Head and neck squamous cell carcinoma (HNSCC), as the most common type (>90%) of head and neck cancer, includes various epithelial malignancies that arise in the nasal cavity, oral cavity, pharynx, and larynx. In 2020, approximately 878 000 new cases and 444 000 deaths linked to HNSCC occurred worldwide (Sung et al., 2021). Due to the associated frequent recurrence and metastasis, HNSCC patients have poor prognosis with a five-year survival rate of 40%–50% (Jou and Hess, 2017). Therefore, novel prognostic biomarkers need to be developed to identify high-risk HNSCC patients and improve their disease outcomes.
The expression of certain genes, especially oncogenes, has been reported to exhibit significant relationships with cancer prognosis, such as genes epidermal growth factor receptor (EGFR) andcyclin-dependent kinase inhibitor 2A(CDKN2A) thathave prognostic value in HNSCC (Polanska et al., 2014). However, thus far, few reported biomarkers have been applied in clinical management because these were usually identified in a small number of patients and lacked independent validation (Kim et al., 2014; Fauzi et al., 2020). With the availability of gene expression profiles and related clinical follow-up information in public databases, such as The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO), the expression profiles of large cohorts could be used for the identification and evaluation of prognostic biomarkers. However, special bioinformatics techniques are needed to analyze and integrate these complex multidimensional omics and clinicopathological data, which has been hindering most researchers without much bioinformatics skills. The application of such bioinformatics tools has shown success in basic and translational medicine studies, especially in the field of biomarker development (Zhang et al., 2019). Unfortunately, a suitable and easy-to-use bioinformatics tool to allow the fast screening and evaluation of prognostic biomarkers in different HNSCC cohorts is still lacking.
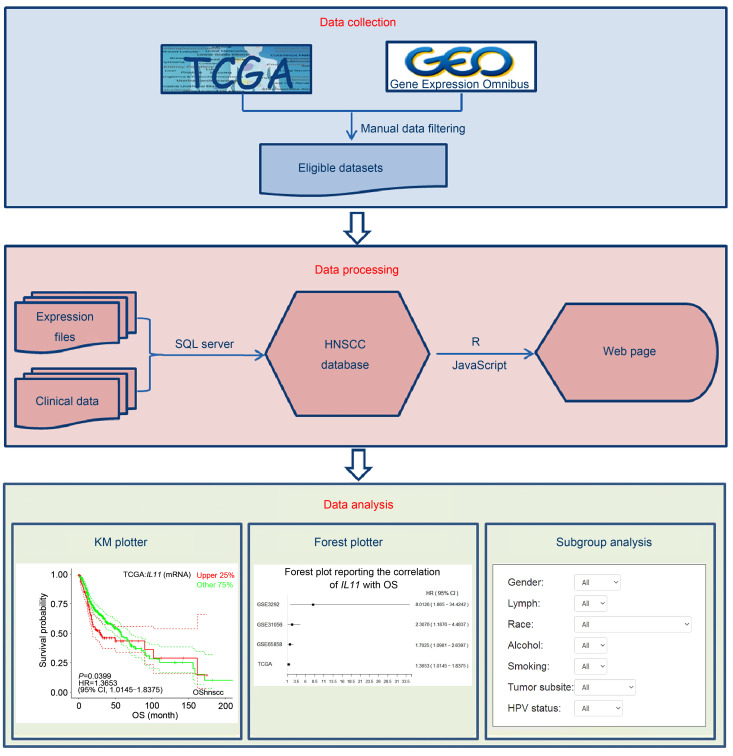
Therefore, in the current study, we developed the Online consensus Survival tool for Head and Neck Squamous Cell Carcinoma (OShnscc), which could offer a new possibility for clinicians and researchers to assess the prognostic value of genes independently in multiple HNSCC cohorts. The development process of OShnscc is presented in Fig. 1. This web-based tool can be accessed at http://bioinfo.henu.edu.cn/HNSC/HNSCList.jsp.
