoding the lysosomal enzyme iduronate-2-sulfatase. IDS is required for the degradation of the glycosaminoglycans (GAGs) heparan sulfate (HS) and dermatan sulfate (DS) [1]. Therefore, the unmetabolized GAGs accumulated within the lysosomes of different tissues, including the central nervous system (CNS), lead to progressive cellular damage [1]. Aggressive phenotypes include clinical manifestations such as cognitive impairment, coarse facies, short stature, skeletal abnormalities, joint stiffness, ocular damage, chronic diarrhea, hearing impairment, and communicating hydrocephalus. Patients with mild phenotypes have the same somatic features with a lower progression rate and without central nervous system involvement [1, 2]. Current treatment alternatives include enzyme replacement therapy (ERT), stem cells, bone marrow, or peripheral blood hematopoietic cells transplantations [3, 4, 5].
Although the storage of HS and DS is considered the primary cause of MPS II, several studies have shown that other signaling and metabolic pathways could also be involved in the disease pathogenesis [2]. For instance, it has been described autophagy impairment, neuroinflammation, dysregulation of calcium homeostasis, synapse, neuroactive ligand-receptor interaction, axon guidance, Wnt signaling and, immune system, among others [6, 7, 8, 9]. Nevertheless, neuropathology causes are not entirely understood, confining the development of novel therapeutic strategies for improving the currently approved. For instance, although ERT improves joint mobility, gait, respiratory function, liver and spleen volumes, and urinary GAGs levels [10, 11], it does not have any impact on CNS alterations since the enzyme cannot cross the blood-brain barrier (BBB) [12]. In addition, although significant advances have been made in the gene therapy field [13] and small molecules [14], these strategies need further studies before they can be approved for MPS II patients. Recently, it was described that mutant IDS is accumulated in the endoplasmic reticulum, which promotes its ubiquitination and degradation. Inhibition of endoplasmic reticulum-associated degradation improved IDS intracellular trafficking and activity [15]. These results show the importance of understanding the IDS maturation process and interactions, as well as the physiopathology of the disease, to identify novel therapeutic targets.
Protein-protein interaction networks are important sources of information on biological processes and complex metabolic functions in living cells [16]. Previously, we demonstrated that proteomics analysis allowed the identification of the IDS interactome, elucidating new physiological roles of this protein through their interaction with proteins involved in different cellular processes such as cell migration, oxidative phosphorylation, redox, immune defense, vesicular and axonal transport [17]. In addition, a bioinformatics protein-protein network analysis led to the identification of proteins with potential physical interactions with IDS [17]. To increase our knowledge of the IDS interactome, in this work we describe the validation of these ten candidates by using the yeast two-hybrid (Y2H) assay, which confirmed the physical interaction of IDS with limbic system-associated membrane protein (LSAMP) and synaptotagmin-1 (SYT1). LSAMP is an immunoglobulin (Ig) superfamily member involved in selective neuronal growth, axon targeting, cellular migration, proliferation, and blood-brain-barrier (BBB) integrity [18, 19]. On the other hand, SYT1 has been proposed to trigger exocytosis and endocytosis through Ca2+-dependent interactions with phosphatidylserine-containing liposomes and SNARE proteins [20]. Although the physiological relevance of these complexes has not been explored, our results suggest that these interactions could provide to IDS a pathway to the specific SYT1-mediated endocytosis during the docking stage, connecting the vesicular membrane with the SNARE fusion complex in the vesicular fusion process; as well as the capability to form a targeting module in the transcytosis process between cellular components from the BBB through the interaction with LSAMP [21, 22].
2. Materials and methods
2.1. Bioinformatic analysis
The protein/gene list of the 187 potential IDS-interacting proteins, previously identified by the affinity purification [17], was used for a global protein-protein interaction network reconstruction with the PPI spider platform [23]. This analysis provided a global picture of protein relationships, extracting the experimental binary interactions deposited in the IntAct database [24]. The dataset was analyzed with geneMANIA [25] at Cytoscape to visualize functional associations between these proteins, based on reported and predicted physical interactions. This last category incorporates functional associations from orthologous organisms to enrich the network [26]. The analysis led to the election of ten proteins directly linked to IDS to be screened by the Y2H system.
2.2. Protein-protein docking
The InterEvScore platform [27] was used to carry out all the protein-protein docking simulations. This software combines evolutionary methods in the docking process, implying that the analysis of heteromeric proteins can be pursued by integrating a residue-based multi-body statistical potential with evolutionary information. The FRODOCK2 was used for the docking search [28]. All the generated models were re-scored with InterEvScore.The SOAP_PP atom-based statistical potential was used to increase the confidence of the outputs [27, 29, 30].
2.3. Mouse brain RNA extraction and PCR amplification
100 mg of brain tissue from 3-month-old male wild-type C57BL/6 mice were resuspended in 1mL of TRIzol reagent and homogenized with a syringe according to the manufacturer's instructions (Sigma-Aldrich). The RNA was treated with 1mL of DNase I per mg of RNA (Invitrogen) for 15 min at room temperature to remove residual DNA contaminants. Subsequently, DNase was inactivated by adding 1 μL of 25 mM EDTA and heating for 10 min at 65 °C. RNA integrity was verified in 1% agarose gel electrophoresis. One-Step RT-PCR was performed in a single tube using gene-specific primers (Table 1), targeting RNAs from total RNA using a mixture of SuperScript® III Reverse Transcriptase and Platinum® Taq DNA polymerase (Invitrogen). Mice were obtained from the Comparative Biology Unit at Pontificia Universidad Javeriana under approved protocols of the Institutional Animal Care and Use Committee.
Table 1.
