Abstract
Macro- and microvascular complications of type 2 diabetes (T2D), obesity, and dyslipidemia share common metabolic pathways. In this study, using a total of 1,300 metabolites from 996 Qatari adults (57% with T2D) and 1,159 metabolites from an independent cohort of 2,618 individuals from the Qatar BioBank (11% with T2D), we identified 373 metabolites associated with T2D, obesity, retinopathy, dyslipidemia, and lipoprotein levels, 161 of which were novel. Novel metabolites included phospholipids, sphingolipids, lysolipids, fatty acids, dipeptides, and metabolites of the urea cycle and xanthine, steroid, and glutathione metabolism. The identified metabolites enrich pathways of oxidative stress, lipotoxicity, glucotoxicity, and proteolysis. Second, we identified 15 patterns we defined as “metabo-clinical signatures.” These are clusters of patients with T2D who group together based on metabolite levels and reveal the same clustering in two or more clinical variables (obesity, LDL, HDL, triglycerides, and retinopathy). These signatures revealed metabolic pathways associated with different clinical patterns and identified patients with extreme (very high/low) clinical variables associated with extreme metabolite levels in specific pathways. Among our novel findings are the role of N-acetylmethionine in retinopathy in conjunction with dyslipidemia and the possible roles of N-acetylvaline and pyroglutamine in association with high cholesterol levels and kidney function.
Introduction
Metabolic association studies have successfully identified pathways perturbed in type 2 diabetes (T2D), a disease with complex etiology that is associated with diverse complications. In our previous study of subjects from the Middle East, an area with a high prevalence of T2D, we analyzed the plasma, saliva, and urine metabolic profiles of >350 individuals and revealed 94 metabolites significantly associated with T2D that were involved in metabolic pathways and different levels of glycemic control (1). We identified pathways involved in kidney function, glycosuria, lipolysis, proteolysis, brain function, and bile acids, among others.
Macro- and microvascular complications resulting from diabetes include cardiovascular conditions—the primary cause of diabetes-related mortality—and diabetic retinopathy. Both have various pathological mechanisms associated with dyslipidemia and abnormal lipoprotein levels. Other diabetes complications are associated with various risk factors, including hyperglycemia, hypertension, and dyslipidemia. Dyslipidemia is considered an independent risk factor for T2D (2): patients with T2D tend to have abnormal plasma lipid and lipoprotein levels, including decreased HDL cholesterol, a predominance of small dense LDL particles, and increased triglycerides (TRI) (3), despite having normal LDL-cholesterol levels. Furthermore, perturbed lipid metabolism and high glucose and insulin levels contribute to the development of atherosclerosis in patients with T2D (4,5). Impaired glucose tolerance and elevated free fatty acid levels in some patients suggest that insulin resistance occurs in those individuals before the onset of hyperglycemia (3,6). Insulin resistance is also associated with smaller and denser LDL particles and decreased HDL levels resulting from the hydrolysis of phospholipids in LDL and HDL particles (3,7–9). Several studies have investigated the levels of serum lipids (LDL, HDL, and TRI) in diabetic retinopathy (10), and increased TRI, total cholesterol, and LDL levels were found in patients with diabetic macular edema.
Most metabolomics studies on T2D have used T2D as an end point, and no studies have investigated correlations between the metabolic pathways involved with T2D and related complications. In this study, we used a recently developed metabolomics platform, the Metabolon DiscoveryHD4, to identify the metabolites in serum samples from 996 Qatari individuals. These high-resolution metabolomics data were combined with clinical data for each individual to investigate the relationships between metabolite levels and T2D, obesity, retinopathy, and dyslipidemia, including independent profiles of LDL, HDL, and TRI. Firstly, we aimed to identify metabolites associated with T2D and its complications and to replicate the findings in an independent set of 2,618 samples from the Qatar Biobank (QBB). Secondly, we clustered patients with T2D to identify groups of metabolites with similar correlations to two or more clinical variables, defining these as “metabo-clinical signatures.”
Research Design and Methods
Study Cohort
A total of 996 Qatari individuals were enrolled with written informed consent at Hamad Medical Corporation (HMC) and HMC Primary Health Care Centers in Doha, Qatar, and the study was approved by the Institutional Review Boards of HMC and Weill Cornell Medicine-Qatar. Of these patients, 574 (57%) had T2D. Individuals were included in the cohort if they were third-generation Qataris (four grandparents born in Qatar). Additional sample demographic characteristics are given in Supplementary Table 1A. The replication cohort (described in Supplementary Table 1A) consisted of 2,618 QBB Qatari subjects, 282 (11%) of whom had T2D (11). These were collected from the QBB and enrolled by written informed consent and were included with the approval of the HMC Institutional Review Board committee.
Metabolomics Data
Metabolomics data were obtained as previously described (12). Briefly, 200 μL of serum was analyzed on the Metabolon DiscoveryHD4 platform, and a total of 1,303 metabolites (including unknowns) were identified. Plasma samples from the QBB replication cohort were collected in BD SST Gel Separator Tubes and stored at −80°C, then aliquoted into 200-μL tubes and sent to the Anti-Doping Lab in Qatar for Metabolon untargeted metabolomic analysis. A total of 1,159 metabolites were profiled for the replication cohort.
Quality control included removal of outlier metabolite measurements (≥3 SDs from the mean) and metabolites missing >20% of the measurements. A total of 826 and 936 metabolites remained for analysis in the discovery and replication cohorts, respectively. A total of 547 metabolites overlapped between the discovery and replication metabolite sets. Metabolite measurements were log-scaled and z-score normalized prior to association analysis.
Metabolomics Association Analysis
To find associations between metabolites and T2D or its complications, metabolite levels were regressed against age, sex, hemolysis, batch effect, cotinine, BMI, T2D, and population stratification estimates using the lm function in the R statistical package (R Bioconductor). Population stratification by ethnicity was estimated by computing principal components (PC1, PC2, and PC3) from genotype data (as described below). A Bonferroni P value threshold was used to report significant associations with phenotypes (P ≤ 0.05/826). LDL, HDL, and TRI levels were analyzed for patients treated with and without statins as an additional covariate. Dyslipidemia was considered present if any of the following criteria were met: LDL >3.4 mmol/L (130 mg/dL), HDL <50 mg/dL (1.3 mmol/L) for women or <40 mg/dL (1.0 mmol/L) for men, TRI >1.7 mmol/L, or taking statins. In the replication cohort, statin information was not available at the time of the study, and dyslipidemia was based on TRI, LDL, and HDL values only.
Metabo-Clinical Signatures Analysis
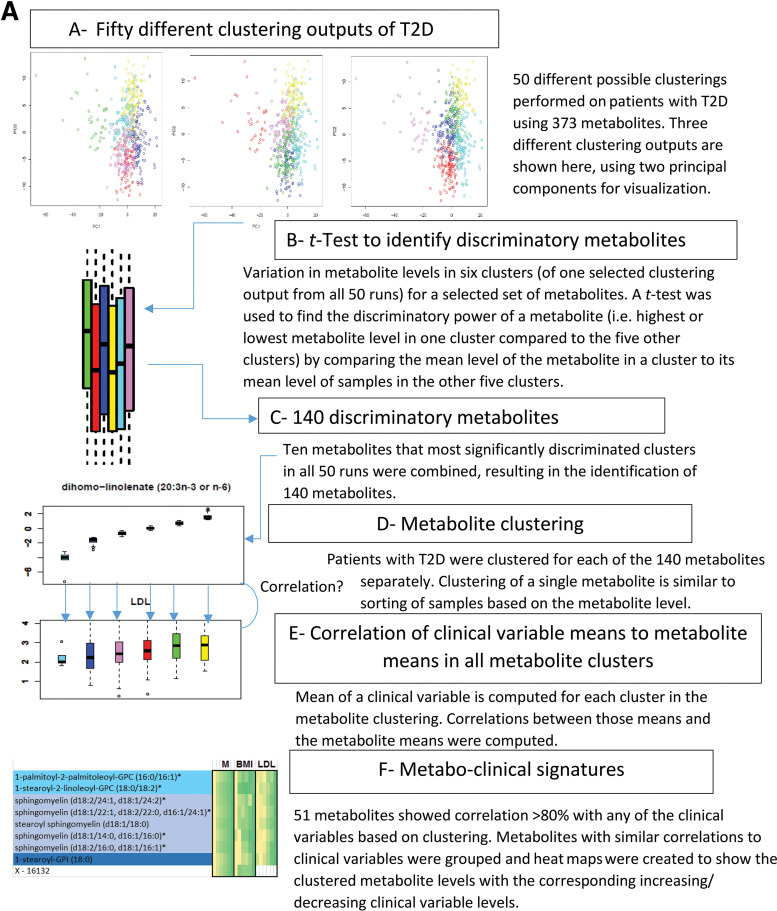
A flow diagram in Fig. 4 illustrates the steps of signature identification, which are explained in different phases below.
Figure 4.
