Summary
Porcine reproductive and respiratory syndrome (PRRS) is a viral disease defined by reproductive problems, respiratory distress and a negative impact on growth rate and general condition. Virulent PRRS virus (PRRSV) strains have emerged in the last years with evident knowledge gaps in their impact on the host immune response. Thus, the present study examines the impact of acute PRRS virus (PRRSV) infection, with two strains of different virulence, on selected immune parameters and on the gut microbiota composition of infected pigs using 16S rRNA compositional sequencing. Pigs were infected with a low virulent (PRRS_3249) or a virulent (Lena) PRRSV‐1 strain and euthanized at 1, 3, 6, 8 or 13 days post‐inoculation (dpi). Faeces were collected from each animal at the necropsy time‐point. Alpha and beta diversity analyses demonstrated that infection, particularly with the Lena strain, impacted the microbiome composition from 6 dpi onwards. Taxonomic differences revealed that infected pigs had higher abundance of Treponema and Methanobrevibacter (FDR < 0.05). Differences were more considerable for Lena‐ than for PRRS_3249‐infected pigs, showing the impact of strain virulence in the intestinal changes. Lena‐infected pigs had reduced abundancies of anaerobic commensals such as Roseburia, Anaerostipes, Butyricicoccus and Prevotella (P < 0.05). The depletion of these desirable commensals was significantly correlated to infection severity measured by viraemia, clinical signs, lung lesions and immune parameters (IL‐6, IFN‐γ and Hp serum levels). Altogether, the results from this study demonstrate the indirect impact of PRRSV infection on gut microbiome composition in a strain virulence‐dependent fashion and its association with selected immune markers.
PRRSV, a porcine respiratory pathogen, indirectly impacts on gut microbiome composition in a strain virulence dependent fashion in association with selected immune markers.

Introduction
Porcine reproductive and respiratory syndrome (PRRS) is one of the infectious diseases with the highest impact on the modern swine industry. The wide diversity among PRRS virus (PRRSV) strains is reflected by the classification of the two genotypes of the virus, PRRSV‐1 and PRRSV‐2, as two independent viral species, Betaarterivirus suid 1 and Betaarterivirus suid 2 respectively (Gorbalenya, 2018). Furthermore, each of these viral species has a broad genetic variability, with strains being assigned to different subtypes, clades and lineages (Shi et al., 2010; Stadejek et al., 2013; Balka et al., 2018).
Since 2006, the heterogeneity among PRRSV strains has been reinforced by the identification of virulent strains in Asia (PRRSV‐2) and Europe (PRRSV‐1) (Tian et al., 2007; Karniychuk et al., 2010). These virulent strains induce high fever and severe respiratory problems complicated with secondary bacterial co‐infections, leading to an increase in morbidity and mortality rates (Tian et al., 2007; Karniychuk et al., 2010; Senthilkumar et al., 2018; Rodríguez‐Gómez et al., 2019). Among several virulent PRRSV‐1 strains, the subtype 3 PRRSV‐1 Lena strain is considered to be the prototype (Karniychuk et al., 2010; Morgan et al., 2013; Frydas et al., 2015; Sinn et al., 2016; Canelli et al., 2017; Stadejek et al., 2017). The emergence of these virulent strains has given more complexity to the control of this disease, which represents a cornerstone in the management of the porcine respiratory disease complex.
PRRSV is characterized by a peak of virus replication in the lung and the onset of fever during the first week post‐infection; with earlier and higher fever when pigs are infected by virulent strains (Karniychuk et al., 2010; Stadejek et al., 2017; Rodríguez‐Gómez et al., 2019). The main clinical signs developed by infected animals consist of dyspnoea and respiratory distress, which may be also associated with poor body condition and diarrhoea according to the virulence of the strain (Sinn et al., 2016; Senthilkumar et al., 2018). Therefore, it is clear that PRRSV, besides affecting the respiratory tract, impacts on the general health status of the animal, which may be reflected in growth retardation and gastroenteric clinical signs.
Despite the gut–lung axis by the interaction of the microbiota and respiratory infections has been already postulated (Budden et al., 2017; Niederwerder, 2017), it is scarcely studied in pigs yet. Recent studies have demonstrated that PRRSV infection, either on its own (Jiang et al., 2019) or together with circovirus infection, modifies the host intestinal microbiota (Niederwerder et al., 2016; Ober et al., 2017). For instance, these studies report a decrease of desirable gut inhabitants such as Prevotella (Jiang et al., 2019) or a lower diversity of the overall faecal microbiome linked to a worse clinical outcome in PRRSV and porcine circovirus type 2 (PCV2) dually infected pigs (Ober et al., 2017). In addition, microbiota changes in dual infections, increased Methanobacteriaceae species and reduced Ruminococcaceae and Streptococcaceae species, were associated with low growth rate pigs (Niederwerder et al., 2016; Ober et al., 2017). These studies should inspire new designs which further explore the changes in the microbiome in the respiratory and intestinal tract after PRRSV infection, including factors such as the immune response, not analysed so far (Jiang et al., 2019). According to the emergence of virulent PRRSV strains, the modulation of the immune response by this virus and the potential interplay between the microbiome and the immune response, there is a clear gap in the scientific literature of previous studies addressing all these issues along PRRSV infection.
In this context, virulent PRRSV strains may directly affect the gut microbiome, leading to a lower diversity than low virulent strains as well as proliferation of pathogenic species, which would relate to the clinical outcome and modulation of different immune mediators. Considering the paucity of studies focused on the role of the faecal microbiome during PRRSV infection as well as the marked interest in deciphering the pathogenesis of PRRSV strains of different virulence, the goal of the present study was to analyse and determine the changes in the composition and diversity of the faecal microbiome in response to infection with two PRRSV‐1 strains of different virulence as well as their association with the host immune response.
