Abstract
Optimal activation of NF-κB signaling is crucial for the initiation of inflammatory responses and eliminating invading bacteria. Bacteria have likewise evolved the ability to evade immunity; however, mechanisms by which bacteria dysregulate host NF-κB signaling are unclear. In this study, we identify eukaryotic translation initiation factor eIF3k, a nonessential member of the eIF3 translation initiation complex, as a suppressor of the NF-κB pathway. Mechanistically, we show that eIF3k expression induced by Vibrio harveyi enhances E3 ligase Nrdp1-mediated K27-linked ubiquitination of MyD88, an upstream regulator of NF-κB pathway activation. Furthermore, we show that eIF3k acts as a bridge linking ubiquitin-tagged MyD88 and ATG5, an important mediator of autophagy. We demonstrate that the MyD88-eIF3k-ATG5 complex is transported to the autophagosome for degradation, and that innate immune signaling is subsequently terminated and does not attack invading V. harveyi. Therefore, our study identifies eIF3k as a specific inhibitor of the MyD88-dependent NF-κB pathway and suggests that eIF3k may act as a selective autophagic receptor that synergizes with ATG5 to promote the autophagic degradation of MyD88, which helps V. harveyi to evade innate immunity. We conclude that V. harveyi can manipulate a host's autophagy process to evade immunity in fish and also provide a new perspective on mammalian resistance to bacterial invasion.
Keywords: eIF3k, MyD88, immune evasion, autophagy, Vibrio harveyi
Abbreviations: 3-MA, 3-methyladenine; ATG, autophagy-related gene; CHX, cycloheximide; Co-IP, coimmunoprecipitation; eIF3k, eukaryotic translation initiation factor 3k; EPC, epithelioma papulosum cyprini; HAM, HEAT analog motif; MIC, M. miiuy intestine cell; MKC, M. miiuy kidney cell; PRR, pattern recognition receptor; SCRV, Siniperca chuatsi rhabdovirus; TIR, Toll/interleukin-1 receptor; TLR, Toll-like receptor
Innate immune system is the host's first line of defense against microbial infection, which can respond quickly to infection. The innate immunity system recognizes the molecular structure of microorganisms through pattern recognition receptors (PRRs), induces the transcription of downstream inflammatory cytokines, thereby triggering innate immunity and inflammatory responses to safeguard the host from infection (1).
Toll-like receptors (TLRs) are the most immemorial PRR, which consist of an ectodomain, a transmembrane, and a Toll/interleukin-1 receptor (TIR). TLRs are mediated by an adaptor molecule containing the TIR domain. TLRs exist on the cell surface and can recognize lipids, carbohydrates, peptides, and nucleic acids (2). With the exception of TLR3, TLRs in all mammals depend, at least in part, on the MyD88 adaptor for signaling (3). TLRs trigger the activation of intracellular signaling via a MyD88-dependent pathway or a MyD88-independent pathway. In the MyD88-dependent pathway, IRAK-1, IRAK-4, and TRAF6 were first enlisted to MyD88. Sequentially, the phosphorylated IRAK and ubiquitinated TRAF6 activate TAK1, ultimately the phosphorylated IKKs promote the transcription of NF-κB and various proinflammatory mediators (4, 5). As a canonical adaptor for inflammatory signaling pathways downstream of TLRs, knockout or knockdown of MyD88 leads to the complete abolishment of LPS-induced NF-κB activation (6). MyD88 deficiency also results in unresponsiveness to bacterial endotoxins in mice (7).
Eukaryotic translation initiation factor 3k (eIF3k) belongs to the eIF3 family, which has been proved to play a critical regulatory role in immune responses (8, 9). In the MAPK/NF-κB signaling pathway, overexpressed eIF3K can downregulate the expression of highly expressed proteins in inflammatory cells (10). eIF3k has two functional domains, a HEAT analog motif (HAM) and a winged-helix (WH), which are both involved in protein–protein interactions (11). In addition, eIF3k promotes apoptosis by enhancing the binding with the keratin in apoptotic cells (12). eIF3k is also a binding chaperone of cyclin D3 and is thought to be involved in the regulation of apoptosis (13). However, whether eIF3k regulates innate immunity in teleost fish remains unknown.
In eukaryotes, autophagy is a conserved process by which abnormal proteins are transported to lysosomes for degradation and plays an important role in innate immune against viral, bacterial, and fungal pathogens (14, 15). Extensive cross talk between autophagy and immune signaling cascades has been well established. In mice, MyD88-dependent induction of autophagy inhibits dissemination of Salmonella enterica subsp (16). Meanwhile, autophagy also participates in the regulation of immune responses. For instance, p62/SQSTM1-mediated autophagy promotes STING degradation to attenuate the activation of cGAS-STING pathway (17). Galectin9 enhances the formation of NLRP3/p62 complex and facilitates p62-dependent autophagic degradation of NLRP3 to avoid excessive activation of NLRP3 inflammasome (18). MyD88 can also mediate autophagy. In the presence of TLR engagement, Beclin1 forms a complex with MyD88 and TRIF to promote autophagy (19). In addition, some pathogens have evolved complex mechanisms to completely inhibit the autophagic process or prevent selective recognition of autophagy or even use autophagy to promote their own replication (20, 21). Francisella tularensis exploits noncanonical autophagy to harvests nutrients to support intracellular growth. After replication, it escapes from autophagosomes to cytoplasm, and ATG5 plays an essential role in this process (22). Autophagy is regulated by different autophagy-related genes (ATGs). mTOR-ULK1 (mammalian target of rapamycin-Unc-51 such as autophagy activating kinase 1) and PI3KC3 (Beclin-1 phosphatidylinositol 3-kinase catalytic subunit type 3) are two important signal pathways to regulate autophagy (23). The complex of ATG5 and ATG12 is a key component of phagophore extension (24). Autophagy is associated with the transformation of LC3B-I to LC3B-II. The ratio of LC3B-II to LC3B-I was also used to measure the degree of autophagy (25, 26). Accumulating evidence suggests that autophagic degradation of proteins is highly selective (27, 28, 29). Selective autophagy packages substrates into the autophagosome for degradation by cargo receptors, which recognize ubiquitinated substrates (30). Cargo receptors, such as p62/SQSTM1 and NBR1, bind directly to ubiquitin and LC3 via the UBA domain and LIR domain, respectively (31, 32). NDP52 does not contain a UBA domain, but is linked to ubiquitin by ubiquitin-binding zinc finger (33). In addition to binding with ubiquitin, ALFY also interacts with autophagosome membranes though phosphatidyl inositol triphosphate (PI3P)-conjugated FYVE domain to mediate selective autophagy (34). Autophagy has been shown to be involved in immunomodulation in mammals. However, the interplay between autophagy and immunomodulation in fish remains to be investigated.
Vibrio harveyi, a Gram-negative bacterium, is the most damaging pathogenic bacteria in aquaculture. V. harveyi infection has been shown to be effective in activating the innate immune response (35, 36). Bacteria have evolved the ability to evade immune attack in their struggle against innate immunity. Shigella achieved successful colonization by inhibiting apoptosis (37). S protein produced by group A streptococcus (GAS) binds to the erythrocyte membrane to evade detection of host immune system (38). During coevolution with the host, these pathogens serve as the host's protective barrier, evade host recognition, or inhibit immune activation.
In this study, we identify that selective autophagy is involved in the regulation of innate immune system in teleost fish, miiuy croaker (Miichthys miiuy). And we identify eIF3k, an eIF3 family member, as a suppressor of NF-κB pathway. eIF3k promotes the E3 ligase Nrdp1-mediated ubiquitination of MyD88 and cooperates with ATG5 to deliver ubiquitinated MyD88 into the autophagosome for degradation, which helps V. harveyi hijack the host's autophagy system to evade innate immune.
Results
eIF3k is upregulated by V. harveyi stimulation
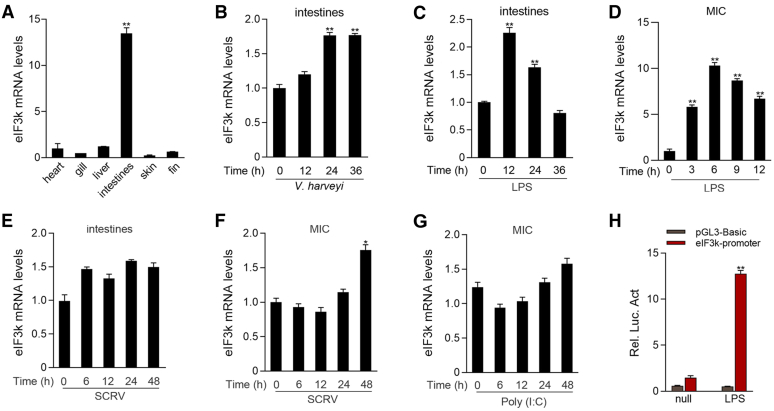
Innate immunity is the first line of defense against bacterial invasion. As mentioned earlier, MyD88 plays a momentous role in the innate immunity. Therefore, we focused on the regulation of MyD88 and performed an endogenous IP experiment with MyD88. We screened out one of the eIF3 members, eIF3k, using IP and mass spectrum analysis (Table S2). To investigate the expression pattern of eIF3k, eIF3k mRNA levels was determined in different tissues by quantitative PCR (qPCR) analysis. eIF3k mRNA was strongly expressed in the intestines (Fig. 1A). The intestine is considered the largest compartment of the immune system. It was previously reported that the intestines can recognize invading pathogens, which is essential for pathogen defense (39). It suggests that eIF3k may involve in immune response. Next, we stimulated M. miiuy with different pathogens to determine the expression of eIF3k. M. miiuy was stimulated with V. harveyi or LPS, and then eIF3k mRNA was measured in the intestines. eIF3k continued to increase after infected with V. harveyi, with a peak at 24 h postinfection (Fig. 1B). Previous studies have demonstrated that LPS is an immune response initiator in M. miiuy (40). eIF3k mRNA levels were significantly enhanced after LPS treatment, with a peak at 12 h (Fig. 1C). The transcription pattern of eIF3k in the M. miiuy intestine cells (MICs) after LPS stimulation was consistent with the results above (Fig. 1D). In addition, eIF3k mRNA was slightly increased in the intestines of Siniperca chuatsi rhabdovirus (SCRV)-infected M. miiuy and in SCRV-infected MIC (Fig. 1, E and F). As an analog of dsRNA, poly(I:C), is also used to mimic viral stimulation in fish (41, 42). eIF3k mRNA was slightly increased in MIC with poly(I:C) stimulation (Fig. 1G). We also found that LPS can markedly induce eIF3k promoter luciferase reporter (Fig. 1H). The results showed that eIF3k mRNA increased to varying degrees after pathogen stimulation, and that it was significantly higher after V. harveyi than viral infection.
