Abstract
A disparate array of plasma/serum markers provides evidence for chronic inflammation in human prediabetes, a condition that is most closely replicated by standard mouse models of obesity and metaflammation. These remain largely nonactionable and contrast with our rich understanding of inflammation in human type 2 diabetes. New data show that inflammatory profiles produced by CD4+ T cells define human prediabetes as a unique inflammatory state. Regulatory T cells (Treg) control mitochondrial function and cytokine production by CD4+ effector T cells (Teff) in prediabetes and type 2 diabetes by supporting T helper (Th)17 or Th1 cytokine production, respectively. These data suggest that Treg control of Teff metabolism regulates inflammation differentially in prediabetes compared with type 2 diabetes. Queries of genes that impact mitochondrial function or pathways leading to transcription of lipid metabolism genes identified the fatty acid importer CD36 as highly expressed in Treg but not Teff from subjects with prediabetes. Pharmacological blockade of CD36 in Treg from subjects with prediabetes decreased Teff production of the Th17 cytokines that differentiate overall prediabetes inflammation. We conclude that Treg control CD4+ T cell cytokine profiles through mechanisms determined, at least in part, by host metabolic status. Furthermore, Treg CD36 uniquely promotes Th17 cytokine production by Teff in prediabetes.
Introduction
Obesity-associated prediabetes is a compromised metabolic state defined mainly by hyperglycemia that falls short of criteria for type 2 diabetes and is less likely to perpetuate complications like renal and cardiovascular diseases. The very large pool of people with prediabetes are at high risk for type 2 diabetes, although prediabetes is very reversible through lifestyle changes, while medications like metformin delay progression to type 2 diabetes (1). A better understanding of prediabetes is an important goal that could prevent a considerable number of type 2 diabetes cases. A disparate array of plasma/serum markers provides evidence for chronic inflammation in prediabetes (2,3) that contrasts with our rich understanding of cellular sources of inflammation in human type 2 diabetes and animal models of insulin resistance (IR) (4).
Myeloid cells dominate inflammation in IR mice, but T cells also influence obesity-associated inflammation, especially the systemic inflammation that provokes numerous obesity comorbidities in people. The T helper (Th)17 subset of CD4+ T cells is a prominent source of obesity-associated inflammation in many human studies (5–8). We identified a Th17 cytokine profile that differentiated peripheral blood mononuclear cells (PBMC) from obese subjects with and without type 2 diabetes and showed that B cells support Th17 inflammation through diabetes-associated mechanisms (9). These studies focused on more extreme ends of the metabolic spectrum of obese subjects rather than prediabetes and concluded that CD4+ T cells were functionally similar in the absence of regulation by support cells (9,10).
The CD4+ T cell subset is a mix of functionally distinct cell types including Th1, Th17, and regulatory T cells (Treg), among others. While Th1 and Th17 along with other T effector subsets like Th2 and Th9 (Teff) promote adipose tissue and systemic inflammation in obesity, Treg counteract inflammation, as demonstrated in IR animals (11,12). Our previous analyses included a mix of functionally contrasting cell types, even within the CD4+ subset, leaving open the possibility that Treg control Teff inflammation during the development of type 2 diabetes.
Treg control Teff, but in turn Treg are controlled through mechanisms that fundamentally differ from those of Teff. One example is a difference in the fuels used to drive effector functions. Although foundational studies concluded that nonmitochondrial glycolysis or fatty oxidation preferentially fuels Teff or Treg, respectively, more nuanced work showed roles for fatty acids and glutamine in Th17 metabolism (13–15) and glycolysis in human Treg function (16,17). Now classical work showed that PPARγ, a transcriptional regulator of many fat metabolism genes, uniquely controls adipose tissue Treg (but not Teff) in obese/IR mice (11). These data raise the possibility that manipulating Treg through the peculiarities of their metabolic machinery to indirectly alter Teff function, as was successful in exploitations of Treg CD36 in cancer to lower Teff suppression (12,18), will provide new avenues for alleviating systemic inflammation in prediabetes to reverse or delay disease progression.
Data herein show that inflammatory profiles produced by purified CD4+ T cells define prediabetes as an inflammatory state that is unique from T cell inflammation in lean/healthy (herein, “lean”) subjects or obese subjects/subjects with type 2 diabetes and that this profile is predominantly generated by the Teff subset. Treg control Teff mitochondrial oxidative phosphorylation (OXPHOS) in prediabetes and type 2 diabetes samples and concomitantly support Th1 or Th17/Treg cytokine production, respectively. These data suggest that Treg-mediated control of Teff metabolism differentially controls inflammation in prediabetes compared with type 2 diabetes. Queries of a range of genes that impact mitochondrial function and/or pathways leading to transcription of lipid metabolism genes identified the long-chain fatty acid importer CD36 as highly expressed in Treg but not Teff of subjects with prediabetes and type 2 diabetes relative to lean subjects. Pharmacological blockade of CD36-mediated long-chain fatty acid import in Treg from subjects with prediabetes decreased production of a subset of Teff cytokines that contribute to prediabetes inflammation. We conclude that Treg control CD4+ T cell cytokine production through multiple mechanisms that are determined, at least in part, by metabolic status of the host. Furthermore, Treg CD36 uniquely promotes Th17 cytokine production by Teff in prediabetes.
Research Design and Methods
Human Subjects
Blood was taken by venipuncture from subjects following informed consent under a protocol approved by University of Kentucky Institutional Review Board, in accordance with the principles in the Declaration of Helsinki. Subject type was defined by American Diabetes Association criteria, and all were recruited from the University of Kentucky Center for Clinical Translational Science. Exclusion criteria included autoimmune disease, recent (within 2 weeks) bacterial/viral infections, cancer <5 years prior, current allergy medications, uncontrolled kidney or cardiovascular disease, insulin >100 units/day, weight change of >5% in the past 3 months, bariatric surgery within 1 year, abnormal thyroid hormone levels, and pregnancy. Subject characteristics can be found in Supplementary Table 1.
Isolation and Culture of Human T Cells
PBMC were isolated from blood using Ficoll-Paque (GE) gradient centrifugation and SepMate tubes (STEMCELL Technologies). CD25+CD4+ Treg and CD25−CD4+ Teff were separated from PBMC with a human CD4+CD25+ Treg isolation kit (Miltenyi Biotec) (Supplementary Fig. 1A) using autoMACS (Miltenyi Biotec). Cells were cryogenically preserved as previously described (19). Cell purity was quantified with CD3 V610, CD4 FITC, CD25 allophycocyanin (APC), and FOXP3 phycoerythrin (PE) with use of a CytoFLEX LX (Beckman Coulter) flow cytometer. Teff and Treg were >90% pure based on CD3+CD4+CD25−FOXP3− or CD3+CD4+CD25+FOXP3+, respectively, in unstimulated samples. Alternatively, Teff and Treg were isolated from frozen PBMC by a Sony SY3200 cell sorter using CD4 PE and CD25 APC antibodies and gating on live CD25−CD4+ and CD25+CD4+ populations, respectively. Results were indistinguishable from magnetic bead–purified populations. Teff were cultured either alone or with Treg at 19:1 (95% Teff:5% Treg) or 4:1 (80% Teff:20% Treg) ratios as previously described (7). Cells from the same donor were combined for cocultures. All cells were stimulated with CD3/CD28 Dynabeads (Thermo Fisher Scientific) for 40 h at a bead:cell ratio of 1:1.
