Abstract
The generation of a comprehensive catalog of null alleles covering all protein-coding genes is the goal of the International Mouse Phenotyping Consortium. Over the past 20 years, significant progress has been made towards achieving this goal through the combined efforts of many large-scale programs that built an embryonic stem cell resource to generate knockout mice and more recently employed CRISPR/Cas9-based mutagenesis to delete critical regions predicted to result in frameshift mutations, thus, ablating gene function. The IMPC initiative builds on prior and ongoing work by individual research groups creating gene knockouts in the mouse. Here, we analyze the collective efforts focusing on the combined null allele resource resulting from strains developed by the research community and large-scale production programs. Based upon this pooled analysis, we examine the remaining fraction of protein-coding genes focusing on clearly defined mouse–human orthologs as the highest priority for completing the mutant mouse null resource. In summary, we find that there are less than 3400 mouse–human orthologs remaining in the genome without a targeted null allele that can be further prioritized to achieve our overall goal of the complete functional annotation of the protein-coding portion of a mammalian genome.
Introduction
Animal models, including mouse knockouts, play an instrumental role in advancing our understanding of how disruption of normal gene function relates to human disease. Traditionally, much of this work focuses on a relatively small number of conserved genes and pathways, reflecting a common tendency for investigators to incrementally build upon existing knowledge. Often these advancements reflect the critical importance of specific genes to human disease (Dolgin 2017); however, the “lamppost effect” greatly limits the opportunity for novel discoveries of gene function (Stoeger et al. 2018), and ultimately restricts the development of new therapeutic avenues (Oprea et al. 2018). This effect is strongly driven by the availability of tools to interrogate gene/pathway functions (Edwards et al. 2011), including mouse mutants (Stoeger et al. 2018). Further, this problem is compounded by poor public availability and/or limited phenotypic characterization of many of these mutant mouse lines. Herein, we summarize our progress towards completion of the mutant mouse null resource that promises to advance our understanding of novel genes and pathways, while reducing the research barriers to entry for individual investigators.
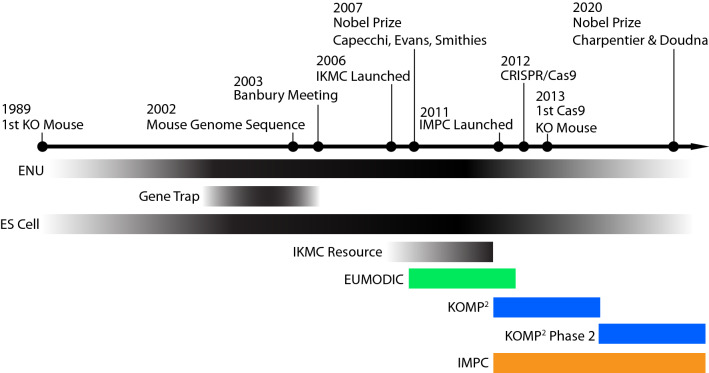
Over the past 20 years, tremendous progress has been made in generating a complete mutant mouse resource covering the roughly 23,000 protein-coding genes in the mouse genome. These efforts, spurred by the publication of the draft sequence of the mouse genome in 2002 (Mouse Genome Sequencing et al. 2002), reflect the consensus view that the mouse is the premier model of mammalian biology and the accumulation of technological innovations in mouse genetics presented a viable path towards functional annotation of the entire mouse genome (Fig. 1; Austin et al. 2004; Auwerx et al. 2004; Birling et al. 2021; Capecchi 1989; de Angelis et al. 2015; International Mouse Knockout et al. 2007; Jinek et al. 2012; Wang et al. 2013). This technological convergence includes the widespread adoption of mouse embryonic stem cells (mESCs) as a platform for gene targeting and innovations in molecular cloning that allowed for high-throughput generation of complex targeting constructs (Angrand et al. 1999; Copeland et al. 2001; Lee et al. 2001; Valenzuela et al. 2003). Early proposals for a more systematic strategy advocated for a hybrid approach of chemical mutagenesis (e.g., ENU), gene trapping, and gene targeting for complete genome coverage and emphasized the need for adopting systematic phenotyping protocols, the development of databases to provide easy access to functional data, and animal repositories to ensure broad access to models and other key resources (Battey et al. 1999; Hrabe de Angelis et al. 2000; Nadeau et al. 2001; Nolan et al. 2000; Skarnes et al. 2004).
Fig. 1.
Timeline highlighting major milestones enabling complete functional annotation of the protein-coding fraction of the mouse genome. Technological advances that made possible the large-scale generation of mutant mouse resources are shown in black. Multi-institutional collaborative programs (e.g., European Mouse Disease Clinic; EUMODIC, and Knockout Mouse Phenotyping Program; KOMP2) implemented these advancements to perform high-throughput animal model production and systematic broad-based phenotyping. This collective work has grown to include additional international sites that have been centralized under the International Mouse Phenotyping Consortium (IMPC) to coordinate animal production, phenotyping, and data dissemination
The Banbury meeting in 2003 further refined this concept to set forth a goal of creating a null, reporter allele for every gene as a foundational starting point for functional annotation (Austin et al. 2004). Despite deliberately flexible in the exact approach, the group adopted several key principles, including the use of a single inbred genetic background, and the open availability of cell, animal, and data resources to the scientific community. Generation of mice from these resources would follow at a pace that reflected both the community demand and capacity for centralized production, ultimately supporting individual investigator research programs. The concept aligned with European efforts, which emphasized the value of generating a conditional version of each allele (Auwerx et al. 2004). Together, these concepts were put into action as the NIH funded Knockout Mouse Project (KOMP), the European Conditional Mouse Mutagenesis Program (EUCOMM), and the Canadian-funded North American Conditional Mouse Mutagenesis Program (NorCOMM), which joined to form a singular effort as the International Knockout Mouse Consortium (IKMC) (Gondo 2008; International Mouse Knockout et al. 2007).
The bold IKMC mission of generating a null or conditional null allele for every gene in the mouse genome was complemented by the more modest goal of turning a small subset of these alleles into mouse strains for systematic characterization (Bradley et al. 2012; Skarnes et al. 2011). Further refinement of standard protocols for mouse phenotyping and the demonstration of the feasibility of large-scale mouse generation and phenotyping (de Angelis et al. 2015; White et al. 2013) underpin the current efforts of the International Mouse Phenotyping Consortium, which are now expanding on the initial vision first proposed at Banbury (Lloyd et al. 2020). The IMPC has generated over 5000 knockout lines from IKMC ES resources (Birling et al. 2021), and the advent of CRISPR/Cas9 technology has further augmented capacity for the generation of mutant mice. To date, the IMPC has generated knockout lines for 7590 genes (Data Release 14, May 7, 2021) and established a robust infrastructure sufficient for completing the draft functional annotation of a mammalian genome. This effort has had an enormous impact on our understanding mammalian biology and human disease (Brown et al. 2018; Meehan et al. 2017). The IMPC has continued to expand the catalog of mammalian essential genes, which are highly enriched in human disease (Cacheiro et al. 2020; Dickinson et al. 2016), while providing novel insights into developmental mechanisms through detailed embryonic phenotyping (Dickinson et al. 2016). The IMPC pipeline has revealed numerous novel gene associations with disease-relevant traits (Bowl et al. 2017; Rozman et al. 2018; Swan et al. 2020), and systematic phenotyping of both sexes reveals widespread sexual dimorphism (Karp et al. 2017).
What was once a long-term aspirational goal of the mouse genetics community is now an achievable milestone? This raises several interesting and important questions: How many more genes remain to be knocked out? How should these genes be prioritized for systematic mutagenesis? Are there other features in the genome (e.g., noncoding RNAs) that merit further consideration as mutagenesis targets? Here, we explore the collective catalog of mutant mouse null alleles generated by the IMPC and the broader scientific community to address these questions and chart a course towards finalizing a blueprint for understanding the activity of the protein-coding fraction of the mammalian genome.
