Abstract
Introduction
Prognosis of uveal melanoma (UM) is assessed using clinical staging or molecular testing. Two modalities often used for prognostication are the American Joint Committee on Cancer (AJCC) staging and a tumor gene expression profile (GEP), the outcomes of which are often discordant. This article discusses a total risk score created to combine the discordant information from both sources.
Methods
A retrospective case series was conducted of all patients presenting with UM over 6 years to 2 referral centers. Each tumor was classified using the AJCC and the GEP. A total risk score was calculated for each patient using results from both AJCC and GEP. Kaplan-Meier analysis of metastasis-free survival was used to compare groups.
Results
A total of 294 patients were included in the study. Kaplan-Meier estimates showed significant curve separation between individual AJCC and GEP risk groups. The combined total risk score provided an accurate estimate of prognosis that incorporated results from both AJCC and GEP.
Conclusions
Clinical staging and molecular prognostication of UM can be discordant. There is important information provided by each system that is not provided by the other. The total risk score provides a simple method to combine information from both AJCC stage and the GEP class in order to provide patients and care teams with a more complete understanding of metastatic risk.
Keywords: Uveal melanoma, Choroidal melanoma, Gene expression profile, Cancer staging, American Joint Committee on cancer, Prognostication, Prognosis
Introduction
Uveal melanoma (UM) is a rare, aggressive primary intraocular tumor. Despite excellent local control of the primary tumor, the 15-year mortality rate of UM patients is almost 50% [1]. As metastases are rarely detectable at the time of diagnosis, great efforts have been directed toward accurate prognostication and identifying high-risk factors for metastasis. For this purpose, several prognostic tests and classification systems have been developed [2, 3, 4]. The American Joint Committee on Cancer (AJCC) tumor, node, metastasis classification is a clinical cancer staging system used across all medical specialties. The AJCC categorization of melanoma involves measurement of tumor size and determination of its anatomic location. The AJCC staging has been shown to correlate with metastasis and death in UM [5]. A gene expression profile (GEP) is a molecular prognostic test using a PCR-based 15-gene test and classifies melanoma into low- or intermediate-risk class 1 (1A and 1B) and high-risk class 2 for metastatic disease [3, 6].
Prognostic information such as the GEP has been shown to alter the surveillance strategies and referral patterns of physicians caring for patients with UM [7]. While it has been shown that higher risk GEP class and higher risk AJCC cancer stage are often correlated at presentation [8], the prognostic outcomes of the 2 entities are not always in agreement. When discordant, it becomes challenging to determine an appropriate surveillance strategy. It has been shown that prognostic accuracy can be improved when the AJCC stage is combined with molecular studies such as alterations in chromosome 3 or 8 [9]. It has also been shown that the largest basal diameter (LBD) of the tumor adds prognostic value when added to the GEP [10, 11]. Recently, Roelofs et al. [12] compared prognostic models combining GEP with various variables including LBD and AJCC classification and showed that a combination of GEP and the LBD was a simple method to include both molecular and clinical prognostic data. In clinical practice at our institutions, AJCC staging and GEP classification are discussed with all patients at the point of care. These variables are also readily understood and often used by colleagues in radiation oncology and medical oncology. We previously published that the combination of GEP classification and AJCC stages enhances the prognostication of each system [13]. The goal of this study was to create a simple prognostic tool that is easy to understand, easy to calculate, and easily combines the discordant information from both GEP molecular status and the AJCC staging so that physicians and patients can better understand prognosis.
Methods
This was a retrospective analysis of patients diagnosed with UM isolated to the choroid who were diagnosed from July 2010 through January 2016 at the Ocular Oncology Clinic at W. K. Kellogg Eye Center at the University of Michigan or at the Ophthalmic Oncology Program at Yale University. Approval for the study was obtained through the Institutional Review Board at both hospitals (Yale Human Investigation Committee #1501015205, Michagan IRB #HUM00046408). All patients were diagnosed with choroidal melanoma by 2 ocular oncologists (H.D. and M.M.) based on history, ophthalmic examination and imaging, or diagnostic fine-needle aspiration biopsy when needed. All patients underwent transscleral or transvitreal fine-needle aspiration biopsy for GEP testing. This fine-needle aspiration biopsy was performed either prior to placement of I-125 plaque radiotherapy or at the time of enucleation. Patients were excluded if they had iris melanomas or iridociliary body melanomas.
