Abstract
This study evaluated the effect of in ovo Bacillus spp. base probiotic (BBP) administration on hatchability, Gram-negative bacteria (GNB) recovery, performance, and microbiota composition in 2 independent trials using a virulent E. coli seeder challenge model. In each trial, one hundred and eighty 18-day-old embryos were allocated into 1 of 2 groups: Control and treated group (inoculated with 107 BBP). On day 19 of embryogenesis, seeder embryos (n = 18) were inoculated with 4.5 × 104E. coli/ mL+ 272 μg/mL tetracycline and segregated into mesh hatching bags. Twelve chicks per group were euthanized at hatch and at day 7 to evaluate the gastrointestinal composition of total GNB or total aerobic pasteurized bacteria. Also, in trial 2, ceca content from five chickens at day 7 were collected to evaluate microbiota composition. Embryos inoculated with BBP showed a significant (P < 0.05) reduction in the total number of GNB at day-of-hatch (DOH) and day 7. Probiotic treatment increased BW at DOH and day 7, and BW gain (days 0 to 7) when compared with Control chickens. Proteobacteria phylum was significantly reduced, while the Firmicutes was significantly increased by the BBP as compared to the Control (P < 0.05). At family level, Enterobacteriaceae was significantly decreased, while the Lachnospiraceae was significantly elevated in the BBP as compared to the Control group (P < 0.05). The genus Oscillospira was significantly enriched in the BBP group, whereas the unidentified genus of family Enterobacteriaceae in the Control group (P < 0.05). The BBP group increased the bacterial species richness, although there was no significant difference between treatments (P > 0.05). Interestingly, beta diversity showed a significant difference in bacterial community structure between Control and BBP groups (P < 0.05). The results of the present study suggest that in ovo administration of a BBP can reduce the severity of virulent E. coli horizontal transmission and infection of broiler chickens during hatch. The reduction in the severity of the transmission and infection by the BPP might be achieved through alterations of microbiota composition and its community structure.
key words: broiler, Escherichia coli, hatchers, in ovo, probiotic
INTRODUCTION
Beneficial bacterial species surpass the number of pathogenic species, complementing the biology and physiology of metazoans (Kikuchi et al., 2009). One example of such beneficial mutualism is found in the gastrointestinal tract (GIT; Xie et al., 2010). The gathering of the gut microbiota is regulated by the elaborate and combinatorial host–microbial and microbial–microbial interactions (Xu and Gordon, 2003). Several studies have described how the microbiota modulates the gut-associated lymphoid tissue (Martin et al., 2010), instructs the immune system (McFall-Ngai, 2007), improves the intestinal integrity (Duerkop et al., 2009), regulates the proliferation and differentiation of the enterocytes (Moran, 2007), regulates blood flow (Sekirov et al., 2010), and activates the enteric nervous system (Tlaskalová-Hogenová et al., 2011). In mammals, colonization of the microbiota initiates at birth and continues throughout life (Di Mauro et al., 2013). Under commercial conditions, chickens hatch in an environment that contains potential pathogens where these pathogens can colonize and shift the microbiota prior to beneficial bacteria colonization. While the natural route of transmission of enteropathogens is fecal-oral (White et al., 1997; Galanis et al., 2006), published studies have also suggested that airborne transmission of enteropathogens in poultry is possible (Kallapura et al., 2014a,b,c). Recently, we demonstrated that the in ovo administration of a probiotic mixed with a Marek's disease (MD) vaccine had no effect on the effectiveness of the MD vaccine, but reduced intestinal Salmonella Enteritidis counts and improved BW and intestinal integrity (Teague et al., 2017). Furthermore, our laboratory has developed a novel in ovo challenge model for virulent Escherichia coli strains (Graham et al., 2019). In that study, we have shown that co-administration of a virulent E. coli strain with tetracycline allows for the hatch of directly challenged chicks and effective horizontal transmission to contact chicks during the hatching process, as evidenced by reduced performance and altered selected enteric bacterial recovery at day 7. Therefore, considering that the 21 d of embryogenesis plus the first 7 d represent 50 to 74% of the commercial life of chickens (Cherian, 2011), in the present study, we evaluated the effect of in ovo administration of a Bacillus base probiotic (BBP) on hatchability, Gram-negative bacteria recovery, performance, and microbiota composition during the first 7 d after hatch, using our published E. coli horizontal infection model in the hatching cabinet in broiler chickens.
MATERIALS AND METHODS
Bacillus Based Probiotic
NorumTM (Eco-Bio/Euxxis Bioscience LLC, Fayetteville, AR) is a Bacillus spore DFM culture, consisting of three isolates: two Ba cillus amyloliquefaciens and one Bacillus subtilis (Latorre et al., 2015b, 2016). The product contains a concentration of stable Bacillus spores (∼3 × 1011 spores/g). Aliquots of vegetative bacterial strains from NorumTM were maintained in 50% glycerol frozen stocks at −80°C. In the present study, 100 µL of a frozen aliquot of each strain were added to 10 mL of tryptic soy broth (TSB, Sigma-Aldrich, St Louis, MO, USA) and incubated at 37°C for 24 h. Then, bacteria were washed 3 times, combined, resuspended in sterile phosphate-buffered saline (PBS) and adjusted to an optical density (OD600) of 0.8 to 0.9. This combination of BBP was diluted in sterile saline to an expected concentration of 5 × 107 cfu/mL for in ovo injection of 0.2 mL into the amnion. Actual colony-forming units from each trial were reported, which were determined retrospectively from spread plating on tryptic soy agar (TSA).
E. coli Culture and Challenge
A lactose negative E. coli strain known to cause respiratory disease and mortality in both chickens and turkeys (Huff et al., 1998, 2002, 2003) was used in the present study. For that, 100 μL of E. coli were removed from a frozen aliquot and added to 10 mL of TSB. The culture was incubated at 37°C for 18 h. Post incubation, bacterial cells were washed with sterile 0.9% saline by centrifugation at 1800 × g for 15 min and reconstituted in saline. The wash procedure was completed three times. E. coli cfu enumeration was determined by serial dilution and plating on MacConkey agar (MacConkey Agar, cat. no. 89,429–342, V.W.R., Suwanee, GA 30,024) to determine the stock concentration and then cells were held overnight at 4°C. Approximately 16 h later, the culture was serially diluted to desired cfu concentration for in ovo challenge (day 19 of embryogenesis) with the selected dose of tetracycline hydrochloride (Tetracycline hydrochloride, cat. no. 64,755, Sigma, St. Louis, MO 63,103). Actual E. coli challenge dose (cfu/mL) was confirmed as described above and reported. Relative minimal inhibitory concentrations of tetracycline were determined in vitro (data not shown) and then adjusted in subsequent trials based on in vivo results.
