The increasing frequency of nosocomial infections due to methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococcus faecium (VRE) and the fear that high-level vancomycin resistance will eventually spread to staphylococci underscore the need for vigilance in the continuing war against pathogenic microbes (18, 39). Current widely used antibiotics are targeted at a surprisingly small number of vital cellular functions: cell wall, DNA, RNA, and protein biosynthesis (Table 1), and instances of resistance to these antibiotics are widespread and well documented (48). Thus, there is little doubt that new antibiotics are needed to combat the growing problem of antibiotic-resistant bacteria, and targeting of new pathways will likely play an important role in discovery of these new antibiotics. In fact, a number of crucial cellular pathways, such as secretion, cell division, and many metabolic functions, remain untargeted today. In the last 3 years, high-throughput automated random genomic DNA sequencing together with robust fragment assembly tools has delivered a wealth of genomic sequence information to assist in the search for new targets. In many cases, entire biochemical pathways can be reconstructed and compared in different pathogens. The purpose of this minireview is to indicate where this information can be found, to outline some of the ways in which it can be used, and to describe new tools to take advantage of genomic sequence information in the drug discovery process.
TABLE 1.
Gene targets of widely used antibiotics
| Target category and gene product | Antibiotic class |
|---|---|
| Protein synthesis | |
| 30S ribosomal subunit | Aminoglycosides, tetracyclines |
| 50S ribosomal subunit | Macrolides, chloramphenicol |
| tRNAIle synthetase | Mupirocin |
| Elongation factor G | Fusidic acid |
| Nucleic acid synthesis | |
| DNA gyrase A subunit; topo-isomerase IV | Quinolones |
| DNA gyrase B subunit | Novobiocin |
| RNA polymerase beta subunit | Rifampin |
| DNA | Metronidazole |
| Cell wall peptidoglycan synthesis | |
| Transpeptidases | Beta-lactams |
| d-Ala-d-Ala ligase substrate | Glycopeptides |
| Antimetabolites | |
| Dihydrofolate reductase | Trimethoprim |
| Dihydropteroate synthesis | Sulfonamides |
| Fatty acid synthesis | Isoniazid |
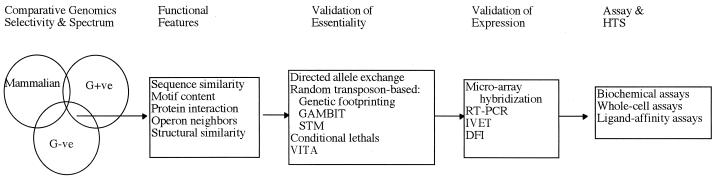
Each potential new antibiotic must meet a number of criteria before it is approved for use, and the choice of an appropriate target is the first step in this process. It is helpful to review the utility of genomic information with regard to some of the key criteria which antimicrobial targets must meet. In general, (i) a target should provide adequate selectivity and spectrum, yielding a drug which is specific or highly selective against the microbe with respect to the human host but also active against the desired spectrum of pathogens; (ii) a target should be essential for growth or viability of the pathogen, at least essential under conditions of infection; and (iii) something about the function of the target should be known so that assays and high-throughput screens can be built. Identification of potential new targets can proceed from any one of these criteria, but ultimately all must be met by a successful target. For example, a variety of methods may be used to find genes which are essential for the survival of an organism under defined conditions or which are necessary for infectivity in an animal model. Comparative genomics may be used to identify potential targets which are shared across multiple microbial species. Several tools, primarily sequence similarity based, may be used to predict the function of most genes so that specific pathways can be targeted. As discussed below, genomic sequence information provides assistance in all of these areas: selectivity, spectrum, functionality, and essentiality (Fig. 1).
FIG. 1.
Schematic view of genomic tools applied to antimicrobial-drug discovery. See the text for details. G+ve and G-ve, gram positive and gram negative, respectively.
CURRENT RESOURCES FOR GENOMIC SEQUENCE AND FUNCTIONALITY INFORMATION
Numerous databases are now available which contain both sequence and functionality information. Most of these are accessible over the Internet through convenient Web browser interfaces. Many also permit downloading of sequence information for use on local servers. Sequence databases now contain the nucleotide and predicted amino acid sequences of virtually every gene in the model microbes Escherichia coli, Bacillus subtilis, and Saccharomyces cerevisiae as well as in a variety of other bacteria (Table 2; a version of this table is updated regularly by The Institute for Genomic Research [TIGR] on their Web site: http://www.tigr.org/tdb/mdb/mdb.html). These databases are the result of extensive analysis of the genomic sequences of those organisms. Open reading frames have been analyzed by sequence comparison and by codon usage to identify those which are most likely to represent transcribed genes. Putative functions have been assigned to slightly more than half of the genes in the model organisms based on sequence comparisons to genes of known function in other organisms, shared sequence motifs, or clustering of sequences into related families. Databases such as EcoCyc, KEGG, and WIT present these data in an organized and useful manner (see Table 3).
TABLE 2.
