Summary
Although the typical genomic and phenotypic changes that characterize the evolution of organisms under the human domestication syndrome represent textbook examples of rapid evolution, the molecular processes that underpin such changes are still poorly understood. Domesticated yeasts for brewing, where short generation times and large phenotypic and genomic plasticity were attained in a few generations under selection, are prime examples. To experimentally emulate the lager yeast domestication process, we created a genetically complex (panmictic) artificial population of multiple Saccharomyces eubayanus genotypes, one of the parents of lager yeast. Then, we imposed a constant selection regime under a high ethanol concentration in 10 replicated populations during 260 generations (6 months) and compared them with propagated controls exposed solely to glucose. Propagated populations exhibited a selection differential of 60% in growth rate in ethanol, mostly explained by the proliferation of a single lineage (CL248.1) that competitively displaced all other clones. Interestingly, the outcome does not require the entire time‐course of adaptation, as four lineages monopolized the culture at generation 120. Sequencing demonstrated that de novo genetic variants were produced in all propagated lines, including SNPs, aneuploidies, INDELs and translocations. In addition, the different propagated populations showed correlated responses resembling the domestication syndrome: genomic rearrangements, faster fermentation rates, lower production of phenolic off‐flavours and lower volatile compound complexity. Expression profiling in beer wort revealed altered expression levels of genes related to methionine metabolism, flocculation, stress tolerance and diauxic shift, likely contributing to higher ethanol and fermentation stress tolerance in the evolved populations. Our study shows that experimental evolution can rebuild the brewing domestication process in ‘fast motion’ in wild yeast, and also provides a powerful tool for studying the genetics of the adaptation process in complex populations.
Here, we propagated panmictic S. eubayanus lines to generate novel strains with greater fermentation capacity. These individuals exhibited different domestication hallmarks, representative of the adaptation process during brewing in lager yeast.
Introduction
Living organisms are continually adapting to changing environments by natural selection, latently harbouring the raw genetic variation required for such responses. When new conditions arise, adaptation to almost every environmental scenario is possible (e.g. temperature, oxygen and nutrients) (Causton et al., 2001; Tamari et al., 2016). In this context, the genomic analysis of human‐made populations (i.e. population genomics of domesticated species) is a relatively new matter and constitutes a promising research approach for the experimental study of evolutionary processes (Sheppard et al., 2018). Nevertheless, studies that search for the causal factors shaping the genetic structure of yeast and fungal populations, such as small nucleotide polymorphisms (SNP), insertions or deletions (INDELS), copy number variation (CNV) and structural variants (SV), are still insufficient to fully characterize the complex genetic process of adaptation to new environments (Peter and Schacherer, 2016).
Adaptive evolution in microorganisms is a process that occurs ubiquitously, including artificial settings where micro‐environments are created, and allows the adaptation of populations to defined conditions, driving the evolution process (domestication) (Doebley et al., 2006). Domestication is a stereotyped adaptive process (a ‘domestication syndrome’; see (Denham et al., 2020; Iqbal et al., 2020)) within a human‐created environment, where several characteristics can be tracked and defined as ‘domestication signatures’. These signatures are present in different fungal species, including Aspergillus oryzae in soya sauce (Gibbons et al., 2012), Penicillium moulds associated with cheese (Bodinaku et al., 2019) and S. cerevisiae (Goncalves et al., 2016; Duan et al., 2018) together with S. pastorianus (Gallone et al., 2018), responsible for beer fermentation. In this context, transcriptional re‐wiring, metabolic remodelling, changes in volatile compound production, faster growth rates and spore production together with viability are considered key traits of microbe domestication. In the case of brewing, the selective environmental pressures, such as high ethanol concentrations, allow to select for fitter individuals utilizing as a resource the new spontaneous mutations generated during cell division and the constant yeast re‐utilization process (Lang et al., 2013). Genomic analysis in beer yeast domesticated strains demonstrated the presence of common genetic patterns, such as large genomic rearrangements, aneuploidies, high heterozygosity levels and infertility, all of which are hallmarks of the adaptation process (Gallone et al., 2016; Goncalves et al., 2016; Gallone et al., 2019; Langdon et al., 2019).
Two main types of yeasts became domesticated under different brewing settings: S. cerevisiae that ferments ale beers at temperatures near 20 °C and S. pastorianus that produce lager beers fermented at lower temperatures (8–15°C) (Bokulich and Bamforth, 2013). S. pastorianus is an interspecific hybrid from the cross between S. cerevisiae and the cryotolerant wild yeast S. eubayanus (Libkind et al., 2011). The hybrid nature of S. pastorianus confers a series of competitive advantages in the fermentation environment, likely due to the combination of performance at relatively cold temperatures, efficient sugar uptake and metabolic switching between sugar sources (Salazar et al., 2019). During an intense domestication process and diversification over approximately 500 years, lager beers have evolved reduced organoleptic complexity, mainly characterized by the presence of ester compounds and the absence of phenolic off‐flavours (Gibson et al., 2017; Gallone et al., 2019). This is reflected in the absence of PAD1 and FDC1 in S. pastorianus, genes which are responsible for the synthesis of such off‐flavours (van den Broek et al., 2015; Diderich et al., 2018) and present in S. eubayanus. Lager yeast domestication is characterized by a reduced lag phase in the switch from glucose to maltose, and regulatory cross‐talk between S. cerevisiae and S. eubayanus sub‐genomes, which complement each other in terms of the genes required for maltose/maltotriose metabolism (Magalhaes et al., 2016; Brouwers et al., 2019a).
Given the recent discovery of S. eubayanus, its puzzling origin and apparently co‐evolutionary association with Nothofagus trees, several authors have analysed the worldwide distribution of S. eubayanus, together with its genetic, phenotypic and fermentative diversity (Eizaguirre et al., 2018; Brouwers et al., 2019a; Langdon et al., 2020; Nespolo et al.,2020b). Patagonian isolates of S. eubayanus exhibit the most extensive genetic diversity, and the presence of the most significant number of lineages compared with Northern Hemisphere populations, including five different lineages and a large group of admixed isolates (Langdon et al., 2020; Nespolo et al., 2020a, 2020b). To date, there is no evidence of S. eubayanus isolates in Europe, where the original S. pastorianus hybrid likely originated. Interestingly, fermentation capacity varies significantly between S. eubayanus isolates, possibly due to differences in maltose consumption and diauxic shift capacity, resulting in two opposite outcomes: successful or stuck fermentations (Nespolo et al., 2020a, 2020b). Patagonian isolates produce fruit and floral flavours in beer (Urbina et al., 2020), but high levels of 4‐vinyl guaiacol considered a phenolic off‐flavour that provides a clove‐like aroma, which is not preferred among consumers (Krogerus et al., 2015; Diderich et al., 2018; Mardones et al., 2020).
Although different reports have provided insights into the genomic and phenotypic changes responsible for the brewing capacity of S. pastorianus, particularly the S. cerevisiae genome portion, we know little about the process of S. eubayanus domestication before or after hybridization. Thus, further evidence is needed to understand the molecular mechanisms underpinning the S. eubayanus fermentative phenotype, which in turn will provide important insights into the inherent evolutionary process represented by directional selection for domestication, and correlated responses. In this study, a genetically complex artificial mixture of 30 different genotypes of S. eubayanus was continually exposed to high ethanol levels, mimicking the domestication process in breweries. We measured their correlated responses including their genomic, transcriptomic and phenotypic changes, and identified candidate genes that confer ethanol tolerance. Our results demonstrate that a single genetic background consistently overcomes the remaining strains, showing greater fermentation performance, but also significantly higher fitness in oxidative and osmotic stress environments. To an extent, we thus recreate the domestication process in the laboratory, showing how this cryotolerant yeast adapted to the competitive beer environment of a human industry and proved that experimental evolution can rebuild the brewing domestication process in S. eubayanus in ‘fast motion’. This provides a powerful tool for disentangling the molecular, physiological and biochemical processes that underlie the domestication of domesticated microorganisms.
Results
S. eubayanus fitness sensitivity under high ethanol conditions
We performed a parallel population assay to obtain high ethanol‐tolerant S. eubayanus individuals (Fig. 1A). For this, thirty S. eubayanus strains belonging to the PB‐2 and PB‐3 lineages, previously isolated in southern Chile (Villarrica, Coyhaique and Puyehue, Table S1; Nespolo et al., 2020a, 2020b), were selected and characterized for microbial growth under different ethanol conditions. These strains were selected aiming to maximize the genetic and phenotypic diversity available at the beginning of this study. Initially, we used micro‐cultures to evaluate biomass generation in 8%, 9% and 10% ethanol. Growth under these conditions showed long lag phases and low growth rates for all strains in concentrations above 9% ethanol (Fig. 1B, Table S2). This growth was significantly lower compared with that of the L299 wine S. cerevisiae control strain (p‐value < 0.05, ANOVA), demonstrating a greater susceptibility of S. eubayanus to high ethanol concentrations (Table S2). Furthermore, for all tested parameters, micro‐culture assays demonstrated significant phenotypic differences between strains (Fig. 1B), representing a genetically and phenotypically heterogeneous group of strains, ideal for the parallel population assay. Based on the above, we chose 9% ethanol as our selective environment for the experimental evolution procedure (from now on referred to as EtOH).
Fig. 1.
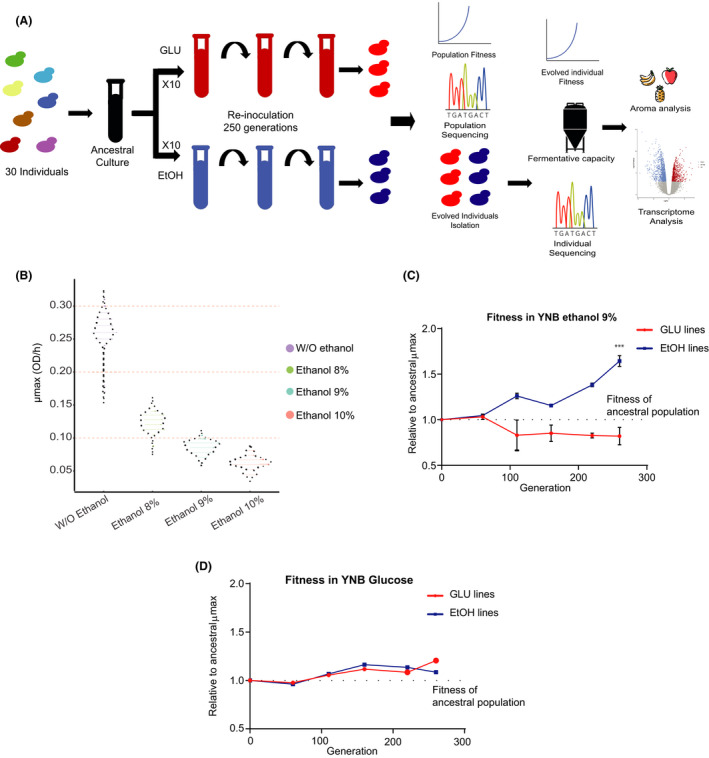
Fitness of the individual and propagated lines under ethanol.
