Abstract
Concussions, both single and repetitive, cause brain and body alterations in athletes during contact sports. The role of the brain-gut connection and changes in the microbiota have not been well established after sports-related concussions or repetitive subconcussive impacts. We recruited 33 Division I Collegiate football players and collected blood, stool, and saliva samples at three time points throughout the athletic season: mid-season, following the last competitive game (post-season), and after a resting period in the off-season. Additional samples were collected from four athletes that suffered from a concussion. 16S rRNA sequencing of the gut microbiome revealed a decrease in abundance for two bacterial species, Eubacterium rectale, and Anaerostipes hadrus, after a diagnosed concussion. No significant differences were found regarding the salivary microbiome. Serum biomarker analysis shows an increase in GFAP blood levels in athletes during the competitive season. Additionally, S100β and SAA blood levels were positively correlated with the abundance of Eubacterium rectale species among the group of athletes that did not suffer a diagnosed concussion during the sports season. These findings provide initial evidence that detecting changes in the gut microbiome may help to improve concussion diagnosis following head injury.
Keywords: Mild traumatic brain injury, Biomarkers, Neuroinflammation, Brain-gut axis, Microbiota
Highlights
-
•
A longitudinal study following college football athletes across a sports season.
-
•
Nanopore 16S rRNA sequencing of gut microbiome reveals changes after head injury.
-
•
Serum biomarker GFAP increased during the competitive period of the season.
-
•
S100β and SAA blood levels were positively correlated with Eubacterium rectale.
-
•
Gut microbiota is suggested as a future biomarker for diagnosis following head injury.
1. Introduction
Concussions (or mild traumatic brain injury [mTBI]) affect 10–20% of athletes in contact sports (Howell et al., 2020). Multiple occurrences of such events are referred to here as repetitive mTBI. A concussion is a complex pathophysiological process affecting the brain, induced by biomechanical forces (McCrory et al., 2013). Sports-related concussion (SRC) is a form of mTBI common in collision sport athletes that has been linked to long-term neurological abnormalities (Conder and Conder, 2015; Jordan, 2013; Li et al., 2021). It is estimated that only 1 in 9 symptom-provoking concussions are reported (Duma et al., 2005; Guskiewicz et al., 2007). Therefore, often these injuries remain undiagnosed and untreated, resulting in implications to brain health that may carry on further on in life (Iverson et al., 2020).
Additionally, athletes in SRC suffer from the cumulative effects of repetitive subconcussive impacts, which are defined as events similar to those giving rise to a concussion but involving insufficient impact forces or accelerations to produce symptoms associated with mTBI (Shuttleworth-Edwards et al., 2008). A recent study that followed college football athletes over five seasons revealed that players sustained a median of 415 recorded head impacts (McCrea et al., 2021). There is an urgent need to understand the mechanism and prevention of concussions and the possible effects of repeated subconcussive impacts. Head impact in sports continues to be an important and growing concern at all levels of football and other sports because of the known adverse outcomes in some cases and the potential for long-term detrimental cognitive effects (Crisco et al., 2010).
Athletes heavily exposed to repetitive mTBI exhibit impaired cognitive function and neuropathological consequences later in life (Manley et al., 2017; Montenigro et al., 2017; Mouzon et al., 2018). Interestingly, the current evidence summarized in a review and meta-analysis refers to professional/elite players only and points towards an association between sustaining a concussion and inferior cognitive function in rugby, American football, and boxing. Because the data included only refers to professional/elite players, it is unclear how this is relevant to college athletes and the general population (Gallo et al., 2020).
The brain inflammation triggered after mTBI plays an essential role in the short and long-term effects of the trauma (Stein et al., 2015). Many neuronal inflammatory markers are substantially altered after concussion compared with non-injured athletes (Smoliga, 2020). However, there are limitations to their use as blood biomarkers since proteins released by the brain are only detectable at low plasma concentrations. They undergo proteolytic degradation in the blood, and their levels are affected by elimination through the liver or kidney (Mondello et al., 2011). Still, developing reliable brain biomarkers (Di Battista, Churchill, Rhind, Richards and Hutchison, 2019) for concussions and repetitive subconcussive hits is critical to improving diagnosis or prognosis in SRC athletes and constitutes the first step to investigate new anti-neuroinflammatory therapeutic approaches.
In addition to central inflammation, concussions are associated with an increase in systemic inflammatory molecules (Oliver et al., 2018). As such, a promising approach for brain recovery is to understand better the mechanisms through which the inflammation generated in the periphery after head injury leads to the development of short- and long-term cognitive deficits. Head trauma leads to both the acute and chronic disruption of the intestinal barrier and the subsequent appearance of bacterial endotoxins in the blood (Mazarati et al., 2021). The gastrointestinal and metabolic disruptions that often accompany brain trauma can be explained by the bidirectional link between the central nervous system (CNS) and the enteric nervous system (e.g., the brain-gut axis). Patients with moderate to severe brain injury often show neurogenic gut dysfunction due to a lack of CNS control over the gastrointestinal tract (Grande et al., 1997), suggesting a possible involvement of the gut microbiota in head injury outcomes. The gut microbiota is an essential neuromodulator of brain-gut signaling and can affect brain inflammation resulting from brain injury (Benakis et al., 2016).
Severe TBI patients treated with Lactobacillus-rich probiotic supplementation have decreased gastrointestinal dysfunction and a shortened time spent in intensive care (Tan et al., 2011). These observed benefits are commonly attributed to probiotic-induced reductions in systemic and central inflammation (Brenner et al., 2017). Similarly, thinking about a post-concussion clinical treatment, novel preventive and therapeutic strategies, including nutritional diets, microbiota manipulations with probiotics or prebiotics, or strengthening the enteric barrier could be applied to modulate the intestinal microbiota and ultimately improve both cognitive and functional TBI health outcomes. In addition, changes in the gut microbiota (or dysbiosis) have previously been correlated with developing certain diseases, including neuropsychiatric disease (Cenit et al., 2017). Dysbiosis of the intestinal microbiota has been described in patients with chronic severe TBI (Arciniegas and McAllister, 2008; McAllister, 2008) and is related to alterations in the permeability of the blood-brain barrier and microglia activation (Hoban et al., 2018), as well as changes in the microbiome composition (Treangen et al., 2018). Although gut dysbiosis after a single, severe TBI has been documented, this has not yet been studied in SRC or associated with subconcussive hits in athletes exposed to repetitive head impacts. Therefore, we suggest that a single diagnosed concussion or multiple subconcussive hits sustained during a collegiate football season may disrupt the gut microbiome.
2. Material and Methods
2.1. Study design and participants
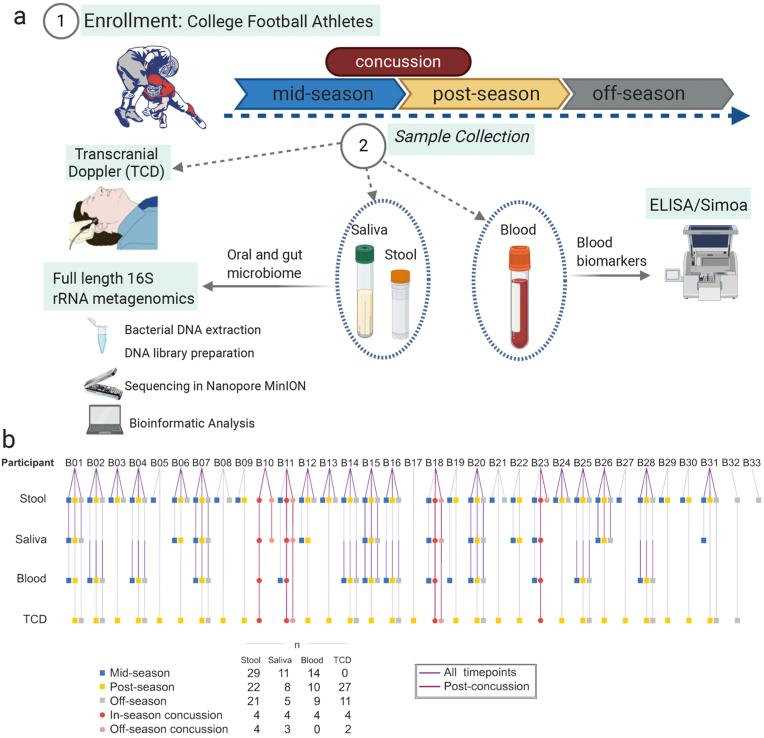
A total of 33 male collegiate football players ages 18–23 were voluntarily recruited to the study from the same Division I university team (Fig. 1). Exclusion criteria included: 1) prior history of a neurological or medical condition that could impact brain and cognition and 2) history of previous psychiatric disorder or substance abuse. The Institutional Review Board (IRB) at Houston Methodist Hospital approved the study protocol and written informed consent was obtained from all participants. To evaluate the time course of microbiota changes in players with a clinical diagnosis of concussion or during the in-season sports activity, samples were collected at three timepoints: mid-season (sports activities start), post-season (last sports activities), and off-season (no sports activities). Since the university's football season went from mid-August 2019 through the end of November 2019, mid-season samples were collected in October 2019, post-season in December 2019, and off-season in February 2020 (Fig. 1). Additional samples were collected within 24–48 h following a diagnosed concussion. For this study, an SRC was defined as an injury resulting from participation in organized intercollegiate practice or completions that requires medical attention by a team-certified athletic trainer or physician following the National Collegiate Athletic Association (NCAA) injury surveillance system definition (Kilcoyne et al., 2014). Demographics of the study cohort are summarized in Table 1.
Fig. 1.
Study Design and participants. (a) Saliva, stool, and blood samples were collected from collegiate football players athletes (n = 33) during the football season. Transcranial doppler imaging measurements were performed, and protein analysis was completed using a commercial ELISA and single molecule array. Bacterial DNA was extracted and 16S rRNA sequencing analysis was performed using the Nanopore platform. (b) Breakdown of samples received from each of the participants in the study at each of the designated timepoints.
Table 1.