Sequenced microbial genomes
| Internet resource | Genome | Strain(s) | Size (Mb) | Institution(s) | Reference |
|---|---|---|---|---|---|
| www.tigr.org/tdb/mdb/hidb/hidb.html | Haemophilus influenzae RD | KW20 | 1.83 | TIGR | 13 |
| www.tigr.org/tdb/mdb/mgdb/mgdb.html | Mycoplasma genitalium | G-37 | 0.58 | TIGR | 15 |
| www.tigr.org/tdb/mdb/mjdb/mjdb.html | Methanococcus jannaschii | DSM 2661 | 1.66 | TIGR | 8 |
| www.kazusa.or.jp/cyano/cyano.html | Synechocystis sp. | PCC 6803 | 3.57 | Kazusa DNA Research Institute | 27 |
| www.zmbh.uni-heidelberg.de/M_pneumoniae/ MP_Home.html | Mycoplasma pneumoniae | M129 | 0.81 | University of Heidelberg | 23 |
| speedy.mips.biochem.mpg.de/mips/yeast/yeast_genome.htmlx or genome-www.stanford.edu/Saccharomyces | Saccharomyces cerevisiae | S288C | 13 | European and North American Consortium | 17 |
| www.tigr.org/tdb/mdb/hpdb/hpdb.html | Helicobacter pylori | 26695 | 1.66 | TIGR | 51 |
| www.genetics.wisc.edu/ | Escherichia coli | K-12 | 4.6 | University of Wisconsin | 7 |
| www.genomecorp.com/gene/sequences/ methanobacter/abstract.html | Methanobacterium thermo-autotrophicum | delta H | 1.75 | Genome Therapeutics and Ohio State University | 43 |
| www.pasteur.fr/Bio/SubtiList.html | Bacillus subtilis | 168 | 4.2 | International Consortium | 31 |
| www.tigr.org/tdb/mdb/afdb/afdb.html | Archaeoglobus fulgidus | VC-16, DSM4304 | 2.18 | TIGR | 29 |
| www.tigr.org/tdb/mdb/bbdb/bbdb.html | Borrelia burgdorferi | B31 | 1.44 | TIGR | 14 |
| www.ncbi.nlm.nih.gov/cgi-bin/Entrez/ framik?db=Genome&gi=133 | Aquifex aeolicus | VF5 | 1.55 | Diversa | 10 |
| www.bio.nite.go.jp/ot3db_index.html | Pyrococcus horikoshii | OT3 | 1.80 | National Institute of Technology and Evaluation | 28 |
| www.sanger.ac.uk/Projects/M_tuberculosis/ | Mycobacterium tuberculosis | H37Rv | 4.40 | Sanger Centre | 9 |
| www.tigr.org/tdb/mdb/tpdb/tpdb.html | Treponema pallidum | Nichols | 1.14 | TIGR and University of Texas | 16 |
| chlamydia-www.berkeley.edu:4231/ | Chlamydia trachomatis | Serovar D (D/UW3/Cx) | 1.05 | University of California at Berke-ley and Stanford University | 46 |
| evolution.bmc.uu.se/∼siv/gnomics/Rickettsia.html | Rickettsia prowazekii | Madrid E | 1.11 | University of Uppsala | 4 |
| www.genomecorp.com/hpylori or www.astra-boston.com/hpylori | Helicobacter pylori | J99 | 1.64 | Genome Therapeutics and Astra AB | 3 |
| www.tigr.org/cig-bin/BlastSearch/blast .cgi?organism=m_tuberculosis | Mycobacterium tuberculosis | CSU#93 | 4.40 | TIGR | Unpublished |
TABLE 3.
Additional Internet resources
Recently, some commercial databases have also become available for nonexclusive use by commercial subscribers. These databases generally also provide sequence information not available in public databases and comparative software and analysis tools for convenient analysis of the data. For example, the results of prerun sequence similarity searches may be stored to provide rapid answers to complex comparative genomic queries by a subscriber. Finally, several Web-accessible sites offer useful tools for sequence analysis via sequence similarity searches, motif searches, and structural comparisons. Examples of relevant Internet sites providing databases of sequence and functionality information and research tools are described in Table 3.
The next advance in microbial genomics will be the availability of the complete genomic sequence from multiple strains of a single bacterial pathogen. The discovery of genes conserved in multiple pathogenic strains or the recognition of genes found only in the most virulent strains are examples of the power such genomic comparisons will provide. Sequence for a second strain of Helicobacter pylori has appeared and sequence for a second strain of Mycobacterium tuberculosis will appear soon (Table 2).
COMPARATIVE GENOMICS TO ASSESS THE SPECTRUM AND SELECTIVITY OF A TARGET
One powerful use of genomic sequence information is to compare all of the identified genes in different bacterial pathogens to determine which genes are, or are not, shared by various species. Indeed, Tatusov et al. (50) have suggested that gene families conserved among bacteria but missing from eukaryotes comprise a pool of potential targets for broad-spectrum antibiotic development. An early step in this direction was taken by Mushegian and Koonin (36), who identified 256 genes shared by the two completely sequenced bacterial genomes at that time, those of Haemophilus influenzae and Mycoplasma genitalium. On the other hand, genes which are apparently unique to a species such as H. pylori might be ideal for targeting that species with a narrow-spectrum antibiotic. As the number of sequenced bacterial and fungal genomes grows, so does the ability to find genes common to most microbial pathogens or truly unique to a particular species. For example, Arigoni et al. (6) identified 26 genes in E. coli, most of which were conserved in the B. subtilis, M. genitalium, H. influenzae, H. pylori, Streptococcus pneumoniae, and Borrelia burgdorferi genomes. They reasoned that this list of genes, which had no predictable function, contained novel targets for broad-spectrum antibiotic development. These analyses can be extended by including sequence comparisons to eukaryotic genomes as a means to examine potential selectivity of a target (50). For example, Arigoni et al. (6) reported that 15 of 26 proteins broadly conserved across bacterial species also exhibited significant sequence similarity to proteins in S. cerevisiae and, therefore, represented targets which, in an assay, might identify compounds that also have human toxicity. While these targets could simply be avoided, it should be noted that the targets of the majority of marketed antimicrobial agents show some conservation with mammalian proteins.