Transformations of Saccharomyces cerevisiae MaV203 strain.
| Test number | Bait plasmid | Prey plasmid | Tested pair | Purpose |
|---|---|---|---|---|
| 0 | None | None | Empty plasmids | Negative Controls |
| 1 | pDEST32/Krev1 | pDEST32/RalGDS-wt | Krev1+RalGDS-wt | Strong positive interaction control |
| 2 | pDEST32/Krev1 | pDEST32/RalGDS-m1 | Krev1+RalGDS-m1 | Weak positive interaction control |
| 3 | pDEST32/Krev1 | pDEST32/RalGDS-m2 | Krev1+RalGDS-m2 | Negative interaction control |
| 4 | pDEST32 | pDEST22 | None (Empty vectors) | Negative activation control |
| 5 | pDEST32/IDS | pDEST22 | IDS | Negative activation control; baseline |
| 6 | pDEST32/IDS | pDEST22/1433G | IDS+1433G | Testing specific two-hybrid interaction |
| 7 | pDEST32 | pDEST22/1433G | 1433G | Negative activation control |
| 8 | pDEST32/IDS | pDEST22/1433Z | IDS+1433Z | Testing specific two-hybrid interaction |
| 9 | pDEST32 | pDEST22/1433Z | 1433Z | Negative activation control |
| 10 | pDEST32/IDS | pDEST22/ALDOA | IDS + ALDOA | Testing specific two-hybrid interaction |
| 11 | pDEST32 | pDEST22/ALDOA | ALDOA | Negative activation control |
| 12 | pDEST32/IDS | pDEST2/ALDOC | IDS + ALDOC | Testing specific two-hybrid interaction |
| 13 | pDEST32 | pDEST22/ALDOC | ALDOC | Negative activation control |
| 14 | pDEST32/IDS | pDEST22/EAA1 | IDS + EAA1 | Testing specific two-hybrid interaction |
| 15 | pDEST32 | pDEST22/EAA1 | EAA1 | Negative activation control |
| 16 | pDEST32/IDS | pDEST22/HSP7C | IDS + HSP7C | Testing specific two-hybrid interaction |
| 17 | pDEST32 | pDEST22/HSP7C | HSP7C | Negative activation control |
| 18 | pDEST32/IDS | pDEST22/LSAMP | IDS + LSAMP | Testing specific two-hybrid interaction |
| 19 | pDEST32 | pDEST22/LSAMP | LSAMP | Negative activation control |
| 20 | pDEST32/IDS | pDEST22/PRDX2 | IDS + PRDX2 | Testing specific two-hybrid interaction |
| 21 | pDEST32 | pDEST22/PRDX2 | PRDX2 | Negative activation control |
| 22 | pDEST32/IDS | pDEST22/SYT1 | IDS + SYT1 | Testing specific two-hybrid interaction |
| 23 | pDEST32 | pDEST22/SYT1 | SYT1 | Negative activation control |
2.4. Construction of plasmids
Cloning procedures were performed using ProQuest Two-Hybrid Technology (Invitrogen), according to the manufacturer's instructions. To generate the bait and prey plasmids, we cloned the ORFs of the selected ten cDNAs and added the att sequences required for site-specific recombination (Table 1. Supporting, Information). The canonical human IDS sequence was used to generate the bait plasmid, and it shares 86% identity with the mouse orthologous protein. The PCRs were performed in 50 μL of Safe-Green™ 2X PCR Bestaq super mix reaction (ABM®) containing 100 ng of cDNA, 0.5 μM of att primers, 1X Safe-Green Bestaq, and nuclease-free H2O. Amplifications were performed as follows: initial denaturation at 95 °C for 3 min, followed by 40 cycles of 30 s at 94 °C, 30 s at the annealing temperature optimal for each primer pair, 1–2 min at 72 °C, and an additional 3 min extension at 72 °C. PCR products were purified using Wizard® SV Gel and PCR Clean-Up System (Promega). 1 μL of each amplification product was loaded onto an agarose gel to check the concentration before BP reactions. Site-specific recombination was performed by BP recombination reaction between a pDONR 201 and 207 and attB PCR products to generate the entry clones containing genes of interest flanked by attL1 and attL2 sites. The desired expression clones were generated by performing an LR recombination reaction between the entry clones and a Gateway™ destination vectors pDESTTM32 and pDESTTM22, respectively, using the Gateway LR Clonase II Enzyme mix (Invitrogen).
2.5. Yeast strain and growth conditions
The Y2H screen was performed by co-transformation of bait and prey plasmids into Saccharomyces cerevisiae MaV203 strain (Invitrogen) (Table 1). The ProQuestTM Two-Hybrid System includes control plasmids based on the interaction of Krev1 (a member of the Ras family of GTP binding proteins) with RalGDS (the Ral guanine nucleotide dissociator stimulator protein). The RalGDS mutants RalGDS-m1 and RalGDS-m2 affect the interaction with Krev1 and were provided by ProQuest™ Control Vectors Box (Invitrogen, Catalog No. PQ10001-01 PQ10002-01). For each transformation, 1 μg plasmid DNA was mixed with 100 μg of denatured sheared salmon sperm DNA and 100 μL of the competent yeast suspension in 2 mL of 1X LiAc/0.5X TE. Cells were incubated at room temperature for 10 min, and 700 μL of 1X LiAc/40% PEG-3350/1X TE was added, mixed, and incubated for 30 min at 30 °C. Furthermore, 88 μL of DMSO was added, and the cell solution was heated at 42 °C for 7 min and centrifuged at 5000 rpm for 10 s. The supernatant was removed and resuspended in 1 mL 1X TE. Finally, the supernatant was removed, and the cells were resuspended in 50 μL of TE to be platelet on a selective plate.
2.6. Characterization of transformants
The identification of transformants was performed by patching and replica plating steps onto the selection/screen plates on dropout SC amino acid mixture formulation (without L-Leucine and L-Tryptophan) for S. cerevisiae growth media (SC-Leu−Trp). Subsequent procedures were performed following the manufacturer's specifications (ProQuest™ Two-Hybrid System, Invitrogen). Interactions were identified by activating both independent reporter genes URA3 and lacZ. To assay, the activation of the HIS3 reporter gene, multiple colonies from each SC-Leu−Trp plate were resuspended in 0.9% NaCl and spotted onto synthetic agar medium containing dropout supplements lacking leucine, tryptophan, and histidine (SC-Leu−Trp−His). Plates were incubated for three days at 30 °C and assessed for growth.
2.7. β-galactosidase activity detection
A β-galactosidase assay was used as a quantitative indicator of the strength of the interaction. The assay was performed following the protocol described in the ProQuest Two-Hybrid System manual (Invitrogen) but with some variations in the cell lysis method. Three colonies containing a pair of proteins previously detected as a positive on HIS3 plates (LSAMP + IDS and SYT1+IDS), as well as both positive and negative controls (Test number 0, 1, 2, 3 4, 5, 19, and 23, Table 1), were growth in 2.5 mL of SC-Leu-Trp medium and incubated for 16 h at 30 °C. Then, 5 mL of yeast extract containing peptone, glucose, and adenine sulfate dihydrate medium (YPAD) was added, and when the cultures reached OD600 = 1.0–1.5; 1.5 mL were centrifuged at 12000 x g for 30 s, and the pellets were washed with 1,5 mL of Z-buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, pH 7.0), resuspended in 300 μL of Z-buffer, and lysed with glass beads (Merck) in 8 cycles of vigorous vertexing during 30 s followed by 30 s on ice. Further, 100 μL of the supernatant were mixed with 700 μL of Z-buffer, 2-mercaptoethanol buffer, and 160 μL of the o-nitrophenyl-β-D-galactopyranoside substrate (ONPG, Thermo Scientific). Samples were incubated for 30 min at 30 °C before stopping with 400 μL of Na2CO3 1M. Finally, the absorbance of each sample was measured at 420 nm. The β-galactosidase activity was calculated using the following equation:
where t = incubation time (min), V = volume of culture used in the assay (mL) = 6mL, OD420 = ONPG reaction absorbance, and OD600 = initial culture density.