Fig. 1. Architecture diagram of the development process of OShnscc. OShnscc: Online consensus Survival tool for Head and Neck Squamous Cell Carcinoma; TCGA: The Cancer Genome Atlas; HNSCC: head and neck squamous cell carcinoma; KM: Kaplan-Meier.
OShnscc contains a total of 1366 clinical cases from nine datasets, including one TCGA dataset and eight GEO datasets. In this tool, six types of survival terms are provided for survival analysis, including overall survival (OS), disease-specific survival (DSS), disease-free survival (DFS), progression-free survival (PFS), disease-free interval (DFI), and progression-free interval (PFI). The survival terms have been described in detail in original studies (Wichmann et al., 2015; Liu et al., 2018). In brief, OS is estimated from the time of diagnosis to the last follow-up, and DSS is similar to OS but only includes patients who died of cancer. DFS is measured from the time of primary recovery to the first observation of relapse or death from any cause by the last follow-up, and DFI is similar to DFS but excludes patients in Stage IV or those who deceased without relapse. PFS is defined from the registration date until the date of detection of definitive disease progression, recurrence, metastasis, new primary tumors in all sites, or death from any cause, and PFI is similar to PFS but excludes patients who died from causes other than cancer. A total of 888 (65%) patients are male, and the median age is 59 years; 731 (54%) patients have OS information, and the median OS time is 24 months. Patients without either clinical follow-up time or gene expression data were removed. The patient clinical characteristics, including tumor node metastasis (TNM) stage, grade, gender, smoking history, lymph invasion, human papilloma virus (HPV) status, and race, were also included and set as confounding clinical factors for subgroup analysis in OShnscc. The clinical characteristics and the detailed items of all datasets in OShnscc are summarized in Tables 1 and S1, respectively.
Table 1.
Clinical characteristics of HNSCC datasets used in OShnscc
| Data source | Platform | Sample size | No. of deaths | Median age(years) |
Median OS (months) |
Male(%) | HPV status (+/-/NA) | StageI/II/III/IV/NA(%) | Smoking (%) | Tumor subsite | Survival term |
|---|---|---|---|---|---|---|---|---|---|---|---|
| TCGA | Illumina HiSeqV2 | 523 | 220 | 61 | 21.50 | 73.04 | 35/239/249 | 5.16/13.96/15.68/51.05/14.15 | 75.14 | OC, OP,HP, L, T | OS, DSS, PFS, DFI, PFI |
| GSE10300 | GPL570 | 44 | 16 | 53 | 79.55 | 2.27/6.82/18.18/43.18/29.55 | 65.91 | OC, OP,HP, L | RFS | ||
| GSE27020 | GPL96 | 109 | 64 | 95.45 | 11.01/16.51/33.03/39.45/0 | 99.08 | L | DFS | |||
| GSE25727 | GPL8432 | 56 | 60 | 92.86 | 87.50 | L | DFS | ||||
| GSE31056 | GPL10526 | 96 | 36 | 59 | 13.54 | 62.50 | 0/20.83/3.13/37.50/38.54 | OC | OS, DFS | ||
| GSE3292 | GPL570 | 33 | 8 | 56 | 24.00 | 81.82 | 6.06/6.06/24.24/57.58/6.06 | OC, OP,HP, L | OS, DFS | ||
| GSE39366 | GPL9053 | 138 | 68 | 57 | 68.84 | 14/82/42 | 5.80/10.14/20.29/60.87/2.90 | 78.26 | OC, OP,HP, L | RFS | |
| GSE41613 | GPL570 | 97 | 51 | 26.30 | 68.04 | 0/97/0 | OC | OS, DSS | |||
| GSE65858 | GPL10588 | 270 | 94 | 59 | 27.90 | 82.59 | 73/196/1 | 6.67/13.70/13.70/65.93/0 | 88.52 | OC, OP,HP, L | OS, PFS |
| Total | 1366 | 493 |
HNSCC: head and neck squamous cell carcinoma; OShnscc: Online consensus Survival tool for Head and Neck Squamous Cell Carcinoma; OS: overall survival; HPV: human papilloma virus; NA: not available; TCGA: The Cancer Genome Atlas; OC: oral cavity; OP: oropharynx; HP: hypopharynx; L: larynx; T: tonsil; DSS: disease-specific survival; PFS: progression-free survival; DFI: disease-free interval; PFI: progression-free interval; RFS: relapse-free survival; DFS: disease-free survival. PFS, DFI, and PFI were defined by Wichmann et al. (2015) and Liu et al. (2018).
