Abstract
We evaluated the combination of immunomagnetic separation (IMS), multiple displacement amplification (MDA), and real-time PCR to detect Salmonella from poultry environmental samples. The limits of detection (LODs) of IMS-MDA real-time PCR with different culture enrichment hours (0, 4, 6, and 8 h) were determined in artificially inoculated litter samples from a specific pathogen-free (SPF) poultry farm. In addition, Salmonella detection rate of IMS-MDA real-time PCR with 8-h culture enrichment was compared with that of conventional real-time PCR and culture-based detection by analyzing 174 poultry environmental samples (boot swabs, drag swabs, and litter), and the levels of Salmonella in the samples were quantified using the most probably number method. The LODs of IMS-MDA real-time PCR with 0, 4 to 6, and 8-h enrichment were 10, 1, and 0.1 CFU/g, respectively. Salmonella was detected in 25 of the 174 environmental samples (14.4%) by IMS-MDA real-time PCR, compared with 24 (13.8%) by conventional real-time PCR and 19 (10.9%) by culturing. Cohen's kappa index indicated strong concordance (0.79) between IMS-MDA real-time PCR and culture detection. We demonstrated the potential of the IMS-MDA real-time PCR assay as a faster and more sensitive alternative to culture-based Salmonella detection from poultry environmental samples.
key words: Salmonella; detection; environmental sample, whole genome amplification; IMS; MDA; real-time PCR
INTRODUCTION
Nontyphoidal Salmonella enterica causes an estimated total of 1.4 million cases of salmonellosis with approximately 600 deaths annually in the United States (Scallan et al., 2011; Lungu et al., 2012). The great majority of human cases of salmonellosis are due to the consumption of contaminated foods of animal origin, especially poultry meat and poultry by-products (Lungu et al., 2012; Berghaus et al., 2013; Soria et al., 2017). According to the Food Safety and Inspection Service (FSIS), the U.S. national prevalence of Salmonella in chicken parts is over 20% (http://www.fsis.usda.gov/shared/PDF/Baseline_Data_Raw_Chicken_Parts.pdf).
Because of the fact that most Salmonella serotypes do not typically cause morbidity or mortality in poultry, environmental samples from poultry farms tend to more readily indicate the presence of Salmonella in the flock than clinical symptoms of birds (Holt et al., 2011; Soria et al., 2017). In addition, environmental monitoring is a useful, effective, and less invasive method to predict potential Salmonella infection of poultry flocks (Waltman and Gast, 2008). Therefore, there is a need for efficient and sensitive detection methods for environmental monitoring of the pathogen in poultry production environments.
According to National Poultry Improvement Plan (NPIP) program standards of U.S. Department of Agriculture (USDA, 2017), culture methods are used to detect Salmonella spp. from poultry environmental samples (https://www.poultryimprovement.org/documents/ProgramStandardsJanuary2017.pdf), and Salmonella detection by culture method takes up to 4 to 6 D. A variety of rapid Salmonella detection assays for poultry environmental samples such as PCR, real-time PCR, and ELISA have been developed to shorten detection time (Charlton et al., 2005; Leon-Velarde et al., 2009; Lungu et al., 2012). However, these methods require at least 24-h culture enrichment due to the often low abundances of target organism and high levels of background flora in the sample matrix (Charlton et al., 2005; Leon-Velarde et al., 2009; Lungu et al., 2012). Furthermore, few validation studies have been conducted using naturally contaminated samples or environmental samples from poultry production.
In our previous study (Hyeon and Deng, 2017), we developed a real-time PCR-based Salmonella detection method that substantially shortened culture enrichment by combining immunomagnetic separation (IMS) for target cell capture and multiple displacement amplification (MDA) for whole genome amplification. MDA is a whole genome amplification technique that utilizes isothermal DNA amplification by highly efficient φ29 polymerase. It has been widely used to generate sufficient quantities of DNA for whole genome and metagenomics sequencing from often small amounts of cells (Hosono et al., 2003; Binga et al., 2008; Rodrigue et al., 2009; Seth-Smith et al., 2013). This method was successfully applied to Salmonella detection from raw chicken breast. IMS-MDA real-time PCR was able to detect 10 CFU/g of Salmonella enterica subsp. enterica serotype Enteritidis (SE) in raw chicken breast without culture enrichment and 0.1 CFU/g of SE with 4-h culture enrichment (Hyeon and Deng, 2017). In the current study, we extended the detection method to poultry environmental samples and validated its performance by comparing it with conventional real-time PCR and culture-based detection.