2.8. Statistical analysis
Data analysis was performed using GraphPad Prism version 8.4.3 (San Diego, CA, United States). One-way with Dunnett's posttests was used to determine the significance of observed differences. Differences were considered statistically significant when p was <0.05.
3. Results
3.1. Bioinformatics analysis of brain IDS-interactome
The mouse brain IDS-interactome was previously isolated and identified using affinity purification and mass spectrometry [17]. The main functional categories among IDS putative interactors were transducers, enzymes, transcriptional activators, transporters, and structural and binding proteins [17]. Here, the interpretation of a set of proteins in the biological context was carried out on PPI Spider and Cytoscape platforms. According to the topological context, the network model of the identified protein list was transferred to a global PPI network. The enrichment took place if missing relevant proteins were included to connect the network. A Monte Carlo simulation was used to obtain the statistical significance of the models. The method has been used as a web-based tool on the PPI spider platform. The re-analysis of the IDS interactome using PPI spider showed that molecular functions related to the binding proteins and metabolic enzymatic activities were predominant in the interaction network (Figure 1). Also, relevant metabolic pathways like ATP-catabolic activity, small GTPase mediated signal transduction, vesicle-mediated transport, hydrogen peroxide catabolic process, tricarboxylic cycle activity, cytoskeleton, response to stress, axogenesis, and positive regulation of cell migration were identified (Figure 2A).
Figure 1.
Functional classification of mouse IDS interactome. Protein analysis through evolutionary relationships algorithm was used for GO term assignment. 187 proteins were classified according to evolutionary groupings, protein class, and functional groupings (gene ontology and pathways). Red lines relate the percentages for each group.
Figure 2.
Protein-protein interaction (PPI) network construction with PPI-Spider. A) D1-3 network model from the input list presented by rectangles. Colors are used to indicate gene function according to the Gene Ontology terms. Proteins from the input list are represented as colored rectangles, where each color represents independent metabolic pathways. The colored rectangles correspond to those proteins that the software finds related to the entry list, but that is not part of it. B) primary IDS network predicted with GeneMANIA selecting physical interactions using query gene-based weighting. Proteins that were selected for Y2H validation are marked with black stars.
As shown in Figure 2B, GeneMANIA analysis represents the association networks between selected proteins built upon physical interactions and predicted network categories to visualize functional relations between genes and proteins by mapping known functional associations across species [17]. Further, considering the coverage sequence percentage and the respective ion score obtained by mass spectrometry (Supplementary information), five proteins were selected to validate their physical predicted interactions from the GeneMANIA network: limbic system-associated membrane protein (LSAMP), CaM kinase-like vesicle-associated protein (CAMKV), synaptotagmin-1 (SYT1), fructose-bisphosphate aldolase C (ALDOC), and excitatory amino acid transporter 1 (EAA1) (Figure 2B), and five proteins from the PPI-Spider network: peroxiredoxin-2 (PRDX2), heat shock cognate 71 kDa protein (HSP7), 14-3-3 protein zeta/delta (1433Z), 14-3-3 protein gamma (1433G) and aldolase A (ALDOA) (Figure 2A).
3.2. Y2H to validate a selection of interactors
The physical interaction of IDS with the selected proteins was validated using the Y2H system. The cDNAs of the selected proteins were amplified by PCR successfully for all proteins, excepting CAMKV, despite making different modifications on PCR conditions (Supplementary data, figure 2). We evaluated the interactions between IDS (bait) and the predicted interactors (prey) through the reconstitution of the active transcription factor, which in turn activated expression of the three reported genes (HIS3, URA3, and lacZ), which one with an independent promoter (Figure 3). The characterization of S. cerevisiae MaV203 transformants cells containing bait and prey interacting proteins as well as positive controls for weak and strong interactions allows the expression of the three reporter genes HIS3, URA3, and lacZ. Positive colonies for HIS3 and URA3 reporters were identified by patching and replica plating steps onto the selection plates (Supplementary data figure 3). In this respect, the cloning of cDNAs by PCR was successful except for CAMKV despite making different modifications to promote clonal amplification (Supplementary data, figure 2).
Figure 3.
Schematic representation of the yeast two-hybrid validation. For the study of the interactions, mouse cDNA of IDS was fused to the DNA-binding domain (DBD) of Gal 4 (Bait vector), and the other proteins were fused with the activation domain (AD) of Gal4 (Prey vector). The yeast strain is transformed with the expression plasmids encoding the bait and prey. If protein IDS interacts with any prey protein in the nucleus, this will bring the activation domain together with the DNA-binding domain to reconstitute transcriptional activation and result in the expression of the reporter genes (HIS3, URA3, and lacZ).
To test the specific interaction with IDS, the transformants were analyzed by plating them on media lacking Leu, Trp, and His. Additionally, the same colonies were grown in a medium lacking uracil (Supporting information figure 3, right panel). Both negative and positive controls were always seeded next to the tests (Supporting information, figure 3, left panel). All the tested colonies grew normally in the non-selective culture medium (SC-Leu-Trp), confirming the presence of both bait and prey vectors (Supplementary Information, figure 3, Assay # 2 and 3, A-C). Colony replating on a selective medium (SC-Leu-Trp-His-Ura) showed differential growth for co-transformed colonies containing IDS + LSAMP and IDS + SYT1 constructs, suggesting that both proteins may interact with IDS (Figure 4). On the other hand, based on the experimental conditions used in this study, colonies containing IDS protein co-transformed with 1433G, ALDOA, ALDOC, EAA1, HSP7C, and PRDX2 did not show growth in patching and replica plating steps onto the selection plates (Supplementary Information, figure 3, Assay # 1B and D). Although the system is designed to reduce false positives because of the uses of low-copy-number vectors and the presence of three different reporter genes with independent promoters, the protein 14-3-3 Z was not considered for further validations because the negative control (pDEST32 + pDEST22/14-3-3 Z) showed that by itself, it could activate the transcription factor. In addition, two-point mutations that probably affect the reading frame were found in the sequence of 14-3-3 Z (Supplementary Information, figure 3, Assay # 1B).
