Abstract
Infectious bursal disease virus (IBDV) is still a vital etiological agent in poultry farms. IBDV outbreaks occasionally occur due to the presence of very virulent, reassortment or variant strains. Vaccine immunization has played crucial roles in IBD control for decades. However, survival pressure of IBDV from the vaccine immunization also increases the reassortments of circulating viruses. In this study, an IBDV strain was isolated from several broiler farms in Henan Province, central part of China, and named IBDV HN strain. Based on the results of RT-PCR, sequencing and phylogenic analyses of VP1 and VP2 genes, the IBDV HN strain is a novel reassortment strain in the Henan region. Segment A of this strain appears to originate from the very virulent IBDV strain, while segment B comes from the other field reassortment strains. This may be the result of natural reassortant of virus circulating in the field. About 60% (6/10) of experimentally infected specific pathogen-free chickens died after 3 to 5 d post-infection with typical symptom and pathological lesions. The IBDV HN strain was prone to horizontal transmission, which poses a serious threat to the chicken industry. Further investigation on the prevalence, virulence, and evolution of HN strain IBDV will provide a foundation for the prevention and control of the disease in this region.
Key words: infectious bursal disease virus, HN strain, phylogenetic analysis, virulence, China
INTRODUCTION
Infectious bursal disease virus (IBDV) is the primary etiological agent of a highly contagious disease among young chickens. The virus damages the immune organs, particularly the bursa of Fabricius, resulting in severe immune suppression and secondary infections with high morbidity and mortality (Rosenberger and Gelb, 1978; Sharma et al., 2000; Ye et al., 2017). As a member of the Birnaviridae family, IBDV contains a double-stranded RNA genome with segments A and B. The 2.8 kb segment B encodes the VP1 protein (879 amino acids (aa)), which is responsible for viral genome replication and RNA synthesis as a RNA-dependent RNA polymerase. The 3.2 kb segment A contains two partially overlapping open reading frames (ORFs), 1 and 2. The smaller ORF1 precedes and overlaps the larger ORF2 and encodes the non-structural protein VP5. The ORF2 encodes a precursor polyprotein (approximately 1012 aa), which is self-processed into three mature proteins: VP2, VP4, and VP3. VP2 is a major structural protein containing the antigen epitope that induces the production of neutralizing antibodies (Alfonso-Morales et al., 2013). Among different strains, the aa sequences of the highly variable region of VP2 are different and commonly affect the virulence of different virus strains (Hoque et al., 2001; Letzel et al., 2007; Lai et al., 2014; Yilmaz et al., 2019). The highly variable region is usually located between the aa residues 206 and 350 and is widely used for molecular epidemiology and phylogenetic studies (Ren et al., 2009; Drissi Touzani et al., 2019).
In China, the first case of IBDV was diagnosed in Guangzhou Province in 1979 (Zhou et al., 1981). Subsequently, the appearances of both hyper-virulent pathogenic and antigenic variants have been reported in China, and IBDV has become an important pathogen, which is responsible for the majority of economic losses in poultry industry (Berg, 2000; Chen et al., 2012; He et al., 2012; Xu et al., 2015). In 1992, the first very virulence IBDV (vvIBDV) strain was isolated from Harbin, Northeast part of China (Wei et al., 2008). Currently, a strict vaccine immunization program is applied to prevent and control IBDV infection. The most common live attenuated vaccine strains prevalent in China are B87, K85, and Gt, whereas the commonly used inactivated vaccine stains include BJQ902, VNJO, SD, and HQ (http://zjs.gov.cn/). These live attenuated and inactivated vaccines play an important role in preventing IBDV outbreaks on farms. However, the widespread use of vaccines exerts high selection pressure on the virus and contributes to the development of genetic diversity of circulating viruses in the field. Despite the implementation of high intensity immunization program in China, outbreaks of IBDV infections occur sporadically in some immunized farms (Sharma et al., 2000; Ren et al., 2009; He et al., 2012; Liu et al., 2013; Xu et al., 2015). Novel or reassortant IBDV strains have been isolated successively (Liu et al., 2002; Jackwood and Sommer-Wagner, 2007; Xia et al., 2008; Chen et al., 2012; He et al., 2012; Cui et al., 2013; Liu et al., 2013; He et al., 2014; Lu et al., 2015; Li et al., 2016; Fan et al., 2019).
In Henan, most of the reports on the IBDV isolates were from south of the Yellow River, such as Xuchang (Wang et al., 2013), Luoyang (Li et al., 2009), Zhengzhou, and other area (Wang et al., 2001; Wang et al., 2013), but there were no reports of IBDV isolates in Xinxiang. In order to clarity the epidemic strain of IBDV in Xinxiang, and provide theoretical basis for its prevention and control of this region, bursa samples from chickens showing clinical signs reminiscent of IBD were collected from four broiler farms of Henan Xinxiang, and one isolate, named IBDV HN strain, was obtained with the analysis of genetic and pathotypic characterization.
MATERIALS AND METHODS
Samples
A total of 20 bursa samples were collected from four broiler farms around Xinxiang, Henan, central part of China. These broilers had been vaccinated with live attenuated B87 strain vaccine at 14 and 28 d of age. After necropsy, severe bleeding was observed in the breast and leg muscles of the birds. Kidneys displayed various degrees of swelling with red and white stripes. Severe bleeding was observed in the bursa of Fabricius of the birds. Bursa samples were collected and stored at −80°C until further analyses. The IBDV strain CJ801, a gifted from Liu Jue, Beijing Institute of Husbandry and Veterinary Research (Beijing, China), was used as classical IBDV control in pathogenicity experimentin.
Virus Isolation
Bursa tissues were homogenized in sterilized phosphate buffered saline (PBS), and the supernatants were harvested by centrifugation at 1500 rpm (4°C; 5 min). The supernatants were then treated with penicillin and streptomycin at a final concentration of 2000 IU/mL at 4°C for 2 h. Then, 200 µ L of the supermatants was inoculated into 10-day-old embryonated specific pathogen-free (SPF) eggs (Merial, Beijing, China) through the chorioallantoic membrane. The embryonated eggs were incubated at 37°C and examined twice daily for viability. Chorioallantoic membranes were harvested for RNA extraction and viral detection after 72 h incubation.
Viral RNA Extraction and RT-PCR
Total RNA was extracted from the chorioallantoic membrane homogenates using E.Z.N.A.® Tissue RNA extraction Kit (Omega, Beijing, China), according to the manufacturer's instructions. The obtained RNA pellets were dissolved in sterile diethyl pyrocarbonate-treated water. The RNA was stored at −80°C until further use.
The total RNA was then reverse transcribed into complementary DNAs (cDNAs) using M-MLV reverse transcriptase (Promega, CA, USA) according to the manufacturer's instructions. Primers were designed based on the conserved region of IBDV VP2, segment A and segment B as shown in Table 1. PCR was conducted in a final reaction volume of 25 µ L containing 2 µ L of cDNA templates, 12.5 µ L of 2 × reaction mix, 1 µ L of each primer (10 µ M), 0.5 µ L of 5 U/µ L Taq DNA polymerase and 8 µ L of nuclease-free water. The reaction conditions were 2 min at 95°C, 30 cycles of denaturation (30 s at 95°C), primer annealing (30 s at 56°C) and extension at 72°C (1 min and 30 s for VP2, 3 min and 20 s for segment A, and 2 min and 45 s for segment B), followed by a final 10-min extension at 72°C. PCR products were examined using agarose gel electrophoresis and visualized with ethidium bromide staining according to standard protocols.
