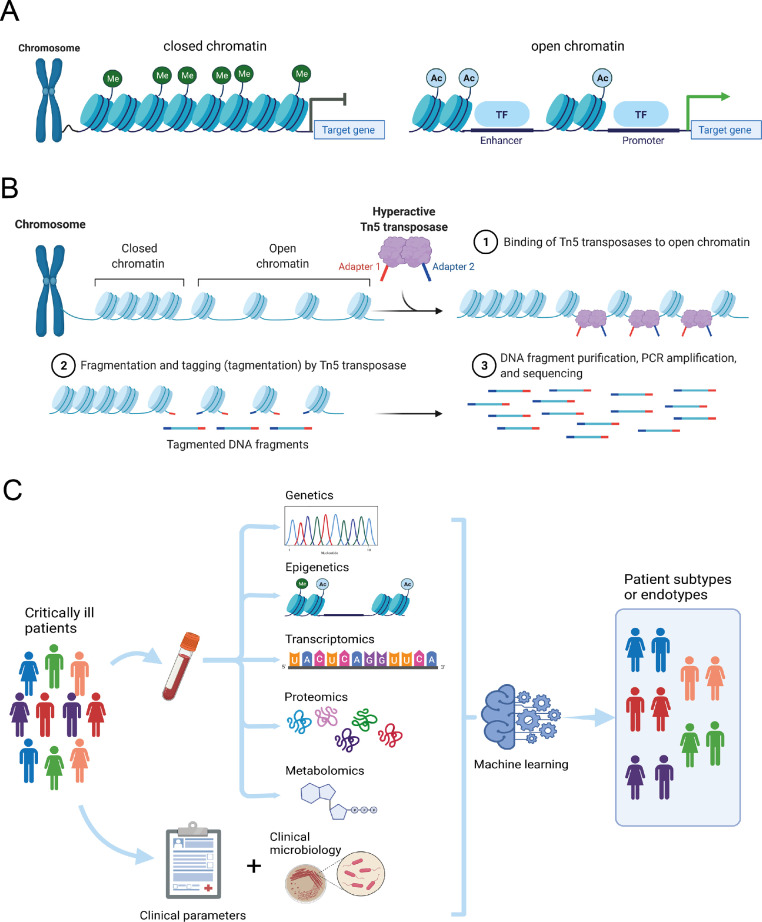
Environmental exposures, including microbes, toxic substances, and vaccines, can significantly impact the functional state of immune cells. It is now accepted that cell identity and functional states are governed by tightly regulated spatiotemporal gene expression programs. Establishing cell type identity depends largely on the coordinated distribution of numerous transcription factors that bind to accessible chromatin regions harbouring cis-regulatory elements, thereby activate or repress gene expression (Figure 1A).1 Therefore, identifying the accessible cis-elements in the genome of specific cell-types is an essential step towards a deeper understanding of health and disease. Considering accessible cis-regulatory elements are typically hypersensitive to nucleases or transposases (whether active or poised), the Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq)2 has been widely used to map chromatin accessibility patterns (Figure 1B). To overcome the limitations of complex tissue samples in resolving cell specific events, methods were recently developed for measuring chromatin accessibility at the level of single cells (scATAC-seq).
Figure 1.
Chromatin accessibility, sequencing and multi-scale modelling in critical illness. (A) Simplified diagrammatic representation of closed or open chromatin conformations that either silence or activate target gene expression by exposing transcriptional enhancer and promoter regions, respectively. Me, methylation; Ac, acetylation; TF, transcription factor. (B) The Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq) method involves Tn5 transposases binding to open chromatin regions, with subsequent tagmentation, fragment purification, amplification and sequencing. (C) Multi-scale modelling of diverse critically ill patient populations integrating different molecular strata, clinical parameters and microbiology in a computational method, e.g. machine learning, to stratify patients into more homogenous groups. Created with BioRender.com.