Data collected included age at diagnosis, sex, the absence or presence of ciliary body involvement or extraocular extension, LBD (mm), tumor thickness (mm), treatment modality (plaque radiotherapy or enucleation), GEP class (1A, 1B or 2), status of metastasis, and follow-up period. The LBD (mm) was estimated based on fundus mapping with indirect ophthalmoscopy and, when possible, by ocular transillumination. Tumor thickness (mm) was measured using B-scan ultrasonography with the calipers extending from just under the retina to the base of the tumor at the inner sclera. Determination of ciliary body involvement was made by clinical examination and ocular transillumination and was confirmed by B-scan ultrasonography and ultrasound biomicroscopy.
The AJCC 8th edition was used, and results were categorized into low risk: stage I (cT1a), intermediate risk: stage II (cT1b–d, cT2a–b, and cT3a), and high risk: stage III (cT2c–d, cT3b–d, and cT4). Patients presenting with stage IV disease at presentation (nodal or metastatic disease) were excluded. Patients were also placed into risk categories based on the following GEP results: low risk (class 1A), intermediate risk (1B), and high risk (class 2). As previous studies have shown that an increasing AJCC stage or increasing GEP class is correlated with higher risk of metastases, and because these scores are already calculated in many patients and understood by many medical specialties, a model was developed to incorporate both of these 2 scores into 1 entity. A risk score was created by applying a point system to the AJCC and GEP result for each patient. For both AJCC and GEP, 1 point was assigned for low risk, 2 points for intermediate risk, and 3 points for high risk (Table 1). The total risk score was calculated by adding the 2 risk scores together. A patient with GEP class 1A and AJCC stage I would have a total risk score of 2. A patient with GEP class 2 and AJCC stage IIIA would have a total risk score of 6.
Table 1.
Total risk score is calculated by adding the points assessed from both AJCC and GEP results for each patient
| Risk category | AJCC | GEP | Points assessed |
|---|---|---|---|
| Low risk | Stage I | Class 1A | 1 |
| Intermediate risk | Stage IIA and IIB | Class 1B | 2 |
| High risk | Stage IIIA, IIIB, and IIIC | Class 2 | 3 |
GEP, gene expression profile; AJCC, American Joint Committee on Cancer.
All patients also underwent complete systemic evaluation regularly at 3-month to yearly intervals with liver ultrasonography or computed tomography or magnetic resonance imaging of the liver. If medical records were not available, the systemic status was updated by contacting the patient or primary care physician. The cause of death was determined by review of medical records and by contacting the patient's family. Statistical analysis was completed using the R Statistical Environment. Kaplan-Meier survival estimates were used to predict and compare survival between groups. Log-rank tests were used to compare Kaplan-Meier curves. p < 0.05 was considered to be statistically significant. A Cox proportional hazard model was created with the final model of total risk scores, and the fit of the model was tested using Bayesian Information Criteria (BIC) and Akaike Information Criteria (AIC).
Results
We identified 296 patients with choroidal melanoma. GEP testing was available in 294 of 296 patients (99%). There were 135 females (46%) and 158 males (54%). The mean patient age at the time of diagnosis was 62 years (SD = 14.5, range 15–93 years). The mean tumor thickness and diameter were 5.1 mm (SD = 3.1) and 12.4 mm (SD = 3.9), respectively. AJCC tumor, node, metastasis staging showed 82 patients (27%) in the low-risk (stage I) group, 178 patients (61%) in the intermediate-risk (stage II) group, and 33 patients (12%) in the high-risk (stage III) group. No patients presented with stage IV disease. GEP testing resulted in 132 patients (45%) with class 1A, 63 patients (22%) with class 1B, and 98 patients (33%) with class 2 status. After a mean follow-up time of 2.81 years (SD = 17.4), 33 (11%) patients developed metastasis with a median time to metastasis of 14 months.