Demographic data in the enrolled participant groups.
| Enrolled Participants | n = 33 |
|---|---|
| Age | |
| Mean (SD) | 19.3 (1.4) |
| Median [Min-Max] | 19 [18–23] |
| Race/ethnicity, n (%) | |
| Black, non-Hispanic | 13 (39.4) |
| White, non-Hispanic | 18 (54.5) |
| Mixed | 2 (6.1) |
| Hispanic | 6 (18.2) |
| Body mass index, mean (SD) | |
| Mean (SD) | 30.2 (4.8) |
| Median [Min-Max] | 29.4 [22.3–41.2] |
| Prior history of concussion, n (%) | 13 (39.4) |
2.2. Sample collection and bacterial DNA extraction
Stool samples were collected using the Stool Nucleic Acid Collection and Preservation System (Norgen, Ontario, Canada). Approximately 2 g of stool was sampled using the sterile spoon attached to the Stool Nucleic Acid Collection and Transport Tube lid. The sample was then suspended in the preservative solution and mixed thoroughly. Samples were kept at room temperature and, upon arrival to the laboratory, further homogenized by vortexing. They were then stored at 4 °C. For DNA extraction, 400 μl of the fecal suspension was transferred to the bead tubes supplied in the QIAamp PowerFecal Pro DNA Kit (Qiagen, Germantown, MD). Bead beating was performed for 1 min at 6.5 m/s on a FastPrep-24 system (MP Biomedicals, Irvine, CA). DNA isolation then continued following the kit manufacturer's instructions. Saliva samples were collected as indicated in the Collection Tube. The Collection Funnel was removed, the contents of the Preservative Ampoule were added and mixed with the collected saliva. Samples were kept at room temperature and then stored at 4 °C until processing. DNA was extracted from the saliva samples using the Norgen Saliva DNA Isolation Reagent Kit (Norgen, Ontario, Canada), per the manufacturer's instructions.
2.3. Long-read 16S rRNA sequencing on the MinION platform
16S rRNA amplicon sequencing was performed on a MinION nanopore sequencer (Oxford Nanopore Technologies, Oxford, UK). The amplicon library was prepared using the 16S Barcoding Kit 1–24 (SQK-16S024, Oxford Nanopore Technologies, Oxford, UK). For the PCR amplification and barcoding, 15 ng of template DNA extracted from fecal samples, and 30 ng in the case of saliva, were added to the LongAmp Hot Start Taq 2X Master Mix (New England Biolabs, Ipswich, MA). Initial denaturation at 95 °C was followed by 35 cycles of 20 s at 95 °C, 30 s at 55 °C, 2 min at 65 °C, and a final extension step of 5 min at 65 °C. The barcoded amplicons were purified using the AMPure XP beads (Beckman Coulter, Brea, CA) as per Nanopore's instructions. Samples were then quantified using a Qubit fluorometer (Life Technologies, Carlsbad, CA) and pooled in an equimolar ratio to a total of 50–100 ng in 10 μl. The pooled library was then loaded into an R9.4.1 flow cell and run per the manufacturer's instructions. MINKNOW software 19.12.5 was used for data acquisition. The provided primer set includes a recently noted issue with the standard ONT forward primer, which contains three mismatching bases to the family Bifidobacteriaceae and thus fails to amplify microbes of these taxa (Fujiyoshi et al., 2020).
2.4. Long-read 16S rRNA sequencing data taxonomic analysis
Raw fastq sequences were base called using Guppy. Sequencing adapters were then removed from sequences with Porechop v0.2.4. The resulting reads were classified taxonomically with Kraken v1.1.1 (Wood et al., 2019). The Pavian web application (https://fbreitwieser.shinyapps.io/pavian/) was utilized to transform Kraken outputs into spreadsheets of taxa counts and relative abundances per sample phylum, family, genus, and species levels. This was completed by: 1) uploading all Kraken, kreport text files for a given sample type, 2) selecting the “Comparison” tab on the web browser, 3) selecting either “read” or “%,” the appropriate taxonomy rank, and “clade” to ensure all bacteria classified at lower taxonomy level were included in the higher-level analysis, and 4) downloading the full tab-separated table. To reduce noise, any bacteria that claimed no more than 2 reads in at least one sample were removed from further analysis. Bacteria classified at the species or strain level that were missing genera or family in their lineage due to NCBI taxonomy were edited to ensure these bacteria were included in the genus- and family-level analysis. This impacted two bacterial species at the genus level: Eubacterium rectale and Ruminococcaceae bacterium CPB6. To match NCBI Taxonomy Database, their listed genera, unclassified Lachnospiraceae, and unclassified Ruminococcaceae, respectively, were treated as classified genera. This also impacted two bacterial species at the family level: Intestinimonas butyriciproducens and Flavonifractor plautii. Both bacteria claim taxonomic family unclassified Clostridiales, which is treated as a classified family for our analysis.
Alpha and beta diversity analyses for microbiome samples were performed with Python 3.8 using the sci-kit bio v0.4.1 diversity package. Alpha diversity values were generated for each sample using the Shannon and Simpson metrics. Beta diversity analyses were observed with the Weighted UniFrac metric. A phylogenetic tree was generated from the Kraken database using the taxdump_to_tree.py python script in biocore, a space for collaboratively developed bioinformatics software. Each branch in the tree was given a length of 1. Beta diversity distance matrices were then generated for each of the two analysis workflows per sample type. To evaluate the statistical difference between groups, p-values were calculated for the entire workflow and each pairwise grouping. This was completed by simulating 9999 ANOSIM permutations for each analysis. To visualize these values, a Matplotlib v.3.3.3 beta diversity scatter plot was generated for each workflow. This was achieved by: generating a principal coordinate analysis table from the appropriate distance matrix, choosing axes that best represented the calculated p-values, and plotting points colored by group. Finally, 95% confidence ellipses were drawn around each group with Matplotlib two-dimensional confidence ellipse source code.
Stacked bar plots were created with GraphPad Prism version 9 to visualize the average relative abundance of bacteria amongst samples in each group for the given workflow. These values were calculated at the phylum, family, and genus level by averaging the appropriate samples using the aligned relative abundance spreadsheet described above. Log2 fold change was calculated between two samples from the same participant by taking the log2 of the percent relative abundance of the later timepoint divided by an earlier timepoint. To avoid mathematical errors, relative abundance values of 0 were transformed to 10−10. Finally, heatmaps were created with the heatmap function in Seaborn v0.11.0 with the calculated fold change values. Scripts for generating diversity plots and heatmaps can be found here: https://gitlab.com/treangenlab/tbi-microbiome-football-2019.
2.5. Prediction of pathway composition by PICRUSt2
Metagenomic functional composition from the taxa abundance was inferred using PICRUSt2 (Douglas et al., 2020). An amplicon sequence variant (ASV) table was generated where the taxonomy of each ASV was assigned by Kraken 2 classification against the Silva database version 138 (Wood et al., 2019). The abundance of MetaCyc pathways was then estimated using PICRUSt2 with default options. Pathways with relative abundance <0.01% were converted to 0 to reduce noise.
2.6. Measurement of serum inflammatory biomarkers
ELISA kits and SIMOA Ultrasensitive assays were performed to measure the candidate blood biomarkers accurately. Venous blood was collected into vacutainer sterile serum tubes and allowed to clot at room temperature for 30 min. After centrifugation at 3000g for 15 min, serum was aliquoted and stored at −80 °C until analysis. Serum NF-L, GFAP, total Tau, and ubiquitin carboxy-terminal hydrolase L1 (UCH-L1) were quantified using the SIMOA Neurology 4-plex assay. According to the manufacturer's protocol, samples were run in duplicates on a HD-X instrument (Quanterix, Lexington, MA). The assay range was calculated from the calibration curve for each analyte. The functional lower limit of quantification was estimated at 1.052 pg/mL for NF-L, 1.728 pg/mL for GFAP, 0.576 pg/mL for Tau, and 13.520 pg/mL for UCH-L1. Measurements for Tau and UCH-L1 were below the limit of quantification for the majority of samples. According to the manufacturer's instructions, serum levels of S100β and SAA were measured using a commercial enzyme-linked immunosorbent assay (ELISA) to verify previous SIMOA data according to the manufacturer's instructions (Human S100B, EZHS100B–33K, Millipore Sigma, Burlington, MA; Human SAA, EL10015L, Anogen, Mississauga, Canada). The lower limit of detection for the S100β and SAA assays were 2.7 pg/mL and 1.1 ng/mL, respectively. When S100β was not detectable, the limit of detection value was assigned to the sample. Each sample was measured in duplicate.
2.7. Optic nerve sheath diameter and velocities with breath-holding
Each participant's optic nerve sheath diameter (ONSD) was measured using a transcranial Doppler (TCD). ONSDs were measured during the post-season and off-season periods. A high-frequency General Electric (Fairfield, CT, US) LOGIQ E9 9 MHz linear probe was utilized for this study. All measurements were performed by a single, experienced, certified neuro-sonographer using the same ultrasound machine to eliminate any operator bias. The probe was placed over the closed eyelid with the patient in the supine position. A small amount of ultrasound gel was used to avoid any discomfort for the patient. The probe was held in a transverse plane with the beam directed posteriorly towards the optic disc complex. The patient was asked to look down while keeping their eyes closed. The ONSD was measured 3 mm behind the globe using an electronic caliper. Each diameter was measured three times, and the mean value was used for the final analysis. Velocities were measured in corresponding locations with temporal (ACA, MCA, PCA), transorbital (ONSD and OA), and suboccipital (VA and BA) windows. BHI and delta velocities were calculated utilizing a 20 s player-patient initiated inhalation and hold.
2.8. Statistical analysis
Statistical analysis of the alpha diversity in the microbiome and serum biomarkers was performed using GraphPad Prism version 9. A Kruskal-Wallis test followed by Dunn's post hoc test for multiple comparisons was performed for the Shannon and Simpson alpha diversity indices. Repeated measures one-way ANOVA was used for the longitudinal analysis of the serum biomarkers. An ordinary one-way ANOVA was performed to find statistical differences between the post-concussion samples and the rest of the groups.