A. Experimental evolution strategy in 10 replicated lines under YNB + glucose (GLU, red tubes) and YNB + GLU + ethanol 9% (EtOH, blue tubes). From every line, individuals were isolated and subjected to phenotyping, fermentation and sequencing analysis.
B. The growth rate (µmax) of the different parental strains used in this study was estimated under ethanol 8%, 9% and 10%. The fitness of the propagated lines under (C) ethanol and (D) glucose.
Our population assay began by mixing the thirty strains in equal proportions and subdividing them into ten mock replicates (YNB–glucose media, from now on referred to as GLU) and ten EtOH lines (Fig. 1A). The ethanol fitness of each propagated line was evaluated at different time points during the progression of the assay (Fig. 1C). After 260 generations (approximately six months), all GLU lines showed a significant decrease in ethanol fitness compared with the ancestral culture (P‐value < 0.05, ANOVA; Fig. 1C). In contrast, the EtOH‐propagated lines showed higher maximum growth rates (µmax) in ethanol compared with the original mixed culture, attaining a 60% greater µmax (P‐value < 0.05, ANOVA). These differences were not observed in glucose micro‐cultures (Fig. 1D), thus demonstrating that the propagated lines performed better in their selective environment compared with the control condition. Interestingly, we did not detect major adverse phenotypic effects in beer wort, suggesting a low accumulation of detrimental mutations (Table S2B).
Genome sequencing reveals consistent strain selection in parallel populations
Three GLU (GLU‐1, GLU‐7 and GLU‐10) and three EtOH lines (EtOH‐2, EtOH‐5 and EtOH‐6) were sequenced at the end of the experiment to identify the genomic changes and the pervasiveness of the different genetic backgrounds across the assay (Table S3). Interestingly, all the EtOH‐sequenced lines showed a sustained prevalence of strain CL248.1 (belonging to PB‐2 and isolated in northern Patagonia), reaching over 95% of the population’s allele frequency by the end of the experimental evolution assay (Fig. 2A). That being said, CL248.1 did not show the highest growth rate (µmax) under ethanol 9% of the S. eubayanus strains considered in this study, suggesting that selection did not occur solely due to ethanol tolerance (Table S2). In contrast, we did not observe a consistent selection in the GLU lines, where different genetic backgrounds were found depending on the propagated line (Fig. 2A). These results likely suggest a milder and different selection pressure in yeast when glucose is used as a selection regime, and a particular competitive fitness advantage of CL248.1 solely under EtOH selection, demonstrating a convergent phenomenon when ethanol and biotic stress are applied together.
Fig. 2.
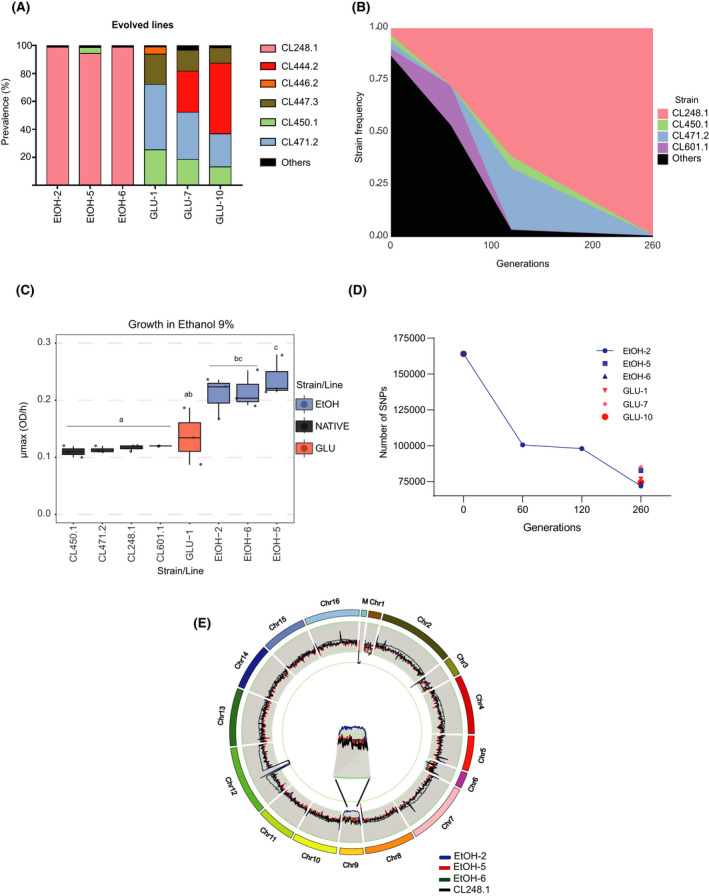
Genomic and phenotypic changes in the propagated lines.
A. The presence of the prevalent genetic backgrounds in three glucose (GLU) and ethanol (EtOH) propagated lines.
B. Prevalence (frequency) of the most prominent genetic backgrounds during the evolution of line EtOH‐2.
C. Ethanol 9% growth rates for the most representative parental strains and propagated lines.
D. Total number of SNPs relative to the CBS12357 reference genome at the beginning and end of the evolution assay for EtOH‐‐5, EtOH‐6 and GLU‐1, GLU‐7 and GLU‐10 lines. In addition, the number of SNPs during the evolution assay is shown for EtOH‐2.
E. Chromosome number estimation across EtOH lines. Only the EtOH‐2 line showed an aneuploidy by the end of the evolution assay (chromosome 9).
Line EtOH‐2 was sequenced at different time points (0, 60, 120 and 260 generations) to identify the genotypic course of the assay and additional genotypes under selection (Fig. 2B). We observed a predominance of CL248.1 and CL601.1 genotypes after 60 generations, demonstrating a competitive displacement of CL248.1 in the culture, together with higher fitness over the other genetic backgrounds (Fig. 2B). Interestingly, after 120 generations, four genotypes monopolized the culture, representing 96.6% of the EtOH‐2 line. Nevertheless, none of these parental genotypes showed high ethanol growth rates compared with the propagated lines (Fig. 2C). A second genotype, CL471.1, reached significant frequencies (maxima 29.3%) during intermediate periods of the evolution assay. However, it was almost absent by the end of the experiment, being detected at a frequency of just 0.15% in the final population. Moreover, over time, we calculated the total number of SNPs in propagated lines against the reference strain CBS12357T. We found a decrease in the number of SNPs over time across all lines relative to the ancestral culture, particularly in EtOH‐2, which exhibited the greatest decay compared with other lines (Fig. 2D).
To identify de novo genetic variants with a potential effect on ethanol tolerance, we used the EtOH‐2 line and compared polymorphisms (SNPs and short INDELs identified using freebayes) before and after selection. We chose this line because it showed the highest homology to a single genetic background (CL248.1), allowing the identification of novel genetic variants over the raw population’s genetic variation. In this way, we arbitrarily selected for polymorphisms with a putative moderate/high impact on the gene function and found 34 impacted genes under these criteria (Table S4). Among others, we found mutations in genes such as YPS6 and IMA1, encoding for a putative GPI‐anchored aspartic protease (Krysan et al., 2005) and a isomaltase (Teste et al., 2010) respectively. We also found a single aneuploidy in the EtOH‐2 line in chromosome IX, where an extra copy was found (Fig. 2E). Altogether, our results demonstrate how ethanol promotes a significant decrease in genetic variability due to genotype selection coupled with the emergence of new adaptive mutations vital for ethanol survival in biological processes such as stress damage and sugar metabolism.
Ethanol‐evolved individuals have greater fermentation capacity and maltotriose consumption
We determined the fitness cost of ethanol adaptation in 24 different environmental conditions for those EtOH‐adapted individuals isolated after 260 generations of selection. For this, we randomly isolated two clones from each EtOH line and estimated growth rates in micro‐cultures considering diverse phenotypic growth conditions, including high temperature, different carbon sources, and oxidative and osmotic stress (Table S5). To control for adaptive mutations in YNB laboratory media, we also isolated two colonies from three GLU lines. In general, individuals from EtOH‐propagated lines showed higher µmax in ethanol (Fig. 3A), and also for a greater number of conditions, compared with GLU‐propagated individuals and the ancestral culture (p‐value < 0.05, ANOVA; Table S5). These conditions included greater growth rates in sources such as glucose, maltose and fructose, together with resistance to oxidative (H2O2) and osmotic stresses (sorbitol 20%) (Fig. 3B), suggesting that selection improved general stress tolerance in these evolved strains. Interestingly, we found that one EtOH‐evolved individual (CLEt5.1, isolate n°1 from the EtOH‐5 line) exhibited greater ethanol tolerance, but a lower growth rate under high temperature (34°C) and an ionic detergent (SDS 0.001%, Fig. 3B), indicating the existence of a trade‐off.
Fig. 3.