Metabolic Analyses
Seahorse was run as we previously described (19), with cells seeded at 2.5 × 105 per well. Data from different Seahorse XF96 plates were combined by the Python-based script SeaHORse Explorer (SHORE) program (19), which uses the median of three or more technical replicate values for each time point to eliminate outliers without bias. Lactate was measured with a colorimetric kit (BioVision). Cell culture supernatants were archived <1 year at −80°C before analysis and then thawed and diluted 50× for lactate concentration measurement.
CD36 Functional Test
Treg were pretreated with 10 μmol/L sulfo-N-succinimidyl oleate (SSO) (Cayman Chemical) or vehicle (1:1,000 DMSO) and stimulated for 4 h in fatty acid–free culture medium (10% fatty acid–free BSA instead of FBS). Treg were washed and then cultured with or without vehicle-treated Teff and restimulated for 40 h. For quantification of impacts of SSO on long-chain fatty acid uptake, cultures restimulated for 40 h were incubated with 1.0 μmol/L BODIPY FL C16 (Thermo Fisher Scientific) for 15 min in PBS at 37°C. Live cells were analyzed by CytoFLEX LX (Beckman Coulter). Mean fluorescence intensity of BODIPY FL C16 was quantified with FlowJo v10.7.1. Supernatants were archived for lactate and cytokine measurements.
Multiplex Cytokine Assay
Cytokines in cell culture supernatants were measured on a Luminex-based FLEXMAP platform (Bio-Rad Laboratories) using Th17 MILLIPLEX kits (Millipore) as previously described (9). Supernatants were diluted 50-fold to stay within the linear range of standard curves as appropriate. For SSO experiments, cytokines were quantified with the HCYTA-60K-04 Human Cytokine/Chemokine/Growth Factor Panel A (IL-17A, IL-17F, IL-10, and IFN-γ; Millipore)
Flow Cytometry
Live cells were identified following staining with either Zombie NIR or Zombie Aqua. For surface staining, CD3 BV605 and CD4 FITC were added at 1:100; CD25 APC was added at 1:50 in staining buffer (1xDPBS, 10% BSA, 1% EDTA). For Foxp3 staining, cells were fixed/permeabilized after surface staining and then FOXP3 PE was added at 1:50 in permeabilization buffer (FOXP3/Transcription Factor Staining Buffer Set; eBiosciences). To differentiate IL-6–positive Teff and Treg, we stained Teff with 1 μmol/L carboxyfluorescein succinimidyl ester (CFSE) and Treg with 5 μmol/L CellTrace Violet for 20 min at room temperature (RT) and then quenched with a 5× volume of cell culture medium. Cells were pelleted and resuspended in prewarmed cell culture medium for 10 min before 40 h activation. To maximize intracellular cytokine signals, we restimulated cells with phorbol myristate acetate (5 ng/mL) and ionomycin (250 ng/mL) during the last 6 h of the culture in the presence of Brefeldin A (3 μg/mL; Thermo Fisher Scientific). For intracellular staining, cells were fixed after surface staining and then treated with permeabilization buffer (Intracellular Fixation & Permeabilization Buffer Set kit; eBiosciences), and IL-6 PE was added at a 1:50 ratio in permeabilization buffer. Stained samples were fixed with 1% paraformaldehyde in PBS before analysis by CytoFLEX LX using CytExpert software (Beckman Coulter). Flow cytometry data were analyzed with FlowJo v10.7.1.
Gene Expression
Whole cell extract was prepared from 1–2 million cells lysed on ice for 20 min with use of 25 μL radioimmunoprecipitation assay buffer/106 cells (Cell Signaling Technologies) supplemented with protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific) and EDTA. Debris was removed by centrifugation at 13,000 rpm for 20 min. Protein was quantified in a Pierce BCA Protein Assay kit (Thermo Fisher Scientific). Protein (15 μg: PPARγ, cat. no. 2430S, Cell Signaling Technologies, and CD36, cat. no. NB400-144, Novus Biologicals; 10 μg: SREBP-1, cat. no. NB600-582, Novus Biologicals, and CPT-1a, cat. no. 12252S, Cell Signaling Technologies) was loaded onto precast gels (4–20%; Bio-Rad Laboratories) and separated at 100 V/20 min and then 140 V/80 min before transfer onto polyvinylidene difluoride membranes at 100 V, 1 h 15 min. Membranes were blocked in 5% dry milk 1× Tris-buffered 0.1% Tween 20 (TBST) for 1 h at RT, followed by three washes with 1× TBST, and then probed with primary antibody (1:1,000 for CD36, SREBP-1, and CPT-1a and 1:500 for PPARγ) overnight at 4°C. After three washes with 1× TBST, membranes were incubated with horseradish peroxidase–conjugated secondary antibody (1:10,000, anti-rabbit or anti-mouse IgG; Cell Signaling Technologies) for 1 h at RT. Ultrasensitive enhanced chemiluminescent (ECL) substrate was used for protein detection. Blots were imaged using automatic exposure with ChemiDock (Bio-Rad Laboratories). Protein expression was quantified by ImageJ.
mRNA Analysis
RNA was isolated with RNeasy (QIAGEN) and then quantified by NanoDrop. cDNA was synthesized with the qScript cDNA Synthesis Kit (Quantabio) before amplification by real-time PCR with SYBR Green master mix (QIAGEN), with 60°C annealing/extension for 40 cycles. For quantification we used the ΔΔCT method. Primers for mRNA quantification can be found in Supplementary Table 2.
Statistical Analysis
Data are presented as mean ± SD unless otherwise indicated. One-way or two-way ANOVAs were conducted in GraphPad Prism 8.0 as appropriate. Alternatively, we fit a mixed model to the log-transformed, normalized values for each individual cytokine with an unstructured correlation matrix. We used fixed effects for cell or clinical group and a random effect for subject. Each model was fit with cell, group, and the statistical interaction between cell and group. If the interaction effect was not significant, it was removed from the model. P values <0.05 were deemed significant for all analyses, and a Tukey adjustment corrected for multiple tests. Cytokine values below the detection limit were replaced by one-half of the minimum log-transformed normalized observed value for the cytokine under consideration. Partial least squares discriminant analysis (PLSDA) of orthogonalized data was completed as previously described (14,20) with work done in R to capture the impact of cytokine interactions that are not captured by the other analytical approaches.