To analyze whether any individual species exhibited a significant difference in relative abundance between timepoints or across health status, we constructed linear models in MaAsLin2 (MaAsLin2 R package version 1.4.0http://huttenhower.sph.harvard.edu/maaslin2, (Mallick et al., 2021). For all models, relative abundance data was calculated for each microbe as a percentage of the total sample, and default transformation, normalization, and outcome distribution options were selected. For the longitudinal analysis among healthy players (n = 17 players x 3 sample times), a random-effects model was selected with the player as the random effect and the fixed effect was specified to be the timepoint (mid/post/off-season); features (i.e., species) were only modeled if they were present with a minimum abundance of 0.05% in at least 10% of the samples (i.e., 6 or more). For the concussion vs. non-concussed samples, models were run separately for in-season (i.e., mid and post-season, combined, n = 51 healthy, 4 concussion) and off-season (n = 21 healthy, 4 concussion). For both models, concussion status was specified as the fixed effect and features were only included if they were present with a relative abundance of at least 1% in a minimum of 10% of the samples. In this case, the higher abundance threshold was due to the small number of cases available, to reduce unnecessary hypothesis tests requiring correction.
To examine the interactions between the serum biomarkers and the gut microbiome composition, Pearson correlations were computed between the concentration of biomarkers throughout the sports season in non-concussed athletes and the relative abundance of species and genus found to be significantly changed in the gut microbiome analyses. Pearson correlation tests followed by false discovery rate (FDR) adjustment were performed in GraphPad v9.
All individual analyses where multiple hypotheses were examined were individually subject to multiple hypothesis correction, although no global correction was applied because each analysis was undertaken with its own motivation rather than planned a priori.
2.9. Data availability
The high-throughput sequence data has been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under the project database with project number PRJNA699316. The authors declare that all other data supporting the findings of this study are available within the article and its Supplementary Information files or from the corresponding author on request.
3. Results
3.1. Demographic and clinical characteristics
This study included 33 male Division I collegiate football players, four of whom suffered from a concussion diagnosed by a medical professional while enrolled in the study. Details of the study population are provided in Table 1 and the Methods section. Three data collection timepoints were established: two during the sports competition (mid- and post-season, following the middle and the last competitive games of the season, respectively) and an off-season collection 86 days after the end of the competition (Fig. 1). Additional data collection was performed for the concussed players within 48 h after diagnosis.
3.2. Changes in the human gut microbiome and corresponding pathways across the football season
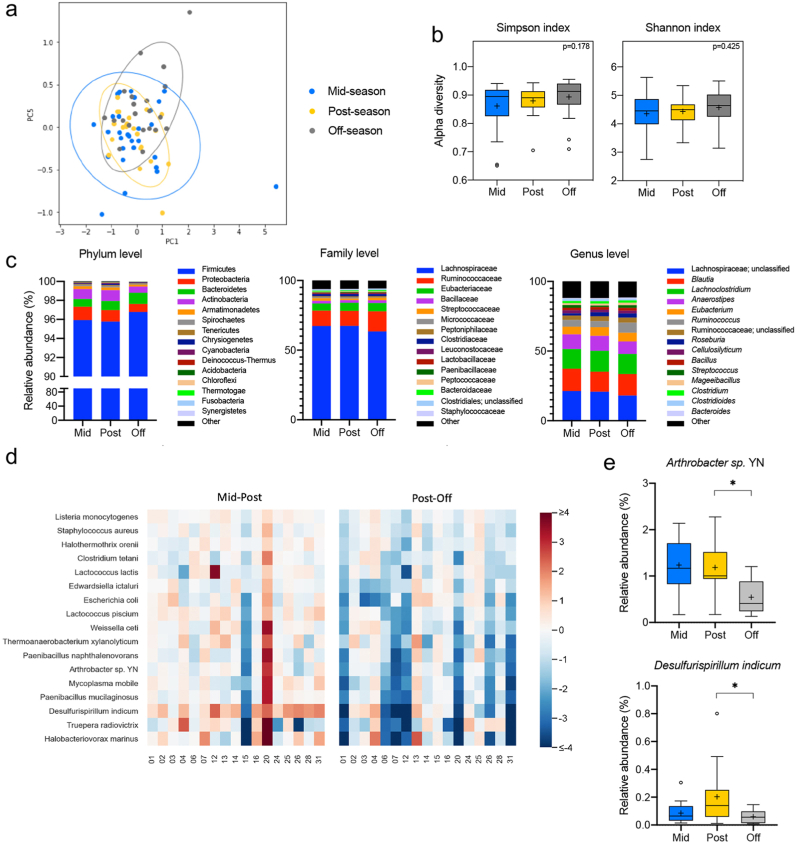
Microbial DNA was extracted from fecal samples, and next-generation 16S rRNA gene sequencing was performed using the Nanopore MinION platform, which has been shown to provide a more accurate taxonomic classification at the species level than Illumina MiSeq (Nygaard et al., 2020). Long-read 16S rRNA amplicons covering the V1–V9 hypervariable region generated reads of approximately 1500 bp. Sequences were then trimmed using Porechop and classified with Kraken (Wood et al., 2019), which yielded 23,249,965 classified reads and led to 1021 species being identified after quality filtering. For the longitudinal analysis of gut microbiota changes across the football season, we excluded any samples collected following a concussion and subsequently grouped all remaining samples according to the three collection timepoints (mid-season, n = 29; post-season, n = 22, and off-season, n = 21). Then, we investigated the differences in gut microbiota community structure across timepoints by calculating beta and alpha diversity metrics. Plotting of weighted UniFrac distances by principal coordinate analysis (PCoA) revealed overlap between the three timepoints, with no statistically significant differences evaluated by analysis of similarity (ANOSIM) (R = 0.0064, p = 0.3622; Fig. 2a). Further, the alpha diversity at the species level based on the Simpson and Shannon metrics did not significantly differ between the time points (p = 0.178 and p = 0.425, respectively) (Fig. 2b). Next, we evaluated the gut microbiota composition at the phylum, family, and genus level for all three-time points. Overall, the relative abundance of prevalent taxa was similar between the mid- and post-seasons. However, some shifts were observed in the off-season compared to the other timepoints (Fig. 2c). A random-effects multivariate analysis that only included data from non-concussed athletes who had provided a sample at all three timepoints (n = 17) was performed using MaAsLin2. Mid-to post-season and post-to off-season were compared in two independent analyses (Supplemental Table 1). This model identified 8 bacteria species with a statistically significant change in abundance between the post- and off-season timepoints (i.e., having an FDR-corrected p-value <0.05). Supplemental Table 1 shows these species but also extends to show all species with an FRD-corrected p-value as high as 0.125. It is noteworthy that of the 17 species shown, all of them had decreased in relative abundance during the off-season (Fig. 2d). Microbial taxa at the family, genus, and species level meeting both statistical significance criteria and expressing prevalent abundance (mean > 1% in at least one group at family or genus level, >0.1% at species level) are represented in Fig. 2e. Of note, the species Anthrobacter sp. YN and Desulfirispirillum indicum decrease in relative abundance during the off-season compared to the post-season (q = 0.027 and 0.039, respectively).
Fig. 2.
Diversity and taxonomic analysis along the sports season in non-concussed football athletes. (a) Plots of Beta diversity analysis of the fecal microbiota from athletes who did not experience a diagnosed concussion during the football season. The Principal Coordinate Analysis (PCoA) ordination plot was based on Weighted UniFrac distances for the mid- (n = 29, blue), post- (n = 22, yellow), and off-season (n = 21, gray) groups. Confidence ellipses represent the 95% confidence interval for each timepoint. Group dissimilarities were evaluated by Analysis of Similarity (ANOSIM). (b) Shannon and Simpson alpha diversity indices at the species level for the mid- (n = 29, blue), post- (n = 22, yellow), and off-season (n = 21, gray) timepoints. In the box and whisker plots, the cross represents the mean. Significance was determined by Kruskal-Wallis followed by Dunn's multiple comparisons test. (c) Relative abundances of the top 15 phyla, families, and genera in the fecal microbiota of the non-concussed athletes through the mid- (n = 29, blue), post- (n = 22, yellow), and off- (n = 21, gray) seasons. (d) Heatmap of the log2 fold changes for the species that are significantly altered across the sport season in the non-concussed athletes. Statistical testing by random-effects multivariate analysis included the samples from athletes who participated in all collection timepoints (n = 17). All statistically significant changes were identified between the post- and off-season. (e) Relative abundances at the mid- (n = 17, blue), post- (n = 17, yellow), and off-season (n = 17, gray) timepoints of the prevalent taxa (abundance >1% in at least one of the groups at the family and genus level, >0.1% at the species level) showing significant alterations in the random-effects multivariate analysis. In the box and whisker plots, the cross represents the mean. ∗q < 0.05; ∗∗∗q < 0.001.
We next inferred the functional composition of the gut microbiota based on the 16S data using PICRUSt2 (Douglas et al., 2020). Changes in abundance of MetaCyc pathways throughout the football season in non-concussed football athletes (n = 17) were identified by two independent MaAsLin2 random-effects multivariate analyses comparing the mid-to post-season and post-to the off-season. Results revealed 251 pathways that were differentially abundant between groups, with all significant changes (q < 0.05) occurring between the post- and off-season (Supplemental Table 3). Identification of the parent classes for the 251 differentially abundant pathways revealed that they are predominantly related to aromatic compound degradation, amino acid biosynthesis and degradation, fatty acid biosynthesis, and biosynthesis of electron carriers, such as quinol and quinone. In particular, the pathways with the highest fold changes between the post- and off-season are most notably involved in the degradation of sugars and aromatic compounds (Fig. 3).
Fig. 3.
Pathway abundance prediction by PICRUSt2 of the fecal microbiota throughout the football season in non-concussed athletes. Log2 fold change of the abundances of the MetaCyc pathways showing significant differences (q < 0.05) by random-effects multivariate analysis of the samples from athletes who provided samples at all three timepoints (n = 17). All statistically significant changes were identified between the post- and off-season. Top 15 log2 fold changes towards increase (red) and reduction (blue) are represented in the bar plot.