Figure 4.
Characterization of transformants based on their phenotypes including self-activation controls and the positive controls for the weak and strong interaction. TPP: Tested proteic pair. NAC: Negative activation control. Krev1+RalGDS-wt: strong positive interaction control; Krev1+RalGDS-m1: weak positive interaction control; Krev1+RalGDS-m2: negative interaction control; MaV203: empty S. cerevisiae Mav203 cells.
LSAMP and SYT1 showed the activation of HIS3 and URA3 reporter genes mediated by a strong interaction with IDS in replica and the clean plates (Figure 5A-B). The negative controls did not grow in the URA3 selective medium (Figure 5C). Also, the negative activation controls (pDEST32/IDS + pDEST22) showed that IDS do not possess domains that promote the activation of the reporter genes (Figure 5, IDS-C). Dilutions for each of the following co-transformants were also evaluated (Figure 5): Krev1-RalGDS-wt for strong interaction control; Krev1-RalGDS-m1 for weak interaction; Krev1-RalGDS-m2, pDEST22-pDEST32 (Empty vectors) for negative interaction. Clones co-expressing pDEST32/IDS and pDEST22/LSAMP, and pDEST32/IDS and pDEST22/SYT1 plasmids grew in URA3 selective medium supplemented with HIS, and total absence of growth was observed for autoactivation controls co-transformed with empty bait plasmid pDEST32 and pDEST22/LSAMP (LSAMP) and pDESTP32 and pDEST/SYT1 (SYT1), respectively (Figure 5C).
Figure 5.
Representative images of yeast growth in media lacking tryptophan, leucine, and histidine for the selection of cells transformed with bait and prey plasmids. The interaction assays between IDS + LSAMP and IDS + SYT were evaluated by the expression of HIS3 and URA3 reporter genes at different dilutions. A. SC-Leu-Trp (+ His + Ura) plates: Positive grow control in the non-selective medium. B. SC-Leu-Trp-His (+ Ura) Plates:HIS3 gene expression in selective medium lacking histidine. C. SC-Leu-Trp-Ura (+ His) plates:URA3 gene expression in selective medium lacking uracil. Krev1+RalGDS-wt; strong positive interaction control, Krev1+RalGDS-m1; weak positive interaction control; Krev1+RalGDS-m2; Negative interaction control; MaV2013; empty cells.
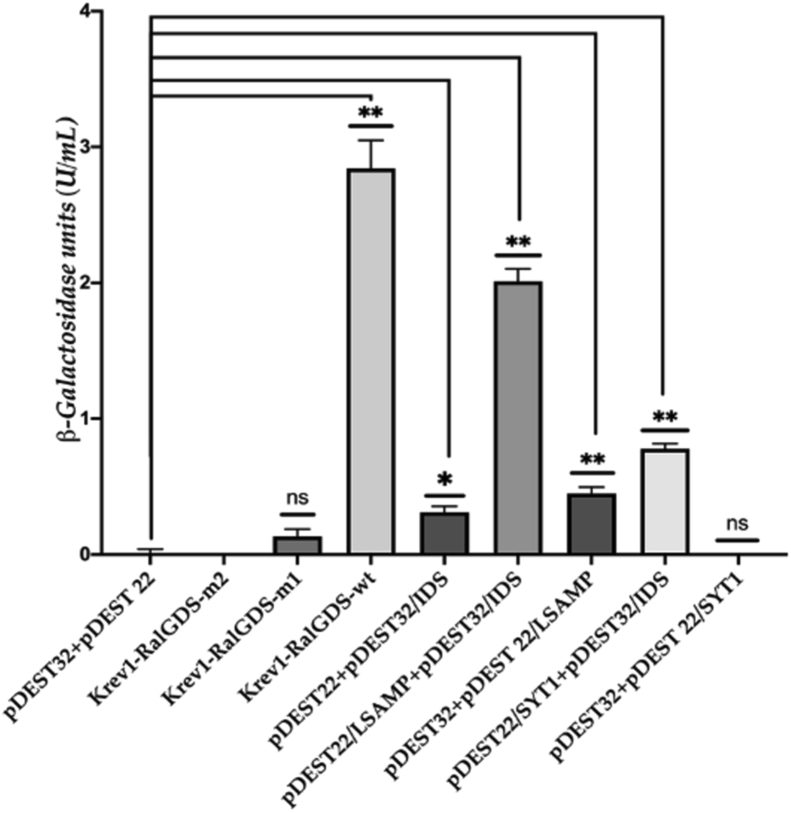
3.3. β-galactosidase activity quantification
The measured β-galactosidase activity reflected the expression level of the reporter gene lacZ. Transformed cells URA3 positive were also analyzed by plating them on media lacking leucine, tryptophan, and URA3. Also, colonies containing positive and negative controls were evaluated. As observed in Figure 6, transformants co-expressing IDS + LSAMP or IDS + SYT1 showed an increase in β-galactosidase activity. IDS + LSAMP showed higher β-galactosidase activity than IDS-SYT1 and all the negative controls (i.e., Krev1-RalGDS-m2, pDEST32 + pDEST22, and pDEST32 + pDEST22/LSAMP), being comparable to that detected for the strong interaction control (Krev1-RalGDS-wt). In contrast, the IDS + SYT1 pair showed a-galactosidase activity close to that observed in the weak interaction control Krev1-RalGDS-m1 (Figure 6). As expected, positive control (Krev1-RalGDS-wt) showed a significantly high β-galactosidase activity; while there was a low or negligible amount of activity for empty plasmids pDEST32 and pDEST22 (negative control) and pDEST32 + pDEST22/SYT1 as compared with the positive interactions. Nevertheless, the cross-activation controls for pDEST32 + pDEST22/LSAMP and pDEST22 + pDEST32/IDS showed that by themselves induce reporter activation.
Figure 6.
β-galactosidase activity. The expression of the lacZ reporter gene was quantified by measuring the β-galactosidase activity in cultures of pDEST22/LSAMP + pDEST32/IDS, pDEST22/SYT1+pDEST32/IDS, positive controls (Krev1-RalGDS-wt; strong interaction, and Krev1-RalGDS-m1; weak interaction), and negative controls (Krev1-RalGDS-m2; negative interaction control, pDEST22 + pDEST32; empty plasmids, pDEST22 + pDEST32/IDS, pDEST32 + pDEST 22/LSAMP and pDEST32 + pDEST22/SYT1; negative activation controls). All values were compared with the mean of the empty vectors (pDEST32 + pDEST22). Differences were considered statistically significant when p was. ∗ = p < 0.05 and ∗∗ = p < 0.001.