The number of patients in low-risk, intermediate-risk, and high-risk groups as determined by both prognostication tools is presented in Table 2. The concordance of risk levels was low with only 36% of patients (105 of 291), demonstrating agreement between classification systems. Overall, 31% of GEP class 1A patients were also AJCC stage I, 61% of GEP class 1B patients were also AJCC stage II, and 27% of GEP class 2 patients were also AJCC stage III.
Table 2.
Concordance of prognostication results of all patients in the study
| GEP class 1A | GEP class 1B | GEP class 2 | |
|---|---|---|---|
| AJCC stage I | 41 | 20 | 21 |
| AJCC stage II | 89 | 38 | 50 |
| AJCC stage III | 2 | 4 | 26 |
GEP, gene expression profile; AJCC, American Joint Committee on Cancer.
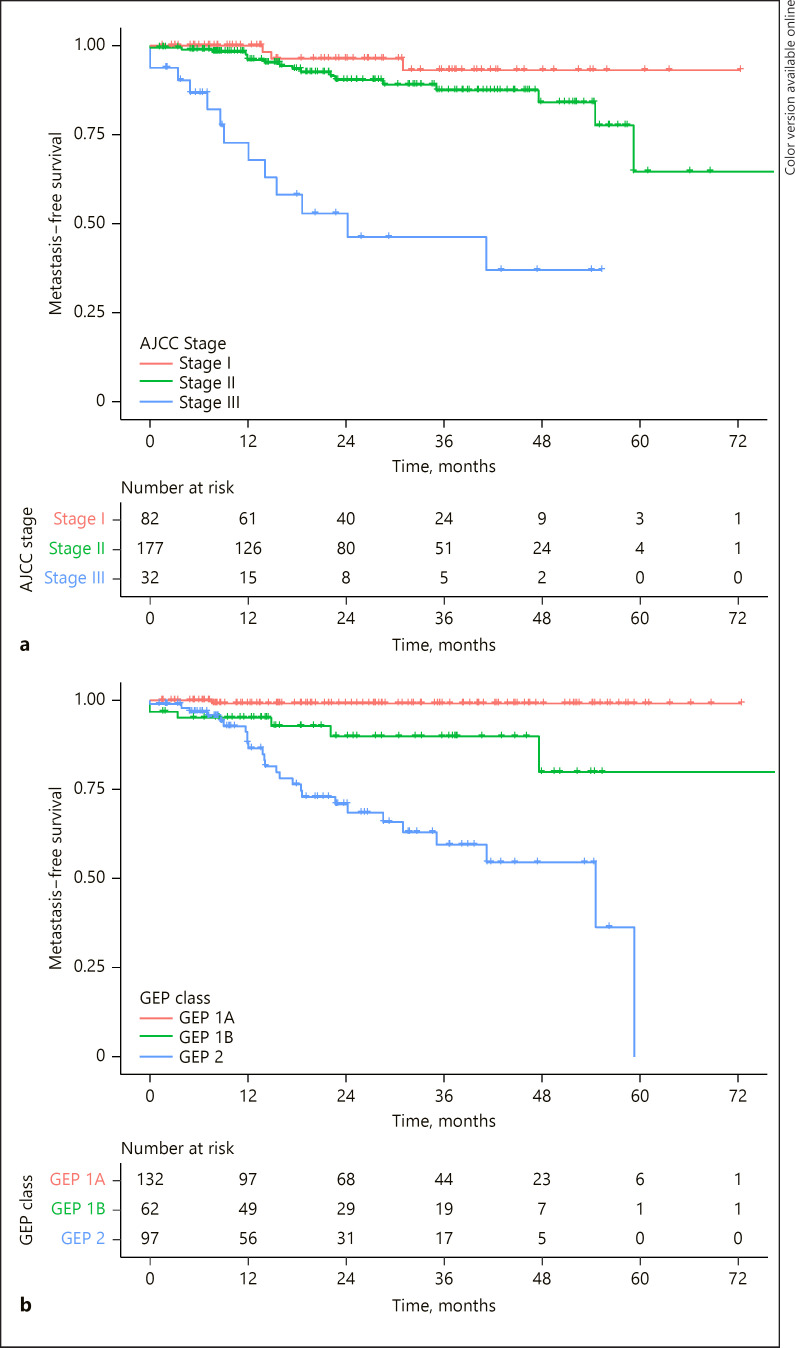
Kaplan-Meier survival curves were used to estimate metastasis-free survival. The 2 prognostication systems performed well independently. The low-, intermediate-, and high-risk groups showed increasing rates of metastasis and demonstrated statistically significant differences in estimated metastasis-free survival between groups within the same prognostic tool (Fig. 1). The comparison of AJCC stage I metastasis-free survival compared to AJCC stage II metastasis-free survival resulted in a p value = 0.09. Otherwise, all pairwise comparisons with the AJCC groups or within the GEP groups resulted in p value <0.01.
