Abstract
To investigate Molybdenum (Mo) and Cadmium (Cd) co-induced the levels of autophagy-related genes via AMPK/mTOR signaling pathway in Shaoxing Duck (Anas platyrhyncha) kidney, 60 healthy 11-day-old ducks were randomly divided into 6 groups, which were treated with Mo or/and Cd at different doses on the basal diet for 120 d. Kidney samples were collected on day 120 to determine the mRNA expression levels of adenosine 5′-monophosphate (AMP)-activated protein kinase α1 (AMPKα1), mammalian target of rapamycin (mTOR), Beclin-1, autophagy-related gene-5 (Atg5), microtubule-associated protein light chain A (LC3A), microtubule-associated protein light chain B (LC3B), sequestosome-1, and Dynein by real-time quantitative polymerase chain reaction. Meanwhile, ultrastructural changes of the kidney were observed. The results indicated that the mTOR and P62 mRNA expression levels were significantly downregulated, but the Atg5 and Beclin-1 mRNA levels were remarkably upregulated in all treated groups compared to control group, and their changes were greater in joint groups. Additionally, compared to control group, the Dynein mRNA expression level was apparently downregulated in co-treated groups, the LC3B, LC3A, and AMPKα1 expression levels were dramatically upregulated in single treated groups and they were not obviously different in co-treated groups. Ultrastructural changes showed that Mo and Cd could markedly increase the number of autophagosomes. Taken together, it suggested that dietary Mo and Cd might induce autophagy via AMPK/mTOR signaling pathway in duck kidney, and it showed a possible synergistic relationship between the 2 elements.
key words: Molybdenum, Cadmium, autophagy, kidney, duck
Highlights
-
1.
The joint toxic effects of Mo and Cd were evaluated in Shaoxing Duck (Anas platyrhyncha).
-
2.
Dietary Mo and Cd could induce autophagy via AMPK/mTOR signaling pathway in Shaoxing Duck (Anas platyrhyncha) kidney.
-
3.
Mo and Cd had a synergistic effect between Mo and Cd induced autophagy in kidney.
INTRODUCTION
Molybdenum (Mo) is an essential trace element for plants, animals, and humans (Bersényi et al., 2008), and it forms the catalytic center of a large variety of enzymes such as nitrogenase, nitrate reductases, sulphite oxidase, and xanthine oxidoreductases (Schwarz et al., 2009). Mo is distributed throughout the environment. It has been widely used for industrial stainless steel, mining, cast iron, fertilizer manufacture, and agricultural activities (Mendel, 2005). A total of 10 million tons of Mo are produced all over the world, 800 tons of which burning into the environment each year. Many studies have reported that improper mining and industrialization can lead to an increase in concentration of Mo in soil, water, and air, then it can be absorbed by aquatic and terrestrial organisms (Davies et al., 2005; Yang et al., 2016). Excessive Mo enters animals and then accumulates in body. High doses of Mo can cause adverse effects on kidney, liver, spleen, and bones in aquatic and terrestrial animals (Abramovich et al., 2011; Cao et al., 2016; Liao et al., 2017). In addition, the kidney is the most important target organ for Mo-induced toxicity. Mo can induce oxidative stress and apoptosis in kidney (Shi et al., 2017), but the relationship between Mo and autophagy is less reported.
Cadmium (Cd) is a toxic heavy metal widely used in industries. It can enter the environment and stay intactly for extremely long biological half-life. Unlike other complex organic pollutants, Cd cannot be degraded by microorganisms, it enters the food chain through contact with Cd in metal industries, paints, fertilizers, automobiles, and batteries, and resulting in Cd accumulation in ecosystems (Luo et al., 2016). It is now well accepted that occupational and environmental pollutant of Cd can accumulate in kidney, liver, pancreas, and testis and so on, and cause adversely effects on the functions and structure of these organs (Brzóska et al., 2003; Satarug et al., 2003; Xu et al., 2013; Liu et al., 2016; Zhang et al., 2017). Exposure to Cd could induce various cellular responses such as apoptosis, autophagy, necrosis, proliferation, and carcinogenesis (Templeton and Liu, 2010; Chen et al., 2011; Lu et al., 2015). The nephrotoxicity of Cd has been extensively studied and reported in literatures, and there are growing evidences that autophagy is the fundamental molecular mechanisms of Cd nephrotoxicity (Chargui et al., 2011; So and Oh, 2016).
Autophagy is an evolutionary conserved catabolic process used by eukaryotic cells for the degradation of damaged or superfluous proteins and organelles (Chen and Klionsky, 2011). It plays important roles in cell survival and maintenance (Parzych and Klionsky, 2014). There are three types of autophagy, namely macro-autophagy, micro-autophagy, and chaperone-mediated autophagy (Wang and Choi, 2014). Among these 3 types of autophagy, macro-autophagy referred as autophagy is the best characterized (Liuzzi et al., 2014). Autophagy plays a role in Cd-induced nephrotoxicity (Thévenod and Lee, 2015). It has been recorded as the initial response of a cell to a toxic metal in a concentration- and time-dependent manner (Yang et al., 2016). At the early stage of exposure, Cd could induce protective autophagy in renal tubular epithelial cells (Liu et al., 2017; Song et al., 2017; Yue et al., 2017). Low doses of Cd can induce autophagy, but if the pressure continues, autophagy eventually fails in maintaining cell viability and in turn induces cell death (Chargui et al., 2011). Various signaling pathways are triggered through cellular proteins and/or protein kinases that can lead to autophagy (Chatterjee et al., 2014). AMPK/mTOR signaling pathway is important in regulating autophagy. Adenosine 5′-monophosphate-activated protein kinase (AMPK) and mammalian target of rapamycin (mTOR) are important regulators of autophagy in cells (Fan et al., 2015). AMPK regulates mTOR activity and plays a central role in controlling cell growth, proliferation and autophagy.
China is rich in tungsten ore. In the mining and screening processes of tungsten ore, due to improper handling of mining and waste, a large number of tailings containing Mo and Cd contaminate soil and water sources, which in turn affect plants and animals, especially waterfowl. Shaoxing duck (Anas platyrhyncha) is the main breed in Southern China, which is also the main breed in Southern Jiangxi province polluted by Mo and Cd. The duck has many advantages such as high laying rate, long egg production peak duration, high feed utilization rate, and strong life force. Therefore it is one of the most popular breeds in China and we used this species in the experiment. Thus, it is necessary to study the combined toxicity of Mo and Cd in view of the current status of combined pollution. Our previous research showed that Mo and Cd presented a synergistic effect on the damage of duck kidney (Xia et al., 2015). However, whether autophagy is involved in Mo and Cd co-induced nephrotoxicity and its underlying mechanism is not well understand. Therefore, in this study, the basis of subchronic Mo or/and Cd poisoning model was carried out to elucidate Mo combined with Cd on renal toxicity of ducks by detecting the mRNA expression levels of AMPKα1, mTOR P62, and Dynein, the autophagy-related genes: Beclin-1, Atg5, LC3A, and LC3B and autophagosomes, thus providing evidence to elucidate the toxicity of Mo or/and Cd on ducks.
