Abstract
Oncogenic stimulation shows a rise in reactive oxygen species (ROS), and ROS can eventually induce carcinogenesis by causing DNA damage. In this context, this study aims to evaluate some biochemical and genotoxic changes in the control of cell death caused by NaBu (Sodium butyrate). treatment in breast cancer cells. NaBu’s impact on cell proliferation was determined via WST-1 assay. The lipid peroxidation (MDA), reduced glutathione (GSH), Nitric Oxide (NO), hydrogen peroxide (H2O2), and superoxide dismutase (SOD) enzyme levels were determined biochemically. NaBu-induced genotoxic damage was estimated via single-cell gel electrophoresis (SCGE). NaBu reduced cell viability and potentially induced GSH, but decreased SOD enzyme activity and the level of MDA and NO decreased also H2O2 decreased at different times and NaBu concentrations. Higher NaBu concentrations amplified DNA damage in MCF-7 cells compared to the control group. NaBu shows anticancer and genotoxic effects, especially through antioxidant enzymes, one of the oxidative stress parameters in breast cancer. However, the anticancer and genotoxic effects of NaBu is changed in the oxidative stress parameters with time and treatment concentration of NaBu in MCF-7 cells. Furthermore, his oxidative stress-dependent effect changes need to be clarified by further evaluation with molecular and more biochemical parameters.
Keywords: Breast cancer, Comet assay, Sodium butyrate, Nitric oxide
1. Introduction
Oncogenic stimulation increases reactive oxygen species (ROS) in cancer cells due to amplified metabolic activity and mitochondrial damage (Khan et al., 2012). The mitochondrial respiratory chain is the primary basis of ROS production in cells, and ROS constructed by cancer cells causes bigger stress (Mansoor et al., 2016, Moloney and Cotter, 2018). ROS causes cytotoxicity at high oxidative stress and can induce apoptosis by inhibiting cell proliferation. In addition, when in a low or moderate oxidative stress state, ROS can eventually induce carcinogenesis by causing DNA damage, cell mutation, inflammation, and cell proliferation (Fang et al., 2009). On the other hand, antioxidants including superoxide dismutase (SOD), catalase (CAT) and glutathione peroxidase are among compounds that can reduce ROS (Atala et al., 2017, Chio and Tuveson, 2017, Yuksel and Deveci Ozkan, 2021). Malondialdehyde (MDA), the major polyunsaturated fatty acid peroxidation, is toxic. Its interaction with DNA and proteins is usually regarded as hypothetically mutagenic atherogenic and is a major contributor to DNA damage and mutation (Del Rio et al., 2005, Ayala et al., 2014). H2O2 is long-lived in plasma and has important effects for selective induction of apoptosis in tumour cells and that cancer cells produce high amounts of H2O2 (López-Lázaro, 2007, Bauer, 2019). Superoxide dismutases (SOD) are essential enzymes that protect cells from damage against free radicals by eliminating superoxide radicals (O-2) (Huang et al., 2000). Studies show that SOD activity and expression are significantly altered in gastric cancer patients (Yi et al., 2010). The key antioxidant enzyme that rummages the superoxide anion radical in mitochondria is manganese superoxide dismutase (MnSOD). MnSOD has a critical function in the growth and progression of cancer (Dhar and Clair, 2012). Reduced glutathione (GSH) plays an essential function in cell biology. It has an important function in most human diseases, such as cancer and cardiovascular diseases, by acting in cellular protection against xenobiotics, free radicals and naturally harmful compounds, including hydroperoxides (Locigno and Castronovo, 2001, Traverso et al., 2013). Since the balance between the number of oxidants and antioxidants affects cancer cell metabolism, it is important to determine these parameters in evaluating the effects of substances with anticancer potential (Isnaini et al., 2018).
The acetylation status of genes controlled via histone acetyltransferases (HAT) and Histone deacetylases (HDAC) acts like a vital regulatory mechanism to control gene expression and chromatin structure (Glozak et al., 2005, Bolden et al., 2006). Sodium butyrate (NaBu), a short-chain fatty acid serving as a histone deacetylase inhibitor (HDACi), is a byproduct of carbohydrate metabolism in the gut and has been reported acting as a possible regulator of cancer cell death (Li et al., 2015, Salimi et al., 2017). It also has important roles in various mechanisms and cellular processes, including cell proliferation, differentiation induced cancer cell growth, DNA double-strand break repair, oxidative stress inhibition and gene expression (Grabarska et al., 2013, Falkenberg and Johnstone, 2014, Eckschlager et al., 2017). It is also known that NaBu induces apoptosis in many cancers (Natoni et al., 2005, Li et al., 2014, Salimi et al., 2017). Moreover, NaBu exhibits good anticancer activity. It can cause relaxation of the chromatin structure and provide improved admission to transcription-related proteins and is therefore widely used in the clinical treatment of many tumours (Jazirehi, 2010).
Breast cancer is the top cancer type in women, other than nonmelanoma skin cancer, and among the cases diagnosed in women, 1 in 4 cancers is breast cancer (Bray et al., 2018, Zendehdel et al., 2018, Waks and Winer, 2019, Sopik, 2021;). Considering the frequency and seriousness of the disease, new therapeutic targets are needed to treat breast cancer. Research into the mechanisms leading to drug resistance in the pathway to treatment success shows that histone deacetylases (HDAC) are promising targets for breast cancer treatment (Hosford and Miller, 2014).
The anti-proliferative and apoptotic effect of NaBu on breast cancer cells is known. In this context, this study aims to evaluate some biochemical and genotoxic changes in the regulation of cell apoptosis caused by NaBu. For this purpose, the effect of NaBu on cell proliferation was determined. Lipid peroxidation (MDA), reduced glutathione (GSH), Nitric Oxide (NO), hydrogen peroxide (H2O2), and superoxide dismutase (SOD) enzyme, which catalyzes superoxide, a product of oxidative stress activities, which are the biochemical parameters of oxidative stress, that has an important role in the apoptotic cell death process, were determined after the most effective dose and time treatment. In addition, oxidative stress parameters of NaBu-induced genotoxic damage were evaluated with single-cell gel electrophoresis (SCGE) analysis, and oxidative stress-induced cell death was evaluated for the first time within the framework of these parameters. Obtained data that may help support the clinical trial of epigenetic treatments for breast cancer.
