Abstract
Methicillin-resistant Staphylococcus aureus (MRSA) are a major concern for public health, and broiler farms are a potential source of MRSA isolates. In this study, a total of 56 MRSA isolates from 15 broiler farms from 4 different counties in Germany were characterised phenotypically and genotypically. Spa types, dru types, SCCmec types, and virulence genes as well as resistance genes were determined by using a DNA microarray or specific PCR assays. In addition, PFGE profiles of isolates were used for analysis of their epidemiological relatedness. While half of the isolates belonged to spa type t011, the other half was of spa types t1430 and t034. On 3 farms, more than 1 spa type was found. The most common dru type was dt10a (n = 19), followed by dt11a (n = 17). Susceptibility testing of all isolates by broth microdilution revealed 21 different resistance phenotypes and a wide range of resistance genes was present among the isolates. Up to 10 different resistance phenotypes were found on individual farms. Resistance to tetracyclines (n = 53), MLSB antibiotics (n = 49), trimethoprim (n = 38), and elevated MICs of tiamulin (n = 29) were most commonly observed. Microarray analysis detected genes for leucocidin (lukF/S), haemolysin gamma (hlgA), and other haemolysines in all isolates. In all t1430 isolates, the egc cluster comprising of genes encoding enterotoxin G, I, M, N, O, U, and/or Y was found. The splitstree analysis based on microarray and PCR gene profiles revealed that all CC9/SCCmec IV/t1430/dt10a isolates clustered apart from the other isolates. These findings confirm that genotypic patterns were specific for clonal lineages rather than for the origin of isolates from individual farms.
Key words: MRSA, broiler, antimicrobial resistance, clonal complex, PCR
INTRODUCTION
Bacterial disease has been life threatening for humans and animals until antibiotics were developed in the early 20th century. Today, this substantial medical invention is threatened by multi-resistant bacteria such as methicillin-resistant Staphylococcus aureus (MRSA). Staphylococcus (S.) aureus colonises the skin and the respiratory system of many animals and humans without causing any symptoms (Devriese, 1990; Vanderhaeghen et al., 2010). However, the capability of some strains to produce toxins and enzymes that interfere with receptor function, damage membranes, and degrade host molecules makes it a potent pathogen (Otto, 2014). MRSA emerge from methicillin-susceptible S. aureus by site-specific integration of the staphylococcal cassette chromosome mec (SCCmec) element (Vanderhaeghen et al., 2010), leading to β-lactam resistance due to an alternative, modified penicillin-binding protein (Monecke et al., 2011). MRSA percentages above 25% in invasive Staphylococcus aureus isolates were reported in one third of European countries in 2017 and serious infections with MRSA remain a problem in all healthcare sectors. Thus, MRSA remains a public health priority in the EU and other countries (ECDC, 2017). Various molecular epidemiological studies have shown that MRSA strains circulating in different reservoirs may be distinct from each other (Graveland et al., 2010; Köck et al., 2013; Monecke et al., 2011). However, most studies focused on transmission of MRSA between different host species or from animals to humans (de Boer et al., 2009). While the focus of initial studies on virulence and antimicrobial resistance of livestock-associated (LA) MRSA was placed mainly on pigs (Vandendriessche et al., 2013), studies on broiler farms often investigated risk factors of MRSA on broiler farms (Geenen et al., 2013) and the risk of people in contact with broilers (Mulders et al., 2010; Wendlandt et al., 2013a, b). Epidemiological data on the transmission of MRSA lineages or strains between poultry farms is therefore still rare (Friese et al., 2013). However, data on the epidemiological diversity of MRSA can provide a better understanding of this issue (Wendlandt et al., 2013a) and a better understanding of factors that contribute to spread and persistence of resistant isolates in poultry production can help to introduce measures preventing broiler farms from becoming a reservoir for MRSA (Crombé et al., 2013; Grontvedt et al., 2016).
Our study investigated the phenotypic and genotypic characteristics of 56 MRSA isolates derived from broiler flocks or the flock environment of 15 farms from 4 different counties in Germany. Different typing techniques were used to characterise the MRSA included in this study and to assess their epidemiological relatedness. To our knowledge, this is one of a few studies with focus on the geographic region of the farms and the distribution of isolates on farm level. Improving the understanding of the transmission routes is crucial for combating transmission and spread of MRSA on broiler farms (Grontvedt et al., 2016).
MATERIALS AND METHODS
Bacterial Isolates and Farms
A total of 56 MRSA isolates from 15 broiler farms were included in this study. Most of the isolates were collected from different farms located in a rural district in northern Germany, in the course of a doctoral research study (n = 48) (Dullweber, 2010). Another 8 isolates were sampled in the same time period by taking pooled samples on different broiler farms from another 3 districts in northern Germany. Detailed information on the source of the MRSA isolates and on flock characteristics is given in Table 1. All sampled broiler houses were thermally insulated, with concrete floor and negative pressure ventilation. Cleaning and disinfection of most plants was done by the farmers using formalin as disinfectant, except for farm number 11 where a mixture of isopropanol 7%, glutaraldehyde 24%, didecyldimethylammoniumchloride 5%, and alkyldimethylammoniumchloride 5% (virocid, CID Lines, Ieper, Belgium) was used as disinfectant. On farm numbers 13 and 15, a third party company did the cleaning and disinfection and information on the disinfectant was not provided. Samples were taken from different locations as shown in Table 1. Environmental samples were collected using gauze pads, which were moistened with isotonic NaCl solution and subsequently autoclaved in sealed plastic bags. At the time of sampling, the bags were opened and pulled back over the gauze pads without touching the pads or the insides of the bag and the swab samples were then taken from a surface of approximately 625 cm². Afterwards, the bag was pulled over the gauze pad again and was subsequently resealed. Samples obtained from live birds consisted of tracheal samples collected with commercially available, sterile swabs (Mast Group Ltd., Brescia, Italy).
Table 1.
Origins of the 56 MRSA isolates included in this study.
| Farm environment |
|||||
|---|---|---|---|---|---|
| Farm | Source of MRSA | Farm size | County | Fattening periodj | Number of isolates per farm |
| 1 | Flock environmenta | Not specified | 1 | Short | 1 |
| 2 | Flock environmenta | Not specified | 2 | Short | 1 |
| 3 | Flock environmenta | Not specified | 3 | Short | 1 |
| 4 | Flock environmenta | Not specified | 3 | Short | 1 |
| 5 | Flock environmenta | Not specified | 3 | Short | 1 |
| 6 | Flock environmenta | Not specified | 2 | Short | 1 |
| 7 | Flock environmenta | Not specified | 3 | Short | 1 |
| 8 | Flock environmenta | Not specified | 4 | Short | 1 |
| 9 | Flock environmentb,c, chicken | >20,000 | 4 | Short | 6 |
| 10 | Flock environmenta,b,c,d,e,f, chicken | >20,000 | 4 | Short | 22 |
| 11 | Flock environmentb,c,h, chicken | >20,000 | 4 | Mid | 4 |
| 12 | Flock environmenta,c,d,f,g, chicken | >20,000 | 4 | Splitting | 8 |
| 13 | Flock environmenta,b,d,f | <20,000 | 4 | Short | 4 |
| 14 | Chicken | >20,000 | 4 | Mid | 3 |
| 15 | Flock environmenti | >100,000 | 4 | Long | 1 |
Pooled sample from flock environment.