Results and Discussion
PRRSV infection contributes to a number of immunological outcomes which alter host health, increasing for instance the susceptibility to secondary infections by opportunistic pathogens (Gómez‐Laguna et al., 2013). Similarly, the infection may be able to alter the gut microbiota, which is essential in host physiological processes (Xiao et al., 2016; Zhang and Wang, 2018; Pearson‐Leary et al., 2019). So far, few studies have analysed the impact of PRRSV on the gut microbiome, and the other way around, (Niederwerder et al., 2016; Ober et al., 2017; Jiang et al., 2019) and none of them have investigated the changes occurring through the infection process from a PRRSV virulence perspective. To assess the goal of this study, an experimental setting was carried out with seventy animals randomly distributed in three experimental groups (Lena‐ and PRRS_3249‐ infected groups and control group) and housed in separate pens at containment facilities.
Clinical signs, gross pathology of the lung and viremia are influenced by the virulence of PRRSV strain
The main clinical signs and pulmonary gross lesions observed in this study are described in detail in Rodríguez‐Gómez et al. (2019). Briefly, pigs infected with the virulent strain Lena showed severe respiratory and systemic signs (i.e. lethargy, anorexia, diarrhoea and pyrexia) as well as marked lung lesions (tan‐mottled areas, atelectasis and areas of consolidation), which occurred earlier and more severely than in pigs from the PRRS_3249 group.
PRRSV was detected by RT‐qPCR in the sera of all Lena‐infected pigs as early as 1 dpi, whereas four out of five PRRS_3249‐infected pigs were simultaneously PRRSV positive. Viraemia was always higher in Lena‐ than PRRS_3249‐infected pigs from 1 to 13 dpi (P < 0.01 at 1, 3, 6 dpi; P < 0.05 at 8 and 13 dpi), reaching the highest level at 6 dpi (1.89 × 107 eq CPD50 per ml) (Fig. 1). PRRSV‐specific antibodies were first detected at 8 dpi in sera from both PRRSV‐infected groups, with slightly higher sample to positive ratio (S/P) values in case of Lena‐infected pigs (File S1).
Fig. 1.
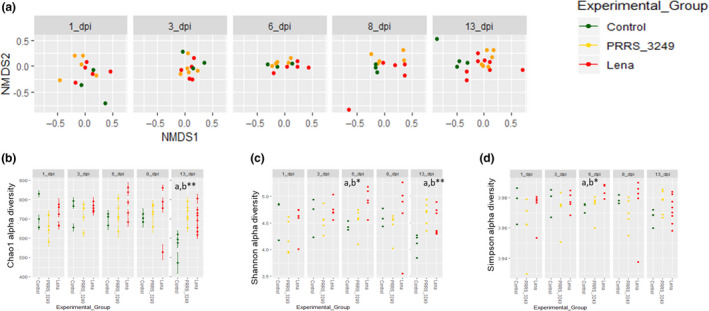
Diversity analyses in faecal samples from PRRSV‐infected pigs.
A. Beta diversity estimated by Bray–Curtis distance and non‐metric multidimensional scaling of samples at each sampling time‐point by experimental group variable.
B–D, Chao1, Shannon and Simpson alpha diversity indexes, respectively, by sampling time‐point. Statistically significant differences were estimated by ANOVA with Tukey correction and were denoted with (*): * P < 0.05; ** P < 0.01; and *** P < 0.001 and ‘a’ Lena vs control, ‘b’ among the three categories.
PRRSV infection alters gut microbiome diversity
Alpha (number of taxa and abundance of these taxa in a particular sample) and beta (differences in taxa between samples) diversity metrics allow evaluating global microbiome composition (Kim and Isaacson, 2015a). Structural analysis of vector spatial distribution regression (P < 0.05) and PERMANOVA (P < 0.001) tests showed the influence of PRRSV strain virulence on the ordination of samples (File S2). A result in line with the observed differences for Chao1 alpha diversity index in the Lena‐infected group compared to the control group (P < 0.05; File S3 and S4). Further analyses were performed after splitting samples by day of sampling, the factor that had the strongest effect on the spatial distribution of samples (P < 0.01), with a greater effect in control pigs compared to infected pigs (Fig. 1A). Interestingly, both diversity metrics were impacted by PRRSV infection, particularly from 6 dpi onwards (Fig. 1A, C and D; File S2, sheet 3), when the peak of viraemia was observed, and the clinical signs were more severe. Thus, significant differences were observed in sample ordination among experimental group categories at 6 dpi (R 2 = 0.25; P < 0.05) and 13 dpi (R 2 = 0.19; P < 0.01) and differences were close to significance for samples collected at 8 dpi (R 2 = 0.22; P = 0.06). Similarly, in alpha diversity analyses, at 6 dpi, there were significant differences in alpha diversity values among experimental groups for Shannon (P < 0.05) and Simpson (P < 0.05) indexes (Fig 1C and D). At 13 dpi, the last time‐point of the study, Shannon and Chao1 indexes differed significantly among experimental group categories (P < 0.01 and P < 0.01 respectively; Fig 1B and C).
The higher diversity of species as well as the increased microbiome evenness and richness in infected pigs, and particularly in the Lena‐infected group, together with the spatial differences between infected and control pigs, which matched with the viremia peak in Lena‐infected group, show that PRRSV infection impairs microbiome composition, allowing the emergence or growth of bacteria present at low proportions in healthy animals, and shifting the gut microbiome composition by strain virulence. Previous microbiome PRRSV studies linked increased diversity to better growth and moderate disease outcomes (Niederwerder et al., 2016; Ober et al., 2017). Differences in study design, such as co‐infection with PCV2 together with the lack of non‐infected pigs to compare with and/or the methodology used to analyse the microbiome, are potential reasons for the observed discrepancies.