Table 1.
Name and sequence of primers used in the manuscript and the amplified product lengths.
| Primer name | Sequence (5′-3′) | Length |
|---|---|---|
| VP2-F | ATGACAAACCTGCAAGATCAAACC | 1356 bp |
| VP2-R | CCTTATGGCCCGGATTATGT | |
| Segment A-F | GGATACGATCGGTCTGAC | 3260 bp |
| Segment A-R | GGGGACCCGCGAACGGATCCAATTTGGGAT | |
| Segment B-F | GGATACGATGGGTCTGAC | 2827 bp |
| Segment B-R | GGGGGCCCCCGCAGGCGAAGGCCGGGGAT |
Sequence Alignment and Phylogenetic Analysis
The purified VP2 PCR product was cloned into a pMD19-T vector. The positive clones and PCR products of segments A and B were further sequenced by a commercial company (Sangon Biotech, Shanghai, China). VP2 nucleotide and deduced aa sequences, segments A and segments B were analyzed using NCBI BLAST and compared with the IBDV reference strains from different regions or countries using DNASTAR 5.0 software (DNASTAR Inc., USA). Phylogenetic analyses based on VP1 and VP2 were performed using the neighbor-joining method in MEGA version 5.05 software (Tamura et al., 2011), respectively. Most of the available IBDV sequences of the very virulent, classical, attenuated, variant and serotype 2 IBDV strains used for further molecular evolution in this study were retrieved from the GenBank database, and their accession numbers are listed in Table 2.
Table 2.
IBDV strains used in the sequence alignment and phylogenetic analysis.
| Accession No. |
||||
|---|---|---|---|---|
| Reference strain | Segment A | Segment B | Origin | Pathotype/serotype |
| Cu-1 | X16107 | AF362775 | German | Attenuated |
| P2 | X84034 | X84035 | German | Attenuated |
| CEF94 | AF194428 | AF194429 | Netherlands | Attenuated |
| B87 | DQ906921 | DQ906922 | China | Attenuated |
| Gt | DQ403248 | DQ403249 | China | Attenuated |
| D78 | AF499929 | EU162090 | Luxembourg | Attenuated |
| 903/78 | JQ411012 | JQ411013 | Hungary | Attenuated |
| Cu1 | D00867 | – | Germany | Attenuated |
| Winterfield-2512 | – | AF083092 | Israel | Attenuated |
| HBDY-1 | KX592158 | KX592159 | China | Classical |
| IM | AY029166 | AY029165 | USA | Classical |
| ZJ2000 | AF321056 | DQ166818 | China | Reassortant |
| TL2004 | DQ088175 | DQ118374 | China | Reassortant |
| HN04 | KC109816 | KC109815 | China | Reassortant |
| CJ801 | AF00670 | – | China | Classical |
| STC | D00499 | JQ619639 | Canada | Variant |
| GLS | AY368653 | AY368654 | USA | Variant |
| Lukert | AY918948 | AY918947 | USA | Classical |
| A-BH83 | JF811920 | JF811921 | Brazil | Classical |
| Faragher 52/70 | HG974565 | HG974566 | UK | Classical |
| Edgar | AY462026 | AY918949 | USA | Classical |
| 9109 | AY462027 | AY459321 | USA | Classical |
| Variant E | AF133904 | AF133905 | USA | Variant |
| OKYM | D49706 | D49707 | Japan | Very virulent |
| Harbin-1 | EF517528 | EF517529 | China | Reassortant |
| Gx | AY444873 | AY705393 | China | Reassortant |
| UK661 | X92760 | X92761 | UK | Very virulent |
| D6948 | AF240686 | AF240687 | Netherlands | Very virulent |
| HK46 | AF092943 | AF092944 | Hong Kong | Very virulent |
| HN | KT884486 | KY948019 | China | Reassortant |
| 0,2015.1 | AJ879932 | AJ880090 | Venezuela | Reassortant |
| T09 | AY099456 | AY099457 | Luxembourg | Very virulent |
| SH99 | LM651365 | LM651366 | China | Very virulent |
| SH95 | AY134874 | AY134875 | China | Reassortant |
| 89,163 | HG974563 | HG974564 | France | Very virulent |
| 94,432 | AM167550 | AM167551 | France | Very virulent |
| TASIK | AF322444 | AF322445 | Australia | Very virulent |
| Cro-lg/02 | EU184685 | EU184686 | Croatia | Very virulent |
| KS | DQ927042 | DQ927043 | Israel | Very virulent |
| NB | EU595667 | EU595673 | China | Very virulent |
| BD/3/99 | AF362776 | AF362770 | Bangladesh | Very virulent |
| HuB-1 | KF569805 | GQ449693 | China | Very virulent |
| SD10LY01 | KF569803 | KF569804 | China | Very virulent |
| CAHFS-785 | JF907702 | JF907705 | USA | Reassortant |
| 2009CAH495 | JF907703 | JF907704 | USA | Reassortant |
| KZC-104 | AB368968 | AB368969 | Zambia | Reassortant |
| 150,124/1.1 | MF969105 | MF969106 | Algeria | Reassortant |
| 150,144/5.1 | MF969115 | MF969116 | Algeria | Reassortant |
| UPM04/190 | KU958716 | KU958717 | Malaysia | Very virulent |
| GX-NN-L | JX134485 | JX134486 | China | Reassortant |
| IBD13HeB01 | KP676467 | KP676468 | China | Reassortant |
| HLJ-0504 | GQ451330 | GQ451331 | China | Reassortant |
| HuN11 | LM651367 | LM651368 | China | Reassortant |
| 88,180 | AM111353 | AM111354 | France | Very virulent |
| 23–82 | AF362773 | AF362774 | German | Serotype 2 |
| OH | M66722 | U30819 | Canada | Serotype 2 |
| Turkey PA/00924/14 | KP642112 | KP642111 | USA | Serotype 2 |
Pathogenicity Experiment
Thirty-six 21-day old SPF chickens (Merial, Beijing, China) were randomly divided into 3 groups. The first group (n = 12) was inoculated with classical IBDV CJ801 strain using as a positive control while the second group (n = 12) were inoculated with HN strain (Liu et al., 2002). The last group (n = 12) was inoculated with PBS as a negative control. Chickens in the infection group were inoculated with 200 µ L of 100 EID50 of the chorioallantoic membrane homogenate via intranasal inoculation and eye drops, respectively. Birds in the control group were treated with the same volume of PBS. After the inoculations, the experimental chickens were observed daily, and clinical symptoms were recorded for 7 d. Three chickens were randomly selected from each group at days 1, 3, 5, and 7 post-infection unless there were died chickens. All the chickens were euthanized and pathological changes were examined after necropsy. The bursa and body weights were weighed and the bursa: body weight was calculated. The bursa samples were further subjected to histopathology or molecular evolution analysis. All animal management and experimental procedures in this study were approved by the Institutional Animal Care and Use Committee at the Henan Institute of Science and Technology (HIST-0,01026).
Histopathological Examination
According to the criteria described by Jackwood et al. (Jackwood et al., 2011), half of the bursa was fixed in 10% formalin, and then embedded in paraffin, sliced and stained with hematoxylin and eosin for further histopathological examination.