Figure 1.
eIF3k is upregulated by V. harveyi stimulation.A, M. miiuy eIF3k mRNAs were determined in different tissues by qPCR. B and C, eIF3k mRNAs in M. miiuy intestines were determined by qPCR after V. harveyi (B) or LPS (C) infection at different times. D, expression of eIF3k mRNAs in MIC was determined by qPCR after LPS stimulation at for different times. E, expression of eIF3k mRNAs was determined in intestines by qPCR after SCRV infection. F and G, expression of eIF3k mRNAs was determined in MIC by qPCR after SCRV or Poly(I:C) stimulation. H, eIF3k was activated by LPS. EPC cells were co-transfected with eIF3k reporter gene plasmid (100 ng) or control vector pGL3-Basic (100 ng). pRL-TK (10 ng) was included to normalize the expression level. Cells were treated with 10 μg/ml LPS for additional 6 h at 24 h posttransfection, followed by detection of luciferase activity. The data are shown as the mean ± SD of three independent experiments. (∗) p< 0.05, (∗∗) p < 0.01 versus the controls. eIF3k, eukaryotic translation initiation factor 3k; EPC, epithelioma papulosum cyprini; MIC, M. miiuy intestine cell.
V. harveyi inhibits NF-κB signaling by inducing eIF3k to increase colonization
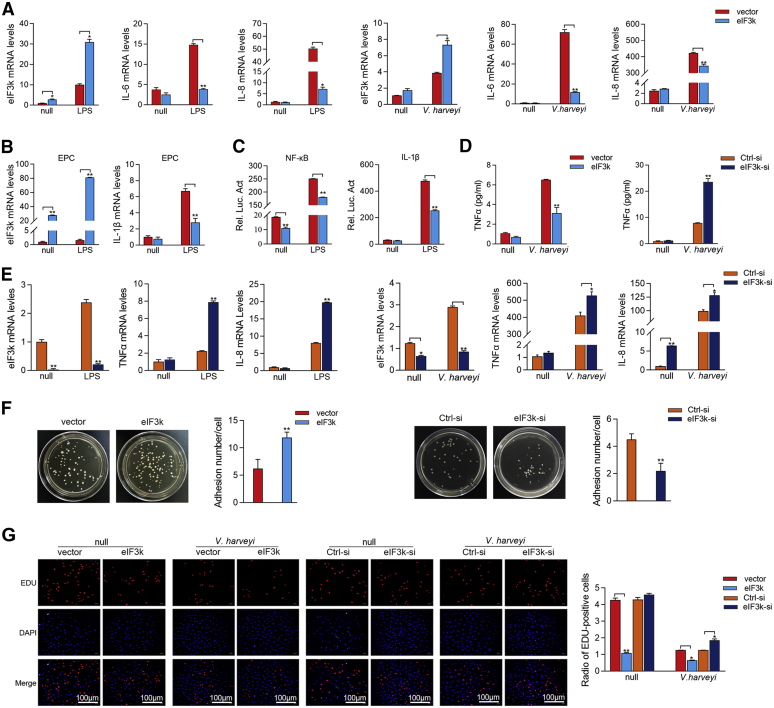
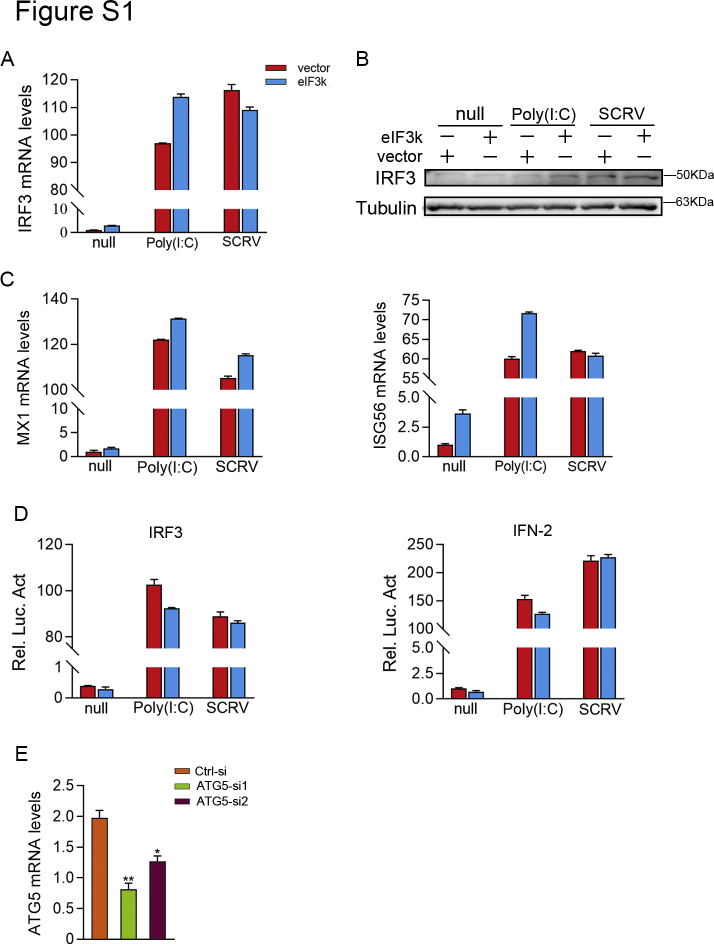
To elucidate whether eIF3k regulates NF-κB signaling, eIF3k was transfected firstly and then the inflammatory cytokines (IL-6 and IL-8) were detected by qPCR. The results indicated that overexpression of eIF3k significantly inhibited LPS- and V. harveyi-triggered transcription of IL-6 and IL-8 in MIC (Fig. 2A). In addition, we found that eIF3k was not only induced by V. harveyi and LPS in M. miiuy (Fig. 2A), but also induced by LPS in epithelioma papulosum cyprini (EPC) cells (Fig. 2B). And eIF3k distinctly inhibited LPS-triggered IL-1β mRNA levels in EPC cells (Fig. 2B). As shown in the luciferase reporter assays, overexpression of eIF3k significantly inhibited LPS-induced activation of NF-κB and IL-1β reporter gene (Fig. 2C). The above results suggested a general inhibitory effect of eIF3k on bacteria-triggered immune responses in fish. Endogenous eIF3k was knocked down by synthesized siRNA to validate its role in the innate immunity (Fig. 2E). We further analyzed the expression of the proinflammatory cytokine TNF-α in V.harveyi infection. eIF3k overexpression significantly reduced TNF-α production, in contrast to eIF3k silencing, which significantly increased TNF-α production (Fig. 2D). The results showed that knockdown of eIF3k significantly enhanced LPS- and V. harveyi-triggered transcription of TNFα and IL-8 (Fig. 2E). Since SCRV and poly(I:C) stimulation also increased eIF3k expression (Fig. 1, E and F), we investigated whether it was also involved in regulating antiviral signaling. Interestingly, we found that overexpression of eIF3k did not effectively inhibit SCRV- and poly(I:C)-triggered IRF-3, MX1, and ISG56 (Fig. S1, A and B). The luciferase reporter assays also showed that overexpression of eIF3k couldn't inhibit poly(I:C)- and SCRV-triggered IRF3 and IFN-2 reporter gene (Fig. S1D). To further explore the role of eIF3k in bacteria invasion, an adhesion assay of V. harveyi was performed. The result showed that eIF3k significantly increased the colony forming unit (cfu) adhering to individual cell (Fig. 2F). Cell proliferation experiment (Fig. 2G) was suggested that both eIF3k and V. harveyi could inhibit MIC proliferation. To sum up, V. harveyi induces eIF3k to inhibit the activation of innate immune signaling during infection for the purpose of immune evasion.
Figure 2.
eIF3k inhibits NF-κB signaling to promote V. haveryi reproduction.A, eIF3k inhibited LPS- and V. harveyi-triggered transcription of pro-inflammatory cytokine in the NF-κB signaling pathway. MIC cells were transfected with eIF3k (500 ng) or control vector pcDNA3.1 (500 ng). Cells were treated with10 μg/ml LPS or V. harveyi for additional 6 h at 36 h post-transfected. eIF3k, IL-6 and IL-8 mRNAs were analyzed by qPCR. B, EPC cells were transfected and stimulate as in (A), eIF3k and IL-1β mRNAs were determined by qPCR. C, eIF3k inhibits LPS-triggered activation of NF-κB and IL-1β reporter gene. EPC cells were transfected with NF-κB, IL-1β reporter gene plasmid, together with pRL-TK Renilla luciferase plasmid. Cells were treated with LPS for an additional 6 h at 24 h post-transfected, followed by detection of luciferase activity. D, TNFα production was detected by ELISA assay in eIF3k overexpression and silenced with V. harveyi infection for 24 h. E, knockdown of eIF3k significantly enhanced LPS- and V. harveyi-triggered transcription of proinflammatory cytokine in MIC cells. Cells were transfected with eIF3k-si, Ctrl-si was used for control. Stimulation with LPS and V. harveyi as before, TNFα and IL-8 mRNAs was determined by qPCR. F, eIF3k increases the adhesion of V. harveyi to MKC cells. Cells were transfected with eIF3k or eIF3k-si, vector and Ctrl-si was used for control. Lysates from V. harveyi infected cells were incubated on LB plates for 12 h, and the colony forming unit (cfu) was counted. G, both bacteria and eIF3k inhibit the proliferation of MKC cells. Cells were transfected with the indicated plasmids for 24 h and infected with V. harveyi for 6 h prior to the cell proliferation assay; Scale bar, 100 μm. The data are shown as the mean ± SD of three independent experiments. (∗) p< 0.05, (∗∗) p< 0.01 versus the controls. eIF3k, eukaryotic translation initiation factor 3k; EPC, epithelioma papulosum cyprini; MIC, M. miiuy intestine cell; MKC, M. miiuy kidney cell.
eIF3k attenuates MyD88-mediated NF-κB signaling
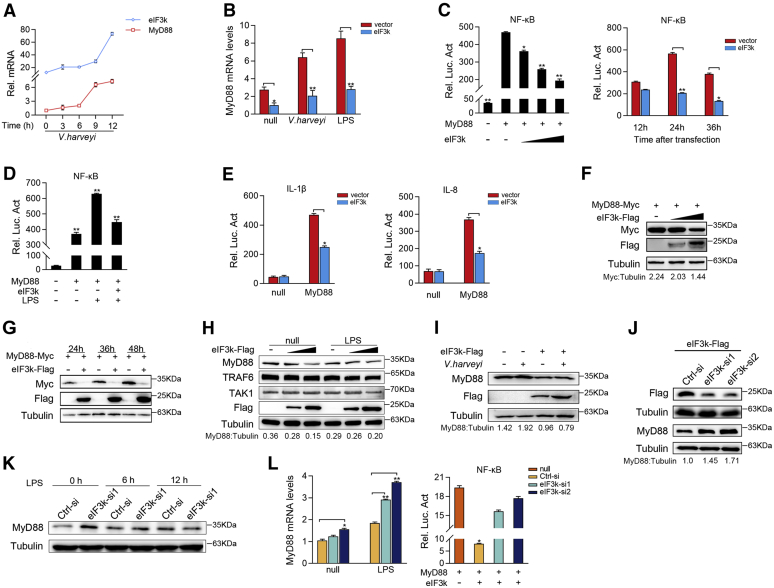
Previous studies have demonstrated that MyD88 is activated and induces inflammatory cytokines to counteract the invasion of Gram-negative bacteria (35, 36). Given that above, we first examined the expression of eIF3k and MyD88 at the time of V. harveyi invasion. There was a significant increase in MyD88 at 6 h after infection; and with the surge of eIF3k, the transcription of MyD88 was repressed (Fig. 3A). Immediately after we overexpressed eIF3k in V. harveyi, LPS stimulated MIC to verify the inhibition of eIF3k on MyD88. We found that eIF3k effectively represses V. harveyi- and LPS-induced transcription of MyD88 (Fig. 3B). The results of luciferase reporter assays showed that overexpression of MyD88 significantly activated NF-κB reporter gene. Nevertheless, coexpression of eIF3k inhibits the activation of NF-κB in a dose-dependent manner and a time-dependent manner (Fig. 3C). Consistent with the above results, eIF3k can also repress the LPS-triggered NF-κB activation (Fig. 3D); coexpression of eIF3k weakened MyD88-induced activation of IL-1β and IL-8 reporter gene (Fig. 3E). To further verify the regulatory of eIF3k on MyD88, immunoblot analysis was performed with cotransfected MyD88 and eIF3k. eIF3k significantly downregulated MyD88 expression in a dose-dependent manner (Fig. 3F) and a time-dependent manner (Fig. 3G). Subsequently, we examined the effects of eIF3k on endogenous MyD88-mediated signaling. Immunoblot analysis shows that eIF3k significantly inhibits endogenous MyD88 regardless of whether the cells were stimulated by LPS or not. However, no significant inhibition of TRAF6 and TAK1 was found (Fig. 3H). We also found that eIF3k obviously inhibited V. harveyi-triggered MyD88 (Fig. 3I).