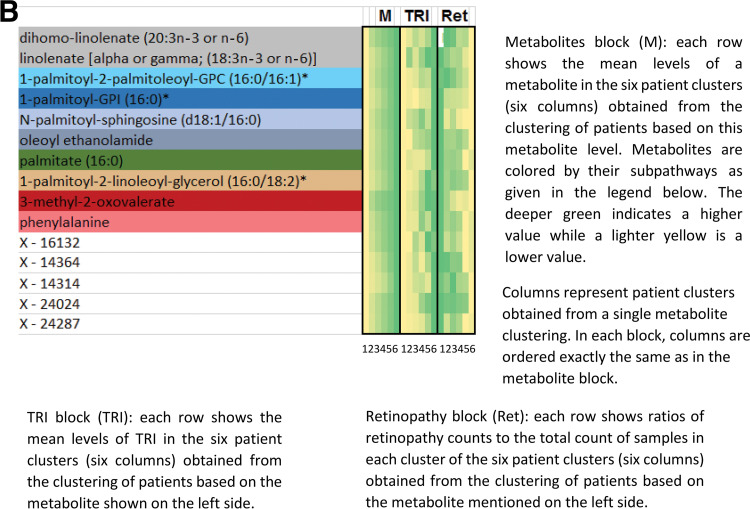
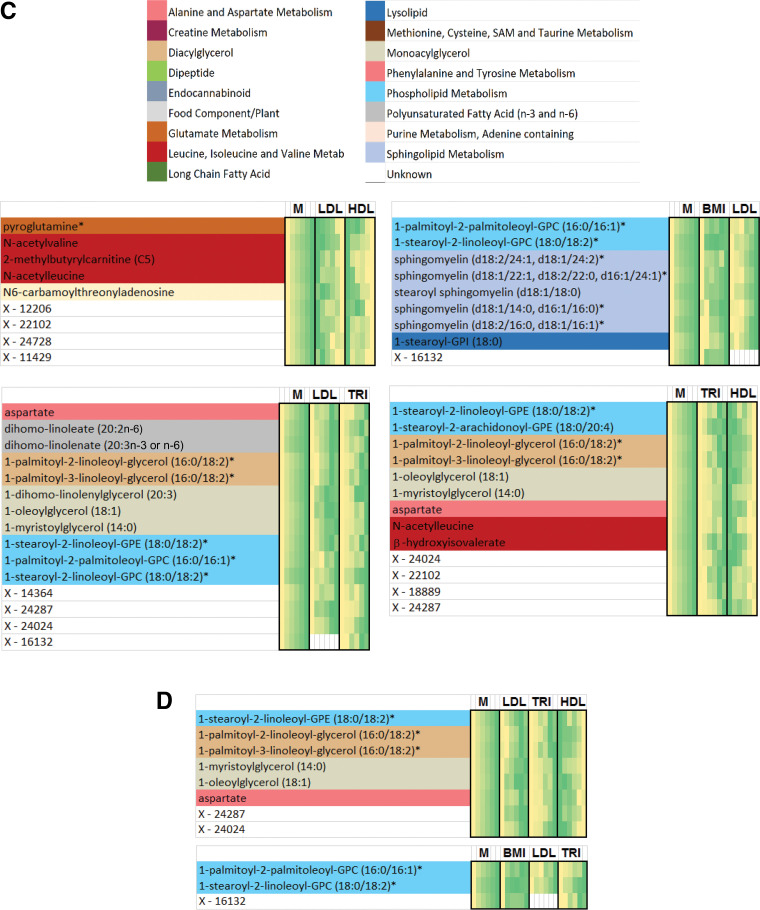
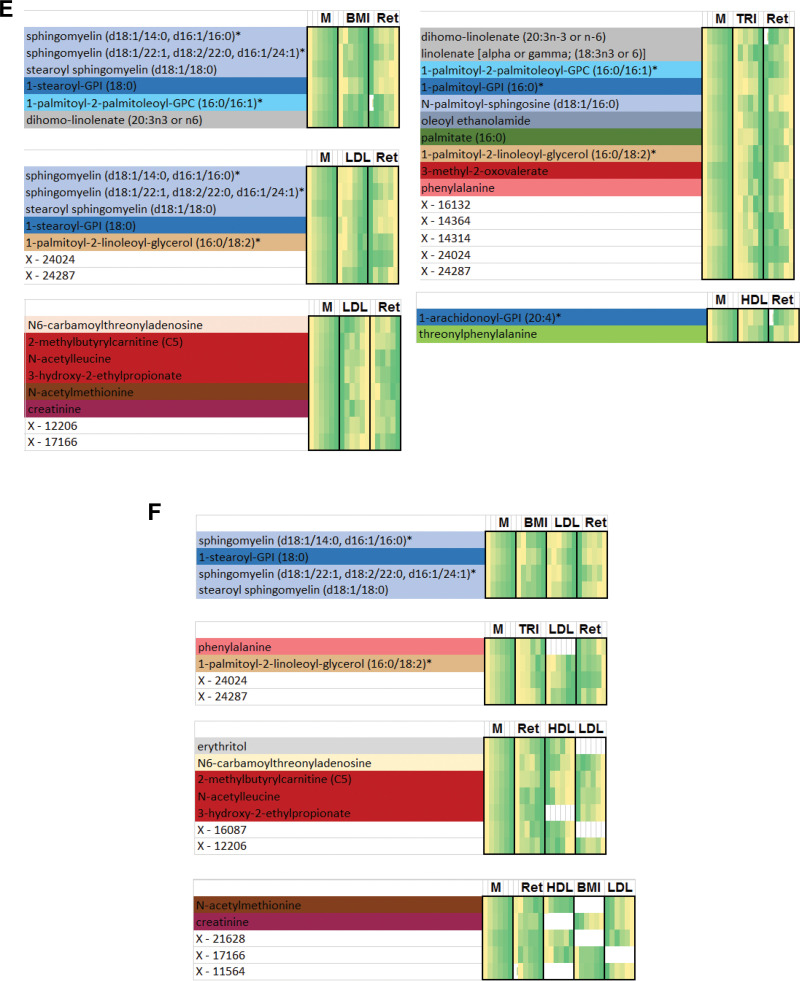
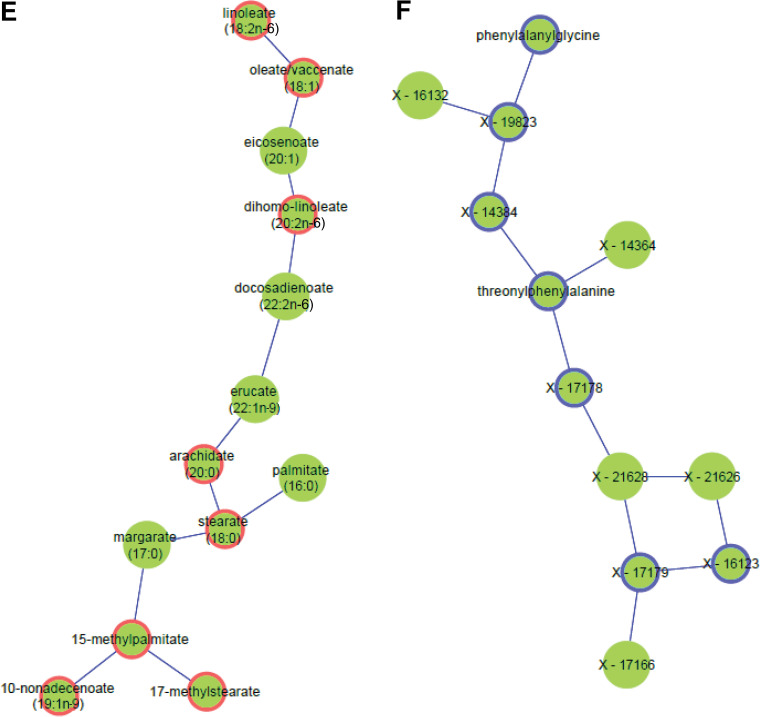
Flow diagram illustrating the identification of metabo-clinical signatures is shown in A. A magnified example metabo-clinical signature shown in B. C–F show all metabo-clinical signatures of patients with T2D for the clinical variables BMI, TRI, LDL, HDL, and retinopathy (see Supplementary Table 12B for magnified detailed views of all signatures). In each heat map, the metabolite block (M) shows the levels of each metabolite in six clusters (metabolites are color coded by their subpathway), followed by a block for each clinical variable to show the signature of that variable as per the clusters of each metabolite in the first block. In each heat map block, clusters are sorted on the means of the metabolite, and the deeper green indicates a higher value, while a lighter yellow is the opposite. Clinical variable names are displayed on the top side of the heat map, where M indicates the metabolites block and Ret indicates retinopathy. Signatures combining BMI, LDL, HDL, and TRI: four identified metabo-clinical signatures of two combined variables (C) and two identified metabo-clinical signatures of three combined clinical variables (D). Signatures combining retinopathy with BMI, LDL, HDL, and TRI: five identified metabo-clinical signatures combining two clinical variables (E) and four identified metabo-clinical signatures of three or more combined clinical variables (F). Rows with empty cells indicate that the correlation of the metabolite with the clinical variable is <80%.
Phase 1: Identification of 140 Discriminatory Metabolites
We ran K-means clustering on patients with T2D and 373 metabolites with 50 random seeds, with the number of clusters (k = 6) chosen as an intermediate number based on visual inspection of the PC analysis (PCA) to avoid overclustering of patients. In each of those 50 clustering results, we identified 10 metabolites that most significantly discriminated samples of each cluster from all the other samples (using P value from a t test to compare metabolite levels in clusters). Collectively, from all 50 runs, we identified 140 metabolites that discriminate the clusters from one another.
Phase 2: Identification of Clinical Variables Correlated With Metabolites Based on Clustering
Patients with T2D were clustered for each of the 140 metabolites using K-means (k = 6) (equivalent to sorting of samples based on the metabolite level). The mean values of metabolite levels and clinical variables (BMI, LDL, HDL, and TRI) and the ratio of patients with retinopathy (the number of retinopathy patients divided by the total number of patients in the cluster) were computed for each cluster. Correlation of the mean values of the metabolite levels with the clinical variables/retinopathy was then computed over the six clusters, and correlations ≥0.8 were selected to identify metabo-clinical signatures.
Phase 3: Heat Maps of Metabo-Clinical Signatures
Metabolites were grouped together based on the similarity of their correlations with clinical variables/retinopathy (Supplementary Tables 11 and 12A). The heat maps that show those similarities were then constructed.
PCA Computation
Genotyping consisted of 996 samples obtained by merging 614 imputed whole-exome sequences with 382 imputed genotype arrays (Illumina 2.5M array), as described in Yousri et al. (12). A total of 1,650,892 single nucleotide polymorphisms remained after removing those with imputation quality R2 < 0.5, minor allele frequency <0.05, genotype call rate <98%, and Hardy-Weinberg P value <10−6. PCA was computed using Plink software (version 1.9).
Gaussian Graphical Models
Partial correlations between two metabolites were calculated using the R package GeneNet, and significant partial correlations were identified as those that achieved a Bonferroni P value ≤0.05 (826 × 825/2).
Data and Resource Availability
Comprehensive data and statistical results that show associations with all metabolites are available in the Supplementary Tables.
Results
Population Characteristics
The 996 Qatari samples were used in the discovery analysis (see Supplementary Table 1A for cohort characteristics) and profiled for 1,300 metabolites (Supplementary Table 1B) (826 metabolites after quality control). Clinical chemistry and medication data (detailed in Research Design and Methods) were available for the majority of this cohort. Another independent set of 2,618 Qatari individuals from the QBB (11) was used to replicate the findings and profiled for 1,159 metabolites (936 after quality control).
Metabolites Associated With T2D
Regression Analysis
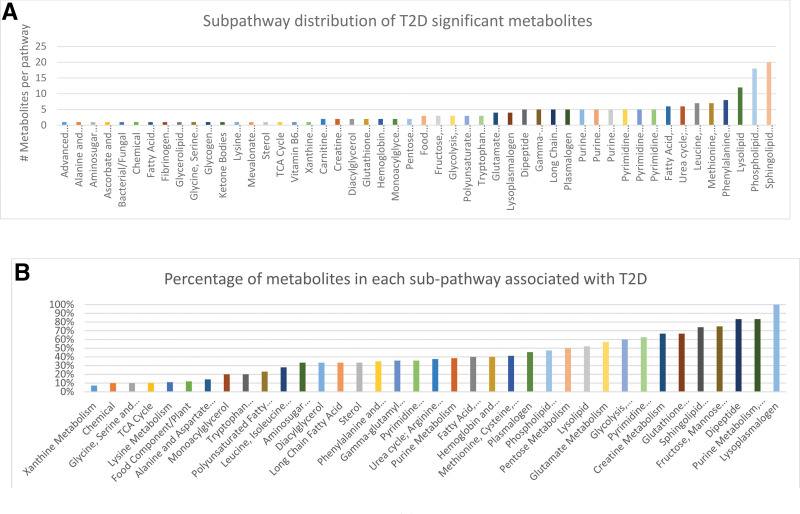
Associations between 826 metabolites and T2D were identified after correcting for age, sex, BMI, population stratification (genotype PCA), and batch effect. A total of 229 metabolites were found to be significantly associated with T2D (Bonferroni P < 0.05/826 = 6.05 × 10−5) (see Table 1 for the top most significant metabolites and Supplementary Tables 7 and 2 for all 229 metabolites and all 826 metabolites, respectively). These associations included 84 lipids, 42 amino acids, 11 carbohydrates, 11 peptides, 10 nucleotides, 6 xenobiotics, 4 cofactors and vitamins, 1 energy metabolite, and 60 unknown metabolites. Fig. 1 shows the pathway distributions of the significant metabolites and their ratios to each pathway size (total number of metabolites detected per pathway). Of the 229 metabolites, 123 had been previously reported in the literature, leaving 106 novel metabolites, of which 50 were replicated in the independent QBB cohort (Supplementary Table 7). Among the novel metabolites were thioproline, 3-methylglutaconate, and the dipeptides leucylalanine, phenylalanylglycine, and threonylphenylalanine. Also included were N-methylproline and N-acetylarginine from the urea cycle; theobromine from xanthine metabolism; and N-acetyltaurine, N-acetylmethionine, and S-methylcysteine from the methionine–taurine–S-adenosylmethionine (SAM) metabolism pathway.
Table 1.