MATERIALS AND METHODS
Animals and Treatments
All animals in this experiment were approved by the Committee of Animal Welfare. Animal care and experimental procedures were also complied with the criteria of the Institutional Animal Care and Use Committee Guidelines at College of Animal Science and Technology, Jiangxi Agricultural University. Hexaammonium molybdate ((NH4)6 Mo7 O24·4H2 O) and Cd sulfate (3CdSO4·8H2 O) were used as sources of Mo and Cd, respectively. A total of 60 healthy 1-day-old Shaoxing ducks (Anas platyrhyncha) were obtained from Zhuji Guowei poultry Co., Ltd., Zhejiang Province. After they were advanced reared for 10 d without abnormality by strict clinical examination, the trail was began. A total of 60 healthy 11-day-old ducks were randomly divided into 6 groups (n = 10 per group). Ducks in each group were fed with basal diet with different concentrations of Mo or/and Cd: control group (0 mg/kg Mo, 0 mg/kg Cd), low dietary intake of Mo group (LMo group, 15 mg/kg Mo), high dietary intake of Mo group (HMo group, 100 mg/kg Mo), Cd group (4 mg/kg Cd), LMoCd group (15 mg/kg Mo, 4 mg/kg Cd), and HMoCd group (100 mg/kg Mo, 4 mg/kg Cd). The basal diet was formulated according to the National Research Council. Ducklings were fed with duckling basal diet and duck basal diet before and after 21 d respectively. The composition of basal diet for ducks and the contents of Mo, Cd in the basal diet and water are shown in Tables 1 and 2. The feeding experiment lasted for 120 d, and ducks were given free access to standard food and water.
Table 1.
Composition and nutrient levels in the basal diet for the ducks.
| Composition of diet |
Content (%) |
Nutrient levels |
Level |
||
|---|---|---|---|---|---|
| Ingredient | 0∼3wk | After 3 wk | Index | 0∼3 wk | After 3 wk |
| Soybean meal | 18.00 | 20.00 | Ca (%) | 0.80 | 2.77 |
| Corn | 59.99 | 44.00 | DE (MJ·kg−1) | 11.93 | 11.44 |
| Wheat bran | 11.00 | 14.40 | Crude protein (%) | 18.03 | 17.63 |
| Rice bran | — | 11.0 | Met+Cys (%) | 0.60 | 0.65 |
| Cottonseed meal | 5.00 | — | Total phosphorous (%) | 0.67 | 0.70 |
| Bone meal | 1.58 | 5.80 | Lys (%) | 0.85 | 0.97 |
| Fish meal | 3.00 | 2.00 | Available phosphorus (%) | 0.35 | 0.40 |
| Salt | 0.37 | 0.30 | |||
| Met | 0.06 | 0.10 | |||
| CaHPO4 | — | 1.40 | |||
| Premix* | 1.00 | 1.00 | |||
| Total | 100.00 | 100.00 | |||
Per kilogram of premix contained the followings: VD3 400 IU, VA 2,500 IU, VB 1,215.0 μg, VK3 0.5 mg, VE 10.0 mg, Riboflavin 4.0 mg, Thiamine 4.0 mg, Nicotinic acid 55.0 mg, Pantothenic acid 11.0 mg, Biotin 0.08 mg, Pyridoxine 2.5 mg, Folic acid 1.0 mg, Choline 1,300.0 mg, Se 0.2 mg, Fe 80.0 mg, Cu 10.0 mg, Zn 60.0 mg, Mn 50.0 mg.
Table 2.
Contents of Mo and Cd in the basal diet and water.
| Contents of trace elements (μgg−1) |
||
|---|---|---|
| Item | Cadmium | Molybdenum |
| Deionized water | 0.0000 | 0.0000 |
| Tap water | 0.0075 | 0.0104 |
| Duckling feed | 0.2471 | 4.1510 |
| Duck feed | 0.4762 | 4.7290 |
Sample Collection
After being treated for 120 d, the kidneys of 10 ducks from each group were randomly removed immediately after sacrificing (After fasting for 12 h) with an overdose intravenous injection of sodium pentobarbital (Nembutal, Abbot Labs, IL, USA; 100 mg/kg). Then kidney tissues were collected and transferred to liquid nitrogen immediately. After that, the samples were stored at −80°C. In addition, the rest of each kidney specimen on the 120th d was collected for ultrastructural studies.
RNA Isolation and Primer Designing
Total RNA was isolated from kidney samples using Trizol reagent (TaKaRa, Dalian, China) according to the manufacturer's instructions and then reverse transcribed. The resultant cDNA was synthesized using a PrimeScriptTM RT reagent Kit with gDNA Eraser (TaKaRa, Dalian, China). The reverse transcription reaction (20 μL) was conducted in a mixture containing 2 μL of 5 × DNA Eraser Buffer, 1 μL of gDNA Eraser, 1 μL of total RNA, and 6 μL of RNase-free ddH2 O and was then incubated for 2 min in a 42°C environment. Next, 4 μL of 5 × Prime Script Buffer 2, 1 μL of Prime Script RT Enzyme Mix I, 1 μL of RT PrimerMix, and 5 μL of RNase-free ddH2 O were added to the reaction solution, and the reaction was run at 37°C for 15 min, 85°C for 5 s, and 4°C for 10 min. The reverse transcription products (cDNA) were stored at −20°C for real-time quantitative polymerase chain reaction (RT-qPCR). We designed primers using Primer Premier Software (PREMIER Biosoft International, CA, USA) according to the Anas platyrhynchos AMPKα1, Beclin-1, Atg5, mTOR, Dynein, LC3A, LC3B and P62 mRNA sequences. The β-actin housekeeping gene was used as an internal reference. The primer sequences are shown in Table 3.
Table 3.