MATERIALS AND METHODS
Microorganism
A SE strain (CFS039 isolated from a poultry source in Georgia) was used for inoculation of litter samples in this study. Cultures of the SE were prepared by growing the stock culture in tryptic soy broth (Difco laboratory, Detroit, MI) overnight at 37°C before inoculation. To obtain viable Salmonella counts, 10-fold serial dilutions of overnight cultures were made in phosphate-buffered saline (PBS, pH 7.2, Amresco, Cleveland, OH) and 100 µL of the dilutions was plated on tryptic soy agar (Difco laboratory). The plates were incubated at 37°C overnight and single colonies were enumerated from the appropriate dilutions.
Inoculation of SE to Litter Samples From a Specific Pathogen-Free Chicken Farm
Salmonella -negative litter samples were collected from a specific pathogen-free (SPF) chicken farm of the U.S. National Poultry Research Center in Athens, Georgia to compare the limits of detection (LOD) of IMS-MDA real-time PCR with those of conventional real-time PCR, IMS real-time PCR, and MDA real-time PCR. In addition, the detection rates of IMS-MDA real-time PCR with different culture enrichment hours were compared. All samples were confirmed Salmonella -negative by culture-based detection before the experiments.
Litter samples (25 g portions) were aseptically placed in sterile Whirl-pak filter bags (Nasco, Fort Atkinson, WI) and inoculated with 2 mL of SE inocula prepared from 10-fold serial dilutions in PBS. Then, the inoculated sample in the Whirl-pak bag was hand massaged for 1 min to enable homogenous distribution of the inoculum. For a negative control, an uninoculated litter sample (25 g) was also prepared. All samples were mixed with 225 mL of buffered peptone water (BPW, Difco Laboratory) by hand massage for 3 min, and 50 mL of homogenate was collected. To compare the detection rates of IMS-MDA real-time PCR after different enrichment hours, 50 mL of enriched BPW was collected after incubation for 4, 6, or 8 h at 37°C. Homogenate or enriched BPW was centrifuged at 100 × g for 10 min to remove solid debris. Then, each supernatant was carefully recovered and centrifuged at 3,000 ×g for 10 min to harvest cell pellet. The pellet was resuspended in 5 mL of BPW, and the suspension was used for DNA extraction or IMS.
Farm Environmental Sample Collection and Preparation
For validation study, environmental samples were collected from the poultry farm of the University of Georgia Poultry Research Center in Athens, Georgia. A total of 174 environmental samples were collected from 46 pen houses (10 to 20 birds/pen), and each sample set consisted of a litter, a drag swab, and a boot swab sample per pen house. The chickens in 4 of the 46 pen houses were infected orally with 107 CFU of Salmonella enterica subsp. enterica serotype Typhimurium (in 1 mL), and the samples were collected at 0, 3, 10, and 17 D post infection.
The environmental samples were collected according to the NPIP procedure. Litter samples were collected in sterile Whirl-pak filter bags and homogenized with 225 mL BPW by hand massage. Drag swab and boot swab samples were prepared using Drag Swabs Poultry Sampling Kits (Solar Biologicals Inc., Newark, DE) and EnviroBootie (Hardy Diagnostics, Santa Maria, CA), respectively. The samples were homogenized with 100 and 150 mL BPW according to the manufacturer's manuals.
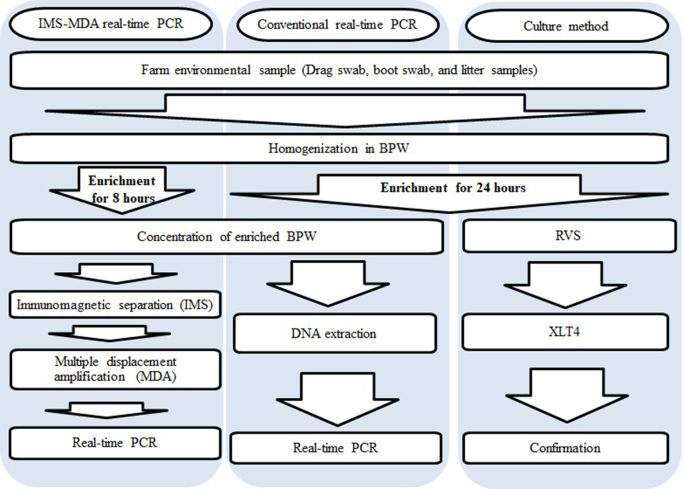
After mixing by hand massage for 3 min, 10, 1, and 0.1 mL of homogenate were used for 3-tube MPN method to enumerate Salmonella in samples according to the USDA-FSIS Microbiology Laboratory Guidebook (MLG) Appendix 2.05 of FSIS, 2014 (https://www.fsis.usda.gov/wps/wcm/connect/8872ec11-d6a3-4fcf-86df-4d87e57780f5/MLG-Appendix-2.pdf?MOD=AJPERES). The remaining samples were incubated at 37°C for 8 h for IMS-MDA real-time PCR or 24 h for conventional real-time PCR and culture method. For conventional real-time PCR and IMS-MDA real-time PCR, 50 mL of enriched BPW was centrifuged as described previously, and the pellet was resuspended in 5 mL of BPW (Figure 1).