Experimental Design
Two independent trials with 18-day-old of embryogenesis Ross 308 embryos were conducted. Embryos were candled and randomly allocated into one of two groups (n = 180 embryos per group) and placed into separate hatchers (G.Q.F. 1550 Digital Cabinet Egg Incubator) based on treatment group, and inoculated via in ovo injection into the amnion with 0.2 mL of sterile PBS or with 5 × 107 cfu/mL (1 × 107 cfu/0.2 mL) of the BBP. Hatchers were housed separately to prevent possible cross-contamination between treatments during the hatch. On day 19 of embryogenesis, seeder embryos (n = 18 seeders/hatcher or 10%/hatcher) were inoculated with E. coli/ tetracycline treatment via in ovo injection into the amnion and segregated into mesh hatching bags (reusable mesh nylon netting, I.D.S., Amazon). Seeder embryos were challenged with a dose of 4.5 × 104 cfu/mL E. coli + 272 μg/mL tetracycline. On day-of-hatch (DOH), dry chicks were removed from hatchers, and hatchability was determined. Then, the contact-challenged chicks were weighed, and 90 chickens were selected to be placed into pens (3pens/group with 30 chicks each). No seeders were placed. The BW on the day of hatching was normalized so that the differences in weight were due to the treatment and not to the higher initial weight of any of the groups. BW allocation was achieved by normalizing the means between all pens and treatments by standard deviation. In each trial, body weight gain (BWG) from d0 to day 7 was determined for the duration of each trial (7-d trial period). Furthermore, 12 chicks per group were euthanized on DOH and day 7 to evaluate the gastrointestinal composition on selective media and enumerate total Gram negative. At day 7, ceca content of five chickens were collected to evaluate microbiota composition. Chickens were provided ad libitum access to water and a balanced, unmedicated corn and soybean diet meeting the nutritional requirements for broilers recommended by Aviagen. This study was carried out in accordance with the recommendations of the Institutional Animal Care and Use Committee (IACUC) at the University of Arkansas, Fayetteville. The IACUC approved protocol #17,073 at the University of Arkansas, Fayetteville for this study.
Enumeration of Bacteria
In both independent trials, the whole gut (ventriculus to the cecum) of 12 chickens per group was aseptically removed and collected into sterile bags as previously described (Tellez et al. 2015). Samples were weighed, homogenized, and 1:4 wt/vol dilutions were made using sterile 0.9% saline. Ten-fold dilutions of each sample, from each group, were made in a sterile 96 well Bacti flat bottom plates and the diluted samples were plated on culture media to evaluate the total number of Gram-negative bacteria on MacConkey agar (MacConkey Agar, cat. no. 89,429–342, V.W.R., Suwanee, GA 30,024). Following heat treatment, 10-fold dilutions of the feed samples were plated on TSA. All plates were incubated at 37°C for 18 h, and bacterial counts were expressed as Log10 cfu/g of sample (Tellez et al. 2015).
Microbiota Analysis
Sample Processing, DNA Extraction, PCR, Library Preparation, and Sequencing
One gram of cecum content from 5 chickens at day 7 was transferred into collection tubes containing a lysis and stabilization buffer. DNA extraction, amplification, and library preparation were performed as described earlier (Almonacid et al., 2017). Briefly, samples were lysed through bead-beating and DNA was extracted by guanidine thiocyanate silica column-based purification method using a liquid-handling robot in a class 1,000 clean room (Cady et al., 2003). The 515F (5'-GTGCCAGCMGCCGCGGTAA) and 806R (5'-GGACTACHVGGGTWTCTAAT) primers that contained Illumina tags and barcodes were used for amplification of the V4 variable region of the 16S rRNA gene (Caporaso et al., 2011). PCR products were then pooled, column-purified, and size-selected through microfluidic DNA fractionation (Minalla et al., 2001). Consolidated libraries were quantified by quantitative real-time PCR using the Kapa Bio-Rad iCycler qPCR kit on a BioRad MyiQ before loading for sequencing. Sequencing was performed in a pair-end modality on the Illumina NextSeq 500 platform rendering 2 × 150 bp pair-end sequences.
16S rRNA Gene Sequences Analysis
After sequencing, demultiplexing of samples was performed using Illumina's BCL2FASTQ algorithm. Forward and reverse reads obtained in each of the 4 lanes per sample were filtered using the following criteria: both forward and reverse reads in a pair must have an average Q-score > 30. Primers and any leading random nucleotides (used to increase the diversity of the library being sequenced) were trimmed, and forward reads were capped at 125 bp and reverse reads are capped at 124 bp. After quality filtering as described above, the Deblur (Amir et al., 2017) workflow was applied for the forward reads to generate a feature table and representative sequences using “qiime deblur denoise-16S” method implemented in QIIME2 version 2019.1 (Bolyen et al., 2018). The features that were present only in a single sample were removed from the feature table. Naive Bayes classifier (Pedregosa et al., 2011) was trained using Green genes 13_8 99% O.T.U.s (DeSantis et al., 2006), where the sequences were trimmed to include only 125 bases from the region of the 16S rRNA gene bound by the 515F/806R primer pair. This pre-trained classifier was used to assign taxonomy to the representative sequences using q2-feature-classifier plugin. Microbial diversity analyses were performed using q2-diversity-plugin of QIIME2 using the even sampling depth of 14,610. The alpha diversity as computed by observed OTUs metric and Shannon's index (Shannon, 1948) and beta diversity as calculated by unweighted UniFrac (Lozupone et al., 2011) distance metrics were reported. All figures were created using ggplot2 packages (Wickham, 2016) on R version 3.5.3.
Statistical Analysis
All data were subjected to one-way analysis of variance as a completely randomized design using the GLM procedure of SAS. (SAS Institute, 2002). Data were expressed as mean ± SE. Significant differences (P < 0.05) among the means were further separated using Duncan's multiple range test for bacterial recovery, BW, and BWG. Hatchability was compared using the chi-squared test of independence to determine the significance for these studies (Zar, 1984). Wilcoxon test was performed for statistical analysis of alpha diversity and bacterial taxonomic groups (phylum, family, and genus) between the 2 treatments. However, PERMANOVA (Anderson, 2001) test was used to calculate significant difference in beta diversity between two treatments.
RESULTS
The results of the effect of in ovo administration of BBP on microbial composition in the GIT of hatching broiler chickens, hatchability and BW in a virulent E. coli horizontal transmission challenge in the hatching cabinet in broiler chickens are summarized in Table 1. In both trials, BBP significantly reduced (P < 0.05) the recovery of Gram-negative bacteria on DOH and day 7 as compared to the in ovo Control group in both trials (Table 1). No significant differences in hatchability were observed in both trials. Nevertheless, in ovo administration of the BBP significantly increased the average BW at DOH and day 7, as well as BWG (days 0 to 7) when compared with Control PBS group (Table 1).
Table 1.