Primers used in this study.
| Gene name | Accession number | Primer sequences (5′ to 3′) |
|---|---|---|
| AMPKα1 | NM_0,010,39603.1 | Forward: TCTCGCCCTCATCCTGAAAG |
| Reverse: CTCGACACACTTCAGCCATG | ||
| mTOR | XM_0,050,21282.3 | Forward: GGAATGAACCGTGATGACCGA |
| Reverse: AGCATTTGACTGAGAGGGCT | ||
| Atg5 | XM_01,310,6467.2 | Forward: GGGAAGCCGAGCCTTACTATT |
| Reverse: TGCACTGTGATGTTCCAAGG | ||
| Beclin-1 | XM_02,127,6709.1 | Forward: TACTGCGAGTTCAAGAGGCA |
| Reverse: CGCGTTGATCTCGTTCCATT | ||
| Dynein | XM_01,528,7795.2 | Forward: ACATCTCTTGAGCTGGCAGT |
| Reverse: AGCCACTTCCTGTTGTCGTA | ||
| LC3A | HM627861.1 | Forward: TCCTTGTCCCAGACCATGTC |
| Reverse: GCCATCCTCATCCTTCTCCT | ||
| LC3B | XM_02,127,1232.1 | Forward: ACAGTACAGACGAGCACCTC |
| Reverse: CCAGAAAACTGTCACACGCA | ||
| P62 | XM_0,050,10532.3 | Forward: CTCTCGCTGGACTCTCTCTG |
| Reverse: ATGCTTGTGACGTGGGTAGA | ||
| β-Actin | EF667345.1 | Forward: ATGTCGCCCTGGATTTCG |
| Reverse: CACAGGACTCCATACCCAAGAA |
Real-Time Quantitative PCR
Gene expression levels were assessed by RT-qPCR. The PCR profiles were as follows: 95°C for 10 min, 40 cycles at 95°C for 15 s, 60°C for 60 s, and extension at 95°C for 15 s. At the end of PCR reactions, melt curve analyses were performed for all genes. All reactions were carried out using the Light Cycler 96 real-time PCR machine (Roche Applied Science, Harbin, China).
Ultrastructure Observation
Transmission electron Microscopy (TEM) studies were performed as previously described (Antúnez et al., 2011). The kidney samples on day 120 were processed and viewed under TEM Zeiss 900 (Zeiss, Germany). The ultrastructural changes in the cells were observed and compared to those of the control group.
Statistical Analysis
Using SPSS version 22.0 (SPSS Inc., Chicago, IL, USA) and GraphPad Prism 5.01 (GraphPad Inc., La Jolla, CA, USA), All data were checked for normal distribution and equal variance. Data were represented as mean ± SD. P < 0.05 means difference significantly, and P < 0.01 means extremely difference significantly. Furthermore, using SPSS, the most important parameters that as pivotal factors for individual variations, were determined by principal component analysis (PCA).
RESULTS
The mRNA Expression Levels of Autophagy-related Genes in Kidney
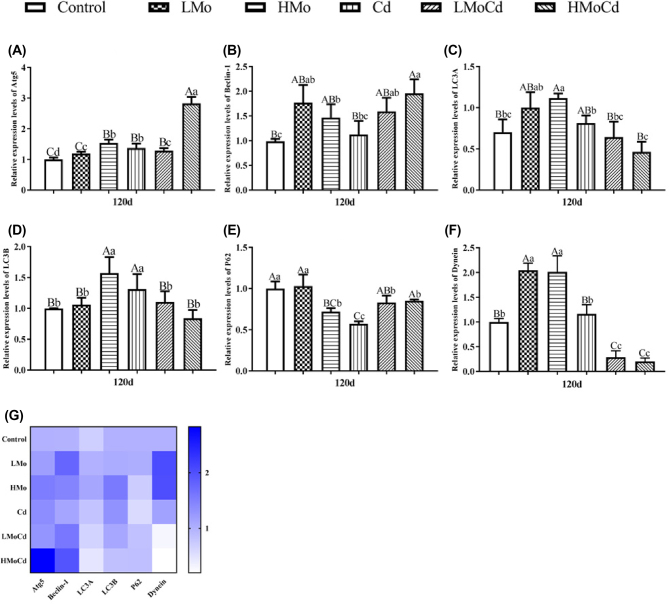
As shown in Figure 1. the Atg5 mRNA expression of kidney was significantly increased (P < 0.05 or P < 0.01) after ducks were treated with Mo and Cd in all exposed groups, and it was remarkably higher in HMoCd group than that in single treated groups (Figure 1 A). The mRNA expression level of Beclin-1 was upregulated (P < 0.05 or P < 0.01) in all exposed groups compared to control group (Figure 1 B). The LC3A mRNA expression was markedly increased (P < 0.05 or P < 0.01) in LMo, HMo, and Cd groups, compared to control group, and it was significantly lower in co-treated groups (P < 0.01) than that in LMo and HMo groups (Figure 1 C). The mRNA level of LC3B was noticeably upregulated (P < 0.01) in HMo and Cd groups compared to control group, it was significantly lower (P < 0.01) in co-treated groups than that in single treated groups (Figure 1 D). Compared to control group, the mRNA expression of P62 was decreased (P < 0.05 or P < 0.01) in all treated groups except for LMo group (Figure 1 E). As shown in Figure 1 F, the mRNA expression level of Dynein was significantly increased (P < 0.01) in LMo and HMo groups compared to control group, it was dramatically lower (P < 0.01) in LMoCd and HMoCd groups than that in control and single treated groups. The Atg5, Beclin-1, LC3A, LC3B, P62, and Dynein mRNA expression levels by Cd and/or Mo exposure were confirmed with heatmap analysis (Figure 1 G).
Figure 1.
Effects of Mo or/and Cd on the mRNA levels of autophagy-related genes in duck kidney on day 120. Panels A-F represent the mRNA levels of Atg5, Beclin-1, LC3A, LC3B, P62, and Dynein, respectively. Statistically significant differences: means with different lowercase letters are significantly different between groups (P < 0.05), means with different uppercase letters are highly significantly different between groups (P < 0.01), and means with common lowercase or uppercase letters are not significantly different between groups (P > 0.05). Each value represented the mean ± SD. The followings present the same. Panel G represents heatmap analysis, bars represent mean ± SD. The followings present the same.
