Abstract
Acinetobacter baumannii has emerged as an important opportunistic pathogen worldwide, being responsible for large outbreaks for nosocomial infections, primarily in intensive care units. A. baumannii ATCC 19606T is the species type strain, and a reference organism in many laboratories due to its low virulence, amenability to genetic manipulation and extensive antibiotic susceptibility. We wondered if frequent propagation of A. baumannii ATCC 19606T in different laboratories may have driven micro- and macro-evolutionary events that could determine inter-laboratory differences of genome-based data. By combining Illumina MiSeq, MinION and Sanger technologies, we generated a high-quality whole-genome sequence of A. baumannii ATCC 19606T, then performed a comparative genome analysis between A. baumannii ATCC 19606T strains from several research laboratories and a reference collection. Differences between publicly available ATCC 19606T genome sequences were observed, including SNPs, macro- and micro-deletions, and the uneven presence of a 52 kb prophage belonging to genus Vieuvirus. Two plasmids, pMAC and p1ATCC19606, were invariably detected in all tested strains. The presence of a putative replicase, a replication origin containing four 22-mer direct repeats, and a toxin-antitoxin system implicated in plasmid stability were predicted by in silico analysis of p1ATCC19606, and experimentally confirmed. This work refines the sequence, structure and functional annotation of the A. baumannii ATCC 19606T genome, and highlights some remarkable differences between domesticated strains, likely resulting from genetic drift.
Keywords: Acinetobacter baumannii ATCC 19606T , genome refinement, native plasmids, Φ19606 phage, strain domestication
Data Summary
The sequence data of the Acinetobacter baumannii ATCC 19606(A) used in this study is freely available from the NCBI BioProject database under accession number PRJNA637288.
Impact Statement.
For decades Acinetobacter baumannii has emerged as a major antibiotic-resistant nosocomial pathogen, and the type strain ATCC 19606T has been used as the reference organism for A. baumannii research in many laboratories. However, frequent subculturing and local differences in culture conditions can result in domestication of laboratory strains, a micro-evolutionary process driven by mutational events at the genome level that can reflect into a variable phenotype. Motivated by the remarkable diversity in publicly available A. baumannii ATCC 19606T whole-genome sequences, we generated an accurately revised whole-genome sequence of ATCC 19606T, which will set a more solid basis for studies of the genetics and genomics of this model organism. Accurate genome assembly made it possible to characterize indigenous plasmids and a new prophage in the A. baumannii ATCC 19606T genome. The remarkable genome diversity among A. baumannii ATCC 19606T strains from different laboratories poses the need for researchers to specify the lineage of the strain used, as local culturing and storage practices may affect strain microevolution.
Introduction
Acinetobacter baumannii is a worldwide-distributed Gram-negative bacterium and a major opportunistic pathogen, especially among critically ill patients in intensive care units [1–3]. Due to extensive antibiotic resistance [4, 5], A. baumannii is on top of the global priority list of pathogens for which new and effective drugs are urgently needed, according to the World Health Organization [6]. While it is likely that infections caused by bacteria now classified as A. baumannii emerged during the 1970s [7], the species was not formally established until 1986, when strain ATCC 19606T was designated as the A. baumannii type strain [8]. A. baumannii ATCC 19606T was isolated in 1948 from the urine of a US patient, and called Bacterium anitratum [9, 10], then filed with the ATCC and renamed A. baumannii in 1986 [8]. More recently, ATCC 19606T was assigned type O by pulsed-field gel electrophoresis and ST52 by multi-locus sequence typing (MLST) [11]. A. baumannii ATCC 19606T has extensively been used as a model strain for research on antimicrobial resistance [12–15], desiccation and osmotic shock tolerance [16], transcriptional regulation and virulence both in vitro and in vivo [17–24], with more than 180 published papers according to ISI Web of Science records (accessed July 2021). The importance of A. baumannii ATCC 19606T as a model organism led us to initiate a whole sequencing and annotation project of its genome. While this work was in progress, three complete genome sequences of ATCC 19606T were released [CP045110.1 [25], hereafter ATCC 19606(H); AP022836 [26], hereafter ATCC 19606(O); CP046654.1 [27], hereafter ATCC 19606(M)], showing remarkable differences in sequence and annotation compared with the A. baumannii ATCC 19606T genome determined in our laboratory. We wondered if the maintenance and propagation of A. baumannii ATCC 19606T in laboratories throughout the world may have entailed micro- and macro-evolutionary events responsible for such differences, and eventually affect the inter-laboratory comparison of genome-based data, as inferred for other laboratory-adapted strains [28–32]. To test this hypothesis, we generated a high-quality complete genome sequence of A. baumannii ATCC 19606T employing Illumina MiSeq and MinION technologies and performed a comparative analysis with previously deposited whole-genome sequences of this strain [25–27]. To gain further insights into inter-laboratory diversification of the A. baumannii ATCC 19606T, the whole-genome sequences of strains maintained in different research institutions throughout Europe and the ATCC (complete genome sequence available at https://www.lgcstandards-atcc.org/Products/All/19606.aspx?geo_country=it#generalinformation) were compared. Numerous single nucleotide polymorphisms (SNPs) and insertions-deletions (INDELs), as well as the occasional loss of a prophage and the invariable presence of two indigenous plasmids were demonstrated, probably reflecting strain domestication. Moreover, the toxin-antitoxin (TA) module and the replicase gene of the indigenous plasmid p1ATCC19606 [25] were in silico characterized and experimentally verified, providing useful information about the mechanisms of plasmid maintenance and replication in Acinetobacter .
Methods
Bacterial strains and culture media
Bacterial strains used in this study (Table 1) were grown in Luria–Bertani (LB) broth or on LB agar (LA) plates at 37 °C. When needed, kanamycin (Km), gentamicin (Gm) or tetracycline (Tc) were added. The Km concentrations used for Escherichia coli DH5α and Acinetobacter baylyi BD413 were 40 µg ml−1 and 15 µg ml−1, respectively. The Gm and Tc concentrations used for E. coli DH5α were 10 µg ml−1 and 12.5 µg ml−1, respectively. Zeocin (Zeo) was added to low-salt LA [33] at 25 µg ml−1 and 250 µg ml−1 for E. coli DH5α and Acinetobacter spp., respectively.
Table 1.
Bacterial strains and plasmids
|
Strain or plasmid |
Relevant characteristics* |
Received year |
Reference and/or source |
|---|---|---|---|
|
Strain | |||
|
|
|||
|
ATCC 19606(A) |
Clinical isolate; type strain |
2014 |
Beate Averhoff collection; genome accession number: CP058289.1 |
|
ATCC 19606(D) |
Clinical isolate; type strain |
2020 |
German Collection of Microorganisms and Cell Cultures, DSMZ GmbH (genome available at https://genomes.atcc.org/genomes/1577c3a70f334038) |
|
ATCC 19606(H) |
Clinical isolate; type strain |
– |
[25]; genome accession number: CP045110.1 |
|
ATCC 19606(M) |
Clinical isolate; type strain |
– |
[27]; genome accession number: CP046654.1 |
|
ATCC 19606(O) |
Clinical isolate; type strain |
– |
|
|
ATCC 19606(S) |
Clinical isolate; type strain |
2019 |
Harald Seifert collection |
|
ATCC 19606(T) |
Clinical isolate; type strain |
2010 |
Kevin Towner collection |
|
ATCC 17978 |
Clinical isolate |
2007 |
[114] |
|
ACICU |
MDR clinical isolate, prototype of the international clonal lineage II |
2007 |
[115] |
|
AB5075 |
MDR and hypervirulent clinical isolate |
2019 |
[116] |
|
A. baylyi BD413 (ADP1) |
Naturally transformable strain |
2017 |
[117] |
|
Member of the ACB complex |
2017 |
[118]; H. Seifert collection |
|
|
Acinetobacter nosocomialis UKK_0361 |
Member of the ACB complex |
2017 |
[66]; H. Seifert collection |
|
Acinetobacter pittii UKK_0145 |
Member of the ACB complex |
2017 |
[66]; H. Seifert collection |
|
E. coli DH5α |
recA1 endA1 hsdR17 supE44 thi-1 gyrA96 relA1 Δ(lacZYA-argF)U169 [ϕ80dlacZΔM15] F- NalR |
– |
[60] |
|
Plasmid |
|
||
|
pCR-Blunt II-TOPO |
E. coli cloning vector; KmR, ZeoR |
– |
ThermoFisher |
|
p1ATCC19606 |
Native plasmid of A. baumannii ATCC 19606T |
– |
[25] |
|
pMAC |
Native plasmid of A. baumannii ATCC 19606T |
– |
[87] |
|
pVRL1 |
E. coli-Acinetobacter species shuttle vector for general cloning purposes; GmR |
– |
[66] |
|
pVRL1ΔTA |
pVRL1 carrying a deletion in the TA system; GmR |
– |
[66] |
|
pVRL2 |
E. coli-Acinetobacter species shuttle vector for arabinose-inducible gene expression; GmR |
– |
[66] |
|
pME6032 |
Broad-host-range shuttle vector for IPTG-inducible gene expression; TcR |
– |
[68] |
|
pCR-p1ATCC19606 |
Full length p1ATCC19606 ligated to pCR-Blunt II-TOPO; KmR, ZeoR |
– |
This work |
|
pCR-p1ATCC19606Δ1 |
Deletion derivative of p1ATCC19606 cloned into pCR-Blunt II-TOPO; KmR, ZeoR |
– |
This work |
|
pCR-p1ATCC19606Δ2 |
Deletion derivative of p1ATCC19606 cloned into pCR-Blunt II-TOPO; KmR, ZeoR |
– |
This work |
|
pCR-p1ATCC19606Δ3 |
Deletion derivative of p1ATCC19606 cloned into pCR-Blunt II-TOPO; KmR, ZeoR |
– |
This work |
|
pCR-p1ATCC19606Δ4 |
Deletion derivative of p1ATCC19606 cloned into pCR-Blunt II-TOPO; KmR, ZeoR |
– |
This work |
|
pCR-p1ATCC19606Δ5 |
Deletion derivative of p1ATCC19606 cloned into pCR-Blunt II-TOPO; KmR, ZeoR |
– |
This work |
|
pCR-p1ATCC19606ΔhigB2A2 |
pCR-p1ATCC19606 carrying a deletion in the TA system; KmR, ZeoR |
– |
This work |
|
pVRL2higA2 |
higA2-like antitoxin promoterless gene cloned into pVRL2; GmR |
– |
This work |
|
pME6032higB2 |
higB2-like toxin promoterless gene cloned into pME6032; TcR |
– |
This work |
*NalR, nalidixic acid resistant; KmR, kanamicin resistant; TcR, tetracycline resistant; ZeoR, zeocin resistant, GmR, gentamicin resistance.
DNA manipulation
Genomic DNA was extracted and purified with the QIAamp DNA minikit (Qiagen), according to the manufacturer’s instructions. Plasmid DNA was extracted from overnight bacterial cultures using the Wizard Plus SV Minipreps DNA Purification System (Promega Corporation), according to the manufacturer’s instructions. PCR reactions were performed using Thermo Scientific Phusion High-Fidelity DNA Polymerase with primers listed in Table S1 (available in the online version of this article). FastDigest restriction enzymes were purchased from Thermo Fisher Scientific. Plasmid DNA sequencing was performed using an ABI3730 Sequencer (service by Bio-fab Research, Rome, IT).
Genome sequencing, assembly and annotation
Whole-genome sequencing of A. baumannii ATCC 19606(A) was performed by GenProbio srl (Parma, Italy) using a MiSeq platform (Illumina, San Diego, CA, USA) according to the supplier’s protocol (Illumina, San Diego, CA, USA). Genomic DNA extracted from A. baumannii ATCC 19606(A) was also subjected to whole-genome sequencing using a MinION (Oxford Nanopore, UK) at GenProbio srl (Parma, Italy) according to the supplier’s protocol (Oxford Nanopore, UK). MinION long reads obtained from genome sequencing runs were used as input for a de novo genome assembly using Canu v1.8 with the estimated parameter ‘genomeSize’ of 4.0 m [34], generating a single complete sequence of the genome. Then, fastq files of Illumina paired-end reads (250 bp) and MinION long reads (ranging from 1000 to 100 065 bp) were used as input for a second genome assembly through the MEGAnnotator pipeline [35]. The SPAdes programme v 3.13.0 was used for the hybrid assembly of the genome sequence with the pipeline option ‘--isolate’, a list of k-mer sizes of 21, 33, 55, 77, 99, 127, and the complete genome sequence obtained through Canu v1.8 for gap closure and repeat resolution using the option ‘--trusted-contigs’ [36]. The chromosome sequence, together with those of plasmids, were then analysed by MEGAnnotator for the prediction of protein-encoding ORFs using Prodigal [37]. Predicted ORFs were functionally annotated by means of RAPSearch2 (Reduced Alphabet based Protein similarity Search) (cutoff E value, 1×10−5; minimum alignment length, 20 aminoacids) by interrogation of the NCBI nr database [38] coupled with hidden Markov model (HMM) profile searches (http://hmmer.org/), performed against the manually curated Pfam-A database (cutoff E value, 1×10−10). tRNA genes were identified using tRNAscan-SE v1.4 [39], while rRNA genes were detected using RNAmmer v1.2 [40]. Before genome submission to NCBI, a protein integrity check that takes neighbouring pairs of proteins and does a blastp analysis on them was performed to detect frameshifts (https://www.ncbi.nlm.nih.gov/genomes/frameshifts/frameshifts.cgi). Pairs of neighbours that hit the same longer protein were annotated as pseudogenes. The presence of genomic islands (GIs) was predicted by IslandViewer 4 [41], which uses SIGI-HMM, IslandPath-DIMOB and IslandPick prediction algorithms to calculate codon usage, dinucleotide bias within a genome, generating a dataset of GIs. Only GIs predicted by at least one of the three algorithms, which do not completely overlap with any predicted phage regions, were considered. Insertion sequences (ISs) were predicted by ISEScan [42]. Prophage sequence prediction and annotation were performed using PHASTER (PHAge Search Tool Enhanced Release) (http://phaster.ca), and only intact (score >90) and questionable (score >70 to 90) phage genomes were considered, whereas incomplete phage genomes (score ≤70) were discarded [43]. The raw read data of the ATCC 19606(M) sequencing project (SRR10295884) were downloaded and screened in search for putative plasmid sequences using both plasmidSPADES [44] and Bowtie 2 [45]. The quality of genome assemblies, namely completeness and contamination percentages were evaluated using CheckM [46].