As in all sequence comparisons, the search parameters and the quality of the input data, e.g., partial human or mammalian sequence information, are critical. Relevant issues which must be addressed include questions such as the following. What degree of sequence similarity to another bacterial genome indicates a shared gene? What degree of sequence similarity to a mammalian gene warns of a possible toxicity problem? Since sequence similarity-searching algorithms allow nearly complete flexibility in the choice of these parameters, some known examples are necessary to calibrate the method. Mushegian and Koonin (36) used a BLASTP score of 90 as the cutoff for defining a biologically relevant relationship between two protein sequences. The appropriate cutoff score for exclusion of genes with apparent mammalian homologs may be more gene specific. Some examples reveal a general trend. Trimethoprim is a highly selective inhibitor of bacterial dihydrofolate reductases (DHFR) despite the fact that the human and E. coli DHFR gene products share 28% amino acid identity over the length of the two proteins (40). Similarly, the quinolones are highly selective against bacterial gyrases despite the fact that the C-terminal domain of human topoisomerase II shares 20% amino acid identity with E. coli gyrase A (25). Fluconazoles are highly selective for fungal lanosterol 14-α demethylases, even though the human and yeast gene products share 37% amino acid identity over their full length (5). These sequence identity percentages translate into BLASTP scores of 132, 125, and 301, respectively, in a search of a large nonredundant protein database comprised of sequences from GenBank, SwissProt, and PIR. Therefore, exclusion of genes having apparent mammalian homologs with scores >150 would likely be suitable for a search of bacterial targets, but the score cutoff would have to be raised to allow identification of the broadest set of antifungal target genes.
IDENTIFICATION OF ESSENTIAL TARGETS EXPERIMENTALLY
Genomic sequence information is not required for discovering essential genes, but such information does facilitate the process. Genes which are essential to pathogenesis and prevent colony formation in a conditional-lethal manner are potential targets for new antimicrobials. This assumes that a small organic molecule which inhibits the activity of an essential gene product would either kill or inhibit the growth of the bacterium which requires that functional protein. Such conditional lethal genes can be discovered through classical mutagenesis techniques. Availability of the sequence of the genome means that the full sequence of each mutated gene, and frequently its cellular role as well, can be gleaned from a short sequence read on a complementing plasmid insert. This additional information accelerates the processing of a mutational study enormously. Depending on the availability of genetic tools for the microbial species in question, a variety of molecular genetic methods can be used to discover essential genes. For example, in E. coli, genes can be placed under control of a regulated promoter by use of an appropriately constructed transposon system (11), or genes can be mutated to a conditional-lethal form. In principle, such conditional mutants can be used in whole-cell screens under moderately suppressing conditions in which the cells may be hypersensitive to drug-like compounds which act against that gene product (see below).
It seems reasonable to assume that most genes which are essential to the cell for growth or viability on laboratory media will also be required for growth or viability in an infected host. Experimentally, media can be varied in order to identify genes which are essential under the widest range of growth conditions and particularly in rich media which may simulate conditions in necrotic tissue of an animal host. Cells carrying auxotrophic mutations may find sufficient nutritional supplement in the host tissues to permit growth or at least survival. Such genes might be poor targets for new antimicrobials unless experiments establish that the particular nutrient is in short supply in the host or that cells are incapable of transporting the nutrient efficiently. In order to establish that a gene target is essential in an infection, a transposon-based gene tagging method called “signature-tagged mutagenesis” (STM) has been used to identify genes which are essential in an animal model (22, 35). However, since cells carrying the disrupted tagged genes must be grown in the laboratory prior to introduction into the animal, the method may be biased against genes which are essential for growth both on laboratory media and in an animal model. Indeed, many of the genes identified by STM appear to encode virulence factors which affect the ability of the pathogen to colonize or damage host tissue rather than the viability of the pathogen. New drugs which intervene in these processes could prove highly selective, and resistance to such drugs might be rare since loss or mutation of the virulence factor would also likely reduce virulence. However, other resistance mechanisms, such as drug modification and efflux pumps, could be problematic. In addition, the absence of a convenient in vitro assay for such drugs would hamper the development, testing, and approval processes. It remains unclear how many important antimicrobial targets would be missed by using as targets for drug discovery only those genes which are essential for growth or viability on laboratory culture media.
A related, important feature of a suitable antimicrobial gene target is its expression pattern in the infection. The absolute level of expression may be less important than information about whether it is expressed at all. A highly expressed, abundant gene product should be no more difficult to inhibit than a low-abundance gene product since an inhibitor with suitably high affinity will be effective in either case unless it is poorly taken up by pathogens. However, if a gene is not expressed at all in an established infection of an animal host, then it will be of no interest as a potential target. A gene already established as being essential for growth or viability in the laboratory by genetic methods obviously must be expressed under these conditions because its failure to be expressed as an active product causes the pathogen to die. Knowledge that such an essential gene is also expressed in an animal model would suggest that it is essential in an infection as well. Two types of methods offer information about gene expression. First, for genes whose sequence is known, reverse transcriptase PCR (RT-PCR) may be used to detect transcripts in cells grown on agar media or in animal infection models (47). Alternatively, for organisms which have been sequenced in their entirety, a whole-genome view of gene expression may be obtained by gridding clones, PCR products, or synthetic oligonucleotides representing every gene onto a solid support. Total RNA may be isolated from cells grown under conditions of interest, labeled, and hybridized to the array (12). While thorough, this type of method suffers from some problems: (i) appropriate controls must be run to eliminate the possibility of bacterial DNA contamination in the RNA preparation, (ii) probes are difficult to prepare because bacterial mRNA is notoriously unstable, and (iii) the whole-genomic scale of the experiments makes the arrayed membranes difficult and expensive to prepare and read. A genetic promoter trap method termed “in vivo expression technology” or IVET may be more feasible for most laboratories (21, 33). In this approach, which has been developed for use in Salmonella typhimurium grown intraperitoneally in BALB/c mice or in cultured macrophages, random DNA fragments are cloned upstream from a gene whose expression is required for growth in an animal host. Cells, which multiply in vivo, are recovered and cloned. The sequences of fragments serving as functional promoters in vivo are then determined. A second, related promoter trap method termed “differential fluorescence induction” (DFI) has been described recently (53). The distinguishing features of this approach are that (i) the gene used for selection encodes a modified green fluorescent protein and (ii) the selection is accomplished with a fluorescence-activated cell sorter. If such methods can be extended to other bacterial species and animal hosts, they will be extremely useful for assessing random genomic fragments or specific genes of interest for expression in vivo.