The mRNA Expression Levels of AMPKα1 and mTOR in Kidney
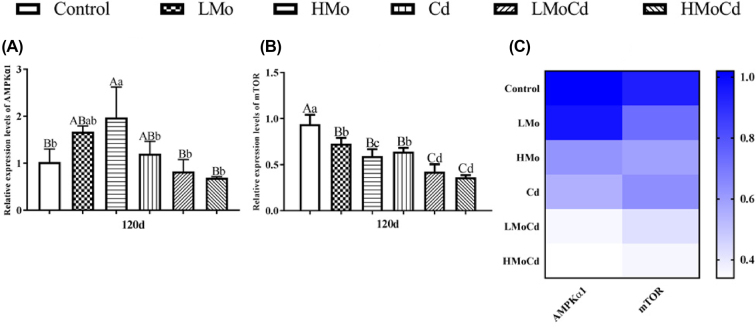
Effects of Mo or/and Cd on the mRNA levels of AMPKα1 and mTOR were presented in Figure 2. On the 120th d, the AMPKα1 expression was increased (P < 0.05 or P < 0.01) in LMo, HMo, and Cd groups compared to control group, but it was lower (P < 0.05) in co-treated groups than that in single treated groups (Figure 2 A). The mTOR mRNA level was dramatically downregulated (P < 0.01) in all treated groups compared to control group, and it was markedly decreased (P < 0.01) in co-treated groups compared to single treated groups (Figure 2 B). The AMPKα1 and mTOR mRNA expression levels were confirmed with heatmap analysis (Figure 2 C).
Figure 2.
Effects of Mo or/and Cd on the mRNA levels of AMPKα1, mTOR, P62, and Dynein in duck kidneys on day 120. Panels A-B represent the mRNA levels of AMPKα1, mTOR, respectively. Panel C represents heatmap analysis, bars represent mean ± SD.
Ultrastructure Observation
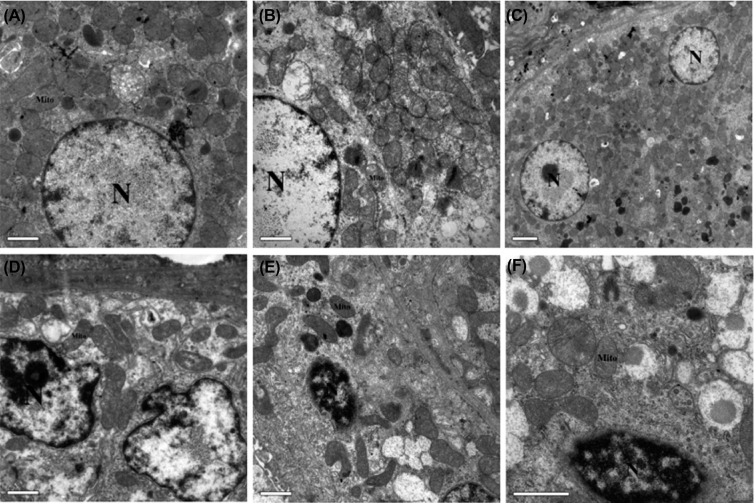
On day 120, ultrastructure of renal cells in duck was observed. The organelle structure and nucleus of renal cells were clear in control group, LMo, and HMo groups (Figure 3 A, B, C). There were a small number of autophagosomes in LMo group, and the number of autophagosomes increased in HMo group (Figure 3 B, C). Partial nucleus was observed to be nuclear pyknosis and there were a small number of autophagosomes in Cd group (Figure 3 D). Nuclear deformation, nuclear pyknosis, and chromatin edge set were observed in LMoCd and HMoCd groups, the mitochondrial cristae were fuzzy (Figure 3 E, F).
Figure 3.
Ultrastructure observation of duck treated with different concentrations of Mo or/and Cd on day 120. A: Control group (×2200); B: LMo group (×2200); C: HMo group (×900); D: Cd group (×2200); E: LMoCd group (×2200); F: HMoCd group (×3900).
Principal Component Analysis
In this study, all parameters measured by using PCA, which the results of measurement were distinguished on the ordination plots, as shown in Figure 4, corresponding to the first, second, and third principal components (51.4, 27.3, and 16.1%, respectively) (Figure 4). The results showed that AMPKα1, mTOR, and LC3B were positively correlated with component 1 while P62, Dynein, Beclin-1, Atg5, and LC3A had negative correlations with component 1. Moreover, AMPKα1, P62, Dynein, Beclin-1, Atg5, LC3A, and LC3B had positive correlations with component 2 except for mTOR, simultaneously, AMPKα1, mTOR, P62, Dynein, Beclin-1, Atg5, and LC3A also had positive correlations with component 3 except for LC3B.
Figure 4.
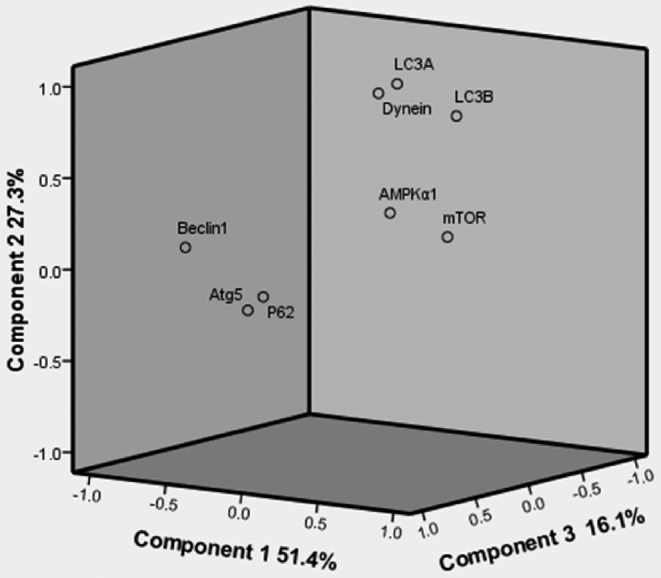
Ordination diagram of the PCA of parameters measured in kidneys exposure to Mo, Cd and a mixture of the two elements. AMPKα1, mTOR, P62, Dynein, ATG5, Beclin-1, LC3A and LC3B are represented in the mRNA levels.
DISCUSSION
The kidney is considered to be the main target of Cd and Mo toxicity (Xiao et al., 2009; Xia et al., 2015; Yang and Shu, 2015). Autophagy is involved in many physiological and pathological processes, and plays a key role in maintaining the metabolic balance and homeostasis of cells (Reggiori, 2006). The autophagy has an important part in the toxic mechanism of heavy metals. It is regulated by a complex signaling network, most of which feed into the AMPK/mTOR pathway (Wang et al., 2016). Our previous results showed the sections of duck kidney tissues treated with Mo or/and Cd showed kidney lesions with swollen epithelial cells of the distal convoluted tubule, granular and blister degeneration, congestion in the renal interstitium, local lymphocytic infiltration and hyperplasia which led to histological changes (Xia et al., 2015), however, it is unclear whether autophagy is involved in kidney damage. Therefore, in this study, Mo and Cd co-induced the levels of autophagy-related genes via AMPK/mTOR signaling pathway in duck kidneys were elucidated.