Comparative genome analyses and mutation detection
ProgressiveMauve [47] was used for pairwise comparison of the A. baumannii ATCC 19606(A) assembled genome with A. baumannii ATCC 19606(D, M, H and O strains) genome assemblies. To detect SNPs and microindels (insertion or deletion events), sequence reads belonging to ATCC 19606(A) were mapped against the ATCC 19606(M) and ATCC 19606(H) genome sequences with BWA mem v0.7.17 [48], using default parameters. A consensus pileup was produced using SAMtools v1.10 [49]. Then, SNPs and microindels were defined using VarScan v2.3.6 [50] with the following parameters: minimum coverage (8), min-reads2 (2), min-avg-qual (15), min-var-freq (0.5), P-value (99×10−2). SNPs and microindels were manually inspected in the output files with Artemis [51]. All discrepancies, i.e. mutations inferred from comparative genome analysis, between ATCC 19606(A), ATCC 19606(M) and ATCC 19606(H) were confirmed by Sanger sequencing of PCR products encompassing the mutated site using an ABI3730 Sequencer (service by Bio-Fab Research, Rome, Italy). For each mutated protein product, a corresponding orthologue from ACICU strain (CP031380.1) was identified using blastp analysis (https://blast.ncbi.nlm.nih.gov/Blast.cgi) with a cutoff value E<10−5 and 50 % identity across at least 80 % of protein sequences. Twenty-two genes showing SNPs and INDELs in pairwise comparisons between ATCC 19606(A), ATCC 19606(H) and ATCC 19606(M) genomes were concatenated, and aligned with mafft v7, selecting the G-INS-i method [52]. A phylogenetic tree based on the concatenated alignment was constructed using the neighbour-joining (NJ) method and visualized using iTOL v6.1.2 [53]. The tree was rooted on ATCC 19606(A).
Detection of phage Φ19606
The presence of phage Φ19606 in A. baumannii ATCC 19606 strains A, D, S and T was verified by PCR using the primer pairs listed in Table S1 and 30 ng of bacterial DNA as a template. To evaluate the loss of Φ19606 during serial propagation steps, ten 1 mm colonies were randomly taken from primary plating on LA of the original A. baumannii ATCC 19606(D) vial and suspended in 500 µl of saline at 4 °C for 30 min with intermittent vortexing to maximize the separation of individual cells. The absence of cell aggregates was verified by bright-field microscopy. Then 1 µl of this suspension was streaked onto an LA plate and incubated at 37 °C for 24 h, while the rest was centrifuged for total DNA extraction from the cell pellet using QIAamp DNA minikit (Qiagen), according to the manufacturer’s instructions. Fourteen serial passages were repeated according to this procedure, for a total of 15 passages including the primary plating from the DSMZ stock vial. For each propagation step of ATCC 19606(D), the presence/absence of Φ19606 was verified by PCR using the primers listed in Table S1 and 30 ng of the purified genome as template.
Phylogenetic analysis of phage Ф19606
The complete genome sequences of Acinetobacter phages belonging to the Siphoviridae family were retrieved from the NCBI database. The resulting dataset contained the genomes of 19 Acinetobacter phages (Table S2), including Ф19606. All pairwise comparisons of the nucleotide sequences were conducted using the Genome-blast Distance Phylogeny (GBDP) method [54] under settings recommended for prokaryotic viruses [55] using VICTOR (https://ggdc.dsmz.de/victor.php). The resulting intergenomic distances were used to infer a balanced minimum evolution tree with branch support via FASTME including SPR postprocessing [56] for the formula D0. Branch support was inferred from 100 pseudo-bootstrap replicates each. The tree was rooted at the midpoint [57] and visualized with iTOL [53]. Taxon boundaries at the genus level were estimated with the optsil programme [58], with the recommended clustering threshold of 0.84 [56] and an F value (fraction of links required for cluster fusion) of 0.5 [59]. The presence of the Ф19606 sequence was searched in all Acinetobacter strains using blastn against the NCBI database with cutoff values of 95 % identity and 85 % coverage of consecutive segments from the query sequence. For each strain containing the Ф19606 sequence, MLST was performed according to the Pasteur scheme (https://pubmlst.org/bigsdb?db=pubmlst_abaumannii_pasteur_seqdef) and the corresponding sequence type (ST) was determined.
Preparation of Acinetobacter spp. and E. coli DH5α competent cells
E. coli competent cells were prepared by the rubidium-calcium chloride method and transformed according to the heat-shock protocol [60]. Electrocompetent cells of A. baumannii were prepared according to Lucidi and coworkers [22]. Plasmid DNA was introduced in Acinetobacter spp. by electroporation as previously described [22]. Naturally competent A. baylyi BD413 was transformed with 150 ng of plasmid, as previously reported [61].
Deletion analysis of p1ATCC19606
DNA fragments encompassing different regions of the p1ATCC19606 plasmid were amplified by PCR using primer pairs listed in Table S1. The amplicons were cloned into pCR-Blunt II-TOPO (pCR, Thermo Fisher Scientific) according to the manufacturer’s instructions, and the resulting constructs (Table 1) were introduced by electroporation into different Acinetobacter spp. The transformation efficiency (TE) was calculated as the ratio between the c.f.u. counts on selective agar plates and the amount of DNA used for transformation and expressed as c.f.u. μg−1 of plasmid DNA.
Homology searches and protein modelling
Putative protein-coding genes from plasmid p1ATCC19606 were manually annotated by integrating the MEGAnnotator [35] and Blast2GO (Basic Local Alignment Search Tool 2 Gene Ontology; [62]) outputs. The putative TA system of p1ATCC19606 was characterized by predicting the protein structures using I-TASSER [63] and SWISS-MODEL [64]. Match marker analyses and superimposition of proteins were performed using the UCSF Chimaera software [65].
Impact of the higB2A2-like toxin-antitoxin system on p1ATCC19606 stability
The p1ATCC19606 plasmid was ligated to the pCR plasmid to enable replication in E. coli , yielding pCR-p1ATCC19606. Subsequently, deletion of the higB2A2-like operon (293 bp) was generated using primers ΔTA FW and ΔTA RV (Table S1) and a Q5 site-directed mutagenesis kit (New England BioLabs). The resulting construct, named pCR-p1ATCC19606ΔhigB2A2, was introduced into E. coli DH5α for plasmid stability testing. Briefly, bacterial strains were preliminarily grown for 18 h in LB with 40 µg ml−1 Km, then washed and diluted 1000-fold in LB (without antibiotic). Bacterial cultures were refreshed (1 : 1000) every 12 h four times, for a total of 48 h. Bacterial colony counts were determined on LA (N0 ) and LA supplemented with 40 µg ml−1 Km (N Ant). Plasmids pVRL1 and pVRL1ΔTA were used as controls for plasmid stability, as reported elsewhere [66]. Plasmid stability is defined by the ratio N Ant /N0 [67].
Construction of plasmids directing the controlled expression of higB2 and higA2 genes
The promoterless higA2-like antitoxin gene was amplified from p1ATCC19606 with primers pVRL2higA2_FW and pVRL2higA2_RV (Table S1), and the 357 bp amplicon was ligated to XhoI and HindIII sites of the pVRL2 vector [66], yielding pVRL2higA2. The promoterless higB2-like toxin gene was amplified by PCR from p1ATCC19606 with primers pME6032higB2_FW and pME6032higB2_RV (Table S1), and the 368 bp amplicon was ligated to the SacI and XhoI sites of the pME6032 vector [68], yielding pME6032higB2. Plasmids pVRL2higA2 and pME6032higB2 were individually introduced into E. coli DH5α, and transformants were selected on LA supplemented with either 10 µg ml−1 Gm or 12.5 µg ml−1 Tc, respectively. Subsequently, pME6032higB2 was introduced in E. coli DH5α(pVRL2higA2) and transformants were routinely maintained on LA supplemented with 10 µg ml−1 Gm, 12.5 µg ml−1 Tc and 0.5 % l-arabinose to ensure plasmid selection and expression of the higA2-like antitoxin gene. To test the neutralizing effect of the HigA2 antitoxin on the HigB2 toxin, E. coli DH5α carrying both pVRL2higA2 and pME6032higB2 was grown in LB supplemented with 10 µg ml−1 Gm, 12.5 µg ml−1 Tc, and 0.5 % l-arabinose, then washed in saline, diluted to OD600=0.001 (corresponding to ca 5×105 c.f.u. ml−1) and dispensed in a microtitre plate containing LB supplemented with 12.5 µg ml−1 Tc and different concentrations of l-arabinose (from 1–0.015 %) and isopropyl-β-D-1-thiogalactopyranoside (IPTG; from 300 to 9.4 µM). After 24 h incubation at 37 °C, the growth was determined by OD600 measurements in a Spark 10M microtitre reader (Tecan). E. coli DH5α(pVRL2higA2 pME6032higB2) was also plated on LA supplemented with 0.5, 0.25, 0.12 and 0 % l-arabinose. Then paper discs containing IPTG (from 5 mM to 0.08 mM) were dispensed on the plate, and the zone of inhibition (ZOI) was determined after 24 h incubation at 37 °C.
Results and discussion
Historical background of A. baumannii ATCC 19606T
In the course of a study on ‘paracolon bacilli’ (old designation for coliform bacteria that do not ferment lactose) carried out by Dr K. Wheeler and Dr C.A. Stuart, several isolates were sent to Dr I.G. Schaub and Dr F.D. Hauber (John Hopkins University, Baltimore, MD, USA) for laboratory identification. Among these, one isolate (no. 81 according to Schaub’s and Hauber’s nomenclature) was reported to be an unidentifiable Bacterium sp. responsible for a urinary tract infection. The isolate was proposed to belong to the species Bacterium anitratum [9]. This isolate was sent to Dr E.O. King (CDC, Atlanta, GA, USA) in 1965, and a descent to Dr R. Hugh (George Washington University, Washington, DC, USA) before being filed with ATCC in 1966, and made publicly available with the ATCC 19606 designation (chain of custody available at https://www.lgcstandards-atcc.org/products/all/19606.aspx#history).
A first taxonomic reassignment of Bacterium anitratum to the genus Acinetobacter was proposed in 1964, but it was not accepted by the scientific community [69]. Thereafter, ATCC 19606 was formally designated as the type strain of B. anitratum [10]. In the 1980s, DNA–DNA hybridization studies reclassified B. anitratum as Acinetobacter calcoaceticus [70] and, subsequently, the definitive name of A. baumannii ATCC 19606T was assigned to the original Schaub and Hauber no. 81 isolate [8].
Genealogy of A. baumannii ATCC 19606T strains from different sources
We wondered if maintenance and sequential propagation of A. baumannii ATCC 19606T in different laboratories may result in genotype differences. To address this issue, four ATCC 19606 strains (A, D, S, T) were obtained from three independent research laboratories in Europe and the German Collection of Microorganisms and Cell Cultures-DSMZ GmbH (Braunschweig, Germany) (Table 1). ATCC 19606(A) was directly purchased from ATCC by microbiologists of the Robert Koch Institute (Wernigerode, Germany), then given to Professor B. Averhoff of the Goethe University (Frankfurt am Main, Germany) before reaching our laboratory. ATCC 19606 strain D was purchased by our laboratory from the DSMZ. ATCC 19606 strains S and T were shared by Dr P.A.D. Grimont of Institute Pasteur (Paris, France) with Professor H. Seifert (Institut Für Medizinische Mikrobiologie Immunologie Und Hygiene, Cologne, Germany) and Professor K.J. Towner (Department of Clinical Microbiology, Nottingham University Hospitals NHS Trust, Queen’s Medical Centre, Nottingham, UK), respectively, then sent to our laboratory.
From the history of the various strains, it emerged that ATCC 19606(A, S, T) were serially propagated several times (personal communications), while ATCC 19606(D) at least five times before ending up in our hands (see previous section Historical background of A. baumannii ATCC 19606T and https://www.lgcstandards-atcc.org/products/all/19606.aspx?geo_country=it#history).