Figure 3.
eIF3k terminates NF-κB signaling by degrading MyD88.A, expression of eIF3k and MyD88 mRNAs in MIC was determined by qPCR after V. harveyi stimulation at for different times. B, eIF3k inhibited the transcription of endogenous MyD88. MIC cells were transfected with the indicated plasmids for 24 h, and then stimulated with V. harveyi and LPS for 6 h. eIF3k and MyD88 mRNAs were determined by qPCR. C–E, eIF3k inhibits myD88-induced NF-κB signaling. EPC cells were transfected with pRL-TK, luciferase reporter genes, together with eIF3k. Luciferase activity was measured and normalized to renilla luciferase activity. F, eIF3k inhibited MyD88 in a dose-dependent manner. EPC cells were co-transfected with MyD88 and eIF3k for 48 h, the expression of MyD88 was determined by immunoblot. G, eIF3k inhibited MyD88 in a schedule-dependent manner. EPC cells were co-transfected with MyD88 and eIF3k; cells were collected at different point for immunoblot. H, eIF3k specifically targets MyD88. MIC cells were transfected with eIF3k or control vector for 48 h. Endogenous proteins were determined by immunoblot. I, eIF3k inhibits V. harveyi-triggered MyD88. MIC cells were infected with V. harveyi for 6 h at post-transfected with eIF3k. Endogenous MyD88 was determined by immunoblot. J–L, knockdown of eIF3k slowed the degradation of endogenous MyD88. MIC cells were transfected with eIF3k-si and Ctrl-si (J), after LPS treatment for 6 h and 12 h (K), the expression of MyD88 was determined by immunoblot and qPCR (L). The data are shown as the mean ± SD of three independent experiments. (∗) p< 0.05, (∗∗) p< 0.01 versus the controls. eIF3k, eukaryotic translation initiation factor 3k; EPC, epithelioma papulosum cyprini; MIC, M. miiuy intestine cell.
Then eIF3k was detected after knockdown with immunoblot, overexpression of eIF3k was significantly increased with eIF3k-si treatment (Fig. 3J, the top panel). It has also come to our notice that knockdown eIF3k could retard the degradation of endogenous MyD88 whether stimulated by LPS or not (Fig. 3, J and K). qPCR results showed that knockdown of eIF3k enhances LPS-triggered MyD88 transcription. In the meantime, loss of eIF3k no longer inhibited MyD88-induced activation of NF-κB (Fig. 3L). The above results indicated that eIF3k has a specifically negative regulation on MyD88.
eIF3k interacts with MyD88
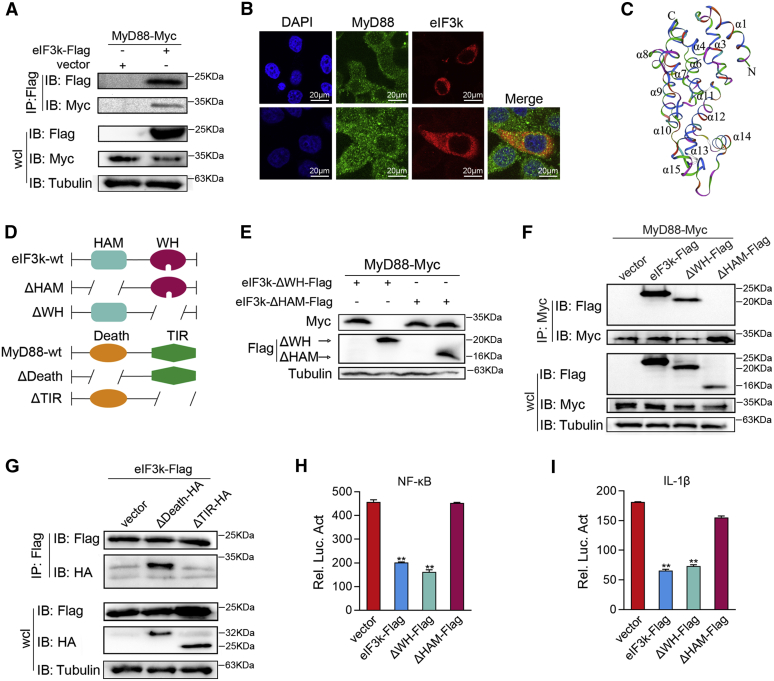
Then the interaction between eIF3k and MyD88 was analyzed. Cells were cotransfected with MyD88-Mycand eIF3k-Flag expression plasmids. As shown in Figure 4A, MyD88-Myc was detected in the anti-Flag immunoprecipitate in coexpression with eIF3k-Flag, but not in negative control pcDNA3-Flag. Subcellular localization of MyD88 and eIF3k was investigated, and the results showed that eIF3k and MyD88 colocalized in the cytoplasm (Fig. 4B). Given this, the results revealed that eIF3k interacted with MyD88. To map the regions of their interaction, we first analyzed the overall structures of eIF3k and MyD88. After alignment with the human eIF3k structure (43), M. miiuy eIF3k was determined to have a HAM domain (composed of 11 α-helices from α1 to α11) and a WH domain (composed of three α-helices structures from α12 to α14) (Fig. 4C). We constructed two truncated mutants of eIF3k (ΔHAM and ΔWH) and two truncated mutants of MyD88 (ΔTIR and ΔDeath) basing on their structures (Fig. 4D). Two truncated mutants of eIF3k and MyD88-wt were cotransfected to verify their effectiveness. We found that only deletion of HAM domain abolished the inhibitory of eIF3k on MyD88 (Fig. 4E). Consistent with the above, the mutant mapping experiments revealed that MyD88 interacts with the HAM domain of eIF3k through its TIR-domain (Fig. 4, F and G). Dual-luciferase activity assay showed the same results; eIF3k-ΔHAM no longer inhibits activation of NF-κB and IL-1β reporter gene (Fig. 4, H and I). Thus, our data raise a possibility that eIF3k targets MyD88 and plays a role in regulating NF-κB signaling.
Figure 4.
Identification of eIF3k as a MyD88-interacting partner.A, eIF3k interacts with MyD88. HEK 293 cells were transfected with the indicated plasmids for 32 h. Co-IP was performed and subjected to immunoblot analysis. B, eIF3k colocalizes with MyD88 in cytoplasm. HeLa cells were transfected with the indicated plasmids for 30 h, then stained with FITC (green) or Cy3 (red). The nuclei were stained by DAPI (blue). Pictures were taken by FCFM. Scale bar, 20 μm; original magnification ×40. C, the overall of eIF3k was predicted by the Swiss model. D, the truncated mutants of MyD88 and eIF3k. E–G, MyD88 associates with eIF3k through its TIR and HAM domain. EPC cells (E) and HEK293 cells (F and G) were transfected with the indicated plasmids for immunoblot (E) and mutant mapping assay. H and I, deletion of the critical domain allows eIF3k no longer inhibit MyD88-induced NF-kB signaling. EPC cells were transfected with the indicated plasmids. The luciferase activity was measured and normalized to renilla luciferase activity. The data are shown as the mean ± SD of three independent experiments. (∗) p< 0.05, (∗∗) p< 0.01 versus the controls. Co-IP, coimmunoprecipitation; eIF3k, eukaryotic translation initiation factor 3k; EPC, epithelioma papulosum cyprini; HAM, HEAT analog motif; TIR, Toll/interleukin-1 receptor.
eIF3k promotes autophagic degradation of MyD88
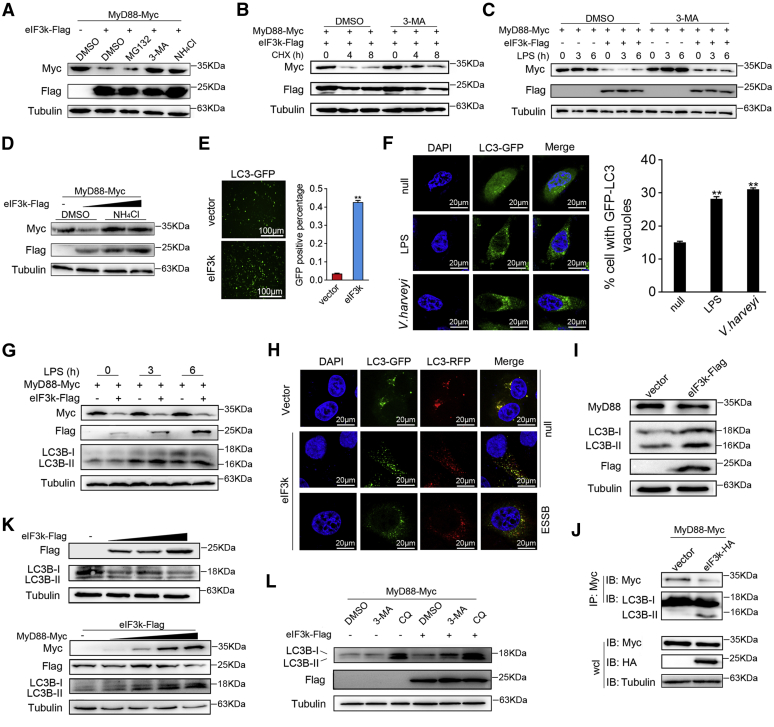
Next, we investigated the mechanism by which eIF3k suppressed MyD88 expression. We found that NH4Cl and 3-Methyladenine (3-MA), the inhibitors of autophagy-lysosome-dependent degradation pathway, both resulted in a significant accumulation of MyD88 in the presence of eIF3k, and MG132 did not affect the degradation of MyD88 (Fig. 5A). Furthermore, the combined cycloheximide (CHX) and DMSO (as control) or 3-MA at 36 h posttransfection were bear out eIF3k mediate MyD88 degradation by autophagy. Since CHX inhibited protein synthesis, 3-MA blocked autophagy at the same time, and the degradation of MyD88 was significantly slowed down (Fig. 5B). The degradation of MyD88 by eIF3k was partially inhibited when 3-MA was present, and this inhibition wasn't relieved by LPS (Fig. 5C). Moreover, similar results showed that overexpression of eIF3k firmly targeted MyD88 for degradation, and the addition of NH4Cl significantly blocked MyD88 degradation in a dose-dependent manner (Fig. 5D). As a result, we speculated that eIF3k may cooperate with the autophagy to mediate MyD88 degradation. More evidence is needed to support our inference. LC3-GFP was cotransfected with eIF3k and MyD88, and the fluorescence assay showed that LC3 puncta in eIF3k-overexpressing were significantly enhanced (Fig. 5E). LPS and V. harveyi stimulation results in the aggregation of LC3 toward the nucleus, whereas it is diffusely distributed in the cytoplasm without stimulation; LPS and V. harveyi stimulation increased the autophagy, as indicated by accumulation of GFP-LC3 puncta (Fig. 5F). eIF3k-mediated degradation of MyD88 was enhanced by LPS-induced activation of autophagy after treatment with LPS for various times (Fig. 5G). However, methods to monitor autophagic activity are complex. Based on the nonquenching characteristics of RFP in the lysosomes, LC3 can be labeled and tracked throughout; LC3-GFP-RFP was used to visually judge the changes in autophagy flux. Compared with the control, yellow puncta decreased and red puncta increased when eIF3k was overexpressed, indicating that autophagy was induced. ESSB treatment reinforced eIF3k-triggered autophagic flux, with more red puncta appearing (Fig. 5H). We found that when eIF3k mediated endogenous MyD88 degradation, the expression of LC3B-II increased (Fig. 5I); and overexpression of eIF3k significantly strengthened the interaction of MyD88 with LC3B-II (Fig. 5J), which further proved that eIF3k mediated MyD88 degradation through autophagy. We found an interesting result that overexpression of eIF3k alone didn’t cause the increase of LC3-II expression. However, after eIF3k was coexpressed with MyD88, LC3B-II was increased in a dose-dependent manner with MyD88 (Fig. 5K). At the same time, we also found that 3-MA, but not CQ, could significantly inhibit eIF3k-induced autophagic flux (Fig. 5L). Taken together, these results indicate that eIF3k mediates the degradation of MyD88 by autophagy, and eIF3k may play a role in the prophase of autophagy.