The formation of autophagosomes is the center in the process of autophagy. In this study, we used TEM to observe autophagosome formation, the results showed that alone Cd and high dose of Mo promoted the formation of autophagosomes in duck kidneys and displayed a synergetic relationship between Cd and Mo. Moreover, the induction of autophagy is supported by detecting the expression levels of autophagy-related genes. Beclin-1 regulates autophagy membrane biosynthesis and it is essential for the initiation of autophagosome formation (Wargasetia et al., 2015). Atg5 plays a vital role in autophagy activation by participating in two essential pathways: Atg12-Atg5 conjugation and LC3 lipidation (Pyo et al., 2013). Atg5 protein in combination with Atg12 and LC3 is involved in the early stages of autophagosome formation (Mizushima et al., 2002). Gao et al. (2013) found that Cd exposure significantly increased the mRNA and protein levels of Beclin-1. It was also found that Cd treatment caused autophagy, and the mRNA expressions of autophagy-related genes Atg5, Atg7, and Beclin-1 increased (Zou et al., 2015). Lysosome plays a vital role in autophagic process, and LC3 is used for autophagic detection as a lysosome-related protein. Microtubule-associated protein light chain A (MAP1LC3A) and MAP1LC3B are structural proteins of autophagosomal membranes, widely used as biomarkers of autophagy (Tsuyuki et al., 2014). P62 is involved in autophagy-dependent elimination of many different cargoes, including ubiquitinated protein aggregates. Due to its interaction with LC3, P62 is continuously degraded by autophagy, and autophagy inhibition leads to accumulation of P62 positive aggregates (Komatsu and Ichimura, 2010). Therefore, the degradation of P62 can be monitored to measure autophagic flux under certain conditions (Pugsley 2017). The inhibition of autophagy flow leads to increased expression of P62 has been reported (Yao et al., 2014). In this study, the results indicated that the Beclin-1 and Atg5 mRNA expression were remarkably upregulated in co-treated groups, however the LC3A and LC3B mRNA expression was decreased in co-treated groups compared to single treated groups, the mRNA expression level of P62 downregulated in all treated groups except for the LMo group, and it was higher in the combined treatment groups, which indicated the impairment of autophagic degradation in Cd and Mo co-treated groups. It was reported that blockage of the degradation step might cause the accumulation of enlarged and unstable autophagic vesicles (Gonzalez et al., 2012; Mena et al., 2012). In the present study, the emergence of large (diameter up to 2 μm) autophagic vesicles in kidney in combined treatment groups under TEM (Figure 3) confirmed that the accumulated autophagosomes were the outcome of impaired autophagic flux. Kimura et al. (2008) found that motor protein was involved in the transportation of autophagosomes. Dynein is a family of cytoskeletal motor proteins, during autophagosome-lysosomal fusion, it has vital part on transporting autophagosomes to lysosomes. The protein expression of Dynein increased under conditions of Cd exposure (Liu et al., 2017). Wang et al. reported Cd could enhance the mRNA expression of the autophagy-related gene Dynein (Wang et al., 2018). Thus, in the study we determined the mRNA level of Dynein, our results showed the Dynein mRNA level significantly upregulated in single Mo and Cd treated groups and markedly downregulated in co-treated groups, which indicated that Mo and Cd could promote the fusion between autophagosome and lysosome and the degradation of damaged organelles in lysosome, but combination of Mo and Cd reduced this effect. Low concentrations of Cd can induce autophagy in a short period of time (Chargui et al., 2011; Son et al., 2011). Depending on the concentration and exposure time, Cd can induce cell survival (for lower exposure) or apoptosis (for elevated exposure) (Chiarelli and Roccheri, 2012). In our results, the expression levels of LC3A, LC3B and Dynein mRNA in combined group were significantly decreased, which might indicate that the autophagy level began to weaken under long time (120 d) and high concentrations of Mo and Cd exposure. Above these results in the study suggested that Mo and Cd could induce autophagy in kidney of ducks and displayed a possible synergistic relationship between the 2 elements.
Many studies have found that autophagy is involved and regulated by multiple signaling pathways (Du et al., 2017; Huang et al., 2017; Luo et al., 2018). ERK1/2 signaling pathway can inhibit Cd-induced apoptosis by activating autophagy (Luo et al., 2018). PI3K/AKT/mTOR signaling pathway regulates the development of autophagy, thereby attenuating Cd-induced nephrotoxicity (Huang et al., 2017). Cd can induce autophagy through the AMPK/mTOR signaling pathwa (Li et al., 2017). AMPK/mTOR signaling pathway is one of the most important pathways regulating autophagy, and it has become a hot topic of research in recent years. AMPK plays a part as a key activator of the signaling network that maintains cellular housekeeping by autophagy (Sengupta et al., 2009). It regulates autophagy through two distinct pathways: one involving the mTOR and the other involving direct activation of unc-51-like kinase 1 (ULk1) (Roach, 2011). Moreover, AMPK usually stimulates autophagy by inhibiting mTOR (Kim et al., 2011). The mTOR is a key regulator of autophagy and mainly contains 2 components, mTORC1 and mTORC2 (Jung et al., 2010). The mTOR plays a negative feedback regulation during autophagy (Pant et al., 2017). The AMPK signaling pathway is upstream of the mTOR signaling pathway (Son et al., 2011). AMPK is activated by stress such as oxidative stress, hypoxia, and nutrient deficiencies (Hardie, 2004). High doses of Mo and Cd can induce oxidative stress, as well as reduce antioxidant enzyme activity and increases free radical accumulation (Raisbeck et al., 2006; Yang et al., 2012). The mTOR mRNA expression level was downregulated in cells exposed to Cd, it is demonstrated that knockdown of AMPKα can inhibit Cd-induced autophagy (Son et al., 2011). In the present study, the AMPKα1 mRNA expression was upregulated in single treated groups and decreased in combination groups, then the mTOR mRNA expression levels significantly downregulated in all treated groups and it was lower in co-treated groups than that in single treated groups. These findings further confirmed that Mo or/and Cd might induce autophagy via AMPK/mTOR signaling pathway in duck kidney, it displayed a possible synergistic relationship between the two elements.