IDENTIFICATION OF ESSENTIAL TARGETS USING DATABASES
Potential gene targets selected from databases can be validated by examining the effect of a gene knockout on cell growth or viability. Recombination is almost exclusively between homologous regions in bacterial genomes, and many common pathogens as well as model bacteria are transformable. Exchange between the chromosomal wild-type allele and a version engineered to carry a deletion and/or an insertion of a drug resistance cassette is generally efficient enough to be practical in the laboratory. Interpreting the results of such an experiment, however, may be difficult for two reasons. First, the frequent occurrence of polycistronic messages in bacteria means that disruption of a gene may have a deleterious effect on expression of a distal neighboring gene, a so-called “polar” effect. In that case, the inviability caused by a gene knockout could be due to loss of expression of a gene other than the one disrupted. Precautions can be taken to reduce these effects by, for example, including a moderate-strength outward reading promoter in the disrupted version of the allele so as to permit expression of the downstream gene(s). Second, the method works better as an exclusionary tool than as an inclusionary one. While success in generating a cell carrying a disrupted allele indicates that the gene is not essential for growth or viability of the cell, failure to generate such an altered cell could be due to any one of multiple causes including polar effects or inefficient recombination in a particular genetic interval.
One solution to this problem is to carry out allele exchange as a two-step process (20, 32). In E. coli, for example, the disrupted allele together with the vector carrying it can be integrated into the genome by means of a single crossover, a so-called “Campbell insertion.” Recombination between homologous regions on the two copies of the allele now on the chromosome will eliminate the vector sequences and either copy of the allele. Which copy is eliminated depends upon which regions of homology were involved in the recombination. Failure to find cells retaining only the disrupted allele strongly suggests that such progeny are inviable. Success in finding cells retaining only the wild-type allele confirms that recombination is efficient in this genetic interval. However, in many naturally competent bacterial species, such as H. influenzae and S. pneumoniae, double-crossover events are extremely efficient, and allele replacement occurs with little or no opportunity to isolate a single crossover intermediate (1). While this complicates evaluation of essential genes in these organisms, it provides a convenient method for disrupting genes under conditions in which they are not essential so that the resulting strains may be examined under a variety of other conditions (e.g., see below).
A new approach promises to accelerate the process of evaluating the essentiality of genes. Smith et al. (44, 45) have described a method for the yeast S. cerevisiae called “genetic footprinting” which makes use of a quasi-random transposable Ty element to generate a rich array of gene knockouts in a population of cells. Further transposition is shut off, and the population is then grown under a variety of conditions. DNA is prepared from cells in the various growth populations, and the DNA is queried by PCR amplification to determine if it will yield PCR products between a gene-specific primer and a transposon-specific primer. Failure to find such PCR products suggests that cells carrying transposons in that gene were inviable under the growth conditions employed. Fluorescent PCR products are viewed on standard sequencing gels by using automated fluorescence sequencing machines and a commercially available software package. An important control in this method is the existence of a gene-to-transposon PCR product in the so-called t0 cell population prior to the shutdown of transposition. This assures the experimenter that this region is not simply a “cold” spot for transposition. The efficiency of this method derives from the use of random transposons to build all necessary gene knockouts rapidly, followed by automated PCR and analysis methods to interpret the results for any given gene of interest.
Recently, a modified version of this method, called “genomic analysis and mapping by in vitro transposition” (GAMBIT), has been applied successfully to two bacterial species (1). In this variation of genetic footprinting, the transposition mutagenesis was done on PCR-amplified genomic segments from H. influenzae or S. pneumoniae in vitro, and the mutations were introduced into these naturally competent host bacteria by transformation. While the method suffers from the absence of a true t0, the focus on 10-kb DNA segments permits near-saturation mutagenesis with the mariner family transposon Himar1, which shows little or no insertion site specificity. These authors identified four essential conserved genes of unknown function from a total of 13 analyzed.
Currently, the main limitation to this method is a requirement for an efficiently transformable host bacterium so that mutations generated in vitro can be evaluated readily in vivo. Other limitations which apply to all genetic footprinting methods include the following: (i) essentiality of the function of a gene that is duplicated or has a functional paralog cannot be analyzed, since footprinting assesses the fitness of a single mutagenized gene; (ii) polarity effects, although not a problem for S. cerevisiae, may lead to misinterpretation of data obtained from bacteria; (iii) the correlation of footprinting data with gene knockout data has not been confirmed in any organism; and (iv) footprinting data are technically difficult to interpret for a variety of reasons, including the facts that some essential genes will tolerate insertions in the C-terminal coding region (e.g., secA [1]) and cells carrying insertions in some genes display an intermediate slow-growth phenotype (e.g., ade2 [44]).
TOOLS FOR PREDICTING THE FUNCTION OF GENE PRODUCTS
Clearly, not all of the predicted functional assignments based on sequence similarities are reliable. In some cases, for example, the function of the closest-related protein has itself been predicted based on its sequence similarity to a gene product of known function. In other cases, the chain of relatedness to a protein of confirmed function may be even longer. About half of the genes in bacterial genomes either lack significant enough sequence similarity to permit functional assignment or have likely homologs whose function is unknown. In neither of these cases can a function be predicted for the gene product. Nevertheless, the results of sequence similarity searches are a useful starting point for further investigation. More sensitive sequence comparison searches may provide a putative function or functional feature such as the presence of a short protein sequence motif. For example, a search against a database of clusters of orthologous groups of genes (COGs [Table 3]) yielded over 100 additional functional predictions for genes in the H. pylori genome (50).
Tools other than sequence similarity have also been useful in a few cases for predicting function of a gene product. For example, a gene product, with no significant sequence relationship to a protein of known function but which is likely to be cotranscribed as part of a polycistronic message with other genes of known function, may play a role in the same pathway with the known gene products. In the E. coli genome, the hypothetical gene yjaF appears to be cotranscribed with the porphyrin biosynthetic gene hemE, and the hypothetical gene yadM appears to be in an operon with the outer-membrane usher protein HtrE, which is involved in transport and binding. It is reasonable to speculate that these genes of unknown function play roles in the same biochemical pathways as their neighboring “known” genes. Of course, experimental evidence would be required to confirm these hypotheses. Methods also exist for identifying likely structural similarity even in the absence of strong primary sequence similarity. As the databases of known structures grow, this will become a powerful approach for assigning likely functions to gene products. For example, the “GenTHREADER” web site (Table 3) presents analysis results from a fast fold recognition program on the predicted open reading frames from three bacterial genomes.