Figure 5.
eIF3k mediates MyD88 degradation through the autophagy pathway.A, 3-MA and NH4Cl blocked eIF3k-mediated MyD88 degradation. EPC cells were transfected with the indicated plasmids for 36 h, cells were stimulated with MG132 (10 μM), 3-MA (10 mM), or NH4Cl (20 mM) for an additional 8 h, DMSO was as a negative control. The level of MyD88 was detected and normalized to Tubulin. B, CHX was jointed use with DMSO or 3-MA to stimulate the EPC cells at 36 h post-transfection. MyD88 level was detected and normalized to Tubulin. C, effects of 3-MA on LPS-induced degradation of MyD88. EPC cells were transfected with eIF3k and MyD88 for 24 h, then cells were pre-treated with 3-MA for 2 h, followed by LPS stimulation for the indicated times. D, NH4Cl blocked eIF3k-mediated MyD88 degradation in a dose-dependent manner. EPC cells were cotransfected with the indicated plasmids for 36 h, NH4Cl was added for an additional 8 h. E, EPC cells were cotransfected with MyD88 and LC3-GFP, together with eIF3k or control vector. The puncta of LC3-GFP were detected by fluorescence microscope. Scale bar, 100 μm. F, effects of LPS and V. harveyi treatment on autophagy. MIC cells were treated with LPS and V. harveyi for 6 h at 24 h posttransfection, and then subjected for fluorescence microscopy. Scale bar, 20 μm; original magnification ×40. LC3-GFP dots were quantified using the Image J software. A minimum of 50 cells were scored for each condition. G, activated autophagy increases eIF3k-mediated MyD88 degradation. EPC cells were treated with LPS at the indicated time at 24 h posttransfected with eIF3k and MyD88. H, effects of EBSS treatment on autophagic flux. HeLa cells were cotransfected with MyD88, eIF3k, and LC3-GFP-RFP for 24 h, and then cells were treated with EBSS for 2 h. The nuclei were stained by DAPI (blue), pictures were taken by FCFM. Scale bar, 20 μm; original magnification ×40. I, eIF3k enhance the autophagy in MIC. MIC was transfected with the indicated plasmids for 48 h before immunoblot. J, eIF3k enhanced the association between MyD88 and LC3B. EPC cells were transfected with the indicated plasmids for 36 h before Co-IP and immunoblot. K, effects of eIF3k on the conversion of LC3B-I to LC3B-II. EPC cells were transfected with the indicated plasmids for 48 h before immunoblot. L, eIF3k plays a role in the prophase of autophagy. EPC cells were co-transfected with MyD88 and eIF3k or control vector for 36 h, cells were stimulated with 3-MA (10 mM) and CQ (50 mM) for an additional 8 h. The level of LC3B was detected and normalized to Tubulin. The data are shown as the mean ± SD of three independent experiments. (∗) p< 0.05, (∗∗) p< 0.01 versus the controls. 3-MA, 3-methyladenine; eIF3k, eukaryotic translation initiation factor 3k; EPC, epithelioma papulosum cyprini; MIC, M. miiuy intestine cell.
eIF3k potentiates Nrdp1-mediated K27-linked ubiquitination and autophagic degradation of MyD88
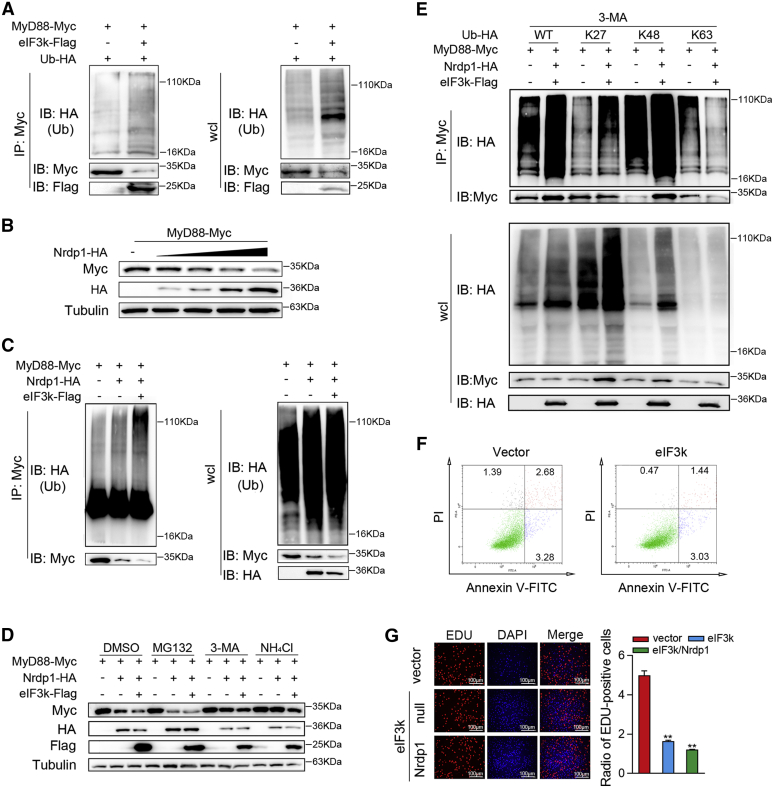
Selective autophagy relies on polyubiquitinated labeling of substrates (30). To determine whether eIF3k regulates the ubiquitination of MyD88, a coimmunoprecipitation (Co-IP) experiment was performed. The result shows that overexpression of eIF3k enhances polyubiquitination of MyD88 (Fig. 6A). Since eIF3k has no E3 ubiquitin ligase activity, we speculate that proteins with E3 ubiquitin ligase activity might be involved. Previous studies demonstrated that Nrdp1 directly binds and polyubiquitinates MyD88, leading to the degradation of MyD88 (44). We found that Nrdp1 significantly downregulated MyD88 in a dose-dependent manner (Fig. 6B). Then we verify whether eIF3k affects Nrdp1-mediated MyD88 ubiquitination. Consistent with previous findings, overexpression of Nrdp1 significantly increased ubiquitination of MyD88. Moreover, Nrdp1-induced ubiquitination of MyD88 is further potentiated when eIF3k is present (Fig. 6C). Surprisingly, Nrdp1-mediated MyD88 degradation was inhibited by 3-MA and NH4Cl, and the inhibition was further weakened when eIF3k was present (Fig. 6D). To confirm the ubiquitinated form of MyD88 that regulated by Nrdp1, we cotransfected ubiquitin-HA or its mutant (with all but one lysine residue simultaneously replaced by Arginine) with MyD88. Overexpression of Nrdp1 and eIF3k significantly enhanced K27-linked ubiquitin of MyD88, slightly increased K48-linked ubiquitin of MyD88, but did not enhance K63-linked ubiquitin of MyD88 (Fig. 6E). An apoptosis analysis was indicated that eIF3k did not significantly reduce the apoptotic (Fig. 6F). The EDU assay has shown that eIF3k counteracts the inflammatory response by inhibiting cell proliferation, and the presence of Nrdp1 enhances its inhibition (Fig. 6G). Collectively, these results indicate that eIF3k enhanced Nrdp1-mediated K27-linked MyD88 ubiquitination and autophagic degradation.
Figure 6.
eIF3k potentiates Nrdp1 ubiquitinate MyD88.A, eIF3k enhances the ubiquitination of MyD88. HEK293 cells were transfected with the indicated plasmids for 32 h, cells were subjected to Co-IP and ubiquitination analysis with the indicated antibodies. B, overexpression of eIF3k decreases the MyD88 protein level. MyD88 were co-transfected with the increased amount of Nrdp1 for 48 h in EPC cells and followed by immunoblot. C, eIF3k potentiates Nrdp1 ubiquitinate MyD88. HEK293 cells were transfected with the indicated plasmids for 32 h; cells were subjected to Co-IP and ubiquitination analysis with the indicated antibodies. D, eIF3k enhances Nrdp1-mediated autophagic degradation of MyD88. EPC cells were transfected with the indicated plasmids for 30 h. Cells were treated with MG132, 3-MA and NH4Cl for an additional 8 h before immunoblot analysis. E, Nrdp1 promote K27-linked ubiquitination of MyD88. HEK293 cells were transfected with the indicated plasmids for 30 h. Cells were treated with 3-MA for 6 h before use in ubiquitination assays. F, apoptosis in eIF3k overexpressing or control MIC cells. Cells were stained with PI and Annexin-V-FITC, and the positive stained cells were counted using FACScan. G, MIC cells were transfected with the indicated plasmids for 36 h before cell proliferation assays. Scale bar, 100 μm. The data are shown as the mean ± SD of three independent experiments. (∗) p< 0.05, (∗∗) p< 0.01 versus the controls. 3-MA, 3-methyladenine; eIF3k, eukaryotic translation initiation factor 3k; EPC, epithelioma papulosum cyprini; MIC, M. miiuy intestine cell.