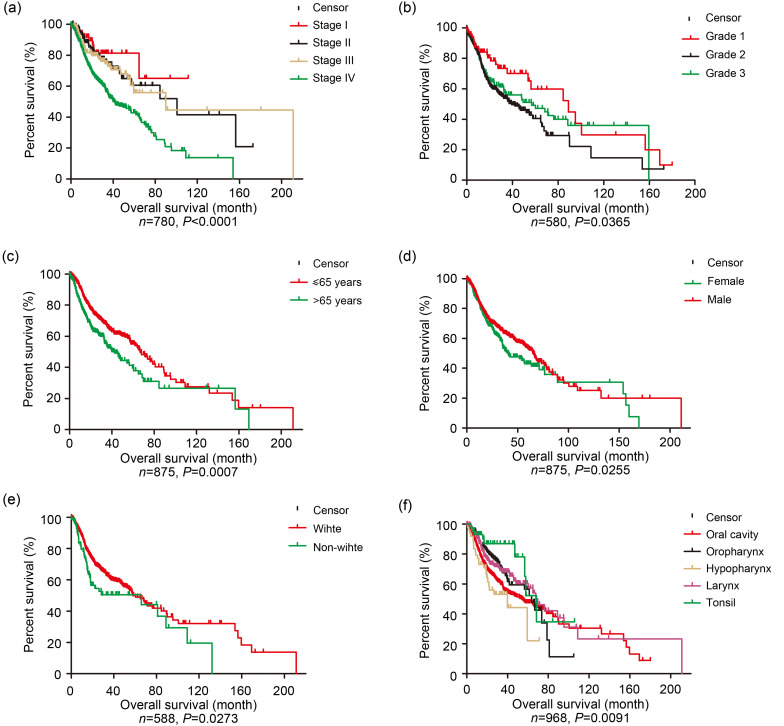
Six clinical characteristics including TNM stage, grade, age, gender, race, and tumor subsite were measured for association with the OS of HNSCC patients in the OShnscc datasets. As shown in Fig. 2, the clinical factors including TNM stage (Fig. 2a), grade (Fig. 2b), age (Fig. 2c), gender (Fig. 2d), race (Fig. 2e), and tumor subsite (Fig. 2f) showed significant associations with the OS of HNSCC patients (P<0.0001, P=0.0365, P=0.0007, P=0.0255, P=0.0273, and P=0.0091, respectively), which was consistent with previous reports (Pai and Westra, 2009; Polanska et al., 2014; Naghavi et al., 2016).
Fig. 2. Survival analysis by clinical factors for all datasets in OShnscc. (a) TNM stage; (b) Grade; (c) Age; (d) Gender; (e) Race; (f) Tumor subsite. OShnscc: Online consensus Survival tool for Head and Neck Squamous Cell Carcinoma; TNM: tumor node metastasis.