Fig. 1.
Kaplan-Meier survival estimates including patients based on their AJCC clinical cancer stage (a) and for patients based on their GEP results (b). GEP, gene expression profile; AJCC, American Joint Committee on Cancer.
Despite the discordance between outcomes in the AJCC and GEP, the risk classifications demonstrated very similar survival estimates. The Kaplan-Meier survival estimates of the 2 low-risk classification (GEP class 1A vs. AJCC stage I) showed no difference between groups (long-rank test, p = 0.20). The same was true for intermediate-risk classifications (GEP 1B vs. AJCC stage II, long-rank test, p = 0.98). There was a statistically significant difference between high-risk classifications (GEP class 2, vs. AJCC stage 3, long-rank test, p = 0.03). While the 2 systems identified different patient cohorts as low, intermediate, and high risk, the different cohorts appeared to have similar prognostic results.
The discordance within the 2 classifications is further evidenced by comparing the survival of patients in a GEP classification based on their AJCC stage. For this purpose, we looked at GEP class 2 patients as there were >20 patients in each AJCC stage within GEP class 2. Within the GEP class 2 group, the survival estimates were very different based on the AJCC classification. For patients with GEP class 2 and AJCC stage I, the Kaplan-Meier estimate of the 3-year survival was 79% (95% confidence interval: 56–100%). For patients with GEP class 2 and AJCC stage II, the 3-year survival estimate was 61% (44–85%). For patients with GEP class 2 and AJCC stage III, the survival estimate was 39% (2–75%). Patients with GEP class 2 and AJCC stage III had significantly worse survival than patients with GEP class 2 and AJCC stage I or II (p = 0.01).
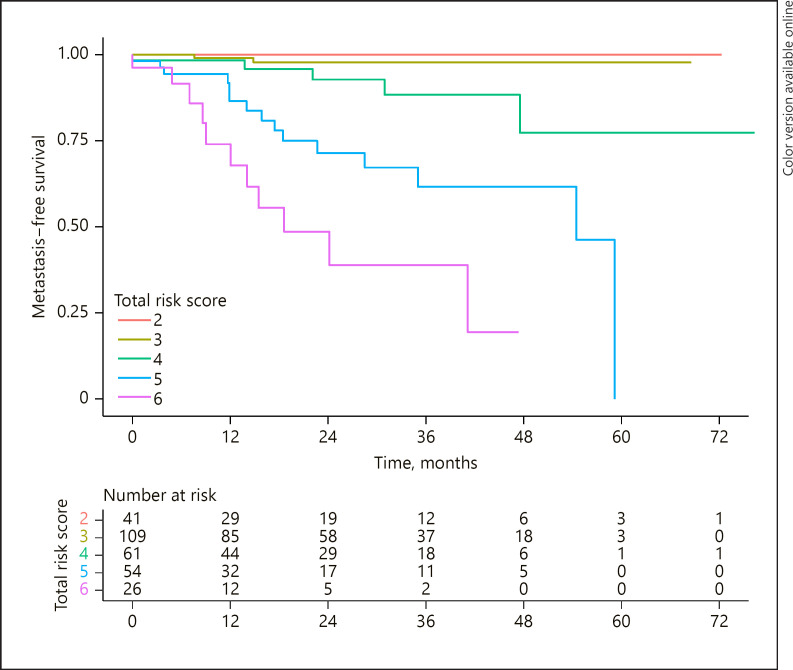
The total risk score was calculated for each patient based on their individual AJCC and GEP results. A total of 291 patients had both AJCC and GEP results that could be combined to form a final total risk score. A Kaplan-Meier survival estimate was calculated for each total -risk-score group (2, 3, 4, 5, and 6). The number of patients, the number of metastases in each group, and the 3-year survival estimate of each group are demonstrated in Table 3. The plotted survival curves demonstrate that as the total risk score increases, the metastasis-free survival decreases (Fig. 2). There was no statistical difference between the survival curves in group 2 versus group 3 (log-rank test, p = 0.40). All other curves were statistically different from all other curves (p < 0.05 in each pairwise comparison). A Cox proportional hazard model was fit using the GEP data only, the AJCC stage data only, and the total risk score of the combined data. The resulting AIC was better for the total risk score model (271) than the model using only the GEP (AIC = 297) and the model using only the AJCC stage (AIC = 320). Similar results were seen when calculating the BIC: total risk score (BIC = 272), GEP (BIC = 298), and AJCC (BIC = 321.8).