Horizontal Transmission Experiment
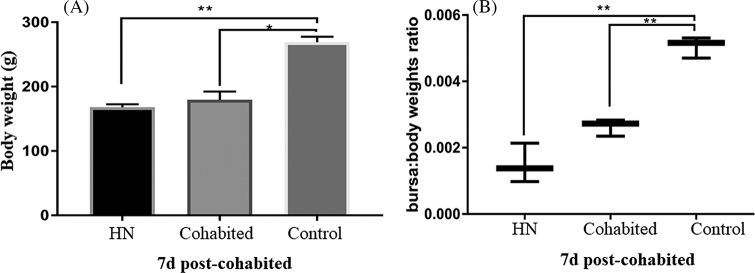
To evaluate the horizontal transmission ability of HN strain, 3 healthy SPF chickens were placed in the same isolator with a group of 10 chickens, which were inoculated with HN. Then, clinical symptoms of chickens were observed daily. At 7-day post-cohabitation, three chickens selected were taken out, euthanized, weighted and the pathological changes were observed, the bursa: body weight was calculated.
Statistical Analysis
T -tests were used to evaluate whether the difference of the mortality, body weights, and bursa: body weights between groups of chickens infected with different IBDV strains was statistically significant. The results withP < 0.05 were considered as statistically significant.
RESULTS
Isolation and Identification of IBDV HN Strain
Four chorioallantoic membrane samples were randomly selected from SPF eggs inoculated with homogenized bursa supernatant to extract total RNA. Conventional RT-PCR was used for amplification and identification based on the conserved region of VP2 gene. Analysis of the RT-PCR revealed presence of the target band in the test samples (data no shown). Then, the target bands were sent for sequencing and the blasting results showed the isolated virus was IBDV and named as IBDV HN strain. Moreover, full-length sequences of segment A and segment B of HN strain were finally obtained by RT-PCR and sequenced. The obtained sequences were submitted to GenBank, under the Accession Numbers KT884486 (segment A) and KY948019 (segment B).
IBDV HN Isolate Evolutionarily Belongs to One Reassortant Strain of the Very Virulent Strain and Unidentified Strain
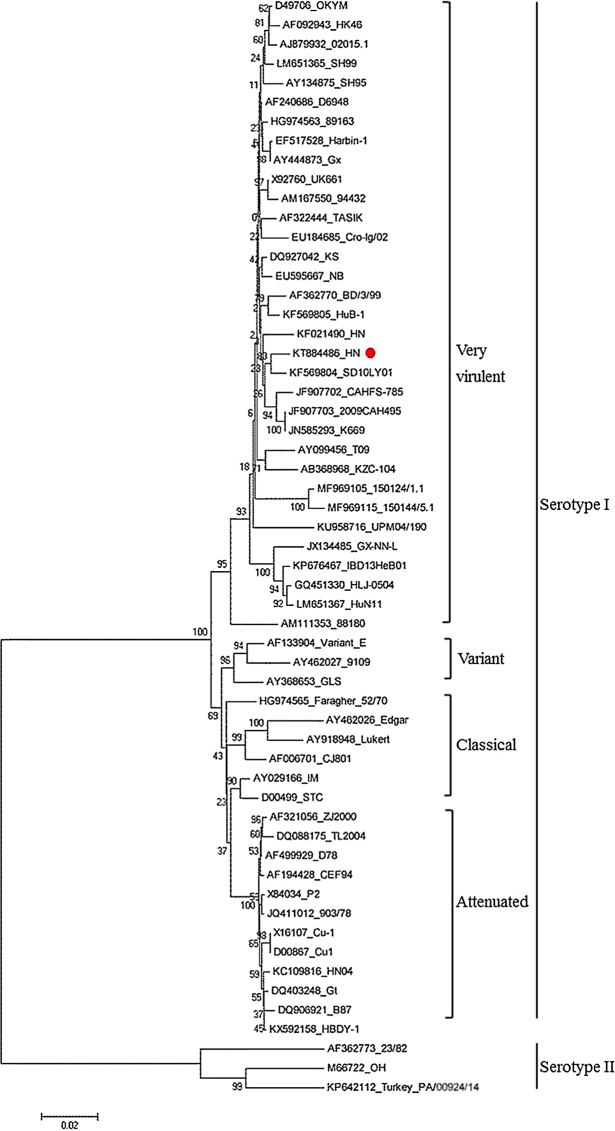
Phylogenetic tree analysis based on the deduced aa sequences of VP2 fragment showed that the IBDV strains were distinctly divided into 5 major branches, including very virulent strains, classical strains, attenuated strains, variant strains, and serotype II strains (Figure 1). The deduced aa sequence of HN strain VP2 fragment exhibited a profile typical for very virulent and clustered in the branch of vvIBDV strains, including UK661, Harbin-1, OKYM, Gx, and D6948, etc. (Figure 1). In the vvIBDV strains branch, HN strain was closely similar to the SD10LY01 strain from Harbin, northeast of China. Compare with other vvIBDV strains in the same branch, the deduced aa sequence of HN strain VP2 was no significant changes, and the aa sequence of the key loci in the hypervariable regions (HVRs), including 222A, 242I, 256I, 294I, and 299S, were in accordance with vvIBDV strains (Table 3). However, there were significant differences in the deduced aa sequence of VP2 between HN strain and vaccine strain B87 and some other attenuated strains, such as CEF94 and variant E. Phylogenetic analysis and the alignment of aa sequence based on VP2 confirmed that segment A of HN strain were from vvIBDV strains.
Figure 1.
Phylogenetic analysis of the nucleotide sequence of VP2 from IBDV isolates identified in this study and IBDV reference strains. The tree was constructed using the neighbor-joining method in MEGA 5 software. Numbers above the branches are bootstrap values computed from 1000 replications. The filled rectangle indicates the IBDV HN strain identified in this study.
Table 3.
Amino acid differences among representative strains.
| VP5 |
VP2 |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IBDV strain | Phenotype | 14 | 76 | 222 | 242 | 253 | 256 | 279 | 284 | 290 | 294 | 299 | 324 | 330 |
| D6948 | Very virulent | E | W | A | I | Q | I | D | A | M | I | S | Q | S |
| UK661 | Very virulent | K | · | · | · | · | · | · | · | · | · | · | · | · |
| Harbin-1 | Reassortant | · | · | · | · | · | · | · | · | · | · | · | K | · |
| HN | Reassortant | K | G | · | · | · | · | · | · | · | · | · | · | · |
| CEF94 | Attenuated | K | · | P | V | H | V | N | T | L | L | N | · | R |
| B87 | Attenuated | K | · | P | V | H | V | N | T | · | L | N | · | R |
| Variant E | Variant | K | · | T | V | · | V | N | · | · | L | N | E | · |
| VP1 |
||||||||||||||
| IBDV strain | Phenotype | 24 | 145 | 146 | 147 | 242 | 390 | 393 | 511 | 515 | 562 | 646 | 687 | 857 |
| D6948 | Very virulent | A | T | D | N | E | M | D | S | E | P | S | P | A |
| UK661 | Very virulent | · | · | · | · | · | · | · | · | · | · | · | · | · |
| Harbin-1 | Reassortant | · | · | E | G | D | L | E | · | · | S | · | · | · |
| HN | Reassortant | T | · | E | G | D | L | E | R | D | S | G | S | V |
| CEF94 | Attenuated | · | N | E | G | D | L | E | R | · | S | G | S | · |
| B87 | Attenuated | · | N | E | G | D | L | E | R | · | S | G | S | · |
| Variant E | Variant | · | N | E | G | D | L | E | R | D | S | G | S | · |
Further alignments results showed that the deduced aa sequences of VP3 and VP4 were no distinct difference from the majority of vvIBDV, which furtherly confirmed that segment A of HN strain was from vvIBDV strains. The deduced aa sequence of VP5 exhibited 2 specific alterations, one in N-terminal domain (K14) and one in the middle of the VP5 sequence (G76). At the position of 14, about half of vvIBDV strains were glutamic acid (E), while the remaining vvIBDV strains and reassortant strains, classical strains, attenuated strains, variant strains, and serotype II strains were lysine (K). But the change at the position of 76 was only occurs in several vvIBDV strains or reassortant strains.