2. Materials and methods
2.1. Cell culture
In this study, MCF-7 breast cancer cells were used. The cells were commercially acquired from the American Type Culture Collection (ATCC). MCF-7 cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Gibco), including 10% heat-inactivated fetal bovine serum (FBS, Sigma Aldrich) and 0.1% penicillin/streptomycin in a humidified incubator at 37 °C with 5% CO2.
2.2. Cell viability assay
The cytotoxic impact of NaBu on MCF-7 cells was determined via WST-1 analysis. For this purpose, the cells were seeded in 96-well plates at 2 × 104 cells/well and incubated for 24 and 48 h with different concentrations of NaBu (1, 2.5, 5 and 10 mM). After incubation, the cells were incubated with WST-1 solution for 45 min. Later, the cells were analyzed using the optical reader (BioTek Instrument ELISA reader) at 450 nm absorbance. MCF-7 cells were incubated in a growth medium without NaBu accepted as a negative control, and the viability of the negative control group was considered 100%. The most effective exposure concentrations and time were selected for further experiments. Each experiment was performed in triplicate.
2.3. Biochemical analyses
2.3.1. Preparation of the cell lysates
To find out the impact of NaBu on oxidative stress parameters in MCF-7 cells, cell lysates were isolated. To this end, the cells (5 × 105) were seeded in 6-well plates and processed with NaBu (1 and 5 mM) for 24 and 48 h. After treatment, cells were treated with RIPA cell lysis buffer (Sigma Aldrich) and centrifuged at 10,000 rpm for 10 min at 4 °C. The supernatant obtained after centrifugation was utilized to analyze antioxidant enzyme activity and oxidative stress analysis.
2.3.2. Determination of malondialdehyde (MDA) level
Malondialdehyde detection was based on the method of Heath and Packer (Heath and Packer, 1968). TCA (Trichloroacetic acid) solution (20%) containing 100 µl cell lysate and 1400 µl of 0.5% TBA (Thiobarbutyric acid) was mixed. The mixture was incubated in an oven at 100 °C for 60 min. Then, the samples were taken on ice, and when they reached room temperature, they were centrifuged at 1500g for 5 min. The supernatant was read at 532 nm and 600 nm in a spectrophotometer and calculated according to the formula of 155 cm−1 M−1 below.
2.3.3. Determination of hydrogen peroxide (H2O2) and nitric oxide (NO) level
The quantity of hydrogen peroxide was calculated based on the study of Alexieva et al., (2001). A total of 1.5 ml of a solution containing 0.25 ml of cell lysate, 0.25 ml of 100 mM potassium phosphate buffer solution (pH 7) and 1 ml of 1 M potassium iodide (KI) (prepared with fresh bidistilled water) was prepared and mixed. The cell lysate was stored in the dark at +4 °C for 1 h to react with KI. The samples were then measured at 390 nm in a spectrophotometer. Nitric oxide (NO) levels were determined with the Nitrite Oxide Colorimetric Assay kit (Elabscience).
2.3.4. Determination of the SOD enzyme activity
As substrate buffer for SOD enzyme determination, 50 mM potassium phosphate buffer (ph 7.8), 9.9 mM L-methionine, 57 mM NBT (Nitroblue tetrazolium chloride), 0.025% (w/v) Triton X-100, 0.0044% (w/v) riboflavin were prepared. The solution was dissolved by stirring in a mixer for 30 min and then incubated at +4 °C (Dixit et al., 2001). The substrate buffer and 0.05 ml of cell lysate were mixed and incubated under a 21-Watt fluorescent lamp for 15 min. The enzyme-free substrate buffer was kept under light for 15 min and used blindly (Tripathi and Gaur, 2004, Mishra et al., 2006). Then, a sample was estimated at 560 nm in a spectrophotometer, and the amount of enzyme was taken as U mg−1.
2.3.5. Determination of the amount of reduced glutathione (GSH)
Determination of the amount of GSH) was based on the study of Kumar et al., (2011). Cell lysate and sulfosalicylic acid (4%) were mixed and incubated at +4 °C for 1 h and centrifuged at 1200 g at +4 °C for 15 min. The supernatant was added to 2.7 ml of potassium phosphate buffer (0.1 M, pH 7.4) and 200 µl of 5.5 dithiobis-(2-nitro benzoic acid) (Ellman's reagent, 0.1 mM, pH 8.0). The samples were measured at 412 nm in a spectrophotometer, and the buffer was used as a blank. GSH level was calculated according to the formula 1.36 × 104 M−1 cm−1 and expressed as micromole per milligram protein.
2.4. Single-cell gel electrophoresis (SCGE) analysis
The cells were seeded in 6-well plates at 1 × 105 cells/well and incubated with different concentrations of NaBu (1 and 5 mM) for 24 and 48 h. After incubation, cells were harvested by trypsinization resuspended in ice-cold PBS. After the slides were first covered with 1% NMP (Normal melting point agar), 75 µl of 1% LMP (Low melting point, prepared with 0.01 M PBS) agar and 50 µl of cell suspension were added and incubated on ice for 15 min. Slides were incubated in lysis solution (2.5 M NaCl, 100 mM Na2EDTA, 10 mM Tris, 1% Triton X-100, and pH 10) for 1 h. Then, they were kept in electrophoresis buffer (300 mM NaOH, 1 mM Na2EDTA, pH > 13) at +4 °C for 15 min. Then, electrophoresis was performed at 25 V, 300 mA for 15 min. After electrophoresis, the slides were kept in cold 0.4 M Tris buffer solution (pH 7.5), distilled water and finally 96% ethanol. The slides were stained with 60 µl (2 µg/ml) Ethidium Bromide, washed twice with distilled water, covered with a coverslip, and examined under a fluorescent microscope (DP71, Olympus) on a 515–560 nm filter. A total of 150 comets were counted, 50 from each slide and my camera was analyzed with CometScore Software.
2.5. Statistical analysis
During statistical analysis, one-way variance analysis (ANOVA) with Tukey's post hoc and Mann‐Whitney U tests were used. Obtained results were reported as a mean ± standard error of the mean (SD), and all experiments were conducted in triplicate. The p < .05 value was statistically significant, and GraphPad Prism 16 was employed while conducting statistical analysis.