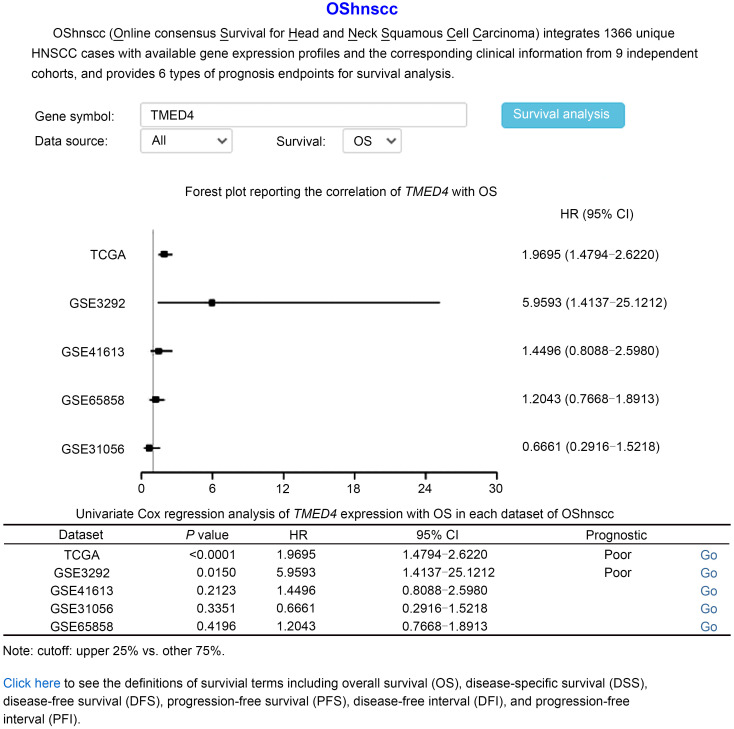
The main function of OShnscc is to evaluate and verify the prognostic value of a given gene in HNSCC. On its home page, OShnscc provides a fast survival analysis model using the default parameter of "Data Source" as "All," the "Survival" term as "OS," and "Split patients by" cutoff as "Upper 25%." The user only needs to input the genes of interest in the term "Gene symbol" and click the "Survival analysis" button. Then, a forest plot and a table that summarize the prognostic values, including hazard ratio (HR) with 95% confidence interval (CI) and the log-rank P value of queried genes in each dataset of OShnscc, will be generated. Taking the transmembrane p24 trafficking protein 4 (TMED4) gene as an example, if the user inputs the gene symbol "TMED4" and then clicks the "Survival analysis" button, a forest plot and a table containing the prognostic value of TMED4 will be presented on the output webpage. The results shown in Fig. 3 indicate that the high expression of TMED4 is associated with the poor OS of HNSCC patients in the dataset of TCGA (n=499, P<0.0001, HR=1.9695, 95% CI=1.4794–2.6220) and GSE3292 (n=33, P=0.0150, HR=5.9593, 95% CI=1.4137–25.1212), while no significant relationship was found between TMED4 expression and the OS of HNSCC patients in the dataset of GSE31056 (n=96, P=0.3351, HR=0.6661, 95% CI=0.2916–1.5218) or GSE65858 (n=270, P=0.4196, HR=1.2043, 95% CI=0.7668–1.8913). When the user query of a gene with a reported prognostic role is submitted, a reminder stating "The query gene has been previously reported as a prognostic biomarker in [Reference]" will be presented underneath the analysis result on the output page. By clicking on the "Go" button on the right side of the table, the user can easily acquire the Kaplan-Meier (KM) plots of TMED4 for individual cohorts such as the TCGA dataset (Fig. 4).
Fig. 3. Summary of prognostic analysis of TMED4 expression in OShnscc using default parameters. TMED4: transmembrane p24 trafficking protein 4 gene; OS: overall survival; TCGA: The Cancer Genome Atlas; HR: hazard ratio; CI: confidence interval.
Fig. 4. Kaplan-Meier curve based on TMED4 expression in OShnscc using default parameters of "TCGA" as data source, "OS" for survival, and "Upper 25%" for patients splitting. TMED4: transmembrane p24 trafficking protein 4 gene; OShnscc: Online consensus Survival tool for Head and Neck Squamous Cell Carcinoma; TCGA: The Cancer Genome Atlas; HNSCC: head and neck squamous cell carcinoma; TNM: tumor node metastasis; mRNA: messenger RNA; HR: hazard ratio; CI: confidence interval; OS: overall survival; HPV: human papilloma virus; DFI: disease-free interval; PFI: progression-free interval.