Phylogenetic tree constructed based on the VP1 aa sequence was distinctly divided into 2 large branches. One branch was vvIBDV strains, and another branch was further divided into classical strains, variant strains, attenuated strains, unidentified reassortment strains, and serotype II strains. HN strain showed higher similarity with the unidentified reassortment strains, but was different from vvIBDV strains (Figure 2). Phylogenetic analysis showed that segment B of HN strain had a different origin with segment A, which confirmed the reassortant nature of the HN isolate. Moreover, the alignment of VP1 deduced aa sequence showed that there were 3 changes between HN strain and other strains in the same branch, S511R, E515D, and S646G. The altered aa were consistent with attenuated strains, variant strains, and some reassortant strains. However, whether these changes were related to virulence has not been reported. Furthermore, there were 2 unique alterations in the deduced aa sequence of VP1 of HN strain. The first changed residue was located within the VP1 N-terminal domain (24T), and the second changed residue was located within the VP1 C-terminal domain (857 V). It is not clear whether the aa changes have an effect on virus replication. In addition, triplet aa TEG were also found to be located at positions 145/146/147 of HN strain VP1, which may affect its virulence.
Figure 2.
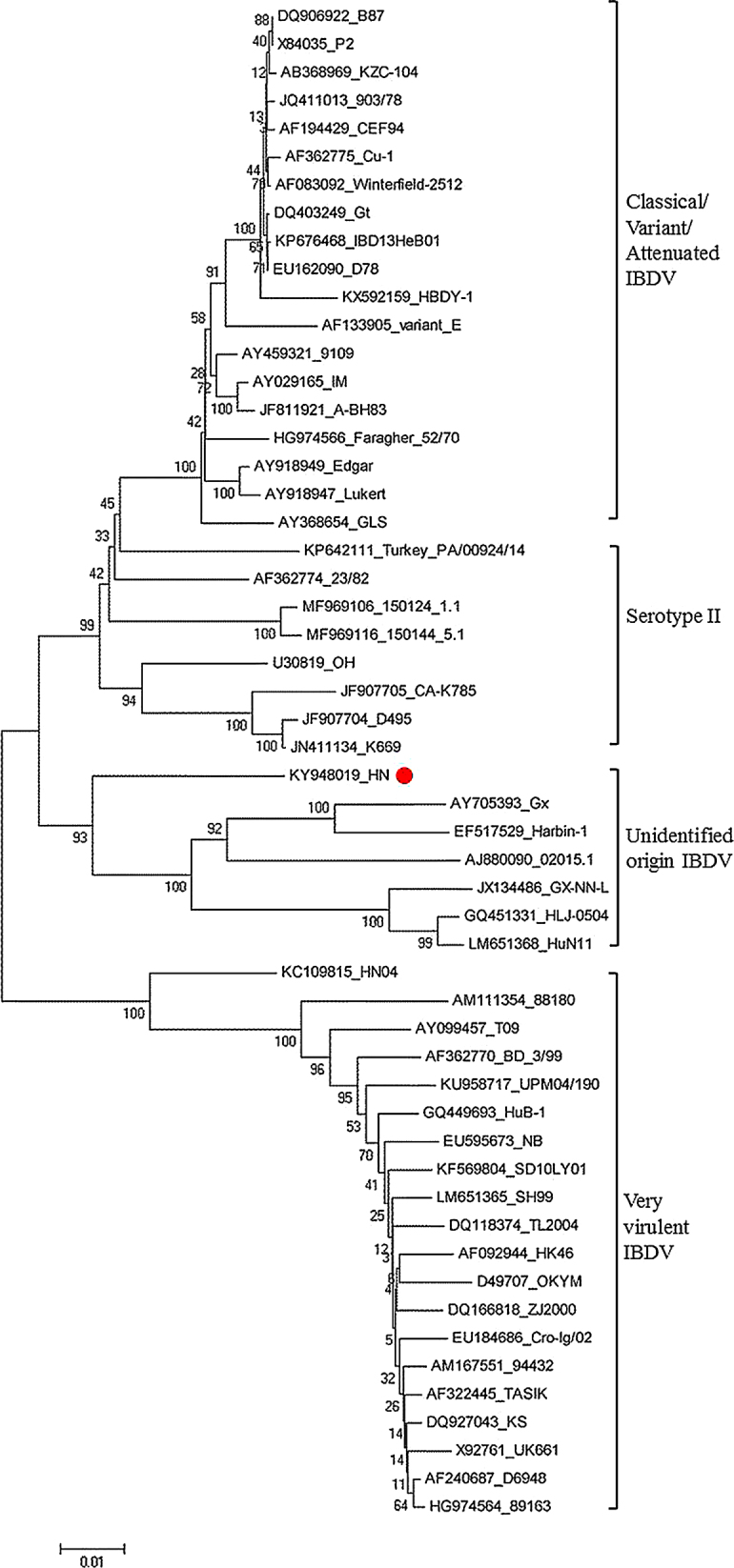
Phylogenetic analysis of the nucleotide sequences of VP1 from IBDV isolates identified in this study and IBDV reference strains. The tree was constructed using the neighbor-joining method in MEGA 5 software. Numbers above the branches are bootstrap values computed from 1,000 replications. The filled rectangle indicates the IBDV HN strain identified in this study.
IBDV HN Isolate has High Pathogenicity to Chickens
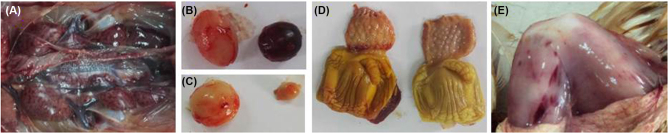
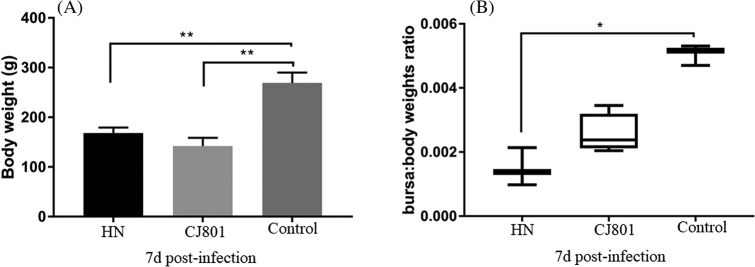
Animal experiments were applied to test the pathogenicity of IBDV HN strain. On 1-day post-infection (dpi) with IBDV HN strain and CJ801 strain, the symptoms of depression and messy feathers were observed in some of the experimental chickens, and some sick chickens even didn't move. On 2 dpi, the sick chickens began to diarrhea. From the third day onwards dpi, some chickens died and the mortality rate was 60% (HN strain, 6/10) and 50% (CJ801 strain, 5/10) (Table 4), respectively. After necropsy, the typical pathological changes were thigh and breast muscle bleeding, border bleeding of proventriculus and kidney swelling and bleeding. Moreover, the bursae obtained from the died chickens between 3 and 6 dpi displayed swelling and bleeding. But at 7 dpi, the bursa exhibited severe atrophy (Figure 3). No clinical symptoms or visible bursa lesions were observed in the PBS control groups. Moreover, the bursa: body weights ratio of the infected chickens in CJ801 group was slightly lower than that of HN strain, but the difference was not statistically significant (P = 0.0568) (Figure 4). Both groups were found to be statistically significantly lower than the normal control groups (P < 0.01). The specific data of pathogenicity experiments are shown in Table 4.