3. Results
3.1. Evaluation of the cytotoxic effect of NaBu on MCF-7 cells
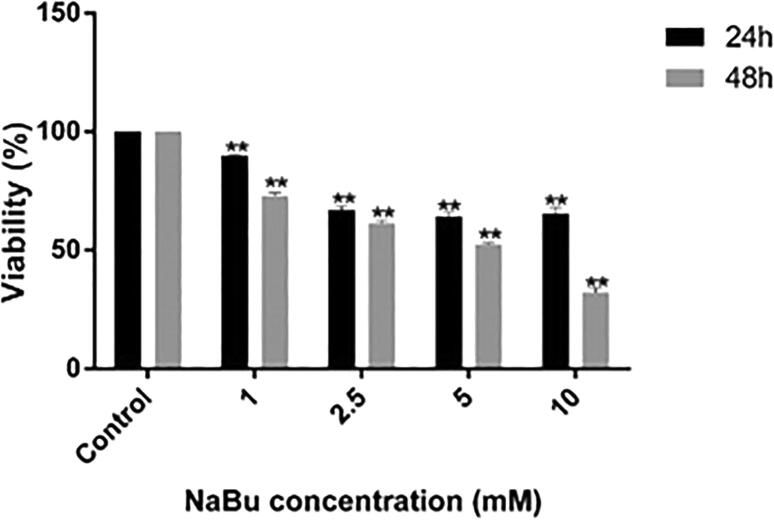
To estimate the most effective exposure concentrations and time of Nabu, WST-1 analysis was conducted. The results showed that NaBu exerted a cytotoxic effect on MCF-7 cells in a dose and time-dependent manner, as shown in Fig. 1. After 24 h of treatment with NaBu (1, 2.5, 5 and 10 mM), the growth rate of MCF-7 cells significantly reduced to 90.5%, 68.1%, 65.5% and 67.8%, respectively (p < .001; Fig. 1). Additionally, the viability of MCF-7 cells significantly reduced to 73,8%, 62,1%, 52,8%, and 33,4 % at concentration of 1, 2.5, 5 and 10 mM NaBu, respectively for 48 h. As a result, our findings showed that the most effective exposure concentrations and time of NaBu were 1 mM and 5 mM and 24 h and 48 h, respectively. Therefore, 1 and 5 mM NaBu treatment were selected at 24 and 48 h time for further experiments.
Fig. 1.
The effects of NaBu on the cell viability of MCF-7 cells were determined by WST-1 analysis for 24 and 48 h (**p < .001).
3.2. The effects of NaBu on the MDA amounts
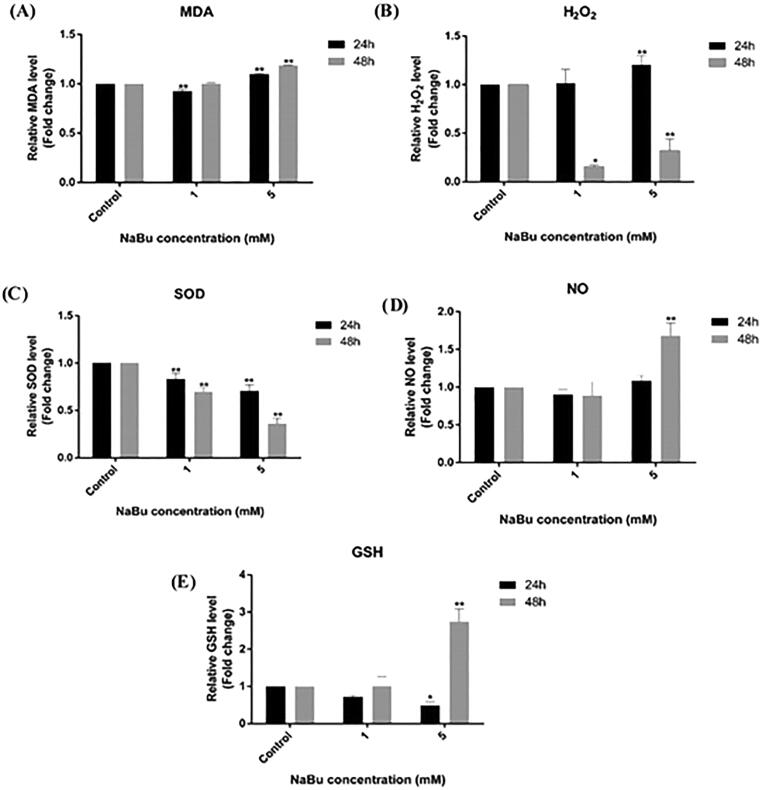
Our results determined that the amount of MDA (2.84 µM) of the cells incubated with 1 mM NaBu for 24 h decreased slightly compared to the control cells that were not treated with NaBu (3.07 µM). On the other hand, the data obtained with 5 mM NaBu showed an increase in the amount of MDA in MCF-7 cells compared to the control group (3.12 µM, 1.09-fold). At the end of the 48 h, the same results were obtained in cells treated with 1 mM NaBu (2.59 µM), compared to the control group, while an increase was observed in 5 mM NaBu treatments (3.085 mM, 1.18-fold) (Fig. 2). The mean values and statistical data of the changes in the amount of MDA in the control group, 1 mM NaBu and 5 mM NaBu treated MCF-7 cells, are given in Table 1.
Fig. 2.
The amounts of (A) MDA, (B) H2O2, (C) SOD, (D) NO and (E) GSH at the end of 24 and 48 h in the control group and NaBu-treated MCF-7 cells. The group without NaBu was determined as the negative control and the changes in the determined parameter (**p < .001).
Table 1.
The mean values (), standard deviations (SD) and statistical data of the changes in the amounts of MDA, H2O2, SOD, NO, GSH in the control group, 1 mM NaBu and 5 mM NaBu treated MCF-7 cells.
| Time (Hours) |
NaBu -Control ± SD |
MDA (µM) ± SD |
H2O2 (µM) ± SD |
SOD U/mg ± SD |
NO (µM) ± SD |
GSH (µM) ± SD |
|---|---|---|---|---|---|---|
| 24 h | Control | 3.07 ± 0.01 | 5.18 ± 0.41 | 10.71 ± 0.73 | 0.92 ± 0.04 | 0.02 ± 0.01 |
| 1 mM | 2.84 ± 0.02** | 5.20 ± 0,25 | 8.92 ± 0.81** | 0.83 ± 0.01 | 0.01 ± 0.02 | |
| 5 mM | 3.12 ± 0.01** | 6.28 ± 0.70** | 6.24 ± 0.13** | 0.90 ± 0.05 | 0.01 ± 0.01* | |
| 48 h | Control | 2.59 ± 0.02 | 8.81 ± 0.47 | 10.94 ± 0.34 | 6.11 ± 0.38 | 0.01 ± 0.02 |
| 1 mM | 2.59 ± 0.09 | 1.34 ± 0.09* | 7.64 ± 0.40** | 5.49 ± 0.16 | 0.01 ± 0.02 | |
| 5 mM | 3.08 ± 0.01** | 0.44 ± 0.16** | 2.77 ± 0.46** | 7.22 ± 0.19** | 0.02 ± 0.01** | |
(*p < .05, ** p < .001).