T2D top most significant metabolites from the total of 229 metabolites, sorted by pathway, excluding unknown metabolites
| Biochemical | Superpathway | Subpathway | N | β (SE) | P value | Replication P value |
|---|---|---|---|---|---|---|
| Aspartate | Amino acid | Alanine and aspartate metabolism | 960 | 0.49 (0.07) | 3.73E-13 | 7.10E-05 |
| Guanidinoacetate | Amino acid | Creatine metabolism | 959 | −0.53 (0.06) | 9.64E-16 | 2.18E-04 |
| Glutamine | Amino acid | Glutamate metabolism | 962 | −0.61 (0.07) | 9.69E-17 | 4.11E-09 |
| Glutamate | Amino acid | Glutamate metabolism | 960 | 0.51 (0.06) | 5.97E-15 | 1.99E-13 |
| Pyroglutamine* | Amino acid | Glutamate metabolism | 961 | −0.51 (0.06) | 8.79E-15 | 3.35E-13 |
| Cysteine-glutathione disulfide | Amino acid | Glutathione metabolism | 902 | −0.58 (0.07) | 1.43E-17 | 6.95E-05 |
| Cys-gly, oxidized | Amino acid | Glutathione metabolism | 877 | −0.55 (0.06) | 5.85E-17 | 2.21E-02 |
| Pipecolate | Amino acid | Lysine metabolism | 944 | 0.52 (0.07) | 2.36E-12 | 3.68E-04 |
| S-adenosylhomocysteine | Amino acid | Methionine, cysteine, SAM, and glyco metabolism | 872 | 0.5 (0.07) | 1.41E-12 | 5.67E-01 |
| N-acetyltaurine | Amino acid | Methionine, cysteine, SAM, and taurine metabolism | 958 | −0.72 (0.07) | 2.88E-23 | NA |
| 2-Hydroxybutyrate/2-hydroxyisobutyrate | Amino acid | Methionine, cysteine, SAM, and taurine metabolism | 959 | 0.59 (0.07) | 4.76E-16 | 6.71E-01 |
| Taurine | Amino acid | Methionine, cysteine, SAM, and taurine metabolism | 962 | −0.57 (0.07) | 2.18E-15 | NA |
| N-acetylmethionine | Amino acid | Methionine, cysteine, SAM, and taurine metabolism | 953 | −0.55 (0.07) | 2.86E-14 | NA |
| 5-Bromotryptophan | Amino acid | Phenylalanine and tyrosine metabolism | 940 | −0.6 (0.07) | 2.62E-16 | 4.26E-03 |
| Citrulline | Amino acid | Urea cycle; arginine and proline metabolism | 964 | −0.74 (0.07) | 2.45E-24 | 1.25E-13 |
| Erythronate* | Carbohydrate | Aminosugar metabolism | 946 | 0.45 (0.07) | 7.29E-11 | 2.43E-03 |
| Mannose | Carbohydrate | Fructose, mannose, and galactose metabolism | 961 | 0.93 (0.06) | 3.63E-49 | NA |
| Fructose | Carbohydrate | Fructose, mannose, and galactose metabolism | 963 | 0.62 (0.07) | 1.27E-17 | NA |
| 1,5-Anhydroglucitol | Carbohydrate | Glycolysis, gluconeogenesis, and pyruvate metabolism | 962 | −1.2 (0.06) | 1.63E-68 | 1.44E-12 |
| Glucose | Carbohydrate | Glycolysis, gluconeogenesis, and pyruvate metabolism | 961 | 0.91 (0.06) | 6.88E-43 | 7.03E-02 |
| Ribonate | Carbohydrate | Pentose metabolism | 949 | 0.75 (0.07) | 2.48E-26 | 5.23E-03 |
| Arabonate/xylonate | Carbohydrate | Pentose metabolism | 950 | 0.73 (0.07) | 1.14E-25 | 4.18E-06 |
| Oxalate (ethanedioate) | Cofactors and vitamins | Ascorbate and aldarate metabolism | 962 | −0.48 (0.07) | 4.44E-13 | NA |
| Malate | Energy | TCA cycle | 949 | 0.52 (0.07) | 9.04E-14 | 1.65E-08 |
| 1-Palmitoyl-2-linoleoyl-glycerol (16:0/18:2)* | Lipid | Diacylglycerol | 963 | 0.49 (0.07) | 3.05E-12 | 1.38E-15 |
| Palmitoylcholine | Lipid | Fatty acid metabolism (acyl choline) | 958 | −0.9 (0.07) | 1.56E-35 | 7.78E-05 |
| 3-Hydroxydecanoate | Lipid | Fatty acid, monohydroxy | 955 | 0.71 (0.07) | 1.09E-22 | 7.77E-01 |
| 3-Hydroxyoctanoate | Lipid | Fatty acid, monohydroxy | 950 | 0.66 (0.07) | 3.76E-19 | 5.21E-01 |
| 3-Hydroxymyristate | Lipid | Fatty acid, monohydroxy | 918 | 0.52 (0.07) | 3.94E-12 | 1.23E-01 |
| 3-Hydroxylaurate | Lipid | Fatty acid, monohydroxy | 954 | 0.51 (0.07) | 6.62E-12 | 5.88E-08 |
| 3-Hydroxybutyrate | Lipid | Ketone bodies | 953 | 0.5 (0.07) | 1.47E-11 | 2.19E-10 |
| 1-Linoleoyl-GPC (18:2) | Lipid | Lysolipid | 964 | −0.6 (0.07) | 2.42E-17 | NA |
| 1-Stearoyl-GPC (18:0) | Lipid | Lysolipid | 964 | −0.56 (0.07) | 9.39E-15 | 8.03E-02 |
| 1-Stearoyl-GPI (18:0) | Lipid | Lysolipid | 959 | −0.55 (0.07) | 1.77E-13 | 7.97E-07 |
| 1-(1-Enyl-palmitoyl)-GPC (P-16:0)* | Lipid | Lysoplasmalogen | 963 | −0.81 (0.07) | 5.36E-30 | 2.50E-09 |
| 1-(1-Enyl-oleoyl)-GPE (P-18:1)* | Lipid | Lysoplasmalogen | 960 | −0.56 (0.07) | 8.18E-15 | 9.98E-05 |
| 1-(1-Enyl-palmitoyl)-GPE (P-16:0)* | Lipid | Lysoplasmalogen | 963 | −0.55 (0.07) | 2.85E-14 | 1.50E-01 |
| Arachidonoylcholine | Lipid | Phospholipid metabolism | 954 | −0.86 (0.07) | 2.32E-31 | 6.05E-09 |
| Oleoylcholine | Lipid | Phospholipid metabolism | 944 | −0.84 (0.07) | 1.46E-30 | 2.62E-01 |
| Dihomo-linolenoyl-choline | Lipid | Phospholipid metabolism | 882 | −0.69 (0.08) | 3.75E-19 | 7.67E-06 |
| 1,2-Distearoyl-GPC (18:0/18:0) | Lipid | Phospholipid metabolism | 923 | −0.66 (0.07) | 7.91E-19 | 4.70E-01 |
| Docosahexaenoylcholine | Lipid | Phospholipid metabolism | 808 | −0.67 (0.08) | 1.25E-16 | 4.93E-03 |
| 1-Palmitoyl-2-arachidonoyl-GPE (16:0/20:4)* | Lipid | Phospholipid metabolism | 962 | 0.5 (0.07) | 1.03E-12 | NA |
| 1-Palmitoyl-2-oleoyl-GPE (16:0/18:1) | Lipid | Phospholipid metabolism | 962 | 0.52 (0.07) | 1.49E-12 | NA |
| GPC | Lipid | Phospholipid metabolism | 957 | −0.5 (0.07) | 2.42E-11 | 4.91E-06 |
| 1-(1-Enyl-palmitoyl)-2-palmitoleoyl-GPC (P-16:0/16:1)* | Lipid | Plasmalogen | 954 | −0.73 (0.07) | 4.42E-24 | 1.00E-15 |
| 1-(1-Enyl-palmitoyl)-2-oleoyl-GPC (P-16:0/18:1)* | Lipid | Plasmalogen | 963 | −0.59 (0.07) | 6.25E-16 | 1.69E-01 |
| 1-(1-Enyl-palmitoyl)-2-linoleoyl-GPC (P-16:0/18:2)* | Lipid | Plasmalogen | 961 | −0.58 (0.07) | 1.17E-15 | 5.70E-05 |
| 1-(1-Enyl-palmitoyl)-2-palmitoyl-GPC (P-16:0/16:0)* | Lipid | Plasmalogen | 962 | −0.55 (0.07) | 5.39E-14 | 2.22E-36 |
| Sphingomyelin (d18:2/14:0, d18:1/14:1)* | Lipid | Sphingolipid metabolism | 963 | −0.7 (0.07) | 4.70E-25 | 1.32E-04 |
| Sphingomyelin (d18:2/24:1, d18:1/24:2)* | Lipid | Sphingolipid metabolism | 964 | −0.7 (0.07) | 3.45E-22 | NA |
| Lactosyl-N-palmitoyl-sphingosine | Lipid | Sphingolipid metabolism | 963 | −0.6 (0.07) | 2.29E-16 | 2.98E-02 |
| Sphingosine | Lipid | Sphingolipid metabolism | 941 | 0.53 (0.07) | 1.75E-14 | 5.36E-02 |
| Sphingomyelin (d18:1/24:1, d18:2/24:0)* | Lipid | Sphingolipid metabolism | 962 | −0.56 (0.07) | 2.21E-14 | 1.23E-02 |
| Sphingomyelin (d18:1/20:1, d18:2/20:0)* | Lipid | Sphingolipid metabolism | 963 | −0.53 (0.07) | 1.43E-13 | 1.56E-06 |
| Sphingomyelin (d18:2/23:0, d18:1/23:1, d17:1/24:1)* | Lipid | Sphingolipid metabolism | 960 | −0.54 (0.07) | 3.04E-13 | 1.77E-01 |
| Sphingomyelin (d18:2/16:0, d18:1/16:1)* | Lipid | Sphingolipid metabolism | 962 | −0.53 (0.07) | 3.66E-13 | 2.23E-03 |
| Sphingomyelin (d18:1/22:1, d18:2/22:0, d16:1/24:1)* | Lipid | Sphingolipid metabolism | 958 | −0.51 (0.07) | 2.21E-12 | 4.33E-10 |
| Palmitoyl sphingomyelin (d18:1/16:0) | Lipid | Sphingolipid metabolism | 961 | −0.51 (0.07) | 8.28E-12 | 1.93E-01 |
| Palmitoyl dihydrosphingomyelin (d18:0/16:0)* | Lipid | Sphingolipid metabolism | 961 | −0.5 (0.07) | 1.26E-11 | 5.55E-01 |
| Inosine | Nucleotide | Purine metabolism, (hypo)xanthine/inosine containing | 963 | −0.64 (0.07) | 1.33E-20 | 3.37E-08 |
| 7-Methylguanine | Nucleotide | Purine metabolism, guanine containing | 957 | −0.62 (0.07) | 9.33E-17 | 2.41E-02 |
| Dihydroorotate | Nucleotide | Pyrimidine metabolism, orotate containing | 847 | −0.48 (0.07) | 3.29E-11 | 5.44E-10 |
| 3-Aminoisobutyrate | Nucleotide | Pyrimidine metabolism, thymine containing | 943 | −0.53 (0.07) | 9.26E-13 | NA |
| 2'-Deoxyuridine | Nucleotide | Pyrimidine metabolism, uracil containing | 922 | −0.59 (0.06) | 1.80E-19 | 1.49E-04 |
| Leucylalanine | Peptide | Dipeptide | 914 | −0.61 (0.07) | 8.44E-17 | 5.14E-11 |
| DSGEGDFXAEGGGVR* | Peptide | Fibrinogen cleavage peptide | 926 | −0.51 (0.07) | 2.57E-13 | 4.44E-13 |
| Gluconate | Xenobiotics | Food component/plant | 944 | 0.85 (0.06) | 1.57E-36 | 4.43E-08 |
See Supplementary Table 7 for additional information.
NA, not available; TCA, tricarboxylic acid. Boldface text indicates a significant replication P value.
indicates compounds that have not been officially confirmed based on a standard, but Metabolon is confident in its identity.
Figure 1.
Subpathway distribution of T2D significant metabolites. A: The number of T2D metabolites in each pathway. B: Percentage of T2D metabolites in each pathway compared with the total number of metabolites in the pathway.