Table 3.
The total risk score accurately represents the complete patient prognosis
| Total risk score | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|
| Patients, n (% of total) | 41 (14) | 109 (37) | 61 (21) | 54 (19) | 26 (9) |
| Metastases, n (% of group) | 0 (0) | 2 (2) | 5 (8) | 15 (28) | 11 (42) |
| 3-Year K-M metastasis-free survival, % | 100 | 98 | 88 | 62 | 39 |
| (95% confidence interval) | (100–100) | (95–100) | (78–100) | (46–83) | (20–76) |
Fig. 2.
Kaplan-Meier survival estimates for patients in each total risk score. There was no statistical difference in these data between the survival curves of group 2 versus group 3. All other survival curves were statistically different.
Discussion
Several prognostication methods have been used in UM. AJCC classification is based on the clinical features of UM including the thickness and diameter of UM, involvement of ciliary body, and extraocular extension. Using AJCC classification 7th edition, Shields and associates reviewed 7,731 patients and reported Kaplan-Meier estimates of metastasis of 5% for stage I, 16% for stage II, and 44% for stage III at 5 years [5]. Similar findings were reported in other studies [14]. Monosomy of chromosome 3 and alterations in chromosome 8 have also been shown to predict mortality in patients with UM [15, 16]. Similar studies using the GEP have shown similar survival results: the time-related probability of metastasis at 3 years was 1% for class 1A, 10% for class 1B, and 40% for class 2 UMs [14]. While gene expression profiling has been shown to be an independent prognostic tool [3, 6], it has also been reported that clinical factors such as the basal diameter of the melanoma can add additional prognostic information beyond the GEP [10, 11, 13].
There is a documented discordance between clinical cancer staging and molecular prognostic results [8]. We observed that 80% of AJCC stage III melanomas were GEP class 2 and half of AJCC stage I and II melanomas were GEP class 1A. However, overall, there was relatively low concordance between the GEP class and AJCC staging. Despite the discordance, both systems provide reliable prognostic forecasting independent of each other, suggesting that there must be prognostic information unique to each system which is not included in the other system. Recent data support this idea, showing that the clinical cancer staging prognosis can be improved upon with additional of molecular data [14, 15]. It is important, if possible, that patients are evaluated for both a clinical staging prognosis as well as a molecular tumor prognosis.
Discerning future prognosis when prognostic tools are discordant can be challenging. The AJCC stage and the GEP result are 2 pieces of information available to many ocular oncologists, patients, and oncology care teams. They are also discussed at the point of care in many situations. In this study, we have developed a simple scoring system, the total risk score, that seamlessly combines the prognostic information from these 2 entities. The scoring system requires a basic calculation and can be done easily in clinic. The results show an increasing rate of metastasis for increases in the total risk score. If patients are at low risk for both AJCC as well as the GEP (total risk score = 2), their overall risk is very low. However, the risk of metastatic disease increases with subsequent increases in the total risk score. In our study, we did not see a significant difference between a total risk score of 2 and 3. However, there were no metastases in the former group and multiple metastases in the latter group. Previous studies have shown that there is a difference between survival in AJCC stage I versus stage II and also in GEP class 1A versus class 1B; therefore, we expect that in larger studies, the difference between the total risk score of 2 and 3 will be significant.