Data and Resource Availability
The data sets generated for the current study are available from the corresponding author upon reasonable request. No new resources were generated during the study.
Results
CD4+ T Cells From Subjects With Prediabetes Produce a Unique Inflammatory Profile
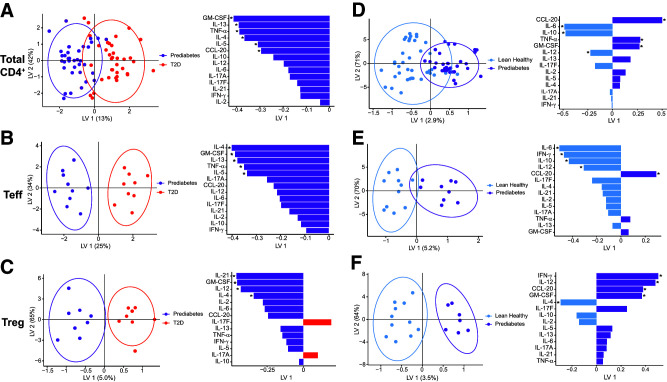
To better define inflammation in prediabetes, we first analyzed T cell (>90% pure based on CD3+CD4+) (Supplementary Fig. 1B) cytokine competence using mixed models that considered each cytokine in isolation. We found numerous differences in cytokine production when compared among cell types (Teff, Treg, or Teff-Treg cocultures) or subject groups (lean or subjects with obesity plus either prediabetes of type 2 diabetes) (Supplementary Table 1), with significant interactions between cell type and subject group for IL-5, -10, and -21 (Supplementary Table 3). To capture potential interactions among cytokines that were not queried in other analyses, we generated combinatorial cytokine profiles (9) from the same cytokine measures. A codominant Th2 (IL-13, IL-4, IL-5)/Th1 (GM-CSF, TNFα) (15,21) cytokine profile delineated prediabetes from type 2 diabetes inflammation in both total CD4 and effector T cells (Teff) (Fig. 1A and B, purple). In contrast, a mixed array of cytokines (IL-21, GM-CSF, IL-12, and IL-4) differentiated Treg in prediabetes (Fig. 1C, purple), although IL-17F and IL-17A were unexpectedly produced at higher concentrations by Treg from subjects with type 2 diabetes (Fig. 1C, red). Consistent with previous literature, purified Treg produced very little/no IL-2 as a functional readout of purity (22) (Supplementary Fig. 1C), recapitulating our mixed-model results for IL-2 (Supplementary Table 3). Comparison of overall inflammation between CD4+ T cells of subjects with prediabetes and of lean subjects showed that a mixed Th1/2/17 profile distinguished prediabetes from lean inflammation (Fig. 1D) (CCL-20, IL-6, IL-10, TNF-α, GM-CSF, and IL-12), while an overlapping but more strictly Th1 profile (IL-6, IFN-γ, and IL-12, with IL-10 as a Th2 cytokine) dominated Teff responses (Fig. 1E). Cytokine profiles from prediabetes relative to lean Treg were similarly dominated by Th1 cytokines like IFN-γ and IL-12 (Fig. 1F). These data support the conclusion that inflammation from CD4+ T cells, and specifically Teff, progressively changes during type 2 diabetes pathogenesis and that prediabetes represents a unique inflammatory state.
Figure 1.
CD4+ T cells from subjects with prediabetes produce a unique cytokine profile. PLSDA of cytokines in supernatants of total CD4+ T cells (A and D), purified Teff (B and E ), or purified Treg (C and F ) stimulated for 40 h with CD3/CD28 Dynabeads. Comparisons are between profiles generated by cells from subjects with prediabetes versus with type 2 diabetes (T2D) (A–C) or subjects with prediabetes and versus lean/healthy (normoglycemic) subjects (D–F). Left panels show distribution of a combination of all cytokines measured, with each dot representing compendium outcomes (“inflammation”) from one sample. Right panels rank cytokines for importance (top = more important; bottom = less important) in differentiating combinatorial inflammatory profiles from the groups indicated, with color fill indicating the clinical group whose cells produced more of that cytokine. *Statistical differences (P < 0.05) in post hoc analysis of ranked cytokines. N = 11, 9, or 9 subjects for lean, prediabetes, or type 2 diabetes, respectively. For analyses in panels A and D we included cytokine profiles from Teff, Treg, and 5% and 20% Treg-Teff cocultures (see Figs. 2 and 3) that together represent a naturally achievable range of Treg-to-Teff ratios and thus total CD4+ T cell inflammation. Each dot is a two-dimensional representation of a multidimensional combination of all cytokines generated from the indicated type of sample(s). Linear combinations of cytokines were visualized as reduced dimensional latent variables (LVs).
Prediabetes Teff Have Higher Mitochondrial Metabolism
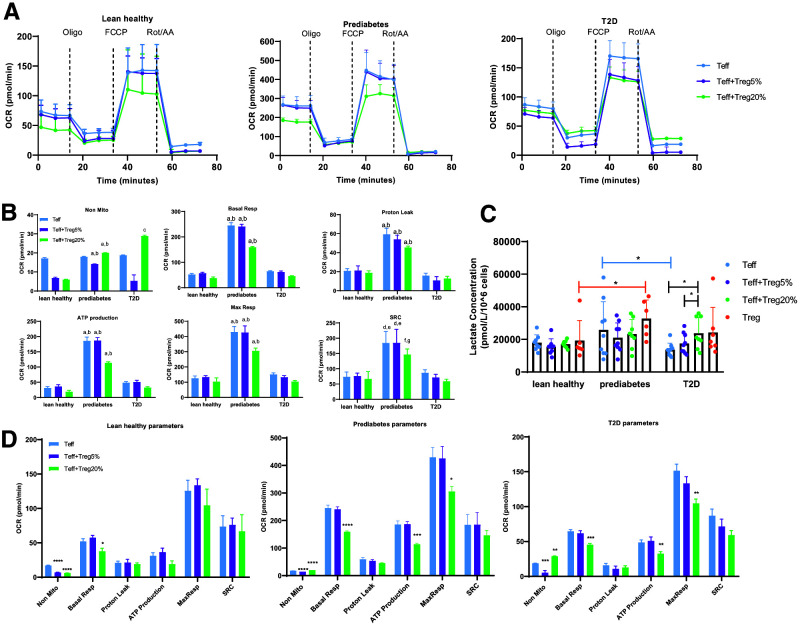
To begin understanding whether prediabetes-associated changes in CD4+ T cell metabolism fuels the unique cytokine profile of prediabetes, we measured mitochondrial function of Teff from lean subjects, or subjects with obesity and either prediabetes or type 2 diabetes, using the Seahorse Mito Stress Test (Fig. 2A). We first analyzed Seahorse-generated oxygen consumption rate (OCR) (a direct measure of OXPHOS) to identify interactions between cell/culture type and disease status (Fig. 2B). This analysis showed that purified Teff from subjects with prediabetes compared with the other cohorts had approximately threefold the OCR under both basal and maximal respiration conditions following CD3/CD28 stimulation (Fig. 2A and B, blue lines/bars). Despite similar nonmitochondrial OXPHOS and higher proton leak, prediabetes Teff had higher ATP production and spare respiratory capacity—the latter a measure of the ability of cells to increase ATP production in response to acute stress (Fig. 2A and B). Prediabetes Teff had higher basal respiration and marginally higher nonmitochondrial glycolysis than Teff from lean subjects or subjects with type 2 diabetes, with the latter measured by lactate production (Fig. 2B and C). We conclude that Teff from prediabetes subjects are more metabolically active than cells from the comparator cohorts, mainly due to higher mitochondrial OXPHOS.