General features of the A. baumannii ATCC 19606(A) genome
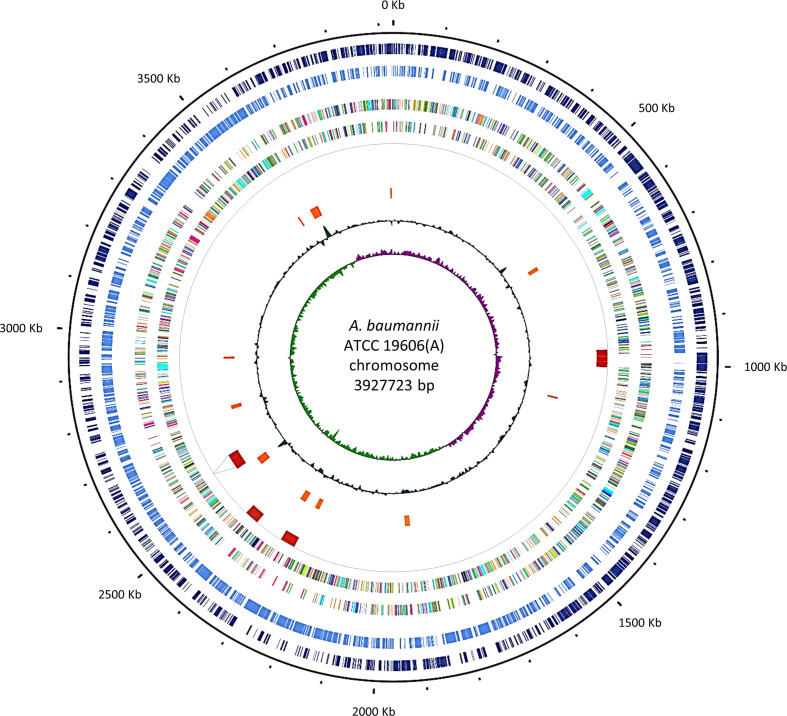
The whole-genome sequence of A. baumannii ATCC 19606(A) was determined in our laboratory using a hybrid de novo assembly combining Illumina paired-end reads and MinION long reads. The resulting A. baumannii ATCC 19606(A) genome sequence represents a 696-fold coverage (426-fold coverage based on short-reads and 270 on long-reads) complete genome with a consensus sequence of 3 927 723 bp and a mean GC content of 39.18 % (Fig. 1). The ATCC 19606(A) genome contains 3618 ORFs, 71 tRNAs and 6 rRNA operons (six copies of 5 s, 16 s and 23 s). Two indigenous plasmids, named p1ATCC19606 and pMAC, were entirely reconstructed, resulting in 9540 and 7655 bp sequences with 11 and 13 predicted ORFs, respectively. The ATCC 19606(A) chromosome and plasmid sequences are now publicly available under GenBank Accession numbers of CP058289, CP058290 and CP058291, respectively. ATCC 19606(A) was assigned to ST52 with MLST Pasteur scheme (cpn60-fusA-gltA-pyrG-recA-rplB-rpoB, 3-2-2-7-9-1-5) [71], in accordance with a previous report [11]. Six ISs were predicted in ATCC 19606(A) genome: five already reported by Zhu and coworkers [27], and an additional IS belonging to the ISL3 family spanning positions 2 528 296–2 531 742 of the chromosome (Table S3). A total of 11 GIs were predicted in the chromosome (Fig. 1, Table S4), many of which are located in the proximity of tRNA genes, which serve as integration sites for exogenous genetic elements [72, 73]. Several genes located in the predicted GI-10 (named GI19606 in [25]) encode proteins involved in arsenic resistance (arsenic resistance protein ArsH, and arsenic reductase and transporter [HTZ92_3354-HTZ92_3356]) and sulfamethoxazole resistance (Sul2 [HTZ92_3349]). Moreover, the gene encoding for a putative chlorhexidine efflux transporter (HTZ92_1759), plausibly implicated in resistance to this disinfectant, was detected in GI-3. In GI-4 two efflux pumps (HTZ92_2076 and HTZ92_2085) were detected, whose role is still unknown. The lprI lipoprotein gene (HTZ92_2140) was detected in GI-5. LprI was previously reported to act as a lysozyme inhibitor [74] and could therefore contribute to elude the host’s innate immunity [75].
Fig. 1.
Chromosome map of A. baumannii ATCC 19606(A). Circular map created by the CGView server. From the outermost to innermost, the tracks show the genes on positive (dark blue) and negative (light blue) strands, ORFs on positive and negative strands (with colours indicating COG classifications; [119]), prophages (red) with dotted lines indicating the excision site of the missing prophage, GIs (orange), GC content (green) and GC skew (purple and light green for positive and negative, respectively). Position 1 in ATCC 19606(A) corresponds to position 3772737 in ATCC 19696(H) and position 1094161 in ATCC 19606(M). Both genomes are in reverse orientation relative to ATCC 19606(A).
Based on the above observations, the GIs predicted in the ATCC 19606(A) genome encode for some functions that could contribute to the general fitness of the bacterium by facilitating its survival under unfavourable environmental conditions.
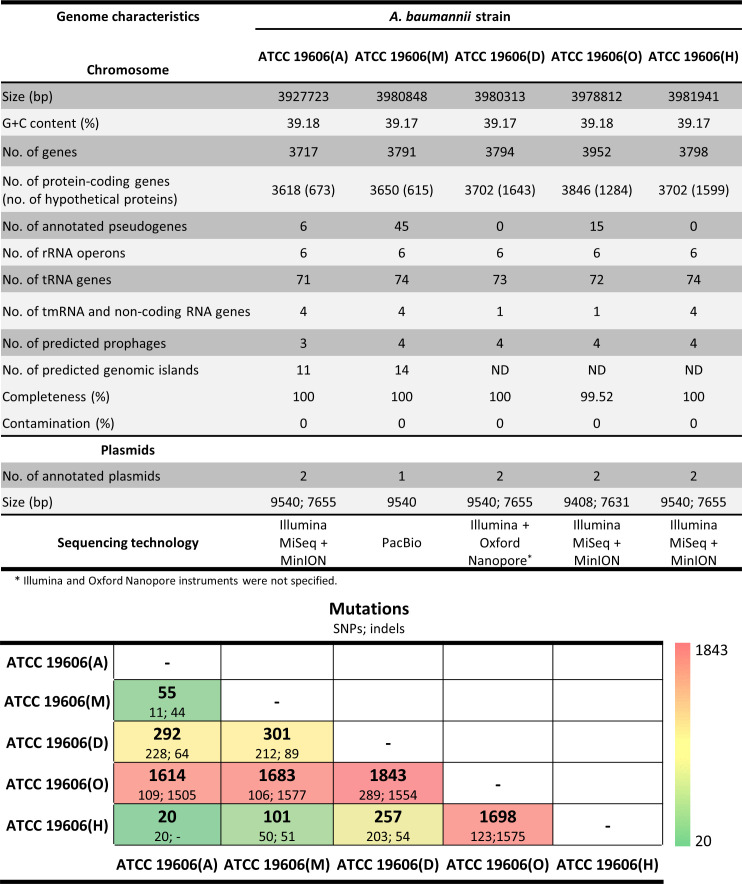
Comparative quality assessment of available ATCC 19606T complete genome sequences
To assess the quality of the assembled ATCC 19606(A) genome sequence, a pairwise comparison between three publicly available complete genome sequences of the same strain retrieved from the NCBI database, i.e. ATCC 19606(M) (CP046654.1 [27]), ATCC 19606(O) (AP022836 [26]) and ATCC 19606(H) (CP045110.1 [25]), and the sequence of ATCC 19606(D) deposited at the ATCC web site (https://genomes.atcc.org/genomes/1577c3a70f334038), was conducted. All genome assemblies showed >99 % completeness and no contamination (Fig. 2). Screening with the MAUVE multiple genome aligner [47] unveiled several SNPs and small INDELs (Fig. 2). ATCC 19606(O) showed the highest number of mutations with respect to ATCC 19606(H), ATCC 19606(M) and ATCC 19606(D), with a total of 1698, 1683 and 1843 SNPs plus INDELs, respectively (Fig. 2). These results are consistent with previous observations by Hamidian and coworkers [25]. Manual screening of the ORFs revealed 263 frameshifted genes in ATCC 19606(O), which also showed the lowest annotation accuracy, with 1284 hypothetical proteins. Although both Racon v1.3.1.1 and Pilon v1.20.1 were used to improve the sequence quality of ATCC 19606(O) [26], the high number of SNPs, INDELs and frameshifted ORFs in its genome suggests they could result from a low-quality sequence. Therefore, further comparative genome analyses were limited to the ATCC 19606(A) and the publicly available genomes with the highest quality represented by ATCC 19606(M) and ATCC 19606(H).
Fig. 2.
Relevant features of genome sequences of different A. baumannii ATCC 19606T strains.
Differential genomic traits of A. baumannii ATCC 19606 strains A, M and H
ATCC 19606(A) and ATCC 19606(M) exhibited similar annotation accuracy (Fig. 2), while ATCC 19606(H) showed lower accuracy, with 1599 hypothetical proteins. Discrepancies were also observed in the number of annotated pseudogenes, specifically six in ATCC 19606(A), none in ATCC 19606(H) and 45 in ATCC 19606(M), including frameshifted, incomplete or internally stopped ORFs [27]. Eleven out of 45 putative pseudogenes annotated in ATCC 19606(M) encode frameshifted proteins, and likely result from DNA sequencing errors; 28 putative pseudogenes shared 100 % sequence identity with ATCC 19606(A) counterparts but were differently annotated in ATCC 19606(M) and ATCC 19606(A) (Table S5). The remaining six putative pseudogenes were detected also in ATCC 19606(A), according to the NCBI annotation pipeline, which interprets pairs of neighbouring proteins that hit the same longer protein in blastp search as encoded by a single pseudogene. This implies that the pair may represent a single gene that has gained frameshift or other mutations along the way. Although the increasing number of pseudogenes is suggestive of genome erosion [76], in this case, the difference in the number of predicted pseudogenes seems to be due to discrepancies in sequencing and annotation.
It was also noticed that ATCC 19606(A) lacks three tRNA genes compared with ATCC 19606 strains M and H (Fig. 2): one tRNA-Gly (GO593_07205) arranged in tandem with another tRNA-Gly and two tRNA-Gln (GO593_10340 and GO593_10345). The absence of these tRNA genes was confirmed in both Illumina and Nanopore assemblies.
SNPs and INDELs
A comparison of ATCC 19606(A), considered as reference, with ATCC 19606(M) and ATCC 19606(H) genome sequences revealed 67 individual mutations: 47 in ATCC 19606(M) only, 12 in ATCC 19606(H) only, and eight in both ATCC 19606(M) and ATCC 19606(H) (Fig. S1). Among the 47 mutations detected in ATCC 19606(M), 13 insertions mapped inside intergenic regions (Table S6) and one SNP resulted in nucleotide substitution in GO593_04005, encoding a tRNA-Arg. None of the intergenic mutations were mapped within putative promoter sequences, as predicted with the BPROM software [77]. The remaining 33 mutations were located within ORFs, causing frameshifts in 12 genes of ATCC 19606(M), compared with ATCC 19606(A) (Table 2). Multiple mutations were detected in GO593_18950 (ten insertions and two SNPs) and in GO593_18955 (11 insertions), predicted to encode a sodium/glutamate symporter and an alpha-beta fold hydrolase, respectively. These mutations were not detected in both ATCC 19606(A) and ATCC 19606(H), in which full-length protein products matched their orthologues in ACICU strain. One insertion in the ATCC 19606(M) GO593_04990 gene (encoding a putative 3′−5′ exonuclease domain-containing protein) produced a shift in the stop codon, leading to an extended protein product.
Table 2.
Comparative analysis of SNPs detected in ATCC 19606(M), ATCC 19606(H) and ATCC 19606(A) genomes
|
Mutation |
Position in ATCC 19606(M) |
Position in ATCC 19606(H) |
Nucleotide change |
Aminoacid change |
Protein length (aa) in ATCC 19606(M)/ ATCC 19606(A)/ ATCC 19606(H) |
Gene designation in ATCC 19606(M)/ ATCC 19606(H) |
Gene designation in ATCC 19606(A) |
Gene product |
Gene designation in ACICU (Protein length) |
|---|---|---|---|---|---|---|---|---|---|
|
|
SNPs between ATCC 19606(M) and ATCC 19606(A) |
||||||||
|
Frameshift |
175 678 |
|
T ->TG |
G80W |
85/513/513 |
GO593_01010/FQU82_02766 |
HTZ92_0804 |
MFS transporter |
DMO12_ 07743 (513) |
|
426 614 |
|
A ->AT |
L235F |
258/548/548 |
GO593_02100/FQU82_02985 |
HTZ92_0587 |
Phospholipid carrier-dependent glycosyltransferase |
DMO12_08688 (548) |
|
|
1 052 567 |
|
G ->GA |
F207I |
222/213/213 |
GO593_04990/FQU82_03557 |
HTZ92_0046 |
3'−5' exonuclease domain-containing protein 2 |
DMO12_10425 (213) |
|
|
1 452 728 |
|
A ->AG |
L101T |
144/250/250 |
GO593_06935/FQU82_00154 |
HTZ92_3299 |
Transcriptional regulator LldL |
DMO12_00333 (250) |
|
|
2 027 426 |
|
A ->AC |
I100Y |
113/462/462 |
GO593_09615/FQU82_00690 |
HTZ92_2784 |
Aminodeoxychorismate synthase component I |
DMO12_02013 (462) |
|
|
2 761 191 |
|
T ->TG |
L96I |
100/209/209 |
GO593_13255/FQU82_01413 |
HTZ92_2145 |
Hypothetical protein |
DMO12_03804 (209) |
|
|
3 154 380 |
|
C ->CT |
N62E |
67/181/181 |
GO593_15095/FQU82_1785 |
HTZ92_1775 |
Acyltransferase |
DMO12_04716 (181) |
|
|
3 271 196 |
|
T ->TG |
R380Q |
381/576/576 |
GO593_15665/FQU82_01898 |
HTZ92_1663 |
Dipeptide ABC transporter ATP-binding protein |
DMO12_05028 (576) |
|
|
3 974 985 |
|
T ->TC |
F157I |
172/212/212 |
GO593_18930/FQU82_02556 |
HTZ92_1005 |
TetR family transcriptional regulator |
DMO12_07332 (212) |
|
|
3 976 370 |
|
G ->GA |
E45R |
75/300/300 |
GO593_18945/FQU82_02559 |
HTZ92_1002 |
ATPase AAA |
DMO12_07341 (300) |
|
|
3 977 672 |
|
T ->TC* |
W9L |
19/411/411 |
GO593_18950/FQU82_02560 |
HTZ92_1001 |
Sodium/glutamate symporter gltS |
DMO12_07344 (411) |
|
|
3 978 830 |
|
A ->ATC† |
21L‡ |
20/382/382 |
GO593_18955/FQU82_02561 |
HTZ92_1000 |
Alpha-beta fold hydrolase |
DMO12_07347 (382) |
|
|
|
SNPs between ATCC 19606(M, H) and ATCC 19606(A) |
||||||||
|
Stop codon |
711 049 |
3 389 623 |
T ->A |
K337‡ |
403/345/403§ |
GO593_03295/FQU82_03219 |
HTZ92_0363 |
Methyltransferase |
DMO12_09396 (286) |
|
Aminoacid change |
261 181 |
2 939 754 |
G ->A |
G40D |
213/213/213 |
GO593_01345/FQU82_02834 |
HTZ92_0737 |
Hypothetical protein |
DMO12_08034 (213) |
|
1 149 064 |
3 827 640 |
A ->T |
V56D |
265/265/265 |
GO593_05440/FQU82_03650 |
HTZ92_3585 |
MBL fold metallo-hydrolase |
DMO12_10692 (265) |
|
|
|
1 168 711 |
3 847 287 |
A ->T |
T36S |
70/70/70 |
GO593_05520/FQU82_03666 |
HTZ92_3569 |
Hypothetical protein |
DMO12_10740 (70) |
|
|
3 331 716 |
2 029 407 |
A ->G |
V94A |
711/711/711 |
GO593_15945/FQU82_01954 |
HTZ92_1607 |
TonB-dependent siderophore receptor |
DMO12_05199 (711) |
|
Synonymous |
20 323 |
2 698 895 |
T ->C |
S117 |
475/478/478 |
GO593_00130/FQU82_02587 |
HTZ92_0978 |
M48 family metalloprotease |
DMO12_07425 (478) |
|
|
2 190 622 |
887 198 |
T ->C |
I319 |
394/394/394 |
GO593_10430/FQU82_00850 |
HTZ92_2632 |
Hypothetical protein |
DMO12_02541 (394) |
|
Intergenic |
1 184 582 |
3 863 158 |
A ->T |
– |
– |
GO593_05600-GO593_05605/FQU82_03682-FQU82_03683 |
HTZ92_3552-HTZ92_3553 |
– |
– |
|
|
SNPs between ATCC 19606(H) and ATCC 19606(A) |
||||||||
|
Stop codon |
|
2 834 189 |
T ->A |
117L‡ |
178/117/178 |
GO593_00915/FQU82_02747 |
HTZ92_0820 |
Peptidase C39 |
DMO12_07686 (178) |
|
Aminoacid change |
|
2 573 642 |
C ->A |
R190L |
201/201/201 |
GO593_18455/FQU82_02459 |
HTZ92_1102 |
Potassium-transporting ATPase C chain |
DMO12_07086 (201) |
|
Synonymous |
|
3 278 427 |
C ->T |
R362|| |
549/549/533¶ |
GO593_02765/FQU82_03113 |
HTZ92_0463 |
Lipid A phosphoethanolamine transferase |
DMO12_09075 (533) |
*Only the first nucleotide change is shown: GO593_18950 contains ten insertions and two SNPs.