To determine the accuracy of using a metabolic profile to classify patients into those with T2D and control subjects, we used an orthogonal partial least square-discriminant analysis method. The accuracy of classification reached 95.6% (specificity 97.6% and sensitivity 94.4%) with a cross-validation Fisher test P value of 4.4 × 10−6 (Supplementary Fig. 1). Loadings (weights) of each metabolite had 80% correlation with their corresponding regression significance values and 82% correlation with their corresponding β-coefficients from the regression analysis. Box plots of the 80 most significant metabolites are shown in Supplementary Fig. 2A. A sensitivity analysis with medication and clinical chemistry was also performed (see Supplementary Material).
Metabolites Associated With Obesity/BMI
We investigated associations of the 826 metabolites with BMI after correcting for T2D, age, sex, population stratification (genotype PCA), and batch effect. We identified 62 significantly associated metabolites (Table 2 and Supplementary Table 2). Of these, 18 were also associated with T2D (i.e., were among the 229 metabolites obtained in the regression analysis) and 39 were replicated in the QBB cohort. Among the 62 metabolites were 5 steroids/sterols, 6 fatty acids, 4 phospholipids, 4 sphingolipids, 3 branched-chain amino acids (BCAAs), 3 lysolipids, 3 phenylalanine and tyrosine metabolites, and 18 unknown metabolites. Out of the 62 total, 34 were reported in previous literature and 28 were considered novel (Supplementary Table 7). Interesting novel metabolites include hydantoin-5-propionic acid from histidine metabolism and several steroids: pregnanediol-3-glucuronide, 5α-androstan-3β, 17α-diol disulfate, etiocholanolone glucuronide, 4-androsten-3β, 17β-diol disulfate (1), and 4-cholesten-3-one.
Table 2.
List of 62 metabolites significantly (Bonferroni) associated with BMI, sorted by pathway
| Metabolite | Superpathway | Subpathway | N | β (SE) | P value | Replication P value |
|---|---|---|---|---|---|---|
| Glutamate | Amino acid | Glutamate metabolism | 960 | 0.022 (0.004) | 5.20E-07 | 2.54487E-51 |
| Glutamine | Amino acid | Glutamate metabolism | 962 | −0.022 (0.005) | 4.67E-06 | 4.6646E-13 |
| Hydantoin-5-propionic acid | Amino acid | Histidine metabolism | 894 | 0.023 (0.005) | 3.60E-06* | NA |
| α-Hydroxyisovalerate | Amino acid | Leucine, isoleucine, and valine metabolism | 959 | 0.023 (0.005) | 2.67E-06* | 4.17463E-13 |
| 3-Methyl-2-oxobutyrate | Amino acid | Leucine, isoleucine, and valine metabolism | 943 | 0.024 (0.005) | 3.32E-06* | 5.8722E-12 |
| Isovalerylcarnitine | Amino acid | Leucine, isoleucine, and valine metabolism | 962 | 0.025 (0.005) | 2.28E-07* | NA |
| 5-Hydroxylysine | Amino acid | Lysine metabolism | 919 | 0.026 (0.005) | 1.95E-07* | 1.53686E-29 |
| 2-Aminoadipate | Amino acid | Lysine metabolism | 937 | 0.021 (0.005) | 8.39E-06* | 4.36112E-19 |
| Tyrosine | Amino acid | Phenylalanine and tyrosine metabolism | 961 | 0.025 (0.005) | 2.73E-07* | 2.16741E-27 |
| 3-(4-Hydroxyphenyl)lactate | Amino acid | Phenylalanine and tyrosine metabolism | 960 | 0.028 (0.005) | 4.24E-09* | 5.96568E-22 |
| N-acetyltyrosine | Amino acid | Phenylalanine and tyrosine metabolism | 918 | 0.023 (0.005) | 5.11E-06 | 1.283E-12 |
| C-glycosyltryptophan | Amino acid | Tryptophan metabolism | 937 | 0.020 (0.005) | 4.61E-05* | 1.74184E-28 |
| Homoarginine | Amino acid | Urea cycle; arginine and proline metabolism | 961 | 0.020 (0.005) | 4.10E-05 | 3.37588E-16 |
| Mannose | Carbohydrate | Fructose, mannose, and galactose metabolism | 961 | 0.016 (0.004) | 6.01E-05 | 1.48241E-18 |
| Pyruvate | Carbohydrate | Glycolysis, gluconeogenesis, and pyruvate metabolism | 956 | 0.024 (0.005) | 8.57E-07* | 2.11799E-08 |
| α-Ketoglutarate | Energy | TCA cycle | 944 | 0.026 (0.005) | 1.22E-07* | 1.11728E-13 |
| 1-Palmitoleoyl-2-oleoyl-glycerol (16:1/18:1)* | Lipid | Diacylglycerol | 882 | 0.027 (0.005) | 5.18E-08* | NA |
| 1-Palmitoleoyl-3-oleoyl-glycerol (16:1/18:1)* | Lipid | Diacylglycerol | 912 | 0.025 (0.005) | 5.97E-07* | NA |
| Propionylglycine | Lipid | Fatty acid metabolism (also BCAA metabolism) | 952 | −0.024 (0.005) | 1.69E-06* | 2.17619E-05 |
| Butyrylcarnitine | Lipid | Fatty acid metabolism (also BCAA metabolism) | 944 | 0.023 (0.005) | 2.40E-06* | NA |
| Palmitoleoylcarnitine* | Lipid | Fatty acid metabolism (acyl carnitine) | 963 | 0.022 (0.005) | 1.44E-05* | NA |
| Dodecanedioate | Lipid | Fatty acid, dicarboxylate | 939 | −0.021 (0.005) | 2.35E-05* | NA |
| Glycerol | Lipid | Glycerolipid metabolism | 962 | 0.033 (0.005) | 2.08E-12 | 1.31193E-17 |
| palmitoleate (16:1n7) | Lipid | LCFA | 962 | 0.021 (0.005) | 3.28E-05* | 5.29171E-09 |
| 1-Linoleoyl-GPC (18:2) | Lipid | Lysolipid | 964 | −0.022 (0.005) | 3.70E-06 | 2.33103E-05 |
| 1-Lignoceroyl-GPC (24:0) | Lipid | Lysolipid | 781 | −0.025 (0.006) | 6.79E-06* | 6.25697E-05 |
| 1-Linoleoyl-GPE (18:2)* | Lipid | Lysolipid | 963 | −0.023 (0.005) | 2.45E-06* | 0.003377949 |
| 1-Palmitoleoylglycerol (16:1)* | Lipid | Monoacylglycerol | 950 | 0.021 (0.005) | 1.79E-05* | 3.72058E-10 |
| 1-Palmitoyl-2-palmitoleoyl-GPC (16:0/16:1)* | Lipid | Phospholipid metabolism | 962 | 0.023 (0.005) | 1.08E-06* | 1.00148E-12 |
| 1,2-Dilinoleoyl-GPC (18:2/18:2) | Lipid | Phospholipid metabolism | 963 | −0.026 (0.005) | 1.21E-07 | 6.59082E-07 |
| 1-Linoleoyl-2-linolenoyl-GPC (18:2/18:3)* | Lipid | Phospholipid metabolism | 947 | −0.025 (0.005) | 3.73E-07* | 0.005034975 |
| 1-Oleoyl-2-linoleoyl-GPC (18:1/18:2)* | Lipid | Phospholipid metabolism | 960 | −0.030 (0.005) | 1.89E-09* | NA |
| Sphingomyelin (d18:2/14:0, d18:1/14:1)* | Lipid | Sphingolipid metabolism | 963 | 0.024 (0.004) | 5.76E-08 | 3.36128E-31 |
| Sphingomyelin (d18:1/18:1, d18:2/18:0) | Lipid | Sphingolipid metabolism | 962 | 0.030 (0.005) | 2.22E-10 | 1.9024E-16 |
| Sphingomyelin (d18:2/16:0, d18:1/16:1)* | Lipid | Sphingolipid metabolism | 962 | 0.020 (0.005) | 2.65E-05 | 3.3749E-10 |
| Stearoyl sphingomyelin (d18:1/18:0) | Lipid | Sphingolipid metabolism | 959 | 0.025 (0.005) | 6.83E-07 | 4.32037E-06 |
| Pregnanediol-3-glucuronide | Lipid | Steroid | 901 | −0.023 (0.005) | 1.54E-05* | 3.40421E-05 |
| 5α-Androstan-3β,17α-diol disulfate | Lipid | Steroid | 775 | −0.027 (0.005) | 9.78E-07* | 5.14421E-05 |
| Etiocholanolone glucuronide | Lipid | Steroid | 906 | −0.022 (0.005) | 1.32E-05* | 0.000123988 |
| 4-Androsten-3β,17β-diol disulfate (1) | Lipid | Steroid | 960 | 0.025 (0.005) | 3.11E-07* | NA |
| 4-Cholesten-3-one | Lipid | Sterol | 945 | 0.021 (0.005) | 2.13E-05* | NA |
| X-12026 | NA | NA | 942 | 0.024 (0.005) | 5.95E-07 | 1.06903E-24 |
| X-23639 | NA | NA | 961 | −0.023 (0.005) | 2.90E-06* | 2.15974E-14 |
| X-11849 | NA | NA | 956 | 0.022 (0.005) | 1.09E-05* | 6.89644E-11 |
| X-13835 | NA | NA | 926 | 0.022 (0.005) | 9.57E-06* | 3.74598E-10 |
| X-24337 | NA | NA | 950 | 0.021 (0.005) | 2.32E-05* | 7.53641E-06 |
| X-16132 | NA | NA | 962 | 0.028 (0.005) | 2.80E-09 | NA |
| X-24287 | NA | NA | 941 | 0.025 (0.005) | 5.15E-07 | NA |
| X-24286 | NA | NA | 920 | 0.023 (0.005) | 3.08E-06 | NA |
| X-11564 | NA | NA | 940 | 0.023 (0.005) | 1.07E-06* | NA |
| X-21785 | NA | NA | 956 | 0.021 (0.005) | 1.16E-05* | NA |
| X-12063 | NA | NA | 955 | 0.033 (0.005) | 1.47E-10* | NA |
| X-15245 | NA | NA | 859 | 0.032 (0.005) | 1.64E-09* | NA |
| X-24054 | NA | NA | 960 | −0.021 (0.005) | 2.00E-05* | NA |
| X-24513 | NA | NA | 943 | 0.029 (0.005) | 2.03E-09* | NA |
| X-23652 | NA | NA | 938 | 0.020 (0.005) | 3.51E-05* | NA |
| X-24125 | NA | NA | 892 | 0.023 (0.005) | 5.03E-06* | NA |
| X-23749 | NA | NA | 955 | −0.021 (0.005) | 6.60E-06* | NA |
| X-17359 | NA | NA | 925 | 0.025 (0.005) | 6.81E-07* | NA |
| Urate | Nucleotide | Purine metabolism, (Hypo)xanthine/inosine containing | 963 | 0.031 (0.005) | 6.35E-11 | 1.924E-48 |
| N6-carbamoylthreonyladenosine | Nucleotide | Purine metabolism, adenine containing | 941 | 0.021 (0.005) | 2.23E-05* | 3.45616E-29 |
| γ-Glutamylglutamine | Peptide | γ-Glutamyl amino acid | 962 | −0.021 (0.005) | 8.29E-06 | 5.86113E-21 |
Table includes 18 metabolites significant to both BMI and T2D and 44 associated only with BMI (P values marked with *).
Boldface text indicates a significant replication P value. NA, not available; TCA, tricarboxylic acid.
Metabolites Associated With Diabetic Retinopathy
Consistent with the high prevalence of retinopathy in Qatar, 123 patients with T2D had retinopathy, while 116 did not, and no information was available for the remaining 335 patients. We identified 28 significantly associated metabolites (Table 3 and Supplementary Table 4), of which 13 were reported in previous studies and 15 were considered novel (Supplementary Table 7). Notably, eight were only associated with retinopathy (did not associate with T2D), namely: xanthine, 1-palmitoyl-GPA (16:0), tryptophan, glycoursodeoxycholate, phenylacetylglutamine, X-23997, X-13729, and 5-methylthioadenosine (MTA). The identified metabolites are from purine, (hypo)xanthine/inosine, phenylalanine and tyrosine, glutathione and sphingolipid metabolism, as well as other pathways, and included 11 unknown metabolites. Interesting novel metabolites associated with retinopathy included 1-palmitoyl-GPA (16:0), glycylvaline, and N-acetylmethionine and oxidized cys-gly and cysteine-glutathione disulfide from glutathione metabolism.