V. harveyi evades immunity through synergistic inhibition of MyD88 by eIF3k and ATG5
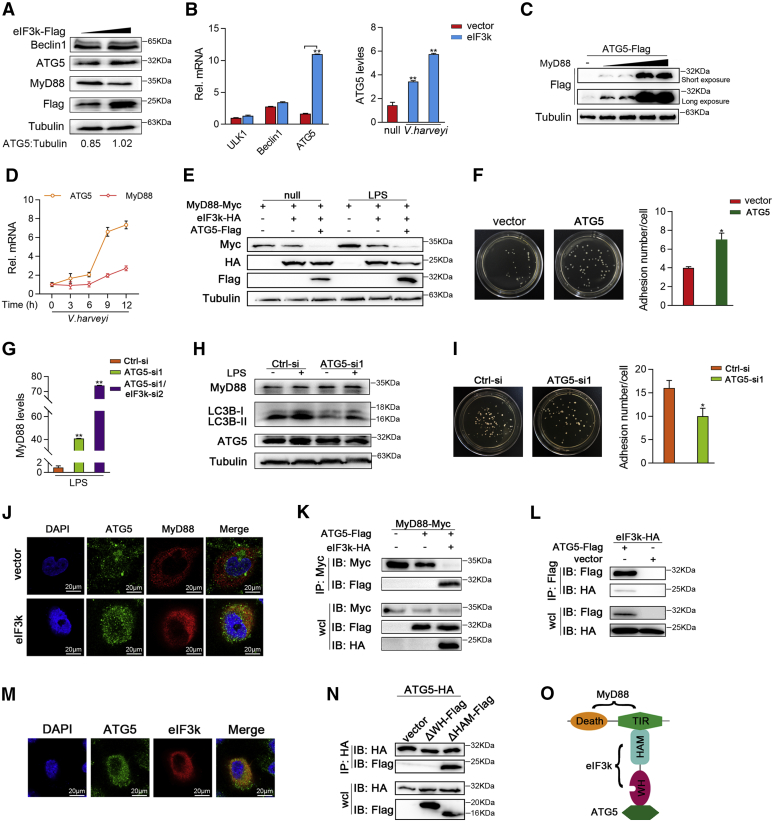
Degradation of abnormal intracellular components by autophagy is an important pathway to maintain cellular homeostasis. However, some bacteria are able to actively use autophagy to promote their own growth and infection, while the replication rate of bacteria is reduced when autophagy is absent. Thus, we next explored the role of eIF3k in the autophagic degradation of MyD88 during V. harveyi infection. Endogenic Beclin1 and ATG5 were detected and the accumulation of ATG5 showed dose-dependent with eIF3k, whereas Beclin1was not increased (Fig. 7A). A qPCR assay showed no significant changes in ULK1 and Beclin1 except for a significant increase in ATG5 upon eIF3k overexpression; and V. harveyi-triggered ATG5 transcription is further enhanced in the presence of eIF3k (Fig. 7B). ATG5 is necessary for autophagy due to its role in autophagosome elongation since the loss of ATG5 completely blocks the autophagy process (45). Overexpression of MyD88 adequately heightened ATG5 in a dose-dependent manner (Fig. 7C). A qPCR assay showed that ATG5 mRNA was time-dependent and positively correlated with MyD88 in V. harveyi persistent infection (Fig. 7D). To validate the role of ATG5 in MyD88 degradation by eIF3k, an immunoblot was performed after coexpressed ATG5, eIF3k, and MyD88. The results indicated that ATG5 significantly enhances the degradation of MyD88 by eIF3k (Fig. 7E). To further confirm the role of ATG5 in the MyD88-mediated signal, we synthesized siRNA for ATG5 (Fig. S1E). The result of qPCR showed that LPS-triggered MyD88 mRNA was significantly increased upon ATG5 knockdown. MyD88 increased more significantly when eIF3k and ATG5 were simultaneously knocked down than knocked down ATG5 alone (Fig. 7G). At the same time, we found that knockdown of ATG5 reduced LPS-triggered autophagic flux (Fig. 7H). Adhesion assay showed that the number of colonies adhering to individual cell increased significantly in the presence of ATG5 (Fig. 7, F and I).
Figure 7.
V. harveyi evades innate immunity through synergistic inhibition of MyD88 by eIF3k and ATG5.A and B, overexpression of eIF3k induces ATG5. MIC cells were transfected the indicated plasmids for 36 h. Endogenous proteins were detected by immunoblot (A) and ATG5 mRNAs was detected by qPCR (B). C, MyD88 induces ATG5 in a dose-dependent manner. EPC cells were transfected with the indicated plasmids for 48 h followed by immunoblot. D, ATG5 mRNAs was induced by V. harveyi. Expression of ATG5 and MyD88 in MIC was determined by qPCR after V. harveyi stimulation at for different times. E, ATG5 enhanced the degradation of MyD88 by eIF3k. EPC cells were transfected with the indicated plasmids for 24 h before LPS stimulation. F, ATG5 increases the adhesion of V. harveyi to MKC cells. Cells were transfected with ATG5, vector was used for control. Lysates from V. harveyi infected cells were incubated on LB plates for 12 h, and the cfu was counted. G and H, MIC cells were transfected with ATG5-si and eIF3k-si. Cells were treatment with LPS for 6 h at 48 h post-transfection. The expression MyD88 were detected by qPCR (G), autophagic flux was analysis by immunoblot (H). I, knocking down ATG5 weakened the adhesion of V. harveyi to MKC cells. Methods were same to (F). J and M, HeLa cells were transfected with the indicated plasmids for 30 h, then stained with FITC (green) or Cy3 (red). The nuclei were stained by DAPI (blue). Pictures were taken by FCFM. Scale bar, 20 μm; original magnification ×40. K, L, and N, HEK293 cells were transfected with the indicated plasmids for 32 h before Co-IP and followed by immunoblot. O, a schematic presentation of interactions among MyD88, eIF3k and ATG5. The data are shown as the mean ± SD of three independent experiments. (∗) p < 0.05, (∗∗) p< 0.01 versus the controls. eIF3k, eukaryotic translation initiation factor 3k; EPC, epithelioma papulosum cyprini; MIC, M. miiuy intestine cell; MKC, M. miiuy kidney cell.
Next, we examined whether eIF3k participates in the regulation of MyD88 by forming a complex with ATG5. We first determine whether there was a colocalization between MyD88 and ATG5. Surprisingly, MyD88 (Red) and ATG5 (Green) were distributed in the cytoplasm but not show colocalization (Fig. 7J). An interesting phenomenon emerged, in the presence of eIF3k, the puncta of ATG5 and MyD88 were overlapped in the cytoplasm, suggesting that the colocalization occurs (Fig. 7J). To test these results, a Co-IP experiment was performed. The result showed no interaction between MyD88 and ATG5 when MyD88 was cotransfected with ATG5 only. However, the interaction of MyD88 with ATG5 was readily detectable when cells were also cotransfected with eIF3k (Fig. 7K). We went on to explore the mechanism for this outcome that intrigued us. It was evident that eIF3k associated with ATG5, eIF3k-HA was detected in the anti-Flag immunoprecipitate in coexpression with ATG5-Flag, but not in the negative control (Fig. 7L). The result of immunofluorescence was also consistent with the Co-IP result, eIF3k and ATG5 were colocalized in the cytoplasm (Fig. 7M). The mutant mapping experiments revealed that ATG5 interacts with the WH domain of eIF3k (Fig. 7N). According to the results, we draw a diagram of eIF3k links MyD88 and ATG5 through diverse domains (Fig. 7O). Taken together, these findings suggest that eIF3k likely works in concert with ATG5 to regulate MyD88, a process that is exploited by V. harveyi to evade immune.
Discussion
In this study, we addressed the role of eIF3k-regulated pathways in a bacteria-triggered inflammatory response. We found that eIF3k inhibits the induction of proinflammatory cytokines by mediating the degradation of MyD88. In mechanism, eIF3k first potentiates Nrdp1-mediated K27-linked ubiquitination of MyD88. Then ubiquitinated MyD88 is linked to ATG5 in the presence of eIF3k to form a MyD88-eIF3k-ATG5 complex, which is delivery to the autophagosome for degradation. In this manner, bacteria-triggered innate signaling is subverted by eIF3k-targeted MyD88 degradation. Meanwhile, considering that LPS can also trigger the upregulation of eIF3k, we believe that eIF3k can improve the tolerance to LPS during bacterial invasion. Our study first confirmed that selective autophagy is involved in innate immune regulation in teleost fish. What's more,it was also the first time to discover that V. harveyi evade innate immune attack by eIF3k and ATG5 synergy to hijacked autophagy. And eIF3k was first found to be associated with innate immunity, which had never been found in mammals and fish before. TLRs recognize pathogen-associated molecular patterns (PAMPs) and represent the first line of immune defense, which is also the first-to-barrier encountered when bacteria invade (1). All TLRs except TLR3 use MyD88 to initiate signal transduction (3). MyD88 signaling is important for hosts to prevent bacterial invasion. Therefore, direct regulation of MyD88 may be most effective. It was found that mice lacking MyD88 were less sensitivity to LPS (46). S-1-propenylcysteine (S1PC) inhibited IL-6 production by inducing the degradation of MyD88 (47). In fish, we reported that IRF3 negatively regulates NF-κB signaling by targeting MyD88 (40), and microRNAs directly target MyD88 and affect the NF-κB signaling through posttranscriptional regulation (35, 36).
The eIF3 complex has dual roles in the initiation of mRNA translation as well as pre- and posttranslational regulation in previous studies (48, 49). Identification of eIF3k, a subunit of the eIF3 complex, as a MyD88 interactor prompted us to investigate the detailed regulatory mechanism between MyD88-mediated signaling and eIF3k. Site-directed mutagenesis and deletion analysis have shown that the TIR domain of MyD88 is essential for Toll and IL-1 activities (50). A TIR domain was also found in plant cytoplasmic proteins implicated in host defense, not only in mammals (51). MyD88s, a splicing mutant of MyD88, that lacks part of between DD and TIR, failed to activate NF-κB signaling (52). And HAM domain of eIF3k had been proposed to mediate protein–protein interactions, which can supply binding surfaces for protein–protein interactions (53). A winged-helix (WH) domain is found in our eIF3k structure, and the WH domain is generally thought to be involved in RNA binding (43). However, not all WH domains have nucleic-acid-binding activity. The interaction among eIF3k, MyD88, and ATG5 was drawn according to the mutant mapping (Fig. 7O). eIF3k, as a bridge, connects MyD88 and ATG5 through different domains. The results also demonstrated that the WH domain of fish eIF3k is involved in protein–protein interaction.
In the early screening process, we also carried out experiments with another Gram-negative bacterium, Vibrio Anguillarum, which also induces eIF3k. Therefore, eIF3k is nonspecific in Gram-negative bacterium invasion. However, V. anguillarum does not significantly alter the expression of MyD88, indicating that V. anguillarum invasion was not achieved by regulating MyD88-mediated NF-κB signaling by eIF3k, so V. harveyi was used as the subject of our study. In vertebrates, bacteria can evade immune attack through a variety of strategies (37, 54). Adhesion is the first step in Vibrio infection, which is important for the bacteria to invade the host cell and act effectively as a toxin. Vibrio adhesion is twofold: on the one hand, it activates the host's immune defenses; on the other hand, it enhances evasion and promotes reproduction. Interference with intracellular signaling pathways and disruption of the host immune response are one of the main ways of bacterial immune evasion (55). In our study, we found a significant increase in eIF3k expression at both the protein and transcriptional levels following V. harveyi infection. Subsequent adhesion assays disproved our hypothesis of eIF3k as an antibacterial molecule. Obviously, V. harveyi induces eIF3k to interfere with innate immune signaling and complete colonization and infection. As a highly conserved survival mechanism, autophagy clears invading pathogens for the host cell (56); substrates are encapsulated into autophagosomes and delivered to lysosomes for degradation (57). In our study, both 3-MA and NH4Cl could rescue eIF3k-induced MyD88 degradation. Overexpression of eIF3k can be found to promote autophagy activation both from fluorescence and immunoblot assay. Interestingly, we found that overexpression of either eIF3k or MyD88 alone did not trigger autophagy. In the presence of both MyD88 and eIF3k, autophagy is activated and autophagic flux was in a dose-dependently manner with MyD88.