Results
Current state of the mutant mouse null allele catalog to assess protein-coding gene function
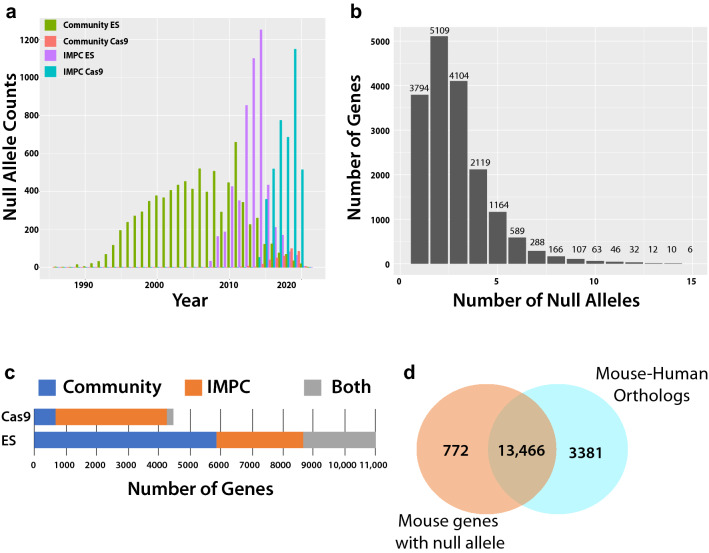
To focus on defining the community-wide effort to mutagenize the mouse genome, we include all null alleles, irrespective of their relationship to systematic production efforts or public availability. We define “IMPC” as all mouse model generation activities of the IKMC or IMPC, and “community” for all other alleles (individual investigator, non-IMPC programs, etc.). For curating null alleles, we used the MouseMine query tool to obtain annotation information detailing the target gene, allele symbol, methodology, and associated publications (Motenko et al. 2015). These queries identified 29,341 unique null alleles corresponding to 13,973 protein-coding genes with the majority of community-derived alleles generated using an ES-based resource and IMPC alleles a mixture of both ES and Cas9 derived. In addition to these targeted null mutations, we identified an additional 265 unique genes as having an annotated null allele resulting from random mutagenesis (spontaneous, ENU or gene trap). While our emphasis here is on the null resource, there are currently over 67,000 mutant mouse alleles documented by Mouse Genome Informatics (http://www.informatics.jax.org) that represent a diverse set of alleles ranging from hypomorphs to dominant negative, and humanized regions as well as others. This set of curated null alleles covers ~ 62% (14,238/23,000) of the protein-coding genes currently cataloged in the mouse genome. A detailed look at the time course for animal model generation highlights community production of null alleles rapidly accelerating in the mid-1990s as the technology spread and was implemented in core facilities to provide stable production of 400–500 new alleles published each year peaking in 2011 (Fig. 2a). About this time, IMPC production of null alleles ramped up significantly (Birling et al. 2021), which has been accompanied by a decline in community production of ESC-based alleles. This reduction may reflect the uptake of IKMC- and IMPC-generated resources by individual investigators, obviating the need to produce their own knockout. In 2015, the IMPC pivoted to the use of CRISPR/Cas9 to generate knockout alleles, which quickly replaced ESC-based animal production resulting in the establishment of ~ 4000 new lines within this 5-year window from 2015 to 2020 (Fig. 2a). Concurrently, there is a lower, but growing rate of community produced null alleles utilizing CRISPR/Cas9. Reflecting challenges in public availability of resources and parallel research aims, multiple null alleles are frequently generated for genes (Fig. 2b), including 150 genes with more than 10 null alleles. Most of community-generated null alleles are ESC based, while the vast majority of Cas9-generated null alleles reported in MGI were generated by the IMPC, due to rapid adoption of the technology as a core mutagenesis method to reduce cost and increase throughput (Fig. 2c).
Fig. 2.
Summary of mouse null allele production and progress towards completing a resource encompassing mouse–human orthologs. a Overview of animal production using either ES-cell based technology or CRISPR/Cas9 within independent research groups representing the Community effort or as part of the International Mouse Phenotyping Consortium. Dates for unique alleles generated by the Community were determined using earliest citation for allele transmission and dates for IMPC alleles were based upon record of successful confirmation of germline line transmission (GLT). b Histogram for null allele counts per gene shows that most genes have 1–3 unique null alleles while another ~ 2000 have 5 or more null alleles. c Contributions of different technology employed by the Community and IMPC to generate the null allele. d Venn diagram comparing all mouse genes with a null allele to the total number of mouse genes with a high-confidence human ortholog
Given the current efforts for complete functional annotation of the mammalian genome, with an overarching goal to understand human biology and disease, we examined how many mouse knockout alleles corresponded to protein-coding genes with high-confidence human orthologs defined by having multiple independent lines of supporting evidence from different resources (Munoz-Fuentes et al. 2018), and how many of these genes remain to be targeted. Of the 14,238 mouse genes that have a null allele, 94% (13,466) have a human ortholog while the remaining 772 lack a clearly identifiable ortholog or are mouse specific (Fig. 2d). Therefore, of the total 16,847 mouse genes with a high-confidence human ortholog, 79.9% have a reported null allele, leaving 3381 genes to complete the mutant mouse null resource.
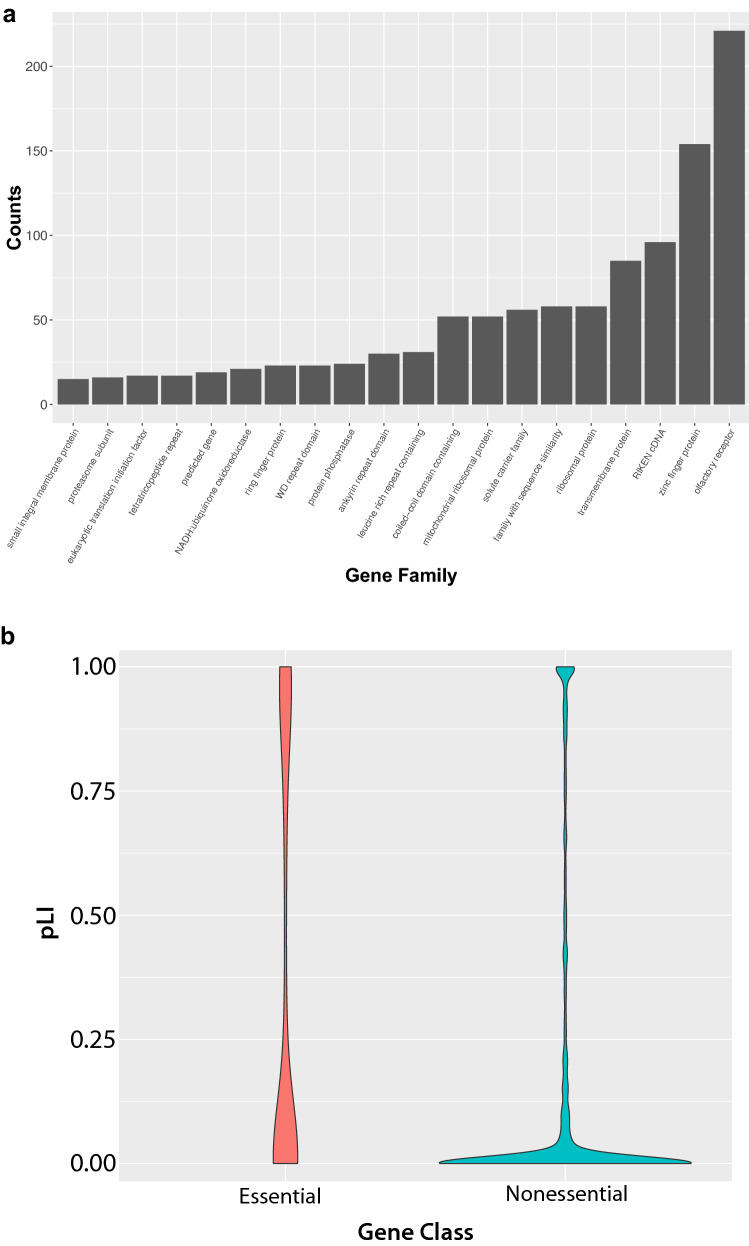
To further develop a prioritization framework, we analyzed the remaining 3381 genes to determine if there were any gene families that were over-represented or if there was evidence of functional constraint on these genes lacking null alleles. Within this set of non-targeted genes, olfactory receptors were the most highly represented class of genes followed by zinc finger domain containing genes and RIKEN cDNA clones (Fig. 3a). This is consistent with large size of these gene families and highlights the barriers to research on genes for which there is little existing functional data (Stoeger et al. 2018). In addition, it is important to consider whether the information generated from knockout mice for a class of genes such as olfactory receptors is merited given the phenotyping tests included in the IMPC, and the expected impact of single gene mutation. Conversely, transmembrane proteins and solute carriers are more likely to be druggable and, thus, could be prioritized as gene families warranting completion. There are also many ribosome-related genes left to be characterized, and ribosomopathies are an emerging disease class that displays a wide range of phenotypes (Kampen et al. 2020). Additionally, genes essential for life are highly enriched for human disease genes (Bartha et al. 2018; Cacheiro et al. 2020; Dickinson et al. 2016; Georgi et al. 2013). Of the 3381 remaining genes, 15% (507/3381) are classified as cell essential based upon CRISPR/Cas9 screens in human cell lines (Cacheiro et al. 2020; Tsherniak et al. 2017). To determine if these genes were under constraint against mutation in the human population, we used the human orthologs to obtain the probability of loss-of-intolerance (pLI) score from the gnomAD database (Karczewski et al. 2020). pLI scores range from 0 to 1 with values closer to 1 indicative of significant constraint in the human genome (Lek et al. 2016). Of the cell essential genes, 144/507 have a human ortholog associated with a pLI score greater than 0.8 further supporting the idea that this set of cell essential genes is under functional constraint. In total, 11% (378/3381) of the genes without a null allele have a pLI score greater than 0.8 (Fig. 3b). Genes that are nonessential in cell lines but have high pLI scores have previously been shown to be enriched for critical developmental regulators that are also associated with human disease (Cacheiro et al. 2020).