3.4. Protein-protein docking
The modeling of protein-protein interactions allows us to elucidate how cellular networks work. To provide a possible visualization of the LSAMP-IDS and SYT1-IDS interaction, the coordinates of their unbound components were used to generate potential predictions from a rigid-body 6D search followed by a second refining round. Then, we performed multiple sequence alignments to identify binding interfaces on the InterEvDock2 server. This is a commonly used platform to study heteromeric proteins, combining a residue-based multi-body statistical potential with evolutionary information derived from the multiple sequence alignments between potential protein partners [27]. The results predicted that the 42 kDa heavy chain of IDS might overlap with the -terminal residues of LSAMP, while the light chain appears to be overlapping with the N-terminal region of SYT1 (Figure 7).
Figure 7.
Residue Interface Consensus. The server runs several steps to propose a selection of the ten most likely models for each score (InterEvScore, SOAP_PP, FRODOCK scores) as well as ten consensus models and five most likely interface residues on each protein. Columns marked in blue show amino acids from either LSAMP or SYT1 that could be interacting with IDS residues (marked in red for each interaction). Figures A and B show the predicted interface for IDS-LSAMP and IDS-SYT1, respectively. C. The Eukaryotic Linear Motifs analysis for the LSAMP sequence reveals the presence of four SH3 domains.
4. Discussion
Recent advances in high-throughput technologies have provided relevant information about the neuronal damage in murine models for MPS II. Alterations of secondary cellular processes have been described as a consequence of the lysosomal accumulation of HS and DS, such as the permeabilization of lysosomal and mitochondrial membranes, and the dysregulation of calcium homeostasis, among others [31]. Also, some proteins have been identified as damage-triggers factors on lipid metabolism, GAGs degradation, glycosphingolipid biosynthesis, and autophagy processes like TFEB, mTOR, and SERBP [32]. Likewise, a systems biology approach was performed to understand the effect of metabolism alterations in MPS through metabolic reconstruction. Silencing MPS-related enzymes also supported severe damage affecting mainly the mitochondrial function and the amino acid and lipid metabolism [32]. New contributions in this field have shown differentially expressed protein profiles in the hippocampus and cerebral cortex from MPS II mouse models through transcriptomic analysis, showing significant alterations in axon guidance, calcium homeostasis, synapse, neuroactive ligand-receptor interaction, neuroinflammation, and the calcium-dependent exocytotic vesicles [33].
On the other hand, functional classification analysis for the specific mouse brain IDS-proteome revealed that most of the proteins identified as direct IDS putative interactors are related to axogenesis, vesicle-mediated intracellular and extracellular transport, and neuron projection [17]. These proteins were previously isolated and identified by affinity chromatography, using a human recombinant IDS as a ligand under high stringency conditions (150 mM NaCl) to favor the selection of the strongest interactors. In addition, we confirmed some of these interactions by immunoprecipitation from mouse brain protein extracts. The antibodies against myelin, 14-3-3 proteins zeta (14-3-3Z), 143-3 gamma (14-3-3G), and aldolase C, were able to co-immunoprecipitated IDS [17]. Although there is no evidence about IDS physiological functions beyond its known catalytic role, these interactions suggest that IDS may be involved in a wide spectrum of cellular functions related to cell growth, enzymatic activity induction, intracellular trafficking, biogenesis, and glycolytic pathways respectively [17]. Additionally, we isolated functional native IDS-complexes in cytosolic fractions from the mouse brain, containing 9 proteins also identified by affinity chromatography (The mitochondrial ATP synatse, glutathione s-transferase, peroxiredoxins (isoforms 2 and 6), creatine kinases, ubiquinol–cytochrome c oxidoreductase, rab GDP dissociation inhibitor alpha, and alfa enolase) [17]. Although these assays allowed us to provide evidence that IDS may form complexes with different proteins, they did not allow us to identify direct physical interactions between these proteins and IDS. In this sense, we decided to use the yeast two-hybrid system to confirm direct-physical protein interactions of some of these putative interactors.
We have applied orthogonal approaches to unravel protein-protein interactions with IDS: protein-protein docking, affinity purification/MS-based analysis for primary identification, and now, targeted Y2H for physical interaction validations. To decrease the rate of false-positive interactions, we used a stringent triple auxotrophic yeast strain with independent promoters for yeast colony analysis. After secondary URA3 screening, candidate colonies were identified as containing IDS-interacting proteins, and their respective autoactivation controls for this marker showed negative growth on the selection plates (Figures 4 and 5). Subsequent quantitative analysis of β-galactosidase to evaluate lacZ gene from URA3 plate colonies showed a positive but residual activity for auto-activation controls (pDEST32/IDS + pDEST22 and pDEST22/LSAMP, Figure 6). Two factors can explain this: first, the URA3 reporter gene is the most stringent reporter in the Y2H system due to the URS1 sequence presence that strongly represses transcription, which means that only solid protein-protein interactions induce this gene sufficiently to allow growth [34, 35], and second, colonies screened on SC-Leu-Trp-Ura plates that did not grow, should not be excluded as true interactors. Furthermore, while showing strong induction of the HIS3 and lacZ reporter genes, specific bait fusions may lead to weak induction of URA3. In addition, it is well known that the system could generate false negatives in some cases because the interaction between test proteins may place the BD and AD in a non-favorable configuration. Since post-translational modifications would be absent in a Y2H assay, interactions that require them will be missed [36]. LSAMP and, to a lesser extent, IDS could have low affinities for many system components, including nuclear signal, activation domain, or the RNA, providing certain false-positive activities. Still, when PDEST32/IDS + PDST22/LSAMP were assayed for lacZ and URA3, the interaction was more robust than the controls (whose activity was abolished entirely in URA3 plates), suggesting that this interaction is undoubtedly strong. On the other hand, the sensitivity of the HIS3 reporter was evident since the yeast strain already expresses a basal level of it. Also, bait proteins often contain a certain level of transcriptional activity that could be enough to initiate the transcription of HIS3, which was undoubtedly confirmed in our experiments using the respective bait controls (Figure 6).