†Only the first nucleotide change is shown: GO593_18955 contains 11 insertions.
‡Indicates a stop codon.
§The predicted ORF in ATCC 19606(A) starts 27 nucleotides upstream, relative to ATCC 19606(M) and ATCC 19606(H).
||Only the first nucleotide change is shown: FQU82_03113 contains ten SNPs.
¶The predicted ORF in ATCC 19606(A) starts 48 nucleotides downstream, relative to ATCC 19606(M) and ATCC 19606(H).
Of the eight mutations shared by both ATCC 19606(M) and ATCC 19606(H) (Table 2), one SNP mapped in an intergenic region, and two SNPs were synonymous mutations. Only one SNP introduced a stop codon in HTZ92_0363, whose predicted product is a methyltransferase, leading to the production of truncated protein form in ATCC 19606(A), compared with the full-length protein predicted for both ATCC 19606(M) and ATCC 19606(H) genomes. Four SNPs detected in both ATCC 19606(M) and ATCC 19606(H) caused aminoacid substitutions relative to corresponding ATCC 19606(A) proteins (Table 2). Twelve additional mutations were detected by direct comparison between ATCC 19606(H) and ATCC 19606(A), taken as reference genome; ten were synonymous changes in the pmrC gene encoding lipid A phosphoethanolamine transferase (FQU2_03113), one caused a non-conservative (R→L) aminoacid substitution, and one generated a stop codon in the peptidase C39 gene (FQU82_02747), resulting in a truncated protein in ATCC 19606(H).
To validate the above observations, Sanger sequencing of the PCR-generated genomic regions encompassing individual SNPs was performed using the ATCC 19606(A) DNA as template. Results confirmed the sequence determined by the hybrid assembly for all SNP-containing regions (100 % identity). Since multiple validations of the reconstructed genome sequence of ATCC 19606(A) made it possible to exclude sequencing errors, it can be argued that ATCC 19606T strains domesticated in different laboratories had diversified their genome sequence.
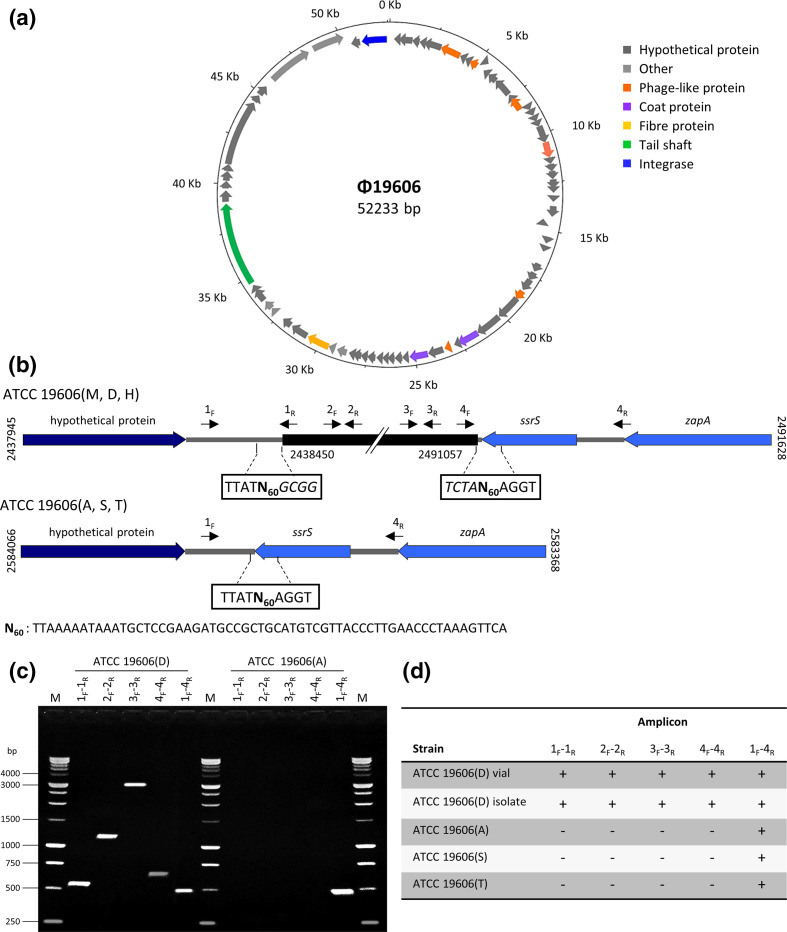
Domesticated A. baumannii ATCC 19606 strains A, S and T lack the Φ19606 prophage
A preliminary comparison between ATCC 19606(A) and ATCC 19606(M) genomes revealed one major structural difference, consisting in the absence of a 52 kb prophage region spanning from GO593_11545 to GO593_11900 in ATCC 19606(M), mapping between a gene coding for a hypothetical protein (HTZ92_2409) and the ssrS gene (HTZ92_ssRs) in ATCC 19606(A). This prophage region was also detected in the ATCC 19606(D) and ATCC 19606(H) genomes and was predicted by PHASTER as an ‘intact’ prophage in the region 2 438 683–2 490 916 of the ATCC 19606(M) genome, with a GC content of 38.22 %. We propose to designate this putative prophage Φ19606 (Fig. 3a). A 60-nucleotide repeat (N60), overlapping the ssrS gene sequence, encoding the 6S regulatory RNA, was detected at both ends of the prophage in ATCC 19606 strains D, H and M. Notably, N60 was present as a single copy in ATCC 19606(A), which lacks Φ19606 (Fig. 3b). Genome inspection of other A. baumannii strains, such as AYE (CU459141.1) and AB307-0294 (CP001172.2), revealed that the ssrS gene is also an insertion site for other phages [78]. No significant homology was observed between Φ19606 and known phage genomes. Integration of Φ19606 in ATCC 19606(M) had occurred immediately downstream of the stop codon of the ssrS gene, causing target site duplication of the N60 sequence. Consequently, N60 could represent a homology region between Φ19606 and ATCC 19606(M) genome, possibly implicated in phage integration. Similar repeats could constitute possible recognition sites for the predicted phage terminase (GO593_11720); at the end of their infection cycle, dsDNA phages generally form concatemers that are cut by the terminase, enabling packaging of the mature phage genome [79].
Fig. 3.
A. baumannii Φ19606 phage. (a) Circular map of the Φ19606 genome drawn with DNAPlotter. The genome map illustrates putative ORFs along with the direction of transcription indicated with arrows. Functional proteins predicted by PHASTER are depicted in different colours. (b) Integration site of Φ19606 (black) into the ATCC 19606(M, D, H) chromosomes (top). The double slash denotes a phage region that is not shown. Positions refer to the ATCC 19606(M) genome sequence. Structure of ATCC 19606(A, S, T) after phage loss (bottom). Positions refer to ATCC 19606(A) genome sequence. Sequences flanking the insertion site are boxed, with predicted phage nucleotides italicized. Primer positions are indicated with black arrows. N60 stands for the 60-nucleotide sequence generated by phage insertion/excision. (c) Agarose gel electrophoresis of the PCR products obtained by using different primer pairs indicated in (b). (d) Presence (+) or absence (-) of amplicons detected in the different A. baumannii ATCC 19606T strains.
Seventy-two ORFs were predicted in the Φ19606 prophage, 48 of which match proteins in the phage protein database, five match bacterial proteins and 19 are annotated as hypothetical proteins (Table S7). According to PHASTER results, prophage Φ19606 showed partial similarity with previously published Acinetobacter siphoviral phages YMC/09/02/B1251_ABA_BP (NC_019541 [80]) and YMC11/11 /R3177 (NC_041866 [81]), with 38 and 31 homologous proteins, respectively. Intriguingly, Φ19606 harbours the GO593_11890 gene, encoding a putative lipid A phosphoethanolamine transferase (eptA), showing 95 % identity with the chromosomal pmrC gene, implicated in colistin resistance [82]. However, colistin susceptibility testing by the broth microdilution method showed similar MIC values (1 µg ml−1 colistin) for both ATCC 19606(A) and ATCC 19606(D) (data not shown).
While the absence of Φ19606 is a distinctive feature of ATCC 19606(A), three prophage regions classified as ‘intact’ and ‘questionable’ by PHASTER, were conserved among ATCC 19606 strains A, D, H and M (Table S8).
Since the whole-genome sequence is unavailable for ATCC 19606 strains S and T, the occurrence of Φ19606 in the genome of these domesticated strains was experimentally tested by PCR, including the ATCC 19606(D) and ATCC 19606(A) DNA as positive and negative control, respectively (Fig. 3c, d). An amplicon of 498 bp was invariably detected in domesticated strains ATCC 19606(A, S, T), indicating the absence of Φ19606 prophage. Intriguingly, the generation of both 560 bp and 498-bp-amplicons from genomic DNA of ATCC 19606(D) suggests that Φ19606 prophage was present in both integrated and episomal form (Fig. 3c). Coherently, genes encoding Cro (GO593_11615) and CI (GO593_11625) repressors, likely implicated in the switch control between the lysogeny and lytic cycle [83, 84], were detected in Φ19606, also presenting with the divergent gene organization typical of the λ-like coliphages.
Since ATCC 19606(A, S, T) were originally filed with ATCC before being distributed to the various laboratories, we hypothesized that multiple propagations from the original vial resulted in Φ19606 loss. To reproduce the in vitro conditions that may have led to the phage loss, serial propagation steps of ATCC 19606(D) from the original vial obtained from DSMZ were conducted. In particular, preliminary viable counts showed that a single ~1 mm colony of A. baumannii ATCC 19606(D) generated after 24 h growth at 37 °C on LA plates is composed of 2.14 (±0.7)×107 c.f.u. Assuming that one colony originates from a single cell, it was calculated that A. baumannii ATCC 19606(D) replicates ca 24 times to produce a ~1 mm colony, with a mean generation time of ~1 h. Therefore, it was estimated that a total of 360 generations were made by A. baumannii ATCC 19606(D) after 15 daily passages on LA plates. For each propagation step, the presence of the prophage was verified by PCR with the primers listed in Table S1, using 30 ng of genomic DNA purified from ten randomly selected colonies as a template for PCR. Amplicons of 560 and 498 bp were detected for all colonies at all passages. In addition, a large screening conducted on 200 randomly selected colonies from the last propagation plate invariably yielded both amplicons, indicating that not even a single cell had lost Φ19606 during 360 generations. Spontaneous prophage induction could be due to both extrinsic and intrinsic factors, or a combination of both [85]. In fact, extrinsic factors, such as pH variations, accumulation of reactive oxygen species, UV radiation or other factors causing DNA damages could have triggered Φ19606 excision. On the other hand, spontaneous activation of genetic circuitry causing prophage excision in single cells of bacterial populations was also observed in the absence of an external trigger, a phenomenon dubbed ‘spontaneous prophage induction’ [85]. Consequently, the events that have led to Φ19606 loss remain unpredictable and difficult to reproduce in vitro, likely resulting from a combination of stochastic and/or ill-defined environmental conditions.