Effect of in ovo administration of Bacillus spp. base probiotic (BBP) on microbial composition in the gastrointestinal tract of hatching broiler chickens, hatchability, body weight (g), and horizontal transmission of virulent E. coli during hatch.
| Treatment | Gram-negative bacterial recovery DOH (Log10 cfu/g) | Gram-negative bacterial recovery day 7 (Log10 cfu/g) | Hatchability (%) | Average BW DOH (CV) | Average BW day 7 (CV) | Average BWG days 0 to 7 (CV) |
|---|---|---|---|---|---|---|
| Trial 1 | ||||||
| In ovo PBS Control | 5.70 ± 0.28a | 7.43 ± 0.12a | 174/180 (96.66%) | 40.03 ± 0.07b (0.089) | 164.56 ± 2.52b (0.120) | 116.93 ± 2.63b (0.144) |
| In ovo 107 cfu/mL BBP | 4.32 ± 0.91b | 4.11 ± 0.47b | 175/180 (97.22%) | 47.77 ± 0.86a (0.086) | 175.15 ± 2.71a (0.097) | 127.38 ± 2.69a (0.112) |
| Trial 2 | ||||||
| In ovo PBS Control | 4.92 ± 0.32a | 6.34 ± 0.33a | 176/180 (97.77%)) | 41.30 ± 0.03b (0.099) | 161.31 ± 2.68b (0.140) | 111.81 ± 1.91b (0.137) |
| In ovo 107 cfu/mL BBP | 3.41 ± 0.81b | 3.89 ± 0.35b | 178/180 (98.88%) | 42.77 ± 0.11a (0.120) | 181.15 ± 2.71a (0.150) | 138.38 ± 3.69a (0.160) |
Hatchability total: hatched chickens/total embryos placed (%). Body weight (BW), n = 30.
Indicates significant difference between columns (P < 0.05).
Bacterial Composition at the Phylum Level
At the phylum level, Firmicutes and Proteobacteria were the only 2 phyla detected from the 2 treatment groups, whose relative abundances are shown in Figure 1. Firmicutes (Control 64.38 ± 5.51%, BBP 79.88 ± 3.74%) was the predominant phylum in both groups followed by the Proteobacteria (Control 35.62 ± 5.51%, BBP 20.12 ± 3.74%). The Proteobacteria was significantly reduced, while Firmicutes was significantly increased in BBP group as compared to the Control (Table 2; Wilcoxon test, P < 0.05).
Figure 1.
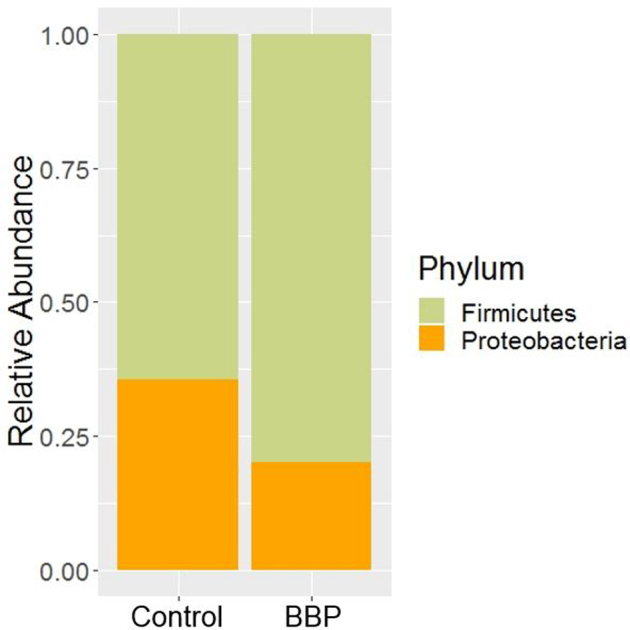
Relative abundance of bacterial phyla recovered from Control and Bacillus base probiotic (BBP) in trial 2.
Table 2.
Differentially abundant bacterial taxa at different levels of taxonomic classification in 2 treatment groups: Control and BBP (Wilcoxon test, P < 0.05).
| Control | BBP | |
|---|---|---|
| Phylum Level | ||
| Proteobacteria | Firmicutes | |
| Family Level | ||
| Enterobacteriaceae | Lachnospiraceae | |
| Genus Level | ||
| Unidentified genus | Oscillospira |
Bacterial Composition at the Family Level
The relative abundance of different bacterial families recovered from Control and BBP groups is shown in Figure 2. In both groups, Lachnospiraceae (Control 40.72 ± 1.84%, BBP 53.30 ± 4.26%) as found the highest percentage followed by Enterobacteriaceae in the Control group (Control; 35.62 ± 5.51%; BBP; 20.11 ± 3.74%), while Ruminococcaceae in the BPP group (Control; 15.01 ± 5.01%, BBP; 20.83 ± 4.44%). Other notable bacterial families were Lactobacillaceae (Control 4.26 ± 2.02%, BBP 1.21 ± 0.60%) and Erysipelotrichaceae (Control 3.73 ± 1.25%, BBP 2.53 ± 0.63%). The Clostridiaceae, Enterococcaceae, and Peptostreptococcaceae were observed as minor members whose average relative abundance was less than 1% in both groups. The statistical analysis revealed that the Enterobacteriaceae was significantly decreased, while the Lachnospiraceae was significantly elevated in BBP as compared to the Control group (Table 2; Wilcoxon test, P < 0.05).
Figure 2.
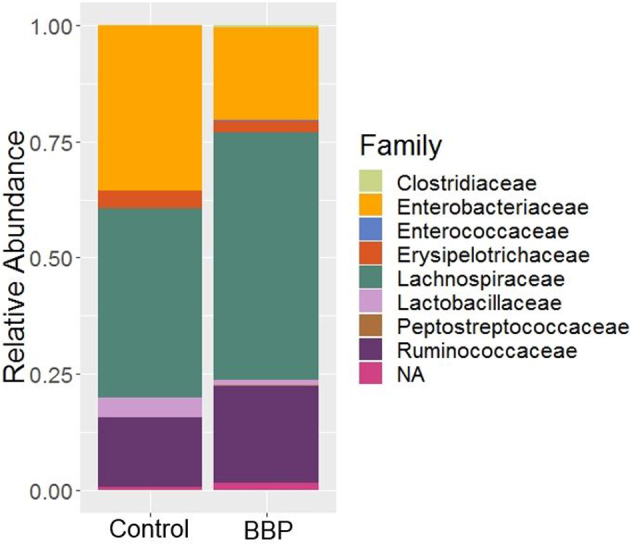
Relative abundance of bacterial families recovered on two treatment groups: Control and Bacillus base probiotic (BBP) in trial 2. NA represent those sequence reads which were not assigned at the family level, however, were assigned at the higher level of taxonomic classification.