CONCLUSION
In summary, Mo and Cd might induce autophagy via AMPK/mTOR signaling pathway in duck kidney, and it showed a possible synergistic relationship between the 2 elements.
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (Grant no. 31260625, Beijing, P.R. China). All authors thank Xiaoquan Guo, Ping Liu, Bing Xia, Yibo Zong, Jin Xiong, Mengmeng Zhang, Zhiyue Liao, and Shuang Yang in the clinical veterinary medicine laboratory in the College of Animal Science and Technology, Jiangxi Agricultural University, for help in the experimental process.
CONFLICT OF INTEREST STATEMENT
The authors declare that there are no conflicts of interest.
Footnotes
All authors have read the manuscript and agreed to submit it in its current form for consideration for publication in the Journal.
Contributor Information
Guoliang Hu, Email: hgljx3818@jxau.edu.cn.
Caiying Zhang, Email: zhangcaiying0916@163.com.
REFERENCES
- Abramovich M., Miller A., Yang H., Friel J.K. Molybdenum content of Canadian and US infant formulas. Biol. Trace Elem. Res. 2011;143:844. doi: 10.1007/s12011-010-8950-4. [DOI] [PubMed] [Google Scholar]
- Antúnez M.I.C., Penco J.M.M., Lozano J.L.A., Macho A.L., Climent V. Influence of intestinal resections on biliary composition and liver ultrastructure. Clin. Nutr. 2011;30:247–251. doi: 10.1016/j.clnu.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Bersényi A., Berta E., Kádár I., Glávits R., Szilágyi M., Fekete S.G. Effects of high dietary molybdenum in rabbits. Acta Vet. Hung. 2008;56:41–55. doi: 10.1556/AVet.56.2008.1.5. [DOI] [PubMed] [Google Scholar]
- Brzóska M.M., Kamiński M., Supernakbobko D., Zwierz K., Moniuszkojakoniuk J. Changes in the structure and function of the kidney of rats chronically exposed to cadmium. I. Biochemical and histopathological studies. Arch Toxicol. 2003;77:344–352. doi: 10.1007/s00204-003-0451-1. [DOI] [PubMed] [Google Scholar]
- Cao H., Zhang M., Xia B., Xiong J., Zong Y., Hu G., Zhang C. Effects of Molybdenum or/and Cadmium on mRNA expression levels of inflammatory Cytokines and HSPs in duck spleens. Biol. Trace Elem. Res. 2016;170:1–8. doi: 10.1007/s12011-015-0442-0. [DOI] [PubMed] [Google Scholar]
- Chargui A., Zekri S., Jacquillet G., Rubera I., Ilie M., Belaid A., Duranton C., Tauc M., Hofman P., Poujeol P. Cadmium-induced autophagy in rat kidney: an early biomarker of subtoxic exposure. Toxicol. Sci. 2011;121:31. doi: 10.1093/toxsci/kfr031. [DOI] [PubMed] [Google Scholar]
- Chatterjee S., Sarkar S., Bhattacharya S. Toxic metals and autophagy. Chem. Res. Toxicol. 2014;27:1887–1900. doi: 10.1021/tx500264s. [DOI] [PubMed] [Google Scholar]
- Chen L., Xu B., Liu L., Luo Y., Zhou H., Chen W., Shen T., Han X., Kontos C.D., Huang S. Cadmium induction of reactive oxygen species activates mTOR pathway, leading to neuronal cell death. Free Radic. Biol. Med. 2011;50:624–632. doi: 10.1016/j.freeradbiomed.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Klionsky D.J. The regulation of autophagy - unanswered questions. J. Cell Sci. 2011;124:161–170. doi: 10.1242/jcs.064576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiarelli R., Roccheri M.C. Heavy metals and metalloids as autophagy inducing agents: focus on cadmium and arsenic. Cells. 2012;1:597. doi: 10.3390/cells1030597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies T.D., Pickard J., Hall K.J. Acute molybdenum toxicity to rainbow trout and other fish. J. Environ. Eng. Sci. 2005;4:481–485. [Google Scholar]
- Du C., Zhang T., Xiao X., Shi Y., Duan H., Ren Y. Protease-activated receptor-2 promotes kidney tubular epithelial inflammation by inhibiting autophagy via the PI3K/Akt/mTOR signalling pathway. Biochem. J. 2017;474:2733–2747. doi: 10.1042/BCJ20170272. [DOI] [PubMed] [Google Scholar]
- Fan X., Wang J., Hou J., Lin C., Bensoussan A., Chang D., Liu J., Wang B. Berberine alleviates ox-LDL induced inflammatory factors by up-regulation of autophagy via AMPK/mTOR signaling pathway. J. Transl. Med. 2015;13:1–11. doi: 10.1186/s12967-015-0450-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D., Xu Z., Kuang X., Qiao P., Liu S., Zhang L., He P., Jadwiga W.S., Wang Y., Min W. Molecular characterization and expression analysis of the autophagic gene beclin 1 from the purse red common carp (Cyprinus carpio) exposed to cadmium. Comparative Biochem. Physiol. Part C. 2013;160:15. doi: 10.1016/j.cbpc.2013.11.004. [DOI] [PubMed] [Google Scholar]
- Gonzalez P., Mader I., Tchoghandjian A., Enzenmüller S., Cristofanon S., Basit F., Debatin K.M., Fulda S. Impairment of lysosomal integrity by B10, a glycosylated derivative of betulinic acid, leads to lysosomal cell death and converts autophagy into a detrimental process. Cell Death Differ. 2012;8:1250–1251. doi: 10.1038/cdd.2012.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie D.G. The AMP-activated protein kinase pathway–new players upstream and downstream. J. Cell Sci. 2004;117:5479–5487. doi: 10.1242/jcs.01540. [DOI] [PubMed] [Google Scholar]
- Huang M., Su L., Yang L., Zhu L., Liu Z., Duan R. Effect of exogenous TGF-β1 on the cadmium-induced nephrotoxicity by inhibiting apoptosis of proximal tubular cells through PI3K-AKT-mTOR signaling pathway. Chem. Biol. Interact. 2017;269:25. doi: 10.1016/j.cbi.2017.03.010. [DOI] [PubMed] [Google Scholar]