Table 4.
Results of in vivo IBDV virulence to SPF chickens
| Group | IBDV strain | Mortality (%) | BBIX at 7 dpi* |
|---|---|---|---|
| 1 | CJ801 | 5/10 (50) | 0.506 ± 0.105 |
| 2 | HN | 6/10 (60) | 0.296 ± 0.095 |
| 3 | Normal control | 0/10 (0) | 1 ± 0.051 |
BBIX = (bursa: body weight ratios)/(bursa: body weight ratios in the negative group).
Figure 3.
Pathological lesions observed in SPF chicken inoculated with the IBDV HN strain. A. Kidney tissue from a bird that died at 4 dpi. B and C. Bursal tissues from SPF chickens inoculated with the HN strain of IBDV (right) or control (left). D. Proventriculus from SPF chickens inoculated with the HN strain of IBDV (right) or control (left). E. Bleeding in leg muscles from a bird that died at 4 dpi.
Figure 4.
Evaluation of the virulence of HN strain IBDV using SPF Chicken. A. The body weight on 7 dpi. B. The bursae/body weight ratio of chickens on 7 dpi.
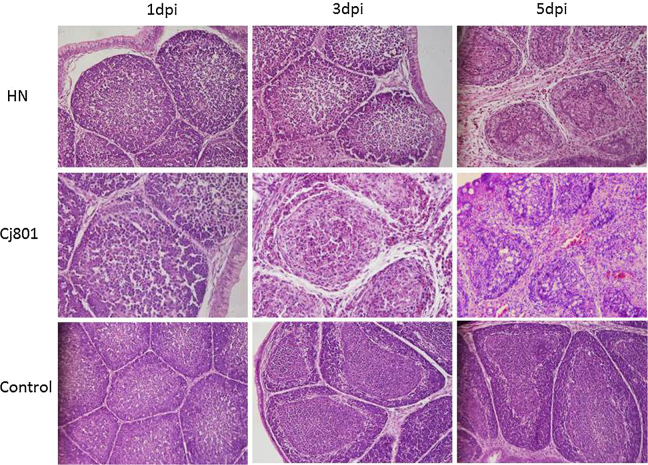
Histopathological examination of the bursa of Fabricius collected from groups 1 and 2 indicated typical bursa lesions and obvious microscopic changes. At 1 dpi, the follicles of the bursa were edematous and the boundary of cortical and medullary was blurred. At 3 dpi, the bursa was infiltrated by inflammatory cells, such as neutrophils and monocytes. Medullary and cortical regions of the lymphatic follicles became depletion or necrosis due to the degeneration of lymphoid cells or pyknosis and karorrhexis of nuclear. At 5 dpi, lymphatic follicles exhibited atrophy and vacuoles, and then was replaced by a large number of connective tissue and reticular cells. Finally lymphocytes in the follicles were marked by necrosis (Figure 5). Bursa tissues microscopic alterations in the HN and CJ801 groups had a similar intensity. The pathological lesion of bursa of HN strain was slighter than that of CJ801 strain, but the difference was not statistically significant. These results indicated that the HN and CJ801 strains have similar virulence. The bursa of fabricius showed normal tissue structure in the simulated inoculated control group, and no obvious changes were observed.
Figure 5.
Differences in histopathological appearance of bursa of Fabricius macroscopic lesions in examined groups (hematoxylin and eosin staining).
Horizontal Transmission Ability of IBDV HN Strain
At 3-day post-cohabitation (dpc), the three chickens that had been exposed to the inoculated birds showed signs of depression. At autopsy, the bursa of the chicken which died at 5 dpc displayed visible swelling and bleeding. At 7 dpc, the bursa from the two remaining chicken revealed mild atrophy. The body weights of all the three chickens decreased (Figure 6 A), and there was no significant difference in the average ratios of bursa: body weight between cohabitation chicken and the infected group (P = 0.25) (Figure 6 B). The bursa samples of three cohabitation chickens were collected and RT-PCR was performed using specific primers of VP2 fragment. Specific bands were observed at expected sizes. These results showed that HN strain had the ability of horizontal transmission and cause bursa damage.
Figure 6.
Evaluation of the horizontal transmission ability of HN strain IBDV using SPF Chicken. A. The body weight on 7 dpi. B. The bursae/body weight ratio of chickens on 7 dpi.
DISCUSSION
With the increasing demand for poultry products, chicken-related food safety issues, including antibiotics residue and various pathogens variants, have drawn increasing attentions. The presence of immunosuppressive diseases increases risks of infection with other pathogens. IBD is an important immunosuppressive disease in poultry, which poses a serious threat to the poultry industry worldwide. In recent years, IBD has occurred occasionally in vaccinated and non-vaccinated farms, causing serious economic losses in some regions of China (He et al., 2012; Liu et al., 2013; He et al., 2014). It may be due to the frequent occurrences of vvIBDV strains (Xia et al., 2008; Ren et al., 2009; Chen et al., 2012; He et al., 2012; Li et al., 2015) or reassortment strains (He et al., 2009; Cui et al., 2013; Lu et al., 2015), or a lack of protection provided by existing vaccines.
As an RNA virus, IBDV has the potential for rapid mutation. In addition, similar to influenza A virus, reassortment of IBDV with segmented genomes is also one of the mechanisms that contribute to the genetic diversity of IBDV. Since the first reassortment strain IBDV was reported (Le Nouën et al., 2006), more and more reassortment IBDV strains have been found (Hon et al., 2006; Wei et al., 2006; Gao et al., 2007; Wei et al., 2008; Jackwood et al., 2011; Chen et al., 2012; Kasanga et al., 2013; He et al., 2014; Lu et al., 2015; He et al., 2016; Patel et al., 2016; Raja et al., 2016; Soubies et al., 2017; Abed et al., 2018; Chen et al., 2018; Pikuła et al., 2018). The virulence of the natural reassortant IBDV reported is mostly between very virulent and classical pathotype strains (Le Nouën et al., 2006; Wei et al., 2006; Wei et al., 2008; Lu et al., 2015).
In this study, aa alignment phylogenetic analysis showed that the segment A of IBDV HN strain was highly homologous with vvIBDV strains, which was significantly different from the HN isolate reported by Cui (Cui et al., 2013). The aa sequence of the key loci in HVRs of HN strain VP2, including 222A, 242I, 253Q, 256I, 294I, and 299S were completely consistent with vvIBDV strains, such as OKYM from Japan, UK661 from Europe and D6984 from the Netherlands. In general, these sites, especially 222A, 294I, and 299S, can change the hydrophilicity of the vvIBDV strains, and may enhance the virulence of virus. In addition, the residue changed within the VP5 N-terminal domain, half of the vvIBDV strains are E14, while HN strain, the classical strains, weakened strains, and variant strains are all K14. The relationship between the change and virulence is currently unknown. However, it is reported that the VP5 N-terminal domain can interact with voltage-dependent anion channel 2 to induce apoptosis (Li et al., 2012), and whether the change of amino acid in this domain will impact the activation of cell apoptosis process is difficult to anticipate and further investigation is required.