Drinking and/or feeding line.
Animal weight scale.
Gas heating.
Temperature sensor.
Air inlet flap.
Air outlet flap.
Barn wall.
Anteroom.
Fattening periods are defined as: short (28 to 30 D), mid (32 to 35 D), long (38 to 42 D), splitting (removal of some animals before the end of the fattening period.
MRSA were isolated and identified as described by Dullweber (2010). Briefly, pre-enrichment was done for 16 to 18 h at 36°C by using Mueller-Hinton Broth (Oxoid, Wesel, Germany) supplemented with 6.5% NaCl. Subsequently, 1 mL was transferred in 9 mL Tryptic Soy Broth (Merck, Germany) supplemented with 75 mg/L aztreonam and 3.5 mg/L cefoxitin. After another 18 h of incubation at 36°C, aliquots were streaked on a selective MRSA agar (BBL CHROMagar MRSA, Becton Dickinson, Heidelberg, Germany). Presumptive MRSA isolates were identified using a triplex PCR, as described previously (Poulsen et al., 2003). For this, the following primers were used: mecup1 (5′-GGGATCATAGCGTCATTATTC-3′) and mecup2 (5′-AACGATTGTGACACGATAGCC-3′), nucPCR1 (5′-TCAGCAAATGCATCACAAACAG-3′) and nucPCR2 (5′-CGTAAATGCACTTGCTTCAGG-3′), and 16Sup1 (5′-GTGCCAGCAGCCGCGGTAA-3′) and 16Sup2 (5′-AGACCCGGGAACGTATTCAC-3′). Thermal cycling parameters consisted of an initial denaturation step of 5 min at 94°C, followed by 30 cycles of 30 s at 94°C, 30 s at 55°C, and 30 s at 72°C, and a final extension step of 2 min at 72°C.
Typing of Isolates
Isolates were spa typed as described by Shopsin et al. (1999) using the primers Spa Seq fw (5′-GACGATCCTTCGGTGAGCAAAG-3′) and Spa Seq rv (5′-CTGTATCACCAGGTTTAACGAC-3′) with the following PCR conditions: initial 10 min at 95°C, 30 cycles of 30 s at 95°C, 30 s at 60°C, 45 s at 72°C, and a final extension at 72°C for 10 min. SCCmec types were determined by 2 multiplex PCRs and 1 single target PCR as described by Zhang et al. (2005), using 14 different primer pairs detecting specific mec complex types, ccr complex types, and SCCmec types. The multiplex PCRs were performed using the following conditions: 94°C for 5 min, followed by 10 cycles of 45 s at 94°C, 45 s at 65°C, and 1.5 min at 72°C, followed by another 25 cycles with an annealing temperature of 55°C, and a final extension step of 10 min at 72°C. The single target PCR targeting type 5 ccr began with 5 min at 94°C, followed by 30 cycles of 1 min at 94°C, 1 min at 50°C, and 2 min at 72°C, ending with a final extension step of 10 min at 72°C. The number of SCCmec -associated direct repeat units (dru) was determined as described previously (Goering et al., 2008) using the primers dru fw (5′-GTTAGCATATTACCTCTCCTTGC-3′) and dru rv (5′-GCCGATTGTGCTTGATGAG-3′) with a PCR protocol consisting of an initial 2 min at 94°C, followed by 30 cycles of 1 min at 94°C, 1 min at 52°C, and 1 min at 72°C. The dru types were assigned using the database at http://dru-typing.org. Macrorestriction analysis was performed according to the HARMONY protocol (Murchan et al., 2003) using a CHEF-DR II system (BioRad, Munich, Germany). However, as CC398 isolates are typically not typeable by Sma I macrorestriction, the restriction enzyme Xma I was used instead. The run time was 10 h with a switch time from 5 to 15 s followed by a run time of 13 h using a switch time from 15 to 60 s.
Susceptibility Testing
All isolates were tested for their antimicrobial susceptibility by broth microdilution method. For determination of the minimum inhibitory concentration (MIC) values, custom-made microtiter plates (Sensititre, East Grinstead, UK) were used. Overall, 30 antimicrobial agents and combinations were tested (test ranges in brackets): Amoxicillin/clavulanic acid (0.03/0.015 to 64/32 µ g/mL), ampicillin (0.03 to 64 µ g/mL), apramycin (0.03 to 64 µ g/mL), ceftiofur (0.03 to 64 µ g/mL), cefotaxime (0.015 to 32 µ g/mL), cefoperazon (0.06 to 32 µ g/mL), cefquinome (0.015 to 32 µ g/mL), cephalothin (0.06 to 128 µ g/mL), chloramphenicol (0.5 to 256 µ g/mL), clindamycin (0.03 to 64 µ g/mL), doxycycline (0.06 to 128 µ g/mL), enrofloxacin (0.008 to 16 µ g/mL), erythromycin (0.015 to 32 µ g/mL), florfenicol (0.12 to 256 µ g/mL), gentamicin (0.12 to 256 µ g/mL), nalidixic acid (0.06 to 128 µ g/mL), oxacillin + 2% NaCl (0.03 to 16 µ g/mL), penicillin G (0.015 to 32 µ g/mL), pirlimycin (0.03 to 64 µ g/mL), quinupristin/dalfopristin (0.008 to 16 µ g/mL), spectinomycin (0.12 to 256 µ g/mL), spiramycin (0.06 to 128 µ g/mL), tetracycline (0.12 to 256 µ g/mL), tiamulin (0.03 to 64 µ g/mL), tilmicosin (0.06 to 128 µ g/mL), trimethoprim (0.06 to 128 µ g/mL), trimethoprim/sulfamethoxazole 1:19 ratio (0.015/0.3 to 32/608 µ g/mL), tylosin (0.03 to 64 µ g/mL), and vancomycin (0.008 to 16 µ g/mL). Susceptibility testing and interpretation was done according to the Clinical and Laboratory Standards Institute (CLSI) documents (CLSI, 2014, 2015). CLSI breakpoints were used for all antibiotics, for which approved breakpoints are available. Staphylococcus aureus ATCC29233 was used for quality control purposes.
Molecular Analyses
All 56 isolates were further subjected to microarray analysis using the S. aureus Genotyping Kit 2.0 (StaphyType, Alere Technologies GmbH, Jena, Germany) (Monecke et al., 2011) which contains more than 300 probes corresponding to approximately 170 distinct genes and their allelic variants. Array experiments were performed according to the manufacturer's instructions. A full list of primer/probe sequences has been published previously (Monecke et al., 2011). Hybridisation patterns of the microarray were also used to infer the clonal complex (CC) of isolates (Monecke et al., 2008).