The scoring system becomes very useful for patients with discordant results. A class 2 GEP result will have a very different prognosis if the tumor is low, intermediate, or high risk in the AJCC classification. In this analysis, the molecular prognostic test used was the GEP. This research was completed prior to the availability of next-generation sequencing, which may provide additional precision to the total risk score. It is likely that other molecular prognostic tests can be adapted to the total risk score.
This study is limited by its small size, limited follow-up in some patients, and retrospective paradigm. However, despite its small size, the survival curves for the total-risk-score groups demonstrated statistically significant differences, signifying that the total risk score accurately stratifies patients. The total risk score estimates a more precise prognosis than either the AJCC or GEP prognosis alone, highlighted by the improved fit of the model using the total risk score compared to the AJCC or GEP alone. If only one of the prognostic tools is used, a large number of patients with discordant prognostic data may be considered higher or lower risk than their true risk. The total risk score is easy to calculate at the point of care, easy to understand, and precisely estimates total metastatic risk. It can be used as a method of risk stratification to more precisely tailor systemic surveillance after primary treatment of choroidal melanoma as well as to identify high-risk patients who may benefit from adjuvant therapies or clinical trials.
Statement of Ethics
The study protocol was approved by the Institutional Review Boards of the University of Michigan Medical School and Yale University Medical School and follows the tenants set forth by the Declaration of Helsinki. Individual consent was not obtained for this retrospective analysis as it is not required by local guidelines.
Conflict of Interest Statement
A.W.S. is a member of the advisory board of Immunocore. V.S.D. has no disclosures. M.M. is a member of the advisory board of Castle Biosciences, a speaker for Carl Zeiss Meditec, and a consultant for Astra Zeneca. H.D. is a member of the advisory boards of Castle Biosciences and Immunocore.
Funding Sources
A.W.S. was supported in part by an unrestricted grant from Research to Prevent Blindness to the Ophthalmology Department at the University of Washington.
Author Contributions
A.W.S. analyzed the data and wrote the manuscript; V.S.D. collected the data and wrote the manuscript; M.M. conceived of the study, collected data, and edited the manuscript; and H.D. conceived of the study, collected data, and edited the manuscript.
Data Availability Statement
The data that support the findings in the manuscript contain dates and variables that are identifiable and are not available publicly. Requests for de-identified summary data can be made to the authors.
References
- 1.Kujala E, Mäkitie T, Kivelä T. Very long-term prognosis of patients with malignant uveal melanoma. Invest Ophthalmol Vis Sci. 2003 Nov;44((11)):4651–4659. doi: 10.1167/iovs.03-0538. [DOI] [PubMed] [Google Scholar]
- 2.Damato B, Eleuteri A, Taktak AF, Coupland SE. Estimating prognosis for survival after treatment of choroidal melanoma. Prog Retin Eye Res. 2011 Sep;30((5)):285–295. doi: 10.1016/j.preteyeres.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 3.Onken MD, Worley LA, Ehlers JP, Harbour JW. Gene expression profiling in uveal melanoma reveals two molecular classes and predicts metastatic death. Cancer Res. 2004 Oct;64((20)):7205–7209. doi: 10.1158/0008-5472.CAN-04-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amin MB, Edge S, Greene F, Byrd DR, Brookland RK, Washington MK, et al. 8th ed. Springer International Publishing: American Joint Commission on Cance; 2017. AJCC cancer staging manual. [Google Scholar]