3.3. Relationship between the gut microbiota and a single concussion
We next evaluated the changes in the gut microbiome associated with concussion. Since mid- and post-season samples proved similar in the longitudinal analysis, both timepoints were combined to form an “in-season” group. Samples were then divided into four groups: in-season (n = 51), in-season concussion (n = 4), off-season (n = 21), and off-season concussion (n = 4) (Fig. 1b). The in-season concussion group consists of the samples from the four athletes who suffered a concussion collected within 24–48 h following the diagnosed concussion. In contrast, the off-season concussion group includes the samples from the same four athletes who received a concussion collected in the later off-season timepoint.
Beta diversity analysis showed significant differences in the gut microbiota structure between groups (R = 0.2309, p < 0.001). Pairwise ANOSIM analysis indicated that samples from concussed and non-concussed players were significantly different for both in- (R = 0.3959, p = 0.0127) and off-season (R = 0.5711, p = 0.0024) comparisons (Fig. 4a). Additionally, greater alpha diversity was observed in the concussed athletes when compared to their non-concussed teammates, for both the in-season (Simpson, p = 0.0082; Shannon p = 0.0043) and off-season (Simpson, p = 0.0435; Shannon p = 0.0329) comparison (Fig. 4b). The abundance of the most prevalent genera and families from the gut microbiota was similar within each group of participants (concussed and non-concussed athletes) between the in- and the off-season timepoints (Fig. 4c). At the phylum level, fecal samples collected post-concussion during the in-season exhibited an overall distinct distribution of the most abundant phyla compared to the rest of the groups represented (Fig. 4c).
Fig. 4.
Diversity and taxonomic analysis in athletes after a single concussion. (a) Beta diversity analysis of fecal microbiota from athletes who experienced a diagnosed concussion during the football season. Principal Coordinate Analysis (PCoA) ordination plot was based on Weighted UniFrac distances for the in-season (n = 51, green), in-season concussion (n = 4, red), off-season (n = 21, gray), and off-season concussion (n = 4, pink) groups. Confidence ellipses represent the 95% confidence interval for each timepoint. Group dissimilarities were evaluated by Analysis of Similarity (ANOSIM). (b) Shannon and Simpson alpha diversity indices at the species level for concussed (n = 4 in-season, red and off-season, pink) and non-concussed (n = 51 in-season, green and n = 21 off-season, gray) football athletes. In the box and whisker plots, the cross represents the mean. Significance was determined by Kruskal-Wallis followed by Dunn's multiple comparisons test. (c) Relative abundances of the top 15 phyla, families, and genera in the fecal microbiota for the in-season (n = 51, green), in-season concussion (n = 4, red), off-season (n = 21, gray) and off-season concussion (n = 4, pink) groups. (d) Relative abundances of the prevalent taxa (abundance >1% in at least one of the groups at the family and genus level, >0.1% at the species level) showing significant alterations in the random-effects multivariate analysis for concussed (n = 4 in-season, red and off-season, pink) and non-concussed (n = 51 in-season, green and n = 21 off-season, gray) athletes. In the box and whisker plots, the cross represents the mean. ∗q < 0.05; ∗∗q < 0.01.
A fixed-effects model in MaAsLin2 was used to identify specific microbial taxa with statistically significant differences in abundance between the concussed and non-concussed players at the in- and off-season timepoints (Supplemental Table 2). The model identified 9 species with a significant difference in the off-season model, while none showed significance in the in-season analysis. The relative abundance of the family Lachnospiraceae was decreased in the concussed athletes compared to their non-concussed teammates (q = 0.003), whereas the families Ruminococcaceae (q = 0.028) and Oscillospiraceae (q = 0.028) were increased (Fig. 4d). At the genus level, Bacillus and Bacteroidetes were increased in the concussion group for the off-season analysis (q = 0.025 and q = 0.023, respectively). The species Eubacterium rectale and Anaerostipes hadrus, both belonging to the Lachnospiraceae family, significantly reduced relative abundance in concussed athletes (q = 0.014 and q = 0.034, respectively). On the contrary, seven bacterial species, including Ruminococcaceae bacterium CPB6 (q = 0.007), Mageeibacillus indolicus (q = 0.021), Ethanoligenes harbinense (q = 0.007), Anaerococcus prevotii (q = 0.032), Bacillus thuringiensis (q = 0.014), Flavonifractor plautii (q = 0.014) and Bacillus cereus (q = 0.017), showed a greater relative abundance in the concussion group.
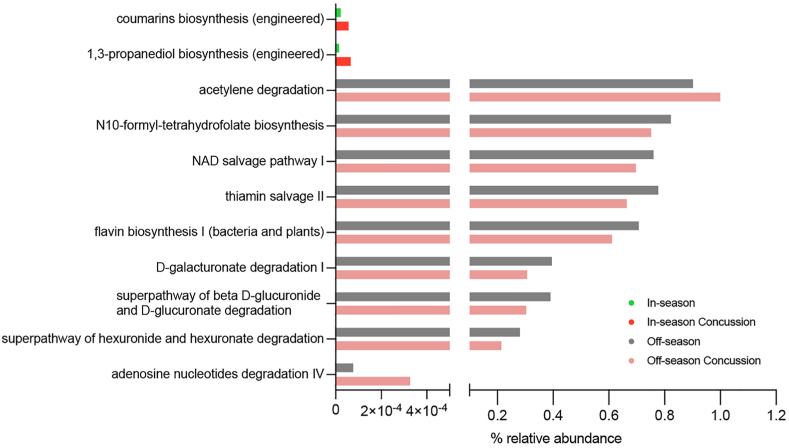
To investigate the functional changes in the gut microbiota following concussion, we examined pathway abundances generated with PICRUSt2. MaAsLin2 multivariate analysis between the concussed and non-concussed groups identified two and eight differentially abundant pathways in the in-season and off-season, respectively (Fig. 5, Supplemental Table 3). These results indicate that the major changes between off-season concussion and non-concussion included: acetylene degradation, N10-formyl-tetrahydrofolate biosynthesis, NAD salvage pathway, thiamin salvage, flavin biosynthesis, D-galacturonate degradation, beta D-glucuronide and D-glucuronate degradation, hexuronide and hexuronate degradation, and adenosine nucleotides degradation. On the other hand, the major changes between the in-season concussion and in-season included: coumarins biosynthesis and 1, 3-propanediol biosynthesis (Fig. 5). Notably, among the pathways altered in the off-season, sugar acid degradation pathways had significantly reduced relative abundances in the concussion group compared to the non-concussed athletes.
Fig. 5.
Pathway abundance prediction by PICRUSt2 of the fecal microbiota following concussion. Abundance of the MetaCyc pathways showing significant differences (q < 0.05) by random-effects multivariate analysis in the in-season (n = 51, green), in-season concussion (n = 4, red), off-season (n = 21, gray), and off-season concussion (n = 4, pink) groups.
3.4. No alterations in the oral microbiota during the longitudinal study or after a concussion
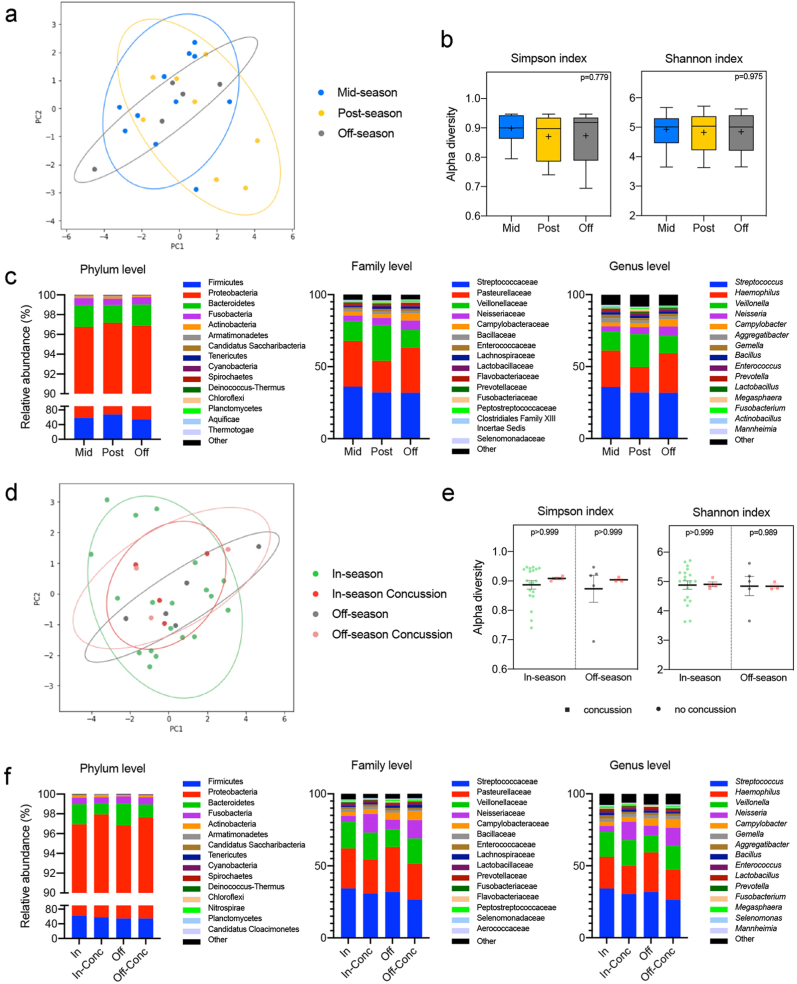
To study the changes in the oral microbiome taking place as the football season progressed, we analyzed 16S rRNA gene sequencing data from the saliva of non-concussed players (mid-season, n = 11; post-season, n = 8; and off-season, n = 5) (Fig. 1b). Nanopore MinION long-read 16S rRNA sequencing yielded 6,967,795 classified reads with 793 species identified after quality filtering. The structure of the saliva microbial community was similar between groups, as indicated by the beta diversity analysis (R = −0.0178, p = 0.5415) (Fig. 6a). Similarly, no significant differences in alpha diversity were observed across the three timepoints analyzed (Simpson, p = 0.779; Shannon, p = 0.975) (Fig. 6b). The overall shifts in the oral microbiota composition at the phylum, family, and genus level do not appear to be consistent across the three timepoints analyzed (Fig. 6c).
Fig. 6.