Relevant resistance determinants that were not included in the microarray were investigated using additional PCR assays. The according protocols and primers were described previously. All isolates were tested for the presence of erm (T) (Fessler et al., 2010), vga (B) (Fessler et al., 2011), vga (C) (Fessler et al., 2010), and vga (E) (Schwendener and Perreten, 2011), while only isolates that were suspected to carry resistance genes due to their phenotype were examined for spc (Fessler et al., 2010), dfrK (Fessler et al., 2010), and tet (L) (Aarestrup et al., 2000).
Data Analysis
Band patterns were analysed using the Bionumerics 7.5 software. A dice coefficient with 0.5% optimization and 1% position tolerance was used for cluster analysis. Genome profiles from micrroarray data and PCR analyses were furthermore clustered using SplitsTree4 software.
RESULTS
Sampling and Isolates
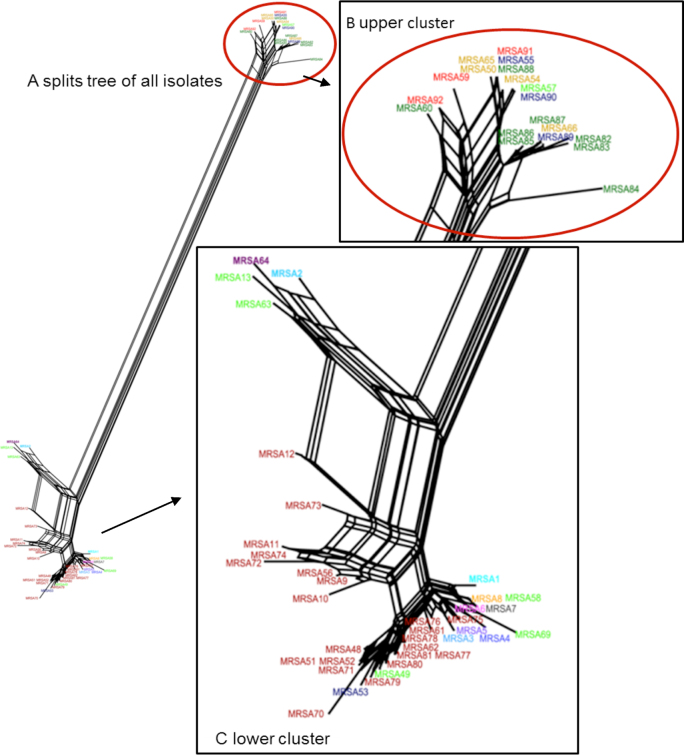
A total of 56 MRSA isolates were collected in the 15 broiler houses. Due to the differences in sample numbers and sample types taken per farm (Table 1), the number of MRSA isolates per farm varied between 1 and 22. While on farm numbers 1 to 8, single isolates were obtained from pooled samples from the flock environment, on farm numbers 9 to 15, MRSA were isolated from broilers as well as from 9 different locations of the broiler houses. As a result, the number of isolates per farm ranged between 0 and 22 MRSA isolates. Comparisons of microarray profiles using the SplitsTree and the BioNumerics software did not result in farm-specific MRSA clusters, as shown in Figure 1.
Figure 1.
Network graph illustrating similarity of the microarray and PCR gene profiles of all 56 MRSA isolates in a SplitsTree analysis. All t1430 isolates form one similarity cluster (marked with red circle). Colors of the isolates´ designation represent their origins from the same (same color) or different farm (different color).
Typing of Isolates
To characterise the isolates and to identify the genetic relationship of the MRSA, all isolates were typed by SCCmec typing, spa - and dru -typing, macrorestriction analysis and microarray analysis.
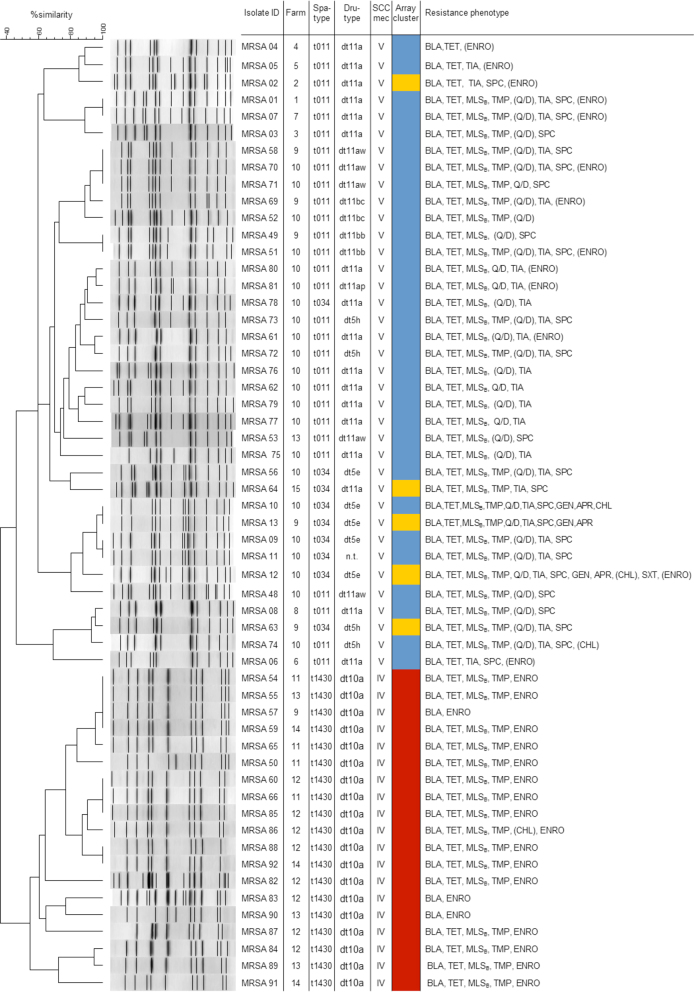
Overall, 3 different spa types were detected in the isolates. A total of 28 isolates belonged to the most common spa type t011 and 9 isolates belonged to t034. Both of these spa types are associated with CC398. The spa type t1430 (CC9) was identified in 19 isolates (Figure 2). On 3 farms (9, 10, and 13), MRSA belonging to more than 1 spa type were detected. Isolates with spa type t1430 were found on 5 farms, 2 of them harbouring also isolates with spa types t011 and t034 (Figure 2). The most common dru type was dt10a (19 isolates), followed by dt11a (17 isolates). All other dru types were found in a few isolates only. All t034 and t011 isolates carried a SCCmec type V cassette with different dru types, while all MRSA of spa type t1430 carried the SCCmec IV cassette and the dru type dt10a. There was 1 t034 isolate that was non-typable by dru typing.
Figure 2.
All 56 isolates were typed by various typing methods and cluster analysis was performed with PFGE and array data. Susceptibility of the isolates to 32 antibiotic agents was analysed by broth microdilution. All t1430 isolates showed high similarity in all typing methods applied. Parentheses indicate that these isolates were classified as intermediate susceptible to the antimicrobial agent according to CLSI standards. Isolates listed as resistant to apramycin, enrofloxacin, spectinomycin or tiamulin showed MIC values of ≥128 µ g/mL, ≥4 µ g/mL, ≥512 µ g/mL, and ≥16 µ g/mL, respectively. An enrofloxacin MIC value of 1 µ g/mL is indicated as intermediate resistance.