Fig. 3.
Analysis of gene family representation and constraint for the 3381 mouse–human orthologs that currently lack a null allele in mouse. a The set of remaining genes is specifically enriched for large gene families and the RIKEN cDNA collections. Olfactory receptors and zinc finger proteins are the most highly represented gene families. b Classification of the remaining genes using human orthologs to assess cell essentiality based upon CRISPR/Cas9 screens in cancer cell lines and functional constraint using probability of loss-of-intolerance (pLI) scores that range from 0 to 1 with values closer to 1 associated with higher level of constraint
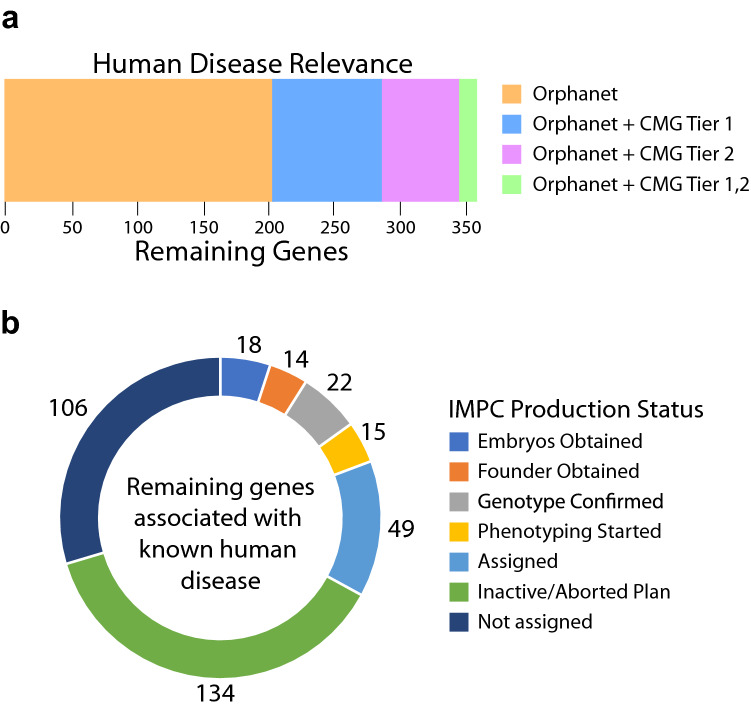
To determine the extent that genes without knockout alleles are related to human disease genes, we examined the overlap with ORPHANET (http://www.orpha.net) and Tier 1 (solved cases) and Tier 2 (unsolved cases) gene lists from the Centers for Mendelian Genomics (CMG; http://mendelian.org/phenotypes-genes) (Posey et al. 2019). This analysis highlighted 358 genes with support from ORPHANET-only or had additional evidence from the CMG (Fig. 4a). While these genes were identified as lacking a knockout mouse, the ongoing production efforts of the IMPC have made progress towards establishing knockouts for 54 genes and begun phenotyping on another 15 genes (Fig. 4b). Production attempts have failed for 134 genes with the remaining 106 yet to be targeted by the IMPC. It will be important to emphasize this set of human disease related genes for generation of mouse knockouts.
Fig. 4.
Potential human disease relevance for genes without a mouse null allele. a Human orthologs were used to query the Orphanet database (https://www.orpha.net/) and the Centers for Mendelian Genomics (CMG) gene lists (http://mendelian.org/phenotypes-genes). CMG genes are classified into Tier 1 and Tier 2 based upon the supporting level of evidence. Tier 1 genes have the highest-level of confidence with multiple levels of supporting evidence and Tier 2 genes are strong candidates but do not meet stringency criteria set for Tier 1. Currently, ~ 10% of the remaining mouse genes are related to human disease genes. b IMPC progress towards making knockouts for genes relevant to human disease. Of these 358 genes, 252 genes have either been previously attempted and failed (Inactive/aborted) or are currently assigned or in progress through the IMPC production and phenotyping pipeline
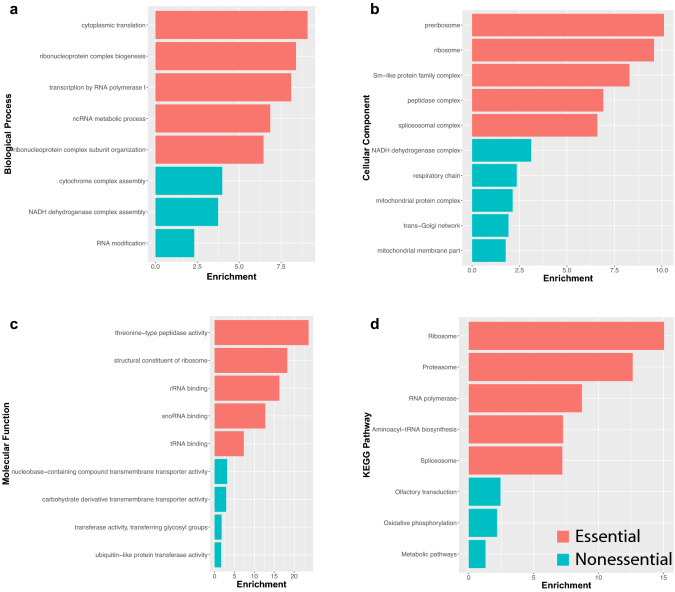
Next, we performed Gene Ontology (GO) term enrichment (Fig. 5a–c) and KEGG pathway analysis (Fig. 5d) on the previously defined cell essential and nonessential gene sets using WebGestalt (Liao et al. 2019). Strikingly, these analyses revealed a stark distinction in the biological processes and pathways associated with these two groups of genes. Cell essential genes were significantly enriched (FDR < 0.05) for terms associated with pathways for mRNA splicing (spliceosome), translation (ribosome), and protein degradation (proteasome; Fig. 5a–d). In contrast, nonessential genes showed enrichment for olfactory transduction and metabolic pathways (Fig. 5d). These findings indicate a clear distinction between core biological functions required for cell survival versus potentially tissue-specific differences in energy requirements or activity of certain cell types. In support of these differences, we used the human orthologs for the remaining genes to identify potential human disease associations within OMIM and observed an enrichment for Mitochondrial Complex I Deficiency (P-value = 2.0673e−11), Alcohol Dependence (P-value = 6.5255e−4), and Mitochondrial Complex IV Deficiency (P-value = 4.4197e−5).
Fig. 5.