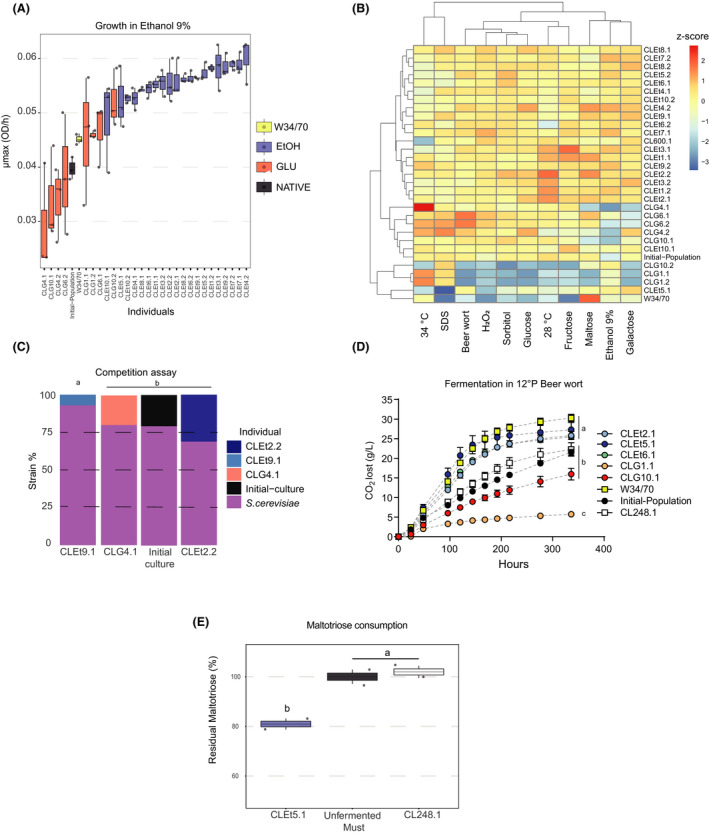
Phenotypic profiling of evolved individuals.
A. Growth rates under YNB–glucose–ethanol 9% of different evolved individuals.
B. Phenotypic heat map based on micro‐culture growth rates of EtOH‐ and GLU‐evolved individuals, evaluated in 11 different conditions.
C. The evolved strains were challenged using a GFP mutant S. cerevisiae in YNB media supplemented with ethanol at 9%. The strain frequency of the evolved individuals was evaluated using flow cytometry.
D. Fermentation in 12 °P beer wort of different evolved individuals. The fermentative capacity was estimated from the CO2 lost at different time points. The statistical differences were calculated after 216 h of fermentation using ANOVA.
E. Maltotriose consumption in YNB maltotriose 2% micro‐cultures.
To determine the relative fitness of EtOH‐ and GLU‐evolved individuals, we carried out a competition assay in YNB–glucose supplemented with 6% ethanol, against a recombinant S. cerevisiae that constitutively expresses GFP (Fig. S1). We observed that all tested strains were unable to outcompete S. cerevisiae; however, significant differences were found in the final proportion of the tested strains at the end of the experiment (p‐value < 0.05, ANOVA). For example, strain CLEt9.1 was almost absent at the end of the competition assay (relative frequency < 0.1), while CLEt2.2 was found to represent 31% of the cells quantified in the final culture (Fig. 3C). These results demonstrate fitness differences between EtOH‐ and GLU‐isolated individuals.
Additionally, we evaluated the fermentative capacity in small‐scale lager wort fermentations at low temperature (12 °C) of three EtOH‐evolved and two GLU‐evolved individuals. The selected strains were monitored for 15 days, and their fermentative capacity was estimated by measuring CO2 loss and sugar consumption throughout the fermentative process (Fig. 3D). Surprisingly, all the EtOH‐evolved individuals showed a similar fermentative profile compared with the commercial strain, where no significant differences were found in terms of total CO2 loss (P‐value < 0.05, ANOVA). Furthermore, the best‐evolved isolate (CLEt5.1) showed a 22.6% increase in loss of CO2 compared with the ancestral culture after 14 days of fermentation (Fig. 3D and Fig. S2A; p‐value > 0.05, ANOVA), and also exceeded the fermentative performance, in terms of fermentation rate, of its parental genetic background CL248.1 (Fig. 3D, S2B and C). Moreover, sugar consumption differed between the W‐34/70 commercial strain and the evolved individuals. Although the isolates were able to consume all the glucose, maltose and fructose found in the wort (Fig. S2D), no maltotriose consumption was observed (p‐value < 0.05, ANOVA; Table S6) in the evolved strains. We only detected maltotriose consumption under fermentation conditions in the lager commercial strain, in agreement with the inability of S. eubayanus to use this carbon source (Fig. S2D (Magalhaes et al., 2016)). To further analyse maltotriose consumption, we quantified the remaining maltotriose concentration after a 5 day incubation period of the evolved individuals in YNB synthetic media supplemented with 2% maltotriose as the sole carbon source (Fig. 3E). Interestingly, we detected 19.1% maltotriose consumption in the evolved strain CLEt5.1, while no consumption was found in CL248.1 (Fig. 3E). These results suggest genomic and molecular changes leading to maltotriose metabolization in this genetic background that only arise when maltotriose is used as the sole carbon source.
Identification of de novo genetic variants in the EtOH‐evolved strain CLEt5.1
The genome of the EtOH‐evolved individual CLEt5.1 was sequenced by coupling Nanopore and Illumina Technologies to elucidate the genetic origin of the phenotypic changes acquired through the evolution process. We obtained a high‐quality assembly and identified 5,946 genes in the final genome annotation, organized in 37 scaffolds (Fig. 4, Table S3 and Table S7A). The completeness analysis using BUSCO showed that the de novo assembly contained almost all the expected set of genes for a member of the Saccharomyces genus (97.5%). By comparing the scaffolds of the assembly against the CBS12357T reference genome, high synteny between genomes was observed, except for an evident translocation between chromosomes IV‐R and XVI‐L (Fig. 4). Therefore, we proceeded to identify structural variants between CLEt5.1 and its parental background (CL248.1) using MUM&Co (O'Donnell and Fischer, 2020). In this way, we identified 100 structural variants (deletions: 47, insertions: 41, duplications: 10, inversions: 0 and translocations: 2; Table S7B), primordially INDELs and confirming the translocation between chromosomes IV‐R and XVI‐L of 980 kb. Additionally, we found a 47 kb deletion in chromosome XII, and two 24 kb and 39 kb duplications in chromosomes VII and IV respectively. Among the genes present in the chromosome VII duplication, we found VID30, which is involved in the regulation of carbohydrate metabolism and the balance of nitrogen metabolism towards glutamate production, and HAP2, a transcription factor, which is predicted to regulate many of the proteins induced during the diauxic shift (Murphy et al., 2015) (Table S7C). SNP calling using freebayes detected 1,006 high‐quality SNPs. Based on these rearrangements, we measured spore viability in 20 tetrads in CLEt5.1 and found that 33% of the spores were viable, compared with 100% in the CL248.1 parental strain.
Fig. 4.
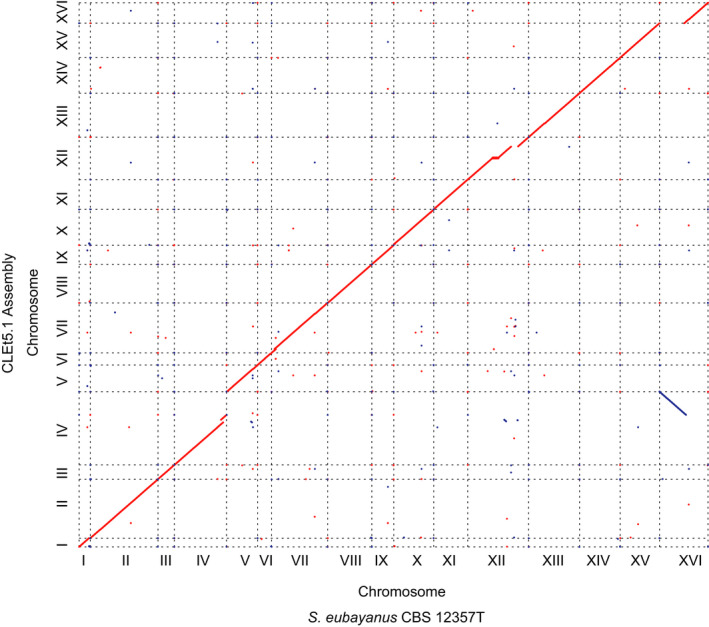
Genome synteny analysis of the EtOH‐evolved CLEt5.1 strain. Dot plot representation of DNA sequence identity between the S. eubayanus CBS12357T strain and the EtOH‐evolved.
To better understand the molecular basis of ethanol adaptation, we searched for polymorphisms across the CLEt5.1 genome that could generate moderate‐ or high‐impact mutations on the gene function (based on snpeff predictions). We found 11 genes with significant polymorphisms between CLEt5.1 and the native CL248.1 strain (Table S7D). For example, we found a missense variant in PUT4, which encodes for a proline permease essential in proline assimilation during fermentation (Long et al., 2018). Similarly, we found a frameshift in IRA2, which encodes for a GTPase‐activating protein, and previously related to high‐temperature fermentation (Wang et al., 2019) and low‐glucose growth defect rescue (Ramakrishnan et al., 2007). These results demonstrate that this relatively short period of ethanol adaptation promoted punctual, small and large rearrangements, which, taken together, may be responsible for the phenotypic differences between the CLEt5.1 and CL248.1 strains.
Transcriptome and organoleptic analysis of the CLEt5.1‐evolved strain under beer fermentation
To determine the impact of genetic changes in metabolic processes during wort fermentation in EtOH‐adapted individuals, we used a transcriptome approach. This allowed us to identify differentially expressed genes (DEGs) between the CLEt5.1 and the CL248.1 parental strain after 24 h of fermentation in a 1.5 l fermenter. Overall, we observed 92 DEGs (fold change > 0.7 and FDR < 0.05; Fig. 5A and Table S8), of which 59 and 33 were up‐ and downregulated in the CLEt5.1 strain respectively. Enrichment analysis of GO terms in upregulated genes revealed that diverse biological and molecular pathways, including sulfur compounds, methionine metabolism and several cellular amino acid metabolic processes, were enriched in the evolved strain (Table S8). In contrast, downregulated genes were significantly enriched in alpha‐amino acid metabolism and pheromone response metabolism, together with cofactor and vitamin‐binding molecular functions (Table S8). Similarly, KEGG enrichment analysis highlighted that genes within several pathways were differentially expressed between genotypes. For example, assimilatory sulfate reduction, cysteine and methionine metabolism, seleno‐compound metabolism and biosynthesis of antibiotics pathways were enriched in the upregulated genes set (Table S8). In contrast, we found a significant enrichment of the amino acid biosynthesis pathway among downregulated genes (p‐value < 0.01, hypergeometric test). Interestingly, these two analyses highlight that several DEGs were related to nitrogen and amino acid uptake, stress tolerance and faster diauxic shift, suggesting that nitrogen uptake and a rapid stress response play important roles during fermentation in this evolved strain.
Fig. 5.