Bacterial Composition at the Genus Level
The relative abundance of dominant bacterial genera in Control and BBP groups is shown in Figure 3. Majority of the reads (>50%) were not assigned at the genus level, however, they were assigned at the higher taxonomic level, and are grouped under NA (Figure 3). Among those identified genera, the genus Ruminococcus that belong to the family Lachnospiraceae was found the highest in both Control (13.25 ± 6.24%) and BBP (15.15 ± 4.82%) groups. This was followed by theRuminococcus (4.67 ± 1.72%) of family Ruminococcaceae in the Control, while the Oscillospira (8.86 ± 2.12%) in the BBP group. The Ruminococcus in the BBP group and the Oscillospira in the Control group were 2.39 ± 0.65% and 3.92 ± 0.74%, respectively. The genus Oscillospira was significantly enriched in the BBP group, whereas the unidentified genus of family Enterobacteriaceae in the Control group (Table 2; Wilcoxon test, P < 0.05). Other important numerical observations were the increase of Lactobacillus and Proteus in the Control group, and Butyricicoccus in the BBP group. Also, the genus Clostridium that belongs to the families Clostridiaceae and Lachnospiraceae was numerically increased in the BBP group while the Clostridium of family Erysipelotrichaceae was elevated in the Control group (Figure 3).
Figure 3.
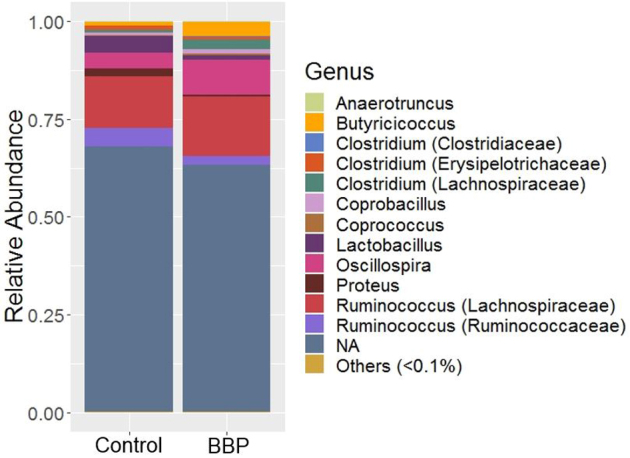
Relative abundance of bacterial genera recovered on2treatment groups: Control and Bacillus base probiotic (BBP) in trial 2. NA represent those sequence reads which were not assigned at the genus level, however, were assigned at the higher level of taxonomic classification. Others represent minor bacterial genera whose average relative abundance across all samples was <0.1%.
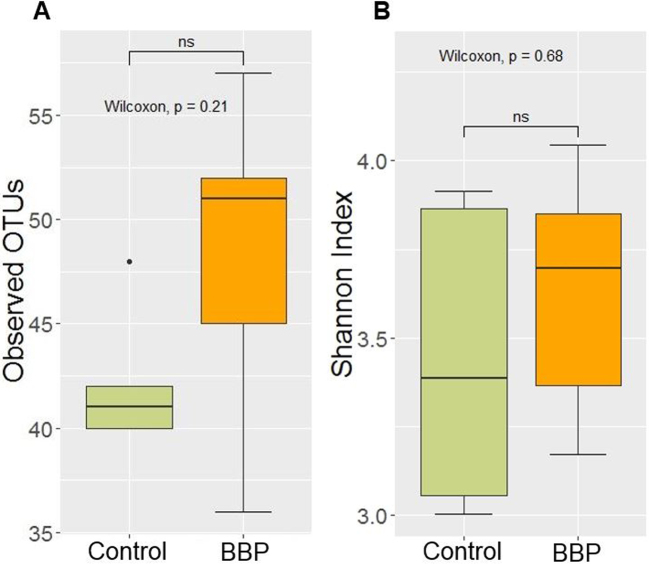
Alpha Diversity
The alpha diversity of Control and BBP group as calculated by observed OTUs metric and Shannon's index are shown in Figure 4 A and 4 B, respectively. Although the species richness was increased by the BBP group (Figure 4 A), there was no significant difference between the 2 groups, as shown in Figure 4 (Wilcoxon test,P > 0.05).
Figure 4.
The alpha diversity of two treatment groups: Control and Bacillus base probiotic (BBP) in trial 2. The alpha diversity was calculated by observed OTUs metric (A) and Shannon's index (B), where the statistical significant difference between treatment groups was calculated by Wilcoxon test. NS represent non-significant difference between treatment groups (P > 0.05).
Beta Diversity
The beta diversity between Control and BBP groups as calculated by unweighted UniFrac distance metric is illustrated in the PCoA plot (Figure 5). As shown in Figure 5, there was a significant difference in bacterial community structure between Control and BBP groups (PERMANOVA, P < 0.05).
Figure 5.
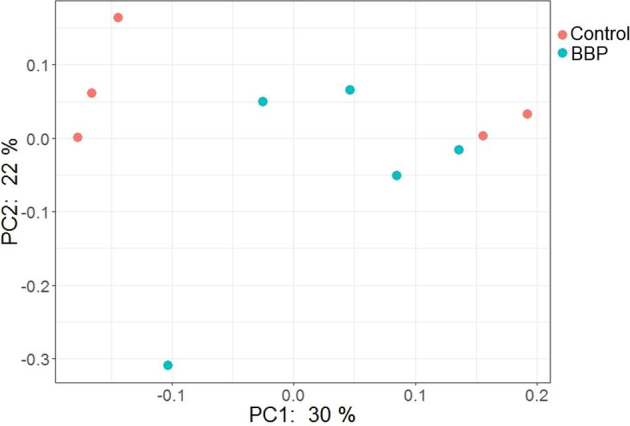
The PCoA plot showing the significant difference in beta diversity between 2 treatment groups: Control and Bacillus base probiotic (BBP) in trial 2. PERMANOVA, P < 0.05.
DISCUSSION
The spread of antibiotic resistance genes has created public and scientific concerns leading to new regulations that limit the use of antibiotics as growth promoters, creating a need to evaluate different alternative products. Hence, the use of probiotics as alternative tools to antibiotic growth promoters has been increasing, and many investigators around the world have demonstrated their efficacy. Probiotics regulate the immune system (Lyte, 2011; Molinaro et al., 2012), exert anti-oxidant properties (Tao et al., 2006), and enhance barrier integrity (Yu et al., 2012). Recent studies published by our laboratory demonstrate that 90% of Bacillus s pp. probiotic spores germinate within 60 min in the crop having full cycle from spores to vegetative cells to spores in different sections of the GIT (Latorre et al., 2014). After spore germination, bacteria become metabolically active to produce enzymes and other compounds that are beneficial to the host.