Figure 1.
Flow diagrams of IMS-MDA real-time PCR, conventional real-time PCR, and culture method for detecting Salmonella in farm environmental samples in this study. “RVS”: Rappaport-Vassiliadis soya broth. “XLT4”: Xylose lysine Tergitol 4 agar.
DNA Preparation
Genomic DNA for PCR reactions was prepared the using PrepSEQ Rapid Spin Sample Preparation Kit (Thermo Fisher Scientific, Waltham, MA). Briefly, each 750 μL aliquot of cells suspended in BPW was pelleted by centrifugation at 16,000 × g for 3 min. After discarding the supernatant, 50 μL of Lysis Buffer was mixed with the pellet and the suspension was incubated at 97 ± 2°C for 12 ± 2 min. Then, the suspension was cooled at the room temperature for 2 min and centrifuged at 16,000 × g for about 1 min. The resulting pellet was resuspended in 250 μL of nuclease-free water, followed by a final round of centrifugation at 16,000 × g for 1 to 2 min. The final supernatant (approximately 200 μL) was collected in a new tube for use as DNA templates.
IMS, MDA, and IMS-MDA Real-Time PCR
IMS, MDA, and real-time PCR were performed as previously described (Hyeon and Deng, 2017) with some modification for IMS real-time PCR and MDA real-time PCR.
For IMS, 1 mL of resuspended cells in BPW was incubated with 20 μL of Dynabeads anti-Salmonella (Thermo Fisher Scientific) at the room temperature using a rotating mixer (Thermo Fisher Scientific) for 30 min. After incubation, the bead-Salmonella complexes were magnetically separated from the suspension using a magnetic particle concentrator (Thermo Fisher Scientific) for 3 min, and then washed 3 times with 1 mL of PBS containing 0.05% (v/v) Tween 20 (Thermo Fisher Scientific) to remove nonspecifically binding bacteria from the complex. At the final washing step, supernatants were discarded and the bead-Salmonella complexes were used for IMS real-time PCR or IMS-MDA real-time PCR.
MDA was performed using the Illustra GenomePhi V2 DNA amplification kit (GE Healthcare Life Sciences, Piscataway, NJ) according to the manufacturer's instruction. The extracted DNA (2 μL) or the bead-Salmonella complexes from IMS were mixed with 9 μL of sample buffer and incubated at 95°C for 3 min for denaturation. After cooling to 4°C on ice, 9 μL of reaction buffer with 1 μL of enzyme mix was combined to each sample on ice. After incubation at 30°C for 1.5 to 2 h for amplification, the samples were heated to 65°C for 10 min to inactivate the enzyme and cooled to 4°C on ice. Then, the final products (approximately 20 μL) were stored at −20°C until use for real-time PCR.
Real-time PCR was performed as previously described (Hyeon and Deng, 2017), and the bead-Salmonella complexes resuspended in 20 μL of PBS (2 μL), MDA products (2 μL), IMS-MDA products (2 μL), and extracted DNA (2 μL) were used as templates for IMS, MDA, IMS-MDA, and conventional real-time PCR, respectively. In addition, internal amplification control (IAC) was tested using Taqman Exogenous Internal Positive Control reagent (Thermo Fisher Scientific) in order to rule out the presence of PCR inhibitor in the samples.
Salmonella Enumeration and Culture-Based Detection
Quantification of Salmonella in poultry environmental samples was performed using a miniature 3-tube most probable number (MPN) procedure according to MLG Appendix 2.05. Culture-based detection for Salmonella was performed following USDA-FSIS MLG 4.10, 2019 (https://www.fsis.usda.gov/wps/wcm/connect/700c05fe-06a2-492a-a6e1-3357f7701f52/MLG-4.pdf?MOD=AJPERES). A 3-tube MPN using 10, 1, and 0.1 mL of homogenate was performed to estimate Salmonella contamination levels of Salmonella -positive samples following the MPN procedure of MLG with the modification that 225 mL of BPW was used. After 24 h pre-enrichment, 0.1 mL of BPW culture was transferred to Rappaport–Vassiliadis soya broth (RVS, Oxoid, Basingstroke, Hampshire, UK) and incubated for 24 h at 42°C. After the selective enrichment, a loopful of each enriched sample was streaked on differential media Xylose lysine Tergitol 4 agar (XLT4, Oxoid). The presumptive Salmonella colonies from the selective agar were confirmed using real-time PCR as previously described (Hyeon and Deng, 2017).