Analysis of the similarities between gene profiles via the SplitsTree software revealed that all CC9/SCCmec IV/t1430/dt10a isolates formed a separate cluster apart from the other isolates. The CC398/t011 isolates and CC398/t034 isolates clustered together (Figures 1 and 2). When using BioNumerics software for unweighted pair group method with arithmetic mean (UPGMA) cluster analysis of pulsed-field gel electrophoresis (PFGE) profiles or microarray data, a separate clustering of CC9/t1430 isolates was confirmed (defined as less than 50% similarity), as indicated in Figure 2.
Susceptibility Testing and Resistance Gene Determination
Overall, susceptibility testing revealed 21 different resistance phenotypes of the MRSA isolates, with most isolates showing resistance to substances from 3 or more classes of antimicrobials and they were thus regarded as multiresistant (Figure 2). Looking at the diversity of phenotypic profiles on farm-level, up to 10 different resistance profiles were found per individual farm (Figure 2). MRSA with 2 different phenotypic resistance profiles were found on 4 farms, while isolates with 5 and 10 phenotypes, respectively, were detected on another 2 farms. Altogether, 7 resistance phenotypes were detected on more than 1 farm (Figure 2).
Isolates of spa type t1430 (CC9) exhibited 3 different resistance phenotypes: (i) β-lactam antibiotics, tetracyclines, macrolides, lincosamides and streptrogramin B antibiotics (MLSB), trimethoprim, and elevated MICs of enrofloxacin (MIC ≥ 4 µ g/mL) (n = 15), (ii) β-lactam antibiotics and elevated MICs of enrofloxacin (n = 3), or (iii) β-lactam antibiotics, tetracyclines, MLSB, trimethoprim, chloramphenicol, and elevated MICs of enrofloxacin (n = 1). These phenotypic profiles differed from t011 and t034 (CC398) isolates. As shown in Figure 2, the most common phenotypic resistance profile of t011 and t034 isolates was resistances to β-lactam antibiotics, tetracyclines, MLSB antibiotics, trimethoprim, quinupristin/dalfopristin as well as elevated MICs of tiamulin (MIC ≥ 16 µ g/mL) and spectinomycin (MIC ≥ 512 µ g/mL) (n = 7).
The most common resistances among the isolates were resistances to tetracyclines (n = 53), MLSB antibiotics (n = 49), and trimethoprim (n = 38). In addition, elevated MICs of tiamulin (MICs of 16–≥ 128 µ g/mL) and enrofloxacin (MICs of 4–16 µ g/mL) were observed in 29 and 19 isolates, respectively. Another 13 MRSA showed slightly elevated MIC values for enrofloxacin (MIC = 1 µ g/mL). Resistance to quinupristin/dalfopristin was observed in 8 MRSA, while another 24 isolates showed intermediate resistance to these substances.
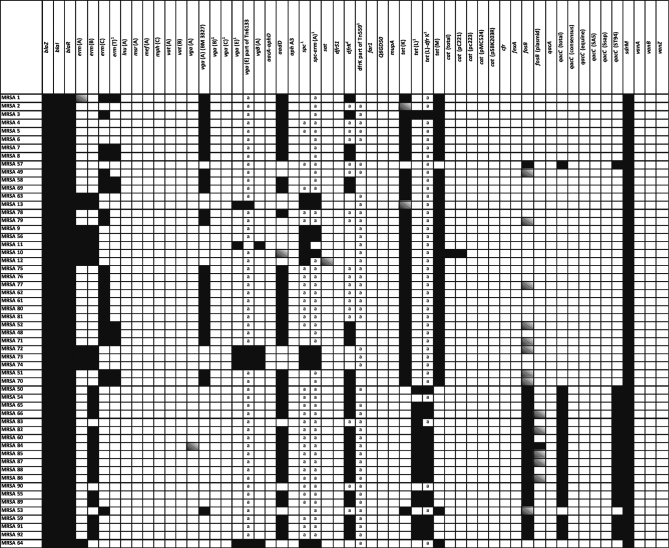
Tetracycline resistance was mediated by the genes tet (K), tet (L), and/or tet (M). Isolates of the spa types t011 and t034 mainly harboured tet (K) and tet (M), while the tet (L) gene was present in most of the t1430 isolates (Figure 3). However, the tet (L) gene was also present in 1 isolate phenotypically susceptible to tetracycline. Resistance to MLSB antibiotics in CC9/t1430 isolates was mediated by the erm (B) gene. All these isolates were phenotypically susceptible to quinupristin/dalfopristin. In all t011 and t034 isolates, at least 1 MLSB or streptogramin resistance determinant [erm (A), erm (B), erm (C), erm (T) vga (A), vga (B), vga (C), or vgb (A)], was present. Although microarray analysis confirmed the presence of the vga (A) allele from strain BM 3327 gene in 26 MRSA, 4 of these isolates were phenotypically susceptible to the combination of quinopristin/dalfopristin. However, a separate streptogramin A compound was not included in the panel of antibiotics tested. The gene dfrK, conferring trimethoprim resistance, was present in 27 of the 38 isolates with MICs above breakpoint level (≥16 µ g/mL). However, the remaining 11 isolates with phenotypic trimethoprim resistance carried neither dfrK nor dfrS1. Elevated MICs of tiamulin (MIC ≥ 16 µ g/mL) were found in 29 MRSA isolates, with vga (A) genes present in 27 of them. Less than half of the examined isolates (n = 24) showed elevated MIC values for spectinomycin (MIC of ≥ 512 µ g/mL), which were mediated by the gene spc in 11 isolates. Only 1 isolate was classified as chloramphenicol-resistant and resistance was found to be mediated by the gene cat (pC221). None of the isolates showed a florfenicol MIC value above 8 µ g/mL. A total of 3 isolates were resistant to gentamicin and also showed apramycin MIC values of ≥ 128 µ g/mL, but neither aacA-aphD nor aphA3 genes were detected. All isolates were susceptible to vancomycin and did not carry vanA, vanB, or vanZ resistance genes.
Figure 3.
Resistance genes of MRSA isolates as determined by microarray or PCR analyses. Isolates from different farms are separated by bold lines. Presence of genes is indicated as black, absence as white, and ambiguous genes as grey boxes.
Virulence Genes
Microarray analysis of the MRSA revealed alleles of staphylococcal superantigen-like proteins 1, 2, 5, 7, and 10, genes for haemolysin gamma (lukF, lukS, and hlgA), leucocidin (lukY 3 isolates ambiguous results, lukX 1 negative, and 3 ambiguous results), and for haemolysine genes (hl, hla, hIIII, and hlb) in all 56 isolates (exceptions as stated in brackets). Furthermore, genes coding for proteases such as aureolysin, glutamyl endopeptidase, and staphopains A and B were detected in all isolates. None of the isolates harboured toxic shock syndrome protein genes or genes coding for the Panton-Valentine leukocidin. Interestingly, alleles present in isolates with spa type t1430 were distinct from the alleles present in the t011 and t034 isolates, although similar virulence genes were detected. In addition, none of the t011 and t034 isolates harboured genes coding for enterotoxins. However, in all t1430 isolates, the enterotoxin G, I, M, N, O, U, and/or Y encoding genes were found as well as genes encoding staphylococcal superantigen-like proteins (ssl 1 additional probes positive, ssl 5, and ssl 9).