- Jung C.H., Ro S.H., Cao J., Otto N.M., Kim D.H. mTOR regulation of autophagy. Febs Lett. 2010;584:1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Kundu M., Viollet B., Guan K.L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S., Noda T., Yoshimori T. Dynein-dependent movement of autophagosomes mediates efficient encounters with lysosomes. Cell Struct. Funct. 2008;33:109. doi: 10.1247/csf.08005. [DOI] [PubMed] [Google Scholar]
- Komatsu M., Ichimura Y. Physiological significance of selective degradation of p62 by autophagy. Febs Lett. 2010;584:1374–1378. doi: 10.1016/j.febslet.2010.02.017. [DOI] [PubMed] [Google Scholar]
- Li R., Luo X., Zhu Y., Zhao L., Li L., Peng Q., Ma M., Gao Y. ATM signals to AMPK to promote autophagy and positively regulate DNA damage in response to cadmium-induced ROS in mouse spermatocytes. Environ. Pollut. 2017;231:1560–1568. doi: 10.1016/j.envpol.2017.09.044. [DOI] [PubMed] [Google Scholar]
- Liao Y., Cao H., Xia B., Xiao Q., Liu P., Hu G., Zhang C. Changes in Trace Element Contents and Morphology in Bones of Duck Exposed to Molybdenum or/and Cadmium. Biol. Trace Elem. Res. 2017;175:449–457. doi: 10.1007/s12011-016-0778-0. [DOI] [PubMed] [Google Scholar]
- Liu F., Wang X.Y., Zhou X.P., Liu Z.P., Song X.B., Wang Z.Y., Wang L. Cadmium disrupts autophagic flux by inhibiting cytosolic Ca(2+)-dependent autophagosome-lysosome fusion in primary rat proximal tubular cells. Toxicology. 2017;383:13–23. doi: 10.1016/j.tox.2017.03.016. [DOI] [PubMed] [Google Scholar]
- Liu W., Dai N., Wang Y., Xu C., Zhao H., Xia P., Gu J., Liu X., Bian J., Yuan Y. Role of autophagy in cadmium-induced apoptosis of primary rat osteoblasts. Sci. Rep. 2016;6 doi: 10.1038/srep20404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liuzzi J.P., Guo L., Yoo C., Stewart T.S. Zinc and autophagy. Biometals. 2014;27:1087–1096. doi: 10.1007/s10534-014-9773-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Zhou Z., Zheng J., Zhang Z., Lu R., Liu H., Shi H., Tu Z. 2D-DIGE and MALDI TOF/TOF MS analysis reveal that small GTPase signaling pathways may play an important role in cadmium-induced colon cell malignant transformation. Toxicol. Appl. Pharmacol. 2015;288:106–113. doi: 10.1016/j.taap.2015.07.020. [DOI] [PubMed] [Google Scholar]
- Luo H.F., Zhang J.Y., Jia W.J., Ji F.M., Yan Q., Xu Q., Ke S., Ke J.S. Analyzing the role of soil and rice cadmium pollution on human renal dysfunction by correlation and path analysis. Environ. Sci. Pollut. Res. Int. 2016;24:1–8. doi: 10.1007/s11356-016-7845-0. [DOI] [PubMed] [Google Scholar]
- Luo T., Zhang H., Yu Q., Liu G., Long M., Zhang K., Liu W., Song R., Bian J., Gu J. ERK1/2 MAPK promotes autophagy to suppress ER stress-mediated apoptosis induced by cadmium in rat proximal tubular cells. Toxicol. In Vitro. 2018;52:60–69. doi: 10.1016/j.tiv.2018.06.001. [DOI] [PubMed] [Google Scholar]
- Mena S., Rodríguez M.L., Ponsoda X., Estrela J.M., Jäättela M., Ortega A.L. Pterostilbene-induced tumor cytotoxicity: a lysosomal membrane permeabilization-dependent mechanism. Plos One. 2012;7 doi: 10.1371/journal.pone.0044524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendel R.R. Molybdenum: biological activity and metabolism. Dalton Trans. 2005;21:3404–3409. doi: 10.1039/b505527j. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Ohsumi Y., Yoshimori T. Autophagosome formation in mammalian cells. Cell Struct. Funct. 2002;27:421–429. doi: 10.1247/csf.27.421. [DOI] [PubMed] [Google Scholar]
- Pant K., Saraya A., Venugopal S.K. Oxidative stress plays a key role in butyrate-mediated autophagy via Akt/mTOR pathway in hepatoma cells. Chem. Biol. Interact. 2017;273:99. doi: 10.1016/j.cbi.2017.06.001. [DOI] [PubMed] [Google Scholar]
- Parzych K.R., Klionsky D.J. An Overview of Autophagy: Morphology, Mechanism, and Regulation. Antioxid Redox Signal. 2014;20:460–473. doi: 10.1089/ars.2013.5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugsley H.R. Assessing Autophagic Flux by Measuring LC3, p62, and LAMP1 Co-localization Using Multispectral Imaging Flow Cytometry. J. Vis. Exp. 2017 doi: 10.3791/55637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyo J.O., Yoo S.M., Ahn H.H., Nah J., Hong S.H., Kam T.I., Jung S., Jung Y.K. Overexpression of Atg5 in mice activates autophagy and extends lifespan. Nat. Commun. 2013;4 doi: 10.1038/ncomms3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisbeck M.F., Siemion R.S., Smith M.A. Modest copper supplementation blocks molybdenosis in cattle. J. Vet. Diagn. Invest. 2006;18:566–572. doi: 10.1177/104063870601800607. [DOI] [PubMed] [Google Scholar]
- Reggiori F. 1. Membrane origin for autophagy. Curr. Top Dev. Biol. 2006;74:1. doi: 10.1016/S0070-2153(06)74001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach P.J. AMPK → ULK1 → autophagy. Mol. Cell. Biol. 2011;31:3082–3084. doi: 10.1128/MCB.05565-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satarug S., Baker J.R., Urbenjapol S., Haswellelkins M., Reilly P.E., Williams D.J., Moore M.R. A global perspective on cadmium pollution and toxicity in non-occupationally exposed population. Toxicol. Lett. 2003;137:65. doi: 10.1016/s0378-4274(02)00381-8. [DOI] [PubMed] [Google Scholar]
- Schwarz G., Mendel R.R., Ribbe M.W. Molybdenum cofactors, enzymes and pathways. Nature. 2009;460:839–847. doi: 10.1038/nature08302. [DOI] [PubMed] [Google Scholar]