Table 3.
List of 28 metabolites significantly (Bonferroni) associated with retinopathy, tested in T2D samples
| Metabolite | Superpathway | Subpathway | N | β | SE | P value |
|---|---|---|---|---|---|---|
| X-14364 | NA | NA | 224 | −0.72 | 0.11 | 4.81E-10 |
| Glycylvaline | Peptide | Dipeptide | 220 | −0.73 | 0.11 | 8.92E-10 |
| Xanthine | Nucleotide | Purine metabolism, (hypo)xanthine/inosine containing | 225 | −0.73 | 0.12 | 4.67E-09* |
| Cysteine-glutathione disulfide | Amino acid | Glutathione metabolism | 215 | 0.73 | 0.12 | 8.35E-09 |
| Sphinganine | Lipid | Sphingolipid metabolism | 214 | −0.64 | 0.12 | 1.44E-07 |
| X-17166 | NA | NA | 226 | 0.67 | 0.12 | 1.75E-07 |
| DSGEGDFXAEGGGVR* | Peptide | Fibrinogen cleavage peptide | 214 | 0.55 | 0.10 | 2.36E-07 |
| X-16134 | NA | NA | 208 | 0.59 | 0.11 | 3.29E-07 |
| Sphingosine | Lipid | Sphingolipid metabolism | 220 | −0.64 | 0.12 | 3.55E-07 |
| X-14095 | NA | NA | 211 | −0.57 | 0.11 | 5.71E-07 |
| Phenylalanine | Amino acid | Phenylalanine and tyrosine metabolism | 226 | −0.55 | 0.11 | 8.47E-07 |
| Aspartate | Amino acid | Alanine and aspartate metabolism | 226 | −0.54 | 0.11 | 1.21E-06 |
| Phenylacetylglutamine | Amino acid | Phenylalanine and tyrosine metabolism | 224 | 0.59 | 0.12 | 1.43E-06* |
| X-14314 | NA | NA | 216 | −0.53 | 0.11 | 1.74E-06 |
| Glutamate | Amino acid | Glutamate metabolism | 226 | −0.53 | 0.11 | 1.76E-06 |
| Inosine | Nucleotide | Purine metabolism, (hypo)xanthine/Inosine containing | 226 | 0.53 | 0.11 | 2.30E-06 |
| 1-Palmitoyl-GPA (16:0) | Lipid | Lysolipid | 222 | −0.59 | 0.12 | 2.76E-06* |
| Cys-gly, oxidized | Amino acid | Glutathione metabolism | 210 | 0.62 | 0.13 | 3.68E-06 |
| X-21628 | NA | NA | 220 | 0.52 | 0.11 | 3.81E-06 |
| Tryptophan | Amino acid | Tryptophan metabolism | 226 | −0.53 | 0.11 | 3.95E-06* |
| X-17167 | NA | NA | 209 | 0.52 | 0.11 | 5.67E-06 |
| X-11381 | NA | NA | 225 | −0.56 | 0.12 | 1.20E-05 |
| X-23997 | NA | NA | 205 | 0.61 | 0.14 | 2.57E-05* |
| X-13729 | NA | NA | 186 | 0.64 | 0.15 | 3.57E-05* |
| 5-MTA | Amino acid | Polyamine metabolism | 207 | 0.53 | 0.13 | 3.95E-05* |
| Glycoursodeoxycholate | Lipid | Secondary bile acid metabolism | 226 | −0.53 | 0.13 | 3.99E-05* |
| X-21626 | NA | NA | 210 | 0.47 | 0.11 | 5.30E-05 |
| N-acetylmethionine | Amino acid | Methionine, cysteine, SAM, and taurine metabolism | 223 | 0.48 | 0.12 | 5.54E-05 |
Negative β value means retinopathy decreased compared with nonretinopathy. Eight metabolites are associated only with retinopathy (P values marked with *).
NA, not available.
Metabolites Associated With Dyslipidemia and Lipid Traits
Among the 621 individuals with dyslipidemia, 458 (74%) had T2D (i.e., 79% of patients with T2D had dyslipidemia). Twenty-four metabolites were associated with dyslipidemia (Bonferroni P < 0.05/826 = 6.05 × 10−5) (Table 4 and Supplementary Tables 5 and 6 report all associations), of which 15 were reported in previous studies and 9 were novel. Nine of the 24 metabolites were replicated in the QBB cohort (Supplementary Table 7). Next, to understand the detailed mechanism of cholesterol metabolism and its relation to each of the studied clinical variables, we identified the metabolites associated with TRI and lipoproteins. The results showed that 143, 52, 27, and 22 metabolites were significantly associated with TRI, LDL, HDL, and LDL/HDL, respectively. Of these, 44, 33, 12, and 14 metabolites, respectively, were considered novel (Table 4 and Supplementary Tables 5, 6, and 8 show replicated, previously reported, and novel metabolites, respectively). These were mostly lysolipids, sphingolipids, phospholipids, fatty acids, and, interestingly, BCAAs. The 24 metabolites associated with dyslipidemia were also associated with either TRI or HDL. Excluding the identified metabolites associated with T2D and BMI, 64 of 143 metabolites were only associated with TRI, 11 of 52 with LDL, and 2 of 27 with HDL (Table 4 and Supplementary Table 5). Of the 44 metabolites associated with BMI but not T2D, 19 were associated with lipoproteins, including 16 with TRI, 6 with HDL, 3 with both HDL and TRI, 1 with the LDL/HDL ratio, and 1 with LDL (both were also associated with TRI) (Supplementary Table 9). The results were similar after adjustment for statins.
Table 4.
Top 10 metabolites most significant to each of the lipids, TRI, and dyslipidemia
| Metabolite | Superpathway | Subpathway | HDL | LDL | TRI | LDL/HDL | Dyslipidemia | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β | P value | β | P value | β | P value | β | P value | β | P value | |||
| 1-Palmitoyl-2-linoleoyl-glycerol (16:0/18:2)* | Lipid | Diacylglycerol | −0.69 | 2.25E-12 | 0.12 | 9.95E-04 | 0.46 | 3.53E-42 | 0.19 | 4.59E-09 | 0.53 | 7.29E-10 |
| 1-Palmitoyl-3-linoleoyl-glycerol (16:0/18:2)* | Lipid | Diacylglycerol | −0.67 | 3.18E-11 | 0.15 | 7.05E-05 | 0.48 | 1.20E-42 | 0.20 | 1.32E-09 | 0.52 | 4.17E-09 |
| 1-Oleoyl-2-linoleoyl-glycerol (18:1/18:2) | Lipid | Diacylglycerol | −0.69 | 3.89E-11 | 0.08 | 3.35E-02 | 0.51 | 4.12E-47 | 0.16 | 4.48E-06 | 0.59 | 9.22E-11 |
| 1-Oleoyl-3-linoleoyl-glycerol (18:1/18:2) | Lipid | Diacylglycerol | −0.67 | 3.76E-10 | 0.11 | 4.41E-03 | 0.50 | 2.08E-42 | 0.17 | 1.07E-06 | 0.58 | 6.08E-10 |
| 1-Palmitoleoylglycerol (16:1)* | Lipid | Monoacylglycerol | −0.48 | 7.09E-06 | 0.07 | 7.24E-02 | 0.48 | 3.09E-35 | 0.13 | 2.00E-04 | 0.51 | 4.44E-08 |
| 1-Oleoylglycerol (18:1) | Lipid | Monoacylglycerol | −0.61 | 3.41E-09 | 0.09 | 1.75E-02 | 0.43 | 5.14E-31 | 0.17 | 4.26E-07 | 0.54 | 2.48E-09 |
| 1-Palmitoyl-2-stearoyl-GPC (16:0/18:0) | Lipid | Phospholipid metabolism | 0.15 | 1.78E-01 | 0.30 | 5.80E-13 | 0.23 | 4.66E-08 | 0.16 | 8.44E-06 | 0.25 | 1.21E-02 |
| 1-Stearoyl-2-arachidonoyl-GPE (18:0/20:4) | Lipid | Phospholipid metabolism | −0.38 | 2.49E-04 | 0.05 | 1.74E-01 | 0.50 | 9.13E-45 | 0.10 | 3.23E-03 | 0.58 | 1.16E-10 |
| 1-(1-Enyl-palmitoyl)-2-oleoyl-GPC (P-16:0/18:1)* | Lipid | Plasmalogen | 0.78 | 7.63E-13 | 0.17 | 2.49E-05 | −0.11 | 7.19E-03 | −0.02 | 6.08E-01 | −0.36 | 1.91E-04 |
| 1-(1-Enyl-palmitoyl)-2-linoleoyl-GPC (P-16:0/18:2)* | Lipid | Plasmalogen | 0.73 | 4.05E-12 | 0.19 | 1.02E-06 | −0.08 | 3.55E-02 | −0.01 | 8.34E-01 | −0.39 | 3.90E-05 |
| 1-(1-Enyl-palmitoyl)-2-palmitoleoyl-GPC (P-16:0/16:1)* | Lipid | Plasmalogen | 0.66 | 2.26E-10 | 0.10 | 1.36E-02 | −0.11 | 3.13E-03 | −0.04 | 1.82E-01 | −0.39 | 1.57E-05 |
| Behenoyl sphingomyelin (d18:1/22:0)* | Lipid | Sphingolipid metabolism | 0.16 | 1.59E-01 | 0.36 | 6.18E-19 | 0.15 | 2.39E-04 | 0.20 | 3.69E-08 | 0.16 | 1.02E-01 |
| Palmitoyl dihydrosphingomyelin (d18:0/16:0)* | Lipid | Sphingolipid metabolism | 0.31 | 6.00E-03 | 0.33 | 1.02E-15 | 0.00 | 9.76E-01 | 0.15 | 1.61E-05 | −0.03 | 7.60E-01 |
| Sphingomyelin (d18:1/20:0, d16:1/22:0)* | Lipid | Sphingolipid metabolism | 0.29 | 9.92E-03 | 0.32 | 8.32E-15 | 0.15 | 1.78E-04 | 0.15 | 2.59E-05 | 0.11 | 2.49E-01 |
| Sphingomyelin (d18:1/22:1, d18:2/22:0, d16:1/24:1)* | Lipid | Sphingolipid metabolism | 0.33 | 2.91E-03 | 0.30 | 2.77E-14 | 0.01 | 7.40E-01 | 0.13 | 1.65E-04 | −0.01 | 9.32E-01 |
| Sphingomyelin (d18:1/17:0, d17:1/18:0, d19:1/16:0) | Lipid | Sphingolipid metabolism | 0.21 | 6.11E-02 | 0.30 | 3.85E-13 | 0.00 | 9.79E-01 | 0.15 | 2.22E-05 | 0.09 | 3.41E-01 |
| Stearoyl sphingomyelin (d18:1/18:0) | Lipid | Sphingolipid metabolism | 0.08 | 4.73E-01 | 0.30 | 7.90E-13 | 0.10 | 1.04E-02 | 0.18 | 4.21E-07 | 0.24 | 1.56E-02 |
| Sphingomyelin (d18:1/21:0, d17:1/22:0, d16:1/23:0)* | Lipid | Sphingolipid metabolism | 0.30 | 7.74E-03 | 0.29 | 8.82E-13 | 0.05 | 2.44E-01 | 0.13 | 3.52E-04 | 0.05 | 6.27E-01 |
| N-palmitoyl-sphingosine (d18:1/16:0) | Lipid | Sphingolipid metabolism | −0.16 | 1.80E-01 | 0.29 | 1.11E-11 | 0.31 | 4.32E-13 | 0.22 | 2.16E-09 | 0.30 | 3.47E-03 |
| N-palmitoyl-sphinganine (d18:0/16:0) | Lipid | Sphingolipid metabolism | −0.14 | 2.22E-01 | 0.27 | 5.13E-10 | 0.29 | 9.55E-11 | 0.19 | 3.39E-07 | 0.30 | 3.15E-03 |
| Cholesterol | Lipid | Sterol | 0.10 | 3.97E-01 | 0.37 | 2.35E-18 | 0.23 | 2.01E-08 | 0.22 | 2.91E-09 | 0.22 | 3.14E-02 |
| X-24286 | NA | NA | −0.64 | 1.08E-09 | 0.08 | 3.75E-02 | 0.45 | 1.36E-35 | 0.17 | 1.13E-06 | 0.57 | 4.50E-10 |
| X-24125 | NA | NA | −0.67 | 1.63E-09 | 0.02 | 5.93E-01 | 0.50 | 1.03E-37 | 0.12 | 6.16E-04 | 0.54 | 1.87E-08 |
| X-24287 | NA | NA | −0.54 | 2.40E-07 | 0.11 | 4.31E-03 | 0.46 | 6.03E-36 | 0.16 | 2.43E-06 | 0.51 | 1.78E-08 |
| X-24263 | NA | NA | −0.37 | 4.35E-04 | 0.00 | 9.89E-01 | 0.40 | 1.89E-27 | 0.04 | 2.97E-01 | 0.54 | 4.54E-09 |
| X-24278 | NA | NA | −0.39 | 3.18E-04 | 0.05 | 1.98E-01 | 0.39 | 2.55E-26 | 0.07 | 4.42E-02 | 0.53 | 1.74E-08 |
indicates compounds that have not been officially confirmed based on a standard, but Metabolon is confident in its identity. NA, not available.