Phylogenetic tree based on the VP1 showed that HN strain was different from vvIBDV strains and was located in a large branch composed of the classical, variant, attenuated and serum type II strains. However, HN strain VP1 differs from B87, D78, K85, Gt, and other commonly used vaccine strains and form an individual small branch with some unidentified origin reassortment strains. The same result was obtained by aligning VP1 amino acid sequences. These results confirm the different origins of both segment sequences from HN strain. It may be related to the natural mutation during the long-term coexistence of vaccine strains and wild strains in the process of natural transmission, or as Xia et al. suggested that the segment B of Harbin-1 strain might have originated from an unidentified ancestral virus maintained in avian or other hosts, but its actual evolutionary process remains to be determined (Xia et al., 2008). The deduced aa sequence of VP1 protein of HN strain exhibits 2 specific alterations. The first changed residue is located at position 24 (A to T) within N-terminal domain and the second change residue is at position 857 (A to V) within C-terminal domain. Whether the change of aa had an effect in viral replication still needs to be confirmed by further experiments. Pikula et al. (Pikuła et al., 2018) reported that one change at position 24(A to V) in the VP1 aa sequence of Bpop/03 reassortant strain was found, but did not provide information about the function of this region of VP1 in viral replication. By sequences alignment analysis and experimental verification, Gao et al. (Gao et al., 2014) found that the triplet amino acids at positions 145/146/147 of VP1 represent a unique characteristic among different branches and played a critical role in the replication and pathogenicity of IBDV. The triplet aa NEG are highly conserved in non-vvIBDV strains, while TDN and TEG triplets are highly conserved in vvIBDV strains. However, substitution of the TDN triplet with TEG or NEG reduced viral replication and pathogenicity of the vvIBDV HuB-1 strain in chickens. It shows that virulence of these isolates with TEG triplet are weaker than those strains with TDN triplet, indicating that IBDV HN strain has a potentially weaken tendency. Distinct difference was observed between the studied isolate and the HN isolates reported by cui et al. (Cui et al., 2013) by sequence alignment. This suggests that the prevalent strain in central and northern of Henan in recent years is a new reassortant strain, which is different from that previously reported strain.
Currently, there have been many reports on IBDV reassortant strains, and most of the reported natural reassortant IBDV have exhibited intermediate virulence between very virulent and classical pathotype strains, such as IBD13HeB01 strain isolated from northern China (Lu et al., 2015), TL2004 strain (Wei et al., 2008) and ZJ2000 strain (Wei et al., 2006) isolated by Wei et al. There are also many reports that the new reassortant IBDV strain inherits high virulence and represents a very virulent phenotype, such as the reassortant strain Bpop/03 isolated from the field of Poland (Pikula et al., 2018), and the reassortant strain GX-NN-L isolated (Chen et al., 2012). In animal experiments, IBDV HN strain could cause obvious clinical symptoms and pathological changes with a mortality rate of 60%, which was more than that of CJ801 strain. The results indicated that the virulence of HN strains was higher than CJ801, and the HN isolate in this study maintains high virulence despite shuffling of genomic segments originating from 2 parental strains with distinct pathotypes. It is well known that Segment A (especially VP2) is the major factor responsible for the virulence of IBDV (Boot et al., 2000; Brandt, et al., 2001; Qi et al., 2009; Qi et al., 2013; Lu et al., 2015). According to the analysis of several reassortant virus reported, segment A of these reassortant strains with reduced virulence commonly originate from vaccine or attenuated strains, while segment B is often derived from virulent strain, but the situations for the reassortant strains with higher virulence are different. There are some reports that segment B also contributes to the virulence of IBDV (Boot et al., 2000; Escaffre et al., 2013; Gao et al., 2014; Lu et al., 2015). TDN triplet aa at positions 145/146/147 of VP1 is highly conserved in the vvIBDV strains, which is crucial for viral replication and pathogenicity (Gao et al., 2014; Abou El-Fetouh and Abdallah, 2018). More investigations, for instance by reverse genetics technology, are needed to explore whether there are other key aa in VP1 that affect viral replication and pathogenicity.
In conclusion, the HN isolate in this study is a novel reassortant strain in China. Although the exact origin is still unclear, segment A appear to be derived from the vvIBDV strains, while segment B is closely related to some other reassortant strains. The HN isolate seem to have retained high virulence and the ability of naturally transmitting to other chickens. IBDV has exacerbated the harm to the breeding industry due to immunosuppression. Therefore, the study on new reassortant IBDV strains is beneficial to monitor the prevalence of IBDV.
ACKNOWLEDGMENTS
This work was supported by grant from the National Natural Science Foundation of China (No. 31402169), key Scientific and Technological Project of Henan Province (182102110301 and 192102110189), Program for Innovative Research Team (in Science and Technology) in University of Henan Province (18IRTSTHN019), Modern agricultural industrial technology system of Henan Province (S2012-06-G02) and the postdoctoral fund of Henan Institute of Science and Technology. We also appreciate Dr. Henry Muriuki Kariithi for manuscript revision.
Contributor Information
Changbo Ou, Email: ouchangbo2004@163.com.
Xingyou Liu, Email: lxingyou@sohu.com.
REFERENCES
- Abou El-Fetouh M.S., Abdallah F.M. Genetic characterization of Infectious Bursal Disease Viruses isolated from the vaccinated broiler chicken flocks in Egypt during 2015–2016. Pol. J. Vet. Sci. 2018;21:581–588. doi: 10.24425/124293. [DOI] [PubMed] [Google Scholar]
- Abed M., Soubies S., Courtillon C., Briand F.X., Allée C., Amelot M., De Boisseson C., Lucas P., Blanchard Y., Belahouel A., Kara R., Essalhi A., Temim S., Khelef D., Eterradossi N. Infectious bursal disease virus in Algeria: Detection of highly pathogenic reassortant viruses. Infect. Genet. Evol. 2018;60:48–57. doi: 10.1016/j.meegid.2018.01.029. [DOI] [PubMed] [Google Scholar]
- Alfonso-Morales A., Martinez-Perez O., Dolz R., Valle R., Perera C.L., Bertran K., Frias M.T., Majo N., Ganges L., Pérez L.J. Spatiotemporal phylogenetic analysis and molecular characterisation of infectious bursal disease viruses based on the vp2 hyper-variable region. PloS One. 2013;8 doi: 10.1371/journal.pone.0065999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg T.P. Acute infectious bursal disease in poultry: a review. Avian. Pathol. 2000;29:175–194. doi: 10.1080/03079450050045431. [DOI] [PubMed] [Google Scholar]