DISCUSSION
In the present study, broiler farms from 4 different counties in Germany were sampled in order to investigate the occurrence of MRSA and to characterise the isolates phenotypically and genotypically. For analysis of typing results, different clustering methods were used. The results from the SplitsTree network graph suggest that genotypic patterns of isolates were rather specific for clonal lineages than for origins of isolates from individual farms, as previously described for virulence genes. This was confirmed by UPGMA analysis with CC9/t1430 isolates clustering together and showing a greater distance (similarity <50%) to CC398/t011 and CC398/t034 isolates (Figure 2). However, the close clustering of t034 and t011 isolates could be expected, as both spa types differ only by the doubling (or deletion) of 2 repeats. It should be noted that isolates of CC398 (similar to CC22) might not be sufficiently discriminated by spa -typing or PFGE. In this case Whole Genome Sequencing and associated typing techniques might be the methods of choice to investigate strain relatedness. MRSA with non-distinguishable PFGE profiles were obtained from up to 4 farms and isolates from a single farm differed in their resistance and virulence patterns (Figure 2). In contrast, in a previous study from Germany all t1430 isolates showed closely related PFGE patterns regardless if the isolates derived from humans or chickens (Wendlandt et al., 2013b).
The detection of up to 3 spa types on a single farm might be a result of the high regional density of poultry farming in some geographic regions in Germany, promoting intense transmission of bacteria between farms (Crombé et al., 2013). Other studies reported up to 2 different spa types occurring on farms in Germany and The Netherlands (Friese et al., 2013; Wendlandt et al., 2013a). However, sources of MRSA are difficult to identify and might include introduction through hatchlings, farm workers, or environmental contamination. A study on broiler flocks in Germany found all flocks to be free from MRSA 1 D after new hatchlings arrived, but increasing numbers of environmental and animal samples were tested positive in the course of the fattening period. The authors therefore concluded that MRSA is not mainly introduced via colonised hatchlings (Friese et al., 2013). However, Broens et al. (2011b) reported that pig farms with a MRSA positive supplier have a 10-fold higher odds ratio for being MRSA positive, underlining the importance of suppliers and animal traffic. Another study has shown that people living on a turkey farm can be positive for MRSA, although turkeys were tested negative. The authors concluded that the farm personnel might have been infected in previous production cycles, but re-infection of turkeys might occur (Richter et al., 2012). Thus, biosecurity and a good hygiene practice of farm workers are highly important to prevent colonization of poultry farms (Friese et al., 2013). However, in the present study, there is no information on the MRSA status of the suppliers of chickens, but a good hygiene practice and a high level of biosecurity was reported (Dullweber, 2010). In addition, several authors reported that the herd size is highly associated with MRSA prevalence in pigs due to multiple risk factors (antimicrobial use, animal trade, and hygiene level) (Alt et al., 2011; Broens et al., 2011a), but for poultry farming data are still rare. Of these risk factors, antimicrobial use deserves special attention because selection pressure imposed by the use of antimicrobials favours resistant phenotypes (Crombé et al., 2013). In the present study the herd size of 5 farms was >20,000 animals, of which 1 farm even had >100,000 animals.
However, it must be pointed out that the present study was neither intended to identify MRSA prevalence nor to determine the frequencies of specific MRSA types on the farms. Differences in isolate numbers mainly result from differences in sample numbers and types taken per farm and the availability of isolates (Dullweber, 2010). Although frequencies could not be determined, the phenotypic and genotypic characteristics of individual isolates provide valuable information about the lineages and properties of isolates present on farms.
Most isolates belonged to CC398, the predominant LA-MRSA in Europe (Ye et al., 2016). As displayed in Figure 2, all CC398 isolates carried a SCCmec type V cassette. This finding is in accordance with previous studies, indicating that CC398-MRSA-V is the major lineage in poultry (El-Adawy et al., 2016). However, CC398-MRSA isolates with unknown or truncated SCCmec elements have also been reported (El-Adawy et al., 2016). The majority of the non-CC398 isolates in the current study belonged to CC9, which is the most prevalent LA-MRSA in Asia (Ye et al., 2016). Genetic variation in MRSA isolates has been reported previously, including a hybrid LA-MRSA CC9/CC398 in isolates from human and animal sources, showing both poultry and human genetic adaptation and consisting of a CC398 chromosomal backbone and a smaller CC9 region (Larsen et al., 2016). This increases the possibility of transfer from animal sources to humans (Larsen et al., 2016). Unlike the isolates in the current study, however, these hybrid isolates belonged to spa type t899. Other CCs commonly associated with poultry, such as CC5, were not detected in the present study (Monecke et al., 2013).
The most common spa type in our study was t011 which is also the most common type in pigs (Van Den Broek et al., 2009) and veal calves (Graveland et al., 2010). All spa types identified in the course of this study (t011, t034, and t1430) were previously detected in poultry and poultry products (Fessler et al., 2011, 2013). However, a single t034 isolate was non-typeable by dru typing. Non-typeability might be attributed to a deletion or truncation in the relevant gene region (the direct-repeat unit adjacent to IS431 in SCCmec), leading to a lack of amplification (El-Adawy et al., 2016).
Among the 56 MRSA isolates included in our study, a high diversity of resistance phenotypes was present and up to 10 different phenotypes were found on a single farm (Figure 2). The CC9/t1430 isolates were all resistant to β-lactams, showed elevated MICs of enrofloxacin and were frequently resistant to MLSB antibiotics and trimethoprim (Figure 2). They exhibited phenotypes that were previously reported in German CC9-poultry isolates (Monecke et al., 2013; Wendlandt et al., 2013b). The phenotypes of t011 and t034 isolates were rather heterogenic, as discussed in other reports (Nemati et al., 2008; Fessler et al., 2011; Richter et al., 2012; Monecke et al., 2013; Wendlandt et al., 2013b).