Laboratory methods can also be invoked to solve questions of unknown gene identities. An unknown gene may be used as the bait in a yeast two-hybrid interaction trap to identify genes whose protein products interact with the unknown protein. The identity of an interacting partner will frequently implicate the unknown in a particular cellular pathway (19). Finally, an unknown gene may be expressed as a tagged fusion, the protein purified by affinity column, and the product tested for categories of activities such as proteolysis, DNA cleavage or binding, ATP or GTP hydrolysis, and binding, to name a few. The probability of successfully identifying an activity of an unknown by the latter method is low, but this method may be warranted if sequence comparisons suggest the presence of a motif associated with an assayable function. An attractive alternative is to focus on assays which do not require knowledge of the cellular function of a gene product (see below).
THE FUTURE: DEALING WITH GENE TARGETS HAVING NO PREDICTABLE FUNCTIONAL FEATURES
The array of tools described so far, including comparative genomic methods for identifying potentially useful gene targets and allele exchange methods for validating the essentiality of those genes, provides both gene targets whose cellular function can be predicted and gene targets for which little or no functional information is available. Targets in the first class may be used immediately to build biochemical assays and high-throughput screens to detect small organic molecules which inhibit the biochemical activity. Typically, the gene sequence is amplified by PCR from genomic DNA of a given bacterium, inserted into an expression vector, and expressed in E. coli sometimes with affinity tags to facilitate purification of the resulting protein product.
It is far less obvious how to proceed with gene targets lacking any functional information. This problem has attracted considerable attention in recent years because of the growing number of such targets known to be shared across many bacterial species (24), some of which are known to be essential in at least one species. As a general guide, about 40% of bacterial genes cannot be assigned a putative function at this time. If 10 to 15% of these genes are essential, then 4 to 6% of the genes in a typical bacterial genome (about 100 genes) represent potential antimicrobial targets which have never been used in screens. Three basic types of approaches seem feasible and have shared some initial success. First, cells expressing higher- or lower-than-normal levels of particular genes have in some cases been shown to be more resistant or more sensitive, respectively, than their wild-type parents to chemical compounds known to inhibit those gene products. For example, overexpression of the yeast ALG7 gene results in cells more resistant than wild-type cells to tunicamycin (38), while reduced activity of the same gene product results in cells more sensitive to the drug (30). Similarly, increased expression of the ERG11 gene in Candida glabrata results in higher levels of resistance to the azole family of drugs which target that enzyme (54). A gene of unknown function could be overexpressed in a host strain, and the resulting assay strain could be tested for increased resistance to a library of compounds. It is clear, however, that many gene targets when overexpressed do not lead to resistance to chemical compounds that are known to bind to the protein product (e.g., gyrA [52]). Furthermore, overexpression of proteins often leads to lethality or growth defects (e.g., kasA [34]). Alternatively, a gene could be underexpressed or crippled by a mutation so that cells might show increased sensitivity to a compound which inhibits the protein product. Scientists at Microcide Pharmaceuticals, Inc., have applied this approach on a large scale using temperature-sensitive mutants grown at intermediate temperatures in order to reduce the level of activity of the target gene product (39a). Of course, it is not clear what fraction of unknown gene products would provide the cell with increased drug resistance or sensitivity when over- or underexpressed in these ways.
The second approach to this problem of assaying gene products of unknown function is probably more generally applicable. Libraries of small molecules are screened for strong binding affinity to proteins of unknown function. This has been achieved with peptides in phage display libraries because binding can be readily detected by elution of bound phage from the protein tethered on a solid support. Proteins of unknown function can be produced easily as affinity fusion products for attachment to solid supports, and a variety of peptide phage display libraries are commercially available. Conformationally constrained disulfide-bonded peptides with affinities in the 100 μM to 100 nM range can be obtained by this approach (55). Of course, not all peptides detected by this approach will bind to sites which inhibit activity, but an elegant new method, called “validation in vivo of targets for anti-infectives” (VITA), has been devised to identify those peptides which inhibit essential cellular functions (49). Potential inhibitory peptides were expressed in a regulated manner within bacterial host cells which were grown either on agar medium or in an animal model of infection. Inhibition of cell growth or viability upon induction of peptide expression validated the peptide-protein interaction as useful for further drug development. While peptides are not ideal drug candidates, a wider array of techniques are applicable after a moderate binder has been obtained. The peptide may be used as a surrogate ligand in a competition assay to identify a small organic compound with higher affinity. Scintillation proximity assays (26) or fluorescence polarization assays (41) may be used in a high-throughput mode to identify compounds in chemical libraries which compete for binding with a labeled peptide. Alternatively, ligand binding assays may be configured to work directly on libraries of unlabeled chemical compounds. Shuker et al. (42) have described a nuclear magnetic resonance-based method capable of a throughput of 1,000 compounds per day. Mass spectrometric methods are also of interest as potentially rapid ways to detect bound ligands from chemical libraries. One concern about these approaches is that proteins may have multiple accessible binding sites, many of which have nothing to do with catalytic activity. It is not clear at this early stage how significant an issue multiple binding sites will be. However, it is worth noting that Shuker et al. (42) took advantage of a second binding site to increase the affinity of an inhibitor for the protein. Ultimately, of course, affinity ligands must be shown to inhibit cell growth, that is, to have antimicrobial activity. Some chemical engineering of the compound may be required to increase microbial uptake.