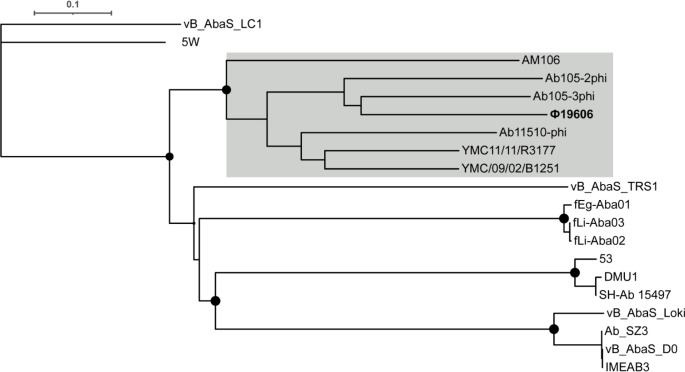
Ф19606 belongs to genus Vieuvirus and its host range is likely restricted to A. baumannii
To address the phylogeny of bacteriophage Ф19606, genome comparisons between Ф19606 and all 19 previously reported Acinetobacter phages belonging to the Siphoviridae family were performed using VICTOR, in agreement with the recommendations of the International Committee on Taxonomy of Viruses [55]. The resulting minimum evolution tree grouped Ф19606 with members of the genus Vieuvirus, with Ab105-3phi and Ab105-2phi resulting the closest relatives (Fig. 4). Therefore, Ф19606 can be classified as a member of the genus Vieuvirus, within the Siphoviridae family.
Fig. 4.
Phylogenetic tree of Acinetobacter phages belonging to the Shiphoviridae family. The tree was generated by VICTOR using the complete genome sequences of the Shiphoviridae family members. Filled circles at the nodes are GBDP pseudo-bootstrap support values >70 % from 100 replications. The scale bar indicates the number of substitutions per variable site. Phages belonging to the genus Vieuvirus are grey-shaded, Ф19606 is in bold. The tree was rooted at the midpoint.
A significant portion (over 85%) of the Ф19606 sequence was detected in 123 non-redundant A. baumannii complete genomes (Table S9). A. baumannii strains harbouring Ф19606 belonged to clonal complex 2 (95 %), including ST2 and a single-locus variant of ST2, and to ST52 and a single-locus variant of ST52 (3 %). In most of the A. baumannii genomes (75 %), Ф19606 was inserted between a homologue of the hypothetical protein coding sequence HTZ92_2409 [according to the ATCC 19606(A) annotation] and the ssrS gene (HTZ92_ssRs), and in six strains an ISAba1 was detected between HTZ92_2409 and Ф19606. In 19 % of A. baumannii genomes, Ф19606 mapped between the HTZ92_2409 homologue and the gene encoding for another hypothetical protein [absent in ATCC 19606(A)], and in 6 % between an integrase and the ssrS genes. Ф19606 was not detected in any organisms other than A. baumannii, in line with previous evidence that Vieuvirus only infect Acinetobacter spp. [86].
A. baumannii ATCC 19606T harbours two plasmids: pMAC and p1ATCC19606
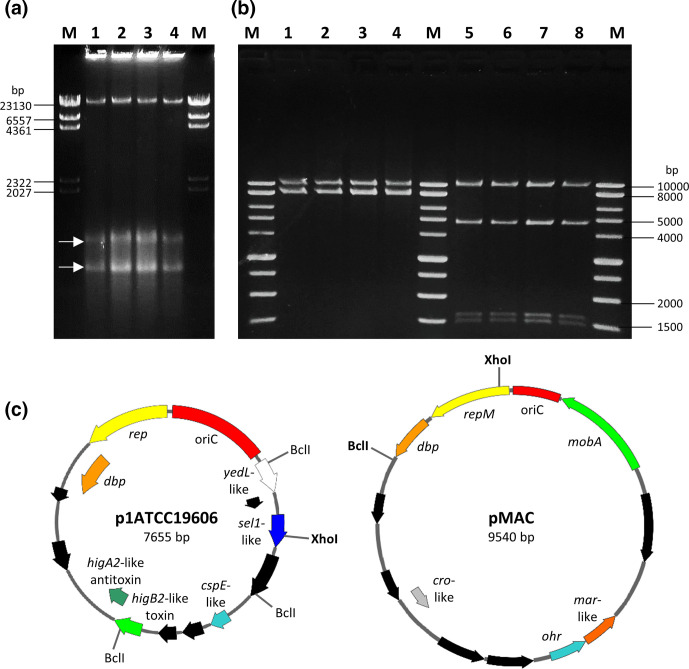
An indigenous plasmid, called pMAC, was previously identified and characterized in A. baumannii ATCC 19606T [87]. pMAC is a 9.5 kb mobilizable episomal element carrying the genetic determinants for resistance to organic peroxides [87]. The existence of an additional replicon was firstly predicted from previous genome assemblies (https://genomes.atcc.org/genomes/1577c3a70f334038; [25, 26]), but no physical evidence of the existence of two different replicons in A. baumannii ATCC 19606T has so far been provided. Moreover, the predicted size of A. baumannii ATCC 19606T plasmids differs from previous studies (Fig. 2). To address these discrepancies, both A. baumannii ATCC 19606T plasmids were isolated and entirely sequenced. The two indigenous plasmids were found to coexist in four different A. baumannii type strains (A, D, S, T), as demonstrated by agarose gel electrophoresis of clear lysates (Fig. 5a). The 7655 bp extrachromosomal element was identified as p1ATCC19606 [25], copurified with pMAC in order to perform a restriction analysis of both replicons (Fig. 5b), and Illumina sequences were confirmed by primer walking and Sanger sequencing with primers listed in Table S1. Fig. 5(c) displays the physical and functional maps of pMAC and p1ATCC19606. The pMAC size (9540 bp) and annotated ORFs exactly match previous data [87]. Inspection of publicly available A. baumannii genomes did not detect p1ATCC19606 and pMAC in strains other than ATCC 19606T. Multiple analyses conducted on the raw sequence read data of ATCC 19606(M) did not detect plasmid p1ATCC19606 (7655 bp). This is because small plasmids are frequently overlooked from long-read-only assemblies [88], likely due to the removal of <~10 kb DNA fragments during the library preparation.
Fig. 5.
Plasmids p1ATCC19606 and pMAC harboured by A. baumannii ATCC 19606T strains. (a) Agarose gel electrophoresis of clear lysates of A. baumannii ATCC 19606(A) (lane 1), ATCC 19606(D) (lane 2), ATCC 19606(S) (lane 3) and ATCC 19606(T) (lane 4). M, Lambda DNA/HindIII marker (ThermoFisher). White arrows indicate the closed circular forms of pMAC (upper band) and p1ATCC19606 (lower band). (b) p1ATCC19606 and pMAC were copurified from A. baumannii strains ATCC 19606(A) (lanes 1 and 5), ATCC 19606(D) (lanes 2 and 6), ATCC 19606(S) (lanes 3 and 7) and ATCC 19606(T) (lanes 4 and 8), and digested with XhoI (lanes 1–4) and BclI (lanes 5–8). M, BenchTop 1 kb DNA Ladder (Promega). (c) Physical and functional maps of the p1ATCC19606 and pMAC plasmids. Restriction sites for the enzymes used to generate the electropherogram in (b) are shown. Unique cutter restriction enzymes are indicated in bold. Nomenclature of p1ATCC19606: rep, putative replicase; dbp, gene encoding a predicted DNA-binding protein; cspE-like, putative cold-shock protein gene; sel1-like, putative gene coding for a Sel1-repeat family protein; yedL-like, gene coding for the putative YedL N-acetyltransferase; oriC, predicted origin of replication. Nomenclature of pMAC: repM, replication protein M; dbp, gene encoding a predicted DNA-binding protein; ohr, gene encoding an organic hydroperoxide resistance protein, mobA, plasmid mobilization protein; oriC, origin of replication. ORFs shown in black are predicted to encode for hypothetical proteins. All genes are reported in scale over the total length of each plasmid. Images were obtained by the use of the SnapGene software (GSL Biotech).
Determination of the minimal self-replicating region of p1ATCC19606
For a preliminary functional characterization of p1ATCC19606, protein-coding genes were annotated by integrating the output of MEGAnnotator and Blast2GO. Thirteen putative ORFs (ORF-1 to ORF-13) were identified (Table 3). Among them, three overlapping ORFs (ORF 10, 11 and 12) were predicted to be involved in plasmid replication and segregation, namely ORF-10 encoding a putative replication protein located downstream the predicted origin of replication, ORF-11 encoding a putative DNA-binding protein, and ORF-12 encoding a putative integral membrane protein, likely involved in plasmid segregation.
Table 3.
Annotation of protein-coding genes of plasmid p1ATCC19606
|
Predicted ORF |
Gene ID |
Position (bp) |
Protein length (aa) |
Blast2GO description (e-value) |
|---|---|---|---|---|
|
ORF-1 |
HTZ92_3642 |
225–497 |
91 |
Helix-turn-helix domain-containing protein (7.34E-59) |
|
ORF-2 |
HTZ92_3643 |
490–810 |
107 |
Type II toxin-antitoxin system RelE/ParE family toxin (4.55E-71) |
|
ORF-3 |
HTZ92_3644 |
997–1194 |
66 |
Hypothetical protein (3.33E-39) |
|
ORF-4 |
HTZ92_3645 |
1261–1488 |
76 |
Hypothetical protein (2.14E-45) |
|
ORF-5 |
HTZ92_3646 |
1586–1801 |
73 |
Cold shock-like protein CspE (1.97E-42) |
|
ORF-6 |
HTZ92_3647 |
2128–2640 |
171 |
Hypothetical protein (6.05E-70) |
|
ORF-7 |
HTZ92_3648 |
2735–3094 |
120 |
Sel1 repeat family protein (6.67E-80) |
|
ORF-8 |
HTZ92_3649 |
3203–3343 |
47 |
Uncharacterized protein (6.73E-25) |
|
ORF-9 |
HTZ92_3650 |
3343–3714 |
124 |
N-acetyltransferase YedL (1.3311E-84) |
|
ORF-10 |
HTZ92_3651 |
4925–5875 |
317 |
Initiator replication family protein (0) |
|
ORF-11 |
HTZ92_3652 |
5868–6443 |
192 |
DNA replication protein (1.73E-140) |
|
ORF-12 |
HTZ92_3653 |
6463–6606 |
48 |
Hypothetical protein - integral component of membrane (5.11E-23) |
|
ORF-13 |
HTZ92_3654 |
7050–7385 |
112 |
Hypothetical protein (2.1E-60) |
The p1ATCC19606 origin of replication (oriC), predicted by using the DoriC 5.0 software [89], consists of a 1072–bp sequence containing four 22-mer direct repeats (5′-GCAAGGTAAACGGTGTCATATT-3′). Similarly arranged iterons of four 21 bp direct repeats were also identified in pMAC [87] and could be implicated in the initiation of plasmid replication and copy-number control [90, 91]. Interrogation of PlasmidFinder (https://cge.cbs.dtu.dk/services/PlasmidFinder/; [92]) did not retrieve any replication class for the p1ATCC19606 oriC region, suggesting that p1ATCC19606 has a narrow-host range, consistent with the absence of predicted mobilization proteins.
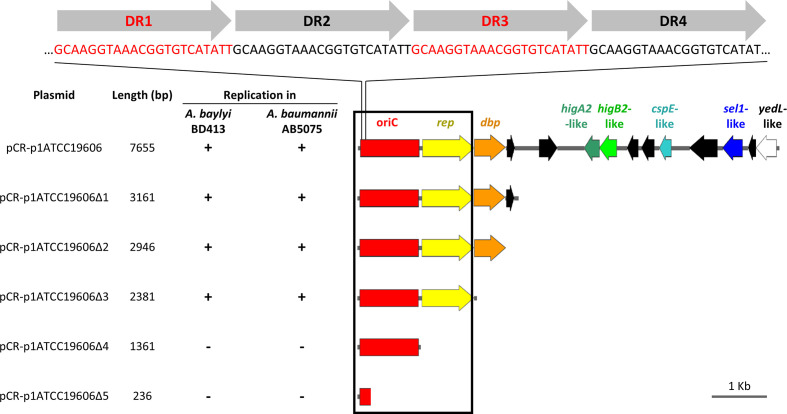
To determine the shortest DNA region enabling self-replication in Acinetobacter spp., deletion analysis of p1ATCC19606 was performed. Overlapping DNA fragments encompassing the oriC region were generated by PCR with the primer pairs listed in Table S1, and the resulting amplicons were cloned into the pCR vector for transformation of E. coli DH5α. After sequence verification, all constructs (designated as pCR-p1ATCC19606 to pCR-p1ATCC19606Δ5) were individually transferred by electroporation in different Acinetobacter spp. to assess self-replication (Fig. 6). Except for the putative replicase gene, all ORFs could be deleted from p1ATCC19606 without affecting the replication of the hybrid constructs in Acinetobacter spp. All p1ATCC19606 deletion derivatives showed similar TEs (Table S10). Intriguingly, the transformation of Acinetobacter pittii UKK_0145 with pCR-p1ATCC19606Δ4, lacking the putative replicase gene, yielded some transformants, though with low efficiency. A blastp search of p1ATCC19606 putative replicase protein sequence in the NCBI nr database of A. pittii proteins retrieved a chromosomally located DNA replication protein (KQF43430.1) sharing 85 % identity with the putative replicase gene of p1ATCC19606. It can therefore be speculated that a chromosomal DNA replicase could act in trans to enable pCR-p1ATCC19606Δ4 replication in A. pittii UKK_0145 (Table S10).
Fig. 6.