Many isolates in our study were resistant to tetracycline (53 isolates), mediated by the genes tet (K), tet (L), and tet (M), and to MLSB antibiotics, mediated by the genes erm (A), erm (B), erm (C), erm (T), vga (A), or vgb (A) (49 isolates). These antimicrobial agents are among the most often used substances in poultry industry and resistances are frequently detected in poultry isolates (Nemati et al., 2008; Mulders et al., 2010; Vanderhaeghen et al., 2010; Fessler et al., 2011; Monecke et al., 2013; Wendlandt et al., 2013b). Studies on MRSA isolates from Belgium (Nemeghaire et al., 2013) and The Netherlands (Wendlandt et al., 2013b) found MLSB resistance rates of up to 50%, although earlier studies detected lower rates (Nemati et al., 2008). A number of 38 MRSA of the present strain collection was resistant to trimethoprim. Although MICs of trimethoprim were ≥256 µ g/mL in 11 of these isolates, no trimethoprim resistance genes were detected. However, point mutations in the dihydrofolate reductase gene might be responsible for elevated MICs (Dale et al., 1997; Vickers et al., 2009). In contrast, all 26 isolates collected from 4 German broiler farms in a previous study were trimethoprim resistant (Wendlandt et al., 2013a). While results from a Dutch study on MRSA in broiler flocks at slaughterhouses detected gentamicin resistance in 22% of the isolates, a lower percentage of gentamicin resistance was found in our study (n = 3.5%), which might be due to different antibiotic breakpoints usage (Mulders et al., 2010). The authors of a study focused on MRSA from German food and food products of poultry origin (Fessler et al., 2011) reported elevated MICs of enrofloxacin in 15.6% of isolates. This was considerably lower than in our study, where 34% of MRSA isolates showed enrofloxacin MICs of ≥4 µ g/mL and another 23% of isolates with MICs of 1 µ g/mL.
Notably, a single isolate with MIC value of tetracycline below the breakpoint carried the resistance gene tet (L), as confirmed by microarray and PCR analysis. However, dysfunctionality of this gene in Gram-positive cocci has been reported previously (Cauwerts et al., 2007).
Only 11 out of 24 isolates with elevated MICs of spectinomycin carried the resistance gene spc. These results suggested that resistance in these isolates might be conferred by other resistance mechanisms or other resistance genes, which were not included in microarray or PCR analyses, such as spd or spw (Jamrozy et al., 2014; Wendlandt et al., 2014). In 11 isolates from 2 farms, the MIC of tiamulin was ≥128 µ g/mL, but only 6 of them carried a vga resistance determinant. Since none of the isolates carried cfr, point mutations in the V domain of the 23S rRNA of the rplC gene encoding the ribosomal protein L3 or the presence of another resistance determinant might be responsible for elevated MICs of tiamulin (van Duijkeren et al., 2014). In contrast, the vga (A) (BM 3327) positive MRSA belonging to CC398/t011 and CC398/t034 exhibited wild-type MICs for MLSB antibiotics and tiamulin. Thus, dysfunctionality of the vga (A) gene might explain the susceptible phenotype, but further studies are needed to elucidate the mechanism.
The occurrence of virulence genes in isolates of our study was similar to previous studies (Fessler et al., 2011; Richter et al., 2012; Monecke et al., 2013; Wendlandt et al., 2013b). Enterotoxin genes of the egc cluster were found only in t1430 isolates, but neither in t011 nor in t034 MRSA (Fessler et al., 2011) and important virulence factors such as genes encoding toxic shock syndrome toxin1, PVL and exfoliative toxins were not identified in any of the examined isolates. This is in accordance with other studies (Vanderhaeghen et al., 2010; Fessler et al., 2011; Wendlandt et al., 2013c), since the presence of the egc is a common feature of certain CCs including CC9 (Fessler et al., 2011; Monecke et al., 2011; Kraushaar et al., 2017).
In conclusion, various resistance phenotypes and resistance genes were present in MRSA isolates from German broiler farms. Isolates present at farm level may contribute to the contamination of all subsequent stages of the food chain (Kraushaar et al., 2017). Therefore, insight into resistance determinants present on farm level and detailed studies on distribution of MRSA along the food production line is necessary (Wendlandt et al., 2013c).
REFERENCES
- Aarestrup F.M., Agersø L.Y., Ahrens P., Jørgensen J.C., Madsen M., Jensen L.B. Antimicrobial susceptibility and presence of resistance genes in staphylococci from poultry. Vet. Microbiol. 2000;74:353–364. doi: 10.1016/s0378-1135(00)00197-8. [DOI] [PubMed] [Google Scholar]
- Alt K., Fetsch A., Schroeter A., Guerra B., Hammerl J.A., Hertwig S., Senkov N., Geinets A., Mueller-Graf C., Braeunig J., Kaesbohrer A., Appel B., Hensel A., Tenhagen B.A. Factors associated with the occurrence of MRSA CC398 in herds of fattening pigs in Germany. BMC Vet. Res. 2011;7:69. doi: 10.1186/1746-6148-7-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broens E.M., Graat E.A., Van der Wolf P.J., van de Giessen A.W., De Jong M.C. Prevalence and risk factor analysis of livestock associated MRSA-positive pig herds in The Netherlands. Prev. Vet. Med. 2011;102:41–49. doi: 10.1016/j.prevetmed.2011.06.005. [DOI] [PubMed] [Google Scholar]
- Broens E.M., Graat E.A., van der Wolf P.J., van de Giessen A.W., van Duijkeren E., Wagenaar J.A., van Nes A., Mevius D.J., de Jong M.C. MRSA CC398 in the pig production chain. Prev. Vet. Med. 2011;98:182–189. doi: 10.1016/j.prevetmed.2010.10.010. [DOI] [PubMed] [Google Scholar]
- Cauwerts K., Decostere A., De Graef E.M., Haesebrouck F., Pasmans F. High prevalence of tetracycline resistance in Enterococcus isolates from broilers carrying the erm(B) gene. Avian Pathol. 2007;36:395–399. doi: 10.1080/03079450701589167. [DOI] [PubMed] [Google Scholar]
- CLSI . Twenty-fourth informational supplement M100-S24. Clinical and Laboratory Standards Institute; Wayne, PA: 2014. Performance standards for antimicrobial susceptibility testing. [Google Scholar]
- CLSI . Third edition supplement VET01S. Clinical and Laboratory Standards Institute; Wayne, PA: 2015. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. [Google Scholar]