Diversity and taxonomic analysis of salivary microbiome throughout the season in football athletes. (a) Beta diversity analysis of the saliva microbiota from athletes who did not experience a diagnosed concussion during the football season. Principal Coordinate Analysis (PCoA) ordination plot was based on Weighted UniFrac distances for the mid- (n = 11, blue), post- (n = 8, yellow), and off-season (n = 5, gray) groups. Confidence ellipses represent the 95% confidence interval for each timepoint. Group dissimilarities were evaluated by Analysis of Similarity (ANOSIM). (b) Shannon and Simpson alpha diversity indices at the species level for the mid- (n = 11, blue), post- (n = 8, yellow), and off-season (n = 5, gray) timepoints. In the box and whisker plots, the cross represents the mean. Significance was determined by Kruskal-Wallis followed by Dunn's multiple comparisons test. (c) Relative abundances of the top 15 phyla, families, and genera in the oral microbiota of the non-concussed athletes through the mid- (n = 11, blue), post- (n = 8, yellow), and off- (n = 5, gray) seasons. (d) Beta diversity analysis of the saliva microbiota from athletes who experienced a diagnosed concussion during the sports season. Principal Coordinate Analysis (PCoA) ordination plot was based on Weighted UniFrac distances for the in-season (n = 19, green), in-season concussion (n = 4, red), off-season (n = 5, gray), and off-season concussion (n = 3, pink) groups. Confidence ellipses represent the 95% confidence interval for each timepoint. Group dissimilarities were evaluated by Analysis of Similarity (ANOSIM). (e) Shannon and Simpson alpha diversity indices at the species level for the concussed (n = 4 in-season, red and n = 3 off-season, pink) and non-concussed (n = 19 in-season, green and n = 5 off-season, gray) football athletes. In the box and whisker plots, the cross represents the mean. Significance was determined by Kruskal-Wallis followed by Dunn's multiple comparisons test. (f) Relative abundances of the top 15 phyla, families and genera in the oral microbiota for the in-season (n = 19, green), in-season concussion (n = 4, red), off-season (n = 5, gray), and off-season concussion (n = 3, pink) groups.
We next assessed whether the oral microbiome was altered in concussed athletes (in-season, n = 4; off-season, n = 3) compared to their non-concussed teammates (in-season, n = 19; off-season n = 5) (Fig. 1b). No significant differences were found in either beta diversity (R = −0.0464; p = 0.6387; Fig. 6d and e) or alpha diversity (Simpson, p > 0.999; Shannon p > 0.999 in-season, p = 0.989 off-season). At the phylum, family, and genus level, the most abundant taxa are overall comparable in abundance between the in- and the off-season within the same group of athletes (concussed and non-concussed) (Fig. 6f). In conclusion, saliva microbiota biomarkers remained close to their baseline values through all timepoints and after a concussion.
3.5. No changes in the optic nerve sheath diameter or cerebral blood flow measurements
There were no significant differences in the optic nerve sheath diameters (ONSD) or transcranial Doppler (TCD) parameters in the right (R) and left (L) cerebral blood flow velocities (CBFV) (velocity without breath-holding (BH)), the velocity with BH, holding time, anterior cerebral artery (ACA), posterior cerebral artery (PCA), internal carotid artery (ICA), L and R PI without BH, and with BH, and diameters of the optic nerve sheath (ONSD) for R and L (diameter and BHI). A single sonographer assessed TCD measurements at the post-season and off-season timepoints (Table 2). No clinical concussion participants underwent clinical TCD testing, and no meaningful differences in velocity or diameter were observed across the timepoints.
Table 2.
Transcranial doppler imaging optic nerve sheath diameter statistics. Right (R) and Left (L), breath-holding (BH), anterior cerebral artery (ACA), posterior cerebral artery (PCA), internal carotid artery (ICA), optic nerve sheath (ONSD).
| R Velocities | |||
|---|---|---|---|
| Group | Off-season | Post-season | P-Value |
| R Velocity without Holding (Mean, SD) | 56.6 ± 12.8 | 57.6 ± 15.1 | 0.88 |
| R Velocity with Holding (Mean, SD) | 70.7 ± 16.7 | 76.4 ± 20.6 | 0.51 |
| R Holding Time (Median, IQR) | 20 [20-20] | 20 [20-20] | 1 |
| R ACA (Mean, SD) | 51.6 ± 11.7 | 55.1 ± 8.2 | 0.5 |
| R PCA (Mean, SD) | 36.9 ± 5.6 | 31.4 ± 5.4 | 0.04 |
| R ICA (Mean, SD) | 31 ± 4.9 | 33.3 ± 4.2 | 0.25 |
| RPI without holding (Mean, SD) | 1 ± 0.1 | 1 ± 0.1 | 0.26 |
|
RPI with holding (Mean, SD) |
0.9 ± 0.1 |
0.8 ± 0.2 |
0.08 |
| L Velocities | |||
|
Group |
Off-season |
Post-season |
P-Value |
| L Velocity without Holding (Mean, SD) | 57.1 ± 12.6 | 59.1 ± 10.2 | 0.71 |
| L Velocity with Holding (Mean, SD) | 74.4 ± 13.1 | 73.3 ± 16.4 | 0.88 |
| L Holding Time (Median, IQR) | 20 [20-20] | 20 [20-20] | 1 |
| L ACA (Mean, SD) | 49 ± 9.2 | 47.6 ± 6.9 | 0.72 |
| L PCA (Mean, SD) | 35.2 ± 5.5 | 34.2 ± 5.7 | 0.68 |
| L ICA (Mean, SD) | 31.9 ± 3.8 | 32.8 ± 5.1 | 0.66 |
| LPI without holding (Mean, SD) | 1 ± 0.1 | 1 ± 0.2 | 0.9 |
|
LPI with holding (Mean, SD) |
0.8 ± 0.1 |
0.8 ± 0.1 |
0.98 |
| Diameters ONSD | |||
|
Group |
Off-season |
Post-season |
P-Value |
| R Diameter (Mean, SD) | 3.6 ± 0.2 | 3.6 ± 0.3 | 0.68 |
| R BHI (Mean, SD) | 1.2 ± 0.5 | 1.4 ± 0.5 | 0.5 |
| L Diameter (Mean, SD) | 3.6 ± 0.3 | 3.7 ± 0.4 | 0.88 |
| L BHI (Mean, SD) | 1.6 ± 0.7 | 1.5 ± 0.5 | 0.59 |
3.6. Changes in blood biomarkers across the football season
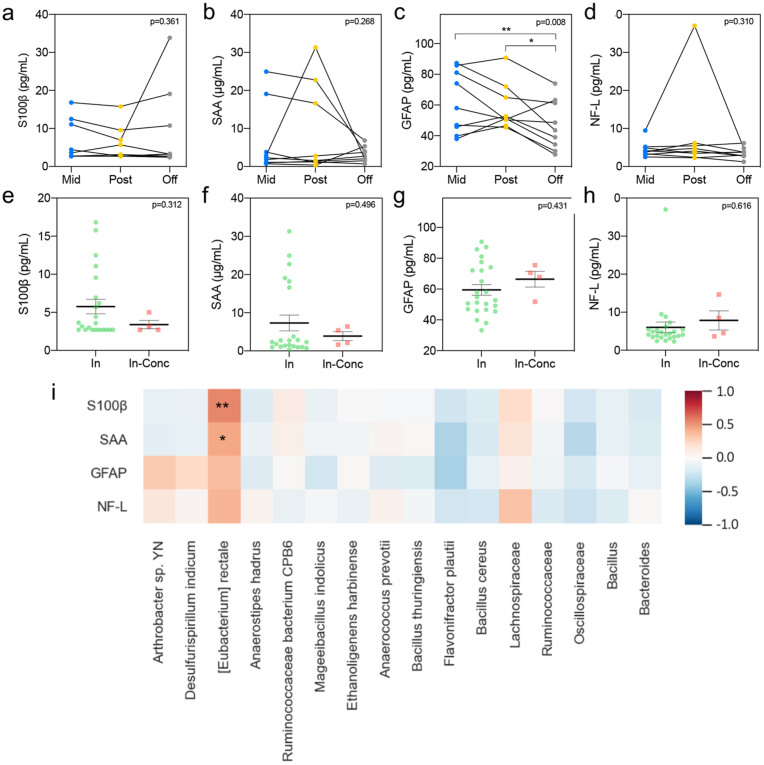
Eleven participants provided blood samples at each of the three timepoints, enabling us to perform a longitudinal analysis of blood serum biomarkers. Specifically, repeated measures ANOVAs were used to determine if the levels of several biomarkers in serum changed across the football season. Results revealed no significant differences for S100β (F (2, 16) = 1.087, p = 0.361), serum amyloid A (SAA) (F (2, 16) = 1.433, p = 0.268) and neurofilament light chain (NF-L) (F (2,16) = 1.262, p = 0.310) (Fig. 7). However, serum glial fibrillary acidic protein (GFAP) was significantly decreased in both the mid-season and the post-season compared to the off-season (p = 0.00813 and p = 0.0437, respectively; F (2,16) = 6.597, p = 0.0081) (Fig. 7c). We also compared the biomarker levels in serum in the four samples collected in-season after a concussion to the samples from the rest of the athletes. There was no statistical difference between the groups tested for S100β, SAA, glial fibrillary acidic protein (GFAP), and neurofilament light (NF-L) (p = 0.312, p = 0.496, p = 0.431, and p = 0.616, respectively) (Fig. 7e–h). However, we did detect a significant correlation between the concentration of the serum biomarkers S100β and SAA in the longitudinal samples of non-concussed athletes with the abundance of the bacterium species Eubacterium rectale (Fig. 7i).
Fig. 7.
Serum levels of biomarkers related to brain injury. (a–d) S100β (a), SAA (b), GFAP (c), and NF-L (d) serum concentrations represented longitudinally for each of the football athletes who did not experience a diagnosed concussion, at mid- (n = 9, blue), post- (n = 9, yellow), and off-season (n = 9, gray). Significance was determined by Repeated Measures ANOVA followed by Tukey multiple comparisons test (∗p < 0.05). (e–h) S100β (e), SAA (f), GFAP (g), and NF-L (h) serum concentrations after diagnosed concussion (n = 4, red) compared to the values in the in-season (n = 24, green) for the rest of the players in the study. Significance was determined by an unpaired T-test. SAA: Serum amyloid A; GFAP: Glial fibrillary acidic protein; NF-L: Neurofilament light. (i) Heatmap of the Pearson's correlation coefficients between the concentration of serum biomarkers throughout the sports season and the relative abundance of species and genus significantly changed in the gut microbiome analysis. ∗p < 0.05; ∗∗p < 0.01.