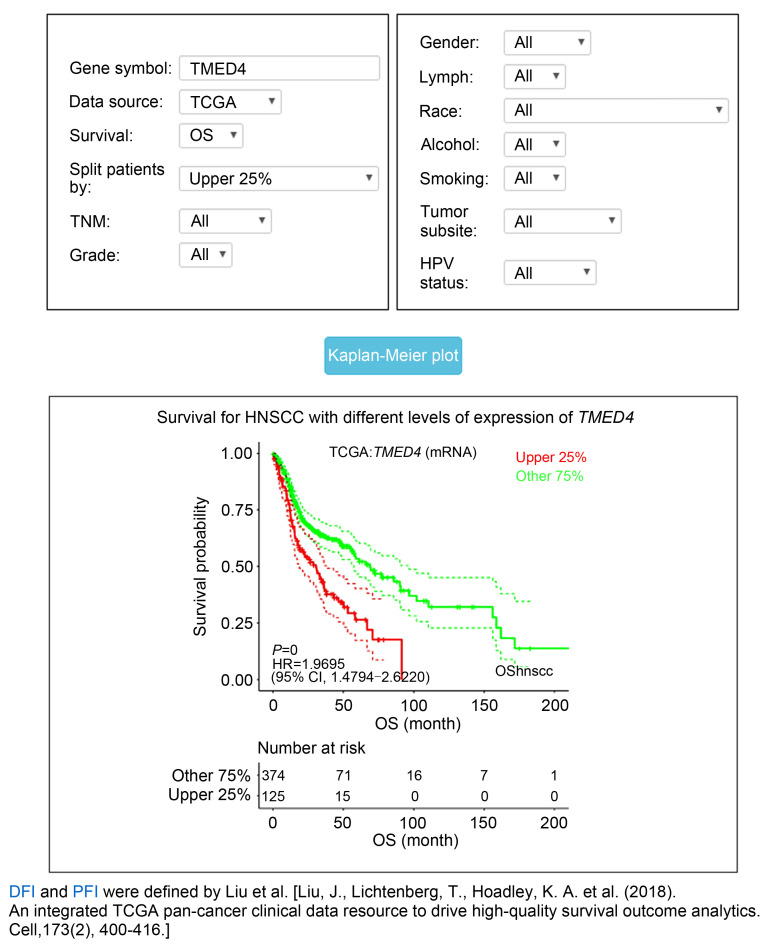
The OShnscc platform also provides diverse options of parameters for survival analysis. Through choosing the datasets in the term "Data source," OShnscc turns to the individual dataset page, and several parameters with 2–5 options are provided. For example, users may want to evaluate the association between the gene of interest and the disease prognosis of patients from the TCGA dataset, and subsequently choose the TCGA dataset in "Data source." Two main parameters including "Survival" and "Split patients by" and nine optional parameters including TNM stage, grade, gender, etc. are displayed on the TCGA webpage (Fig. 5). Users only need to type the gene symbol, select the prognostic endpoint in "Survival" and the cutoff value in "Split patients by," and then click the "Kaplan-Meier plot" button for the survival curve with HR, 95% CI, and log-rank P value to be graphically displayed on the webpage. In addition, optional parameters including TNM stage, HPV status, smoking history, gender, lymph, histological type, and race are also provided for subgroup analysis to limit the prognosis in a subgroup for the intended clinical factor of HNSCCs.
Fig. 5. Options of main input parameters and clinical factors provided in OShnscc. (a) Survival term; (b) Split patients by; (c) TNM stage; (d) Grade; (e) Gender; (f) Lymph invasion; (g) Race; (h) Alcohol; (i) Smoking; (j) Tumor subsite; (k) HPV status. OShnscc: Online consensus Survival tool for Head and Neck Squamous Cell Carcinoma; TNM: tumor node metastasis; HPV: human papilloma virus.
In order to measure the performance of OShnscc and the repeatability of prognostic biomarkers reported in the literature, a total of 39 prognostic genes reported by 36 studies were collected for testing in OShnscc. As shown in Tables S2 and S3, 38 out of 39 previously reported genes (97%) have been confirmed for their prognostic values in OShnscc, and 16 out of 39 genes (41%) showed significant association with HNSCC outcomes for at least two independent datasets in OShnscc. However, one gene (SRY-box transcription factor 4 (SOX4)) did not show any significance for prognosis in any cohort from OShnscc. A total of 39 previously reported prognostic genes, except SOX4, were confirmed in at least one of the HNSCC cohorts in OShnscc. The discrepancy of the prognostic performance of SOX4 between OShnscc and the literature is likely due to race (Naghavi et al., 2016). The race of cohorts reported in the literature was Asian (Korea), while the cohort included in OShnscc is mostly the Caucasian population. The survival analysis of previously reported prognostic biomarkers in OShnscc demonstrated that OShnscc performs well in evaluating the prognosis of genes and can be a highly useful tool for screening and evaluating prognostic biomarkers for HNSCC patients.