GO term enrichment and pathway analysis on the set of 3381 genes classified into cell essential and nonessential categories. GO terms for a Biological process, b Cellular component, and c Molecular function highlight a role for essential genes in core biological processes including transcription, splicing, and translation, and a role for nonessential genes in metabolic processes and mitochondrial function. d KEGG pathway analysis supports the distinction between cell essential and nonessential genes and associated biological function. Enrichment values were determined using WebGestalt with FDR < 0.5
Overview of noncoding transcripts annotated in the mouse genome
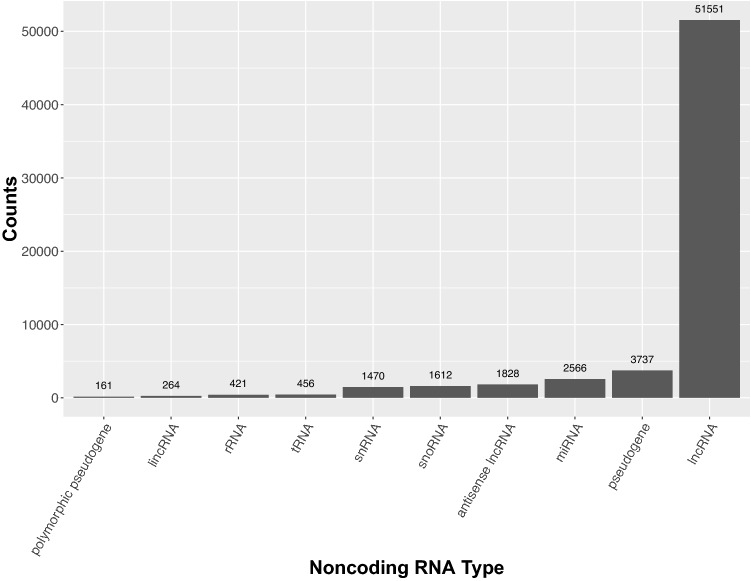
Despite the intense focus on the protein-coding portion of the genome, there has been a long-standing appreciation for the role of noncoding transcripts for regulating diverse cellular events such as modulating chromatin structure (e.g., Xist), targeting genes to regulate expression (e.g., miRNA), and building the translational machinery (e.g., rRNA and tRNA). While these functions are known, the number of annotated noncoding transcripts continues to grow and now exceeds the number of protein-coding genes with the vast majority generically referred to as long noncoding RNA’s (lncRNAs) with unknown function (Fig. 6). To gain a greater understanding of how noncoding transcripts impact biology, we will need to invest and explore in alternative targeting strategies to determine if the noncoding transcript itself is functional or if it is merely the act of transcription that is required. Functional testing of the transcripts can be achieved using whole gene ablation strategies with CRISPR/Cas9; however, while large deletions (> 10 kb) are feasible with CRISPR/Cas9 these are often accompanied with other structural variants such as inversions and duplications which would require additional screening (Birling et al. 2017). Additionally, the removal of large genomic regions has the potential to impact the expression of neighboring genes by altering the cis-regulatory landscape. An alternative approach is to introduce a poly-adenylation sequence to terminate transcription (Engreitz et al. 2016) or knockdown the target the transcript by introducing a degradation sequence or using Cas13a (Gao et al. 2020). These approaches would keep the locus intact but prevent the transcript from accumulating; and thereby, determine if the transcript itself is functional or is it merely the engagement of RNA polymerase II that is required. There is also a pre-existing set of ESC-based resources available for studying noncoding RNA function (Hansen et al. 2021). Our current animal production methodologies are well suited to be scaled to address these questions and can readily implement all of these approaches.
Fig. 6.
Top 10 noncoding RNA sequence features annotated in the mouse genome. The number of known long noncoding RNA’s (lncRNAs) currently exceeds the number of protein-coding genes and the vast majority remain to be studied in depth
Conclusions
In summary, extraordinary progress has been made towards generating the first complete functional annotation of the protein-coding fraction of the mammalian genome. This celebratory milestone is rapidly approaching thanks to the combined efforts of the research community and the ongoing IMPC initiative. For the remaining genes, strategic implementation of different prioritization strategies will allow us to focus on sets of genes that have the potential to inform on human disease, increase our understanding of the broad biological function of large gene families, and shed light on the darkest parts of the genome. Thus far, in IMPC-generated strains, the rate of gene essentiality has remained constant at ~ 35% where 25% of lines are classified as lethal and an additional 10% are classified as subviable (Cacheiro et al. 2020; Dickinson et al. 2016). It will be informative to determine whether these rates will remain stable for this gene set given the relative dearth of functional information available for these genes. Elucidating the full spectrum of gene essentiality within mammals has the potential to further support ongoing human disease gene discovery efforts and to provide new insights into the underlying mechanisms of congenital birth defects and later onset diseases that have a developmental etiology. Our initial characterization of the production attempts for these remaining genes suggests that some may be refractory to our current methodologies possibly due to gene essentiality or haploinsufficiency and, thus, may require a more nuanced approach. Conditional alleles could provide a means to circumvent production challenges and would provide useful tools for further mechanistic investigation of essential gene function. Alternatively, novel approaches such as the implementation of auxin-inducible protein degradation may help to overcome null allele production efforts (Yesbolatova et al. 2020). Further, the classification of a null allele is based upon the assumption that frameshift mutations will result in mRNA degradation via the nonsense-mediated decay (NMD) pathway. While the IMPC design principles adhere to well-established rules of NMD, recent work has shown that there are a number of exceptions to the canonical rules, and that roughly 25% of the variance from expected NMD remains unexplained (Lindeboom et al. 2016). Moreover, there are reported cases of exon skipping as well as the use of alternate downstream translation start sites that allow for some level of activity (Makino et al. 2016). The extent to which these apply to the current catalog will require further bioinformatic and experimental determination. As mentioned above, the rate of gene essentiality has remained constant for IMPC-generated lines regardless of the technology used to generate the allele. Thus, as the foundation for a phenotype-driven screen, the mutant alleles characterized to date significantly disrupt gene function.
In addition, the generation of alleles for precision medicine corresponding to patient-specific mutations will undoubtedly be a critical next step to further our understanding of the molecular basis of human disease. Beyond the coding genome, the vast majority of noncoding RNAs have yet to be characterized in mutant mice. The collective effort to generate a complete null resource will provide a strong foundation to support these future initiatives, expanding our understanding of genome function, which holds great promise for improving human health and personalized medicine.