Changes in relative abundance are associated with the infection process and PRRSV strain virulence
16S rRNA compositional sequencing allows the estimation of relative abundance of different groups of bacteria (Kim et al., 2015b). By detailed analysis of differences in abundance, we aimed to decipher the particular groups of bacteria involved in the shift observed in the microbiome after PRRSV infection (Fig. 2A; File S5). Changes at taxonomic level were influenced by strain virulence and particularly linked to Lena strain infection. This infected group showed an increased abundance of genera Treponema and Methanobrevibacter, as was the case for Treponema in comparison with PRRS_3249 group. In contrast, the genera Anaerostipes, Prevotella and Butyricicoccus abundance in Lena‐infected pigs were significantly lower compared to the other two groups. We finally observed a decreased abundance of Roseburia in Lena‐infected pigs as well as a decrease in Mycoplasma abundance in both Lena‐ and PRRS_3249‐ infected pigs compared to controls. The increased abundance of Treponema and Methanobrevibacter, a bacterium of the phylum Euryarchaeota, herein observed has been previously reported in low growth PRRSV‐infected pigs (Ober et al., 2017). The role of this sort of methanogens in health and disease is still under discussion (Chaudhary et al., 2018) and may require specific designed amplicons for an accurate characterization (Klindworth et al., 2013).
Fig. 2.
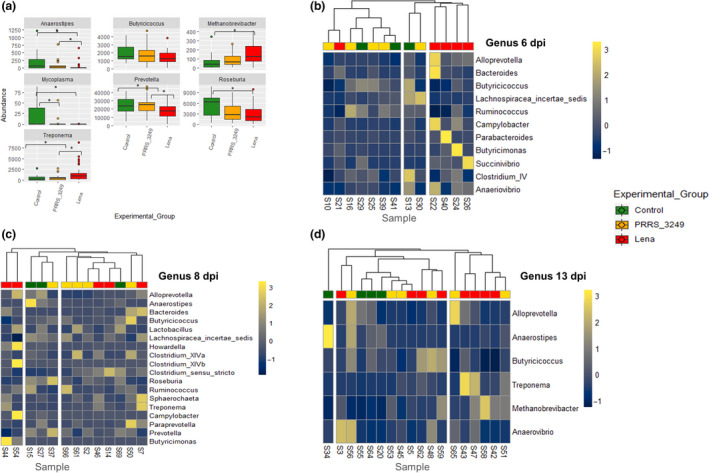
Genera abundance analyses in experimental groups.
A. differentially abundant genera established by fitzig function in Metagenomeseq package among Lena, PRRS_3249 and control groups. Whiskers denote the significant differences among experimental groups and * denotes FDR < 0.05.
B–D. represent heatmaps of the log‐transformed relative temporal abundance of genera for which P < 0.01 (Shade et al., 2013) illustrating the relative counts of genera in samples from 6 dpi (B), 8 dpi (C) and 13 dpi (D). The dark blue colour indicates low relative abundance taxa, while light yellow represents those at high relative abundance. The dendrogram was built using Euclidean distances and Ward’s method with red colour for pig from Lena‐infected group, yellow for PRRS_3249‐infected group and green for control group respectively.
The analysis of differential abundance by sampling day (File S5) revealed a higher abundance of Succinivibrio in infected pigs compared to controls at 6 dpi, coinciding with the viraemia peak. At 8 dpi, a higher abundance of Treponema and Bacteroides and lower abundance of two Prevotellacea, the genera Prevotella and Alloprevotella, and two Clostridiales, Roseburia and Clostridium cluster XIVa, was observed. Finally, at 13 dpi, we also observed a higher abundance of Treponema in both infected groups. The clustering of samples by relative abundance of genera at targeted days post‐infection (6, 8 and 13 dpi) revealed an influence of PRRSV experimental group variable in pig clustering by taxa abundance (Figs. 2B–D). The observed decrease in abundance of anaerobe saccharolytic inhabitants of the distal gut (Anaerostipes, Prevotella and Alloprevotela) as well as in other members of the class Clostridia such as Roseburia and Clostridium cluster XIVa, both from the family Lachnospiraceae, and Butyricicoccus (Clostridiaceae) illustrates the displacement of anaerobes from the gut in Lena‐infected pigs. Furthermore, increased abundance of Proteobacteria and Spirochaetes in faeces from Lena‐infected pigs is indicator of potential favourable conditions for pig enterobacteria such as E. coli, Salmonella or the two pathogenic spirochaetes Brachyspira hyodysenteriae and B. pilosicoli. This finding may explain the on‐farm link between PRRSV infection and enteric diseases and highlights the relevance of PRRSV control for successful treatment and control of other diseases, including enteric pathogens (Beloeil et al., 2007).
Association between factors linked to PRRSV disease and microbiome abundance
In this study, respiratory and systemic clinical signs, lung lesions, viraemia and several immune markers were measured and correlated to abundance of gut bacteria to establish potential indirect relationships between infection and microbial abundance. An immunologic link between the gastrointestinal tract and respiratory tract mucosa is postulated via mesenteric lymph nodes and thoracic duct connection (He et al., 2017). Following this premise, we looked for potential correlations among studied immune factors or acute‐phase proteins and gut microbiome abundance.
Significant correlations between host disease analyses and taxa abundance are detailed in table 2 at phylum, family and genus levels. The relative abundance of six genera was significantly correlated with changes in variables under study (Fig. 3; File S6). Clinical signs and cumulative gross lesions score were positively correlated with the relative abundance of Methanobrevibacter and Spirochaeta, result which was also observed for the family Spirochaetaceae and the phylum Spirochaetes (Table 1). In contrast, the score of these two variables was negatively correlated with the relative abundance of genera from the phylum Firmicutes, including two Lachnospiraceae (Blautia and Roseburia) and Faecalibacterium as well as the Prevotella from the phylum Bacteroides. These correlations demonstrate the link between disease severity and depletion of anaerobic commensals. A similar depletion of anaerobes from class Clostridia is observed in Salmonella infection as a consequence of mucosal inflammation (Rivera‐Chávez and Bäumler, 2015; Argüello et al., 2018). Accordingly, to the virulence of PRRSV strain and the severity of clinical signs, including marked inappetence and severe respiratory signs, anorexia or fasting may be the principal motive of the microbial changes observed. Indeed, a study performed in fasted healthy adult horses describes a reduction of class Clostridia as occurs in our study (Schoster et al., 2016). Other correlate of disease, such as the viraemia, was strongly correlated with the relative abundance of Micrococcaceae and to a lesser extent with the relative abundance of Spirochaetaceae, while we observed a negative correlation to the phylum Firmicutes (Table 1). In this sense, the depletion observed in strict anaerobes may provide an opportunity for other families, such as Spirochaeta, Helicobacteriaceae or Campylobacteriaceae to proliferate, as observed here. As above mentioned, these groups include pathogenic bacteria which may co‐infect PRRSV‐infected pigs.