4. Discussion
The results of our preliminary study highlight alterations to the gut microbiome profiles in collegiate student-athletes who have suffered a single concussion, as well as in the active period of the football season (in-season) relative to the off-season. Currently, collegiate athletes are removed from play if signs and symptoms of traumatic head or cervical injury or concussion are observed by the sideline medical staff. However, since an asymptomatic player may still have a mild brain injury, indicative biomarkers corroborating a brain injury diagnosis are needed. This study identifies specific bacteria that decrease in abundance after concussion (Eubacterium rectale and Aerostipes hadrus). Even subtle changes in gut microbiota, and the metabolites that are produced in microbial communities, may affect gut health and athletic performance (Han et al., 2020) and exercise intensity (Ribeiro et al., 2021), and the microbiome of professional athletes was previously shown to differ from than of sedentary individuals (Barton et al., 2018).
Changes in the gut microbiota in response to the inflammatory stage have been previously observed in severe TBI patients (Nicholson et al., 2019; Urban et al., 2020). It has been shown that concussion can trigger long-term delayed changes in the colon and that subsequent bacterial infections in the gastrointestinal tract can increase post-traumatic brain inflammation and neurodegeneration in mice (Ma et al., 2017). Our previous studies have established a relationship between TBI and ensuing changes in the gut microbiome of mice (Treangen et al., 2018), providing the potential for a direct connection between brain-gut-microbiota and TBI. This raises the question of whether the same phenomenon occurs in humans, although collecting data appropriate to test this hypothesis presents obvious ethical and logistical challenges. We tested this hypothesis in our study by following a cohort of high-risk subjects through multiple timepoints, which yields useful longitudinal data from a homogenous population at the expense of comparatively low sample size among cases (in this case, n = 4). We observe noticeable and, in some cases, statistically significant differences in the microbiome of subjects according to a concussion status and across time among those without a diagnosed concussion. These two effects, timepoint, and concussion status warrant individual discussion.
4.1. Time-point comparison
Although the primary hypothesis motivating this study is the effect of a diagnosed concussion on the human microbiome in the 24–48 h following the event, a secondary hypothesis is that the chronic subconcussive impact experienced by most football players has a comparable but less pronounced effect which subsides sometime after the season. Although not statistically significant in the classical sense, the ANOSIM analysis discussed above suggests that there may be a material change in the gut microbiome for this group happening between the post-season and off-season. The comparison between timepoints for these athletes is shown in Fig. 2. For this group, the study design uses the homogenous nature of the cohort and multiple timepoints to boost its statistical power. Still, it naturally implies that differences between timepoints (particularly in and out of season) are associated with a substantial number of common environmental changes beyond frequent hits. We cannot associate this change with a single factor in the absence of a well-chosen control group.
One thing we can examine are specific microorganisms that show significant per-sample change in abundance based on a linear random-effects model. These microbes are presented in Supplemental Table 1, which includes the model-indicated fold-change in abundance from the post-season to the off-season. The Supplemental Table 1 is sorted according to p-value after multiple-hypothesis correction, with a cutoff of p < 0.125, and includes a note about the organism's known environment. Two observations are possible from this table. First, all the fold-change values are negative. The difference in communities is characterized by a common set of organisms that declined in abundance, but not by a common set of organisms that increased to make up the difference. In other words, the samples diverged. This is not particularly surprising since the athlete's behavior, schedule, diet, and physical location likely also diverged as they transitioned from the rigidly structured football season to their schedules, habits, diets, and locations in the off-season.
Second, the known environment of these specific microbes suggests a possible narrative of the change. Of those with decreased average abundance, three are commonly known environmental pathogens (one of which, S. aureus, is known to be higher risk specifically among football teams) and the other 14 are primarily associated with soil, water, or both. All but one are under 0.5% average abundance in the postseason samples. Given these associations and low abundances the most likely explanation seems to be that these are microbes the players are exposed to regularly in the football environment, are transmitted to the gut inadvertently during the season, represent environmental contaminants, or potential misclassifications. The Fig. 2d shows the per-subject abundance change for each bacterium; the panel on the right shows that the post-to-off change for this set of bacteria appears to be highly correlated across players. Namely, about half the subjects show a clear drop in abundance across all these organisms, while the other half show small changes with no particular pattern.
4.2. Concussion-status comparison
For the four players who were diagnosed with a concussion during the season, both alpha and beta diversity analyses show a significant difference between the post-concussion microbial community and that of other players during the season. The difference appears to be present in the off-season samples as well. This study is underpowered for this analysis due to the small number of cases and the fact a random-effects model is not available. Nonetheless, the comparison of subjects by concussion status and timepoint is shown in Fig. 3. The differences by concussion status are visible even in the simple stacked bar charts in Fig. 3c.
Beyond the alpha and beta diversity comparison, the multivariate analysis indicated what species are contributing most to these differences. As discussed above, the in-season model did not yield any statistically significant alterations, although the off-season model gave 9 with a p-value below 0.05, listed in Supplemental Table 2. The table also includes the coefficients and p-values for the same organisms from the in-season model, and it is worth noting that the in- and off-season coefficients are generally within a standard error of each other. It is important to re-emphasize the low statistical power in this analysis because the multiple-hypothesis correction reduces it even further. Given the difficulty of gathering a fecal sample immediately following a TBI event, the data should be considered valuable but taken lightly.
Two bacterial species were significantly depleted in relative abundance in the gut microbiota from players diagnosed with a concussion. One of the organisms identified is Eubacterium rectale, which was lower relative abundance (q < 0.05) in the post-concussion samples. Eubacterium rectale is a ubiquitous member of the human gut microbiome and an anti-inflammatory microbe (Mukherjee et al., 2020). A decrease in Eubacterium rectale has been previously correlated with peripheral inflammation in patients suffering from cognitive impairment and brain amyloidosis (Cattaneo et al., 2017). Also, Eubacterium rectale was found less frequently in children with neurodevelopmental disorders such as autism (Bojovic et al., 2020). The second bacterium, Anaerostipes hadrus, which is a common member of the healthy human gut microbiome with known anti-inflammatory properties (Brereton et al., 2021), showed similar depletion in the post-concussion group. Together, they hint at a narrative of the injury triggering pro-inflammatory pathways leading to a depletion in these two microbes. This is constant with effects observed in mice and underscores the need for a much larger study in the future with a large enough sample size that can provide more robust conclusions.
Next, we will review the additional results presented in the study: (i) biomarker analysis, (ii) pathway analysis, and (iii) salivary microbiome analysis.
4.2.1. Biomarker analysis
There are two main limitations to the development of reliable blood TBI biomarkers: 1) Highly expressed proteins within the CNS are only detectable at low plasma concentrations, and 2) Potential biomarkers undergo proteolytic degradation in the blood, such that their levels are affected by elimination through the liver or kidney14. Here, we used ELISA kits for blood protein detection, in addition to the ultrasensitive multiplex array (Simoa)15. S100β has been proposed as a fluid biomarker following brain injury. Blood levels of S100β are elevated after mTBI and have been associated with the presence of intracranial lesions, which lead to clinical studies for the use of this biomarker as a predictor of computed tomography (CT) scan findings (Bazarian et al., 2013; Zongo et al., 2012). As for SRC, serum S100β levels have been reported to be increased in athletes at 3 h following the injury but went back to baseline levels within 2 days post-concussion (Kiechle et al., 2014). A more recent study provided evidence of elevated S100β levels at 6 h after a concussion followed by a recovery to preseason levels in the 24–48 h window (Meier et al., 2017). High serum levels of SAA have been detected in children with mild head trauma (Gao et al., 2021), infants with white matter injury (Leviton et al., 2011), neonates with hypoxic-ischemic encephalopathy (Aly et al., 2011) and adult TBI patients (Hergenroeder et al., 2008). However, we did not find changes in serum S100β levels across the sports season or the post-concussion athletes compared to their non-concussed teammates. But notably, both serum blood biomarkers, S100β and SAA, were positively correlated with the abundance of the Eubacterium rectale species. Therefore, we report that there may be a relationship between inflammatory markers in the blood and the abundance of this bacterium among football players over the course of the year for athletes experiencing repetitive subconcussive impacts. A limitation of this analysis is the high degree of variability between the biomarkers, especially for S100β, SAA, and GFAP. We could not structure the limited group of players by the position they occupy to see if this can account for the variability observed. Future studies with a larger number of participants will include the risk of receiving concussions and subconcussive hits depending on the position (e.g., lower risk for non-contact positions such as Kicker or players that do not play all the games) as a variable to be evaluated.
4.2.2. Pathway analysis
Prediction of the pathway abundances associated with the fecal microbiota changes from the post to off-season indicated that processes related to the synthesis and degradation of sugars and aromatic compounds were among those showing the most significant fold changes. Interestingly, both bacteria species significantly decreased in abundance in players diagnosed with a concussion, E. rectale, and A. hadrus are major butyrate producers, with the former associated with insulin metabolism (Venkataraman et al., 2016). Butyrate producers are garnering renewed attention as “next-generation” probiotics for increasing colon butyrate production and their role in countering disease-associated alterations to the microbiome (Boesmans et al., 2018). Metabolomic analysis of the fecal samples in future studies will help characterize these changes further and elucidate their role in brain damage.
4.2.3. Salivary microbiome
We did not identify any observable changes in salivary microbiota, dampening the potential utility of this sample type as a biomarker of concussions. Despite this, a recent large prospective observational study of non-invasive concussion biomarkers has shown changes in salivary small non-coding RNAs (sncRNAs) in saliva in rugby players diagnosed with concussion (Di Pietro et al., 2021). Therefore, using RNA identifiers in saliva that correlate with biomarkers of the gut microbiota could represent a complete panel of concussion profiles that should be analyzed going forward.