3.3. The effects of NaBu on the H2O2 amounts
The H2O2 amounts of cells treated with 1 mM NaBu and 5 mM NaBu for 24 h increased compared to the control group (5.18 µM) (5.20 µM, 6.28 µM; 1.01 and 1.2-fold, respectively) (Fig. 2). In the data we received at the end of 48 h, a very serious decrease was observed in the amount of H2O2. As a result of 1 mM, NaBu and 5 mM, NaBu treatments compared to the control group (8.81 µM), 1.34 µM and 0.44 µM were determined in MCF-7 cells, respectively. The mean values and statistical data of changes in H2O2 amounts in MCF-7 cells treated with the control group, 1 mM NaBu and 5 mM NaBu are given in Table 1.
3.4. The effects of NaBu on the SOD activity
In our studies to determine the SOD enzyme activity, it was determined that the activities of the SOD enzyme decreased with increasing NaBu concentration compared to the control cells in both the 24-hour and 48-hour groups. In NaBu treatments, MCF-SOD enzyme activities were determined as 8.92 U/mg and 6.24 U/mg, respectively. After 48 h, 10.94 U/mg activity in the control group, 7.64 U/mg in 1 mM NaBu treatments, and 2.77 U/mg in 5 mM NaBu treatments were detected (Table 1). The mean values and statistical data of the changes in SOD activities in MCF-7 cells treated with 1 mM NaBu and 5 mM NaBu in the control group are given in Table 1. In contrast, the fold change in enzyme activities is given in Fig. 2.
3.5. The effects of NaBu on the NO levels
It was determined that there were no changes in nitric oxide (NO) amounts in the experimental group for 24 h compared to the control (0.92 µM) (0.83 µM and 0.92 µM) (Table 1). In the 48-hour experimental group, a slight decrease was observed in the 1 mM NaBu treatments (5.49 µM) compared to the control group (6.11 µM), while a 1.3-fold increase was observed in the 5 mM NaBu (7.22 µM) treatment (Fig. 2). The mean values and statistical data of the changes in NO amounts in the control group, 1 mM NaBu and 5 mM NaBu treated MCF-7 cells, are given in Table 1.
3.6. The effects of NaBu on the GSH amount
In our GSH data, there was a decrease in NaBu treatments in our 24-hour experimental group when the results were compared with those of the control group. Moreover, there was a 2.7-fold increase in the amount of GSH in the 5 mM NaBu treatments in our experimental group at the end of 48 h compared to the control group (Fig. 2). The mean values and statistical data of the changes in the amount of GSH in the control group, MCF-7 cells treated with 1 mM NaBu and 5 mM NaBu, are given in Table 1.
3.7. Evaluation of the genotoxic effects of NaBu
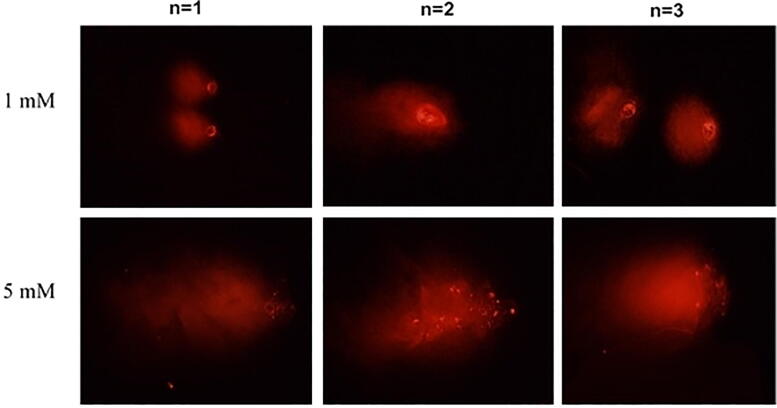
DNA damage (head DNA (HDNA%), tail DNA (TDNA%), tail moment, olive tail moment (OTM) data in the control group, MCF-7 cells treated with 1 mM NaBu and 5 mM NaBu at the end of 48 h given in Table 2. Since there is no effective result after 24 h, only 48-hour results are given. According to these data, OTM increases with 1 mM NaBu and 5 mM NaBu concentrations (25.63 μm; 37.26 μm, respectively) compared to the control group (23.39 μm) at the end of the 48th hour. In TDNA%, an increase was detected compared to the control groups in both NaBu treatments at the end of 48 h (Table 2). In contrast to the increase seen in TDNA% and OTM, a decrease was detected in the HDNA% value compared to the control group (20.99 μm), 1 mM NaBu and 5 mM NaBu (15.95 μm and 13.17 μm) treatment (Fig. 3).
Table 2.
DNA damage (head DNA (%), tail DNA (%), tail moment, olive tail moment) in the MCF-7 cells treated with 1 mM NaBu and 5 mM NaBu compared to the control group for 48 h.
| Time (Hours) |
NaBu-Control | Head DNA (%) ± SD |
Tail DNA (%) ± SD |
Tail moment (μm) ± SD |
Olive tail moment (μm) ± SD |
|---|---|---|---|---|---|
| 48 h | Control | 20.99 ± 2.48 | 79.01 ± 2.48 | 29.65 ± 3.24 | 23.39 ± 3.08 |
| 1 mM | 15.95 ± 2.79 | 84.05 ± 2.79 | 30.23 ± 0.51 | 25.63 ± 0.40 | |
| 5 mM | 13.17 ± 6.46 | 86.83 ± 6.46 | 42.66 ± 5.91 | 37.26 ± 7.00 |
Fig. 3.
The images of NaBu induced genotoxic effect determined by comet assay in MCF-7 cells after 48 h. The images were shown in triplicate (n = 3).