Metabolic Associations Shared Among Clinical Variables
In total, 373 metabolites were significant to any studied clinical variable. Of these, 161 were novel (86 known metabolites and 75 unknowns, out of which 50 were replicated in QBB), while 212 were previously reported (T2D, obesity, their complications, or dyslipidemia) in 78 unique references (Supplementary Table 10), including our previous study (1) and other studies (13–17).
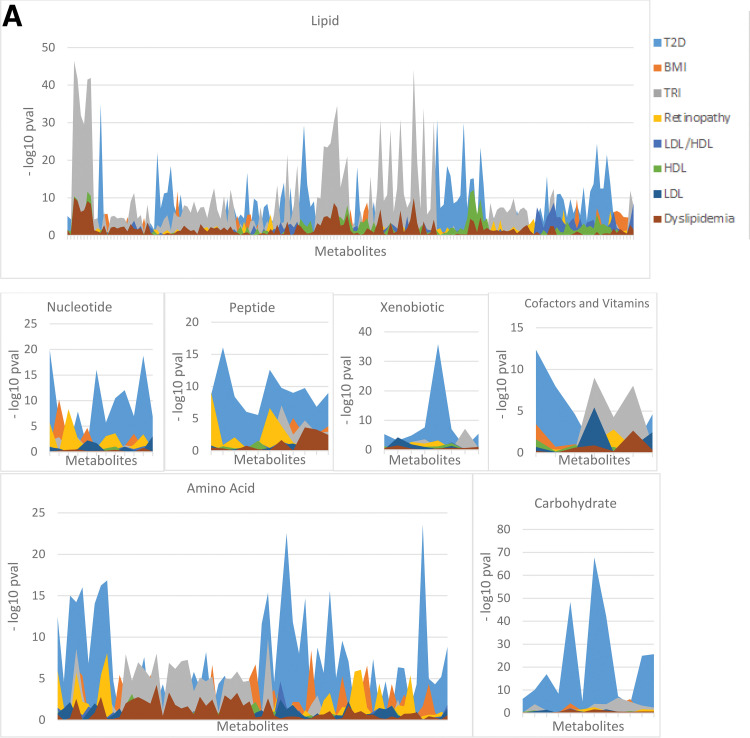
Metabolites were either only significant to one clinical variable (n = 231 metabolites, including 122 to T2D, 25 to BMI, 8 to retinopathy, 3 to HDL, 6 to LDL, and 67 to TRI) or shared by two or more phenotypes (n = 142 metabolites) (Supplementary Table 9). Fig. 2A compares the significance of metabolites for the eight clinical variables in each of the superpathways. T2D has the highest significance of associations in most of the pathways, while both T2D and TRI show the highest significance with lipids. Fig. 2B shows the heat map of associations of the clinical variables with 373 metabolites. Supplementary Fig. 3 shows the pairwise correlations of clinical variables based on their metabolic associations, while Supplementary Fig. 4 shows box plots of metabolites shared between clinical variables (Supplementary Fig. 4A and B) and associated with a single clinical variable (Supplementary Fig. 4C–H).
Figure 2.
A: Pathway associations with clinical variables. The y-axis shows the –log10 P value for all metabolites in each pathway, and the x-axis represents the metabolites in this pathway. The peaks present the −log10 P values, while the areas under the peaks are only used for a better visualization of the P values. B: Pathway associations with eight clinical variables. Clustered heat map of (Sign[β] ∗ −log10 [P value]) of metabolite associations with the clinical variables displaying 17 subpathways that have >5 metabolites from the 373 identified metabolites. Deep blue indicates the highest positive association, and deep red indicates the lowest negative association.
Interestingly, only two metabolites were shared among retinopathy and clinical outcomes other than T2D: glutamate, which was associated with T2D, BMI, retinopathy, and TRI; and N-acetylmethionine, associated with the LDL/HDL ratio. Among the many novel shared metabolites identified in our study were the following interesting findings: T2D and retinopathy shared associations with oxidized cys-gly and cysteine-glutathione disulfide from glutathione metabolism and with N-acetylmethionine (also shared with LDL/HDL) and glycylvaline. BMI, LDL, and TRI shared associations with the novel metabolite 4-cholesten-3-one. Novel metabolites that only associated with a single clinical variable included 1-palmitoyl-GPA (16:0), associated with retinopathy, and hydantoin-5-propionic acid, which was associated with BMI. Interestingly, three BCAAs—N-acetylvaline, β-hydroxyisovalerate, and N-acetylisoleucine—among other metabolites in fatty acid and lipid metabolism, were novel and associated only with TRI.
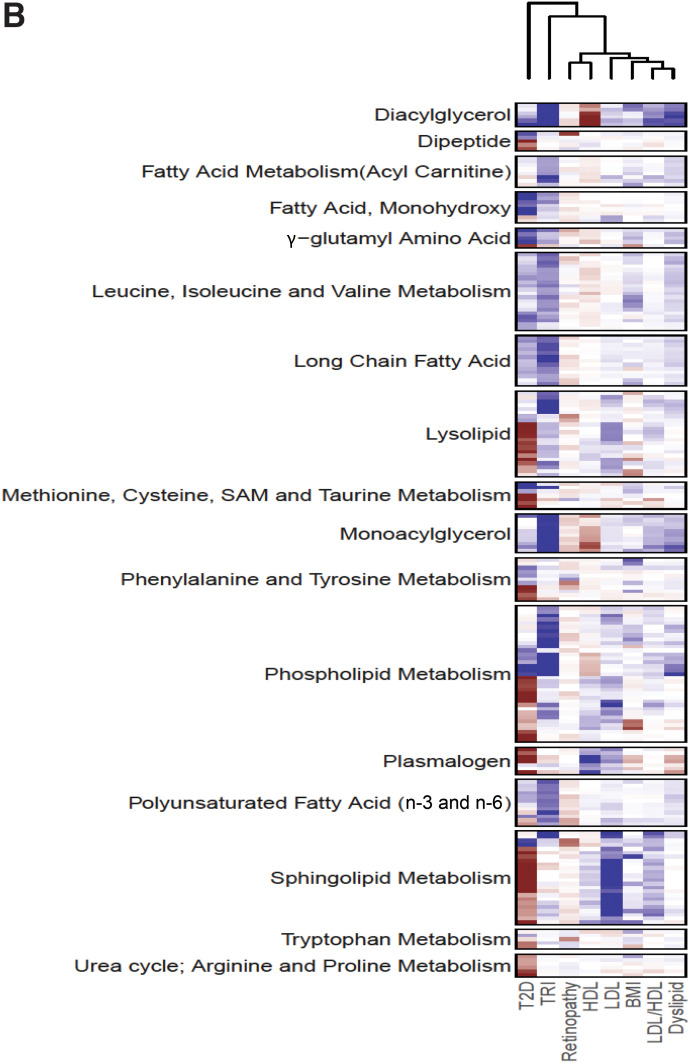
Metabolic Interactions and Gaussian Graphical Models
To investigate potential metabolic pathways associated with the clinical variables, Gaussian graphical models (GGMs) were constructed based on significant partial correlations among all 826 metabolites and by selecting only edges (i.e., significant partial correlations) connecting the 373 clinically significant metabolites. A total of 27 subnetworks were identified with ≥3 metabolites. The largest 11 subnetworks (≥6 metabolites) are split between Fig. 3 and Supplementary Fig. 5. Metabolites significant with T2D only or with TRI only are highlighted, showing several subnetworks linking those metabolites together. The largest (the lipid subnetwork) connected 65 metabolites, mostly palmitoyls/stearoyl/arachidonoyl/oleoyl/linoleoyl-glycerophosphorylcholine (GPC)/glycerophosphatidylethanolamine (GPE)/glycerophosphatidylinositol (GPI) lipids (Supplementary Fig. 5A), including 25 phospholipids, 19 lysolipids, and 5 plasmalogens. Fifteen metabolites of this subnetwork were only associated with TRI, 6 were only associated with T2D, and 37 were common between two or more phenotypes.
Figure 3.
A selected set of subnetworks from the largest 11 GGM subnetworks constructed from significant partial correlation of 373 metabolites associated with any of the 8 clinical variables. The larger the node, the more phenotypes are associated with the metabolite (up to six phenotypes can be associated with metabolites). If four or more metabolites are associated only with a single clinical variable, the border of the node is colored: a red border indicates association with TRI only, and a blue border indicates association with T2D only. Monoacylglycerol/phospholipid/PUFAs (17 metabolites) (A), monohydroxy fatty acids (B), carbohydrates/sugars (C), fungal/xenobiotic/unknowns (D), fatty acid subnetwork (LCFA/PUFAs) (13 metabolites) (E), and dipeptides and unknowns (F).
Metabo-Clinical Signatures of Patients With T2D
We clustered patients with T2D using each of 140 metabolites separately (see Research Design and Methods and Fig. 4A) to: 1) identify metabolites that correlate – based on their cluster means, with two or more clinical variables, and 2) identify groups of metabolites that have similar correlations to those variables and present them together with their correlated clinical variables in what we defined as a “metabo-clinical signature.”
Out of the 140 metabolites, 51 correlated with the mean levels of two or more clinical variables across the clusters at >80% (see research design and methods and Fig. 4). Supplementary Table 11 shows the correlations. Accordingly, we identified six metabo-clinical signatures when considering obesity (BMI), LDL, HDL, and TRI and identified nine metabo-clinical signatures after adding retinopathy to those variables. Fig. 4 (detailed in Supplementary Table 12A and B) shows the flow diagram of identifying the signatures and the resulting heat maps of the identified metabo-clinical signatures.
Among the observed signatures for cholesterols is a group of patients with simultaneous highest HDL levels and highest LDL levels (Fig. 4C, top left corner) having the lowest values of BCAA metabolism (N-acetylvaline, N-acetylleucine, and 2-methylbutyrylcarnitine), pyroglutamine, and N6-carbamoylthreonyladenosine or vice versa (lowest HDL and LDL levels and highest metabolite levels). Another signature (Fig. 4C, bottom left corner) showed patients with the highest LDL and TRI, associated with extreme levels of aspartate, phospholipids, and mono- and diacylglycerols.
Metabo-clinical signatures that included retinopathy (Fig. 4E and F) showed that clusters with the highest number of patients with retinopathy are the least obese patients and associated mostly with sphingolipids and creatinine. Interestingly, retinopathy and LDL had two signatures (Fig. 4E, bottom left corner): showing the largest portion of patients with retinopathy with the lowest LDL levels associated with the highest levels of BCAAs and with the lowest levels of sphingolipids. The largest portion of patients with retinopathy with the lowest TRI levels (Fig. 4E, top right corner) associated with 3-methyl-2-oxovalerate and phenylalanine, among other lipids. In contrast, another signature (Fig. 4F, third panel from top) showed their association with the lowest HDL and LDL levels and the highest values of N-acetylleucine and 2-methylbutyrylcarnitine.