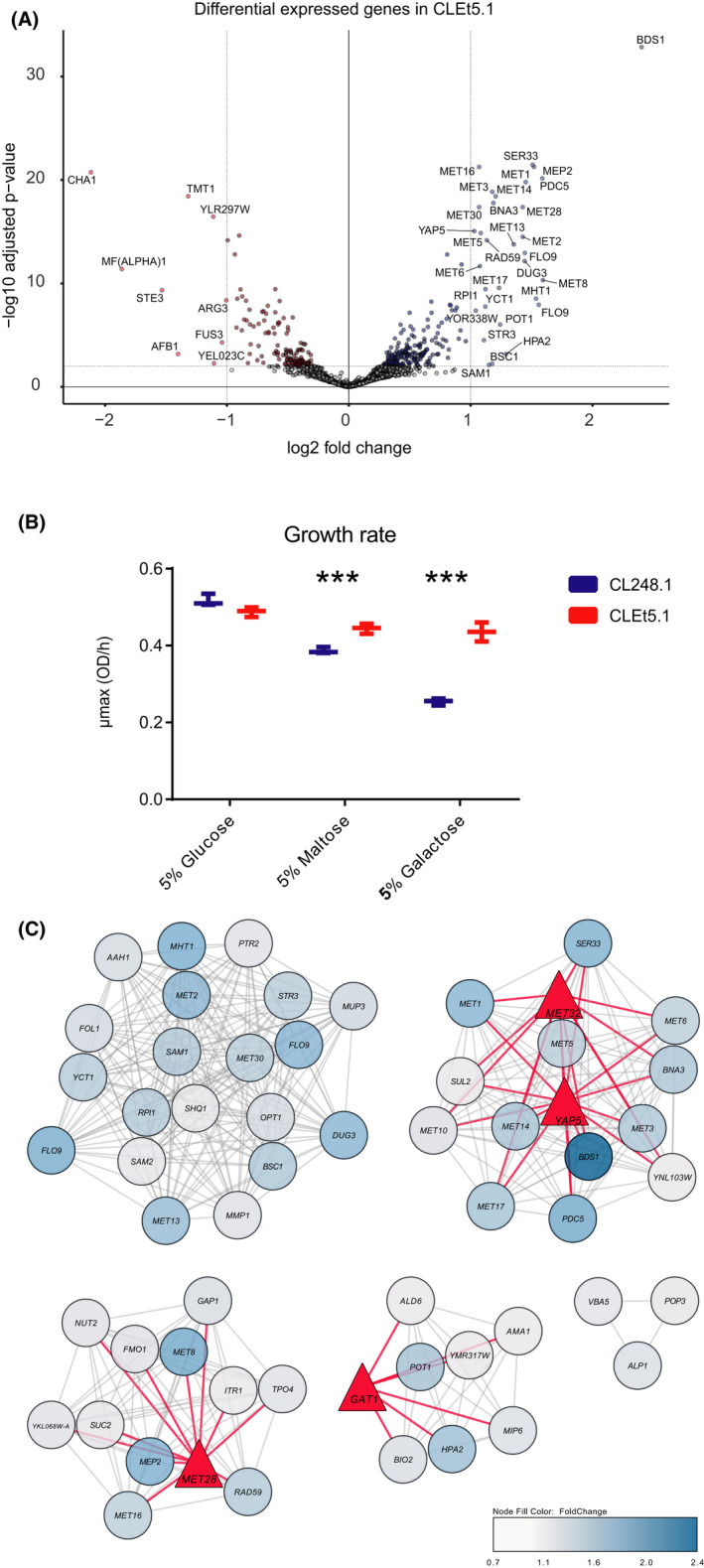
Differential gene expression analysis between the EtOH‐evolved CLEt5.1 strain and its native parental strain under beer wort fermentation conditions. The transcriptome of the EtOH‐evolved CLEt5.1 strain was evaluated and compared against the CL248.1 native strain under beer wort fermentation conditions.
A. The volcano plot depicts differentially expressed genes between CLEt5.1 and CL248.1.
B. Relative growth rates of CLEt5.1 and CL248.1 strains shifted from two 24 hours 5% glucose pre‐cultures to 5% maltose and 5% galactose media.
C. Network analysis in upregulated genes in CLEt5.1 depicting the most relevant hubs differently regulating genes between CLEt5.1 and CL248.1. Transcription factors are shown in red triangles, while TF‐gene connections are shown in red lines.
To evaluate the fast diauxic shift and the capacity of these two strains to switch from glucose to other disaccharides, we estimated their growth capacity under maltose and galactose after two 24 h pre‐cultures in 5% glucose. In agreement with our transcriptome results, the evolved strain showed a significantly greater growth rate compared with CL248.1 under 6% maltose and 6% galactose concentrations after long glucose incubation periods (Fig. 5B).
Additionally, to identify possible common regulatory elements of the upregulated genes, we analysed their promoter sequences (500 bp upstream of the transcription start site) and found a significant enrichment of transcription factor‐binding sites (p‐value < 0.05, Fisher's exact test) for transcription co‐activators of the Cbf1‐Met4‐Met28p complex (methionine metabolism), Dal80p and Uga3p (activators of nitrogen metabolism), Tye7p (glycolytic genes activator) and Sfl1p (repression of flocculation‐related genes and activation of stress‐responsive genes; Table S9). Additionally, we used Cytoscape to visualize the resulting network predicting regulatory interactions from the set of upregulating genes (Fig. 5C). According to our network model, we found four transcription factors: Met28p, Met32p, Gatp and Yap5p modulating the expression of these upregulated genes in CLEt5.1. Interestingly, Yap5p is known to be involved in the diauxic shift (Zampar et al., 2013). These results highlight a transcriptional re‐wiring in CLEt5.1 for genes related to nutrient acquisition, stress tolerance and methionine metabolism during the evolution of tolerance to fermentation stress.
During the fermentation process, we subjectively perceived that the organoleptic properties of the beers produced by the evolved strain differed from those of the parental native strain. Therefore, to determine how the transcriptional re‐wiring and genomic changes impacted the production of volatile compounds and the beer profile in the CLEt5.1‐evolved strain, we quantified volatile compound production using HS‐SPME‐GC/MS at the end of fermentation (day 15). As expected, we found significant differences in the composition of volatile compounds produced in beer between the evolved and parental strains (p‐value < 0.05, paired t‐test; Fig. 6A, Table S10). In general, the evolved clone showed lower levels of ester compounds, such as isoamyl acetate and ethyl octanoate (p‐value < 0.05, ANOVA). Additionally, we detected high levels of benzaldehyde 4‐methyl (aromatic aldehyde) and ethyl hexadecanoate in the evolved strain compared with the native genetic background, which could confer a fruity aroma to the beer similar to those found in lager beers. The most interesting differences were found in terms of off‐flavours. We detected a significantly lower production of 2‐methoxy‐4‐vinylphenol (4‐vinyl guaiacol) in the evolved strain, likely reducing its clove‐like flavour, which is typically found in fermented beverages by wild strains (p‐value < 0.05, ANOVA; Fig. 6A). Interestingly, we did not find mutations in the FDC1‐ and PAD1‐coding regions, or a significant difference in gene expression for FDC1 (log2FC = −0.038, p‐value‐adjusted = 0.838) and PAD1 (log2FC = −0.0095, p‐value‐adjusted = 0.965) between both strains. However, a series of mutations in the regulatory regions of both genes were found in CLEt5.1, which could alter expression levels in later fermentation stages. These results suggest that the evolution process significantly impacted the volatile compound profile of beers produced by CLEt5.1, emulating the domestication process that modified several commercial yeasts.
Fig. 6.
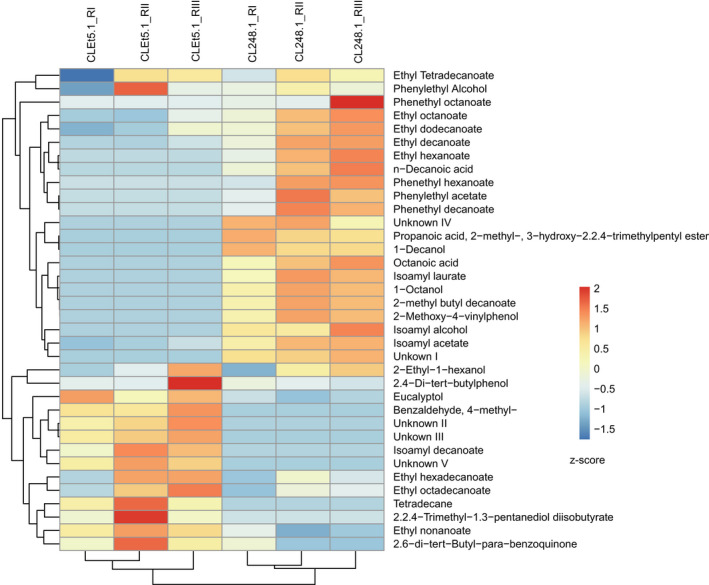
Volatile compound production on beer wort. The final beer from EtOH‐evolved CLEt5.1 and its parental strain CL248.1 was analysed using HS‐SPME‐GC/MS. The relative abundance of each compound detected was evaluated, and a heat map was constructed. The compounds were grouped in accordance with their relative abundance.
Discussion
Human‐driven selection associated with yeast domestication in fermentative environments has been extensively reported in S. cerevisiae and related hybrids (Gallone et al., 2019; Langdon et al., 2019). However, the genetic basis and molecular changes in other Saccharomyces genomes associated with alcoholic beverages are still unclear. In our study, we have reconstructed the putative domestication history of the yeast S. eubayanus under biotic and abiotic stresses, using a panmictic founding population that simulated the natural process of adaptive evolution together with strain sorting, and using an ethanol environment as the selective agent. We used dozens of wild genotypes in a single culture, in order to replicate the natural genetic variability of these organisms and promote strain sorting. After selection, we observed a decrease in the genetic diversity of the EtOH‐propagated lines and that a single genetic background, CL248.1, systematically outcompetes the others, acquiring de novo mutations and improving basal ethanol tolerance. Interestingly, the time‐course of this competitive displacement was complex, involving genotype selection and innovations throughout the assay (key adaptive mutations) that were constantly replaced by others during the ‘fast‐motion’ evolution time‐course. Thus, the evolved lineages derived from our founding genetic background exhibited higher ethanol growth rates compared with their ancestors, demonstrating a rapid response to selection, and so adapted successfully to their new environment. However, CL248.1 was not the best ethanol‐tolerant strain, suggesting that pre‐existing variants, together with de novo mutations, combined to positively affect fitness in this strain. In this sense, it has been demonstrated that pre‐existing and de novo genetic variants can both drive long‐term adaptation to environmental changes in yeast (Vázquez‐García et al., 2017). This indicates that not only a fitness advantage related to a given environmental selection pressure is essential for directional selection to occur in populations (Hoekstra et al., 2001), but also that a combination of standing genetic variation with some genomic plasticity for beneficial mutations is essential (Elena et al., 1996). In this way, the success of an individual is established in such a competitive environment (Dragosits et al., 2013). Our results show that both pre‐existing genetic variation and de novo mutations of a range of effects were important in explaining rapid evolution in this ecological context (Thompson, 1998; Callahan et al., 2014). Importantly, the Saccharomyces ‘make‐accumulate‐consume (ethanol)’ life strategy is fundamental for withstanding the antimicrobial effects of ethanol in a complex population (Hagman et al., 2013; Nespolo et al., 2020a, 2020b). Thanks to this, multiple Saccharomyces genotypes were selected, domesticated and used over centuries in the beer industries, including the S. pastorianus hybrid (Gallone et al., 2019; Langdon et al., 2019).