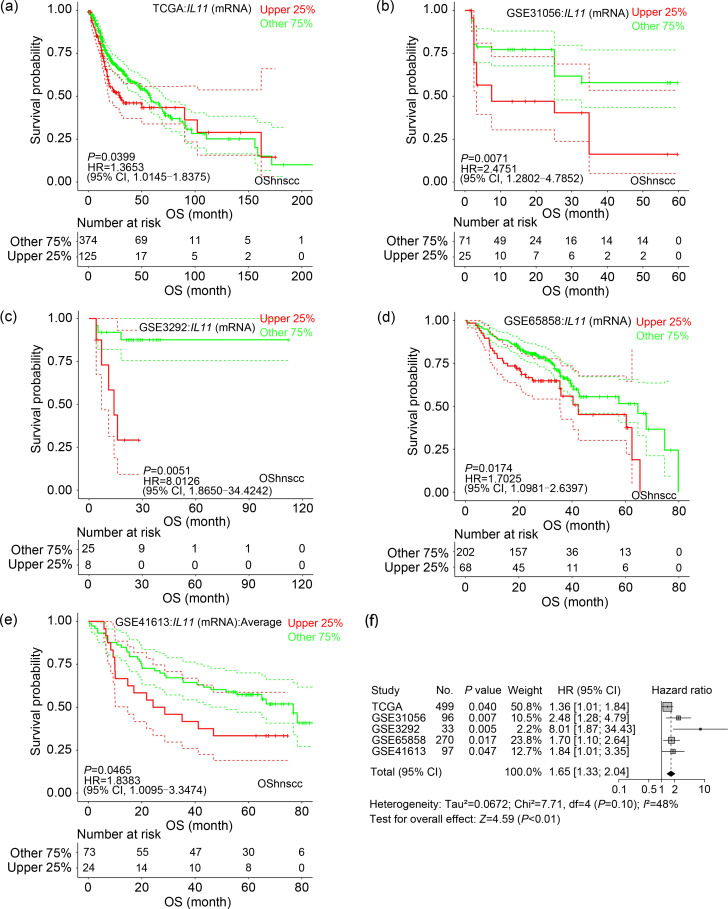
OShnscc can be used to facilitate discovering new prognostic biomarkers. Through OShnscc, we found that interleukin 11 (IL11) has a significant association with the OS of HNSCC patients in five OShnscc datasets (Figs. 6a–6e). Highly expressed IL11 is significantly associated with poor OS in the HNSCC datasets including TCGA (n=499, P=0.040, HR=1.365, 95% CI=1.015–1.838), GSE31056 (n=96, P=0.007, HR=2.475, 95% CI=1.280–4.785), GSE3292 (n=33, P=0.005, HR=8.013, 95% CI=1.865–34.424), GSE65858 (n=270, P=0.017, HR=1.703, 95% CI=1.098–2.640), and GSE41613 (n=97, P=0.047, HR=1.838, 95% CI=1.010–3.347). Moreover, HNSCC patients with high IL11 expression levels had shorter OS (n=995, HR=1.64, 95% CI=1.32–2.03) based on the results of meta-analysis (Fig. 6f). To our knowledge, IL11 is a member of glycoprotein-130 (GP-130) cytokines, and overexpressed IL11 has been reported in tumorigenesis through the Janus kinase (JAK)-signal transducers and activators of transcription 3 (STAT3) pathway in cancers, including breast cancer, colorectal cancer, and gastric cancer (Xu et al., 2016). In addition, the upregulation of IL11 has been shown to be associated with shortened survival in clear cell renal cell carcinoma (Pan et al., 2015), non-small cell lung cancer (Zhao et al., 2018), and breast cancer (Hanavadi et al., 2006). This gene has not been previously reported as a prognostic biomarker in HNSCC, and thus it should be validated in a clinical study for novel prognostic biomarker development. Based on the above data, we hypothesize that IL11 could be a potential candidate for further experimental verification.