Discussion
T2D affects half a billion people and is a leading cause of morbidity and mortality worldwide (18). In Qatar, >15% of the population has T2D and >20% are obese, leading to chronic health conditions that require public health intervention and better predictive and prognostic biomarkers. Macro- and microvascular complications resulting from diabetes are associated with hyperglycemia, hypertension, dyslipidemia, and other risk factors. Thus, studying the underlying metabolic mechanisms can reveal shared metabolic pathways that better explain the pathogenesis of T2D and its complications.
Major Perturbed Metabolic Pathways
Analysis of seven major metabolic pathways (Fig. 2A) showed that compared with the other clinical variables, T2D and TRI contributed to the highest significant perturbations. The most significant perturbations in the amino acids, carbohydrates, nucleotides, peptides, and xenobiotics pathways were observed in T2D, and the most significant perturbations in lipid pathways were observed in TRI. Based on associations with the 373 metabolites, T2D was positively correlated with BMI and TRI and negatively correlated with HDL, reflecting the interplay between dyslipidemia and T2D. Moreover, the phospholipid and lysolipid pathways contained the largest number of metabolites affected in T2D, BMI, LDL, and TRI (Fig. 2B). Sphingolipids were also perturbed in T2D, BMI, and LDL. Lipid metabolic pathways that were highly correlated with TRI in comparison with other clinical variables include phospholipids, fatty acids, long-chain fatty acids (LCFAs), polyunsaturated fatty acids (PUFAs), and monoacylglycerols, except for sphingolipids, which were not as highly correlated with TRI as with other clinical outcomes. The GGM subnetworks linked together metabolites only associated with TRI with those only associated with T2D, further confirming interactions between their pathways (Fig. 3A and C).
Perturbations in Lipid Pathways and Links to Oxidative Stress
As expected, the largest set of perturbed metabolites shared among the complications and HDL, LDL, and TRI are in the lipid pathways, including phospholipids, sphingolipids, lysolipids, fatty acids, PUFAs, and LCFAs. Notably, 36 metabolites, including 15 phospholipids, 12 lysolipids, and 3 lysoplasmalogens, were also nominally significant in a previous study that compared the metabolic profiles of obesity with and without metabolic syndrome and nonobesity (13). One of the three largest GGM subnetworks reflected pathways showing correlations with phospholipids, lysolipids, and plasmalogens, some of which were only associated with T2D or TRI. Another subnetwork contained a set of correlated sphingomyelins associated with more than one clinical variable, and a third correlated metabolites only associated with T2D or TRI with monoacylglycerol/phospholipid/PUFA pathways.
Sphingomyelins were previously associated with insulin resistance (19) and with diabetic nephropathy pathogenesis (20). Lysolipids were decreased with T2D in our cohort, in agreement with previous studies (21,22), but increased with high TRI levels. However, phospholipids, which are central to the pathogenesis of metabolic diseases (19) and associated with insulin resistance and sensitivity (13), are significantly increased with TRI and T2D. Plasmalogen and lysoplasmalogen, which are enriched in PUFA (arachidonate), were decreased in T2D, consistent with a decrease in PUFAs. Decreased concentrations of plasmalogens indicate higher oxidative stress (23). The large number of perturbed lipids and fatty acids identified in our study highlights pathways involved in oxidative stress and lipotoxicity. Oxidative stress is a major cause of insulin resistance, β-cell dysfunction, glucose intolerance, and T2D micro- and macrovascular dysfunction (24) (see Supplementary Material for extended discussion).
Perturbations in Amino Acid Pathways and Links to Oxidative Stress and Lipotoxicity
Many amino acids from BCAAs, phenylalanine, tyrosine, methionine, cysteine, and the urea cycle pathways were perturbed in T2D, and BCAAs were increased in T2D, TRI, and BMI. Perturbed levels of circulating BCAAs, leucine, isoleucine, valine, 3-hydroxyisobutyric acid (3-HIB), and the aromatic amino acids tryptophan, phenylalanine, and tyrosine have been repeatedly associated with T2D (1). Elevated BCAA levels indicate proteolysis in insulin resistance (25), which promotes oxidative stress (26). Increased 3-HIB levels are associated with obesity-related insulin resistance and with the future development of T2D (27). 3-HIB mechanistic links explain how increased BCAA catabolic flux causes diabetes (28). Glutathione and taurine perturbations observed in this study are also linked to oxidative stress (29,30) (see Supplementary Material).
T2D Metabolites and Links to Oxidative Stress
Metabolites associated only with T2D are involved in kidney function (1,5-anhydroglucitol), creatine metabolism (creatinine and guanidinoacetate) (31), and glutamate metabolism (N-acetylglutamine and pyroglutamine). Pseudouridine, tartronate (novel to T2D), and 4-hydroxychlorothalonil were also associated only with T2D (32,33). Proline is known to protect mammalian cells against oxidative stress (34), and the novel associations of thioproline, an antioxidant that protects the body from oxidative stress (35), and N-methylproline are only associated with T2D. N-acetylarginine, which affects oxidative stress in rats (36), and dopamine sulfate were also novel and only associated with T2D. Similarly, N6-carboxymethyl-lysine has been associated with an increased risk for cardiovascular disease and other diabetic complications (37,38), renal function (39), and diabetic retinopathy (40). It is an advanced glycation end product metabolite and has roles in reactive oxygen species generation, oxidative stress, increased inflammation, and increased vascular smooth muscle apoptosis, leading to atherosclerosis (37,41) (Supplementary Material).
Diabetic Retinopathy
Xanthine, inosine, and adenosine were reported to be higher in patients with diabetic retinopathy compared with those with diabetes in a small cohort (42). We found lower xanthine levels and higher inosine and 5-MTA levels in diabetic retinopathy compared with T2D. Metabolites in the retinopathy cluster that showed altered metabolite levels compared with the other patients with T2D included X-17166, DSGEGDFXAEGGGVR, inosine, N-acetylmethionine, oxidized cys-gly, cysteine-glutathione disulfide, aspartate, glutamate, X-14364, and sphingosine. Inosine is known to affect sucrose and has a role in diabetes prevention; it was decreased in those with T2D versus control subjects, and its increased levels in retinopathy compared with nonretinopathy may indicate that it has been controlled for the patients with retinopathy. Notably, pyroglutamate promotes the survival of retinal ganglion cells (43), consistent with the observed decrease in glutamate in retinopathy. The aspartate and glutamate metabolic pathways have been previously implicated in retinopathy (44,45). Importantly, glutamate and N-acetylmethionine were also perturbed in association with TRI and the LDL/HDL ratio and thus might be involved in a common pathological mechanism in retinopathy and dyslipidemia. The perturbation of N-acetylmethionine with retinopathy, a novel finding in our study, may be indicative of the role of an imbalance between HDL and LDL in diabetic retinal damage. N-acetylmethionine targets rhodopsin, a photoreceptor required for image-forming vision at low light intensities (46).
Metabo-Clinical Signatures
Metabo-clinical signatures enabled the identification of diverse clinical patterns in patients with T2D that strongly correlate with specific metabolites and also identified groups that show multiple extreme clinical variables at extreme metabolite levels. Such metabolites that gradually change in tandem with clinical variables may help in identifying possible treatment pathways. One of the observed signatures had the highest LDL and HDL levels associated with the lowest values of N-acetylvaline, N-acetylleucine, 2-methylbutyrylcarnitine, pyroglutamine, and N6-carbamoylthreonyladenosine. N-acetylleucine and 2-methylbutyrylcarnitine are both associated with renal function (47,48). The association between the lowest levels of those metabolites and the highest LDL and HDL levels possibly hints on their involvement in the pathway linking high cholesterol with renal failure. It also reveals the possible roles of N-acetylvaline and pyroglutamine in association with high cholesterol and kidney function, which have not been previously reported. It is important to note in this study that correlations of creatinine to N-acetylvaline and pyroglutamine based on the metabo-clinical signature clusters are 0.81 and 0.85, respectively, again hinting on their link to kidney function. The involvement of N-acetylleucine in renal function may highlight some common pathways between retinopathy and nephropathy that are associated with dyslipidemia. Similarly, the association of 3-hydroxy-2-ethylpropionate with muscle mass (49) and its association with the largest portion of patients with retinopathy with the lowest LDL levels may indicate the role of LDL in muscle mass loss in people with diabetes and possible involvement in vision impairment. The highest aspartate levels were observed in patients exhibiting the highest TRI and LDL and lowest HDL levels and were also inversely associated with retinopathy in the regression results. This highlights the involvement of aspartate with cholesterols and TRI and their link to retinopathy. As previously indicated in this study, N-acetylmethionine is also found at its highest levels in patients with retinopathy with the highest HDL and lowest LDL levels, confirming our previous claim above on its possible role in linking retinopathy with dyslipidemia. See the Supplementary Material for further interpretation of results.
Metabolic Interactions and Interplay Between Different Pathways
Metabolites are interdependent, making the identification of metabolic networks important for our understanding of biomarker interactions. Interestingly, the identified GGMs show the strongest correlations between metabolites from one or more subpathways of the same super pathway or of different superpathways. Figure 3A links together phospholipids, PUFAs, and monoacylglycerols, while Fig. 3E shows LCFAs and PUFAs connected together. More remarkable are metabolites that could possibly indicate the metabolic flux as a result of a metabolic reaction. This is seen in metabolites of the same subnetwork in which one is positively correlated with a clinical variable and another is negatively correlated with it. For example, Fig. 3A links the PUFA metabolite docosatrienoate (22:3n-3), which is increased in those with T2D compared with control subjects, and dihomo-linolenoyl-choline, which is decreased. Such investigations are beyond the scope of this study but could lead to many interesting findings on the interactions of different biomarkers of T2D and its complications. Metabolites involved in lipotoxicity and glucotoxicity were also mapped onto Kyoto Encyclopedia of Genes and Genomes pathways and found in six interlinked pathways (Supplementary Fig. 6).
Limitations of the Study
Limitations of the study include factors (e.g., sample handling) affecting the samples that could impact metabolite levels. We have included hemolysis as a covariate to correct for one of the major factors affecting sample quality. The number of overlapping metabolites between the discovery and replication cohort was 547 metabolites, so we could not replicate some of the metabolites. However, we could also confirm many findings from literature. We could not replicate the retinopathy findings in the replication cohort as that information was not available, yet some of the metabolites confirmed previous findings in other studies. Although association studies are observational and could not be used to explain causality, the identification of shared metabolic pathways between T2D and complications paves the way to understanding the common underlying mechanisms of those clinical end points.
Conclusions
We performed high-resolution metabolomics profiling of 996 Qataris, of whom 57% had T2D, and obtained replication data from 2,618 individuals, of whom 11% had T2D. A total of 373 metabolites associated with T2D and its clinical variables were identified. Of these, 161 were novel and highlighted several pathways linked to oxidative stress and lipotoxicity, among other mechanisms. We identified metabo-clinical signatures of clusters of patients with T2D that highlighted patients with extreme values in more than two clinical variables aligned with extreme values in a group of metabolites. This highlighted metabolites that may mark the progression of complications in patents. Among our findings are the role of N-acetylmethionine in retinopathy in conjunction with dyslipidemia and the possible roles of N-acetylvaline and pyroglutamine in association with high cholesterol levels linked to kidney function.
To our knowledge, this is the first large-scale study to look at shared metabolic signatures of T2D and complications in an untargeted manner using >3,500 individuals from a population with a high prevalence of diabetes and metabolic disorders. We believe that the results provide the research community with a comprehensive reference of metabolic perturbations in T2D and complications, which may help further studies investigating personalized therapies and drug pathways affected by metabolic mechanisms.