4. Discussion
Regulation of the homeostatic mechanism of cancer cell formation and metastasis depends on the balance between cancer cell growth and death (Mirzaei et al., 2016, Vafaei et al., 2019). In this context, some biochemical and genotoxic changes in the regulation of cell apoptosis caused by NaBu were evaluated together with this study. Our results demonstrated that NaBu reduced cell viability dose and time-dependent. Additionally, NaBu potentially induced GSH but decreased SOD enzyme activity at higher concentrations, but the level of MDA and NO decreased at a low concentration of NaBu. Also, H2O2 decreased after incubation with higher and lower concentrations of NaBu for only 48 h treatment, not 24 h in MCF-7 cells. Our comet assay results determined that higher concentrations of NaBu treatments increased DNA damage in MCF-7 cells compared to the control group, inconsistent with the biochemical parameter results. NaBu shows its anticancer and genotoxic effects in breast cancer, especially through antioxidant enzymes, one of the oxidative stress parameters. However, the anticancer and genotoxic effects of NaBu are changes in the oxidative stress parameters with time and treatment concentration of NaBu in MCF-7 cells.
Mutation in DNA repair mechanisms due to any factor can cause MDA-DNA adducts mutations (dot and frameshift), strand breaks, cell cycle arrest, and induction of apoptosis (Valko et al., 2007, Sies, 2015, Liguori et al., 2018). In recent studies, it has been shown by various researchers that the amount of MDA increased compared to the control group (Gonenc et al., 2001, Chole et al., 2010, Rašić et al., 2018). In the data we obtained, a slight decrease in the amount of MDA in the cells incubated with low-dose NaBu at the end of 24 h is an indication that NaBu reduces lipid peroxidation against cancer cells, while detection of a very slight increase in high-dose NaBu treatments at the end of 48 h supports related data.
An upsurge in cellular levels of H2O2 has been associated with some major changes in cancer, such as DNA alterations, cell proliferation, metastasis, angiogenesis, and hypoxia-inducible factor 1 (HIF-1) activation (Graves, 2012, Pizzino et al., 2017). In contrast, it was also observed that H2O2 could selectively stimulate apoptosis in cancer cells and partially mediated the activity of several anticancer drugs frequently employed in clinical practice (Li et al., 2017, Rahman et al., 2019). Consistent with the literature, the very significant decrease in H2O2 amounts following NaBu treatment in the data we obtained at the end of 48 h reveal the H2O2 mediated anticancer activity of NaBu.
While many cancer cells have low levels of MnSOD proteins and enzymatic activity, some cancer cells have been reported to have high levels of MnSOD expression and activity (Dhar and Clair, 2012). Extracellular SOD (EcSOD) expression levels are significantly reduced in most cancers, including breast, head and neck, lung and sarcoma (Teng et al., 2012, Griess et al., 2017). Despite the marked decrease in EcSOD expression in breast carcinomas, an inverse correlation has been reported between EcSOD mRNA expression levels and breast cancer stage (Hubackova et al., 2012, Teoh-Fitzgerald et al., 2014). However, the Determination of the SOD level by specific oncogenic separators or by the general state of the entire redox system can make the situation more complex. Papa et al., (2014) showed in their study that decreased Cu/ZnSOD expression in some breast cancer cells caused a compensatory increase in MnSOD. In the data we obtained from our study, the activities of the SOD enzyme decreased with increasing NaBu concentration. As stated in the literature, this decrease, contrary to expectations, shows that there are significant changes in the level of SOD among cancer cells, indicating that there is differential regulation of SOD in cancer cells, and this regulation may be associated with a typical stage of cancer development.
Nitric oxide (NO) is a short-lived gas that is produced endogenously by Nitric oxide synthase (NOS) enzymes in the body and acts as a key signalling molecule in various physiological processes (Choudhari et al., 2013, Galadari et al., 2017). While nitric oxide (NO) plays a role as a mediator of the cancer phenotype, in some cases, it is also being investigated for therapeutic purposes due to its tumour suppressor properties (Choudhari et al., 2013, Li et al., 2017, Smeda et al., 2018). In the data we obtained, an increase was detected in the amount of NO in high dose NaBu treatment for 48 h, while a decrease was observed after low dose NaBu treatment after 24 h of treatment. The chemical and biochemical properties of NO, its interactions with cellular targets differ greatly due to the heterogeneity of tumours (Somasundaram et al., 2019, Khan et al., 2020). Understanding the complexity of NO's role in cancer will contribute to cancer treatment processes. NO seems to have a stimulating or inhibitory role on cancer depending on various factors such as time and dose. The molecular events associated with these differences need to be studied extensively.
GSH has an important impact in the detoxification of carcinogens and appears as a target in developing new strategies to improve cancer treatment (Estrela et al., 2006, Ballatori et al., 2009). In our data, a significant increase in the amount of GSH, especially following high-dose NaBu treatment, reveals the GSH-mediated anticancer activity of NaBu in breast cancer cells.
DNA damages occur due to acute outcomes stemming from impaired DNA metabolism, stopping cell cycle progression or causing cell death (Liao et al., 2009). Single Cell Gel electrophoresis – i.e., a comet assay – is a method employed to determine DNA damages (Collins, 2004, Møller et al., 2020.). The induction of DNA damage in cancer cells is a famous therapeutic technique for killing cancer (Liao et al., 2009). Researchers have reported that treatment with NaBu increases human endometrial cancer cell line intracellular ROS production and DNA damage response signals and induces DNA damage (Kato et al., 2011, Pant et al., 2017). Our results determined that NaBu treatments increased DNA damage in MCF-7 cells compared to the control group in Comet assay data. HDAC inhibitors can be used as antitumor therapy according to their inhibitory impact on cancer cells.
5. Conclusions
The data we obtained revealed that NaBu shows its anticancer and genotoxic effects in breast cancer, especially through antioxidant enzymes, one of the oxidative stress parameters. However, this oxidative stress-dependent effect changes depending on dose and time, and this change needs to be clarified by further evaluation with molecular and more biochemical parameters. It is supported by the data we obtained that the detection of additional targets in breast cancer is highly encouraging. Prospective therapy developments will involve HDAC inhibitors and combination therapy using chemotherapy or other inhibitors.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
Special thanks to Dr Ozlem AKSOY from the Department of Biology of Kocaeli University for providing the laboratory facilities.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Footnotes
Peer review under responsibility of King Saud University.