Deletion analysis of p1ATCC19606 to determine the minimal region required for autonomous plasmid replication in Acinetobacter spp. Deletion fragments of p1ATCC19606 were generated by PCR amplification with primers listed in Table S1 and cloned into pCR. The resulting p1ATCC19606 deletion derivatives were introduced in A. baylyi BD413 and A. baumannii AB5075 to map the minimal self-replicating region (black box). Relevant coding regions are indicated with colours: red, predicted minimal origin of replication (oriC); yellow, putative replicase (rep); orange, gene encoding a predicted DNA-binding protein (dbp); dark green, putative higA2-like antitoxin gene; light green, putative higB2-like toxin gene; cyan, putative cold-shock protein gene (cspE); blue, gene coding for putative a Sel1-repeat family protein (sel1); white, gene coding for the putative YedL N-acetyltransferase (yedL). Four copies of the 22-mer direct repeat (DR1–DR4) in the predicted origin of replication are shown on top. ORFs in black are predicted to encode for hypothetical proteins. All genes are reported in scale over the total length of the plasmid. Images were obtained by the use of the SnapGene software (GSL Biotech).
The p1ATCC19606 predicted replicase was modelled on the E. coli π initiator protein–iteron complex of plasmid R6K (2NRA [93]). Both SWISS-MODEL and I-TASSER provided highly superimposable models (Fig. S2), with maximum superimposition at the level of the replicase DNA-binding domain. This prediction suggests that the p1ATCC19606 replicase could interact with the four 22-mer direct repeats identified in oriC, similar to R6K replicase. However, R6K replication also requires iterons located outside the predicted origin of replication [93, 94], which are not detectable in p1ATCC19606, arguing for a different replication mechanism.
A HigB2-HigA2-like TA system accounts for p1ATCC19606 plasmid stability
ORF-1 and ORF-2 of p1ATCC19606 (Table 3) were predicted to encode a putative antitoxin and a toxin, respectively, possibly constituting a type-II TA module [95–97]. TA modules are implicated in plasmid maintenance since they encode a poisonous toxin, and a neutralizing antitoxin [98–100].
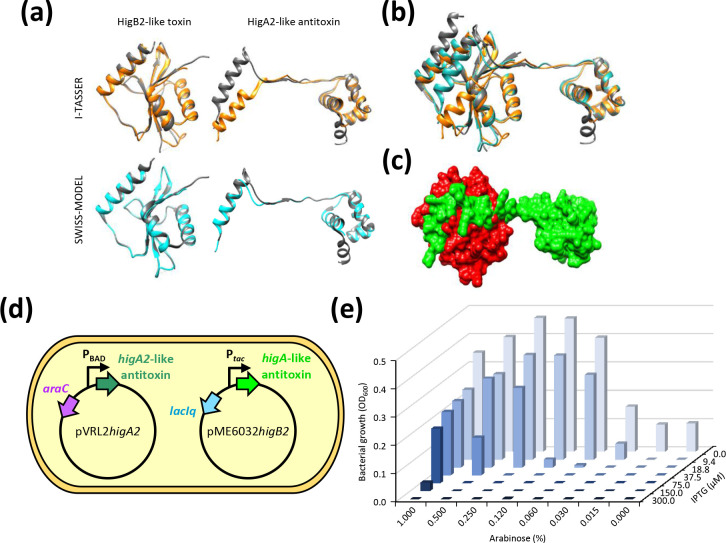
Homology modelling of ORF-2 and superimposition to the HigB2 toxin from V. cholerae (5JA8; [101]), using both I-TASSER and SWISS-MODEL, provided high scores for template modelling (TM) and Qualitative Model Energy ANalysis (QMEAN) (Table S11; Fig. 7a). ORF-1 was modelled by I-TASSER and SWISS-MODEL on the HigA2 antitoxin cocrystallized with its cognate HigB2 toxin (5JAA; [101]) (Fig. 7a). The predicted HigB2A2-like module was substantiated by the unusual genetic organization of this TA system: although the antitoxin gene is usually located upstream of the toxin gene, higB2A2 module has reverse gene organization [102–104]. Although I-TASSER and SWISS-MODEL software employed the same protein template to model the putative HigA2-like antitoxin, SWISS-MODEL provided higher modelling scores compared with I-TASSER (Table S11), probably because antitoxin proteins show considerable structural flexibility, which limits the superimposition of the ORF-1 product on the template structure, compared with the HigA2 template (Fig. 7a). After structure editing of the I-TASSER-derived antitoxin model, consisting of the torsion angle modification of five residues (from 26 to 30), both SWISS-MODEL and I-TASSER models became superimposable onto the HigB2A2 crystal structure complex (5JAA; [101]) (Fig. 7b), enabling to predict a 3D model of the interaction between the p1ATCC19606 putative toxin and antitoxin proteins (Fig. 7c).
Fig. 7.
HigB2-like and HigA2-like components the TA system of p1ATCC19606. (a) Superimposition of the HigBA2-like TA complex on the Vibrio cholerae HigBA2 TA crystal structure (5JAA). The query structure is shown in grey, while the structural analogue is displayed in orange or cyan for I-TASSER- and SWISS-MODEL-based models, respectively. Only the first-ranked model predicted by I-TASSER and SWISS-MODEL for each query is shown. Torsion angles of amminoacid residues 26–30 of the I-TASSER-based model of the predicted HigA2-like antitoxin were modified to orient the α-helix involved in the interaction with HigB2-like toxin. (b) Superimposition of the predicted p1ATCC19606 TA complex models (I-TASSER, orange; SWISS-MODEL, cyan) over the crystal structure of HigB2-HigA2 (grey; 5JAA). (c) GRASP surface representation of the HigB2-like toxin (red)-HigA2-like antitoxin (green) complex based on the SWISS-MODEL predictions, displaying the interaction between the putative toxin and antitoxin proteins. The images shown in (a–c) were obtained using UCSF Chimaera. (d) Schematic illustration of HigB2-like toxin neutralization by the HigA2-like antitoxin. The arabinose-inducible expression of the higA2-like antitoxin gene provided in trans from pVRL2 allows the growth of E. coli DH5α expressing the IPTG-inducible higB2-like toxin gene from plasmid pME6032higB2. (e) Bacterial growth assessed after 24 h incubation at 37 °C in LB supplemented with the appropriate antibiotic concentration. To induce the expression of the higA2-like antitoxin gene from the arabinose-inducible PBAD promoter and of the higB2-like toxin gene from the IPTG-inducible P tac promoter, the medium was supplemented with the indicated arabinose and IPTG concentrations, respectively. OD600 values are representative of three independent experiments giving similar results.
To demonstrate the involvement of the predicted TA system in p1ATCC19606 stability, the higB2A2-like gene system was deleted from pCR-p1ATCC19606, yielding pCR-p1ATCC19606ΔhigB2A2. The stability of p1ATCC19606ΔhigB2A2 in E. coli DH5α after 48 h growth in the absence of antibiotic selection (N Ant /N0 ratios) was reduced by ca 99 % compared with the parent pCR-p1ATCC19606 plasmid (Table S12). A comparable reduction of plasmid stability was also observed for the TA deletion derivative of pVRL1 (i.e. pVRL1ΔTA; [66]), used as a control. Therefore, the deletion of the TA module dramatically reduces p1ATCC19606 stability.
To provide direct evidence of the toxicity of the HigB2-like toxin, E. coli DH5α was transformed with both pVRL2higA2 and pME6032higB2 plasmids, directing the arabinose- and IPTG-inducible expression of higA2 and higB2 genes, respectively (Fig. 7d). Assuming that the HigB2-like protein is a toxin and the HigA2-like protein is the cognate antitoxin, cells should be viable only upon expression of the higA2-like antitoxin gene, i.e. in the presence of arabinose. On this assumption, the growth of E. coli DH5α carrying both pVRL2higA2 and pME6032higB2 was determined after 24 h incubation at 37 °C in LB supplemented with 10 µg ml−1 Tc and different concentration of IPTG and arabinose. Results clearly demonstrate that IPTG-inducible expression of the higB2-like toxin gene abrogates E. coli growth unless compensated by arabinose-inducible expression of the higA2-like antitoxin gene (Fig. 7e). Indeed, bacterial growth increased with increasing arabinose concentration and decreased with increasing IPTG concentrations. A similar growth inhibition profile of E. coli DH5α carrying both pVRL2higA2 and pME6032higB2 was observed around paper discs soaked with increasing IPTG concentrations and applied to LA plates supplemented with arabinose (Fig. S3).
The peculiar gene architecture together with the structural and functional similarities between the p1ATCC19606 TA system and the HigB2A2 complex of Enterobacteriaceae suggest that the A. baumannii HigB2-like toxin acts as a translating mRNA ribonuclease, causing a stall in protein synthesis in plasmid-free daughter cells and cell death [105]. The activity of the HigB2-like toxin is neutralized by the cognate HigA2-like antitoxin, securing the survival of the A. baumannii cells that inherit the p1ATCC19606 plasmid.
Conclusion
For decades A. baumannii ATCC 19606T has been the reference strain for A. baumannii research. However, it is known that frequent subculturing and local differences in culture conditions can result in the domestication of laboratory strains, a micro-evolutionary process driven by mutational events at the genome level that could even reflect into variable phenotype [28–32, 106]. Motivated by the remarkable diversity in publicly available A. baumannii ATCC 19606T genome sequences, we generated an accurately revised genome sequence of ATCC 19606T, which will hopefully set a more solid basis for studies of the genetics and genomics of this model organism. Previous long-read sequence data, such as those generated by PacBio and Oxford Nanopore technologies [26, 27], allowed for complete assembly of the ATCC 19606T genomic sequence without manual intervention. However, assembling long-read data alone can result in sequencing errors [107], and in the case of ATCC 19606T failed to detect both plasmids [25]. To overcome these limitations, we combined deep Illumina short-read with MinION long-read and Sanger technologies to generate a high-quality genome sequence. Subsequent annotation made it possible to lower the number of genes encoding hypothetical proteins as well as of pseudogenes, compared with formerly released ATCC 19606T genome sequences. Indeed, comparative analysis of our genomic sequence with previously published ones [25–27] highlighted a high number of SNPs and INDELs, and a difference in the annotation of putative pseudogenes. Sanger sequencing of the genomic regions encompassing individual SNPs confirmed the sequence determined by the hybrid assembly of ATCC 19606(A), allowing us to exclude sequencing errors. Therefore, the confirmed SNPs and INDELs could result from micro-evolutionary events of individual strains during domestication, whereas differences in pseudogene number are suggestive of sequencing and/or annotation errors, rather than genome erosion events.
Accurate genome assembly also made it possible to characterize the indigenous plasmid p1ATCC19606, whose presence and size were undefined in previous versions of the A. baumannii ATCC 19606T genome sequence (Fig. 2). The HigB2A2-like TA system and the minimal self-replicating region of p1ATCC19606 were characterized both in silico and in vitro, providing insights into the mechanisms of plasmid maintenance and replication, respectively. Of note, HigB2A2-like modules are the most prevalent plasmid-borne TA systems in A. baumannii [100, 108]. Coherently, the invariable presence of p1ATCC19606 in all tested ATCC 19606 strains A, D, S and T denotes intrinsic stability, which can be ascribed to a very efficient maintenance system rather than a selective advantage, e.g. antibiotic resistance, conferred by plasmid carriage. However, the function of p1ATCC19606 remains so far elusive.
Prophages are important sources of new genetic information, having the potential of transferring virulence and antibiotic resistance genes [109–111]. Prophages belonging to the Siphoviridae and Myoviridae families were the most frequently discovered in A. baumannii genomes [109, 112]. Here, we show that a remarkable difference among domesticated A. baumannii ATCC 19606T strains was the uneven presence of a 52 kb region, which we identified as the siphoviral Φ19606 prophage. This genetic element was only detected in A. baumannii ATCC 19606T stocks directly originating from ATCC, as inferred from genome analysis of strains D, H, M and O, but not in strains A, S and T, which were passed from lab to lab since the 1980s. Φ19606 belongs to the Vieuvirus genus and was not detected in species other than A. baumannii, showing high prevalence among strains belonging to the successful clonal complex 2 [113]. In A. baumannii ATCC 19606(D) both integrated and episomal forms of Φ19606 were experimentally proven to coexist during serial passages under non-curing conditions (ca 360 generations on LA plates), denoting substantial phage stability. Therefore, at which stage of the of A. baumannii ATCC 19606 A, S and T evolutionary history Φ19606 was lost remains an open question.
The genetic drift resulting from laboratory domestication of reference strains can reflect phenotypic variability, and genome-level differences among laboratory-adapted strains were observed to affect the inter-laboratory experimental reproducibility in the case of Pseudomonas aeruginosa PAO1 lineages [28, 30]. Our studies on A. baumannii ATCC 19606T strains from different laboratories highlighted remarkable diversity at the genome level. This poses the need for researchers to specify the lineage of the strain used, as individual culturing and storage practices may affect micro-evolution, and should encourage the storage of the strains in a single glycerol stock, revitalizing an aliquot when necessary, without recurring to subculturing. Stringent quality controls and strain assessments, including sequencing and the use of low-passage cultures, will help ensure the reproducibility and consistency of A. baumannii research.
Supplementary Data
Funding information
This work was supported by the Excellence Departments grant (art. 1, commi 314-337 Legge 232/2016) to the Department of Science, Roma Tre University, and grant PRIN 2017 (Prot. 20177J5Y3P) to P.V., both from the Italian Ministry of Education, University and Research (MIUR), and by grants no. 85-2017-13763 ANTIMET and no. A0375-2020-36558 GAVAP, both from Regione Lazio. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Acknowledgements
We are grateful to Professor Beate Averhoff (Department of Molecular Microbiology and Bioenergetics, Johann Wolfgang Goethe University Frankfurt am Main, Germany), Professor Harald Seifert (Institute for Medical Microbiology, Immunology and Hygiene, University of Cologne, Cologne, Germany) and Professor Kevin J. Towner (Department of Clinical Microbiology, Nottingham University Hospitals NHS Trust, Nottingham, United Kingdom) for providing A. baumannii ATCC 19606(A), A. baumannii ATCC 19606(S) and A. baumannii ATCC 19606(T), respectively.