A significant limitation of this study is that we were following a cohort of high-risk subjects across multiple timepoints, yielding valuable longitudinal data from a homogeneous population at the expense of a comparatively low sample size among concussion cases. Missing samples and data points, which are common in clinical studies due to participant dropout, constitute a further caveat for the analysis performed here. Since the study has low statistical power, some observations may be considered suggestive and worthy of additional study but should not be taken as conclusive. For example, we need to address the history of prior concussion in future studies, incorporate the measurement of head impacts and head impact exposure over time, a depth diet analysis, or the position they occupy in the game related to the risk of receiving more exposures to hits. These and other variables can be assumed if we increase the number of participants in future studies. Another limitation of this study is the lack of a baseline sample from each player taken prior to the start of the football season, which was not possible due mainly to the coincident timing of students returning to campus after the summer. As an alternative, we considered the off-season as a resting period of the season to be used for comparison with the in-season timepoints. However, the inclusion of a baseline control would more forcefully illustrate which changes are associated specifically with the onset of football season. We plan to expand this study to increase the cohort pool during the football season in the following years to address these limitations.
5. Conclusions
In summary, we report alterations in the gut microbiome in college football players after a single concussion and during seasonal sports activity. To our knowledge, this is the first study to highlight possible changes in the relative abundance of key gut bacteria during the football season. Specifically, Eubacterium rectale was associated with serum inflammatory biomarkers, which could indicate that it represents a microbial biomarker of SRC. Our study suggests that assessing species-specific changes in the gut microbiota after a concussion may represent a valuable tool in the clinic and will allow designing new evaluation and management of concussions in these athletes. Future research focused on in-depth multi-omics analyses in a more significant number of athletes who practice contact sports will be required to identify more reliable microbiome biomarkers and to identify concussions and subconcussive hits of difficult clinical diagnosis.
Funding
This work was supported by the National Institute for Neurological Disorders and Stroke (NINDS) (grant number R21NS106640 (SV)), Institute of Biosciences and Bioengineering (IBB) Hamill Innovation Award (TT), National Institute of Allergy and Infectious Diseases (NIAID) (grant number P01AI152999-01 (TT)), and funds from Houston Methodist Research Institute (SV).
Author contributions
S.Sc., S.Sa., R.G., K.P., G.B., T.T., and S.V., designed the study, S.Sa., A.C., and S.Sc. coordinated the participants enrollment, J.W., A.C., S.So. collected the samples, R.K. performed TCD analysis, S.So. performed laboratory analysis, K.C., Q.W., M.N., E.R., and T.T. performed bioinformatic and statistical analysis, and S.V., T.T., S.So., K.C., and M.N. wrote the manuscript, S.V., T.T., R.G., S.So., K.C., and M.N. analyzed the data, S.V. and T.T. coordinated the study. All authors reviewed and revised the final manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We would like to thank all of the college athletes who participated in this study. The authors are indebted to Dr. Gillian Hamilton for editing. Fig. 1 was created using Biorender.com.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbih.2022.100438.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Aly H., Hamed Z., Mohsen L., Ramy N., Arnaoot H., Lotfy A. Serum amyloid A protein and hypoxic ischemic encephalopathy in the newborn. J. Perinatol. 2011;31(4):263–268. doi: 10.1038/jp.2010.130. [DOI] [PubMed] [Google Scholar]
- Arciniegas D.B., McAllister T.W. Neurobehavioral management of traumatic brain injury in the critical care setting. Crit. Care Clin. 2008;24(4):737–765. doi: 10.1016/j.ccc.2008.06.001. viii. [DOI] [PubMed] [Google Scholar]
- Barton W., Penney N.C., Cronin O., Garcia-Perez I., Molloy M.G., Holmes E.…O'Sullivan O. The microbiome of professional athletes differs from that of more sedentary subjects in composition and particularly at the functional metabolic level. Gut. 2018;67(4):625–633. doi: 10.1136/gutjnl-2016-313627. [DOI] [PubMed] [Google Scholar]
- Bazarian J.J., Blyth B.J., He H., Mookerjee S., Jones C., Kiechle K.…Khan J. Classification accuracy of serum Apo A-I and S100B for the diagnosis of mild traumatic brain injury and prediction of abnormal initial head computed tomography scan. J. Neurotrauma. 2013;30(20):1747–1754. doi: 10.1089/neu.2013.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benakis C., Brea D., Caballero S., Faraco G., Moore J., Murphy M.…Anrather J. Commensal microbiota affects ischemic stroke outcome by regulating intestinal gammadelta T cells. Nat. Med. 2016;22(5):516–523. doi: 10.1038/nm.4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boesmans L., Valles-Colomer M., Wang J., Eeckhaut V., Falony G., Ducatelle R.…Verbeke K. Butyrate producers as potential next-generation probiotics: safety assessment of the administration of butyricicoccus pullicaecorum to healthy volunteers. mSystems. 2018;3(6) doi: 10.1128/mSystems.00094-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojovic K., Ignjatovic Eth I., Sokovic Bajic S., Vojnovic Milutinovic D., Tomic M., Golic N., Tolinacki M. Gut microbiota dysbiosis associated with altered production of short chain fatty acids in children with neurodevelopmental disorders. Front. Cell. Infect. Microbiol. 2020;10:223. doi: 10.3389/fcimb.2020.00223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner L.A., Stearns-Yoder K.A., Hoffberg A.S., Penzenik M.E., Starosta A.J., Hernandez T.D.…Lowry C.A. Growing literature but limited evidence: a systematic review regarding prebiotic and probiotic interventions for those with traumatic brain injury and/or posttraumatic stress disorder. Brain Behav. Immun. 2017;65:57–67. doi: 10.1016/j.bbi.2017.06.003. [DOI] [PubMed] [Google Scholar]
- Brereton N.J.B., Pitre F.E., Gonzalez E. Reanalysis of the Mars500 experiment reveals common gut microbiome alterations in astronauts induced by long-duration confinement. Comput. Struct. Biotechnol. J. 2021;19:2223–2235. doi: 10.1016/j.csbj.2021.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattaneo A., Cattane N., Galluzzi S., Provasi S., Lopizzo N., Festari C.…Group I.-F. Association of brain amyloidosis with pro-inflammatory gut bacterial taxa and peripheral inflammation markers in cognitively impaired elderly. Neurobiol. Aging. 2017;49:60–68. doi: 10.1016/j.neurobiolaging.2016.08.019. [DOI] [PubMed] [Google Scholar]
- Cenit M.C., Sanz Y., Codoner-Franch P. Influence of gut microbiota on neuropsychiatric disorders. World J. Gastroenterol. 2017;23(30):5486–5498. doi: 10.3748/wjg.v23.i30.5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conder R.L., Conder A.A. Sports-related concussions. N. C. Med. J. 2015;76(2):89–95. doi: 10.18043/ncm.76.2.89. [DOI] [PubMed] [Google Scholar]
- Crisco J.J., Fiore R., Beckwith J.G., Chu J.J., Brolinson P.G., Duma S.…Greenwald R.M. Frequency and location of head impact exposures in individual collegiate football players. J. Athl. Train. 2010;45(6):549–559. doi: 10.4085/1062-6050-45.6.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Battista A.P., Churchill N., Rhind S.G., Richards D., Hutchison M.G. Evidence of a distinct peripheral inflammatory profile in sport-related concussion. J. Neuroinflammation. 2019;16(1):17. doi: 10.1186/s12974-019-1402-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pietro V., O'Halloran P., Watson C.N., Begum G., Acharjee A., Yakoub K.M.…Belli A. Unique diagnostic signatures of concussion in the saliva of male athletes: the Study of Concussion in Rugby Union through MicroRNAs (SCRUM) Br. J. Sports Med. 2021 doi: 10.1136/bjsports-2020-103274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas G.M., Maffei V.J., Zaneveld J.R., Yurgel S.N., Brown J.R., Taylor C.M.…Langille M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020;38(6):685–688. doi: 10.1038/s41587-020-0548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duma S.M., Manoogian S.J., Bussone W.R., Brolinson P.G., Goforth M.W., Donnenwerth J.J.…Crisco J.J. Analysis of real-time head accelerations in collegiate football players. Clin. J. Sport Med. 2005;15(1):3–8. doi: 10.1097/00042752-200501000-00002. [DOI] [PubMed] [Google Scholar]
- Fujiyoshi S., Muto-Fujita A., Maruyama F. Evaluation of PCR conditions for characterizing bacterial communities with full-length 16S rRNA genes using a portable nanopore sequencer. Sci. Rep. 2020;10(1):12580. doi: 10.1038/s41598-020-69450-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo V., Motley K., Kemp S.P.T., Mian S., Patel T., James L.…McElvenny D. Concussion and long-term cognitive impairment among professional or elite sport-persons: a systematic review. J. Neurol. Neurosurg. Psychiatry. 2020;91(5):455–468. doi: 10.1136/jnnp-2019-321170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Boryczka J., Zheng P., Kasani S., Yang F., Engler-Chiurazzi E.B.…Wu N. A "hot Spot"-Enhanced paper lateral flow assay for ultrasensitive detection of traumatic brain injury biomarker S-100beta in blood plasma. Biosens. Bioelectron. 2021;177:112967. doi: 10.1016/j.bios.2021.112967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grande P.O., Asgeirsson B., Nordstrom C. Aspects on the cerebral perfusion pressure during therapy of a traumatic head injury. Acta Anaesthesiol. Scand. Suppl. 1997;110:36–40. doi: 10.1111/j.1399-6576.1997.tb05493.x. [DOI] [PubMed] [Google Scholar]
- Guskiewicz K.M., Mihalik J.P., Shankar V., Marshall S.W., Crowell D.H., Oliaro S.M.…Hooker D.N. Measurement of head impacts in collegiate football players: relationship between head impact biomechanics and acute clinical outcome after concussion. Neurosurgery. 2007;61(6):1244–1252. doi: 10.1227/01.neu.0000306103.68635.1a. discussion 1252-1243. [DOI] [PubMed] [Google Scholar]