Domestication signatures in yeast, as a result of the human domestication syndrome, included genomic changes in the S. cerevisiae and S. eubayanus genomic portions leading to faster fermentation rates under low temperatures, a more moderate organoleptic complexity and the absence of off‐flavours in beers (Gibson et al., 2017; Gallone et al., 2019). Under the premise that evolutionary experiments can lead to unexpected and somewhat counterintuitive results (Van den Bergh et al., 2018), we evaluated the beer fermentation performance of S. eubayanus‐evolved individuals. Interestingly, evolved individuals exhibited a similar fermentation performance compared with lager yeast, suggesting in turn that ethanol, together with competitive displacement, could be the leading drivers of yeast domestication in brewing environments. This persistent directional selection involved correlated selection of other traits, such as osmotic stress tolerance and efficient nitrogen uptake (Gibson et al., 2007). In general, domesticated fungi used in fermented foods exhibit genomic rearrangements, fewer spores and produce desirable volatile compounds (Bodinaku et al., 2019). These domestication signatures have been reported in other systems, such as Aspergillus and Penicillium, where a transition to environments rich in carbon and nitrogen sources led to extensive metabolism remodelling when used to produce cheese (Gibbons et al., 2012; Bodinaku et al., 2019). Indeed, the CLEt5.1‐evolved individual showed a lower spore viability compared with the CL248.1 parental strain, representing another domestication hallmark.
Ethanol‐evolved individuals presented a series of genomic changes related to yeast domestication, such as aneuploidy, chromosomal rearrangements and lower spore viability (Gallone et al., 2016). Furthermore, signatures of trait domestication are evident in evolved individuals showing improved stress resistance, fast fermentation rates, lower organoleptic complexity and a lower production of phenolic off‐flavours (Gallone et al., 2019). S. cerevisiae beer strains are characterized by strong domestication signatures in their genomes, including polyploidies, the decay of sexual reproduction and maltotriose consumption (Gallone et al., 2016). Interestingly, one of our strains evidenced maltotriose consumption, which is another key domestication hallmark. This phenomenon was solely observed in CLEt5.1 after five days, when maltotriose was the only carbon source and no consumption was detected under fermentation conditions. Previous studies in S. eubayanus demonstrated that improved maltotriose consumption was possible due to the non‐reciprocal translocations between genes encoding maltose transporters (Baker and Hittinger, 2019; Brouwers et al., 2019a, 2019b). In our case, we could not identify large rearrangements or other mutations in maltose transporters that could lead to maltotriose consumption, suggesting that further experiments should be performed to identify the molecular basis of this phenotype.
In terms of the molecular mechanisms that explain their increased fermentative capacity, we observed that some stress response genes were either mutated or upregulated in the ethanol‐evolved line compared with its parental genetic background. In this way, the mutations and genomic rearrangements found in the CLEt5.1‐evolved individual could explain the transcriptional re‐wiring and improved fermentative profile. Indeed, ethanol exposure leads to the recruitment of error‐prone DNA polymerases, causing DNA replication stress and increased mutation rates (Voordeckers et al., 2020). Accordingly, we found that RAD59 (involved in DNA double‐strand break repair) was overexpressed in the evolved strain CLEt5.1, likely indicative of a mechanism that counteracts the mutagenic effect of ethanol (Davis and Symington, 2001). Other overexpressed genes could also be directly related to an increased fermentative capacity, such as SUC2, YAP5 and MET, which could promote glucose uptake, a dynamic diauxic shift and the accumulation of S‐adenosyl‐L‐methionine respectively (Outten and Albetel, 2013; Mohandesi et al., 2016). In this context, genomic rearrangements, such as the duplication found in chromosome VII containing HAP2, which is involved in promoting the diauxic shift, are in agreement with these findings. Furthermore, previous reports in lager yeast demonstrated that the accumulation and exogenous supplementation of S‐adenosyl‐L‐methionine promote an increase in the fermentative capacity of yeast under high‐gravity wort (Oomuro et al., 2018).
Concluding remarks
In summary, the results found in our study could be applied to determine the domestication dynamics of the S. eubayanus genomic portion in the lager strain, given the occurrence of similar desirable traits for beer. Based on multiple analyses, we provide evidence of the intermediate evolutionary changes in S. eubayanus, which have direct implications in the generation of novel yeasts for the industry. In this way, genomic changes promote a transcriptional re‐wiring that induces a favourable response in a fermentative environment. For the first time, these findings provide novel insights into the genomic and phenomic changes in wild S. eubayanus leading to faster wort fermentation rates and desirable organoleptic complexity, demonstrating its broad feasible use in the beer industry.
Experimental procedures
Microorganisms and culture media
Thirty S. eubayanus strains isolated from bark samples obtained from Nothofagus pumilio trees in south Chile were utilized for the experimental evolution assay, as listed in Table S1. These strains were previously reported and belong to the Patagonia B cluster (Nespolo et al., 2020a, 2020b). S. cerevisiae L299 (Martinez et al., 2007) and MTF2444 (EC1118 hsp12::GFP) (Tesniere et al., 2013) strains were used as growth control and in the competition assays respectively. Additionally, we used the S. pastorianus Saflager W‐34/70 (Fermentis, France) strain as a lager fermentation control. All isolates were maintained in YPD agar media (yeast extract 1%, peptone 2%, glucose 2% and agar 2%) and stored at −80°C in 20% glycerol stocks.
Experimental evolution
Initially, one colony from each S. eubayanus strain was cultured in 0.67% yeast nitrogen base (YNB) media (Difco, France) with 2% glucose at 20°C (hereinafter referred to as GLU) and 150 rpm orbital shaking. Later, each pre‐inoculum was utilized to prepare a co‐culture in a single 250 mL flask to obtain a final concentration of 1x106 cells ml‐1 of each strain. Ten replicates were set up (parallel populations) in 5 mL GLU and ten supplemented with 0.67% YNB media, 2% glucose and 9% ethanol (hereinafter referred to as EtOH). The inoculum was resuspended and transferred to the 20 replicates to obtain a final concentration of 1x106 cells ml‐1 (Fig. 1A). The adaptive evolution assays were performed at 20°C at 150 rpm for 72 h. Subsequently, the cultures were used to inoculate fresh 5 mL cultures at an inoculum density of 1 x 106 cell ml‐1, and this procedure was sequentially repeated. The number of generations was estimated using the ‘generations = log (final cells ‐ log initial cells)/log2’ formula, summing up the number of cells per ml doublings between every culture transfer during the adaptive evolution process.
Phenotyping assay
The phenotyping assay was performed as previously described (Nespolo et al., 2020a, 2020b). Briefly, isolates were pre‐cultivated in 200 µl 0.67% YNB medium supplemented with glucose 2% for 48 h at 25°C. Next, strains were inoculated to an optical density (OD) of 0.03–0.1 (wavelength 630 nm) in 200 µl growth media, where the following carbon sources were considered: glucose 2%, fructose 2%, maltose 2%, galactose 2% and 12 °Plato (°P) pilsner beer wort and incubated without agitation at 20°C for 24 h using a Tecan Sunrise absorbance microplate reader. Additionally, several environmental stressors were assessed, including ethanol 9%, sorbitol 20%, H2O2 3 mM, SDS 0.001% and high temperature (28 and 34°C) during 48 h. For ethanol 9%, experiments were carried out for 96 h. The OD was measured every 30 minutes using a 630 nm filter. Each experiment was performed in triplicate. Maximum growth rate, lag time and OD max parameters were obtained for each strain using the GrowthRates software with default parameters (Hall et al., 2014).
Growth curves incorporating carbon source switching from glucose to maltose and galactose were determined under micro‐cultivation conditions in YP (1% yeast extract, 2% peptone) media including either 5% glucose, 5% maltose or 5% galactose at 25°C for 48 h. Pre‐cultures were grown in YP with 5% glucose medium at 25°C for 24 h. Cultures were then diluted to an initial OD600nm of 0.1 in fresh YP 5% glucose medium for an extra overnight growth. The next day, cultures were used to inoculate a 96‐well plate with a final volume of 200 μL YP with the disaccharide source at an initial OD600nm of 0.1. The growth curves were monitored by measuring the OD600nm every 30 min as previously mentioned. All experiments were performed in triplicate. Lag phase and maximum specific growth rate (μmax) were estimated as previously described (Perez‐Samper et al., 2018) using the R software version 3.6.3.
Fermentations in beer wort
Fermentations were carried out as previously described (Mardones et al., 2020; Urbina et al., 2020). Briefly, fermentations were performed in at least three biological replicates, depending on the experiment, in 12 °P using a BrewFerm Pilsner Commercial Beer Kit (Beringen, Belgium). For this, a colony was transferred to 5 mL 6 °P pilsner beer wort supplemented with 0.3 ppm ZnCl2 and incubated at 20°C with orbital shaking at 150 rpm for 24 h. Then, the complete pre‐inoculum was transferred to 50 mL 12 °P pilsner beer wort and incubated in similar conditions for 24 h. Cells were utilized to inoculate 50 mL fresh 12 °P pilsner beer wort to a final concentration of 1.8 x 107 cell ml‐1. Cultures were maintained at 12°C for 14 days without agitation and weighed every day to calculate the CO2 released.