Fig. 3.

Correlation matrix between abundance of the main genera and the PRRSV infection parameters under study. Spearman correlations among significant genera abundance and disease parameters.
Table 1.
Significant correlations observed between taxa relative abundance and host parameters along PRRSV infection.
| Taxa | Host factors | |||||
|---|---|---|---|---|---|---|
| Clinical signs | Gross lesions | Viraemia | IFN‐γ | IL‐6 | Hp | |
| Phyla | ||||||
| Firmicutes | –0.69* | –0.78** | –0.70* | –0.69* | NS | NS |
| Spirochaetes | NS | 0.67* | NS | NS | NS | NS |
| Proteobacteria | NS | 0.71** | NS | NS | NS | NS |
| Familiae | ||||||
| Spirochaetaceae | 0.72** | 0.78** | NS | NS | NS | NS |
| Helicobacteriaceae | 0.61* | 0.71** | NS | NS | NS | NS |
| Campylobacteriaceae | 0.52* | 0.64* | NS | NS | NS | NS |
| Clostridiaceae | 0.57* | NS | NS | NS | NS | NS |
| Lachnospiraceae | –0.77** | –0.82*** | NS | NS | NS | NS |
| Prevotellaceae | –0.56* | –0.60* | NS | NS | NS | NS |
| Micrococcaceae | NS | NS | 0.80*** | NS | 0.63* | NS |
| Spirochaetaceae | NS | NS | 0.52* | NS | NS | NS |
| Lachnospiraceae | NS | NS | NS | NS | NS | 0.54* |
| Streptococcaceae | NS | NS | NS | –0.55* | NS | NS |
| Genera | ||||||
| Methanobrevibacter | 0.67* | 0.72* | NS | NS | NS | NS |
| Spirochaeta | 0.72** | 0.84*** | NS | NS | NS | NS |
| Blautia | –0.73** | –0.74** | NS | NS | NS | NS |
| Faecalibacterium | –0.80** | –0.73** | NS | NS | NS | NS |
| Prevotella | –0.78** | –0.79** | NS | NS | NS | NS |
| Roseburia | –0.77** | –0.73** | NS | NS | NS | NS |
IFN, interferon; IL, interleukin; Hp, haptoglobin.
P‐values and Rho values were established by Spearman rank correlation (P < 0.05); *(P < 0.05); **(P < 0.01); and ***(P < 0.001).
From the set of immunity markers tested, serum level of IL‐6 in PRRS_3249 group showed maximum levels at 6 dpi (mean of 350 ± SD pg ml‐1), dropping at 8 dpi, whereas the Lena group reached the highest IL‐6 level at 8 dpi (mean of 480 ± SD pg ml‐1). Serum IL‐6 level was correlated with the relative abundance of Micrococcaceae. A significant increase in IFN‐γ serum levels was detected at 6 and 8 dpi in Lena‐infected pigs (maximum mean level of 234 ± SD pg ml‐1 at 6 dpi) compared to control (P < 0.05) and PRRS_3249 groups (P < 0.01). Serum concentration of IFN‐γ was negatively correlated with the relative abundance of Streptococcaceae (Table 1). Finally, for IL‐10, Hp and LBP, no differences between groups were observed throughout the study. Among these immune markers, Hp levels were positively correlated with the relative abundance of Lachnospiraceae and negatively correlated with the relative abundance of Helicobacteriaceae (File S6).
The correlations observed between immune mediators, such as IFN‐γ, IL‐6 or Hp, and the changes in the gut microbiome, highlight the potential impact of the immune modifications prompted by PRRSV infection in the lung (Gómez‐Laguna et al., 2013; Rodríguez‐Gómez et al., 2019) on the intestine. Ultimately, the gut‐lung immune response axis is complex (He et al., 2017) and further research is needed to determine the exact mechanisms that result in shifts in the gut microbiome.
Altogether, the results from this study demonstrate the impact of PRRSV infection on gut microbiome composition. Microbiome changes were clearly influenced by strain virulence and were more evident from viremia peak onwards. PRRSV infection, principally associated with Lena strain, displaced desirable gut anaerobes and allowed colonization by microbial groups such as Proteobacteria or Spirochaetes which are indicative of favourable conditions for usual enteric pathogens. These microbial changes were associated with disease severity, but further research is needed to disclose the gut‐lung axis relationships in PRRSV infection or secondary factors that prompt the observed changes.
Experimental procedures
Ethics statement
All the animal experiments described in the present study were conducted in strict accordance with the guidelines of the European Union (Directive 2010/63/EU). All animal studies were conducted under protocols approved by the IRTA Ethics Committee and by the Catalan Autonomous Government (Project 3647; FUE‐2017‐00533413). All efforts were made to minimize suffering and ensure the highest ethical and humane standards. Accordingly, euthanasia of the animals at the different time‐points was performed by initial sedation with azaperone (4 mg kg‐1; Stresnil, Ecuphar Veterinaria, S.L.U., Barcelona, Spain), followed by anaesthesia with ketamine (15 mg kg‐1; Ketaset, Zoetis Spain S.L., Madrid, Spain) and a lethal dose of 5 % sodium thiopental (Tiobarbital, Braun Vet Care S.A., Barcelona, Spain).