- Boot H.J., ter Huurne A.A., Hoekman A.J., Peeters B.P., Gielkens A.L. Rescue of very virulent and mosaic infectious bursal disease virus from cloned cDNA: VP2 is not the sole determinant of the very virulent phenotype. J. Virol. 2000;74:6701–6711. doi: 10.1128/jvi.74.15.6701-6711.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt M., Yao K., Liu M., Heckert R.A., Vakharia V.N. Molecular determinants of virulence, cell tropism, and pathogenic phenotype of infectious bursal disease virus. J. Virol. 2001;75:11974–11982. doi: 10.1128/JVI.75.24.11974-11982.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Liu J., Yan Z., Liu D., Ji J., Qin J., Li H., Ma J., Bi Y., Xie Q. Complete genome sequence analysis of a natural reassortant infectious bursal disease virus in China. J. Virol. 2012;86:11942–11943. doi: 10.1128/JVI.02043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., He X., Yang L., Wei P. Antigenicity characterization of four representative natural reassortment IBDVs isolated from commercial three-yellow chickens from Southern China reveals different subtypes co-prevalent in the field. Vet. Microbiol. 2018;219:183–189. doi: 10.1016/j.vetmic.2018.04.024. [DOI] [PubMed] [Google Scholar]
- Cui P., Ma S.J., Zhang Y.G., Li X.S., Gao X.Y., Cui B.A., Chen H.Y. Genomic sequence analysis of a new reassortant infectious bursal disease virus from commercial broiler flocks in Central China. Arch. Virol. 2013;158:1973–1978. doi: 10.1007/s00705-013-1682-y. [DOI] [PubMed] [Google Scholar]
- Drissi Touzani C., Fellahi S., Gaboun F., Fassi Fihri O., Baschieri S., Mentag R., El Houadf M. Molecular characterization and phylogenetic analysis of very virulent infectious bursal disease virus circulating in Morocco during 2016-2017. Arch. Virol. 2019;164:381–390. doi: 10.1007/s00705-018-4076-3. [DOI] [PubMed] [Google Scholar]
- Escaffre O., Le Nouen C., Amelot M., Ambroggio X., Ogden K.M., Guionie O., Toquin D., Muller H., Islam M.R., Eterradossi N. Both genome segments contribute to the pathogenicity of very virulent infectious bursal disease virus. J. Virol. 2013;87:2767–2780. doi: 10.1128/JVI.02360-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L., Wu T., Hussain A., Gao Y., Zeng X., Wang Y., Gao L., Li K., Wang Y., Liu C., Cui H., Pan Q., Zhang Y., Liu Y., He H., Wang X., Qi X. Novel variant strains of infectious bursal disease virus isolated in China. Vet. Microbiol. 2019;230:212–220. doi: 10.1016/j.vetmic.2019.01.023. [DOI] [PubMed] [Google Scholar]
- Gao H.L., Wang X.M., Gao Y.L., Fu C.Y. Direct evidence of reassortment and mutant spectrum analysis of a very virulent infectious bursal disease virus. Avian Dis. 2007;51:893–899. doi: 10.1637/7626-042706R1.1. [DOI] [PubMed] [Google Scholar]
- Gao L., Li K., Qi X., Gao H., Gao Y., Qin L., Wang Y., Shen N., Kong X., Wang X. Triplet amino acids located at positions 145/146/147 of the RNA polymerase of very virulent infectious bursal disease virus contribute to viral virulence. J. Gen. Virol. 2014;95:888–897. doi: 10.1099/vir.0.060194-0. [DOI] [PubMed] [Google Scholar]
- He C.Q., Ma L.Y., Wang D., Li G.R., Ding N.Z. Homologous recombination is apparent in infectious bursal disease virus. Virology. 2009;384:51–58. doi: 10.1016/j.virol.2008.11.009. [DOI] [PubMed] [Google Scholar]
- He X., Chen G., Yang L., Xuan J., Long H., Wei P. Role of naturally occurring genome segment reassortment in the pathogenicity of IBDV field isolates in Three-Yellow chickens. Avian Pathol. 2016;45:178–186. doi: 10.1080/03079457.2016.1139687. [DOI] [PubMed] [Google Scholar]
- He X., Wei P., Yang X., Guan D., Wang G., Qin A. Molecular epidemiology of infectious bursal disease viruses isolated from Southern China during the years 2000–2010. Virus genes. 2012;45:246–255. doi: 10.1007/s11262-012-0764-3. [DOI] [PubMed] [Google Scholar]
- He X., Xiong Z., Yang L., Guan D., Yang X., Wei P. Molecular epidemiology studies on partial sequences of both genome segments reveal that reassortant infectious bursal disease viruses were dominantly prevalent in southern China during 2000–2012. Arch Virol. 2014;159:3279–3292. doi: 10.1007/s00705-014-2195-z. [DOI] [PubMed] [Google Scholar]
- Hon C.C., Lam T.Y., Drummond A., Rambaut A., Lee Y.F., Yip C.W., Zeng F., Lam P.Y., Ng P.T., Leung F.C. Phylogenetic analysis reveals a correlation between the expansion of very virulent infectious bursal disease virus and reassortment of its genome segment B. J. Virol. 2006;80:8503–8509. doi: 10.1128/JVI.00585-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoque M.M., Omar A.R., Chong L.K., Hair-Bejo M., Aini I. Pathogenicity of SspI-positive infectious bursal disease virus and molecular characterization of the VP2 hypervariable region. Avian Pathol. 2001;30:369–380. doi: 10.1080/03079450120066377. [DOI] [PubMed] [Google Scholar]
- Jackwood D.J., Sommer-Wagner S. Genetic characteristics of infectious bursal disease viruses from four continents. Virology. 2007;365:369–375. doi: 10.1016/j.virol.2007.03.046. [DOI] [PubMed] [Google Scholar]
- Jackwood D.J., Sommer-Wagner S.E., Crossley B.M., Stoute S.T., Woolcock P.R., Charlton B.R. Identifcation and pathogenicity of a natural reassortant between a very virulent serotype 1 infectious bursal disease virus (IBDV) and a serotype 2 IBDV. Virology. 2011;420:98–105. doi: 10.1016/j.virol.2011.08.023. [DOI] [PubMed] [Google Scholar]
- Kasanga C.J., Yamaguchi T., Munan'andu H.M., Ohya K., Fukushi H. Genomic sequence of an infectious bursal disease virus isolate from Zambia: classical attenuated segment B reassortment in nature with existing very virulent segment A. Arch. Virol. 2013;158:685–689. doi: 10.1007/s00705-012-1531-4. [DOI] [PubMed] [Google Scholar]
- Lai S.Y., Chang G.R., Yang H.J., Lee C.C., Lee L.H., Vakharia V.N., Wang M.Y. A single amino acid in VP2 is critical for the attachment of infectious bursal disease subviral particles to immobilized metal ions and DF-1 cells. Biochim. Biophys. Acta. 2014;1844:1173–1182. doi: 10.1016/j.bbapap.2014.04.004. [DOI] [PubMed] [Google Scholar]
- Le Nouën C., Rivallan G., Toquin D., Darlu P., Morin Y., Beven V., Boisseson C., Cazaban C., Comte S., Gardin Y., Eterradossi N. Very virulent infectious bursal disease virus: reduced pathogenicity in a rare natural segment-B-reassorted isolate. J. Gen. Virol. 2006;87:209–216. doi: 10.1099/vir.0.81184-0. [DOI] [PubMed] [Google Scholar]
- Letzel T., Coulibaly F., Rey F.A., Delmas B., Jagt E., van Loon A.A., Mundt E. Molecular and structural bases for the antigenicity of VP2 of infectious bursal disease virus. J. Virol. 2007;81:12827–12835. doi: 10.1128/JVI.01501-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Courtillon C., Guionie O., Allee C., Amelot M., Qi X., Gao Y., Wang X., Eterradossi N. Genetic, antigenic and pathogenic characterization of four infectious bursal disease virus isolates from China suggests continued evolution of very virulent viruses. Infect. Genet. Evol. 2015;30:120–127. doi: 10.1016/j.meegid.2014.12.016. [DOI] [PubMed] [Google Scholar]