Larger volume fermentations for RNA extraction and metabolite production analysis were carried out in 1.5 l 12 °P beer wort for 14 days at 12°C. At the end of the fermentation, metabolites such as glucose, fructose, maltose, maltotriose, ethanol and glycerol were estimated using HPLC (Nespolo et al., 2020a, 2020b). Volatile compounds were detected using HS‐SPME‐GC‐MS as previously described (Urbina et al., 2020).
Competition assays
A total of 1 x 106 cells ml‐1 of the evolved and S. cerevisiae MTF2444 (EC1118 hsp12::GFP) strains were separately pre‐incubated in 5 ml YNB media supplemented with 2% glucose for 24 h. Evolved individuals were mixed in equal proportions with the S. cerevisiae MTF2444 GFP‐expressing mutant strain at a final concentration of 2 x 106 cell ml‐1 in YNB media supplemented with 2% glucose and 6% ethanol. Cultures were incubated in an orbital shaker at 20°C and 150 rpm during 72 h, and 100 µl samples from each culture were extracted every 24 h. Aliquots were washed twice in PBS and stored in the same buffer. Cultures were then analysed in a BD FACSCanto II Cytometer (Biosciences, San Jose, CA, USA). Finally, the proportion of non‐fluorescent/GFP‐fluorescent cells was estimated. Experiments were performed in triplicate.
Sequencing of the propagated lines and identification of mutations
DNA extraction was performed as previously described (Mardones et al., 2020; Nespolo et al., 2020a, 2020b). Sequencing of three parallel populations at final and intermediate stages of the evolution process was performed using the Illumina HiSeq X Ten Platform (BGI sequencing, China). Overall, approximately 45 million reads (paired‐end) were obtained for each evolved line. The raw reads were processed to remove adaptor sequences using the Fastp tool and filtered considering a 20 phred score cut‐off (Chen et al., 2018). Reads were aligned against the S. eubayanus CBS12357T reference genome (Brickwedde et al., 2018) using the Burrows–Wheeler Aligner (Li and Durbin, 2009). Overall, 99% of the reads were aligned, obtaining a mean coverage of 980X. Genome sequences of 27 parental strains were previously sequenced (Nespolo et al., 2020a, 2020b), from which a list of SNPs that were unique for each of those sequenced strains was obtained, using a custom R script. To estimate the proportion of the parental genetic backgrounds in every propagated line, the alternative genotype coverage at each unique SNP coordinate was obtained using bcftools mpileup (Li, 2011) (McKenna et al., 2010). De novo SNP calling in the propagated lines was performed using freebayes v 1.3.0 (https://github.com/ekg/freebayes). The total number of SNPs was calculated using Freebayes (Garrison and Marth, 2012), and the effect of each SNP was predicted with SnpEff (Cingolani et al., 2012) and the S. eubayanus CBS12357T reference genome (Brickwedde et al., 2018). Reads are available in the Biosample Database Project PRJNA666059.
Genome reconstruction of the CLEt5.1 mutant
The genome of the CLEt5.1 mutant was reconstructed using Nanopore sequencing coupled with Illumina sequencing. Nanopore sequencing was performed using a minION system (Oxford Nanopore, Oxford, UK). For this, DNA extraction and sequencing proceeded as previously described (Mardones et al., 2020). Overall, 26.1 million reads for Illumina and 96,000 reads for Nanopore were obtained (Table S2). The raw fast5 files were transformed to fastq files and debarcoded using Guppy 2.3.5 (Ueno et al., 2003). Barcode and adapter sequences were trimmed using Porechop (https://github.com/rrwick/Porechop) and filtered with Filtlong (https://github.com/rrwick/Filtlong) using a Phred score of 30. Genome assembly was performed with Canu (https://github.com/marbl/canu) using default settings. Additionally, two rounds of nanopolish (https://github.com/jts/nanopolish) and pilon (https://github.com/broadinstitute/pilon) were carried out. Moreover, the raw assembly was polished using the Illumina reads filtered with a Phred score of 20 (Burrows–Wheeler Aligner). The genome assembly was annotated with the pipeline LRSDAY (Yue and Liti, 2018) using the S. eubayanus CBS12357T reference genome as model for training AUGUSTUS (Stanke and Morgenstern, 2005), supported by the transcriptome assembly produced by TRINITY (Grabherr et al., 2011). The completeness of the genome assembly was evaluated using BUSCO (Simao et al., 2015). The assembly was compared with CBS12357T using nucmer (Marçais et al, 2018) to evaluate the synteny, while specific structural variants (SVs) were identified using MUM&Co (O'Donnell and Fischer, 2020). All the parameters of the pipeline were set up as default. The enrichment analysis of Gene Ontology (GO) terms and KEGG pathways was performed using METASCAPE (Zhou et al., 2019). The identification of transcription factor‐binding sites in the regulatory region 500 bp upstream of the upregulated genes of the evolved strain was performed using CiiDER (Gearing et al., 2019). Reads are available in the Biosample Database Project PRJNA666059.
RNA sequencing and differential expression analysis
RNA was extracted using the E.Z.N.A.® Total RNA Kit I (Omega Bio‐Tek, Waltham, MA, USA). RNA was DNase I‐treated (Thermo Fisher, USA) and purified using the RNeasy MinElute Cleanup Kit (Qiagen, Hilden, Germany). The Illumina libraries and sequencing were performed as previously described (Mardones et al., 2020) in the BGI facilities (Hong Kong, China). Briefly, RNA integrity was confirmed using a Fragment Analyzer (Agilent, Santa Clara, CA, USA). The RNA‐seq libraries were constructed using the TruSeq RNA Sample Prep Kit v2 (Illumina, San Diego, CA, USA). The sequencing was conducted using paired‐end 100 bp reads on an Illumina HiSeq X Ten in a single lane for the six samples. Reads are available in the Biosample Database Project PRJNA666059. Reads were mapped to the S. eubayanus CBS12357T reference genome using RNASTAR ver. 2.7.3 (Dobin et al., 2013) and analysed using featureCounts in R (Liao et al., 2014). Differential expression was analysed statistically using DESeq2 package in R (Love et al., 2014). Genes showing an adjusted P‐value of 0.05 or less were considered as differentially expressed genes (DEGs). Analysis of GO term enrichment was performed with the R package enrichGO (https://www.rdocumentation.org/packages/clusterProfiler/versions/3.0.4/topics/enrichGO). Cytoscape was used to visualize transcription factor regulatory networks (Shannon et al., 2003).
Conflicts of interest
The authors declare that they have no conflicts of interest with the content of this article.
Supporting information
Fig. S1. Competition assay of evolved individual in ethanol 9%.
Fig. S2. Fermentative capacity of the evolved individuals. (A) The fermentative capacity is indicated as a percentage of the capacity of the S. pastorianus control strain (W34/70) at 7 days. The fermentative capacity was estimated from the loss of CO2 over time. All assays were performed in triplicate. (B) The fermentative capacity was also determined at 14 days. (C) The velocity of the fermentation was estimated and (D) the residual sugars and metabolites in the wort were evaluated using HPLC.
Table S1. Native S. eubayanus strains used in the experimental evolution assay. The strain ID and the location of isolation site are indicated.
Table S2. Growth kinetic parameters in glucose and ethanol of the native parental strains used for the ancestral culture. Growth parameters µmax (OD/hr), OD max (OD) and lag phase (1/hr).
Table S3. Bioinformatics Summary statistics.
Table S4. SNP effect analysis of the novel polymorphisms in EtOH‐2. Snpeffect analysis of the novel/fixed polymorphisms in EtOH‐2 after 260 generations
Table S5. Phenotype data of evolved individuals. The data shows the average µmax across three replicates and the standard deviation (SD) for diverse growth conditions, including high temperature (28°C and 34°C), different carbon sources (glucose, fructose, maltose, galactose, xylose), and oxidative (ethanol 9%, 3 mM H2O2) and osmotic stress (beer wort, SDS 0.001%, Sorbitol 20%).
Table S6. Sugar consumption and metabolite production of the evolved individuals from fermentations in beer wort. Sugar consumption (g/L) and metabolite production (g/L) are informed.
Table S7. Structural variants identified in CLEt5.1 using MUM&Co. A. CLEt5.1 genome assembly and annotation statistics. The genome assembly of CLEt5.1 using Nanopore and Illumina sequencing technology was used to calculate several assembly statistics. B. All structural variants. C. Duplicated genes present in the chromosome IV – chromosome XVI duplication in CLEt5.1. D. High/moderate SNPeff prediction of SNPs and short INDELs in CLEt5.1
Table S8. Differential gene expression between CL248.1 and CLEt5.1 under beer wort. A. Gene expression results. B. Upregulated and C. Downregulated genes in CLEt5.1. R1, R2 and R3 represent the three biological replicates for each genotype.
Table S9. Enrichment analysis of Transcription Factor binding sites in regulatory regions of upregulated genes using CiiDER.
Table S10. Volatile compound production in CL248.1 and CLEt5.1 in beer wort.
Acknowledgements
This research is supported to FC by Comisión Nacional de Investigación Científica y Tecnológica CONICYT FONDECYT [1180161] and Millennium Institute for Integrative Biology (iBio). WM is supported by CONICYT FONDECYT [Grant No. 3190532]. CV is supported by CONICYT FONDECYT [Grant No. 3170404]. JM is supported by ANID FONDECYT POSTDOCTORADO [Grant No. 3200545]. RN is supported by FIC ‘Transferencia Levaduras Nativas para Cerveza Artesanal’ and Fondecyt Grant [No. 1180917]. We thank Michael Handford (Universidad de Chile) for language support.