Transcriptional profiling of blood leukocytes obtained from critically ill patients due to severe trauma and burn injuries revealed substantial alterations in gene expression that were associated with extent of organ dysfunction and outcomes.3 Applying single cell RNA-sequencing to peripheral blood mononuclear cells (PBMCs) obtained from trauma patients, revealed gene expression patterns in myeloid cells (mainly monocytes) contributed markedly to departure from steady state.4 Notably, gene expression changes in key transcription factors could be attributed to the myeloid progenitor pool. Whether those trauma-induced gene expression changes reflected chromatin accessibility patterns was an open question. In this issue of eBioMedicine, Chen and colleagues addressed this question by testing chromatin accessibility using scATAC-seq in PBMCs obtained from trauma patients.5 As expected, gene expression alterations were largely concomitant with open chromatin conformations, which the authors stratified as either “focal” loci, associated with well-characterized inflammatory and immune suppressive gene sets, or “global” loci characterized by gene expression changes attuned to pathways unspecific to the immune response, including de-repression of Polycomb targets, suppression of genes involved in DNA repair and RNA processing. This “global epigenetic signature” was subsequently explored in large datasets of whole-blood leukocyte transcriptomes and the associated clinical outcomes from trauma, severe burns,3 and sepsis6,7 cohorts. Using a gene filtering bioinformatics strategy, the authors found that, firstly, combining the “global” epigenetic alterations (designated EG subtypes) with an inflammatory response transcriptomic signature (termed SG subtypes), which was previously derived by the same group, provided better resolution of patient heterogeneity in critical illness due to trauma than using either EG or SG subtypes in isolation. Moreover, and intriguingly, higher expression of genes associated with the “global” epigenetic group was associated with worse outcomes in critical illness due to burns or sepsis. The potential prognostic value of the epigenetic group subtypes was independent of transcriptional changes of genes involved in typical immune response pathways, including pro-, anti-inflammatory signalling, antigen presentation, and interferon signalling pathways, which were associated with transcriptomic SG subtypes in the group's previous study, and also defined by other investigators in sepsis utilizing blood transcriptomics. Though direct scATAC-seq measurements in independent trauma and sepsis patients will be needed to ascertain the robustness of the “global” epigenetic group to predict outcomes of critical illness, the author's findings raise interesting possibilities. In particular, chromatin accessibility signals at gene regions involved in key cell identity, development and maintenance, may represent a common theme in poor prognosis critical illness, that is not exclusive to trauma, burns, sepsis or any other aetiology of critical illness – a common sickness response. Overall, the authors are to be congratulated for providing a benchmark in chromatin accessibility dynamics in circulating mononuclear cells obtained from trauma patients, demonstrating that combining different molecular strata can provide more precision to patient stratification efforts. Whether adding epigenomic features into a critically ill patient subtyping strategy could assist in resolving patient heterogeneity will require direct investigations in larger patient cohorts.
Classification of critically ill patients as blood transcriptomic endotypes has been earmarked as an invaluable tool to identify patients who would benefit, or be harmed, from specific interventions. Several researchers have successfully used blood transcriptomes to identify patient subtypes with clinical implications. While myself, and other investigators, would argue that critical care medicine is in particular need for this new approach in patient stratification,8,9 the question whether molecular subtypes determined from the systemic compartment represent dominant immunopathologies versus epiphenomena remains outstanding. Going forward necessitates a more holistic approach by integrating different molecular strata, including genetic, epigenetic, transcriptomic, proteomic and metabolomic, as well as clinical parameters and microbiology (Fig. 1C), requiring multidisciplinary collaborations among various research groups, and validation in both available data sets, as well as new prospective multi-scale studies.
Declaration of interests
The author declares no conflict of interest.
Footnotes
Article type: Commentary on article entitled "The independent prognostic value of global epigenetic alterations: An analysis of single-cell ATAC-seq of circulating leukocytes from trauma patients followed by validation in whole blood leukocyte transcriptomes across three etiologies of critical illness ."
References
- 1.Shen Y., et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xiao W., et al. A genomic storm in critically injured humans. J Exp Med. 2011;208:2581–2590. doi: 10.1084/jem.20111354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen T., et al. A road map from single-cell transcriptome to patient classification for the immune response to trauma. JCI Insight. 2021 doi: 10.1172/jci.insight.145108. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen T., et al. The independent prognostic value of global epigenetic alterations: an analysis of single-cell ATAC-seq of circulating leukocytes from trauma patients followed by validation in whole blood leukocyte transcriptomes across three etiologies of critical illness. EBioMedicine. 2022 doi: 10.1016/j.ebiom.2022.103860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scicluna B.P., et al. Classification of patients with sepsis according to blood genomic endotype: a prospective cohort study. Lancet Respir Med. 2017;5:816–826. doi: 10.1016/S2213-2600(17)30294-1. [DOI] [PubMed] [Google Scholar]
- 7.Davenport E.E., et al. Genomic landscape of the individual host response and outcomes in sepsis: a prospective cohort study. Lancet Respir Med. 2016;4:259–271. doi: 10.1016/S2213-2600(16)00046-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeMerle K.M., et al. Sepsis subclasses: a framework for development and interpretation. Crit Care Med. 2021;49:748–759. doi: 10.1097/CCM.0000000000004842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matthay M.A., et al. Acute respiratory distress syndrome. Nat Rev Dis Prim. 2019;5:18. doi: 10.1038/s41572-019-0069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]