Article Information
Acknowledgments. The authors thank the QBB for the support in obtaining the replication data.
Funding. This study was made possible by National Priorities Research Program grants from the Qatar National Research Fund (a member of the Qatar Foundation) (NPRP11S-0114-180299, NPRP09-740-3-192, and NPRP09-741-3-793). This work was also supported by the Bioinformatics Core and Biomedical Research Program in Weill Cornell Medicine-Qatar, funded by the Qatar Foundation.
The findings achieved in this article are solely the responsibility of the authors.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. N.A.Y., K.S., S.C.H., R.G.C., and K.A.F. were responsible for study design. N.A.Y. performed computational and statistical analysis. N.A.Y. was responsible for manuscript preparation. N.A.Y. and E.Y. performed the literature review. A.A., A.R., O.C., and K.A.F. were responsible for sample collection. M.E. contributed to the classification of retinopathy samples. N.A.Y., K.S., S.C.H., R.G.C., and K.A.F. revised and edited the manuscript. N.A.Y. is the guarantor of this work and, as such, had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains supplementary material online at https://doi.org/10.2337/figshare.16896382.
References
- 1. Yousri NA, Mook-Kanamori DO, Selim MM, et al. A systems view of type 2 diabetes-associated metabolic perturbations in saliva, blood and urine at different timescales of glycaemic control. Diabetologia 2015;58:1855–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wilson PW, Meigs JB, Sullivan L, Fox CS, Nathan DM, D’Agostino RB Sr. Prediction of incident diabetes mellitus in middle-aged adults: the Framingham Offspring Study. Arch Intern Med 2007;167:1068–1074 [DOI] [PubMed] [Google Scholar]
- 3. Krauss RM. Lipids and lipoproteins in patients with type 2 diabetes. Diabetes Care 2004;27:1496–1504 [DOI] [PubMed] [Google Scholar]
- 4. Ng TW, Khan AA, Meikle PJ. Investigating the pathogenesis and risk of type 2 diabetes: clinical applications of metabolomics. Clin Lipidol 2012;7:641–659 [Google Scholar]
- 5. Tomkin GH, Owens D. Diabetes and dyslipidemia: characterizing lipoprotein metabolism. Diabetes Metab Syndr Obes 2017;10:333–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blüher M, Kratzsch J, Paschke R. Plasma levels of tumor necrosis factor-α, angiotensin II, growth hormone, and IGF-I are not elevated in insulin-resistant obese individuals with impaired glucose tolerance. Diabetes Care 2001;24:328–334 [DOI] [PubMed] [Google Scholar]
- 7. Tan CE, Foster L, Caslake MJ, et al. Relations between plasma lipids and postheparin plasma lipases and VLDL and LDL subfraction patterns in normolipemic men and women. Arterioscler Thromb Vasc Biol 1995;15:1839–1848 [DOI] [PubMed] [Google Scholar]
- 8. Watson TD, Caskake MJ, Freeman DJ, et al. Determinants of LDL subfraction distribution and concentrations in young normolipidemic subjects. Arterioscler Thromb 1994;14:902–910 [DOI] [PubMed] [Google Scholar]
- 9. Zambon A, Austin MA, Brown BG, Hokanson JE, Brunzell JD. Effect of hepatic lipase on LDL in normal men and those with coronary artery disease. Arterioscler Thromb 1993;13:147–153 [DOI] [PubMed] [Google Scholar]
- 10. Zhou Y, Wang C, Shi K, Yin X. Relationship between dyslipidemia and diabetic retinopathy: a systematic review and meta-analysis. Medicine (Baltimore) 2018;97:e12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Al Thani A, Fthenou E, Paparrodopoulos S, et al. Qatar Biobank cohort study: study design and first results. Am J Epidemiol 2019;188:1420–1433 [DOI] [PubMed] [Google Scholar]
- 12. Yousri NA, Fakhro KA, Robay A, et al. Whole-exome sequencing identifies common and rare variant metabolic QTLs in a Middle Eastern population. Nat Commun 2018;9:333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Candi E, Tesauro M, Cardillo C, et al. Metabolic profiling of visceral adipose tissue from obese subjects with or without metabolic syndrome. Biochem J 2018;475:1019–1035 [DOI] [PubMed] [Google Scholar]
- 14. Palmer ND, Okut H, Hsu FC, et al. Metabolomics identifies distinctive metabolite signatures for measures of glucose homeostasis: the Insulin Resistance Atherosclerosis Family Study (IRAS-FS). J Clin Endocrinol Metab 2018;103:1877–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cirulli ET, Guo L, Leon Swisher C, et al. Profound perturbation of the metabolome in obesity is associated with health risk. Cell Metab 2019;29:488–500.e2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Urpi-Sarda M, et al. Metabolomics for biomarkers of type 2 diabetes mellitus: advances and nutritional intervention trends. Curr Cardiovasc Risk Rep 2015;9:12 [Google Scholar]
- 17. Vangipurapu J, Fernandes Silva L, Kuulasmaa T, Smith U, Laakso M. Microbiota-related metabolites and the risk of type 2 diabetes. Diabetes Care 2020;43:1319–1325 [DOI] [PubMed] [Google Scholar]
- 18. Kaiser AB, Zhang N, Van Der Pluijm W. Global prevalence of type 2 diabetes over the next ten years (2018-2028). Diabetes 2018;67(Suppl. 1):202 [Google Scholar]
- 19. Chang W, Hatch GM, Wang Y, Yu F, Wang M. The relationship between phospholipids and insulin resistance: from clinical to experimental studies. J Cell Mol Med 2019;23:702–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhu D, Sreekumar PG, Hinton DR, Kannan R. Expression and regulation of enzymes in the ceramide metabolic pathway in human retinal pigment epithelial cells and their relevance to retinal degeneration. Vision Res 2010;50:643–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang SJ, Kwak SY, Jo G, Song TJ, Shin MJ. Serum metabolite profile associated with incident type 2 diabetes in Koreans: findings from the Korean Genome and Epidemiology Study. Sci Rep 2018;8:8207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Razquin C, Toledo E, Clish CB, et al. Plasma lipidomic profiling and risk of type 2 diabetes in the PREDIMED Trial. Diabetes Care 2018;41:2617–2624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Braverman NE, Moser AB. Functions of plasmalogen lipids in health and disease. Biochim Biophys Acta 2012;1822:1442–1452 [DOI] [PubMed] [Google Scholar]
- 24. Wright E Jr, Scism-Bacon JL, Glass LC. Oxidative stress in type 2 diabetes: the role of fasting and postprandial glycaemia. Int J Clin Pract 2006;60:308–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kahl S, Roden M. Amino acids - lifesaver or killer in patients with diabetes? Nat Rev Endocrinol 2018;14:449–451 [DOI] [PubMed] [Google Scholar]
- 26. Zhenyukh O, Civantos E, Ruiz-Ortega M, et al. High concentration of branched-chain amino acids promotes oxidative stress, inflammation and migration of human peripheral blood mononuclear cells via mTORC1 activation. Free Radic Biol Med 2017;104:165–177 [DOI] [PubMed] [Google Scholar]
- 27. Mardinoglu A, Gogg S, Lotta LA, et al. Elevated plasma levels of 3-hydroxyisobutyric acid are associated with incident type 2 diabetes. EBioMedicine 2018;27:151–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jang C, Oh SF, Wada S, et al. A branched-chain amino acid metabolite drives vascular fatty acid transport and causes insulin resistance. Nat Med 2016;22:421–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lutchmansingh FK, Hsu JW, Bennett FI, et al. Glutathione metabolism in type 2 diabetes and its relationship with microvascular complications and glycemia. PLoS One 2018;13:e0198626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ribeiro RA, Bonfleur ML, Batista TM, Borck PC, Carneiro EM. Regulation of glucose and lipid metabolism by the pancreatic and extra-pancreatic actions of taurine. Amino Acids 2018;50:1511–1524 [DOI] [PubMed] [Google Scholar]
- 31. Li M, Gu L, Yang J, Lou Q. Serum uric acid to creatinine ratio correlates with β-cell function in type 2 diabetes. Diabetes Metab Res Rev 2018;34:e3001. [DOI] [PubMed] [Google Scholar]
- 32. Rebholz CM, Surapaneni A, Levey AS, et al. The serum metabolome identifies biomarkers of dietary acid load in 2 studies of adults with chronic kidney disease. J Nutr 2019;149:578–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Luo S, Coresh J, Tin A, et al.; Chronic Kidney Disease Biomarkers Consortium Investigators . Serum metabolomic alterations associated with proteinuria in CKD. Clin J Am Soc Nephrol 2019;14:342–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Krishnan N, Dickman MB, Becker DF. Proline modulates the intracellular redox environment and protects mammalian cells against oxidative stress. Free Radic Biol Med 2008;44:671–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ham Y-H, Jason Chan KK, Chan W. Thioproline serves as an efficient antioxidant protecting human cells from oxidative stress and improves cell viability. Chem Res Toxicol 2020;33:1815–1821 [DOI] [PubMed] [Google Scholar]
- 36. Sasso S, Dalmedico L, Delwing-Dal Magro D, Wyse AT, Delwing-de Lima D. Effect of N-acetylarginine, a metabolite accumulated in hyperargininemia, on parameters of oxidative stress in rats: protective role of vitamins and L-NAME. Cell Biochem Funct 2014;32:511–519 [DOI] [PubMed] [Google Scholar]
- 37. Fishman SL, Sonmez H, Basman C, Singh V, Poretsky L. The role of advanced glycation end-products in the development of coronary artery disease in patients with and without diabetes mellitus: a review. Mol Med 2018;24:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hegab Z, Gibbons S, Neyses L, Mamas MA. Role of advanced glycation end products in cardiovascular disease. World J Cardiol 2012;4:90–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wagner Z, Wittmann I, Mazák I, et al. N(epsilon)-(carboxymethyl)lysine -levels in patients with type 2 diabetes: role of renal function. Am J Kidney Dis 2001;38:785–791 [DOI] [PubMed] [Google Scholar]
- 40. Mishra SK, Jha N, Shankar PR, Dahal PK, Khatiwada B, Sapkota YD. An assessment of diabetic retinopathy and diabetes management system in Nepal. J Nepal Health Res Counc 2016;14:104–110 [PubMed] [Google Scholar]
- 41. Rhee SY, Kim YS. The role of advanced glycation end products in diabetic vascular complications. Diabetes Metab J 2018;42:188–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xia J, Wang Z, Zhang F. Association between related purine metabolites and diabetic retinopathy in type 2 diabetic patients. Int J Endocrinol 2014;2014:651050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Oono S, et al. Pyroglutamic acid enhances survival of axotomized retinal ganglion cells in adult rats. Invest Ophthalmol Vis Sci 2008;49:5507 [Google Scholar]
- 44. Jin H, Zhu B, Liu X, Jin J, Zou H. Metabolic characterization of diabetic retinopathy: an 1H-NMR-based metabolomic approach using human aqueous humor. J Pharm Biomed Anal 2019;174:414–421 [DOI] [PubMed] [Google Scholar]
- 45. Rhee SY, Jung ES, Park HM, et al. Plasma glutamine and glutamic acid are potential biomarkers for predicting diabetic retinopathy. Metabolomics 2018;14:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wishart DS, Knox C, Guo AC, et al. Drugbank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res 2006;34(Database issue):D668–D672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gagnebin Y, Jaques DA, Rudaz S, de Seigneux S, Boccard J, Ponte B. Exploring blood alterations in chronic kidney disease and haemodialysis using metabolomics. Sci Rep 2020;10:19502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Devi S, Nongkhlaw B, Limesh M, et al. Acyl ethanolamides in diabetes and diabetic nephropathy: novel targets from untargeted plasma metabolomic profiles of South Asian Indian men. Sci Rep 2019;9:18117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lustgarten MS, Price LL, Chale A, Phillips EM, Fielding RA. Branched chain amino acids are associated with muscle mass in functionally limited older adults. J Gerontol A Biol Sci Med Sci 2014;69:717–724 [DOI] [PMC free article] [PubMed] [Google Scholar]