- Crombé F., Argudín M.A., Vanderhaeghen W., Hermans K., Haesebrouck F., Butaye P. Transmission dynamics of methicillin-resistant Staphylococcus aureus in pigs. Front. Microbiol. 2013;4:57. doi: 10.3389/fmicb.2013.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale G.E., Broger C., D'Arcy A., Hartman P.G., DeHoogt R., Jolidon S., Kompis I., Labhardt A.M., Langen H., Locher H., Page M.G., Stüber D., Then R.L., Wipf B., Oefner C. A single amino acid substitution in Staphylococcus aureus dihydrofolate reductase determines trimethoprim resistance. J. Mol. Biol. 1997;266:23–30. doi: 10.1006/jmbi.1996.0770. [DOI] [PubMed] [Google Scholar]
- de Boer E., Zwartkruis-Nahuis J.T., Wit B., Huijsdens X.W., de Neeling A.J., Bosch T., van Oosterom R.A., Vila A., Heuvelink A.E. Prevalence of methicillin-resistant Staphylococcus aureus in meat. Int. J. Food Microbiol. 2009;134:52–56. doi: 10.1016/j.ijfoodmicro.2008.12.007. [DOI] [PubMed] [Google Scholar]
- Devriese L.A. Staphylococci in healthy and diseased animals. Soc. Appl. Bacteriol. Symp. Ser. 1990;19:71S–80S. doi: 10.1111/j.1365-2672.1990.tb01799.x. [DOI] [PubMed] [Google Scholar]
- Dullweber A. Stiftung Tierärztliche Hochschule Hannover; Hannover: 2010. Occurence of methicillin-resistant Staphylococcus aureus (MRSA) in poultry flocks. Doctoral Thesis. [Google Scholar]
- ECDC . European Centre for Disease Prevention and Control (ECDC); Stockholm: 2017. Summary of the latest data on antibiotic resistance in the European Union: EARS-Net surveillance data, November 2017. [Google Scholar]
- El-Adawy H., Ahmed M., Hotzel H., Monecke S., Schulz J., Hartung J., Ehricht R., Neubauer H., Hafez H.M. Characterization of methicillin-resistant Staphylococcus aureus isolated from healthy turkeys and broilers using DNA microarrays. Front. Microbiol. 2016;7 doi: 10.3389/fmicb.2016.02019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fessler A., Scott C., Kadlec K., Ehricht R., Monecke S., Schwarz S. Characterization of methicillin-resistant Staphylococcus aureus ST398 from cases of bovine mastitis. J. Antimicrob. Chemother. 2010;65:619–625. doi: 10.1093/jac/dkq021. [DOI] [PubMed] [Google Scholar]
- Fessler A.T., Kadlec K., Hassel M., Hauschild T., Eidam C., Ehricht R., Monecke S., Schwarz S. Characterization of methicillin-resistant Staphylococcus aureus isolates from food and food products of poultry origin in Germany. Appl. Environ. Microbiol. 2011;77:7151–7157. doi: 10.1128/AEM.00561-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friese A., Schulz J., Zimmermann K., Tenhagen B.A., Fetsch A., Hartung J., Rösler U. Occurrence of livestock-associated methicillin-resistant Staphylococcus aureus in turkey and broiler barns and contamination of air and soil surfaces in their vicinity. Appl. Environ. Microbiol. 2013;79:2759–2766. doi: 10.1128/AEM.03939-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geenen P.L., Graat E.A., Haenen A., Hengeveld P.D., Van Hoek A.H., Huijsdens X.W., Kappert C.C., Lammers G.A., Van Duijkeren E., van De Giessen A.W. Prevalence of livestock-associated MRSA on Dutch broiler farms and in people living and/or working on these farms. Epidemiol. Infect. 2013;141:1099–1108. doi: 10.1017/S0950268812001616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goering R.V., Morrison D., Al-Doori Z., Edwards G.F., Gemmell C.G. Usefulness of mec-associated direct repeat unit (dru) typing in the epidemiological analysis of highly clonal methicillin-resistant Staphylococcus aureus in Scotland. Clin. Microbiol. Infect. 2008;14:964–969. doi: 10.1111/j.1469-0691.2008.02073.x. [DOI] [PubMed] [Google Scholar]
- Graveland H., Wagenaar J.A., Heesterbeek H., Mevius D., van Duijkeren E., Heederik D. Methicillin resistant Staphylococcus aureus ST398 in veal calf farming: human MRSA carriage related with animal antimicrobial usage and farm hygiene. PLoS ONE. 2010;5 doi: 10.1371/journal.pone.0010990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grontvedt C.A., Elstrom P., Stegger M., Skov R.L., Skytt Andersen P., Larssen K.W., Urdahl A.M., Angen O., Larsen J., Amdal S., Lotvedt S.M., Sunde M., Bjornholt J.V. Methicillin-resistant Staphylococcus aureus CC398 in humans and pigs in Norway: a “One Health” perspective on introduction and transmission. Clin. Infect. Dis. 2016;63:1431–1438. doi: 10.1093/cid/ciw552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamrozy D.M., Coldham N.G., Butaye P., Fielder M.D. Identification of a novel plasmid-associated spectinomycin adenyltransferase gene spd in methicillin-resistantStaphylococcus aureus ST398 isolated from animal and human sources. J. Antimicrob. Chemother. 2014;69:1193–1196. doi: 10.1093/jac/dkt510. [DOI] [PubMed] [Google Scholar]
- Köck R., Schaumburg F., Mellmann A., Köksal M., Jurke A., Becker K., Friedrich A.W. Livestock-associated methicillin-resistant Staphylococcus aureus (MRSA) as causes of human infection and colonization in Germany. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0055040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraushaar B., Ballhausen B., Leeser D., Tenhagen B.A., Käsbohrer A., Fetsch A. Antimicrobial resistances and virulence markers in methicillin-resistant Staphylococcus aureus from broiler and turkey: a molecular view from farm to fork. Vet. Microbiol. 2017;200:25–32. doi: 10.1016/j.vetmic.2016.05.022. [DOI] [PubMed] [Google Scholar]
- Larsen J., Stegger M., Andersen P.S., Petersen A., Larsen A.R., Westh H., Agersø Y., Fetsch A., Kraushaar B., Käsbohrer A., Febetaler A.T., Schwarz S., Cuny C., Witte W., Butaye P., Denis O., Haenni M., Madec J.Y., Jouy E., Laurent F., Battisti A., Franco A., Alba P., Mammina C., Pantosti A., Monaco M., Wagenaar J.A., de Boer E., van Duijkeren E., Heck M., Domínguez L., Torres C., Zarazaga M., Price L.B., Skov R.L. Evidence for human adaptation and foodborne transmission of livestock-associated methicillin-resistant Staphylococcus aureus. Clin. Infect. Dis. 2016;63:1349–1352. doi: 10.1093/cid/ciw532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monecke S., Coombs G., Shore A.C., Coleman D.C., Akpaka P., Borg M., Chow H., Ip M., Jatzwauk L., Jonas D., Kadlec K., Kearns A., Laurent F., O'Brien F.G., Pearson J., Ruppelt A., Schwarz S., Scicluna E., Slickers P., Tan H.L., Weber S., Ehricht R. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0017936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monecke S., Ruppelt A., Wendlandt S., Schwarz S., Slickers P., Ehricht R., Jäckel S.C. Genotyping of Staphylococcus aureus isolates from diseased poultry. Vet. Microbiol. 2013;162:806–812. doi: 10.1016/j.vetmic.2012.10.018. [DOI] [PubMed] [Google Scholar]
- Monecke S., Slickers P., Ehricht R. Assignment of Staphylococcus aureus isolates to clonal complexes based on microarray analysis and pattern recognition. FEMS Immunol. Med. Microbiol. 2008;53:237–251. doi: 10.1111/j.1574-695X.2008.00426.x. [DOI] [PubMed] [Google Scholar]