A third approach for assaying gene products of unknown function relies on the complex gene expression regulatory network found in many bacteria. Expression levels of genes in metabolic pathways are often regulated in response to the amounts of intermediates in the cell. For example, disruption of the general secretory pathway in E. coli by mutation results in dramatic up-regulation of secA gene expression (37). Alksne et al. (2) took advantage of this fact to build a strain of E. coli carrying a secA-lacZ fusion as a detectable reporter. Several synthetic compounds and natural products were identified by their ability to induce expression of the reporter. Many of these exhibited antimicrobial activity and reduced the secretion of Staphylococcus aureus toxin 1. Similarly, Mdluli et al. (34) have reported that sublethal concentrations of isoniazid lead to upregulation of the kasA and acpM genes. This group has initiated a whole-cell, high-throughput screen of chemical compounds which induce expression of a luciferase reporter fused to a gene in this regulated pathway. Screens of this type, which take advantage of the bacterial gene regulatory network, are inherently less specific than the two other types described here. In addition, they suffer from the basic limitation of all whole-cell screens: compounds must be capable of entering the cell in order to be detected. However, these types of screens offer the potential advantage of identifying compounds which act at any of several points in a pathway.
CONCLUSIONS
The availability of genomic sequence information for all or nearly all of several different bacterial species provides important new advantages for target discovery. First, it permits use of a comparative genomic analysis to identify potential new targets shared across several bacterial species or particular to a single species. In this manner, it is possible to generate lists of genes which represent potential targets for broad-spectrum or highly focused narrow-spectrum antibiotics. Sequence comparisons can also provide some assurance against mammalian toxicity if proteins of similar sequence do not exist in mammalian sequence databases. Second, sequence similarity provides some insights into putative functions for most gene products. Finally, availability of the entire sequence of the gene target of interest permits rapid construction of gene knockouts to validate the utility of the target and facile construction of expression plasmids for production of protein and development of assays. The fact that bacterial and fungal genes can be assessed rapidly for their relevance as potential antibiotic targets by determining the effect of knocking out the gene and the fact that their genomes are small enough to be sequenced in their entirety are compelling reasons that the field of genomics will likely find its first real utility in the development of new antimicrobials.
ACKNOWLEDGMENTS
We thank our colleagues at Genome Therapeutics Corporation and the Schering-Plough Research Institute for helpful discussions about genomic approaches to drug discovery. In particular, Skip Shimer, Brad Guild, and Lucy Ling were instrumental in the analysis of the approaches summarized here. We thank Douglas Smith of Genome Therapeutics Corporation for the compilation of Internet resources presented in Table 3.
REFERENCES
- 1.Akerley B J, Rubin E J, Camilli A, Lampe D J, Robertson H M, Mekalanos J J. Systematic identification of essential genes by in vitro mariner mutagenesis. Proc Natl Acad Sci USA. 1998;95:8927–8932. doi: 10.1073/pnas.95.15.8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alksne L E, Burgio P, Bradford P, Feld B, Hu W, Labthavikul P, McGlynn M, Petersen P J, Tuckman M, Projan S. Abstracts of the 38th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1998. Identification of inhibitors of bacterial secretion by using a SecA reporter system; p. 272. [Google Scholar]
- 3.Alm R A, Ling L L, Moir D T, King B L, Brown E D, Doig P C, Smith D R, Noonan B, Guild B C, deJonge B L, Carmel G, Tummino P J, Caruso A, Uria-Nickelsen M, Mills D M, Ives C, Gibson R, Merberg D, Mills S D, Jiang Q, Taylor D E, Vovis G F, Trust T J. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 4.Andersson S G E, Zomorodipour A, Andersson J O, Sicheritz-Ponten T, Alsmark U C M, Podowski R M, Naeslund A K, Eriksson A-S, Winkler H H, Kurland C G. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 5.Aoki Y, Yoshihara F, Kondoh M, Nakamura Y, Nakayama N, Arisawa M. Ro 09-1470 is a selective inhibitor of P-450 lanosterol C-14 demethylase of fungi. Antimicrob Agents Chemother. 1993;37:2662–2667. doi: 10.1128/aac.37.12.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arigoni F, Talabot F, Peitsch M, Edgerton M D, Meldrum E, Allet E, Fish R, Jamotte T, Curchod M-L, Loferer H. A genome-based approach for the identification of essential bacterial genes. Nat Biotechnol. 1998;16:851–856. doi: 10.1038/nbt0998-851. [DOI] [PubMed] [Google Scholar]
- 7.Blattner F R, Plunkett G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 8.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 9.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail M A, Rajandream M-A, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston J E, Taylor K, Whitehead S, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 10.Deckert G, Warren P V, Gaasterland T, Young W G, Lenox A L, Graham D E, Overbeek R, Snead M A, Keller M, Aujay M, Huberk R, Feldman R A, Short J M, Olsen G J, Swanson R V. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;393:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 11.de Lorenzo V, Eltis L, Kessler B, Timmis K N. Analysis of Pseudomonas gene products using lacIq/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene. 1993;123:17–24. doi: 10.1016/0378-1119(93)90533-9. [DOI] [PubMed] [Google Scholar]