Figure 2.
Treg control Teff metabolism. A: OCR (a direct measurement of mitochondrial OXPHOS) of CD3/CD28-stimulated Teff from lean subjects, subjects with prediabetes, and subjects with type 2 diabetes in the presence or absence of Treg as indicated in the key and analyzed as we previously described (19). OCR following treatment with the ATP synthase inhibitor oligomycin (oligo), the mitochondrial uncoupler trifluoromethoxy carbonlycyanide phenylhydrazone (FCCP), or mitochodrial complex I/III inhibitors rotenone and antimycin A (Rot/AA) are shown. B: Quantification of Mito Stress test OCR (i.e., OXPHOS) of Teff in the presence/absence of Treg included nonmitochondrial oxygen consumption (Non Mito), basal respiration (Basal Resp), proton leak, ATP production, maximal respiration (Max Resp), and spare respiratory capacity (SRC) from panel A. Analysis takes into account interactions between cell/culture type and disease status of cell donor as labeled on x-axis. Differences are indicated as follows: a, prediabetes vs. lean, P < 0.001; b, prediabetes vs. type 2 diabetes, P < 0.001; c, lean vs. type 2 diabetes, P < 0.001; d, prediabetes vs. lean, P < 0.01; e, prediabetes vs. type 2 diabetes, P < 0.01; f, prediabetes vs. lean, P < 0.05; g, prediabetes vs. type 2 diabetes, P < 0.05. C: Lactate concentrations in culture supernatants of stimulated Teff ± Treg as indicated in key. N = 9 for each cohort. *P < 0.05. D: Reanalysis of data to focus on cell type differences independent from interactions with disease status. Data were analyzed with one-way ANOVA, Kruskal-Wallis test. *P < 0.05, **P < 0.01, ***P < 0.005, ****P < 0.001, compared with Teff alone. For panels A, B, and D: N = 8, 7, and 8 for lean, prediabetes, and type 2 diabetes samples, respectively. For panels B–D, data were analyzed with SHORE (19) and are displayed as median and range. T2D, type 2 diabetes.
Treg Control CD4+ T Cell Metabolism and Function Through Disease-Associated Mechanisms
Treg frequencies differ in lean compared with mice and humans with IR/type 2 diabetes amid modest changes in Teff frequencies (7,11,12,14), raising the possibility that changes in Teff-to-Treg ratios, and thus cross talk, impact Teff metabolism and cytokine production in prediabetes. We repeated OCR measures in Teff cocultured with 5% or 20% syngeneic Treg to mimic the broad range of Teff-to-Treg ratios identified previously (23). We found no evidence that Treg impacted Teff-Treg coculture OCR through disease-associated mechanisms (Fig. 2B; blue, purple and green bars within each subject cohort are similar). However, analysis of culture-specific differences by subject group showed that 20% Treg lowered basal OCR in cells from all three subject types and decreased ATP production and maximal respiration only in Teff from subjects with obesity, regardless of metabolic health (Fig. 2D). Treg lowered nonmitochondrial respiration in Teff from subjects who were lean (at 1:19 ratios), but at 1:4 ratios, Treg increased nonmitochondrial respiration in Teff from subjects with type 2 diabetes (Fig. 2D). Treg increased lactate production only in cocultures from subjects with type 2 diabetes (Fig. 2C). The 12,500–50,000 Treg added to Teff consume immeasurable oxygen using this method (data not shown), although we cannot entirely rule out the possibility that Teff activate measurable oxygen consumption by Treg in cocultures. CFSE dilution assays to measure Treg blockade of Teff proliferation showed that, as expected, Treg did not affect Teff proliferation in short-term cocultures (Supplementary Fig. 1D and E), eliminating one potential confounder of the outcomes. We conclude that Treg change basal and maximal mitochondrial function, nonmitochondrial OXPHOS, ATP production, and nonmitochondrial glycolysis of T cells (largely Teff) through obesity- and/or type 2 diabetes–associated mechanisms.