Acknowledgements
We would like to thank members of the International Mouse Phenotyping Consortium for all of their feedback and input over the years on these analyses. In particular, Violetta Munoz-Fuentes and Pilar Cacheiro for their assistance in defining mouse–human orthologs and essential gene classification. Additionally, this analysis would not have been possible without the contributions of all the individual research labs around the world and the curational efforts of the MGI team. This work was supported by the NIH Common Fund and National Institutes of Health Office of the Director UM1 OD023222 (S.A.M.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Kevin A. Peterson, Email: kevin.peterson@jax.org
Stephen A. Murray, Email: steve.murray@jax.org
References
- Angrand PO, Daigle N, van der Hoeven F, Scholer HR, Stewart AF. Simplified generation of targeting constructs using ET recombination. Nucleic Acids Res. 1999;27:e16. doi: 10.1093/nar/27.17.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin CP, Battey JF, Bradley A, Bucan M, Capecchi M, Collins FS, Dove WF, Duyk G, Dymecki S, Eppig JT, Grieder FB, Heintz N, Hicks G, Insel TR, Joyner A, Koller BH, Lloyd KC, Magnuson T, Moore MW, Nagy A, Pollock JD, Roses AD, Sands AT, Seed B, Skarnes WC, Snoddy J, Soriano P, Stewart DJ, Stewart F, Stillman B, Varmus H, Varticovski L, Verma IM, Vogt TF, von Melchner H, Witkowski J, Woychik RP, Wurst W, Yancopoulos GD, Young SG, Zambrowicz B. The knockout mouse project. Nat Genet. 2004;36:921–924. doi: 10.1038/ng0904-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auwerx J, Avner P, Baldock R, Ballabio A, Balling R, Barbacid M, Berns A, Bradley A, Brown S, Carmeliet P, Chambon P, Cox R, Davidson D, Davies K, Duboule D, Forejt J, Granucci F, Hastie N, de Angelis MH, Jackson I, Kioussis D, Kollias G, Lathrop M, Lendahl U, Malumbres M, von Melchner H, Muller W, Partanen J, Ricciardi-Castagnoli P, Rigby P, Rosen B, Rosenthal N, Skarnes B, Stewart AF, Thornton J, Tocchini-Valentini G, Wagner E, Wahli W, Wurst W. The European dimension for the mouse genome mutagenesis program. Nat Genet. 2004;36:925–927. doi: 10.1038/ng0904-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartha I, di Iulio J, Venter JC, Telenti A. Human gene essentiality. Nat Rev Genet. 2018;19:51–62. doi: 10.1038/nrg.2017.75. [DOI] [PubMed] [Google Scholar]
- Battey J, Jordan E, Cox D, Dove W. An action plan for mouse genomics. Nat Genet. 1999;21:73–75. doi: 10.1038/5012. [DOI] [PubMed] [Google Scholar]
- Birling MC, Schaeffer L, Andre P, Lindner L, Marechal D, Ayadi A, Sorg T, Pavlovic G, Herault Y. Efficient and rapid generation of large genomic variants in rats and mice using CRISMERE. Sci Rep. 2017;7:43331. doi: 10.1038/srep43331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birling MC, Yoshiki A, Adams DJ, Ayabe S, Beaudet AL, Bottomley J, Bradley A, Brown SDM, Burger A, Bushell W, Chiani F, Chin HG, Christou S, Codner GF, DeMayo FJ, Dickinson ME, Doe B, Donahue LR, Fray MD, Gambadoro A, Gao X, Gertsenstein M, Gomez-Segura A, Goodwin LO, Heaney JD, Herault Y, de Angelis MH, Jiang ST, Justice MJ, Kasparek P, King RE, Kuhn R, Lee H, Lee YJ, Liu Z, Lloyd KCK, Lorenzo I, Mallon AM, McKerlie C, Meehan TF, Fuentes VM, Newman S, Nutter LMJ, Oh GT, Pavlovic G, Ramirez-Solis R, Rosen B, Ryder EJ, Santos LA, Schick J, Seavitt JR, Sedlacek R, Seisenberger C, Seong JK, Skarnes WC, Sorg T, Steel KP, Tamura M, Tocchini-Valentini GP, Wang CL, Wardle-Jones H, Wattenhofer-Donze M, Wells S, Wiles MV, Willis BJ, Wood JA, Wurst W, Xu Y, International Mouse Phenotyping C, Teboul L, Murray SA A resource of targeted mutant mouse lines for 5061 genes. Nat Genet. 2021;53:416–419. [Google Scholar]
- Bowl MR, Simon MM, Ingham NJ, Greenaway S, Santos L, Cater H, Taylor S, Mason J, Kurbatova N, Pearson S, Bower LR, Clary DA, Meziane H, Reilly P, Minowa O, Kelsey L, International Mouse Phenotyping C, Tocchini-Valentini GP, Gao X, Bradley A, Skarnes WC, Moore M, Beaudet AL, Justice MJ, Seavitt J, Dickinson ME, Wurst W, de Angelis MH, Herault Y, Wakana S, Nutter LMJ, Flenniken AM, McKerlie C, Murray SA, Svenson KL, Braun RE, West DB, Lloyd KCK, Adams DJ, White J, Karp N, Flicek P, Smedley D, Meehan TF, Parkinson HE, Teboul LM, Wells S, Steel KP, Mallon AM, Brown SDM A large scale hearing loss screen reveals an extensive unexplored genetic landscape for auditory dysfunction. Nat Commun. 2017;8:886. doi: 10.1038/s41467-017-00595-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley A, Anastassiadis K, Ayadi A, Battey JF, Bell C, Birling MC, Bottomley J, Brown SD, Burger A, Bult CJ, Bushell W, Collins FS, Desaintes C, Doe B, Economides A, Eppig JT, Finnell RH, Fletcher C, Fray M, Frendewey D, Friedel RH, Grosveld FG, Hansen J, Herault Y, Hicks G, Horlein A, Houghton R, Hrabe de Angelis M, Huylebroeck D, Iyer V, de Jong PJ, Kadin JA, Kaloff C, Kennedy K, Koutsourakis M, Lloyd KC, Marschall S, Mason J, McKerlie C, McLeod MP, von Melchner H, Moore M, Mujica AO, Nagy A, Nefedov M, Nutter LM, Pavlovic G, Peterson JL, Pollock J, Ramirez-Solis R, Rancourt DE, Raspa M, Remacle JE, Ringwald M, Rosen B, Rosenthal N, Rossant J, Ruiz Noppinger P, Ryder E, Schick JZ, Schnutgen F, Schofield P, Seisenberger C, Selloum M, Simpson EM, Skarnes WC, Smedley D, Stanford WL, Stewart AF, Stone K, Swan K, Tadepally H, Teboul L, Tocchini-Valentini GP, Valenzuela D, West AP, Yamamura K, Yoshinaga Y, Wurst W. The mammalian gene function resource: the International Knockout Mouse Consortium. Mamm Genome. 2012;23:580–586. doi: 10.1007/s00335-012-9422-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SDM, Holmes CC, Mallon AM, Meehan TF, Smedley D, Wells S. High-throughput mouse phenomics for characterizing mammalian gene function. Nat Rev Genet. 2018;19:357–370. doi: 10.1038/s41576-018-0005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacheiro P, Munoz-Fuentes V, Murray SA, Dickinson ME, Bucan M, Nutter LMJ, Peterson KA, Haselimashhadi H, Flenniken AM, Morgan H, Westerberg H, Konopka T, Hsu CW, Christiansen A, Lanza DG, Beaudet AL, Heaney JD, Fuchs H, Gailus-Durner V, Sorg T, Prochazka J, Novosadova V, Lelliott CJ, Wardle-Jones H, Wells S, Teboul L, Cater H, Stewart M, Hough T, Wurst W, Sedlacek R, Adams DJ, Seavitt JR, Tocchini-Valentini G, Mammano F, Braun RE, McKerlie C, Herault Y, de Angelis MH, Mallon AM, Lloyd KCK, Brown SDM, Parkinson H, Meehan TF, Smedley D, Genomics England Research Consortium, International Mouse Phenotyping Consortium Human and mouse essentiality screens as a resource for disease gene discovery. Nat Commun. 2020;11:655. doi: 10.1038/s41467-020-14284-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capecchi MR. Altering the genome by homologous recombination. Science. 1989;244:1288–1292. doi: 10.1126/science.2660260. [DOI] [PubMed] [Google Scholar]
- Copeland NG, Jenkins NA, Court DL. Recombineering: a powerful new tool for mouse functional genomics. Nat Rev Genet. 2001;2:769–779. doi: 10.1038/35093556. [DOI] [PubMed] [Google Scholar]