Animals and experimental design
Animals and samples used in this study were part of a project to elucidate the pathogenesis of PRRSV strains of different virulence (Rodríguez‐Gómez et al., 2019). Seventy piglets (Landrace × Large White, four‐week‐old) were randomly distributed in three different groups and housed in separate pens at the Biosafety Level 3 containment facilities of Centre de Recerca en Sanitat Animal (IRTA‐CReSA, Bellaterra, Barcelona, Spain). Pigs were obtained from a high health status farm, historically negative for PRRSV and also confirmed as negative for PCV2 and Mycoplasma hyopneumoniae [IDEXX PRRS X3 Ab Test; (Mattsson et al., 1995; Sibila et al., 2004)]. After a 7‐day acclimation period, pigs were intranasally inoculated as follows: (i) PRRS_3249 group, 26 pigs were inoculated with 2 ml (1.0 ml per nostril) of 1 × 105 TCID50 of the subtype 1 PRRSV‐1 3249 strain; (ii) Lena group, 28 pigs were inoculated at the same conditions with the subtype 3 PRRSV‐1 Lena strain; and (iii) control group, 16 pigs were inoculated with porcine alveolar macrophages supernatant diluted in RPMI at the same conditions. Three control pigs and five infected pigs from each group were euthanized on days 1, 3, 6 and 8 post‐inoculation (dpi). At 13 dpi, four control pigs, six pigs from PRRS_3249 group and eight pigs from the Lena group were humanely killed.
Clinical signs and samples collection
Rectal temperature and clinical signs, such as abnormal behaviour, anorexia, dyspnoea, cough, skin lesions, presence of diarrhoea and faeces consistency, were daily monitored along the study. Blood samples were collected at 0, 1, 3, 6, 8 and 13 dpi and routinely processed to analyse the viraemia and perform serological studies. At necropsy, immediately after the euthanasia, rectal faeces from each animal were aseptically collected into sterile containers and freeze at −80°C until processing. Gross lung lesions were recorded as previously reported (Rodríguez‐Gómez et al., 2019).
Viremia, detection of antibodies and serological analyses
Sera were collected to evaluate the viraemia, the presence of specific antibodies and the serum concentration of the cytokines IFN‐γ, IL‐6 and IL‐10 as well as the acute‐phase proteins haptoglobin (Hp) and lipopolysaccharide‐binding protein (LBP). RNA was purified from sera using NucleoSpin RNA virus (Macherey‐Nagel, Düren, Germany) according to manufacturer’s instructions. Viraemia for either PRRS_3249 strain or Lena strain was quantified by RT‐qPCR using LSI™ VetMAX™ PRRSV EU/NA 2.0 kit (Life Technologies, Carlsbad, CA, USA). Serial dilutions of PRRS_3249 strain or Lena strain (known viral titres) were used to determine genome quantification. RT‐qPCR reactions were carried out with the QuantStudio 5 Real‐time PCR System (Life Technologies, Carlsbad, CA, USA) for 5 min (min) at 50°C, 10 min at 95°C followed by 40 cycles of 3 s (s) at 95°C and 30 s at 60 °C. Viraemia results are showed in equivalent (eq) cytopathic dose 50 (CPD50) per mL. PRRSV‐specific antibodies were detected using IDEXX PRRS X3 ELISA test (IDEXX laboratories, Barcelona, Spain) following manufacturer’s instructions. For cytokines and acute‐phase protein detection, different commercially available ELISA and colorimetric assays were used in accordance with manufacturer’s guidelines (IFN‐γ, IL‐6, IL‐10 [Invitrogen, Carlsbad, CA, USA]; LBP, [Hycult Biotech, Uden, Netherlands] and Hp, [Tridelta Development Limited, Kildare, Ireland]). The results were expressed in pg/mL for IFN‐γ, IL‐6 and IL‐10; ng ml‐1 for LPB, and mg ml‐1 for Hp. The minimum detectable concentrations were 2 pg ml‐1 for IFN‐γ, 45 pg ml‐1 for IL‐6, 3 pg ml‐1 for IL‐10, 1.6 ng ml‐1 for LBP and 0.005 mg ml‐1 for Hp.
16S rRNA sequencing and bioinformatic processing analysis
The microbiota composition of faecal samples was established by amplicon sequencing. The V3‐V4 variable region of the 16S rRNA gene was amplified from each extracted DNA sample according to the 16S metagenomic sequencing library protocol (Illumina Inc., San Diego, CA, USA), following manufacturer’s instructions and subsequently sequenced on the Illumina MiSeq platform using v3 sequencing chemistry with 2 × 3000 pb paired‐end reads according to the manufacturer’s guidelines. The Illumina reads were filtered on the basis of quality (removal of low quality nucleotides at the 3' end, and remove windows 20 nt with a low average quality) and length (removal of sequences with <300 pb) with prinseq (Schmieder and Edwards, 2011), and the paired‐end reads with a minimum overlap of 20 bp were joined using Fastq‐join (Aronesty, 2013). Finally, all single files were processed to a final filtering sequence mean quality score > 25. In addition, the sequences were cleaned of dereplicates, elimination of unique sequences and chimeras against gold database using UPARSE‐OTU algorithm with Usearch v8.0 algorithm (Edgar, 2010). The resulting sequences were clustered with 97 % identity to obtain operative taxonomic unit (OTUs) using UPARSE‐OTU algorithm with Usearch v8.0 algorithm (Edgar, 2010). The taxonomic assignment of these OTUs was obtained against the Ribosomal Database Project (RDP) (Cole et al., 2017).