Moreover, the inclusion of this selected Bacillus- DFM (NorumTM) that produce a different set of extracellular enzymes using different poultry diets, significantly reduce both viscosity and C. perfringens proliferation (Latorre et al., 2015b). Further studies confirmed that this multiple enzymes producing Bacillus- based DFM improved growth performance, digesta viscosity, bacterial translocation, microbiota composition, and bone mineralization in broiler chickens fed with a rye-based diet (Latorre et al., 2015a) as well as mitigate the negative impacts of necrotic enteritis in broiler chickens (Hernandez-Patlan et al., 2019). In the present study, we evaluated for the first time, thein ovo application of the vegetative Bacillus spp. strains contained in NorumTM against experimental horizontal infection of E. coli in the hatching cabinet. Virulent E. coli strains can invade the host via the respiratory tract, leading to septicemia and airsacculitis (Dziva and Stevens, 2008). Under commercial conditions, chicks may be exposed to virulent E. coli strains during the hatch, indicating a need for a laboratory model allowing for evaluation of the effects of exposure during the hatching process.
In chickens, metagenome sequencing has shown that there are 4 major microbial phyla (Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria), which represent 99% of the intestinal microbiota. These phyla fall into 3 main groups of strict extremophile anaerobes: Bacteroides, Clostridium cluster XIVa, and Clostridium cluster IV (Oakley and Kogut, 2016). Clostridium and Bacillus are both in the phylum Firmicutes, but they are in different classes, orders, and families. Also, Clostridium is distinguished from Bacillus by being strict anaerobes. Clostridium genus contains over 100 commensal species, but only a few of them such as Clostridium perfringens and Clostridium tetani are pathogenic and produce some of the most potent toxins known in nature. Interestingly, most of Clostridia have a remarkable commensal and central relationship with their metazoan host, playing decisive roles in the physiology, immunology, and even cognitive activities as some of the essential butyric acid-producing bacteria of the GIT (Lopetuso et al., 2013; Liu et al., 2015; Zhong et al., 2015).
In both trials of the present study, hatchability was not affected by the treatment. However, embryos inoculated with BBP showed a significant reduction in the total number of Gram-negative bacteria in the GIT on DOH and day 7. Probiotic treatment increased BW DOH, BW day 7, and days 0 to 7 BWG when compared with control chickens. Moreover, the bacterial composition at the phylum level revealed that Proteobacteria was significantly higher in the PBS Control, while Firmicutes was significantly higher in the BBP. The gut microbiota complements the biology of metazoans playing important roles in animal overall health and productiveness (Wei et al., 2013; Tellez, 2014). Several studies have documented some mechanisms by which BBP can balance the gut microbiota and improve performance (Latorre et al., 2015a; Qin et al., 2018).
At the family level, in ovo administration of the BBP increased the presence of Lachnospiraceae but showed a significant reduction of Enterobacteriaceae when compared with the PBS Control group. Lachnospiraceae (phylum Firmicutes, class Clostridia) is abundant in the digestive tracts of many mammals, are also important within the group because of the production of butyric acid (Meehan and Beiko, 2014; Schnabl and Brenner, 2014). In this context, at the genus level, BBP group also showed a significant increase in Oscillospira and a significant reduction in unidentified genus of family Enterobacteriaceae when compared with the PBS Control group. Even though alpha diversity richness was increased by the BBP group, it was not significant when compared with the control group. Interestingly, beta diversity showed a significant difference in bacterial community structure between Control and BBP groups. Oscillospira is an anaerobic bacterial genus from Clostridial cluster IV, and this genus was found to reduce significantly in the gut of human having enteric inflammation, whereas positively correlate with the leanness (Konikoff and Gophna, 2016; Gophna et al., 2017). In chickens, Clostridium and Ruminococcus are some of the most predominant genera found in the cecum (Wei et al., 2013).
The Clostridium genus, along with Oscillospira and Coprococcus, also encompasses bacteria capable of producing butyrate (Yang et al., 2017). Butyrate has been demonstrated to have a decisive role on growth performance, intestinal villus structure, and pathogen control, as well as anti-inflammatory properties (Onrust et al., 2015). Furthermore, Ruminococcus genus can also produce other short-chain fatty acids, such as acetic and succinic acid (Flint et al., 2008). It is well known that short chain fatty acids are an essential source of energy for enterocytes and are vital for intestinal health (Biasato et al., 2018). The large intestine is abundant in Clostridium clusters IV and XIVa of the phylum Firmicutes that produce butyric acid (Onrust et al., 2015). Some strategies are available to stimulate butyrate production in the distal gut. These include delivery of prebiotic, probiotic, or symbiotic products (Tellez et al., 2006; Plöger et al., 2012). Members of Clostridium clusters XIVa and IV such as Lachnospiraceae, Ruminococcus, and Roseburia are depleted continuously in humans with intestinal inflammation disorders, suggesting that these organisms play a vital role in preserving gastrointestinal homeostasis (Kabeerdoss et al., 2013). Hence, it is essential to distinguish the presence of beneficial clostridial groups from opportunistic pathogenic strains such as Clostridium perfringens and Clostridium difficile (Honneffer et al., 2014). Clostridiales-OTUs in cecal digesta may have triggered a higher mucosal tolerance towards the commensal microbiota by increasing the expression levels of IL10 and TGFB1 at the cecal mucosa (Wahl et al., 2004). The results of the present study suggest that in ovo administration of a BBP can reduce the severity of virulent E. coli horizontal transmission and infection of broiler chickens in the hatching cabinet. The reduction in the severity of the transmission and infection were associated with significant changes in beta diversity induced by the BBP, suggesting that the BBP treatment may drive large-scale changes in the microbial community structure and composition, which in turn provided protection against the pathogenic effects of the E. coli infection. This has been shown previously; for example treatment with Bifidobacterium protected against the virulence of E. coli toxins through the production of butyrate in the mouse gut (Fukuda et al., 2011, 2012). We hypothesize that a similar mechanism may explain the results presented here, with the consequent improvement in the health and productivity of broiler chickens.
ACKNOWLEDGEMENTS
The authors would like to thank A. M. Donoghue, Poultry Production and Product Safety Research Unit, USDA ARS, for providing the isolate used in these experiments.
Footnotes
Supplementary data are available at Poultry Science online.