- de Angelis MH, Nicholson G, Selloum M, White J, Morgan H, Ramirez-Solis R, Sorg T, Wells S, Fuchs H, Fray M, Adams DJ, Adams NC, Adler T, Aguilar-Pimentel A, Ali-Hadji D, Amann G, Andre P, Atkins S, Auburtin A, Ayadi A, Becker J, Becker L, Bedu E, Bekeredjian R, Birling MC, Blake A, Bottomley J, Bowl M, Brault V, Busch DH, Bussell JN, Calzada-Wack J, Cater H, Champy MF, Charles P, Chevalier C, Chiani F, Codner GF, Combe R, Cox R, Dalloneau E, Dierich A, Di Fenza A, Doe B, Duchon A, Eickelberg O, Esapa CT, El Fertak L, Feigel T, Emelyanova I, Estabel J, Favor J, Flenniken A, Gambadoro A, Garrett L, Gates H, Gerdin AK, Gkoutos G, Greenaway S, Glasl L, Goetz P, Da Cruz IG, Gotz A, Graw J, Guimond A, Hans W, Hicks G, Holter SM, Hofler H, Hancock JM, Hoehndorf R, Hough T, Houghton R, Hurt A, Ivandic B, Jacobs H, Jacquot S, Jones N, Karp NA, Katus HA, Kitchen S, Klein-Rodewald T, Klingenspor M, Klopstock T, Lalanne V, Leblanc S, Lengger C, le Marchand E, Ludwig T, Lux A, McKerlie C, Maier H, Mandel JL, Marschall S, Mark M, Melvin DG, Meziane H, Micklich K, Mittelhauser C, Monassier L, Moulaert D, Muller S, Naton B, Neff F, Nolan PM, Nutter LM, Ollert M, Pavlovic G, Pellegata NS, Peter E, Petit-Demouliere B, Pickard A, Podrini C, Potter P, Pouilly L, Puk O, Richardson D, Rousseau S, Quintanilla-Fend L, Quwailid MM, Racz I, Rathkolb B, Riet F, Rossant J, Roux M, Rozman J, Ryder E, Salisbury J, Santos L, Schable KH, Schiller E, Schrewe A, Schulz H, Steinkamp R, Simon M, Stewart M, Stoger C, Stoger T, Sun M, Sunter D, Teboul L, Tilly I, Tocchini-Valentini GP, Tost M, Treise I, Vasseur L, Velot E, Vogt-Weisenhorn D, Wagner C, Walling A, Weber B, Wendling O, Westerberg H, Willershauser M, Wolf E, Wolter A, Wood J, Wurst W, Yildirim AO, Zeh R, Zimmer A, Zimprich A, Consortium E, Holmes C, Steel KP, Herault Y, Gailus-Durner V, Mallon AM, Brown SD Analysis of mammalian gene function through broad-based phenotypic screens across a consortium of mouse clinics. Nat Genet. 2015;47:969–978. doi: 10.1038/ng.3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson ME, Flenniken AM, Ji X, Teboul L, Wong MD, White JK, Meehan TF, Weninger WJ, Westerberg H, Adissu H, Baker CN, Bower L, Brown JM, Caddle LB, Chiani F, Clary D, Cleak J, Daly MJ, Denegre JM, Doe B, Dolan ME, Edie SM, Fuchs H, Gailus-Durner V, Galli A, Gambadoro A, Gallegos J, Guo S, Horner NR, Hsu CW, Johnson SJ, Kalaga S, Keith LC, Lanoue L, Lawson TN, Lek M, Mark M, Marschall S, Mason J, McElwee ML, Newbigging S, Nutter LM, Peterson KA, Ramirez-Solis R, Rowland DJ, Ryder E, Samocha KE, Seavitt JR, Selloum M, Szoke-Kovacs Z, Tamura M, Trainor AG, Tudose I, Wakana S, Warren J, Wendling O, West DB, Wong L, Yoshiki A, International Mouse Phenotyping Consortium, Jackson Laboratory, Infrastructure Nationale Phenomin Institut Clinique de la Souris, Charles River Laboratories, Harwell MRC, Toronto Centre for Phenogenomics, Wellcome Trust Sanger Institute, Riken BioResource Center, MacArthur DG, Tocchini-Valentini GP, Gao X, Flicek P, Bradley A, Skarnes WC, Justice MJ, Parkinson HE, Moore M, Wells S, Braun RE, Svenson KL, de Angelis MH, Herault Y, Mohun T, Mallon AM, Henkelman RM, Brown SD, Adams DJ, Lloyd KC, McKerlie C, Beaudet AL, Bucan M, Murray SA High-throughput discovery of novel developmental phenotypes. Nature. 2016;537:508–514. [Google Scholar]
- Dolgin E. The most popular genes in the human genome. Nature. 2017;551:427–431. doi: 10.1038/d41586-017-07291-9. [DOI] [PubMed] [Google Scholar]
- Edwards AM, Isserlin R, Bader GD, Frye SV, Willson TM, Yu FH. Too many roads not taken. Nature. 2011;470:163–165. doi: 10.1038/470163a. [DOI] [PubMed] [Google Scholar]
- Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, McDonel PE, Guttman M, Lander ES. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539:452–455. doi: 10.1038/nature20149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Cai Y, Kapranov P, Xu D. Reverse-genetics studies of lncRNAs-what we have learnt and paths forward. Genome Biol. 2020;21:93. doi: 10.1186/s13059-020-01994-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgi B, Voight BF, Bucan M. From mouse to human: evolutionary genomics analysis of human orthologs of essential genes. PLoS Genet. 2013;9:e1003484. doi: 10.1371/journal.pgen.1003484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gondo Y. Trends in large-scale mouse mutagenesis: from genetics to functional genomics. Nat Rev Genet. 2008;9:803–810. doi: 10.1038/nrg2431. [DOI] [PubMed] [Google Scholar]
- Hansen J, von Melchner H, Wurst W. Mutant non-coding RNA resource in mouse embryonic stem cells. Dis Model Mech. 2021 doi: 10.1242/dmm.047803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrabe de Angelis MH, Flaswinkel H, Fuchs H, Rathkolb B, Soewarto D, Marschall S, Heffner S, Pargent W, Wuensch K, Jung M, Reis A, Richter T, Alessandrini F, Jakob T, Fuchs E, Kolb H, Kremmer E, Schaeble K, Rollinski B, Roscher A, Peters C, Meitinger T, Strom T, Steckler T, Holsboer F, Klopstock T, Gekeler F, Schindewolf C, Jung T, Avraham K, Behrendt H, Ring J, Zimmer A, Schughart K, Pfeffer K, Wolf E, Balling R. Genome-wide, large-scale production of mutant mice by ENU mutagenesis. Nat Genet. 2000;25:444–447. doi: 10.1038/78146. [DOI] [PubMed] [Google Scholar]
- International Mouse Knockout Consortium. Collins FS, Rossant J, Wurst W. A mouse for all reasons. Cell. 2007;128:9–13. doi: 10.1016/j.cell.2006.12.018. [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampen KR, Sulima SO, Vereecke S, De Keersmaecker K. Hallmarks of ribosomopathies. Nucleic Acids Res. 2020;48:1013–1028. doi: 10.1093/nar/gkz637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alfoldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, Gauthier LD, Brand H, Solomonson M, Watts NA, Rhodes D, Singer-Berk M, England EM, Seaby EG, Kosmicki JA, Walters RK, Tashman K, Farjoun Y, Banks E, Poterba T, Wang A, Seed C, Whiffin N, Chong JX, Samocha KE, Pierce-Hoffman E, Zappala Z, O'Donnell-Luria AH, Minikel EV, Weisburd B, Lek M, Ware JS, Vittal C, Armean IM, Bergelson L, Cibulskis K, Connolly KM, Covarrubias M, Donnelly S, Ferriera S, Gabriel S, Gentry J, Gupta N, Jeandet T, Kaplan D, Llanwarne C, Munshi R, Novod S, Petrillo N, Roazen D, Ruano-Rubio V, Saltzman A, Schleicher M, Soto J, Tibbetts K, Tolonen C, Wade G, Talkowski ME, Genome Aggregation Database Consortium, Neale BM, Daly MJ, MacArthur DG The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp NA, Mason J, Beaudet AL, Benjamini Y, Bower L, Braun RE, Brown SDM, Chesler EJ, Dickinson ME, Flenniken AM, Fuchs H, Angelis MH, Gao X, Guo S, Greenaway S, Heller R, Herault Y, Justice MJ, Kurbatova N, Lelliott CJ, Lloyd KCK, Mallon AM, Mank JE, Masuya H, McKerlie C, Meehan TF, Mott RF, Murray SA, Parkinson H, Ramirez-Solis R, Santos L, Seavitt JR, Smedley D, Sorg T, Speak AO, Steel KP, Svenson KL, International Mouse Phenotyping C, Wakana S, West D, Wells S, Westerberg H, Yaacoby S, White JK Prevalence of sexual dimorphism in mammalian phenotypic traits. Nat Commun. 2017;8:15475. doi: 10.1038/ncomms15475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O'Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, Kosmicki JA, Duncan LE, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Berghout J, Cooper DN, Deflaux N, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki MI, Moonshine AL, Natarajan P, Orozco L, Peloso GM, Poplin R, Rivas MA, Ruano-Rubio V, Rose SA, Ruderfer DM, Shakir K, Stenson PD, Stevens C, Thomas BP, Tiao G, Tusie-Luna MT, Weisburd B, Won HH, Yu D, Altshuler DM, Ardissino D, Boehnke M, Danesh J, Donnelly S, Elosua R, Florez JC, Gabriel SB, Getz G, Glatt SJ, Hultman CM, Kathiresan S, Laakso M, McCarroll S, McCarthy MI, McGovern D, McPherson R, Neale BM, Palotie A, Purcell SM, Saleheen D, Scharf JM, Sklar P, Sullivan PF, Tuomilehto J, Tsuang MT, Watkins HC, Wilson JG, Daly MJ, MacArthur DG, Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Wang J, Jaehnig EJ, Shi Z, Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47:W199–W205. doi: 10.1093/nar/gkz401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeboom RG, Supek F, Lehner B. The rules and impact of nonsense-mediated mRNA decay in human cancers. Nat Genet. 