References
- Alexieva V., Sergiev I., Mapelli S., Karanov E. Drought and ultraviolet radiation affect growth and stress markers in pea and wheat. Plant Cell and Environ. 2001;24:1337–1344. doi: 10.1046/j.1365-3040.2001.00778.x. [DOI] [Google Scholar]
- Atala E., Fuentes J., Wehrhahn M.J., Speisky H. Quercetin and related flavonoids conserve their antioxidant properties despite undergoing chemical or enzymatic oxidation. Food Chem. 2017;234:479–485. doi: 10.1016/j.foodchem.2017.05.023. [DOI] [PubMed] [Google Scholar]
- Ayala A., Muñoz M.F., Argüelles S. Lipid peroxidation: production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxid. Med. Cell. Longevity. 2014;2014:1–31. doi: 10.1155/2014/360438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballatori N., Krance S.M., Notenboom S., Shi S., Tieu K., Hammond C.L. Glutathione dysregulation and the etiology and progression of human diseases. Biol. Chem. 2009;390:191–214. doi: 10.1515/BC.2009.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer G. The synergistic effect between hydrogen peroxide and nitrite, two long-lived molecular species from cold atmospheric plasma, triggers tumor cells to induce their own cell death. Redox Biol. 2019;26:101291. doi: 10.1016/j.redox.2019.101291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolden J.E., Peart M.J., Johnstone R.W. Anticancer activities of histone deacetylase inhibitors. Nat. Rev. Drug. Discov. 2006;5(9):769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- Chio I.I.C., Tuveson D.A. ROS in cancer: the burning question. Trends. Mol. Med. 2017;23(5):411–429. doi: 10.1016/j.molmed.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chole R.H., Patil R.N., Basak A., Palandurkar K., Bhowate R. Estimation of serum malondialdehyde in oral cancer and precancer and its association with healthy individuals, gender, alcohol, and tobacco abuse. J. Cancer Res. Ther. 2010;6:487–491. doi: 10.4103/0973-1482.77106. [DOI] [PubMed] [Google Scholar]
- Choudhari S.K., Chaudhary M., Bagde S., Gadbail A.R., Joshi V. Nitric oxide and cancer: a review. World J. Surg. Oncol. 2013;11:118. doi: 10.1186/1477-7819-11-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins A.R. The comet assay for DNA damage and repair: principles, applications, and limitations. Mol. Biotechnol. 2004;26(3):249–261. doi: 10.1385/MB:26:3:249. [DOI] [PubMed] [Google Scholar]
- Del Rio D., Stewart A.J., Pellegrini N. A review of recent studies on malondialdehyde as toxic molecule and biological marker of oxidative stress. Nutr. Metab. Cardiovasc. Dis. 2005;15:316–328. doi: 10.1016/j.numecd.2005.05. [DOI] [PubMed] [Google Scholar]
- Dhar S.K., St. Clair D.K. Manganese superoxide dismutase regulation and cancer. Free Radic. Biol. Med. 2012;52(11-12):2209–2222. doi: 10.1016/j.freeradbiomed.2012.03.009. [DOI] [PubMed] [Google Scholar]
- Dixit V., Pandey V., Shyam R. Differential antioxidative responses to cadmium in roots and leaves of pea (Pisum sativum L. cv. Azad) J. Exp. Bot. 2001;52:1101–1109. doi: 10.1093/jexbot/52.358.1101. [DOI] [PubMed] [Google Scholar]
- Eckschlager T., Plch J., Stiborova M., Hrabeta J. Histone deacetylase inhibitors as anticancer drugs. Int. J. Mol. Sci. 2017;18:1414. doi: 10.3390/ijms18071414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrela J.M., Ortega A., Obrador E. Glutathione in cancer biology and therapy. Crit. Rev. Clin. Lab. Sci. 2006;43(2):143–181. doi: 10.1080/10408360500523878. [DOI] [PubMed] [Google Scholar]
- Falkenberg K.J., Johnstone R.W. Histone deacetylases and their inhibitors in cancer, neurological diseases, and immune disorders. Nat. Rev. Drug. Discov. 2014;13(9):673–691. doi: 10.1038/nrd4360. [DOI] [PubMed] [Google Scholar]
- Fang J., Seki T., Maeda H. Therapeutic strategies by modulating oxygen stress in cancer and inflammation. Adv. Drug. Deliv. Rev. 2009;61(4):290–302. doi: 10.1016/j.addr.2009.02.005. [DOI] [PubMed] [Google Scholar]
- Galadari S., Rahman A., Pallichankandy S., Thayyullathil F. Reactive oxygen species and cancer paradox: to promote or to suppress? Free Radical Bio. and Med. 2017;104:144–164. doi: 10.1016/j.freeradbiomed.2017.01.004. [DOI] [PubMed] [Google Scholar]
- Glozak M.A., Sengupta N., Zhang X., Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Gonenc A., Ozkan Y., Torun M., Simsek B. Plasma malondialdehyde (MDA) levels in breast and lung cancer patients. J. Clin. Pharm. Ther. 2001;26(2):141–144. doi: 10.1046/j.1365-2710.2001.00334.x. [DOI] [PubMed] [Google Scholar]
- Grabarska A., Dmoszyńska-Graniczka M., Nowosadzka E., Stepulak A. Histone deacetylase inhibitors - molecular mechanisms of actions and clinical applications. Postepy Hig. Med. Dosw. 2013;67:722–735. doi: 10.5604/17322693.1061381. [DOI] [PubMed] [Google Scholar]
- Graves D.B. The emerging role of reactive oxygen and nitrogen species in redox biology and some implications for plasma treatments to medicine and biology. J. Phys. D. 2012;45:263001. [Google Scholar]
- Griess B., Tom E., Domann F., Teoh-Fitzgerald M. Extracellular superoxide dismutase and its role in cancer. Free Radic. Biol. Med. 2017;112:464–479. doi: 10.1016/j.freeradbiomed.2017.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath R.L., Packer L. Photoperoxidation in isolated chloroplasts. I. Kinetics and stoichiometry of fatty acid peroxidation. Arch. Biochem. Biophys. 1968;125(1):189–198. doi: 10.1016/0003-9861(68)90654-1. [DOI] [PubMed] [Google Scholar]