The relevance of these interactions in the pathogenesis of MPS II would be the next issue to investigate. Under physiological conditions, the nature of these interactions remains unclear. However, a functional relationship between IDS and SYT1 could underlie pathways of exocytosis and endocytosis of enzymes dependent on SNARE complexes such as IDS. In this regard, it is well known that the extracellular presence of lysosomal enzymes may be due to a mannose 6-phosphate targeting signal (M6P-dependent pathway). Likewise, a new vesicular M6P-independent mechanism of newly synthesized lysosomal enzymes to lysosomes has been reported [37, 38]. ERT for MPS II takes advantage of exposed M6P residues present in the recombinant IDS used for this purpose (i.e., idursulfase and idursulfase-beta), which target cell surface M6P receptors and promote the internalization via clathrin-coated vesicles [39]. On the other hand, in addition to the degradative functions, lysosomes are also capable of extracellularly releasing their contents by lysosomal exocytosis via a Ca2+ dependent process [37]. The dynamics include the peripheral transport along microtubules, the presence of vesicle-associated membrane protein 7 (VAMP7) on the surface of lysosomes interacting with syntaxin-4 and with synaptosome-associated protein of 23 kDa (SNAP23) on the plasma membrane, and a local Ca2+ sensor like synaptotagmin 7 to facilitate the cytoplasmic membrane fusion [40]. In this regard, the synaptotagmin family comprises 17 members, many of which trigger exocytosis in response to calcium, such as SYT1, which also functions as a primary calcium sensor in the synchronous neurotransmitter release synaptic vesicles (SV) [41]. SYT1 is a membrane protein with conserved Ca2+ sensors capable of initiating SNARE-dependent vesicle fusion during endo-exocytosis of synaptic vesicles and hormone stimulation [42].
Interestingly, our docking studies allowed us to predict possible interactions between SYT1 domains, specifically C2A, and the C-terminal light chain of IDS. It has been well known that SYT1 interacts with a clathrin adaptor protein (AP-2), suggesting a role in the clathrin-mediated endocytosis [43]. SYT1 is one of the main modules for exocytosis and endocytosis Ca2+ dependent [44]. Nevertheless, most studies have exposed a compensatory modulation of endocytosis switching from Ca2+−dependent to Ca2+ independent in the absence of SYT1 [45]. In addition, experiments with SYT1 knockout neurons revealed a slower endocytosis rate than wild-type neurons. These results were consistent when the SYT1 native function was abolished from Drosophila neuromuscular junction, rising impaired synaptic vesicle endocytosis [46]. Clathrin-coated vesicles are formed by their interaction with adaptor membrane proteins capable of recruiting transmembrane receptors into the fledgling vesicle [47]. Then endosomal trafficking takes place when the enzyme–receptor complex dissociates [48, 49]. An instance of the mechanisms, receptors of transferrin, insulin, low-density lipoprotein, growth factors, and numerous other ligands that have been used to internalize lysosomal enzymes such as IDS [50, 51]. This recruitment mechanism can be independent of ligand attachment or specific to the spatial conformation of a ligand-receptor complex. Hence, the clathrin pathway will be attractive as a flexible tool for drug design, enabling the uptake of selective drugs or enzymes conjugated to antibodies, peptides, and natural sensors like SYT1 that target receptors in clathrin-coated vesicles [51].
LSAMP is a critical member of the IgLONs protein family, encoded as a preproprotein proteolytically processed to generate a neuronal surface glycoprotein. IgLONs proteins are known to form homophilic and heterophilic complexes along the cell surface [52]. Furthermore, this protein may act as a selective homophilic adhesion molecule during axon guidance and neuronal growth in the developing limbic system but may also behave as a tumor suppressor [53]. Structurally, LSAMP is an immunoglobulin superfamily member with 3 Ig-like C2 domains, mediating cellular interactions [18]. According to our protein-protein docking, it was predicted that the interaction between the IDS-heavy chain and LSAMP could take place on the Ig-like C2–N-terminal domain from LSAMP (Figure 7, left panel). This family of cell adhesion molecules has been involved in neurite outgrowth regulation, dendritic arborization, and synapse formation [18]. Recently, functional studies have shown that LSAMP can promote and inhibit neurite outgrowth [54]. Recent advances in molecular evolution using bioinformatics allowed the identification of a KVTVNYP motif on LSAMP that serves as the SH3 recognition domain. SH3 domains are short sequences that seem to play an essential role in the BBB integrity on the level of cellular adhesion between astrocytes, endothelial cells, and leukocytes. These domains are typically expressed on scaffolding proteins localizing to the tight junctions and endothelial cells [18, 55]. In this regard, LSAMP possesses several small SH3 modules (Figure 7C) that could be influencing the BBB integrity [56, 57]. Hence, the interaction between IDS and LSAMP is attractive in the way that it could be used as a targeting module between cellular components from BBB. However, the interaction of LSAMP within the respective components of the BBB is still to be uncovered [58].
5. Conclusions
This study has identified LSAMP and SYT1 as novel interactors of human IDS. These results highlight the importance of proteomics and interactomics studies to continue understanding the role of IDS in the brain. The biological relevance of the interaction of IDS with LSAMP and SYT1 may underlie the intracellular trafficking pathways, which could involve the formation of multiprotein complexes for translocation of ligands, i.e., from the BBB to the brain, and the clathrin-endocytic mechanism mediated by SYT1, respectively. These results may help generate novel hypotheses to increase our knowledge regarding the molecular and cellular bases of MPS II and to design novel therapeutic alternatives.
Declarations
Author contribution statement
Eliana Benincore-Flórez: Performed the experiments; Analyzed and interpreted the data.
Jorge El-Azaz: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Gabriela Alejandra Solarte, Alexander Rodríguez: Performed the experiments.
Luis H. Reyes: Contributed reagents, materials, analysis tools or data.
Carlos Javier Alméciga-Díaz: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Carolina Cardona-Ramírez: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Funding statement
This work was supported by Pontificia Universidad Javeriana (Project ID 6699), Grupo de Investigaciones Biomédicas y de Genética Humana Aplicada GIBGA, Facultad de Ciencias de la Salud, Universidad de Ciencias Aplicadas y Ambientales U.D.C.A, Bogotá, Colombia (Project ID 15402038), the Ministry of Science, Technology and Innovation (Grant No. FP44842-233-2015). EBF received a master's scholarship from Faculty of Science of Pontificia Universidad Javeriana. CJAD was also supported by Pontificia Universidad Javeriana (Grant ID 3964, 5537, and 7204) and COLCIENCIAS (Grant ID 5174, contract No. 120356933205; and Grant ID 5170, contract No. 120356933427).
Data availability statement
Data included in article/supplementary material/referenced in article.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Contributor Information
Carlos Javier Alméciga-Díaz, Email: cjalmeciga@javeriana.edu.co.