Supplementary Material
REFERENCES
- Almonacid D.E., Kraal L., Ossandon F.J., Budovskaya Y.V., Cardenas J.P., Bik E.M., Goddard A.D., Richman J., Apte Z.S. 16S rRNA gene sequencing and healthy reference ranges for 28 clinically relevant microbial taxa from the human gut microbiome. PLoS One. 2017;14 doi: 10.1371/journal.pone.0176555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir A., McDonald D., Navas-Molina J.A., Kopylova E., Morton J.T., Xu Z.Z., Kightley E.P., Thompson L.R., Hyde E.R., Gonzalez A., Knight R. Deblur rapidly resolves single-nucleotide community sequence patterns. MSystems. 2017;2:e00191. doi: 10.1128/mSystems.00191-16. e00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson M.J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26:32–46. [Google Scholar]
- Biasato I., Ferrocino I., Biasibetti E., Grego E., Dabbou S., Sereno A., Gai F., Gasco L., Schiavone A., Cocolin L., Capucchio M.T. Modulation of intestinal microbiota, morphology and mucin composition by dietary insect meal inclusion in free-range chickens. BMC Vet. Res. 2018;14:383. doi: 10.1186/s12917-018-1690-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C., Al-Ghalith G.A., Alexander H., Alm E.J., Arumugam M., Asnicar F., Bai Y., Bisanz J.E., Bittinger K., Brejnrod A., Brislawn C.J., Brown C.T., Callahan B.J., Caraballo-Rodríguez A.M., Chase J., Cope E., Da Silva R., Dorrestein P.C., Douglas G.M., Durall D.M., Duvallet C., Edwardson C.F., Ernst M., Estaki M., Fouquier J., Gauglitz J.M., Gibson D.L., Gonzalez A., Gorlick K., Guo J., Hillmann B., Holmes S., Holste H., Huttenhower C., Huttley G., Janssen S., Jarmusch A.K., Jiang L., Kaehler B., Kang K.B., Keefe C.R., Keim P., Kelley S.T., Knights D., Koester I., Kosciolek T., Kreps J., Langille M.G., Lee J., Ley R., Liu Y., Loftfield E., Lozupone C., Maher M., Marotz C., Martin B.D., McDonald D., McIver L.J., Melnik A.V., Metcalf J.L., Morgan S.C., Morton J., Naimey A.T., Navas-Molina J.A., Nothias L.F., Orchanian S.B., Pearson T., Peoples S.L., Petras D., Preuss M.L., Pruesse E., Rasmussen L.B., Rivers A., Robeson M.S., II, Rosenthal P., Segata N., Shaffer M., Shiffer A., Sinha R., Song S.J., Spear J.R., Swafford A.D., Thompson L.R., Torres P.J., Trinh P., Tripathi A., Turnbaugh P.J., Ul-Hasan S., van der Hooft J.J., Vargas F., Vázquez-Baeza Y., Vogtmann E., von Hippel M., Walters W., Wan Y., Wang M., Warren J., Weber K.C., Williamson C.H., Willis A.D., Xu Z.Z., Zaneveld J.R., Zhang Y., Zhu Q., Knight R., Caporaso J.G. QIIME 2: Reproducible, interactive, scalable, and extensible microbiome data science. PeerJ. Preprints. 2018;6 doi: 10.7287/peerj.preprints.27295v2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cady N.C., Stelick S., Batt C.A. Nucleic acid purification using microfabricated silicon structures. Biosens. Bioelectron. 2003;19:59–66. doi: 10.1016/s0956-5663(03)00123-4. [DOI] [PubMed] [Google Scholar]
- Caporaso J.G., Lauber C.L., Walters W.A., Berg-Lyons D., Lozupone C.A., Turnbaugh P.J., Fierer N., Knight R. Proc. of the National Academy of Sciences of the USA. National Academy of Sciences; St. Louis, MO: 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherian G. Essential fatty acids and early life programming in meat-type birds. World's Poult. Sci. J. 2011;67:599–614. [Google Scholar]
- DeSantis T.Z., Hugenholtz P., Larsen N., Rojas M., Brodie E.L., Keller K., Huber T., Dalevi D., Hu P., Andersen G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Mauro A., Neu J., Riezzo G., Raimondi F., Martinelli D., Francavilla R., Indrio F. Gastrointestinal function development and microbiota. Ital. J. Pediatr. 2013;39:15. doi: 10.1186/1824-7288-39-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerkop B.A., Vaishnava S., Hooper L.V. Immune responses to the microbiota at the intestinal mucosal surface. Immunity. 2009;31:368–376. doi: 10.1016/j.immuni.2009.08.009. [DOI] [PubMed] [Google Scholar]
- Dziva F., Stevens M.P. Colibacillosis in poultry: unravelling the molecular basis of virulence of avian pathogenic Escherichia coli in their natural hosts. Avian Pathol. 2008;37:355–366. doi: 10.1080/03079450802216652. [DOI] [PubMed] [Google Scholar]
- Flint H.J., Bayer E.A., Rincon M.T., Lamed R., White B.A. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat. Rev. Microbiol. 2008;6:121–131. doi: 10.1038/nrmicro1817. [DOI] [PubMed] [Google Scholar]
- Fukuda S., Toh H., Hase K., Oshima K., Nakanishi Y., Yoshimura K., Tobe T., Clarke J.M., Topping D.L., Suzuki T., Taylor T.D., Itoh K., Kikuchi J., Morita H., Hattori M., Ohno H. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- Fukuda S., Toh H., Taylor T.D., Ohno H., Hattori M. Acetate-producing bifidobacteria protect the host from enteropathogenic infection via carbohydrate transporters. Gut Microbes. 2012;3:449–454. doi: 10.4161/gmic.21214. [DOI] [PubMed] [Google Scholar]
- Galanis E., Lo Fo Wong D.M., Patrick M.E., Binsztein N., Cieslik A., Chalermchikit T., Aidara-Kane A., Ellis A., Angulo F.J., Wegener H.C. Web-based surveillance and global Salmonella distribution, 2000–2002. Emerg. Infect. Dis. 2006;12:381–388. doi: 10.3201/eid1203.050854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gophna U., Konikoff T., Nielsen H.B. Oscillospira and related bacteria-From metagenomic species to metabolic features. Environ. Microbiol. 2017;19:835–841. doi: 10.1111/1462-2920.13658. [DOI] [PubMed] [Google Scholar]
- Graham B.D., Selby C.M., Teague K.D., Graham L.E., Vuong C.N., Latorre J.D., Tellez G., Hargis B.M. Development of a novel in ovo challenge model for virulent Escherichia coli strains. Poult. Sci. 2019;0:1–6. doi: 10.3382/ps/pez321. [DOI] [PubMed] [Google Scholar]
- Hernandez-Patlan D., Solis-Cruz B., Pontin K.P., Hernandez-Velasco X., Merino-Guzman R., Adhikari B., Lopez-Arellano R., Kwon Y.M., Hargis B.M., Arreguin-Nava M., Tellez G. Impact of a Bacillus direct-fed microbial on growth performance, intestinal barrier integrity, necrotic enteritis lesions and ileal microbiota in broiler chickens using a laboratory challenge model. Front. Vet. Sci. 2019;6:108. doi: 10.3389/fvets.2019.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honneffer J.B., Minamoto Y., Suchodolski J.S. Microbiota alterations in acute and chronic gastrointestinal inflammation of cats and dogs. World J. Gastroenterol. 2014;20:16489–16497. doi: 10.3748/wjg.v20.i44.16489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huff G.R., Huff W.E., Balog J.J.M., Rath N. The effects of dexamethasone immunosuppression on turkey osteomyelitis complex in an experimental Escherichia coli respiratory infection. Poult. Sci. 1998;77:654–661. doi: 10.1093/ps/77.5.654. [DOI] [PubMed] [Google Scholar]