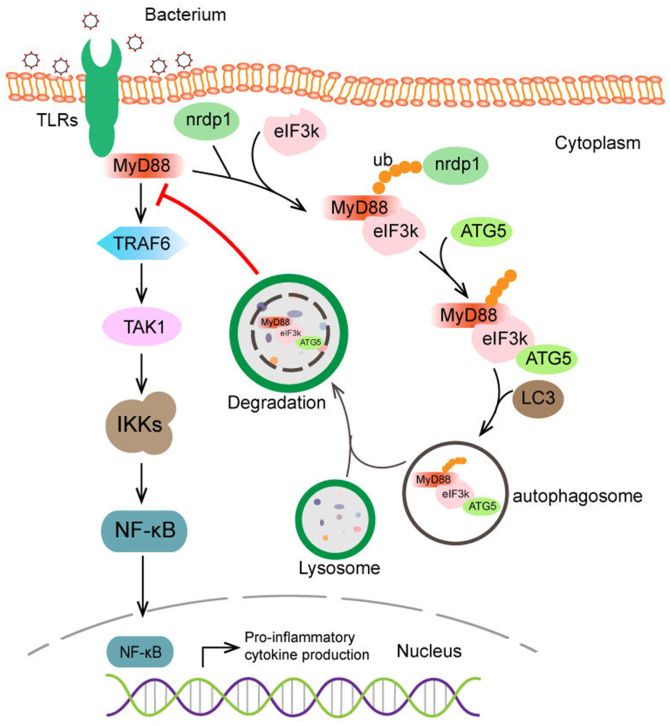
Accumulating evidence suggests that autophagy is highly selective (27, 28, 29). Selective autophagy is achieved through selective autophagic receptors. Ubiquitination, one of the most common posttranslational modifications, mediates the regulation of protein homeostasis in vivo. Selective autophagy receptors recognize ubiquitinated proteins and transported them to lysosomes for degradation (57). In this study, eIF3k markedly enhanced Nrdp1-mediated k27-linked ubiquitination of MyD88. K27-linked ubiquitination-mediated autophagy plays a major role in preserve innate immune homeostasis. K27-linked ubiquitination of TRIM23 triggers autophagy to resist the viral infection (58); and it can also negatively regulate immune responses through selective autophagy. NDP52 recognizes K27-linked ubiquitination of MAVS and mediates MAVS targeting into the autophagosome, which ultimately inhibits the immune processes (59). Upon Salmonella infection, the ubiquitin ligase LRSAM1 forms K27 ubiquitin chains and recruits the cargo receptor NDP52 to mediate autophagy (60). K48-linked ubiquitination primarily mediates proteasome degradation, and K63-linked ubiquitination mainly mediates nonprotein degradation signals. However, unlike the classical theory that K48-linked ubiquitination only targets cargo for proteasome degradation, OPTN proteins are degraded mainly through autophagy after HACE1-mediated modification of K48-linked ubiquitination (61). As previously stated, some bacteria actively use autophagy to promote reproduction and infection. ATG5 is a key molecule involved in autophagosome membrane extension, and it plays an important role in bacterial immune evasion (22). ATG5-ATG12 complex can interact with the RIG-I and MAVS to inhibit the production of IFN-1 (24). Overexpressing ATG5 not only repressed the expression of MyD88-mediated proinflammatory cytokine, but also enhanced V. harveyi adhesion to cells. We also found that ATG5 and eIF3k cooperate to induce autophagic degradation of MyD88. A novel autophagy receptor, Erp1, was identified in S. pombe. The main function of Epr1 in ER autophagy is to bridge Atg8 and VAP (62). Similarly, we found that eIF3k plays a role as a bridge linking ubiquitinated MyD88 and ATG5. Thus, we speculate that eIF3k may also act as a selective autophagic receptor and promote the selective autophagic degradation of MyD88. Based on our data, a working model of eIF3k in MyD88-mediated innate immunity was drawn (Fig. 8). When the innate immune system suffers bacterial invasion, TRAF6 and downstream components are activated and recruit to MyD88, inducing the production of proinflammatory cytokine to identify invasive pathogens. To evade immunity, eIF3k induced by V. harveyi enhanced Nrdp1-mediated K27-linked ubiquitination of MyD88. Subsequently, eIF3k acts as a bridge linking ubiquitin-tagged MyD88 and ATG5, then the eIF3k-MyD88-ATG5 complex was transported into the autophagosome to fuse with lysosomes for MyD88 degradation, contributing to the termination of MyD88-mediated innate immune and inflammatory responses.
Figure 8.
A working model of eIF3k negatively regulates MyD88 in fish.
In summary, our results reveal a novel mechanism of V. harveyi immune evasion and LPS tolerance in fish. Fish eIF3k induced by V. harveyi invasion may act as a selective autophagic receptor to promote the selective autophagic degradation of MyD88. V. harveyi manipulated autophagy to evade innate immunity was first observed, which provide new insights into understanding the effects of autophagy on host–bacterium interaction in teleost fish. And it also provides a new perspective on mammalian resistance to bacterial invasion.
Experimental procedures
Animals and challenge
M. miiuy was injected with V. harveyi or LPS as described previously (35). SCRV or poly(I:C) challenge was performed as described previously (64). RNA was extracted from the intestinal at different times after challenge. All animal experimental procedures were performed following the National Institutes of Health’s Guide for the Care and Use of Laboratory Animals, and the experimental protocols were approved by the Research Ethics Committee of Shanghai Ocean University (No. SHOU-DW-2018-047).
Cell culture and treatment
MICs and M. miiuy kidney cells (MKCs) were cultured in L-15 medium (HyClone) supplemented with 15% fetal bovine serum (FBS, Gibco), 100 U/ml penicillin, and 100 μg/ml streptomycin at 26 °C. (EPC) cells were maintained in medium 199 (Invitrogen) supplemented with 10% FBS, 1% penicillin, and streptomycin at 26 °C with 5% CO2 (41). HEK293 cells and HeLa cells were cultured in high-glucose DMEM (Hyclone) supplemented with 10% FBS and 1% penicillin and grown in a humidified incubator at 37 °C with 5% CO2. For stimulation experiments, MIC cells were stimulated with SCRV, Poly(I:C) (42), and LPS (36) at different time, and the cells were used for qPCR analysis and immunoblot analysis.
Plasmids and antibodies
The CDS of eIF3k, ATG5, LC3B, and Nrdp1 were amplified from the M.miiuy cDNA through standard PCR methods, and then the expression plasmids were cloned into pcDNA3.1 with Myc, HA, and Flag tag, respectively. The fusion plasmids MyD88-GFP, LC3-GFP, LC3-GFP-RFP were cloned into GFP or GFP-RFP vector. All the recombinant plasmid was confirmed by Western blot and Sanger sequencing. The expression plasmids ubiquitin-HA, ubiquitin-k27-HA, ubiquitin-k48-HA, ubiquitin-k63-HA were obtained from Addgene. The encoding constructs of MyD88 and eIF3k were confirmed with uniprot database, and their domain was predicted through a Simple Modular Architecture Research Tool (SMART, http://smart.embl-heidelberg.de/), two deletion mutants of MyD88 and eIF3k were amplified by PCR with specific primers based on the recombinant plasmid, named MyD88-ΔT for deletion of TIR domain, MyD88-ΔD for deletion of Death domain, eIF3k-ΔHAM for deletion of HAM domain, and eIF3k-ΔWH for deletion of WH domain. Luciferase reporter plasmids NF-κB, IL-1β, IL6, IRF3, and IFN-2 were characterized previously (35, 63).
The antibodies used in this study were as follows: anti-Flag, anti-HA, anti-Myc were purchased from Sigma; anti-GFP, anti-tubulin, Cy3-labeled goat anti-mouse IgG, and FITC-labeled goat anti-rabbit IgG were purchased from Beyotime. Anti-Myc, anti-HA, and anti-Flag for Coimmunoprecipitation were purchased from Abbkin. Endogenous antibodies, anti-MyD88, anti-TRAF6, anti-TAK1, anti-ATG5, anti-Beclin1, and anti-LC3B were purchased from Boster Biological Technology, the second antibody against mouse or rabbit was purchased from Beyotime.
RNA extraction, cDNA synthesis, and quantitative real-time PCR
Total RNA extraction and cDNA synthesis were performed as described previously (42). ChamQ Universal SYBR qPCR Master Mix (Vazyme) was used for quantitative real-time PCR (qPCR) according to the manufacturer’s protocol, all samples were analyzed in triplicate, and the expression values, unless indicated, were normalized to β-actin. Primers used for qPCR analysis are listed in Table S1. The procedure is as follows: 95 °C for 3 min, 40 cycles of 95 °C for 10 s, 60 °C for 10 s, and 95 °C for 15 s.
Transfection and luciferase activity assays
To determine the functional regulation of eIF3k to MyD88, the plasmids constructed above were transfected by Lipofectamin 3000 (Thermo Fisher) according to the manufacturer’s protocol. The 6-well plate was used for Co-IP, the 24-well plate was used for WB, and 48-well plate was used for luciferase activity assays. Luciferase activity assays were performed by dual-luciferase reporter assay system (Promega) according to the manufacturer’s protocol. Renilla luciferase reporter plasmid (pRL-TK) (10 ng/well) was used as the internal control. Cells were lysed at 24 h posttransfection, and the supernatant was analyzed. Besides, the LPS challenge experiment was performed with LPS at 24 h posttransfection.
Western blotting
Cells were washed using phosphate-buffered saline (PBS) twice to remove the dead cells. NP40 was used to obtain whole cell lysate. The lysate was centrifuged at 12,000 rpm for 4 °C, 3 min, and then heated at 95 °C, 5 min. For SDS-PAGE, equal amounts of protein were loaded into a gel, and then it was electrophoretically transferred onto PVDF membranes with a semidry process (Bio-Rad TransBlot Turbo System) after electrophoresis. The PVDF membranes were blocked with 5% skim milk (dissolved with 1% TBST) for 90 min and then used the corresponding antibody to bind overnight at 10 rpm, 4 °C. After that, the second antibody was incubated after washing PVDF with 1% TBST 3 times at 25 °C. The protein was detected by CCD system (Tanon).
RNA interference
Two eIF3k-targeted small interfering RNAs and ATG5-targeted small interfering RNAs were designed by the online website Thermo Fisher Scientific (http://www.thermofisher.com). The siRNAs and a nontargeting control siRNA (NC) were purchased from Gene Pharma. RNA interference for eIF3k is as follows: 5′-ACCGACTTCACTCTCTGCAAGTGCA-3′ (eIF3k-si1) and 5′-AGGTGAAGGTTTGGATGAATAAGTA-3′ (eIF3k-si2). RNA interference for ATG5 is as follows: 5′-GG AGTATCACTGTGCACTT-3′ (ATG5-si1) and 5′-GGAATATTCCACCGAGGAA-3′ (ATG5-si2). Lipofectamine RNAi MAX (Thermo Fisher) was used to transfected RNAi oligos. The knockdown efficiency of eIF3k was determined by qPCR and immunoblotting.