Statistical Analysis
The mean Ct values of each trial were analyzed to compare conventional real-time PCR, IMS real-time PCR, MDA real-time PCR, and IMS-MDA real-time PCR. The statistical difference between the number of Salmonella -positive samples of different detection methods was analyzed with GraphPad Software version 3.05 (San Diego, CA). The difference was considered statistically significant when P ≤ 0.05. Fisher's exact probability tests for 2 × 2 contingency tables were applied to compare IMS-MDA real-time PCR, conventional real-time PCR, and culture method. To validate IMS-MDA real-time PCR with 8-h enrichment, Cohen's Kappa index values were calculated by comparison with the culture method (Cohen, 1960; ISO:16140,2003).
The threshold cycle (Ct), which is the intersection between each fluorescence curve and a threshold line, was calculated using Applied Biosystems 7500 real-time PCR system software version 2.0.6 (Thermo Fisher Scientific). Negative results correspond to Ct values ≥40 or sample with Ct values higher than that of negative control.
RESULTS AND DISCUSSION
LOD of IMS-MDA Real-Time PCR for Salmonella Detection From Litter
The LODs of SE in litter samples by conventional real-time PCR, IMS real-time PCR, MDA real-time PCR, and IMS-MDA real-time PCR are shown in Table 1. IMS and MDA were separately coupled with real-time PCR to evaluate their individual contribution to improving real-time PCR-based Salmonella detection.
Table 1.
Detection limits of conventional real-time PCR, immunomagnetic separation (IMS) real-time PCR, and immunomagnetic separation-multiple displacement amplification (IMS-MDA) real-time PCR to detect Salmonella Enteritidis inoculated on litter samples from specific-pathogen-free chicken farm.
| Ct1 values ± S.D (detection rate %, n = 8) |
||||
|---|---|---|---|---|
| SE inoculum (Log CFU/g) | Conventional real-time PCR | IMS real-time PCR | MDA real-time PCR | IMS-MDA real-time PCR |
| 5 | 25.90 ± 2.87 (100) | 25.05 ± 0.43 (100) | 28.79 ± 0.40 (100) | 23.36 ± 1.35 (100) |
| 4 | 29.29 ± 2.04 (100) | 27.59 ± 1.08 (100) | 29.99 ± 3.99(100) | 27.16 ± 3.89 (100) |
| 3 | 33.10 ± 2.32 (100) | 31.94 ± 1.74 (100) | 30.45 ± 3.5 (100) | 29.39 ± 2.54 (100) |
| 2 | ND2 | 33.44 ± 2.26 (100) | 30.66 ± 1.05 (25) | 31.69 ± 1.71 (100) |
| 1 | ND | ND | ND | 34.36 ± 1.06 (100) |
The threshold cycle (Ct), which is the intersection between each fluorescence curve and a threshold line, was calculated using Applied Biosystems 7500 real-time PCR system software version 2.0.6 (Thermo Fisher Scientific, Waltham, MA). Negative results correspond to Ct values ≥40 or sample with Ct values higher than that of negative control.
ND, not detected.
The use of IMS has been shown to decrease LOD by selectively concentrating target bacteria (Leon-Velarde et al., 2009; Tatavarthy et al., 2009; Zheng et al., 2014; Hyeon and Deng, 2017). In this study, the LOD by IMS real-time PCR (102 CFU/g) was 1 log lower than that by conventional real-time PCR (103 CFU/g), while the LODs by MDA real-time PCR and conventional real-time PCR were similar. Although the use of MDA alone did not lower the LOD, the combination of IMS and MDA showed a synergistic effect in improving detection sensitivity. The LOD by IMS-MDA real-time PCR (10 CFU/g) was remarkably lower than that by conventional real-time PCR and those by previously reported methods that incorporated IMS in Salmonella detection, such as 104– 106 CFU/mL from environmental swabs using IMS-enzyme immunoassay (Leon-Velarde et al., 2009) and 104 to 105 CFU/g from food samples using IMS real-time PCR assays (Wang et al., 2007; Zheng et al., 2014, 2016). This result was comparable to that of our previous study (Hyeon and Deng, 2017) on SE detection in raw chicken meat homogenate using the same IMS-MDA real-time PCR method (10 CFU/g with 100% detection rate). The litter samples used in this study were collected from an SPF chicken farm, which may contain less competing flora than litter from conventional farms. This difference may also contribute to the lower LODs obtained in this study.