Carolina Cardona-Ramírez, Email: clacardona@udca.edu.co.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Tomatsu S., et al. Mucopolysaccharidoses Update (2 Volume Set) Nova Science Publishers, Inc.; Hauppauge, NY: 2018. Mucopolysaccharidosis type II: clinical features, biochemistry, diagnosis, genetics and treatment; pp. 165–209. [Google Scholar]
- 2.D'Avanzo F., et al. Mucopolysaccharidosis type II: one hundred years of research, diagnosis, and treatment. Int. J. Mol. Sci. 2020;21(4) doi: 10.3390/ijms21041258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sohn Y.B., et al. Safety and efficacy of enzyme replacement therapy with idursulfase beta in children aged younger than 6 years with Hunter syndrome. Mol. Genet. Metabol. 2015;114(2):156–160. doi: 10.1016/j.ymgme.2014.08.009. [DOI] [PubMed] [Google Scholar]
- 4.Muenzer J., et al. Long-term, open-labeled extension study of idursulfase in the treatment of Hunter syndrome. Genet. Med. : Off. J. Am. Coll. Med. Genet. 2011;13(2):95–101. doi: 10.1097/GIM.0b013e3181fea459. [DOI] [PubMed] [Google Scholar]
- 5.Scarpa M., et al. Mucopolysaccharidosis type II: European recommendations for the diagnosis and multidisciplinary management of a rare disease. Orphanet J. Rare Dis. 2011;6:72. doi: 10.1186/1750-1172-6-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maeda M., et al. Autophagy in the central nervous system and effects of chloroquine in mucopolysaccharidosis type II mice. Int. J. Mol. Sci. 2019;20(23) doi: 10.3390/ijms20235829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fusar P.E., et al. Murine neural stem cells model Hunter disease in vitro: glial cell-mediated neurodegeneration as a possible mechanism involved. Cell Death Dis. 2013;4:e906. doi: 10.1038/cddis.2013.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gleitz H.F., et al. Brain-targeted stem cell gene therapy corrects mucopolysaccharidosis type II via multiple mechanisms. EMBO Mol. Med. 2018;10(7) doi: 10.15252/emmm.201708730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salvalaio M., et al. Brain RNA-seq profiling of the mucopolysaccharidosis type II mouse model. Int. J. Mol. Sci. 2017;18(5) doi: 10.3390/ijms18051072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wraith J.E., et al. Enzyme replacement therapy in patients who have mucopolysaccharidosis I and are younger than 5 years: results of a multinational study of recombinant human alpha-L-iduronidase (laronidase) Pediatrics. 2007;120(1):e37–46. doi: 10.1542/peds.2006-2156. [DOI] [PubMed] [Google Scholar]
- 11.Muenzer J., et al. Idursulfase treatment of Hunter syndrome in children younger than 6 years: results from the Hunter Outcome Survey. Genet. Med. 2011;13(2):102–109. doi: 10.1097/GIM.0b013e318206786f. [DOI] [PubMed] [Google Scholar]
- 12.Cho S.Y., et al. Effect of systemic high dose enzyme replacement therapy on the improvement of CNS defects in a mouse model of mucopolysaccharidosis type II. Orphanet J. Rare Dis. 2015;10:141. doi: 10.1186/s13023-015-0356-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zapolnik P., Pyrkosz A. Gene therapy for mucopolysaccharidosis type II-A review of the current possibilities. Int. J. Mol. Sci. 2021;22(11) doi: 10.3390/ijms22115490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoshina H., et al. Chaperone effect of sulfated disaccharide from heparin on mutant iduronate-2-sulfatase in mucopolysaccharidosis type II. Mol. Genet. Metabol. 2018;123(2):118–122. doi: 10.1016/j.ymgme.2017.12.428. [DOI] [PubMed] [Google Scholar]
- 15.Osaki Y., et al. Shutdown of ER-associated degradation pathway rescues functions of mutant iduronate 2-sulfatase linked to mucopolysaccharidosis type II. Cell Death Dis. 2018;9(8):808. doi: 10.1038/s41419-018-0871-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barabasi A.L., Oltvai Z.N. Network biology: understanding the cell's functional organization. Nat. Rev. Genet. 2004;5(2):101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 17.Cardona C., et al. Identification of the iduronate-2-sulfatase proteome in wild-type mouse brain. Heliyon. 2019;5(5):e01667. doi: 10.1016/j.heliyon.2019.e01667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kubick N., Brosamle D., Mickael M.E. Molecular evolution and functional divergence of the IgLON family. Evol. Bioinform. 2018;14 doi: 10.1177/1176934318775081. 1176934318775081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fierro H., et al. [Polyradiculoneuritis. Increase of the occurrence of the severe forms] Bol. Med. Hosp. Infant. Mex. 1978;35(4):665–672. [PubMed] [Google Scholar]
- 20.Wu Z., et al. Phosphatidylserine-containing liposomes inhibit the differentiation of osteoclasts and trabecular bone loss. J. Immunol. 2010;184(6):3191–3201. doi: 10.4049/jimmunol.0803609. [DOI] [PubMed] [Google Scholar]
- 21.van Breevoort D., et al. STXBP1 promotes Weibel-Palade body exocytosis through its interaction with the Rab27A effector Slp4-a. Blood. 2014;123(20):3185–3194. doi: 10.1182/blood-2013-10-535831. [DOI] [PubMed] [Google Scholar]
- 22.Pulgar V.M. Transcytosis to cross the blood brain barrier, new advancements and challenges. Front. Neurosci. 2018;12:1019. doi: 10.3389/fnins.2018.01019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Antonov A.V., et al. PPI spider: a tool for the interpretation of proteomics data in the context of protein-protein interaction networks. Proteomics. 2009;9(10):2740–2749. doi: 10.1002/pmic.200800612. [DOI] [PubMed] [Google Scholar]
- 24.Schweingruber C., et al. Identification of interactions in the NMD complex using proximity-dependent biotinylation (BioID) PLoS One. 2016;11(3):e0150239. doi: 10.1371/journal.pone.0150239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warde-Farley D., et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–W220. doi: 10.1093/nar/gkq537. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shannon P., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andreani J., Faure G., Guerois R. InterEvScore: a novel coarse-grained interface scoring function using a multi-body statistical potential coupled to evolution. Bioinformatics. 2013;29(14):1742–1749. doi: 10.1093/bioinformatics/btt260. [DOI] [PubMed] [Google Scholar]
- 28.Garzon J.I., et al. FRODOCK: a new approach for fast rotational protein-protein docking. Bioinformatics. 2009;25(19):2544–2551. doi: 10.1093/bioinformatics/btp447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramirez-Aportela E., Lopez-Blanco J.R., Chacon P. FRODOCK 2.0: fast protein-protein docking server. Bioinformatics. 2016;32(15):2386–2388. doi: 10.1093/bioinformatics/btw141. [DOI] [PubMed] [Google Scholar]