- 12.DeRisi J L, Iyer V R, Brown P O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 13.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, McKenney K, Sutton G, FitzHugh W, Fields C, Gocayne J D, Scott J, Shirley R, Liu L, Glodek A, Kelley J M, Weldman J F, Phillips C A, Spriggs T, Hedblom E, Cotton M D, Utterback T R, Hanna M C, Nguyen D T, Saudek D M, Brandon R C, Fine L D, Fritchman J L, Fuhmann J L, Geoghagen N S M, Gnehm C L, McDonald L A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 14.Fraser C M, Casjens S, Huang W M, Sutton G G, Clayton R, Lathigra R, White O, Ketchum K A, Dodson R, Hickey E K, Gwinn M, Dougherty B, Tomb J-F, Fleischmann R D, Richardson D, Peterson J, Kerlavage A R, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams M D, Gocayne J, Weidman J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fujii C, Cotton M D, Horst K, Roberts K, Hatch B, Smith H O, Venter J C. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 15.Fraser C M, Gocayne J D, White O, Adams M D, Clayton R A, Fleischmann R D, Bult C J, Kerlavage A R, Sutton G, Kelley J M, Fritchman J L, Weidman J F, Small K V, Sandusky M, Fuhrmann J, Nguyen D, Utterback T R, Saudek D M, Phillips C A, Merrick J M, Tomb J-F, Daugherty B A, Bott K F, Hu P-C, Lucier T S, Peterson S N, Smith H O, Hutchison III C A, Venter J C. The minimal gene complement of Mycoplasma genitalium. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 16.Fraser C M, Norris S J, Weinstock G M, White O, Sutton G G, Dodson R, Gwinn M, Hickey E K, Clayton R, Ketchum K A, Sodergren E, Hardham J M, McLeod M P, Salzberg S, Peterson J, Khalak H, Richardson D, Howell J K, Chidambaram M, Utterback T, McDonald L, Artiach P, Bowman C, Cotton M D, Venter J C, et al. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science. 1998;281:375–388. doi: 10.1126/science.281.5375.375. [DOI] [PubMed] [Google Scholar]
- 17.Goffeau A, Barrell B G, Bussey H, Davis R W, Dujon B, Feldmann H, Galibert F, Hoheisel J D, Jacq C, Johnston M, Louis E J, Mewes H W, Murakami Y, Philippsen P, Tettelin H, Oliver S G. Life with 6000 genes. Science. 1996;274:546–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 18.Gold H S, Moellering R C. Antimicrobial-drug resistance. N Engl J Med. 1996;335:1445–1453. doi: 10.1056/NEJM199611073351907. [DOI] [PubMed] [Google Scholar]
- 19.Gyuris J, Golemis E, Chertkov H, Brent R. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 20.Hamilton C M, Aldea M, Washburn B K, Babitzke P, Ku S R. New method for generating deletions and gene replacements in Escherichia coli. J Bacteriol. 1989;171:4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heithoff D M, Conner C P, Hanna P C, Julio S M, Hentschel U, Mahan M J. Bacterial infection as assessed by in vivo gene expression. Proc Natl Acad Sci USA. 1997;94:934–939. doi: 10.1073/pnas.94.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hensel M, Shea J E, Gleeson C, Jones M D, Dalton E, Holden D W. Simultaneous identification of bacterial virulence genes by negative selection. Science. 1995;269:400–403. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- 23.Himmelreich R, Hilbert H, Plagens H, Pirkl E, Li B C, Herrmann R. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 1996;24:4420–4449. doi: 10.1093/nar/24.22.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hinton J C D. The Escherichia coli genome sequence: the end of an era or the start of the FUN? Mol Microbiol. 1997;26:417–422. doi: 10.1046/j.1365-2958.1997.6371988.x. [DOI] [PubMed] [Google Scholar]
- 25.Hoshino K, Sato K, Une T, Osada Y. Inhibitory effects of quinolones on DNA gyrase of Escherichia coli and topoisomerase II of fetal calf thymus. Antimicrob Agents Chemother. 1989;33:1816–1818. doi: 10.1128/aac.33.10.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jenh C H, Zhang M, Wiekowski M, Tan J C, Fan X D, Hegde V, Patel M, Bryant R, Narula S K, Zavodny P J, Chou C C. Development of a CD28 receptor binding-based screen and identification of a biologically active inhibitor. Anal Biochem. 1998;256:47–55. doi: 10.1006/abio.1997.2495. [DOI] [PubMed] [Google Scholar]
- 27.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 28.Kawarabayasi Y, Sawada M, Horikawa H, Haikawa Y, Hino Y, Yamamoto S, Sekine M, Baba S, Kosugi H, Hosoyama A, Nagai Y, Sakai M, Ogura K, Otsuka R, Nakazawa H, Takamiya M, Ohfuku Y, Funahashi T, Tanaka T, Kudoh Y, Yamazaki J, Kushida N, Oguchi A, Aoki K, Kikuchi H. Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 1998;5(Suppl.):147–155. doi: 10.1093/dnares/5.2.147. [DOI] [PubMed] [Google Scholar]
- 29.Klenk H-P, Clayton R A, Tomb J-F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, Richardson D L, Kerlavage A R, Graham D E, Kyrpides N C, Fleischmann R D, Quackenbush J, Lee N H, Sutton G G, Gill S, Kirkness E F, Dougherty B A, McKenney K, Adams M D, Loftus B, Peterson S, Reich C I, McNeil L K, Badger J H, Glodek A, Zhou L, Overbeek R, Gocayne J D, Weidman J F, McDonald L, Utterback T, Cotton M D, Spriggs T, Artiach P, Kaine B P, Sykes S M, Sadow P W, D’Andrea K P, Bowman C, Fujii C, Garland S A, Mason T M, Olsen G J, Fraser C M, Smith H O, Woese C R, Venter J C. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 30.Kukuruzinska M A, Lennon K. Diminished activity of the first N-glycosylation enzyme, dolichol-P-dependent N-acetylglucosamine-1-P transferase (GPT), gives rise to mutant phenotypes in yeast. Biochim Biophys Acta. 1995;1247:51–59. doi: 10.1016/0167-4838(94)00201-q. [DOI] [PubMed] [Google Scholar]