Author contributions
P.V. supervised the work and revised the manuscript. I.A. performed genomic analysis with assistance from G.A.L. and M.V., and drafted part of the manuscript. M.L. carried experimental work with the assistance of G.C. and D.V. and drafted part of the manuscript. All authors interpreted the results, contributed to the editing of the manuscript, and read and approved the final version of the manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: Blast2GO, Basic Local Alignment Search Tool 2 Gene Ontology; DR, direct repeat; GBDP, Genome-BLAST Distance Phylogeny; GIs, genomic islands; Gm, gentamicin; HMM, hidden Markov model; INDELs, insertions-deletions; IPTG, isopropyl-β-D-1-thiogalactopyranoside; ISs, insertion sequences; Km, kanamycin; LA, LB agar; LB, Luria–Bertani; MLST, multi-locus sequence typing; NJ, neighbour-joining; pCR, pCR-Blunt II-TOPO; PHASTER, PHAge Search Tool Enhanced Release; QMEAN, Qualitative Model Energy ANalysis; RAPSearch2, Reduced Alphabet based Protein similarity Search; SNPs, single nucleotide polymorphisms; ST, sequence type; TA, toxin-antitoxin; Tc, tetracycline; TE, transformation efficiency; TM, template modelling; Zeo, zeocin; ZOI, zone of inhibition.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Twelve supplementary tables and three supplementary figures are available with the online version of this article.
References
- 1.Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008;21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Visca P, Seifert H, Towner KJ. Acinetobacter infection--an emerging threat to human health. IUBMB Life. 2011;63:1048–1054. doi: 10.1002/iub.534. [DOI] [PubMed] [Google Scholar]
- 3.Antunes LCS, Visca P, Towner KJ. Acinetobacter baumannii: evolution of a global pathogen. Pathog Dis. 2014;71:292–301. doi: 10.1111/2049-632X.12125. [DOI] [PubMed] [Google Scholar]
- 4.Harding CM, Hennon SW, Feldman MF. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat Rev Microbiol. 2018;16:91–102. doi: 10.1038/nrmicro.2017.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee C-R, Lee JH, Park M, Park KS, Bae IK, et al. Biology of Acinetobacter baumannii: pathogenesis, antibiotic resistance mechanisms, and prospective treatment options. Front Cell Infect Microbiol. 2017;7:55. doi: 10.3389/fcimb.2017.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.WHO Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. World Health Organisation. 2017 [Google Scholar]
- 7.Bergogne-Bérézin E, Towner KJ. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin Microbiol Rev. 1996;9:148–165. doi: 10.1128/CMR.9.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouvet PJM, Grimont PAD. Taxonomy of the genus Acinetobacter with the recognition of Acinetobacter baumannii sp. nov., Acinetobacter haemolyticus sp. nov., Acinetobacter johnsonii sp. nov., and Acinetobacter junii sp. nov. and emended descriptions of Acinetobacter calcoaceticus and Acinetobacter lwoffii . Int J Syst Bacteriol. 1986;36:228–240. doi: 10.1099/00207713-36-2-228. [DOI] [Google Scholar]
- 9.Schaub IG, Hauber FD. A biochemical and serological study of a group of identical unidentifiable gram-negative bacilli from human sources. J Bacteriol. 1948;56:379–385. doi: 10.1128/jb.56.4.379-385.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hugh R, Reese R. Designation of the type strain for Bacterium anitratum Schaub and Hauber 1948. Int J Syst Bacteriol. 1967;17:245–254. doi: 10.1099/00207713-17-3-245. [DOI] [Google Scholar]
- 11.Giannouli M, Antunes LCS, Marchetti V, Triassi M, Visca P, et al. Virulence-related traits of epidemic Acinetobacter baumannii strains belonging to the international clonal lineages I-III and to the emerging genotypes ST25 and ST78. BMC Infect Dis. 2013;13:282. doi: 10.1186/1471-2334-13-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moffatt JH, Harper M, Harrison P, Hale JDF, Vinogradov E, et al. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob Agents Chemother. 2010;54:4971–4977. doi: 10.1128/AAC.00834-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krizova L, Poirel L, Nordmann P, Nemec A. TEM-1 β-lactamase as a source of resistance to sulbactam in clinical strains of Acinetobacter baumannii . J Antimicrob Chemother. 2013;68:2786–2791. doi: 10.1093/jac/dkt275. [DOI] [PubMed] [Google Scholar]
- 14.Malone L, Kwon DH. Carbapenem-associated multidrug-resistant Acinetobacter baumannii are sensitised by aztreonam in combination with polyamines. Int J Antimicrob Agents. 2013;41:70–74. doi: 10.1016/j.ijantimicag.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamidian M, Hall RM. Acinetobacter baumannii ATCC 19606 carries GIsul2 in a genomic island located in the chromosome. Antimicrob Agents Chemother. 2017;61:e01991-16. doi: 10.1128/AAC.01991-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeidler S, Müller V. Coping with low water activities and osmotic stress in Acinetobacter baumannii: significance, current status and perspectives. Environ Microbiol. 2019;21:2212–2230. doi: 10.1111/1462-2920.14565. [DOI] [PubMed] [Google Scholar]
- 17.Pachón-Ibáñez ME, Docobo-Pérez F, López-Rojas R, Domínguez-Herrera J, Jiménez-Mejias ME, et al. Efficacy of rifampin and its combinations with imipenem, sulbactam, and colistin in experimental models of infection caused by imipenem-resistant Acinetobacter baumannii . Antimicrob Agents Chemother. 2010;54:1165–1172. doi: 10.1128/AAC.00367-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaddy JA, Arivett BA, McConnell MJ, López-Rojas R, Pachón J, et al. Role of acinetobactin-mediated iron acquisition functions in the interaction of Acinetobacter baumannii strain ATCC 19606T with human lung epithelial cells, Galleria mellonella caterpillars, and mice. Infect Immun. 2012;80:1015–1024. doi: 10.1128/IAI.06279-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henry R, Crane B, Powell D, Deveson Lucas D, Li Z, et al. The transcriptomic response of Acinetobacter baumannii to colistin and doripenem alone and in combination in an in vitro pharmacokinetics/pharmacodynamics model. J Antimicrob Chemother. 2015;70:1303–1313. doi: 10.1093/jac/dku536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo L, Wu L, Xiao Y, Zhao D, Chen Z, et al. Enhancing pili assembly and biofilm formation in Acinetobacter baumannii ATCC19606 using non-native acyl-homoserine lactones. BMC Microbiol. 2015;15:62. doi: 10.1186/s12866-015-0397-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alkasir R, Ma Y, Liu F, Li J, Lv N, et al. Characterization and transcriptome analysis of Acinetobacter baumannii persister cells. Microb Drug Resist. 2018;24:1466–1474. doi: 10.1089/mdr.2017.0341. [DOI] [PubMed] [Google Scholar]
- 22.Lucidi M, Visaggio D, Prencipe E, Imperi F, Rampioni G, et al. New shuttle vectors for real-time gene expression analysis in multidrug-resistant Acinetobacter species: in vitro and in vivo responses to environmental stressors. Appl Environ Microbiol. 2019;85:e01334-19. doi: 10.1128/AEM.01334-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Runci F, Gentile V, Frangipani E, Rampioni G, Leoni L, et al. Contribution of active iron uptake to Acinetobacter baumannii pathogenicity. Infect Immun. 2019;87:e00755-18. doi: 10.1128/IAI.00755-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu Y, Zhao J, Maifiah MHM, Velkov T, Schreiber F, et al. Metabolic responses to polymyxin treatment in Acinetobacter baumannii ATCC 19606: integrating transcriptomics and metabolomics with genome-scale metabolic modeling. mSystems. 2019;4:e00157-18. doi: 10.1128/mSystems.00157-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamidian M, Blasco L, Tillman LN, To J, Tomas M, et al. Analysis of complete genome sequence of Acinetobacter baumannii strain ATCC 19606 reveals novel mobile genetic elements and novel prophage. Microorganisms. 2020;8:1851. doi: 10.3390/microorganisms8121851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsubouchi T, Suzuki M, Niki M, Oinuma K-I, Niki M, et al. Complete genome sequence of Acinetobacter baumannii ATCC 19606T, a model strain of pathogenic bacteria causing nosocomial infection. Microbiol Resour Announc. 2020;9:e00289-20. doi: 10.1128/MRA.00289-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu Y, Lu J, Zhao J, Zhang X, Yu HH, et al. Complete genome sequence and genome-scale metabolic modelling of Acinetobacter baumannii type strain ATCC 19606. Int J Med Microbiol. 2020;310:151412. doi: 10.1016/j.ijmm.2020.151412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klockgether J, Munder A, Neugebauer J, Davenport CF, Stanke F, et al. Genome diversity of Pseudomonas aeruginosa PAO1 laboratory strains. J Bacteriol. 2010;192:1113–1121. doi: 10.1128/JB.01515-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Draper JL, Hansen LM, Bernick DL, Abedrabbo S, Underwood JG, et al. Fallacy of the unique genome: sequence diversity within single Helicobacter pylori strains. mBio. 2017;8:e02321-16. doi: 10.1128/mBio.02321-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chandler CE, Horspool AM, Hill PJ, Wozniak DJ, Schertzer JW, et al. Genomic and phenotypic diversity among ten laboratory isolates of Pseudomonas aeruginosa PAO1. J Bacteriol. 2019;201:e00595-18. doi: 10.1128/JB.00595-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pascoe B, Williams LK, Calland JK, Meric G, Hitchings MD, et al. Domestication of Campylobacter jejuni NCTC 11168. Microb Genom. 2019;5 doi: 10.1099/mgen.0.000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dorman MJ, Thomson NR. “Community evolution” - laboratory strains and pedigrees in the age of genomics. Microbiology (Reading) 2020;166:233–238. doi: 10.1099/mic.0.000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luna BM, Ulhaq A, Yan J, Pantapalangkoor P, Nielsen TB, et al. Selectable markers for use in genetic manipulation of extensively drug-resistant (XDR) Acinetobacter baumannii HUMC1. mSphere. 2017;2:e00140-17. doi: 10.1128/mSphere.00140-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lugli GA, Milani C, Mancabelli L, van Sinderen D, Ventura M. MEGAnnotator: a user-friendly pipeline for microbial genomes assembly and annotation. FEMS Microbiol Lett. 2016;363:fnw049. doi: 10.1093/femsle/fnw049. [DOI] [PubMed] [Google Scholar]
- 36.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hyatt D, Chen G-L, Locascio PF, Land ML, Larimer FW, et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao Y, Tang H, Ye Y. RAPSearch2: a fast and memory-efficient protein similarity search tool for next-generation sequencing data. Bioinformatics. 2012;28:125–126. doi: 10.1093/bioinformatics/btr595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chan PP, Lowe TM. tRNAscan-SE: Searching for tRNA Genes in Genomic Sequences. Methods Mol Biol. 2019;1962:1–14. doi: 10.1007/978-1-4939-9173-0_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lagesen K, Hallin P, Rødland EA, Staerfeldt H-H, Rognes T, et al. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bertelli C, Laird MR, Williams KP, Simon Fraser University Research Computing Group. Lau BY, et al. IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017;45:W30–W35. doi: 10.1093/nar/gkx343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xie Z, Tang H. ISEScan: automated identification of insertion sequence elements in prokaryotic genomes. Bioinformatics. 2017;33:3340–3347. doi: 10.1093/bioinformatics/btx433. [DOI] [PubMed] [Google Scholar]
- 43.Arndt D, Grant JR, Marcu A, Sajed T, Pon A, et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44:W16–21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Antipov D, Raiko M, Lapidus A, Pevzner PA. Plasmid detection and assembly in genomic and metagenomic data sets. Genome Res. 2019;29:961–968. doi: 10.1101/gr.241299.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Langmead B, Wilks C, Antonescu V, Charles R. Scaling read aligners to hundreds of threads on general-purpose processors. Bioinformatics. 2019;35:421–432. doi: 10.1093/bioinformatics/bty648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Darling ACE, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H. Aligning sequence reads, clone sequences and assembly contigs with bwa-mem. arXiv:13033997 [q-bio] 2021 [Google Scholar]
- 49.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koboldt DC, Chen K, Wylie T, Larson DE, McLellan MD, et al. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics. 2009;25:2283–2285. doi: 10.1093/bioinformatics/btp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carver T, Harris SR, Berriman M, Parkhill J, McQuillan JA. Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics. 2012;28:464–469. doi: 10.1093/bioinformatics/btr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 53.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meier-Kolthoff JP, Auch AF, Klenk H-P, Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meier-Kolthoff JP, Göker M. VICTOR: genome-based phylogeny and classification of prokaryotic viruses. Bioinformatics. 2017;33:3396–3404. doi: 10.1093/bioinformatics/btx440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lefort V, Desper R, Gascuel O. FastME 2.0: A comprehensive, accurate, and fast distance-based phylogeny inference program. Mol Biol Evol. 2015;32:2798–2800. doi: 10.1093/molbev/msv150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Farris JS. Estimating phylogenetic trees from distance matrices. The American Naturalist. 1972;106:645–668. doi: 10.1086/282802. [DOI] [Google Scholar]
- 58.Göker M, García-Blázquez G, Voglmayr H, Tellería MT, Martín MP. Molecular taxonomy of phytopathogenic fungi: a case study in Peronospora. PLoS One. 2009;4:e6319. doi: 10.1371/journal.pone.0006319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meier-Kolthoff JP, Hahnke RL, Petersen J, Scheuner C, Michael V, et al. Complete genome sequence of DSM 30083(T), the type strain (U5/41(T)) of Escherichia coli, and a proposal for delineating subspecies in microbial taxonomy. Stand Genomic Sci. 2014;9:2. doi: 10.1186/1944-3277-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. Molecular cloning: a laboratory manual. 1989 [Google Scholar]