Microb. Biotechnol. (2022) 15(00), 967–984
References
- Baker, E.P. , and Hittinger, C.T. (2019) Evolution of a novel chimeric maltotriose transporter in Saccharomyces eubayanus from parent proteins unable to perform this function. PLoS Genet 15: e1007786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodinaku, I. , Shaffer, J. , Connors, A. B. , Steenwyk, J. L. , Biango‐Daniels, M. N. , Kastman, E. K. , et al. (2019) Rapid phenotypic and metabolomic domestication of wild Penicillium molds on cheese. mBio 10(5): e02445‐19. https://10.1128/mBio.02445‐19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich, N.A. , and Bamforth, C.W. (2013) The microbiology of malting and brewing. Microbiol Mol Biol Rev 77: 157–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickwedde, A. , Brouwers, N. , van den Broek, M. , Gallego Murillo, J.S. , Fraiture, J.L. , Pronk, J.T. , and Daran, J.G. (2018) Structural, physiological and regulatory analysis of maltose transporter genes in Saccharomyces eubayanus CBS 12357(T). Front Microbiol 9: 1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwers, N. , Brickwedde, A. , Gorter de Vries, A. R. , van den Broek, M. , Weening, S. M. , van den Eijnden, L. , et al. (2019a) Himalayan Saccharomyces eubayanus genome sequences reveal genetic markers explaining heterotic maltotriose consumption by saccharomyces pastorianus hybrids. Appl Environ Microbiol 85: e01516‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwers, N. , Gorter de Vries, A.R. , van den Broek, M. , Weening, S.M. , Elink Schuurman, T.D. , Kuijpers, N.G.A. , et al. (2019b) In vivo recombination of Saccharomyces eubayanus maltose‐transporter genes yields a chimeric transporter that enables maltotriose fermentation. PLoS Genet 15: e1007853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan, B.J. , Fukami, T. , and Fisher, D.S. (2014) Rapid evolution of adaptive niche construction in experimental microbial populations. Evolution 68: 3307–3316. [DOI] [PubMed] [Google Scholar]
- Causton, H.C. , Ren, B. , Koh, S.S. , Harbison, C.T. , Kanin, E. , Jennings, E.G. , et al. (2001) Remodeling of yeast genome expression in response to environmental changes. Mol Biol Cell 12: 323–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S. , Zhou, Y. , Chen, Y. , and Gu, J. (2018) fastp: an ultra‐fast all‐in‐one FASTQ preprocessor. Bioinformatics 34: i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani, P. , Platts, A. , le Wang, L. , Coon, M. , Nguyen, T. , Wang, L. , et al. (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso‐2; iso‐3. Fly 6: 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, A.P. , and Symington, L.S. (2001) The yeast recombinational repair protein Rad59 interacts with Rad52 and stimulates single‐strand annealing. Genetics 159: 515–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denham, T. , Barton, H. , Castillo, C. , Crowther, A. , Dotte‐Sarout, E. , Florin, S.A. , et al. (2020) The domestication syndrome in vegetatively propagated field crops. Ann Bot 125: 581–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diderich, J.A. , Weening, S.M. , van den Broek, M. , Pronk, J.T. , and Daran, J.G. (2018) Selection of Pof(‐)Saccharomyces eubayanus variants for the construction of S. cerevisiae x S. eubayanus hybrids with reduced 4‐vinyl guaiacol formation. Front Microbiol 9: 1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin, A. , Davis, C.A. , Schlesinger, F. , Drenkow, J. , Zaleski, C. , Jha, S. , et al. (2013) STAR: ultrafast universal RNA‐seq aligner. Bioinformatics 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley, J.F. , Gaut, B.S. , and Smith, B.D. (2006) The molecular genetics of crop domestication. Cell 127: 1309–1321. [DOI] [PubMed] [Google Scholar]
- Dragosits, M. , Mozhayskiy, V. , Quinones‐Soto, S. , Park, J. , and Tagkopoulos, I. (2013) Evolutionary potential, cross‐stress behavior and the genetic basis of acquired stress resistance in Escherichia coli . Mol Syst Biol 9: 643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, S.‐F. , Han, P.‐J. , Wang, Q.‐M. , Liu, W.‐Q. , Shi, J.‐Y. , Li, K. , et al. (2018) The origin and adaptive evolution of domesticated populations of yeast from Far East Asia. Nat Commun 9: 2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eizaguirre, J.I. , Peris, D. , Rodriguez, M.E. , Lopes, C.A. , De Los Rios, P. , Hittinger, C.T. , and Libkind, D. (2018) Phylogeography of the wild Lager‐brewing ancestor (Saccharomyces eubayanus) in Patagonia. Environ Microbiol 18: 1137–1147. [DOI] [PubMed] [Google Scholar]
- Elena, S.F. , Cooper, V.S. , and Lenski, R.E. (1996) Punctuated evolution caused by selection of rare beneficial mutations. Science 272: 1802–1804. [DOI] [PubMed] [Google Scholar]
- Gallone, B. , Mertens, S. , Gordon, J.L. , Maere, S. , Verstrepen, K.J. , and Steensels, J. (2018) Origins, evolution, domestication and diversity of Saccharomyces beer yeasts. Curr Opin Biotechnol 49: 148–155. [DOI] [PubMed] [Google Scholar]
- Gallone, B. , Steensels, J. , Prahl, T. , Soriaga, L. , Saels, V. , Herrera‐Malaver, B. , et al. (2016) Domestication and divergence of Saccharomyces cerevisiae beer yeasts. Cell 166(1397–1410): e1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallone, B. , Steensels, J. , Mertens, S. , Dzialo, M.C. , Gordon, J.L. , Wauters, R. , et al. (2019) Interspecific hybridization facilitates niche adaptation in beer yeast. Nat Ecol Evol 3: 1562–1575. [DOI] [PubMed] [Google Scholar]
- Garrison, E. , and Marth, G. (2012) Haplotype‐based variant detection from short‐read sequencing. arXiv 1207. [Google Scholar]
- Gearing, L.J. , Cumming, H.E. , Chapman, R. , Finkel, A.M. , Woodhouse, I.B. , Luu, K. , et al. (2019) CiiiDER: a tool for predicting and analysing transcription factor binding sites. PLoS One 14: e0215495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons, J.G. , Salichos, L. , Slot, J.C. , Rinker, D.C. , McGary, K.L. , King, J.G. , et al. (2012) The evolutionary imprint of domestication on genome variation and function of the filamentous fungus Aspergillus oryzae . Curr Biol 22: 1403–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson, B. , Geertman, J. A. , Hittinger, C. T. , Krogerus, K. , Libkind, D. , Louis, E. J. , et al. (2017) New yeasts‐new brews: modern approaches to brewing yeast design and development. FEMS Yeast Res 17: fox038. [DOI] [PubMed] [Google Scholar]
- Gibson, B.R. , Lawrence, S.J. , Leclaire, J.P.R. , Powell, C.D. , and Smart, K.A. (2007) Yeast responses to stresses associated with industrial brewery handling. FEMS Microbiol Rev 31: 535–569. [DOI] [PubMed] [Google Scholar]
- Goncalves, M. , Pontes, A. , Almeida, P. , Barbosa, R. , Serra, M. , Libkind, D. , et al. (2016) Distinct domestication trajectories in top‐fermenting beer yeasts and wine yeasts. Curr Biol 26: 2750–2761. [DOI] [PubMed] [Google Scholar]
- Grabherr, M.G. , Haas, B.J. , Yassour, M. , Levin, J.Z. , Thompson, D.A. , Amit, I. , et al. (2011) Full‐length transcriptome assembly from RNA‐Seq data without a reference genome. Nat Biotechnol 29: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagman, A. , Sall, T. , Compagno, C. , and Piskur, J. (2013) Yeast "make‐accumulate‐consume" life strategy evolved as a multi‐step process that predates the whole genome duplication. PLoS One 8: e68734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, B.G. , Acar, H. , Nandipati, A. , and Barlow, M. (2014) Growth rates made easy. Mol Biol Evol 31: 232–238. [DOI] [PubMed] [Google Scholar]
- Hoekstra, H.E. , Hoekstra, J.M. , Berrigan, D. , Vignieri, S.N. , Hoang, A. , Hill, C.E. , et al. (2001) Strength and tempo of directional selection in the wild. Proc Natl Acad Sci USA 98: 9157–9160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal, M.M. , Erskine, W. , Berger, J.D. , and Nelson, M.N. (2020) Phenotypic characterisation and linkage mapping of domestication syndrome traits in yellow lupin (Lupinus luteus L.). Theor Appl Genet 133: 2975–2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogerus, K. , Magalhaes, F. , Vidgren, V. , and Gibson, B. (2015) New lager yeast strains generated by interspecific hybridization. J Ind Microbiol Biotechnol 42: 769–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan, D.J. , Ting, E.L. , Abeijon, C. , Kroos, L. , and Fuller, R.S. (2005) Yapsins are a family of aspartyl proteases required for cell wall integrity in Saccharomyces cerevisiae . Eukaryot Cell 4: 1364–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang, G.I. , Parsons, L. , and Gammie, A.E. (2013) Mutation rates, spectra, and genome‐wide distribution of spontaneous mutations in mismatch repair deficient yeast. Genes|Genomes|Genetics 3(9): 1453–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdon, Q.K. , Peris, D. , Baker, E.P. , Opulente, D.A. , Nguyen, H.V. , Bond, U. , et al. (2019) Fermentation innovation through complex hybridization of wild and domesticated yeasts. Nat Ecol Evol 3: 1576–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdon, Q.K. , Peris, D. , Eizaguirre, J.I. , Opulente, D.A. , Buh, K.V. , Sylvester, K. , et al. (2020) Postglacial migration shaped the genomic diversity and global distribution of the wild ancestor of lager‐brewing hybrids. PLoS Genet 16: e1008680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. (2011) A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27: 2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , and Durbin, R. (2009) Fast and accurate short read alignment with Burrows‐Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, Y. , Smyth, G.K. , and Shi, W. (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930. [DOI] [PubMed] [Google Scholar]
- Libkind, D. , Hittinger, C.T. , Valerio, E. , Goncalves, C. , Dover, J. , Johnston, M. , et al. (2011) Microbe domestication and the identification of the wild genetic stock of lager‐brewing yeast. Proc Natl Acad Sci USA 108: 14539–14544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, D. , Wilkinson, K.L. , Taylor, D.K. , and Jiranek, V. (2018) Novel wine yeast for improved utilisation of proline during fermentation. Fermentation 4: 10. [Google Scholar]