Optimization of Enrichment Time for Detection of Salmonella From Litter Using IMS-MDA Real-Time PCR
To further lower the LOD of IMS-MDA real-time PCR, a short period (4, 6, or 8 h) of culture enrichment was added before IMS (Table 2). This method allowed consistent detection of 0.1 CFU/g of SE with 100% detection rate after 8-h enrichment. It is longer than that of previous reports with a comparable LOD by IMS real-time PCR, such as 6 h in pork cutlet (Notzon et al.,2006) and 7 h in raw duck wing (Zheng et al., 2014), but shorter than 10 h in ground beef (Mercanoglu and Griffiths, 2005) to detect 1 to 10 CFU/g of Salmonella. It is also longer than the 4-h culture enrichment that allowed the detection of 0.1 CFU/g of SE in raw chicken meat by IMS-MDA real-time PCR (Hyeon and Deng, 2017). The longer enrichment time required for litter samples was likely due to the interference of Salmonella growth by high levels of competitive flora in samples. The high abundances of background microflora might decrease the efficiency of IMS by cross-reactivity and nonspecific binding (Fu et al., 2005; Tatavarthy et al., 2009; Zheng et al., 2014).
Table 2.
Comparison of enrichment hours for detecting Salmonella Enteritidis in inoculated litter using immunomagnetic separation-multiple displacement amplification (IMS-MDA) real-time PCR.
| Salmonella Enteritidis inoculum (Av. ± S.D. CFU/g) | Ct1 values ± S.D. (detection rate %, n = 8) |
||
|---|---|---|---|
| 4 h | 6 h | 8 h | |
| 10.5 ± 1.4 | 28.7 ± 2.2 (100) | 29.01 ± 1.9 (100) | 28.9 ± 1.3 (100) |
| 1.05 ± 0.1 | 32.8 ± 0.7 (100) | 32.4 ± 1.2 (100) | 32.1 ± 1.7 (100) |
| 0.11 ± 0.01 | 32.5 ± 1.1 (50) | 33.4 ± 1.2 (50) | 32.9 ± 1.2 (100) |
The threshold cycle (Ct), which is the intersection between each fluorescence curve and a threshold line, was calculated using Applied Biosystems 7500 real-time PCR system software version 2.0.6 (Thermo Fisher Scientific, Waltham, MA). Negative results correspond to Ct values ≥40 or sample with Ct values higher than that of negative control.
Equivalence Assessment of IMS-MDA Real-Time PCR, Conventional Real-Time PCR, and Culture Method
To validate the IMS-MDA real-time PCR method with optimized enrichment time (8 h), a total of 174 poultry environmental samples, including litter, boot swab, and drag swab, were analyzed with culture method, conventional real-time PCR, and IMS-MDA real-time PCR for Salmonella detection (Table 3).
Table 3.
Comparison of the numbers of positive and negative samples in immunomagnetic separation-multiple displacement amplification (IMS-MDA) real-time PCR, conventional real-time PCR, and culture method in detection of Salmonella spp. in boot swab, drag swab, and litter samples from a chicken farm.
| Real-time PCR1 (24 h enrichment) |
IMS-MDA real-time PCR1 (8 h enrichment) |
||||
|---|---|---|---|---|---|
| Sample type | Culture method | Positive | Negative | Positive | Negative |
| Boot swab (n = 58) | Positive (n = 8) | 7 | 1 | 7 | 1 |
| Negative (n = 50) | 2 | 48 | 3 | 47 | |
| Drag swab (n = 58) | Positive (n = 2) | 2 | 0 | 2 | 0 |
| Negative (n = 56) | 3 | 53 | 3 | 53 | |
| Litter (n = 58) | Positive (n = 9) | 9 | 0 | 9 | 0 |
| Negative (n = 49) | 1 | 48 | 1 | 48 | |
| Total (n = 174) | Positive (n = 19) | 18 | 1 | 18 | 1 |
| Negative (n = 155) | 6 | 149 | 7 | 148 | |
P value is 0.51 and Cohen's Kappa index is 0.82 between real-time PCR and culture method, and 0.42 and 0.79 between IMS-MDA real-time PCR and culture method. P value < 0.05 indicates a statistically significant difference with culture method, and Cohen's Kappa index values of between 0.41 and 0.60 indicate clear concordance, those between 0.61 and 0.80 indicated strong concordance, and those between 0.81 and 1.00 indicated nearly complete concordance.