Treg Impact CD4+ T Cell Function in Prediabetes and Type 2 Diabetes
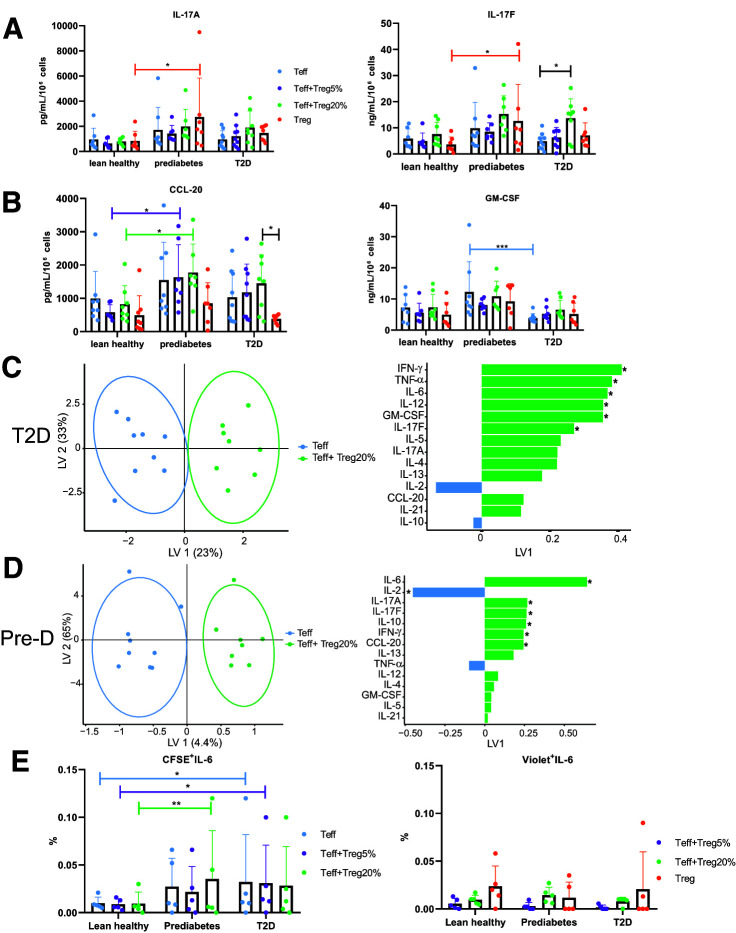
To determine whether disease-associated changes in metabolism of Teff/Treg cocultures regulate the Th17 and Th2 cytokines that define the unique inflammatory profile of Teff in prediabetes (Fig. 1), we quantified cytokines produced by Teff alone, Treg alone, or Teff-Treg cocultures from replicates of samples analyzed with Seahorse (Fig. 2). Supplementary Table 3 highlights significant differences in single cytokines based on cell type, including differences between Teff and Treg in IL-2, -4, -6, and -17A and CCL20. ANOVA showed that Treg did not impact production of multiple cytokines traditionally characterized as Th17 or Th1 (Fig. 3A and B, respectively) in samples from the cohorts of lean subjects or subjects with prediabetes, although higher IL-17A and IL-17F production by Treg from subjects with prediabetes compared with lean subjects may contribute to Th17 profiles in prediabetes (Figs. 1F and 3A). However, Treg significantly increased IL-17F production by CD4+ T cells from subjects with type 2 diabetes (Fig. 3A). This approach cannot entirely rule out the possibility that Teff triggers robust Treg cytokine production in coculture, although adding 20% of the cytokines produced by Treg would not account for the highlighted difference. With PLSDA we confirmed that IL-17F production differentiated cytokine profiles of Teff produced in the presence versus absence of Treg from subjects with type 2 diabetes, along with an ability of Treg to induce a comprehensive Th1 profile (IFN-γ, TNF-α, IL-6, IL-12, GM-CSF) (Fig. 3C). In contrast, PLSDA analysis of cytokine profiles produced by Teff compared with Teff-Treg cocultures from subjects with prediabetes revealed a mixed Th17-supportive (IL-17A and -17F/CCL-20)/Treg-supportive (IL-2 and -10) profile generated in the presence of Treg (Fig. 3D). Comparison of profiles produced by Treg and Teff-Treg 20% cocultures showed that cytokines well known to be produced by Teff but not Treg (IL-2 and IFN-γ, among others) were important for differentiating culture types similarly in cells from subjects with prediabetes or type 2 diabetes (Fig. 3C and D), indicating that Teff cytokine production almost exclusively accounts for disease-associated differences in coculture inflammation. Models comparing Teff and Teff-Treg 5% profiles in cells from all three cohorts or Teff and Teff-Treg 20% profiles from lean subjects showed no differences (not shown), indicating modest if any impact of Treg on CD4+ T cell cytokine production under these conditions. Models comparing Treg with Teff-Treg 20% profile showed that a highly overlapping set of cytokines differentiates function of these two culture types (IL-12 and -21, IFN-γ, etc.) irrespective of donor group, consistent with the interpretation that Treg cytokine production contributes modestly to the disease-associated differences in cytokine production by Teff versus Teff-Treg cocultures. (Supplementary Fig. 2). Intracellular staining for IL-6, a cytokine that distinguished function of Teff versus Teff-Treg cultures from subjects with type 2 diabetes and prediabetes (but not lean subjects) (Fig. 3C and D), supported the conclusion that a higher frequency of CFSE-stained Teff in the Teff-Treg cocultures produced IL-6 in prediabetes (or type 2 diabetes) compared with lean subjects’ cells (Fig. 3E, left). IL-6+ violet-stained Treg were similarly frequent among subject types (Fig. 3E, right). The finding of similarities in IL-6 production by Teff and Teff-Treg cultures in flow analyses is consistent with our understanding that multivariate analysis (per Fig. 3C and D) can identify differences that fail to meet standard statistical criteria for differences. We conclude that the ability of Treg to change Teff metabolism (Fig. 2) associates with cohort-determined differences in CD4+ T cell cytokine production.
Figure 3.
Treg change CD4+ T cell cytokine profiles produced by cells from subjects with prediabetes and subjects with type 2 diabetes. Concentrations of IL-17A and IL-17F (A) or CCL-20 and GM-CSF (B) in supernatants of Teff stimulated ± Treg as indicated. Purified Treg cytokine production is shown to suggest that diluting Treg cytokines by 1:4 or 1:19 in cocultures is unlikely to explain differences. Data are from lean subjects (n = 8), subjects with prediabetes (n = 8), and subjects with type 2 diabetes (n = 9). For panels A and B, data show mean ± SD. Two-way ANOVA identified differences as *P < 0.05, ***P < 0.005. C and D: PLSDA of 25 measured cytokines in supernatants of Teff stimulated alone (blue) or in the presence of 20% Treg (green). Left panels show distribution of a combination of all cytokines measured, with each dot representing compendium outcomes (“inflammation”) from one sample. Right panels rank cytokines for importance (top = more important; bottom = less important) in differentiating combinatorial inflammatory profiles from the groups indicated, with color fill indicating the clinical group whose cells produced more of that cytokine. Cells were from subjects with type 2 diabetes (C) or subjects with prediabetes (Pre-D) (D). *indicates statistical differences (P < 0.05) in post hoc analysis of ranked cytokines. Linear combinations of analytes were visualized as reduced dimensional latent variables (LVs). E: Intracellular staining to identify cell type(s) producing IL-6. Frequency of Teff (labeled with CSFE [left]) or Treg (labeled with CellTrace Violet [right]) that costained for IL-6 following culture as indicated in the key. Differences as identified by two-way ANOVA are indicated as *P < 0.05 or **P < 0.01. N = 5.
