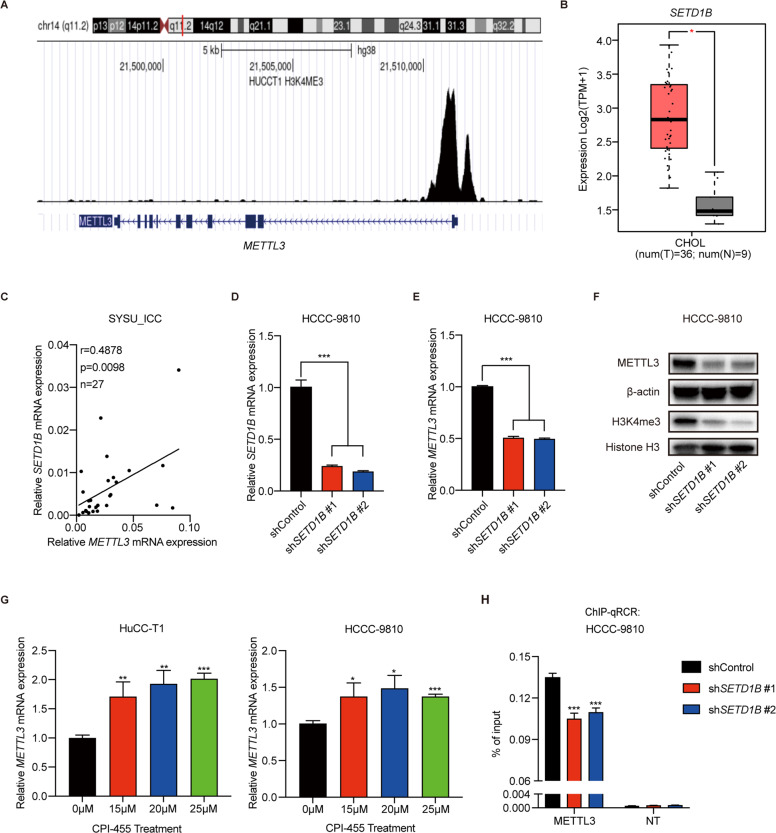
Fig. 2. H3K4me3 activates METTL3 transcription in ICC.
A Analysis of H3K4me3 modification in the METTL3 locus in HuCC-T1 cell (datasets from the Cistrome Data Browser). B SETD1B expression in ICC tumors (n = 36) and normal tissues (n = 9) in the GEPIA2 database. C Correlation analysis of METTL3 expression with SETD1B expression in our ICC cohort. D The mRNA levels of SETD1B in SETD1B-KD and control HCCC-9810 cells were confirmed by RT-qPCR. E The mRNA levels of METTL3 in SETD1B-KD and control HCCC-9810 cells were confirmed by RT-qPCR. F The protein levels of METTL3 and H3K4me3 modification after SETD1B silencing in HCCC-9810 cells were confirmed by western blotting. G The mRNA level of METTL3 after CPI-455 treatment for 72 h in HuCC-T1 and HCCC-9810 cells was confirmed by RT-qPCR. H Chromatin immunoprecipitation-qPCR analysis of H3K4me3 enrichment in the METTL3 locus or a flank region with no signal in SETD1B-KD or control HCCC-9810 cells. NT = A flank region with no signal was used as a negative control. The results are presented as mean ± SD of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, according to Student’s t test.