- Sengupta A., Molkentin J.D., Yutzey K.E. FoxO transcription factors promote autophagy in cardiomyocytes. J. Biol. Chem. 2009;284 doi: 10.1074/jbc.M109.024406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L., Cao H., Luo J., Liu P., Wang T., Hu G., Zhang C. Effects of molybdenum and cadmium on the oxidative damage and kidney apoptosis in Duck. Ecotoxicol. Environ. Saf. 2017;145:24–31. doi: 10.1016/j.ecoenv.2017.07.006. [DOI] [PubMed] [Google Scholar]
- So K.Y., Oh S.H. Cadmium-induced Heme-Oxygenase-1 expression plays dual roles in autophagy and apoptosis and is regulated by both PKC-δ and PKB/Akt activation in NRK52E kidney cells. Toxicology. 2016;370:49–59. doi: 10.1016/j.tox.2016.09.010. [DOI] [PubMed] [Google Scholar]
- Son Y.O., Wang X., Hitron J.A., Zhang Z., Cheng S., Budhraja A., Ding S., Lee J.C., Shi X. Cadmium induces autophagy through ROS-dependent activation of the LKB1-AMPK signaling in skin epidermal cells. Toxicol. Appl. Pharmacol. 2011;255:287–296. doi: 10.1016/j.taap.2011.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X.B., Liu G., Liu F., Yan Z.G., Wang Z.Y., Liu Z.P., Wang L. Autophagy blockade and lysosomal membrane permeabilization contribute to lead-induced nephrotoxicity in primary rat proximal tubular cells. Cell Death Dis. 2017;8 doi: 10.1038/cddis.2017.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton D.M., Liu Y. Multiple roles of cadmium in cell death and survival. Chem. Biol. Interact. 2010;188:267–275. doi: 10.1016/j.cbi.2010.03.040. [DOI] [PubMed] [Google Scholar]
- Thévenod F., Lee W.K. Live and let die: roles of autophagy in cadmium nephrotoxicity. Toxics. 2015;3:130. doi: 10.3390/toxics3020130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuyuki S., Takabayashi M., Kawazu M., Kudo K., Watanabe A., Nagata Y., Kusama Y., Yoshida K. Detection of WIPI1 mRNA as an indicator of autophagosome formation. Autophagy. 2014;10:497–513. doi: 10.4161/auto.27419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Qi Q., Feng Z., Zhang X., Huang B., Chen A., Prestegarden L., Li X., Wang J. Berberine induces autophagy in glioblastoma by targeting the AMPK/mTOR/ULK1-pathway. Oncotarget. 2016;7:66944–66958. doi: 10.18632/oncotarget.11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Xu Z., Yin H., Min Y., Li S. Alleviation mechanisms of selenium on cadmium-spiked in chicken ovarian tissue: perspectives from autophagy and energy metabolism. Biol. Trace Elem. Res. 2018;186:521–528. doi: 10.1007/s12011-018-1341-y. [DOI] [PubMed] [Google Scholar]
- Wang Z., Choi M.E. Autophagy in kidney health and disease. Antioxid. Redox. Signal. 2014;20:519–537. doi: 10.1089/ars.2013.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wargasetia T.L., Shahib N., Martaadisoebrata D., Dhianawaty D., Hernowo B. Characterization of apoptosis and autophagy through Bcl-2 and Beclin-1 immunoexpression in gestational trophoblastic disease. Iran J. Reprod. Med. 2015;13:413–420. [PMC free article] [PubMed] [Google Scholar]
- Xia B., Cao H., Luo J., Liu P., Guo X., Hu G., Zhang C. The Co-induced effects of Molybdenum and Cadmium on antioxidants and heat shock proteins in duck kidneys. Biol. Trace Elem. Res. 2015;168:261–268. doi: 10.1007/s12011-015-0348-x. [DOI] [PubMed] [Google Scholar]
- Xiao J., Cui H.M., Peng X., Cui Y. Effect of dietary high Molybdenum on the cell cycle and apoptosis of kidney in broilers. Biol. Trace Elem. Res. 2009;142:523–531. doi: 10.1007/s12011-010-8772-4. [DOI] [PubMed] [Google Scholar]
- Xu S., Pi H., Chen Y., Zhang N., Guo P., Lu Y., He M., Xie J., Zhong M., Zhang Y. Cadmium induced Drp1-dependent mitochondrial fragmentation by disturbing calcium homeostasis in its hepatotoxicity. Cell Death Dis. 2013;4:e540. doi: 10.1038/cddis.2013.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Zhang C., Zhuang Y., Gu X., Xiao Q., Guo X., Hu G., Cao H. Oxidative stress and cell apoptosis in caprine liver induced by Molybdenum and Cadmium in combination. Biol. Trace Elem. Res. 2016;173:1–8. doi: 10.1007/s12011-016-0633-3. [DOI] [PubMed] [Google Scholar]
- Yang H., Shu Y. Cadmium Transporters in the kidney and Cadmium-induced nephrotoxicity. Int. J. Mol. Sci. 2015;16:1484–1494. doi: 10.3390/ijms16011484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Zhang Z., He J., Li J., Zhang J.L., Xing H., Xu S. Ovarian toxicity induced by dietary Cadmium in hen. Biol. Trace Elem. Res. 2012;148:53. doi: 10.1007/s12011-012-9343-7. [DOI] [PubMed] [Google Scholar]
- Yao X.F., Cao J., Xu L.M., Sun X.C., Kang J., Yang G., Jiang L.P., Geng C.Y., Gao C.Z., Zhong L.F., Ma Y.F. Perfluorooctane sulfonate blocked autophagy flux and induced lysosome membrane permeabilization in HepG2 cells. Food Chem. Toxicol. 2014;67:96–104. doi: 10.1016/j.fct.2014.02.017. [DOI] [PubMed] [Google Scholar]
- Yue T., Gu L., Chen D., Wu W., Liu H., Hu H., Wan Y., Sun W. Rhein Inhibits Autophagy in Rat Renal Tubular Cells by Regulation of AMPK/mTOR Signaling. Sci. Rep. Uk. 2017;7 doi: 10.1038/srep43790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Xing S., Li Z. Antagonistic effects of lycopene on cadmium-induced hippocampal dysfunctions in autophagy, calcium homeostatis and redox. Oncotarget. 2017;8:44720–44731. doi: 10.18632/oncotarget.18249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H., Zhuo L., Han T., Hu D., Yang X., Wang Y., Yuan Y., Gu J., Bian J., Liu X. Autophagy and gap junctional intercellular communication inhibition are involved in cadmium-induced apoptosis in rat liver cells. Biochem. Biophys. Res. Commun. 2015;459:713–719. doi: 10.1016/j.bbrc.2015.03.027. [DOI] [PubMed] [Google Scholar]