Prediabetes Changes Metabolism-Associated Proteins in Treg and Teff
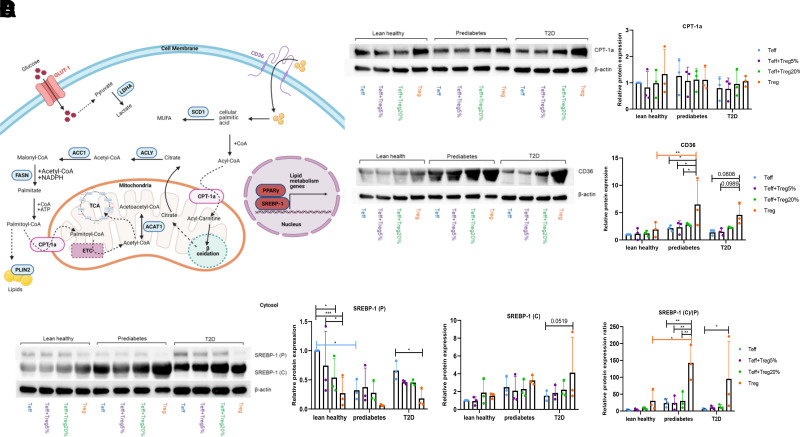
Mitochondrial OXPHOS is one important outcome of coordination among numerous lipid-related genes and pathways, outlined in Fig. 4A. Given the role long-chain fatty acids play in T cell cytokine production in type 2 diabetes (7,13,14), we quantified proteins responsible for fatty acid metabolism/flux to determine whether similar changes may play roles in the mixed Th17/Th2 profile produced by CD4+ cells in prediabetes. mRNA quantification showed that GLUT1, ACC1, LDHa, ACLY, FASN, ACAT1, SCD1, PLIN2, and CPT-1a had disease-independent expression, although some of these mRNAs were expressed more in Teff or cocultures than Treg (Supplementary Fig. 3A). In contrast to the higher CPT-1a expression by PBMC from subjects with obesity/type 2 diabetes compared with subjects with obesity/normoglycemia (14), T cells from cohorts analyzed herein expressed similar amounts of CPT-1a protein (Fig. 4B). We preliminarily conclude that proteins involved in mitochondrial fatty acid processing modestly differ in T cells from comparator cohorts herein. In contrast, the fatty acid importer CD36 was expressed more highly in Treg from subjects with obesity (Fig. 4C). Although Western blots for PPARγ, a transcription factor downstream of CD36, were inconclusive (Supplementary Fig. 3B), mRNA analysis suggested that Treg express more PPARγ than Teff from all cohorts (Supplementary Fig. 3C). Similarly, the cleaved-to-precursor ratio of the transcription factor SREBP-1 was highest in Treg from both obese cohorts (Fig. 4D). We conclude that key transcriptional mediators that are regulated by long-chain fatty acid signaling pathways, and control lipid metabolism genes, are altered through cell type and, for CD36 and SREBP, disease-associated mechanisms.
Figure 4.
More CD36 in Treg from subjects with prediabetes associated with higher expression of downstream transcriptional regulators but not mitochondrial genes. A: Schema of CD36-downstream transcriptional regulators and mitochondrial genes. GLUT1 and LDH (lactate dehydrogenase) regulate glucose metabolism; pyruvate entry into mitochondrial metabolism is not shown. CD36 (fatty acid translocase) mediates fatty acid uptake. SCD1 (stearoyl-CoA desaturase 1), ACLY (ATP-citrate lyase), ACC1 (acetyl-CoA carboxylase 1), FASN (fatty acid synthase), and SREBP-1 (sterol regulatory element binding protein 1) regulate fatty acid synthesis. CPT-1a (carnitine-palmitate transferase 1a) and ACAT1 (acetyl-CoA acetyltransferase 1) regulate fatty acid oxidation. PPARγ (peroxisome proliferator–activated receptor γ) regulates lipid metabolism and inflammation. PLIN2 (perilipin 2) regulates lipid droplet formation. ETC designates the mitochondrial electron transport chain CPT-1a (B), CD36 (C), or SREBP-1 (D) (precursor [P] or cleaved [C]) protein in stimulated T cell cultures as indicated. Panels B, C, and D show a representative Western blot; bar graphs show quantification of biological replicates. For all panels, N = 3 for each subject group and data (mean ± SD) were analyzed with two-way ANOVA. *P < 0.05; **P < 0.01; ***P < 0.005.
Treg CD36 Promotes Teff Cytokine Production in Prediabetes
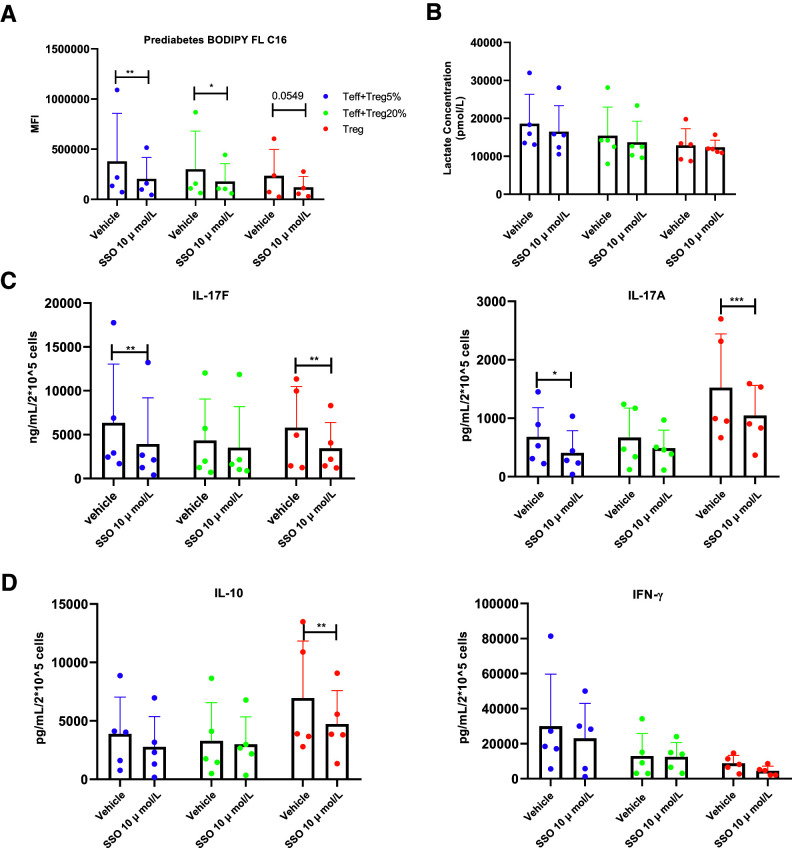
Given that 1) Treg upregulate CD36 in adipose tissue of obese subjects among other pathogenic environments (11,18), 2) Treg CD36 is highest in subjects with prediabetes but CD36 is expressed similarly by Teff from all subject types (Fig. 4C), 3) CD36 imports long-chain fatty acids that promote T cell inflammation in type 2 diabetes (14), and 4) CD36 can be manipulated through approaches consistent with limited Treg numbers, we questioned the functional importance of higher Treg CD36 on T cell inflammation in prediabetes. Because the number of Treg we recover from each subject is insufficient for genetic manipulation per our previous approaches (14), we instead relied on an irreversible and specific CD36 inhibitor, SSO, that blunts long-chain fatty acid uptake without impacting glucose uptake or fatty acid metabolism (24). As expected, SSO inhibited BODIPY FL C16 uptake by T cells (Fig. 5A) and had no effect on nonmitochondrial glycolysis as indicated by lactate production (Fig. 5B). SSO lowered production of the Th17 cytokines IL-17A and IL-17 in both purified Treg and in cocultures of 95% Teff + 5% Treg (Fig. 5C). IL-10 and IFN-γ production were unchanged in these cocultures, although Treg IL-10 was modestly lower following SSO treatment (Fig. 5D). Mechanisms that explain the lack of an effect of SSO on the cocultures with 80% Teff + 20% Treg remain unknown (Fig. 5C). Studies with cells from N = 3 individuals processed as in Fig. 1 versus Fig. 5 confirmed that absolute differences in cytokine production were due to protocol differences (not shown). Overall, these data support the conclusion that Treg CD36 supports CD4+-generated inflammation in prediabetes.