- 30.Dong G.Q., et al. Optimized atomic statistical potentials: assessment of protein interfaces and loops. Bioinformatics. 2013;29(24):3158–3166. doi: 10.1093/bioinformatics/btt560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ballabio A., Gieselmann V. Lysosomal disorders: from storage to cellular damage. Biochim. Biophys. Acta. 2009;1793(4):684–696. doi: 10.1016/j.bbamcr.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 32.Salazar D.A., et al. Systems biology study of mucopolysaccharidosis using a human metabolic reconstruction network. Mol. Genet. Metabol. 2016;117(2):129–139. doi: 10.1016/j.ymgme.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 33.Salvalaio M., et al. Brain RNA-seq profiling of the mucopolysaccharidosis type II mouse model. Int. J. Mol. Sci. 2017;18(5) doi: 10.3390/ijms18051072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Das S., Kalpana G.V. Reverse two-hybrid screening to analyze protein-protein interaction of HIV-1 viral and cellular proteins. Methods Mol. Biol. 2009;485:271–293. doi: 10.1007/978-1-59745-170-3_19. [DOI] [PubMed] [Google Scholar]
- 35.Kuroda H., et al. A comprehensive analysis of interaction and localization of Arabidopsis SKP1-like (ASK) and F-box (FBX) proteins. PLoS One. 2012;7(11):e50009. doi: 10.1371/journal.pone.0050009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galletta B.J., Rusan N.M. A yeast two-hybrid approach for probing protein-protein interactions at the centrosome. Methods Cell Biol. 2015;129:251–277. doi: 10.1016/bs.mcb.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nathan D.G., Orkin S.H. Musings on genome medicine: enzyme-replacement therapy of the lysosomal storage diseases. Genome Med. 2009;1(12):114. doi: 10.1186/gm114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marques A.R.A., Saftig P. Lysosomal storage disorders - challenges, concepts and avenues for therapy: beyond rare diseases. J. Cell Sci. 2019;132(2) doi: 10.1242/jcs.221739. [DOI] [PubMed] [Google Scholar]
- 39.Muro S. Challenges in design and characterization of ligand-targeted drug delivery systems. J. Contr. Release. 2012;164(2):125–137. doi: 10.1016/j.jconrel.2012.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.MacDougall D.D., et al. The high-affinity calcium sensor synaptotagmin-7 serves multiple roles in regulated exocytosis. J. Gen. Physiol. 2018;150(6):783–807. doi: 10.1085/jgp.201711944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maximov A., et al. Genetic analysis of synaptotagmin-7 function in synaptic vesicle exocytosis. Proc. Natl. Acad. Sci. U. S. A. 2008;105(10):3986–3991. doi: 10.1073/pnas.0712372105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y.C., et al. Synaptotagmin-1- and synaptotagmin-7-dependent fusion mechanisms target synaptic vesicles to kinetically distinct endocytic pathways. Neuron. 2017;93(3):616–631 e3. doi: 10.1016/j.neuron.2016.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang J.Z., et al. Synaptotagmin I is a high affinity receptor for clathrin AP-2: implications for membrane recycling. Cell. 1994;78(5):751–760. doi: 10.1016/s0092-8674(94)90442-1. [DOI] [PubMed] [Google Scholar]
- 44.Wu L.G., et al. Exocytosis and endocytosis: modes, functions, and coupling mechanisms. Annu. Rev. Physiol. 2014;76:301–331. doi: 10.1146/annurev-physiol-021113-170305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yao J., et al. Uncoupling the roles of synaptotagmin I during endo- and exocytosis of synaptic vesicles. Nat. Neurosci. 2011;15(2):243–249. doi: 10.1038/nn.3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nicholson-Tomishima K., Ryan T.A. Kinetic efficiency of endocytosis at mammalian CNS synapses requires synaptotagmin I. Proc. Natl. Acad. Sci. U. S. A. 2004;101(47):16648–16652. doi: 10.1073/pnas.0406968101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mousavi S.A., et al. Clathrin-dependent endocytosis. Biochem. J. 2004;377(Pt 1):1–16. doi: 10.1042/BJ20031000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chung Y.K., et al. A biochemical and physicochemical comparison of two recombinant enzymes used for enzyme replacement therapies of hunter syndrome. Glycoconj. J. 2014;31(4):309–315. doi: 10.1007/s10719-014-9523-0. [DOI] [PubMed] [Google Scholar]
- 49.Neufeld E.F. The uptake of enzymes into lysosomes: an overview. Birth Defects Orig. Artic. Ser. 1980;16(1):77–84. [PubMed] [Google Scholar]
- 50.McMahon H.T., Boucrot E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 2011;12(8):517–533. doi: 10.1038/nrm3151. [DOI] [PubMed] [Google Scholar]
- 51.Chen C.H., et al. Aptamer-based endocytosis of a lysosomal enzyme. Proc. Natl. Acad. Sci. U. S. A. 2008;105(41):15908–15913. doi: 10.1073/pnas.0808360105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McNamee C.J., Youssef S., Moss D. IgLONs form heterodimeric complexes on forebrain neurons. Cell Biochem. Funct. 2011;29(2):114–119. doi: 10.1002/cbf.1730. [DOI] [PubMed] [Google Scholar]
- 53.Hua X., et al. LSAMP-AS1 binds to microRNA-183-5p to suppress the progression of prostate cancer by up-regulating the tumor suppressor DCN. EBioMedicine. 2019;50:178–190. doi: 10.1016/j.ebiom.2019.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Philips M.A., et al. Lsamp is implicated in the regulation of emotional and social behavior by use of alternative promoters in the brain. Brain Struct. Funct. 2015;220(3):1381–1393. doi: 10.1007/s00429-014-0732-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pawson T., Schlessingert J. SH2 and SH3 domains. Curr. Biol. 1993;3(7):434–442. doi: 10.1016/0960-9822(93)90350-w. [DOI] [PubMed] [Google Scholar]
- 56.Bauer H.C., et al. "You Shall Not Pass"-tight junctions of the blood brain barrier. Front. Neurosci. 2014;8:392. doi: 10.3389/fnins.2014.00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stamatovic S.M., et al. Junctional proteins of the blood-brain barrier: new insights into function and dysfunction. Tissue Barriers. 2016;4(1):e1154641. doi: 10.1080/21688370.2016.1154641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martin-Blondel G., et al. Migration of encephalitogenic CD8 T cells into the central nervous system is dependent on the alpha4beta1-integrin. Eur. J. Immunol. 2015;45(12):3302–3312. doi: 10.1002/eji.201545632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in article/supplementary material/referenced in article.