ELISA
Cells were transfected with eIF3k or eIF3k-si, pcDNA3.1 and Ctrl-si were used for control. Cells were stimulated with V. harveyi for addition 6 h at 12 h posttransfection, and then cells were collected and lysed. Then an ELISA assay was performed with fish TNFα ELISA kit (MAISHA Industries) according with the manufacturer's guidelines.
Coimmunoprecipitation and detection of ubiquitin-modified proteins
ForCo-IP, the cells were collected with RIPA (Beyotime) at 32 h posttransfection. Fifty microliters of agarose protein A/G (Sigma) was washed three times with 1 ml RIPA. After incubating agarose protein A/G with primary antibody for 2 h, the supernatant of cell lysates was added. These complexes were placed on a turntable mixer at 10 rpm and incubated overnight at 4 °C. After that, the beads were washed with cold PBS three times; all the steps need to be on the ice.
Mass spectrum analysis
The anti-MyD88 was used in the endogenous Co-IP experiment. Then the agarose-MyD88 complex was eluted with NP40, and the resulting solution was digested with trypsin. The digested solution was analyzed by Q-Exactive (Thermo), and the results were qualitatively and quantitatively analyzed by mascot software.
Immunofluorescence
Cells were grown on glass coverslips in 24-well plates and transfected. The glass coverslips are soaked with polylysine for 4 h in advance, dried, and sterilized afterward. At 32 h posttransfection, cells were washed with PBS three times and then fixed with 4% paraformaldehyde. After blocking with 0.5% FBS for 1 h, the primary antibody was added to react for 6 h. Cells were washed three times with PBS and then incubated with a secondary antibody for 1 h. Finally, after counterstaining with DAPI staining solution for 10 min, the coverslips were moved out and mounted with an antifluorescence quenching agent. Fluorescence signals were assessed by confocal microscopy (Leica).
Cell apoptosis and proliferation
Cell apoptosis was analyzed by flow cytometry at 48 h posttransfection, plasmids (500 ng/well) were transfected in a 24-well plate. Hundred microliters of Trypsin was added for 20 s, and then cells were collected and prepared with Annexin V-FITC apoptotic Kit according to the manufacturer’s protocol. In brief, cells were stained with PI and Annexin-V-FITC, and the positive stained cells were counted using FACScan. Cell proliferation assays were performed with BeyoClickEdU cell Proliferation Kit with AlexaFluor 488 (Beyotime) following the manufacturer’s instructions. All the experiments were performed in triplicate.
The adhesion of V. harveyi
After the cells were infected with V. harveyi for 2 h, the culture medium was sucked, washed with PBS for four times, digested with trypsin for 2 to 3 min, discarded the trypsin, blown down the cells with PBS, centrifuged in a centrifuge at 1200g for 5 min, removed the supernatant after centrifugation, added the cell lysate and mixed evenly, stood for 30 min, diluted 1000 times and coated on the LB plate, cultured the plate at 37 °C for 12 h and counted.
Statistical analysis
The statistical analysis was performed using Student’s two-tailed t test. The data are represented as the mean ± SD of n independent experiments. Value of p < 0.05 was considered significant, p < 0.01 was considered highly significant.
Data availability
All data are contained within the manuscript.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflict of interest with the contents of this article.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (31822057).
Author contributions
T. X. conceptualization; Y. C., W. Z., Y. S., and T. X. formal analysis; Y. C. and B. C. investigation; T. X. methodology; Y. C. and T. X. resources; Y. C. and T. X. writing—original draft.
Edited by Peter Cresswell
Supporting information
Supplemental Figure S1.
eIF3k is not involved in the regulation of antiviral signaling.A and B, MIC cells were transfected with eIF3k or control vector. Cells were treatment with SCRV or Poly(I:C) for an additional 12 h at posttransfection. IRF3 mRNAs, and IRF3 protein were detected with qPCR and immunoblot. C, MX1 mRNAs and ISG56 mRNAs were determined by qPCR after treatment with SCRV or Poly(I:C) for an additional 12 h at post-transfection. D, EPC cells were transfected with the indicated plasmids for 24 h, SCRV or Poly(I:C) treatment was as before, followed by detection of luciferase activity. E, MIC cells were transfected with ATG5-si and Ctrl-si; and the qPCR was used to detect the knockdown efficiency. The data are shown as the mean ± SD of three independent experiments. (∗) p< 0.05, (∗∗) p< 0.01 versus the controls.
References
- 1.Wolf A.J., Underhill D.M. Peptidoglycan recognition by the innate immune system. Nat. Rev. Immunol. 2018;18:243–254. doi: 10.1038/nri.2017.136. [DOI] [PubMed] [Google Scholar]
- 2.Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 3.Takeda K., Kaisho T., Akira S. Toll-like receptors. Annu. Rev. Immunol. 2003;21:335–376. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 4.Cao Z., Xiong J., Takeuchi M., Kurama T., Goeddel D.V. TRAF6 is a signal transducer for interleukin-1. Nature. 1996;383:443–446. doi: 10.1038/383443a0. [DOI] [PubMed] [Google Scholar]
- 5.Emmerich C.H., Ordureau A., Strickson S., Arthur J.S., Pedrioli P.G., Komander D., Cohen P. Activation of the canonical IKK complex by K63/M1-linked hybrid ubiquitin chains. Proc. Natl. Acad. Sci. U. S. A. 2013;110:15247–15252. doi: 10.1073/pnas.1314715110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamamoto M., Sato S., Hemmi H., Hoshino K., Kaisho T., Sanjo H., Takeuchi O., Sugiyama M., Okabe M., Takeda K., Akira S. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 2003;301:640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 7.Kawai T., Adachi O., Ogawa T., Kiyoshi S.A. Unresponsiveness of MyD88-deficient mice to endotoxin. Immunity. 1999;11:115–122. doi: 10.1016/s1074-7613(00)80086-2. [DOI] [PubMed] [Google Scholar]
- 8.Albornoz A., Carletti T., Corazza G., Marcello A. The stress granule component TIA-1 binds tick-borne encephalitis virus RNA and is recruited to perinuclear sites of viral replication to inhibit viral translation. J. Virol. 2014;88:6611–6622. doi: 10.1128/JVI.03736-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cattie D.J., Richardson C.E., Reddy K.C., Ness-Cohn E.M., Droste R., Thompson M.K., Gilbert K.D. Mutations in nonessential eIF3k and eIF3l genes confer Lifespan extension and enhanced resistance to ER stress in Caenorhabditis elegans. PLoS Genet. 2016;12 doi: 10.1371/journal.pgen.1006326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yao L., Xu B., Li X. Neisseria gonorrhoeae-induced salpingitis is targeted by circular RNA EIF3K via miR-139-5p and regulating MAPK/NF-κB signaling pathway to promotes apoptosis and autophagy bacterial cells. Microb. Pathog. 2020;142:104051. doi: 10.1016/j.micpath.2020.104051. [DOI] [PubMed] [Google Scholar]
- 11.Dong Z., Zhang J.T. Initiation factor eIF3 and regulation of mRNA translation, cell growth, and cancer. Crit. Rev. Oncol. Hematol. 2006;59:169–180. doi: 10.1016/j.critrevonc.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 12.Lin Y.M., Chen Y.R., Lin J.R., Wang W.J., Inoko A., Inagaki M., Wu Y.C., Chen R.H. eIF3k regulates apoptosis in epithelial cells by releasing caspase 3 from keratin-containing inclusions. J. Cell Sci. 2008;121:2382–2393. doi: 10.1242/jcs.021394. [DOI] [PubMed] [Google Scholar]
- 13.Shen X., Yang Y., Liu W., Sun M., Jiang J., Zong H., Gu J. Identification of the p28 subunit of eukaryotic initiation factor 3(eIF3k) as a new interaction partner of cyclin D3. FEBS Lett. 2004;573:139–146. doi: 10.1016/j.febslet.2004.07.071. [DOI] [PubMed] [Google Scholar]
- 14.Hurley J.H., Young L.N. Mechanisms of autophagy initiation. Annu. Rev. Biochem. 2017;86:225–244. doi: 10.1146/annurev-biochem-061516-044820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He X., Zhu Y., Zhang Y., Geng Y., Gong J., Geng J., Zhang P., Zhang X., Liu N., Peng Y., Wang C., Wang Y., Liu X., Wan L., Gong F., et al. RNF34 functions in immunity and selective mitophagy by targeting MAVS for autophagic degradation. EMBO J. 2019;38 doi: 10.15252/embj.2018100978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benjamin J.L., Sumpter R., Levine B., Hooper L.V. Intestinal epithelial autophagy is essential for host defense against invasive bacteria. Cell Host Microbe. 2013;13:723–734. doi: 10.1016/j.chom.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prabakaran T., Bodda C., Krapp C., Zhang B., Christensen M.H., Sun C., Reinert L., Cai Y., Jensen S.B., Skouboe M.K. Attenuation of cGAS-STING signaling is mediated by a p62/SQSTM1-dependent autophagy pathway activated by TBK1. EMBO J. 2018;37 doi: 10.15252/embj.201797858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang W., Qin Y., Song H., Wang L., Jia M., Zhao C., Gong M., Zhao W. Galectin-9 targets NLRP3 for autophagic degradation to limit inflammation. J. Immunol. 2021;206:2692–2699. doi: 10.4049/jimmunol.2001404. [DOI] [PubMed] [Google Scholar]
- 19.Shi C.S., Kehrl J.H. MyD88 and Trif target beclin 1 to trigger autophagy in macrophages. J. Biol. Chem. 2008;283:33175–33182. doi: 10.1074/jbc.M804478200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Case E.D., Chong A., Wehrly T.D., Hansen B., Child R., Hwang S., Virgin H.W., Celli J. The Francisella O-antigen mediates survival in the macrophage cytosol via autophagy avoidance. Cell Microbiol. 2014;16:862–877. doi: 10.1111/cmi.12246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi J.H., Cheong T.C., Ha N.Y., Ko Y., Cho C.H., Jeon J.H., So I., Kim I.K., Choi M.S., Kim I.S., Cho N.H. Orientia tsutsugamushi subverts dendritic cell functions by escaping from autophagy and impairing their migration. PLoS Negl. Trop. Dis. 2013;7 doi: 10.1371/journal.pntd.0001981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steele S., Brunton J., Ziehr B., Taft-Benz S., Moorman N., Kawula T. Francisella tularensis harvests nutrients derived via ATG5-independent autophagy to support intracellular growth. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee H.K., Lund J.M., Ramanathan B., Mizushima N., Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315:1398–1401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- 24.Jounai N., Takeshita F., Kobiyama K., Sawano A., Miyawaki A., Xin K.Q., Ishii K.J., Kawai T., Akira S., Suzuki K., Okuda K. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc. Natl. Acad. Sci. U. S. A. 2007;104:14050–14055. doi: 10.1073/pnas.0704014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaaf M., Keulers T.G., Vooijs M.A., Rouschop K. LC3/GABARAP family proteins: Autophagy-(un)related functions. FASEB J. 2016;30:3961–3978. doi: 10.1096/fj.201600698R. [DOI] [PubMed] [Google Scholar]