Fig. 6. Survival analysis of IL11 gene in OShnscc. (a‒e) Kaplan-Meier survival curves of IL11 with OS in the data source of TCGA (a), GSE31056 (b), GSE3292 (c), GSE65858 (d), GSE41613 (e); (f) Forest plots for the meta-analysis of the prognostic values of IL11 gene for OS analysis in OShnscc. IL11: interleukin 11; OShnscc: Online consensus Survival tool for Head and Neck Squamous Cell Carcinoma; TCGA: The Cancer Genome Atlas; OS: overall survival; HR: hazard ratio; CI: confidence interval; mRNA: messenger RNA.
Currently, several messenger RNA (mRNA)-focused survival analysis tools exist, such as Gene Expression Profiling Interactive Analysis (GEPIA) (Tang et al., 2017), KM plotter (Nagy et al., 2018), and UALCAN (Xu et al., 2021). Nevertheless, most of these tools are mainly or exclusively based on datasets collected from the TCGA database, have limited number of HNSCC cases, and lack individual cohorts for independent validation. In comparison to these tools, OShnscc has several unique features: (1) it is an online survival assessment tool specifically for HNSCC, and contains by far the largest number of HNSCC cases (1366 cases) for prognostic analysis; (2) it enables users to evaluate the prognostic value of the queried gene independently in individual cohorts, which could generate more robust results for prognostic analysis; (3) it allows users to perform survival analysis for subgroups by filtering HNSCC patients through the use of different terms of clinical confounding factors. However, OShnscc has limitations in survival analysis. For example, although it features the largest number of HNSCC patients, the corresponding case numbers for the prognostic analysis of recurrence and metastasis are still low. Nevertheless, we aim to expand the number of HNSCC cases for OShnscc once the novel relative HNSCC datasets become available.
In summary, OShnscc is proposed as a publicly available, free online web tool for the rapid analysis of the prognostic values of genes in HNSCC. This tool is user-friendly, allowing researchers with limited bioinformatics background to screen and evaluate the prognostic values of genes for different cohorts of HNSCC, which may accelerate the clinical development of prognostic biomarkers in HNSCC. Upon the release of additional HNSCC datasets, we will keep updating the database of OShnscc to perform prognostic analysis for HNSCC patients at a large scale.
Materials and methods
Detailed methods are provided in the electronic supplementary materials of this paper.
Supplementary information
Funding Statement
This work was supported by the National Natural Science Foundation of China (No. U2004136), the Supporting Program for Central Plain Young Top Talents (No. ZYQR201912176), the Program for Innovative Talents of Science and Technology in Henan Province (No. 18HASTIT048), the Program for Science and Technology Development in Henan Province (Nos. 212102310150, 202102310063, and SBGJ2018041), the Kaifeng Science and Technology Project (No. 1908001), and the Supporting Grant of Henan University (Nos. 2018YLJC01, 2019YLXKJC01, and 2020YLZDYJ14), China.
Author contributions
Guosen ZHANG performed data collection, developed the web tool, and wrote and edited the manuscript. Qiang WANG and Xinlei QI performed data collection and developed the web tool. Huimin YANG, Xiaodong SU, Manman YANG, Chao JIANG, and Yang AN contributed to data collection and analysis. Hong ZHENG, Lu ZHANG, and Wan ZHU performed data analysis and paper writing. Jiancheng GUO and Xiangqian GUO contributed to the study design, data analysis, writing and editing of the manuscript. All authors have read and approved the final manuscript, and therefore, have full access to all the data in the study and take responsibility for the integrity and security of the data.