Statistical analysis
Statistical analyses were performed in R v3.4.2. Microbiota, and study variables (infection, experimental group and sampling day) were included in the estimation of alpha diversity richness (Shannon, Simpson and Chao1 indexes) by the Vegan and Phyloseq R packages (McMurdie and Holmes, 2013). For diversity values, assumption of normality was checked using the Shapiro–Wilk test. Potential differences in richness of factors included in the study were estimated by repeated measures analysis of variance (ANOVA), using sampling time‐point as a co‐factor and a Tukey multiple comparison test. Dissimilarities in beta diversity between pairs of samples were therefore estimated with the Bray–Curtis dissimilarity index and weighted unifrac index and analysed with mon‐metric multidimensional scaling (NMDS) in Vegan (Bray and Curtis, 1957; Lozupone et al., 2007). The Vegan envfit function, which fits environmental vectors or factors onto an ordination, was used to evaluate whether the factors sampling day and infection status were associated with the (NMDS) ordinations; the significance of the fitted factors was estimated using 999 permutations. Analysis of similarities (Vegan anosim function) and Permutational Multivariate Analysis of Variance Using Distance Matrices (Vegan adonis function) were also used to establish the influence of the variables under study into the sample ordination. Differences in taxa abundance for 33 experimental groups (Control vs Infected, Control vs Lena/3249 or Lena‐infected vs 3249‐infected) were analysed using a zero‐inflated gaussian model by the fitZig function of the metagenomeSeq R package (Paulson et al., 2013), and significance was established with a false discovery rate (FDR) correction with a threshold of 0.05. Heatmaps of genera for which P < 0.01 were built using their relative temporal abundance as previously recommended (Shade, et al., 2013), clustered using Euclidean distances and Ward’s method using Pheatmap package. Spearman correlations were performed among animal parameters and microbial relative abundance using R core functions and represented by Corrplot and Performance Analytics packages. For viraemia and serological parameters, statistical analyses were performed in GraphPad Prism 7 software (GraphPad software Inc., La Jolla, CA, USA) using the Mann–Whitney Test (P < 0.05).
Sequences accession number
The full data sets have been submitted under BioProject accession number PRJNA596172.
Funding Information
J. Gómez‐Laguna is supported by a ‘Ramón y Cajal’ contract of the Spanish Ministry of Economy and Competitiveness (RYC‐2014‐16735). Hector Argüello is supported by the ‘Beatriz Galindo’ Programme from the Spanish Ministry of Education (BEAGAL‐18‐106). This work was partially supported by the Spanish Ministry of Education and Science (AGL2016‐76111‐R). Research in the Cotter laboratory is funded by Science Foundation Ireland in the form of a centre grants (APC Microbiome Ireland, Grant Number SFI/12/RC/2273, and Vistamilk, Grant Number SFI/16/RC/3835) and by the European Commission under the Horizon 2020 programme under grant number 818368 (MASTER).
Conflict of interests
The authors declare no conflict of interest regarding the results of this research.
Ethical approval
All the animal experiments described in the present study were conducted in strict accordance with the guidelines of the European Union (Directive 2010/63/EU). All animal studies were conducted under protocols approved by the IRTA Ethics Committee and by the Catalan Autonomous Government (Project 3647; FUE‐2017‐00533413).
Supporting information
File S1. PRRSV viral load and antibodies in sera from infected groups. PRRSV genome load was quantified in sera by RT‐qPCR (primary axis). Antibodies against PRRSV‐N‐protein were detected in sera using IDEXX PRRS X3 ELISA test (secondary axis). All data are reported as the median with range of results obtained for the pigs in the Lena group (filled red diamonds for viremia; unfilled red diamonds for antibodies) or in PRRS_3249 group (filled green triangles for viremia; unfilled green triangles for antibodies). Statistically significant differences are indicated with (*): * P < 0.05; ** P < 0.01 (Mann‐Whitney Test; GraphPad Prism 7 software, GraphPad software Inc., La Jolla, CA, USA). S/P: Sample to positive ratio. ELISA cut‐off value of 0.4 was applied according to manufacturer’s instructions.
File S2. Analysis (envfit function of Vegan) of the influence of different factors on the ordination of samples. Significance was established at α = 0.05. Results of the permutation multivariate ANOVA test performed in the ordination analysis.
File S3. Alpha diversity values in fecal samples from PRRSV‐infected pigs. Figure 3A, Chao1 index values in fecal samples by experimental groups Lena, PRRS_3249 and control.
File S4. Alpha‐diversity estimations (Observed OTUs, Shannon, inverted Simpson, Simpson, Chao1) and P‐values obtained when alpha‐diversity by the estimators selected was compared by the factors under study were compared.
File S5. Differentially abundant genus among pigs infected with differentially virulent PRRSV strains. Difference in mean of log2 normalized counts among Lena‐infected, PRRS_3249‐infected and control groups.
File S6. Correlation matrix between abundance of the main genera and the PRRSV infection parameters under study. Spearman correlations among significant phyla (A) and familiae (B) abundance and disease parameters.