- Hosford S.R., Miller T.W. Clinical potential of novel therapeutic targets in breast cancer: CDK4/6, Src, JAK/STAT, PARP, HDAC, and PI3K/AKT/mTOR pathways. Pharmgenomics Pers. Med. 2014;7:203–215. doi: 10.2147/PGPM.S52762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P., Feng L.i., Oldham E.A., Keating M.J., Plunkett W. Superoxide dismutase as a target for the selective killing of cancer cells. Nature. 2000;407(6802):390–395. doi: 10.1038/35030140. [DOI] [PubMed] [Google Scholar]
- Hubackova M., Vaclavikova R., Ehrlichova M., Mrhalova M., Kodet R., Kubackova K., Vrána D., Gut I., Soucek P. Association of superoxide dismutases and NAD(P)H quinone oxidoreductases with prognosis of patients with breast carcinomas. Int. J. Cancer. 2012;130(2):338–348. doi: 10.1002/ijc.26006. [DOI] [PubMed] [Google Scholar]
- Isnaini I., Permatasari N., Mintaroem K., Prihardina B., Widodo M.A. Oxidants-antioxidants profile in the breast cancer cell line MCF-7. Asian Pac. J. Cancer Prev. 2018;19:3175–3178. doi: 10.31557/APJCP.2018.19.11.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazirehi A.R. Regulation of apoptosis-associated genes by histone deacetylase inhibitors: implications in cancer therapy. Anticancer Drugs. 2010;21:805–813. doi: 10.1097/CAD.0b013e32833dad91. [DOI] [PubMed] [Google Scholar]
- Kato K., Kuhara A., Yoneda T., Inoue T., Takao T., Ohgami T., Dan L.i., Kuboyama A., Kusunoki S., Takeda S., Wake N. Sodium butyrate inhibits the self-renewal capacity of endometrial tumor side-population cells by inducing a DNA damage response. Mol. Cancer. Ther. 2011;10(8):1430–1439. doi: 10.1158/1535-7163.MCT-10-1062. [DOI] [PubMed] [Google Scholar]
- Khan M.I., Mohammad A., Patil G., Naqvi S.A.H., Chauhan L.K.S., Ahmad I. Induction of ROS, mitochondrial damage and autophagy in lung epithelial cancer cells by iron oxide nanoparticles. Biomater. 2012;33(5):1477–1488. doi: 10.1016/j.biomaterials.2011.10.080. [DOI] [PubMed] [Google Scholar]
- Khan F.H., Dervan E., Bhattacharyya D.D., McAuliffe J.D., Miranda K.M., Glynn S.A. The role of nitric oxide in cancer: master regulator or NOt? Int. J. Mol. Sci. 2020;21:9393. doi: 10.3390/ijms21249393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar B., Kuhad A., Chopra K. Neuropsychopharmacological effect of sesamol in unpredictable chronic mild stress model of depression. Behav. Biochem. Evidence. 2011;214:819–828. doi: 10.1007/s00213-010-2094-2. [DOI] [PubMed] [Google Scholar]
- Li L., Sun Y., Liu J., Wu X., Chen L., Ma L.i., Wu P. Histone deacetylase inhibitor sodium butyrate suppresses DNA double-strand break repair induced by etoposide more effectively in MCF-7 cells than in HEK293 cells. BMC Biochem. 2015;16(1):2. doi: 10.1186/s12858-014-0030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Ho Kang M., Sup Uhm H., Joon Lee G., Ha Choi E., Han I. Effects of atmospheric-pressure non-thermal bio-compatible plasma and plasma-activated nitric oxide water on cervical cancer cells. Sci. Rep. 2017;7:45781. doi: 10.1038/srep45781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Wang Y., Yuan C., Zhu Y., Qiu J., Zhang W., Qi B., Wu H., Ye J., Jiang H., Yang J., Cheng J. Oncogenic roles of Bmi1 and its therapeutic inhibition by histone deacetylase inhibitor in tongue cancer. Lab. Invest. 2014;94(12):1431–1445. doi: 10.1038/labinvest.2014.123. [DOI] [PubMed] [Google Scholar]
- Liao W., McNutt M.A., Zhu W.-G. The comet assay: a sensitive method for detecting DNA damage in individual cells. Methods. 2009;48(1):46–53. doi: 10.1016/j.ymeth.2009.02.016. [DOI] [PubMed] [Google Scholar]
- Liguori I., Russo G., Curcio F. Oxidative stress, aging, and diseases. Clin. Interv Aging. 2018;13:757–772. doi: 10.2147/CIA.S158513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locigno R., Castronovo V. Reduced glutathione system: role in cancer development, prevention and treatment. Inter. J. Oncol. 2001;19:221–236. doi: 10.3892/ijo.19.2.221. [DOI] [PubMed] [Google Scholar]
- López-Lázaro M. Dual role of hydrogen peroxide in cancer: possible relevance to cancer chemoprevention and therapy. Cancer Lett. 2007;252(1):1–8. doi: 10.1016/j.canlet.2006.10.029. [DOI] [PubMed] [Google Scholar]
- Shazia M., Imran K., Jasmine F., Mohd S., Huma M. Anti-bacterial, anti-oxidant and cytotoxicity of aqueous and organic extracts of Ricinus communis. African J. of Microbiology Res. 2016;10(8):260–270. doi: 10.5897/AJMR2015.7397. [DOI] [Google Scholar]
- Mirzaei A., Tavoosidana G., Rad A.A., Rezaei F., Tavakoli-Yaraki M., Kadijani A.A., Khalili E., Madjd Z. A new insight into cancer stem cell markers: Could local and circulating cancer stem cell markers correlate in colorectal cancer? Tumour Biol. 2016;37(2):2405–2414. doi: 10.1007/s13277-015-3989-7. [DOI] [PubMed] [Google Scholar]
- Mishra S., Srivastava S., Tripathi R.D., Govindarajan R., Kuriakose S.V., Prasad M.N. Phytochelatin synthesis and response of antioxidants during cadmium stress in Bacopa monnieri L. Plant Physiol. Biochem. 2006;44:25–37. doi: 10.1016/j.plaphy.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Moloney J.N., Cotter T.G. ROS signalling in the biology of cancer. Semin. Cell Dev. Biol. 2018;80:50–64. doi: 10.1016/j.semcdb.2017.05.023. [DOI] [PubMed] [Google Scholar]
- Møller P., Stopper H., Collins A.R. Measurement of DNA damage with the comet assay in high-prevalence diseases: current status and future directions. Mutagenesis. 2020;35:5–18. doi: 10.1093/mutage/gez018. [DOI] [PubMed] [Google Scholar]