- Mulders M.N., Haenen A.P., Geenen P.L., Vesseur P.C., Poldervaart E.S., Bosch T., Huijsdens X.W., Hengeveld P.D., Dam-Deisz W.D., Graat E.A., Mevius D., Voss A., van de Giessen A.W. Prevalence of livestock-associated MRSA in broiler flocks and risk factors for slaughterhouse personnel in The Netherlands. Epidemiol. Infect. 2010;138:743–755. doi: 10.1017/S0950268810000075. [DOI] [PubMed] [Google Scholar]
- Murchan S., Kaufmann M.E., Deplano A., de Ryck R., Struelens M., Zinn C.E., Fussing V., Salmenlinna S., Vuopio-Varkila J., El Solh N., Cuny C., Witte W., Tassios P.T., Legakis N., van Leeuwen W., van Belkum A., Vindel A., Laconcha I., Garaizar J., Haeggman S., Olsson-Liljequist B., Ransjo U., Coombes G., Cookson B. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J. Clin. Microbiol. 2003;41:1574–1585. doi: 10.1128/JCM.41.4.1574-1585.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemati M., Hermans K., Lipinska U., Denis O., Deplano A., Struelens M., Devriese L.A., Pasmans F., Haesebrouck F. Antimicrobial resistance of old and recent Staphylococcus aureus isolates from poultry: first detection of livestock-associated methicillin-resistant strain ST398. Antimicrob. Agents Chemother. 2008;52:3817–3819. doi: 10.1128/AAC.00613-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeghaire S., Roelandt S., Argudín M.A., Haesebrouck F., Butaye P. Characterization of methicillin-resistant Staphylococcus aureus from healthy carrier chickens. Avian Pathol. 2013;42:342–346. doi: 10.1080/03079457.2013.805183. [DOI] [PubMed] [Google Scholar]
- Otto M. Staphylococcus aureus toxins. Curr. Opin. Microbiol. 2014;17:32–37. doi: 10.1016/j.mib.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen A.B., Skov R., Pallesen L.V. Detection of methicillin resistance in coagulase-negative staphylococci and in staphylococci directly from simulated blood cultures using the EVIGENE MRSA Detection Kit. J. Antimicrob. Chemother. 2003;51:419–421. doi: 10.1093/jac/dkg084. [DOI] [PubMed] [Google Scholar]
- Richter A., Sting R., Popp C., Rau J., Tenhagen B.A., Guerra B., Hafez H.M., Fetsch A. Prevalence of types of methicillin-resistant Staphylococcus aureus in turkey flocks and personnel attending the animals. Epidemiol. Infect. 2012;140:2223–2232. doi: 10.1017/S095026881200009X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwendener S., Perreten V. New transposon Tn6133 in methicillin-resistant Staphylococcus aureus ST398 contains vga(E), a novel streptogramin A, pleuromutilin, and lincosamide resistance gene. Antimicrob. Agents Chemother. 2011;55:4900–4904. doi: 10.1128/AAC.00528-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shopsin B., Gomez M., Montgomery S.O., Smith D.H., Waddington M., Dodge D.E., Bost D.A., Riehman M., Naidich S., Kreiswirth B.N. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 1999;37:3556–3563. doi: 10.1128/jcm.37.11.3556-3563.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Broek I.V.F., Cleef B.A., Haenen A., Broens E.M., Van Der Wolf P.J., van den Broek M.J., Huijsdens X.W., Kluytmans J.A., Van De Giessen A.W., Tiemersma E.W. Methicillin-resistant Staphylococcus aureus in people living and working in pig farms. Epidemiol. Infect. 2009;137:700–708. doi: 10.1017/S0950268808001507. [DOI] [PubMed] [Google Scholar]
- van Duijkeren E., Greko C., Pringle M., Baptiste K.E., Catry B., Jukes H., Moreno M.A., Pomba M.C., Pyörälä S., Rantala M., Ružauskas M., Sanders P., Teale C., Threlfall E.J., Torren-Edo J., Törneke K. Pleuromutilins: use in food-producing animals in the European Union, development of resistance and impact on human and animal health. J. Antimicrob. Chemother. 2014;69:2022–2031. doi: 10.1093/jac/dku123. [DOI] [PubMed] [Google Scholar]
- Vandendriessche S., Vanderhaeghen W., Soares F.V., Hallin M., Catry B., Hermans K., Butaye P., Haesebrouck F., Struelens M.J., Denis O. Prevalence, risk factors and genetic diversity of methicillin-resistant Staphylococcus aureus carried by humans and animals across livestock production sectors. J. Antimicrob. Chemother. 2013;68:1510–1516. doi: 10.1093/jac/dkt047. [DOI] [PubMed] [Google Scholar]
- Vanderhaeghen W., Hermans K., Haesebrouck F., Butaye P. Methicillin-resistant Staphylococcus aureus (MRSA) in food production animals. Epidemiol. Infect. 2010;138:606–625. doi: 10.1017/S0950268809991567. [DOI] [PubMed] [Google Scholar]
- Vickers A.A., Potter N.J., Fishwick C.W., Chopra I., O'Neill A.J. Analysis of mutational resistance to trimethoprim in Staphylococcus aureus by genetic and structural modelling techniques. J. Antimicrob. Chemother. 2009;63:1112–1117. doi: 10.1093/jac/dkp090. [DOI] [PubMed] [Google Scholar]
- Wendlandt S., Fessler A.T., Kadlec K., van Duijkeren E., Schwarz S. Identification of the novel spectinomycin resistance gene spd in a different plasmid background among methicillin-resistant Staphylococcus aureus CC398 and methicillin-susceptible S. aureus ST433. J. Antimicrob. Chemother. 2014;69:2000–2003. doi: 10.1093/jac/dku067. [DOI] [PubMed] [Google Scholar]
- Wendlandt S., Kadlec K., Fessler A.T., Mevius D., van Essen-Zandbergen A., Hengeveld P.D., Bosch T., Schouls L., Schwarz S., van Duijkeren E. Transmission of methicillin-resistant Staphylococcus aureus isolates on broiler farms. Vet. Microbiol. 2013;167:632–637. doi: 10.1016/j.vetmic.2013.09.019. [DOI] [PubMed] [Google Scholar]
- Wendlandt S., Kadlec K., Fessler A.T., Monecke S., Ehricht R., van de Giessen A.W., Hengeveld P.D., Huijsdens X., Schwarz S., van Duijkeren E. Resistance phenotypes and genotypes of methicillin-resistant Staphylococcus aureus isolates from broiler chickens at slaughter and abattoir workers. J. Antimicrob. Chemother. 2013;68:2458–2463. doi: 10.1093/jac/dkt239. [DOI] [PubMed] [Google Scholar]
- Wendlandt S., Schwarz S., Silley P. Methicillin-resistant Staphylococcus aureus: a food-borne pathogen? Ann. Rev. Food Sci. Technol. 2013;4:117–139. doi: 10.1146/annurev-food-030212-182653. [DOI] [PubMed] [Google Scholar]
- Ye X., Wang X., Fan Y., Peng Y., Li L., Li S., Huang J., Yao Z., Chen S. Genotypic and phenotypic markers of livestock-associated methicillin-resistant Staphylococcus aureus CC9 in humans. Appl. Environ. Microbiol. 2016;82:3892–3899. doi: 10.1128/AEM.00091-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., McClure J.A., Elsayed S., Louie T., Conly J.M. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 2005;43:5026–5033. doi: 10.1128/JCM.43.10.5026-5033.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]