As shown in Table 3, IMS-MDA real-time PCR detected Salmonella in more samples (n = 25) than culturing (n = 19) and conventional real-time PCR (n = 24) with the shortest enrichment time (8 h). However, few culture-positive samples were not detected by conventional real-time PCR (n = 1) and IMS-MDA real-time PCR (n = 1), including a false-negative result from a boot swab sample by both real-time PCR assays (Table 3). The differences between the culture method and 2 real-time PCR assays were not significant (P > 0.05) in all sample types (Table 3). Cohen's Kappa index indicated nearly complete concordance (0.82) of conventional real-time PCR and strong concordance (0.79) of IMS-MDA real-time PCR with the culture method (Table 2) (Cohen, 1960; ISO:16140, 2003). Real-time PCR-based methods were reportedly more sensitive than culturing alone for detecting Salmonella in poultry-related samples in most previous studies. For example, from 422 boot and drag swabs, Lungu et al. (2012) detected Salmonella in 271 samples using real-time PCR compared with 201 using the culture method. Similarly, Charlton et al. (2005) identified 43 Salmonella -positive samples from 942 drag swabs by real-time PCR compared with 34 by the culture method.
In the current study, the discrepancy in detection between culture and real-time PCR-based methods may be due to the abundance of competing flora in environmental samples, low levels of Salmonella in the samples, and insufficient enrichment time for adequate resuscitation and growth of stressed and injured bacterial cells.
While both culture and real-time PCR detection methods include pre-enrichment where competition between Salmonella and background flora would equally affect both methods, culture-based detection requires an additional round of culturing on agar media that can further subject Salmonella to growth competition with background flora still present in the enrichment culture. If the flora overgrows and outcompetes Salmonella on agar plates, sensitivity of detection can decrease.
Real-time PCR, on the other hand, may detect Salmonella DNA from the dead bacterial cells in the sample. However, compared with conventional real-time PCR, selective capture of culture-enriched Salmonella cells by IMS and subsequent washing of bead-Salmonella complexes may help reduce the detection of soluble DNA from dead cells in the samples. The false-negative results of the real-time PCR assays appeared to be due to low levels of Salmonella in the samples. As shown in Table 4, both conventional and IMS-MDA real-time PCR assays did not detect Salmonella from the sample of the lowest MPN of 0.03 (Table 4). In addition to that, the interference of the interaction between the target pathogen and magnetic beads by the sample matrix and competing flora may also lead to false-negative detection by the IMS-MDA real-time PCR assays. The false-negative results caused by PCR inhibitors have been excluded by an IAC which was detected between 26 and 33 cycles (data not shown).
Table 4.
Most probable number (MPN) and the number of positive samples by immunomagnetic separation-multiple displacement amplification (IMS-MDA) real-time PCR, conventional real-time PCR, and culture method in detection of Salmonella spp. in boot swab, drag swab, and litter samples from chicken farm (n = 174).
| Number of positive samples |
|||
|---|---|---|---|
| MPN (mpn/g) | Culture method | Real-time PCR | IMS-MDA real-time PCR |
| Negative | 0 | 6 | 7 |
| 0.03 | 2 | 1 | 1 |
| 0.036 | 3 | 3 | 3 |
| 0.062 | 2 | 2 | 2 |
| 0.094 | 2 | 2 | 2 |
| 0.29 | 1 | 1 | 1 |
| 0.36 | 2 | 2 | 2 |
| 11 | 1 | 1 | 1 |
| >11 | 6 | 6 | 6 |
Among 3 different sample types, boot swabs and litters had significantly greater incidence of Salmonella than drag swabs by all 3 detection methods (Table 3). For IMS-MDA real-time PCR, Salmonella was detected in 10 of 58 boot swab samples, 10 of 58 litter samples, and 5 of 58 drag swab samples (Table 3). Similar results have been reported in previous studies, and boot swabbing was the most sensitive poultry environmental sampling method (McCrea et al., 2005; Buhr et al., 2007; Lungu et al., 2012). In Buhr et al's study (2007), Salmonella -positive samples were detected in 10 of 36 (28%) fecal samples, 20 of 36 (56%) litter grab samples, 14 of 36 (39%) drag swab samples, and 26 of 36 (72%) sock samples in challenge pens. Lungu et al. (2012) also demonstrated a higher sensitivity of boot swab sampling (67.1%) in comparison to drag swab sampling (61.3%) for detecting Salmonella using real-time PCR.
In conclusion, IMS-MDA real-time PCR was shown to substantially shorten culture enrichment for Salmonella detection from poultry environmental samples and yielded concordant results with those of conventional real-time PCR and the culture method. Therefore, the IMS-MDA real-time PCR method provides a viable alternative for monitoring Salmonella in poultry production environments.