Figure 5.
CD36 mediates long-chain fatty acid uptake in Treg and total CD4+ T cell cytokine production by cells from subjects with prediabetes. A: BODIPY uptake in prediabetes Treg and stimulated Treg-Teff cocultures following pretreatment of Treg with the irreversible CD36 inhibitor SSO as indicated. Uptake is quantified based on mean fluorescence intensity of BODIPY FL C16 as quantified with flow cytometry. N = 4. B: Impact of Treg pretreatment with SSO on lactate production by CD4+ T cells from subjects with prediabetes. Examples of Th17 (C) or non-Th17 (D) cytokine production by CD4+ T cells as indicated after Treg pretreatment with SSO. For panels B–D, N = 5. All data are displayed as mean ± SD, with analysis by two-way ANOVA. *P < 0.05; **P < 0.01; ***P < 0.005.
Discussion
Our data support the conclusion that CD4+ T cell inflammation in subjects with prediabetes differs from that of either lean subjects or subjects with type 2 diabetes largely due to the Teff subset, which in turn is regulated by CD36 on Treg. These data thus highlight nonlinear changes in T cell function during disease development that culminate in the systemic Th17 profile of type 2 diabetes (9). Our findings fundamentally differ from previous demonstrations of slow, steady increases in single inflammatory markers, sometimes limited to specific points of disease progression (i.e., lean → prediabetes; prediabetes → type 2 diabetes), in studies with greater numbers of subjects (2,25). This new appreciation of metaflammation suggests new targets for reversing prediabetes that will complement recommendations for glycemic control and body weight management. Examples include derivatives from type 2 immunity/Th2/Treg-supportive helminths, shown to ameliorate obesity in mouse models (26,27).
Our demonstration that Treg control Teff mitochondrial respiration from subjects with obesity introduces a previously unappreciated mechanism of Treg action. Treg-limited OCR associated with disease-determined cytokine profiles, but these data also justify studies on other outcomes of Teff metabolism, like proliferation or migration (28), to pinpoint the functional significance of this obesity-associated Treg action. The lack of disease-associated differences in mRNAs that encode mitochondrial mediators of T cells suggests that complementary tools like metabolomics will be needed to identify mechanisms that Treg use to alter Teff mitochondrial function uncovered by our work. Regardless, the ability of Treg to shift T cell inflammatory profiles concurrent with lower T cell OXPHOS in disease-associated patterns expands our previous demonstration that B cells support T cell cytokine production in diabetes and add nuance to our original conclusion that total CD4+ T cells are functionally similar in obese/normoglycemic and diabetes-afflicted individuals (9,10). Our focus on prediabetes as a separate metabolic category, combined with more complex analytical tools, implies that follow-up analysis on cells from four cohorts (lean/normoglycemic, obese/normoglycemic, obese/prediabetes, and obese/type 2 diabetes) will be needed to unify data on purified CD4+ T cells herein with outcomes from PBMC of obese/normoglycemic subjects and subjects with obesity/type 2 diabetes (9,14). This comparison will be essential in generating new strategies for the timing of immunomodulatory drugs to prevent disease progression by targeting mediators like Treg CD36, as successfully targeted in cancer (18). Our data also hint that the mechanistic foundation of T cell function significantly differs in prediabetes compared with type 2 diabetes, perhaps in part due to significantly higher mitochondrial function in the former. Although the subjects with type 2 diabetes in our study used glycemic control drugs, our previous work showed that metformin, the most common glycemic control drug for type 2 diabetes, did not alter mitochondrial function in PBMC from subjects with prediabetes (14). Although metformin may not be critical for the metabolic impacts of Treg-Teff cross talk, additional analyses are needed to quantify the impact of metformin on the differences between type 2 diabetes and prediabetes inflammation.
Our data are consistent with the interpretation that transcriptional mediators of lipid metabolism genes downstream of CD36 and its associated fatty acid signaling pathways (29) are altered by cell type and disease-associated mechanisms. Although CD36 on Treg from mouse visceral adipose tissue is modestly increased in IR (11) compared with the increases we see in Treg from subjects with prediabetes, the parallel increases in mouse but not human CPT-1a, PLIN2, and other genes involved in CD4+ T cell lipid metabolism suggest fundamentally different functional outcomes of elevated Treg CD36 in the two models of obesity-associated inflammation. Although higher CD36 on murine adipose Treg was attributed to decreased PPARγ (11,30–32), higher expression of human Treg CD36 seemed largely independent of PPARγ. A comprehensive dissection of lipid metabolism alongside transcription of lipid regulatory genes remains difficult in primary human Treg but might better be achieved in cell lines once we identify an animal model that recapitulates our findings. The challenging questions remain: how does a disease alter immune cell metabolism to regulate inflammation, and how can we translate these new concepts from bench to bedside?
Article Information
Acknowledgments. The authors thank Dr. Anat Erdreich-Epstein from the Departments of Pediatrics and Pathology, Children’s Hospital Los Angeles, Norris Comprehensive Cancer Center, and the Keck School of Medicine, University of Southern California, Los Angeles, CA, for important suggestions that contributed to rigor of results.
Funding. This work was funded by National Institute of Diabetes and Digestive and Kidney Diseases (grant R01DK108056 to D.A.L., P.A.K., and B.S.N.), the University of Kentucky Pharmaceutical Sciences PhD Program (R.L.), the Shared Resource Facility of the University of Kentucky Markey Cancer Center (P30 CA 177558), Barnstable Brown Diabetes and Obesity Center (B.S.N.), National Institutes of Health National Center for Advancing Translational Sciences UL1TR001998 (P.A.K.), and the Army Institute for Collaborative Biotechnologies Cooperative Agreement W911NF-19-2-0026 from the Army Research Office (D.A.L.). All statistics and bioinformatics analysis was funded by collaborative grants or support from the authors’ institutions.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. R.L. researched data and wrote the manuscript. G.H.P., E.T., and K.T. researched data. K.T. and D.A.L. researched data and wrote the manuscript. P.A.K. provided clinical expertise and human samples and wrote the manuscript. B.S.N. researched data and wrote the manuscript. All authors edited the manuscript and contributed to discussion. All statistics and bioinformatics analysis was done collaboratively by R.L., E.T., K.T., and D.A.L. B.S.N. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains supplementary material online at https://doi.org/10.2337/figshare.16915180.