Microb. Biotechnol. (2022) 15(00), 1007–1016
References
- Argüello, H. , Estellé, J. , Zaldívar‐López, S. , Jiménez‐Marín, Á. , Carvajal, A. , López‐Bascón, M.A. , et al. (2018) Early Salmonella Typhimurium infection in pigs disrupts microbiome composition and functionality principally at the ileum mucosa. Sci Rep 8: 7788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronesty, E. (2013) Comparison of sequencing utility programs. Open bioinforma J 7: 1–8. [Google Scholar]
- Balka, G. , Podgórska, K. , Brar, M.S. , Bálint, Á. , Cadar, D. , Celer, V. , et al. (2018) Genetic diversity of PRRSV 1 in Central Eastern Europe in 1994–2014: origin and evolution of the virus in the region. Sci Rep 8: 7811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beloeil, P.A. , Chauvin, C. , Proux, K. , Fablet, C. , Madec, F. , and Alioum, A. (2007) Risk factors for Salmonella seroconversion of fattening pigs in farrow‐to‐finish herds. Vet Res 38: 835–848. [DOI] [PubMed] [Google Scholar]
- Bray, R.J. , and Curtis, J.T. (1957) An ordination of upland forest communities of southern Wisconsin. Ecol Monogr 27: 325–349. [Google Scholar]
- Budden, K.F. , Gellatly, S.L. , Wood, D.L. , Cooper, M.A. , Morrison, M. , Hugenholtz, P. , and Hansbro, P.M. (2017) Emerging pathogenic links between microbiota and the gut‐lung axis. Nat Rev Microbiol 15: 55–63. [DOI] [PubMed] [Google Scholar]
- Canelli, E. , Catella, A. , Borghetti, P. , Ferrari, L. , Ogno, G. , De Angelis, E. , et al. (2017) Phenotypic characterization of a highly pathogenic Italian porcine reproductive and respiratory syndrome virus (PRRSV) type 1 subtype 1 isolate in experimentally infected pigs. Vet Microbiol 210: 124–133. [DOI] [PubMed] [Google Scholar]
- Chaudhary, P.P. , Conway, P.L. , and Schlundt, J. (2018) Methanogens in humans: potentially beneficial or harmful for health. Appl Microbiol Biotechnol 102: 3095–3104. [DOI] [PubMed] [Google Scholar]
- Cole, B.J. , Feltcher, M.E. , Waters, R.J. , Wetmore, K.M. , Mucyn, T.S. , Ryan, E.M. , et al. (2017) Genome‐wide identification of bacterial plant colonization genes. PLoS Biol 15: e2002860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, R.C. (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461. [DOI] [PubMed] [Google Scholar]
- Frydas, I.S. , Trus, I. , Kvisgaard, L.K. , Bonckaert, C. , Reddy, V.R. , Li, Y. , et al. (2015) Different clinical, virological, serological and tissue tropism outcomes of two new and one old Belgian type 1 subtype 1 porcine reproductive and respiratory virus (PRRSV) isolates. Vet Res 46: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez‐Laguna, J. , Salguero, F.J. , Pallarés, F.J. , and Carrasco, L. (2013) Immunopathogenesis of porcine reproductive and respiratory syndrome in the respiratory tract of pigs. Vet J 195: 148–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya . (2018). Riboviria: establishing a single taxon that comprises RNA viruses. URL https://talk.ictvonline.org/taxonomy/p/taxonomyhistory?taxnode_id=20186087 [Google Scholar]
- He, Y. , Wen, Q. , Yao, F. , Xu, D. , Huang, Y. , and Wang, J. (2017) Gut‐lung axis: the microbial contributions and clinical implications. Crit Rev Microbiol 43: 81–95. [DOI] [PubMed] [Google Scholar]
- Jiang, N. , Liu, H. , Wang, P. , Huang, J. , Han, H. , and Wang, Q. (2019) Illumina MiSeq sequencing investigation of microbiota in bronchoalveolar lavage fluid and cecum of the swine infected with PRRSV. Curr Microbiol 76: 222–230. [DOI] [PubMed] [Google Scholar]
- Karniychuk, U.U. , Geldhof, M. , Vanhee, M. , Van Doorsselaere, J. , Saveleva, T.A. , and Nauwynck, H.J. (2010) Pathogenesis and antigenic characterization of a new East European subtype 3 porcine reproductive and respiratory syndrome virus isolate. BMC Vet Res 6: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H.B. , and Isaacson, R.E. (2015a) The pig gut microbial diversity: understanding the pig gut microbial ecology through the next generation high throughput sequencing. Vet Microbiol 177: 242–251. [DOI] [PubMed] [Google Scholar]
- Kim, J. , Nguyen, S.G. , Guevarra, R.B. , Lee, I. , and Unno, T. (2015b) Analysis of swine fecal microbiota at various growth stages. Arch Microbiol 197: 753–759. [DOI] [PubMed] [Google Scholar]
- Klindworth, A. , Pruesse, E. , Schweer, T. , Peplies, J. , Quast, C. , Horn, M. , and Glöckner, F.O. (2013) Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next‐generation sequencing‐based diversity studies. Nucleic Acids Res 41: e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone, C.A. , Hamady, M. , Kelley, S.T. , and Knight, R. (2007) Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol 73: 1576–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattsson, J.G. , Bergström, K. , Wallgren, P. , and Johansson, K.E. (1995) Detection of Mycoplasma hyopneumoniae in nose swabs from pigs by in vitro amplification of the 16S rRNA gene. J Clin Microbiol 33: 893–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie, P.J. , and Holmes, S. (2013) phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8: e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan, S.B. , Graham, S.P. , Salguero, F.J. , Sánchez Cordón, P.J. , Mokhtar, H. , Rebel, J.M. , et al. (2013) Increased pathogenicity of European porcine reproductive and respiratory syndrome virus is associated with enhanced adaptive responses and viral clearance. Vet Microbiol 163: 13–22. [DOI] [PubMed] [Google Scholar]
- Niederwerder, M.C. (2017) Role of the microbiome in swine respiratory disease. Vet Microbiol 209: 97–106. [DOI] [PubMed] [Google Scholar]
- Niederwerder, M.C. , Jaing, C.J. , Thissen, J.B. , Cino‐Ozuna, A.G. , McLoughlin, K.S. , and Rowland, R.R. (2016) Microbiome associations in pigs with the best and worst clinical outcomes following co‐infection with porcine reproductive and respiratory syndrome virus (PRRSV) and porcine circovirus type 2 (PCV2). Vet Microbiol 88: 1–11. [DOI] [PubMed] [Google Scholar]