ACKNOWLEDGMENTS
The authors would like to thank the U.S. National Poultry Research Center for kindly providing litter samples were from a SPF chicken farm and Dr. Mark Harrison and Gwen Hirsch of the University of Georgia for kindly providing the bacterial strain and other support to this study. This work was supported in part by the USDA National Institute of Food and Agriculture Hatch project 1006141.
REFERENCES
- Berghaus R.D., Thayer S.G., Law B.F., Mild R.M., Hofacre C.L., Singer R.S. Enumeration of Salmonella and Campylobacter spp. in environmental farm samples and processing plant carcass rinses from commercial broiler chicken flocks. Appl. Environ. Microbiol. 2013;79:4106–4114. doi: 10.1128/AEM.00836-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binga E.K., Lasken R.S., Neufeld J.D. Something from (almost) nothing: the impact of multiple displacement amplification on microbial ecology. ISME J. 2008;2:233–241. doi: 10.1038/ismej.2008.10. [DOI] [PubMed] [Google Scholar]
- Buhr R.J., Richardson L.J., Cason J.A., Cox N.A., Fairchild B.D. Comparison of four sampling methods for the detection of Salmonella in broiler litter. Poult. Sci. 2007;86:21–25. doi: 10.1093/ps/86.1.21. [DOI] [PubMed] [Google Scholar]
- Charlton B.R., Walker R.L., Kinde B.H., Bauer C.R., Channing-Santiago S.E., Farver T.B. Comparison of a Salmonella Enteritidis-specific polymerase chain reaction assay to delayed secondary enrichment culture for the detection of Salmonella Enteritidis in environmental drag swab samples. Avian Dis. 2005;49:418–422. doi: 10.1637/7283-092404R1.1. [DOI] [PubMed] [Google Scholar]
- Cohen J. A coefficient of agreement for nominal scales. Educ. Psychol. Meas. 1960;20:37–46. [Google Scholar]
- Fu Z., Rogelj S., Kieft T.L. Rapid detection of Escherichia coli O157:H7 by immunomagnetic separation and real-time PCR. Int. J. Food Microbiol. 2005;99:47–57. doi: 10.1016/j.ijfoodmicro.2004.07.013. [DOI] [PubMed] [Google Scholar]
- Holt P.S., Davies R.H., Dewulf J., Gast R.K., Huwe J.K., Jones D.R., Waltman D., Willian K.R. The impact of different housing systems on egg safety and quality. Poult. Sci. 2011;90:251–262. doi: 10.3382/ps.2010-00794. [DOI] [PubMed] [Google Scholar]
- Hosono S., Faruqi A.F., Dean F.B., Du Y., Sun Z., Wu X., Du J., Kingsmore S.F., Egholm M., Lasken R.S. Unbiased whole-genome amplification directly from clinical samples. Genome Res. 2003;13:954–964. doi: 10.1101/gr.816903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyeon J.Y., Deng X. Rapid detection of Salmonella in raw chicken breast using real-time PCR combined with immunomagnetic separation and whole genome amplification. Food Microbiol. 2017;63:111–116. doi: 10.1016/j.fm.2016.11.007. [DOI] [PubMed] [Google Scholar]
- International Standard Organization . International Standard Organization; 2003. Microbiology of food and animal feeding stuffs—Protocol for the validation of alternative methods. 16140. [Google Scholar]
- Leon-Velarde C.G., Zosherafatein L., Odumeru J.A. Application of an automated immunomagnetic separation-enzyme immunoassay for the detection of Salmonella enterica subspecies enterica from poultry environmental swabs. J. Microbiol. Methods. 2009;79:13–17. doi: 10.1016/j.mimet.2009.07.003. [DOI] [PubMed] [Google Scholar]
- Lungu B., Waltman W.D., Berghaus R.D., Hofacre C.L. Comparison of a real-time PCR method with a culture method for the detection of Salmonella enterica serotype enteritidis in naturally contaminated environmental samples from integrated poultry houses. J. Food Prot. 2012;75:743–747. doi: 10.4315/0362-028X.JFP-11-297. [DOI] [PubMed] [Google Scholar]
- McCrea B.A., Norton R.A., Macklin K.S., Hess J.B., Bilgili S.F. Recovery and genetic similarity of Salmonella from broiler house drag swabs versus surgical shoe covers. J. Appl. Poult. Res. 2005;14:694–699. [Google Scholar]