- 5.Shields CL, Kaliki S, Furuta M, Fulco E, Alarcon C, Shields JA. American Joint Committee on cancer classification of uveal melanoma (anatomic stage) predicts prognosis in 7731 patients: the 2013 Zimmerman lecture. Ophthalmology. 2015 Jun;122((6)):1180–1186. doi: 10.1016/j.ophtha.2015.01.026. [DOI] [PubMed] [Google Scholar]
- 6.Onken MD, Worley LA, Char DH, Augsburger JJ, Correa ZM, Nudleman E, et al. Collaborative Ocular Oncology Group report number 1: prospective validation of a multi-gene prognostic assay in uveal melanoma. Ophthalmology. 2012 Aug;119((8)):1596–1603. doi: 10.1016/j.ophtha.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plasseraud KM, Cook RW, Tsai T, Shildkrot Y, Middlebrook B, Maetzold D, et al. Clinical performance and management outcomes with the decisionDx-UM gene expression profile test in a prospective multicenter study. J Oncol. 2016;2016:5325762. doi: 10.1155/2016/5325762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berry DE, Schefler AC, Seider MI, Materin M, Stinnett S, Mruthyunjaya P, et al. Correlation of gene expression profile status and American Joint Commission on cancer stage in uveal melanoma. Retina. 2020 Feb;40((2)):214–224. doi: 10.1097/IAE.0000000000002385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dogrusöz M, Bagger M, van Duinen SG, Kroes WG, Ruivenkamp CAL, Böhringer S, et al. The prognostic value of AJCC staging in uveal melanoma is enhanced by adding chromosome 3 and 8q status. Invest Ophthalmol Vis Sci. 2017 Feb;58((2)):833–842. doi: 10.1167/iovs.16-20212. [DOI] [PubMed] [Google Scholar]
- 10.Corrêa ZM, Augsburger JJ. Independent prognostic significance of gene expression profile class and largest basal diameter of posterior uveal melanomas. Am J Ophthalmol. 2016 Feb;162:20–7.e1. doi: 10.1016/j.ajo.2015.11.019. [DOI] [PubMed] [Google Scholar]
- 11.Walter SD, Chao DL, Feuer W, Schiffman J, Char DH, Harbour JW. Prognostic implications of tumor diameter in association with gene expression profile for uveal melanoma. JAMA Ophthalmol. 2016 Jul;134((7)):734–740. doi: 10.1001/jamaophthalmol.2016.0913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roelofs KA, Grewal P, Lapere S, Larocque M, Murtha A, Weis E. Optimising prediction of early metastasis-free survival in uveal melanoma using a four-category model incorporating gene expression profile and tumour size. Br J Ophthalmol. 2021 Feb; doi: 10.1136/bjophthalmol-2020-317714. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 13.Demirci H, Niziol LM, Ozkurt Z, Slimani N, Ozgonul C, Liu T, et al. Do largest basal tumor diameter and the American Joint Committee on cancer's cancer staging influence prognostication by gene expression profiling in choroidal melanoma. Am J Ophthalmol. 2018;195:83–92. doi: 10.1016/j.ajo.2018.07.033. [DOI] [PubMed] [Google Scholar]
- 14.AJCC Ophthalmic Oncology Task Force International validation of the American Joint Committee on cancer's 7th edition classification of uveal melanoma. JAMA Ophthalmol. 2015 Apr;133((4)):376–383. doi: 10.1001/jamaophthalmol.2014.5395. [DOI] [PubMed] [Google Scholar]
- 15.Shields CL, Ganguly A, Bianciotto CG, Turaka K, Tavallali A, Shields JA. Prognosis of uveal melanoma in 500 cases using genetic testing of fine-needle aspiration biopsy specimens. Ophthalmology. 2011 Feb;118((2)):396–401. doi: 10.1016/j.ophtha.2010.05.023. [DOI] [PubMed] [Google Scholar]
- 16.Shields CL, Say EAT, Hasanreisoglu M, Saktanasate J, Lawson BM, Landy JE, et al. Cytogenetic abnormalities in uveal melanoma based on tumor features and size in 1059 patients: the 2016 W. Richard Green Lecture. Ophthalmology. 2017;124((5)):609–618. doi: 10.1016/j.ophtha.2016.12.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings in the manuscript contain dates and variables that are identifiable and are not available publicly. Requests for de-identified summary data can be made to the authors.