2016;48:1112–1118. doi: 10.1038/ng.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd KCK, Adams DJ, Baynam G, Beaudet AL, Bosch F, Boycott KM, Braun RE, Caulfield M, Cohn R, Dickinson ME, Dobbie MS, Flenniken AM, Flicek P, Galande S, Gao X, Grobler A, Heaney JD, Herault Y, de Angelis MH, Lupski JR, Lyonnet S, Mallon AM, Mammano F, MacRae CA, McInnes R, McKerlie C, Meehan TF, Murray SA, Nutter LMJ, Obata Y, Parkinson H, Pepper MS, Sedlacek R, Seong JK, Shiroishi T, Smedley D, Tocchini-Valentini G, Valle D, Wang CL, Wells S, White J, Wurst W, Xu Y, Brown SDM. The deep genome project. Genome Biol. 2020;21:18. doi: 10.1186/s13059-020-1931-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S, Fukumura R, Gondo Y. Illegitimate translation causes unexpected gene expression from on-target out-of-frame alleles created by CRISPR-Cas9. Sci Rep. 2016;6:39608. doi: 10.1038/srep39608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehan TF, Conte N, West DB, Jacobsen JO, Mason J, Warren J, Chen CK, Tudose I, Relac M, Matthews P, Karp N, Santos L, Fiegel T, Ring N, Westerberg H, Greenaway S, Sneddon D, Morgan H, Codner GF, Stewart ME, Brown J, Horner N, International Mouse Phenotyping C, Haendel M, Washington N, Mungall CJ, Reynolds CL, Gallegos J, Gailus-Durner V, Sorg T, Pavlovic G, Bower LR, Moore M, Morse I, Gao X, Tocchini-Valentini GP, Obata Y, Cho SY, Seong JK, Seavitt J, Beaudet AL, Dickinson ME, Herault Y, Wurst W, de Angelis MH, Lloyd KCK, Flenniken AM, Nutter LMJ, Newbigging S, McKerlie C, Justice MJ, Murray SA, Svenson KL, Braun RE, White JK, Bradley A, Flicek P, Wells S, Skarnes WC, Adams DJ, Parkinson H, Mallon AM, Brown SDM, Smedley D Disease model discovery from 3328 gene knockouts by The International Mouse Phenotyping Consortium. Nat Genet. 2017;49:1231–1238. [Google Scholar]
- Motenko H, Neuhauser SB, O'Keefe M, Richardson JE. MouseMine: a new data warehouse for MGI. Mamm Genome. 2015;26:325–330. doi: 10.1007/s00335-015-9573-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium. Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J, Baertsch R, Bailey J, Barlow K, Beck S, Berry E, Birren B, Bloom T, Bork P, Botcherby M, Bray N, Brent MR, Brown DG, Brown SD, Bult C, Burton J, Butler J, Campbell RD, Carninci P, Cawley S, Chiaromonte F, Chinwalla AT, Church DM, Clamp M, Clee C, Collins FS, Cook LL, Copley RR, Coulson A, Couronne O, Cuff J, Curwen V, Cutts T, Daly M, David R, Davies J, Delehaunty KD, Deri J, Dermitzakis ET, Dewey C, Dickens NJ, Diekhans M, Dodge S, Dubchak I, Dunn DM, Eddy SR, Elnitski L, Emes RD, Eswara P, Eyras E, Felsenfeld A, Fewell GA, Flicek P, Foley K, Frankel WN, Fulton LA, Fulton RS, Furey TS, Gage D, Gibbs RA, Glusman G, Gnerre S, Goldman N, Goodstadt L, Grafham D, Graves TA, Green ED, Gregory S, Guigo R, Guyer M, Hardison RC, Haussler D, Hayashizaki Y, Hillier LW, Hinrichs A, Hlavina W, Holzer T, Hsu F, Hua A, Hubbard T, Hunt A, Jackson I, Jaffe DB, Johnson LS, Jones M, Jones TA, Joy A, Kamal M, Karlsson EK, Karolchik D, Kasprzyk A, Kawai J, Keibler E, Kells C, Kent WJ, Kirby A, Kolbe DL, Korf I, Kucherlapati RS, Kulbokas EJ, Kulp D, Landers T, Leger JP, Leonard S, Letunic I, Levine R, Li J, Li M, Lloyd C, Lucas S, Ma B, Maglott DR, Mardis ER, Matthews L, Mauceli E, Mayer JH, McCarthy M, McCombie WR, McLaren S, McLay K, McPherson JD, Meldrim J, Meredith B, Mesirov JP, Miller W, Miner TL, Mongin E, Montgomery KT, Morgan M, Mott R, Mullikin JC, Muzny DM, Nash WE, Nelson JO, Nhan MN, Nicol R, Ning Z, Nusbaum C, O'Connor MJ, Okazaki Y, Oliver K, Overton-Larty E, Pachter L, Parra G, Pepin KH, Peterson J, Pevzner P, Plumb R, Pohl CS, Poliakov A, Ponce TC, Ponting CP, Potter S, Quail M, Reymond A, Roe BA, Roskin KM, Rubin EM, Rust AG, Santos R, Sapojnikov V, Schultz B, Schultz J, Schwartz MS, Schwartz S, Scott C, Seaman S, Searle S, Sharpe T, Sheridan A, Shownkeen R, Sims S, Singer JB, Slater G, Smit A, Smith DR, Spencer B, Stabenau A, Stange-Thomann N, Sugnet C, Suyama M, Tesler G, Thompson J, Torrents D, Trevaskis E, Tromp J, Ucla C, Ureta-Vidal A, Vinson JP, Von Niederhausern AC, Wade CM, Wall M, Weber RJ, Weiss RB, Wendl MC, West AP, Wetterstrand K, Wheeler R, Whelan S, Wierzbowski J, Willey D, Williams S, Wilson RK, Winter E, Worley KC, Wyman D, Yang S, Yang SP, Zdobnov EM, Zody MC, Lander ES. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Munoz-Fuentes V, Cacheiro P, Meehan TF, Aguilar-Pimentel JA, Brown SDM, Flenniken AM, Flicek P, Galli A, Mashhadi HH, Hrabe de Angelis M, Kim JK, Lloyd KCK, McKerlie C, Morgan H, Murray SA, Nutter LMJ, Reilly PT, Seavitt JR, Seong JK, Simon M, Wardle-Jones H, Mallon AM, Smedley D, Parkinson HE, The International Mouse Phenotyping Consortium I The International Mouse Phenotyping Consortium (IMPC): a functional catalogue of the mammalian genome that informs conservation. Conserv Genet. 2018;19:995–1005. doi: 10.1007/s10592-018-1072-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau JH, Balling R, Barsh G, Beier D, Brown SD, Bucan M, Camper S, Carlson G, Copeland N, Eppig J, Fletcher C, Frankel WN, Ganten D, Goldowitz D, Goodnow C, Guenet JL, Hicks G, Hrabe de Angelis M, Jackson I, Jacob HJ, Jenkins N, Johnson D, Justice M, Kay S, Kingsley D, Lehrach H, Magnuson T, Meisler M, Poustka A, Rinchik EM, Rossant J, Russell LB, Schimenti J, Shiroishi T, Skarnes WC, Soriano P, Stanford W, Takahashi JS, Wurst W, Zimmer A, International Mouse Mutagenesis Consortium Sequence interpretation. Functional annotation of mouse genome sequences. Science. 2001;291:1251–1255. doi: 10.1126/science.1058244. [DOI] [PubMed] [Google Scholar]
- Nolan PM, Peters J, Strivens M, Rogers D, Hagan J, Spurr N, Gray IC, Vizor L, Brooker D, Whitehill E, Washbourne R, Hough T, Greenaway S, Hewitt M, Liu X, McCormack S, Pickford K, Selley R, Wells C, Tymowska-Lalanne Z, Roby P, Glenister P, Thornton C, Thaung C, Stevenson JA, Arkell R, Mburu P, Hardisty R, Kiernan A, Erven A, Steel KP, Voegeling S, Guenet JL, Nickols C, Sadri R, Nasse M, Isaacs A, Davies K, Browne M, Fisher EM, Martin J, Rastan S, Brown SD, Hunter J. A systematic, genome-wide, phenotype-driven mutagenesis programme for gene function studies in the mouse. Nat Genet. 2000;25:440–443. doi: 10.1038/78140. [DOI] [PubMed] [Google Scholar]
- Oprea TI, Bologa CG, Brunak S, Campbell A, Gan GN, Gaulton A, Gomez SM, Guha R, Hersey A, Holmes J, Jadhav A, Jensen LJ, Johnson GL, Karlson A, Leach AR, Ma'ayan A, Malovannaya A, Mani S, Mathias SL, McManus MT, Meehan TF, von Mering C, Muthas D, Nguyen DT, Overington JP, Papadatos G, Qin J, Reich C, Roth BL, Schurer SC, Simeonov A, Sklar LA, Southall N, Tomita S, Tudose I, Ursu O, Vidovic D, Waller A, Westergaard D, Yang JJ, Zahoranszky-Kohalmi G. Unexplored therapeutic opportunities in the human genome. Nat Rev Drug Discov. 2018;17:317–332. doi: 10.1038/nrd.2018.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanet: an online database of rare diseases and orphan drugs. Copyright, INSERM 1997. Available at http://www.orpha.net. Accessed 27 June 2021.