- Mercanoglu B., Griffiths M.W. Combination of immunomagnetic separation with real-time PCR for rapid detection of Salmonella in milk, ground beef, and alfalfa sprouts. J. Food Prot. 2005;68:557–561. doi: 10.4315/0362-028x-68.3.557. [DOI] [PubMed] [Google Scholar]
- Notzon A., Helmuth R., Bauer J. Evaluation of an immunomagnetic separation-real-time PCR assay for the rapid detection of Salmonella in meat. J. Food Prot. 2006;69:2896–2901. doi: 10.4315/0362-028x-69.12.2896. [DOI] [PubMed] [Google Scholar]
- Rodrigue S., Malmstrom R.R., Berlin A.M., Birren B.W., Henn M.R., Chisholm S.W. Whole genome amplification and de novo assembly of single bacterial cells. PLoS One. 2009;4 doi: 10.1371/journal.pone.0006864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scallan E., Hoekstra R.M., Angulo F.J., Tauxe R.V., Widdowson M.A., Roy S.L., Jones J.L., Griffin P.M. Foodborne illness acquired in the United States–major pathogens. Emerg. Infect. Dis. 2011;17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth-Smith H.M., Harris S.R., Skilton R.J., Radebe F.M., Golparian D., Shipitsyna E., Duy P.T., Scott P., Cutcliffe L.T., O'Neill C., Parmar S., Pitt R., Baker S., Ison C.A., Marsh P., Jalal H., Lewis D.A., Unemo M., Clarke I.N., Parkhill J., Thomson N.R. Whole-genome sequences of Chlamydia trachomatis directly from clinical samples without culture. Genome Res. 2013;23:855–866. doi: 10.1101/gr.150037.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soria M.C., Soria M.A., Bueno D.J., Godano E.I., Gomez S.C., ViaButron I.A., Padin V.M., Roge A.D. Salmonella spp. contamination in commercial layer hen farms using different types of samples and detection methods. Poult. Sci. 2017;96:2820–2830. doi: 10.3382/ps/pex053. [DOI] [PubMed] [Google Scholar]
- Tatavarthy A., Peak K., Veguilla W., Cutting T., Harwood V.J., Roberts J., Amuso P., Cattani J., Cannons A. An accelerated method for isolation of Salmonella enterica serotype Typhimurium from artificially contaminated foods, using a short preenrichment, immunomagnetic separation, and xylose-lysine-desoxycholate agar (6IX method) J. Food Prot. 2009;72:583–590. doi: 10.4315/0362-028x-72.3.583. [DOI] [PubMed] [Google Scholar]
- U.S. Department of Agriculture-Food Safety and Inspection Service Microbiology Laboratory Guidebook 4.10. 2019. http://www.fsis.usda.gov/wps/wcm/connect/700c05fe-06a2-492a-a6e1-3357f7701f52/MLG-4.pdf?MOD=AJPERES
- U.S. Department of Agriculture-Food Safety and Inspection Service Microbiology Laboratory Guidebook. 2014. https://www.fsis.usda.gov/wps/wcm/connect/8872ec11-d6a3-4fcf-86df-4d87e57780f5/MLG-Appendix-2.pdf?MOD=AJPERES Appendix 2.05.
- U.S. Department of Agriculture The National Poultry Improvement Plan (NPIP) program standards. 2017. https://www.poultryimprovement.org/documents/ProgramStandardsJanuary2017.pdf
- Waltman D.W., Gast R.K. 5th ed. American Association of Avian Pathologists; Athens, Georgia: 2008. Salmonellosis. [Google Scholar]
- Wang L., Li Y., Mustaphai A. Rapid and simultaneous quantitation of Escherichia coli 0157:H7, Salmonella, and Shigella in ground beef by multiplex real-time PCR and immunomagnetic separation. J. Food Prot. 2007;70:1366–1372. doi: 10.4315/0362-028x-70.6.1366. [DOI] [PubMed] [Google Scholar]
- Zheng Q., Miks-Krajnik M., Yang Y., Lee S.M., Lee S.C., Yuk H.G. Evaluation of real-time PCR coupled with immunomagnetic separation or centrifugation for the detection of healthy and sanitizer-injured Salmonella spp. on mung bean sprouts. Int. J. Food Microbiol. 2016;222:48–55. doi: 10.1016/j.ijfoodmicro.2016.01.013. [DOI] [PubMed] [Google Scholar]
- Zheng Q., Miks-Krajnik M., Yang Y., Xu W., Yuk H.G. Real-time PCR method combined with immunomagnetic separation for detecting healthy and heat-injured Salmonella Typhimurium on raw duck wings. Int. J. Food Microbiol. 2014;186:6–13. doi: 10.1016/j.ijfoodmicro.2014.06.005. [DOI] [PubMed] [Google Scholar]