References
- 1. Diabetes Prevention Program Research Group; Knowler WC, Fowler SE, Hamman RF, et al. 10-year follow-up of diabetes incidence and weight loss in the Diabetes Prevention Program Outcomes Study. Lancet 2009;374:1677–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Grossmann V, Schmitt VH, Zeller T, et al. Profile of the immune and inflammatory response in individuals with prediabetes and type 2 diabetes. Diabetes Care 2015;38:1356–1364 [DOI] [PubMed] [Google Scholar]
- 3. Brahimaj A, Ligthart S, Ghanbari M, et al. Novel inflammatory markers for incident pre-diabetes and type 2 diabetes: the Rotterdam Study. Eur J Epidemiol 2017;32:217–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu R, Nikolajczyk BS. Tissue immune cells fuel obesity-associated inflammation in adipose tissue and beyond. Front Immunol 2019;10:1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Touch S, Clément K, André S. T cell populations and functions are altered in human obesity and type 2 diabetes. Curr Diab Rep 2017;17:81. [DOI] [PubMed] [Google Scholar]
- 6. Dalmas E, Venteclef N, Caer C, et al. T cell-derived IL-22 amplifies IL-1β-driven inflammation in human adipose tissue: relevance to obesity and type 2 diabetes. Diabetes 2014;63:1966–1977 [DOI] [PubMed] [Google Scholar]
- 7. Jagannathan-Bogdan M, McDonnell ME, Shin H, et al. Elevated proinflammatory cytokine production by a skewed T cell compartment requires monocytes and promotes inflammation in type 2 diabetes. J Immunol 2011;186:1162–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fabbrini E, Cella M, McCartney SA, et al. Association between specific adipose tissue CD4+ T-cell populations and insulin resistance in obese individuals. Gastroenterology 2013;145:366–374.e1-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ip B, Cilfone NA, Belkina AC, et al. Th17 cytokines differentiate obesity from obesity-associated type 2 diabetes and promote TNFα production. Obesity (Silver Spring) 2016;24:102–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. DeFuria J, Belkina AC, Jagannathan-Bogdan M, et al. B cells promote inflammation in obesity and type 2 diabetes through regulation of T-cell function and an inflammatory cytokine profile. Proc Natl Acad Sci U S A 2013;110:5133–5138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cipolletta D, Feuerer M, Li A, et al. PPAR-γ is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature 2012;486:549–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Feuerer M, Herrero L, Cipolletta D, et al. Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat Med 2009;15:930–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Endo Y, Asou HK, Matsugae N, et al. Obesity drives Th17 cell differentiation by inducing the lipid metabolic kinase, ACC1. Cell Rep 2015;12:1042–1055 [DOI] [PubMed] [Google Scholar]
- 14. Nicholas DA, Proctor EA, Agrawal M, et al. Fatty acid metabolites combine with reduced β oxidation to activate Th17 inflammation in human type 2 diabetes. Cell Metab 2019;30:447–461.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Johnson MO, Wolf MM, Madden MZ, et al. Distinct regulation of Th17 and Th1 cell differentiation by glutaminase-dependent metabolism. Cell 2018;175:1780–1795.e19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. De Rosa V, Galgani M, Porcellini A, et al. Glycolysis controls the induction of human regulatory T cells by modulating the expression of FOXP3 exon 2 splicing variants. Nat Immunol 2015;16:1174–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Procaccini C, Carbone F, Di Silvestre D, et al. The proteomic landscape of human ex vivo regulatory and conventional T cells reveals specific metabolic requirements. Immunity 2016;44:406–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang H, Franco F, Tsui YC, et al. CD36-mediated metabolic adaptation supports regulatory T cell survival and function in tumors. Nat Immunol 2020;21:298–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nicholas D, Proctor EA, Raval FM, et al. Advances in the quantification of mitochondrial function in primary human immune cells through extracellular flux analysis. PLoS One 2017;12:e0170975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bharath LP, Agrawal M, McCambridge G, et al. Metformin enhances autophagy and normalizes mitochondrial function to alleviate aging-associated inflammation. Cell Metab 2020;32:44–55.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cait A, Hughes MR, Antignano F, et al. Microbiome-driven allergic lung inflammation is ameliorated by short-chain fatty acids. Mucosal Immunol 2018;11:785–795 [DOI] [PubMed] [Google Scholar]
- 22. Chinen T, Kannan AK, Levine AG, et al. An essential role for the IL-2 receptor in Treg cell function. Nat Immunol 2016;17:1322–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Laparra A, Tricot S, Le Van M, et al. The frequencies of immunosuppressive cells in adipose tissue differ in human, non-human primate, and mouse models. Front Immunol 2019;10:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Coort SL, Willems J, Coumans WA, et al. Sulfo-N-succinimidyl esters of long chain fatty acids specifically inhibit fatty acid translocase (FAT/CD36)-mediated cellular fatty acid uptake. Mol Cell Biochem 2002;239:213–219 [PubMed] [Google Scholar]
- 25. Luc K, Schramm-Luc A, Guzik TJ, Mikolajczyk TP. Oxidative stress and inflammatory markers in prediabetes and diabetes. J Physiol Pharmacol 2019;70:809–824 [DOI] [PubMed] [Google Scholar]
- 26. Hussaarts L, García-Tardón N, van Beek L, et al. Chronic helminth infection and helminth-derived egg antigens promote adipose tissue M2 macrophages and improve insulin sensitivity in obese mice. FASEB J 2015;29:3027–3039 [DOI] [PubMed] [Google Scholar]
- 27. Su CW, Chen CY, Li Y, et al. Helminth infection protects against high fat diet-induced obesity via induction of alternatively activated macrophages. Sci Rep 2018;8:4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kishore M, Cheung KCP, Fu H, et al. Regulatory T cell migration is dependent on glucokinase-mediated glycolysis. Immunity 2017;47:875–889.e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Glatz JF, Luiken JJ, Bonen A. Membrane fatty acid transporters as regulators of lipid metabolism: implications for metabolic disease. Physiol Rev 2010;90:367–417 [DOI] [PubMed] [Google Scholar]
- 30. Angela M, Endo Y, Asou HK, et al. Fatty acid metabolic reprogramming via mTOR-mediated inductions of PPARγ directs early activation of T cells. Nat Commun 2016;7:13683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gogg S, Nerstedt A, Boren J, Smith U. Human adipose tissue microvascular endothelial cells secrete PPARγ ligands and regulate adipose tissue lipid uptake. JCI Insight 2019;4:e125914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Marx N, Kehrle B, Kohlhammer K, et al. PPAR activators as antiinflammatory mediators in human T lymphocytes: implications for atherosclerosis and transplantation-associated arteriosclerosis. Circ Res 2002;90:703–710 [DOI] [PMC free article] [PubMed] [Google Scholar]