- Huff W.E., Huff G.R., Rath N.C., Balog J.M., Donoghue A.M. Prevention of Escherichia coli infection in broiler chickens with a bacteriophage aerosol spray. Poult. Sci. 2002;81:1486–1491. doi: 10.1093/ps/81.10.1486. [DOI] [PubMed] [Google Scholar]
- Huff W.E., Huff G.R., Rath N.C., Balog J.M., Donoghue A.M. Evaluation of aerosol spray and intramuscular injection of bacteriophage to treat an Escherichia coli respiratory infection. Poult. Sci. 2003;82:1108–1112. doi: 10.1093/ps/82.7.1108. [DOI] [PubMed] [Google Scholar]
- Kabeerdoss J., Sankaran V., Pugazhendhi S., Ramakrishna B.S. Clostridium leptum group bacteria abundance and diversity in the fecal microbiota of patients with inflammatory bowel disease: a case-control study in India. BMC Gastroenterol. 2013;13:20. doi: 10.1186/1471-230X-13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallapura G., Kogut M.H., Morgan M., Pumford N., Bielke L.R., Wolfenden A.D., Faulkner O.B., Latorre J.D., Menconi A., Hernandez-Velasco X., Kuttappan V.A., Hargis B.M., Tellez G. Fate of Salmonella Senftenberg in broiler chickens evaluated by challenge experiments. Avian Pathol. 2014;43:305–309. doi: 10.1080/03079457.2014.923554. [DOI] [PubMed] [Google Scholar]
- Kallapura G., Morgan M., Pumford N.R., Bielke L.R., Wolfenden A., Faulkner O.B., Latorre J.D., Menconi A., Hernandez-Velasco X., Kuttappan V.A., Hargis B.M., Tellez G. Evaluation of the respiratory route as a viable portal of entry for Salmonella in poultry via intratracheal challenge of Salmonella Enteritidis and Salmonella Typhimurium. Poult. Sci. 2014;93:340–346. doi: 10.3382/ps.2013-03602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallapura G., Botero A., Layton S., Bielke L.R., Latorre J.D., Menconi A., Hernandez-Velasco X., Bueno D., Hargis B.M., Tellez G. Evaluation of recovery of Salmonella from trachea and ceca in commercial poultry. J. Appl. Poult. Res. 2014;23:132–136. [Google Scholar]
- Kikuchi Y., Hosokawa T., Nikoh N., Meng X.-Y., Kamagata Y., Fukatsu T. Host-symbiont co-speciation and reductive genome evolution in gut symbiotic bacteria of acanthosomatid stinkbugs. BMC Biol. 2009;7:2. doi: 10.1186/1741-7007-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konikoff T., Gophna U. Oscillospira: a central, enigmatic component of the human gut microbiota. Trends Microbiol. 2016;24:523–524. doi: 10.1016/j.tim.2016.02.015. [DOI] [PubMed] [Google Scholar]
- Latorre J., Hernandez-Velasco X., Kallapura G., Menconi A., Pumford N.R., Morgan M.J., Layton S., Bielke L.R., Hargis B.M., Téllez G. Evaluation of germination, distribution, and persistence of Bacillus subtilis spores through the gastrointestinal tract of chickens. Poult, Sci. 2014;93:1793–1800. doi: 10.3382/ps.2013-03809. [DOI] [PubMed] [Google Scholar]
- Latorre J., Hernandez-Velasco X., Bielke L., Vicente J., Wolfenden R., Menconi A., Hargis B., Tellez G. Evaluation of a Bacillus direct-fed microbial candidate on digesta viscosity, bacterial translocation, microbiota composition and bone mineralisation in broiler chickens fed on a rye-based diet. Br. Poult. Sci. 2015;56:723–732. doi: 10.1080/00071668.2015.1101053. [DOI] [PubMed] [Google Scholar]
- Latorre J.D., Hernandez-Velasco X., Kuttappan V.A., Wolfenden R.E., Vicente J.L., Wolfenden A.D., Bielke L.R., Prado-Rebolledo O.F., Morales E., Hargis B.M., Tellez G. Selection of Bacillus spp. for cellulase and xylanase production as direct-fed microbials to reduce digesta viscosity and Clostridium perfringens proliferation using an in vitro digestive model in different poultry diets. Front. Vet. Sci. 2015;2:25. doi: 10.3389/fvets.2015.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latorre J.D., Hernandez-Velasco X., Wolfenden R.E., Vicente J.L., Wolfenden A.D., Menconi A., Bielke L.R., Hargis B.M., Tellez G. Evaluation and selection of Bacillus species Based on enzyme production, antimicrobial activity, and biofilm synthesis as direct-fed microbial candidates for poultry. Front. Vet. Sci. 2016;3:95. doi: 10.3389/fvets.2016.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Cao S., Zhang X. Modulation of gut microbiota-brain axis by probiotics, prebiotics, and diet. J. Agric. Food Chem. 2015;63:7885–7895. doi: 10.1021/acs.jafc.5b02404. [DOI] [PubMed] [Google Scholar]
- Lopetuso L.R., Scaldaferri F., Petito V., Gasbarrini A. Commensal clostridia: leading players in the maintenance of gut homeostasis. Gut Pathog. 2013;5:23. doi: 10.1186/1757-4749-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C., Lladser M.E., Knights D., Stombaugh J., Knight R. UniFrac: an effective distance metric for microbial community comparison. ISME J. 2011;5:169–172. doi: 10.1038/ismej.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyte M. Probiotics function mechanistically as delivery vehicles for neuroactive compounds: microbial endocrinology in the design and use of probiotics. Bioessays. 2011;33:574–581. doi: 10.1002/bies.201100024. [DOI] [PubMed] [Google Scholar]
- Martin R., Nauta A., Ben Amor K., Knippels L.M., Knol J., Garssen J. Early life: gut microbiota and immune development in infancy. Benef. Microbes. 2010;1:367–382. doi: 10.3920/BM2010.0027. [DOI] [PubMed] [Google Scholar]
- McFall-Ngai M. Adaptive immunity: care for the community. Nature. 2007;445:153. doi: 10.1038/445153a. [DOI] [PubMed] [Google Scholar]
- Meehan C.J., Beiko R.G. A phylogenomic view of ecological specialization in the Lachnospiraceae, a family of digestive tract-associated bacteria. Genome Biol. Evol. 2014;6:703–713. doi: 10.1093/gbe/evu050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minalla A. R., Dubrow R., Bousse L. J. 2001. Feasibility of high-resolution oligonucleotide separation on a microchip. Proc. SPIE 4560, Microfluidics and BioMEMS. San Francisco, CA. Micromachining and Microfabrication.