Compliance with ethics guidelines
Guosen ZHANG, Qiang WANG, Xinlei QI, Huimin YANG, Xiaodong SU, Manman YANG, Chao JIANG, Yang AN, Hong ZHENG, Lu ZHANG, Wan ZHU, Jiancheng GUO, and Xiangqian GUO declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- Fauzi FH, Hamzan NI, Rahman NA, et al. , 2020. Detection of human papillomavirus in oropharyngeal squamous cell carcinoma. J Zhejiang Univ-Sci B (Biomed & Biotechnol), 21(12): 961-976. 10.1631/jzus.B2000161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanavadi S, Martin TA, Watkins G, et al. , 2006. Expression of interleukin 11 and its receptor and their prognostic value in human breast cancer. Ann Surg Oncol, 13(6): 802-808. 10.1245/ASO.2006.05.028 [DOI] [PubMed] [Google Scholar]
- Jou A, Hess J, 2017. Epidemiology and molecular biology of head and neck cancer. Oncol Res Treat, 40(6): 328-332. 10.1159/000477127 [DOI] [PubMed] [Google Scholar]
- Kim KY, McShane LM, Conley BA, 2014. Designing biomarker studies for head and neck cancer. Head Neck, 36(7): 1069-1075. 10.1002/hed.23444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JF, Lichtenberg T, Hoadley KA, et al. , 2018. An integrated TCGA Pan-Cancer Clinical Data Resource to drive high-quality survival outcome analytics. Cell, 173(2): 400-416.e11. 10.1016/j.cell.2018.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naghavi AO, Echevarria MI, Strom TJ, et al. , 2016. Treatment delays, race, and outcomes in head and neck cancer. Cancer Epidemiol, 45: 18-25. 10.1016/j.canep.2016.09.005 [DOI] [PubMed] [Google Scholar]
- Nagy Á, Lánczky A, Menyhárt O, et al. , 2018. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep, 8: 9227. 10.1038/s41598-018-27521-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai SI, Westra WH, 2009. Molecular pathology of head and neck cancer: implications for diagnosis, prognosis, and treatment. Annu Rev Pathol, 4: 49-70. 10.1146/annurev.pathol.4.110807.092158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D, Xu L, Liu HO, et al. , 2015. High expression of interleukin-11 is an independent indicator of poor prognosis in clear-cell renal cell carcinoma. Cancer Sci, 106(5): 592-597. 10.1111/cas.12638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polanska H, Raudenska M, Gumulec J, et al. , 2014. Clinical significance of head and neck squamous cell cancer biomarkers. Oral Oncol, 50(3): 168-177. 10.1016/j.oraloncology.2013.12.008 [DOI] [PubMed] [Google Scholar]
- Sung H, Ferlay J, Siegel RL, et al. , 2021. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin, 71(3): 209-249. 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- Tang ZF, Li CW, Kang BX, et al. , 2017. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res, 45(W1): W98-W102. 10.1093/nar/gkx247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichmann G, Rosolowski M, Krohn K, et al. , 2015. The role of HPV RNA transcription, immune response-related gene expression and disruptive TP53 mutations in diagnostic and prognostic profiling of head and neck cancer. Int J Cancer, 137(12): 2846-2857. 10.1002/ijc.29649 [DOI] [PubMed] [Google Scholar]
- Xu DH, Zhu ZW, Wakefield MR, et al. , 2016. The role of IL-11 in immunity and cancer. Cancer Lett, 373(2): 156-163. 10.1016/j.canlet.2016.01.004 [DOI] [PubMed] [Google Scholar]
- Xu N, Dong RN, Lin TT, et al. , 2021. Development and validation of novel biomarkers related to M2 macrophages infiltration by weighted gene co-expression network analysis in prostate cancer. Front Oncol, 11: 634075. 10.3389/fonc.2021.634075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang GS, Wang Q, Yang MS, et al. , 2019. OSblca: a web server for investigating prognostic biomarkers of bladder cancer patients. Front Oncol, 9: 466. 10.3389/fonc.2019.00466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Liu Y, Liu R, et al. , 2018. Upregulation of IL-11, an IL-6 family cytokine, promotes tumor progression and correlates with poor prognosis in non-small cell lung cancer. Cell Physiol Biochem, 45(6): 2213-2224. 10.1159/000488166 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.