- Love, M.I. , Huber, W. , and Anders, S. (2014) Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalhaes, F. , Vidgren, V. , Ruohonen, L. , and Gibson, B. (2016) Maltose and maltotriose utilisation by group I strains of the hybrid lager yeast Saccharomyces pastorianus . FEMS Yeast Res 16: fow053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardones, W. , Villarroel, C.A. , Krogerus, K. , Tapia, S.M. , Urbina, K. , Oporto, C.I. , et al. (2020) Molecular profiling of beer wort fermentation diversity across natural Saccharomyces eubayanus isolates, Microb . Biotechnol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais, G. , Delcher, A. L. , Phillippy, A. M. , Coston, R. , Salzberg, S. L. , and Zimin, A. (2018) MUMmer4: a fast and versatile genome alignment system. PLoS Comput Biol 14: e1005944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, C. , Cosgaya, P. , Vasquez, C. , Gac, S. , and Ganga, A. (2007) High degree of correlation between molecular polymorphism and geographic origin of wine yeast strains. J Appl Microbiol 103: 2185–2195. [DOI] [PubMed] [Google Scholar]
- McKenna, A. , Hanna, M. , Banks, E. , Sivachenko, A. , Cibulskis, K. , Kernytsky, A. , et al. (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next‐generation DNA sequencing data. Genome Res 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohandesi, N. , Siadat, S. , Haghbeen, K. , and Hesampour, A. (2016) Cloning and expression of Saccharomyces cerevisiae SUC2 gene in yeast platform and characterization of recombinant enzyme biochemical properties. 3 Biotech 6: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, J.P. , Stepanova, E. , Everley, R.A. , Paulo, J.A. , and Gygi, S.P. (2015) Comprehensive temporal protein dynamics during the diauxic shift in Saccharomyces cerevisiae . Mol Cell Proteomics 14: 2454–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nespolo, R.F. , Solano‐Iguaran, J.J. , Paleo‐Lopez, R. , Quintero‐Galvis, J.F. , Cubillos, F.A. , and Bozinovic, F. (2020a) Performance, genomic rearrangements, and signatures of adaptive evolution: lessons from fermentative yeasts. Ecol Evol 10: 5240–5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nespolo, R.F. , Villarroel, C.A. , Oporto, C.I. , Tapia, S.M. , Vega‐Macaya, F. , Urbina, K. , et al. (2020b) An Out‐of‐Patagonia migration explains the worldwide diversity and distribution of Saccharomyces eubayanus lineages. PLoS Genet 16: e1008777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell, S. , and Fischer, G. (2020) MUM&Co: accurate detection of all SV types through whole‐genome alignment. Bioinformatics 36: 3242–3243. [DOI] [PubMed] [Google Scholar]
- Oomuro, M. , Watanabe, D. , Sugimoto, Y. , Kato, T. , Motoyama, Y. , Watanabe, T. , and Takagi, H. (2018) Accumulation of intracellular S‐adenosylmethionine increases the fermentation rate of bottom‐fermenting brewer's yeast during high‐gravity brewing. J Biosci Bioeng 126: 736–741. [DOI] [PubMed] [Google Scholar]
- Outten, C.E. , and Albetel, A.N. (2013) Iron sensing and regulation in Saccharomyces cerevisiae: Ironing out the mechanistic details. Curr Opin Microbiol 16: 662–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez‐Samper, G. , Cerulus, B. , Jariani, A. , Vermeersch, L. , Barrajon Simancas, N. , Bisschops, M.M.M. , et al. (2018) The crabtree effect shapes the Saccharomyces cerevisiae lag phase during the switch between different carbon sources. mBio 9: e01331‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter, J. , and Schacherer, J. (2016) Population genomics of yeasts: towards a comprehensive view across a broad evolutionary scale. Yeast 33: 73–81. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan, V. , Theodoris, G. , and Bisson, L.F. (2007) Loss of IRA2 suppresses the growth defect on low glucose caused by the snf3 mutation in Saccharomyces cerevisiae . FEMS Yeast Res 7: 67–77. [DOI] [PubMed] [Google Scholar]
- Salazar, A.N. , Gorter de Vries, A.R. , van den Broek, M. , Brouwers, N. , de la Torre Cortes, P. , Kuijpers, N.G.A. , et al. (2019) Chromosome level assembly and comparative genome analysis confirm lager‐brewing yeasts originated from a single hybridization. BMC Genom 20: 916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon, P. , Markiel, A. , Ozier, O. , Baliga, N.S. , Wang, J.T. , Ramage, D. , et al. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard, S.K. , Guttman, D.S. , and Fitzgerald, J.R. (2018) Population genomics of bacterial host adaptation. Nat Rev Genet 19: 549–565. [DOI] [PubMed] [Google Scholar]
- Simao, F.A. , Waterhouse, R.M. , Ioannidis, P. , Kriventseva, E.V. , and Zdobnov, E.M. (2015) BUSCO: assessing genome assembly and annotation completeness with single‐copy orthologs. Bioinformatics 31: 3210–3212. [DOI] [PubMed] [Google Scholar]
- Stanke, M. , and Morgenstern, B. (2005) AUGUSTUS: a web server for gene prediction in eukaryotes that allows user‐defined constraints. Nucleic Acids Res 33: W465–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamari, Z. , Yona, A.H. , Pilpel, Y. , and Barkai, N. (2016) Rapid evolutionary adaptation to growth on an 'unfamiliar' carbon source. BMC Genom 17: 674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesniere, C. , Delobel, P. , Pradal, M. , and Blondin, B. (2013) Impact of nutrient imbalance on wine alcoholic fermentations: nitrogen excess enhances yeast cell death in lipid‐limited must. PLoS One 8: e61645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teste, M.A. , Francois, J.M. , and Parrou, J.L. (2010) Characterization of a new multigene family encoding isomaltases in the yeast Saccharomyces cerevisiae, the IMA family. J Biol Chem 285: 26815–26824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.N. (1998) Rapid evolution as an ecological process. Trends Ecol Evol 13: 329–332. [DOI] [PubMed] [Google Scholar]
- Ueno, Y. , Arita, M. , Kumagai, T. , and Asai, K. (2003) Processing sequence annotation data using the Lua programming language. Genome Inform 14: 154–163. [PubMed] [Google Scholar]
- Urbina, K. , Villarreal, P. , Nespolo, R. F. , Salazar, R. , Santander, R. , and Cubillos, F. A. (2020) Volatile compound screening using HS‐SPME‐GC/MS on Saccharomyces eubayanus strains under low‐temperature pilsner wort fermentation. Microorganisms 8: 755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Bergh, B. , Swings, T. , Fauvart, M. , and Michiels, J. (2018) Experimental design, population dynamics, and diversity in microbial experimental evolution. Microbiol Mol Biol Rev 82: e00008–00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Broek, M. , Bolat, I. , Nijkamp, J.F. , Ramos, E. , Luttik, M.A. , Koopman, F. , et al. (2015) Chromosomal copy number variation in Saccharomyces pastorianus is evidence for extensive genome dynamics in industrial lager brewing strains. Appl Environ Microbiol 81: 6253–6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez‐García, I. , Salinas, F. , Li, J. , Fischer, A. , Barré, B. , Hallin, J. , et al. (2017) Clonal heterogeneity influences the fate of new adaptive mutations. Cell Rep 21: 732–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voordeckers, K. , Colding, C. , Grasso, L. , Pardo, B. , Hoes, L. , Kominek, J. , et al. (2020) Ethanol exposure increases mutation rate through error‐prone polymerases. Nat Commun 11: 3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z. , Qi, Q. , Lin, Y. , Guo, Y. , Liu, Y. , and Wang, Q. (2019) QTL analysis reveals genomic variants linked to high‐temperature fermentation performance in the industrial yeast. Biotechnol Biofuels 12: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue, J.X. , and Liti, G. (2018) Long‐read sequencing data analysis for yeasts. Nat Protoc 13: 1213–1231. [DOI] [PubMed] [Google Scholar]
- Zampar, G.G. , Kümmel, A. , Ewald, J. , Jol, S. , Niebel, B. , Picotti, P. , et al. (2013) Temporal system‐level organization of the switch from glycolytic to gluconeogenic operation in yeast. Mol Syst Biol 9: 651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y. , Zhou, B. , Pache, L. , Chang, M. , Khodabakhshi, A.H. , Tanaseichuk, O. , et al. (2019) Metascape provides a biologist‐oriented resource for the analysis of systems‐level datasets. Nat Commun 10: 1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Competition assay of evolved individual in ethanol 9%.
Fig. S2. Fermentative capacity of the evolved individuals. (A) The fermentative capacity is indicated as a percentage of the capacity of the S. pastorianus control strain (W34/70) at 7 days. The fermentative capacity was estimated from the loss of CO2 over time. All assays were performed in triplicate. (B) The fermentative capacity was also determined at 14 days. (C) The velocity of the fermentation was estimated and (D) the residual sugars and metabolites in the wort were evaluated using HPLC.
Table S1. Native S. eubayanus strains used in the experimental evolution assay. The strain ID and the location of isolation site are indicated.
Table S2. Growth kinetic parameters in glucose and ethanol of the native parental strains used for the ancestral culture. Growth parameters µmax (OD/hr), OD max (OD) and lag phase (1/hr).
Table S3. Bioinformatics Summary statistics.
Table S4. SNP effect analysis of the novel polymorphisms in EtOH‐2. Snpeffect analysis of the novel/fixed polymorphisms in EtOH‐2 after 260 generations
Table S5. Phenotype data of evolved individuals. The data shows the average µmax across three replicates and the standard deviation (SD) for diverse growth conditions, including high temperature (28°C and 34°C), different carbon sources (glucose, fructose, maltose, galactose, xylose), and oxidative (ethanol 9%, 3 mM H2O2) and osmotic stress (beer wort, SDS 0.001%, Sorbitol 20%).
Table S6. Sugar consumption and metabolite production of the evolved individuals from fermentations in beer wort. Sugar consumption (g/L) and metabolite production (g/L) are informed.
Table S7. Structural variants identified in CLEt5.1 using MUM&Co. A. CLEt5.1 genome assembly and annotation statistics. The genome assembly of CLEt5.1 using Nanopore and Illumina sequencing technology was used to calculate several assembly statistics. B. All structural variants. C. Duplicated genes present in the chromosome IV – chromosome XVI duplication in CLEt5.1. D. High/moderate SNPeff prediction of SNPs and short INDELs in CLEt5.1
Table S8. Differential gene expression between CL248.1 and CLEt5.1 under beer wort. A. Gene expression results. B. Upregulated and C. Downregulated genes in CLEt5.1. R1, R2 and R3 represent the three biological replicates for each genotype.
Table S9. Enrichment analysis of Transcription Factor binding sites in regulatory regions of upregulated genes using CiiDER.
Table S10. Volatile compound production in CL248.1 and CLEt5.1 in beer wort.