- Molinaro F., Paschetta E., Cassader M., Gambino R., Musso G. Probiotics, prebiotics, energy balance, and obesity: mechanistic insights and therapeutic implications. Gastroenterol. Clin. North Am. 2012;41:843–854. doi: 10.1016/j.gtc.2012.08.009. [DOI] [PubMed] [Google Scholar]
- Moran N.A. Symbiosis as an adaptive process and source of phenotypic complexity. Proc. Natl. Acad. Sci. U.S.A. 2007;104:8627–8633. doi: 10.1073/pnas.0611659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley B.B., Kogut M.H. Spatial and temporal changes in the broiler chicken cecal and fecal microbiomes and correlations of bacterial taxa with cytokine gene expression. Front. Vet. Sci. 2016;3:11. doi: 10.3389/fvets.2016.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onrust L., Ducatelle R., Van Driessche K., De Maesschalck C., Vermeulen K., Haesebrouck F., Eeckhaut V., Van Immerseel F. Steering endogenous butyrate production in the intestinal tract of broilers as a tool to improve gut health. Front. Vet. Sci. 2015;2:75. doi: 10.3389/fvets.2015.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V., Vanderplas J., Passos A., Cournapeau D., Brucher M., Perrot M., Duchesnay E. Scikit-learn: machine learning in python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- Plöger S., Stumpff F., Penner G.B., Schulzke J.D., Gäbel G., Martens H., Shen Z., Günzel D., Aschenbach J.R. Microbial butyrate and its role for barrier function in the gastrointestinal tract. Ann. N. Y. Acad. Sci. 2012;1258:52–59. doi: 10.1111/j.1749-6632.2012.06553.x. [DOI] [PubMed] [Google Scholar]
- Qin C., Gong L., Zhang X., Wang Y., Wang Y., Wang B., Li Y., Li W. Effect of Saccharomyces boulardii and Bacillus subtilis B10 on gut microbiota modulation in broilers. Anim. Nutr. 2018;4:358–366. doi: 10.1016/j.aninu.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAS Institute . SAS Institute Inc.; Cary, NC: 2002. SAS User Guide. Version 9.1. [Google Scholar]
- Schnabl B., Brenner D.A. Interactions between the intestinal microbiome and liver diseases. Gastroenterology. 2014;146:1513–1524. doi: 10.1053/j.gastro.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekirov I., Russell S.L., Antunes L.C.M., Finlay B.B. Gut microbiota in health and disease. Physiol. Rev. 2010;90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- Shannon C.E. A mathematical theory of communication. Bell Syst. Tech. J. 1948;27:379–423. [Google Scholar]
- Tao Y., Drabik K.A., Waypa T.S., Musch M.W., Alverdy J.C., Schneewind O., Chang E.B., Petrof E.O. Soluble factors from Lactobacillus GG activate MAPKs and induce cytoprotective heat shock proteins in intestinal epithelial cells. Am. J. Physiol. Cell Physiol. 2006;290:C1018–C1030. doi: 10.1152/ajpcell.00131.2005. [DOI] [PubMed] [Google Scholar]
- Teague K., Graham L., Dunn J., Cheng H., Anthony N., Latorre J., Menconi A., Wolfenden R., Wolfenden A., Mahaffey B., Baxter M., Hernandez -Velasco X., Merino-Guzman R., Bielke L., Hargis B., Tellez G. In ovo evaluation of FloraMax®-B11 on Marek' s disease HVT vaccine protective efficacy, hatchability, microbiota composition, morphometric analysis, and Salmonella Enteritidis infection in broiler chickens. Poult. Sci. 2017;96:2074–2082. doi: 10.3382/ps/pew494. [DOI] [PubMed] [Google Scholar]
- Tellez G., Higgins S., Donoghue A., Hargis B. Digestive physiology and the role of microorganisms. J. Appl. Poult. Res. 2006;15:136–144. [Google Scholar]
- Tellez G. Prokaryotes versus Eukaryotes: who is hosting whom? Front. Vet. Sci. 2014;1:3. doi: 10.3389/fvets.2014.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellez G., Latorre J.D., Kuttappan V.A., Hargis B.M., Hernandez-Velasco X. Rye affects bacterial translocation, intestinal viscosity, microbiota composition and bone mineralization in turkey poults. PLoS One. 2015;10 doi: 10.1371/journal.pone.0122390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tlaskalová-Hogenová H., Stěpánková R., Kozáková H., Hudcovic T., Vannucci L., Tučková L., Rossmann P., Hrnčíř T., Kverka M., Zákostelská Z., Klimešová K., Přibylová J., Bártová J., Sanchez D., Fundová P., Borovská D., Srůtková D., Zídek Z., Schwarzer M., Drastich P., Funda D.P. The role of gut microbiota (commensal bacteria) and the mucosal barrier in the pathogenesis of inflammatory and autoimmune diseases and cancer: contribution of germ-free and gnotobiotic animal models of human diseases. Cell. Mol. Immunol. 2011;8:110–120. doi: 10.1038/cmi.2010.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl S.M., Swisher J., McCartney-Francis N., Chen W. TGF-beta: the perpetrator of immune suppression by regulatory T cells and suicidal T cells. J. Leukoc. Biol. 2004;76:15–24. doi: 10.1189/jlb.1103539. [DOI] [PubMed] [Google Scholar]
- Wei S., Morrison M., Yu Z. Bacterial census of poultry intestinal microbiome. Poult. Sci. 2013;92:671–683. doi: 10.3382/ps.2012-02822. [DOI] [PubMed] [Google Scholar]
- White P.L., Baker A.R., James W.O. Strategies to control Salmonella and Campylobacter in raw poultry products. Rev. Sci. Tech. 1997;16:525–541. doi: 10.20506/rst.16.2.1046. [DOI] [PubMed] [Google Scholar]
- Wickham H. Springer; Houston, TX: 2016. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- Xie J., Vilchez I., Mateos M. Spiroplasma bacteria enhance survival of Drosophila hydei attacked by the parasitic wasp Leptopilina heterotoma. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Gordon J.I. Honor thy Symbionts. Proc. Nat. Acad. Sci. 2003;100:10452–10459. doi: 10.1073/pnas.1734063100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Liu S., Ding J., Dai R., He C., Xu K., Honaker C.F., Zhang Y., Siegel P., Meng H. Gut microbiota co-microevolution with selection for host humoral immunity. Front- Microbiol. 2017;8 doi: 10.3389/fmicb.2017.01243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q., Zhu L., Wang Z., Li P., Yang Q. Lactobacillus delbrueckii ssp. lactis R4 prevents Salmonella typhimurium SL1344-induced damage to tight junctions and adherens junctions. J. Microbiol. 2012;50:613–617. doi: 10.1007/s12275-012-1596-5. [DOI] [PubMed] [Google Scholar]
- Zar J. 2nd ed. Prentice Hall; Upper Saddle River, NJ: 1984. Biostatistical Analysis. [Google Scholar]
- Zhong Y., Teixeira C., Marungruang N., Sae-Lim W., Tareke E., Andersson R., Fåk F., Nyman M. Barley malt increases hindgut and portal butyric acid, modulates gene expression of gut tight junction proteins and Toll-like receptors in rats fed high-fat diets, but high advanced glycation end-products partially attenuate the effects. Food Funct. 2015;6:3165–3176. doi: 10.1039/c5fo00150a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.