- Natoni F., Diolordi L., Santoni C., Gilardini Montani M.S. Sodium butyrate sensitizes human pancreatic cancer cells to both the intrinsic and the extrinsic apoptotic pathways. Biochim. Biophys. Acta. 2005;1745:318–329. doi: 10.1016/j.bbamcr.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Pant K., Yadav A.K., Gupta P., Islam R., Saraya A., Venugopal S.K. Butyrate induces ROS-mediated apoptosis by modulating miR-22/SIRT-1 pathway in hepatic cancer cells. Redox Bio. 2017;12:340–349. doi: 10.1016/j.redox.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa L., Hahn M., Marsh E.L., Evans B.S., Germain D. SOD2 to SOD1 switch in breast cancer. J. Biol. Chem. 2014;289(9):5412–5416. doi: 10.1074/jbc.C113.526475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzino G., Irrera N., Cucinotta M., Pallio G., Mannino F., Arcoraci V., Squadrito F., Altavilla D., Bitto A. Oxidative stress: harms and benefits for human health. Oxid Med. Cell. Longev. 2017;2017:1–13. doi: 10.1155/2017/8416763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman A., Pallichankandy S., Thayyullathil F., Galadari S. Critical role of H2O2 in mediating sanguinarine-induced apoptosis in prostate cancer cells via facilitating ceramide generation, ERK1/2 phosphorylation, and Par-4 cleavage. Free Radical Bio. and Med. 2019;134:527–544. doi: 10.1016/j.freeradbiomed.2019.01.039. [DOI] [PubMed] [Google Scholar]
- Rašić I., Rašić A., Akšamija G., Radović S. The relationship between serum level of malondialdehyde and progression of colorectal cancer. Acta Clin. Croat. 2018;57:411–416. doi: 10.20471/acc.2018.57.03.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salimi V., Shahsavari Z., Safizadeh B., Hosseini A., Khademian N., Tavakoli-Yaraki M. Sodium butyrate promotes apoptosis in breast cancer cells through reactive oxygen species (ROS) formation and mitochondrial impairment. Lipids Health Dis. 2017;16:208. doi: 10.1186/s12944-017-0593-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sies H. Oxidative stress: a concept in redox biology and medicine. Redox Biol. 2015;4:180–183. doi: 10.1016/j.redox.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeda M., Kieronska A., Adamski M.G., Proniewski B., Sternak M., Mohaissen T., Przyborowski K., Derszniak K., Kaczor D., Stojak M., Buczek E., Jasztal A., Wietrzyk J., Chlopicki S. Nitric oxide deficiency and endothelial-mesenchymal transition of pulmonary endothelium in the progression of 4T1 metastatic breast cancer in mice. Breast Cancer Res. 2018;20(1) doi: 10.1186/s13058-018-1013-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somasundaram V., Basudhar D., Bharadwaj G., No J.H., Ridnour L.A., Cheng R.Y.S., Fujita M., Thomas D.D., Anderson S.K., McVicar D.W., Wink D.A. Molecular mechanisms of nitric oxide in cancer progression, signal transduction, and metabolism. Antioxid. Redox. Signal. 2019;30(8):1124–1143. doi: 10.1089/ars.2018.7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopik V. International variation in breast cancer incidence and mortality in young women. Breast Cancer Res. Treat. 2021;186(2):497–507. doi: 10.1007/s10549-020-06003-8. [DOI] [PubMed] [Google Scholar]
- Teng Y.-F., Aquino R.S., Park P.W. Molecular functions of syndecan-1 in disease. Matrix bio. 2012;31(1):3–16. doi: 10.1016/j.matbio.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teoh-Fitzgerald M.L., Fitzgerald M.P., Zhong W., Askeland R.W., Domann F.E. Epigenetic reprogramming governs EcSOD expression during human mammary epithelial cell differentiation, tumorigenesis and metastasis. Oncogene. 2014;33(3):358–368. doi: 10.1038/onc.2012.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traverso N., Ricciarelli R., Nitti M., Marengo B., Furfaro A.L., Pronzato M.A., Marinari U.M., Domenicotti C. Role of glutathione in cancer progression and chemoresistance. Oxid. Med. Cell. Longevity. 2013;2013:1–10. doi: 10.1155/2013/972913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi B.N., Gaur J.P. Relationship between copper- and zinc- induced oxidative stress and proline accumulation in Scenedesmus sp. Planta. 2004;219:397–404. doi: 10.1007/s00425-004-1237-2. [DOI] [PubMed] [Google Scholar]
- Valko M., Leibfritz D., Moncol J., Cronin M.T.D., Mazur M., Telser J. Free radicals and antioxidants in normal physiological functions and human disease. Int. J. Biochem. Cell Biol. 2007;39(1):44–84. doi: 10.1016/j.biocel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Waks A.G., Winer E.P. Breast cancer treatment: a review. Jama. 2019;321(3):288. doi: 10.1001/jama.2018.19323. [DOI] [PubMed] [Google Scholar]
- Vafaei S., Fattahi F., Ebrahimi M., Janani L., Shariftabrizi A., Madjd Z. Common molecular markers between circulating tumor cells and blood exosomes in colorectal cancer: a systematic and analytical review. Cancer Manage. Res. 2019;11:8669. doi: 10.2147/CMAR.S219699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi J.F., Li Y.M., Liu T., He W.T., Li X., Zhou W.C., Kang S.L., Zeng X.T., Zhang J.Q. Mn-SOD and CuZn-SOD polymorphisms and interactions with risk factors in gastric cancer. World J. Gastroenterol. 2010;16:4738–4746. doi: 10.3748/wjg.v16.i37.4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuksel B., Deveci Ozkan A. The role of citrus nobiletin on oxidative stress levels and superoxide dismutase activities in metastatic castration-resistant prostate cancer. Commagene Biol. 2021;5:84–89. doi: 10.31594/commagene.895415. [DOI] [Google Scholar]
- Zendehdel M., Niakan B., Keshtkar A., Rafiei E., Salamat F. Subtypes of benign breast disease as a risk factor for breast cancer: a systematic review and meta-analysis protocol. Iranian J. Med. Sci. 2018;43:1–8. [PMC free article] [PubMed] [Google Scholar]