- Ober, R.A. , Thissen, J.B. , Jaing, C.J. , Cino‐Ozuna, A.G. , Rowland, R.R.R. , and Niederwerder, M.C. (2017) Increased microbiome diversity at the time of infection is associated with improved growth rates of pigs after co‐infection with porcine reproductive and respiratory syndrome virus (PRRSV) and porcine circovirus type 2 (PCV2). Vet Microbiol 208: 203–211. [DOI] [PubMed] [Google Scholar]
- Paulson, J.N. , Stine, O.C. , Bravo, H.C. , and Pop, M. (2013) Differential abundance analysis for microbial marker‐gene surveys. Nat Meth 10: 1200–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson‐Leary, J. , Zhao, C. , Bittinger, K. , Eacret, D. , Luz, S. , Vigderman, A.S. , et al. (2019) The gut microbiome regulates the increases in depressive‐type behaviors and in inflammatory processes in the ventral hippocampus of stress vulnerable rats. Mol Psychiatry 25: 1068–1079. [DOI] [PubMed] [Google Scholar]
- Rivera‐Chávez, F. , and Bäumler, A.J. (2015) The pyromaniac inside you: Salmonella metabolism in the host gut. Annu Rev Microbiol 69: 31–48. [DOI] [PubMed] [Google Scholar]
- Rodríguez‐Gómez, I.M. , Sánchez‐Carvajal, J.M. , Pallarés, F.J. , Mateu, E. , Carrasco, L. , and Gómez‐Laguna, J. (2019) Virulent Lena strain induced an earlier and stronger downregulation of CD163 in bronchoalveolar lavage cells. Vet Microbiol 235: 101–109. [DOI] [PubMed] [Google Scholar]
- Schmieder, R. , and Edwards, R. (2011) Quality control and preprocessing of metagenomic datasets. Bioinformatics 27: 863–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoster, A. , Mosing, M. , Jalali, M. , Staempfli, H.R. , and Weese, J.S. (2016) Effects of transport, fasting and anaesthesia on the faecal microbiota of healthy adult horses. Equine Vet J 48: 595–602. [DOI] [PubMed] [Google Scholar]
- Senthilkumar, D. , Rajukumar, K. , Sen, A. , Kumar, M. , Shrivastava, D. , Kalaiyarasu, S. , et al. (2018) Pathogenic characterization of porcine reproductive and respiratory syndrome virus of Indian origin in experimentally infected piglets. Transbound Emerg Dis 65: 1522–1536. [DOI] [PubMed] [Google Scholar]
- Shade, A. , McManus, P. , and Handelsman, J. (2013) Unexpected diversity during community succession in the apple. MBio 4: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, M. , Lam, T.T. , Hon, C.C. , Hui, R.K. , Faaberg, K.S. , Wennblom, T. , et al. (2010) Molecular epidemiology of PRRSV: a phylogenetic perspective. Virus Res 154: 7–17. [DOI] [PubMed] [Google Scholar]
- Sibila, M. , Calsamiglia, M. , Segalés, J. , Blanchard, P. , Badiella, L. , Le Dimna, M. , et al. (2004) Use of a polymerase chain reaction assay and an ELISA to monitor porcine circovirus type 2 infection in pigs from farms with and without postweaning multisystemic wasting syndrome. Am J Vet Res 65: 88–92. [DOI] [PubMed] [Google Scholar]
- Sinn, L.J. , Klingler, E. , Lamp, B. , Brunthaler, R. , Weissenböck, H. , Rümenapf, T. , and Ladinig, A. (2016) Emergence of a virulent porcine reproductive and respiratory syndrome virus (PRRSV) 1 strain in Lower Austria. Porcine Health Manag 2: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadejek, T. , Larsen, L.E. , Podgórska, K. , Bøtner, A. , Botti, S. , Dolka, I. , et al. (2017) Pathogenicity of three genetically diverse strains of PRRSV Type 1 in specific pathogen free pigs. Vet Microbiol 209: 13–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadejek, T. , Stankevicius, A. , Murtaugh, M.P. , and Oleksiewicz, M.B. (2013) Molecular evolution of PRRSV in Europe: current state of play. Vet Microbiol 165: 21–28. [DOI] [PubMed] [Google Scholar]
- Tian, K. , Yu, X. , Zhao, T. , Feng, Y. , Cao, Z. , Wang, C. , et al. (2007) Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS One 2: e526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, L. , Estellé, J. , Kiilerich, P. , Ramayo‐Caldas, Y. , Xia, Z. , Feng, Q. , et al. (2016) A reference gene catalogue of the pig gut microbiome. Nat Microbiol 1: 16161. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , and Wang, R. (2018) Gut microbiota modulates drug pharmacokinetics. Drug Metab Rev 50: 357–368. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
File S1. PRRSV viral load and antibodies in sera from infected groups. PRRSV genome load was quantified in sera by RT‐qPCR (primary axis). Antibodies against PRRSV‐N‐protein were detected in sera using IDEXX PRRS X3 ELISA test (secondary axis). All data are reported as the median with range of results obtained for the pigs in the Lena group (filled red diamonds for viremia; unfilled red diamonds for antibodies) or in PRRS_3249 group (filled green triangles for viremia; unfilled green triangles for antibodies). Statistically significant differences are indicated with (*): * P < 0.05; ** P < 0.01 (Mann‐Whitney Test; GraphPad Prism 7 software, GraphPad software Inc., La Jolla, CA, USA). S/P: Sample to positive ratio. ELISA cut‐off value of 0.4 was applied according to manufacturer’s instructions.
File S2. Analysis (envfit function of Vegan) of the influence of different factors on the ordination of samples. Significance was established at α = 0.05. Results of the permutation multivariate ANOVA test performed in the ordination analysis.
File S3. Alpha diversity values in fecal samples from PRRSV‐infected pigs. Figure 3A, Chao1 index values in fecal samples by experimental groups Lena, PRRS_3249 and control.
File S4. Alpha‐diversity estimations (Observed OTUs, Shannon, inverted Simpson, Simpson, Chao1) and P‐values obtained when alpha‐diversity by the estimators selected was compared by the factors under study were compared.
File S5. Differentially abundant genus among pigs infected with differentially virulent PRRSV strains. Difference in mean of log2 normalized counts among Lena‐infected, PRRS_3249‐infected and control groups.
File S6. Correlation matrix between abundance of the main genera and the PRRSV infection parameters under study. Spearman correlations among significant phyla (A) and familiae (B) abundance and disease parameters.