- Posey JE, O'Donnell-Luria AH, Chong JX, Harel T, Jhangiani SN, Coban Akdemir ZH, Buyske S, Pehlivan D, Carvalho CMB, Baxter S, Sobreira N, Liu P, Wu N, Rosenfeld JA, Kumar S, Avramopoulos D, White JJ, Doheny KF, Witmer PD, Boehm C, Sutton VR, Muzny DM, Boerwinkle E, Gunel M, Nickerson DA, Mane S, MacArthur DG, Gibbs RA, Hamosh A, Lifton RP, Matise TC, Rehm HL, Gerstein M, Bamshad MJ, Valle D, Lupski JR, Centers for Mendelian Genomics Insights into genetics, human biology and disease gleaned from family based genomic studies. Genet Med. 2019;21:798–812. doi: 10.1038/s41436-018-0408-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozman J, Rathkolb B, Oestereicher MA, Schutt C, Ravindranath AC, Leuchtenberger S, Sharma S, Kistler M, Willershauser M, Brommage R, Meehan TF, Mason J, Haselimashhadi H, Consortium I, Hough T, Mallon AM, Wells S, Santos L, Lelliott CJ, White JK, Sorg T, Champy MF, Bower LR, Reynolds CL, Flenniken AM, Murray SA, Nutter LMJ, Svenson KL, West D, Tocchini-Valentini GP, Beaudet AL, Bosch F, Braun RB, Dobbie MS, Gao X, Herault Y, Moshiri A, Moore BA, Kent Lloyd KC, McKerlie C, Masuya H, Tanaka N, Flicek P, Parkinson HE, Sedlacek R, Seong JK, Wang CL, Moore M, Brown SD, Tschop MH, Wurst W, Klingenspor M, Wolf E, Beckers J, Machicao F, Peter A, Staiger H, Haring HU, Grallert H, Campillos M, Maier H, Fuchs H, Gailus-Durner V, Werner T, Hrabe de Angelis M Identification of genetic elements in metabolism by high-throughput mouse phenotyping. Nat Commun. 2018;9:288. doi: 10.1038/s41467-017-01995-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarnes WC, von Melchner H, Wurst W, Hicks G, Nord AS, Cox T, Young SG, Ruiz P, Soriano P, Tessier-Lavigne M, Conklin BR, Stanford WL, Rossant J, International Gene Trap Consortium A public gene trap resource for mouse functional genomics. Nat Genet. 2004;36:543–544. doi: 10.1038/ng0604-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, Jackson D, Severin J, Biggs P, Fu J, Nefedov M, de Jong PJ, Stewart AF, Bradley A. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeger T, Gerlach M, Morimoto RI, Nunes Amaral LA. Large-scale investigation of the reasons why potentially important genes are ignored. PLoS Biol. 2018;16:e2006643. doi: 10.1371/journal.pbio.2006643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan AL, Schutt C, Rozman J, Del Mar Muniz Moreno M, Brandmaier S, Simon M, Leuchtenberger S, Griffiths M, Brommage R, Keskivali-Bond P, Grallert H, Werner T, Teperino R, Becker L, Miller G, Moshiri A, Seavitt JR, Cissell DD, Meehan TF, Acar EF, Lelliott CJ, Flenniken AM, Champy MF, Sorg T, Ayadi A, Braun RE, Cater H, Dickinson ME, Flicek P, Gallegos J, Ghirardello EJ, Heaney JD, Jacquot S, Lally C, Logan JG, Teboul L, Mason J, Spielmann N, McKerlie C, Murray SA, Nutter LMJ, Odfalk KF, Parkinson H, Prochazka J, Reynolds CL, Selloum M, Spoutil F, Svenson KL, Vales TS, Wells SE, White JK, Sedlacek R, Wurst W, Lloyd KCK, Croucher PI, Fuchs H, Williams GR, Bassett JHD, Gailus-Durner V, Herault Y, Mallon AM, Brown SDM, Mayer-Kuckuk P, Hrabe de Angelis M, Consortium I Mouse mutant phenotyping at scale reveals novel genes controlling bone mineral density. PLoS Genet. 2020;16:e1009190. doi: 10.1371/journal.pgen.1009190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, Gill S, Harrington WF, Pantel S, Krill-Burger JM, Meyers RM, Ali L, Goodale A, Lee Y, Jiang G, Hsiao J, Gerath WFJ, Howell S, Merkel E, Ghandi M, Garraway LA, Root DE, Golub TR, Boehm JS, Hahn WC. Defining a cancer dependency map. Cell. 2017;170:564–576 e516. doi: 10.1016/j.cell.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela DM, Murphy AJ, Frendewey D, Gale NW, Economides AN, Auerbach W, Poueymirou WT, Adams NC, Rojas J, Yasenchak J, Chernomorsky R, Boucher M, Elsasser AL, Esau L, Zheng J, Griffiths JA, Wang X, Su H, Xue Y, Dominguez MG, Noguera I, Torres R, Macdonald LE, Stewart AF, DeChiara TM, Yancopoulos GD. High-throughput engineering of the mouse genome coupled with high-resolution expression analysis. Nat Biotechnol. 2003;21:652–659. doi: 10.1038/nbt822. [DOI] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JK, Gerdin AK, Karp NA, Ryder E, Buljan M, Bussell JN, Salisbury J, Clare S, Ingham NJ, Podrini C, Houghton R, Estabel J, Bottomley JR, Melvin DG, Sunter D, Adams NC, Sanger Institute Mouse Genetics P, Tannahill D, Logan DW, Macarthur DG, Flint J, Mahajan VB, Tsang SH, Smyth I, Watt FM, Skarnes WC, Dougan G, Adams DJ, Ramirez-Solis R, Bradley A, Steel KP Genome-wide generation and systematic phenotyping of knockout mice reveals new roles for many genes. Cell. 2013;154:452–464. doi: 10.1016/j.cell.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yesbolatova A, Saito Y, Kitamoto N, Makino-Itou H, Ajima R, Nakano R, Nakaoka H, Fukui K, Gamo K, Tominari Y, Takeuchi H, Saga Y, Hayashi KI, Kanemaki MT. The auxin-inducible degron 2 technology provides sharp degradation control in yeast, mammalian cells, and mice. Nat Commun. 2020;11:5701. doi: 10.1038/s41467-020-19532-z. [DOI] [PMC free article] [PubMed] [Google Scholar]