- Han M., Yang K., Yang P., Zhong C., Chen C., Wang S.…Ning K. Stratification of athletes' gut microbiota: the multifaceted hubs associated with dietary factors, physical characteristics and performance. Gut Microb. 2020;12(1):1–18. doi: 10.1080/19490976.2020.1842991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergenroeder G., Redell J.B., Moore A.N., Dubinsky W.P., Funk R.T., Crommett J.…Dash P.K. Identification of serum biomarkers in brain-injured adults: potential for predicting elevated intracranial pressure. J. Neurotrauma. 2008;25(2):79–93. doi: 10.1089/neu.2007.0386. [DOI] [PubMed] [Google Scholar]
- Hoban A.E., Stilling R.M., Moloney G., Shanahan F., Dinan T.G., Clarke G., Cryan J.F. The microbiome regulates amygdala-dependent fear recall. Mol. Psychiatr. 2018;23(5):1134–1144. doi: 10.1038/mp.2017.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell D.R., Kirkwood M.W., Laker S., Wilson J.C. Collision and contact sport participation and quality of life among adolescent athletes. J. Athl. Train. 2020;55(11):1174–1180. doi: 10.4085/1062-6050-0536.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iverson G.L., Williams M.W., Gardner A.J., Terry D.P. Systematic review of preinjury mental health problems as a vulnerability factor for worse outcome after sport-related concussion. Orthop. J. Sports Med. 2020;8(10) doi: 10.1177/2325967120950682. 2325967120950682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan B.D. The clinical spectrum of sport-related traumatic brain injury. Nat. Rev. Neurol. 2013;9(4):222–230. doi: 10.1038/nrneurol.2013.33. [DOI] [PubMed] [Google Scholar]
- Kiechle K., Bazarian J.J., Merchant-Borna K., Stoecklein V., Rozen E., Blyth B.…Biberthaler P. Subject-specific increases in serum S-100B distinguish sports-related concussion from sports-related exertion. PLoS One. 2014;9(1) doi: 10.1371/journal.pone.0084977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilcoyne K.G., Dickens J.F., Svoboda S.J., Owens B.D., Cameron K.L., Sullivan R.T., Rue J.P. Reported concussion rates for three division I football programs: an evaluation of the new NCAA concussion policy. Sport Health. 2014;6(5):402–405. doi: 10.1177/1941738113491545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leviton A., Kuban K.C., Allred E.N., Fichorova R.N., O'Shea T.M., Paneth N., Investigators E.S. Early postnatal blood concentrations of inflammation-related proteins and microcephaly two years later in infants born before the 28th post-menstrual week. Early Hum. Dev. 2011;87(5):325–330. doi: 10.1016/j.earlhumdev.2011.01.043. [DOI] [PubMed] [Google Scholar]
- Li A.Y., Schupper A.J., Quinones A., Shuman W.H., Ali M., Hannah T.C.…Choudhri T.F. Sport contact level affects post-concussion neurocognitive performance in young athletes. Arch. Clin. Neuropsychol. 2021 doi: 10.1093/arclin/acab021. [DOI] [PubMed] [Google Scholar]
- Ma E.L., Smith A.D., Desai N., Cheung L., Hanscom M., Stoica B.A.…Faden A.I. Bidirectional brain-gut interactions and chronic pathological changes after traumatic brain injury in mice. Brain Behav. Immun. 2017;66:56–69. doi: 10.1016/j.bbi.2017.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallick H, Rahnavard A, McIver LJ, Siyuan M, Yangcong Z, Nguyen LH, Tickle TL, et al. Multivariable association discovery in population-scale meta-omics studies. PLoS Comput Biol; 2021;e1009442 doi: 10.1371/journal.pcbi.1009442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley G., Gardner A.J., Schneider K.J., Guskiewicz K.M., Bailes J., Cantu R.C.…Iverson G.L. A systematic review of potential long-term effects of sport-related concussion. Br. J. Sports Med. 2017;51(12):969–977. doi: 10.1136/bjsports-2017-097791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazarati A., Medel-Matus J.S., Shin D., Jacobs J.P., Sankar R. Disruption of intestinal barrier and endotoxemia after traumatic brain injury: implications for post-traumatic epilepsy. Epilepsia. 2021;62(6):1472–1481. doi: 10.1111/epi.16909. [DOI] [PubMed] [Google Scholar]
- McAllister T.W. Neurobehavioral sequelae of traumatic brain injury: evaluation and management. World Psychiatr. 2008;7(1):3–10. doi: 10.1002/j.2051-5545.2008.tb00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCrea M.A., Shah A., Duma S., Rowson S., Harezlak J., McAllister T.W.…Stemper B.D. Opportunities for prevention of concussion and repetitive head impact exposure in college football players: a concussion assessment, research, and education (care) consortium study. JAMA Neurol. 2021;78(3):346–350. doi: 10.1001/jamaneurol.2020.5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCrory P., Meeuwisse W., Aubry M., Cantu B., Dvorak J., Echemendia R.…Turner M. Consensus statement on concussion in sport - the 4th international conference on concussion in sport held in zurich, november 2012. Phys. Ther. Sport. 2013;14(2):e1–e13. doi: 10.1016/j.ptsp.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Meier T.B., Nelson L.D., Huber D.L., Bazarian J.J., Hayes R.L., McCrea M.A. Prospective assessment of acute blood markers of brain injury in sport-related concussion. J. Neurotrauma. 2017;34(22):3134–3142. doi: 10.1089/neu.2017.5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondello S., Muller U., Jeromin A., Streeter J., Hayes R.L., Wang K.K. Blood-based diagnostics of traumatic brain injuries. Expert Rev. Mol. Diagn. 2011;11(1):65–78. doi: 10.1586/erm.10.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montenigro P.H., Alosco M.L., Martin B.M., Daneshvar D.H., Mez J., Chaisson C.E.…Tripodis Y. Cumulative head impact exposure predicts later-life depression, apathy, executive dysfunction, and cognitive impairment in former high school and college football players. J. Neurotrauma. 2017;34(2):328–340. doi: 10.1089/neu.2016.4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouzon B.C., Bachmeier C., Ojo J.O., Acker C.M., Ferguson S., Paris D.…Crawford F. Lifelong behavioral and neuropathological consequences of repetitive mild traumatic brain injury. Ann. Clin. Transl. Neurol. 2018;5(1):64–80. doi: 10.1002/acn3.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A., Lordan C., Ross R.P., Cotter P.D. Gut microbes from the phylogenetically diverse genus Eubacterium and their various contributions to gut health. Gut Microb. 2020;12(1):1802866. doi: 10.1080/19490976.2020.1802866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson S.E., Watts L.T., Burmeister D.M., Merrill D., Scroggins S., Zou Y.…Schwacha M.G. Moderate traumatic brain injury alters the gastrointestinal microbiome in a time-dependent manner. Shock. 2019;52(2):240–248. doi: 10.1097/SHK.0000000000001211. [DOI] [PubMed] [Google Scholar]
- Nygaard A.B., Tunsjo H.S., Meisal R., Charnock C. A preliminary study on the potential of Nanopore MinION and Illumina MiSeq 16S rRNA gene sequencing to characterize building-dust microbiomes. Sci. Rep. 2020;10(1):3209. doi: 10.1038/s41598-020-59771-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver J.M., Anzalone A.J., Stone J.D., Turner S.M., Blueitt D., Garrison J.C.…Jagim A.R. Fluctuations in blood biomarkers of head trauma in NCAA football athletes over the course of a season. J. Neurosurg. 2018:1–8. doi: 10.3171/2017.12.JNS172035. [DOI] [PubMed] [Google Scholar]
- Ribeiro F.M., Petriz B., Marques G., Kamilla L.H., Franco O.L. Is there an exercise-intensity threshold capable of avoiding the leaky gut? Front Nutr. 2021;8:627289. doi: 10.3389/fnut.2021.627289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuttleworth-Edwards A.B., Smith I., Radloff S.E. Neurocognitive vulnerability amongst university rugby players versus noncontact sport controls. J. Clin. Exp. Neuropsychol. 2008;30(8):870–884. doi: 10.1080/13803390701846914. [DOI] [PubMed] [Google Scholar]
- Smoliga J.M. Interpreting biomarker data after concussion and repeated subconcussive head impacts: challenges in evaluating brain protection. JAMA Neurol. 2020;77(12):1477–1478. doi: 10.1001/jamaneurol.2020.3467. [DOI] [PubMed] [Google Scholar]
- Stein T.D., Alvarez V.E., McKee A.C. Concussion in chronic traumatic encephalopathy. Curr. Pain Headache Rep. 2015;19(10):47. doi: 10.1007/s11916-015-0522-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M., Zhu J.C., Du J., Zhang L.M., Yin H.H. Effects of probiotics on serum levels of Th1/Th2 cytokine and clinical outcomes in severe traumatic brain-injured patients: a prospective randomized pilot study. Crit. Care. 2011;15(6):R290. doi: 10.1186/cc10579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treangen T.J., Wagner J., Burns M.P., Villapol S. Traumatic brain injury in mice induces acute bacterial dysbiosis within the fecal microbiome. Front. Immunol. 2018;9:2757. doi: 10.3389/fimmu.2018.02757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban R.J., Pyles R.B., Stewart C.J., Ajami N., Randolph K.M., Durham W.J.…Sheffield-Moore M. Altered fecal microbiome years after traumatic brain injury. J. Neurotrauma. 2020;37(8):1037–1051. doi: 10.1089/neu.2019.6688. [DOI] [PubMed] [Google Scholar]
- Venkataraman A., Sieber J.R., Schmidt A.W., Waldron C., Theis K.R., Schmidt T.M. Variable responses of human microbiomes to dietary supplementation with resistant starch. Microbiome. 2016;4(1):33. doi: 10.1186/s40168-016-0178-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood D.E., Lu J., Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20(1):257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zongo D., Ribereau-Gayon R., Masson F., Laborey M., Contrand B., Salmi L.R.…Lagarde E. S100-B protein as a screening tool for the early assessment of minor head injury. Ann. Emerg. Med. 2012;59(3):209–218. doi: 10.1016/j.annemergmed.2011.07.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The high-throughput sequence data has been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under the project database with project number PRJNA699316. The authors declare that all other data supporting the findings of this study are available within the article and its Supplementary Information files or from the corresponding author on request.