- 31.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessières P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S-K, Codani J-J, Connerton I F, Cummings N J, Daniel R A, Denizot F, Devine K M, Düsterhöft A, Ehrlich S D, Emmerson P T, Entian K D, Errington J, Fabret C, Ferrari E, Foulger D, Fritz C, Fujita M, Fujita Y, Fuma S, Galizzi A, Galleron N, Ghim S-Y, Glaser P, Goffeau A, Golightly E J, Grandi G, Guiseppi G, Guy B J, Haga K, Haiech J, et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 32.Link A J, Phillips D, Church G M. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J Bacteriol. 1997;179:6228–6237. doi: 10.1128/jb.179.20.6228-6237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahan M J, Tobias J W, Slauch J M, Hanna P C, Collier R J, Mekalanos J J. Antibiotic-based selection for bacterial genes that are specifically induced during infection of a host. Proc Natl Acad Sci USA. 1995;92:669–673. doi: 10.1073/pnas.92.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mdluli K, Slayden R A, Zhu Y-Q, Ramaswamy S, Pan X, Mead D, Crane D D, Musser J M, Barry C E. Inhibition of a Mycobacterium tuberculosis β-ketoacyl ACP synthase by isoniazid. Science. 1998;280:1607–1610. doi: 10.1126/science.280.5369.1607. [DOI] [PubMed] [Google Scholar]
- 35.Mei J M, Nourbakhsh F, Ford C W, Holden D W. Identification of Staphylococcus aureus virulence genes in a murine model of bacteraemia using signature-tagged mutagenesis. Mol Microbiol. 1997;26:399–407. doi: 10.1046/j.1365-2958.1997.5911966.x. [DOI] [PubMed] [Google Scholar]
- 36.Mushegian A R, Koonin E V. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc Natl Acad Sci USA. 1996;93:10268–10273. doi: 10.1073/pnas.93.19.10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Riggs P D, Derman A I, Beckwith J. A mutation affecting the regulation of a secA-lacZ fusion defines a new sec gene. Genetics. 1988;118:571–579. doi: 10.1093/genetics/118.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rine J. Gene overexpression in studies of Saccharomyces cerevisiae. Methods Enzymol. 1991;194:239–251. doi: 10.1016/0076-6879(91)94019-9. [DOI] [PubMed] [Google Scholar]
- 39.Salyers A A, Amabile-Cuevas C F. Why are antibiotic resistance genes so resistant to elimination? Antimicrob Agents Chemother. 1997;41:2321–2325. doi: 10.1128/aac.41.11.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39a.Schmid, M. Personal communication.
- 40.Schweitzer B I, Dicker A P, Bertino J R. Dihydrofolate reductase as a therapeutic target. FASEB J. 1990;4:2441–2452. doi: 10.1096/fasebj.4.8.2185970. [DOI] [PubMed] [Google Scholar]
- 41.Seethala R, Menzel R. A homogeneous, fluorescence polarization assay for src-family tyrosine kinases. Anal Biochem. 1997;253:210–218. doi: 10.1006/abio.1997.2365. [DOI] [PubMed] [Google Scholar]
- 42.Shuker S B, Hajduk P J, Meadows R P, Fesick S W. Discovering high-affinity ligands for proteins: SAR by NMR. Science. 1996;274:1531–1534. doi: 10.1126/science.274.5292.1531. [DOI] [PubMed] [Google Scholar]
- 43.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicaire R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Safer H, Patwell D, Prabhakar S, McDougall S, Shimer G, Goyal A, Pietrokovski S, Church G M, Daniels C J, Mao J-i, Rice P, Noelling J, Reeve J N. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith V, Botstein D, Brown P O. Genetic footprinting: a genomic strategy for determining a gene’s function given its sequence. Proc Natl Acad Sci USA. 1995;92:6479–6483. doi: 10.1073/pnas.92.14.6479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith V, Chou K N, Lashkari D, Botstein D, Brown P O. Functional analysis of the genes of yeast chromosome V by genetic footprinting. Science. 1996;274:2069–2074. doi: 10.1126/science.274.5295.2069. [DOI] [PubMed] [Google Scholar]
- 46.Stephens R S, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov R L, Zhao Q, Koonin E V, Davis R W. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 47.Swartley J S, Liu L-J, Miller Y K, Martin L E, Edupuganti S, Stephens D S. Characterization of the gene cassette required for biosynthesis of the (α1→6)-linked N-acetyl-d-mannosamine-1-phosphate capsule of serogroup A Neisseria meningitidis. J Bacteriol. 1998;180:1533–1539. doi: 10.1128/jb.180.6.1533-1539.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swartz M N. Hospital-acquired infections: diseases with increasingly limited therapies. Proc Natl Acad Sci USA. 1994;91:2420–2427. doi: 10.1073/pnas.91.7.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tao J, Li T, Connelly G, Shen X, Silverman J, Houman F, Wendler P, Tally F P. Abstracts of the 38th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1998. VITA: validation in vivo of targets and assays for anti-infectives; p. 274. [Google Scholar]
- 50.Tatusov R L, Koonin E V, Lipman D J. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 51.Tomb J-F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenney K, Fitzegerald L M, Lee N, Adams M D, Hickey E K, Berg D E, Gocayne J D, Utterback T R, Peterson J D, Kelley J M, Cotton M D, Weidman J M, Fujii C, Bowman C, Watthey L, Wallin E, Hayes W S, Borodovsky M, Karp P D, Smith H O, Fraser C M, Venter J C. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–542. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 52.Truong Q C, Nguyen Van J C, Shlaes D, Gutmann L, Moreau N J. A novel, double mutation in DNA gyrase A of Escherichia coli conferring resistance to quinolone antibiotics. Antimicrob Agents Chemother. 1997;41:85–90. doi: 10.1128/aac.41.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Valdivia R H, Falkow S. Fluorescence-based isolation of bacterial genes expressed within host cells. Science. 1997;277:2007–2011. doi: 10.1126/science.277.5334.2007. [DOI] [PubMed] [Google Scholar]
- 54.Vandenbossche H, Marichal P, Odds F C, Lejeune L, Coene M C. Characterization of an azole-resistant Candida glabrata isolate. Antimicrob Agents Chemother. 1992;36:2602–2610. doi: 10.1128/aac.36.12.2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wrighton N C, Farrell F X, Chang R, Kashyap A K, Barbone F P, Mulcahy L S, Johnson D L, Barrett R W, Jolliffe L K, Dover W J. Small peptides as potent mimetics of the protein hormone erythropoietin. Science. 1996;273:458–463. doi: 10.1126/science.273.5274.458. [DOI] [PubMed] [Google Scholar]