- 26.Li P., He J., Yang Z., Ge S., Zhang H., Zhong Q., Fan X. ZNNT1 long noncoding RNA induces autophagy to inhibit tumorigenesis of uveal melanoma by regulating key autophagy gene expression. Autophagy. 2019;16:1186–1199. doi: 10.1080/15548627.2019.1659614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Q., Liu T.T., Lin H., Zhang M., Wei J., Luo W.W., Hu Y.H., Zhong B., Hu M.M., Shu H.B. TRIM32-TAX1BP1-dependent selective autophagic degradation of TRIF negatively regulates TLR3/4-mediated innate immune responses. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeng Y., Xu S., Wei Y., Zhang X., Wang Q., Jia Y., Wang W., Han L., Chen Z., Wang Z., Zhang B., Chen H., Qi C., Zhu Q. The PB1 protein of influenza A virus inhibits the innate immune response by targeting MAVS for NBR1-mediated selective autophagic degradation. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang R., Varela M., Vallentgoed W., Forn-Cuni G., Vaart M., Meijer A.H. The selective autophagy receptors Optineurin and p62 are both required for zebrafish host resistance to mycobacterial infection. PLoS Pathog. 2019;15 doi: 10.1371/journal.ppat.1007329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu Y., Jin S., Liu Q., Zhang Y., Ma L., Zhao Z., Yang S., Li Y.,P., Cui J. Selective autophagy controls the stability of transcription factor IRF3 to balance type I interferon production and immune suppression. Autophagy. 2021;17:1379–1392. doi: 10.1080/15548627.2020.1761653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeong S.J., Zhang X., Rodriguez-Velez A., Evans T.D., Razani B. p62/SQSTM1 and selective autophagy in cardiometabolic diseases. Antioxid. Redox Signal. 2019;31:458–471. doi: 10.1089/ars.2018.7649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirkin V., Lamark T., Sou Y.S., Bjørkøy G., Nunn J.L., Bruun J.A., Shvets E., McEwan D.G., Clausen T.H., Wild P., Bilusic I., Theurillat J.P., Øvervatn A., Ishii T., Elazar Z., et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell. 2009;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 33.Vonmuhlinen N., Akutsu M., Ravenhill B., Foeglein G., Bloor S., Rutherford T., Freund S.V., Komander D., Randow F. LC3C, bound selectively by a noncanonical LIR motif in NDP52, is required for antibacterial autophagy. Mol. Cell. 2012;48:329–342. doi: 10.1016/j.molcel.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clausen T.H., Lamark T., Isakson P., Finley K., Larsen K.B., Brech A., Øvervatn A., Stenmark H., Bjørkøy G., Simonsen A., Terje J. p62/SQSTM1 and ALFY interact to facilitate the formation of p62 bodies/ALIS and their degradation by autophagy. Autophagy. 2010;6:330–344. doi: 10.4161/auto.6.3.11226. [DOI] [PubMed] [Google Scholar]
- 35.Chu Q., Sun Y.N., Cui J.X., Xu T.J. Inducible microRNA-214 contributes to the suppression of NF-κB-mediated inflammatory response via targeting myd88 gene in fish. J. Biol. Chem. 2017;292:5282–5290. doi: 10.1074/jbc.M117.777078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chu Q., Sun Y.N., Cui J.X., Xu T.J. MicroRNA-3570 modulates the NF-κB pathway in teleost fish by targeting MyD88. J. Immunol. 2017;198:3274–3282. doi: 10.4049/jimmunol.1602064. [DOI] [PubMed] [Google Scholar]
- 37.Günther S.D., Fritsch M., Seeger J.M., Schiffmann L.M., Snipas S.J., Coutelle M., Kufer T.A., Higgins P.G., Hornung V., Bernardini M.L., Hamid K. Cytosolic Gram-negative bacteria prevent apoptosis by inhibition of effector caspases through lipopolysaccharide. Nat. Microbiol. 2020;5:354–367. doi: 10.1038/s41564-019-0620-5. [DOI] [PubMed] [Google Scholar]
- 38.Wierzbicki I.H., Campeau A., Dehaini D., Holay M., Wei X., Greene T., Ying M., Sands J.S., Lamsa A., Zuniga E., David J.G. Group A streptococcal S protein utilizes red blood cells as immune camouflage and is a critical determinant for immune evasion. Cell Rep. 2019;29:2979–2989.e15. doi: 10.1016/j.celrep.2019.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mowat A.M., Agace W.W. Regional specialization within the intestinal immune system. Nat. Rev. Immunol. 2014;14:667–685. doi: 10.1038/nri3738. [DOI] [PubMed] [Google Scholar]
- 40.Yan X.-L., Zhao X.-Y., Huo R.-X., Xu T.-J. IRF3 and IRF8 regulate NF-κB signaling by targeting MyD88 in teleost fish. Front. Immunol. 2020;11:606. doi: 10.3389/fimmu.2020.00606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chu Q., Xu T.-J., Zheng W.W., Chang R.-J., Zhang L. Long noncoding RNA MARL regulates antiviral responses through suppression miR-122-dependent MAVS downregulation in lower vertebrates. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng W.W., Chu Q., Yang L.-Y., Sun L.-P., Xu T.-J. Circular RNA circDtx1 regulates IRF3-mediated antiviral immune responses through suppression of miR-15a-5p-dependent TRIF downregulation in teleost fish. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei Z., Zhang P., Zhou Z., Cheng Z., Wan M., Gong W. Crystal structure of human eIF3k, the first structure of eIF3 subunits. J. Biol. Chem. 2004;279:34983–34990. doi: 10.1074/jbc.M405158200. [DOI] [PubMed] [Google Scholar]
- 44.Wang C., Chen T., Zhang J., Yang M., Li N., Xu X., Cao X. The E3 ubiquitin ligase Nrdp1 “preferentially” promotes TLR-mediated production of type I interferon. Nat. Immunol. 2009;10:744–752. doi: 10.1038/ni.1742. [DOI] [PubMed] [Google Scholar]
- 45.Zhao Y., Sun X., Nie X., Sun L., Tang T.S., Chen D., Sun Q., Iwasaki A. COX5B regulates MAVS-mediated antiviral signaling through interaction with ATG5 and repressing ROS production. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1003086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arbour N.C., Lorenz E., Schutte B.C., Zabner J., Kline J.N., Jones M., Frees K., Watt J.L., Schwartz D.A. TLR4 mutations are associated with endotoxin hyporesponsiveness in humans. Nat. Genet. 2000;25:187–191. doi: 10.1038/76048. [DOI] [PubMed] [Google Scholar]
- 47.Suzuki J.I., Kodera Y., Miki S., Ushijima M., Takashima M., Matsutomo T., Morihara N. Anti-inflammatory action of cysteine derivative S-1-propenylcysteine by inducing MyD88 degradation. Sci. Rep. 2018;8:14148. doi: 10.1038/s41598-018-32431-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi J., Kahle A., Hershey J., Honchak B.M., Warneke J.A., Leong S., Nelson M.A. Decreased expression of eukaryotic initiation factor 3f deregulates translation and apoptosis in tumor cells. Oncogene. 2006;25:4923–4936. doi: 10.1038/sj.onc.1209495. [DOI] [PubMed] [Google Scholar]
- 49.Lee A., Kranzusch P.J., Cate J.H. eIF3 targets cell-proliferation messenger RNAs for translational activation or repression. Nature. 2015;522:111–114. doi: 10.1038/nature14267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Di Padova F., Quesniaux V.F.J., Ryffel B. MyD88 as a therapeutic target for inflammatory lung diseases. Expert Opin. Ther. Targets. 2018;22:401–408. doi: 10.1080/14728222.2018.1464139. [DOI] [PubMed] [Google Scholar]
- 51.Van der Biezen E.A., Jones J.D. Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem. Sci. 1998;23:454–456. doi: 10.1016/s0968-0004(98)01311-5. [DOI] [PubMed] [Google Scholar]
- 52.Mendoza-Barberá E., Corral-Rodríguez M.A., Soares-Schanoski A., Velarde M., Macieira S., Messerschmidt A., López-Collazo E., Fuentes-Prior P. Contribution of globular death domains and unstructured linkers to MyD88.IRAK-4 heterodimer formation: An explanation for the antagonistic activity of MyD88s. Biochem. Biophys. Res. Commun. 2009;380:183–187. doi: 10.1016/j.bbrc.2009.01.069. [DOI] [PubMed] [Google Scholar]
- 53.Andrade M.A., Perez-Iratxeta C., Ponting C.P. Protein repeats: Structures, functions, and evolution. J. Struct. Biol. 2001;134:117–131. doi: 10.1006/jsbi.2001.4392. [DOI] [PubMed] [Google Scholar]
- 54.Underhill D.M., Ozinsky A. Phagocytosis of microbes: Complexity in action. Annu. Rev. Immunol. 2017;20:825–852. doi: 10.1146/annurev.immunol.20.103001.114744. [DOI] [PubMed] [Google Scholar]
- 55.Levine B., Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Glick D., Barth S., Macleod K.F. Autophagy: Cellular and molecular mechanisms. J. Pathol. 2010;221:3–12. doi: 10.1002/path.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zinngrebe J., Montinaro A., Peltzer N., Walczak H. Ubiquitin in the immune system. EMBO Rep. 2014;15:28–45. doi: 10.1002/embr.201338025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sparrer K.M.J., Gableske S., Zurenski M.A., Parker Z.M., Full F., Baumgart G.J., Kato J., Pacheco-Rodriguez G., Liang C., Pornillos O., Michaela G. TRIM23 mediates virus-induced autophagy via activation of TBK1. Nat. Microbiol. 2017;2:1543–1557. doi: 10.1038/s41564-017-0017-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jin S., Tian S., Luo M., Xie W., Liu T., Duan T., Wu Y., Cui J. Tetherin suppresses type I interferon signaling by targeting MAVS for NDP52-mediated selective autophagic degradation in human cells. Mol. Cell. 2017;68:308–322. doi: 10.1016/j.molcel.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 60.Huett A., Heath R., Begun J., Sassi S., Baxt L., Vyas J., Goldberg M., Xavier R. The LRR and RING domain protein LRSAM1 is an E3 ligase crucial for ubiquitin-dependent autophagy of intracellular Salmonella typhimurium. Cell Host Microbe. 2012;12:778–790. doi: 10.1016/j.chom.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu Z., Chen P., Gao H., Gu Y., Yang J., Peng H., Xu X., Wang H., Yang M., Liu X., Hu R. Ubiquitylation of autophagy receptor Optineurin by HACE1 activates selective autophagy for tumor suppression. Cancer Cell. 2014;26:106–120. doi: 10.1016/j.ccr.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao D., Zou C.-X., Liu X.-M., Jiang Z.-D., Yu Z.-Q., Suo F., Du T.-Y., Dong M.-Q., He W., Du L.-L. A UPR-induced soluble ER-phagy receptor acts with VAPs to confer ER stress resistance. Mol. Cell. 2020;79:963–977. doi: 10.1016/j.molcel.2020.07.019. [DOI] [PubMed] [Google Scholar]
- 63.Zheng W., Chu Q., Xu T.-J. The Long noncoding RNA NARL regulates immune responses via microRNA-mediated NOD1 downregulation in teleost fish. J. Biol. Chem. 2021;296:100414. doi: 10.1016/j.jbc.2021.100414. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the manuscript.