- Li Y., Wu T., Cheng X., Zhang C. Molecular characteristic of VP2 gene of infectious bursal disease viruses isolated from a farm in two decades. Virus Genes. 2009;38:408–413. doi: 10.1007/s11262-009-0356-z. [DOI] [PubMed] [Google Scholar]
- Li Y., Cui S., Cui Z., Chang S., Zhao P. Genome analysis and pathogenicity of reticuloendotheliosis virus isolated from a contaminated vaccine seed against infectious bursal disease virus: first report in China. J. Gen. Virol. 2016;97:2809–2815. doi: 10.1099/jgv.0.000588. [DOI] [PubMed] [Google Scholar]
- Li Z., Wang Y., Xue Y., Li X., Cao H., Zheng S.J. Critical role for voltage-dependent anion channel 2 in infectious bursal disease virus-induced apoptosis in host cells via interaction with VP5. J. Virol. 2012;86:1328–1338. doi: 10.1128/JVI.06104-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Zhang X.B., Yan Z.Q., Chen F., Ji J., Qin J.P., Li H.Y., Lu J.P., Xue Y., Liu J.J., Xie Q.M., Ma J.Y., Xue C.Y., Bee Y.Z. Molecular characterization and phylogenetic analysis of infectious bursal disease viruses isolated from chicken in South China in 2011. Trop. Anim. Health. Prod. 2013;45:1107–1112. doi: 10.1007/s11250-012-0333-8. [DOI] [PubMed] [Google Scholar]
- Liu J., Zhou J., Kwang J. Antigenic and molecular characterization of recent infectious bursal disease virus isolates in China. Virus Genes. 2002;24:135–147. doi: 10.1023/a:1014568532292. [DOI] [PubMed] [Google Scholar]
- Lu Z., Zhang L., Wang N., Chen Y., Gao L., Wang Y., Gao H., Gao Y., Li K., Qi X., Wang X. Naturally occurring reassortant infectious bursal disease virus in northern China. Virus Res. 2015;203:92–95. doi: 10.1016/j.virusres.2015.04.003. [DOI] [PubMed] [Google Scholar]
- Patel A.K., Pandey V.C., Pal J.K. Evidence of genetic drift and reassortment in infectious bursal disease virus and emergence of outbreaks in poultry farms in India. Virusdisease. 2016;27:161–169. doi: 10.1007/s13337-016-0306-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikuła A., Lisowska A., Jasik A., Śmietanka K. Identifcation and assessment of virulence of a natural reassortant of infectious bursal disease virus. Vet. Res. 2018;49:89–99. doi: 10.1186/s13567-018-0586-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X., Gao H., Gao Y., Qin L., Wang Y., Gao L., Wang X. Naturally occurring mutations at residues 253 and 284 in VP2 contribute to the cell tropism and virulence of very virulent infectious bursal disease virus. Antiviral Res. 2009;84:225–233. doi: 10.1016/j.antiviral.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Qi X., Zhang L., Chen Y., Gao L., Wu G., Qin L., Wang Y., Ren X., Gao Y., Gao H., Wang X. Mutations of residues 249 and 256 in VP2 are involved in the replication and virulence of infectious bursal disease virus. PLoS One. 2013;8 doi: 10.1371/journal.pone.0070982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja P., Senthilkumar T.M., Parthiban M., Thangavelu A., Gowri A.M., Palanisammi A., Kumanan K. Complete genome sequence analysis of a naturally reassorted infectious bursal disease virus from India. Genome Announc. 2016;4:e00709–e00716. doi: 10.1128/genomeA.00709-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Xue C., Zhang Y., Chen F., Cao Y. Genomic analysis of one Chinese strain YS07 of infectious bursal disease virus reveals unique genetic diversity. Virus genes. 2009;39:246–248. doi: 10.1007/s11262-009-0379-5. [DOI] [PubMed] [Google Scholar]
- Rosenberger J.K., Gelb J.J. Response to several avian respiratory viruses as affected by infectious bursal disease virus. Avian Dis. 1978;22:95–105. [PubMed] [Google Scholar]
- Sharma J.M., Kim I.J., Rautenschlein S., Yeh H.Y. Infectious bursal disease virus of chickens: pathogenesis and immunosuppression. Dev. Comp. Immunol. 2000;24:223–235. doi: 10.1016/s0145-305x(99)00074-9. [DOI] [PubMed] [Google Scholar]
- Soubies M.S., Courtilion C., Briand F.X., Queguiner-Leroux M., Coutois D., Amelot M., Grousson K., Morillon P., Herin J.B., Eterradossi N. Identifcation of a European interserotypic reassortant strain of IBDV. Avian Pathol. 2017;46:19–27. doi: 10.1080/03079457.2016.1200010. [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.Q., Qiao H.X., Liu H.Y., Fang X.Z., Gong X., Luo J., Lu J.F., Chen D.S., Chen P.Y., Zheng H.J. Proceedings of the 2nd International Symposium on Infectious Bursal Disease and Chicken Infectious Anaemia. 2001. Isolation and characterization of vvIBDV from vaccinated chickens with atypical signs of IBD in Henan Province, China; pp. 204–214. Giessen, Germany. [Google Scholar]
- Wang S.T., Chang H.T., Li H., Li W.H., Yan H., Wang C.Q., Yao H.X., Liu H.Y., Wang X.W. Isolation and identification of four infectious bursal disease virus strains and sequence analysis of VP2 gene. J. Yangzhou University (Agr. Life. Sci. Edit.). 2013;34:5–9. [Google Scholar]
- Wei Y., Li J., Zheng J., Xu H., Li L., Yu L. Genetic reassortment of infectious bursal disease virus in nature. Biochem. Biophys. Res. Commun. 2006;350:277–287. doi: 10.1016/j.bbrc.2006.09.040. [DOI] [PubMed] [Google Scholar]
- Wei Y., Yu X., Zheng J., Chu W., Xu H., Yu X., Yu L. Reassortant infectious bursal disease virus isolated in China. Virus Res. 2008;131:279–282. doi: 10.1016/j.virusres.2007.08.013. [DOI] [PubMed] [Google Scholar]
- Xia R.X., Wang H.Y., Huang G.M., Zhang M.F. Sequence and phylogenetic analysis of a Chinese very virulent infectious bursal disease virus. Arch. Virol. 2008;153:1725–1729. doi: 10.1007/s00705-008-0140-8. [DOI] [PubMed] [Google Scholar]
- Xu M.Y., Lin S.Y., Zhao Y., Jin J.H., Tang N., Zhang G.Z. Characteristics of very virulent infectious bursal disease viruses isolated from Chinese broiler chickens (2012-2013) Acta Trop. 2015;141:128–134. doi: 10.1016/j.actatropica.2014.10.003. [DOI] [PubMed] [Google Scholar]
- Ye C., Han X., Yu Z., Zhang E., Wang L., Liu H. Infectious bursal disease virus activates c-Src To promote alpha4beta1 integrin-dependent viral entry by modulating the downstream Akt-RhoA GTPase-actin rearrangement cascade. J. Virol. 2017;91:e01891. doi: 10.1128/JVI.01891-16. e01816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz A., Turan N., Bayraktar E., Gurel A., Cizmecigil U.Y., Aydin O., Bamac O.E., Cecchinato M., Franzo G., Tali H.E., Cakan B., Savic V., Richt J.A., Yilmaz H. Phylogeny and evolution of infectious bursal disease virus circulating in Turkish broiler flocks. Poult. Sci. 2019;98:1976–1984. doi: 10.3382/ps/pey551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Liu F., Tao S., Ai G. Isolation of pathogen of infectious bursal disease in Beijing area. Chin. J. Vet. Med. 1981;8:25–26. [Google Scholar]