- 61.Renda BA, Chan C, Parent KN, Barrick JE. Emergence of a competence-reducing filamentous phage from the genome of Acinetobacter baylyi ADP1. J Bacteriol. 2016;198:3209–3219. doi: 10.1128/JB.00424-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 63.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 66.Lucidi M, Runci F, Rampioni G, Frangipani E, Leoni L, et al. New shuttle vectors for gene cloning and expression in multidrug-resistant acinetobacter species. Antimicrob Agents Chemother. 2018;62:e02480-17. doi: 10.1128/AAC.02480-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hunger M, Schmucker R, Kishan V, Hillen W. Analysis and nucleotide sequence of an origin of DNA replication in Acinetobacter calcoaceticus and its use for Escherichia coli shuttle plasmids. Gene. 1990;87:45–51. doi: 10.1016/0378-1119(90)90494-c. [DOI] [PubMed] [Google Scholar]
- 68.Heeb S, Itoh Y, Nishijyo T, Schnider U, Keel C, et al. Small, stable shuttle vectors based on the minimal pVS1 replicon for use in gram-negative, plant-associated bacteria. Mol Plant Microbe Interact. 2000;13:232–237. doi: 10.1094/MPMI.2000.13.2.232. [DOI] [PubMed] [Google Scholar]
- 69.Steel KJ, Cowan ST. Le rattachement de bacterium antratum, moraxella lwoffii, bacillus mallei et haemophilus parapertussis au genre acinetobacter brisou et prevot. Ann Inst Pasteur. 1964;106:479–483. doi: 10.1099/00207713-17-3-245. [DOI] [PubMed] [Google Scholar]
- 70.Johnson JL, Anderson RS, Ordal EJ. Nucleic acid homologies among oxidase-negative Moraxella species. J Bacteriol. 1970;101:568–573. doi: 10.1128/jb.101.2.568-573.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Karah N, Jolley KA, Hall RM, Uhlin BE. Database for the ampC alleles in Acinetobacter baumannii . PLoS One. 2017;12:e0176695. doi: 10.1371/journal.pone.0176695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reiter WD, Palm P, Yeats S. Transfer RNA genes frequently serve as integration sites for prokaryotic genetic elements. Nucleic Acids Res. 1989;17:1907–1914. doi: 10.1093/nar/17.5.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Williams KP. Integration sites for genetic elements in prokaryotic tRNA and tmRNA genes: sublocation preference of integrase subfamilies. Nucleic Acids Res. 2002;30:866–875. doi: 10.1093/nar/30.4.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sethi D, Mahajan S, Singh C, Lama A, Hade MD, et al. Lipoprotein LprI of Mycobacterium tuberculosis Acts as a Lysozyme Inhibitor. J Biol Chem. 2016;291:2938–2953. doi: 10.1074/jbc.M115.662593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Callewaert L, Aertsen A, Deckers D, Vanoirbeek KGA, Vanderkelen L, et al. A new family of lysozyme inhibitors contributing to lysozyme tolerance in gram-negative bacteria. PLoS Pathog. 2008;4:e1000019. doi: 10.1371/journal.ppat.1000019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mira A, Ochman H, Moran NA. Deletional bias and the evolution of bacterial genomes. Trends Genet. 2001;17:589–596. doi: 10.1016/s0168-9525(01)02447-7. [DOI] [PubMed] [Google Scholar]
- 77.Solovyev V, Salamov A. Automatic Annotation of Microbial Genomes and Metagenomic Sequences. in Metagenomics and Its Applications in Agriculture, Biomedicine and Environmental Studies. Nova Science Publishers; 2011. pp. 61–78. [Google Scholar]
- 78.Ramisetty BCM, Sudhakari PA. Bacterial “Grounded” prophages: hotspots for genetic renovation and innovation. Front Genet. 2019;10:65. doi: 10.3389/fgene.2019.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Black LW. DNA packaging in dsDNA bacteriophages. Annu Rev Microbiol. 1989;43:267–292. doi: 10.1146/annurev.mi.43.100189.001411. [DOI] [PubMed] [Google Scholar]
- 80.Jeon J, Kim J, Yong D, Lee K, Chong Y. Complete genome sequence of the podoviral bacteriophage YMC/09/02/B1251 ABA BP, which causes the lysis of an OXA-23-producing carbapenem-resistant Acinetobacter baumannii isolate from a septic patient. J Virol. 2012;86:12437–12438. doi: 10.1128/JVI.02132-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jeon J, D’Souza R, Pinto N, Ryu C-M, Park J, et al. Complete genome sequence of the siphoviral bacteriophage Βϕ-R3177, which lyses an OXA-66-producing carbapenem-resistant Acinetobacter baumannii isolate. Arch Virol. 2015;160:3157–3160. doi: 10.1007/s00705-015-2604-y. [DOI] [PubMed] [Google Scholar]
- 82.Potron A, Vuillemenot J-B, Puja H, Triponney P, Bour M, et al. ISAba1-dependent overexpression of eptA in clinical strains of Acinetobacter baumannii resistant to colistin. J Antimicrob Chemother. 2019;74:2544–2550. doi: 10.1093/jac/dkz241. [DOI] [PubMed] [Google Scholar]
- 83.Ackers GK, Johnson AD, Shea MA. Quantitative model for gene regulation by lambda phage repressor. Proc Natl Acad Sci U S A. 1982;79:1129–1133. doi: 10.1073/pnas.79.4.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Atsumi S, Little JW. Regulatory circuit design and evolution using phage lambda. Genes Dev. 2004;18:2086–2094. doi: 10.1101/gad.1226004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nanda AM, Thormann K, Frunzke J. Impact of spontaneous prophage induction on the fitness of bacterial populations and host-microbe interactions. J Bacteriol. 2015;197:410–419. doi: 10.1128/JB.02230-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.López-Leal G, Reyes-Muñoz A, Santamaria RI, Cevallos MA, Pérez-Monter C, et al. A novel vieuvirus from multidrug-resistant Acinetobacter baumannii . Arch Virol. 2021;166:1401–1408. doi: 10.1007/s00705-021-05010-4. [DOI] [PubMed] [Google Scholar]
- 87.Dorsey CW, Tomaras AP, Actis LA. Sequence and organization of pMAC, an Acinetobacter baumannii plasmid harboring genes involved in organic peroxide resistance. Plasmid. 2006;56:112–123. doi: 10.1016/j.plasmid.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 88.Wick RR, Judd LM, Wyres KL, Holt KE. Recovery of small plasmid sequences via Oxford Nanopore sequencing. Microb Genom. 2021;7 doi: 10.1099/mgen.0.000631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gao F, Luo H, Zhang C-T. DoriC 5.0: an updated database of oriC regions in both bacterial and archaeal genomes. Nucleic Acids Res. 2013;41:D90–3. doi: 10.1093/nar/gks990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kües U, Stahl U. Replication of plasmids in gram-negative bacteria. Microbiol Rev. 1989;53:491–516. doi: 10.1128/mr.53.4.491-516.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chattoraj DK. Control of plasmid DNA replication by iterons: no longer paradoxical. Mol Microbiol. 2000;37:467–476. doi: 10.1046/j.1365-2958.2000.01986.x. [DOI] [PubMed] [Google Scholar]
- 92.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Swan MK, Bastia D, Davies C. Crystal structure of pi initiator protein-iteron complex of plasmid R6K: implications for initiation of plasmid DNA replication. Proc Natl Acad Sci U S A. 2006;103:18481–18486. doi: 10.1073/pnas.0609046103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Abhyankar MM, Reddy JM, Sharma R, Büllesbach E, Bastia D. Biochemical investigations of control of replication initiation of plasmid R6K. J Biol Chem. 2004;279:6711–6719. doi: 10.1074/jbc.M312052200. [DOI] [PubMed] [Google Scholar]
- 95.Goeders N, Van Melderen L. Toxin-antitoxin systems as multilevel interaction systems. Toxins (Basel) 2014;6:304–324. doi: 10.3390/toxins6010304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Page R, Peti W. Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat Chem Biol. 2016;12:208–214. doi: 10.1038/nchembio.2044. [DOI] [PubMed] [Google Scholar]
- 97.Unterholzner SJ, Poppenberger B, Rozhon W. Toxin-antitoxin systems: Biology, identification, and application. Mob Genet Elements. 2013;3:e26219. doi: 10.4161/mge.26219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chan WT, Espinosa M, Yeo CC. Keeping the wolves at bay: antitoxins of prokaryotic type II toxin-antitoxin systems. Front Mol Biosci. 2016;3:9. doi: 10.3389/fmolb.2016.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rocker A, Meinhart A. Type II toxin: antitoxin systems. More than small selfish entities? Curr Genet. 2016;62:287–290. doi: 10.1007/s00294-015-0541-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Armalytė J, Jurėnas D, Krasauskas R, Čepauskas A, Sužiedėlienė E. The higBA toxin-antitoxin module from the opportunistic pathogen Acinetobacter baumannii - regulation, activity, and evolution. Front Microbiol. 2018;9:732. doi: 10.3389/fmicb.2018.00732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hadži S, Garcia-Pino A, Haesaerts S, Jurenas D, Gerdes K, et al. Ribosome-dependent Vibrio cholerae mRNAse HigB2 is regulated by a β-strand sliding mechanism. Nucleic Acids Res. 2017;45:4972–4983. doi: 10.1093/nar/gkx138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hurley JM, Woychik NA. Bacterial toxin HigB associates with ribosomes and mediates translation-dependent mRNA cleavage at A-rich sites. J Biol Chem. 2009;284:18605–18613. doi: 10.1074/jbc.M109.008763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yamaguchi Y, Park J-H, Inouye M. MqsR, a crucial regulator for quorum sensing and biofilm formation, is a GCU-specific mRNA interferase in Escherichia coli . J Biol Chem. 2009;284:28746–28753. doi: 10.1074/jbc.M109.032904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Heaton BE, Herrou J, Blackwell AE, Wysocki VH, Crosson S. Molecular structure and function of the novel BrnT/BrnA toxin-antitoxin system of Brucella abortus . J Biol Chem. 2012;287:12098–12110. doi: 10.1074/jbc.M111.332163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Christensen-Dalsgaard M, Gerdes K. Two higBA loci in the Vibrio cholerae superintegron encode mRNA cleaving enzymes and can stabilize plasmids. Mol Microbiol. 2006;62:397–411. doi: 10.1111/j.1365-2958.2006.05385.x. [DOI] [PubMed] [Google Scholar]
- 106.Graña-Miraglia L, Lozano LF, Velázquez C, Volkow-Fernández P, Pérez-Oseguera Á, et al. Rapid gene turnover as a significant source of genetic variation in a recently seeded population of a healthcare-associated pathogen. Front Microbiol. 2017;8:1817. doi: 10.3389/fmicb.2017.01817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Watson M, Warr A. Errors in long-read assemblies can critically affect protein prediction. Nat Biotechnol. 2019;37:124–126. doi: 10.1038/s41587-018-0004-z. [DOI] [PubMed] [Google Scholar]
- 108.Jurenaite M, Markuckas A, Suziedeliene E. Identification and characterization of type II toxin-antitoxin systems in the opportunistic pathogen Acinetobacter baumannii . J Bacteriol. 2013;195:3165–3172. doi: 10.1128/JB.00237-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Costa AR, Monteiro R, Azeredo J. Genomic analysis of Acinetobacter baumannii prophages reveals remarkable diversity and suggests profound impact on bacterial virulence and fitness. Sci Rep. 2018;8:15346. doi: 10.1038/s41598-018-33800-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wachino J-I, Jin W, Kimura K, Arakawa Y. Intercellular transfer of chromosomal antimicrobial resistance genes between Acinetobacter baumannii strains mediated by prophages. Antimicrob Agents Chemother. 2019;63:e00334-19. doi: 10.1128/AAC.00334-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.López-Leal G, Santamaria RI, Cevallos MÁ, Gonzalez V, Castillo-Ramírez S. Letter to the editor: prophages encode antibiotic resistance genes in Acinetobacter baumannii . Microb Drug Resist. 2020;26:1275–1277. doi: 10.1089/mdr.2019.0362. [DOI] [PubMed] [Google Scholar]
- 112.Loh B, Chen J, Manohar P, Yu Y, Hua X, et al. A biological inventory of prophages in A. baumannii genomes reveal distinct distributions in classes, length, and genomic positions. Front Microbiol. 2020;11:579802. doi: 10.3389/fmicb.2020.579802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hamidian M, Nigro SJ. Emergence, molecular mechanisms and global spread of carbapenem-resistant Acinetobacter baumannii . Microb Genom. 2019;5:e000306. doi: 10.1099/mgen.0.000306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Smith MG, Gianoulis TA, Pukatzki S, Mekalanos JJ, Ornston LN, et al. New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev. 2007;21:601–614. doi: 10.1101/gad.1510307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Iacono M, Villa L, Fortini D, Bordoni R, Imperi F, et al. Whole-genome pyrosequencing of an epidemic multidrug-resistant Acinetobacter baumannii strain belonging to the European clone II group. Antimicrob Agents Chemother. 2008;52:2616–2625. doi: 10.1128/AAC.01643-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jacobs AC, Thompson MG, Black CC, Kessler JL, Clark LP, et al. AB5075, a highly virulent isolate of Acinetobacter baumannii, as a model strain for the evaluation of pathogenesis and antimicrobial treatments. mBio. 2014;5:e01076–14. doi: 10.1128/mBio.01076-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Vaneechoutte M, Young DM, Ornston LN, De Baere T, Nemec A, et al. Naturally transformable Acinetobacter sp. strain ADP1 belongs to the newly described species Acinetobacter baylyi . Appl Environ Microbiol. 2006;72:932–936. doi: 10.1128/AEM.72.1.932-936.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cosgaya C, Marí-Almirall M, Van Assche A, Fernández-Orth D, Mosqueda N, et al. Acinetobacter dijkshoorniae sp. nov., a member of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex mainly recovered from clinical samples in different countries. Int J Syst Evol Microbiol. 2016;66:4105–4111. doi: 10.1099/ijsem.0.001318. [DOI] [PubMed] [Google Scholar]
- 119.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.